Summary
Colletotrichum graminicola causes maize anthracnose, an agronomically important disease with a worldwide distribution. We have identified a fungalysin metalloprotease (Cgfl) with a role in virulence. Transcriptional profiling experiments and live cell imaging show that Cgfl is specifically expressed during the biotrophic stage of infection. To determine whether Cgfl has a role in virulence, we obtained null mutants lacking Cgfl and performed pathogenicity and live microscopy assays. The appressorium morphology of the null mutants is normal, but they exhibit delayed development during the infection process on maize leaves and roots, showing that Cgfl has a role in virulence. In vitro chitinase activity assays of leaves infected with wild‐type and null mutant strains show that, in the absence of Cgfl, maize leaves exhibit increased chitinase activity. Phylogenetic analyses show that Cgfl is highly conserved in fungi. Similarity searches, phylogenetic analysis and transcriptional profiling show that C. graminicola encodes two LysM domain‐containing homologues of Ecp6, suggesting that this fungus employs both Cgfl‐mediated and LysM protein‐mediated strategies to control chitin signalling.
Keywords: anthracnose, chitinase, Colletotrichum graminicola, fungalysin, host–pathogen interaction, protease
Introduction
Maize (Zea mays L.) is one of the most important crops worldwide (Wu and Guclu, 2013). In addition to its agronomic importance, maize has been a classic model organism for fundamental research for nearly a century (Strable and Scanlon, 2009). Colletotrichum graminicola (Ces.) G.W. Wils. causes anthracnose stalk rot and anthracnose leaf blight of maize (Bergstrom and Nicholson, 1999; Jamil and Nicholson, 1991), producing annual yield losses of more than one billion dollars in the US alone (Frey et al., 2011). The incidence of maize anthracnose is likely to increase as a result of climate change and changes in agriculture practices, threatening global food security (Frey et al., 2011; Munkvold, 2002).
Plants possess a variety of mechanisms to defend themselves against pathogens. One mechanism involves the secretion of hydrolytic enzymes, such as chitinases, which target components of the fungal cell wall. These enzymes can degrade structural components of the cell wall, such as chitin, rendering the cell wall unstable. Many chitinases are classified as pathogenesis‐related (PR) proteins, as they are induced in response to infection by pathogens (Kasprzewska, 2003; van Loon et al., 2006).
Chitinases also have the secondary effect of releasing chitin, which acts as a pathogen‐ or microbe‐associated molecular pattern (PAMP/MAMP). Because of the crucial role of chitin for the activation of the plant immune system, fungi have evolved mechanisms to suppress this signal (Bolton et al., 2008; Marshall et al., 2011; Mentlak et al., 2012). A broad diversity of plant‐associated fungi secrete LysM domain‐containing proteins in order to stop the chitin‐mediated activation of the plant immune system. LysM motifs, also known as CBM50 carbohydrate‐binding modules, bind to chitin and peptidoglycans, thereby masking their recognition by plant receptors (de Jonge and Thomma, 2009). In this context, the C. graminicola and C. higginsianum genomes encode more proteins with CBM50 modules than all other fungi examined (O'Connell et al., 2012). The tomato leaf mould pathogen Cladosporium fulvum secretes apoplastic chitin‐binding effectors, such as Ecp6. This protein contains three LysM domains which bind chitin fragments released from degraded cell walls by host chitinases in order to suppress the activation of plant defence receptors (Bolton et al., 2008; de Jonge et al., 2012).
In addition to LysM‐containing proteins, fungi may possess other mechanisms for the control of chitin‐mediated activation of plant defences (Naumann et al., 2011). Recently, our group has identified several putative effector proteins in C. graminicola (Rech et al., 2014; Vargas et al., 2015), and we are actively studying their roles in causing plant disease. One of the putative effectors encodes a metalloprotease belonging to the M36 family of fungalysins. Fungalysins are a group of metalloproteases that belong to MEROPS family M36 (http://merops.sanger.ac.uk/).
The first fungalysin was described from the opportunistic human pathogen Aspergillus fumigatus, which causes aspergillosis by the degradation of lung tissue through proteolytic activity (Monod et al., 1993). Fungalysins are reported to be potential virulence factors in pathogenic dermatophytes that colonize keratinized structures (Brouta et al., 2002; Jousson et al., 2004). Recently, a fungalysin from Fusarium verticillioides has been shown to cleave within a conserved sequence in class IV plant chitinases Chit A and Chit B, thereby establishing the biochemical function for this protein (Naumann et al., 2011). Subsequently, researchers have shown that fungalysins from a number of pathogens of monocot and dicot plants have the same function, truncating these chitinases and producing the same degradation products (Karimi Jashni et al., 2015; Naumann and Price, 2012; Naumann & Wicklow, 2013).
There are several examples of metalloproteases from plant pathogens that have roles in virulence (Dow et al., 2000; Jia et al., 2000; Zhang et al., 1999). Perhaps the best known example is Avr‐Pita, a zinc (Zn) metalloprotease from the rice blast fungus Magnaporthe oryzae. Avr‐Pita is recognized by a cytoplasmic resistance (R) gene‐encoded protein that triggers a signalling cascade leading to resistance (Jia et al., 2000). The C. graminicola gene Cgfl (GLRG_06543) was originally identified by Vargas et al. (2012) as a secreted fungalysin that is strongly up‐regulated during the early stages of infection. Subsequent transcriptional profiling experiments using RNA‐Seq confirmed its expression at the early stages of infection (O'Connell et al., 2012). Together, the contribution of metalloproteases to pathogenicity in other plant pathogens, the involvement of fungalysins in the pathogenicity of animal pathogens (Mathy et al., 2010) and the characteristic expression pattern in C. graminicola suggest that fungalysins play an important role in infection and anthracnose development.
In the present study, we show that fungalysins are a conserved family of metalloproteases found in a broad range of fungi. The C. graminicola fungalysin encoding gene Cgfl is only expressed in biotrophic hyphae during colonization of the plant. Pathogenicity and live cell imaging carried out with Cgfl null mutants show that this gene has an important role in virulence on maize leaves and roots. Using in vitro assays, we also show that Cgfl null mutants exhibit decreased proteolytic activity. Maize leaves inoculated with the Cgfl null mutants exhibit increased chitinase activity, consistent with the role of fungalysins as chitinase‐degrading enzymes. Together, our evidence shows that Cgfl plays an important role in the plant–fungus interaction and highlights its importance for successful plant colonization.
Results
GLRG_06543 is predicted to encode a chitin‐degrading fungalysin metalloprotease, Cgfl
Analysis of the GLRG_06543 amino acid sequence with InterProScan shows that it encodes an 18‐amino‐acid signal peptide, a fungalysin/thermolysin propeptide (FTP) domain (Markaryan et al., 1994), which is predicted to be cleaved to generate the active protein, and the fungalysin M36 peptidase domain. The M36 peptidase domain is responsible for catalytic activity and contains the HEXXH motif characteristic of Zn metalloproteases (Fig. 1a). Analysis of the predicted mature protein with the Phyre2 protein fold recognition server shows that 389 residues (95%) are modelled with 100% confidence using the structure of mep, a fungalysin from A. fumigatus, as a template. We aligned the mature protein to fungalysins of fungal pathogens that have been shown experimentally to degrade Z. mays class IV A and B chitinases (Naumann and Wicklow, 2010, 2013; Naumann et al., 2011). The alignment reveals strong sequence conservation in the catalytic HEEXH motif, and the residues implicated in the protein–protein interaction (Fig. 1b), suggesting that Cgfl functions in a similar manner to the other fungalysins.
Figure 1.
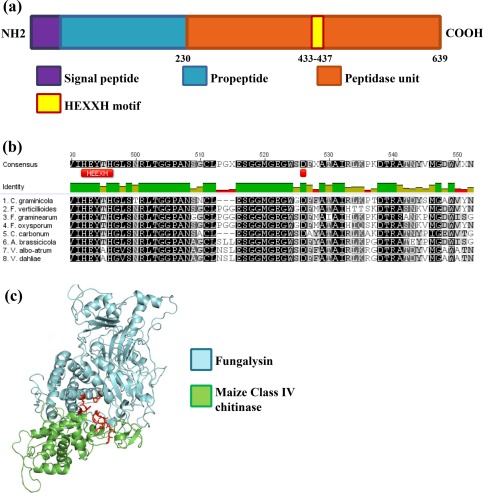
Structure of Cgfl. (a) Schematic view of Cgfl protein domains. (b) A portion of a multiple sequence alignment of fungalysins from diverse pathogenic fungi that have been shown previously to degrade chitinases. The conserved catalytic site HEXXH is present in all of them, as are the residues implicated in catalytic activity, Glu465 and His524. (c) Predicted interaction model between Colletotrichum graminicola fungalysin (blue) and Zea mays class IV chitinase ACX37090.1 (green) made with the Phyre2 server. The residues implicated in catalytic activity are shown in red.
Members of the M36 fungalysin family in other filamentous fungi are known to degrade chitinases (Karimi Jashni et al., 2015; Naumann and Price, 2012; Naumann and Wicklow, 2010, 2013; Naumann et al., 2009, 2011). To determine whether GLRG_06543 encodes a chitinase‐binding protein, we predicted the three‐dimensional structure of Z. mays class IV chitinase ACX37090.1 using the Phyre2 server (Kelley and Sternberg, 2009) and compared its structure with that of Cgfl. The predicted model shows that three residues are important in the fungalysin–chitinase interaction: His433, Glu465 and His524 (Fig. 1c). Based on the strong sequence and structural similarity of GLRG_06543 to known chitinase‐degrading fungalysins, we conclude that this locus encodes a fungalysin and named the protein Cgfl (C. graminicola fungalysin).
