Summary
Fungal plant pathogens, such as Zymoseptoria tritici (formerly known as Mycosphaerella graminicola), secrete repertoires of effectors to facilitate infection or trigger host defence mechanisms. The discovery and functional characterization of effectors provides valuable knowledge that can contribute to the design of new and effective disease management strategies. Here, we combined bioinformatics approaches with expression profiling during pathogenesis to identify candidate effectors of Z. tritici. In addition, a genetic approach was conducted to map quantitative trait loci (QTLs) carrying putative effectors, enabling the validation of both complementary strategies for effector discovery. In planta expression profiling revealed that candidate effectors were up‐regulated in successive waves corresponding to consecutive stages of pathogenesis, contrary to candidates identified by QTL mapping that were, overall, expressed at low levels. Functional analyses of two top candidate effectors (SSP15 and SSP18) showed their dispensability for Z. tritici pathogenesis. These analyses reveal that generally adopted criteria, such as protein size, cysteine residues and expression during pathogenesis, may preclude an unbiased effector discovery. Indeed, genetic mapping of genomic regions involved in specificity render alternative effector candidates that do not match the aforementioned criteria, but should nevertheless be considered as promising new leads for effectors that are crucial for the Z. tritici–wheat pathosystem.
Keywords: bioinformatics, effectors, knock‐out, QTL mapping, septoria tritici blotch, wheat, Zymoseptoria tritici
Introduction
Zymoseptoria tritici (Desm.) Quaedvlieg & Crous (Quaedvlieg et al., 2011), the causal agent of septoria tritici blotch (STB) disease, is a major threat for global wheat production (Eyal, 1999). This foliar blight frequently occurs in many countries throughout the world, but particularly in regions with high rainfall and moderate temperatures, where the disease is responsible for significant yield losses, causing very large direct and indirect costs, representing millions of euros for disease control (Eyal, 1987). Over the last decade, Z. tritici has emerged as a genetic model for the Dothideales (Goodwin et al., 2004) as a result of its completed genome sequence (Goodwin et al., 2011) and detailed genetic studies (Kema et al., 1996c, 2002; Linde et al., 2002; Mirzadi Gohari et al., 2014; Wittenberg et al., 2009), and was recently placed in the top ten of the most important global plant pathogens (Dean et al., 2012).
Zymoseptoria tritici has a hemibiotrophic lifestyle with two distinct colonization phases (a stealthy biotrophic phase and a ramifying necrotrophic phase) in which various aspects of growth and differentiation can be studied in detail using a range of biological and molecular tools. Following stomatal penetration, the initial symptomless biotrophic phase, in which hyphae colonize the extracellular space, lasts for about 10 days post‐infection (dpi). The transition to necrotrophy coincides with the formation of small chlorotic lesions that gradually expand and coalesce into large necrotic blotches bearing abundant pycnidia, the asexual fructifications that contain the splash‐borne pycnidiospores (Orton et al., 2011). An array of pathogen‐derived toxic compounds have been suggested to be actively secreted into the apoplast, but the accurate events and mechanisms underlying this complex phase are poorly understood (Cohen and Eyal, 1993; Kema et al., 1996d). The genetic diversity in natural populations of the fungus is driven by the sexual process, which comprises several cycles within a single growing season and results in extraordinary diverse airborne inoculum (Chen and McDonald, 1996; Wittenberg et al., 2009; Zhan et al., 2003). In addition, Z. tritici produces splash‐dispersed asexual conidia during the growing season which disseminate over shorter distances and result in largely clonal foci (Hunter et al., 1999; Kema et al., 1996c; Suffert and Sache, 2011; Zhan et al., 2003).
Zymoseptoria tritici is pathogenic on both hexaploid bread wheat (Triticum aestivum L., AABBDD, 2n = 42) and tetraploid durum wheat [T. turgidum L. (Thell.) ssp. durum L., AABB, 2n = 28] (Kema et al., 1996a), as well as various grass species (Stukenbrock et al., 2007). Interestingly, isolates of Z. tritici exhibit a high degree of host species specificity and cultivar specificity (Kema et al., 1996a, b; Kema and van Silfhout, 1997). These are hierarchical levels of pathogenicity. Host species specificity in Z. tritici refers to avirulence to the vast majority of wheat cultivars of a wheat species. Thus, the majority of durum wheat cultivars are highly resistant to the majority of Z. tritici isolates originating from bread wheat, whereas the majority of bread wheat cultivars are highly resistant to the majority of Z. tritici isolates originating from durum wheat. Cultivar specificity is at a lower hierarchical level and refers to avirulence on particular cultivars within these wheat species (Eyal et al., 1973; Saadaoui, 1987; Van Ginkel and Scharen, 1988). A gene‐for‐gene interaction for cultivar specificity of Z. tritici in bread wheat has been proven, in which host resistance and pathogenicity are controlled by complementary single genetic loci (Brading et al., 2002; Kema et al., 2002). However, the genes controlling host species and cultivar specificity have thus far not been identified.
Fungal effector molecules are small secreted proteins (SSPs) that modulate physiological and morphological processes in plant hosts, thus promoting infection or triggering defence responses (Rep, 2005). This dual biological activity of effectors, which can function as virulence or avirulence factors, has been widely accepted to determine the eventual outcome of interactions between pathogens and their associated hosts (Bent and Mackey, 2007; Stergiopoulos and de Wit, 2009). Hence, the discovery and functional characterization of effectors can principally yield valuable knowledge that will eventually contribute to the design of new and effective disease management and resistance breeding strategies (Vleeshouwers and Oliver, 2014). The majority of characterized effectors in plant‐pathogenic fungi share similar structural features that can be used for their identification. Candidate effectors are usually small proteins (less than 300 amino acids) containing cysteine residues and an N‐terminus signal peptide that is required for extracellular secretion, here collectively called SSPs. It is well documented that some effectors are broadly present in different taxa, such as Ecp6 (de Jonge and Thomma, 2009), whereas others are unique and specific to an individual fungal species, such as AVRPiz‐t of Magnaporthe oryzae (Park et al., 2012). Despite their polymorphism, homologues of some effector proteins, such as Ecp6, Ecp2 and Avr4 (SSPs of the tomato pathogen Cladosporium fulvum), have been found in Z. tritici as well as the banana black leaf streak pathogen Mycosphaerella fijiensis (Bolton et al., 2008; Stergiopoulos et al., 2010, 2012, 2014). Nevertheless, the identification of fungal effectors through homology analyses is complicated because of their low conservation when compared with the identification of resistance gene analogues (Chisholm et al., 2006; Dangl and Jones, 2001). Hence, several complementary approaches have been employed to successfully identify functional SSPs in plant‐pathogenic fungi, including genetic analyses, bioinformatics cataloguing and functional genomics. For instance, a combined bioinformatics and RNA sequencing approach resulted in the discovery of Avr5 in the fungal tomato pathogen C. fulvum (Mesarich et al., 2014). In other cases, map‐based strategies have been used to clone effector genes, such as AvrLm1, AvrLm6 and AvrLm11 in the oilseed rape pathogen Leptosphaeria maculans (Balesdent et al., 2013; Fudal et al., 2007; Gout et al., 2006; Van de Wouw et al., 2014).
In Z. tritici, two effectors were identified by the functional analyses of homologues of the well‐known effector proteins MgNLP and Mg3LysM, which were functionality analysed by knock‐out and heterologous protein expression strategies. MgNLP belongs to the necrosis and ethylene‐inducing peptide 1 (Nep1)‐like protein family (NLP), but is not instrumental for the virulence of Z. tritici. However, its expressed protein in Pichia pastoris triggers cell death and the activation of defence‐related genes in Arabidopsis leaves (Motteram et al., 2009). Mg3LysM plays an essential role in the establishment of the initial symptomless biotrophic phase of Z. tritici (Marshall et al., 2011).
Previously, we have developed a robust protocol to cross Z. tritici isolates, providing an excellent tool for the generation of mapping populations and their analysis and deployment in genome assembly (Goodwin et al., 2011; Wittenberg et al., 2009). Here, we report the cataloguing of SSPs, subsequent expression profiling during pathogenesis and, eventually, a complementary quantitative trait locus (QTL) mapping approach to identify whether candidate effectors map to these regions on the Z. tritici genome. The analyses have resulted in a list of promising SSPs that remain to be explored in future studies. However, they also indicate an intriguing ambiguity between bioinformatics‐ and expression profiling‐driven SSP identification and characterization versus map‐based strategies, thereby questioning the potential of unbiased sequence‐based strategies for effector discovery.
Results
Identification of candidate effectors
In order to build a comprehensive list of conceivable SSPs, we followed two strategies. First, we mined the genome of Z. tritici, which resulted in the identification of 266 secreted proteins with a size of ≤300 amino acids and with four or more cysteine residues. Twenty‐four of these were predicted to possess transmembrane (TM) domains outside the signal peptide sequence, and were therefore excluded from the list. Subsequently, the expressed sequence tag (EST) database, which is accessible via the JGI genome browser (http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html), was used to further narrow down the list to 68 SSPs with transcript support (Table 1 ). Second, we used the in vitro secretome of Z. tritici (Mirzadi Gohari et al., 2014) for another round of independent SSP identification. This resulted in the identification of 114 extracellular proteins, 94 of which were supported by EST analyses. Eventually, after using the above‐mentioned criteria, we narrowed this number down to 30 candidates. Interestingly, both strategies resulted in two largely complementary sets of candidates, as the overlap was only nine SSPs. Eventually, we selected the entire set of 68 candidates from the in silico bioinformatics approach and supplemented this with 10 randomly selected candidates from the secretome analysis, resulting in a total of 78 SSPs for expression profiling (Table 1).
