Summary
Only few fungal effectors have been described to be delivered into the host cell during obligate biotrophic interactions. RTP1p, from the rust fungi Uromyces fabae and U. striatus, was the first fungal protein for which localization within the host cytoplasm could be demonstrated directly. We investigated the occurrence of RTP1 homologues in rust fungi and examined the structural and biochemical characteristics of the corresponding gene products. The analysis of 28 homologues showed that members of the RTP family are most likely to occur ubiquitously in rust fungi and to be specific to the order Pucciniales. Sequence analyses indicated that the structure of the RTPp effectors is bipartite, consisting of a variable N‐terminus and a conserved and structured C‐terminus. The characterization of Uf‐RTP1p mutants showed that four conserved cysteine residues sustain structural stability. Furthermore, the C‐terminal domain exhibits similarities to that of cysteine protease inhibitors, and it was shown that Uf‐RTP1p and Us‐RTP1p are able to inhibit proteolytic activity in Pichia pastoris culture supernatants. We conclude that the RTP1p homologues constitute a rust fungi‐specific family of modular effector proteins comprising an unstructured N‐terminal domain and a structured C‐terminal domain, which exhibit protease inhibitory activity possibly associated with effector function during biotrophic interactions.
Introduction
During invasion of their hosts, plant pathogens release numerous effector molecules which enable infection processes and allow the pathogen to overcome plant defences. The modulation of plant defence mechanisms is especially important for biotrophic and hemibiotrophic pathogens which establish a close interaction with their hosts and depend on a persisting continuation of this interaction. The manipulation of host metabolism and defence reactions by the secretion of effector proteins is a common strategy of many plant pathogens (Panstruga and Dodds, 2009). These effectors can act in the plant apoplast or may be translocated to the host cytoplasm following secretion. The transfer of effectors to the host cytoplasm is well known in pathogenic bacteria, which use an elaborate secretion apparatus for the delivery of a wide repertoire of proteins, called type III effectors (Hann et al., 2010). Similar to pathogenic bacteria, plant pathogenic oomycetes and fungi possess a repertoire of effectors which are transferred to the host cytoplasm. Oomycete effectors contain N‐terminal host translocation signals, such as the LXLFLAK, RXLR and CHXC motifs (Birch et al., 2006; Kemen et al., 2011; Schornack et al., 2010; Whisson et al., 2007). Functional variants have also been described for some fungal effectors (Kale et al., 2010; Rafiqi et al., 2010). Although many effectors of oomycete and fungal plant pathogens have been discovered, little is known about their functions. Most of these effectors were identified because they elicit plant defence reactions by interacting with plant resistance proteins, and were therefore termed avirulence (Avr) proteins (Catanzariti et al., 2006; Orbach et al., 2000). However, it can be assumed that these proteins also have positive effects for pathogen virulence in the absence of the corresponding R protein, as shown for the Avr3a protein from Phytophthora infestans, which is able to inhibit infestin 1 (INF1)‐induced cell death by stabilizing a host E3 ubiquitin ligase required for programmed cell death (PCD) (Bos et al., 2010). The functions of several apoplastic effector proteins have been reported, e.g. for EPIC1 and EPIC2B from P. infestans and Avr2 from Cladosporium fulvum. These effectors inhibit the tomato cysteine protease Rcr3, which is involved in basal defence reactions, and thus contribute to successful infection of the host plant (van Esse et al., 2008; Tian et al., 2007). The inhibition of plant proteases may be a critical factor for most pathogens, as many apoplastic and cytosolic plant proteases are involved in pathogen recognition and defence reactions during the infection process (D'Silva et al., 1998; van der Hoorn, 2008; Solomon et al., 1999).
Rust transferred protein 1 (RTP1p) from the bean rust fungus Uromyces fabae and its homologue from U. striatus were the first fungal proteins for which haustorium‐specific expression and transfer to the host cytoplasm during the biotrophic interaction could be shown directly (Kemen et al., 2005). After the initial identification of RTP1p in U. fabae and U. striatus (Kemen et al., 2005), recently released expressed sequence tag (EST) and genome sequence data of a range of rust fungi revealed the existence of further RTP1 homologues, some of which could be shown to be expressed in biotrophic structures (Duplessis et al., 2011; Fernandez et al., 2012; Puthoff et al., 2008). These findings suggested that RTP1 homologues might be widespread in the order Pucciniales, and therefore might be crucial for the biotrophic lifestyle.
The aim of this study was to further investigate the role of RTP1p and its homologues during the biotrophic interaction of rust fungi and their host plants. We show that RTP1p is a member of a new rust‐specific family of secreted effectors. Members of this protein family are divided into a variable, unstructured N‐terminus and a conserved and highly structured C‐terminal domain, which shows similarity to the C‐terminal domain of cysteine protease inhibitors.
Results
Identification of RTP1 homologues from different genera of rust fungi
Based on the sequences of Uf‐RTP1 and Us‐RTP1 (Kemen et al., 2005) and additional RTP sequences from Puccinia graminis f.sp. tritici and U. appendiculatus, published by Puthoff et al. (2008), Uneven‐ and SiteFinding‐polymerase chain reaction (PCR) (Chen and Wu, 1997; Tan et al., 2005), as well as blast searches (Altschul et al., 1997), were performed to identify further homologues from different rust genera. We identified and sequenced RTP1 homologues from Gymnosporangium sabinae, Hemileia vastatrix and Puccinia coronata. blast searches of publicly available genome sequences of P. graminis f.sp. tritici, P. triticina, Melampsora larici‐populina and Phakopsora pachyrhizi revealed the existence of five further RTP1 homologues in P. graminis f.sp. tritici, three homologues in M. larici‐populina and one homologue in P. pachyrhizi. Two further sequences from U. appendiculatus and P. pachyrhizi (Ua‐RTP9 and Pp‐RTP2) were kindly provided by T. Link (Universität Hohenheim, Stuttgart, unpublished results). For further analysis, we additionally used RTP1 homologous sequences from Melampsora occidentalis, Melampsora medusae f.sp. tremuloides, Melampsora medusae f.sp. deltoides and Puccinia striiformis, identified by Feau et al. (2007), Joly et al. (2010) and Ma et al. (2009).
The lengths of the thus identified 28 complete genomic sequences ranged between 1005 bp (Mlp‐RTP2) and 1380 bp (Gs‐RTP1). All genes contained five highly conserved exons encoding the RTPp C‐termini (exons 2–6, Fig. 1). The region encoding the N‐termini of the proteins was highly variable and consisted of one to three exons (exons 1A–C, Fig. 1). The length of the amino acid sequences ranged between 199 and 265 residues. SignalP 3.0 analysis (Bendtsen et al., 2004) identified an N‐terminal leader peptide in all RTPps, indicating secretion of all homologues. The theoretical molecular weight varied in the range 22.0–28.9 kDa for the unprocessed polypeptides and 19.6–25.7 kDa for mature proteins. Alignment of the RTP amino acid sequences confirmed the previous finding that RTP1p homologues are divided into a divergent N‐terminal part, encoded by the variable exon 1A–C, and a highly conserved C‐terminal region starting at position 94 referring to the Uf‐RTP1p sequence, encoded by exons 2–6 (Figs 1 and S1, see Supporting Information). The smaller N‐terminal part comprised 71–113 residues, and the C‐terminal part contained 125–155 residues. These findings suggest a bipartite structure of RTP1p homologues, comprising variable N‐terminal and conserved C‐terminal domains.
Figure 1.