Cgfl is evolutionarily conserved in fungi
In the MEROPS peptidase database, fungalysins are classified as family M36 and examination of this family reveals that members are widely distributed in both fungi and bacteria. A search of the OrthoDB database (Waterhouse et al., 2013) revealed that Cgfl belongs to group EOG7RV9QR (OrthoDV V7). This group has an evolutionary rate of 1.01, approximately the average of all of the clusters in the database. Previously, we have performed a comparative analysis of the genomes of seven C. graminicola isolates (Rech et al., 2014) and a re‐examination of the supplementary data revealed that the fungalysins of all seven isolates show 100% identity. Taken together, these data show that fungalysins do not have an unusually fast rate of evolution, as has been revealed in some effector coding gene families. The study by Rech et al. also revealed that both the 5′ and 3′ regions of Cgfl contain substitutions that are consistent with positive Darwinian selection. Both the 5′ and 3′ flanking regions are known to be important for transcriptional regulation and transcript stability, suggesting that, instead of selection for mutations that affect the protein, there has been selection for mutations that affect the transcription of the gene.
We constructed a multi‐gene phylogeny of several representative species in the Sordariomycetes (Fig. 2). The phylogeny reveals at least four losses of fungalysins during the evolution of the Sordariomycetes. One gene loss event is within a lineage of fungi that includes Neurospora, Myceliophthora and others. All of the species in this lineage are saprophytes, suggesting a link between the presence of the fungalysin and the ability to infect plants. Two other species, Grossmania clavigera and Cryphonectria parasitica, lack fungalysins, both of which are tree pathogens. Several species, including C. gloeosporioides and two Verticillium spp., contain two fungalysin genes, indicating that a gene duplication occurred during the evolution of these fungi. We constructed a gene tree of the fungalysin family within the Sordariomycetes (Fig. S1, see Supporting Information). The phylogeny reveals evidence that the gene duplication probably occurred prior to the appearance of the Sordariomycetes, with numerous losses of the second gene copy in most of the family.
Figure 2.
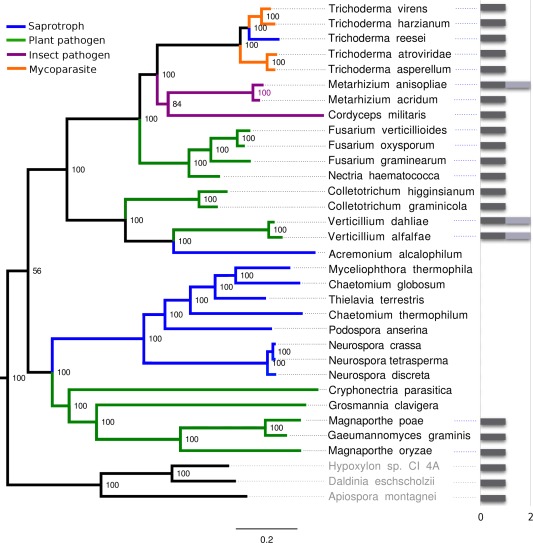
Phylogenetic tree of fungalysins in the Sordariomycetes. Grey bars represent the gene copy number in each species. Most members of the Sordariomycetes contain a single fungalysin, but gene losses and duplications have occurred. Most gene losses are in saprotrophs, with a few exceptions. Internal node labels indicate percentages of bootstrap support.
Cgfl is expressed in primary hyphae
The infection of maize leaves by C. graminicola begins with a biotrophic phase in which the fungus develops primary hyphae that are irregularly shaped, relatively wide in diameter and that colonize host tissues without causing cell death. The biotrophic hyphae colonize several host cells and, after approximately 48–60 h, the biotrophic hyphae begin to give rise to narrow, necrotrophic secondary hyphae that eventually cause cell death. By 72 h post‐infection (hpi) (Vargas et al., 2012), most of the hyphae in the central part of the lesion are necrotrophic. As the lesion expands, the leading hyphae are biotrophic and, behind the advancing hyphae, the necrotrophic hyphae colonize plant cells (O'Connell et al., 2012).
To analyse the expression of Cgfl during the infection process, we performed reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assays using maize leaves inoculated with C. graminicola M1.001. The expression of Cgfl was quantified at 12‐h intervals from 12 to 72 hpi, with final samples taken at 8 days post‐infection (dpi). Mycelium grown on potato dextrose broth (PDB) and conidia were also included. No Cgfl gene expression was evident in in vitro‐cultured mycelium or in conidia. During the infection process, gene expression is evident at 36 hpi and reaches a peak at 60 hpi when the majority of the hyphae are biotrophic. After 60 hpi, the level of expression is reduced, but expression still occurs at low levels up to 8 dpi (Fig. 3a). There is a correlation between the expression profile of Cgfl and the relative abundance of biotrophic hyphae in colonized plant tissue.
Figure 3.
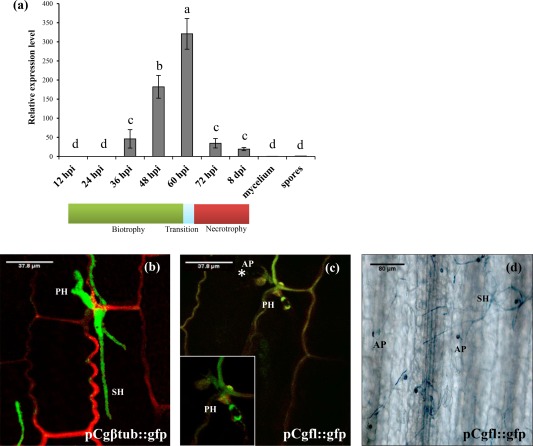
Analysis of Cgfl expression during anthracnose development. (a) Quantitative real‐time polymerase chain reaction analysis of Cgfl expression during anthracnose development. RNA samples were extracted from leaves infected with the wild‐type (WT) strain M1.001. The transcript levels of Cgfl were normalized to the constitutively expressed Colletotrichum graminicola histone H3 gene. Bars ± standard deviation (SD) denote mean expression values of three biological replicates, and different letters indicate significant differences according to Tukey's honestly significant difference (HSD) analysis (P < 0.05). A 29‐Cycle Threshold (Ct) cycle was used as the maximum limit of detection and the spore value was denoted as reference for the analysis. hpi, hours post‐infection; dpi, days post‐infection. (b) Confocal image of a maize leaf infected with PCgfl‐tubulin::gfp, used to visualize biotrophic and necrotrophic hyphae. (c) To further study the expression pattern of Cgfl, the reporter gene gfp was fused to the promoter region of fungalysin PCgβ‐tu::gfp. Fluorescence was only detected in primary hyphae at 60 h post‐infection (hpi), when the fungus switches from biotrophic to necrotrophic growth. (d) Bright field image and lactophenol blue staining denote the presence of secondary hyphae in the same site of infection as previously observed with confocal microscopy. AP, appressorium; PH, primary (biotrophic) hyphae; SH, secondary (necrotrophic) hyphae.
To further study Cgfl expression in planta, we engineered a strain (PCgfl::gfp) expressing a Cgfl promoter–green fluorescent protein (GFP) fusion construct. We also visualized the development of biotrophic and necrotrophic hyphae in wild‐type (WT) C. graminicola M1.001 by constructing a transformant (PCgβ‐tubulin::gfp) expressing a β‐tubulin promoter–GFP fusion construct (Fig. 3b). Maize leaves were inoculated with both strains and infection sites were observed by confocal microscopy. The PCgβ‐tubulin::gfp transformant fluoresced in both biotrophic and necrotrophic hyphae, enabling the visualization of both structures in planta (Fig. 3b). At early stages (24 and 48 hpi), no GFP fluorescence was detected in the strain carrying the PCgfl::gfp construct (data not shown); however, fluorescence caused by the activation of the Cgfl promoter was observed at 60 hpi, specifically in biotrophic hyphae, the same time point at which Cgfl is strongly up‐regulated in planta (Fig. 3c). In contrast, no fluorescence was seen in secondary hyphae at 60 hpi or at later time points, such as 96 hpi. To confirm the presence of secondary hyphae in tissues at 60 and 96 hpi, excised infection sites were stained with lactophenol blue. The staining revealed secondary hyphae penetrating the infected epidermal tissue, which is characteristic of typical anthracnose development (Fig. 3d), and ensured that infection was progressing normally. From these experiments, we conclude that Cgfl is specifically expressed in biotrophic hyphae.
ΔCgfl mutants are severely compromised in virulence on maize leaves and roots
To characterize the role of Cgfl during the infection process, targeted gene deletion mutants were constructed using the DelsGate method (García‐Pedrajas et al., 2008). The native Cgfl gene was replaced by the vector pKW1, which contains the gfp reporter gene and the hygromycin phosphotransferase gene (hph) (Fig. S2a, see Supporting Information). Approximately 200 transformants were analysed, two of which contained the expected gene deletion and showed GFP fluorescence (ΔCgfl36 and ΔCgfl88, Fig. S2b). Integration of the deletion construct by homologous recombination, replacing the gene, was confirmed by PCR and Southern blot analysis (Fig. S2c,d). Growth rate assays, sporulation rate and appressorium formation assays showed no statistical differences between the ΔCgfl mutants and a gfp‐tagged version of the WT strain, M1.001 BH‐GFP (Sukno et al., 2008), indicating that these phenotypes are not affected by the loss of Cgfl (Fig. S3, see Supporting Information), further supporting a role for this protein only in planta, during the interaction of the fungus with the plant.
