Summary
Plants have developed diverse mechanisms to fine tune defence responses to different types of enemy. Cross‐regulation between signalling pathways may allow the prioritization of one response over another. Previously, we identified SUPPRESSOR OF rps4‐RLD 1 (SRFR 1) as a negative regulator of ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1)‐dependent effector‐triggered immunity against the bacterial pathogen P seudomonas syringae pv. tomato strain DC3000 expressing avr R ps4. The use of multiple stresses is a powerful tool to further define gene function. Here, we examined whether SRFR 1 also impacts resistance to a herbivorous insect in leaves and to a cyst nematode in roots. Interestingly, srfr1‐1 plants showed increased resistance to herbivory by the beet army worm S podoptera exigua and to parasitism by the cyst nematode H eterodera schachtii compared with the corresponding wild‐type Arabidopsis accession RLD. Using quantitative real‐time PCR (qRT‐PCR) to measure the transcript levels of salicylic acid (SA) and jasmonate/ethylene (JA/ET) pathway genes, we found that enhanced resistance of srfr1‐1 plants to S. exigua correlated with specific upregulation of the MYC2 branch of the JA pathway concurrent with suppression of the SA pathway. In contrast, the greater susceptibility of RLD was accompanied by simultaneously increased transcript levels of SA, JA and JA/ET signalling pathway genes. Surprisingly, mutation of either SRFR 1 or EDS 1 increased resistance to H . schachtii, indicating that the concurrent presence of both wild‐type genes promotes susceptibility. This finding suggests a novel form of resistance in Arabidopsis to the biotrophic pathogen H . schachtii or a root‐specific regulation of the SA pathway by EDS1, and places SRFR 1 at an intersection between multiple defence pathways.
Keywords: Arabidopsis thaliana, beet armyworm, beet cyst nematode, jasmonic acid, salicylic acid, signalling
Introduction
A variety of perception mechanisms, inducible signal cascades and transcriptional regulation of many defence‐ and stress‐related genes enable plants to survive and reproduce in challenging environments (Van Poecke, 2007). In general, salicylic acid (SA)‐dependent responses are triggered by and active against biotrophic pathogens, whereas resistance to necrotrophs and insect herbivores is usually associated with the jasmonic acid (JA)/ethylene (ET) pathway. Predominantly, these two defence pathways have been observed to act antagonistically, which allows plants to prioritize their response to a prevailing threat (Glazebrook, 2005; Kunkel and Brooks, 2002; Pieterse et al., 2012; Zander et al., 2014). However, in a network analysis, both pathways contributed to resistance to biotrophs and necrotrophs (Tsuda et al., 2009), consistent with considerable overlap in the transcriptional response to these hormones (Schenk et al., 2000) and hormone concentration‐dependent SA and JA/ET synergism or antagonism in plant resistance (Mur et al., 2006). Although these responses have been studied extensively in shoots, less is known about the contribution of these pathways to the resistance of roots to pests and pathogens.
The Arabidopsis accession RLD is defective in RPS4 and is fully susceptible to DC3000 and DC3000 carrying the effector avrRps4 (Gassmann et al., 1999; Hinsch and Staskawicz, 1996). Mutations in SUPPRESSOR OF rps4‐RLD1 (SRFR1) in RLD or combined with a mutation in the RPS6 resistance gene in the RLD background result in resistance to DC3000(avrRps4) or DC3000(hopA1), respectively, whereas mutants remain fully susceptible to DC3000 (Kim et al., 2009b; Kwon et al., 2009). Specific enhancement of these effector‐triggered immunity pathways in srfr1 mutants is most likely a consequence of SRFR1 interaction with the positive immune regulator ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) (Bhattacharjee et al., 2011), which, in turn, was found to be guarded by the Toll/interleukin‐1 receptor–nucleotide binding–leucine‐rich repeat (TNL) resistance proteins RPS4 and RPS6, and targeted by the effectors AvrRps4 and HopA1 (Bhattacharjee et al., 2011; Heidrich et al., 2011). Recently, the RPS4 co‐resistance protein RRS1 (Birker et al., 2009; Narusaka et al., 2009, 2013) has been identified as an additional target of AvrRps4 (Sarris et al., 2015; Williams et al., 2014), although it is not yet clear whether this interaction is direct or mediated by a common factor. Like EDS1, SRFR1 may have additional functions beyond specific effector‐triggered immunity pathways. To address this question, we used multiple stresses as a valuable tool to further elucidate gene function.
The recessive srfr1 phenotype and the similarity of SRFR1 to transcriptional repressors of Saccharomyces cerevisiae and Caenorhabditis elegans led to the hypothesis that it functions as a negative transcriptional regulator (Kwon et al., 2009), which is consistent with the observed upregulation of defence genes in srfr1 plants (Kim et al., 2009a) and interaction with TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors (Kim et al., 2014). The upregulation of both SA and JA/ET defence pathway genes in srfr1 mutants suggests that SRFR1 may also influence resistance in Arabidopsis to pests and pathogens, such as the sugar beet cyst nematode Heterodera schachtii and the beet army worm Spodoptera exigua.
The chewing insect S. exigua is of significant agricultural interest and feeds on the leaves of more than 50 plant species worldwide, including many crops and vegetables (Smits et al., 1987). When its larvae feed on plants, wounding and probably also larvae‐derived elicitors trigger the biosynthesis of JA, ET and JA derivatives, leading to defence activation (Bonaventure et al., 2011; Wu and Baldwin, 2010). However, it has also been reported that S. exigua can manipulate plant defences to activate the SA pathway and counteract the JA/ET pathway (Diezel et al., 2009).
Heterodera schachtii, a sedentary, obligate endoparasite, also infects many economically important crops, including sugar beet, cabbage and broccoli, and causes extensive worldwide agricultural and horticultural crop losses (Evans and Rowe, 1998). Second‐stage infective juveniles (J2) hatch from eggs in the soil and infect the roots of host plants using a stylet to puncture the cell wall and to secrete enzymes that degrade plant cell walls and effectors that modify host cells and processes (Hewezi and Baum, 2013; Mitchum et al., 2013). The fusing of the initial feeding cell with neighbouring cells forms a syncytium that feeds the sedentary nematode as it develops into a reproductive male or female adult. Adult males leave the root to mate with sedentary females exposed on the root surface. The dead female body becomes the cyst which protects the eggs until conditions are favourable for hatching.
Although recent advances have been made in the identification of nematode effectors and corresponding resistance genes (Cook et al., 2012; Hewezi and Baum, 2013; Liu et al., 2012; Mitchum et al., 2013), plant resistance mechanisms that limit nematode reproduction are not well understood (Kandoth and Mitchum, 2013). Heterodera schachtii parasitism upregulates PR1 (PATHOGENESIS‐RELATED GENE 1) expression in shoots and roots of infected Arabidopsis, and overexpression of PR1 reduces susceptibility of Arabidopsis to H. schachtii (Hamamouch et al., 2011; Wubben et al., 2008). The JA pathway has been reported to be important in inducing resistance to root‐knot nematode infection in rice (Cooper et al., 2005; Nahar et al., 2011); however, a tomato mutant impaired in JA perception is more resistant (Bhattarai et al., 2008).
Here, we show in feeding experiments that the Arabidopsis srfr1‐1 mutant exhibits significantly enhanced resistance to S. exigua feeding and H. schachtii parasitism when compared with RLD in leaves and roots, respectively. We further present evidence that constitutive JA, JA‐Ile (JA conjugated with the amino acid isoleucine) and SA levels are not responsible for these differences. In insect experiments, higher transcript levels of both SA and JA pathway genes in mock‐treated srfr1‐1 leaves than in RLD indicate a low level of generalized constitutive defences. Following insect herbivory, concurrent induction of the JA pathway and repression of the JA/ET and SA pathways in srfr1‐1 is consistent with the finding that the JA pathway is mainly responsible for insect resistance in plants (Wasternack and Hause, 2013). In contrast, simultaneously increased levels of transcripts in all three pathways following insect herbivory appear to correlate with the greater susceptibility of RLD. In nematode‐parasitized roots, neither activation of the SA nor JA pathway appears to correlate with greater srfr1‐1 resistance. Instead, increased levels of EDS1 transcript correlate with RLD susceptibility. Correspondingly, we showed that eds1‐2 mutants were more resistant than wild‐type Col‐0 to H. schachtii infection. Mutations in PAD4 (PHYTOALEXIN DEFICIENT4), which encodes an EDS1‐related lipase‐like protein that forms heterodimers with EDS1 and is required for SA‐mediated defences (Feys et al., 2001; Rietz et al., 2011), or in SRFR1, did not add to eds1‐2 resistance. Indeed, only genotypes that had a functional SRFR1 gene together with a functional EDS1 gene were fully susceptible, suggesting a novel form of cyst nematode resistance in srfr1 roots.
