Summary
Oxalate oxidases (OxO) catalyse the degradation of oxalic acid (OA). Highly resistant transgenic soybean carrying an OxO gene and its susceptible parent soybean line, AC Colibri, were tested for genome‐wide gene expression in response to the necrotrophic, OA‐producing pathogen Sclerotinia sclerotiorum using soybean cDNA microarrays. The genes with changed expression at statistically significant levels (overall F‐test P‐value cut‐off of 0.0001) were classified into functional categories and pathways, and were analysed to evaluate the differences in transcriptome profiles. Although many genes and pathways were found to be similarly activated or repressed in both genotypes after inoculation with S. sclerotiorum, the OxO genotype displayed a measurably faster induction of basal defence responses, as observed by the differential changes in defence‐related and secondary metabolite genes compared with its susceptible parent AC Colibri. In addition, the experiment presented provides data on several other transcripts that support the hypothesis that S. sclerotiorum at least partially elicits the hypersensitive response, induces lignin synthesis (cinnamoyl CoA reductase) and elicits as yet unstudied signalling pathways (G‐protein‐coupled receptor and related). Of the nine genes showing the most extreme opposite directions of expression between genotypes, eight were related to photosynthesis and/or oxidation, highlighting the importance of redox in the control of this pathogen.
Keywords: defence, microarray, necrotroph, photosynthesis, transcriptomics, white mould
Introduction
White mould disease, caused by the filamentous ascomycete fungus Sclerotinia sclerotiorum (Lib.) de Bary, can be a devastating disease of soybean and many other plant species. The disease is difficult to control and no fully resistant soybean genotype has been found to date. Sclerotinia sclerotiorum is a necrotrophic pathogen and its main virulence factor has been shown to be oxalic acid (OA) (Godoy et al., 1990). The secretion of OA from growing mycelia, in contact with the plant surface, conditions the tissue for infection and enhances the formation of lesions. The dying tissue is penetrated by invading hyphae, which presumably release OA continuously, allowing further ingression throughout the plant.
Several mechanisms of action for secreted OA have been proposed. Marciano et al. (1983) proposed that OA chelates Ca2+ ions in the plant middle lamellae, decreasing Ca2+‐mediated cross‐linking of pectin, thereby facilitating the degradation of the plant cell wall by pathogen‐released pectin‐degrading enzymes. Sperry and Tyree (1988) found that the decrease in conductivity of the host xylem, assumed to be caused by the occlusion of vessels by the fungus, is a result of the induction of embolism directed by OA in combination with endogenous calcium. Studies in soybean and tobacco have indicated that OA can act independently of calcium to inhibit the defence‐associated oxidative burst, presumably by restraining the production of hydrogen peroxide (Cessna et al., 2000). Various exo‐ and endopolygalacturonases have been found to show increased expression and activity in the presence of OA (Favaron et al., 2004; Riou et al., 1991). Guimaraes and Stotz (2004) demonstrated that OA can act at a cellular level to regulate stomata by disrupting abscisic acid (ABA)‐dependent stomatal closure, thus leaving them more easily penetrable by fungal mycelia. Kim et al. (2008) provided evidence that OA induces cell death, and that the pathogen may make use of the initial dying cells to initiate infection of the tissue. Cell death induced by OA may be related to its ability to chelate iron, as suggested in a recent high‐throughput gene expression study in soybean (Calla et al., 2014).
Oxalate oxidases (OxO) (EC 1.2.3.4) are enzymes that catalyse a reaction that converts OA into hydrogen peroxide (H2O2) and carbon dioxide (CO2). Several species of plant, fungi and bacteria produce OxO enzymes. Enzymes with OxO activity include the true‐cereal‐only germins and some other germin‐like proteins which belong to the cupin superfamily (van Loon et al., 2006). Germins have long been known, and so named, by their presence in germinating wheat embryos and by having strong resistance to degradation by proteases and sodium dodecylsulphate (SDS) (Thompson et al., 1995; Thompson and Lane, 1980). These proteins possess an N‐terminal secretory signal that targets them to the apoplast. Several lines of evidence have indicated a role for germins not only during germination, but also during defence responses. For example, the presence of germins is proposed to be a defining factor in the domestication of wheat (Triticum aestivum), which would have been largely selected based on its resistance to pathogens (Lane, 1994). It has been proposed previously that germins give the plant an antimicrobial capability because of their catalytic production of H2O2. In the specific case of OA‐producing pathogens, germins also confer plants with the ability to degrade the secreted OA (Lane, 2002; Thompson and Lane, 1980).
Several crops that lack OA‐degrading capacity have been transformed with OxO genes. As early as 1995, an oilseed rape (Brassica napus) was transformed with an OxO gene from barley (Hordeum vulgare) (Thompson et al., 1995). The leaves of transformed plants showed resistance to wilting caused by dipping excised leaves in OA solution. However, resistance to pathogens was not reported. Later, a transgenic OxO soybean was developed by Donaldson et al. (2001). This transgenic plant, 80(30)‐1, was obtained by Agrobacterium‐mediated transformation of the Canadian‐adapted soybean cultivar AC Colibri with the wheat germin gene gf‐2.8. The 80(30)‐1 transgenic plants showed enhanced resistance to S. sclerotiorum after inoculation of cotyledons in young plants (VC stage), and after stem inoculation of V1–V2 stage plants (Cober et al., 2003). In addition, the plants showed enhanced field resistance against S. sclerotiorum relative to the parent line, and an equivalent resistance level when compared with the most resistant short‐season cultivars. The same gf‐2.8 from wheat was later introduced into sunflower (Helianthus annuus) (Hu et al., 2003). Increased OxO activity, elevated levels of H2O2 and induction of the defence signalling molecule salicylic acid (SA) were reported in the transformed sunflower leaves without pathogen inoculation. In addition, three defence‐related genes were reported to be induced in the transgenic sunflower leaves and stems relative to the non‐transformed tissues, namely pathogenesis‐related 5‐1 (PR5‐1), defensin and sunflower carbohydrate oxidase (SCO). Sunflower is also a very susceptible host of S. sclerotiorum and the gf‐2.8‐expressing plants showed increased resistance to direct inoculation with the pathogen. More recently, another oilseed rape line was transformed to express an unspecified ‘wheat OxO’ (Dong et al., 2008). The stable transgenic plants showed increased OxO activity, H2O2 production and resistance to S. sclerotiorum which was documented in the laboratory using detached leaves and in field studies (Dong et al., 2008). Livingstone et al. (2005) transformed peanut plants (Harachis hypoagea) with barley OxO using particle bombardment of embryos. Peanut is an important crop that is susceptible to Sclerotinia minor, a related OA‐producing fungus and causal agent of Sclerotinia blight of peanut. These transformants showed the same positive defensive effects as the other OxO transgenic plant species.
This work evaluates the changes in global transcript abundances resulting from the infection of soybean leaves with S. sclerotiorum, and compares the differences in these changes between the OxO‐expressing transgenic soybean [80(30)‐1], referred to in this work as ‘OxO’, and its parent AC Colibri, referred to as ‘AC’. From the expression data, it appears that the OxO transgenic soybean line is more resistant by initiating a stronger and more rapid defence response than does AC. In addition, this study identifies several specific genes and gene families that may play a role in response to OA and may serve as the basis for further studies that may help us to gain better insights into the mechanism of pathogenesis and to develop more resistant soybean genotypes.
Results and Discussion
Two time‐course microarray experiments were carried out: one with resistant transgenic OxO plants and one with the susceptible parent AC plants. Each experiment contained four time points ranging from 0 to 36 h post‐inoculation (hpi) (Fig. 1), and was analysed independently. At 12 hpi, both OxO and AC plants showed no visible symptoms and, by 24 and 36 hpi, both genotypes began to show browning and possible necrosis. After 36 hpi, the fungus and symptoms continued to spread in AC, but clearly slowed at this stage in OxO. After microarray image acquisition, data normalization and statistical analysis of the 36 700 different cDNAs represented on the microarrays, 1748 cDNA spots showed differential expression levels in AC and 1425 in OxO at an overall F‐test P‐value cut‐off of <0.0001. The large number of statistically significant cDNA spots at this very stringent cut‐off indicates that the data were of high quality because of the low error‐derived variability. The data were further filtered in both experiments by selecting only features with a log2 intensity ratio of ≥2 or ≤−2 for at least one of the six possible pair‐wise sample comparisons (12 hpi vs. 0 hpi, 24 hpi vs. 0 hpi, 36 hpi vs. 0 hpi, 36 hpi vs. 24 hpi, 36 hpi vs. 12 hpi and 24 hpi vs. 12 hpi) for each genotype. This more stringent filtering identified 724 differential cDNA spots in the AC experiment and 578 in the OxO experiment as having the most confidence of being differentially expressed, and these cDNAs are the focus of this article (Table S1, see Supporting Information). These cDNAs identified by stringent filtration were annotated by blasting the sequences against the National Center for Biotechnology Information non‐redundant (NCBI NR) database, The Arabidopsis Information Resource (TAIR) database and the predicted gene set of the soybean genome (Glyma1.0) available on Phytozome v7.0 (Schmutz et al., 2010), and a best annotation and functional category assignment was obtained. The genes were sorted by category and displayed as heat maps to aid in the identification of gross physiological differences and interesting patterns of expression (Fig. 2). In addition, the gene lists from each genotype were merged by retrieving data for all spots that were significant in at least one of the studies, resulting in a combined list of 1041 genes (pre‐sorted by difference in expression ratios at 12 or 36 hpi between OxO and AC) that were clustered (Fig. 3), again with the goal of facilitating the identification of expression patterns of interest. It is clear from these heat map images (Fig. 3) that most of the 1041 genes of the combined list had the same direction of differential expression across the time points and genotypes. To aid in the identification of cDNAs that varied in expression levels between the two genotypes, expression data were clustered for cDNAs that increased at least two‐fold or decreased at least 1.5‐fold between OxO and AC (Fig. S1, see Supporting Information).
