Summary
Brown spot disease, caused by C ochliobolus miyabeanus, is currently considered to be one of the most important yield reducers of rice (O ryza sativa L.). Despite its agricultural importance, little is known about the virulence mechanisms deployed by the fungus. Therefore, we set out to identify novel virulence factors with a role in disease development. This article reports, for the first time, the production of tentoxin by C . miyabeanus as a virulence factor during brown spot disease and the identification of the non‐ribosomal protein synthetase (NRPS) CmNps3, responsible for tentoxin biosynthesis. We compared the chemical compounds produced by C . miyabeanus strains differing in virulence ability using ultra‐high‐performance liquid chromatography (UHPLC) coupled to high‐resolution Orbitrap mass spectrometry (HRMS). The production of tentoxin by a highly virulent strain was revealed by principal component analysis of the detected ions and confirmed by UHPLC coupled to tandem‐quadrupole mass spectrometry (MS/MS). The corresponding NRPS was identified by in silico genome analysis and confirmed by gene deletion. Infection tests with wild‐type and C mnps3 mutants showed that tentoxin acts as a virulence factor and is correlated with chlorosis development during the second phase of infection. Although rice has previously been classified as a tentoxin‐insensitive plant species, our data demonstrate that tentoxin production by C . miyabeanus affects symptom development.
Keywords: Bipolaris oryzae, brown spot disease, Cochliobolus miyabeanus, Oryza sativa L., plant–pathogen interactions, rice, tentoxin
Introduction
Brown spot disease, caused by Cochliobolus miyabeanus, is currently considered to be one of the most important yield reducers of rice (Oryza sativa L.) (Savary et al., 2012). Despite its agricultural importance, little is known about the virulence mechanisms deployed by the fungus. Cochliobolus miyabeanus is a filamentous ascomycete belonging to the Pleosporales order of the class Dothideomycetes. Many species belonging to this order are notorious for the production of phytotoxins (Condon et al., 2013; Panaccione, 1993; Stergiopoulos et al., 2013). Phytotoxins can be either host specific (HST) or non‐host specific (non‐HST), and are mainly small secondary metabolites, such as terpenes, non‐ribosomal peptides, polyketides, alkaloids or metabolites of mixed origin, that ultimately cause plant cell death. HSTs often act as pathogenicity determinants and determine the host range of a pathogen (Berestetskiy, 2008; Stergiopoulos et al., 2013). Although the existence of a C. miyabeanus HST has been proposed, such a compound has never been characterized (Vidhyasekaran et al., 1986). Instead, C. miyabeanus seems to rely mostly on an array of non‐host‐specific virulence factors, which is reflected in its host range (Chakrabarti, 2001).
Cochliobolus miyabeanus produces multiple ophiobolins, a family of sesterterpenoid non‐host‐specific phytotoxins (Nakamura and Ishibashi, 1958; Orsenigo, 1957; Xiao et al., 1991; Yun et al., 1988). The highly bioactive analogue ophiobolin A contributes significantly and quantitatively to the virulence level of C. miyabeanus (L. De Bruyne, unpublished results). Although, to date, no other virulence factors have been characterized for C. miyabeanus, their existence is suggested by the remaining virulence ability in an ophiobolin‐negative background. This idea is also supported by the identification of 11 non‐ribosomal protein synthetase (NRPS) and 21 polyketide synthetase (PKS) genes in the C. miyabeanus genome, although none are unique for the species (Condon et al., 2013). Small peptides and polyketides produced via multimodular megasynthases (NRPSs and PKSs, respectively) comprise a group of secondary metabolites with diverse bioactivity, including phytotoxicity (Stergiopoulos et al., 2013). They are good virulence factor candidates.
In an attempt to identify unknown virulence factors produced by C. miyabeanus during brown spot disease, we compared the chemical compounds produced by a highly virulent strain with those of less virulent strains. We used ultra‐high‐performance liquid chromatography coupled to high‐resolution Orbitrap mass spectrometry (UHPLC‐Orbitrap‐HRMS). A compound characteristic to the most virulent strain was selected and analysed for involvement in disease development. In addition, a genetic analysis was performed to identify the genes involved in its biosynthesis.
Results
UHPLC‐Orbitrap‐HRMS analysis of liquid culture filtrates
We compared the metabolites produced by four C. miyabeanus strains with variable virulence abilities in a chemical screening. The highly virulent strain Cm988 and the less virulent strains WK1C and G513 were grown in liquid Fries medium, known to stimulate toxin production in vitro. Non‐inoculated medium was used as a control to distinguish between medium components and fungal secretions. Metabolites were extracted from the crude culture filtrate using acetonitrile–ethylacetate in an attempt to collect as many compounds as possible. The extracted metabolites were screened using liquid chromatography coupled to an Orbitrap mass spectrometer (UHPLC‐Orbitrap‐HRMS) in the positive (ESI+) and negative (ESI−) electrospray ionization mode. Different metabolites were separated into frames based on retention times, and accurate mass data were generated by Orbitrap HRMS using Sieve™ software. The 5000 frames with the highest detection level in ESI+ mode were selected for analysis. A principal component analysis was performed on the integrated intensities of the selected compounds using SPSS Statistics 21 software (IBM Corp; Armonk, NY, US) to identify compounds that correlate with the virulence level of the investigated strains.
The first three calculated principal components explained 92% of the total variance in the dataset (Table S1, see Supporting Information). PC1 and PC2 clearly divided control and fungal samples, enabling a separation between medium compounds and metabolites. PC3 distinguished between strains differing in virulence level and provided an insight into possible virulence factors (Fig. S1, see Supporting Information). A combination of PC2 and PC3 gave the best separation of the different strains. One ion correlated strongly with PC3 and was characteristic of the most virulent strain (Fig. 1).
Figure 1.
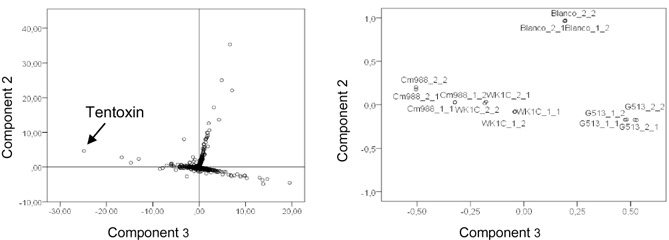
Score and loading plots representing how 5000 ions detected in a liquid culture filtrate correlate with different C ochliobolus miyabeanus strains after separation according to principal components 2 and 3. Tentoxin is characteristic of the highly virulent strain Cm988.
This compound was selected as a candidate virulence factor and analysed further. The compound eluted at 14.12 min and the Orbitrap measured an m/z value of 415.2329 for the precursor ion generated in ESI+ mode, corresponding to a chemical compound with a molecular weight (MW) of 414.50 and a monoisotopic mass of 414.2267. Based on the Orbitrap accurate mass data, an elemental composition of C22H30N4O4 was proposed with a mass error Δppm of 2.68. A second minor peak with the same mass characteristics and a Δppm of 1.65 eluted 1.27 min earlier (at 12.85 min). A KEGG database search via the ChemSpider website (www.chemspider.com) based on the elemental composition returned tentoxin (C08441) as the only known metabolite in the KEGG metabolic database with the same chemical composition (Fig. 2a). The compound was also detected by UHPLC‐Orbitrap‐HRMS analysis in ESI− mode (data not shown).
Figure 2.
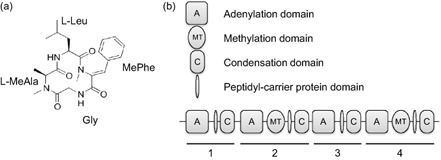
(a) Chemical structure of tentoxin. Gly, glycine; Leu, leucine; MeAla, methylalanine; MePhe, methylphenylalanine. (b) Module and domain architecture of the non‐ribosomal protein synthetase CmNps3, responsible for tentoxin biosynthesis in C ochliobolus miyabeanus. Modules (1–4) and the corresponding domains were predicted using antiSMASH.
Confirmation of tentoxin based on UPLC‐MS/MS analysis
To confirm the identity of the selected compound, we compared the retention time and fragmentation pattern with those of a commercial tentoxin reference standard using a UPLC‐MS/MS system. The retention time and multiple reaction monitoring (MRM) spectra were indistinguishable. Both compounds had a retention time of 12.79 min and precursor ion of m/z 413.3 in negative mode. The difference of 1.33 min in retention time compared with the peak in the UPLC‐Orbitrap system (14.12 min) was caused by a different configuration of the UPLC system.