Table 1.
Overall characterization of the Zymoseptoria tritici small secreted proteins (SSPs) used in this study
| Protein | Protein ID | EST support | Location on chromosome | Length | No. Cys | PFAM domain | Z. tritici specific | Approach to identify |
|---|---|---|---|---|---|---|---|---|
| SSP1 | 73448 | Y | 6 | 180 | 8 | N | Y | Bioinformatics analysis |
| SSP2 | 79161 | Y | 1 | 69 | 7 | N | Y | Bioinformatics analysis |
| SSP3 | 79286 | Y | 2 | 64 | 8 | N | Y | Bioinformatics analysis |
| SSP4 | 80332 | Y | 4 | 77 | 6 | N | Y | Bioinformatics analysis |
| SSP5 | 82029 | Y | 9 | 67 | 7 | N | Y | Bioinformatics analysis |
| SSP6 | 82925 | Y | 12 | 58 | 6 | N | Y | Bioinformatics analysis |
| SSP7 | 83081 | Y | 13 | 54 | 6 | N | Y | Bioinformatics analysis |
| SSP8 | 87205 | Y | 8 | 56 | 9 | N | Y | Bioinformatics analysis |
| SSP9 | 98580 | Y | 1 | 271 | 8 | CFEM domain | N | Bioinformatics analysis |
| SSP10 | 99124 | Y | 2 | 113 | 7 | N | Y | Bioinformatics analysis |
| SSP11 | 99161 | Y | 2 | 165 | 18 | N | N | Bioinformatics analysis |
| SSP12 | 99676 | Y | 3 | 169 | 5 | N | N | Bioinformatics analysis |
| SSP13 | 100649 | Y | 7 | 76 | 8 | N | Y | Bioinformatics analysis |
| SSP14 | 102617 | Y | 1 | 159 | 13 | N | Y | Bioinformatics analysis |
| SSP15 | 102792 | Y | 1 | 115 | 8 | N | Y | Bioinformatics analysis |
| SSP16 | 102849 | Y | 1 | 286 | 6 | N | N | Bioinformatics analysis |
| SSP17 | 102996 | Y | 1 | 164 | 8 | N | Y | Bioinformatics analysis |
| SSP18 | 103572 | Y | 3 | 67 | 7 | N | Y | Bioinformatics analysis |
| SSP19 | 103713 | Y | 3 | 104 | 4 | N | Y | Bioinformatics analysis |
| SSP20 | 103792 | Y | 3 | 197 | 16 | PAN domain | N | Bioinformatics analysis |
| SSP21 | 104000 | Y | 4 | 181 | 6 | N | N | Bioinformatics analysis |
| SSP22 | 104383 | Y | 5 | 75 | 8 | N | Y | Bioinformatics analysis |
| SSP23 | 104444 | Y | 5 | 80 | 10 | N | Y | Bioinformatics analysis |
| SSP24 | 104758 | Y | 6 | 119 | 10 | N | Y | Bioinformatics analysis |
| SSP25 | 105182 | Y | 7 | 144 | 6 | N | N | Bioinformatics analysis |
| SSP26 | 105826 | Y | 10 | 99 | 8 | N | Y | Bioinformatics analysis |
| SSP27 | 106125 | Y | 11 | 71 | 6 | N | Y | Bioinformatics analysis |
| SSP28 | 106127 | Y | 11 | 98 | 6 | N | Y | Bioinformatics analysis |
| SSP29 | 106260 | Y | 12 | 110 | 16 | N | N | Bioinformatics analysis |
| SSP30 | 106445 | Y | 13 | 120 | 10 | N | Y | Bioinformatics analysis |
| SSP31 | 106502 | Y | 13 | 90 | 8 | N | Y | Bioinformatics analysis |
| SSP32 | 107286 | Y | 1 | 117 | 6 | N | Y | Bioinformatics analysis |
| SSP33 | 108877 | Y | 3 | 112 | 6 | N | Y | Bioinformatics analysis |
| SSP34 | 110220 | Y | 7 | 132 | 6 | N | Y | Bioinformatics analysis |
| SSP35 | 110386 | Y | 8 | 195 | 10 | LysM domain | N | Bioinformatics analysis |
| SSP38 | 103900 | Y | 3 | 130 | 8 | N | Y | Bioinformatics analysis |
| SSP39 | 104283 | Y | 4 | 227 | 12 | N | N | Bioinformatics analysis |
| SSP40 | 104404 | Y | 5 | 180 | 4 | Ecp2 | N | Bioinformatics analysis |
| SSP41 | 104697 | Y | 5 | 150 | 4 | N | N | Bioinformatics analysis |
| SSP42 | 104794 | Y | 6 | 158 | 4 | N | N | Bioinformatics analysis |
| SSP43 | 104867 | Y | 6 | 171 | 4 | N | N | Bioinformatics analysis |
| SSP44 | 105223 | Y | 7 | 189 | 5 | N | N | Bioinformatics analysis |
| SSP45 | 105265 | Y | 7 | 200 | 10 | N | N | Bioinformatics analysis |
| SSP46 | 105478 | Y | 8 | 136 | 9 | N | N | Bioinformatics analysis |
| SSP47 | 105487 | Y | 8 | 98 | 4 | LysM domain | N | Bioinformatics analysis |
| SSP48 | 105659 | Y | 9 | 183 | 4 | N | Y | Bioinformatics analysis |
| SSP49 | 105677 | Y | 9 | 199 | 4 | N | N | Bioinformatics analysis |
| SSP50 | 106106 | Y | 11 | 157 | 6 | N | N | Bioinformatics analysis |
| SSP51 | 106335 | Y | 12 | 239 | 5 | N | N | Bioinformatics analysis |
| SSP52 | 106345 | Y | 12 | 164 | 4 | N | N | Bioinformatics analysis |
| SSP53 | 111505 | Y | 12 | 198 | 4 | N | N | Bioinformatics analysis |
| SSP54 | 108976 | Y | 4 | 201 | 6 | N | N | Bioinformatics analysis |
| SSP55 | 109137 | Y | 4 | 155 | 4 | N | N | Bioinformatics analysis |
| SSP56 | 107904 | Y | 2 | 171 | 6 | Ecp2 | N | Bioinformatics analysis |
| SSP57 | 108329 | Y | 2 | 193 | 6 | N | N | Bioinformatics analysis |
| SSP58 | 108482 | Y | 3 | 109 | 10 | N | N | Bioinformatics analysis |
| SSP59 | 110756 | Y | 9 | 247 | 7 | Cerato‐platanin | N | Bioinformatics analysis |
| SSP60 | 109710 | Y | 6 | 201 | 5 | SCP domain | N | Bioinformatics analysis |
| SSP61 | 111636 | Y | 13 | 160 | 4 | Ecp2 | N | Bioinformatics analysis |
| SSP62 | 111352 | Y | 11 | 165 | 6 | N | Y | Bioinformatics analysis |
| SSP63 | 111008 | Y | 10 | 220 | 4 | N | N | Bioinformatics analysis |
| SSP64 | 111027 | Y | 10 | 152 | 4 | N | N | Bioinformatics analysis |
| SSP65 | 67799 | Y | 2 | 273 | 7 | N | N | Bioinformatics analysis |
| SSP66 | 70376 | Y | 3 | 203 | 4 | N | N | Bioinformatics analysis |
| SSP67 | 76021 | Y | 10 | 280 | 7 | Peptidase‐M43 | N | Bioinformatics analysis |
| SSP68 | 71724 | Y | 5 | 274 | 6 | Glyco‐hydro | N | Bioinformatics analysis |
| SSP69 | 111203 | Y | 11 | 155 | 6 | N | Y | Bioinformatics analysis |
| SSP70 | 85504 | N | 4 | 150 | 4 | Cerato‐platanin | N | Bioinformatics analysis |
| SSP126 | 110887 | Y | 9 | 284 | 14 | N | Y | Secretome analysis |
| SSP128 | 86778 | Y | 7 | 132 | 5 | N | Y | Secretome analysis |
| SSP134 | 106446 | Y | 13 | 120 | 10 | N | Y | Secretome analysis |
| SSP138 | 100429 | Y | 6 | 164 | 1 | N | Y | Secretome analysis |
| SSP139 | 41491 | N | 5 | 82 | 8 | N | N | Secretome analysis |
| SSP140 | 104441 | Y | 5 | 285 | 6 | N | Y | Secretome analysis |
| SSP142 | 103877 | Y | 3 | 169 | 5 | N | N | Secretome analysis |
| SSP146 | 91995 | N | 3 | 168 | 5 | N | N | Secretome analysis |
| SSP147 | 96868 | N | 11 | 157 | 4 | N | N | Secretome analysis |
| SSP160 | 92365 | N | 4 | 107 | 8 | PI‐PLC‐X | N | Secretome analysis |
| SSP71 | 90699 | N | 2 | 154 | 6 | N | N | QTL analysis |
| SSP74 | 103274 | N | 2 | 177 | 3 | N | Y | QTL analysis |
| SSP77 | 79484 | Y | 2 | 73 | 4 | N | Y | QTL analysis |
| SSP84 | 102982 | Y | 1 | 107 | 2 | N | Y | QTL analysis |
| SSP85 | 102983 | Y | 1 | 55 | 0 | N | Y | QTL analysis |
| SSP89 | 41315 | Y | 4 | 196 | 6 | N | N | QTL analysis |
| SSP91 | 71216 | Y | 4 | 197 | 1 | N | N | QTL analysis |
| SSP100 | 104754 | Y | 6 | 164 | 1 | N | Y | QTL analysis |
| SSP101 | 93741 | N | 6 | 160 | 0 | N | N | QTL analysis |
| SSP103 | 72923 | Y | 6 | 113 | 0 | N | N | QTL analysis |
| SSP112 | 94597 | N | 7 | 163 | 1 | N | N | QTL analysis |
| SSP114 | 83064 | Y | 13 | 75 | 6 | N | Y | QTL analysis |
| SSP116 | 97500 | Y | 13 | 138 | 10 | N | Y | QTL analysis |
| SSP118 | 97526 | N | 13 | 206 | 12 | N | Y | QTL analysis |
| SSP125 | 93501 | N | 5 | 106 | 0 | N | N | QTL analysis |
| SSP150 | 71681 | Y | 5 | 410 | 4 | Cellulase | N | QTL analysis |
| SSP151 | 58567 | Y | 5 | 251 | 2 | Adh‐short | N | QTL analysis |
| SSP152 | 41969 | N | 5 | 545 | 11 | PLA2B | N | QTL analysis |
| SSP153 | 104341 | Y | 5 | 146 | 1 | N | Y | QTL analysis |
| SSP154 | 92954 | N | 5 | 363 | 2 | N | N | QTL analysis |
| SSP155 | 42164 | Y | 5 | 302 | 8 | HRXXH | N | QTL analysis |
| SSP156 | 100094 | Y | 5 | 303 | 0 | ZIP | N | QTL analysis |
Cys, cysteine; EST, expressed sequence tag; QTL, quantitative trait locus.