Results
Enhanced resistance of srfr1‐1 to S . exigua and H . schachtii
To examine resistance to insect herbivory by S. exigua, two first instar larvae were confined to and allowed to feed on a 4‐week‐old plant for 7 days. Each replicate consisted of 26–30 plants per genotype. Leaves of srfr1‐1 were more resistant than RLD leaves, as shown by significant differences in the proportion of leaf eaten and in the lower mean larval weight per plant in two of three experiments (Fig. 1). In a two‐choice assay, in which srfr1‐1 and RLD were grown in the same pot and larvae were allowed to select a plant for feeding, larvae showed a significant difference in the choice of RLD leaves over srfr1‐1 leaves in two of four experiments (Fig. S1, see Supporting Information). In this assay, smaller plants, shorter feeding times and a larger variation in the beginning of feeding time probably contributed to the greater variability.
Figure 1.
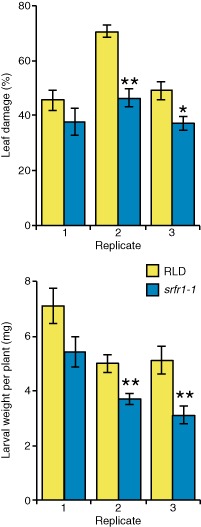
srfr1‐1 plants are more resistant than RLD wild‐type plants to feeding by S podoptera exigua. Digitally quantified leaf damage proportions of RLD and srfr1‐1 (top) and the weight of caterpillars feeding on these plants (bottom) were determined in no‐choice assays. Two first instar larvae confined to a single 4‐week‐old plant were allowed to feed for 7 days. The value shown for a genotype is the average for 26–30 plants. Error bars represent the standard error. Asterisks represent significance as determined by Student's t‐test (*P < 0.01, **P < 0.005).
To evaluate the resistance of srfr1‐1 and RLD to nematode infection by H. schachtii, 14‐day‐old Arabidopsis seedlings (20–33 seedlings of each genotype per replicate) were inoculated with approximately 200 J2s per plant, and the number of fourth‐stage infective juveniles (J4s) and adult female nematodes were determined at 14 and 30 days post‐inoculation. The results showed that srfr1‐1 was 33%–55% more resistant than RLD and that resistance had occurred by day 14 (Fig. 2). We next determined whether the smaller number of nematodes per srfr1‐1 plant was a result of decreased penetration. The number of nematodes that had penetrated the roots at 3 days post‐inoculation was similar for both genotypes (Fig. S2, see Supporting Information), indicating that the difference between RLD and srfr1‐1 resistance to nematode infection most probably results from an altered defence response in srfr1‐1.
Figure 2.
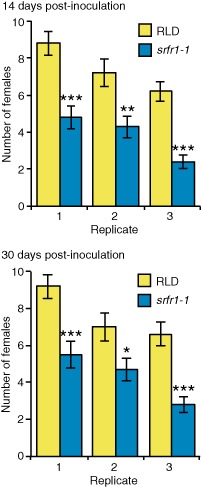
srfr1‐1 is more resistant than RLD to H eterodera schachtii infection. Number of fourth‐stage infective juvenile (J4) and adult females per plant at 14 days (top) and 30 days (bottom) post‐inoculation. Error bars represent standard error, and the averages shown represent 20–33 plants for each genotype per replicate. Asterisks indicate significant differences determined by Student's t‐test (*P < 0.05, **P < 0.005, ***P < 0.0005).
One explanation for the greater resistance of srfr1‐1 relative to RLD in our insect feeding and nematode infection experiments and the previously reported constitutively upregulated PR1, PR2 and PDF1.2 (PLANT DEFENSIN1.2) defence gene expression in srfr1‐1 plants (Kim et al., 2009a) could be higher endogenous levels of JA, JA‐Ile and SA in srfr1‐1 leaves and roots. However, no significant differences in defence hormone levels between RLD and srfr1‐1 were found in untreated shoot or root tissue (Fig. S3, see Supporting Information). Therefore, we reasoned that the greater resistance to S. exigua feeding and H. schachtii infection may result from altered sensitivity to defence hormones in srfr1‐1, which would manifest itself as changes in constitutive or induced SA pathway or JA pathway gene expression levels in srfr1‐1.
Differential induction of SA‐ and JA‐responsive transcripts in srfr1‐1 and RLD leaves
We examined S. exigua‐induced changes in transcript levels of a subset of SA and JA response marker genes (Fig. S4, see Supporting Information) in local and systemic srfr1‐1 and RLD leaves using quantitative real‐time PCR (qRT‐PCR). Because the pressure of the cage on the leaves can cause thigmotropic stimuli that may contribute to elevated gene expression (Rehrig et al., 2014), we also measured transcript levels in mock‐treated leaves on which cages were applied without insects. At 24 h, SALICYLIC ACID INDUCTION DEFICIENT2 (SID2) in the SA pathway and transcript levels of all analysed genes in the JA and JA/ET pathways, except JASMONATE‐ZIM‐DOMAIN PROTEIN1 (JAZ1), showed consistently higher average levels of expression in mock‐treated srfr1‐1 leaves relative to RLD, although not all differences were supported statistically (Fig. S4). Overall, this indicates a generalized constitutive defence or induction by thigmotropic stimuli in srfr1‐1, consistent with the reported constitutive upregulation of SA‐ and JA‐responsive defence genes in srfr1 plants (Kim et al., 2009a).
In size‐matched insect‐damaged leaves at 24 h, there were differences between srfr1‐1 and RLD in terms of SID2 levels and JA marker genes (Fig. S4). Transcript levels of genes in the MYC2 branch [LOX2 (LIPOXYGENASE2), COI1 (CORONATINE INSENSITIVE1), MYC2 and VSP2 (VEGETATIVE STORAGE PROTEIN2)] were higher in srfr1‐1 than in RLD (Fig. S4). In contrast, SID2, OCTADECANOID‐RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59), PDF1.2 and JAZ1 were expressed to a lower level in insect‐damaged srfr1‐1 leaves than in RLD (Fig. S4). We did not observe the co‐regulation of MYC3 with MYC2 in srfr1‐1 plants, and MYC4 expression was very low (Fig. S4). Younger leaves on insect‐eaten plants were also harvested to measure systemically induced gene expression. Induction in srfr1‐1 of the MYC2 branch of the JA pathway was not as comprehensive in systemic leaves as in insect‐damaged leaves: LOX2, COI1 and VSP2 transcript levels were higher than in mock‐treated leaves, whereas MYC2 levels were not (Fig. S4). In systemic leaves of RLD, as in insect‐damaged leaves, the expression of all genes, except MYC4, was higher than in mock‐treated leaves, again suggesting generalized defence responses of RLD as a result of insect herbivory (Fig. S4).
These consistent patterns in expression between RLD and srfr1‐1 responses to herbivory were analysed statistically using Wilcoxon signed rank tests on pair‐wise treatment comparisons of changes in mRNA levels mapped onto a simplified network (Figs 3 and 4; Table S1, see Supporting Information). Comparing transcript levels in RLD insect‐damaged or systemic leaves with those in mock‐treated leaves, the patterns show a statistically significant (α < 0.01) generalized herbivory‐induced upregulation of SA‐, JA‐ and JA/ET‐responsive genes (Fig. 3). In contrast, a comparison of levels of gene expression in insect‐damaged or systemic leaves with mock‐treated srfr1‐1 leaves yielded no statistically significant upregulation over all genes. Instead, the patterns suggest that srfr1‐1 resistance to S. exigua correlates with significant (α = 0.01) herbivory‐induced upregulation specifically of MYC2 branch JA‐responsive genes, leading to VSP2 upregulation (‘MYC2 branch’ genes) in insect‐damaged leaves (Fig. 3). This contrast is more easily discerned when expression levels in different leaf tissues are compared between RLD and srfr1‐1 (Fig. 4). In these analyses, all genes as a group are significantly higher (α < 0.01) in mock‐treated leaves of srfr1‐1 compared with RLD. In contrast, insect‐damaged srfr1‐1 leaves show a significant (α = 0.01) upregulation of MYC2 branch genes, concomitant with suppression of remaining SA‐ and JA/ET‐responsive genes compared with RLD (Fig. 4). In systemic srfr1‐1 leaves, upregulation of MYC2 branch genes is not observed (α > 0.1), whereas the suppression of the remaining genes is still significant (α = 0.02; Fig. 4, Table S1).
Figure 3.
