Summary
Smut fungi are biotrophic plant pathogens that exhibit a very narrow host range. The smut fungus S porisorium reilianum exists in two host‐adapted formae speciales: S . reilianum f. sp. reilianum (SRS), which causes head smut of sorghum, and S . reilianum f. sp. zeae (SRZ), which induces disease on maize. It is unknown why the two formae speciales cannot form spores on their respective non‐favoured hosts. By fungal DNA quantification and fluorescence microscopy of stained plant samples, we followed the colonization behaviour of both SRS and SRZ on sorghum and maize. Both formae speciales were able to penetrate and multiply in the leaves of both hosts. In sorghum, the hyphae of SRS reached the apical meristems, whereas the hyphae of SRZ did not. SRZ strongly induced several defence responses in sorghum, such as the generation of H2O2, callose and phytoalexins, whereas the hyphae of SRS did not. In maize, both SRS and SRZ were able to spread through the plant to the apical meristem. Transcriptome analysis of colonized maize leaves revealed more genes induced by SRZ than by SRS, with many of them being involved in defence responses. Amongst the maize genes specifically induced by SRS were 11 pentatricopeptide repeat proteins. Together with the microscopic analysis, these data indicate that SRZ succumbs to plant defence after sorghum penetration, whereas SRS proliferates in a relatively undisturbed manner, but non‐efficiently, on maize. This shows that host specificity is determined by distinct mechanisms in sorghum and maize.
Keywords: defence responses, fluorescence microscopy, host specificity, phytopathogenic fungi, RNA sequencing, Sporisorium reilianum, transcriptome
Introduction
The smut fungus Sporisorium reilianum causes head smut disease of maize and sorghum, two of the most important commercial cereals in the world. Although the fungus is a biotroph, the disease is devastating for the plant because it leads to complete harvest loss of the affected individual. In the field, the main sources of inoculum are soil‐borne diploid teliospores which, under favourable conditions, undergo meiosis, germinate and give rise to haploid sporidia of different mating types (Halisky, 1963; Martinez et al., 2002). Compatible sporidia recognize each other through a pheromone–pheromone receptor system and form conjugation hyphae that grow towards each other and fuse at their tips (Schirawski et al., 2005). From then on, the fungus grows as a dikaryotic filament that penetrates and colonizes the plant without causing obvious symptoms (Martinez et al., 1999; Prom et al., 2011; Schirawski et al., 2010). When the fungus invades the undifferentiated floral tissue, the emerging inflorescence or flowers are replaced by sori containing black masses of teliospores (Wilson and Frederiksen, 1970). In this way, sexual proliferation of the fungus depends on successful host plant colonization.
Sporisorium reilianum exists in two formae speciales with different host preferences (Halisky, 1963; Zuther et al., 2012). Sporisorium reilianum f. sp. reilianum (SRS) is highly virulent on sorghum, but does not produce spores on maize, whereas S. reilianum f. sp. zeae (SRZ) is virulent on maize, but does not lead to head smut of sorghum. The inoculation of sorghum with SRZ induces the generation of the sorghum‐specific phytoalexins luteolinidin and apigeninidin, of which luteolinidin has been shown to inhibit the vegetative growth of both formae speciales of S. reilianum in vitro (Zuther et al., 2012). This suggests that the formation of phytoalexins inhibits the spread of SRZ in sorghum. In contrast, no information is available regarding the inability of SRS to cause smut disease on maize.
New fungal diseases emerge as a result of host switching events and host range extensions of phytopathogenic fungi (Friesen et al., 2006; Giraud et al., 2010). It is thought that the original host of S. reilianum was sorghum and that the disease only later spread to maize (Halisky, 1963). The mechanism and conditions that allowed this host jump are unknown. For a few fungi, the mechanisms of host adaptation have been elucidated. In the necrotroph Alternaria alternata, virulence is dependent on the production of particular toxins, and the transfer of biosynthesis genes from one pathovar to another alters the host range (Akagi et al., 2009; Masunaka et al., 2005; Miyashita et al., 2001; Morisseau et al., 1999; Walton, 2000), whereas, in the hemibiotroph Fusarium oxysporum, lineage‐specific regions containing four chromosomes occur (Ma et al., 2010). In F. oxysporum f. sp. lycopersici, host specificity has been associated with the presence of SIX (secreted‐in‐xylem) proteins that are secreted by the fungus into the plant xylem (Lievens et al., 2009; Takken and Rep, 2010). In Magnaporthe oryzae and Phytophthora infestans, host specificity has been related to PWL (pathogenicity on weeping lovegrass) and RXLR (arginine, any amino acid, leucine, arginine) effector proteins, respectively (Kang et al., 1995; Lee et al., 2014; Sweigard et al., 1995). Strains that are unable to cause disease on a particular plant typically induce a variety of plant defence responses, which include the generation of reactive oxygen species, the reinforcement of the cell wall and the generation or mobilization of antimicrobial substances (Ahuja et al., 2012; Heller and Tudzynski, 2011; Underwood, 2012). The accumulation of hydrogen peroxide (H2O2) can lead to cell wall strengthening, signal transduction or programmed cell death (Gadjev et al., 2008; Gilchrist, 1998; Hückelhoven, 2007; Kuźniak and Urbanek, 2000). In addition, the deposition of callose and lignin at sites of fungal penetration and cell‐to‐cell crossings acts as a physical barrier to block fungal entry and spread (Bhuiyan et al., 2009; Luna et al., 2011). The biosynthesis of antimicrobial compounds, such as phytoalexins, is also a very efficient strategy, protecting plant tissues against pathogen spread (Hammerschmidt, 1999).
The factors involved in the host specificity of S. reilianum are yet undiscovered. For the SRS–sorghum interaction, at least six different races are known (Prom et al., 2011), indicating the possible presence of a gene‐for‐gene resistance mechanism. A gene‐for‐gene resistance mechanism does not seem to be active in the SRZ–maize interaction, as no fully resistant maize lines have been described (Lübberstedt et al., 1999). The SRZ genome shows a marked paucity of polyketide synthase (PKS) and non‐ribosomal peptide synthetase (NRPS) genes potentially involved in the production of host‐specific toxins (Schirawski et al., 2010). Instead, genome sequencing reveals the presence of hundreds of small secreted proteins suggested to contribute to host selection (Laurie et al., 2012; Schirawski et al., 2010; Wollenberg and Schirawski, 2014).
In this study, we investigated the differences between the two formae speciales of S. reilianum during plant infection. Using fluorescence microscopy and the quantification of fungal DNA, we showed that both SRS and SRZ are able to spread in both hosts, but to different extents. On sorghum, SRZ is strongly challenged by multiple plant defence compounds, whereas these compounds are only marginally induced by SRS. On maize, neither SRS nor SRZ induced strong plant defences, and both formae speciales proliferated well. Whole transcriptome analysis of colonized maize leaves revealed that SRZ induced more plant defence genes than did SRS. This excludes a mechanism of plant defence in the leaf for the inability of SRS to sporulate on maize and indicates that the host specificity of S. reilianum is achieved by different mechanisms in maize and sorghum.
Results
SRS reaches the apical meristems in maize, whereas SRZ does not enter sorghum nodes
To identify the exact point at which SRS and SRZ cease to proliferate in the non‐favoured host plant, we macro‐ and microscopically investigated maize and sorghum plants after inoculation with SRS or SRZ. We observed that SRS induced chlorosis on sorghum leaves, sori replacing apical meristems and spore‐filled sori replacing inflorescences (Fig. S1A–C, see Supporting Information) in all investigated plants (n = 19). In contrast, SRZ induced phytoalexin deposition in sorghum leaves, whereas the apical meristems and inflorescences of all samples analysed (n = 20) were healthy (Fig. S1D–F). On maize, SRS induced chlorosis on leaves, and young cobs showed phyllody (Ghareeb et al., 2011) on about 70% of evaluated plants (n = 20), but there were no spores and the tassels were healthy (Fig. S1G–I). SRZ induced chlorosis and necrosis on maize leaves and resulted in about 60% of the young cobs carrying fungal spores and some tassels showing phyllody and spores (Fig. S1J–L).
Fluorescence microscopy of samples stained with wheat germ agglutinin (WGA)‐Alexafluor revealed that both formae speciales efficiently colonized sorghum leaves at 4 days after inoculation (dai) (n = 12, Fig. 1A, B). However, whereas SRS clearly seemed to prefer bundle sheath cells (Fig. 1A), the hyphae of SRZ did not show such a preference and were abundant in mesophyll areas (Fig. 1B). At 9 dai, hyphae of SRS colonized bundle sheath cells and vascular bundles in inoculated leaves (Fig. 1C), whereas hyphae of SRZ were found in bundle sheath cells, but rarely in vascular cells (Fig. 1D). When the stem tissue was investigated at 15 dai, hyphae of SRS were readily visible in vascular bundles near the growing point of the plant (Fig. 1E), whereas the stem tissues of plants inoculated with SRZ were completely healthy (Fig. 1F).
Figure 1.