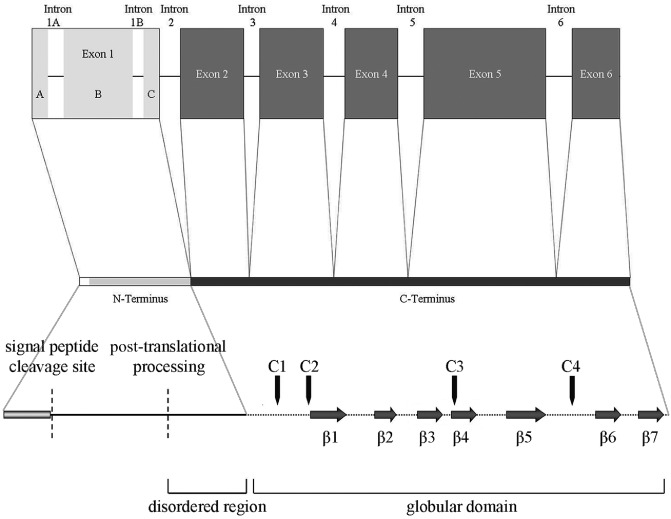
Schematic diagram of the RTP exon/intron structure and domain structure of the rust transferred proteins (RTPps). The variable exon 1 may be interrupted by additional introns (A, B) and codes for the structurally disordered N‐terminus. Exons 2–6 code for the C‐terminal globular domain. Black arrows indicate the positions of the four conserved cysteine residues (C1–C4). Grey arrows represent the seven β strands (β1–β7) of the globular domain.
Phylogenetic analysis of the RTPp family
In order to investigate the evolutionary relationship among RTPp family members, we carried out OrthoMCL analyses using the parameters defined by Moustafa et al. (2009) and the genomes of 12 eukaryotes to verify RTPp candidates and to identify possible candidates outside the rust fungi. We were unable to identify any RTP homologues outside the Pucciniales, suggesting that RTPp homologues constitute a family of effectors specific to rust fungi. Phylogenetic analyses of RTPp N‐ and C‐termini suggest that the clades of the N‐terminal and C‐terminal domains largely show co‐evolution, whereas three genes showed a diverging distribution within these clades (Fig. 2). These findings were supported by TribeMCL analyses of RTPp N‐ and C‐terminal domains, which showed that both the RTPp N‐terminal domains and the RTPp C‐terminal domains could be clustered into six tribes, five of which were consistent, whereas the N‐terminal domains of tribe one showed a higher degree of divergence (Fig. S2, see Supporting Information).
Figure 2.
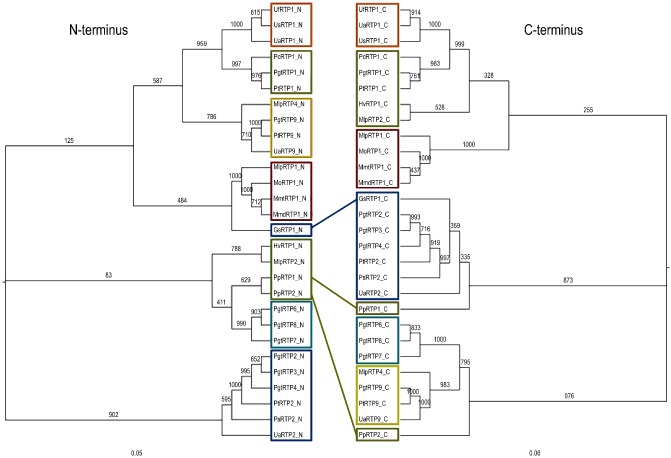
Phylogenetic tree of the N‐ versus C‐terminus of the rust transferred proteins (RTPps). Colours are selected according to TribeMCL analyses. Clades of RTPp N‐termini and C‐termini show co‐evolution, except for three genes that show divergence between N‐ and C‐termini (indicated by blue connection bars). Tree based on a neighbour‐joining (NJ) analysis. All bootstrap counts refer to 1000 replications.
Sequence analyses of the RTPp N‐ and C‐terminal domains
In silico structural analyses using DisEMBLTM, GLOBPLOT2 and PONDR‐FIT algorithms (Linding et al., 2003a, b; Xue et al., 2010) supported the previous finding that RTP1p homologues consist of two distinct domains corresponding to the variable and conserved regions described above (Fig. 1). Accordingly, the N‐terminus is structurally disordered, whereas the C‐terminus consists of a globular domain, which indicates that RTPp domains are structurally distinct. Secondary structure prediction based on different algorithms was performed for each sequence. As expected, no conserved secondary structural motif could be identified within the N‐terminal region. By contrast, all algorithms consistently indicated the presence of seven highly conserved β strands within the C‐terminal domain, which were interrupted by six equally conserved coil or loop regions (Fig. 1). The identified secondary structural motifs of the C‐terminal domain appear to be more strongly conserved than the amino acid sequences, indicating a crucial function for these β sheets. Searches for conserved motifs within the N‐terminal RTPp sequences revealed a dibasic cleavage site for post‐translational processing in the N‐terminus, which resembles NEC1/NEC2 or subtilisin/kexin processing sites (Fig. S1). N‐terminal sequencing of Uf‐RTP1p and Us‐RTP1p expressed in Pichia pastoris showed that the sequences of the mature peptides started with Glu55 and Asp64, respectively, indicating additional processing of the proteins after cleavage of the N‐terminal leader peptide. First analyses of Uf‐RTP1p isolated from infected Vicia faba leaves suggest that an identical N‐terminal processing also occurs in rust fungi. Corresponding to the findings of Puthoff et al. (2008) a nuclear localization signal (NLS), initially postulated for Uf‐RTP1p (Kemen et al., 2005), could not be detected in any of the homologous sequences, indicating that the primary target for RTPps might be different from the host cell nucleus.
The RTP1p C‐terminal domains show signs of purifying selection
Selection analyses of the variable N‐terminal sequences and the conserved C‐terminal sequences showed that a large number of residues in the C‐terminal domains are under purifying selection, whereas no significant signs of selection could be detected within the N‐terminal domains (Fig. 3). Twenty‐six residues in the C‐terminal part of RTPps (amino acid positions 94–220) show strong signatures of purifying selection. In particular, four conserved cysteine residues (C1–C4, matching Uf‐RTP1p residues Cys104, Cys117, Cys147 and Cys179) and two conserved glycine residues (Gly188 and Gly189) underlie purifying selection (Fig. 3).
Figure 3.
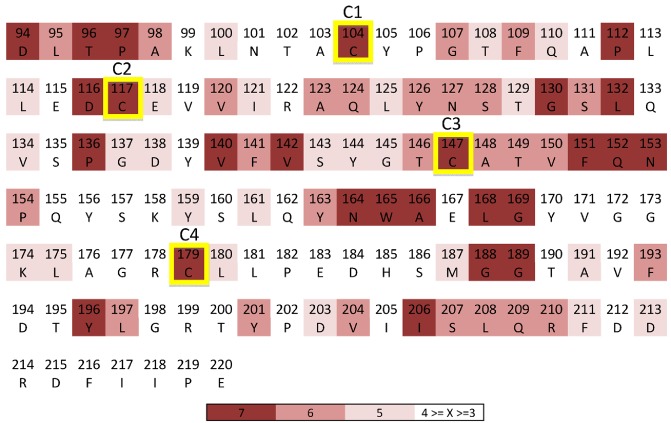
Rust transferred proteins (RTPps) show signs of purifying selection. Numerous C‐terminal residues of RTPs show signatures of purifying selection. In particular, conserved cysteine residues (C1–C4) are under purifying selection. X, value on Selecton selection scale, where ‘1’ indicates positive selection and ‘7’ indicates purifying selection.
Importance of disulphide bridges for the stabilization of the globular domain
The localization of the four highly conserved cysteine residues which show signs of purifying selection within the C‐terminal globular domain of RTPps indicated the importance of disulphide bridge formation for the stability of this domain. According to DiANNA1.1 and DISULFIND predictions (Ferre and Clote, 2005; Vullo and Frasconi, 2004), the formation of disulphide bridges between C1 and C2, as well as C3 and C4, is the most probable combination. Furthermore, additional cysteine residues occurring in some homologues were predicted to be linked by another disulphide bond. The substitution of the cysteine residues by serine residues resulted in reduced or completely absent secretion, as well as an alteration of the molecular weight and N‐glycosylation state of the mutated proteins (Fig. 4). Endoglycosidase Hf treatment of the mutated proteins showed that the variation of the molecular weight originated from altered N‐glycosylation. The failure of secretion and aberrant post‐translational modifications can be considered as an effect of protein misfolding and destabilization, as described, for example, for the GPHα protein (Darling et al., 2000). Therefore, we assume that the formation of disulphide bridges is essential for the structural integrity of the globular domain.