Zymoseptoria tritici SSPs show expression profiles that correspond with infection stages
Ninety‐three per cent of the selected SSP‐encoding genes were supported by EST data generated under either in vitro or in planta conditions (Kema et al., 2008; Keon et al., 2005). Here, we used reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis to determine their in planta expression profiles during pathogenesis. All genes, including those without previous EST support, were transcribed in planta and their profiles corresponded remarkably well with: (i) the early stage of infection or biotrophic phase (2 and 4 dpi); (ii) the transition from biotrophy to necrotrophy (8 dpi); and (iii) the necrotrophic phase (>8 dpi) (Fig. S1, see Supporting Information). For example, SSP42 was strongly induced (around 20‐fold) during biotrophy at 2 dpi (Fig. 1a). The SSPs specifically expressed during the transition phase, usually accompanied by early macroscopic chlorosis, included SSP15, whose expression at 8 dpi was 120‐fold (Fig. 1b). The group expressed during necrotrophy included, for instance, SSP44. Its expression started at 8 dpi, peaked at 12 dpi and subsequently dropped off until 20 dpi (Fig. 2a), whereas the expression of other SSPs peaked at 8 dpi and then steadily decreased until 20 dpi (Fig. 2b). In addition to these profiles, only four SSPs were particularly up‐regulated at the very end of pathogenesis (20 dpi), a phase that is characterized by the development of abundant pycnidia, the asexual fructifications of Z. tritici (Fig. S2, see Supporting Information). In summary, 42% of the selected SSPs were specifically expressed during necrotrophy [Figs 2, 3 and S3 (see Supporting Information)] and 21% were expressed during biotrophy and necrotrophy [Figs 3 and S4 (see Supporting Information)]. Finally, 18% of the selected SSPs were transcribed at low levels (less than one‐fold) throughout the entire infection process [Figs 3 and S5 (see Supporting Information)].
Figure 1.
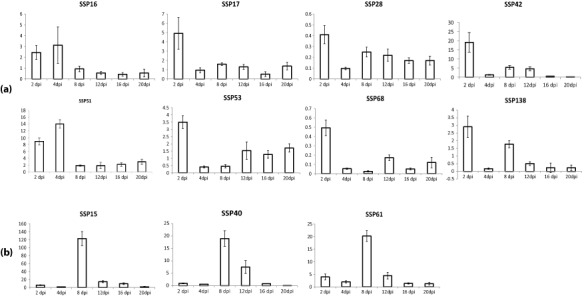
(a) Relative in planta expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) that are up‐regulated during biotrophy. (b) The transcription levels of the Z. tritici SSPs that are exclusively up‐regulated during the transition to necrotrophy. dpi, days post‐infection.
Figure 2.
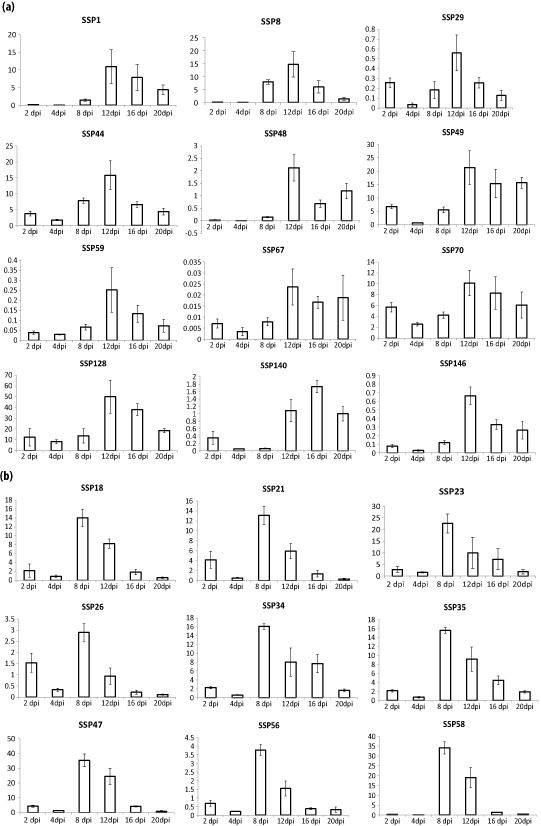
Relative in planta expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) that are specifically up‐regulated during necrotrophy: (a) SSPs that peak at 12 days post‐infection (dpi); (b) SSPs that peak at 8 dpi and subsequently decrease steadily towards 20 dpi.
Figure 3.
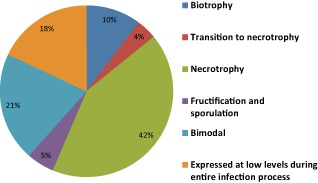
The majority of studied Zymoseptoria tritici small secreted proteins (SSPs) were highly expressed during necrotrophy. Each section corresponds to the percentage of studied SSPs that are up‐regulated during biotrophy [0–4 days post‐infection (dpi)], the transition phase to necrotrophy (4–8 dpi) and necrotrophy (>8 dpi). Also shown are sections with SSPs particularly important during fructification and sporulation (20 dpi), with a bimodal expression pattern (during biotrophy and necrotrophy), and SSPs that are expressed at low levels throughout the entire infection process.
Functional analyses of Z. tritici SSP15 and SSP18 reveal their dispensability for pathogenicity
As SSP15 and SSP18 were highly expressed at 8 dpi (120‐fold and 14‐fold, respectively), we generated three independent knock‐out mutants for each gene by homologous recombination to determine their role during pathogenesis. At 21 dpi, the developed phenotypes of the knock‐out strains and the wild‐type (WT) were similar on each evaluated wheat cultivar, indicating that SSP15 and SSP18 are dispensable for pathogenesis (Fig. S6, see Supporting Information). However, daily comparison of symptom development between the SSP15ΔIPO323 and WT strains showed a slight delay in pathogenesis in wheat accession FD3 between 14 and 16 dpi (necrosis and pycnidia differences), suggesting a quantitative effect of SSP15 during the late phase of infection, but only in this specific wheat accession, which is also used as a parent in the Stb mapping population (Goudemand et al., 2013).
An unbiased QTL approach results in SSPs that are expressed at low levels during pathogenesis
As the bioinformatics‐ and proteomics‐driven SSP approach did not reveal SSPs with a clear function in pathogenesis, despite their unique expression profiles, we alternatively considered a genetic approach to map candidate effectors. An existing mapping population from a cross between the Z. tritici reference strain IPO323 and the Algerian durum wheat field strain IPO95052 (Goodwin et al., 2011; Wittenberg et al., 2009) resulted in a progeny of 163 isolates that were phenotyped on a suite of durum wheat and bread wheat cultivars (Table S1, see Supporting Information). Isolate IPO323 developed less than 1% leaf area covered by pycnidia (P p) on the durum wheat cultivars, but was highly pathogenic on the bread wheat cultivars (P p values between 56% and 71%). As expected, the other parental isolate IPO95052 showed opposite responses, as it was highly pathogenic on the durum wheat cultivars (P p values between 49% and 62%), but avirulent on the bread wheat cultivars (P p values of less than 1%; Fig. 4a). Although both parental isolates were avirulent on cv. Shafir, we included this cultivar in the phenotyping to study the independent segregation of host species and cultivar specificity. The progeny strains clearly showed a highly diverse range of pathogenicity, with large qualitative and quantitative differences for host species and cultivar specificity (Fig. 4b). Among 163 progeny, 150 isolates represented recombinant phenotypes (for examples, see Fig. 4b), including isolates that were virulent or avirulent on all tested cultivars. Interestingly, nine isolates were virulent on bread wheat cv. Shafir despite the avirulence of both parental isolates on this cultivar.