Tentoxin was fragmented into the deprotonated dipeptides [M − H]− Leu‐Me(Δ)Phe and [M − H]− Gly‐MeAla (Gly, glycine; Leu, leucine; MeAla, methylalanine; MePhe, methylphenylalanine) with m/z 271.2 and m/z 141.0, respectively (Liu and Rychlik, 2013). Hence, we used the MRM mode with ion transitions 413.3 → 271.2 and 413.3 → 141.0 in ESI− mode to analyse our sample. The m/z 413.3 [M − H]− ion in the sample produced the same fragment ions as tentoxin, and their integrated peak intensity ratio ([m/z 271.2]/[m/z 141]) was 0.5 for both the sample and the standard (Fig. 3). Similar to our observations in the UHPLC‐Orbitrap‐HRMS system, a second smaller peak with the same chemical characteristics was detected at 11.57 min, 1.22 min before the main peak. We believe that this peak might represent the tentoxin isomer, isotentoxin (Horiuchi et al., 2003; Liebermann et al., 1996).
Figure 3.
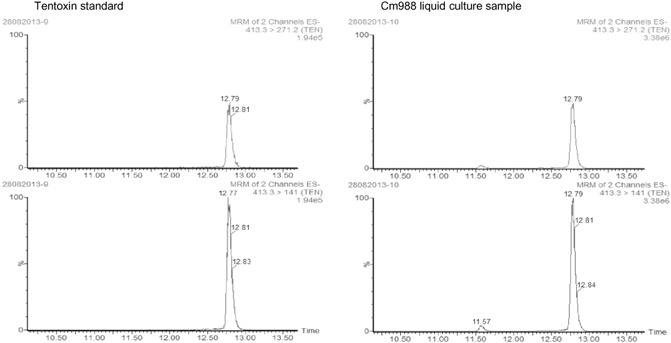
Verification of the identity of the compound produced by C ochliobolus miyabeanus, based on two multiple reaction monitoring (MRM) ion transitions characteristic of tentoxin: m/z 413.3 → 271.2 and m/z 413.3 → 141.0. The left side of the figure shows the peaks generated from a pure tentoxin standard; the right side of the figure shows the compound detected in a liquid culture extract of C . miyabeanus strain Cm988 grown in Fries medium for 7 days.
Tentoxin synthase gene identification and deletion
Tentoxin is a cyclic tetrapeptide with the structure cyclo(Gly‐l‐MeAla‐l‐Leu‐MePhe[(Z)Δ]), produced by an NRPS (Fig. 2a). The corresponding NRPS has not been identified previously, but, based on the peptide structure, is probably constructed of four modules, featuring two methylation domains. The genome of C. miyabeanus features 11 NRPS encoding genes (Condon et al., 2013). The web‐based application antiSMASH was used to identify the different modules in the predicted NRPS genes. We used NRPSPredictor2 for fungal sequences to predict the likely physico‐chemical properties of the monomers of the resulting NRP based on a support vector machine (SVM) method, and compared them with the Stachelhaus code (Table 1). antiSMASH returned three predicted C. miyabeanus NRPSs harbouring four modules, but only one featured methylation domains in two of its modules. The C. miyabeanus NPS3 homologue n8391 from C. miyabeanus strain WK1C (designated CmNps3), with Joint Genome Institute (JGI) protein ID 98843, met the structural requirements (Fig. 2b). CmNps3 is 5158 amino acids long, corresponding to a 574.7‐kDa protein. The encoding gene is 15.477 kb long and does not contain introns in its coding sequence. Of all the NRPS genes detected in the C. miyabeanus genome, CmNPS3 is the most likely candidate involved in tentoxin production, although the monomer predictions were not completely consistent with the known tentoxin structure (Table 1).
Table 1.
Monomer specificity predictions of CmNps3 adenylation domains compared with actual tentoxin monomers
| A‐domain | Modification |
NRPSPredictor2*
Prediction—Score |
Stachelhaus code Prediction—Score | Tentoxin |
|---|---|---|---|---|
| 1 | — | Hydrophobic‐aliphatic—0.6995 | Gly—90% | Gly |
| 2 | Me | Hydrophobic‐aliphatic—1.1171 | Val—70% | MeAla |
| 3 | — | Hydrophobic‐aromatic—0.0578 | Phe—70% | Leu |
| 4 | Me | Hydrophobic‐aliphatic—1.07 | Phe—60% | MePhe |
A‐domain, adenylation domain; Gly, glycine; Leu, leucine; MeAla, methylalanine; MePhe, methylphenylalanine; Phe, phenylalanine; Val, valine.
*Rausch et al. (2005).
The SVM‐based method predicted that the first two monomers are hydrophobic‐aliphatic amino acids with SVM scores of 0.70 and 1.12, respectively. The second module features a methylation domain. Therefore, these monomers could correspond to Gly and MeAla in tentoxin. The prediction of the last two monomers was less consistent with the tentoxin sequence. Although tentoxin features Leu on position 3, NRPSPredictor2 predicts a hydrophobic‐aromatic monomer for this module, but the low SVM score (score = 0.06) could indicate an inconsistent prediction. Although the last module harbours a methylation domain corresponding to MePhe in tentoxin, NRPSPredictor2 predicts a hydrophobic‐aliphatic monomer, with a high score of 1.07. Based on the Stachelhaus code, the monomer sequence would be Gly‐MeVal‐Phe‐MePhe (MeVal, methylvaline). Gly was confirmed as the first monomer with 90% identity to a known Stachelhaus code. As Stachelhaus predictions at 70% or lower are less reliable, we cannot trust the last three monomer predictions (Rausch et al., 2005). Because this NRPS was still the best candidate for a tentoxin synthase in C. miyabeanus, it was selected for further analysis.
To determine the phenotype of CmNPS3, the gene was deleted from two C. miyabeanus field strains, WK1C and Cm988, using homologous recombination via protoplast transformation. The candidate transformants were purified by single spore selection, and exchange of the CmNPS3 gene for the hygromycin B resistance gene (HygB) was confirmed by polymerase chain reaction (PCR) verification and fungal growth on HygB+ medium (Fig. S2, see Supporting Information). One confirmed knockout mutant in each background was selected for subsequent phenotyping: Cm988ΔCmnps3 and WK1CΔCmnps3.
The production of tentoxin by the wild‐type (WT) and mutant strains was measured using UPLC‐MS/MS (Fig. 4). Cm988 WT strain produced tentoxin in vitro and in planta, whereas WK1C only produced tentoxin in planta. After 7 days of growth in Fries medium, we extracted an average of 2121 ng tentoxin/g dry weightfungus (DWfungus) from the WT Cm988 cultures, whereas tentoxin production by the mutant strain was completely abolished. Similarly, Cmnps3 mutants of both WT strains could no longer produce tentoxin in planta at 48 h post‐inoculation (hpi), whereas WT Cm988 produced an average of 38.8 ng tentoxin/g fresh weightleaf (FWleaf) after spore inoculation and WK1C produced 2.28 ng tentoxin/g FWleaf after mycelium inoculation. These results confirm the involvement of CmNps3 in tentoxin biosynthesis.
Figure 4.
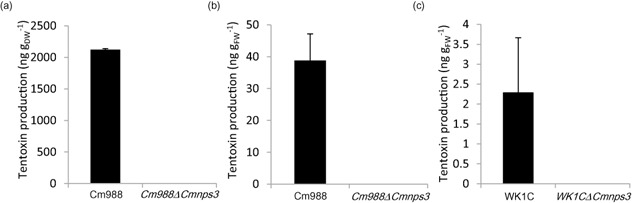
Quantification of tentoxin production by C ochliobolus miyabeanus strains Cm988 and WK1C using ultra‐high‐performance liquid chromatography‐tandem mass spectrometry (UPLC‐MS/MS). (a) Tentoxin production in liquid culture. The wild‐type (WT) and C m988Δ C mnps3 mutant were grown in liquid Fries medium for 7 days; tentoxin was extracted from the culture medium and divided by the total fungal dry weight. (b) Tentoxin production in planta. Rice (O ryza sativa) leaf pieces were spray inoculated with a spore solution (1 × 104 spores/mL) containing Cm988 WT or C m988Δ C mnps3 spores. Tentoxin was extracted from the infected leaf tissue at 48 h post‐inoculation (hpi) and divided by the total leaf fresh weight. (c) Tentoxin production in planta. Rice leaf pieces were inoculated with a mycelial suspension [5 × 104 colony‐forming units (CFU)/mL] containing WK1C WT or WK 1 C Δ C mnps3 mycelium. Tentoxin was extracted from the infected leaf tissue at 48 hpi and divided by the total leaf fresh weight. Results are means ± standard error (SE) of at least three biological repeats.
