Summary
The tomato I ‐3 and I ‐7 genes confer resistance to F usarium oxysporum f. sp. lycopersici (F ol) race 3 and were introgressed into the cultivated tomato, S olanum lycopersicum, from the wild relative S olanum pennellii. I‐3 has been identified previously on chromosome 7 and encodes an S‐receptor‐like kinase, but little is known about I ‐7. Molecular markers have been developed for the marker‐assisted breeding of I ‐3, but none are available for I ‐7. We used an RNA‐seq and single nucleotide polymorphism (SNP) analysis approach to map I ‐7 to a small introgression of S . pennellii DNA (c. 210 kb) on chromosome 8, and identified I ‐7 as a gene encoding a leucine‐rich repeat receptor‐like protein (LRR‐RLP), thereby expanding the repertoire of resistance protein classes conferring resistance to F ol. Using an eds1 mutant of tomato, we showed that I‐7, like many other LRR‐RLPs conferring pathogen resistance in tomato, is EDS1 (Enhanced Disease Susceptibility 1) dependent. Using transgenic tomato plants carrying only the I ‐7 gene for F ol resistance, we found that I ‐7 also confers resistance to F ol races 1 and 2. Given that F ol race 1 carries A vr1, resistance to F ol race 1 indicates that I‐7‐mediated resistance, unlike I‐2‐ or I‐3‐mediated resistance, is not suppressed by Avr1. This suggests that Avr1 is not a general suppressor of F ol resistance in tomato, leading us to hypothesize that Avr1 may be acting against an EDS1‐independent pathway for resistance activation. The identification of I ‐7 has allowed us to develop molecular markers for marker‐assisted breeding of both genes currently known to confer F ol race 3 resistance (I ‐3 and I ‐7). Given that I‐7‐mediated resistance is not suppressed by Avr1, I ‐7 may be a useful addition to I ‐3 in the tomato breeder's toolbox.
Keywords: Fusarium oxysporum f. sp. lycopersici, Fusarium wilt, leucine‐rich repeat, plant disease resistance gene, receptor‐like protein, Solanum lycopersicum, Solanum pennellii
Introduction
The fungus Fusarium oxysporum causes devastating wilt diseases of many important crop plants, including banana/plantain, cotton, potato, tomato, capsicum, beans, peas, chickpeas and melons. However, individual pathogenic isolates of F. oxysporum are highly specific for a particular species of host plant. For example, F. oxysporum f. sp. cubense (Foc) is specific for banana/plantain, F. oxysporum f. sp. vasinfectum (Fov) is specific for cotton and F. oxysporum f. sp. lycopersici (Fol) is specific for tomato. In contrast with Fusarium wilt of banana or cotton, Fusarium wilt of tomato (caused by Fol) has been managed for many years by the breeding of disease‐resistant cultivars, but this approach is not possible for all crop plants. Either breeding for resistance is very difficult, as is the case for banana, or no major genes for resistance have so far been identified, as is the case for cotton. Therefore, in addition to its importance as a crop pathosystem, the tomato–Fol pathosystem has become an important, well‐established model system for the study of plant–F. oxysporum interactions at a genetic and molecular level.
Fol infection involves the germination of soil‐borne spores in the vicinity of growing roots, attachment to the root surface, penetration of the root cortex and proliferation of hyphae within the root vasculature. Eventually, Fol invades and colonizes the parenchyma of the dying tomato plant and sporulates on the plant surface (Michielse and Rep, 2009). The disease is characterized by yellowing, wilting and browning of the leaves, stunted growth and, eventually, death of the plant. These symptoms result from the obstruction of water and nutrient flow caused by hyphae within the xylem vessels, as well as tyloses, callose, gums and gels produced by the host plant.
Three races of Fol have been reported and named races 1, 2 and 3 in order of appearance. Fol races 1 and 2 were discovered over 70 years ago (Alexander and Tucker, 1945; Bohn and Tucker, 1939) and are distributed almost worldwide. Fol race 3 was first discovered in Queensland (Australia) in 1978 (Grattidge and O'Brien, 1982) and by the 1980s had caused great yield losses (McGrath et al., 1987). Fol race 3 was later reported in the USA (Volin and Jones, 1982) and subsequently Mexico (Valenzuela‐Ureta et al., 1996), Brazil (Reis et al., 2005) and many other parts of the world. The use of resistant cultivars remains the most practical, cost‐effective and environmentally safe method for the control of Fusarium wilt in tomato.
Wilt‐resistant cultivars have been generated by the introgression of genes for Fol resistance, designated Immunity genes, into the cultivated tomato Solanum lycopersicum, from wild relatives. The I gene was identified in the wild tomato Solanum pimpinellifolium (accession PI79532) and confers resistance against Fol race 1 (Bohn and Tucker, 1939). However, the rapid emergence of race 2, virulent on tomato cultivars carrying the I gene, soon necessitated the search for a new source of resistance. The I‐2 gene was identified in an S. lycopersicum × S. pimpinellifolium hybrid (accession PI126915; Stall and Walter, 1965) and confers resistance to Fol race 2. I‐2 encodes a coiled‐coil, nucleotide‐binding, leucine‐rich repeat (CC‐NB‐LRR) resistance protein (Simons et al., 1998). The eventual emergence of race 3, virulent on tomato cultivars carrying the I‐2 gene, necessitated another search for a new source of resistance. Two genes for resistance to Fol race 3 were identified in Solanum pennellii, one from accession LA716 (Scott and Jones, 1989) and the other from accession PI414773 (McGrath et al., 1987). Originally, both genes were designated I‐3, but gene‐mapping work by Lim et al. (2006) revealed that the two genes were not the same. The I‐3 gene from S. pennellii LA716 encodes an S‐receptor‐like kinase (SRLK) protein (Catanzariti et al., 2015; Lim et al., 2008) and, together with the rice Pi‐d2 gene for resistance to Magnaporthe grisea (Chen et al., 2006) and the Arabidopsis thaliana RFO3 gene for resistance to F. oxysporum (Cole and Diener, 2013), forms the basis for a new class of plant disease resistance genes. However, at the time of commencing the work described here, nothing was known about the resistance gene from S. pennellii PI414773, renamed I‐7, except that it was not located in the I‐3 region on chromosome 7 (Lim et al., 2006).
Here, we describe an RNA‐seq analysis of tomato cultivars, differing by the presence or absence of I‐7, which not only enabled us to identify genes expressed in roots (where I‐7 would be expected to act), but also the introgressed region of S. pennellii DNA carrying I‐7. This enabled a candidate gene to be identified and complementation tests to be carried out confirming the identification of I‐7.
Results
Single nucleotide polymorphism (SNP) analyses of Tristar and M82 RNA‐seq data reveal a cluster of SNPs on Tristar chromosome 8
An RNA‐seq analysis of root transcripts was carried out on the tomato cultivars Tristar (carrying I‐7) and M82 (lacking I‐7). More than 169 million reads for Tristar and 152 million reads for M82 (Table S1, see Supporting Information) were mapped against the tomato reference transcriptome and analysed for SNPs, and, in both cases, more than 55,000 SNPs were identified (Table S1). SNPs supported by at least 75% of the mapped reads were extracted: 30,069 for Tristar and 31,391 for M82. Then, 20,450 SNPs common to Tristar and M82 were discarded. The remaining unique polymorphisms were used to calculate the SNP frequency for each transcript (number of SNPs per transcript divided by the length of the transcript). The SNP frequency was then plotted against the corresponding gene positions on each tomato chromosome. We reasoned that an introgressed region of DNA from a tomato relative should generate a cluster of genes showing a higher frequency of unique SNPs compared with the corresponding region of S. lycopersicum DNA or surrounding S. lycopersicum sequences. We also reasoned that the S. pennellii introgression carrying I‐7 should be relatively small, given the failure of an extensive genome‐wide marker survey that attempted to identify the I‐7 introgression (Gonzalez‐Cendales et al., 2014). Consistent with these expectations, a small cluster of genes with a higher frequency of SNPs in Tristar relative to M82 and to adjacent Tristar genes was identified on chromosome 8 (Fig. 1).
Figure 1.
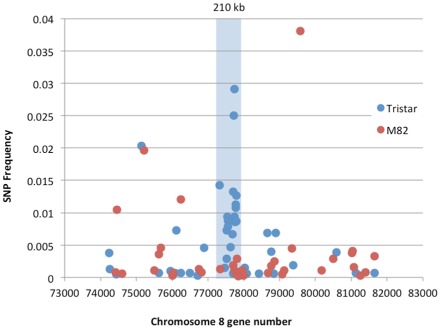
Plot of single nucleotide polymorphism (SNP) frequencies for Tristar and M82 transcripts relative to the tomato reference transcriptome for genes on the long arm of chromosome 8. The SNP cluster corresponding to the introgressed region of S olanum pennellii DNA on Tristar chromosome 8 is highlighted in blue. SNP frequencies were calculated as the number of SNPs per transcript unique to Tristar or M82 divided by the length of the transcript.
Identification of I ‐7 as the Tristar allele of S olyc08g77740
Cleaved amplified polymorphic sequence (CAPS) markers were designed for several genes in the SNP cluster found on chromosome 8 (Table S2, see Supporting Information) based on restriction site polymorphisms between Tristar and M82. To determine whether these markers were linked to I‐7, marker analysis was performed on 15 Tristar × M82 F3 or F4 lines scored as homozygous resistant and 16 scored as homozygous susceptible. The results showed complete correspondence between marker genotype and resistance (Fig. S1, see Supporting Information), indicating that we had correctly identified the S. pennellii introgression carrying I‐7.