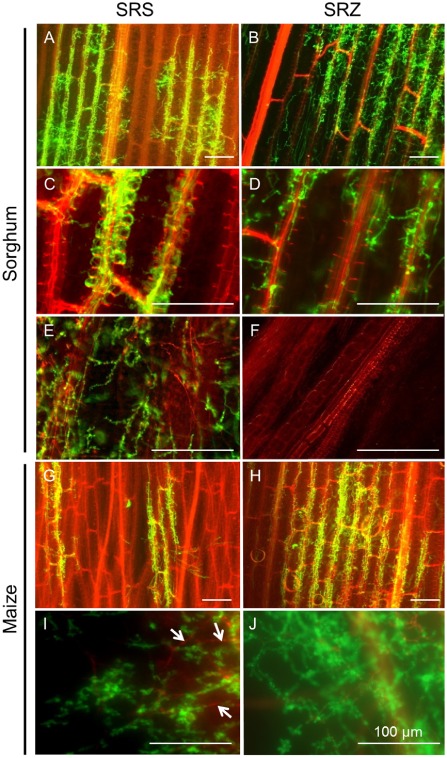
Microscopic characterization of sorghum and maize infection by S porisorium reilianum. Sorghum (A–F) and maize (G–J) seedlings were syringe inoculated with S . reilianum f. sp. reilianum (SRS) (left) or S . reilianum f. sp. zeae (SRZ) (right). Samples were collected at 4 days after inoculation (dai) (A, B, G, H), 9 dai (C, D) or 15 dai (E, F, I, J). Plant material and dead hyphae were stained with propidium iodide and appear red; fungal hyphae were stained with wheat germ agglutinin (WGA)‐Alexafluor‐488 and appear green. Sorghum samples inoculated with SRS show hyphae colonizing leaf tissues (A) and presenting a preference for vascular bundles (C), later reaching the nodes and apical meristems (E). SRZ infects leaves (B) and leaf sheaths (D), without showing such a preference for vascular bundles, and without reaching the nodes and apical meristems (F). Maize plants inoculated with SRS show hyphae colonizing leaf tissues (G) and reaching the nodes and apical meristem (I), where some dead hyphae are also observed (white arrows). Leaves inoculated with SRZ show extensive fungal growth (H) and the pathogen reaches the nodes and apical meristem (J). Bars, 100 μm; n = 12 plant samples.
Inoculated maize leaves showed colonization by SRS and SRZ at 4 dai (n = 12, Fig. 1G, H). However, in samples infected with SRS, apparently smaller amounts of hyphae were observed (Fig. 1G) when compared with SRZ (Fig. 1H). In maize stems, fungal proliferation was observed for both SRZ (Fig. 1J) and SRS (Fig. 1I), and SRS‐infected stems showed several hyphae stained by propidium iodide (Fig. 1I, white arrows), indicating that they were dead or damaged (Hickey et al., 2004). These results indicate that both strains are able to proliferate from the inoculated leaves up to the nodes and meristems in maize, whereas only SRS can do so in sorghum.
SRS and SRZ show better proliferation in their compatible hosts
To confirm and quantify the differences observed by microscopy, we analysed the content of fungal genomic DNA relative to total plant DNA at 9 dai by quantitative polymerase chain reaction (qPCR). In sorghum, a similar quantity of fungal DNA was observed at the inoculation site, whereas a lower quantity of SRZ relative to SRS was measured in the ligules, leaf sheaths and stems (Fig. 2A). The relative amount of genomic DNA of SRZ decreased with increasing distance from the inoculation site, thus being undetectable in the stems (Fig. 2A). Therefore, the proliferation of SRZ seems to be effectively blocked during its spread in the inoculated leaf, and the fungus does not reach the nodes or apical meristems of sorghum.
Figure 2.
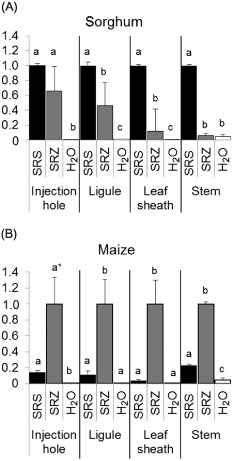
Quantification of fungal biomass in planta by quantitative polymerase chain reaction (qPCR). Fungal DNA was quantified relative to plant DNA and relative to S porisorium reilianum f. sp. reilianum (SRS) DNA (A) or to S . reilianum f. sp. zeae (SRZ) DNA (B). Samples from the inoculation site, ligule, leaf sheath and stems were collected at 9 days after inoculation (dai) from sorghum and maize. (A) In sorghum, a greater amount of DNA from SRS than from SRZ was detected in ligules, leaf sheaths and stems. (B) In maize, SRZ was dominant in ligules, leaf sheaths and stems. The presence of SRS was observed in maize stems, whereas SRZ could not be detected in sorghum stems. The experiment was performed in three biological replicates of 10 plants each that were inoculated with SRS, SRZ or water. Error bars represent the standard error of the mean (SEM). Different letters above the bars indicate a significant difference (t‐test with P ≤ 0.05) within one tissue. *P value comparing the data of SRS and SRZ at the injection hole is slightly above 0.05.
In maize, the reverse result was obtained: although the level of genomic DNA was not statistically significantly different at the inoculation site, the relative amount of SRZ DNA was higher at all other sites analysed. However, SRS DNA could still be detected in maize stems (Fig. 2B), indicating a relatively small, but clearly observable, proliferation of SRS in maize, confirming the microscopic analysis of the nodes (Fig. 1I) and the visual observations of the meristems (Fig. S1H). Taken together, fungal DNA quantification in maize and sorghum indicates that both SRS and SRZ can grow in both plants, with a clear proliferation preference for the compatible host. In contrast with SRS on maize, SRZ is unable to spread systemically in sorghum.
SRS and SRZ show differences in the plant–fungus interaction zones
To determine whether the differences in the colonization of SRS and SRZ in maize and sorghum were reflected by variations in the regions of fungal–plant interaction, we observed the ultrastructure of fungal cell walls (FCWs) and interfacial matrix (IM) in sorghum and maize leaves by transmission electron microscopy (TEM). Most fungal occurrences were characterized by a dark layer of FCW that was surrounded by a grey sheath of IM (Yi and Valent, 2013), which separated the fungal hyphae from the plant plasma membrane (Fig. 3). In sorghum, IM was roughly twice as thick as FCW for both SRS and SRZ, and no obvious differences between FCWs could be observed (Fig. 3A, C, E). IM appeared to be very similar for the hyphae of SRS that grew in maize (Fig. 3B). In contrast, the hyphae of SRZ had a thicker FCW when compared with SRS, and had an extremely thin or seemingly lacking IM which, on average, only reached about half the thickness of FCW (Fig. 3D, F). This indicates that the contact zones between fungi and hosts differ between maize and sorghum for SRZ and differ between SRS and SRZ in maize.
Figure 3.
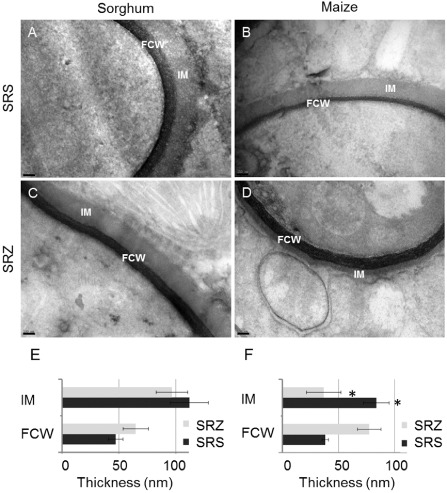
Electron micrographs of sections obtained from sorghum (A, C) and maize (B, D) infected with S porisorium reilianum f. sp. reilianum (SRS) (A, B) or S . reilianum f. sp. zeae (SRZ) (C, D). Photographs show transverse cuts of fungal hyphae with black fungal cell wall (FCW) and grey interfacial matrix (IM). Measurements were performed for FCW and IM in sorghum (E, n = 10) and maize (F, n = 8). Error bars give the standard error of the mean (SEM); size bars, 100 nm.
H2O2 occurs more often at penetration sites of SRZ in sorghum
Sorghum responds to SRZ infection by the generation of phytoalexins at 3 dai (Poloni and Schirawski, 2014). To investigate the occurrence of earlier plant defence responses, we evaluated the production of H2O2 at penetration sites. On sorghum, 20% of the appressoria of SRS showed the accumulation of H2O2 at 1 dai (Fig. 4B). Unexpectedly, for SRZ on sorghum, we observed a strong accumulation of H2O2 at the majority (61%) of penetration sites at 1 dai (Fig. 4B). Interestingly, in spite of a strong H2O2 response, the hyphae of SRZ continued to spread in sorghum leaves after penetration (Fig. 1B).
Figure 4.
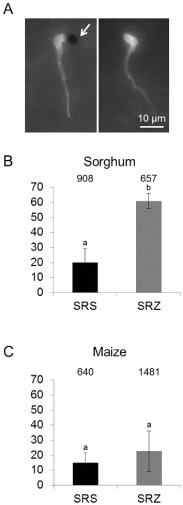
H2O2 accumulation at S porisorium reilianum infection sites in maize and sorghum. Samples were collected at 1 day after inoculation (dai) and were stained with 3,3′‐diaminobenzidine (DAB) and calcofluor. (A) Examples of appressoria showing H2O2 deposition (left, white arrow) and without H2O2 (right). (B, C) Percentage of appressoria with H2O2 deposition for sorghum (B) and maize (C). Three biological replicates containing three to four plants each were analysed. Data given are the mean values and standard deviations. The total number of counted appressoria is indicated above the bars. Different letters above the columns indicate significant differences (t‐test with P ≤ 0.05).