Fungal growth rate and conidiation
To determine whether CmNps3 plays a role in normal fungal development, we investigated the effect of CmNPS3 deletion on fungal growth and conidiation (Fig. 5).
Figure 5.
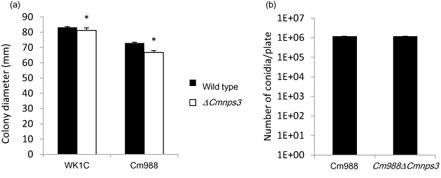
Effect of CmNPS 3 deletion on fungal growth and conidiation of C ochliobolus miyabeanus. (a) Fungal growth rate on potato dextrose agar (PDA) at 28 °C in the dark of two different field strains: Cm988 and WK1C. Black bars, wild‐type; white bars, C mnps3 mutant. (b) Conidiation on PDA plates measured after 14 days of growth at 28 °C in the dark. Data presented are means ± standard error (SE). *Significantly different from respective wild‐type strain (Kruskal–Wallis rank test, α = 0.05).
Although the colony diameter of tentoxin knockout strains was significantly smaller at 7 days when compared with the respective WT strains (P < 0.05), the differences were small. Furthermore, Cm988ΔCmnps3 produced the same amount of spores as the parental strain during 14 days of growth on potato dextrose agar (PDA). The WK1C strain sporulates very poorly under laboratory conditions.
Tentoxin production is related to the severity of disease symptoms
In order to determine whether tentoxin could play a role during infection, we compared symptom development after infection of rice leaves by WT and tentoxin‐deficient strains. We compared the number of lesions and the total affected leaf surface (Fig. 6). Plants were inoculated with a spore suspension of Cm988 or the corresponding mutant, and with a mycelium suspension of WK1C or the respective mutant. Because of this difference in inoculation method, the results cannot be compared between strains. The average number of lesions present at 48 hpi was not significantly different between Cm988 and Cm988ΔCmnps3 (53 and 50, respectively; P > 0.05). However, the amount of leaf surface affected differed significantly. Although Cm988 caused, on average, damage to 29% of the leaf surface at 72 hpi, only 10% of the leaf surface was affected by Cm988ΔCmnps3 infection. This difference is mainly attributable to the development of chlorosis during WT infection, which was mostly absent during infection with tentoxin‐deficient mutants. No significant differences were observed in the number of lesions (80 and 81, respectively; P > 0.05) or affected leaf area (24% and 21%, respectively; P > 0.05) during infection by the WK1C WT and WK1CΔCmnps3 mutant, respectively.
Figure 6.
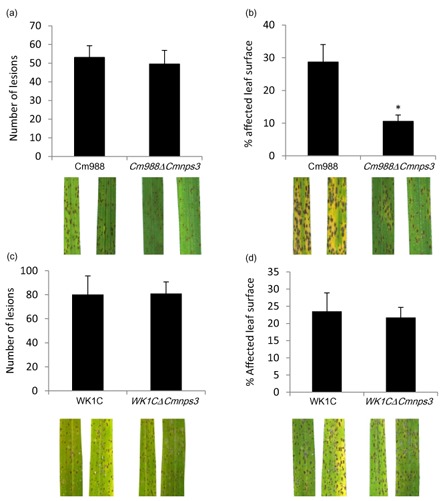
C ochliobolus miyabeanus infection assays on rice (O ryza sativa) leaf pieces. (a, b) Leaf pieces of 5‐week‐old rice plants were spray inoculated with a spore solution (1 × 104 conidia/mL) of C . miyabeanus Cm988 wild‐type (WT) or C m988Δ C mnps3. Lesions were counted per leaf piece at 48 h post‐inoculation (hpi) (a) and the percentage affected leaf area was measured at 72 hpi (b). (c, d) Leaf pieces of 5‐week‐old plants were spray inoculated with a mycelial suspension [5 × 104 colony‐forming units (CFU)/mL] of WK1C WT or WK1C Δ C mnps3 mutant. Lesions were counted per leaf piece at 48 hpi (c) and the percentage affected leaf area was measured at 72 hpi (d). Photographs show representative symptoms at the respective time points. Results are means ± standard error (SE). *Significant differences (Kruskal–Wallis rank test, α = 0.05).
To test the effect of pure tentoxin on mature rice leaves, we punctured leaf pieces and applied two concentrations of pure tentoxin (Fig. 7). The control leaves and leaves treated with 50 μm tentoxin did not show any symptoms. However, treatment with 500 μm tentoxin caused a necrotic ring around the puncture wound in 33% of treatments. The other 66% of leaf pieces did not show any symptoms.
Figure 7.
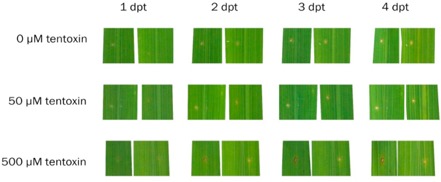
Effect of pure tentoxin on mature rice (O ryza sativa) leaves. Leaf pieces of 5‐week‐old plants were punctured and treated with 10 μL of pure tentoxin at the indicated concentrations. Symptoms were monitored during 4 days. Photographs represent the same replicate at four different time points after treatment. dpt, days post‐treatment.
Furthermore, we quantified the amount of tentoxin produced during infection by field strains with different virulence abilities (Fig. 8a). All strains started to produce tentoxin at 24 hpi, and production increased whilst the disease progressed. Interestingly, the highly virulent Cm988 strain produced very large amounts of tentoxin. Tentoxin production by the less virulent strains G513 and WK1C, and the very weak strain S4, was two‐fold lower, and amounts were not significantly different from each other.
Figure 8.
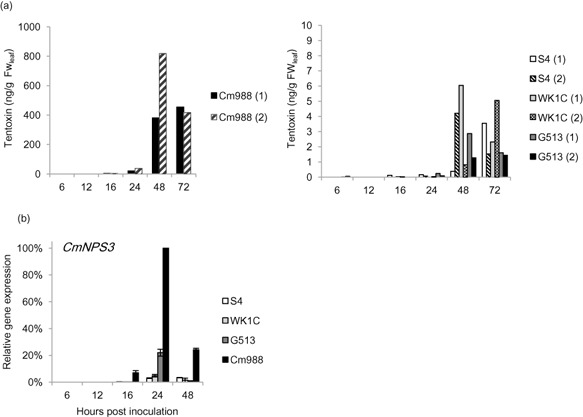
Tentoxin production during infection of rice (O ryza sativa) leaves is correlated with disease severity caused by different C ochliobolus miyabeanus wild‐type strains. Cm988 is highly virulent, G513 is intermediate in virulence and WK1C and S4 are weakly virulent. (a) Tentoxin was extracted from infected leaf pieces at the indicated time points after mycelium inoculation and quantified using ultra‐high‐performance liquid chromatography‐tandem mass spectrometry (UPLC‐MS/MS). The results of two independent biological repeats are presented. (b) The amount of tentoxin produced by different C ochliobolus miyabeanus wild‐type strains is correlated with CmNPS 3 gene expression during infection of rice (O ryza sativa) leaves. Gene expression levels were normalized using C m A ctin as an internal reference and expressed relative to maximal gene expression (Cm988 at 24 h post‐inoculation). Results represent means [±standard error (SE)] of two independent biological and two technical repeats.
To determine whether the difference in tentoxin production between the strains could be caused by a difference in gene expression, we measured the relative expression level of CmNPS3 at different time points during infection (Fig. 8b). Fold induction followed a similar pattern to tentoxin production, with a much higher relative expression in the highly virulent Cm988 WT strain compared with the less virulent strains. Fold induction reached a maximum at 24 hpi in all strains, preceding maximal tentoxin production at 48 hpi.