Figure 4.
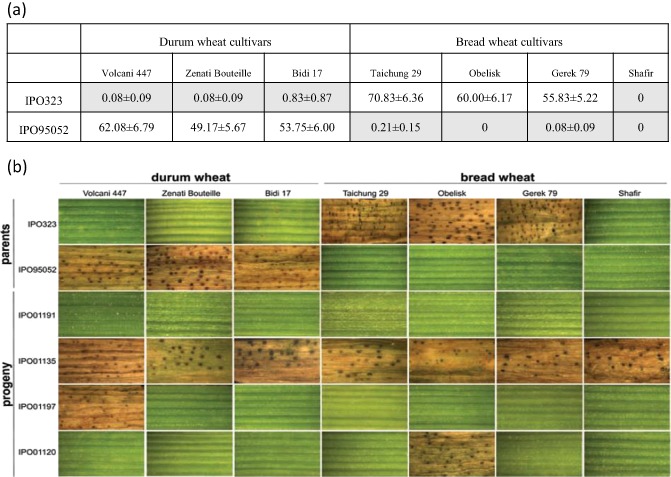
Phenotyping of Zymoseptoria tritici on wheat. (a) Percentages of primary leaf area covered with pycnidia averaged over 11 individual experiments. Grey highlighting indicates incompatible interactions. (b) Screening of the parental Z. tritici isolates IPO323 and IPO95052 and four progeny isolates from a cross between these strains on three durum wheat and four bread wheat cultivars.
Data analyses revealed a major QTL on chromosome 5 [logarithm of the odds (LOD) = 17.56], covering genes controlling specificity for the durum wheat cultivars Volcani 447, Zenati Bouteille and Bidi 17, as well as for the bread wheat cultivar Shafir, which explained up to 47% of the observed variation on these cultivars (Fig. 5, Table S1). In addition, eight additional QTLs with lower, although significant, LOD values were detected on chromosomes 2, 3, 4, 5, 6, 7 and 13 (Table 2, Fig. S7, see supporting information), see Supporting Information). Five of these eight QTLs control specificity for bread wheat cultivars and three for durum wheat cultivars (Table S1). None of the identified QTLs mapped to the dispensable chromosomes 14–21. Finally, we mapped the identified QTLs to the Z. tritici genome sequence and determined that they covered a total of 2795 genes (Table 2), comprising 64 secreted proteins containing signal peptides. Eventually, 15 SSPs were excluded because they contained glycosylphosphatidylinositol (GPI) anchors (two SSPs) and TM domain(s) outside of the signal peptide (13 SSPs), resulting in a final number of 49 SSPs under the identified QTLs that were partially supported by ESTs (http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html). We therefore performed an RT‐PCR of these SSPs in pooled cDNA at several days post‐infection to check for in planta expression (data not shown), and finally selected 22 SSPs as candidate effectors for further analyses (Table 1). Overall, the protein length for the selected SSPs ranged from 55 to 545 amino acids and the number of cysteine residues varied between 0 and 12. Twelve SSPs have a predicted role associated with polysaccharide degradation, seven with protein modification and two with lipid degradation (Table S2, see Supporting Information). Expression profiling of these SSPs surprisingly revealed that all SSPs were expressed less than one‐fold throughout pathogenesis, except for SSP114, which was ≤35‐fold expressed during necrotrophy [Figs 6 and S8 (see Supporting Information)].
Figure 5.
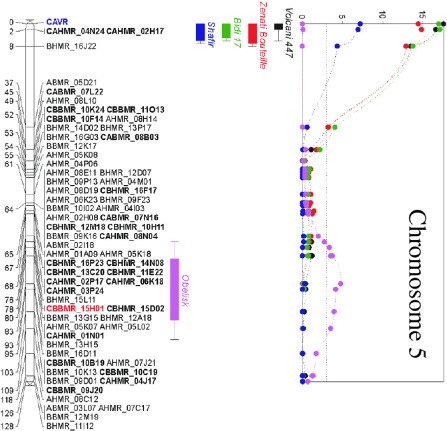
Quantitative trait loci (QTLs) for Zymoseptoria tritici pathogenicity measured by the percentage of foliage covered by pycnidia, the asexual fructifications that are positioned in the substomatal cavities, mapped on chromosome 5. The subtelomeric region carries a QTL with a logarithm of the odds (LOD) value of 17.56 that covers genes involved in cultivar or host specificity.
Table 2.
Overview and characterization of mapped quantitative trait loci for P parameters (leaf area covered by pycnidia) in Z ymoseptoria tritici with their physical boundaries on the genome and the protein families covered
| Chromosome number | QTL locus | QTL specificity and LOD value | QTL explained variance (%) | QTL position* | Number of predicted genes |
|---|---|---|---|---|---|
| Chromosome 2 | QTL‐P1 | BW Taichung 29 (LOD ≥ 5.70) | 15.1 | 1468524–3025189 | 586 |
| 1703574–2109386 (LOD ≥ 4.6) | 154 | ||||
| Chromosome 3 | QTL‐P2 | BW Shafir (LOD ≥ 3.22) | 8.6 | 3313278–3502029 | 69 |
| Chromosome 4 | QTL‐P3 | DW Volcani 447 (LOD ≥ 7.64) | 12.4 | 703405–2293727 | 581 |
| 1913310–2242122 (LOD ≥ 6) | 120 | ||||
| Chromosome 5 | QTL‐P4.1 | BW Shafir (LOD ≥ 7.20) | 23.5 | 146–252047 | 84 |
| QTL‐P4.2 | DW Zenati Bouteille (LOD ≥ 14.83) | 45.2 | 146–252047 | 84 | |
| QTL‐P4.3 | DW Volcani (LOD ≥ 16.60) 447 | 47.9 | 146–252047 | 84 | |
| QTL‐P4.4 | DW Bidi 17 (LOD ≥ 17.12) | 48.5 | 146–252047 | 84 | |
| QTL‐P5 | BW Obelisk (LOD ≥ 4.80) | 16.7 | 2308292–2709039 | 140 | |
| Chromosome 6 | QTL‐P6 | DW Bidi 17 (LOD ≥ 3.33) | 6.2 | 527868–942291 | 158 |
| Chromosome 7 | QTL‐P7 | BW Obelisk (LOD ≥ 3.76) | 10.9 | 977135–2038435 | 391 |
| QTL‐P8.1 | DW Bidi 17 (LOD ≥ 2.91) | 5.4 | 37079–95788 | 24 | |
| QTL‐P8.2 | DW Zenati Bouteille (LOD ≥ 3.89) | 7.7 | 37079–95788 | 24 | |
| Chromosome 13 | QTL‐P9.1 | BW Gerek (LOD ≥ 3.06) | 14.1 | 901651–1184505 | 106 |
| QTL‐P9.2 | BW Taichung 29 (LOD ≥ 7.61) | 28.3 | 901651–1184505 | 106 |
LOD, logarithm of the odds; QTL, quantitative trait locus.
*Sequence markers above the LOD threshold of 3.0 were used to determine the boundaries. If no sequences were available which mapped in the LOD > 3 interval, the first flanking marker with a mapped sequence was taken to determine the chromosomal location.
Figure 6.
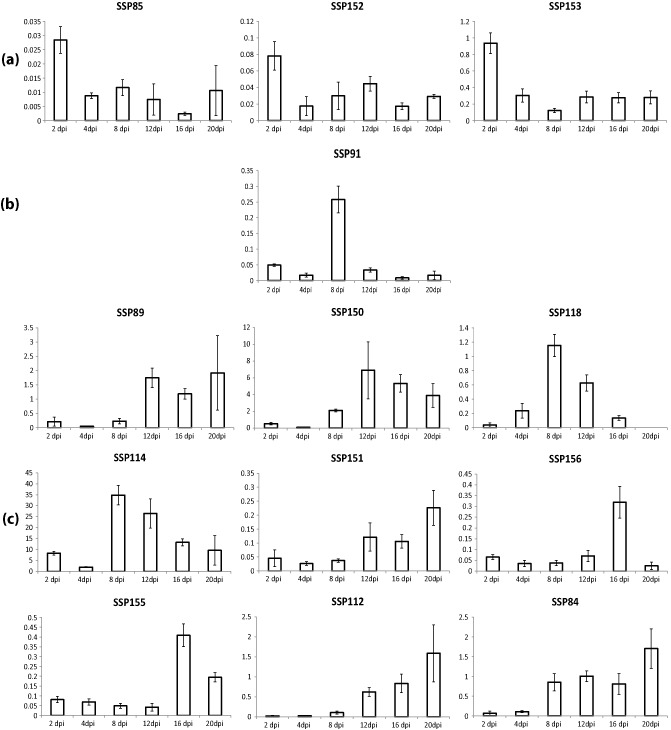
Relative in planta expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) that are positioned under the mapped quantitative trait loci (QTLs) on the Zymoseptoria tritici genome. (a) SSPs up‐regulated during biotrophy. (b) SSPs expressed during the transition to necrotrophy. (c) SSPs primarily expressed during necrotrophy, fructification and sporulation. dpi, days post‐infection.