Maize leaves were inoculated with spore suspensions of M1.001 BH‐GFP, ΔCgfl36 and ΔCgfl88 (Fig. 4a). Control plants were inoculated with water. No differences between inoculations were seen at early points and, at 72 hpi, leaves inoculated by both WT and mutant strains exhibited visible lesions (not shown). At 96 hpi, the necrotic lesions on leaves inoculated with the ΔCgfl strains were significantly smaller than those on the leaves inoculated with the WT strain (Fig. 4b). These results show that Cgfl is important for host colonization and disease development.
Figure 4.
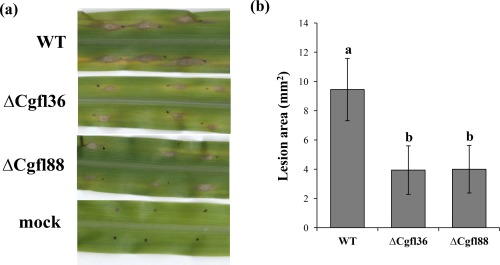
Anthracnose leaf blight assays at 96 h post‐infection (hpi) performed with wild‐type (WT) and ΔCgfl transformants. (a) Lesions on maize leaves at 96 hpi inoculated with spore suspensions of M1.001 BH‐GFP and ΔCgfl. Two independent transformants are shown. Mock‐inoculated leaves were inoculated with water. Disease symptoms were reduced on plants inoculated with the ΔCgfl mutants. (b) Mean lesion size was significantly reduced in ΔCgfl mutant infections compared with M1.001 BH‐GFP. Error bars denote ± standard deviation (SD). Bars with different letters indicate significant differences according to Tukey's honestly significant difference (HSD) test (P < 0.001). Three independent experiments were performed. Black dots indicate inoculation points.
Fungal infection was monitored by laser scanning confocal microscopy at 24, 48 and 60 hpi, which corresponds to the time point of maximum Cgfl expression in planta, and 96 hpi, when the central zone of the lesions comprises secondary, necrotrophic hyphae. No differences between inoculations were seen at 24 and 48 hpi (data not shown). Both the WT and ΔCgfl strains contained a constitutively expressed gfp construct, enabling us to quantify the differences in tissue colonization by measuring GFP fluorescence at lesion borders and lesion central zones. Examination of the central zone and lesion borders using confocal microscopy showed that, at 60 hpi, the infection process proceeded more slowly with the ΔCgfl36 strain than with the WT strain, indicating that ΔCgfl36 reduced the ability to colonize maize cells (Fig. 5a,b,d,e). The analysis of the fluorescence intensity in images taken at the lesion border and the lesion central zone showed that lesions of the ΔCgfl36 strain were significantly less fluorescent than those of the WT, consistent with reduced colonization of the host tissue (Fig. 5c,f). At 96 hpi, there were no significant differences in the levels of fluorescence in the central zones of lesions produced by the WT and ΔCgfl36 strain (Fig. 5g–i). However, the lesion borders still contained significantly reduced numbers of hyphae, based on visual inspection and quantification of fluorescence (Fig. 5j–l). These results suggest that the ΔCgfl36 strain has a reduced capacity to invade plant tissues during the early stages of colonization, coinciding with the biotrophic stage of development, and at the borders of the lesions which predominantly comprise biotrophic hyphae. Necrotrophic development appears not to be impeded in the ΔCgfl36 mutant.
Figure 5.
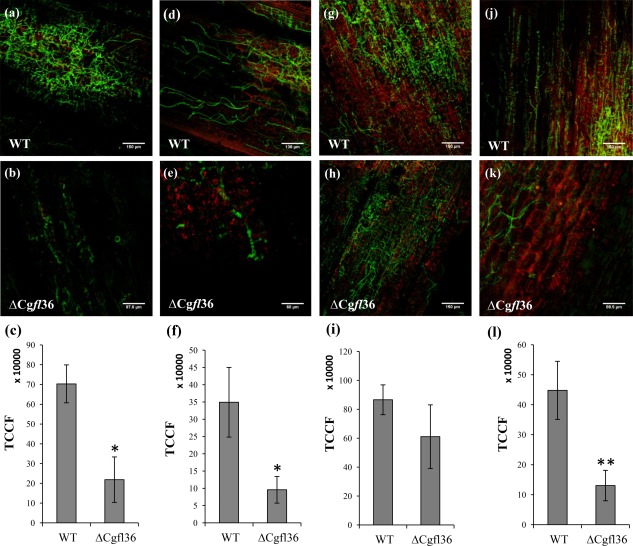
Live cell confocal imaging of maize leaves infected with wild‐type (WT) Colletotrichum graminicola and the ΔCgfl transformants. (a) Fungal infection in maize leaves was monitored by confocal microscopy at 60 h post‐infection (hpi) and 96 hpi. A detailed examination of the lesions at 60 hpi showed that the infection proceeded more slowly in the ΔCgfl36 strain relative to the WT strain, indicating that the fungus had reduced ability to colonize host maize cells in the central zone (a, b) and in the borders of the lesion (d, e) with respect to the WT strain. Differences in fungal growth were quantified by measuring green fluorescent protein (GFP) fluorescence (c, f) using the software ImageJ. The values shown in the graph represent averages of total corrected cellular fluorescence (TCCF) of two independent experiments with standard deviation (SD) bars. Asterisk denotes samples that were significantly different from the WT (P < 0.05). At 96 hpi, both WT and ΔCgfl36 strains showed similar growth in the central zone of lesions (g, h) but, in the lesion borders, the ΔCgfl36 mutant exhibited delayed development (j, k). Fungal growth was quantified by measuring GFP fluorescence, and TCCF values are represented in graphs (i) and (l). Double asterisks denote samples that were significantly different from the WT (P < 0.001).
We also tested the ability of strain ΔCgfl36 to infect maize roots by performing pathogenicity assays with the gfp‐tagged WT and ΔCgfl36 strains. At 6 dpi, colonization of the epidermis and first cell layers of the cortex by the WT strain was visible (Fig. 6a). In contrast, the epidermal cells of the roots infected with ΔCgfl36 showed reduced colonization and, instead, extensive colonization of the exterior of the root (Fig. 6b). At 12 dpi, roots infected with the WT strain were extensively colonized by intra‐ and intercellular hyphae, which extended from the cortex to the endodermis (Fig. 6d). Roots infected with ΔCgfl36 showed significantly less colonization of the cortex. Strain ΔCgfl36 colonized the epidermal layer and cortex, but did not reach the endodermis (Fig. 6e). Quantification of fluorescence revealed significantly reduced fluorescence at both 6 and 12 dpi (Fig. 6c,f). These observations show that Cgfl contributes to virulence in maize roots.
Figure 6.
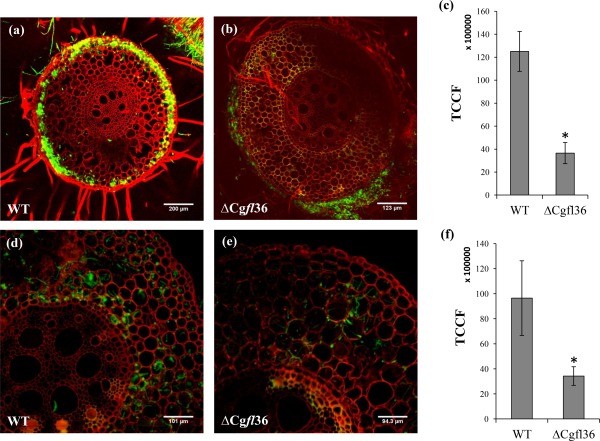
Live cell confocal imaging of inoculated maize roots using wild‐type (WT) and ΔCgfl36 transformants. Confocal analysis of ΔCgfl36 revealed that it showed reduced root colonization relative to WT at 6 and 12 days post‐infection (dpi) (a–c). At 6 dpi, ΔCgfl36 remained mostly outside of the root epidermis, whereas WT colonized epidermal cells and parts of the cortex (d–f). At 12 dpi, ΔCgfl36 colonized less root tissue than WT. Quantification of the green fluorescent protein (GFP) fluorescence signal was made using ImageJ software, and the values shown in the graphs are averages of the total corrected cellular fluorescence (TCCF) of two independent inoculations ± standard deviation (SD). Significant differences (P < 0.05) between WT and ΔCgfl36 are indicated by asterisks.
We also constructed a strain constitutively expressing Cgfl by ligating the gpda promoter to the Cgfl coding sequence. First, we tested several types of liquid medium for their capacity to induce Cgfl transcription in the WT strain in vitro. Maize leaf extract (LE) medium was also used to mimic the plant environment, thereby transcriptionally activating genes that are usually only expressed in planta (Krijger et al., 2008). RT‐PCR assays confirmed that, in the WT strain, Cgfl is expressed in vitro in maize LE medium, but not in minimal medium, and that Pgpda::Cgfl is expressed in both media (Fig. 7a). In vitro assays of growth rate, sporulation and appressorium development showed that Pgpda::Cgfl is not affected in any of these phenotypes (Fig. S3). Pathogenicity assays also failed to detect any differences between Pgpda::Cgfl and the WT strain (Fig. S6, see Supporting Information).