These results suggest that tentoxin production is not necessary for pathogenicity and initiation of the infection, but contributes to disease severity during later phases.
Tentoxin synthase gene conservation
CmNPS3 is an orthologue of C. heterostrophus NPS3 (ChNPS3) (Condon et al., 2013; Turgeon et al., 2008). Comparison of the two genomic sequences (C. miyabeanus strain WK1C and C. heterostrophus strain C4) revealed the presence of many single nucleotide polymorphisms (SNPs). Furthermore, an intron spanning 51 bp in the adenylation (A)‐domain of the third module of ChNPS3 caused a loss of 17 amino acids in ChNps3 relative to CmNps3. These changes could indicate a change or loss of function of ChNps3, although the overall protein similarity is still 95%. Therefore, we screened C. heterostrophus strain C4 for tentoxin production in vitro and in planta during infection of maize leaves. We could not detect any tentoxin (data not shown), suggesting that ChNps3 function may indeed be different from CmNps3. Because tentoxin has been reported previously in Alternaria alternata, we also screened the genome of the A. alternata strain SRC1lrK2f for orthologues. We found an NRPS with 85% similarity to CmNps3: AaNps3 (JGI protein ID 1065195) (Table 2). Like ChNPS3, AaNPS3 features an intron in the A‐domain of the third module resulting in a loss of 13 amino acids, but at a different site from the ChNps3 intron (Fig. S3 and Fig. S4, see Supporting Information). Functional analysis of the gene should shed light on a possible involvement in A. alternata tentoxin production.
Table 2.
Protein sequence similarities between Nps3 homologues of A lternaria alternata, C ochliobolus miyabeanus and C . heterostrophus
| AaNps3 | 100% | ||
| CmNps3 | 85% | 100% | |
| ChNps3 | 85% | 95% | 100% |
| AaNps3 | CmNps3 | ChNps3 |
Discussion
The aim of this study was to characterize new disease‐associated metabolites in C. miyabeanus using a comparative chemical screening. We have shown that the non‐HST tentoxin is a virulence factor in C. miyabeanus. To our knowledge, this is the first report of tentoxin production by a non‐Alternaria plant pathogen. Tentoxin biosynthesis was induced at 24 hpi and was correlated with chlorosis development. This finding reinforces the hypothesis that C. miyabeanus produces other virulence factors in addition to ophiobolins, and relies mainly on non‐host‐specific phytotoxins.
Although tentoxin production by Alternaria has been known for a long time, the NRPS responsible for its synthesis has not been characterized previously. NRPSs are multimodular megasynthases which catalyse the biosynthesis of small peptides independent of the ribosomes (Stachelhaus et al., 1999). Each module represents a single step in chain elongation and is composed of separate domains catalysing the selection, activation and modification of its amino acid substrate (Stein et al., 1996). Monomer specificity is conferred by the A‐domain. Based on the amino acids lining the substrate‐binding pocket, predictions can be made about substrate specificity. Several algorithms have been developed to aid in monomer prediction of each module. We used antiSMASH to predict the modular backbone and monomer specificity of C. miyabeanus NRPS, based on the Stachelhaus code and NRPSPredictor2 (Medema et al., 2011; Rausch et al., 2005; Röttig et al., 2011). One NRPS, n8391, stood out because of the tetramodular structure, including methylation domains in the second and fourth modules. Because n8391 is an orthologue of C. heterostrophus NPS3, we designated the enzyme CmNps3. Deletion of CmNPS3 from two C. miyabeanus strains confirmed its involvement in the biosynthesis of tentoxin.
Although the first two predicted monomers of CmNPS3 were consistent with the known amino acid sequence of tentoxin (Gly, MeAla, Leu, MePhe), the last two monomers were not. It is not clear whether the A‐domain specificity prediction is erroneous or whether the biosynthesis of tentoxin does not follow the collinearity rule. The currently available amount of training data for NRPS prediction algorithms is relatively low; there are still a large number of sequences for which prediction is uncertain or impossible, and this is especially true for fungal sequences. Most of the characterized A‐domains that serve as a basis for specificity predictions have a bacterial origin, and this, plus data indicating that eukaryotic A‐domains might have developed alternative substrate binding patterns, makes predictions difficult (Kalb et al., 2013; Rausch et al., 2005; Stack et al., 2007). In addition, eukaryotic NRPS elongation does not always follow the collinearity rule, which states that the module sequence of the NRPS reflects the monomer sequence in the resulting peptide (Marahiel et al., 1997). Thus, it is not surprising to find discrepancies between expected and predicted monomer specificity. Furthermore, it is unclear whether the peptide elongation process proceeds stepwise starting from glycine or by the binding of two dipeptides Gly‐MeAla and Leu‐MePhe (Ramm et al., 1994). The latter case would explain the switched positions of the predicted monomers, but cannot explain the methylation of an aliphatic amino acid. To resolve this, the module specificity needs to be confirmed by an adenosine triphosphate–pyrophosphate (ATP–PPi) exchange reaction (Lee and Lipmann, 1975; Phelan et al., 2009).
CmNPS3 is an orthologue of ChNPS3 and is also conserved in C. sativus (Condon et al., 2013; Lee et al., 2005). Although other sequenced Cochliobolus sp. do not have a full NPS3 orthologue, they all have additional NRPS proteins constructed from separate NPS1/NPS3 modules. The seemingly volatile nature of NPS1/NPS3 modules could lead to new proteins with new functionalities, and eventually new virulence factors (Condon et al., 2013). This would imply that tentoxin is only one possible Nps3 product, resulting from a specific combination of four modules. Mining of the genome of the tentoxin producer A. alternata revealed the presence of another NPS3 orthologue (AaNPS3) which could be involved in tentoxin production in this genus. Although the same monomer specificity is predicted for ChNps3, AaNps3 and CmNps3, the functionality of the ChNPS3 gene can be questioned. The C. heterostrophus NPS3 region shows many SNPs (only 90% sequence identity) when compared with CmNPS3. In addition, part of the DNA sequence changed to an intron, resulting in the loss of a stretch of 17 amino acids in the A‐domain of the third module. Cochliobolus heterostrophus strain C4 showed minimal expression of ChNPS3 after 24 h of culture in a nitrogen‐poor medium (Lee et al., 2005). We could not detect any tentoxin present in the supernatant under these conditions, nor during infection of maize leaves (data not shown). Similarly, a screening of C. heterostrophus NRPS mutants did not reveal a phenotype for Chnps3, although ChNPS3 was expressed in liquid culture (Lee et al., 2005; Turgeon et al., 2008). The reported production of tentoxin by A. alternata suggests that AaNPS may function in tentoxin biosynthesis, despite missing 13 amino acids in the third A‐domain compared with CmNps3. Further investigations are needed to verify this claim.
Several phytotoxic effects have been reported for tentoxin, but the main mode of action is the induction of chlorosis via the inhibition of photophosphorylation in sensitive plant species. In vitro, tentoxin binds to chloroplast coupling factor (CF1)‐ATPase of sensitive plant species, thereby blocking ATP synthesis (Groth, 2002; Meiss et al., 2008; Pavlova et al., 2004; Steele et al., 1976). Although ATPase inhibition alone cannot explain the chlorosis effects, it could deplete the ATP necessary for the incorporation of polyphenol oxidase (PPO) into thylakoids (Holland et al., 1997; Sommer et al., 1994). Loss of plastidic PPO can stop the electron flow, leading to the over‐energization of thylakoid membranes and the production of reactive oxygen species, which ultimately results in chlorosis (Holland et al., 1997; Vaughn and Duke, 1981, 1984). Other phytotoxic effects include interference with membrane functions responsible for ion transport and stomatal closure (Durbin et al., 1973; Erdei and Klotz, 1988; Heitz et al., 1986; Klotz, 1988; Klotz and Erdei, 1988). Although rice seedlings have been classified as being insensitive to the chlorosis effects of tentoxin (Durbin and Uchytil, 1977), this study showed a correlation between tentoxin production and chlorosis during C. miyabeanus infection of fully developed rice leaves of 5‐week‐old plants. Furthermore, treatment of mature rice leaves with 500 μm tentoxin resulted in necrotic symptoms 33% of the time, indicating that tentoxin does not affect only seedlings of species with a sensitive CF1‐ATPase. Interestingly, photophosphorylation was also inhibited in chloroplasts of insensitive plant species when high concentrations of tentoxin were applied. This photophosphorylation inhibition was independent of ATPase inhibition, suggesting the existence of additional tentoxin effects on energy transfer (Steele et al., 1976). Further investigation should shed light on the tentoxin sensitivity of rice, in particular the involvement of photophosphorylation.