The extent of the introgressed region was determined by looking for root‐expressed genes showing a higher frequency of SNPs in Tristar than in M82 relative to the reference transcriptome, and for flanking root‐expressed genes lacking SNPs relative to the reference transcriptome (Table S3, see Supporting Information). Based on this analysis, the introgressed region was found to be about 210 kb and to carry 29 genes (Solyc08g077520 to Solyc08g077800). The Tristar RNA‐seq data showed that 11 of these 29 genes were not expressed in roots, as determined by no or very few mapped reads (Table S3). I‐7 should be expressed in roots because that is where it acts to limit Fol infection. Therefore, annotations of the 18 root‐expressed genes (Table S3) were examined for members of the major plant disease resistance gene classes. One gene, Solyc08g077740, was found to encode an extracellular LRR receptor‐like protein (LRR‐RLP), thus resembling a resistance gene. Of the remaining 17 root‐expressed genes, 16 encoded proteins with no resemblance to known resistance proteins and one encoded a protein of unknown function, leaving Solyc08g077740 as the sole candidate for I‐7.
To test this possibility, the Solyc08g077740 genes from Tristar and M82 were first polymerase chain reaction (PCR) amplified from genomic DNA using cultivar‐specific LIC7774 primers designed using Tristar and M82 consensus sequences for Solyc08g077740 assembled from the RNA‐seq data (Table S4, see Supporting Information). The resulting PCR products of approximately 2.9 kb were cloned via ligation‐independent cloning (LIC) into the binary LIC vector pL2. The resulting plasmids pL2‐Tristar and pL2‐M82 were used for the transformation of tomato cultivars Moneymaker and M82 in the case of pL2‐Tristar and Moneymaker alone in the case of pL2‐M82. Six independent transgenic Moneymaker T1 plants (MM‐7774Tristar) and 12 independent transgenic M82 T1 plants (M82‐7774Tristar) were obtained with the Tristar gene. Seven independent transgenic Moneymaker T1 plants (MM‐7774M82) were obtained with the M82 gene. The presence of the transgene in T1 plants was confirmed by PCR as described below.
T2 seedlings from two independent MM‐7774Tristar lines and one M82‐7774Tristar line were screened for the presence of the transgene by multiplex PCR using the CAPS7774 marker (Table S2, Fig. S2, see Supporting Information), which amplified a product from the endogenous Solyc08g077740 gene, and 7774F4 (Table S4) and M13 reverse primers, which amplified a product from the Solyc08g077740 transgene (Fig. S2). Seedlings carrying the transgene from each line were inoculated with Fol race 3. All were found to show little or no sign of disease, like Tristar their transgene donor, but unlike Moneymaker or M82, their progenitors, which were severely diseased (Fig. 2). Pathogen tests were also carried out on two additional MM‐7774Tristar lines and four MM‐7774M82 lines without first screening for seedlings carrying the transgene, i.e. on plants segregating for the transgene. Both MM‐7774Tristar lines segregated for plants showing little or no sign of disease and a small number of severely diseased plants (Fig. S3, see Supporting Information), whereas the MM‐7774M82 lines showed diseased plants with symptoms ranging from severe to mild (Fig. 3), the latter indicating that overexpression of the M82 gene may reduce susceptibility. Together, these data indicate that the Solyc8g077740 gene from Tristar confers resistance to Fol race 3 and corresponds to I‐7, whereas the Solyc8g077740 gene from M82 confers no or little resistance to Fol race 3 and is here designated i‐7.
Figure 2.
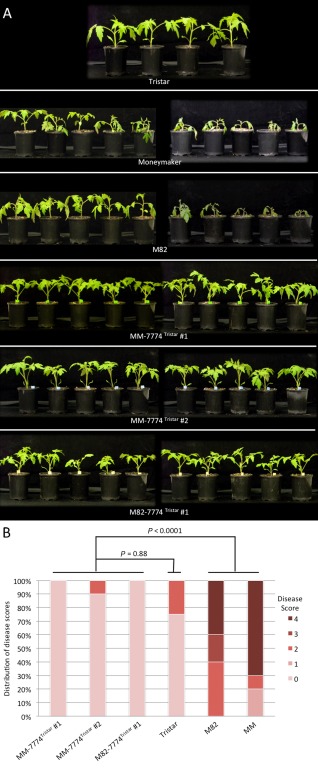
(A) Disease assays on T2 plants carrying the S olyc08g077740 gene from Tristar in a Moneymaker (MM) or M82 background. T2 plants, together with Tristar (resistant), Moneymaker (susceptible) and M82 (susceptible) control plants, were inoculated with F ol race 3. Photographs were taken at 18 days post‐inoculation. (B) Disease scores for the T2 plants shown in (A). Probability values were obtained using the non‐parametric Kruskal–Wallis test to determine significant differences in disease scores between plant lines.
Figure 3.
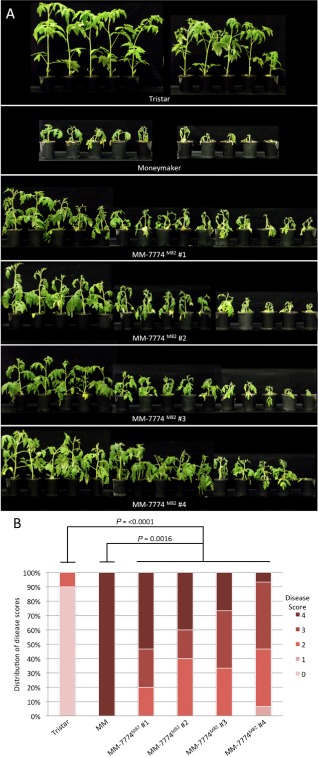
(A) Disease assays on T2 plants carrying the S olyc08g077740 gene from M82 in a Moneymaker (MM) background. T2 plants, together with Tristar (resistant) and Moneymaker (susceptible) control plants, were inoculated with F ol race 3. Photographs were taken at 21 days post‐inoculation. (B) Disease scores for the T2 plants shown in (A). Probability values were obtained using the non‐parametric Mann–Whitney test to determine significant differences in disease scores between plant lines.
I ‐7 confers resistance to F ol races 1 and 2
Although I‐7 confers resistance to Fol race 3, it was not known whether it also confers resistance to Fol races 1 and 2 because Tristar also carries the I gene for race 1 resistance and the I‐2 gene for race 2 resistance. Therefore, T2 seedlings from MM‐7774Tristar line #1, which lacks I and I‐2, were screened by multiplex PCR as described above to identify seedlings carrying the I‐7 transgene, and then inoculated with Fol races 1 or 2. These seedlings showed little or no sign of disease with either race, like Tristar, but unlike Moneymaker, which was severely diseased (Figs S4 and S5, see Supporting Information). These results show that I‐7 confers resistance to Fol races 1, 2 and 3.
I ‐7 resistance is dependent on EDS 1
Some LRR‐RLPs, including Cf‐9 and Ve1, have been shown to depend on EDS1 (Enhanced Disease Susceptibility 1), a lipase‐like protein involved in defence activation (Hu et al., 2005). To assess whether I‐7‐mediated resistance is dependent on EDS1, an F2 family segregating for both I‐7 versus i‐7 and EDS1 versus eds1 was obtained from a cross between Tristar and the sun1‐1 (eds1) mutant of tomato (Hu et al., 2005). Around 50 Tristar × sun1‐1 F2 seedlings were genotyped for EDS1 and eds1 by multiplex PCR using a single reverse primer (CCTGCTGCACGAAGACACAG; Hu et al., 2005), annealing downstream of the deletion in sun1‐1, and two forward primers, one (TGCTTGCTCCTCTTTCATCA) annealing to the region of EDS1 deleted in sun1‐1 plants and the other (GTAGGGGTGTTCGTGATTCGATTTG; Hu et al., 2005) annealing upstream of the deletion in sun1‐1 (Fig. S6A, see Supporting Information). Based on these results, two sets of F2 seedlings were selected for a disease assay: those homozygous for the eds1 mutation (eds1/eds1) and those that were heterozygous (EDS1/eds1). These plants, together with the parent lines, Tristar and sun1‐1, were inoculated with Fol race 3, and DNA samples used to screen for the presence of eds1 were screened for the presence of I‐7 using the CAPS7774 marker (Fig. S6B). Although resistance was slightly reduced in some EDS1/eds1 plants carrying I‐7 compared with Tristar, resistance was lost altogether in eds1/eds1 plants carrying I‐7 (Fig. 4). These results show clearly that EDS1 is required for the resistance response mediated by I‐7.
Figure 4.
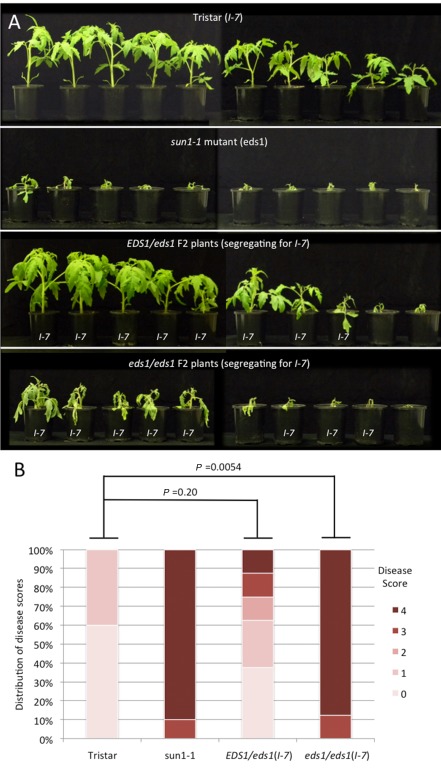
(A) Disease assays on Tristar × sun1‐1 EDS 1/eds1 and eds1/eds1 F2 plants (segregating for I ‐7) and parent lines inoculated with F ol race 3. F2 plants were genotyped for I ‐7 by cleaved amplified polymorphic sequence (CAPS) marker analysis and plants carrying I ‐7 are indicated. Photographs were taken at 21 days post‐inoculation. (B) Disease scores for the T2 plants shown in (A), excluding F2 plants not carrying I ‐7. Probability values were obtained using the non‐parametric Mann–Whitney test to determine significant differences in disease scores between plant lines.