Figure 7.
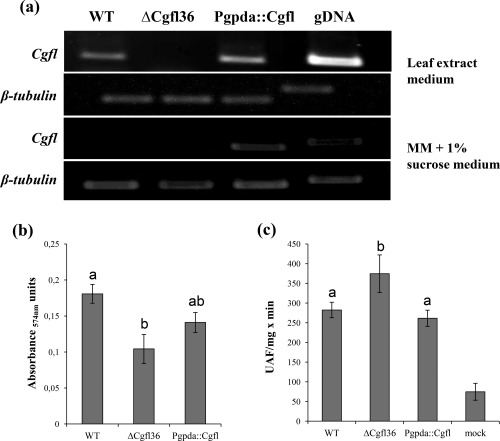
In vitro expression, proteolytic and chitinolytic activity assays. (a) Reverse transcription‐polymerase chain reaction (RT‐PCR) assays showed that fungalysin is differentially expressed using in vitro cultures. Maize leaf extract (LE) medium induced Cgfl expression, whereas minimal medium (MM) did not. (b) Secreted endoproteolytic activity assay revealed reduced absorbance in ΔCgfl36 with respect to the wild‐type (WT) in LE medium, in which Cgfl is expressed. Two independent experiments were performed with duplicates each time. Error bars represent ± standard deviation (SD). Bars with different letters differ significantly according to Tukey's honestly significant difference (HSD) analysis at the 5% level. (c) Chitinase activity assay with protein extractions of maize leaves inoculated with Colletotrichum graminicola WT, ΔCgfl and Pgpda::Cgfl strains, showing significant differences (P < 0.05) according to Tukey's HSD test. Three independent experiments with duplicates each time were performed. UAF, Units of Arbitrary Fluorescence.
The ΔCgfl mutant shows reduced proteolytic activity
We confirmed the endoproteolytic activity of Cgfl using an in vitro protease activity assay of strains WT, ΔCgfl36 and the constitutively expressed strain Pgpda::Cgfl. Cgfl transcripts of the WT strain were only detected in LE medium (Fig. 7a). Therefore, LE medium was used for the protease assays. The fungal strains were grown in LE medium and culture filtrates were tested for their capacity to degrade resorufin‐labelled casein, a substrate for the detection of endoproteolytic activity. The absorbance of ΔCgfl36 was significantly reduced relative to the WT and Pgpda::Cgfl strains, indicating a reduced proteolytic activity in this strain (Fig. 7b). To evaluate whether Cgfl plays a role in plant chitinase degradation during the establishment of infection, we performed an in vitro chitinase activity assay with protein extracts of infected maize leaves, testing for differences in total chitinase activity present in leaves inoculated with WT and ΔCgfl strains. The artificial substrate 4‐methylumbelliferyl‐β‐d‐N,N ′,N ″‐triacetylchitotrioside hydrate was used, which, when cleaved by endochitinases, releases a fluorescent product. Samples from plants inoculated with ΔCgfl showed significantly elevated fluorescence, reflecting increased chitinase activity, compared with those inoculated with the WT strain (Fig. 7c). No significant differences were observed between the WT and constitutively expressed Pgpda::Cgfl strains. Taken together, these results show that, in the absence of Cgfl, maize leaves show increased chitinase activity. This result is consistent with the role of fungalysins in the degradation of chitinases in other fungi (Karimi Jashni et al., 2015; Naumann and Wicklow, 2013; Naumann et al., 2011).
The C. graminicola genome encodes two Ecp6 homologues
Bioinformatic analysis shows that Cgfl is a fungalysin, a family that is known to degrade chitinases produced by plants. Fungalysins may play a role in the mediation of chitin‐triggered immunity (CTI) by degrading the enzymes needed by the plant to activate CTI. Another mechanism by which fungi mediate CTI is through chitin‐binding proteins, such as Ecp6 from Passalora fulva (Cl. fulvum). Homologues of the gene encoding Ecp6 have been identified in several other plant pathogens, including Slp1 from M. oryzae and Mg3LysM from Zymoseptoria tritici. Many species that contain Ecp6 homologues also possess copies of Cgfl, suggesting the presence of two complementary mechanisms for the mediation of CTI. To determine whether C. graminicola also contains Ecp6 homologues, we performed a blast search using Ecp6 as a query sequence vs. a blast database of C. graminicola proteins, and identified two proteins with high similarity to Ecp6: GLRG_07767 and GLRG_02947. The structural similarity of the two proteins was analysed using the Phyre2 web server and, in both cases, 90% of the amino acids in both proteins were aligned at over 90% confidence to the Ecp6 structure. We performed another blast search, this time using the non‐redundant (nr) database and Ecp6 as a query sequence, in order to identify more Ecp6 homologues in other fungi. A phylogenetic analysis of the two C. graminicola Ecp6 homologues and the blast hits revealed two clades that contain proteins that have been experimentally proven to interact with chitin and that modulate plant immunity (Fig. S4, see Supporting Information). Both C. graminicola homologues are within the clade that includes M. oryzae Slp1 and Slp2, C. lindemuthianum Cih1 and C. higginsianum ChELP1 and ChELP2 (Kombrink, 2014; Mentlak et al., 2012).
We also performed RT‐qPCR experiments to determine whether the Ecp6 homologues are expressed during the infection of maize leaves (Fig. S5, see Supporting Information). Both genes show peak expression during the biotrophic stage, after which the expression levels continue to decrease. No transcripts of either gene were detected in in vitro‐cultured mycelium or in germinating spores.
Discussion
In recent years, an increasing number of effectors have been identified, but, for many, their biological function and plant targets remain elusive (Oliva et al., 2010; Rafiqi et al., 2012; Stotz et al., 2014). In this work, we describe a fungalysin metalloprotease that plays a role in the virulence and colonization of maize leaves and roots. The biological activity of fungalysins has been demonstrated recently through biochemical analysis in several plant‐pathogenic fungi, including C. higginsianum; they have been shown to specifically degrade chitinases produced by plants, suggesting that they play a role in chitin‐mediated plant defence (Karimi Jashni et al., 2015; Naumann and Wicklow, 2013; Naumann et al., 2011). The C. graminicola Cgfl protein represents another mechanism by which fungi control chitin‐mediated activation of plant defences.
We have shown that fungalysins are highly conserved enzymes, and are present in most fungi. Most members of the Sordariomycetes have a single fungalysin gene, although some species carry two copies, which may have arisen though an ancient gene duplication event or through horizontal gene transfer. Vertebrate pathogens of the Onygenales, an order of dermatophytic fungi, have duplications of genes encoding fungalysins, and it has been proposed that these gene family expansions play a role in host specialization (Jousson et al., 2004; Li and Zhang, 2014). The same may be true in plant pathogens, and we hypothesize that, in species with gene duplications, both copies play a role in pathogenesis. Our phylogenetic analysis also revealed that some members of the Sordariomycetes lack a fungalysin gene, including a lineage that contains species that are typically regarded as saprophytes, suggesting that a loss of the fungalysin gene may be associated with a loss of pathogenicity. The plant pathogens within the Sordariomycetes have a variety of lifestyles, including those that specialize in the colonization of above‐ground plant parts and those that specialize in the colonization of roots. This suggests that fungalysins are important for fungi with a diversity of different pathogenic lifestyles.
Previous studies have revealed that C. graminicola effector gene expression is highly dynamic and is induced in successive waves during specific stages of the infection process (O'Connell et al., 2012). During the biotrophic phase, transcriptional profiling has revealed that secondary metabolites and effector proteins are preferentially secreted in order to establish and facilitate host infection, whereas CAZymes, proteases and transporters are expressed at the switch to the necrotrophic stage. Previously, we have identified a fungalysin in C. graminicola that is strongly up‐regulated during the early stages of infection (Vargas et al., 2012). Genome‐wide expression profiling by RNA‐Seq also revealed that Cgfl is up‐regulated during the biotrophic phase and is among the 100 most highly expressed genes induced in biotrophy (O'Connell et al., 2012). In this study, biotrophy‐associated gene expression was confirmed by RT‐qPCR and live cell imaging of a WT strain carrying a Cgfl promoter‐gfp fusion, indicating that it is specifically expressed in biotrophic hyphae during the late biotrophic phase of the infection process. This highly modulated pattern of expression is consistent with other effectors and suggests that Cgfl is not needed during necrotrophic growth.
Interestingly, constitutive expression of Cgfl has no effect on virulence or any of the other studied phenotypes. Several cases have been reported in the literature in which the constitutive expression of defence‐associated genes has no effect on pathogenicity or other phenotypes (Baeza‐Montañez et al., 2015; Yu et al., 2015). It is well known that transcript abundance levels may not be correlated with protein levels and/or enzymatic activity because of the regulation of translation, protein post‐translational modifications or turnover (Baeza‐Montañez et al., 2015; Yang et al., 2013). Thus, it is possible that, even though the gene is constitutively expressed, the protein is not present or is not in a mature and fully functional state. Alternatively, if Cgfl specifically targets one or a few plant chitinases, its expression in conditions and at time points at which the chitinases are not present would not result in a change in phenotype. Future studies aimed at the study of maize chitinase expression during the infection process may reveal whether Cgfl is expressed specifically and only when maize chitinases are present.