Chlorosis development during the second phase of brown spot disease shows high biological variability. The amount of tentoxin measured for different biological replicates of one strain is similarly highly variable. When CmNPS3 was deleted, the variability in disease symptoms decreased considerably. It is not clear why tentoxin production is highly variable, but the analysis of CmNPS3 gene expression shows that the difference in tentoxin production between strains can be explained, at least partially, by a difference in relative gene expression. Strong induction of CmNPS3 gene expression and tentoxin production at 24 h after inoculation suggests the need for an extracellular stimulus. This might be an infection‐specific trigger originating from the plant or fungus, the strength of which could correlate with the amount of tentoxin produced. This could also explain the difference in gene expression between different strains, as less virulent strains cause less disease symptoms during the first phase of infection. Furthermore, NRPSs are generally organized in gene clusters, sometimes comprising specific transcription factors or regulatory proteins. Both specific transcription factors as well as global transcription factors can regulate the expression of secondary metabolism genes (Wu et al., 2012; Yu and Keller, 2005). As no specific transcription factors have been predicted in the near vicinity of CmNPS3 in the WK1C genome published on the JGI MycoCosm website, further research should shed light on how tentoxin biosynthesis is triggered and regulated.
During C. miyabeanus infection, rice physiology is subjected to radical changes, accompanied by a decrease in photosynthetic processes and the remobilization of nitrogen, characterized by the induction of glutamine synthetases (Van Bockhaven et al., 2015). Fungi rely almost exclusively on the plant for their nitrogen source (Seifi et al., 2013). The availability of the amino acids necessary to produce tentoxin (Gly, Ala, Leu and Phe), as well as a methionine as source for the methylation reactions, could determine the level of tentoxin produced. Apart from possible phytotoxic effects, tentoxin could enhance disease by using plant amino acids for its production. In this way, valuable nitrogen sources may be drained away from the plant and lost for recycling to other plant parts.
Tentoxin has mainly been studied as a mycotoxin produced by A. alternata, often in the context of food contamination. Although the risk of tentoxin to human health has not been investigated adequately, detection strategies have been developed to detect tentoxin in food matrices (Walravens et al., 2014). Often, tentoxin detection is seen as an indicator of Alternaria sp. Our results show that tentoxin production could also be correlated with the presence of Cochliobolus sp.
To summarize, we have shown that, in addition to ophiobolins, C. miyabeanus also produces the non‐specific toxin tentoxin as a virulence factor. This supports the idea that broad‐host‐range necrotrophs rely on an array of non‐specific virulence factors. Each factor typically targets a general plant target or suppresses defence responses, adding quantitatively to the overall virulence. The identification of new virulence factors can complement breeding efforts, as it provides additional tools to screen for more resistant genotypes.
Experimental Procedures
Biological material and growth conditions
Cochliobolus miyabeanus wild‐type strains Cm988 (De Vleesschauwer et al., 2010) and WK1C (Condon et al., 2013) were used. CmNPS3 (n8391) was deleted from each strain, resulting in the tentoxin‐deficient mutants Cm988Δnps3 and WK1CΔnps3, respectively. Cm988 and Cm988Δnps3 were grown on PDA under dark conditions at 28 °C, and WK1C and WK1CΔnps3 were grown on complete medium with xylose (CMX) (Inderbitzin et al., 2010) under constant light at 23 °C, for optimal conidiation.
The O. sativa japonica cultivar Nipponbare was grown in commercial potting soil in a growth room under 12 h light/12 h dark at 28 °C, as described previously (De Vleesschauwer et al., 2010).
Sample preparation for chemical analysis
Liquid culture extracts were collected as follows. Three mycelium plugs per strain were grown in 100‐mL Erlenmeyer flasks containing 30 mL of Fries medium (Pringle and Braun, 1957) supplemented with 0.1% yeast extract and 1 mL of plant extract at 28 °C in the dark on a rotary shaker (150 rpm). The plant extract was prepared by extracting 4 g of ground fresh rice leaves with 50 mL of double‐distilled water, and filter sterilized using a syringe‐driven 0.22‐μm Millex‐HV filter unit (Merck Millipore, Cork, Ireland) before use. After 3 days, the culture was blended and 1 mL was transferred to fresh medium and incubated for another 7 days under the same circumstances. The supernatant was filtered through two layers of cheese cloth and one layer of filter paper (Grade 3hw, Sartorius Stedim Biotech S.A., Goettingen, Germany), and sterilized using a syringe‐driven 0.45‐μm Millex‐HV filter unit (Merck Millipore). The mycelium was collected from the filter and dried at 80 °C for 16 h before measurement of the dry weight. The total volume of the crude culture filtrate was measured and a 4‐mL aliquot was used for metabolite extraction.
Samples for in planta tentoxin quantification were prepared as follows. Leaf pieces were inoculated as described for the virulence assays. Infected leaf samples were crushed using liquid N2 and stored at −80 °C until chemical extraction. A total of approximately 0.5 g of leaf material was used for toxin extraction.
The samples (either crude extract or leaf material) were extracted with 10 mL of ethylacetate–acetonitrile (1:1, v/v) after the addition of 1.8 g of MgSO4. The sample was vigorously shaken on an end‐over‐end shaker (Agitelec, J. Toulemonde & Cie, Paris, France) for 30 min and then centrifuged at 4000 g for 15 min. The organic upper phase was evaporated to dryness under an N2 flow at 40 °C. The dry residue was redissolved in 100 μL of mobile phase A : B (60:40, v/v) and filter centrifuged in an Ultrafree‐MC centrifugal filter unit (Merck Millipore, Bedford, MA, USA) for 5 min at 14 000 g.
UHPLC‐Orbitrap‐HRMS conditions
An Accela™ High Speed LC (UHPLC) system (Thermo Fisher Scientific, San José, CA, USA), coupled to an Orbitrap Exactive™ mass analyser (Thermo Fisher Scientific), was used to analyse the samples. The Orbitrap was equipped with an ESI interface, which was operated in alternating positive and negative ionization mode. The following conditions were used: spray voltage, 4.5 kV; sheath, auxiliary and sweep gas flow rates, 45, 10 and 5 arbitrary units, respectively; capillary and heater temperature, 250 °C; capillary, tube lens and skimmer voltages (V), 45 (−45), 125 (−125) and 22 (−23), respectively, for positive (negative) mode. The instrument was operated in full‐scan mode at a resolution of 100 000 full width at half‐maximum (FWHM) height with an m/z scan range of 100–1000. The maximum injection time was 250 ms and the automatic gain control (AGC) target was set at ultimate mass accuracy (5 × 105 ions).
The column used was a Zorbax Eclipse C18 (2.1 mm × 100 mm inside diameter, 1.8 μm) (Agilent, Santa Clara, CA, USA). Mobile phase A consisted of H2O–methanol (95:5, v/v) and mobile phase B of methanol–H2O (95:5, v/v). To facilitate ionization, 0.1% formic acid and 1 mm ammonium formate were added to both mobile phases. The gradient elution programme for LC‐Orbitrap MS analyses was applied as follows: 0–0.5 min, 0% B; 0.5–20 min, 0–99.1% B; 20–21 min, 99.1% B; 21–24 min, 99.1–0% B; 24–28 min, 0% B. The flow rate was 0.4 mL/min. The column temperature was set to 30 °C and the temperature of the autosampler was 10 °C.
Instrument control and data processing were carried out using Xcalibur™ 2.1 (Thermo Fisher Scientific). Sieve™ software (Thermo Fisher Scientific) was used for peak picking according to the following settings: positive charge; m/z range, 100–1000 Da; m/z width, 10 ppm; retention time range, 5.0–25.0 min; peak intensity threshold, 105 000 arbitrary units; maximum peak width, 0.5 min; maximum number of 5000 frames. The peak intensities of the 5000 selected frames were further analysed using principal component analysis in SPSS Statistics 21.