Figure 4.
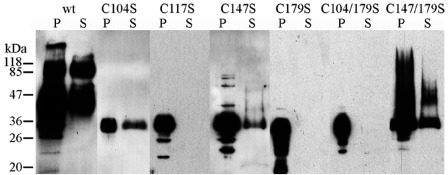
Western blot analysis of Uf‐RTP1p cysteine mutants. Mutated proteins were heterologously expressed in Pichia pastoris. P, pellet; S, culture supernatant; wt, wild‐type Uf‐RTP1p. Uf‐RTP1p cysteine residues C104, C117, C147 and C179, corresponding to the conserved cysteine residues C1, C2, C3 and C4, respectively, were replaced by serine residues. Differences in the molecular weight of the Uf‐RTP1p variants originate from altered N‐glycosylation of the mutated proteins.
Similarities of RTPps to cysteine protease inhibitors
As the secondary structure of the RTPps appears to be highly conserved, and accurate folding of the secondary structural motifs was found to be critical for protein integrity, we searched for structural homologues of RTPps. Two families of cysteine protease inhibitors, the cystatins (MEROPS family I25) and the chagasin‐like inhibitors (MEROPS family I42), showed similarities with respect to secondary structure, as members of both families are characterized by β sheet structures (Fig. 5) (Alvarez‐Fernandez et al., 2005; Smith et al., 2006). The cystatins contain a five‐stranded antiparallel β sheet and one α helix; type 2 cystatins, which are secreted cystatins, additionally contain two conserved disulphide bridges (Alvarez‐Fernandez et al., 2005). Members of the chagasin‐like inhibitor family are characterized by an immunoglobulin‐like β sandwich fold composed of seven essential β strands (Smith et al., 2006). Phylogenetic analyses based on secondary structure alignments show a relationship between members of the RTPp family and cystatin‐like proteins (Fig. S3, see Supporting Information). The RTP1p C‐terminal domains and cystatin‐ and chagasin‐like proteins show distinct similarities with regard to the position of the β strands as well as some amino acid positions (Fig. 5). Analysis of the RTPp sequences corresponding to the protease interacting loop sequences of the chagasin‐like proteins revealed similarities between the protease interacting motifs, especially with regard to two highly conserved glycine residues that correspond to the second chagasin loop motif (Fig. 5).
Figure 5.
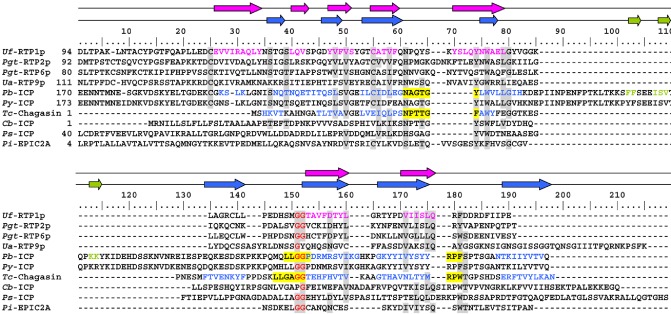
Alignment of selected rust transferred protein (RTPp) globular domain sequences and sequences of eukaryotic and bacterial cysteine protease inhibitors. Pb‐ICP, Py‐ICP, Tc‐ICP (chagasin), Cb‐ICP and Ps‐ICP are chagasin‐like cysteine protease inhibitors, whereas Pi‐EPIC2A is cystatin‐like. Cb, Coxiella burnetti; Pb, Plasmodium berghei; Pgt, Puccinia graminis f.sp. tritici; Pi, Phytophthora infestans; Ps, Pseudomonas syringae; Py, Plasmodium yoelii; Tc, Trypanosoma cruzi; Ua, Uromyces appendiculatus; Uf, Uromyces fabae. Pink arrows and sequences mark the positions of the seven β strands of Uf‐RTPp. The positions of β sheets of Tc‐ICP and Pb‐ICP according to Rennenberg et al. (2010) are marked by blue arrows and sequences. Green arrows, positions of β sheets specific for Pb‐ICP; yellow, sequence motifs that are known to be involved in cysteine protease binding (Rennenberg et al., 2010); red, conserved glycine residues.
Inhibition of proteases by Uf‐RTP1p and Us‐RTP1p
As RTPp homologues exhibit certain structural similarities with cysteine protease inhibitors, we investigated possible protease inhibitory effects of Uf‐RTP1p and Us‐RTP1p. As the yeast P. pastoris is known to show proteolytic activity within the culture medium (Sinha et al., 2005), we tested the protease activity of supernatants from different strains containing Uf‐RTP1p, Us‐RTP1p, a U. fabae invertase (Voegele et al., 2006) or no heterologous protein. We detected phenylmethanesulfonyl fluoride (PMSF) inhibitable proteolytic activity corresponding to a molecular mass of about 100 kDa in P. pastoris strains not expressing any heterologous protein or expressing a U. fabae invertase. However, this proteolytic activity was absent in P. pastoris strains expressing Uf‐RTP1p or Us‐RTP1p (Fig. 6, lanes 1–4). The addition of Uf‐RTP1p or Us‐RTP1p abolished the proteolytic activity within the supernatants of control strains, suggesting that the observed inhibition of the proteolytic activity can be attributed to RTP1p (Fig. 6, lanes 11–14). We therefore conclude that RTPps are able to inhibit protease activity and may act as cysteine protease inhibitors within the host plant.
Figure 6.
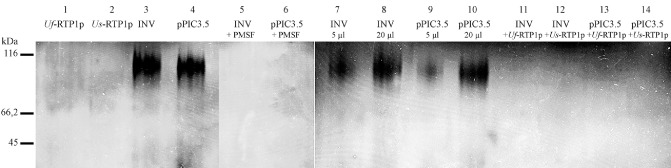
Inhibition of proteolytic activity by Uf‐RTP1p and Us‐RTP1p. Gelatin‐based zymograms show proteolytic activity in Pichia pastoris supernatants from strains expressing Uf‐RTP1p, Us‐RTP1p, U. fabae invertase (INV) or no heterologous protein (pPIC3.5). Supernatant (20 μL) was loaded onto lanes 1–6. The addition of phenylmethanesulfonyl fluoride (PMSF) (10 mm), Uf‐RTP1p or Us‐RTP1p (equal amount as sample volume) to supernatants from INV and pPIC3.5 strains abolished proteolytic activity (lanes 5 and 6, and 11–14, respectively). The intensity of the proteolytic activity depends on the amount of the loaded supernatant (lanes 7–10).
Discussion
The analysis of 28 complete RTPp sequences from 13 rust species emanating from six genera showed that RTPs constitute a new family of effector proteins presumably occurring ubiquitously in the order Pucciniales. Since the identification of RTP1 in U. fabae and U. striatus (Kemen et al., 2005), a number of RTP1 homologues have been identified in various genera within all suborders of the Pucciniales, including phylogenetically distant groups, such as the genus Hemileia (Duplessis et al., 2011; Fernandez et al., 2012; Puthoff et al., 2008). The discovery of homologues in such a wide spectrum of rust fungi suggests that RTP1 homologues are conserved in all rust species. RTP genes lack homology to sequences from species outside this order. This indicates that the RTPp family is confined to rust fungi and may therefore be associated with their obligate biotrophic lifestyle. Similarly, a restricted occurrence of effector protein families possibly involved in biotrophic interactions has been described recently for powdery mildews (Spanu et al., 2010). The RTPp family shows a distribution into several subgroups, with most species containing members from different subgroups. As some of the subgroups are conserved between genera, it seems likely that these subgroups evolved early in the Pucciniales, which is further supported by the fact that members of different subgroups have been described recently for H. vastatrix by Fernandez et al. (2012). Some groups, however, are likely to have evolved from gene duplication events in one species.