Figure 1.
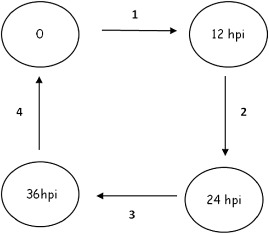
Microarray slide hybridization layout. Loop design used in the 36‐h time course experiment. Circles represent samples of either AC (parent line) or OxO (transgenic line). Arrows represent microarray slides hybridized with samples at the arrowhead and tail. ‘0’ represents the mock‐inoculated T0 sample; hpi, hours post‐inoculation. Three independent biological replications were performed.
Figure 2.
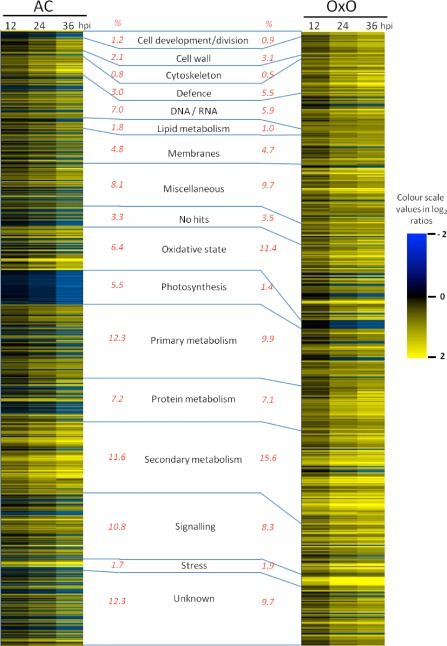
Statistically significant genes. Genes differentially expressed in susceptible AC Colibri (AC, 724 genes) or the highly resistant oxalate oxidase transgenic line 80(30)‐1 (OxO, 578 genes). Each row represents a single gene, and the blue colour depicts genes whose expression decreased at 12, 24 or 36 h post‐inoculation (hpi) compared with the mock‐inoculated T0 control. Yellow colour depicts increased abundance in gene expression. The percentage of the total number of significant genes that fall into each category is given in red to the left (AC%) or right (OxO%) of the category name.
Figure 3.
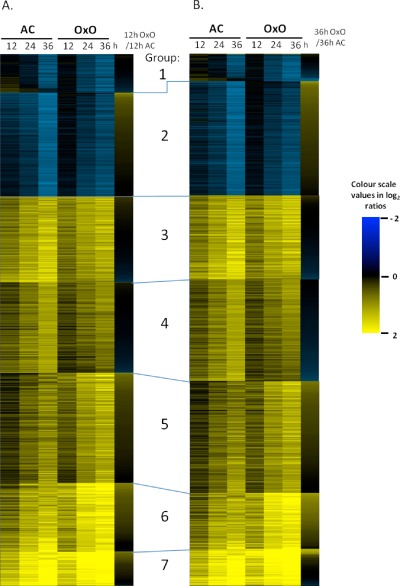
K‐means clustering of the differential expression of all genes deemed significant in at least one genotype. (A) Genes initially sorted by 12 h differential expression, OxO (highly resistant oxalate oxidase transgenic line) over AC (AC Colibri). (B) Genes initially sorted by 36 h differential expression, OxO over AC. Group 1, mostly down‐regulated in both genotypes, but some up in AC and down or non‐responsive in OxO at 12 and 24 hpi. Group 2, down‐regulated in both, but more so in AC. Group 3, both up‐regulated about equally, but some slightly more strongly expressed in AC. Group 4, similar to Group 3, but slightly weaker differential expression. Groups 5, 6 and 7, all induced, but slightly stronger in OxO; differential expression stronger in 7 than 6, and 6 stronger than 5.
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) confirms the microarray results
qRT‐PCR was used to verify the validity of the microarray results. Eight up‐regulated genes and eight down‐regulated genes were randomly selected from the list of significantly differentially regulated genes from the microarray study for each of the soybean lines, and their expression was tested using qRT‐PCR. For all the genes tested, the qRT‐PCR expression result was similar to that of the microarray study, with transcripts showing the same direction of expression (Fig. 4), validating the microarray results.
Figure 4.
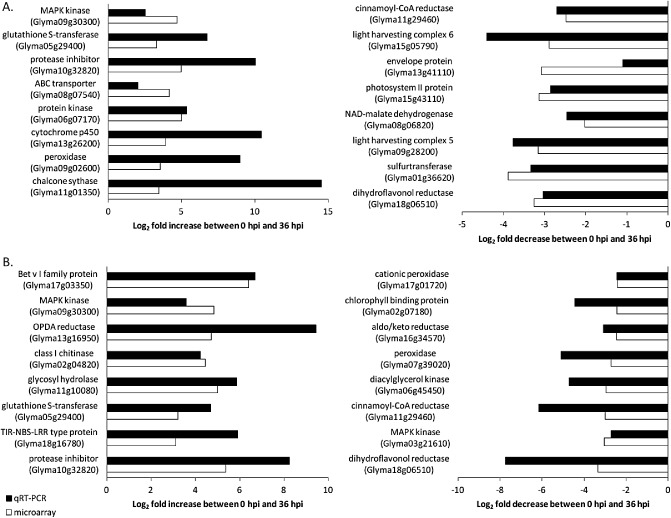
Genes showing increased or decreased expression between 0 and 36 h post‐inoculation with Sclerotinia sclerotiorum in the parent line AC Colibri (A) and the transgenic line OxO (B).
Additional confirmation of the microarray data came from qRT‐PCR of PR5. As in previous results from our group, which showed that PR5 coding sequences were highly up‐regulated in soybean stems that were infected with S. sclerotiorum (Calla et al., 2009), both the OxO and AC microarray experiments showed that PR5 was up‐regulated on infection. To confirm and quantify the expression of PR5, qRT‐PCR analysis was performed from two genomic loci (Glyma01g42660.1 and Glyma01g42670.1) represented by four different cDNA microarray spots that showed induction. The qRT‐PCR analyses showed an increase in expression for both loci in AC and OxO leaves following inoculation with S. sclerotiorum, confirming that PR5 is induced on S. sclerotiorum infection, thus adding additional validity to the microarray results (Fig. S2, see Supporting Information).
Reduced expression of transcripts in AC and OxO over time
One of the most obvious differences in how AC and OxO responded to S. sclerotiorum was in the number of significantly down‐regulated genes. The AC experiment identified 240 down‐regulated cDNAs (32% of the significantly differentially regulated spots), whereas the OxO experiment showed only 51 significantly down‐regulated genes (8.8% of the selected differentially expressed genes) within the 36‐h time course. Many of the down‐regulated genes in the AC parental line (40 cDNAs, accounting for 5.5% of total) showed homology to components of photosynthesis, including photosystem I, photosystem II and chlorophyll biosynthesis, but only eight photosynthetic‐related spots were present in the set of highly significant down‐regulated genes in the OxO transgenic line (Fig. 2 and Table S1). Most of the photosynthesis‐related cDNAs that showed a significant change in expression in at least one study were reduced in both genotypes, but to a greater extent in AC (Fig. S3, see Supporting Information). Of the 44 photosynthesis‐related cDNAs that showed differential expression in at least one study, 75% of the genes were more reduced in AC than OxO. Down‐regulation of photosynthesis‐related transcripts is not surprising, as this response has been documented as being common in plants undergoing biotic stress (Berger et al., 2007; Bilgin et al., 2010; Zou et al., 2005).
Looking at the expression differences over time, 17 cDNAs showed a statistically significant reduction in expression over time that was also significant in both genotypes. Several of these reduced genes were oxidases and primary metabolism‐related genes. Five belong to the photosynthesis process. The repression pattern of the 17 genes was notably different between AC and OxO; whereas AC showed mild down‐regulation in the first 12–24 hpi and a spike of down‐regulation between the 24‐ and 36‐hpi time points, OxO showed a steady and more pronounced repression of genes during the first 24 h of infection and a milder repression between 24 and 36 hpi (Fig. 5), consistent with a resistance response that effectively limits colonization.