On maize, the penetrating hyphae of SRZ exhibited the accumulation of similar small amounts of H2O2 to SRS on sorghum (Fig. 4B, C). In contrast, the penetrating hyphae of SRS showed a much lower production of H2O2 at penetration sites in maize than did SRS in sorghum (Fig. 4B, C). This indicates that the H2O2 deposition response of sorghum to SRZ is distinct from and much stronger than that of maize to SRS.
Callose deposition occurs more frequently and more strongly at cell‐to‐cell crossings of SRZ in sorghum
We determined the occurrence of callose at attempted hyphal cell‐to‐cell crossings in maize and sorghum at 2 dai. In sorghum leaves, for about 6% of the cell‐to‐cell crossings, the hyphae of SRS induced callose deposition that was localized at the tip of the crossing hyphae (Fig. 5A, E). In contrast, about 25% of the hyphal cell‐to‐cell crossings of SRZ induced callose deposition that was stronger—often affecting the complete cell wall of the responding cell (Fig. 5B, E).
Figure 5.
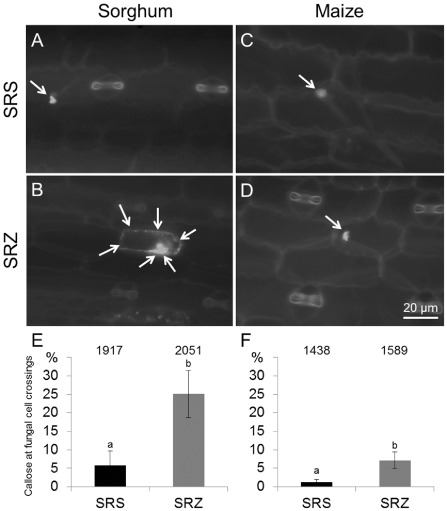
Callose deposition in infected sorghum and maize. Sorghum (A, B, E) and maize (C, D, F) leaves were collected at 2 days after inoculation (dai). Sorghum infected with S porisorium reilianum f. sp. reilianum (SRS) showed weak and localized callose deposition (A, E), whereas stronger and more frequent levels of callose were observed for S . reilianum f. sp. zeae (SRZ) (B, E). In maize, callose was observed in small and localized amounts for SRS (C) and SRZ (D), and was slightly more frequent for SRZ (F). (E) and (F) show the percentage of occurrence of callose at hyphal cell‐to‐cell crossings in sorghum and maize, respectively. Hyphal cell‐to‐cell crossings were counted for three biological replicates consisting of two leaves of two plants each. Data given are the mean values and standard deviations. The total number of counted cell‐to‐cell crossings is given above the bars. Different letters above the columns indicate significant differences (t‐test with P ≤ 0.05).
In maize, cell‐to‐cell crossings of SRZ hyphae induced callose deposition with the same low percentage and localized response as the hyphae of SRS in sorghum (Fig. 5C, E, F). In contrast, the hyphae of SRS showed much weaker signs of callose deposition in maize than did SRZ in sorghum (Fig. 5D, E, F). This indicates that the callose deposition response of sorghum to SRZ is distinct from and much stronger than that of maize to SRS.
Expression of marker genes reveals differential plant responses in maize and sorghum
To investigate the occurrence of additional defence responses, we determined the expression of several plant defence marker genes in inoculated leaves at 3 dai using quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR). In sorghum, we selected the gene Sb03g030800 as a marker for callose deposition, as it is the best homologue of the Arabidopsis thaliana glucan synthase‐like 5 (GSL5, Jacobs et al., 2009). In addition, we selected the genes Sb03g030100, encoding a chitinase, Sb01g037970, encoding the pathogenesis‐related protein 10 (PR10), SbDFR3 (Sb02g000220), encoding the enzyme dihydroflavonol 4‐reductase (DFR3) involved in phytoalexin biosynthesis in sorghum (Liu et al., 2010), Sb05g018800, encoding a leucine‐rich repeat (LRR)‐containing extracellular glycoprotein precursor, and Sb08g022440, potentially encoding a thaumatin‐like protein. Of these, genes encoding the putative chitinase, DFR3, LRR‐containing protein and thaumatin‐like protein all showed specific up‐regulation in SRZ‐colonized sorghum leaf samples, when compared with SRS‐infected samples (Fig. 6A).
Figure 6.
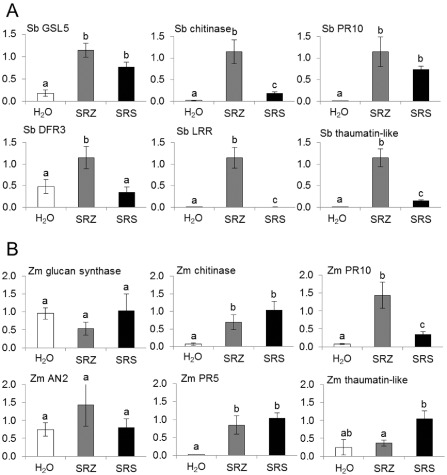
Quantification of defence gene expression in sorghum and maize inoculated with S porisorium reilianum f. sp. reilianum (SRS) (black), S . reilianum f. sp. zeae (SRZ) (grey) or water (H2O, white). Changes in transcript levels of marker genes of sorghum (Sb) are shown in (A), whereas maize (Zm) genes are shown in (B). Mean values and standard deviations of three biological replicates containing eight plants each are shown. Different letters above the columns show significant differences (t‐test with P ≤ 0.05). AN2, involved in kauralexin biosynthesis; DFR3, dihydroflavonol 4‐reductase; GSL5, glucan synthase‐like 5; LRR, leucine‐rich repeat; PR5, pathogenesis‐related protein 5; PR10, pathogenesis‐related protein 10.
In maize, we investigated the expression of the gene GRMZM2G430680, encoding a 1,3‐β‐glucan synthase, a putative chitinase (GRMZM2G005633), a putative PR10 (GRMZM2G112538), the gene AN2 (GRMZM2G044481) involved in kauralexin biosynthesis, the putative PR5 gene (GRMZM2G402631) and a gene putatively encoding a thaumatin‐like protein (GRMZM2G149809). Of these six genes, the glucan synthase and AN2 gene did not show differences in expression among H2O, SRZ and SRS, whereas the chitinase and PR5 gene were similarly up‐regulated by both SRZ and SRS (Fig. 6B). In contrast, PR10 was significantly up‐regulated in maize leaves colonized by SRZ in comparison with SRS, whereas the gene encoding the thaumatin‐like protein was up‐regulated only in SRS‐colonized maize leaves (Fig. 6B). In summary, a large variety of plant defence marker genes are up‐regulated in sorghum colonized with SRZ, whereas, in maize, both strains induce particular plant defence genes, indicating a mechanistic difference in host response to the two formae speciales of S. reilianum in maize and sorghum.
Transcriptome analysis reveals distinct gene sets induced by SRS and SRZ in maize
As no clear differences were observed in maize with regard to the evaluated plant defence responses, we analysed the gene expression profile of infected maize leaves by Illumina RNA sequencing. Sporisorium reilianum‐inoculated samples were compared with water‐inoculated samples (Zm‐SRS vs. Zm‐H2O, and Zm‐SRZ vs. Zm‐H2O) and with each other (Zm‐SRS vs. Zm‐SRZ). In SRS‐inoculated samples compared with control (Zm‐SRS vs. Zm‐H2O), 1450 maize genes were differentially regulated, 1279 (88%) of which showed up‐regulation and only 171 (12%) of which were down‐regulated (Fig. 7A). In SRZ‐infected samples compared with control (Zm‐SRZ vs. Zm‐H2O), 1563 genes showed differential regulation, 1443 (92%) of which were up‐regulated and 120 (8%) of which were down‐regulated (Fig. 7A). Among the differentially regulated genes, 698 were present in both SRS‐ and SRZ‐infected samples compared with control. Of these, 670 were up‐regulated and 28 were down‐regulated in plants infected with SRZ compared with control plants. This was very similar for SRS, and only three genes (GRMZM2G113378, GRMZM2G502350 and GRMZM2G333022) that were up‐regulated in SRZ‐infected samples showed down‐regulation for plants infected with SRS. Therefore, SRZ infection led to the regulation of an exclusive set of 865 genes, 773 (89%) of which were up‐regulated and only 92 (11%) of which were down‐regulated. Infection with SRS led to the regulation of a different set of 752 genes, 612 (81%) of which were up‐regulated and 140 (19%) of which were down‐regulated (Fig. 7A). This shows that, although the presence of both SRS and SRZ resulted in a large set of genes commonly regulated, the presence of each forma specialis induced or decreased an even larger set of distinct genes.
Figure 7.