We showed that members of the RTPp family are characterized by a bipartite structure that comprises two domains which differ in sequence conservation and secondary structure. Although, in most cases, both parts have evolved in congruence, it is possible that at least three RTPs are the product of recombination and therefore do not follow the expected phylogeny. This domain organization is reflected at the genomic level, with the highly diverse exon 1 coding for the variable N‐terminal domain and the invariable exons 2–6 coding for the conserved C‐terminal domain. A modular protein structure has been described previously for oomycete effectors, where the domains adopt different functions during the interaction with the host plant (Win et al., 2007). In these cytoplasmic effector proteins, the N‐terminal region is involved in secretion and translocation into the host cell, whereas the C‐terminal domain can be linked to effector activity (Schornack et al., 2009). Both domains are under different selection pressure, which is consistent with their distinct function (Schornack et al., 2009; Win et al., 2007). In contrast with these oomycete effectors, we were unable to detect such evolutionary processes for the RTPp domains.
The highly variable RTPp N‐terminal domain does not contain any structural motifs, except for the N‐terminal leader peptide, and probably consists of a disordered region. These flexible regions within proteins are often involved in specific but transient protein interactions (Gsponer et al., 2008), which may be advantageous for possible protein interactions during the translocation process or within the target compartments. Only one conserved sequence motif could be identified in the N‐terminal region of RTP1p homologues. This motif forms a dibasic cleavage site for post‐translational processing at which the proteins are cleaved during secretion. Translocation motifs of oomycete RXLR and CRN effectors, as well as some fungal effector proteins, are located in the N‐termini, about 30–60 amino acids downstream of the signal peptides (Bhattacharjee et al., 2006; Haas et al., 2009; Kale et al., 2010; Rafiqi et al., 2010). The oomycete RXLR motif, as well as its functional variants in fungal effectors, resembles the Pexel motif of translocated proteins from the malaria pathogen Plasmodium spp. and is interchangeable with this motif (Grouffaud et al., 2008; Hiller et al., 2004; Kale et al., 2010). These proteins are cleaved at the Pexel motif during secretion from the parasite and enter the erythrocyte through a pathogen‐derived translocon (Chang et al., 2008; de Koning‐Ward et al., 2009). As the cleavage site of RTPp N‐termini is located within a similar region and represents the sole conserved sequence within this highly variable domain, one could speculate that this motif is involved in the transfer of RTP1p homologues, and the observed cleavage may be a mechanism associated with the translocation process, although it is not yet known whether all RTPp homologues are actually located within the host cell cytoplasm, as recent studies by Hacquard et al. (2012) have suggested a diverse localization of different RTPp homologues. Therefore, proteolytic processing of the RTPp N‐terminus may be necessary for the release or activation of the C‐terminal protease inhibitor domain.
Unlike the N‐terminal region, the RTPp C‐terminal domain is highly conserved in terms of sequence, as well as secondary structural motifs, in particular a β aggregation domain described for Uf‐RTP1p and Us‐RTP1p by E. Kemen et al. (Max Planck Institute for Plant Breeding Research, Cologne, unpublished results), which mediates amyloid‐like characteristics and can be found in all RTP1p homologues. This high degree of structural as well as sequence conservation indicates an association with effector function inside the host cell. All our analyses indicate that the overall structure of the C‐terminal domain is most likely globular and ordered, which is consistent with other intracellular effector proteins, particularly AvrM and AvrL567 from flax rust, which also possess highly structured C‐terminal domains (Catanzariti et al., 2010; Wang et al., 2007). Site‐directed mutagenesis of four highly conserved cysteine residues in the C‐terminal region indicates that the formation of disulphide bridges is required for correct folding and stability of the RTPp C‐terminal domain. A range of effector proteins are known to be stabilized by disulphide bridges, among them Avr2 and Avr4 from C. fulvum and Pep1 from Ustilago maydis (van den Burg et al., 2003; Doehlemann et al., 2009; Van't Klooster et al., 2011). These disulphide bridges sustain the globular structure in the apoplastic compartment and thus are indispensable for effector function (Jones and Catanzariti, 2010). Similarly, disulphide bridges may stabilize the RTPp C‐terminal domain within the extrahaustorial matrix and during transfer into the plant cell.
As consistently predicted by secondary structure analyses, the RTPp globular domain is likely to contain seven entirely conserved β strands, suggesting a crucial role for protein structure or secondary structural motifs. As the structure of the RTPp C‐terminal domain appears to be a critical factor, we attempted to identify proteins characterized by a similar structural motif. We showed that members of the RTPp family share similarities with chagasin‐ and cystatin‐like proteins, two families of cysteine protease inhibitors. These findings were supported by phylogenetic analyses showing the relationship between members of the RTPp family and cysteine protease inhibitors. Interestingly, some members of the chagasin family are secreted by the malaria pathogen Plasmodium spp. and enter the host cell cytoplasm, where they are involved in the suppression of host cell death (Rennenberg et al., 2010). The N‐terminal region of these proteins undergoes post‐translational processing, which leads to the release of the C‐terminal chagasin‐like domain that is active inside the host cell (Rennenberg et al., 2010). The RTPp C‐terminal domains and chagasin‐like inhibitors exhibit a similar molecular weight and resemble, in number and position, conserved secondary structure elements. Members of the chagasin‐like inhibitor family exhibit low sequence similarity and are characterized by an immunoglobulin‐like β sandwich fold composed of seven essential β strands (Casados‐Vazquez et al., 2011; Ljunggren et al., 2007; Smith et al., 2006). Three of the loop regions between the β strands form a wedge‐like structure that mediates interaction with the proteases (Rigden et al., 2001). Regarding these structural similarities to chagasin‐like protease inhibitors, it is possible that the RTPp C‐terminal domain adopts a similar overall structure. Interestingly, the loop regions of the chagasin‐like proteins can be associated with the three protease interacting regions of the cystatins (Redzynia et al., 2009). One of these motifs comprising two glycine residues can also be found in the RTPps (Fig. 5), and our selection analyses revealed that these residues are under purifying selection, supporting their functional relevance (Fig. 3).
We observed the inhibition of proteolytic activity within P. pastoris culture supernatants from strains expressing Uf‐RTP1p and Us‐RTP1p. This suggests that Uf‐RTP1p and Us‐RTP1p may be able to inhibit proteases. Little is known about the proteases which are active in P. pastoris supernatants, although the degradation of many heterologously expressed proteins is a frequently reported problem (Cereghino and Cregg, 2000; Sinha et al., 2005). Currently, it is not yet possible to define the type of inhibited proteases, although the inhibition of proteolytic activity suggests an involvement of serine proteases or papain‐like cysteine proteases.
Several apoplastic effector proteins of phytopathogenic oomycetes and fungi have been shown to act as protease inhibitors, among them EPIC1 and EPIC2B from P. infestans and Avr2 from C. fulvum, which are inhibitors of the tomato cysteine protease Rcr3 that is involved in the hypersensitive response (van Esse et al., 2008; Tian et al., 2007). As plant proteases are active in numerous pathogen recognition processes and defence reactions (van der Hoorn, 2008), these proteases represent essential targets for effector protein activity. Some studies have shown stress‐ and PCD‐associated activity of cysteine proteases in the plant cytosol (D'Silva et al., 1998; Solomon et al., 1999). Such cytoplasmic cysteine proteases may be inhibited by members of the RTPp effector protein family during the infection process, but this may not be the only function of the RTPps.
As described by E. Kemen et al. (Max Planck Institute for Plant Breeding Research, Cologne, unpublished results), Uf‐RTP1p and Us‐RTP1p show characteristics of amyloid‐like proteins and are able to form filamentous structures. It is conceivable that the RTPps exhibit variable functions during different stages of infection or within different compartments passed during the course of secretion and translocation to the host cytoplasm. The inhibition of plant cysteine proteases within the extrahaustorial matrix or the plant cytoplasm may therefore be an essential functional feature of the RTPp effector family that contributes to the establishment and maintenance of the biotrophic interaction.
Experimental Procedures
Isolation of fungal DNA
DNA of different rust fungi, required for the identification of RTP1 homologues by Uneven‐PCR (Chen and Wu, 1997) and SiteFinding‐PCR (Tan et al., 2005), was obtained from infected host plants. Hemileia vastatrix isolate 1427 DNA was isolated from infected leaves of Coffea arabica ‘Catuai’ (IICT/CIFC Centro Investigação das Ferrungens do Cafeeiro, Oeiras, Portugal) using the ZR Plant/Seed DNA Kit (Zymo Research Corporation, Orange, CA, USA). Gymnosporangium sabinae DNA was isolated in a similar fashion from field samples of infected Pyrus communis leaves from a pear orchard near Lake Constance, Germany. Puccinia coronata DNA was kindly provided by Dr Les Szabo (US Department of Agriculture‐Agricultural Research Service, St. Paul, MN, USA).