Recently, we have performed a genome‐wide survey of natural selection acting on protein coding and non‐coding DNA sequences in C. graminicola (Rech et al., 2014). The fungalysin‐encoding gene Cgfl was among a group of genes for which there is evidence for positive selection in the 3′‐untranslated region (3′‐UTR) and downstream region of the gene. There is increasing evidence that this region of the gene plays an important role in gene transcriptional regulation. We speculate that there may be strong selective pressure for the fungus to modulate the expression of Cgfl to coincide with the expression and secretion of the plant's chitinases.
To validate the role of Cgfl during pathogenesis, we developed C. graminicola null mutants lacking the Cgfl gene and tested their ability to infect maize leaves and roots. Infection assays on maize leaves with Cgfl null mutants showed that Cgfl is required for full virulence on maize plants. Confocal microscopic analysis revealed that ΔCgfl mutants, during the early stages of infection and later at lesion margins, which mainly comprise primary hyphae, show reduced capacity to proliferate through leaf tissue. These results reinforce the idea that Cgfl suppresses the host immune system during the biotrophic phase of the infection and that it has no role during necrotrophic growth. Infection assays carried out on maize roots showed that ΔCgfl mutants had reduced capacity to colonize the cortex, revealing that Cgfl also plays a role in chitin‐mediated signalling in roots. In Fusarium oxysporum f.sp. lycopersici, Karimi Jashni et al. (2015) found that a null mutant of a fungalysin (FoMep1) had no effect on virulence; however, when both FoMep1 and a serine protease (FoSep1) were deleted in the same strain, a reduction in virulence was observed. As the ΔCgfl mutants show a clear phenotype in C. graminicola, we conclude that fungalysin‐mediated chitinase degradation plays a more important role in the virulence of C. graminicola than in F. oxysporum f.sp. lycopersici. However, the C. graminicola genome encodes two serine proteases with high similarity to FoSep1, and so we cannot discount the possibility that they may also contribute to chitinase degradation.
Many fungi that interact with plants secrete proteins that contain LysM motifs, which enable the proteins to interact with, and sequester, chitin molecules, or to protect chitin from degradation by plant hydrolytic enzymes (Gust et al., 2012; Kombrink et al., 2011). Several fungal effectors of Cl. fulvum have been reported to interfere with chitin perception (Bolton et al., 2008; de Jonge et al., 2012). Interestingly, LysM effectors are not restricted to Cl. fulvum, and orthologues can be found in a wide range of fungal pathogens (Bolton et al., 2008; Marshall et al., 2011; Mentlak et al., 2012; Stergiopoulos et al., 2010), including Colletotrichum spp., such as C. lindemuthianum (Takahara et al., 2009), C. higginsianum and C. graminicola (O'Connell et al., 2012). One of these genes, Ecp6, has been shown to be important in virulence in Cl. fulvum (Bolton et al., 2008) by sequestering chitin oligomers and preventing chitin‐triggered microbe‐/pathogen‐triggered immunity (M/PTI) in plants (de Jonge et al., 2011). Homologues of Ecp6 have been identified in Z. tritici, M. oryzae, C. lindemuthianum and C. higginsianum. Some of these species carry two Ecp6 homologues, but, in each case, at least one of the copies has been demonstrated to suppress chitin‐induced M/PTI (Marshall et al., 2011; Mentlak et al., 2012). The C. graminicola genome encodes two genes (GLRG_02947 and GLRG_07767) with homology to Ecp6. The similarities in domain structure, phylogenetic relationships and gene expression patterns are all consistent with the hypothesis that these two C. graminicola genes have functions similar to Ecp6. Both genes are expressed specifically during the infection process and have a peak of expression at 36 and 48 hpi, respectively, before the expression peak of Cgfl, suggesting that C. graminicola employs both strategies to control CTI, but not at the same time. Like Ecp6, fungalysins are broadly distributed in fungi, but, unlike Ecp6, the fungalysins have highly conserved amino acid sequences, suggesting that their functions have remained essentially unchanged despite hundreds of millions of years of evolution in distinct fungal lineages. We propose that fungalysins are an ancient mechanism by which fungi control CTI, and that this mechanism is present in a wide diversity of fungi that interact with plants.
In Z. tritici, evidence of positive Darwinian selection was detected in Ecp6 homologues (Marshall et al., 2011). Interestingly, our previous genome‐wide survey of selection in C. graminicola did not detect positive selection in the coding sequence of these genes. Instead, we found evidence of positive selection in the flanking non‐coding parts of the gene, suggesting selection for the transcriptional profile (Rech et al., 2014).
The arms race model is often used to describe the evolution of plant–fungal interactions. Chitin signalling is a good example of the evolution of plant–fungal cross‐talk. Chitin is a key component of fungal cell walls and plants have evolved receptors to detect it and activate plant defences. One of these defences is the production of chitinases, which degrade the fungal cell wall, releasing chitin monomers that improve the plant's ability to detect the presence of fungi. Chitinases therefore represent an evolutionarily conserved mechanism to degrade fungal cell walls and aid in their detection. Fungi have evolved several mechanisms to mitigate the chitin‐mediated defence responses. One of these is chitin deacetylation, converting chitin to chitosan, which has a significantly reduced capacity relative to chitin to elicit a response (El Gueddari et al., 2002; Vander et al., 1998). Fungi also secrete chitin‐binding proteins to protect themselves from chitin‐mediated activation of the plant immune system. In addition, fungi have evolved proteases, such as fungalysin, which degrade chitinases directly. Recently, a further step in this model of plant–pathogen co‐evolutionary competition has been described. Hevein‐like antimicrobial peptides that block fungalysin activity have been described in wheat (Slavokhotova et al., 2014).
Our discovery of the precise transcriptional regulation of Cgfl during the first hours of infection and our subsequent demonstration of its importance in virulence demonstrate that secreted proteases represent an important class of effectors that target specific proteins involved in the plant immune system. In conclusion, this work supports the hypothesis that Cgfl is a fungalysin metalloprotease, which is important during the establishment of infection and contributes to virulence. This protein is highly conserved among fungi, suggesting that chitinase degradation is a characteristic of many fungal species. Fungalysin‐mediated degradation of chitinases is another mechanism by which fungi may attenuate chitin‐mediated activation of the plant's immune system.
Experimental Procedures
Phylogenetic analysis
To identify homologues of GLRG_06543, we performed a blast search against the complete fungal proteomes available in UniProt (www.uniprot.org), GenBank (http://www.ncbi.nlm.nih.gov/genbank/), Joint Genome Institute (http://genome.jgi.doe.gov/programs/fungi/index.jsf) and Broad Institute (http://www.broadinstitute.org/) databases. The homologous proteins were aligned with MAFFT (Katoh et al., 2002). MODELGENERATOR (Keane et al., 2006) was used to identify the best‐fitting substitution model and PhyML (Guindon et al., 2010) to reconstruct the phylogeny. A bootstrap test with 100 replicates was also performed with PhyML. The sequence alignments and phylogenetic analysis were performed with Geneious (http://www.geneious.com).
The complete proteomes of the selected species were analysed with Mirlo (https://github.com/mthon/mirlo) to identify conserved single‐copy protein families. Five protein families were aligned with MAFFT and the alignments were concatenated. The phylogenetic tree was constructed using PhyML.
Homologues of Passalora fulva Ecp6 were identified by performing a blastp search to the nr database. The top 100 hits to members of the Fungi were kept for the analysis. The proteins were reviewed to remove duplicate sequences and were aligned with MAFFT. The alignment was edited to remove the highly variable parts of the sequences, leaving only two conserved LysM domains. The phylogenetic tree was constructed with RAxML using the GAMMA JTT substitution model. The blast search, alignment and other operations were completed using Geneious.
Prediction of the fungalysin structure and interaction model
The signal peptide and cleavage site were identified with SignalP (Petersen et al., 2011). InterProScan was used to identify the protein domains. The three‐dimensional structures of Cgfl and Z. mays class IV chitinase ACX37090.1 were constructed using the Phyre 2 server (Kelley and Sternberg, 2009). We manipulated the three‐dimensional models with PyMOL (Version 1.2r3pre, Schrödinger, LLC, München, Germany).
Fungal strains and culture conditions
Colletotrichum graminicola M1.001 BH WT strain and its derivative GFP‐tagged strain M1.001 BH‐GFP (Sukno et al., 2008) were used. Cultures were maintained as described previously (Sukno et al., 2008) and grown in solid medium, i.e. potato dextrose agar (PDA) (Difco, Becton, Dickinson and Company, Sparks, MD, USA), minimal medium or oatmeal agar (Difco). To prepare vegetative mycelium, cultures were grown in potato dextrose broth (PDB) (Difco), LE medium (Krijger et al., 2008) or minimal medium supplemented with 1% sucrose, and incubated at 25 °C and 180 rpm. In vitro growth rate assays and in vitro appressorium formation rate assay were performed as described previously (Fang et al., 2002; Thon et al., 2002).