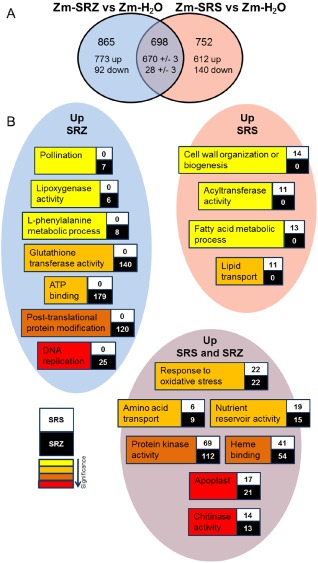
Comparison of transcriptome analysis of maize leaves infected with S porisorium reilianum f. sp. reilianum (SRS) and S . reilianum f. sp. zeae (SRZ) vs. water control samples. (A) Venn diagram showing the distribution of genes differentially regulated (P ≤ 0.05) in the comparison of SRS‐infected samples vs. water‐inoculated control samples (Zm‐SRS vs. Zm‐H2O) and SRZ‐infected samples vs. water‐inoculated control samples (Zm‐SRZ vs. Zm‐H2O). (B) Examples of gene ontology (GO) terms up‐regulated by SRS, SRZ or both fungi when compared with water‐inoculated control samples. Boxes contain the number of genes induced by SRS (white box) or by SRZ (black box) belonging to each associated GO term. Background colours of the GO terms indicate significance according to the key.
To determine which functional processes were activated by each strain, the 1279 and 1443 genes up‐regulated by SRS and SRZ, respectively, were subjected to a gene ontology (GO) term enrichment analysis using the Singular Enrichment Analysis (SEA) tool of the agriGO platform (Zhou et al., 2010). This analysis revealed 62 and 100 GO terms as significantly enriched for SRS and SRZ, respectively, which were reduced to 55 and 78 GO terms using REVIGO (Supek et al., 2011; Tables S1, S2, see Supporting Information). GO terms enriched for both SRS and SRZ included apoplast, chitinase activity, heme binding, response to oxidative stress, nutrient reservoir activity and amino acid transport (Fig. 7B). The GO terms protein kinase activity and protein amino acid phosphorylation were up‐regulated by both SRS and SRZ, but almost twice as many genes were present for SRZ compared with control (Fig. 7B, Tables S1, S2). GO terms specifically enriched in SRZ‐induced genes included DNA replication, post‐translational protein modification, glutathione transferase, ATP binding, l‐phenylalanine metabolic process, lipoxygenase activity and pollination. GO terms specifically enriched for SRS‐induced genes included lipid transport, fatty acid metabolic process, acetyltransferase activity and cell wall organization or biogenesis (Fig. 7B).
The comparison of SRS‐infected samples with SRZ‐infected samples (Zm‐SRS vs. Zm‐SRZ) revealed 500 differentially regulated maize genes. Of these, 270 and 230 genes were up‐regulated in SRZ‐ and SRS‐infected samples, respectively (Fig. 8A). The logarithmic fold changes of these genes were used for Parametric Analysis of Gene Set Enrichment (PAGE) from agriGO (Zhou et al., 2010). Of the 500 genes, only 242 (48%) were annotated and appeared in the query list. Of these, 171 (63% of 270 genes) were up‐regulated in SRZ‐inoculated samples and 71 (31% of 230 genes) in SRS‐inoculated samples. The analysis resulted in 55 GO terms that were all significantly up‐regulated in maize colonized with SRZ relative to SRS, 18 of which belonged to the main ontology ‘molecular function’, 37 to ‘biological process’ and none to ‘cellular components’ (Fig. 8B; Table S3, see Supporting Information). The genes strongly up‐regulated by SRZ relative to SRS belonged to functionally connected GO terms that culminated in the four GO terms ‘regulation of transcription’, ‘protein amino acid phosphorylation’, ‘ATP binding’ and ‘protein serine/threonine kinase activity’.
Figure 8.
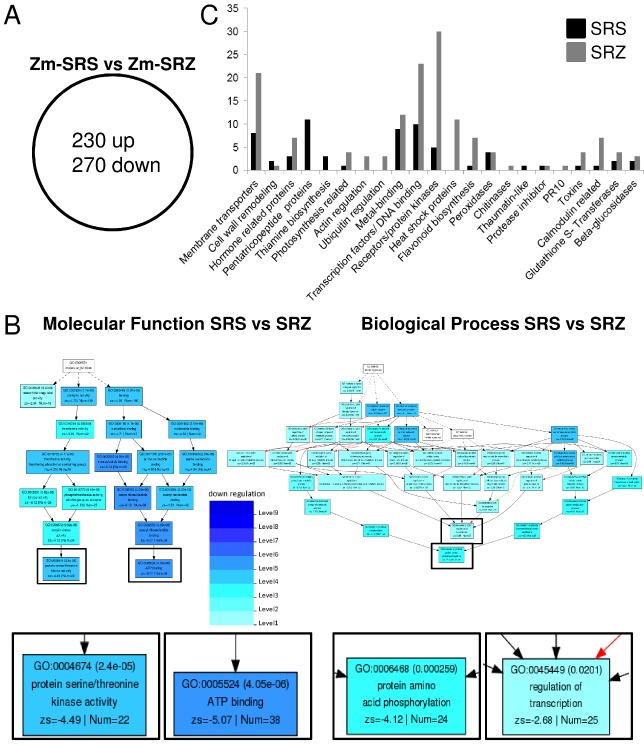
Comparison of transcriptome analysis of maize leaves infected with S porisorium reilianum f. sp. reilianum (SRS) relative to S . reilianum f. sp. zeae (SRZ)‐infected samples. (A) Venn diagram showing the distribution of genes differentially regulated (P ≤ 0.05) in the comparison of SRS‐ with SRZ‐infected samples (Zm‐SRS vs. Zm‐SRZ). (B) Pathway mapping of the significantly enriched gene ontology (GO) terms in the comparison of Zm‐SRS vs. Zm‐SRZ showing the categories ‘Molecular function’ and ‘Biological process’. Four culminating GO terms are enlarged below for better readability. Boxes with GO terms have the GO ID, the false discovery rate (FDR), the GO term, the z score and the number of submitted maize genes belonging to the respective GO term. Background colours of GO terms indicate significance according to the key. (C) Summary of gene identity of differentially up‐regulated annotated genes of (A) in samples infected with SRS (black) and SRZ (grey). The graph shows the annotation and number of genes belonging to each function (212 genes are represented; complete gene lists are available in Tables S2 and S3, see Supporting Information).
As, in agriGO, annotation was available for only one‐half of the differentially regulated genes, we manually annotated each of the 500 genes by comparison with the non‐redundant databases using blast (Altschul et al., 1990). Among the 270 genes induced by SRZ, at least 70 were predicted to be involved in defence responses, and included four peroxidases, one chitinase, one PR10, three β‐glucosidases, seven genes related to calmodulin, one protease inhibitor, four glutathione‐S‐transferases, three genes involved in the biosynthesis of toxins and seven genes associated with flavonoid biosynthesis (Fig. 8C). Moreover, 30 genes coding for plant receptors or protein kinases were up‐regulated, as well as 11 heat shock proteins mainly belonging to the group of Hsp70. DNA binding or transcription factors comprised 23 genes, five of which belonged to the WRKY transcription factor group. Other genes included 21 membrane transporters, seven hormone‐related genes, 12 genes associated with metal binding, four genes related to photosynthesis, three genes involved in the regulation of actin and three genes involved in the regulation of ubiquitin (Fig. 8C; Table S4, see Supporting Information).
Of the 230 maize genes induced by SRS, at least 30 were potentially implicated in plant defences (Fig. 8C, Table S5, see Supporting Information). These genes included four peroxidases, one thaumatin‐like gene, one calmodulin‐related gene, one protease inhibitor, two β‐glucosidases, one toxin and two genes encoding glutathione‐S‐transferases. Receptors and protein kinases comprised five genes. Furthermore, eight membrane transporters, three hormone‐related genes, nine genes associated with metal binding, two genes involved in cell wall remodelling and 10 genes representing DNA‐binding proteins and transcription factors were up‐regulated. Interestingly, 11 pentatricopeptide repeat (PPR) proteins and three genes involved in thiamine biosynthesis were induced (Fig. 8C). Transcriptome analysis demonstrated that the presence of either SRS or SRZ in maize leaves led to a clear transcriptional response in maize which involved a greater number of genes in response to SRZ than to SRS. Defence genes were induced in both cases, but, here also, a larger number of genes were induced by SRZ.
Discussion
The smut fungus S. reilianum exists in two host‐adapted formae speciales that infect sorghum (SRS) or maize (SRZ) (Fig. S1). We found that both formae speciales could colonize both plants, with SRS being more proliferative on sorghum and SRZ on maize. SRS was able to colonize maize and reach meristematic tissues, whereas the proliferation of SRZ was restricted to the inoculated leaves in sorghum (Figs 1, 2). The proliferation behaviour correlated with the observed levels of plant defence responses that were strong only for SRZ in sorghum (Figs 5, 6), and included H2O2 formation at 1 dai, callose deposition at 2 dai and phytoalexin induction at 3 dai. Obviously, discrete mechanisms exist that determine the host specificity of SRS and SRZ. In sorghum, SRZ is recognized at least as soon as it penetrates, and multiple successive layers of defence responses were shown to be induced during the first 3 dai. Together, these defences are effective in restricting the growth of SRZ to the inoculated leaf, and the prevention of node and meristem colonization is efficient in inhibiting smut disease.