Figure 5.
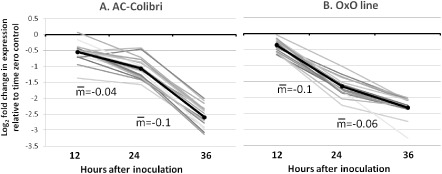
Seventeen transcripts reduced in abundance in both the parent line AC Colibri (AC) and the transgenic line (OxO) inoculated with Sclerotinia sclerotiorum. Each line represents a transcript.
Increased expression of transcripts in AC and OxO over time
Although AC showed more significantly reduced genes than OxO in response to S. sclerotiorum, the OxO line had a greater number of significantly induced genes. The microarray analysis identified 484 (of 724, 66.9%) up‐regulated cDNAs in AC and 527 (of 578, 91.2%) in OxO. Many categories (Fig. 2) were fairly consistent in the percentage of identified spots for both lines (e.g. signalling, primary metabolism, membrane transport genes and the phenylpropanoid pathway), indicating that the general plant defence response pathways were activated in both genotypes, but were stronger in OxO. In addition, 20 transcription factors were up‐regulated in the AC line and 19 in OxO, including WRKYs, MYB domains and zinc finger domain‐containing proteins. Seven of these transcription factors showed induction in both genotypes, suggesting an important role in the Sclerotinia–soybean interaction (Table 1).
Table 1.
Transcription factors showing statistically significant induction in response to Sclerotinia sclerotiorum inoculation: (A) in both genotypes tested (AC and OxO); (B) in AC only; and (C) in OxO only
| (A) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Microarray spot ID | GlymaID | AC | OxO | Transcription factor | ||||
| 12 hpi | 24 hpi | 36 hpi | 12 hpi | 24 hpi | 36 hpi | |||
| BE020308 | Glyma02g42140.2 | 2.16 | 2.63 | 3.35 | 2.12 | 2.25 | 2.84 | MYB |
| AW459137 | Glyma03g28050.1 | 0.96 | 1.83 | 2.64 | 1.05 | 2.18 | 2.46 | MYB |
| AW423555 | Glyma13g27460.1 | 2.09 | 2.52 | 3.29 | 2.06 | 2.58 | 3.05 | RAU1 |
| AW423555 | Glyma13g27460.1 | 1.69 | 2.04 | 2.50 | 2.06 | 2.58 | 3.05 | RAU1 |
| AW309165 | Glyma07g36180.1 | 0.16 | 0.70 | 3.01 | 0.07 | 1.58 | 3.03 | WRKY33 |
| AW185876 | Glyma10g43080.1 | 1.39 | 2.32 | 2.82 | 1.15 | 1.99 | 2.44 | Jumonji (jmjC) |
| AW133440 | Glyma02g39870.1 | 1.40 | 1.54 | 2.08 | 2.15 | 2.17 | 2.25 | WRKY33 |
| AW432526 | Glyma09g00820.1 | 1.45 | 1.81 | 2.20 | 1.99 | 2.20 | 2.51 | WRKY1 |
| AW432526 | Glyma09g00820.1 | 1.62 | 1.91 | 2.13 | 1.99 | 2.20 | 2.51 | WRKY |
| (B) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Microarray spot ID | GlymaID | 12 hpi | 24 hpi | 36 hpi | Transcription factor | |||
| BE021564 | Glyma13g27460.1 | 1.69 | 2.04 | 2.50 | RAU1 | |||
| BE020282 | Glyma09g00820.1 | 1.62 | 1.91 | 2.13 | WRKY | |||
| AW277496 | Glyma10g03690.1 | −0.81 | −1.48 | −2.89 | Basic helix–loop–helix (bHLH) | |||
| AW307328 | Glyma16g32330.1 | 1.29 | 2.09 | 3.00 | CBF‐like protein | |||
| BE022247 | Glyma05g26990.1 | −1.13 | −1.59 | −2.21 | DNA bi zinc finger | |||
| BE020866 | Glyma06g08390.2 | 1.64 | 2.12 | 2.54 | Basic leucine zipper 9 | |||
| AW279451 | Glyma04g02860.1 | 0.39 | 1.31 | 2.23 | Basic pentacysteine 6 | |||
| AW432784 | Glyma12g10790.1 | 2.42 | 2.78 | 3.10 | MYB | |||
| AI442281 | Glyma04g42800.1 | 1.31 | 1.78 | 2.42 | NAC/NAM | |||
| AI494927 | Glyma13g35550.1 | 2.28 | 2.85 | 3.38 | NAC/NAM | |||
| AW278647 | Glyma01g02250.1 | −0.36 | −1.05 | −2.43 | Phaseolin G‐box binding protein PG1 | |||
| AW156772 | Glyma11g27640.1 | 0.93 | 1.30 | 2.06 | PHD finger | |||
| AW507617 | Glyma13g25100.1 | −0.61 | −1.21 | −2.37 | Plus‐3 domain | |||
| AI496441 | Glyma17g03930.1 | 0.55 | 1.33 | 2.21 | Undefined transcription factor | |||
| AW308923 | Glyma06g23400.2 | −1.57 | −1.72 | −3.44 | Transcription initiation factor IIE | |||
| AI442350 | Glyma08g15430.1 | −1.25 | −1.75 | −2.08 | Transcription initiation factor IIS‐associated | |||
| AW132302 | Glyma16g04930.1 | −0.30 | −0.80 | −2.04 | Transducin/WD40 repeat‐like superfamily | |||
| AI748342 | Glyma20g26260.1 | 1.15 | 1.67 | 2.39 | Transducin/WD40 repeat‐like superfamily | |||
| AW432634 | Glyma13g17250.1 | 2.21 | 3.22 | 3.65 | Undefined transcription factor | |||
| AW101413 | Glyma08g23380.4 | 1.79 | 2.11 | 2.21 | WRKY and harpin induced | |||
| (C) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Microarray spot ID | GlymaID | 12 hpi | 24 hpi | 36 hpi | Transcription factor | |||
| AW831622 | Glyma08g26110.1 | 0.63 | 1.84 | 2.42 | bHLH transcription factor | |||
| BG510547 | Glyma18g52660.1 | 2.71 | 4.26 | 5.09 | F box/zinc finger | |||
| AW782286 | Glyma06g45940.1 | 1.44 | 3.18 | 3.67 | MYB | |||
| BG882487 | Glyma16g02570.1 | 1.89 | 3.53 | 3.90 | MYB | |||
| BG509767 | Glyma05g35050.1 | 1.93 | 2.54 | 3.18 | MYB | |||
| BF009475 | Glyma10g04350.1 | 1.56 | 2.50 | 2.68 | NAC/NNAM/CUC2‐like protein | |||
| AW781918 | Glyma07g30140.1 | 1.42 | 2.15 | 1.74 | TIFY2A | |||
| AW760574 | Glyma11g08780.1 | 0.33 | 1.44 | 2.08 | Transcription factor IIIC, subunit 5 | |||
| BF324626 | Glyma09g00820.1 | 1.14 | 1.55 | 2.10 | WRKY1 | |||
| AW306758 | Glyma02g39870.1 | 2.15 | 2.17 | 2.25 | WRKY33 | |||
| BI893195 | Glyma14g38010.1 | 2.06 | 2.01 | 2.18 | WRKY39 | |||
| BM177031 | Glyma11g29720.1 | 2.15 | 2.89 | 2.95 | WRKY | |||
AC, parent AC Colibri; Oxo, highly resistant oxalate oxidase transgenic line.
Many genes that are defined as being actively involved in defence were significantly expressed in the transgenic OxO line. Five resistance (R) genes were identified with high induction in OxO, but not in AC (Table S1). In addition, the OxO plants showed twice as many PR proteins with a significant change in expression relative to AC (Table S1). At our statistical cut‐off of significance, nine cDNAs that putatively encode PR proteins were found in AC: a PR2, six PR5s and two PR4s. In OxO, 16 spotted cDNAs with homology to PR protein coding genes were up‐regulated. In addition to PR5s and PR4s, the OxO line also showed a statistically significant induction of cDNAs corresponding to PR1, PR3 and PR10 homologues (Table S1). cDNAs corresponding to starvation‐associated message 22 (SAM22), which may be considered as a PR10 homologue, were annotated as ‘stress related’ and were not included in the accounting above. Of the 40 defence‐related genes that were differentially expressed in the studies, 75% were clearly more induced in OxO than in AC (Fig. S3 and Table S1).
There were 244 cDNA spots that showed up‐regulation in both genotypes that mirrored the response noted for the 17 down‐regulated sequences (Fig. 5). The expression of these transcripts suggested that a more pronounced defensive response occurred in the transgenic OxO line during the initial 24 hpi. A comparison of the 49 secondary metabolism‐related genes with significant induction in both AC and OxO showed a higher level of induction after infection in OxO relative to AC at 24 hpi, which then tapered off at 36 hpi for OxO (Fig. 6), again suggesting that the OxO line had the pathogen effectively contained by 36 hpi, whereas AC continued to mount a defence response, as reflected in the continued increased expression of these genes in AC (Fig. 6).