Pathogenicity assays
In vivo leaf blight assays were performed with susceptible maize inbred line Mo940 (Warren et al., 1975) as described previously (Vargas et al., 2012). Infected leaves were excised from the plant, scanned using a flat‐bed scanner and lesion areas were measured using the imaging processing software Paint.NET. Differences among average lesion sizes were tested using a completely randomized analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) test. Maize root infection assays were performed as described previously (Sukno et al., 2008). Fluorescence intensities were quantified using ImageJ software. Three independent experiments were performed and, in each experiment, four images were used to calculate the total corrected cellular fluorescence (TCCF). TCCF was calculated using the formula: TCCF = integrated density – (area of selection × mean fluorescence of background readings).
Gene deletion
The Cgfl gene deletion construct was prepared using the DelsGate System (García‐Pedrajas et al., 2008). The 5′ and 3′ regions flanking the GLRG_06543 coding sequence were amplified by PCR using the primer pairs 6543‐1–6543‐2 and 6543‐3–6543‐4. The pKW1 vector, which contains a gfp cassette and a hygromycin B phosphotransferase cassette (hph), was used as the pDONR vector to clone the Cgfl flanking regions using Gateway technology (Invitrogen, Carlsbad, CA, USA). The construct was confirmed by sequencing. The construct was linearized with SceI (Roche Diagnostics, Rotkreuz, Switzerland) and transformed into C. graminicola M1.001 as described previously (Sukno et al., 2008; Thon et al., 2000). Transformants containing the desired gene deletion were identified by PCR amplification of the internal region using two sets of PCR primers (Table S1, see Supporting Information).
Genomic DNA extraction and Southern blot analysis
Genomic DNA was extracted from mycelium using the protocol adapted from Baek and Kenerley (1998). For Southern blot analysis, 10 µg of genomic DNA were digested with NcoI (Roche Diagnostics, Basel, Switzerland), separated in 0.8% (w/v) agarose gel, denatured and transferred to nitrocellulose membrane (Amersham HyBond‐N, GE‐HealthCare, Little Chalfont, UK). A digoxigenin (DIG) dUTP‐labelled probe (the 3′ flanking region of the Cgfl coding sequence) was generated by PCR using a DIG DNA labelling kit (Roche Diagnostics) and primers 6543‐3 and 6543‐4, and C. graminicola M1.001 genomic DNA as the template. The membranes were hybridized and washed under high stringency as described by Sambrook and Russell (2001).
RNA extraction and RT‐qPCR assays
Total RNA was purified from liquid cultures as described previously (Vargas et al., 2012). To confirm the absence of genomic DNA, RT‐PCR and RT‐qPCR assays were performed without reverse transcriptase. qPCR was carried out using a KAPA SYBR Green qPCR Master Mix (KAPA Biosystems, Wilmington, MA, USA), and 1 µL of the reverse transcription reaction. Amplifications were performed in a StepOnePlus™ Real‐Time PCR system (Applied Biosystems, Foster City, CA, USA) using the following program: 40 cycles at 95 ºC for 30 s and 60 ºC for 30 s. The accuracy of SYBR‐Green detection was verified by checking the melting curves for the presence of only one specific peak. The PCR product size was checked by electrophoresis in 2.5% (w/v) agarose gel stained with ethidium bromide. Samples were collected from three different plant infection assays, and an independent set of cDNAs was prepared for each biological replicate. The gene‐specific primers used in qPCRs are listed in Table S1. All primer pairs showed efficiencies greater than 95%. The relative transcript abundance was normalized using the reference gene histone H3 (Krijger et al., 2008), whose expression was confirmed to be stable in all of the conditions tested. Relative expression levels were calculated using the comparative Ct method (Livak et al., 1995) All Ct calculations and statistical analyses were performed using StepOne™ software V2.2.2.
Secreted endoprotease activity assays
Liquid fungal cultures were grown at 25 ºC and 150 rpm for 4 days in different media (PDB medium, Fries medium, minimal medium supplemented with 1% sucrose or maize LE medium). To minimize the effect of endogenous proteases that are present in LEs, the medium was heated to 65 °C for 5 min (Sambrook and Russell, 2001). Supernatants were recovered by filtration through Miracloth and mycelia were frozen in liquid nitrogen and used for RT‐PCR assays. Resorufin‐labelled casein (Roche Diagnostics) was used for the detection of proteolytic activity following the protocol of Jousson et al. (2004). The experiments were performed twice and in duplicate each time. The results were tested with a one‐way ANOVA followed by Tukey's HSD test.
Total chitinase activity assay
Densely inoculated maize leaves were collected at 60 hpi and frozen in liquid nitrogen. Proteins were extracted according to Lange et al. (1996). 4‐Methylumbelliferyl‐β‐d‐N,N ′,N ″‐triacetylchitotrioside hydrate (Sigma‐Aldrich, St. Louis, MO, USA) was used as an artificial substrate (Ren et al., 2000). Maize protein extracts were incubated with the substrate and sodium acetate buffer (50 mm, pH 5.0) for 1 h at 37 ºC, and the reaction was stopped with 0.1 m NaOH. Fluorescence was measured in a Synergy 2 Multi‐Mode Microplate Reader (BioTek, Winooski, VT, USA) with an excitation wavelength of 355 nm and emission wavelength at 460 nm. One unit of activity was defined as a change in fluorescence of five absorbance units per minute. The protein concentration in extracts was measured according to the method of Bradford with bovine serum albumin (BSA) as a standard.
Construction of the fungalysin::gfp transcriptional fusion
To construct the PCgfl::gfp transcriptional fusion, the Cgfl gene promoter was predicted to be 1.4 kb upstream of the Cgfl start codon. Primers 6543FTr Fw and 6543FTr Rv (Table S1) were designed to amplify this region from genomic DNA. The fragment was cloned using pEntr/D‐TOPO vector (Invitrogen), confirmed by sequencing and recombined into pKW3 destination vector, which contains a gfp cassette (Vargas et al., 2015). The construct was confirmed by sequencing, and PCgfp::gfp transformants were confirmed by PCR with primers 6543FTr Fw and GFP Rv. A construct fusing the β‐tubulin promoter to the gfp coding sequence was prepared to visualize the WT infection process following the same strategy.
A strain constitutively expressing Cgfl was obtained by amplifying the Cgfl coding sequence with primers 6543OE Fw and 6543OE Rv, and ligating into plasmid pRF‐HUE, containing the Gpda promoter, by the USER friendly cloning reaction (Frandsen et al., 2008). The resulting vector was transformed into C. graminicola M1.001 protoplasts, and transformants with ectopic integrations of the plasmid were selected. The construct and transformants were confirmed by PCR using primers gpdA Seq F1 and 6543OE Rv.
Microscopy and image processing
Confocal images were taken on a TCS/SP2 Leica Laser Scanning spectral confocal microscope (Leica, Bensheim, Germany) using an argon laser at 488 nm, and GFP fluorescence was detected at 490–520 nm. A range of 595–680 nm was used to eliminate plant autofluorescence. For fluorescence and bright field microscopy, a Leica DMLB fluorescence microscope, Leica DG300F digital camera and Leica IM1000 software were used. Projections were generated from adjusted individual channels in the image stacks using Leica Confocal Software.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Phylogenetic analysis of Cgfl homologues in the Sordariomycetes. Two species of Dothidiomycetes were used as outgroup.
Fig. S2 (a) Schematic representation of targeted gene replacement using the DelsGate method. pKW1, which is a modified version of pDNOR‐A‐Hyg and contains a constitutively expressed gfp cassette, was used as the donor vector. Arrows indicate the primer pairs used to prepare the construct. (b) Conidia of the ΔCgfl36 transformant showing green fluorescent protein (GFP) fluorescence. (c) Polymerase chain reaction (PCR) of wild‐type (WT) and ΔCgfl transformants with 6543KO F and GFP Rv primers. Bands show the replacement of Cgfl with pKW1. (d) Southern blot analysis of WT strain and the ΔCgfl transformants. The genomic DNA was digested with NcoI and hybridized with the digoxigenin‐labelled 3′ Cgfl flank probe.
Fig. S3 In vitro radial growth rate (a) and appressorium formation assays (b) with wild‐type (WT) and Cgfl transformants. No difference was observed between the WT and Cgfl transformants. Error bars represent ± standard deviation (SD) of three independent replicates. PDA, potato dextrose agar.
Fig. S4 Phylogenetic analysis of Ecp6 homologues. The sequences were aligned with MAFFT and a central region containing two LysM motifs (although several proteins have only one motif) was used for construction of the phylogenetic tree using RAxML. Two clades that include proteins that have been experimentally shown to bind chitin and modulate plant immunity are indicated in green and blue. The proteins with experimental evidence to support their function are shown in orange. The two Colletotrichum graminicola homologues are shown in red.
Fig. S5 Transcriptional profiles of Ecp6 homologues GLRG_02947 and GLRG_07767 and Cgfl using reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Transcript levels were normalized to the constitutively expressed Colletotrichum graminicola histone H3 gene. The graph shows the values of two biological replicates with two technical replicates. Error bars denote standard deviation (SD).
Fig. S6 Anthracnose leaf blight assay of the Pgpda::Cgfl transformant. No differences in lesion size were seen between maize plants inoculated with the wild‐type and Pgpda::Cgfl transformant at 96 h post‐infection (hpi). Bars represent means + standard deviations based on three independent replicates.
Table S1. Primers used in this study.