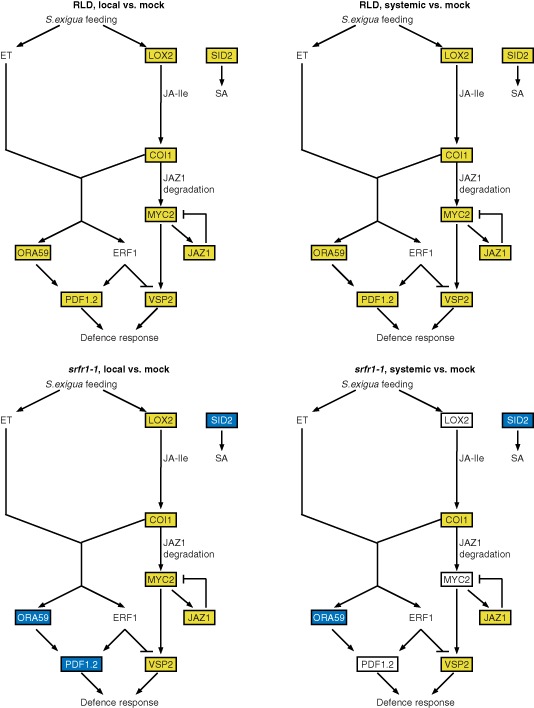
RLD responds to S podoptera exigua feeding with generalized gene induction, whereas, in srfr1‐1, specifically jasmonic acid (JA)‐responsive genes are upregulated. Schematics of simplified JA/ethylene (ET) and salicylic acid (SA) pathways. Marker gene mRNA levels were taken from the data presented in Fig. S4. Genes that were upregulated in both replicates of the indicated pair‐wise comparisons are shown in a yellow box, whereas downregulated genes are shown in a blue box (empty box: no consistent change). See Table S1 for Wilcoxon signed‐rank test statistics.
Figure 4.
srfr1‐1 specifically upregulates the MYC2 branch of the jasmonic acid (JA) response pathway on S podoptera exigua feeding compared with RLD. Schematics of simplified JA/ethylene (ET) and salicylic acid (SA) pathways. Marker gene mRNA levels were taken from the data presented in Fig. S4. Genes that were upregulated in both replicates of the indicated pair‐wise comparisons are shown in a yellow box, whereas downregulated genes are shown in a blue box (empty box: no consistent change). See Table S1 for Wilcoxon signed‐rank test statistics.
Results at later time points suggest that insect feeding‐induced changes in transcript levels are not likely to persist after 24 h. In insect‐damaged and systemic leaves of both genotypes, LOX2 and VSP2 expression was greatly reduced between 24 and 48 h after treatment (Figs S4 and S5, see Supporting Information). Although COI1 continued to be expressed at a level similar to that at 24 h, there was no difference between srfr1‐1 and RLD. Only the PDF1.2 transcript was higher in srfr1‐1 systemic leaves than in RLD at 48 h (Figs S4 and S5).
Thus, as summarized in Figs 3 and 4, the SA, JA and JA/ET defence pathways appear to be primed in srfr1‐1 relative to RLD. In contrast, 24 h after insect feeding, srfr1‐1 showed specifically increased transcript levels in the pathway most likely to be effective against S. exigua, the MYC2 branch of the JA pathway, whereas a generalized induction of defence pathways was seen in RLD.
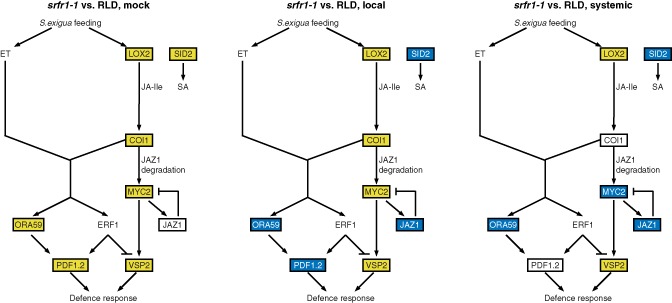
Differential induction of SA‐ and JA‐responsive transcripts in srfr1‐1 and RLD roots and leaves
To evaluate the contribution of the SA and JA signalling pathways to the increased resistance of srfr1‐1 to H. schachtii, transcript levels of SA‐responsive (EDS1, PAD4 and PR1) and JA‐responsive (MYC2 and JAZ1) genes were determined in 14‐day‐old Arabidopsis seedlings at 3 days post‐infection and compared with transcript levels in mock‐treated seedlings. In seedling shoots of mock‐treated srfr1‐1, levels of the SA‐responsive transcripts PAD4 and PR1 were elevated compared with RLD, whereas no differences were evident for the JA‐responsive transcripts (Fig. 5). After nematode infection, levels of SA‐responsive transcripts were reduced in srfr1‐1 seedling shoots and remained low in RLD, suggesting suppression of systemic SA‐mediated defence and resulting in little difference between the two, except for PAD4 (Fig. 5). JA‐responsive transcripts increased in shoots of both RLD and srfr1‐1 (Fig. 5). Thus, srfr1‐1 shoots appeared to be primed in the SA pathway relative to RLD; however, soon after nematode infection, this difference was largely lost.
Figure 5.
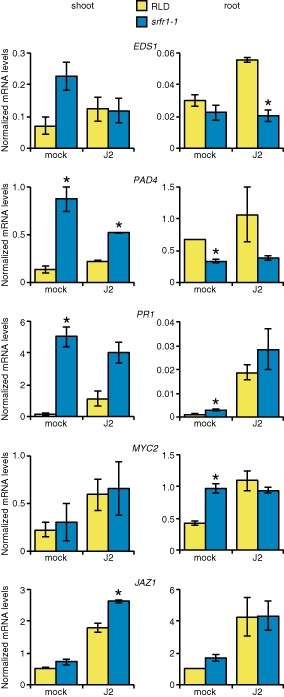
Downregulation of ENHANC EDS 1 in srfr1‐1 roots relative to RLD after H eterodera schachtii infection. Transcript levels of the indicated salicylic acid (SA) and jasmonic acid (JA) pathway genes in shoot (left) and root (right) tissue of RLD and srfr1‐1 at 3 days post‐inoculation of roots with agarose (mock) or H . schachtii (second‐stage juveniles, J2s) as measured by quantitative real‐time PCR (qRT‐PCR) with normalization using SAND gene (At2g28390) mRNA levels as an internal standard. Values represent averages from two biological replicates, with error bars showing the standard error. Asterisks denote significant differences between srfr1‐1 and RLD expression levels in a treatment (mock or J2) as determined by Student's t‐test (P < 0.05).
In roots, we observed that, in both genotypes, the levels of EDS1 and PR1 transcripts were much lower than systemic shoot levels (Fig. 5). Lower PR1 expression in roots than shoots has been reported previously (Seo et al., 2008). In mock‐treated roots, there were no clear patterns in the comparison of mRNA levels of SA‐responsive and JA‐responsive genes in srfr1‐1 and RLD (Fig. 5). The transcript level of PAD4 was higher in RLD, whereas MYC2 and PR1 were higher in srfr1‐1, but there was little difference in EDS1 and JAZ1 transcript levels (Fig. 5). Comparing mock treatment with nematode infection within a genotype suggested that both RLD and srfr1‐1 increased or maintained high levels of MYC2 and JAZ1 transcripts after infection with H. schachtii (Fig. 5), resulting in no difference between srfr1‐1 and RLD after J2 treatment. In the SA pathway, both genotypes increased PR1 transcript levels and maintained PAD4 transcript levels, whereas EDS1 transcript levels increased only in RLD (Fig. 5). Thus, after infection with H. schachtii J2s, the lower EDS1 transcript level in srfr1‐1 roots than in RLD suggests that increased EDS1 transcript levels in roots may be associated with increased susceptibility to nematodes.
To test the significance of this surprising finding, we performed infection assays with the corresponding mutants in the Col‐0 background, which is also susceptible to H. schachtii. Because srfr1 mutants in the Col‐0 background are severely stunted (Kim et al., 2010; Li et al., 2010), they could not be used for cyst nematode infection assays; however, growth of the double mutant eds1‐2 srfr1‐4 is normal (Bhattacharjee et al., 2011). No difference in root morphology was observed in all tested genotypes. Both eds1‐2 and eds1‐2 srfr1‐4 showed significantly enhanced resistance relative to wild‐type Col‐0 at 14 and 30 days post‐inoculation in two of three replications (Fig. 6), confirming that a functional EDS1 gene correlates with increased susceptibility. The enhanced resistance of eds1‐2 srfr1‐4 and eds1‐2 was not significantly different, indicating that srfr1‐4 and eds1‐2 do not have additive effects. In another set of experiments that included the pad4‐1 mutant, enhanced resistance was found in all tested eds1‐2 single and double mutants relative to wild‐type Col‐0 (Fig. 7), again indicating that functional EDS1 promotes parasitism by H. schachtii. The absence of PAD4 did not enhance resistance in eds1‐2 pad4‐1 double mutants relative to eds1‐2, and the susceptibility of pad4‐1 to H. schachtii was not statistically significantly different from that of Col‐0 (Fig. 7). This suggests that PAD4 does not play a role in susceptibility or resistance to this nematode. Surprisingly, therefore, within these tested genotypes, maximal susceptibility was observed when plants possessed both a functional SRFR1 and EDS1 gene.