Analysis of the protein sequence encoded by I‐7
The I‐7 gene [National Center for Biotechnology Information (NCBI) accession KT185194] encodes a typical LRR‐RLP of 966 amino acids (Fig. 5). The first 25 amino acids are predicted to function as a signal peptide directing protein secretion (von Heijne, 1986). Amino acids 26–85 constitute the mature N‐terminus comprising an N‐terminal LRR‐flanking domain containing four conserved cysteines likely to be involved in disulfide bond formation. The N‐terminal end is followed by a typical extracellular LRR region consisting of 30 repeats (from amino acids 86 to 851) separated into two blocks (26 and four repeats) by a loop out region (amino acids 709–755). Amino acids 852–875 constitute a C‐terminal LRR‐flanking region containing two conserved cysteines potentially involved in disulfide bond formation. Amino acids 876–906 comprise a very acidic domain with 13 negatively charged residues and only one positively charged residue. Amino acids 907–929 comprise a predicted transmembrane domain. Amino acids 930–966 comprise a very basic cytosolic C‐terminal domain, with 13 positively charged residues and only two negatively charged residues. The cytosolic domain also includes a putative YXXø endocytosis motif (YVKF), where ø represents an amino acid with a bulky hydrophobic side chain (Bonifacino and Traub, 2003). The predicted LRR region contains 21 potential N‐glycosylation sites matching the consensus motif Nx(S/T)x (where x is any amino acid except P; Gavel and von Heijne, 1990).
Figure 5.
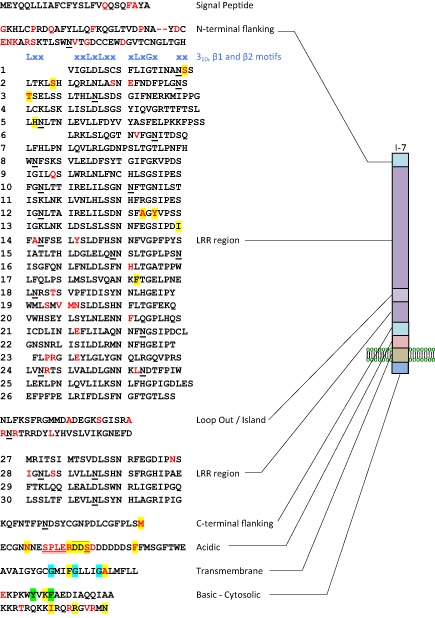
Sequence and graphic representation of the predicted I‐7 protein divided into eight domains: signal peptide, N‐terminal flanking region, typical extracellular leucine‐rich repeat (LRR) region (with LRRs numbered 1–30), divided into two blocks by a loop out region or island domain, C‐terminal LRR‐flanking region, acidic domain, transmembrane domain and basic cytosolic domain. The locations of predicted 310 (xxLxx), concave β1 (xxLxLxx) and convex β2 (xLxGx) motifs are shown in blue above the LRR domain. The positions of amino acid differences between I‐7 and i‐7M82 are shown in red, whereas those between I‐7 and i‐7LA716 are highlighted in yellow. Putative N‐glycosylation sites are underlined and a putative endocytosis motif is highlighted in green. A possible SOBIR1‐dimerization motif (GxxxGxxxG; Bi et al., 2015) is highlighted in light blue. Amino acids present in I‐7, but missing from i‐7M82, owing to deletions in i‐7 relative to I ‐7, are double underlined, whereas those missing from i‐7LA716 are overlined.
Compared with I‐7, i‐7 encodes a slightly shorter 963‐amino‐acid protein with 93.4% identity to I‐7. I‐7 and i‐7 differ by 68 polymorphic residues (Fig. 5). The N‐terminal flanking domain contains a high proportion of the polymorphisms comprising only 6.6% of the protein, but containing 23.5% (16) of the polymorphisms. The acidic domain is also fairly polymorphic; it comprises only 3.2% of the protein, but possesses 13.2% (nine) of the polymorphisms. The LRR domain is relatively conserved between the two proteins compared with the non‐LRR regions. Surprisingly, the predicted solvent‐exposed residues in the β‐sheet region of the LRR domain, thought to determine recognitional specificity in other LRR‐RLPs, show very few polymorphisms between I‐7 and i‐7, with only five polymorphic residues out of 150 (3.3%).
I ‐7 is a member of a small gene family
A blastn search (http://www.sgn.cornell.edu/tools/blast/; default settings) of the S. lycopersicum Heinz 1706 genome sequence reveals an I‐7 orthologue (i.e. i‐7) on chromosome 8 (Solyc08g077740; 96% nucleotide identity) and a paralogue on chromosome 6 (Solyc06g033920; 86% nucleotide identity). Similarly, a blastn search of the S. pennellii LA716 genome sequence reveals an orthologue on chromosome 8 (Sopen08g026260; 99% nucleotide identity) and a paralogue on chromosome 6 (Sopen06g014090; 86% nucleotide identity). However, this search also revealed a paralogue on chromosome 8 (Sopen08g026250; 89% nucleotide identity) adjacent to the I‐7 orthologue. This raised the possibility that the S. pennellii introgression in Tristar also carried a paralogue adjacent to I‐7. Further examination of the RNA‐seq data described above revealed transcripts corresponding to a homologue present in Tristar, but absent in M82. The coding sequence for this homologue, apart from the first 52 bases, was assembled from the Tristar RNA‐seq data (NCBI accession KT185195). This homologue showed higher nucleotide identity to the I‐7 paralogue on chromosome 8 of LA716 (99% nucleotide identity) than I‐7 (88% nucleotide identity), the I‐7 orthologue on chromosome 8 of LA716 (89% nucleotide identity) or the paralogue on chromosome 6 of LA716 (90% nucleotide identity). From these data, we infer the presence of an I‐7 paralogue adjacent to the I‐7 gene in Tristar.
Alignment of the predicted protein sequences (Fig. 6) clearly shows that the chromosome 6 and 8 paralogues are more closely related to one another than they are to I‐7 or its orthologues. The alignment also shows that there are 78 variable positions with three or more different amino acids represented among the seven sequences: 56 within the LRR domain and 21 at predicted solvent‐exposed positions in the β‐sheet region of the LRR domain. Almost one‐half (25) of these positions and two‐thirds of those predicted to be solvent exposed are clustered within LRRs 14–19. In contrast, only seven of the variable positions are present in LRRs 20–30 and none of these is predicted to be solvent exposed. The alignment also shows extensive polymorphism for the presence/absence of putative N‐glycosylation sites among the gene family, with 18 of 33 positions being polymorphic, and one site unique to I‐7. This alignment also shows that there are only 21 amino acid differences between I‐7 and its orthologue in LA716.
Figure 6.
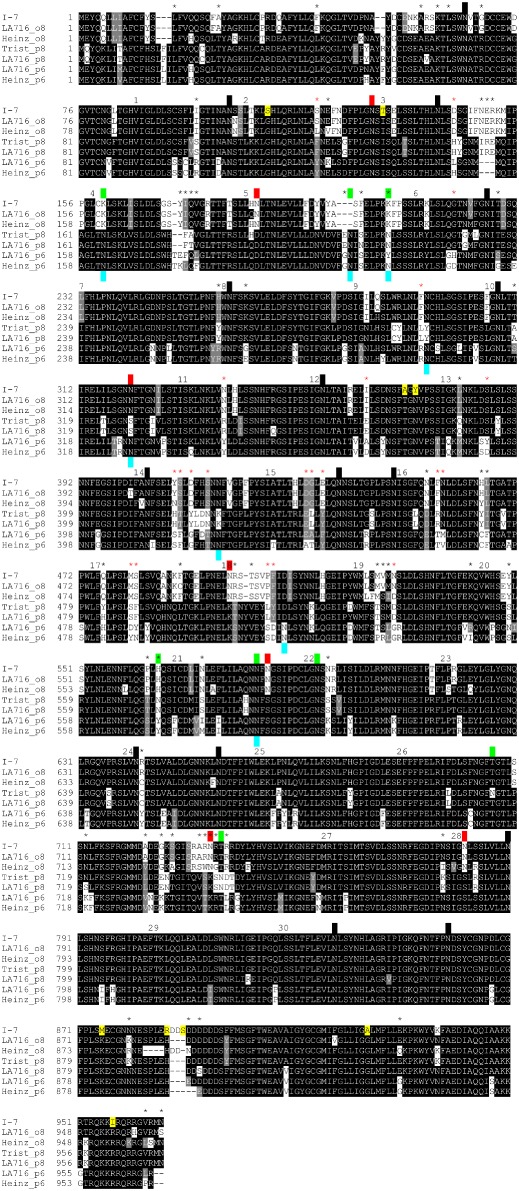
Alignment of I‐7 and orthologous and paralogous proteins from Tristar, Heinz 1706 and LA716. Sequences were aligned using the EMBL‐EBI MAFFT server at http://www.ebi.ac.uk/Tools/msa/mafft/, and the alignment was shaded using the ExPASy BoxShade server at http://www.ch.embnet.org/software/BOX_form.html. Amino acid identities are highlighted in black. Amino acid similarities are highlighted in grey. Putative N‐glycosylation sites encoded by orthologous and paralogous genes on chromosome 8 (labelled o8 and p8, respectively) and paralogous genes on chromosome 6 (labelled p6) are indicated by black boxes above the sequence alignment. Putative N‐glycosylation sites encoded by genes at only one or two of these three loci are indicated by red boxes (o8) or green boxes (p8) above the alignment or blue boxes (p6) below the alignment. Residues unique to I‐7 are highlighted in yellow. Numbers above the alignment indicate the leucine‐rich repeat (LRR) number, and asterisks indicate variable sites with three or more different residues, with red asterisks indicating those in predicted solvent‐exposed residues in the β‐strand motif of the LRR. It should be noted that the first 18 amino acids of the Trist_p8 sequence are inferred from the LA716_p8 sequence.
To test whether the LA716 orthologue of I‐7 also confers Fol resistance, three LA716 introgression lines, IL8‐1, IL8‐2 and IL8‐3, which together span S. pennellii chromosome 8 (Eshed and Zamir, 1995), were tested for resistance to Fol race 3. None of the three lines, including IL8‐2, which carries the LA716 orthologue of I‐7, showed resistance to Fol race 3 (Fig. S7, see Supporting Information). This result is consistent with previous work showing that the IL8 lines do not confer resistance to Fol races 1 or 2 (Sela‐Buurlage et al., 2001). Thus, the LA716 orthologue of I‐7 is also an i‐7 allele. We have designated this gene i‐7LA716 to distinguish it from the M82 allele, which we have now designated i‐7M82. Nine of the amino acid differences between I‐7 and i‐7M82 or i‐7LA716 are unique to I‐7 (Fig. 6) and, although four of these nine (S110, T131, A171 and Y173) occur in the LRR region, none are at solvent‐exposed positions within the β‐sheet region of the LRR domain (Fig. 5).