Figure 6.
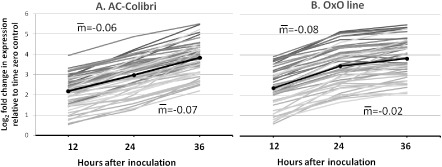
Forty‐nine defence‐related transcripts that increased in abundance in both the parent line AC Colibri (AC) and the transgenic line (OxO) inoculated with Sclerotinia sclerotiorum. Each line represents a transcript.
Two classes of genes that were strongly induced in both genotypes, but more strongly in OxO, in addition to the 40 categorized as defence related (Fig. S3), were matrix metalloproteinases (MMPs) and SAM22s. MMPs are named after their location within the matrix of mammalian cells, and have been implicated in plant defence in soybean (Liu et al., 2001; Zou et al., 2005) and tobacco (Schiermeyer et al., 2009). SAM22, one of the main allergens in soybean, has homology to both the major birch tree allergen ‘Bet v 1’ and PR10, which was shown to have ribonuclease activity (Bufe et al., 1996). As these two gene classes were amongst the strongest induced genes of this study, as well as in other soybean–pathogen studies involving Pseudomonas syringae, S. sclerotinia and Fusarium virguliforme (Calla et al., 2009; Radwan et al., 2013; Zou et al., 2005), it would be of value to better understand the actual role of these classes of proteins during defence.
Genes with different direction of expression between AC and OxO
The expression patterns (Fig. 3, S1 and S3) show that nearly all the genes that were differentially expressed at a statistically significant level in one genotype were also differentially expressed in the same direction in the other genotype, even across all time points. This observation suggests that, independent of shared statistical significance, many genes respond similarly to infection in both genotypes. The main difference in the expression of genes between AC and OxO was the robustness or strength of the differential expression. However, a handful of genes showed an opposite differential expression between AC and OxO (Fig. 3, bottom rows of Group 1, and Figs S1, S3 and S4, see Supporting Information). Of the 10 genes with the strongest opposite expression difference between AC and OxO, eight were related to photosynthesis or the redox state of the cells. These genes were all up‐regulated in AC at 12 and/or 24 hpi, but generally unchanged or down‐regulated in expression in OxO in response to S. sclerotiorum. This is perhaps most clearly observed in Fig. S4, where the cDNAs that showed this pattern the strongest (located at the bottom of the clusters in Fig. S4) were nearly all related to photosynthesis or the redox state of the cell. One of the genes with the strongest opposite differential expression was Glyma12g35050.3, a putative protochlorophyllide oxidoreductase. A down‐regulation of this gene would presumably lead to less of the protein gene product and, in this case, to a possible buildup of NADPH:protochlorophyllide, which, in the presence of light, leads to rapid singlet oxygen production, a trigger for defences and cell death. One could speculate that this difference in AC and OxO (up in AC, unchanged or down in OxO) could lead to a more rapid defence response in OxO, and may be a factor contributing to OxO resistance.
Other genes of interest
Peroxidases
Other cDNAs highly down‐regulated in OxO plants were seven peroxidases. Peroxidases oxidize various molecules in the presence of H2O2, and convert H2O2 to water. They have been implicated in lignin biosynthesis and in defence to pathogens, and they can remove harmful excess H2O2 in the cell environment (Caruso et al., 2001; Moy et al., 2004; Yoshida et al., 2003). They are also known to function in the oxidation of indole acetic acid (IAA), and have been demonstrated to show antifungal properties because of their ability to inhibit germ‐tube elongation in Botrytis cinerea (a close relative of S. sclerotiorum), Fusarium culmorum and Trichoderma viride (Caruso et al., 2001). Several studies have been undertaken to differentiate the roles of anionic versus cationic peroxidases. It has been postulated that anionic peroxidases are involved in lignin biosynthesis and that cationic peroxidases are involved in IAA catabolism (Castillo and Greppin, 1986), but these hypotheses have not been confirmed. A study on the kinetics and reactive forms produced during the oxidation of IAA by anionic and cationic peroxidases showed that both were able to oxidize IAA, but with some differences in intermediate forms (Gazaryan et al., 1999).
The statistically significant peroxidase response differed in the AC and OxO lines. AC showed 15 up‐regulated and no down‐regulated peroxidases, and OxO showed 17 up‐regulated and six down‐regulated peroxidases; however, three of these cDNAs appear to be from the same gene: Glyma17g01720.1. The down‐regulation of peroxidase transcripts in response to pathogens does not agree with several studies, which reported an increase in abundance of peroxidase gene expression on infection with pathogens (Lagrimini et al., 1987; van Loon et al., 2006). Peroxidase transcripts have also been reported to increase in abundance in plants following S. sclerotiorum stem inoculation (Calla et al., 2009; Yang et al., 2007); the protein levels have also been observed to increase (Liang et al., 2008). It is noteworthy, however, that the repressed peroxidases identified in the OxO experiment of this study were only of the ‘cationic peroxidase’ type. Interestingly, the up‐regulated and down‐regulated peroxidase gene expression patterns matched very closely those observed in avirulent P. syringae‐challenged soybean leaves (Zou et al., 2005). A comparison of the expression patterns of all peroxidases in response to S. sclerotiorum and P. syringae showed that the same genes tended to have the same direction of modulation to both pathogens (summarized by GlymaID in Fig. 7 and listed by individual cDNAs in Fig. S5, see Supporting Information), suggesting a hypersensitive response (HR)‐like reaction (or at least some aspects of the HR) as part of the initial response to S. sclerotiorum. The triggering of HR‐related programmed cell death (PCD) seems to be a beneficial strategy taken by some necrotrophs, as the prevention of PCD can lead to enhanced resistance to this class of pathogen (Lincoln et al., 2002).
Figure 7.
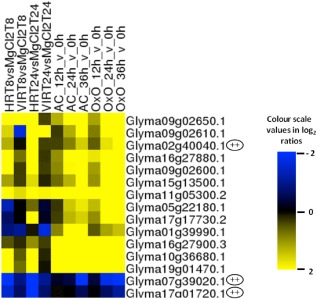
Hierarchical clustering of comparative peroxidase genes represented on the microarray set assayed. HR and VIR samples were from leaves inoculated with Pseudomonas syringae with or without avrB (Zou et al., 2005). AC and OxO are the parent and transgenic lines inoculated with Sclerotinia. Putative cationic peroxidases are indicated with dual pluses.
Lignins
Lignins are biopolymers derived from the oxidative polymerization of hydroxycinnamyl alcohols or monolignols, and are very important for structural support to the cell wall (Lacombe et al., 1997). In addition, a direct correlation between lignin synthesis and defence against pathogens has been reported (Dixon and Paiva, 1995; Hammerschmidt and Kuc, 1982; Vance et al., 1980; Zabala et al., 2006). The biosynthesis of lignin is initiated off the phenylpropanoid pathway, where enzymes such as phenylalanine ammonia lyase (PAL) and 4‐coumarate‐CoA ligase (4CL) are common to the multitude of branches, leading to the synthesis of diverse phenolic compounds. Cinnamoyl CoA esters are the last common precursor shared between these phenolic pathways, and they are substrates of both chalcone synthase (CHS) and cinnamoyl CoA reductase (CCR). CHS is the enzyme leading to the synthesis of chalcone and subsequent anthocyanins, flavonoid and isoflavonoid derivatives, whereas CCR represents the first committed step in the lignin pathway. Subsequent steps in the lignin pathway are carried out by caffeic acid O‐methyltransferase (COMT) and cinnamyl alcohol dehydrogenase (CAD) (Lagrimini et al., 1987).
The strongest down‐regulated genes in both the AC and OxO lines showed homology to both CCR and dihydroflavonol‐4‐reductase (DFR), two enzymes of different downstream branches of the phenylpropanoid pathway: CCR initiating lignin formation and DFR involved in the synthesis of anthocyanins. The ambiguous CCR/DFR annotation in the public databases is a result of the homology between the peptide sequences of the enzymes, with the N‐terminal domain having the strongest conservation (Lacombe et al., 1997). For this reason, the annotation of DFR and CCR enzymes using sequence homology between 5′ expressed sequence tags (ESTs) from different species is not accurate. Table 2 lists 11 ESTs corresponding to seven Glyma models with ambiguous CCR/DFR annotations in the public databases. To clarify our results, the peptide sequences of the seven predicted genes were aligned with the well‐characterized CCR sequences of Eucalyptus gunii (Lacombe et al., 1997) using ClustalW (Fig. S6, see Supporting Information). All the sequences showed high homology to known CCRs along the entire peptide; however, the internal amino acid sequence, known to code for the most conserved region and putative catalytic site of CCRs (NWYCY) (Barakat et al., 2011), was only fully present in one of the sequences: Glyma13g44700.1. Gene Glyma12g12230.1 contains no similarity to this putative active site, and the other genes showed only partial conservation of these residues. Together, the evidence indicates that most of the sequences ambiguously annotated as CCR/DFR and differentially expressed in this study are more likely to be DFRs, and only one, Glyma13g44700.1, is likely to be a CCR. Therefore, for this study, judging by the behaviour of Glyma13g44700.1, a CCR gene that showed strong induction in AC inoculated with S. sclerotiorum at the three time points tested relative to time zero (Table 2), we can conclude that the lignin pathway is induced in leaves on infection with S. sclerotiorum. Supporting this conclusion, two CAD enzymes were also found to be strongly induced in AC plants. Because OA (degraded in OxO) is higher in AC, it is likely that OA is at least partly responsible for the induction of the lignin pathway.