Figure 6.
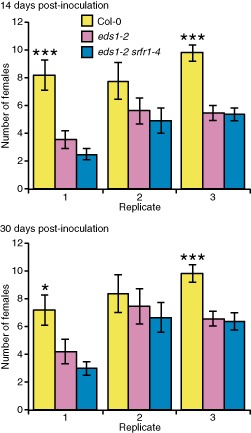
eds1‐2 plants are more resistant than Col‐0 plants to nematode infection. The number of fourth‐stage juvenile (J4) and adult females per plant at 14 days (top) and 30 days (bottom) post‐inoculation obtained from 22–35 samples per genotype. Error bars represent the standard error. Asterisks indicate a significant difference determined by Student's t‐test (*P < 0.05, ***P < 0.0005).
Figure 7.
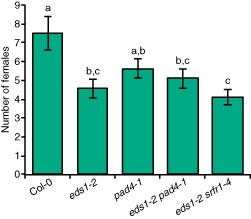
EDS 1 and SRFR 1, but not PAD 4, contribute to nematode susceptibility. The number of fourth‐stage juvenile (J4) and adult females per plant at 30 days post‐inoculation obtained from 20–30 samples per genotype. Error bars represent standard error, with different letters denoting significant differences (P < 0.05) as determined by Student's t‐test. This experiment was repeated once with similar results.
Discussion
Here, we have shown that mutations in SRFR1 lead to reduced leaf herbivory by S. exigua and to reduced development of H. schachtii. We have also shown that there were marked differences in leaf and root transcript level patterns between genotypes and in the responses to insect feeding and nematode parasitism. Insect‐damaged srfr1‐1 leaves 24 h after feeding showed specifically increased JA‐responsive and concomitantly suppressed SA‐ and JA/ET‐responsive transcript levels. The importance of the JA‐mediated response to wounding and herbivory has been well documented in transcriptome analyses and in studies with mutants impaired in the perception or synthesis of JA (Reymond et al., 2004; Song et al., 2014; Verhage et al., 2011; Wu and Baldwin, 2010). Consistent with our gene expression results, in a recent feeding study, Pieris rapae and S. exigua larvae gained most weight when feeding on myc2 myc3 myc4 mutants, intermediate weight on Col‐0 and least weight on ein3 eil1 mutants (Song et al., 2014). Correspondingly, the application of methyl jasmonate resulted in reduced levels of VSP2 transcripts in myc2 myc3 myc4 plants relative to Col‐0, but increased levels in ein3 eil1 plants.
In contrast with srfr1‐1, the generalized increase in all analysed defence marker transcripts in RLD and the pronounced increase in the JAZ1 transcript, which can inhibit JA pathway induction, correlated with greater leaf damage by S. exigua. In a recent report, MYC2, ORA59, PDF1.2, PR3 and PR4 transcript levels were elevated in Col‐0 local leaves 6 h after S. exigua feeding, suggesting a similar generalized upregulated defence in another susceptible accession (Rehrig et al., 2014). In addition, JA, JA‐Ile and ET were increased by 30 min and remained higher 6 h after initiation of S. exigua feeding, confirming that the larvae activated both JA and ET pathways. In that study, transcript levels had decreased by 24 h, except for PR3, perhaps because the feeding time was shorter and there was less damage. Nevertheless, transcript levels of 23 transcription factors in 10 classes were elevated in the local leaf 24 h after feeding (Rehrig et al., 2014).
Although SRFR1 has a well‐described function in effector‐triggered immunity as a negative regulator of TNL‐mediated EDS1‐dependent resistance (Gassmann and Bhattacharjee, 2012), we consider it unlikely that alteration of this sector of the defence network can explain the increased resistance to insect feeding displayed by srfr1‐1. Instead, srfr1‐1 leaves appear to respond more strongly in either the JA or SA pathway, depending on the stimulus, placing SRFR1 as a negative regulator at a cross‐point between these two pathways. We propose that the recently described interaction of SRFR1 with class I TCP transcription factors (Kim et al., 2014) may form the basis for these observations. SRFR1 interacts most strongly with TCP8, TCP14 and TCP15, and less strongly with TCP20, TCP22 and TCP23. Because a tcp8 tcp14 tcp15 triple mutant was compromised in effector‐triggered immunity to DC3000 expressing the effectors AvrRps4, HopA1, AvrRpm1 or AvrRpt2, it appears that these TCPs positively regulate general defence genes active against DC3000, and that SRFR1 functions as a co‐repressor by interacting with TCPs (Bhattacharjee et al., 2013; Kim et al., 2014).
Interestingly, the class I TCPs TCP20 and TCP9 have been reported recently to negatively regulate LOX2 expression in developing leaves, whereas the class II TCP TCP4 induces LOX2 (Danisman et al., 2012; Schommer et al., 2008). If this repressive function depends on SRFR1 as a co‐repressor, one would predict an upregulation of LOX2 in srfr1 mutants. This upregulation may be partly constitutive, but mainly more pronounced after a stimulus, which matches our observations. A connection of TCPs to insect defences would underscore the emerging importance of TCPs in plant biotic stress responses to a wide range of organisms (Kim et al., 2014; Mukhtar et al., 2011; Sugio et al., 2011; Weßling et al., 2014). Ultimately, induction of LOX2 and the JA pathway would result in the accumulation of VSP2 (Berger et al., 2002; Liu et al., 2005; Reymond et al., 2004) and probably other insect deterrents, such as glucosinolates (Mewis et al., 2005; Rehrig et al., 2014; Schweizer et al., 2013; Textor and Gershenzon, 2009). Alternatively, SRFR1's role in herbivory responses may depend on its interaction with the receptor co‐chaperone constituent SGT1b (Li et al., 2010), which, together with heat shock proteins HSP90 and HSP70, stabilizes the JA receptor COI1 (Zhang et al., 2015). Consistent with this, SGT1 in Nicotiana attenuata has been shown to positively regulate JA accumulation and resistance to Manduca sexta, a specialist herbivore (Meldau et al., 2011).
Comparing the results obtained from leaf bacteria and insects with those from nematodes feeding on RLD and srfr1‐1 indicates that defence signalling pathways in shoots and roots differ. Because H. schachtii parasitism was inhibited by SA treatment of wild‐type Col‐0, was increased in SA‐deficient NahG‐expressing plants and the sid2‐1 mutant (Wubben et al., 2008), and was reduced by overexpression of PR1 (Hamamouch et al., 2011), we expected increased parasitism in eds1‐2 plants. However, in our experiments, increased levels of EDS1 transcript in RLD relative to srfr1‐1 suggested that induction of the SA pathway correlated with susceptibility. Mutation of either SRFR1 or EDS1 led to similar levels of resistance, and the simultaneous presence of EDS1 and SRFR1 appeared to correlate with increased susceptibility to nematode parasitism, suggesting a novel form of resistance to nematodes or a different regulation of the SA pathway by EDS1 in roots.
It is possible that EDS1 constitutes an important virulence target for a nematode effector. Because the eds1‐2 mutant was more resistant to H. schachtii, the function of such an effector would probably not consist of the blocking or degradation of EDS1, but rather of the hyperactivation of EDS1's negative effect on root nematode resistance. In this interaction, EDS1 in roots does not appear to occupy as central and positive a role in plant resistance and SA pathway regulation as in shoots. It was shown that SA is required for Mi‐1‐mediated resistance to root‐knot nematodes; in contrast, a functional JA pathway was required for nematode parasitism, perhaps via antagonism of the SA pathway (Bhattarai et al., 2008). Future research will therefore need to examine whether, in Arabidopsis roots, the SA pathway is still fully active against H. schachtii in the absence of EDS1.
Heterodera effectors that impact plant processes, such as signalling, defence and cell wall remodelling, are beginning to be identified (Hewezi and Baum, 2013; Mitchum et al., 2013). Nematode infection is a complex process requiring cell cycle alteration, reprogramming of root cells that form the feeding site, and altered plant metabolism. A time course study of infection between 3 days post‐infection, when penetration was assayed, and day 14, when resistance was observed, could narrow down when the responses in srfr1‐1 and eds1‐2 roots begin to differ from those in wild‐type plants and uncover the affected processes. In addition, further studies need to determine whether full susceptibility requires EDS1 and SRFR1 expression locally (in the developing syncytium) or in the surrounding root tissue.