Discussion
Prior to this work, the only genes for resistance to Fusarium wilt that had been cloned from tomato were I‐2, encoding a CC‐NB‐LRR protein (Simons et al., 1998), and I‐3, encoding an SRLK protein (Catanzariti et al., 2015). Now, through the identification of I‐7, which encodes an LRR‐RLP, another class of R gene has been added to this repertoire. The existence of a diverse group of resistance genes against just one pathogen is unusual. In most plant–pathogen systems, genes conferring resistance to different isolates of the same pathogen belong to just one or two classes. The fact that resistance to Fol is provided by genes in three different classes suggests that pathogen recognition and defence activation are more complex in the tomato–Fol pathosystem than in other pathosystems. Such complexity may also be a feature of resistance in other plant–F. oxysporum pathosystems, given that Arabidopsis genes for quantitative resistance to F. oxysporum show similar diversity, with RFO1 encoding a wall‐associated kinase (a non‐LRR receptor‐like kinase; Diener and Ausubel, 2005), RFO2 encoding an LRR‐RLP (Shen and Diener, 2013), like I‐7, and RFO3 encoding an SRLK (Cole and Diener, 2013), like I‐3. It is possible that the complexity of the F. oxysporum infection process (penetration of the root epidermis, growth across the root cortex, colonization of the root xylem and a switch from biotrophy to necrotrophy) provides multifarious opportunities for recognition that might result in a more complex array of plant resistance mechanisms that counter infection. Moreover, the fact that tomato uses both cytosolic (I‐2) and surface (I‐3 and, most likely, I‐7) receptors to detect Fol effectors suggests that Fol secretes effectors that act both inside and outside the plant cell.
Many resistance genes occur in gene clusters. For example, the I‐2 gene is one of seven homologous genes clustered at the I‐2 locus (Simons et al., 1998), and I‐3 is one of six homologous genes clustered at the I‐3 locus. I‐7 belongs to a small gene family in tomato that includes a homologue on chromosome 6 and a homologue on chromosome 8 adjacent to I‐7. Although it is clear that the homologue on chromosome 6 does not confer resistance to Fol, we cannot rule out the possibility that the chromosome 8 homologue in Tristar also confers resistance to Fol. However, given its amino acid sequence divergence from I‐7, it would be unlikely to confer the same specificity and, given that I‐7 can function in its absence, it is unlikely to be a co‐receptor.
I‐7 encodes a protein with a domain architecture typical of LRR‐RLPs (Fig. 5). Other LRR‐RLPs identified in tomato and tobacco include disease resistance proteins, such as the tomato Cf‐2 and Cf‐9 proteins, conferring resistance to Cladosporium fulvum (Dixon et al., 1996; Jones et al., 1994), and pattern recognition receptors (PRRs), including the tomato Ve1 protein, which mediates resistance against Verticillium dahliae (Kawchuk et al., 2001) through recognition of the Ave1 protein present in various pathogenic fungi (de Jonge et al., 2012), the tomato Eix1 and Eix2 receptors, which mediate perception of the cell wall‐derived ethylene‐inducing xylanase (EIX) from Trichoderma species (Ron and Avni, 2004), and the tobacco EILP protein, which responds to a cell wall elicitor from Phytophthora infestans (Takemoto et al., 2000). The LRR domains of these proteins differ in repeat number and the sequence of non‐LRR residues within the LRR motif. Conversely, these proteins show conserved N‐ and C‐terminal regions encompassing three N‐ and 11 C‐terminal LRRs that contain conserved cysteine residues likely to be involved in disulfide bonds, two or four at the N‐terminus, two in adjacent LRRs and two in the C‐terminal LRR‐flanking domain (Fig. 7). C‐terminal conservation, including the 11 C‐terminal LRRs, C‐terminal LRR‐flanking domain and transmembrane GxxxGxxxG motif (Fig. 5), is thought to reflect interaction with a downstream signalling partner, possibly SOBIR1 (Bi et al., 2015; Liebrand et al., 2013) and/or BAK1 (Postma et al., 2015). The intervening LRRs, which vary in number and sequence between LRR‐RLPs, are often involved in the recognition of microbial proteins, including specific avirulence proteins (Chakrabarti et al., 2009; van der Hoorn et al., 2001; Thomas et al., 1997, 1998; Wulff et al., 2009). Accordingly, I‐7 might recognize an Avr7 effector protein yet to be identified, possibly corresponding to one of the SIX (secreted in xylem) proteins produced by Fol during tomato root colonization (Rep, 2005; Schmidt et al., 2013).
Figure 7.
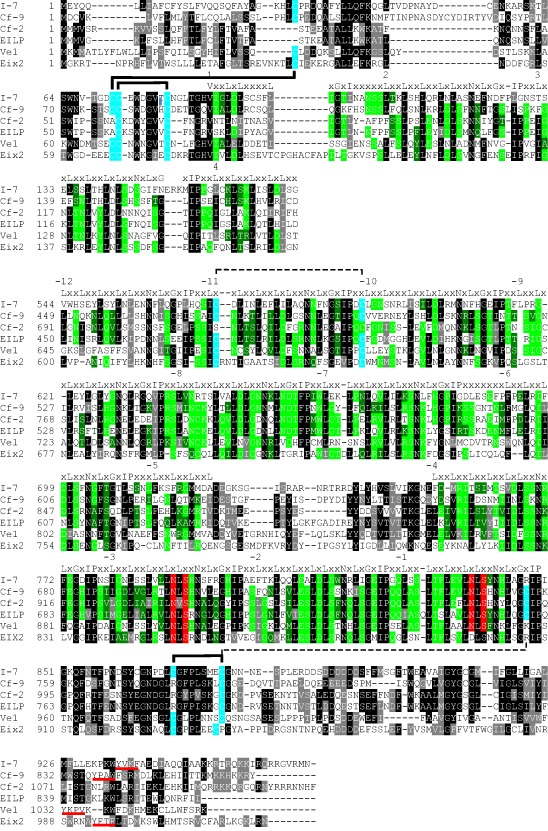
Alignment of N‐ and C‐terminal regions conserved between I‐7 and other tomato and tobacco leucine‐rich repeat receptor‐like proteins (LRR‐RLPs). Sequences were aligned using the EMBL‐EBI MAFFT server at http://www.ebi.ac.uk/Tools/msa/mafft/, and the alignment was shaded using the ExPASy BoxShade server at http://www.ch.embnet.org/software/BOX_form.html. Amino acid identities are highlighted in black. Amino acid similarities are highlighted in grey. In LRR regions, the consensus sequence is shown above the alignment, and amino acid identities for non‐consensus residues are highlighted in bright green and amino acid similarities for non‐consensus residues are highlighted in dark green. Conserved cysteine residues likely to be involved (full connecting lines) or potentially involved (broken connecting lines) in disulfide bonding, based on structural studies of other plant extracellular LRR proteins (Di Matteo et al., 2003; Hothorn et al., 2011), are highlighted in blue. Conserved N‐glycosylation motifs are highlighted in red. LRR‐RLP sequences were derived from GenBank accessions AAA65235 (Cf‐9), AAC15779 (Cf‐2), BAA88636 (EILP), ACR33106 (Ve1) and AAR28378 (Eix2).
Most of the amino acid differences between I‐7 and its homologues are among predicted solvent‐exposed β‐strand residues clustered within LRRs 14–19 (Fig. 6). Plant extracellular LRR domains generally fold into superhelical structures with parallel β‐sheets on the concave side and parallel β‐sheets and α‐ or 310‐helices on the convex side (Di Matteo et al., 2003; Hothorn et al., 2011; She et al., 2011; Sun et al., 2013), providing a structural framework for the presentation of amino acids involved in protein–protein interactions (Kobe and Kajava, 2001). Solvent‐exposed residues on the conserved concave surface β‐strand/β‐turn structural motif are often determinants of recognitional specificity in plant extracellular LRR proteins (Benedetti et al., 2011; Dunning et al., 2007; van der Hoorn et al., 2001; Leckie et al., 1999; Spinelli et al., 2009; Wulff et al., 2009). However, none of the amino acid substitutions that distinguish I‐7 from i‐7LA716 occur in the concave β‐strand/β‐turn region of I‐7. Interestingly, the only residues in central LRRs that distinguish I‐7 from both i‐7M82 and i‐7LA716 are two solvent‐exposed residues (A171 and Y173) in the predicted convex β‐strand region (xLxGx) of LRR12 (Fig. 5), suggesting that I‐7 could function differently from other LRR‐RLPs involved in microbial protein recognition. Other differences that distinguish I‐7 from both i‐7M82 and i‐7LA716 include three residues (S104, S110 and T131) in or near the predicted 310‐helix region (xxLxx) of LRRs 1, 2 and 3 (Fig. 5), which might also be involved in recognition. T131 replaces the conserved leucine of the xxLxx motif, suggesting that it could potentially disrupt 310‐helix formation and therefore also affect protein structure and stability. The remaining differences that distinguish I‐7 from both i‐7M82 and i‐7LA716 are clustered in the non‐LRR C‐terminus of the protein (Fig. 5), suggesting that they could play a role in protein stability or signalling rather than recognition.