Cultivation of microorganisms
Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA, USA) was grown in Luria–Bertani medium (Sambrook and Russell, 2001) supplemented with an adequate concentration of antibiotic, isopropyl‐β‐d‐thiogalactopyranoside and 5‐bromo‐4‐chloro‐indolyl‐β‐D‐galactopyranoside for blue/white screening following transformation. Pichia pastoris strain KM71 was grown at 30 °C and 360 rpm in minimal glycerol medium or minimal methanol medium for induction, according to the manufacturer's instructions (Multi‐Copy Pichia Expression Kit, Invitrogen).
Nucleic acid manipulations
Molecular procedures were performed according to standard protocols (Sambrook and Russell, 2001). Escherichia coli strain DH5α was used for plasmid propagation. Pichia pastoris was transformed by electroporation according to the manufacturer's instructions (Invitrogen). Plasmids were linearized with SalI prior to P. pastoris transformation. Sequencing was performed by GATC Biotech, Konstanz, Germany. Sequencing data were evaluated and analysed using BioEdit software (Hall, 1999). Uneven‐PCR (Chen and Wu, 1997) and SiteFinding‐PCR (Tan et al., 2005) were used to amplify unknown sequence fragments of the Gs‐RTP1, Hv‐RTP1 and Pc‐RTP1 genes (Table S1, see Supporting Information). Substitutions of cysteine residues by serine were introduced to Uf‐RTP1 containing a C‐terminal 6 × His cluster by PCR with overlapping mutagenic primers (Table S2, see Supporting Information).
Plasmid constructions
Products of Uneven‐ and SiteFinding‐PCR were ligated into the pDRIVE vector from the Qiagen PCR cloning kit according to the manufacturer's instructions (Qiagen GmbH, Hilden, Germany). For expression in Pi. pastoris, cysteine mutants were amplified during mutagenic PCR with primers PIG7‐full5 (5′‐CGTAGAATTCCATTATGTCAAACCTTCGCTTAC‐3′) and Pig7‐f‐his3′ (5′‐GCCGCCCTAGGTCAGTGGTGGTGGTGGTGG‐3′), introducing EcoRI and AvrII restriction sites, respectively (italic type). PCR products were digested with EcoRI and AvrII successively, and ligated into EcoRI and AvrII digested and dephosphorylated vector pPIC3.5 (Invitrogen). The accuracy of the plasmid constructs was confirmed by sequence analysis.
Homology searches, sequence alignments and phylogenetic analysis
Homologous sequences were obtained from public databases of the Broad Institute FGI (Fungal Genome Initiative), DOE Joint Genome Institute and National Center for Biotechnology Information (NCBI) GenBank websites by blast search (Altschul et al., 1997) using blastn, blastp, blastx, tblastn and tblastx algorithms (Table S3, see Supporting Information). RTPp sequences were aligned using the ClustalW algorithm (Thompson et al., 1994). To identify RTP1 orthologues within the Pucciniales, in more distant fungi and other Eukaryotes, we performed OrthoMCL analyses using the genomes of Blumeria graminis, M. larici‐populina, P. graminis, P. striiformis, U. maydis, Sporisorium reilianum, Fusarium oxysporum, Saccharomyces cerevisiae, Tuber melanosporum, Laccaria bicolor, Hyaloperonospora arabidopsidis and Albugo laibachii.
We used default parameters as described by Moustafa et al. (2009). All alignments were performed using e‐values of 1E‐3 and 1E‐5. Tribe analysis was performed using hierarchical clustering with average linkage implemented in the clusterx program (Nepusz et al., 2010). For selection analyses, transcribed RTPp sequences were aligned using ClustalW2 (Thompson et al., 1994). The alignment was transferred onto the nucleotide sequence using the Pythonscript revtrans.py (version 1.4, http://www.cbs.dtu.dk/services/RevTrans/download.php) and analysed using the selecton algorithm with default parameters. Selecton was downloaded from http://selecton.tau.ac.il/source.html and run locally as described by ‘selecton‐h’. As input tree, a tree based on the ClustalW2 alignment using 1000 optimization steps was used.
In silico analyses of amino acid sequences and structural cluster analyses
Searches for conserved functional motifs were performed using the ELM server (Puntervoll et al., 2003). The presence of signal peptides was verified with the SignalP 3.0 program (Bendtsen et al., 2004). N‐Glycosylation sites were predicted with the Net N‐Glyc 1.0 algorithm (Blom et al., 2004). Prediction of cysteine disulphide bonding was performed using the DISULFIND and DiANNA 1.1 servers (Ferre and Clote, 2005; Vullo and Frasconi, 2004). Analyses of protein domain structure were carried out using the DisEMBLTM, GLOBPLOT2 and PONDR‐FIT algorithms (Linding et al., 2003a, b; Xue et al., 2010). Protein secondary structure was predicted with the Porter, Sable and PsiPred algorithms (Adamczak et al., 2004; McGuffin et al., 2000; Pollastri and McLysaght, 2005).
Sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and immunoblot analysis
Protein samples were separated on 12% SDS‐PAGE gels (Laemmli, 1970) and subsequently transferred to polyvinylidene difluoride (PVDF) membranes. Immunoblots were performed according to Towbin et al. (1979). Purified guinea pig anti‐Uf‐RTP1p serum S746p or rabbit anti‐Uf‐RTP1p serum S844p (Kemen et al., 2005) were used as primary antibodies. Peroxidase‐coupled goat–anti‐guinea pig or goat–anti‐rabbit secondary antibodies (Sigma‐Aldrich, Taufkirchen, Germany) and ECL Western Blot detection reagent (GE Healthcare, Munich, Germany) were used for detection. For analyses of heterologously expressed proteins, pellet fractions of the appropriate Pi. pastoris cultures were dissolved in sample buffer after centrifugation. Cell‐free culture supernatants were mixed with sample buffer following filtration through a 0.2‐μm pore size filter.
N‐terminal sequencing
Protein samples from 30‐fold concentrated P. pastoris culture supernatant were separated on 12% SDS‐PAGE gels and transferred to PVDF membranes. Membranes were stained with 0.2% (w/v) Serva Blue R250, 40% methanol and destained with 50% methanol. Edman sequencing of the excised bands was performed by TOPLAB GmbH, Munich, Germany.
Gelatin zymography
Gelatin zymography was performed on 7% SDS‐PAGE gels containing 0.1% gelatin. Pichia pastoris culture supernatant (20 μL) was mixed with sample buffer without reducing agent and loaded onto the gel without previous heating. Gels were incubated in 2.5% Triton X‐100 for 1 h at room temperature after electrophoresis, followed by incubation overnight at 37 °C in 10 mm Tris, 20 mm NaCl, 5 mm cysteine, pH 5.0. Proteolytic activity was detected as unstained bands after staining with 45% (v/v) methanol, 10% (v/v) acetic acid and 0.25% (v/v) Serva Blue R, and destaining with 35% (v/v) methanol, 10% (v/v) acetic acid and 0.3% (v/v) glycerol. For analysis of inhibitory effects, samples were pretreated with 10 mm PMSF or mixed with an equal amount of P. pastoris culture supernatant containing Uf‐RTP1p or Us‐RTP1p, and incubated for 20 min at room temperature before electrophoresis.
Supporting information
Fig. S1 Alignment of the 28 complete rust transferred protein 1 (RTP1p) homologues. Conserved residues are shaded grey. Blue, signal peptide; green, cleavage site for post‐translational processing; yellow, highly conserved cysteine residues. Arrows mark the positions of the conserved β strands.