In summary, the results here expand on and provide first insights into the heightened resistance of srfr1‐1 to a wide spectrum of biotic stresses, and demonstrate the value of using multiple stresses to better understand gene function. A more complete analysis of gene expression and defence hormone level changes in response to Pseudomonas syringae, S. exigua and H. schachtii will provide insights into the molecular mechanism of system‐wide defence regulation by SRFR1. In addition, the effects of mutation of SRFR1 on the yield and resistance of food crops need to be tested to determine whether decreasing expression of this regulator could be an asset in disease and pest control.
Experimental Procedures
Insect bioassays and plant growth
Eggs of S. exigua were obtained from Benzon Research (Carlisle, PA, USA) and larvae reared on artificial diet (Bioserv, Frenchtown, NJ, USA). First instar caterpillars were transferred to RLD and srfr1‐1 plants 1 day before the experiments for acclimation as described by Rehrig et al. (2014). All seeds were germinated in half‐strength Murashige and Skoog (MS) medium containing 1% agar and sown into 6 × 5‐cm2 pots containing sterile Metromix 200 soil (Sun Gro Horticulture Ltd., Agawam, MA, USA) and grown in chambers at 22 °C, 65% ± 5% relative humidity and light intensity of 200 μmol/m2/s with a short‐day photoperiod (8 h light : 16 h dark), as described by Rehrig et al. (2014). Each of three bioreplications consisted of 30 4–5‐week‐old plants of each genotype and 10 mock‐treated plants of each genotype.
To estimate plant performance in response to herbivore feeding, no‐choice assays and two‐choice assays were conducted. In the no‐choice assay, RLD and srfr1‐1 were grown in separate pots, and two first‐instar larvae were fed for 1 week on each plant. The plant rosettes were caged by transparent Mylar cylinders (diameter, 5 cm; height, 9 cm) with fine mesh gauze tops (<0.01‐mm mesh) to maintain air exchange. In the two‐choice assay, both RLD and srfr1‐1 were grown in the same pot caged by a similar cylinder, and one second‐instar caterpillar was placed between the two plants for free selection during a 16‐h feeding period. Camel hair brushes were used to catch and move the caterpillars. Mock treatments were caged plants without a caterpillar, and all plants were returned to the growth chamber during the caterpillar feeding time.
To investigate early gene expression events, one third‐instar caterpillar was forced to feed on a single seventh fully expanded mature leaf that had half its area caged by a clip with fine mesh gauze (<0.01‐mm mesh). The caterpillar was allowed to feed for 3–5 h until 40%–50% of the leaf area was removed. Cages without insects were placed on size‐matched leaves on mock‐treated plants. Once sufficient damage was achieved, the caterpillar was removed and the plant was returned to the growth chamber. Caterpillar‐damaged leaves and two systemic leaves, younger leaves on both sides of the damaged leaf, were harvested for gene expression analysis at 24 and 48 h after feeding. Caged leaves on mock‐treated plants were harvested at the same time.
Quantification of eaten leaf area and caterpillar weight measurement
Photographs of RLD and srfr1‐1 leaves before feeding (0 days) and after feeding (7 days) were taken with a 10‐megapixel Canon (Melville, NY, USA) Rebel digital camera in a customized stand. They were analysed with a computer algorithm that automatically corrects image size, setting the green areas to 10 000 pixels/cm2, and then masks the leaf images to calculate the pixels of leaf tissues (Green et al., 2012). Untreated plants were used to determine the increases in leaf area resulting from growth, and the growth rates of each genotype were factored into calculations of the final area of leaf tissue removed by a caterpillar.
The caterpillar weight at the beginning of a feeding assay was assumed to be zero. After feeding, caterpillars in the no‐choice assay were weighed with an analytical balance (Mettler Toledo, LLC, OH, USA) and the average growth during a 1‐week feeding period was calculated. The 16‐h feeding period in the choice assay was too short for meaningful measurement of caterpillar growth.
Plant material and growth conditions for nematode assays
All Arabidopsis plants were grown from surface‐sterilized seed planted on modified Knops nutrient medium, pH 6.4, with 0.8% Daishin agar (Brunschwig Chemie, Amsterdam, the Netherlands) (Sijmons et al., 1991) on 12‐well plates or 9‐cm2 vertical plates (Falcon Brand; BD, Franklin Lakes, NJ, USA) in a growth chamber under 16‐h light : 8‐h dark at 25 °C (Replogle et al., 2013). Using eds1‐2 plants as a recipient, eds1‐2 srfr1‐4 and eds1‐2 pad4‐1 double knockout plants were generated and genotyped by polymerase chain reaction (PCR) with the primers, as described previously (Bartsch et al., 2006; Kim et al., 2010; Ng et al., 2011).
Cyst nematode infection assays
Heterodera schachtii eggs were collected and surface sterilized in 0.02% sodium azide for 20 min, washed with tap water and set up to hatch at 27 °C on a modified Baermann pan for 2–3 days in antibiotic solution containing 1.5 μg/mL gentamicin and 0.05 μg/mL nystatin. Freshly hatched infective J2s were collected and surface sterilized for 8 min in solution containing 0.04% mercuric chloride, 0.04% sodium azide and 0.002% Triton X‐100, washed four times in sterile water, resuspended in sterile 0.1% agarose with rotation for 60 min and diluted to a concentration of approximately 200 J2s/25 μL resuspended in sterile 0.1% agarose (Replogle et al., 2013). Each 14‐day‐old Arabidopsis seedling was inoculated with 25 μL of this J2 suspension. The numbers of J4 and adult females that developed on each plant by 14 and 30 days post‐inoculation were counted under a dissecting microscope. The experimental design for the assessment of mutant susceptibility to H. schachtii employed randomly arranged mutant and wild‐type seeds within 12‐well plates, with one seed per well. Each individual mutant was analysed in at least three independent experiments with a minimum of 20 plants per experiment. Data were analysed using Student's t‐test.
Penetration assay
Fourteen‐day‐old RLD and srfr1‐1 plants grown on 12‐well plates were each inoculated with 25 μL of agarose containing approximately 200 J2s. Acid fuchsin staining was performed at 3 days post‐inoculation as described by Ithal et al. (2007) with some modifications. The whole intact root in each well was gently separated from agarose medium, and transferred to a new 12‐well plate with water to wash out agarose. The roots in each well were incubated in 10% bleach for 7 min, followed by 20‐min incubation in tap water. Fifty millilitres of water were brought to the boil in a glass beaker to which 1 mL of acid fuchsin solution (0.35 g acid fuchsin in 25 mL of acetic acid) was added. Three millilitres of boiled acid solution were added to each root twice, 2 min apart. The roots were then incubated in acid fuchsin for 7–10 min. Root segments were rinsed with water and stored in 1.5‐mL Eppendorf tubes containing 100% ethanol. Before observations under a dissecting microscope, the roots were briefly rinsed in water and placed on a slide containing 50% glycerine. Photographs were taken using an Olympus (Center Valley, PA, USA) camera.
RNA extraction and qRT‐PCR
For insects, the transcript abundance of investigated genes was analysed by qRT‐PCR performed on RNA samples that were generated from Arabidopsis leaf tissue with or without S. exigua treatments. For nematodes, plants were grown on vertical plates containing modified Knop's medium, with about 10 plants per plate. Each 8‐day‐old seedling was inoculated with 25 μL of agarose containing about 200 J2s for infected samples or no J2s for mock‐treated samples. At 3 days post‐inoculation, root and shoot tissues from a minimum of 10 plants per genotype were collected.
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and treated with Turbo DNase (Ambion, Austin, TX, USA). For qRT‐PCR, first‐strand cDNA was synthesized from 2 μg of total RNA using an oligo(dT)15 primer and Moloney murine leukaemia virus (MMLV) reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer's instructions. qRT‐PCR was performed using an ABI 7500 system and a SYBR Green qPCR Master Mix (Applied Biosystems, Warrington, UK). For insect experiments, a reaction volume of 20 μL was used with the following cycling conditions: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 59 °C for 1 min. For nematode experiments, a reaction volume of 10 μL was used with the following cycling conditions: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 65 °C for 1 min, followed by a dissociation step of 95 °C for 15 s. For a given cDNA sample, reactions were run in duplicate or triplicate (technical replicates), each for two biological replicates.