Functional annotation of 100 ZtSSPs
Gene ontology analyses of the 100 studied ZtSSPs provided an overview of their putative biological and molecular functions. This revealed that the in planta‐expressed ZtSSPs are not clearly attributed to a specific biological process (Fig. S9a, see Supporting Information), whereas more than 50% are specifically considered to be involved in either catalytic activity or hydrolase activity (Fig. S9b).
Discussion
Septoria leaf blotch caused by Z. tritici is one of the most economically devastating wheat diseases around the world and globally impacts food security. The Z. tritici–wheat pathosystem currently represents a model to investigate the molecular mechanisms involved in pathogenicity and the infection process of hemibiotrophic fungal plant pathogens (Brading et al., 2002; Goodwin et al., 2004; Kellner et al., 2014; Rudd et al., 2015; Yang et al., 2013). The identification and characterization of effectors have contributed significantly to our understanding of the molecular mechanisms underlying pathogenesis, which are increasingly being used in the design of effector‐driven breeding programmes (Rietman et al., 2012; Vleeshouwers and Oliver, 2014; Vleeshouwers et al., 2008). To date, several strategies, including biochemical (Rose et al., 2002) and genetic (Fudal et al., 2007; Gout et al., 2006) approaches have been implemented to discover effector genes from plant pathogens, but primarily next‐generation sequencing (NGS) technologies have provided broad sets of genome sequence data from diverse microorganisms, including oomycete and fungal plant pathogens, that have resulted in a massive amount of putative effector genes (Dean et al., 2005; Haas et al., 2009; Kämper et al., 2006; Lévesque et al., 2010; Rouxel et al., 2011). The application of high‐performance bioinformatics tools, such as pexfinder (Torto et al., 2003), has enabled the mining of fungal genome sequences for effector proteins (Orsomando et al., 2001; Stergiopoulos et al., 2012) and, indeed, has resulted in the identification of key effectors playing major roles in host–pathogen interactions and other biological processes (de Jonge et al., 2012).
Despite the functional analyses of a range of genes that play key roles during pathogenesis in the Z. tritici—wheat pathosystem, key effectors have thus far not been identified (Cousin et al., 2006; Marshall et al., 2011; Mehrabi et al., 2006a, b; Motteram et al., 2009; Rudd et al., 2015). Here, we focused on a wide approach to identify and functionally analyse putative effector proteins by taking advantage of the completed genome sequence of Z. tritici (Goodwin et al., 2011). We primarily identified SSPs through a funnel strategy, starting with bioinformatics cataloguing, followed by expression profiling and functional analyses and, eventually, the short‐listing of candidates by linkage mapping.
First, we built a comprehensive list of 78 SSPs from in silico and in vitro proteomic analyses (Mirzadi Gohari et al., 2014) adopting protein size (less than 300 amino acids) and protein secretion as key qualifiers for candidate discovery (Rep, 2005). In addition, we included cysteine richness, the presence of GPI anchors and TM domains as important characteristics (Rep, 2005; Stergiopoulos and de Wit, 2009). Finally, we analysed expression profiles, enabling the identification of SSP15 and SSP18, two top candidate SSPs that were uniquely expressed during a specific and defined phase of pathogenesis. However, subsequent functional analyses revealed that these SSPs were dispensable for pathogenicity. We therefore concluded that an unbiased approach indeed results in a range of effector candidates, but does not suffice in the conclusive identification of key effectors. Moreover, each and every candidate needs to be functionally analysed, which is very time consuming. Hence, discovery criteria should be redefined or, alternatively, additional strategies should support candidate discovery. For instance, recently, MoCDIP1, a secreted protein of Magnaporthe oryzae, was discovered which encodes a homologue of ricin B lectin inducing cell death in both monocot and dicot species (Songkumarn, 2013). This protein has a size of 355 amino acids, suggesting that a borderline of 300 amino acids for SSPs is too narrow a qualification. Furthermore, recent transcriptome analysis of Colletotrichum higginsianum candidate effector (ChEC) genes has revealed successive waves of expression that correspond with the phases of infection. The first wave included genes that were particularly expressed in appressoria before penetration, and the second wave contained genes transcribed before and during penetration (O'Connell et al., 2012). Giraldo and Valent (2013) also suggested that plant pathogens express effector proteins during distinct stages of pathogenesis, probably based on delicately regulated and fine‐tuned requirements. Hence, sample preparations at defined phases of pathogenesis—such as, in our case, at 2, 4, 8, 12, 16 and 20 dpi—can still miss crucial SSPs with unique expression profiles. Our expression analysis of ZtSSPs is generally consistent with the above‐mentioned considerations, as we identified SSPs with distinct and/or high expression profiles (e.g. SSP42, SSP39 and SSP15), as well as those with similar expression profiles throughout pathogenesis (e.g. SSP2 and SSP6) or with remarkably low expression profiles (e.g. SSP5 and SSP20). In general, we observed three major in planta SSP expression waves: at biotrophy, when Z. tritici commences the invasion of the extracellular space of the mesophyll tissue; at the transition to necrotrophy, which is accompanied by the sudden appearance of chlorosis and necrosis and; at necrotrophy, when the largest group of candidate SSPs is up‐regulated, apparently to facilitate the further destruction of host cells and the access to nutrients supporting fructification. Indeed, these observations could suggest that Z. tritici deploys distinct SSPs at different pathogenic stages, e.g. to suppress host defences, facilitate colonization and, finally, induce host necrosis and survival by massive fructification (Kema et al., 1996d). Previous histological analyses have suggested that the switch from biotrophy to necrotrophy is associated with massive changes in mesophyll cell content, which are actually already underway from the moment Z. tritici hyphae colonize the apoplast at early phases of infection (Kema et al., 1996d). Therefore, we were particularly interested in SSPs with distinct expression patterns during this switch. Nevertheless, the functional analyses of two of the most highly expressed SSPs (SSP15 and SSP18) showed their dispensability for Z. tritici pathogenicity. Intriguingly, Rudd et al. (2015), following a RNAseq strategy, selected five candidate SSPs, including those functionally analysed in this study, and concluded that all of these candidates were dispensable for the pathogenicity of Z. tritici in wheat.
A search of the Z. tritici genomic database also resulted in the discovery of homologues of well‐known effector proteins, including two LysM effectors (SSP35 and SSP47) which have been functionally characterized previously (Marshall et al., 2011) and are highly transcribed during necrotrophy. This result accords with a recent transcriptomic analysis of the wheat—Z. tritici interaction by Yang et al. (2013). It has also been suggested that LysM effector proteins play a role in the suppression of chitin‐mediated wheat defence responses during the entire infection cycle, and may be essential for disease development because of the enhanced level of chitin/biomass during the necrotrophic phase (Lee et al., 2013; Marshall et al., 2011). We also identified cerato‐platanin, a protein that has been implicated as a phytotoxin or pathogen‐associated molecular pattern (PAMP) triggering host defence mechanisms, such as the salicylic acid (SA) pathway (Frías et al., 2011, 2013; Yang et al., 2009). Expression analysis of the Z. tritici cerato‐platanin protein, which we designate as ZtCP (SSP70), revealed that it is highly expressed during the necrotrophic phase. This suggests a possible role during the transition phase to necrotrophy or in ascertaining fungal proliferation during the end phase of pathogenesis, as has been proposed in the necrotrophic plant pathogen Botrytis cinerea (Frías et al., 2013). We therefore generated a knock‐out strain of ZtCP and tested its pathogenicity on 20 wheat cultivars. However, none of these IPO323ΔZtCP strains were attenuated in pathogenicity or virulence (data not shown). Hence, the exact biological role of ZtCP in the Z. tritici–wheat interaction remains to be elucidated in subsequent studies.
Eventually, essentially as a result of the failure of the aforementioned approaches to identify SSPs that are crucial for pathogenesis, we decided to explore the map‐based identification of SSP candidates. We mapped nine QTLs explaining between 5.4% and 47.9% of the observed variation on durum wheat cultivars Volcani 447, Zenati Bouteille and Bidi 17, as well as the bread wheat cultivars Shafir, Taichung 29, Gerek 79 and Obelisk. Subsequently, we catalogued SSPs that were placed under the mapped QTLs and showed that none of these overlapped with any of the SSPs identified by the other strategies. We subsequently determined their expression profiles and observed that all of these SSPs, except SSP114, were expressed at low levels throughout pathogenesis. This was contrary to our expectations and may indicate that an unbiased map‐based approach for effector discovery is the way forward to uncover essential components in the host–pathogen interaction. For instance, we identified SSP115 (302 amino acids) positioned under QTL5 with a LOD value of 17.56 explaining 47% of the observed variation, which is a homologue of BEC1019, recently characterized as a new class of biotrophic fungal effectors notably present in 97 of 271 sequenced fungal genomes (Whigham, 2013).
In summary, our data show that predetermined key qualifiers, including protein size, cysteine residues and expression patterns or magnitudes (Rep, 2005), so far have not revealed any useful links in the Z. tritici—wheat pathosystem. This corroborates the recent findings of Rudd et al. (2015). Clearly, any sampling strategy will exclude candidates that peak (transiently) at other stages of pathogenesis. Moreover, redundancy may also significantly affect the determination of individual SSPs and their role in pathogenesis, which is experimentally complicated to address, requiring double or triple knock‐out strains (Bakkeren and Valent, 2014; Gijzen et al., 2014). Furthermore, incomplete or incorrect annotations of the genomic stretches carrying the QTLs significantly hampers the discovery of essential SSPs. Therefore, we will pursue our strategy to fine map QTL windows in the regions of interest, followed by functional analyses of the QTL‐based SSPs, in order to determine their contribution to cultivar and wheat species specificity, which is crucial for effector‐driven wheat breeding programmes.