Protein sequence analysis also revealed that more than 50% of the putative N‐glycosylation sites showed polymorphisms for presence/absence among I‐7 and its orthologues and paralogues (Fig. 6). Plant LRR domains are often characterized by large numbers of putative N‐glycosylation sites. For example, Cf‐9 contains 22 putative N‐linked glycosylation sites in its extracellular domain, 19 of which are evenly distributed over different LRRs (van der Hoorn et al., 2003). Functional and structural studies have shown that putative N‐linked glycosylation sites not only contribute to Cf‐9 activity, but are essential when located in the putative helical regions of the LRR domain (van der Hoorn et al., 2005). More recently, a functional study of the interactions between N‐glycosylation mutants of the Arabidopsis LRR receptor kinases (EFR and FLS2) and their bacterial interactors (elf18 from elongation factor Tu and flg22 from flagellin) has shown that a single N‐glycan plays a crucial role in receptor abundance and ligand recognition (Haweker et al., 2010). Given the important role of N‐glycosylation sites in the function of various LRR‐containing proteins, it is possible that polymorphisms for the presence/absence of putative N‐glycosylation sites among I‐7 family members might have an impact on the abundance and activity of these proteins. In addition to its possible disruptive effect on 310‐helix formation, the T131 residue also converts N129 into a putative glycosylation site unique to I‐7, so that this residue could potentially have a significant impact on I‐7 structure and function.
Several LRR‐RLPs contain a potential endocytosis motif, e.g. a di‐leucine E/DXXXLø (in Ve1) or tyrosine YXXø (in Ve1, Eix1 and Eix2) motif, where ø is any amino acid with a bulky hydrophobic side chain (Fradin et al., 2014; Kawchuk et al., 2001; Ron and Avni, 2004) (Fig. 7). Like Ve1, Eix1 and Eix2, I‐7 possesses a potential endocytosis motif (YVKF) in its cytosolic tail. However, the motif is not well conserved in sequence or position among LRR‐RLPs, and mutation experiments have shown that the E/DXXXLø and YXXø motifs of Ve1 are not required for function (Zhang et al., 2014). Similarly, A. Chakrabarti et al. (unpublished results) have shown that mutation of the Y or W of the YXXø motif (YAPW) of Cf‐9 does not affect Avr9‐dependent necrosis. In contrast, mutation of the Y in the YXXø motif of Eix2 abolishes the EIX response (Ron and Avni, 2004). The role of the YVKF motif in I‐7 resistance remains to be determined experimentally.
Avr1, present in Fol race 1, suppresses both I‐2‐ and I‐3‐mediated resistance (Houterman et al., 2008). We have shown that I‐7 confers resistance against Fol race 1, indicating that Avr1 does not suppress I‐7‐mediated resistance and is therefore not a general suppressor of resistance. It is possible that the action of Avr1 is limited to Avr2 and Avr3 through a functional relationship between the three effectors, e.g. Avr1 might direct the degradation of a host protein (or proteins) targeted for modification by Avr2 and Avr3, but not by Avr7, to circumvent recognition of the modified host protein(s) by I‐2 and I‐3. Alternatively, the failure of Avr1 to suppress resistance triggered by Avr7 could indicate that Avr1 is a pathway‐specific effector, acting on or after a point of convergence in I‐2 (CC‐NB‐LRR) and I‐3 (SRLK) signalling, but not I‐7 (LRR‐RLP) signalling. EDS1 is required for resistance mediated by LRR‐RLPs and Toll‐Interleukin 1 Receptor (TIR)‐NB‐LRRs, basal defences and systemic acquired resistance, but not resistance mediated by CC‐NB‐LRRs (Aarts et al., 1998; Breitenbach et al., 2014; Fradin et al., 2009; Hu et al., 2005; Parker et al., 1996). We have shown that EDS1 is required for I‐7 resistance, and it can be assumed that EDS1 is not required for I‐2 resistance because I‐2 is a CC‐NB‐LRR protein. If I‐3 is found to be EDS1 independent, it is plausible that I‐7 operates through a different signalling pathway to I‐2 and I‐3. Significantly, Avr1 triggers defence signalling through recognition by I, which is also EDS1 dependent (Hu et al., 2005), indicating that the I gene most probably encodes a TIR‐NB‐LRR or LRR‐RLP that signals through the same pathway as I‐7. Together, these observations are consistent with the idea that Avr1 might act to suppress signalling through a non‐EDS1‐dependent pathway.
The identification of I‐7 has allowed us to develop a CAPS marker to the gene itself (CAPS7774; see Table S2 and Fig. S2), thereby providing plant breeders with perfect genetic markers for the selection and breeding of elite tomato cultivars carrying either or both genes currently known to confer Fol race 3 resistance (see Catanzariti et al., 2015 for I‐3 markers). Given that I‐7‐mediated resistance is not suppressed by Avr1, I‐7 may be a useful backup to I‐3 in the plant breeder's toolbox.
Experimental Procedures
Plant and fungal material
Tomato cultivars Tristar (carrying I‐7; resistant to Fol race 3; McGrath, 1988) and M82 (lacking I‐7; susceptible to Fol race 3) were crossed, and F3 and F4 lines were produced by recurrent selfing. Homozygous resistant or susceptible F3 or F4 lines were identified by screening with Fol race 3, as described below. Tristar was also crossed to the sun1‐1 mutant (homozygous for a mutation of the tomato EDS1 gene; Hu et al., 2005) of the tomato cultivar VF36 (lacking I‐7; susceptible to Fol race 3) and an F2 line produced by selfing. F2 plants were scored for I‐7‐dependent resistance by screening with Fol race 3, as described below. Tomato cultivars Moneymaker (no resistance to Fol) and M82 (lacking I‐7, but carrying the I gene conferring resistance to Fol race 1) were used as Fol race 3‐susceptible lines for plant transformation.
Fol race 1 (Fol004 carrying Avr1, Avr2 and Avr3) and race 2 (Fol007 carrying Avr2 and Avr3) were provided by Dr Martijn Rep (University of Amsterdam, the Netherlands). Fol race 3 isolate #1943 (carrying Avr3) was provided by Mr Des McGrath (Agri‐Science Queensland, Australia).
Pathogen tests for Fusarium wilt resistance
Tomato seeds were sown in seed‐raising mix (Martins Fertilizers, Yass, NSW, Australia), and seedlings were grown for 10–13 days in a temperature‐controlled glasshouse with a maximum day temperature of 25 °C and minimum night temperature of 18 °C prior to fungal inoculation. Fol cultures were grown in potato dextrose broth (Difco, Detroit, MI, USA) for 5 days on a rotary shaker (200 rpm) at 25 °C, and conidia were collected by filtering the cultures through four layers of miracloth. Conidia were washed twice by centrifugation at 6000 g for 10 min and resuspension in deionized water. Conidial concentrations were adjusted to 5 × 106 or 1 × 107 conidia/mL after the final resuspension. Seedlings were removed from the seed‐raising mix and their roots were washed with deionized water, trimmed and then dipped in a conidial suspension for 3 min before replanting in potting mix (Martins Fertilizers). Roots of control plants were dipped in deionized water for the purposes of mock inoculation to ensure that disease phenotypes were a consequence of Fol infection, rather than the inoculation process per se (results not shown apart from the example in Fig. S7). After inoculation, plants were kept in a controlled‐environment growth room with a 16 h, 25 °C day/8 h, 20 °C night cycle. After 18 or 21 days, wilt symptoms and vascular browning were recorded and used to calculate disease scores according to the following criteria described by Rep et al. (2005): 0, no reaction, healthy plant; 1, slightly swollen or bent hypocotyl; 2, one or two brown vascular bundles in hypocotyl; 3, at least two brown vascular bundles and growth distortion; 4, all vascular bundles are brown; plant either dead or very small and wilted.
RNA‐seq analysis
Ten‐day‐old seedlings of tomato cultivars Tristar and M82 were inoculated with Fol race 3 or mock inoculated for the purposes of conducting an RNA‐seq experiment looking at transcriptional responses to Fol infection. Only the data obtained for the mock‐inoculated plants were used for the work described in this paper. Roots of 15 plants from each cultivar were collected 2 days after mock inoculation, washed with sterile deionized water, pooled and frozen in liquid nitrogen for RNA extraction.
Frozen root samples were ground in liquid nitrogen, and total RNA was isolated using a Plant RNeasy kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Extracted RNA (5 μg per sample) was sent to The Ramaciotti Centre (University of New South Wales, Sydney, Australia) for cDNA synthesis, library preparation and the generation of 100‐base paired‐end reads using the HiSeq2000 sequencing platform (Illumina, San Diego, CA, USA). The RNA‐seq experiment was repeated three times to generate biological replicates, but, for the purposes of the analysis described in this paper, data from the replicates were pooled.
The RNA‐seq data were analysed using CLC Genomics Workbench 4.0 (http://www.clcbio.com/). Sequence reads were trimmed using default parameters and then mapped against the tomato (cv. Heinz 1706) reference transcriptome (file ITAG2.3_cdna_alignments.gff3) obtained from the SOL genomics network (http://solgenomics.net/organism/Solanum_lycopersicum/genome). Mapping was performed using the read mapper tool with global alignment (mismatch cost = 2; insertion cost = 3; deletion cost = 3; length fraction = 0.5; similarity = 0.8). The RNA‐seq analysis tool was used to analyse the transcript data (maximum pair distance = 500; minimum paired distance = 1; maximum mismatches = 2; minimum length fraction = 0.97; minimum similarity fraction = 0.8) to determine which genes were expressed in roots. Expression levels were recorded as reads per kilobase per million (RPKM) mapped reads (Mortazavi et al., 2008).
SNP analysis
To identify SNPs between mapped reads and the reference transcriptome, the CLC Genomics Workbench 4.0 SNP detection tool was used with default parameters, and a minimum coverage of 10 and a minimum variant frequency of 35. The SNP data were then transferred into an Excel spreadsheet, and SNPs with a read frequency of ≥75% were selected for further analysis using the Filter function in Excel. SNPs that were common to Tristar and M82 relative to the reference transcriptome were identified using the VLOOKUP function in Excel, and discarded. The remaining polymorphisms were used to calculate the frequency of SNPs in each transcript (number of SNPs per transcript divided by the length of transcript) that were unique to Tristar or M82. The SNP frequency was plotted against the corresponding tomato gene positions on each tomato chromosome to identify genes with a higher frequency of SNPs in Tristar compared with M82 or with surrounding Tristar genes.