Fig. S2 TribeMCL analysis of rust transferred protein (RTPp) N‐ and C‐termini. Cluster analyses of conserved RTPp termini were performed using the TribeMCL method. The network structure highlights the major relationships between members of a tribe. (a) RTPp C‐termini can be clustered into six tribes. (b) RTPp N‐termini also cluster into six tribes. Colours are selected according to C‐termini clustering and reveal that the N‐terminus of tribe 1 shows a higher degree of divergence than those of tribes 2–6.
Fig. S3 Phylogenetic analyses based on secondary structure prediction using PsiPred. Analyses were conducted using sequences of rust transferred proteins (RTPps), cystatin‐ and chagasin‐like cysteine protease inhibitors and Kunitz‐type serine protease inhibitors as outgroup. Cb, Coxiella burnetti; Gg, Gallus gallus; Gm, Glycine max; Hs, Homo sapiens; Mlp, Melampsora larici‐populina; Mm, Mus musculus; Mmd, Melampsora medusae f.sp. deltoides; Mmt, Melampsora medusae f.sp. tremuloides; Mo, Melampsora occidentalis; Pb, Plasmodium berghei; Pc, Puccinia coronata; Pgt, Puccinia graminis f.sp. tritici; Pi, Phytophthora infestans; Pn, Populus nigra; Ps, Puccinia striiformis; Psa, Pisum sativum; Pt, Puccinia triticina; Py, Plasmodium yoelii; Tc, Trypanosoma cruzi; Ua, Uromyces appendiculatus; Uf, Uromyces fabae; Us, Uromyces striatus.
Table S1 Uneven‐ and SiteFinding‐PCR primers for Gs‐RTP1, Pc‐RTP1 and Hv‐RTP1 sequencing.
Table S2 Mutagenic primers for substitution of Uf ‐RTP1p amino acid residues.
Table S3 Accession numbers of sequences used in this study.
Acknowledgements
We are grateful to Tobias Link (Fachgebiet Phytopathologie, Institut für Phytomedizin, Fakultät Agrarwissenschaften, Universität Hohenheim, Stuttgart, Germany) for providing additional RTP sequences. This work was supported by grants provided by the Deutsche Forschungsgemeinschaft to Kurt Mendgen (ME523/25‐1) and Ralf Voegele (VO595/2‐2).
References
- Adamczak, R. , Porollo, A. and Meller, J. (2004) Accurate prediction of solvent accessibility using neural networks‐based regression. Proteins, 56, 753–767. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schaeffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Fernandez, M. , Liang, Y.‐H. , Abrahamson, M. and Su, X.‐D. (2005) Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile. J. Biol. Chem. 280, 18 221–18 228. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , von Nielsen, H., Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: signalp 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. , Hiller, N.L. , Liolios, K. , Win, J. , Kanneganti, T.D. , Young, C. , Kamoun, S. and Haldar, K. (2006) The malarial host‐targeting signal is conserved in the Irish potato famine pathogen. Plos Pathog. 2, e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch, P.R. , Rehmany, A.P. , Pritchard, L. , Kamoun, S. and Beynon, J.L. (2006) Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol. 14, 8–11. [DOI] [PubMed] [Google Scholar]
- Blom, N. , Sicheritz‐Ponten, T. , Gupta, R. , Gammeltoft, S. and Brunak, S. (2004) Prediction of post‐translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics, 4, 1633–1649. [DOI] [PubMed] [Google Scholar]
- Bos, J.I. , Armstrong, M.R. , Gilroy, E.M. , Boevink, P.C. , Hein, I. , Taylor, R.M. , Zhendong, T. , Engelhardt, S. , Vetukuri, R.R. and Harrower, B. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA, 107, 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg, H.A. , Westerink, N. , Francoijs, K.‐J. , Roth, R. , Woestenenk, E. , de Boeren, S., Wit, P.J.G.M. , Joosten, M.H.A.J. and Vervoort, J. (2003) Natural disulfide bond‐disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf‐4‐mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278, 27 340–27 346. [DOI] [PubMed] [Google Scholar]
- Casados‐Vazquez, L.E. , Lara‐Gonzalez, S. and Brieba, L.G. (2011) Crystal structure of the cysteine protease inhibitor 2 from Entamoeba histolytica: functional convergence of a common protein fold. Gene, 471, 45–52. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.‐M. , Dodds, P.N. , Ve, T. , Kobe, B. , Ellis, J.G. and Staskawicz, B.J. (2010) The AvrM effector from flax rust has a structured C‐terminal domain and interacts directly with the M resistance protein. Mol. Plant–Microbe Interact. 23, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino, J.L. and Cregg, J.M. (2000) Heterologous protein expression in the methylotrophic yeast Pichia pastoris . FEMS Microbiol. Rev. 24, 45–66. [DOI] [PubMed] [Google Scholar]
- Chang, H.H. , Falick, A.M. , Carlton, P.M. , Sedat, J.W. , DeRisi, J.L. and Marletta, M.A. (2008) N‐terminal processing of proteins exported by malaria parasites. Mol. Biochem. Parasitol. 160, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. and Wu, R. (1997) Direct amplification of unknown genes and fragments by Uneven polymerase chain reaction. Gene, 185, 195–199. [DOI] [PubMed] [Google Scholar]
- Darling, R.J. , Ruddon, R.W. , Perini, F. and Bedows, E. (2000) Cystine knot mutations affect the folding of the glycoprotein hormone alpha‐subunit. Differential secretion and assembly of partially folded intermediates. J. Biol. Chem. 275, 15 413–15 421. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. , van der Linde, K. , Aβmann, D. , Schwammbach, D. , Hof, A. , Mohanty, A. , Jackson, D. and Kahmann, R. (2009) Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. Plos Pathog. 5, e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva, I. , Poirier, G.G. and Heath, M.C. (1998) Activation of cysteine proteases in cowpea plants during the hypersensitive response—a form of programmed cell death. Exp. Cell Res. 245, 389–399. [DOI] [PubMed] [Google Scholar]
- Duplessis, S. , Cuomo, C.A. , Lin, Y.‐C. , Aerts, A. , Tisserant, E. , Veneault‐Fourrey, C. , Joly, D.L. , Hacquard, S. , Amselem, J. , Cantarel, B.L. , Chiu, R. , Coutinho, P.M. , Feau, N. , Field, M. , Frey, P. , Gelhaye, E. , Goldberg, J. , Grabherr, M.G. , Kodira, C.D. , Kohler, A. , Kües, U. , Lindquist, E.A. , Lucas, S.M. , Mago, R. , Mauceli, E. , Morin, E. , Murat, C. , Pangilinan, J.L. , Park, R. , Pearson, M. , Quesneville, H. , Rouhier, N. , Sakthikumar, S. , Salamov, A.A. , Schmutz, J. , Selles, B. , Shapiro, H. , Tanguay, P. , Tuskan, G.A. , Henrissat, B. , Van de Peer, Y. , Rouzé, P. , Ellis, J.G. , Dodds, P.N. , Schein, J.E. , Zhong, S. , Hamelin, R.C. , Grigoriev, I.V. , Szabo, L.J. and Martin, F. (2011) Obligate biotrophy features unravelled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA, 108, 9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse, H.P. , Van't Klooster, J.W. , Bolton, M.D. , van Yadeta, K.A., Baarlen, P. , Boeren, S. , de Vervoort, J., Wit, P.J. and Thomma, B.P. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell, 20, 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feau, N. , Bergeron, M.‐J. , Joly, D.L. , Roussel, F. and Hamelin, R.C. (2007) Detection and validation of EST‐derived SNPs for poplar leaf rust Melampsora medusae f. sp. deltoidae . Mol. Ecol. Notes, 7, 1222–1228. [Google Scholar]
- Fernandez, D. , Tisserant, E. , Talhinhas, P. , Azinheira, H. , Vieira, A.N.A. , Petitot, A.‐S. , Loureiro, A. , Poulain, J. , Da Silva, C. , Silva, M.D.O.C. and Duplessis, S. (2012) 454‐pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix reveals in planta‐expressed pathogen‐secreted proteins and plant functions in a late compatible plant–rust interaction. Mol. Plant Pathol. 13, 17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre, F. and Clote, P. (2005) DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 33, 230–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grouffaud, S. , van West, P. , Avrova, A.O. , Birch, P.R.J. and Whisson, S.C. (2008) Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans . Microbiology, 154, 3743–3751. [DOI] [PubMed] [Google Scholar]