The specific recognition of SRZ in sorghum may be induced by microbe‐associated molecular patterns (MAMPs), such as chitin and β‐1,3‐glucan, which are known to induce H2O2, callose, phytoalexins and PR proteins. Although SRS is also expected to contain these MAMPs, recent studies have shown that plant pathogens can hide or escape from MAMP recognition through various mechanisms (De Jonge et al., 2010; El Gueddari et al., 2002; Fujikawa et al., 2009, 2012; Oliveira‐Garcia and Deising, 2013; Van den Burg et al., 2006), some or all of which could also be used by S. reilianum. Alternatively, the distinct responses encountered by SRS and SRZ in sorghum may indicate the existence of specific avirulence effectors in SRZ that are recognized by resistance receptors in sorghum and trigger defence reactions, as described for other plant pathogens, such as Melampsora lini, M. oryzae and Cladosporium fulvum (Ellis et al., 2007; Sweigard et al., 1995; Tosa et al., 2005; Van der Does and Rep, 2007).
In the compatible interaction of Ustilago maydis and maize, microarray analysis of plant genes showed a differential regulation of 575 genes at 2 dai, including genes associated with stress, secondary metabolism, cell wall metabolism, protein, transcription and RNA processing (Doehlemann et al., 2008). In addition, U. maydis induced Bax‐inhibitor 1 and several cystatin genes expected to be involved in cell death suppression (Doehlemann et al., 2008). These genes were not among the genes induced by SRZ in maize (Table S4). A different maize response to the two compatible maize pathogens might reflect the different lifestyle of the fungi. In the leaf, SRZ proliferates without causing symptoms, whereas U. maydis induces tumours to assist spore formation.
In maize, SRS does not proliferate to the same level as SRZ (Fig. 2), which could be caused by compatibility issues, such as the lack of virulence factors or the misexpression of factors needed for growth, or to the presence of enhanced plant resistance. However, no or only very weak phenotypic defence responses were elicited by either fungus, and the weak defence responses observed were similar or even stronger for SRZ than for SRS on maize (Figs 4, 5). Analysis of maize gene expression largely corroborated a weaker defence gene expression response for SRS (Fig. 8). Transcriptome analysis revealed that the presence of both fungi is clearly detected by the plant, and commonly induced a large set of genes (Fig. 7). However, each forma specialis also induced a specific set of genes that could be involved in the observed differences in fungal proliferation in maize leaves. Notably, a group of 11 PPR proteins was up‐regulated for SRS‐infected plants. In Arabidopsis and soybean, specific PPR proteins have been shown to be involved in resistance against various pathogens (Laluk et al., 2011; Park et al., 2014; Wong et al., 2014) and have been reported to function in RNA binding (Fujii and Small, 2011; Lurin et al., 2004; Saha et al., 2007; Yagi et al., 2014). As the function of PPR proteins is still unknown in maize, further work is required to determine whether they are involved in slowing the growth of SRS in maize. Interestingly, hyphae of SRZ that colonize maize leaves show a thinner IM (Fig. 3). Possibly, a thinner IM is necessary for efficient colonization of maize. If this is so, the lack of efficient proliferation of SRS in maize might be a result of compatibility issues caused by an inconveniently thick IM.
It is interesting that the transcriptional plant responses of maize to SRZ involve many more genes than its response to SRS (Fig. 7), including a much larger number of defence‐related genes (Fig. 8C). This corroborates the suggestion that SRS is not successful on maize because of incompatibility rather than increased plant defence. Such a scenario would also be in support of a proposed host jump from sorghum to maize. If ancient SRS strains colonized maize, they would not have met with a strong defence response, allowing persistent colonization and spread into the meristematic tissues, which, under certain environmental conditions, might have allowed spore formation on maize and the subsequent evolution of strains specific for maize. A host jump of SRZ strains from maize to sorghum would not have been possible because of excessive plant defence responses met by SRZ strains in sorghum.
In this study, we performed a detailed analysis of the fungal growth behaviour and plant responses induced by the two formae speciales of S. reilianum on maize and sorghum. We showed that host specificity is determined after plant penetration, and different plant responses and fungal growth patterns are observed in each pathosystem. In sorghum, the spread of SRZ is inhibited by the induction of plant defence responses that limit fungal growth to the inoculated leaf. In maize, SRS does not meet with stronger defence responses than SRZ and seems to show an altered thickness of the plant–fungus interaction zone, which indicates the existence of more subtle compatibility issues. It would be interesting to compare the gene expression profiles of SRS and SRZ in maize in order to identify compatibility‐related fungal genes.
Experimental Procedures
Plant material, fungal strains and seedling inoculation
Seeds of Zea mays Gaspe Flint and Sorghum bicolor Tall Polish were grown for 7 and 14 days, respectively, under glasshouse conditions of 15 h daylight, at 28 °C and 50% relative humidity, and 9 h night, at 22 °C and 60% relative humidity.
The compatible wild‐type Sporisorium reilianum strains SRZ1_5‐2 (a1b1) and SRZ2_5‐1 (a2b2), originally isolated from maize (Schirawski et al., 2010), and SRS1_H2‐8 (a1b1) and SRS2_H2‐7 (a2b6), isolated from sorghum (Zuther et al., 2012), were used in this study. Fungal strains were inoculated in 2 mL of YEPS light medium (1% tryptone, 1% yeast extract and 1% sucrose) and incubated at 28 °C with shaking at 200 rpm for 8 h. The cultures were used to inoculate 50 mL of potato dextrose broth (BD, Heidelberg, Germany), and were incubated at 28 °C overnight, until they reached an optical density at 600 nm (OD600) of 0.6–0.8. The cultures were centrifuged at 5700 g for 5 min in a multifuge X3R centrifuge (Thermo Fischer Scientific, Dreieich, Germany), and the cell pellets were suspended in water to reach an OD600 of 2.0. Cell suspensions were mixed at a 1 : 1 ratio and mixtures of SRZ1_5‐2 and SRZ2_5‐1 (SRZ), or SRS1_H2‐8 and SRS2_H2‐7 (SRS), were used for syringe inoculation of leaf whorls of maize or sorghum seedlings.
Macroscopic and microscopic characterization of plant infection
Leaf blades, ligules, leaf sheaths of inoculated leaves and stems containing the nodes and floral meristems were used for microscopic analysis. The tissues were collected at several time points between 4 and 75 dai. Samples were used directly for light microscopy, or were stained with propidium iodide and WGA‐Alexafluor‐488 prior to analysis by fluorescence microscopy, as described by Ghareeb et al. (2011).
Sample preparation and TEM
Infected leaves were cut into small pieces (0.5–1 cm), fixed in a solution of glutaraldehyde 2.5% and then kept in a solution of osmium tetroxide (1%) for 90 min. Samples were washed in water and dehydrated in six successive rinsing concentrations of ethanol for 30 min each. Samples were incubated in LR White (SPI Supplies, West Chester, PA, USA)–ethanol (2 : 1) for 2 h and embedded in pure LR White resin overnight. After polymerization at 50 °C for 24 h, ultra‐thin cuts of 90 nm were prepared and stained with 4% uranyl acetate.
Quantification of fungal genomic DNA
Sorghum and maize plants inoculated with SRS, SRZ or water were harvested at 9 dai. Samples were obtained from the leaf blade, ligule and leaf sheath of the inoculated leaf, and from the inner stem containing nodes and meristems of the inoculated plant. Ten plants were collected and pooled, and the experiment was repeated three times. The plant material was frozen and ground in liquid nitrogen until a fine powder was obtained, and DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Oligonucleotides oSP101 (GTGCATCAACTGCCAGAAGG) and oSP102 (TCGTAGCCGTAGTACCAAGC) were used to amplify a 396‐bp fragment of S. reilianum genomic DNA derived from the gene sr16559. As reference genes, the sorghum actin gene SbActin was amplified using oligonucleotides HEL794 and HEL795 (Du et al., 2010), whereas, in maize, the actin gene ZmActin was amplified with primers oBH73 (ACCTCACCGACCACCTAATG) and oBH74 (ACCTGACCATCAGGCATCTC). A 25‐μL reaction mixture was composed of 1 × NH4 reaction buffer, 2 mm MgCl2, 0.25 U BIOTaq DNA polymerase, 100 μm deoxynucleoside triphosphates (dNTPs) (all from Bioline, Luckenwalde, Germany), 0.2 × SYBR Green solution (Invitrogen, Karlsruhe, Germany), 0.25 μm of each primer (Sigma‐Aldrich, Taufkirchen, Germany) and 1 μL of template DNA. PCR amplification was performed in a CFX Connect cycler (Bio‐Rad Laboratories, München, Germany) with an initial denaturation of 95 °C for 6 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min, followed by a plate reading step. Finally, a product melting curve was performed at 55–95 °C. Expression ratios in samples of inoculated plants compared with water‐inoculated plants were calculated using CFX Manager 3.0 (Bio‐Rad Laboratories).
Staining of plant material for H2O2 and callose
For the detection of H2O2, leaf samples were collected at 1 and 2 dai, vacuum infiltrated for 3 min in 3,3′‐diaminobenzidine solution (DAB, 1 mg/mL, Sigma Aldrich, Taufkirchen, Germany; D3939) and stained for 6 h in the dark. Sections were destained in ethanol and soaked for 30 s in a solution of calcofluor white (10 mg/mL, Fluorescence Brightener 28, Sigma). The generation of H2O2 was visualized as a reddish‐brown coloration, whereas fungal surface structures were fluorescent under UV illumination using a fluorescence microscope (DM6000B, Leica Bensheim, Germany) and a DAPI (4′,6‐diamidino‐2‐phenylindole) filter (EX BP 365, BS FT 395, EM BP 445/50). For the detection of callose, infected leaves were collected at 1 and 2 dai, soaked overnight in ethanol and incubated for 1 h in a staining solution containing aniline blue (0.005%) in 50 mm sodium phosphate buffer (pH 8.2). The material was analysed by fluorescence microscopy (DM6000B, Leica) using a DAPI filter set.