Once such cluster of genes was identified. CAPS markers were designed targeting restriction site polymorphisms associated with the SNPs in these genes. Markers were then used to screen plants from 15 homozygous resistant and 16 homozygous susceptible Tristar × M82 F3 or F4 lines to identify correlation between marker genotypes and resistance or susceptibility to Fol race 3.
Generation of transgenic tomato lines for complementation testing
The I‐7 candidate gene from Tristar and the corresponding gene from M82 were PCR amplified from genomic DNA using LIC7774 forward and reverse primers (Table S4), designed for ligation‐independent cloning (LIC), and iProof high‐fidelity DNA polymerase (BioRad Laboratories, Hercules, CA, USA), according to the manufacturer's instructions. PCR products were cloned via LIC into pL2, a binary vector based on pGreenII (Hellens et al., 2000), containing an nptII gene for the selection of transformed plants and a LIC site flanked by a cauliflower mosaic virus (CaMV) 35S promoter and terminator (Fig. S8, see Supporting Information). Purified PCR product was treated with T4 DNA polymerase (NEB, Ipswich, MA, USA) in 1 × Buffer 2 (NEB) and 2.5 mm dCTP at 22 °C for 30 min. To prepare the vector, pL2 was digested with SnaBI, purified and treated with T4 DNA polymerase in 1 × Buffer 2 and 2.5 mm dGTP at 22 °C for 30 min. PCR products (75 ng) and vector (50 ng) were incubated for 20 min at 75 °C, cooled at 4 °C, mixed together, the volume adjusted to 10 μL with water, and incubated at 65 °C for 1 min, and then 22 °C for 20 min, to anneal the complementary vector and insert overhangs prior to heat shock transformation of Escherichia coli Mach1 cells. Transformants were screened by colony PCR using insert‐specific primers, and insert integrity was confirmed by sequencing.
Binary vectors were extracted from E. coli and transformed together with pSoup, required for the replication of pGreenII‐based vectors in Agrobacterium (Hellens et al., 2000), into Agrobacterium tumefaciens GV3101 (pMP90) by electroporation. Transformants were screened by colony PCR using insert‐specific primers, and positive transformants were confirmed by plasmid rescue (plasmid extraction and re‐transformation of E. coli). Agrobacterium transformants were then used to transform tomato cultivars M82 (susceptible to Fol races 2 and 3, but resistant to race 1) and Moneymaker (susceptible to all three races) using the standard Agrobacterium co‐cultivation technique, essentially as described by McCormick (1991). Transgenic plants were selected using 100 mg/L kanamycin and 200 mg/L timentin, and the presence of the transgene was confirmed by PCR analysis of plant DNA extracts using a transgene‐specific forward primer and a T‐DNA‐specific (M13) reverse primer.
Supporting information
Fig. S1 Marker analysis on Tristar, M82 and 15 homozygous‐resistant and 16 homozygous‐susceptible Tristar × M82 F3 or F4 lines using CAPS7774 primers (Table S2) targeting the Solyc08g077740 gene. Polymerase chain reaction (PCR) amplification generates an 808‐bp product with all genomic DNA templates (not shown) and, after digestion with AgeI, Tristar shows digested products of 612 and 196 bp, whereas M82 products remain undigested. Resistant lines all show the Tristar banding pattern, whereas susceptible lines all show the M82 banding pattern, indicating complete linkage between I‐7 and the CAPS7774 marker.
Fig. S2 Polymerase chain reaction (PCR) screening of MM‐7774Tristar T2 plants from two lines for the presence of the Solyc08g077740 transgene from Tristar using 7774F4 (Table S4) and M13 reverse primers to detect the transgene (top band) and CAPS7774 primers (Table S2) to detect the transgene and the endogenous Solyc08g077740 gene in transgenic plants, or the endogenous Solyc08g077740 gene alone in non‐transgenic sibs (bottom band). Asterisks indicate plants carrying the transgene. Controls included a genomic DNA sample from Moneymaker (MM), a no tempate control (Water) and plasmid DNA containing the transgene (pL2‐Tristar).
Fig. S3 (A) Disease assays on T2 plants from two additional lines segregating for the Solyc08g077740 gene from Tristar in a Moneymaker (MM) background. T2 plants, together with Tristar (resistant) and Moneymaker (susceptible) control plants, were inoculated with Fol race 3. Photographs were taken at 21 days post‐inoculation (dpi). (B) Disease scores for the plants shown in (A).
Fig. S4 (A) Disease assays on MM‐7774Tristar #1 T2 plants carrying the Solyc08g077740 gene from Tristar in a Moneymaker (MM) background. T2 plants, together with Tristar (resistant) and Moneymaker (susceptible) control plants, were inoculated with Fol race 1. Photographs were taken at 20 days post‐inoculation (dpi). (B) Disease scores for the T2 plants shown in (A).
Fig. S5 (A) Disease assays on MM‐7774Tristar #1 T2 plants carrying the Solyc08g077740 gene from Tristar in a Moneymaker (MM) background. T2 plants, together with Tristar (resistant) and Moneymaker (susceptible) control plants, were inoculated with Fol race 2. Photographs were taken at 20 days post‐inoculation (dpi). (B) Disease scores for the T2 plants shown in (A).
Fig. S6 (A) Polymerase chain reaction (PCR) screening for the presence of EDS1 and the eds1 mutation in Tristar × sun1‐1 F2 plants using the primers as described in the main text. Plants containing the EDS1 gene produce a PCR product of 644 bp, whereas plants carrying the eds1 mutation produce a PCR product of about 850 bp. The presence of the EDS1 gene is represented by a capital E and the presence of the eds1 allele by a lower case e. Plants heterozygous for eds1 (with an Ee genotype shown in white) and homozygous for eds1 (with an ee genotype shown in red) were inoculated with Fol race 3 and screened for the presence of I‐7 as shown in (B). (B) PCR screening for the presence of I‐7 among the Ee and ee plant DNA samples shown in (A) using the CAPS7774 primers (Table S2). I‐7 generates 612‐ and 196‐bp products after digestion with AgeI, whereas the i‐7 allele from sun1‐1 generates an 808‐bp product that remains undigested. Plants carrying I‐7 are indicated. In both (A) and (B), the PCR products amplified from genomic DNA of the parent lines (Tristar and sun1‐1) are shown (boxed).
Fig. S7 (A) Authentication of IL8‐1, IL8‐2 and IL8‐3 lines using chromosome 8 cleaved amplified polymorphic sequence (CAPS) and sequence characterised amplified region (SCAR) markers diagnostic for each line (Table S5, see Supporting Information). (B) Analysis of the Tristar, M82, IL8‐1, IL8‐2 and IL8‐3 lines using the CAPS7774 marker (Table S2), showing that IL8‐2 carries the LA716 allele (i‐7LA716) of the I‐7 gene. (C) Disease assays on IL8‐1, IL8‐2 and IL8‐3 plants inoculated with Fol race 3 or mock inoculated. Photographs were taken at 21 days post‐inoculation (dpi).
Fig. S8 Ligation‐independent cloning (LIC) using the in‐house binary vector pL2 derived from pGreenII showing: (A) steps in the LIC process; (B) a map of pL2.
Table S1 RNA‐seq output, mapping and single nucleotide polymorphism (SNP) analysis data
Table S2 Cleaved amplified polymorphic sequence (CAPS) markers designed for genes within the cluster of Tristar single nucleotide polymorphisms (SNPs) on chromosome 8
Table S3 Genes present in the introgressed and neighbouring regions of Tristar chromosome 8. The introgressed region encompasses genes Solyc08g077520 to Solyc08g077800, genes which show a high frequency of single nucleotide polymorphisms (SNPs) unique to Tristar.
Table S4 Primers used for amplification, cloning and sequencing of Solyc08g077740 genes from Tristar and M82.
Table S5 Cleaved amplified polymorphic sequence (CAPS) and sequence characterised amplified region (SCAR) markers used to authenticate Solanum pennellii LA716 chromosome 8 introgression lines IL8‐1, IL8‐2 and IL8‐3.
Acknowledgements
This work was funded by an Australian Research Council Linkage Project grant (LP100100172) awarded to DAJ and DJM, which, in turn, funded an Australian Postgraduate Award (Industry) scholarship awarded to YG‐C. A‐MC was funded by an Australian Research Council postdoctoral fellowship (DP1095157). We are grateful to the Queensland Department of Agriculture and Fisheries for their financial contribution to the ARC Linkage Project as industry partner. We are also grateful to ANU Plant Culture staff for their assistance.