Acknowledgements
This research was supported by grants AGL2011‐29446 and AGL2012‐34139 (Ministerio de Economía y Competitividad (MINECO), Spain) and grant SA‐165U13 (Junta de Castilla y León). J.M.S.‐M. was supported by a graduate fellowship (AP2009‐2656), W.A.V. was supported by a fellowship (JCI‐2009‐05364) and J.R.P.‐A. was supported by a fellowship (237402, Consejo Nacional de Ciencia y Tecnología (CONACyT), Mexico). We would like to thank the staff at the Plataforma Andaluza de Bioinformática of the University of Málaga, Spain, for providing computer resources and technical support.
References
- Baek, J.‐M. and Kenerley, C.M. (1998) The arg2 gene of Trichoderma virens: cloning and development of a homologous transformation system. Fungal Genet. Biol. 23, 34–44. [DOI] [PubMed] [Google Scholar]
- Baeza‐Montañez, L. , Gold, S.E. , Espeso, E.A. and García‐Pedrajas, M.D. (2015) Conserved and distinct functions of the “Stunted” (StuA)‐homolog Ust1 during cell differentiation in the corn smut fungus Ustilago maydis . Mol. Plant–Microbe Interact. 28, 86–102. [DOI] [PubMed] [Google Scholar]
- Bergstrom, G.C. and Nicholson, R.L. (1999) The biology of corn anthracnose: knowledge to exploit for improved management. Plant Dis. 83, 596–608. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , van Esse, H.P. , Vossen, J.H. , de Jonge, R. , Stergiopoulos, I. , Stulemeijer, I. J. E. , van den Berg, G. C. M. , Borrás‐Hidalgo, O. , Dekker, H. L. , de Koster, C. G. , de Wit, Pierre, J. G. M. , Joosten, M. H. A. J. , and Thomma, B. P. H. J. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69, 119–136. [DOI] [PubMed] [Google Scholar]
- Brouta, F. , Descamps, F. , Monod, M. , Vermout, S. , Losson, B. and Mignon, B. (2002) Secreted metalloprotease gene family of Microsporum canis . Infect. Immun. 70, 5676–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow, M. , Newman, M.‐A. and Roepenack, E. von (2000) The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261. [DOI] [PubMed] [Google Scholar]
- El Gueddari, N.E. , Rauchhaus, U. , Moerschbacher, B.M. and Deising, H.B. (2002) Developmentally regulated conversion of surface‐exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156, 103–112. [Google Scholar]
- Fang, G.‐C. , Hanau, R.M. and Vaillancourt, L.J. (2002) The SOD2 gene, encoding a manganese‐type superoxide dismutase, is up‐regulated during conidiogenesis in the plant‐pathogenic fungus Colletotrichum graminicola . Fungal Genet. Biol. 36, 155–165. [DOI] [PubMed] [Google Scholar]
- Frandsen, R.J. , Andersson, J.A. , Kristensen, M.B. and Giese, H. (2008) Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, T.J. , Weldekidan, T. , Colbert, T. , Wolters, P.J.C.C. and Hawk, J.A. (2011) Fitness evaluation of Rcg1, a locus that confers resistance to Colletotrichum graminicola (Ces.) G.W. Wils. using near‐isogenic maize hybrids. Crop Sci. 51, 1551. [Google Scholar]
- García‐Pedrajas, M.D. , Nadal, M. , Kapa, L.B. , Perlin, M.H. , Andrews, D.L. and Gold, S.E. (2008) DelsGate, a robust and rapid gene deletion construction method. Fungal Genet. Biol. 45, 379–388. [DOI] [PubMed] [Google Scholar]
- Guindon, S. , Dufayard, J.‐F. , Lefort, V. , Anisimova, M. , Hordijk, W. and Gascuel, O. (2010) New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Gust, A.A. , Willmann, R. , Desaki, Y. , Grabherr, H.M. and Nürnberger, T. (2012) Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci. 17, 495–502. [DOI] [PubMed] [Google Scholar]
- Jamil, F.F. and Nicholson, R.L. (1991) Response of sorghum lines of different ages to Colletotrichum graminicola isolates from shattercane, sorghum and corn. Pak. J. Phytopathol. 3, 12–18. [Google Scholar]
- Jia, Y. , McAdams, S.A. , Bryan, G.T. , Hershey, H.P. and Valent, B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. and Thomma, B.P.H.J. (2009) Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 17, 151–157. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , Bolton, M.D. and Thomma, B.P. (2011) How filamentous pathogens co‐opt plants: the ins and outs of fungal effectors. Curr. Opin. Plant Biol. 14, 400–406. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M. K. , Zhang, Z. , Usami, T. , Lievens, B. , Subbarao, K.V. and Thomma, B.P.H.J. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousson, O. , Léchenne, B. , Bontems, O. , Capoccia, S. , Mignon, B. , Barblan, J. , Quadroni, M. and Monod, M. (2004) Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum . Microbiology, 150, 301–310. [DOI] [PubMed] [Google Scholar]
- Karimi Jashni, M. , Dols, I.H.M. , Iida, Y. , Boeren, S. , Beenen, H.G. , Mehabi, R. , Collemare, J. and De Wit, P.J.G.M. (2015) Synergistic action of serine‐ and metallo‐proteases from Fusarium oxysporum f. sp. lycopersici cleaves chitin‐binding tomato chitinases, reduces their antifungal activity and enhances fungal virulence. Mol. Plant–Microbe Interact. 28, 996–1008. [DOI] [PubMed] [Google Scholar]
- Kasprzewska, A. (2003) Plant chitinases–regulation and function. Cell. Mol. Biol. Lett. 8, 809–824. [PubMed] [Google Scholar]
- Katoh, K. , Misawa, K. , Kuma, K. and Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane, T.M. , Creevey, C.J. , Pentony, M.M. , Naughton, T.J. and Mclnerney, J.O. (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 6, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L.A. and Sternberg, M.J.E. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Kombrink, A. (2014) Functional analysis of LysM effectors secreted by fungal plant pathogens. PHD thesis, Wageningen University.
- Kombrink, A. , Sánchez‐Vallet, A. and Thomma, B.P.H.J. (2011) The role of chitin detection in plant–pathogen interactions. Microbes Infect. 13, 1168–1176. [DOI] [PubMed] [Google Scholar]
- Krijger, J.‐J. , Horbach, R. , Behr, M. , Schweizer, P. , Deising, H.B. and Wirsel, S.G.R. (2008) The yeast signal sequence trap identifies secreted proteins of the hemibiotrophic corn pathogen Colletotrichum graminicola . Mol. Plant–Microbe Interact. 21, 1325–1336. [DOI] [PubMed] [Google Scholar]
- Lange, J. , Mohr, U. , Wiemken, A. , Boller, T. and Vogeli‐Lange, R. (1996) Proteolytic processing of class IV chitinase in the compatible interaction of bean roots with Fusarium solani . Plant Physiol. 111, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. and Zhang, K.‐Q. (2014) Independent expansion of zinc in metalloproteinases in Onygenales fungi may be associated with their pathogenicity. PLoS One, 9, e90225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. , Flood, S.J. , Marmaro, J. , Giusti, W. and Deetz, K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4, 357–362. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Markaryan, A. , Morozova, I. , Yu, H. and Kolattukudy, P.E. (1994) Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect. Immun. 62, 2149–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, R. , Kombrink, A. , Motteram, J. , Loza‐Reyes, E. , Lucas, J. , Hammond‐Kosack, K.E. , Thomma, B.P.H.J. and Rudd, J.J. (2011) Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 156, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy, A. , Baldo, A. , Schoofs, L. , Cambier, L. , Defaweux, V. , Tabart, J. , Maréchal, F. , Symoens, F. and Mignon, B. (2010) Fungalysin and dipeptidyl‐peptidase gene transcription in Microsporum canis strains isolated from symptomatic and asymptomatic cats. Vet. Microbiol. 146, 179–182. [DOI] [PubMed] [Google Scholar]
- Mentlak, T.A. , Kombrink, A. , Shinya, T. , Ryder, L.S. , Otomo, I. , Saitoh, H. , Terauchi, R. , Nishizawa, Y. , Shibuya, N. , Thomma, B.P.H.J. and Talbot, N.J. (2012) Effector‐mediated suppression of chitin‐triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell, 24, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod, M. , Paris, S. , Sanglard, D. , Jaton‐Ogay, K. , Bille, J. and Latge, J.P. (1993) Isolation and characterization of a secreted metalloprotease of Aspergillus fumigatus . Infect. Immun. 61, 4099–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold, G.P. (2002) Managing anthracnose stalk rot: a moving target. In Proc. 56th Annu. Corn & Sorghum Res. Conf., pp. 196–206. Chicago, IL.