UPLC‐MS/MS conditions
We confirmed tentoxin via analysis of a pure standard and its corresponding fragmentation pattern. A Waters Acquity UPLC system, coupled to a Waters Quattro Premier XE tandem‐quadrupole mass spectrometer (Waters, Milford, MA, USA), was used to confirm the identity of tentoxin based on the fragmentation patterns of a pure commercial tentoxin standard (Biopure Referenzsubstanzen GmbH, Tulln/Donau, Austria). Data processing was performed with Masslynx software v4.1 (Waters). The column and LC conditions were the same as described for the Accela™ High Speed LC (UHPLC) system discussed in the section ‘UHPLC‐Orbitrap‐HRMS conditions’.
The mass spectrometer was operated in the ESI− mode. The capillary voltage was 3.5 kV, and nitrogen was used as the desolvation gas. The source and desolvation temperatures were set at 120 and 300 °C, respectively. Tentoxin was analysed using the selected reaction monitoring (MRM) mode with the following ion transitions: 413.3 → 271.2 (cone, 44 V; collision energy, 18 eV); 413.3 → 141.0 (cone, 44 V; collision energy, 24 eV). The ion transition 413.3 → 141.0 was used for quantification; the ion transition 413.3 → 271.2 was used as qualifier.
Quanlynx (Waters) was used for the quantification of tentoxin in the samples by generating a calibration curve using different concentrations of a tentoxin standard. Tentoxin production per gram of mycelium dry weight or per gram of leaf fresh weight was calculated for in vitro and in planta production, respectively.
Gene identification and gene knockout
The NRPSs present in the C. miyabeanus genome, as identified by Condon et al. (2013), were screened using the program antiSMASH (Medema et al., 2011). One candidate tentoxin synthase was withheld [NODE 8391 (Condon et al., 2013), JGI protein ID 98843].
The selected gene was deleted using homologous recombination, as described by Bi et al. (2013). The entire selection marker fragment ‘hygB’, including the promoter, was amplified from plasmid pUCATPH using M13R and M13F primers, and fused to the 5′ and 3′ flanking fragments of the target CmNPS3 gene. PCR amplification of transformation constructs was carried out with phusion high‐fidelity DNA polymerase (Thermo Fisher Scientific, Aalst, Belgium) following the manufacturer's instructions.
Transformation of C. miyabeanus was carried out as described previously (Inderbitzin et al., 2010) with slight modifications. The Cm988 strain was cultured on PDA for 7 days in the dark, and WK1C on CMX for 14 days under continuous light. For protoplasting, conidia were transferred to 300 mL of complete medium (CM) in a 1‐L flask and shaken at 100 rpm for 16 h in the dark at 24 °C. Protoplasts were harvested and transformed as described previously (Catlett et al., 2003). Candidate transformants were single conidiated and transferred to PDA or CMX containing hygromycin B (100 μg/mL) to confirm sensitivity to the drug, as described previously (Oide, 2006). Integration of the constructs at the intended sites was confirmed by diagnostic PCRs, as described previously (Inderbitzin et al., 2010; Wu et al., 2012). All PCRs were conducted with GoTaq (Promega, Leiden, The Netherlands). Single‐conidiated, confirmed transformants were selected and scanned for tentoxin production using UPLC‐MS/MS. The primer sequences used for gene deletion are listed in Table 3.
Table 3.
Primer sequences used in this study. Sequences in italics are complementary to M13R and M13F sequences, respectively
| Primer name | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| NLC38 | CGTTGCAAGACCTGCCTGAA |
Forward primer for HygB
Catlett et al. (2003) |
| NLC37 | GGATGCCTCCGCTCGAAGTA |
Reverse primer for HygB
Catlett et al. (2003) |
| M13F | CGCCAGGGTTTTCCCAGTCACGAC |
Reverse primer to amplify HygB
Catlett et al. (2003) |
| M13R | AGCGGATAACAATTTCACACAGGA |
Forward primer to amplify HygB
Catlett et al. (2003) |
| CmNODE8391_UF1 | GCCGTTTTGCTGCTATCTT | Forward primer of 5′ flank for CmNPS3 deletion |
| CmNODE8391_UR1 | TCCTGTGTGAAATTGTTATCCGCTAAGGTGTCAGAACGCTACG | Reverse primer of 5′ flank for CmNPS3 deletion, complementary to M13R |
| CmNODE8391_DF1 | GTCGTGACTGGGAAAACCCTGGCGGGATGAGGACAAGGAAAGG | Forward primer of 3′ flank for CmNPS3 deletion, complementary to M13F |
| CmNODE8391_DR1 | GCGGGTGTGTGAATAAAGA | Reverse primer of 3′ flank for CmNPS3 deletion |
| CmNODE8391_amp1F1 | AGAAGTCATCAACAGCAGCA | Internal forward primer for CmNPS3; deletion verification |
| CmNODE8391_amp1R1 | CCTGACTGAAACCCCTGAC | Internal reverse primer for CmNPS3; deletion verification |
| 8391_UpVER1 | TGGTTAGACGCACCCATACA | Forward primer outside of 5′ flank for CmNPS3 deletion; deletion verification |
| 8391_DownVER1 | AGTTGTGCACTCCTCTCTCC | Reverse primer outside of 3′ flank for CmNPS3 deletion; deletion verification |
| CmNPS3_F | GTCGACATCTTGAACGCAGA | Forward primer for CmNPS3 mRNA; qRT‐PCR fungal gene expression analysis |
| CmNPS3_R | CTCCCAGGAAGATCCATCAA | Reverse primer for CmNPS3 mRNA; qRT‐PCR fungal gene expression analysis |
| CmAct1.2_F | AAGTCGCAGCTCTCGTCATC | Forward primer for CmActin; normalizer for qRT‐PCR fungal gene expression analysis |
| CmAct1.2_R | TGAGTCCTTCTGGCCCATAC | Reverse primer for CmActin; normalizer for qRT‐PCR fungal gene expression analysis |
qRT‐PCR, quantitative reverse transcription‐polymerase chain reaction.
Fungal growth rate and conidiation
A 6‐mm‐diameter plug from a 7‐day‐old colony of the respective strain was placed in the centre of a PDA plate and incubated at 28 °C in the dark. Fungal growth was measured as the average colony diameter after 3 or 7 days of growth. After 7 days of growth, the conidia from each plate were suspended in 15 mL of double‐distilled water and counted using a Bürker counting chamber (Marienfield, Lauda‐Königshofen, Germany). The experiment was repeated once using three PDA plates per strain.
Infection assays
Disease severity was assessed using leaf piece infection assays. For Cm988 and Cm988Δnps3 inoculum preparation, conidia were harvested as described by Thuan et al. (2006) and resuspended in 0.02% Tween‐20 to a final density of 1 × 104 conidia/mL.
For all other strains (and also for Cm988 in the infection assay in which tentoxin production by different field strains was compared), a mycelial suspension was used as inoculum. Five mycelium plugs per strain were grown in 100‐mL Erlenmeyer flasks containing 30 mL of potato dextrose broth (PDB) at 28 °C in the dark on a rotary shaker (150 rpm). After 48 h of growth, the mycelium was collected and blended in 15 mL of double‐distilled water. The resulting mycelial suspension was counted and diluted to 5 × 104 colony‐forming units (CFU)/mL in 0.02% Tween‐20.
The youngest fully developed leaves of 5‐week‐old rice plants were cut into 7‐cm pieces and kept in square Petri dishes (9 cm × 9 cm, Greiner bio one, Wemmel, Belgium) lined with moistened tissue. Leaf pieces were spray inoculated with 0.5 mL of inoculum per Petri dish. The leaf pieces were kept in the dark at 28 °C and moved to growth chamber conditions after 16 h. The number of lesions was counted at 48 hpi using APS Assess Software (the American Phytopathological Society, St. Paul, MN, USA). Disease symptoms were quantified as the percentage affected leaf surface at 3 dpi using APS Assess Software. Statistical analysis was performed using non‐parametric Kruskal–Wallis rank test (α = 0.05).