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis . Proc. Natl. Acad. Sci. USA, 95, 10 306–10 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, L.J. and Tucker, C.M. (1945) Physiological specialization in the Tomato wilt fungus Fusarium oxysporum f. sp. lycopersici . J. Agric. Res. 70, 303–313. [Google Scholar]
- Benedetti, M. , Leggio, C. , Federici, L. , De Lorenzo, G. , Pavel, N.V. and Cervone, F. (2011) Structural resolution of the complex between a fungal polygalacturonase and a plant polygalacturonase‐inhibiting protein by small‐angle X‐ray scattering. Plant Physiol. 157, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, G. , Liebrand, T.W.H. , Bye, R.R. , Postma, J. , van der Burgh, A.M. , Robatzek, S. , Xu, X. and Joosten, M.H.A.J. (2015) SOBIR1 requires the GxxxG dimerization motif in its transmembrane domain to form constitutive complexes with receptor‐like proteins. Mol. Plant Pathol. doi: 10.1111/mpp.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn, G. and Tucker, C. (1939) Immunity to Fusarium wilt in the tomato. Science, 89, 603–604. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J.S. and Traub, L.M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447. [DOI] [PubMed] [Google Scholar]
- Breitenbach, H.H. , Wenig, M. , Wittek, F. , Jordá, L. , Maldonado‐Alconada, A.M. , Sarioglu, H. , Colby, T. , Knappe, C. , Bichlmeier, M. , Pabst, E. , Mackey, D. , Parker, J.E. and Vlot, A.C. (2014) Contrasting roles of the apoplastic aspartyl protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1‐DEPENDENT1 and LEGUME LECTIN‐LIKE PROTEIN1 in Arabidopsis systemic acquired resistance. Plant Phys. 165, 791–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Lim, G.T. and Jones, D.A. (2015) The tomato I‐3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207, 106–118. [DOI] [PubMed] [Google Scholar]
- Chakrabarti, A. , Panter, S.N. , Harrison, K. , Jones, J.D.G. and Jones, D.A. (2009) Regions of the Cf‐9B disease resistance protein able to cause spontaneous necrosis in Nicotiana benthamiana lie within the region controlling pathogen recognition in tomato. Mol. Plant–Microbe Interact. 22, 1214–1226. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Shang, J. , Chen, D. , Lei, C. , Zou, Y. , Zhai, W. , Liu, G. , Xu, J. , Ling, Z. , Cao, G. , Ma, B. , Wang, Y. , Zhao, X. , Li, S. and Zhu, L. (2006) A B‐lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804. [DOI] [PubMed] [Google Scholar]
- Cole, S.J. and Diener, A.C. (2013) Diversity in receptor‐like kinase genes is a major determinant of quantitative resistance to Fusarium oxysporum f.sp. matthioli . New Phytol. 200, 172–184. [DOI] [PubMed] [Google Scholar]
- Di Matteo, A. , Federici, L. , Mattei, B. , Salvi, G. , Johnson, K.A. , Savino, C. , De Lorenzo, G. , Tsernoglou, D. and Cervone, F. (2003) The crystal structure of polygalacturonase‐inhibiting protein (PGIP), a leucine‐rich repeat protein involved in plant defense. Proc. Natl. Acad. Sci. USA, 100, 10 124–10 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, A.C. and Ausubel, F.M. (2005) Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease‐resistance gene, is not race specific. Genetics, 171, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S. , Jones, D.A. , Keddie, J.S. , Thomas, C.M. , Harrison, K. and Jones, J.D.G. (1996) The tomato Cf‐2 disease resistance locus comprises two functional genes encoding leucine‐rich repeat proteins. Cell, 84, 451–459. [DOI] [PubMed] [Google Scholar]
- Dunning, F.M. , Sun, W. , Jansen, K.L. , Helft, L. and Bent, A.F. (2007) Identification and mutational analysis of Arabidopsis FLS2 leucine‐rich repeat domain residues that contribute to flagellin perception. Plant Cell, 19, 3297–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y. and Zamir, D. (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield‐associated QTL. Genetics, 141, 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Juarez Ayala, J.C. , Castroverde, C.D.M. , Nazar, R.N. , Robb, J. , Liu, C.‐M. and Thomma, B.P.H.J. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1 . Plant Physiol. 150, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Rovenich, H. , Song, Y. , Liebrand, T.W.H. , Masini, L. , van den Berg, G.C.M. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2014) Functional analysis of the tomato immune receptor Ve1 through domain swaps with its non‐functional homolog Ve2. PLoS ONE, 9, e88208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel, Y. and von Heijne, G. (1990) Sequence differences between glycosylated and non‐glycosylated Asn‐X‐Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Cendales, Y. , Do, H.T.T. , Lim, G.T.T. , McGrath, D.J. , Catanzariti, A.‐M. and Jones, D.A. (2014) Application of CAPS markers to the mapping and marker‐assisted breeding of genes for resistance to Fusarium wilt in the tomato In: Cleaved Amplified Polymorphic Sequences (CAPS) Markers in Plant Biology (Shavrukov Y., ed.), pp. 91–108, New York: Nova Science Publishers. [Google Scholar]
- Grattidge, R. and O'Brien, R.G. (1982) Occurrence of a third race of Fusarium wilt of tomatoes in Queensland. Plant Dis. 66, 165–166. [Google Scholar]
- Haweker, H. , Rips, S. , Koiwa, H. , Salomon, S. , Saijo, Y. , Chinchilla, D. , Robatzek, S. and von Schaewen, A. (2010) Pattern recognition receptors require N‐glycosylation to mediate plant immunity. J. Biol. Chem. 285, 4629–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1986) A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14, 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P. , Edwards, E.A. , Leyland, N.R. , Bean, S. and Mullineaux, P.M. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium‐mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. , Roth, R. and De Wit, P.J.G.M. (2001) Identification of distinct specificity determinants in resistance protein Cf‐4 allows construction of a Cf‐9 mutant that confers recognition of avirulence protein Avr4. Plant Cell, 13, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. , Rivas, S. , Wulff, B.B.H. , Jones, J.D.G. and Joosten, M.H.A.J. (2003) Rapid migration in gel filtration of the Cf‐4 and Cf‐9 resistance proteins is an intrinsic property of Cf proteins and not because of their association with high‐molecular‐weight proteins. Plant J. 35, 305–315. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. , Wulff, B.B.H. , Rivas, S. , Durrant, M.C. , van der Ploeg, A. , de Wit, P.J.G.M. and Jones, J.D.G. (2005) Structure–function analysis of Cf‐9, a receptor‐like protein with extracytoplasmic leucine‐rich repeats. Plant Cell, 17, 1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn, M. , Belkhadir, Y. , Dreux, M. , Dabi, T. , Noel, J.P. , Wilson, I.A. and Chory, J. (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature, 474, 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman, P.M. , Cornelissen, B.J.C. and Rep, M. (2008) Suppression of plant resistance gene‐based immunity by a fungal effector. PLoS Pathol. 4, e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, G. , deHart, A.K. , Li, Y. , Ustach, C. , Handley, V. , Navarre, R. , Hwang, C.‐F. , Aegerter, B.J. , Williamson, V.M. and Baker, B. (2005) EDS1 in tomato is required for resistance mediated by TIR‐class R genes and the receptor‐like R gene Ve . Plant J. 42, 376–391. [DOI] [PubMed] [Google Scholar]
- Jones, D.A. , Thomas, C.M. , Hammond‐Kosack, K.E. , Balint‐Kurti, P.J. and Jones, J.D.G. (1994) Isolation of the tomato Cf‐9 gene for resistance to Cladosporium fulvum by transposon tagging. Science, 266, 789–793. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M.K. , Zhang, Z. , Usami, T. , Lievens, B. , Subbarao, K.V. and Thomma, B.P.H.J. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk, L.M. , Hachey, J. , Lynch, D.R. , Kulcsar, F. , van Rooijen, G. , Waterer, D.R. , Robertson, A. , Kokko, E. , Byers, R. , Howard, R.J. , Fischer, R. and Prüfer, D. (2001) Tomato Ve disease resistance genes encode cell surface‐like receptors. Proc. Natl. Acad. Sci. USA, 98, 6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B. and Kajava, A.V. (2001) The leucine‐rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732. [DOI] [PubMed] [Google Scholar]
- Leckie, F. , Mattei, B. , Capodicasa, C. , Hemmings, A. , Nuss, L. , Aracri, B. , De Lorenzo, G. and Cervone, F. (1999) The specificity of polygalacturonase inhibiting protein (PGIP): a single amino acid substitution in the solvent‐exposed β‐strand/β‐turn region of the leucine‐rich repeats (LRRs) confers a new recognition capability. EMBO J. 18, 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W.H. , van den Berg, G.C.M. , Zhang, Z. , Smit, P. , Cordewener, J.H.G. , America, A.H.P. , Sklenard, J. , Jones, A.M.E. , Tameling, W.I.L. , Robatzek, S. , Thomma, B.P.H.J. and Joosten, M.H.A.J. (2013) Receptor‐like kinase SOBIR1/EVR interacts with receptor‐like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA, 110, 10 010–10 015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, G.T.T. , Wang, G.P. , Hemming, M.N. , Basuki, S. , McGrath, D.J. , Carroll, B.J. and Jones, D.A. (2006) Mapping the I‐3 gene for resistance to Fusarium wilt in tomato: application of an I‐3 marker in tomato improvement and progress towards the cloning of I‐3 . Australas. Plant Pathol. 35, 671–680. [Google Scholar]
- Lim, G.T.T. , Wang, G.P. , Hemming, M.N. , McGrath, D.J. and Jones, D.A. (2008) High resolution genetic and physical mapping of the I‐3 region of tomato chromosome 7 reveals almost continuous microsynteny with grape chromosome 12 but interspersed microsynteny with duplications on Arabidopsis chromosomes 1, 2 and 3. Theor. Appl. Genet. 118, 57–75. [DOI] [PubMed] [Google Scholar]
- McCormick, S. (1991) Transformation of tomato with Agrobacterium tumefaciens In: Plant Tissue Culture Manual (Lindsey K., ed.), pp. 1–9. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- McGrath, D.J. (1988) Fusarium wilt resistant tomatoes. Queensland Agric. J. 114, 290. [Google Scholar]
- McGrath, D. , Gillespie, D. and Vawdrey, L. (1987) Inheritance of resistance to Fusarium oxysporum f. sp. lycopersici races 2 and 3 in Lycopersicon pennellii . Aust. J. Agric. Res. 38, 729–733. [Google Scholar]