RNA extraction and cDNA synthesis
Sorghum and maize leaf pieces of about 3 cm inoculated with SRS, SRZ or water control were collected at 3 dai in three biological replicates containing eight plants each. Samples were macerated in liquid nitrogen, and 100 mg of the resulting powder was used for RNA extraction using Trizol (Sigma). A clean‐up step was performed using a Qiagen RNeasy Plus Mini Kit. The final RNA concentration was determined using a NanoDrop Spectrophotometer (Peqlab, Erlangen, Gemany), and RNA integrity was confirmed through denaturating agarose gel electrophoresis. One microgram of total RNA was subjected to cDNA synthesis using oligo(dT)18 oligonucleotides (First Strand cDNA Synthesis Kit, Fermentas, St. Leon‐Rot, Germany) following the manufacturer's protocol.
Expression of plant defence marker genes by qRT‐PCR
Real‐time PCR was performed in a CFX 96 Thermal Cycler (Bio‐Rad Laboratories) in a 25‐μL reaction mixture composed of 1 × NH4 reaction buffer (Bioline), 3 mm MgCl2, 100 μm dNTPs, 0.4 μm of each gene‐specific primer, 0.25 units BIOTaq DNA polymerase (Bioline), 1 μL of cDNA and 100 000 times diluted SYBR Green I solution (Cambrex, Wiesbaden, Germany). Primers used for the amplification of marker genes in maize and sorghum are listed in Table S6 (see Supporting Information). As reference genes in sorghum, actin, ubiquitin and glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) genes were selected, whereas, for maize, ubiquitin, GAPDH and tubulin were used. The PCR conditions consisted of an initial denaturation at 95 °C for 6 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min, followed by a plate reading step and a final product melting curve at 55–95 °C. Expression ratios in samples of inoculated plants compared with water‐inoculated plants were calculated using CFX Manager 3.0 (Bio‐Rad), and statistical calculations were performed using Graph Pad software.
Transcriptomic analysis
Total RNA was extracted as described above from maize leaves inoculated with SRS, SRZ or water (three biological replicates containing samples of 10 plants each were pooled prior to sequencing). Illumina sequencing was performed by Beckman Coulter Genomics (Danvers, MA, USA) using the paired‐end sequencing protocol. Read files were imported into CLC genomics workbench version 6 and trimmed by ambiguous bases, quality and Illumina adapter sequences. The resulting reads were mapped to the reference genomes of Zea mays (maize B73; Schnable et al., 2009). Logarithmic fold changes, P values and false discovery rates (FDRs) were calculated with the extension package edgeR 1.6.5. Comparisons were made between three groups of samples: SRS‐ and SRZ‐infected plants and water‐inoculated control samples. Genes with a corrected P ≤ 0.05 were considered to be significantly regulated. Gene annotation was obtained from maize GDB (http://www.maizegdb.org/), Plant GDB (http://www.plantgdb.org/ZmGDB/) or Phytosome (http://www.phytozome.net), when available. Genes that did not present annotation were subjected to blast search at the National Center for Biotechnology Information (NCBI) (Altschul et al., 1990; http://blast.ncbi.nlm.nih.gov/Blast.cgi). GO enrichment was performed for genes that showed significant expression change using the agriGO tool kit (http://bioinfo.cau.edu.cn/agriGO/), and redundant GO terms were reduced using the REVIGO tool (http://revigo.irb.hr/).
Supporting information
Fig. S1 Macroscopic symptoms on sorghum and maize after inoculation with Sporisorium reilianum. Seedlings of sorghum (A–F) or maize (G–L) were syringe inoculated with Sporisorium reilianum f. sp. reilianum (SRS) (A–C, G–I) or S. reilianum f. sp. zeae (SRZ) (D–F, J–L). Samples of plants were analysed at 4 days after inoculation (dai) (A, D, G, J), 21 dai (I, L), 35 dai (B, E), 50 dai (H, K) or 75 dai (C, F). Sorghum plants inoculated with SRS showed only chlorosis on leaves (A), but resulted in meristems wrapped in a white peridium (B) and spore formation in the apical inflorescences (C). Penetrating hyphae were colourless (A, inset). However, SRZ‐inoculated sorghum plants displayed red phytoalexin accumulation in leaves (D), and phytoalexins also stained fungal hyphae red (D, inset). SRZ‐inoculated sorghum plants developed healthy meristems (E), and inflorescences containing normal seeds (F) were produced. In maize, SRS‐inoculated plants showed weak chlorosis on leaves (G) and leaf‐like structures in cobs (H), but tassels were healthy (I). SRZ‐inoculated maize presented strong chlorosis on leaves (J), and produced spores and leaf‐like structures in cobs (K) and tassels (L). Bars: 1 cm in A–L, 20 μm in the insets. Insets in (A) and (D) are bright field micrographs of untreated leaf samples. Arrows in the insets of (A) and (D) point to fungal hyphae. Insets in (H) and (I) represent inflorescences of mock‐inoculated plants.
Table S1 List of significant gene ontology (GO) terms in Sporisorium reilianum f. sp. zeae (SRZ)‐infected samples vs. water control samples.
Table S2 List of significant gene ontology (GO) terms in Sporisorium reilianum f. sp. reilianum (SRS)‐infected samples vs. water control samples.
Table S3 List of significant gene ontology (GO) terms in Sporisorium reilianum f. sp. reilianum (SRS)‐infected samples vs. S. reilianum f. sp. zeae (SRZ)‐infected samples.
Table S4 List of maize genes significantly up‐regulated in the comparison Zm‐SRZ vs. Zm‐SRS. SRS, Sporisorium reilianum f. sp. reilianum; SRZ, S. reilianum f. sp. zeae; Zm, Zea mays.
Table S5 List of maize genes significantly up‐regulated in the comparison Zm‐SRS vs. Zm‐SRZ. SRS, Sporisorium reilianum f. sp. reilianum; SRZ, S. reilianum f. sp. zeae; Zm, Zea mays.
Table S6 List of oligonucleotides used for quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR).
Acknowledgements
We thank Theresa Wollenberg (Aachen) for help with mapping of the transcriptomic data, Steven Stadler for statistical transcriptomic analysis, Annie Minkyung Cho for proofreading, Michael Hoppert (Göttingen) for help with sample preparation for electron microscopy and for operating the electron microscope, and Susanne Mester (Göttingen) for help with plant care. We thank Ivo Feussner (Göttingen) for use of the glasshouse and RWTH Aachen University for institutional funding. This work was supported by a grant from the German Academic Exchange Service (DAAD) to Alana Poloni, and by the German Initiative of Excellence (Deutsche Forschungsgemeinschaft grant no. ZUK45/1) to Jan Schirawski.