- Naumann, T.A. and Price, N.P.J. (2012) Truncation of class IV chitinases from Arabidopsis by secreted fungal proteases. Mol. Plant Pathol. 13, 1135–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann, T.A. and Wicklow, D.T. (2010) Allozyme‐specific modification of a maize seed chitinase by a protein secreted by the fungal pathogen Stenocarpella maydis . Phytopathology, 100, 645–654. [DOI] [PubMed] [Google Scholar]
- Naumann, T.A. and Wicklow, D.T. (2013) Chitinase modifying proteins from phylogenetically distinct lineages of Brassica pathogens. Physiol. Mol. Plant Pathol. 82, 1–9. [Google Scholar]
- Naumann, T.A. , Wicklow, D.T. and Kendra, D.F . (2009) Maize seed chitinase is modified by a protein secreted by Bipolaris zeicola. Physiol. Mol. Plant Pathol. 74, 134–141. [Google Scholar]
- Naumann, T.A. , Wicklow, D.T. and Price, N.P.J. (2011) Identification of a chitinase‐modifying protein from Fusarium verticillioides truncation of a host resistance protein by a fungalysin metalloprotease. J. Biol. Chem. 286, 35 358–35 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, R.J. , Thon, M.R. , Hacquard, S. , Amyotte, S.G., Kleemann, J., Torres, M.F., Damm, U., Buiate, E.A., Epstein, L., Alkan, N., Altmüller, J., Alvarado‐Balderrama, L., Bauser, C.A., Becker, C., Birren, B.W., Chen, Z., Choi, J., Crouch, J.A., Farman, M.A., Gan, P., Heiman, D., Henrissat, B., Howard, R.J., Kabbage, M., Koch, C., Kracher, B., Kubo, Y., Law, A.D., Lebrun, M.H., Lee, Y.H., Miyara, I., Moore, N., Neumann, U., Nordström, K., Panaccione, D.G., Panstruga, R., Place, M., Proctor, R.H., Prusky, D., Rech, G., Reinhardt, R., Rollins, J.A., Rounsley, S., Schardl, C.L., Schwartz, D.C., Shenoy, N., Shirasu, K., Sikhakolli, U.R., Stüber, K., Sukno, S.A., Sweigard, J.A., Takano, Y., Takahara, H., Trail, F., van der Does, H.C., Voll, L.M., Will, I., Young, S., Zeng, Q., Zhang, J., Zhou, S., Dickman, M.B., Schulze‐Lefert, P., Ver Loren van Themaat, E., Ma, L.J. and Vaillancourt, M.J. (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44, 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva, R. , Win, J. , Raffaele, S. , et al (2010) Recent developments in effector biology of filamentous plant pathogens. Cell. Microbiol. 12, 705–715. [DOI] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , Heijne, G. von and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Rafiqi, M. , Ellis, J.G. , Ludowici, V.A. , Hardham, A.R. and Dodds, P.N. (2012) Challenges and progress towards understanding the role of effectors in plant–fungal interactions. Curr. Opin. Plant Biol. 15, 477–482. [DOI] [PubMed] [Google Scholar]
- Rech, G.E. , Sanz‐Martín, J.M. , Anisimova, M. , Sukno, S.A. and Thon, M.R. (2014) Natural selection on coding and noncoding DNA sequences is associated with virulence genes in a plant pathogenic fungus. Genome Biol. Evol. 6, 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y. , Wee, K.E. and Chang, F.N. (2000) Deficiency of current methods in assaying endochitinase activity. Biochem. Biophys. Res. Commun. 268, 302–305. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual. CSHL Press. Cold Spring Harbor, NY. [Google Scholar]
- Slavokhotova, A.A. , Naumann, T.A. , Price, N.P.J. , Rogozhin, E.A. , Andreev, Y.A. , Vassilevski, A.A. and Odintsova, T.I. (2014) Novel mode of action of plant defense peptides – hevein‐like antimicrobial peptides from wheat inhibit fungal metalloproteases. FEBS J. 281, 4754–4764. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Burg, H.A. , van den, Ökmen, B. , Beenen, H.G. , Liere, S. , van, Kema, G.H.J. , and Wit, P.J.G.M. de (2010) Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA 107, 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz, H.U. , Mitrousia, G.K. , de Wit, P.J.G.M. and Fitt, B.D.L. (2014) Effector‐triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 19, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strable, J. and Scanlon, M.J. (2009) Maize (Zea mays): a model organism for basic and applied research in plant biology. Cold Spring Harb. Protoc. 2009, pdb.emo132. [DOI] [PubMed] [Google Scholar]
- Sukno, S.A. , García, V.M. , Shaw, B.D. and Thon, M.R. (2008) Root infection and systemic colonization of maize by Colletotrichum graminicola . Appl. Environ. Microbiol. 74, 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara, H. , Dolf, A. , Endl, E. and O'Connell, R. (2009) Flow cytometric purification of Colletotrichum higginsianum biotrophic hyphae from Arabidopsis leaves for stage‐specific transcriptome analysis. Plant J. 59, 672–683. [DOI] [PubMed] [Google Scholar]
- Thon, M.R. , Nuckles, E.M. and Vaillancourt, L.J. (2000) Restriction enzyme‐mediated integration used to produce pathogenicity mutants of Colletotrichum graminicola . Mol. Plant–Microbe Interact. 13, 1356–1365. [DOI] [PubMed] [Google Scholar]
- Thon, M.R. , Nuckles, E.M. , Takach, J.E. and Vaillancourt, L.J. (2002) CPR1: a gene encoding a putative signal peptidase that functions in pathogenicity of Colletotrichum graminicola to maize. Mol. Plant–Microbe Interact. 15, 120–128. [DOI] [PubMed] [Google Scholar]
- Vander, P. , Vårum, K.M. , Domard, A. , Gueddari, N.E.E. and Moerschbacher, B.M. (1998) Comparison of the ability of partially N‐acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol. 118, 1353–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, W.A. , Martín, J.M.S. , Rech, G.E. , Rivera, L.P. , Benito, E.P. , Díaz‐Mínguez, J.M. , Thon, M.R. and Sukno, S.A. (2012) Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotrichum graminicola in maize. Plant Physiol. 158, 1342–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, W.A. , Sanz‐Martin, J.M. , Rech, G.E. , Armijos‐Jaramillo, V.D. , Rivera, L.P. , Echeverria, M.M. , Díaz‐Mínguez, J.M. , Thon, M. and Sukno, S. (2015) A fungal effector with host nuclear localization and DNA‐binding properties is required for maize anthracnose development. Mol. Plant–Microbe Interact. (in press, DOI: 10.1094/MPMI-09-15-0209-R). [DOI] [PubMed] [Google Scholar]
- Warren, H.L. , Nicholson, R.L. and Turner, M.T. (1975) Field reaction of corn inbreds to Colletotrichum graminicola . Plant Dis. 59, 767–769. [Google Scholar]
- Waterhouse, R.M. , Tegenfeldt, F. , Li, J. , Zdobnov, E.M. and Kriventseva, E.V. (2013) OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 41, D358–D365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. and Guclu, H. (2013) Global maize trade and food security: implications from a social network model. Risk Anal. 33, 2168–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F. , Li, W. and Jørgensen, H.J.L. (2013) Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PLoS One, 8, e81606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Lee, K.‐M. , Son, M. and Kim, K.‐H. (2015) Effects of the deletion and over‐expression of Fusarium graminearum gene FgHal2 on host response to mycovirus Fusarium graminearum virus 1. Mol. Plant Pathol. 16, 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Bak, D.D. , Heid, H. and Geider, K. (1999) Molecular characterization of a protease secreted by Erwinia amylovora . J. Mol. Biol. 289, 1239–1251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Phylogenetic analysis of Cgfl homologues in the Sordariomycetes. Two species of Dothidiomycetes were used as outgroup.
Fig. S2 (a) Schematic representation of targeted gene replacement using the DelsGate method. pKW1, which is a modified version of pDNOR‐A‐Hyg and contains a constitutively expressed gfp cassette, was used as the donor vector. Arrows indicate the primer pairs used to prepare the construct. (b) Conidia of the ΔCgfl36 transformant showing green fluorescent protein (GFP) fluorescence. (c) Polymerase chain reaction (PCR) of wild‐type (WT) and ΔCgfl transformants with 6543KO F and GFP Rv primers. Bands show the replacement of Cgfl with pKW1. (d) Southern blot analysis of WT strain and the ΔCgfl transformants. The genomic DNA was digested with NcoI and hybridized with the digoxigenin‐labelled 3′ Cgfl flank probe.
Fig. S3 In vitro radial growth rate (a) and appressorium formation assays (b) with wild‐type (WT) and Cgfl transformants. No difference was observed between the WT and Cgfl transformants. Error bars represent ± standard deviation (SD) of three independent replicates. PDA, potato dextrose agar.
Fig. S4 Phylogenetic analysis of Ecp6 homologues. The sequences were aligned with MAFFT and a central region containing two LysM motifs (although several proteins have only one motif) was used for construction of the phylogenetic tree using RAxML. Two clades that include proteins that have been experimentally shown to bind chitin and modulate plant immunity are indicated in green and blue. The proteins with experimental evidence to support their function are shown in orange. The two Colletotrichum graminicola homologues are shown in red.
Fig. S5 Transcriptional profiles of Ecp6 homologues GLRG_02947 and GLRG_07767 and Cgfl using reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Transcript levels were normalized to the constitutively expressed Colletotrichum graminicola histone H3 gene. The graph shows the values of two biological replicates with two technical replicates. Error bars denote standard deviation (SD).
Fig. S6 Anthracnose leaf blight assay of the Pgpda::Cgfl transformant. No differences in lesion size were seen between maize plants inoculated with the wild‐type and Pgpda::Cgfl transformant at 96 h post‐infection (hpi). Bars represent means + standard deviations based on three independent replicates.
Table S1. Primers used in this study.