Primers used in qRT‐PCR experiments are listed in Table S2 (see Supporting Information). Transcript levels of a target gene were normalized using the SAND gene (Czechowski et al., 2005). This gene showed no significant transcript level differences between mock‐treated and wounded plants (Rehrig et al., 2011). In the present experiments, the cycle threshold (Ct) values for SAND were similar between mock‐treated and nematode‐infected plants in roots and shoots. Quantitative RT‐PCR data were analysed as described previously (Kwon et al., 2009). PCR efficiency was calculated using linear regression in the LinRegPCR program (Ramakers et al., 2003), and was determined to be >1.68 for all samples with R 2 > 0.9975. The Ct value for SAND was subtracted from the Ct value for target genes. Expression values (Exp) for target genes were calculated as: Exp = (PCR efficiency)−ΔCt.
For insect experiments, Wilcoxon signed‐rank tests (Sokal and Rohlf, 1995) were performed on groups of genes to analyse the statistical significance of differences in pathway regulations between two samples. For example, to compare expression levels in RLD insect‐damaged leaves with those in mock‐treated leaves, the ratio of expression values (RLD local/RLD mock) of a given gene from a given biological replicate was determined and log2‐transformed. Absolute log2‐transformed values of groups of genes from both biological replicates of the same treatment were ranked from lowest to highest values. The test statistics W+ and W − were calculated by adding rank values with positive and negative signs, respectively, and compared with a table of critical W values. For nematode experiments, differences in expression levels between RLD and srfr1‐1 were analysed using Student's t‐test, with P < 0.05 used as a threshold for statistical significance.
JA, JA‐Ile and SA quantification
SA, JA and JA‐Ile levels were measured in untreated leaf and root tissues of RLD and srfr1‐1. Leaf tissue was harvested from plants grown as described for insect feeding experiments, and root tissue from seedlings as grown for nematode infection assays. SA, JA and JA‐Ile were extracted and measured by ultra performance liquid chromatography‐electrospray ionization‐tandem mass spectrometry (UPLC‐ESI‐MS/MS) as described previously (Chung et al., 2008; Koo et al., 2009; Smith et al., 2014).
Statistical analyses
All statistical analyses were performed using the program Excel (Microsoft, Redmond, WA, USA).
Accession numbers
COI1 (At2g39940), EDS1 (At3g48090), JAZ1 (At1g19180), LOX2 (At3g45140), MYC2 (At1g32640), MYC3 (At5g46760), ORA59 (At1g06160), PAD4 (At3g52430), PDF1.2 (At5g44420), PR1 (At2g14610), SID2 (At1g74710), SRFR1 (At4g37460), VSP2 (At5g24770).
Supporting information
Fig. S1 Spodoptera exigua larvae tend to prefer RLD over srfr1‐1 in a two‐choice assay. Single RLD and srfr1‐1 plants were grown in the same pot and S. exigua was allowed to feed for 16 h when plants were 3 weeks old. The digitally quantified leaf area eaten (%) was determined for each plant, and damage on srfr1‐1 was subtracted from that on RLD for each pair. Positive values indicate more damage on RLD, and negative values indicate more damage on srfr1‐1. Differences were statistically significant (P < 0.01) by the Wilcoxon signed rank test in two of four replicates.
Fig. S2 Heterodera schachtii penetration assay in RLD and srfr1‐1 roots. A minimum of 20 infected roots of 14‐day‐old seedlings in 12‐well plates were stained with acid fuchsin at 3 days post‐inoculation. No statistically significant difference between RLD and srfr1‐1 was detected at P < 0.05 as analysed by Student's t‐test.
Fig. S3 Endogenous defence hormone levels in RLD and srfr1‐1 do not differ. Total resting‐state salicylic acid (SA) (top), jasmonic acid (JA) (middle) and JA‐Ile (JA conjugated with the amino acid isoleucine) (bottom) levels in leaf tissue of RLD and mutant srfr1‐1 soil‐grown plants (left) or roots of plate‐grown seedlings (right) were quantified by ultra performance liquid chromatography‐electrospray ionization‐tandem mass spectrometry (UPLC‐ESI‐MS/MS). Tissue was harvested at the same stage as used for insect feeding and nematode infection experiments, respectively. Error bars represent standard errors of three to four samples. There were no significant differences between RLD and srfr1‐1 as determined by Student's t‐test (P > 0.05). Similar results were found in a second experiment.
Fig. S4 Differences in jasmonic acid/ethylene (JA/ET) and salicylic acid (SA) pathway gene regulation in srfr1‐1 and RLD in response to Spodoptera exigua feeding. mRNA levels of the indicated SA and JA/ET pathway genes in Arabidopsis leaf tissue were determined 24 h after S. exigua feeding in mock‐treated (mock), insect‐damaged (local) and systemic (systemic) leaves. Expression levels were quantified by quantitative real‐time PCR (qRT‐PCR) and normalized using SAND gene (At2g28390) mRNA levels as an internal standard. Values represent averages from two biological replicates, with error bars showing the standard error.
Fig. S5 Dampened mRNA level changes 48 h after S. exigua feeding. mRNA levels of the indicated JA/ET pathway genes in Arabidopsis leaf tissue were determined in mock‐treated (mock), insect‐damaged (local) and systemic (systemic) leaves. Expression levels were quantified by quantitative real‐time PCR (qRT‐PCR) and normalized using SAND gene (At2g28390) mRNA levels as an internal standard. Values represent averages from two biological replicates, with error bars showing the standard error. For ease of comparison, the same scales were used for the genes shown in both Figs 3 and 4.
Table S1 W test statistic values for Wilcoxon signed‐rank tests on the data in Figs 3 and 4.
Table S2 Forward (F) and reverse (R) qRT‐PCR primers.
Acknowledgements
We thank Dean E. Bergstrom, Sree D. Gopaluni, Caitlin Vore and Abigail Ferrieri (Schultz/Appel Laboratory, University of Missouri, Columbia, MO, USA) for technical assistance with insect experiments. We thank Dr Jeanne D. Mihail (University of Missouri, Columbia, MO, USA) for advice on statistical analyses, Dr Jane E. Parker (Max Planck Institute, Köln, Germany) for providing the eds1‐2 line, and Dr Jane Glazebrook (University of Minnesota, St Paul, MN, USA) for pad4‐1. Initial data for this manuscript were obtained by PDTN in a class project for PLNT_S 8530, Research with Plant Stress Agents, supervised by MGM and HMA. This work was supported by graduate fellowships from the Vietnam Overseas Scholarship Program and the Daniel F. Millikan Graduate Endowment, Division of Plant Sciences (to PDTN). The authors have no conflicts of interest to declare.
The copyright line for this article was changed on 7 April 2016 after original online publication.