References
- Ahuja, I. , Kissen, R. and Bones, A.M. (2012) Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 73–90. [DOI] [PubMed] [Google Scholar]
- Akagi, Y. , Akamatsu, H. , Otani, H. and Kodama, M. (2009) Horizontal chromosome transfer, a mechanism for the evolution and differentiation of a plant‐pathogenic fungus. Eukaryot. Cell, 8, 1732–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bhuiyan, N.H. , Selvaraj, G. , Wei, Y. and King, J. (2009) Role of lignification in plant defense. Plant Signal. Behav. 4, 158–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge, R. , van Esse, H.P. , Kombrink, A. , Shinya, T. , Desaki, Y. , Bours, R. , van der Krol, S. , Shibuya, N. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2010) Conserved fungal LysM effector Ecp6 prevents chitin‐triggered immunity in plants. Science, 329, 953–955. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. , Wahl, R. , Horst, R.J. , Voll, L.M. , Usadel, B. , Poree, F. , Stitt, M. , Pons‐Kühnemann, J. , Sonnewald, U. , Kahmann, R. and Kämper, J. (2008) Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis . Plant J. 56, 181–195. [DOI] [PubMed] [Google Scholar]
- Du, Y. , Chu, H. , Wang, M. , Chu, I. and Lo, C. (2010) Identification of flavone phytoalexins and a pathogen‐inducible flavone synthase II gene (SbFNSII) in sorghum. J. Exp. Bot. 61, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gueddari, N.E. , Rauchhaus, U. , Moerschbacher, B.M. and Deising, H.B. (2002) Developmentally regulated conversion of surface‐exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156, 103–112. [Google Scholar]
- Ellis, J.G. , Lawrence, G.J. and Dodds, P.N. (2007) Further analysis of gene‐for‐gene disease resistance specificity in flax. Mol. Plant Pathol. 8, 103–109. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Stukenbrock, E.H. , Liu, Z. , Meinhardt, S. , Ling, H. , Faris, J.D. , Rasmussen, J.B. , Solomon, P.S. , McDonald, B.A. and Oliver, R.P. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. [DOI] [PubMed] [Google Scholar]
- Fujii, S. and Small, I. (2011) The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191, 37–47. [DOI] [PubMed] [Google Scholar]
- Fujikawa, T. , Kuga, Y. , Yano, S. , Yoshimi, A. , Tachiki, T. , Abe, K. and Nishimura, M. (2009) Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 73, 553–570. [DOI] [PubMed] [Google Scholar]
- Fujikawa, T. , Sakaguchi, A. , Nishizawa, Y. , Kouzai, Y. , Minami, E. , Yano, S. , Koga, H. , Meshi, T. and Nishimura, M. (2012) Surface α‐1,3‐glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 8, e1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev, I.Z. , Stone, J.M. and Gechev, T.S. (2008) Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. Int. Rev. Cell Mol. Biol. 270, 87–144. [DOI] [PubMed] [Google Scholar]
- Ghareeb, H. , Becker, A. , Iven, T. , Feussner, I. and Schirawski, J. (2011) Sporisorium reilianum infection changes inflorescence and branching architectures of maize. Plant Physiol. 156, 2037–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist, D.G. (1998) Programmed cell death in plant disease: the purpose and promise of cellular suicide. Annu. Rev. Phytopathol. 36, 393–414. [DOI] [PubMed] [Google Scholar]
- Giraud, T. , Gladieux, P. and Gavrilets, S. (2010) Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol. Evol. 25, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halisky, P.M. (1963) Head smut of sorghum, sudan grass, and corn, caused by Sphacelotheca reiliana (Kühn) Clint. Hilgardia, 34, 287–304. [Google Scholar]
- Hammerschmidt, R. (1999) Phytoalexins: what have we learned after 60 years? Annu. Rev. Phytopathol. 37, 285–306. [DOI] [PubMed] [Google Scholar]
- Heller, J. and Tudzynski, P. (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390. [DOI] [PubMed] [Google Scholar]
- Hickey, P.C. , Swift, S.R. , Roca, G.M. and Read, N.D. (2004) Live‐cell imaging of filamentous fungi using vital fluorescent dyes and confocal microscopy. Methods Microbiol. 34, 63–87. [Google Scholar]
- Hückelhoven, R. (2007) Cell wall‐associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. [DOI] [PubMed] [Google Scholar]
- Jacobs, A. , Lipka, V. , Burton, R. , Panstruga, R. , Strizhov, N. , Schulze‐Lefert, P. and Fincher, G. (2009) An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell, 15, 2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S. , Sweigard, J.A. and Valent, B. (1995) The PWL host specificity gene family in the blast fungus Magnaporthe grisea . Mol. Plant–Microbe Interact. 8, 939–948. [DOI] [PubMed] [Google Scholar]
- Kuźniak, E. and Urbanek, H. (2000) The involvement of hydrogen peroxide in plant responses to stresses. Acta Physiol. Plant. 22, 195–203. [Google Scholar]
- Laluk, K. , Abuqamar, S. and Mengiste, T. (2011) The Arabidopsis mitochondria‐localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 156, 2053–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie, J.D. , Ali, S. , Linning, R. , Mannhaupt, G. , Wong, P. , Güldener, U. , Münsterkötter, M. , Moore, R. , Kahmann, R. , Bakkeren, G. and Schirawski, J. (2012) Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species‐specific presence of transposable elements. Plant Cell, 24, 1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.A. , Kim, S.Y. , Oh, S.K. , Yeom, S.I. , Kim, S.B. , Kim, M.S. , Kamoun, S. and Choi, D. (2014) Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans . New Phytol. 203, 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens, B. , Houterman, P.M. and Rep, M. (2009) Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other forma speciales. FEMS Microbiol. Lett. 300, 201–215. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Du, Y. , Chu, H. , Shih, C.H. , Wong, Y.W. , Wang, M. , Chu, I.K. , Tao, Y. and Lo, C. (2010) Molecular dissection of the pathogen‐inducible 3‐deoxyanthocyanidin biosynthesis pathway in sorghum. Plant Cell Physiol. 51, 1173–1185. [DOI] [PubMed] [Google Scholar]
- Luna, E. , Pastor, V. , Robert, J. , Flors, V. , Mauch‐Mani, B. and Ton, J. (2011) Callose deposition: a multifaceted plant defence response. Mol. Plant–Microbe Interact. 24, 183–193. [DOI] [PubMed] [Google Scholar]
- Lurin, C. , Andrés, C. , Aubourg, S. , Bellaoui, M. , Bitton, F. , Bruyère, C. , Caboche, M. , Debast, C. , Gualberto, J. , Hoffmann, B. , Lecharny, A. , Le Ret, M. , Martin‐Magniette, M.L. , Mireau, H. , Peeters, N. , Renou, J.P. , Szurek, B. , Taconnat, L. and Small, I. (2004) Genome‐wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell, 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübberstedt, T. , Xia, X.C. , Tan, G. , Liu, X. and Melchinger, A.E. (1999) QTL mapping of resistance to Sporisorium reiliana in maize. Theor. Appl. Genet. 99, 593–598. [DOI] [PubMed] [Google Scholar]
- Ma, L.J. , van der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.J. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. , Houterman, P.M. , Kang, S. , Shim, W.B. , Woloshuk, C. , Xie, X. , Xu, J.R. , Antoniw, J. , Baker, S.E. , Bluhm, B.H. , Breakspear, A. , Brown, D.W. , Butchko, R.A. , Chapman, S. , Coulson, R. , Coutinho, P.M. , Danchin, E.G. , Diener, A. , Gale, L.R. , Gardiner, D.M. , Goff, S. , Hammond‐Kosack, K.E. , Hilburn, K. , Hua‐Van, A. , Jonkers, W. , Kazan, K. , Kodira, C.D. , Koehrsen, M. , Kumar, L. , Lee, Y.H. , Li, L. , Manners, J.M. , Miranda‐Saavedra, D. , Mukherjee, M. , Park, G. , Park, J. , Park, S.Y. , Proctor, R.H. , Regev, A. , Ruiz‐Roldan, M.C. , Sain, D. , Sakthikumar, S. , Sykes, S. , Schwartz, D.C. , Turgeon, B.G. , Wapinski, I. , Yoder, O. , Young, S. , Zeng, Q. , Zhou, S. , Galagan, J. , Cuomo, C.A. , Kistler, H.C. and Rep, M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, C. , Roux, C. and Dargent, R. (1999) Biotrophic development of Sporisorium reilianum zeae in maize shoot apex. Phytopathology, 89, 247–253. [DOI] [PubMed] [Google Scholar]
- Martinez, C. , Roux, C. , Jauneau, A. and Dargent, R. (2002) The biological cycle of Sporisorium reilianum f.sp. zeae: an overview using microscopy. Mycologia, 94, 505–514. [PubMed] [Google Scholar]
- Masunaka, A. , Ohtani, K. , Peever, T.L. , Timmer, L.W. , Tsuge, T. , Yamamoto, M. , Yamamoto, H. and Akimitsu, K. (2005) An isolate of Alternaria alternata that is pathogenic to both tangerines and rough lemon and produces two host‐selective toxins, ACT‐ and ACR‐toxins. Phytopathology, 95, 241–247. [DOI] [PubMed] [Google Scholar]
- Miyashita, M. , Nakamori, T. , Murai, T. , Yonemoto, T. , Miyagawa, H. , Akamatsu, M. and Ueno, T. (2001) Structure–activity relationship study of host‐specific phytotoxins (AM‐toxin analogs) using a new assay method with leaves from apple meristem culture. Z. Naturforsch. [C], 56, 1029–1037. [DOI] [PubMed] [Google Scholar]
- Morisseau, C. , Ward, B.L. , Gilchrist, D.G. and Hammock, B.D. (1999) Multiple epoxide hydrolases in Alternaria alternata f. sp. lycopersici and their relationship to medium composition and host‐specific toxin production. Appl. Environ. Microbiol. 65, 2388–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira‐Garcia, E. and Deising, H.B. (2013) Infection structure‐specific expression of β‐1,3‐glucan synthase is essential for pathogenicity of Colletotrichum graminicola and evasion of β‐glucan‐triggered immunity in maize. Plant Cell, 25, 2356–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y.J. , Lee, H.J. , Kwak, K.J. , Lee, K. , Hong, S.W. and Kang, H. (2014) MicroRNA400‐guided cleavage of pentatricopeptide repeat protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol. 55, 1660–1668. [DOI] [PubMed] [Google Scholar]