Table 2.
Differential expressiona of cDNAs coding for ambiguously annotated homologues to cinnamoyl CoA reductase (CCR) or dihydroflavonol‐4‐reductase (DFR) in four soybean genotypes inoculated with S clerotinia sclerotiorum
| GlymaID | EST ID | Will82c | PI194639 | AC Colibrib | OxO transgenicb | ||||
|---|---|---|---|---|---|---|---|---|---|
| T14/T8 | T14/T8 | T12 | T24 | T36 | T12 | T24 | T36 | ||
| Glyma11g29460.3 | AI496533 | −1.35 | −1.04 | 0.13 | −0.67 | −2.48 | — | — | — |
| AW132589 | −1.15 | −0.86 | — | — | — | — | — | — | |
| AW156703 | −1.55 | −1.34 | — | — | — | −0.35 | −1.31 | −2.62 | |
| Glyma12g12230.1 | BI320600 | −0.90 | −0.92 | — | — | — | — | — | — |
| Glyma18g06510.1 | BF071487 | −1.46 | −1.16 | — | — | — | −0.10 | −1.70 | −3.34 |
| AW101559 | −1.27 | −1.01 | 0.34 | −0.96 | −3.25 | — | — | — | |
| AI901207 | — | — | −0.16 | −0.99 | −2.97 | 0.24 | −1.79 | −3.26 | |
| Glyma19g00980.1 | BF009613 | −0.44 | −0.39 | — | — | — | — | — | — |
| Glyma13g44700.1 | AI522858 | — | — | 1.22 | 2.10 | 2.08 | — | — | — |
| Glyma12g02250.1 | AI441276 | — | — | 0.89 | 1.70 | 2.46 | — | — | — |
| Glyma09g33820.2 | AI437595 | — | — | 0.80 | 1.33 | 2.19 | 0.82 | 2.05 | 2.10 |
Differential expression calculated as log2 fold change.
Expression values in AC Colibri and OxO are relative to time zero.
Williams82.
AC, parent AC Colibri; Oxo, highly resistant oxalate oxidase transgenic line.
Will82 and PI194639 data from infected stems (Calla et al., 2009).
AC Colibri and OxO data from infected leaves.
In the case of inoculated stems (Calla et al., 2009), an additional enzyme in the lignin pathway, COMT, was found to be down‐regulated after infection, suggesting that, perhaps, the down‐regulation of this pathway is a tissue‐specific event occurring in stems, where the plant may be diverting substrates from the lignin pathway to the flavonoid and isoflavonoid pathway to increase the production of defensive substrates when challenged with the fungus. That node and internode levels of lignin were negatively correlated with resistance (Peltier et al., 2009) may support this hypothesis; however, the study of Peltier et al. (2009) did not address the induction of genes after inoculation, but, instead, the levels of lignins in resistant and susceptible soybean cultivars without infection.
G proteins
Our analysis of gene expression in response to S. sclerotiorum detected differential expression of a diversity of signalling‐related genes. As expected for a necrotrophic pathogen, cDNAs related to jasmonic acid and ethylene signalling were induced, as were several auxin‐related cDNAs (Fig. S1). Interestingly, the data here indicate some involvement of a G‐protein signalling pathway in response to S. sclerotiorum, as several cDNAs showed differential expression in the studies (Fig. S3). The component of G‐protein signal transduction that showed the strongest induction in response to infection over time was a G‐protein‐coupled receptor (GPCR) gene, Glyma10g05370.1. The gene product for GPCR may sense a signal from the pathogen to enhance the defence response. We are currently investigating the role of G‐protein‐related signalling in plant defence.
Experimental Procedures
Plant growth and inoculation with S . sclerotiorum
Soybean plants of the OxO resistant transgenic line (Donaldson et al., 2001) and plants of the susceptible parent (AC) were grown in environmentally controlled cabinets at 26/24 °C with a 13‐h photoperiod, watered every day and fertilized with a 20‐20‐20 (N‐P‐K) nutrient solution twice per week. At the V4 stage, when the trifoliate leaflets were fully expanded (21–28 days), they were transferred to a mist chamber (22/18 °C), 7 h prior to inoculation.
Inoculum was prepared using soybean flowers. Newly opened soybean flowers of X5 plants (AAFC breeding line X2650‐7‐2‐3) were inoculated between the standard and wing petals with 10 μL of ascospores suspended in 0.006% Triton X‐100 and incubated at room temperature in humid Petri dishes for 3 days. Twelve random plants (six pots) were inoculated at a time. To inoculate the OxO and AC plants, the infected X5 flowers were placed behind the first lateral vein on the central leaflet of the V4 trifoliate of each plant. Tissue samples were collected at three disease stages: stage 1, infection cushions appeared following contact with the plant cell wall, 10–12 hpi (referred to as the 12‐hpi sample); stage 2, hyphae penetrated the lateral vascular system (early vascular entry), 18–24 hpi (24‐hpi sample); stage 3, infection present in the main vein of the leaflet (36‐hpi sample). To ensure the correct stage of the disease at stage 1, the inoculated leaflets were quickly observed under a dissecting microscope just prior to sampling. The two lateral leaflets were removed and the inoculated leaflet was cut from the plant together with the petiole. The leaflet was cut transversely 3 cm beyond its base and immediately frozen in liquid nitrogen. For each time point replicate, a total of 10 plants was sampled and pooled. Plant growth, inoculation and sampling were repeated three times to obtain three independent biological replications that were also independent experimental replications.
RNA extraction, purification and quantification
Total RNA was isolated using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA). Tubes containing Phase Lock Gel™ (Brinkmann Instruments, Inc., Westbury, NY, USA) were used to improve the organic/inorganic phase separation. After extraction, RNA concentrations were estimated using a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For microarray experiments, RNA was further purified through Qiagen RNeasy columns (Qiagen, Valencia, CA, USA) and final quality was determined by a combination of spectrophotometry (NanoDrop ND‐1000) and gel electrophoresis (BioAnalyzer 2100, Agilent Technologies, Santa Clara, CA, USA).
Microarrays
Microarray hybridization procedures followed closely that described by Zou et al. (2005). Three independent biological/experimental replications were analysed. RNA was reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Invitrogen) in the presence of aminoallyl‐dUTP. Purified cDNA was quantified and coupled to either Amersham™ Cyanine 3 or Cyanine 5 mono‐reactive fluorescent dyes (GE Healthcare UK Ltd., Amersham, Buckinghamshire, UK), mixed in pairs according to a loop design and applied onto corresponding microarray slides. Loop designs for microarrays are basically incomplete block designs; after normalizing fluorescence intensities across all samples of the loop and calculating variances, they allow for the comparison between all samples independent of whether or not they were hybridized on the same slide (Kerr and Churchill, 2001) (Fig. 1). Statistical analysis of the expression data was performed in a manner similar to that described previously (Calla et al., 2009). For each of the genotypes, an analysis of variance (ANOVA) was run with array as a random effect and time as a fixed effect. The interaction between time and array was tested and proven to be non‐significant; for this reason, it was further removed from the model. The experiment used soybean cDNA‐based microarrays 18k‐A and 18k‐B, which represent approximately 36 700 predicted genes (Vodkin et al., 2004, 2007). Hierarchical and K‐means clustering were performed with Cluster v3.0 (Eisen et al., 1998), and were visualized as a heat map using TreeView v1.60 (http://rana.lbl.gov/EisenSoftware.htm).
qRT‐PCR
RNA from infected leaves was extracted with the RNeasy plant mini kit (Qiagen), DNase treated with Turbo DNA‐free (Invitrogen or Ambion, Austin, TX, USA) according to the manufacturer's protocol, and quantified with a NanoDrop ND‐1000 spectrophotometer. cDNA was synthesized using an iScript cDNA synthesis kit (Bio‐Rad, Hercules, CA, USA) containing 0.5 μg RNA, 8 μL of 5 × iScript reaction mix and 2 μL of iScript reverse transcriptase in a volume of 40 μL. The reaction was incubated at 25°C for 5 min, 42°C for 35 min and 85°C for 5 min in an Eppendorf Mastercycler. PCR was performed with 1 μL of cDNA template, 7.5 μL of SsoFast EvaGreen Supermix (Bio‐Rad), 0.6 μL of 10 μm forward and reverse primer mix in a volume of 15 μL. The reaction was conducted in a Bio‐Rad CFX96 C1000 system. The constitutive control cons15 (Libault et al., 2008) was used to normalize expression. Primers (Table 3) were designed with either Primer Express software v. 3.0 (2004, Applied Biosystems) or Primer3 software v. 0.4.0 (2006, Whitehead Institute for Biomedical Research, Cambridge, MA, USA).