Experimental Procedures
Identification and bioinformatics analyses of SSPs
The genome of Z. tritici, which is publicly available at the JGI website (http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html), was mined and all SSPs ≤ 300 amino acids were retrieved. We followed the same pipeline as do Amaral et al. (2012) to predict the secretome of Z. tritici IPO323 with minor modifications. In silico prediction of extracellular proteins was performed using SignalP (v3.0; http://www.cbs.dtu.dk/services/SignalP/) to determine the presence of signal peptides. The number of cysteine residues inside the mature proteins was manually computed and the number of selected SSPs was narrowed down to those with four or more cysteine residues. The TargetP program (v1.1; http://www.cbs.dtu.dk/services/TargetP) was used to identify and remove SSPs with either a chloroplast transit peptide or a mitochondrial targeting peptide. tmhmm software was then utilized to identify and remove the TM‐containing SSPs, except those with one TM in the N‐terminal signal peptide. The remaining SSPs were screened for the presence of GPI anchor proteins using big‐PI (http://mendel.imp.ac.at/gpi/fungi_server.html). The EST databases (http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html) were used to further narrow down the SSPs to those that were transcribed in at least one of the three in planta or seven in vitro EST libraries. Finally, the selected SSPs were subjected to PFAM analysis using the PFAM database (ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/). In addition, BlastP analysis was conducted to determine whether SSPs were either conserved or unique for Z. tritici (e‐value of 10−6). Finally, we performed gene ontology analyses using Blast2go (Conesa et al., 2005).
Inoculations, RNA isolation and SSP expression profiling during pathogenesis in wheat
Wheat cv. Obelisk was used throughout these experiments. Ten‐day‐old plants were inoculated with a spore suspension of Z. tritici IPO323 using previously reported protocols (Mehrabi et al., 2006a). The infected leaves were collected in three biological replications and flash frozen in liquid nitrogen and ground manually using a mortar and pestle. Total RNA was isolated with the RNeasy plant mini kit (Qiagen, Valencia, CA, USA) and residual DNA was subsequently removed using the DNAfree kit (Ambion, Huntingdon, Cambridgeshire, UK). First‐strand cDNA was obtained from 2 μg of total RNA primed with oligo(dT) using SuperScript III (Invitrogen, Carlsbad, CA 92008, USA) according to the manufacturer's instructions. Expression analyses of selected SSPs were performed using RT‐qPCR. One microlitre of the resulting cDNA was used in a 25‐μL PCR employing a QuantiTect SYBR Green PCR Kit (Applied Biosystems, Warrington, UK), and run and analysed using an ABI 7500 Real‐Time PCR machine (Applied Biosystems, Foster City, CA 94404, USA). The relative expression levels of each gene were initially normalized by the constitutively expressed Z. tritici β‐tubulin gene (Keon et al., 2007; Motteram et al., 2009) and then calculated based on the comparative C(t) method described previously (Schmittgen and Livak, 2008).
Functional analyses
The gene deletion constructs, pΔMgSSP15 and pΔMgSSP18, were generated using the USER friendly cloning method with minor modifications, as described previously (Mirzadi Gohari et al., 2014). The constructs were then transformed into Agrobacterium tumefaciens strain LBA1100 via electroporation and, subsequently, A. tumefaciens‐mediated transformation (ATMT) was performed to delete MgSSP15 and MgSSP18 in Z. tritici IPO323, as described previously (Mehrabi et al., 2006a). All knock‐out strains and the WT strain Z. tritici IPO323 were compared for pathogenicity on 12 wheat cultivars (Table S3, see Supporting Information) following regular protocols (Mehrabi et al., 2006a).
Zymoseptoria tritici crosses, and selection and analyses of mapping populations
We used the Z. tritici reference IPO323 and isolate IPO95052, an Algerian field strain originating from durum wheat, for in planta crosses (Kema et al., 1996c). Two F1 progenies were generated on either the bread wheat cv. Obelisk (n = 103) or the durum wheat cv. Inbar (n = 60) which, after initial molecular and in planta analyses, were not significantly different and therefore bulked for further analyses. Each progeny isolate was used to inoculate (30 mL of 107 spores/mL) a set of bread and durum wheat cultivars (Table S1) in at least two biological repetitions, and disease severity was scored as the percentage primary leaf area covered by pycnidia (P p) at 21 dpi following the protocols and conditions reported previously (Kema et al., 1996c). Histograms with frequency distributions of progeny using log‐transformed P p data were generated for each cultivar using bins (classes) in intervals of 0.1 after logarithmic transformation to evaluate segregation distributions. The P p scores from 147 of the 163 progeny were used to map QTLs on a genetic linkage map previously generated with this population (Goodwin et al., 2011; Wittenberg et al., 2009). This subset includes 23 twin pairs of isolates for which P p data were merged after averaging. These twin pairs are genetically identical isolates resulting from mitosis after meiosis in an ascus, leading to four pairs of genetically identical ascospores (Wittenberg et al., 2009). Because histograms did not reveal normal distributions of virulence data, QTL mapping based on a continuous scale of P p was necessary. Mapping with the average of the log‐transformed P p data [log(average P p) + 1] from each isolate–cultivar result, versus mapping with the average raw P p data, yielded higher LOD values; hence, we therefore continued with log‐transformed data. The software program MapQTL 5.0 (van Ooijen, 1992) was used to detect QTLs with both the interval mapping (Lander and Botstein, 1989) and Multiple‐QTL Mapping (MQM) mapping (Jansen, 1994) methods. First, interval mapping was performed to detect QTLs. Subsequently, co‐factors were determined using the automatic co‐factor selection option, followed by MQM mapping of the same trait with the selected co‐factor(s) to identify new QTLs. The LOD profiles and the percentage of explained variance were obtained with the MQM mapping approach when co‐factors were selected. When only one QTL was detected, the LOD profile of the interval mapping procedure was shown. Permutation tests were performed to determine QTL significance, which resulted in a genome‐wide significance threshold of LOD = 3.0 for all traits. LOD profiles were graphically displayed using MapChart version 2.2 (Voorrips, 2002), including the LOD − 1/LOD − 2 support interval to approximate a 95% confidence interval (van Ooijen, 1992).
Supporting information
Fig. S1 Schematic diagram showing different infection processes of Zymoseptori tritici.
Fig. S2 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) up‐regulated during fructification/sporulation. dpi, days post‐infection.
Fig. S3 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) up‐regulated during the transition/necrotrophic phase of pathogenesis. dpi, days post‐infection.
Fig. S4 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) with a bimodal pattern. dpi, days post‐infection.
Fig. S5 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) expressed at low levels throughout pathogenesis. dpi, days post‐infection.
Fig. S6 Disease development after inoculation with the Zymoseptoria tritici wild‐type (WT) strain IPO323 and the IPO323ΔSSP15 and IPO323ΔSSP18 knock‐out strains in 12 wheat cultivars.
Fig. S7 Mapped quantitative trait loci involved in (a)virulence of Zymoseptoria tritici to seven wheat cultivars.
Fig. S8 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) positioned under mapped quantitative trait loci (QTLs) and expressed at low levels throughout pathogenesis.
Fig. S9 Gene ontology (GO) analyses of 100 Zymoseptoria tritici small secreted proteins (SSPs).
Table S1 Mapped quantitative trait loci involved in (a)virulence in progeny isolates of Zymoseptoria tritici from a cross between isolates IPO323 and IPO95052 tested for virulence on the durum wheat cultivars Volcani 447, Zenati Bouteille and Bidi 17 and the bread wheat cultivars Taichung 29, Obelisk, Gerek 79 and Shafir.
Table S2 Zymoseptoria tritici small secreted proteins (SSPs) positioned under the quantitative trait loci (QTLs) involved in the degradation of polysaccharides, proteins and lipids.
Table S3 Cultivars used in this study.
Acknowledgements
A. Mirzadi Gohari was financially supported by the Ministry of Research and Technology of Iran. We would like to thank Bertus vander Laan at Unifarm of Wageningen University for maintaining the glasshouse in an excellent condition. This research was partially supported by a grant from the French ‘Fonds de Soutien à l'Obtention Végétale’ (FSOV) within the program FSOV 2010K ‘Développement d'un nouvel outil d'aide à la sélection de variétés de blé résistant à la septoriose’.