- Poloni, A. and Schirawski, J. (2014) Red card for pathogens: phytoalexins in sorghum and maize. Molecules, 19, 9114–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prom, L.K. , Perumal, R. , Erattaimuthu, S.R. , Erpelding, J.E. , Montes, N. , Odvody, G.N. , Greenwald, C. , Jin, Z. , Frederiksen, R. and Magill, C.W. (2011) Virulence and molecular genotyping studies of Sporisorium reilianum isolates in sorghum. Plant Dis. 95, 523–529. [DOI] [PubMed] [Google Scholar]
- Saha, D. , Prasad, A.M. and Srinivasan, R. (2007) Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 45, 521–534. [DOI] [PubMed] [Google Scholar]
- Schirawski, J. , Heinze, B. , Wagenknecht, M. and Kahmann, R. (2005) Mating type loci of Sporisorium reilianum: novel pattern with three a and multiple b specificities. Eukaryot. Cell, 4, 1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski, J. , Mannhaupt, G. , Münch, K. , Brefort, T. , Schipper, K. , Doehlemann, G. , Di Stasio, M. , Rössel, N. , Mendoza‐Mendoza, A. , Pester, D. , Müller, O. , Winterberg, B. , Meyer, E. , Ghareeb, H. , Wollenberg, T. , Münsterkötter, M. , Wong, P. , Walter, M. , Stuckenbrock, E. , Güldener, U. and Kahmann, R. (2010) Pathogenicity determinants in smut fungi revealed by genome comparison. Science, 330, 1546–1548. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S. , Ware, D. , Fulton, R.S. , Stein, J.C. , Wei, F. , Pasternak, S. , Liang, C. , Zhang, J. , Fulton, L. , Graves, T.A. , Minx, P. , Reily, A.D. , Courtney, L. , Kruchowski, S.S. , Tomlinson, C. , Strong, C. , Delehaunty, K. , Fronick, C. , Courtney, B. , Rock, S.M. , Belter, E. , Du, F. , Kim, K. , Abbott, R.M. , Cotton, M. , Levy, A. , Marchetto, P. , Ochoa, K. , Jackson, S.M. , Gillam, B. , Chen, W. , Yan, L. , Higginbotham, J. , Cardenas, M. , Waligorski, J. , Applebaum, E. , Phelps, L. , Falcone, J. , Kanchi, K. , Thane, T. , Scimone, A. , Thane, N. , Henke, J. , Wang, T. , Ruppert, J. , Shah, N. , Rotter, K. , Hodges, J. , Ingenthron, E. , Cordes, M. , Kohlberg, S. , Sgro, J. , Delgado, B. , Mead, K. , Chinwalla, A. , Leonard, S. , Crouse, K. , Collura, K. , Kudrna, D. , Currie, J. , He, R. , Angelova, A. , Rajasekar, S. , Mueller, T. , Lomeli, R. , Scara, G. , Ko, A. , Delaney, K. , Wissotski, M. , Lopez, G. , Campos, D. , Braidotti, M. , Ashley, E. , Golser, W. , Kim, H. , Lee, S. , Lin, J. , Dujmic, Z. , Kim, W. , Talag, J. , Zuccolo, A. , Fan, C. , Sebastian, A. , Kramer, M. , Spiegel, L. , Nascimento, L. , Zutavern, T. , Miller, B. , Ambroise, C. , Muller, S. , Spooner, W. , Narechania, A. , Ren, L. , Wei, S. , Kumari, S. , Faga, B. , Levy, M.J. , McMahan, L. , Van Buren, P. , Vaughn, M.W. , Ying, K. , Yeh, C.T. , Emrich, S.J. , Jia, Y. , Kalyanaraman, A. , Hsia, A.P. , Barbazuk, W.B. , Baucom, R.S. , Brutnell, T.P. , Carpita, N.C. , Chaparro, C. , Chia, J.M. , Deragon, J.M. , Estill, J.C. , Fu, Y. , Jeddeloh, J.A. , Han, Y. , Lee, H. , Li, P. , Lisch, D.R. , Liu, S. , Liu, Z. , Nagel, D.H. , McCann, M.C. , San Miguel, P. , Myers, A.M. , Nettleton, D. , Nguyen, J. , Penning, B.W. , Ponnala, L. , Schneider, K.L. , Schwartz, D.C. , Sharma, A. , Soderlund, C. , Springer, N.M. , Sun, Q. , Wang, H. , Waterman, M. , Westerman, R. , Wolfgruber, T.K. , Yang, L. , Yu, Y. , Zhang, L. , Zhou, S. , Zhu, Q. , Bennetzen, J.L. , Dawe, R.K. , Jiang, J. , Jiang, N. , Presting, G.G. , Wessler, S.R. , Aluru, S. , Martienssen, R.A. , Clifton, S.W. , McCombie, W.R. , Wing, R.A. and Wilson, R.K. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science, 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Supek, F. , Bošnjak, M. , Škunca, N. and Šmuc, T. (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE, 6 (7), e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard, J.A. , Carroll, A.M. , Kang, S. , Farrall, L. , Chumley, F.G. and Valent, B. (1995) Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell, 7, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken, F. and Rep, M. (2010) The arms race between tomato and Fusarium oxysporum . Mol. Plant Pathol. 11, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosa, Y. , Osue, J. , Eto, Y. , Oh, H.‐S. , Nakayashiki, H. , Mayama, S. and Leong, S.A. (2005) Evolution of an avirulence gene, AVR1‐CO39, concomitant with the evolution and differentiation of Magnaporthe oryzae . Mol. Plant–Microbe Interact. 18, 1148–1160. [DOI] [PubMed] [Google Scholar]
- Underwood, W. (2012) The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci. 3, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Burg, H.A. , Harrison, S.J. , Joosten, M.H.A.J. , Vervoort, J. and de Wit, P.J.G.M. (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant–Microbe Interact. 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Van der Does, H.C. and Rep, M. (2007) Virulence genes and the evolution of host specificity in plant‐pathogenic fungi. Mol. Plant–Microbe Interact. 20, 1175–1182. [DOI] [PubMed] [Google Scholar]
- Walton, J.D. (2000) Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet. Biol. 30, 167–171. [DOI] [PubMed] [Google Scholar]
- Wilson, J.M. and Frederiksen, R.A. (1970) Histopathology of the interaction of Sorghum bicolor and Sphacelotheca reiliana . Phytopathology, 60, 828–833. [Google Scholar]
- Wollenberg, T. and Schirawski, J. (2014) Comparative genomics of plant fungal pathogens: the Ustilago–Sporisorium paradigm. PLoS Pathog. 10 (7), e1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J. , Gao, L. , Yang, Y. , Zhai, J. , Arikit, S. , Yu, Y. , Duan, S. , Chan, V. , Xiong, Q. , Yan, J. , Li, S. , Liu, R. , Wang, Y. , Tang, G. , Meyers, B.C. , Chen, X. and Ma, W. (2014) Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 79, 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi, Y. , Nakamura, T. and Small, I. (2014) The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 78, 772–782. [DOI] [PubMed] [Google Scholar]
- Yi, M. and Valent, B. (2013) Communication between filamentous pathogens and plants at the biotrophic interface. Annu. Rev. Phytopathol. 51, 587–611. [DOI] [PubMed] [Google Scholar]
- Zhou, D. , Xin, Z. , Yi, L. , Zhenhai, Z. and Zhen, S. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther, K. , Kahnt, J. , Utermark, J. , Imkampe, J. , Uhse, S. and Schirawski, J. (2012) Host specificity of Sporisorium reilianum is tightly linked to generation of the phytoalexin luteolinidin by Sorghum bicolor . Mol. Plant–Microbe Interact. 25, 1230–1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Macroscopic symptoms on sorghum and maize after inoculation with Sporisorium reilianum. Seedlings of sorghum (A–F) or maize (G–L) were syringe inoculated with Sporisorium reilianum f. sp. reilianum (SRS) (A–C, G–I) or S. reilianum f. sp. zeae (SRZ) (D–F, J–L). Samples of plants were analysed at 4 days after inoculation (dai) (A, D, G, J), 21 dai (I, L), 35 dai (B, E), 50 dai (H, K) or 75 dai (C, F). Sorghum plants inoculated with SRS showed only chlorosis on leaves (A), but resulted in meristems wrapped in a white peridium (B) and spore formation in the apical inflorescences (C). Penetrating hyphae were colourless (A, inset). However, SRZ‐inoculated sorghum plants displayed red phytoalexin accumulation in leaves (D), and phytoalexins also stained fungal hyphae red (D, inset). SRZ‐inoculated sorghum plants developed healthy meristems (E), and inflorescences containing normal seeds (F) were produced. In maize, SRS‐inoculated plants showed weak chlorosis on leaves (G) and leaf‐like structures in cobs (H), but tassels were healthy (I). SRZ‐inoculated maize presented strong chlorosis on leaves (J), and produced spores and leaf‐like structures in cobs (K) and tassels (L). Bars: 1 cm in A–L, 20 μm in the insets. Insets in (A) and (D) are bright field micrographs of untreated leaf samples. Arrows in the insets of (A) and (D) point to fungal hyphae. Insets in (H) and (I) represent inflorescences of mock‐inoculated plants.
Table S1 List of significant gene ontology (GO) terms in Sporisorium reilianum f. sp. zeae (SRZ)‐infected samples vs. water control samples.
Table S2 List of significant gene ontology (GO) terms in Sporisorium reilianum f. sp. reilianum (SRS)‐infected samples vs. water control samples.
Table S3 List of significant gene ontology (GO) terms in Sporisorium reilianum f. sp. reilianum (SRS)‐infected samples vs. S. reilianum f. sp. zeae (SRZ)‐infected samples.
Table S4 List of maize genes significantly up‐regulated in the comparison Zm‐SRZ vs. Zm‐SRS. SRS, Sporisorium reilianum f. sp. reilianum; SRZ, S. reilianum f. sp. zeae; Zm, Zea mays.
Table S5 List of maize genes significantly up‐regulated in the comparison Zm‐SRS vs. Zm‐SRZ. SRS, Sporisorium reilianum f. sp. reilianum; SRZ, S. reilianum f. sp. zeae; Zm, Zea mays.
Table S6 List of oligonucleotides used for quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR).