- Gsponer, J. , Futschik, M.E. , Teichmann, S.A. and Babu, M.M. (2008) Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science, 322, 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. , Torto‐Alalibo, T. , Bozkurt, T.O. , Ah‐Fong, A.M. , Alvarado, L. , Anderson, V.L. , Armstrong, M.R. , Avrova, A. , Baxter, L. , Beynon, J. , Boevink, P.C. , Bollmann, S.R. , Bos, J.I. , Bulone, V. , Cai, G. , Cakir, C. , Carrington, J.C. , Chawner, M. , Conti, L. , Costanzo, S. , Ewan, R. , Fahlgren, N. , Fischbach, M.A. , Fugelstad, J. , Gilroy, E.M. , Gnerre, S. , Green, P.J. , Grenville‐Briggs, L.J. , Griffith, J. , Grunwald, N.J. , Horn, K. , Horner, N.R. , Hu, C.H. , Huitema, E. , Jeong, D.H. , Jones, A.M. , Jones, J.D. , Jones, R.W. , Karlsson, E.K. , Kunjeti, S.G. , Lamour, K. , Liu, Z. , Ma, L. , Maclean, D. , Chibucos, M.C. , McDonald, H. , McWalters, J. , Meijer, H.J. , Morgan, W. , Morris, P.F. , Munro, C.A. , O'Neill, K. , Ospina‐Giraldo, M. , Pinzon, A. , Pritchard, L. , Ramsahoye, B. , Ren, Q. , Restrepo, S. , Roy, S. , Sadanandom, A. , Savidor, A. , Schornack, S. , Schwartz, D.C. , Schumann, U.D. , Schwessinger, B. , Seyer, L. , Sharpe, T. , Silvar, C. , Song, J. , Studholme, D.J. , Sykes, S. , van de Thines, M., Vondervoort, P.J. , Phuntumart, V. , Wawra, S. , Weide, R. , Win, J. , Young, C. , Zhou, S. , Fry, W. , van Meyers, B.C., West, P. , Ristaino, J. , Govers, F. , Birch, P.R. , Whisson, S.C. , Judelson, H.S. and Nusbaum, C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hacquard, S. , Joly, D.L. , Lin, Y.C. , Tisserant, E. , Feau, N. , Delaruelle, C. , Legue, V. , Kohler, A. , Tanguay, P. , Petre, B. , Frey, P. , Van de Peer, Y. , Rouze, P. , Martin, F. , Hamelin, R.C. and Duplessis, S. (2012) A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici‐populina (poplar leaf rust). Mol. Plant–Microbe Interact. 25, 279–293. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hann, D.R. , Gimenez‐Ibanez, S. and Rathjen, J.P. (2010) Bacterial virulence effectors and their activities. Curr. Opin. Plant Biol. 13, 388–393. [DOI] [PubMed] [Google Scholar]
- Hiller, N.L. , van Bhattacharjee, S., Ooij, C. , Liolios, K. , Harrison, T. , Lopez‐Estrano, C. and Haldar, K. (2004) A host‐targeting signal in virulence proteins reveals a secretome in malarial infection. Science, 306, 1934–1937. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59, 191–223. [DOI] [PubMed] [Google Scholar]
- Joly, D.L. , Feau, N. , Tanguay, P. and Hamelin, R.C. (2010) Comparative analysis of secreted protein evolution using expressed sequence tags from four poplar leaf rusts (Melampsora spp). BMC Genomics, 11, 422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A. and Catanzariti, A.M. (2010) Effector proteins of extracellular fungal plant pathogens that trigger host resistance. Funct. Plant Biol. 37, 901–906. [Google Scholar]
- Kale, S.D. , Gu, B. , Capelluto, D.G. , Dou, D. , Feldman, E. , Rumore, A. , Arredondo, F.D. , Hanlon, R. , Fudal, I. and Rouxel, T. (2010) External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell, 142, 284–295. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Kemen, A.C. , Rafiqi, M. , Hempel, U. , Mendgen, K. , Hahn, M. and Voegele, R.T. (2005) Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant–Microbe Interact. 18, 1130–1139. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Gardiner, A. , Schultz‐Larsen, T. , Kemen, A.C. , Balmuth, A.L. , Robert‐Seilaniantz, A. , Bailey, K. , Holub, E. , Studholme, D.J. , MacLean, D. and Jones, J.D.G. (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana . Plos Biol. 9, e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning‐Ward, T.F. , Gilson, P.R. , Boddey, J.A. , Rug, M. , Smith, B.J. , Papenfuss, A.T. , Sanders, P.R. , Lundie, R.J. , Maier, A.G. , Cowman, A.F. and Crabb, B.S. (2009) A newly discovered protein export machine in malaria parasites. Nature, 459, 945–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Linding, R. , Jensen, L.J. , Diella, F. , Bork, P. , Gibson, T.J. and Russell, R.B. (2003a) Protein disorder prediction, implications for structural proteomics. Structure, 11, 1453–1459. [DOI] [PubMed] [Google Scholar]
- Linding, R. , Russell, R.B. , Neduva, V. and Gibson, T.J. (2003b) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31, 3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljunggren, A. , Redzynia, I. , Alvarez‐Fernandez, M. , Abrahamson, M. , Mort, J.S. , Krupa, J.C. , Jaskolski, M. and Bujacz, G. (2007) Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease. J. Mol. Biol. 371, 137–153. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Huang, X. , Wang, X. , Chen, X. , Qu, Z. , Huang, L. and Kang, Z. (2009) Identification of expressed genes during compatible interaction between stripe rust (Puccinia striiformis) and wheat using a cDNA library. BMC Genomics, 10, 586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin, L.J. , Bryson, K. and Jones, D.T. (2000) The PSIPRED protein structure prediction server. Bioinformatics, 16, 404–405. [DOI] [PubMed] [Google Scholar]
- Moustafa, A. , Beszteri, B. , Maier, U.G. , Bowler, C. , Valentin, K. and Bhattacharya, D. (2009) Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science, 324, 1724–1726. [DOI] [PubMed] [Google Scholar]
- Nepusz, T. , Sasidharan, R. and Paccanaro, A. (2010) SCPS: a fast implementation of a spectral method for detecting protein families on a genome‐wide scale. BMC Bioinformatics, 11, 120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach, M.J. , Farrall, L. , Sweigard, J.A. , Chumley, F.G. and Valent, B. (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi‐ta . Plant Cell, 12, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga, R. and Dodds, P.N. (2009) Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science, 324, 748–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri, G. and McLysaght, A. (2005) Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics, 21, 1719–1720. [DOI] [PubMed] [Google Scholar]
- Puntervoll, P. , Linding, R. , Gemund, C. , Chabanis‐Davidson, S. , Mattingsdal, M. , Cameron, S. , Martin, D.M. , Ausiello, G. , Brannetti, B. and Costantini, A. (2003) ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31, 3625–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthoff, D.P. , Neelam, A. , Ehrenfried, M.L. , Scheffler, B.E. , Ballard, L. , Song, Q. , Campbell, K.B. , Cooper, B. and Tucker, M.L. (2008) Analysis of expressed sequence tags from Uromyces appendiculatus hyphae and haustoria and their comparison to sequences from other rust fungi. Phytopathology, 98, 1126–1135. [DOI] [PubMed] [Google Scholar]
- Rafiqi, M. , Gan, P.H. , Ravensdale, M. , Lawrence, G.J. , Ellis, J.G. , Jones, D.A. , Hardham, A.R. and Dodds, P.N. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell, 22, 2017–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redzynia, I. , Ljunggren, A. , Bujacz, A. , Abrahamson, M. , Jaskolski, M. and Bujacz, G. (2009) Crystal structure of the parasite inhibitor chagasin in complex with papain allows identification of structural requirements for broad reactivity and specificity determinants for target proteases. FEBS J. 276, 793–806. [DOI] [PubMed] [Google Scholar]