Table 3.
Primers used for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR)
| Microarray spot ID | GlymaID | Short annotation | Forward primer | Reverse primer |
|---|---|---|---|---|
| B020384 | Glyma01g42660 | PR5 | aggcatcagggcacctctcctt | ttcctggcggctgcaacaac |
| AI494656 | Glyma01g42670 | PR5 | taagggcacctgggggatgcaa | cgaccttgtagttagagccagcgg |
| AI855519 | Glyma01g36620 | Sulphurtransferase | ttgatgtctctgccacttgc | aacttgggtggaacagcaac |
| BI424301 | Glyma02g04820 | Class I chitinase | ggctctttcaatggctttgg | acccacctgtggtttcatgag |
| BQ297212 | Glyma02g07180 | Chlorophyll‐binding protein | ctggtggtgcttttgatcct | cacaaaagccaacaaagcaa |
| AW458674 | Glyma03g21610 | MAPK kinase | agagcttggggatggttctt | ttgaccgcgacaatctcata |
| BE020966 | Glyma05g29400 | Glutathione S‐transferase | tcgatacagataattgacccattga | cttgatcacaggatggctaagaaa |
| BI786214 | Glyma05g29400 | Glutathione S‐transferase | tcgatacagataattgacccattga | cttgatcacaggatggctaagaaa |
| AI442353 | Glyma06g07170 | Serine–threonine protein kinase | gccttgctttgttcttccac | tcaggcaacctttgctttct |
| BG550911 | Glyma06g45450 | Diacylglycerol kinase | catggaactcgtcttgctca | aaggttccccatcaatcctc |
| AI441522 | Glyma07g39020 | Peroxidase | gccctggagttgtttcatgt | accacatcagctctgctcct |
| AW132407 | Glyma08g06820 | NAD‐malate dehydrogenase | tttccaggaagggagtgatg | gcaaagaaagggattgacca |
| AI939107 | Glyma08g07540 | ABC transporter | aaccaggatgctgatgaagg | tcttcttcttcccacatgcac |
| AI496388 | Glyma09g02600 | Peroxidase | gcgtggtttcttgtgctgat | caagggttcgatttgctgtt |
| AW133469 | Glyma09g28200 | Light‐harvesting complex 5 | gatgggggtacgctgaacta | gcacctcccacaagaacaat |
| AI441060 | Glyma09g30300 | MAPK kinase | cgccggagtttcacgatt | acctctccccggactcctt |
| AI443042 | Glyma10g32820 | Protease inhibitor | ggtaagagctcatggcctgagt | aaaccctatcacatcggaaatca |
| AI441937 | Glyma11g01350 | Chalcone synthase | cacatgaccgagctcaaaga | catgtcttgcctagcatcca |
| BI972951 | Glyma11g10080 | Glycosyl hydrolase | ccagcacttgaaaacattcagaa | tgtctattgctgttgacaccttcat |
| AI496533 | Glyma11g29460 | Cinnamoyl CoA reductase | gactttgcgaaggagacagg | taggtctctttgcccccttt |
| AW570395 | Glyma11g29460 | Cinnamoyl CoA reductase | gactttgcgaaggagacagg | taggtctctttgcccccttt |
| BG550897 | Glyma13g16950 | OPDA reductase | caggtagaggcatggagaaatgta | aacatgccagagttgacagaagata |
| AI443120 | Glyma13g26200 | Cytochrome p450 | ggagaggttgttgagggtga | agtctgttcctgccgagaaa |
| AW507727 | Glyma13g41110 | Envelope protein | gctggcgtttatgttggaat | ttgttgctggctgcagatac |
| AI736217 | Glyma15g05790 | Light‐harvesting complex 6 | ctgctgcaccaaagaagtca | ccaaggcctagtggatcaaa |
| AW397279 | Glyma15g43110 | Photosystem II protein | tcctctggtccattctttgg | ttggcagcattcactacgag |
| AW761087 | Glyma16g34570 | Aldo/keto reductase | aaggcaaaagggaagaccat | ggttctgcttcatcctctcg |
| BI788114 | Glyma17g01720 | Cationic peroxidase | gccagagatggcattgtttc | cattgtggtctgggaggaac |
| AW761241 | Glyma17g03350 | Bet v I family protein | catcccaaaggctgtcgaa | gccaccgttcccctcaa |
| AW101559 | Glyma18g06510 | Dihydroflavonol reductase | ttatgccaagccctaactgg | caaccccttctgtttgcagt |
| AI901207 | Glyma18g06510 | Dihydroflavonol reductase | ttatgccaagccctaactgg | caaccccttctgtttgcagt |
| BQ080089 | Glyma18g16780 | TIR‐NBS‐LRR‐type protein | agggagcaatccaatcgtaaga | ggccacaatttgttacggttaca |
| AW396185* | Glyma10g38456 | Cons15 | taaagagcaccatgcctatcc | tggttatgtgagcagatgcaa |
*Constitutive control cons15 used for data normalization (Libault et al., 2008).
LRR, leucine‐rich repeat; MAPK, mitogen‐activated protein kinase; NBS, nucleotide‐binding site; OPDA, 12‐oxophytodienoate; PR5, pathogenesis‐related 5; TIR, Toll/interleukin‐1 receptor.
qRT‐PCR for PR5
PR5 expression was analysed separately from the other genes studied by qRT‐PCR. Total RNA was purified from S. sclerotiorum‐inoculated leaves at 0, 12 and 24 hpi with the RNeasy Mini Kit (Qiagen), as described above with the addition of DNAse I treatment. First‐strand cDNA was synthesized from 2 μg of total RNA by reverse transcription. One microlitre of 10 mm deoxynucleoside triphosphates (dNTPs) and 1 μL of oligo dT (25‐mer) were added to each RNA sample. RNA was then denatured at 65 °C for 5 min, chilled on ice for 10 min and quickly centrifuged. Ten microlitres of a master mix were added to each reaction containing 4 μL of 5 × First Strand Buffer, 1 μL of RNAse OUT, 2 μL of 10 mm MgCl2, 2 μL of 0.1 mm dithiothreitol (DTT) and 1 μL (200 units) of SuperScript III (all from Invitrogen) per sample. The reaction was incubated at 50 °C for 1 h, followed by 15 min at 70 °C to inactivate the enzyme. Following cDNA synthesis, RNA was degraded by the addition of 1 μL (2 units) of RNAse H (Invitrogen) and incubation at 37 °C for 20 min. Samples were adjusted to 30 ng/μL with nuclease‐free water and PCR was run with 2 μL of cDNA template, 7.5 μL of water, 0.8 μL of each primer at 4 μm and 8.9 μL of SYBR Green master mix (Agilent Technologies) with ROX as the reference dye. The reaction (20 μL) was used to quantify cDNA by amplification employing the MX3000P QPCR system (Stratagene, Kirkland, WA, USA). The soybean constitutive gene cons15 (Libault et al., 2008) was used to normalize expression. The experiment was performed with three biological replications and three technical replications employing different samples from those collected for the microarray experiments. Data analysis was performed using the ΔΔCt method (Pfaffl, 2001).
Conclusions
Soybean plants with and without OxO activity (i.e. OxO transgenic, the resistant line, and its parent AC Colibri, the susceptible line, respectively) were challenged with S. sclerotiorum. Gene expression profiling allowed for the identification of genes that are modulated during infection and those that may be a direct consequence of OA secreted by the pathogen into the plant. In general, OxO and AC plants seemed to respond to fungal attack with many of the same set of genes that are primarily known protagonists of the basal defence response: the induction of cell wall‐related transcripts, ethylene and jasmonate pathways. The phenylpropanoid pathway, an identified source of antimicrobial phytoalexins, UV protectants and antioxidants, was also induced. The induction of cytochrome p450s and PR proteins (including PR4, PR5 and PR10) was detected as a common response in both genotypes. Peroxidases play an important role during the plant response to S. sclerotiorum, probably because of the large oxidative state modification undergone by the plant.
Although most genes in both the transgenic and parent plants showed the same direction of differential expression, a remarkable difference between the degrees of up‐ and down‐regulation was evident: the response from AC was clearly milder than that from OxO during the first 24 h, followed by an accelerated change in expression during the period between 24 and 36 hpi. The OxO line, in turn, showed a stronger initial change from 12 to 24 hpi, followed by a dampening of expression of defence‐related genes between 24 and 36 hpi. These differences in defence responses between plants able and unable to degrade OA indicate that OA might quell the defence response. However, it is also probable that the OxO plants show a strengthened defence response by the production of H2O2, a well‐characterized inducer of plant defence responses, from the degradation of OA. If OA is indeed capable of attenuating the defence response independently of its pH and other factors present during the first hours of infection, it would support the hypothesis of Williams et al. (2011) that OA is able to suppress reactive oxygen species production to initially manipulate the cellular redox environment.