References
- Bartsch, M. , Gobbato, E. , Bednarek, P. , Debey, S. , Schultze, J.L. , Bautor, J. and Parker, J.E. (2006) Salicylic acid‐independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell, 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S. , Mitchell‐Olds, T. and Stotz, H.U. (2002) Local and differential control of vegetative storage protein expression in response to herbivore damage in Arabidopsis thaliana . Physiol. Plant. 114, 85–91. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. , Halane, M.K. , Kim, S.H. and Gassmann, W. (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science, 334, 1405–1408. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. , Garner, C.M. and Gassmann, W. (2013) New clues in the nucleus: transcriptional reprogramming in effector‐triggered immunity. Front. Plant Sci. 4, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai, K.K. , Xie, Q.G. , Mantelin, S. , Bishnoi, U. , Girke, T. , Navarre, D.A. and Kaloshian, I. (2008) Tomato susceptibility to root‐knot nematodes requires an intact jasmonic acid signaling pathway. Mol. Plant–Microbe Interact. 21, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Birker, D. , Heidrich, K. , Takahara, H. , Narusaka, M. , Deslandes, L. , Narusaka, Y. , Reymond, M. , Parker, J.E. and O'Connell, R. (2009) A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 60, 602–613. [DOI] [PubMed] [Google Scholar]
- Bonaventure, G. , VanDoorn, A. and Baldwin, I.T. (2011) Herbivore‐associated elicitors: FAC signaling and metabolism. Trends Plant Sci. 16, 294–299. [DOI] [PubMed] [Google Scholar]
- Chung, H.S. , Koo, A.J. , Gao, X. , Jayanty, S. , Thines, B. , Jones, A.D. and Howe, G.A. (2008) Regulation and function of Arabidopsis JASMONATE ZIM‐domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, D.E. , Lee, T.G. , Guo, X. , Melito, S. , Wang, K. , Bayless, A.M. , Wang, J. , Hughes, T.J. , Willis, D.K. , Clemente, T.E. , Diers, B.W. , Jiang, J. , Hudson, M.E. and Bent, A.F. (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science, 338, 1206–1209. [DOI] [PubMed] [Google Scholar]
- Cooper, W.R. , Jia, L. and Goggin, L. (2005) Effects of jasmonate‐induced defenses on root‐knot nematode infection of resistant and susceptible tomato cultivars. J. Chem. Ecol. 31, 1953–1967. [DOI] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. , van der Wal, F. , Dhondt, S. , Waites, R. , de Folter, S. , Bimbo, A. , van Dijk, A.D. , Muino, J.M. , Cutri, L. , Dornelas, M.C. , Angenent, G.C. and Immink, R.G. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezel, C. , von Dahl, C.C. , Gaquerel, E. and Baldwin, I.T. (2009) Different lepidopteran elicitors account for cross‐talk in herbivory‐induced phytohormone signaling. Plant Physiol. 150, 1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, K. and Rowe, J. (1998) Distribution and economic importance In: The Cyst Nematodes (Sharma S.B., ed.), pp. 1–30, Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Feys, B.J. , Moisan, L.J. , Newman, M.A. and Parker, J.E. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann, W. and Bhattacharjee, S. (2012) Effector‐triggered immunity signaling: from gene‐for‐gene pathways to protein–protein interaction networks. Mol. Plant–Microbe Interact. 25, 862–868. [DOI] [PubMed] [Google Scholar]
- Gassmann, W. , Hinsch, M.E. and Staskawicz, B.J. (1999) The Arabidopsis RPS4 bacterial‐resistance gene is a member of the TIR‐NBS‐LRR family of disease‐resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Green, J.M. , Appel, H. , Rehrig, E.M. , Harnsomburana, J. , Chang, J.F. , Balint‐Kurti, P. and Shyu, C.R. (2012) PhenoPhyte: a flexible affordable method to quantify 2D phenotypes from imagery. Plant Methods, 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamouch, N. , Li, C. , Seo, P.J. , Park, C.M. and Davis, E.L. (2011) Expression of Arabidopsis pathogenesis‐related genes during nematode infection. Mol. Plant Pathol. 12, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich, K. , Wirthmueller, L. , Tasset, C. , Pouzet, C. , Deslandes, L. and Parker, J.E. (2011) Arabidopsis EDS1 connects pathogen effector recognition to cell compartment‐specific immune responses. Science, 334, 1401–1404. [DOI] [PubMed] [Google Scholar]
- Hewezi, T. and Baum, T.J. (2013) Manipulation of plant cells by cyst and root‐knot nematode effectors. Mol. Plant–Microbe Interact. 26, 9–16. [DOI] [PubMed] [Google Scholar]
- Hinsch, M. and Staskawicz, B.J. (1996) Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi . Mol. Plant–Microbe Interact. 9, 55–61. [DOI] [PubMed] [Google Scholar]
- Ithal, N. , Recknor, J. , Nettleton, D. , Hearne, L. , Maier, T. , Baum, T.J. and Mitchum, M.G. (2007) Parallel genome‐wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Mol. Plant–Microbe Interact. 20, 293–305. [DOI] [PubMed] [Google Scholar]
- Kandoth, P.K. and Mitchum, M.G. (2013) War of the worms: how plants fight underground attacks. Curr. Opin. Plant Biol. 16, 457–463. [DOI] [PubMed] [Google Scholar]
- Kim, S.H. , Kwon, S.I. , Bhattacharjee, S. and Gassmann, W. (2009a) Regulation of defense gene expression by Arabidopsis SRFR1 . Plant Signal. Behav. 4, 149–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , Kwon, S.I. , Saha, D. , Anyanwu, N.C. and Gassmann, W. (2009b) Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR‐NBS‐LRR protein RPS6 and is enhanced by mutations in SRFR1 . Plant Physiol. 150, 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , Gao, F. , Bhattacharjee, S. , Adiasor, J.A. , Nam, J.C. and Gassmann, W. (2010) The Arabidopsis resistance‐like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 6, e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , Son, G.H. , Bhattacharjee, S. , Kim, H.J. , Nam, J.C. , Nguyen, P.D.T. , Hong, J.C. and Gassmann, W. (2014) The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector‐triggered immunity. Plant J. 78, 978–989. [DOI] [PubMed] [Google Scholar]
- Koo, A.J. , Gao, X. , Jones, A.D. and Howe, G.A. (2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59, 974–986. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Kwon, S.I. , Kim, S.H. , Bhattacharjee, S. , Noh, J.J. and Gassmann, W. (2009) SRFR1, a suppressor of effector‐triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J. 57, 109–119. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Li, S. , Bi, D. , Cheng, Y.‐T. , Li, X. and Zhang, Y. (2010) SRFR1 negatively regulates plant NB‐LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 6, e1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kandoth, P.K. , Warren, S.D. , Yeckel, G. , Heinz, R. , Alden, J. , Yang, C. , Jamai, A. , El‐Mellouki, T. , Juvale, P.S. , Hill, J. , Baum, T.J. , Cianzio, S. , Whitham, S.A. , Korkin, D. , Mitchum, M.G. and Meksem, K. (2012) A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature, 492, 256–260. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Ahn, J.E. , Datta, S. , Salzman, R.A. , Moon, J. , Huyghues‐Despointes, B. , Pittendrigh, B. , Murdock, L.L. , Koiwa, H. and Zhu‐Salzman, K. (2005) Arabidopsis vegetative storage protein is an anti‐insect acid phosphatase. Plant Physiol. 139, 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldau, S. , Baldwin, I.T. and Wu, J. (2011) SGT1 regulates wounding‐ and herbivory‐induced jasmonic acid accumulation and Nicotiana attenuate's resistance to the specialist lepidopteran herbivore Manduca sexta . New Phytol. 189, 1143–1156. [DOI] [PubMed] [Google Scholar]
- Mewis, I. , Appel, H.M. , Hom, A. , Raina, R. and Schultz, J.C. (2005) Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem‐feeding and chewing insects. Plant Physiol. 138, 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum, M.G. , Hussey, R.S. , Baum, T.J. , Wang, X. , Elling, A.A. , Wubben, M. and Davis, E.L. (2013) Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 199, 879–894. [DOI] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. , Tasan, M. , Galli, M. , Hao, T. , Nishimura, M.T. , Pevzner, S.J. , Donovan, S.E. , Ghamsari, L. , Santhanam, B. , Romero, V. , Poulin, M.M. , Gebreab, F. , Gutierrez, B.J. , Tam, S. , Monachello, D. , Boxem, M. , Harbort, C.J. , McDonald, N. , Gai, L. , Chen, H. , He, Y. , Vandenhaute, J. , Roth, F.P. , Hill, D.E. , Ecker, J.R. , Vidal, M. , Beynon, J. , Braun, P. and Dangl, J.L. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Atzorn, R. , Miersch, O. and Wasternack, C. (2006) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar, K. , Kyndt, T. , De Vleesschauwer, D. , Höfte, M. and Gheysen, G. (2011) The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 157, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka, M. , Shirasu, K. , Noutoshi, Y. , Kubo, Y. , Shiraishi, T. , Iwabuchi, M. and Narusaka, Y. (2009) RRS1 and RPS4 provide a dual Resistance‐gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. [DOI] [PubMed] [Google Scholar]
- Narusaka, M. , Kubo, Y. , Hatakeyama, K. , Imamura, J. , Ezura, H. , Nanasato, Y. , Tabei, Y. , Takano, Y. , Shirasu, K. and Narusaka, Y. (2013) Interfamily transfer of dual NB‐LRR genes confers resistance to multiple pathogens. PLoS ONE, 8, e55954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, G. , Seabolt, S. , Zhang, C. , Salimian, S. , Watkins, T.A. and Lu, H. (2011) Genetic dissection of salicylic acid‐mediated defense signaling networks in Arabidopsis . Genetics, 189, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Ramakers, C. , Ruijter, J.M. , Deprez, R.H.L. and Moorman, A.F.M. (2003) Assumption‐free analysis of quantitative real‐time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Rehrig, E.M. , Appel, H.M. and Schultz, J.C. (2011) Measuring ‘normalcy’ in plant gene expression after herbivore attack. Mol. Ecol. Resour. 11, 294–304. [DOI] [PubMed] [Google Scholar]