- Rennenberg, A. , Lehmann, C. , Heitmann, A. , Witt, T. , Hansen, G. , Nagarajan, K. , Deschermeier, C. , Turk, V. , Hilgenfeld, R. and Heussler, V.T. (2010) Exoerythrocytic Plasmodium parasites secrete a cysteine protease inhibitor involved in sporozoite invasion and capable of blocking cell death of host hepatocytes. Plos Pathog. 6, e1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden, D.J. , Monteiro, A.C. and Grossi de Sa, M.F. (2001) The protease inhibitor chagasin of Trypanosoma cruzi adopts an immunoglobulin‐type fold and may have arisen by horizontal gene transfer. FEBS Lett. 504, 41–44. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schornack, S. , Huitema, E. , Cano, L.M. , Bozkurt, T.O. , Oliva, R. , Van Damme, M. , Schwizer, S. , Raffaele, S. , Chaparro‐Garcia, A. , Farrer, R. , Segretin, M.E. , Bos, J. , Haas, B.J. , Zody, M.C. , Nusbaum, C. , Win, J. , Thines, M. and Kamoun, S. (2009) Ten things to know about oomycete effectors. Mol. Plant Pathol. 10, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , van Damme, M. , Bozkurt, T.O. , Cano, L.M. , Smoker, M. , Thines, M. , Gaulin, E. , Kamoun, S. and Huitema, E. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA, 107, 17 421–17 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, J. , Plantz, B.A. , Inan, M. and Meagher, M.M. (2005) Causes of proteolytic degradation of secreted recombinant proteins produced in methylotrophic yeast Pichia pastoris: case study with recombinant bovine interferon‐tau. Biotechnol. Bioeng. 89, 102–112. [DOI] [PubMed] [Google Scholar]
- Smith, B.O. , Picken, N.C. , Westrop, G.D. , Bromek, K. , Mottram, J.C. and Coombs, G.H. (2006) The structure of Leishmania mexicana ICP provides evidence for convergent evolution of cysteine peptidase inhibitors. J. Biol. Chem. 281, 5821–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, M. , Belenghi, B. , Delledonne, M. , Menachem, E. and Levine, A. (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell, 11, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu, P.D. , Abbott, J.C. , Amselem, J. , Burgis, T.A. , Soanes, D.M. , Stüber, K. , Loren van Themaat, E.V. , Brown, J.K.M. , Butcher, S.A. , Gurr, S.J. , Lebrun, M.‐H. , Ridout, C.J. , Schulze‐Lefert, P. , Talbot, N.J. , Ahmadinejad, N. , Ametz, C. , Barton, G.R. , Benjdia, M. , Bidzinski, P. , Bindschedler, L.V. , Both, M. , Brewer, M.T. , Cadle‐Davidson, L. , Cadle‐Davidson, M.M. , Collemare, J. , Cramer, R. , Frenkel, O. , Godfrey, D. , Harriman, J. , Hoede, C. , King, B.C. , Klages, S. , Kleemann, J. , Knoll, D. , Koti, P.S. , Kreplak, J. , López‐Ruiz, F.J. , Lu, X. , Maekawa, T. , Mahanil, S. , Micali, C. , Milgroom, M.G. , Montana, G. , Noir, S. , O'Connell, R.J. , Oberhaensli, S. , Parlange, F. , Pedersen, C. , Quesneville, H. , Reinhardt, R. , Rott, M. , Sacristán, S. , Schmidt, S.M. , Schön, M. , Skamnioti, P. , Sommer, H. , Stephens, A. , Takahara, H. , Thordal‐Christensen, H. , Vigouroux, M. , Weßling, R. , Wicker, T. and Panstruga, R. (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science, 330, 1543–1546. [DOI] [PubMed] [Google Scholar]
- Tan, G. , Gao, Y. , Shi, M. , Zhang, X. , He, S. , Chen, Z. and An, C. (2005) SiteFinding‐PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Res. 33, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M. , Win, J. , van der Song, J., van der Hoorn, R., Knaap, E. and Kamoun, S. (2007) A Phytophthora infestans cystatin‐like protein targets a novel tomato papain‐like apoplastic protease. Plant Physiol. 143, 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H. , Staehelin, T. and Gordon, J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Klooster, J.W. , Van Der Kamp, M.W. , Vervoort, J. , Beekwilder, J. , Boeren, S. , Joosten, M.H.A. , Thomma, B.P.H.J. and De Wit, P.J.G.M. (2011) Affinity of Avr2 for tomato cysteine protease Rcr3 correlates with the Avr2‐triggered Cf‐2‐mediated hypersensitive response. Mol. Plant Pathol. 12, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T. , Wirsel, S. , Möll, U. , und Lechner, M. and Mendgen, K. (2006) Cloning and characterization of a novel invertase from the obligate biotroph Uromyces fabae and analysis of expression patterns of host and pathogen invertases in the course of infection. Mol. Plant–Microbe Interact. 19, 625–634. [DOI] [PubMed] [Google Scholar]
- Vullo, A. and Frasconi, P. (2004) Disulfide connectivity prediction using recursive neural networks and evolutionary information. Bioinformatics, 20, 653–659. [DOI] [PubMed] [Google Scholar]
- Wang, C.I. , Guncar, G. , Forwood, J.K. , Teh, T. , Catanzariti, A.M. , Lawrence, G.J. , Loughlin, F.E. , Mackay, J.P. , Schirra, H.J. , Anderson, P.A. , Ellis, J.G. , Dodds, P.N. and Kobe, B. (2007) Crystal structures of flax rust avirulence proteins AvrL567‐A and ‐D reveal details of the structural basis for flax disease resistance specificity. Plant Cell, 19, 2898–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , van Grouffaud, S., West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Win, J. , Morgan, W. , Bos, J. , Krasileva, K.V. , Cano, L.M. , Chaparro‐Garcia, A. , Ammar, R. , Staskawicz, B.J. and Kamoun, S. (2007) Adaptive evolution has targeted the C‐terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell, 19, 2349–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, B. , Dunbrack, R.L. , Williams, R.W. , Dunker, A.K. and Uversky, V.N. (2010) PONDR‐FIT: a meta‐predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta, 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Alignment of the 28 complete rust transferred protein 1 (RTP1p) homologues. Conserved residues are shaded grey. Blue, signal peptide; green, cleavage site for post‐translational processing; yellow, highly conserved cysteine residues. Arrows mark the positions of the conserved β strands.
Fig. S2 TribeMCL analysis of rust transferred protein (RTPp) N‐ and C‐termini. Cluster analyses of conserved RTPp termini were performed using the TribeMCL method. The network structure highlights the major relationships between members of a tribe. (a) RTPp C‐termini can be clustered into six tribes. (b) RTPp N‐termini also cluster into six tribes. Colours are selected according to C‐termini clustering and reveal that the N‐terminus of tribe 1 shows a higher degree of divergence than those of tribes 2–6.
Fig. S3 Phylogenetic analyses based on secondary structure prediction using PsiPred. Analyses were conducted using sequences of rust transferred proteins (RTPps), cystatin‐ and chagasin‐like cysteine protease inhibitors and Kunitz‐type serine protease inhibitors as outgroup. Cb, Coxiella burnetti; Gg, Gallus gallus; Gm, Glycine max; Hs, Homo sapiens; Mlp, Melampsora larici‐populina; Mm, Mus musculus; Mmd, Melampsora medusae f.sp. deltoides; Mmt, Melampsora medusae f.sp. tremuloides; Mo, Melampsora occidentalis; Pb, Plasmodium berghei; Pc, Puccinia coronata; Pgt, Puccinia graminis f.sp. tritici; Pi, Phytophthora infestans; Pn, Populus nigra; Ps, Puccinia striiformis; Psa, Pisum sativum; Pt, Puccinia triticina; Py, Plasmodium yoelii; Tc, Trypanosoma cruzi; Ua, Uromyces appendiculatus; Uf, Uromyces fabae; Us, Uromyces striatus.
Table S1 Uneven‐ and SiteFinding‐PCR primers for Gs‐RTP1, Pc‐RTP1 and Hv‐RTP1 sequencing.
Table S2 Mutagenic primers for substitution of Uf ‐RTP1p amino acid residues.
Table S3 Accession numbers of sequences used in this study.