Toxicity test
Tentoxin (Sigma‐Aldrich, Diegem, Belgium) was dissolved in 10% ethanol to a final stock concentration of 1.2 mm. The youngest fully developed leaves of 5‐week‐old rice plants were cut into 7‐cm pieces and kept in square Petri dishes (9 cm × 9 cm) lined with moistened tissue. Each leaf piece was punctured at three different places and treated with 10 μL of tentoxin solution at the indicated concentrations, or a control. All solutions contained 4% ethanol. Symptom development was monitored at 1–4 dpi.
RNA extraction and quantitative reverse transcription‐polymerase chain reaction (RT‐PCR)
Total RNA was extracted from infected leaf material using TRIZOL reagent (Invitrogen, Merelbeke, Belgium) and subsequently treated with Turbo DNase (Ambion, ThermoFisher Scientific, Leusden, Netherlands) to remove genomic DNA. First‐strand cDNA was synthesized from 2 μg of total RNA using Multiscribe reverse transcriptase (Applied Biosystems, ThermoFisher Scientific, Leusden, Netherlands) and random primers following the manufacturer's instructions. qPCR amplifications were conducted in optical 96‐well plates with the Mx3005P real‐time PCR detection system (Stratagene, Agilent technologies, Santa Clara, CA, USA), using GoTaq qPCR Master Mix containing the CXR reference dye (Promega, Leiden, The Netherlands) to monitor double‐stranded DNA synthesis. The expression of each gene was assayed in duplicate in a total volume of 25 μL according to the manufacturer's instructions (Promega). The thermal profile used consisted of an initial denaturation step at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 59 °C for 30 s and 72 °C for 30 s. To verify amplification specificity, a melting curve analysis was included according to the thermal profile suggested by the manufacturer (Stratagene). The amount of CmNPS3 mRNA was normalized using CmActin as internal control for fungal gene expression. Statistical analysis was performed using MxPro software (Stratagene). The primer sequences used for gene expression analysis are listed in Table 3.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Score plots representing how the first three principal components help to differentiate between samples, based on differences in compounds produced in liquid Fries medium by different Cochliobolus miyabeanus strains.
Fig. S2 Illustration of the strategy used to confirm successful deletion of the target gene and specific integration of the transformation construct at the target site.
Fig. S3 Pairwise alignment of the nucleotide sequences of CmNPS3 from Cochliobolus miyabeanus, ChNPS3 from Cochliobolus heterostrophus and AaNPS3 from Alternaria alternata.
Fig. S4 Pairwise alignment of the amino acid sequences of CmNPS3 from Cochliobolus miyabeanus, ChNPS3 from Cochliobolus heterostrophus and AaNPS3 from Alternaria alternata.
Table S1 Total variance explained by the first three principal components based on differences in 5000 ions detected in culture filtrates of three different Cochliobolus miyabeanus wild‐type strains grown in liquid Fries medium.
Acknowledgements
Lieselotte De Bruyne was supported by a personal PhD Grant for Strategic Basic Research provided by the Agency for Innovation by Science and Technology (IWT), Belgium. Nur Ain Izzati Mohd Zainudin was supported by Universiti Putra Malaysia (UPM) and the Ministry of Higher Education (MOHE). We thank the Department of Energy Joint Genome Institute (DOE JGI) for making the sequence data of C. miyabeanus strain WK1C publicly available on their website (http://www.jgi.doe.gov).
References
- Berestetskiy, A.O. (2008) A review of fungal phytotoxins: from basic studies to practical use. Appl. Biochem. Microbiol. 44, 453–465. [PubMed] [Google Scholar]
- Bi, Q. , Wu, D. , Zhu, X. and Turgeon, B.G. (2013) Cochliobolus heterostrophus Llm1—a Lae1‐like methyltransferase regulates T‐toxin production, virulence, and development. Fungal Genet. Biol. 51, 21–33. [DOI] [PubMed] [Google Scholar]
- Catlett, N.L. , Lee, B.‐N. , Yoder, O.C. and Turgeon, B.G. (2003) Split‐marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 50, 9–11. [Google Scholar]
- Chakrabarti, N.K. (2001) Epidemiology and disease management of brown spot of rice in India In: Major Fungal Diseases of Rice (Sreenivasaprasad S. and Johnson R., eds), pp. 293–305, Dordrecht: Springer. [Google Scholar]
- Condon, B.J. , Leng, Y. , Wu, D. , Bushley, K.E. , Ohm, R.A. , Otillar, R. , Martin, J. , Schackwitz, W. , Grimwood, J. , MohdZainudin, N. , Xue, C. , Wang, R. , Manning, V.A. , Dhillon, B. , Tu, Z.J. , Steffenson, B.J. , Salamov, A. , Sun, H. , Lowry, S. , Labutti, K. , Han, J. , Copeland, A. , Lindquist, E. , Barry, K. , Schmutz, J. , Baker, S.E. , Ciuffetti, L.M. , Grigoriev, I.V. , Zhong, S. and Turgeon, B.G. (2013) Comparative genome structure, secondary metabolite, and effector coding capacity across Cochliobolus pathogens. PLoS Genet. 9, e1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D., Yang Y., Cruz C.V. and Höfte M. (2010) Abscisic acid‐induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP Kinase‐mediated repression of ethylene signaling. Plant Physiol. 152, 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin, R.D. and Uchytil, T.F. (1977) A survey of plant insensitivity to tentoxin. Phytopathology, 67, 602–603. [Google Scholar]
- Durbin, R.D. , Uchytil, T.F. and Sparapano, L. (1973) The effect of tentoxin on stomatal aperture and potassium content of guard cells. Phytopathology, 63, 1077–1078. [Google Scholar]
- Erdei, L. and Klotz, M.G. (1988) Growth and internal ion concentrations in seedlings of winter wheat are affected by 1 pM tentoxin. Physiol. Plant. 73, 295–298. [Google Scholar]
- Groth, G. (2002) Structure of spinach chloroplast F1‐ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc. Natl. Acad. Sci. USA, 99, 3464–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, F. , Jacquier, R. , Kaddari, F. and Verducci, J. (1986) Aggregation and ion transfer induced by tentoxin. Biophys. Chem. 23, 245–249. [DOI] [PubMed] [Google Scholar]
- Holland, N. , Evron, Y. , Jansen, M. , Edelman, M. and Pick, U. (1997) Involvement of thylakoid overenergization in tentoxin‐induced chlorosis in Nicotiana spp. Plant Physiol. 114, 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi, M. , Akimoto, N. , Ohnishi, K. , Yamashita, M. and Maoka, T. (2003) Rapid and simultaneous determination of tetra cyclic peptide phytotoxins, tentoxin, isotentoxin and dihydrotentoxin, from Alternaria porri by LC/MS. Chromatography, 24, 109–116. [Google Scholar]
- Inderbitzin, P. , Asvarak, T. and Turgeon, B.G. (2010) Six new genes required for production of T‐toxin, a polyketide determinant of high virulence of Cochliobolus heterostrophus to maize. Mol. Plant–Microbe Interact. 23, 458–472. [DOI] [PubMed] [Google Scholar]
- Kalb, D. , Lackner, G. and Hoffmeister, D. (2013) Fungal peptide synthetases: an update on functions and specificity signatures. Fungal Biol. Rev. 27, 43–50. [Google Scholar]
- Klotz, M.G. (1988) Action of tentoxin on membrane processes in plants. Physiol. Plant. 74, 575–582. [Google Scholar]
- Klotz, M.G. and Erdei, L. (1988) Effect of tentoxin on K+ transport in winter wheat seedlings of different K+ status. Physiol. Plant. 72, 298–304. [Google Scholar]
- Lee, B. , Kroken, S. , Chou, D.Y.T. , Robbertse, B. , Yoder, O.C. and Turgeon, B.G. (2005) Functional analysis of all non‐ribosomal peptide synthetases in Cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryot. Cell, 4, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.G. and Lipmann, F. (1975) Volume 43: antibiotics In: Methods in Enzymology (Hash J.H., ed.), pp. 585–602, New York: Academic Press. [DOI] [PubMed] [Google Scholar]
- Liebermann, B. , Ellinger, R. and Pinet, E. (1996) Isotentoxin, a conversion product of the phytotoxin tentoxin. Phytochemistry, 42, 1537–1540. [Google Scholar]