References
- do Amaral, A.M. , Antoniw, J. , Rudd, J.J. and Hammond‐Kosack, K.E. (2012) Defining the predicted protein secretome of the fungal wheat leaf pathogen Mycosphaerella graminicola . PLoS ONE, 7, e49904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkeren, G. and Valent, B. (2014) Do pathogen effectors play peek‐a‐boo? Front. Plant Sci. 5, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balesdent, M.H. , Fudal, I. , Ollivier, B. , Bally, P. , Grandaubert, J. , Eber, F. , Chèvre, A.M. , Leflon, M. and Rouxel, T. (2013) The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa . New Phytol. 198, 887–898. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , Van Esse, H.P. , Vossen, J.H. , De Jonge, R. , Stergiopoulos, I. , Stulemeijer, I.J. , Van Den Berg, G. , Borrás‐Hidalgo, O. , Dekker, H.L. and De Koster, C.G. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69, 119–136. [DOI] [PubMed] [Google Scholar]
- Brading, P.A. , Verstappen, E.C. , Kema, G.H.J. and Brown, J.K. (2002) A gene‐for‐gene relationship between wheat and Mycosphaerella graminicola, the Septoria tritici blotch pathogen. Phytopathology, 92, 439–445. [DOI] [PubMed] [Google Scholar]
- Chen, R.‐S. and McDonald, B.A. (1996) Sexual reproduction plays a major role in the genetic structure of populations of the fungus Mycosphaerella graminicola . Genetics, 142, 1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cohen, L. and Eyal, Z. (1993) The histology of processes associated with the infection of resistant and susceptible wheat cultivars with Septoria tritici. Plant Pathol. 42, 737–743. [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J.M. , Terol, J. , Talón, M. and Robles, M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cousin, A. , Mehrabi, R. , Guilleroux, M. , Dufresne, M. , Van der Lee, T. , Waalwijk, C. , Langin, T. and Kema, G.H.J. (2006) The MAP kinase‐encoding gene MgFus3 of the non‐appressorium phytopathogen Mycosphaerella graminicola is required for penetration and in vitro pycnidia formation. Mol. Plant Pathol. 7, 269–278. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. and Ellis, J. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R.A. , Talbot, N.J. , Ebbole, D.J. , Farman, M.L. , Mitchell, T.K. , Orbach, M.J. , Thon, M. , Kulkarni, R. , Xu, J.‐R. and Pan, H. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature, 434, 980–986. [DOI] [PubMed] [Google Scholar]
- Eyal, Z. (1987) The Septoria Diseases of Wheat: Concepts and Methods of Disease Management. Mexico: International Maize and Wheat Improvement Center (CIMMYT). [Google Scholar]
- Eyal, Z. (1999) The Septoria tritici and Stagonospora nodorum blotch diseases of wheat. Eur. J. Plant Pathol. 105, 629–641. [Google Scholar]
- Eyal, Z. , Amiri, Z. and Wahl, I. (1973) Physiologic specialization of Septoria tritici. Phytopathology, 63, 91. [Google Scholar]
- Frías, M. , González, C. and Brito, N. (2011) BcSpl1, a cerato‐platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192, 483–495. [DOI] [PubMed] [Google Scholar]
- Frías, M. , Brito, N. and González, C. (2013) The Botrytis cinerea cerato‐platanin BcSpl1 is a potent inducer of systemic acquired resistance (SAR) in tobacco and generates a wave of salicylic acid expanding from the site of application. Mol. Plant Pathol. 14, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudal, I. , Ross, S. , Gout, L. , Blaise, F. , Kuhn, M. , Eckert, M. , Cattolico, L. , Bernard‐Samain, S. , Balesdent, M. and Rouxel, T. (2007) Heterochromatin‐like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map‐based cloning of AvrLm6. Mol. Plant–Microbe Interact. 20, 459–470. [DOI] [PubMed] [Google Scholar]
- Gijzen, M. , Ishmael, C. and Shrestha, S.D. (2014) Epigenetic control of effectors in plant pathogens. Front. Plant Sci. 5, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo, M.C. and Valent, B. (2013) Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. [DOI] [PubMed] [Google Scholar]
- Goodwin, S.B. , Waalwijk, C. and Kema, G.H.J. (2004) Genetics and genomics of Mycosphaerella graminicola: a model for the dothideales. Appl. Mycol. Biotechnol. 4, 315–330. [Google Scholar]
- Goodwin, S.B. , M'Barek, S.B. , Dhillon, B. , Wittenberg, A.H. , Crane, C.F. , Hane, J.K. , Foster, A.J. , Van der Lee, T.A. , Grimwood, J. and Aerts, A. (2011) Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7, e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudemand, E. , Laurent, V. , Duchalais, L. , Ghaffary, S.M.T. , Kema, G.H.J. , Lonnet, P. , Margalé, E. and Robert, O. (2013) Association mapping and meta‐analysis: two complementary approaches for the detection of reliable Septoria tritici blotch quantitative resistance in bread wheat (Triticum aestivum L.). Mol. Breed. 32, 563–584. [Google Scholar]
- Gout, L. , Fudal, I. , Kuhn, M.L. , Blaise, F. , Eckert, M. , Cattolico, L. , Balesdent, M.H. and Rouxel, T. (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans . Mol. Microbiol. 60, 67–80. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. and Torto‐Alalibo, T. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hunter, T. , Coker, R. and Royle, D. (1999) The teleomorph stage, Mycosphaerella graminicola, in epidemics of septoria tritici blotch on winter wheat in the UK. Plant Pathol. 48, 51–57. [Google Scholar]
- Jansen, R.C. (1994) Controlling the type I and type II errors in mapping quantitative trait loci. Genetics, 138, 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. and Thomma, B.P. (2009) Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 17, 151–157. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M.K. , Zhang, Z. , Usami, T. , Lievens, B. and Subbarao, K.V. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper, J. , Kahmann, R. , Bölker, M. , Ma, L.‐J. , Brefort, T. , Saville, B.J. , Banuett, F. , Kronstad, J.W. , Gold, S.E. and Müller, O. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature, 444, 97–101. [DOI] [PubMed] [Google Scholar]
- Kellner, R. , Bhattacharyya, A. , Poppe, S. , Hsu, T.Y. , Brem, R.B. and Stukenbrock, E.H. (2014) Expression profiling of the wheat pathogen Zymoseptoria tritici reveals genomic patterns of transcription and host‐specific regulatory programs. Genome Biol. 6, 1353–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kema, G.H.J. and van Silfhout, C.H. (1997) Genetic variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem III. Comparative seedling and adult plant experiments. Phytopathology, 87, 266–272. [DOI] [PubMed] [Google Scholar]
- Kema, G.H.J. , Annone, J.G. , Sayoud, R. , Van Silfhout, C.H. , Van Ginkel, M. and De Bree, J. (1996a) Genetic variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem I. Interactions between pathogen isolates and host cultivars. Phytopathology, 86, 200–212. [Google Scholar]
- Kema, G.H.J. , Sayoud, R. , Annone, J. and Van Silfhout, C. (1996b) Genetic variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem. II: analysis of interactions between pathogen isolates and host cultivars. Phytopathology, 86, 213–220. [Google Scholar]
- Kema, G.H.J. , Verstappen, E.C. , Todorova, M. and Waalwijk, C. (1996c) Successful crosses and molecular tetrad and progeny analyses demonstrate heterothallism in Mycosphaerella graminicola . Curr. Genet. 30, 251–258. [DOI] [PubMed] [Google Scholar]
- Kema, G.H.J. , Yu, D. , Rijkenberg, F. , Shaw, M. and Baayen, R. (1996d) Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology, 86, 777–786. [Google Scholar]
- Kema, G.H.J. , Goodwin, S.B. , Hamza, S. , Verstappen, E.C. , Cavaletto, J.R. , Van der Lee, T.A. , de Weerdt, M. , Bonants, P.J. and Waalwijk, C. (2002) A combined amplified fragment length polymorphism and randomly amplified polymorphism DNA genetic linkage map of Mycosphaerella graminicola, the Septoria tritici leaf blotch pathogen of wheat. Genetics, 161, 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kema, G.H.J. , van der Lee, T.A. , Mendes, O. , Verstappen, E.C. , Lankhorst, R.K. , Sandbrink, H. , van der Burgt, A. , Zwiers, L.‐H. , Csukai, M. and Waalwijk, C. (2008) Large‐scale gene discovery in the septoria tritici blotch fungus Mycosphaerella graminicola with a focus on in planta expression. Mol. Plant–Microbe Interact. 21, 1249–1260. [DOI] [PubMed] [Google Scholar]
- Keon, J. , Antoniw, J. , Rudd, J. , Skinner, W. , Hargreaves, J. and Hammond‐Kosack, K. (2005) Analysis of expressed sequence tags from the wheat leaf blotch pathogen Mycosphaerella graminicola (anamorph Septoria tritici). Fungal Genet. Biol. 42, 376–389. [DOI] [PubMed] [Google Scholar]
- Keon, J. , Antoniw, J. , Carzaniga, R. , Deller, S. , Ward, J.L. , Baker, J.M. , Beale, M.H. , Hammond‐Kosack, K. and Rudd, J.J. (2007) Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant–Microbe Interact. 20, 178–193. [DOI] [PubMed] [Google Scholar]
- Lander, E.S. and Botstein, D. (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics, 121, 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.‐S. , Rudd, J.J. , Hammond‐Kosack, K.E. and Kanyuka, K. (2013) Mycosphaerella graminicola LysM effector‐mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant–Microbe Interact. 27, 236–243. [DOI] [PubMed] [Google Scholar]
- Lévesque, C.A. , Brouwer, H. , Cano, L. , Hamilton, J.P. , Holt, C. , Huitema, E. , Raffaele, S. , Robideau, G.P. , Thines, M. and Win, J. (2010) Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde, C. , Zhan, J. and McDonald, B.A. (2002) Population structure of Mycosphaerella graminicola: from lesions to continents. Phytopathology, 92, 946–955. [DOI] [PubMed] [Google Scholar]