Disclaimer
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Supporting information
Fig. S1 Heat map clusters of differentially expressed genes selected by ratio OxO (transgenic expressing oxalate oxidase enzyme) at T12 [12 h post‐inoculation (hpi)] over AC (parent AC Colibri) at T12 greater than two‐fold (A) or less than 1.5‐fold (B), and by ratio OxO at T36 (36 hpi) over AC at T36 greater than two‐fold (C) or less than 1.5‐fold (D). After selection of gene lists, the genes were sorted alphabetically by functional category. Yellow indicates log2 ratios above zero, and blue below zero; colours were more intense with increased distance from zero.
Fig. S2 Comparison of quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) results and microarray in two cDNAs coding for pathogenesis‐related 5 (PR5) protein in two lines, the parent AC Colibri and OxO (transgenic expressing oxalate oxidase enzyme), at two time points: 12 and 24 h post‐inoculation.
Fig. S3 Some subclusters of interest. (A) Defence‐related genes. (B) Photosynthesis‐related genes. (C) Matrix metalloproteinases (MMPs). (D) Lignin/lignin‐related genes. (E) G‐protein signalling‐related genes. (F) Starvation‐associated message 22s (SAM22s). Log2 expression comparisons were at 12, 24 and 36 h post‐inoculation (hpi) compared with time zero (T0) for either the parent AC Colibri (AC) or the oxalate oxidase‐expressing transgenic (OxO). The last two columns show the expression comparisons of OxO/AC at either 12 or 36 hpi. Yellow represents increased expression, blue reduced expression.
Fig. S4 Top 25 cDNAs whose expression was higher in the parent AC Colibri (AC) than in the oxalate oxidase‐expressing transgenic (OxO) at T12 [12 h post‐inoculation (hpi)] (A) and/or T36 (36 hpi) (B). Yellow indicates an increase in expression after inoculation, and blue a decrease in expression.
Fig. S5 Hierarchical clustering of all cDNA spots assayed in the study that are predicted to be peroxidases, compared with their expression in the soybean response to Pseudomonas syringae with (HR) or without (VIR) avrB (Zou et al., 2005).
Fig. S6 ClustalW alignment of sequences annotated as cinnamoyl CoA reductase (CCR) or CCR‐like with known CCRs from Eucalyptus gunii. The alignment shows that only one of the soybean sequences contains the internal peptide that is presumed to be the catalytic site of CCRs (boxed).
Table S1 Statistics, ratios and descriptions of all genes discussed in the study.
Acknowledgements
The authors would like to thank the United States Department of Agriculture (USDA) National Sclerotinia Initiative and the Ontario Soybean Board for generous financial support, as well as Sheryl Hubbard and Suqin Zheng for technical assistance.
References
- Barakat, A. , Yassin, N.B.M. , Park, J.S. , Choi, A. , Herr, J. and Carlson, J.E. (2011) Comparative and phylogenomic analyses of cinnamoyl‐CoA reductase and cinnamoyl‐CoA‐reductase‐like gene family in land plants. Plant Sci. 181, 249–257. [DOI] [PubMed] [Google Scholar]
- Berger, S. , Benediktyová, Z. , Matouš, K. , Bonfig, K. , Mueller, M.J. , Nedbal, L. and Roitsch, T. (2007) Visualization of dynamics of plant–pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. syringae and of oxylipins on A. thaliana . J. Exp. Bot. 58, 797–806. [DOI] [PubMed] [Google Scholar]
- Bilgin, D.D. , Zavala, J.A. , Zhu, J.I.N. , Clough, S.J. , Ort, D.R. and DeLucia, E.H. (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 33, 1597–1613. [DOI] [PubMed] [Google Scholar]
- Bufe, A. , Spangfort, M.D. , Kahlert, H. , Schlaak, M. and Becker, W.‐M. (1996) The major birch pollen allergen, Bet v 1, shows ribonuclease activity. Planta, 199, 413–415. [DOI] [PubMed] [Google Scholar]
- Calla, B. , Vuong, T. , Radwan, O. , Hartman, G.L. and Clough, S.J. (2009) Gene expression profiling soybean stem tissue early response to Sclerotinia sclerotiorum and in silico mapping in relation to resistance markers. Plant Genome, 2, 149–166. [Google Scholar]
- Calla, B. , Blahut‐Beatty, L. , Koziol, L. , Simmonds, D. and Clough, S.J. (2014) Transcriptome analyses suggest a disturbance of iron homeostasis in soybean leaves during white mould disease establishment. Mol. Plant Pathol. 15, 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, C. , Chilosi, G. , Leonardi, L. , Bertini, L. , Magro, P. , Buonocore, V. and Caporale, C. (2001) A basic peroxidase from wheat kernel with antifungal activity. Phytochemistry, 58, 743–750. [DOI] [PubMed] [Google Scholar]
- Castillo, F.J. and Greppin, H. (1986) Balance between anionic and cationic extracellular peroxidase activities in Sedum album leaves after ozone exposure. Analysis by high‐performance liquid chromatography. Physiol. Plant. 68, 201–208. [Google Scholar]
- Cessna, S.G. , Sears, V.E. , Dickman, M.B. and Low, P.S. (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell, 12, 2191–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cober, E.R. , Rioux, S. , Rajcan, I. , Donaldson, P.A. and Simmonds, D.H. (2003) Partial resistance to white mold in a transgenic soybean line. Crop Sci. 43, 92–95. [Google Scholar]
- Dixon, R.A. and Paiva, N.L. (1995) Stress‐induced phenylpropanoid metabolism. Plant Cell Online, 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, P.A. , Anderson, T. , Lane, B.G. , Davidson, A.L. and Simmonds, D.H. (2001) Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf‐2.8 (germin) gene are resistant to the oxalate‐secreting pathogen Sclerotina sclerotiorum . Physiol. Mol. Plant Pathol. 59, 297–307. [Google Scholar]
- Dong, X. , Ji, R. , Guo, X. , Foster, S. , Chen, H. , Dong, C. , Liu, Y. , Hu, Q. and Liu, S. (2008) Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta, 228, 331–340. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B. , Spellman, P.T. , Brown, P.O. and Botstein, D. (1998) Cluster analysis and display of genome‐wide expression patterns. Proc. Natl. Acad. Sci. USA, 95, 14 863–14 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaron, F. , Sella, L. and D'Ovidio, R. (2004) Relationships among endo‐polygalacturonase, oxalate, pH, and plant polygalacturonase‐inhibiting protein (PGIP), in the interaction between Sclerotinia sclerotiorum and soybean. Mol. Plant–Microbe Interact. 17, 1402–1409. [DOI] [PubMed] [Google Scholar]
- Gazaryan, I.G. , Chubar, T.A. , Mareeva, E.A. , Lagrimini, L.M. , Van Huystee, R.B. and Thorneley, R.N.F. (1999) Aerobic oxidation of indole‐3‐acetic acid catalysed by anionic and cationic peanut peroxidase. Phytochemistry, 51, 175–186. [Google Scholar]
- Godoy, G. , Steadman, J.R. , Dickman, M.B. and Dam, R. (1990) Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris . Physiol. Mol. Plant Pathol. 37, 179–191. [Google Scholar]
- Guimaraes, R.L. and Stotz, H.U. (2004) Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 136, 3703–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt, R. and Kuc, J. (1982) Lignification as a mechanism for induced systemic resistance in cucumber. Physiol. Plant Pathol. 20, 61–71. [Google Scholar]
- Hu, X. , Bidney, D.L. , Yalpani, N. , Duvick, J.P. , Crasta, O. , Folkerts, O. and Lu, G. (2003) Overexpression of a gene encoding hydrogen peroxide‐generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol. 133, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, M.K. and Churchill, G.A. (2001) Experimental design for gene expression microarrays. Biostatistics, 2, 183–201. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. , Min, J.‐Y. and Dickman, M.B. (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant–Microbe Interact. 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Lacombe, E. , Hawkins, S. , Van Doorsselaere, J. , Piquemal, J. , Goffner, D. , Poeydomenge, O. , Boudet, A.‐M. and Grima‐Pettenati, J. (1997) Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 11, 429–441. [DOI] [PubMed] [Google Scholar]
- Lagrimini, L.M. , Burkhart, W. , Moyer, M. and Rothstein, S. (1987) Molecular cloning of complementary DNA encoding the lignin‐forming peroxidase from tobacco: molecular analysis and tissue‐specific expression. Proc. Natl. Acad. Sci. USA, 84, 7542–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, B.G. (2002) Oxalate, germins, and higher‐plant pathogens. IUBMB Life V53, 67–75. [DOI] [PubMed] [Google Scholar]