- Liu, Y. and Rychlik, M. (2013) Development of a stable isotope dilution LC‐MS/MS method for the Alternaria toxins tentoxin, dihydrotentoxin, and isotentoxin. J. Agric. Food Chem. 61, 2970–2978. [DOI] [PubMed] [Google Scholar]
- Marahiel, M.A. , Stachelhaus, T. and Mootz, H.D. (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674. [DOI] [PubMed] [Google Scholar]
- Medema, M.H. , Blin, K. , Cimermancic, P. , de Jager, V. , Zakrzewski, P. , Fischbach, M.A. , Weber, T. , Takano, E. and Breitling, R. (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiss, E. , Konno, H. , Groth, G. and Hisabori, T. (2008) Molecular processes of inhibition and stimulation of ATP synthase caused by the phytotoxin tentoxin. J. Biol. Chem. 283, 24594–24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, M. and Ishibashi, K. (1958) On the new antibiotic ‘ophiobolin’ produced by Ophiobolus miyabeanus . J. Agric. Chem. Soc. Jpn. 32, 739–744. [Google Scholar]
- Oide S., Moeder W., Krasnoff S., Gibson D., Haas H., Yoshioka K. and Turgeon, B.G. (2006). NPS6, encoding a nonribosomal peptide synthetase involved in siderophore‐mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. The Plant Cell 18, 2836–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsenigo, M. (1957) Toxin production by Helminthosporium oryzae . Phytopathol. Z. 29, 189–196. [Google Scholar]
- Panaccione, D.G. (1993) The fungal genus Cochliobolus and toxin‐mediated plant disease. Trends Microbiol. 1, 14–20. [DOI] [PubMed] [Google Scholar]
- Pavlova, P. , Shimabukuro, K. , Hisabori, T. , Groth, G. , Lill, H. and Bald, D. (2004) Complete inhibition and partial re‐activation of single F1‐ATPase molecules by tentoxin: new properties of the re‐activated enzyme. J. Biol. Chem. 279, 9685–9688. [DOI] [PubMed] [Google Scholar]
- Phelan, V.V. , Du, Y. , McLean, J.A. and Bachmann, B.O. (2009) Adenylation enzyme characterization using gamma‐18O4‐ATP pyrophosphate exchange. Chem. Biol. 16, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle R.B. and Armin C.B. (1957) The isolation of the toxin of Helminthosporium victoriae. Phytopathology 47, 369–371. [Google Scholar]
- Ramm, K. , Ramm, M. , Liebermann, B. and Reuter, G. (1994) Studies of the biosynthesis of tentoxin by Alternaria alternata . Microbiology, 140, 3257–3266. [DOI] [PubMed] [Google Scholar]
- Rausch, C. , Weber, T. , Kohlbacher, O. , Wohlleben, W. and Huson, D.H. (2005) Specificity prediction of adenylation domains in non‐ribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33, 5799–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röttig, M. , Medema, M.H. , Blin, K. , Weber, T. , Rausch, C. and Kohlbacher, O. (2011) NRPSPredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary, S. , Ficke, A. , Aubertot, J.‐N. and Hollier, C. (2012) Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 4, 519–537. [Google Scholar]
- Seifi, H.S. , Van Bockhaven, J. , Angenon, G. and Höfte, M. (2013) Glutamate metabolism in plant disease and defense: friend or foe? Mol. Plant–Microbe Interact. 26, 475–485. [DOI] [PubMed] [Google Scholar]
- Sommer, A. , Néeman, E. , Steffens, J.C. , Mayer, A.M. and Harel, E. (1994) Import, targeting, and processing of a plant polyphenol oxidase. Plant Physiol. 105, 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachelhaus, T. , Mootz, D. and Marahiel, M.A. (1999) The specificity‐conferring code of adenylation domains in non‐ribosomal peptide synthetases. Chem. Biol. 6, 493–505. [DOI] [PubMed] [Google Scholar]
- Stack, D. , Neville, C. and Doyle, S. (2007) Non‐ribosomal peptide synthesis in Aspergillus fumigatus and other fungi. Microbiology, 153, 1297–1306. [DOI] [PubMed] [Google Scholar]
- Steele, J.A. , Uchytil, T.F. , Durbin, R.D. , Bhatnagar, P. and Rich, D.H. (1976) Chloroplast coupling factor 1: a species‐specific receptor for tentoxin. Proc. Natl. Acad. Sci. USA, 73, 2245–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, T. , Vater, J. , Kruft, V. , Otto, A. , Wittmann‐Liebold, B. , Franke, P. , Panico, M. , McDowell, R. and Morris, H.R. (1996) The multiple carrier model of non‐ribosomal peptide biosynthesis at modular multienzymatic templates. J. Biol. Chem. 271, 15428–15435. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Collemare, J. , Mehrabi, R. and De Wit, P.J.G.M. (2013) Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol. Rev. 37, 67–93. [DOI] [PubMed] [Google Scholar]
- Thuan, T.N.N. , Bigirimana, J. , Roumen, E. , Van Der Straeten, D. and Höfte, M. (2006) Molecular and pathotype analysis of the rice blast fungus in North Vietnam. Eur. J. Plant Pathol. 114, 381–396. [Google Scholar]
- Turgeon, B.G. , Oide, S. and Bushley, K. (2008) Creating and screening Cochliobolus heterostrophus non‐ribosomal peptide synthetase mutants. Mycol. Res. 112, 200–206. [DOI] [PubMed] [Google Scholar]
- Van Bockhaven, J. , Steppe, K. , Bauweraerts, I. , Kikuchi, S. , Asano, T. , Höfte, M. and De Vleesschauwer, D. (2015) Primary metabolism plays a central role in moulding silicon‐inducible brown spot resistance in rice. Mol. Plant Pathol. 16, 811–824. doi: 10.1111/mpp.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, K.C. and Duke, S.O. (1981) Tentoxin‐induced loss of plastidic polyphenol oxidase. Physiol. Plant. 53, 421–428. [Google Scholar]
- Vaughn, K.C. and Duke, S.O. (1984) Tentoxin stops the processing of polyphenol oxidase into an active protein. Physiol. Plant. 60, 257–261. [Google Scholar]
- Vidhyasekaran, P. , Borromeo, E.S. and Mew, T.W. (1986) Host‐specific toxin production by Helminthosporium oryzae . Phytopathology, 76, 261–266. [Google Scholar]
- Walravens, J. , Mikula, H. , Rychlik, M. , Asam, S. , Ediage, E.N. , Diana Di Mavungu, J. , Van Landschoot, A. , Vanhaecke, L. and De Saeger, S. (2014) Development and validation of an ultra‐high‐performance liquid chromatography tandem mass spectrometric method for the simultaneous determination of free and conjugated Alternaria toxins in cereal‐based foodstuffs. J. Chromatogr. A, 1372C, 91–101. [DOI] [PubMed] [Google Scholar]
- Wu, D. , Oide, S. , Zhang, N. , Choi, M.Y. and Turgeon, B.G. (2012) ChLae1 and ChVel1 regulate T‐toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus . PLoS Pathog. 8, e1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J.Z. , Tsuda, M. , Doke, N. and Nishimura, S. (1991) Phytotoxins produced by germinating spores of Bipolaris oryzae . Physiol. Biochem. 81, 58–64. [Google Scholar]
- Yu, J.H. and Keller, N. (2005) Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43, 437–458. [DOI] [PubMed] [Google Scholar]
- Yun, C.‐H. , Sugawara, F. and Strobel, G.A. (1988) The phytotoxic ophiobolins produced by Drechslera oryzae, their structures and biological activity on rice. Plant Sci. 54, 237–243. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Score plots representing how the first three principal components help to differentiate between samples, based on differences in compounds produced in liquid Fries medium by different Cochliobolus miyabeanus strains.
Fig. S2 Illustration of the strategy used to confirm successful deletion of the target gene and specific integration of the transformation construct at the target site.
Fig. S3 Pairwise alignment of the nucleotide sequences of CmNPS3 from Cochliobolus miyabeanus, ChNPS3 from Cochliobolus heterostrophus and AaNPS3 from Alternaria alternata.
Fig. S4 Pairwise alignment of the amino acid sequences of CmNPS3 from Cochliobolus miyabeanus, ChNPS3 from Cochliobolus heterostrophus and AaNPS3 from Alternaria alternata.
Table S1 Total variance explained by the first three principal components based on differences in 5000 ions detected in culture filtrates of three different Cochliobolus miyabeanus wild‐type strains grown in liquid Fries medium.