- Marshall, R. , Kombrink, A. , Motteram, J. , Loza‐Reyes, E. , Lucas, J. , Hammond‐Kosack, K.E. , Thomma, B.P. and Rudd, J.J. (2011) Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 156, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi, R. , van der Lee, T. , Waalwijk, C. and Kema, G.H.J. (2006a) MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant–Microbe Interact. 19, 389–398. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , Zwiers, L.‐H. , de Waard, M.A. and Kema, G.H.J. (2006b) MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola . Mol. Plant–Microbe Interact. 19, 1262–1269. [DOI] [PubMed] [Google Scholar]
- Mesarich, C.H. , Griffiths, S.A. , van der Burgt, A. , Ökmen, B. , Beenen, H.G. , Etalo, D.W. , Joosten, M.H.A.J. and de Wit, P.J.G.M. (2014) Transcriptome sequencing uncovers the Avr5 avirulence gene of the tomato leaf mold pathogen Cladosporium fulvum . Mol. Plant–Microbe Interact. 27, 846–857. [DOI] [PubMed] [Google Scholar]
- Mirzadi Gohari, A. , Mehrabi, R. , Robert, O. , Ince, I.A. , Boeren, S. , Schuster, M. , Steinberg, G. , Wit, P.J. and Kema, G.H.J. (2014) Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici . Mol. Plant Pathol. 15, 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motteram, J. , Küfner, I. , Deller, S. , Brunner, F. , Hammond‐Kosack, K.E. , Nürnberger, T. and Rudd, J.J. (2009) Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain‐containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola . Mol. Plant–Microbe Interact. 22, 790–799. [DOI] [PubMed] [Google Scholar]
- O'Connell, R.J. , Thon, M.R. , Hacquard, S. , Amyotte, S.G. , Kleemann, J. , Torres, M.F. , Damm, U. , Buiate, E.A. , Epstein, L. and Alkan, N. (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44, 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen, J.W. (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor. Appl. Genet. 84, 803–811. [DOI] [PubMed] [Google Scholar]
- Orsomando, G. , Lorenzi, M. , Raffaelli, N. , Dalla Rizza, M. , Mezzetti, B. and Ruggieri, S. (2001) Phytotoxic protein PcF, purification, characterization, and cDNA sequencing of a novel hydroxyproline‐containing factor secreted by the strawberry pathogen Phytophthora cactorum . J. Biol. Chem. 276, 21578–21584. [DOI] [PubMed] [Google Scholar]
- Orton, E.S. , Deller, S. and Brown, J.K. (2011) Mycosphaerella graminicola: from genomics to disease control. Mol. Plant Pathol. 12, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.‐H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. , Ning, Y. , Wang, R. and Bellizzi, M. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell Online, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg, W. , Kema, G.H.J. , Groenewald, J. , Verkley, G. , Seifbarghi, S. , Razavi, M. , Gohari, A.M. , Mehrabi, R. and Crous, P. (2011) Zymoseptoria gen. nov.: a new genus to accommodate Septoria‐like species occurring on graminicolous hosts. Persoonia, 26, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep, M. (2005) Small proteins of plant‐pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253, 19–27. [DOI] [PubMed] [Google Scholar]
- Rietman, H. , Bijsterbosch, G. , Cano, L.M. , Lee, H.‐R. , Vossen, J.H. , Jacobsen, E. , Visser, R.G. , Kamoun, S. and Vleeshouwers, V.G. (2012) Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Mol. Plant–Microbe Interact. 25, 910–919. [DOI] [PubMed] [Google Scholar]
- Rose, J.K. , Ham, K.‐S. , Darvill, A.G. and Albersheim, P. (2002) Molecular cloning and characterization of glucanase inhibitor proteins: coevolution of a counterdefense mechanism by plant pathogens. Plant Cell, 14, 1329–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouxel, T. , Grandaubert, J. , Hane, J.K. , Hoede, C. , van de Wouw, A.P. , Couloux, A. , Dominguez, V. , Anthouard, V. , Bally, P. and Bourras, S. (2011) Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat‐induced point mutations. Nat. Commun. 2, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, J. , Kanyuka, K. , Hassani‐Pak, K. , Derbyshire, M. , Devonshire, J. , Saqi, M. , Desai, N. , Powers, S. , Hooper, J. and Ambroso, L. (2015) Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat (Triticum aestivum) reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions, and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 167, 1158–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadaoui, E. (1987) Physiologic specialization of Septoria tritici in Morocco. Plant Dis. 71, 153–155. [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative CT method. Nat. Protocols, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Songkumarn, P. (2013) Identification and Characterization of In‐Planta Expressed Secreted Effector Proteins from Magnaporthe oryzae. Columbus: The Ohio State University. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. and de Wit, P.J.G.M. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , van den Burg, H.A. , Ökmen, B. , Beenen, H.G. , van Liere, S. , Kema, G.H.J. and de Wit, P.J.G.M. (2010) Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA, 107, 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Kourmpetis, Y.A. , Slot, J.C. , Bakker, F.T. , De Wit, P.J.G.M. and Rokas, A. (2012) In silico characterization and molecular evolutionary analysis of a novel superfamily of fungal effector proteins. Mol. Biol. Evol. 29, 3371–3384. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Cordovez, V. , Ökmen, B. , Beenen, H.G. , Kema, G.H.J. and Wit, P.J.G.M. (2014) Positive selection and intragenic recombination contribute to high allelic diversity in effector genes of Mycosphaerella fijiensis, causal agent of the black leaf streak disease of banana. Mol. Plant Pathol. 15, 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock, E.H. , Banke, S. , Javan‐Nikkhah, M. and McDonald, B.A. (2007) Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol. Biol. Evol. 24, 398–411. [DOI] [PubMed] [Google Scholar]
- Suffert, F. and Sache, I. (2011) Relative importance of different types of inoculum to the establishment of Mycosphaerella graminicola in wheat crops in north‐west Europe. Plant Pathol. 60, 878–889. [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A. , van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wouw, A.P. , Lowe, R.G.T. , Elliott, C.E. , Dubois, D.J. and Howlett, B.J. (2014) An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol. Plant Pathol. 15, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ginkel, M. and Scharen, A. (1988) Host–pathogen relationships of wheat and Septoria tritici. Phytopathology, 78, 762–766. [Google Scholar]
- Vleeshouwers, V.G. and Oliver, R.P. (2014) Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant–Microbe Interact. 27, 196–206. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S.‐K. , Wang, M. , Bouwmeester, K. , Vosman, B. and Visser, R.G. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE, 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips, R. (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. [DOI] [PubMed] [Google Scholar]
- Whigham, E.L. (2013) Characterization of a cell death suppressing effector broadly conserved across the fungal kingdom. Graduate Thesis and Dissertation, Iowa State University.
- Wittenberg, A.H. , van der Lee, T.A. , M'Barek, S.B. , Ware, S.B. , Goodwin, S.B. , Kilian, A. , Visser, R.G. , Kema, G.H.J. and Schouten, H.J. (2009) Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola . PLoS ONE, 4, e5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F. , Li, W. and Jørgensen, H.J. (2013) Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PLoS ONE, 8, e81606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Zhang, H. , Li, G. , Li, W. , Wang, X. and Song, F. (2009) Ectopic expression of MgSM1, a cerato‐platanin family protein from Magnaporthe grisea, confers broad‐spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 7, 763–777. [DOI] [PubMed] [Google Scholar]
- Zhan, J. , Pettway, R. and McDonald, B.A. (2003) The global genetic structure of the wheat pathogen Mycosphaerella graminicola is characterized by high nuclear diversity, low mitochondrial diversity, regular recombination, and gene flow. Fungal Genet. Biol. 38, 286–297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Schematic diagram showing different infection processes of Zymoseptori tritici.
Fig. S2 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) up‐regulated during fructification/sporulation. dpi, days post‐infection.
Fig. S3 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) up‐regulated during the transition/necrotrophic phase of pathogenesis. dpi, days post‐infection.
Fig. S4 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) with a bimodal pattern. dpi, days post‐infection.
Fig. S5 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) expressed at low levels throughout pathogenesis. dpi, days post‐infection.
Fig. S6 Disease development after inoculation with the Zymoseptoria tritici wild‐type (WT) strain IPO323 and the IPO323ΔSSP15 and IPO323ΔSSP18 knock‐out strains in 12 wheat cultivars.
Fig. S7 Mapped quantitative trait loci involved in (a)virulence of Zymoseptoria tritici to seven wheat cultivars.
Fig. S8 Expression profiling of Zymoseptoria tritici small secreted proteins (SSPs) positioned under mapped quantitative trait loci (QTLs) and expressed at low levels throughout pathogenesis.
Fig. S9 Gene ontology (GO) analyses of 100 Zymoseptoria tritici small secreted proteins (SSPs).
Table S1 Mapped quantitative trait loci involved in (a)virulence in progeny isolates of Zymoseptoria tritici from a cross between isolates IPO323 and IPO95052 tested for virulence on the durum wheat cultivars Volcani 447, Zenati Bouteille and Bidi 17 and the bread wheat cultivars Taichung 29, Obelisk, Gerek 79 and Shafir.
Table S2 Zymoseptoria tritici small secreted proteins (SSPs) positioned under the quantitative trait loci (QTLs) involved in the degradation of polysaccharides, proteins and lipids.
Table S3 Cultivars used in this study.