- Lane, B.G. (1994) Oxalate, germin, and the extracellular matrix of higher plants. FASEB J. 8, 294–301. [DOI] [PubMed] [Google Scholar]
- Liang, Y. , Srivastava, S. , Rahman, M.H. , Strelkov, S.E. and Kav, N.N.V. (2008) Proteome changes in leaves of Brassica napus L. as a result of Sclerotinia sclerotiorum challenge. J. Agric. Food Chem. 56, 1963–1976. [DOI] [PubMed] [Google Scholar]
- Libault, M. , Thibivilliers, S. , Bilgin, D.D. , Radwan, O. , Benitez, M. , Clough, S.J. and Stacey, G. (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome, 1, 44–54. [Google Scholar]
- Lincoln, J.E. , Richael, C. , Overduin, B. , Smith, K. , Bostock, R. and Gilchrist, D.G. (2002) Expression of the antiapoptotic baculovirus p35 gene in tomato blocks programmed cell death and provides broad‐spectrum resistance to disease. Proc. Natl. Acad. Sci. USA, 99, 15 217–15 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Dammann, C. and Bhattacharyya, M.K. (2001) The matrix metalloproteinase gene GmMMP2 is activated in response to pathogenic infections in soybean. Plant Physiol. 127, 1788–1797. [PMC free article] [PubMed] [Google Scholar]
- Livingstone, D.M. , Hampton, J.L. , Phipps, P.M. and Grabau, E.A. (2005) Enhancing resistance to Sclerotinia minor in peanut by expressing a barley oxalate oxidase gene. Plant Physiol. 137, 1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Marciano, P. , Di Lenna, P. and Magro, P. (1983) Oxalic acid, cell wall‐degrading enzymes and pH in pathogenesis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiol. Plant Pathol. 22, 339–345. [Google Scholar]
- Moy, P. , Qutob, D. , Chapman, B.P. , Atkinson, I. and Gijzen, M. (2004) Patterns of gene expression upon infection of soybean plants by Phytophthora sojae . Mol. Plant–Microbe Interact. 17, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Peltier, A.J. , Hatfield, R.D. and Grau, C.R. (2009) Soybean stem lignin concentration relates to resistance to Sclerotinia sclerotiorum . Plant Dis. 93, 149–154. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan, O. , Li, M. , Calla, B. , Li, S. , Hartman, G.L. and Clough, S.J. (2013) Effect of Fusarium virguliforme phytotoxin on soybean gene expression suggests a role in multidimensional defence. Mol. Plant Pathol. 14, 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou, C. , Freyssinet, G. and Fevre, M. (1991) Production of cell wall‐degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum . Appl. Environ. Microbiol. 57, 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiermeyer, A. , Hartenstein, H. , Mandal, M.K. , Otte, B. , Wahner, V. and Schillberg, S. (2009) A membrane‐bound matrix‐metalloproteinase from Nicotiana tabacum cv. BY‐2 is induced by bacterial pathogens. BMC Plant Biol. 9, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , Hyten, D.L. , Song, Q. , Thelen, J.J. , Cheng, J. , Xu, D. , Hellsten, U. , May, G.D. , Yu, Y. , Sakurai, T. , Umezawa, T. , Bhattacharyya, M.K. , Sandhu, D. , Valliyodan, B. , Lindquist, E. , Peto, M. , Grant, D. , Shu, S. , Goodstein, D. , Barry, K. , Futrell‐Griggs, M. , Abernathy, B. , Du, J. , Tian, Z. , Zhu, L. , Gill, N. , Joshi, T. , Libault, M. , Sethuraman, A. , Zhang, X.C. , Shinozaki, K. , Nguyen, H.T. , Wing, R.A. , Cregan, P. , Specht, J. , Grimwood, J. , Rokhsar, D. , Stacey, G. , Shoemaker, R.C. and Jackson, S.A. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Sperry, J.S. and Tyree, M.T. (1988) Mechanism of water stress‐induced xylem embolism. Plant Physiol. 88, 581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, C. , Dunwell, J.M. , Johnstone, C.E. , Lay, V. , Ray, J. , Schmitt, M. , Watson, H. and Nisbet, G. (1995) Degradation of oxalic acid by transgenic oilseed rape plants expressing oxalate oxidase. Euphytica, 85, 169–172. [Google Scholar]
- Thompson, E.W. and Lane, B.G. (1980) Relation of protein synthesis in imbibing wheat embryos to the cell‐free translational capacities of bulk mRNA from dry and imbibing embryos. J. Biol. Chem. 255, 5965–5970. [PubMed] [Google Scholar]
- Vance, C.P. , Kirk, T.K. and Sherwood, R.T. (1980) Lignification as a mechanism of disease resistance. Annu. Rev. Phytopathol. 18, 259–288. [Google Scholar]
- Vodkin, L.O. , Khanna, A. , Shealy, R. , Clough, S.J. , Gonzalez, D.O. , Philip, R. , Zabala, G. , Thibaud‐Nissen, F. , Sidarous, M. , Stromvik, M.V. , Shoop, E. , Schmidt, C. , Retzel, E. , Erpelding, J. , Shoemaker, R.C. , Rodriguez‐Huete, A.M. , Polacco, J.C. , Coryell, V. , Keim, P. , Gong, G. , Liu, L. , Pardinas, J. and Schweitzer, P. (2004) Microarrays for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. BMC Genomics, 5, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin, L.O. , Thibaud‐Nissen, F. , Gonzalez, D.O. , Zabala, G. , Clough, S.J. and Shealy, R. (2007) An update on soybean functional genomics and microarray resources for gene discovery and crop improvement. PETRIA, 17, 43–53. [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.‐J. , Britt, R. and Dickman, M.B. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Srivastava, S. , Deyholos, M.K. and Kav, N.N.V. (2007) Transcriptional profiling of canola (Brassica napus L.) responses to the fungal pathogen Sclerotinia sclerotiorum . Plant Sci. 173, 156–171. [Google Scholar]
- Yoshida, K.Y. , Kaothien, P.K. , Matsui, T.M. , Kawaoka, A.K. and Shinmyo, A.S. (2003) Molecular biology and application of plant peroxidase genes. Appl. Microbiol. Biotechnol. 60, 665–670. [DOI] [PubMed] [Google Scholar]
- Zabala, G. , Zou, J. , Tuteja, J. , Gonzalez, D.O. , Clough, S.J. and Vodkin, L.O. (2006) Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol. 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Rodriguez‐Zas, S. , Aldea, M. , Li, M. , Zhu, J. , Gonzalez, D.O. , Vodkin, L.O. , DeLucia, E. and Clough, S.J. (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense‐related genes and rapid HR‐specific downregulation of photosynthesis. Mol. Plant–Microbe Interact. 18, 1161–1174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Heat map clusters of differentially expressed genes selected by ratio OxO (transgenic expressing oxalate oxidase enzyme) at T12 [12 h post‐inoculation (hpi)] over AC (parent AC Colibri) at T12 greater than two‐fold (A) or less than 1.5‐fold (B), and by ratio OxO at T36 (36 hpi) over AC at T36 greater than two‐fold (C) or less than 1.5‐fold (D). After selection of gene lists, the genes were sorted alphabetically by functional category. Yellow indicates log2 ratios above zero, and blue below zero; colours were more intense with increased distance from zero.
Fig. S2 Comparison of quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) results and microarray in two cDNAs coding for pathogenesis‐related 5 (PR5) protein in two lines, the parent AC Colibri and OxO (transgenic expressing oxalate oxidase enzyme), at two time points: 12 and 24 h post‐inoculation.
Fig. S3 Some subclusters of interest. (A) Defence‐related genes. (B) Photosynthesis‐related genes. (C) Matrix metalloproteinases (MMPs). (D) Lignin/lignin‐related genes. (E) G‐protein signalling‐related genes. (F) Starvation‐associated message 22s (SAM22s). Log2 expression comparisons were at 12, 24 and 36 h post‐inoculation (hpi) compared with time zero (T0) for either the parent AC Colibri (AC) or the oxalate oxidase‐expressing transgenic (OxO). The last two columns show the expression comparisons of OxO/AC at either 12 or 36 hpi. Yellow represents increased expression, blue reduced expression.
Fig. S4 Top 25 cDNAs whose expression was higher in the parent AC Colibri (AC) than in the oxalate oxidase‐expressing transgenic (OxO) at T12 [12 h post‐inoculation (hpi)] (A) and/or T36 (36 hpi) (B). Yellow indicates an increase in expression after inoculation, and blue a decrease in expression.
Fig. S5 Hierarchical clustering of all cDNA spots assayed in the study that are predicted to be peroxidases, compared with their expression in the soybean response to Pseudomonas syringae with (HR) or without (VIR) avrB (Zou et al., 2005).
Fig. S6 ClustalW alignment of sequences annotated as cinnamoyl CoA reductase (CCR) or CCR‐like with known CCRs from Eucalyptus gunii. The alignment shows that only one of the soybean sequences contains the internal peptide that is presumed to be the catalytic site of CCRs (boxed).
Table S1 Statistics, ratios and descriptions of all genes discussed in the study.