- Michielse, C.B. and Rep, M. (2009) Pathogen profile update: Fusarium oxysporum . Mol. Plant Pathol. 10, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi, A. , Williams, B.A. , McCue, K. , Schaeffer, L. and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Parker, J.E. , Holub, E.B. , Frost, L.N. , Falk, A. , Gunn, N.D. and Daniels, M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell, 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma, J. , Liebrand, T.W. , Bi, G. , Evrard, A. , Bye, R.R. , Mbengue, M. , Joosten, M.H.A.J. and Robatzek, S. (2015) The Cf‐4 receptor‐like protein associates with the BAK1 receptor‐like kinase to initiate receptor endocytosis and plant immunity. bioRχiv, 019471. [DOI] [PubMed]
- Reis, A. , Costa, H. , Boiteux, L.S. and Lopes, C.A. (2005) First report of Fusarium oxysporum f. sp. lycopersici race 3 on tomato in Brazil. Fitopatol. Bras. 30, 426–428. [Google Scholar]
- Rep, M. (2005) Small proteins of plant‐pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253, 19–27. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Meijer, M. , Houterman, P.M. , van der Does, H.C. and Cornelissen, B.J.C. (2005) Fusarium oxysporum evades I‐3‐mediated resistance without altering the matching avirulence gene. Mol. Plant–Microbe Interact. 18, 15–23. [DOI] [PubMed] [Google Scholar]
- Ron, M. and Avni, A. (2004) The receptor for the fungal elicitor ethylene‐inducing xylanase is a member of a resistance‐like gene family in tomato. Plant Cell, 16, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, S.M. , Houterman, P.M. , Schreiver, I. , Ma, L. , Amyotte, S. , Chellappan, B. , Sjef Boeren, S. , Takken, F.L.W. and Rep, M. (2013) MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum . BMC Genomics, 14, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, J.W. and Jones, J.P. (1989) Monogenic resistance in tomato to Fusarium oxysporum f. sp. lycopersici race 3. Euphytica, 40, 49–53. [Google Scholar]
- Sela‐Buurlage, M.B. , Budai‐Hadrian, O. , Pan, Q. , Carmel‐Goren, L. , Vunsch, R. , Zamir, D. and Fluhr, R. (2001) Genome‐wide dissection of Fusarium resistance in tomato reveals multiple complex loci. Mol. Genet. Genomics, 265, 1104–1111. [DOI] [PubMed] [Google Scholar]
- She, J. , Han, Z. , Kim, T.W. , Wang, J. , Cheng, W. , Chang, J. , Shi, S. , Wang, J. , Yang, M. , Wang, Z.‐Y. and Chai, J. (2011) Structural insight into brassinosteroid perception by BRI1. Nature, 474, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. and Diener, A.C. (2013) Arabidopsis thaliana resistance to Fusarium oxysporum 2 implicates tyrosine‐sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet. 9, e1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, G. , Groenendijk, J. , Wijbrandi, J. , Reijans, M. , Groenen, J. , Diergaarde, P. , Van der Lee, T. , Bleeker, M. , Onstenk, J. , de Both, M. , Haring, M. , Mes, J. , Cornelissen, B. , Zabeau, M. and Vos, P. (1998) Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell, 10, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli, F. , Mariotti, L. , Mattei, B. , Salvi, G. , Cervone, F. and Caprari, C. (2009) Three aspartic acid residues of polygalacturonase‐inhibiting protein (PGIP) from Phaseolus vulgaris are critical for inhibition of Fusarium phyllophilum PG. Plant Biol. 11, 738–743. [DOI] [PubMed] [Google Scholar]
- Stall, R. and Walter, J. (1965) Selection and inheritance of resistance in tomato to isolates of races 1 and 2 of the Fusarium wilt organism. Phytopathology, 55, 1213–1215. [Google Scholar]
- Sun, Y. , Li, L. , Macho, A.P. , Han, Z. , Hu, Z. , Zipfel, C. , Zhou, J.‐M. and Chai, J. (2013) Structural basis for flg22‐induced activation of the Arabidopsis FLS2‐BAK1 immune complex. Science, 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Takemoto, D. , Hayashi, M. , Doke, N. , Mishimura, M. and Kawakita, K. (2000) Isolation of the gene for EILP, an elicitor‐inducible LRR receptor‐like protein, from tobacco by differential display. Plant Cell Physiol. 41, 458–464. [DOI] [PubMed] [Google Scholar]
- Thomas, C.M. , Jones, D.A. , Parniske, M. , Harrison, K. , Balint‐Kurti, P.J. , Hatzixanthis, K. and Jones, J.D.G. (1997) Characterization of the tomato Cf‐4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf‐4 and Cf‐9 . Plant Cell, 9, 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M. , Dixon, M.S. , Parniske, M. , Golstein, C. and Jones, J.D.G. (1998) Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum . Philos. Trans. R. Soc. London, B: Biol. Sci. 353, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela‐Ureta, J. , Lawn, D. , Heisey, R.F. and Zamudio‐Guzman, V. (1996) First report of Fusarium wilt race 3, caused by Fusarium oxysporum f. sp. lycopersici, of tomato in Mexico. Plant Dis. 80, 105. [Google Scholar]
- Volin, R.B. and Jones, J.P. (1982) A new race of Fusarium wilt of tomato in Florida and sources of resistance. Proc. Florida State Hort. Soc. 95, 268–270. [Google Scholar]
- Wulff, B.B.H. , Heese, A. , Tomlinson‐Buhot, L. , Jones, D.A. , de la Pena, M. and Jones, J.D.G. (2009) The major specificity‐determining amino acids of the tomato Cf‐9 disease resistance protein are at hypervariable solvent‐exposed positions in the central leucine‐rich repeats. Mol. Plant–Microbe Interact. 22, 1203–1213. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Song, Y. , Liu, C.M. and Thomma, B.P.H.J. (2014) Mutational analysis of the Ve1 immune receptor that mediates Verticillium resistance in tomato. PLoS ONE, 9, e99511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Marker analysis on Tristar, M82 and 15 homozygous‐resistant and 16 homozygous‐susceptible Tristar × M82 F3 or F4 lines using CAPS7774 primers (Table S2) targeting the Solyc08g077740 gene. Polymerase chain reaction (PCR) amplification generates an 808‐bp product with all genomic DNA templates (not shown) and, after digestion with AgeI, Tristar shows digested products of 612 and 196 bp, whereas M82 products remain undigested. Resistant lines all show the Tristar banding pattern, whereas susceptible lines all show the M82 banding pattern, indicating complete linkage between I‐7 and the CAPS7774 marker.
Fig. S2 Polymerase chain reaction (PCR) screening of MM‐7774Tristar T2 plants from two lines for the presence of the Solyc08g077740 transgene from Tristar using 7774F4 (Table S4) and M13 reverse primers to detect the transgene (top band) and CAPS7774 primers (Table S2) to detect the transgene and the endogenous Solyc08g077740 gene in transgenic plants, or the endogenous Solyc08g077740 gene alone in non‐transgenic sibs (bottom band). Asterisks indicate plants carrying the transgene. Controls included a genomic DNA sample from Moneymaker (MM), a no tempate control (Water) and plasmid DNA containing the transgene (pL2‐Tristar).
Fig. S3 (A) Disease assays on T2 plants from two additional lines segregating for the Solyc08g077740 gene from Tristar in a Moneymaker (MM) background. T2 plants, together with Tristar (resistant) and Moneymaker (susceptible) control plants, were inoculated with Fol race 3. Photographs were taken at 21 days post‐inoculation (dpi). (B) Disease scores for the plants shown in (A).
Fig. S4 (A) Disease assays on MM‐7774Tristar #1 T2 plants carrying the Solyc08g077740 gene from Tristar in a Moneymaker (MM) background. T2 plants, together with Tristar (resistant) and Moneymaker (susceptible) control plants, were inoculated with Fol race 1. Photographs were taken at 20 days post‐inoculation (dpi). (B) Disease scores for the T2 plants shown in (A).
Fig. S5 (A) Disease assays on MM‐7774Tristar #1 T2 plants carrying the Solyc08g077740 gene from Tristar in a Moneymaker (MM) background. T2 plants, together with Tristar (resistant) and Moneymaker (susceptible) control plants, were inoculated with Fol race 2. Photographs were taken at 20 days post‐inoculation (dpi). (B) Disease scores for the T2 plants shown in (A).
Fig. S6 (A) Polymerase chain reaction (PCR) screening for the presence of EDS1 and the eds1 mutation in Tristar × sun1‐1 F2 plants using the primers as described in the main text. Plants containing the EDS1 gene produce a PCR product of 644 bp, whereas plants carrying the eds1 mutation produce a PCR product of about 850 bp. The presence of the EDS1 gene is represented by a capital E and the presence of the eds1 allele by a lower case e. Plants heterozygous for eds1 (with an Ee genotype shown in white) and homozygous for eds1 (with an ee genotype shown in red) were inoculated with Fol race 3 and screened for the presence of I‐7 as shown in (B). (B) PCR screening for the presence of I‐7 among the Ee and ee plant DNA samples shown in (A) using the CAPS7774 primers (Table S2). I‐7 generates 612‐ and 196‐bp products after digestion with AgeI, whereas the i‐7 allele from sun1‐1 generates an 808‐bp product that remains undigested. Plants carrying I‐7 are indicated. In both (A) and (B), the PCR products amplified from genomic DNA of the parent lines (Tristar and sun1‐1) are shown (boxed).
Fig. S7 (A) Authentication of IL8‐1, IL8‐2 and IL8‐3 lines using chromosome 8 cleaved amplified polymorphic sequence (CAPS) and sequence characterised amplified region (SCAR) markers diagnostic for each line (Table S5, see Supporting Information). (B) Analysis of the Tristar, M82, IL8‐1, IL8‐2 and IL8‐3 lines using the CAPS7774 marker (Table S2), showing that IL8‐2 carries the LA716 allele (i‐7LA716) of the I‐7 gene. (C) Disease assays on IL8‐1, IL8‐2 and IL8‐3 plants inoculated with Fol race 3 or mock inoculated. Photographs were taken at 21 days post‐inoculation (dpi).
Fig. S8 Ligation‐independent cloning (LIC) using the in‐house binary vector pL2 derived from pGreenII showing: (A) steps in the LIC process; (B) a map of pL2.
Table S1 RNA‐seq output, mapping and single nucleotide polymorphism (SNP) analysis data
Table S2 Cleaved amplified polymorphic sequence (CAPS) markers designed for genes within the cluster of Tristar single nucleotide polymorphisms (SNPs) on chromosome 8
Table S3 Genes present in the introgressed and neighbouring regions of Tristar chromosome 8. The introgressed region encompasses genes Solyc08g077520 to Solyc08g077800, genes which show a high frequency of single nucleotide polymorphisms (SNPs) unique to Tristar.
Table S4 Primers used for amplification, cloning and sequencing of Solyc08g077740 genes from Tristar and M82.
Table S5 Cleaved amplified polymorphic sequence (CAPS) and sequence characterised amplified region (SCAR) markers used to authenticate Solanum pennellii LA716 chromosome 8 introgression lines IL8‐1, IL8‐2 and IL8‐3.