- Rehrig, E.M. , Appel, H.M. , Jones, A.D. and Schultz, J.C. (2014) Roles for jasmonate‐ and ethylene‐induced transcription factors in the ability of Arabidopsis to respond differentially to damage caused by two insect herbivores. Front. Plant Sci. 5, 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle, A. , Wang, J. , Paolillo, V. , Smeda, J. , Kinoshita, A. , Durbak, A. , Tax, F.E. , Wang, X. , Sawa, S. and Mitchum, M.G. (2013) Synergistic interaction of CLAVATA1, CLAVATA2, and RECEPTOR‐LIKE PROTEIN KINASE 2 in cyst nematode parasitism of Arabidopsis . Mol. Plant–Microbe Interact. 26, 87–96. [DOI] [PubMed] [Google Scholar]
- Reymond, P. , Bodenhausen, N. , Van Poecke, R.M. , Krishnamurthy, V. , Dicke, M. and Farmer, E.E. (2004) A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell, 16, 3132–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz, S. , Stamm, A. , Malonek, S. , Wagner, S. , Becker, D. , Medina‐Escobar, N. , Corina Vlot, A. , Feys, B.J. , Niefind, K. and Parker, J.E. (2011) Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119. [DOI] [PubMed] [Google Scholar]
- Sarris, P.F. , Duxbury, Z. , Huh, S.U. , Ma, Y. , Segonzac, C. , Sklenar, J. , Derbyshire, P. , Cevik, V. , Rallapalli, G. , Saucet, S.B. , Wirthmueller, L. , Menke, F.L. , Sohn, K.H. and Jones, J.D. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell, 161, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M. , Kazan, K. , Wilson, I. , Anderson, J.P. , Richmond, T. , Somerville, S.C. and Manners, J.M. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA, 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer, C. , Palatnik, J.F. , Aggarwal, P. , Chételat, A. , Cubas, P. , Farmer, E.E. , Nath, U. and Weigel, D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6, e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, F. , Fernández‐Calvo, P. , Zander, M. , Diez‐Diaz, M. , Fonseca, S. , Glauser, G. , Lewsey, M.G. , Ecker, J.R. , Solano, R. and Reymond, P. (2013) Arabidopsis basic helix‐loop‐helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell, 25, 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, P.J. , Lee, A.K. , Xiang, F. and Park, C.M. (2008) Molecular and functional profiling of Arabidopsis pathogenesis‐related genes: insights into their roles in salt response of seed germination. Plant Cell Physiol. 49, 334–344. [DOI] [PubMed] [Google Scholar]
- Sijmons, P.C. , Grundler, F.M.W. , von Mende, N. , Burrows, P.R. and Wyss, U. (1991) Arabidopsis thaliana as a new model host for plant‐parasitic nematodes. Plant J. 1, 245–254. [Google Scholar]
- Smith, J.M. , Leslie, M.E. , Robinson, S.J. , Korasick, D.A. , Zhang, T. , Backues, S.K. , Cornish, P.V. , Koo, A.J. , Bednarek, S.Y. and Heese, A. (2014) Loss of Arabidopsis thaliana dynamin‐related protein 2B reveals separation of innate immune signaling pathways. PLoS Pathog. 10, e1004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, P.H. , van Velden, M.C. , van de Vrie, M. and Vlak, J.M. (1987) Feeding and dispersion of Spodoptera exigua larvae and its relevance for control with a nuclear polyhedrosis virus. Entomol. Exp. Appl. 43, 67–72. [Google Scholar]
- Sokal, R.R. and Rohlf, F.J. (1995) Biometry: The Principles and Practice of Statistics in Biological Research, New York: W. H. Freeman and Company. [Google Scholar]
- Song, S. , Huang, H. , Gao, H. , Wang, J. , Wu, D. , Liu, X. , Yang, S. , Zhai, Q. , Li, C. , Qi, T. and Xie, D. (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis . Plant Cell, 26, 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. , Kingdom, H.N. , MacLean, A.M. , Grieve, V.M. and Hogenhout, S.A. (2011) Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA, 108, E1254–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor, S. and Gershenzon, J. (2009) Herbivore induction of the glucosinolate‐myrosinase defense system: major trends, biochemical bases and ecological significance. Phytochem. Rev. 8, 149–170. [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri, F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poecke, R.M. (2007) Arabidopsis–insect interactions. Arabidopsis Book 5, e0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage, A. , Vlaardingerbroek, I. , Raaymakers, C. , Van Dam, N.M. , Dicke, M. , Van Wees, S.C. and Pieterse, C.M. (2011) Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. and Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany . Ann. Bot. 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling, R. , Epple, P. , Altmann, S. , He, Y. , Yang, L. , Henz, S.R. , McDonald, N. , Wiley, K. , Bader, K.C. , Gläßer, C. , Mukhtar, M.S. , Haigis, S. , Ghamsari, L. , Stephens, A.E. , Ecker, J.R. , Vidal, M. , Jones, J.D. , Mayer, K.F. , Ver Loren van Themaat, E. , Weigel, D. , Schulze‐Lefert, P. , Dangl, J.L. , Panstruga, R. and Braun, P. (2014) Convergent targeting of a common host protein‐network by pathogen effectors from three kingdoms of life. Cell Host Microbe, 16, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S.J. , Sohn, K.H. , Wan, L. , Bernoux, M. , Sarris, P.F. , Segonzac, C. , Ve, T. , Ma, Y. , Saucet, S.B. , Ericsson, D.J. , Casey, L.W. , Lonhienne, T. , Winzor, D.J. , Zhang, X. , Coerdt, A. , Parker, J.E. , Dodds, P.N. , Kobe, B. and Jones, J.D. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science, 344, 299–303. [DOI] [PubMed] [Google Scholar]
- Wu, J. and Baldwin, I.T. (2010) New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24. [DOI] [PubMed] [Google Scholar]
- Wubben, M.J. , Jin, J. and Baum, T.J. (2008) Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA‐independent pathogenesis‐related gene expression in roots. Mol. Plant–Microbe Interact. 21, 424–432. [DOI] [PubMed] [Google Scholar]
- Zander, M. , Thurow, C. and Gatz, C. (2014) TGA transcription factors activate the salicylic acid‐suppressible branch of the ethylene‐induced defense program by regulating ORA59 expression. Plant Physiol. 165, 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.‐C. , Millet, Y.A. , Cheng, Z. , Bush, J. and Ausubel, F.M. (2015) Jasmonate signalling in Arabidopsis involves SGT1b–HSP70–HSP90 chaperone complexes. Nat. Plants, 1, 15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Spodoptera exigua larvae tend to prefer RLD over srfr1‐1 in a two‐choice assay. Single RLD and srfr1‐1 plants were grown in the same pot and S. exigua was allowed to feed for 16 h when plants were 3 weeks old. The digitally quantified leaf area eaten (%) was determined for each plant, and damage on srfr1‐1 was subtracted from that on RLD for each pair. Positive values indicate more damage on RLD, and negative values indicate more damage on srfr1‐1. Differences were statistically significant (P < 0.01) by the Wilcoxon signed rank test in two of four replicates.
Fig. S2 Heterodera schachtii penetration assay in RLD and srfr1‐1 roots. A minimum of 20 infected roots of 14‐day‐old seedlings in 12‐well plates were stained with acid fuchsin at 3 days post‐inoculation. No statistically significant difference between RLD and srfr1‐1 was detected at P < 0.05 as analysed by Student's t‐test.
Fig. S3 Endogenous defence hormone levels in RLD and srfr1‐1 do not differ. Total resting‐state salicylic acid (SA) (top), jasmonic acid (JA) (middle) and JA‐Ile (JA conjugated with the amino acid isoleucine) (bottom) levels in leaf tissue of RLD and mutant srfr1‐1 soil‐grown plants (left) or roots of plate‐grown seedlings (right) were quantified by ultra performance liquid chromatography‐electrospray ionization‐tandem mass spectrometry (UPLC‐ESI‐MS/MS). Tissue was harvested at the same stage as used for insect feeding and nematode infection experiments, respectively. Error bars represent standard errors of three to four samples. There were no significant differences between RLD and srfr1‐1 as determined by Student's t‐test (P > 0.05). Similar results were found in a second experiment.
Fig. S4 Differences in jasmonic acid/ethylene (JA/ET) and salicylic acid (SA) pathway gene regulation in srfr1‐1 and RLD in response to Spodoptera exigua feeding. mRNA levels of the indicated SA and JA/ET pathway genes in Arabidopsis leaf tissue were determined 24 h after S. exigua feeding in mock‐treated (mock), insect‐damaged (local) and systemic (systemic) leaves. Expression levels were quantified by quantitative real‐time PCR (qRT‐PCR) and normalized using SAND gene (At2g28390) mRNA levels as an internal standard. Values represent averages from two biological replicates, with error bars showing the standard error.
Fig. S5 Dampened mRNA level changes 48 h after S. exigua feeding. mRNA levels of the indicated JA/ET pathway genes in Arabidopsis leaf tissue were determined in mock‐treated (mock), insect‐damaged (local) and systemic (systemic) leaves. Expression levels were quantified by quantitative real‐time PCR (qRT‐PCR) and normalized using SAND gene (At2g28390) mRNA levels as an internal standard. Values represent averages from two biological replicates, with error bars showing the standard error. For ease of comparison, the same scales were used for the genes shown in both Figs 3 and 4.
Table S1 W test statistic values for Wilcoxon signed‐rank tests on the data in Figs 3 and 4.
Table S2 Forward (F) and reverse (R) qRT‐PCR primers.


