Summary
Membrane trafficking is vital to plant development and adaptation to the environment. It is suggested that post‐Golgi vesicles and multivesicular bodies are essential for plant defence against directly penetrating fungal parasites at the cell wall. However, the actual plant proteins involved in membrane transport for defence are largely unidentified. We applied a candidate gene approach and single cell transient‐induced gene silencing for the identification of membrane trafficking proteins of barley involved in the response to the fungal pathogen Blumeria graminis f.sp. hordei. This revealed potential components of vesicle tethering complexes [putative exocyst subunit HvEXO70F‐like and subunits of the conserved oligomeric Golgi (COG) complex] and Golgi membrane trafficking (COPIγ coatomer and HvYPT1‐like RAB GTPase) as essential for resistance to fungal penetration into the host cell.
Introduction
Plant cells constantly survey their surface for nonself molecules or for signs of nonself activity. Conserved microbe‐associated molecular patterns (MAMPs), which are absent from plants, can be recognized as nonself molecules. Damage‐associated molecular patterns (DAMPs) are released from the plant itself on microbial activity that harms plant cell integrity. DAMPs can be fragments of cell wall polymers or molecules derived from damaged proteins or membranes. Pattern recognition receptors bind MAMPs or DAMPs and mediate cross‐membrane signal transduction to symplastic signalling for subsequent immune responses (Boller and Felix, 2009). In addition, mechanosensitive plasma membrane proteins may contribute to defence signalling (Hématy et al., 2009). The plant immune response includes a transcriptional and metabolic reprogramming of the plant cells that are in contact with the microbe, and can additionally lead to systemic priming of the plant for future pathogen challenge (Spoel and Dong, 2012). The efficacy of the plant defence response may differ greatly depending on its spatiotemporal accuracy. Membrane transport is probably pivotal for the spatial control of plant defence that is focused to a particular area of the plant cell or apoplast. This is demonstrated by mutants that are affected in factors of polar membrane trafficking and thus defective in different types of immunity to adapted or nonadapted pathogens. Pathogens also secrete virulence effectors into plant host cells for direct manipulation of host membrane trafficking (for excellent reviews, see Frei dit Frey and Robatzek, 2009; Lipka et al., 2010).
The plant's secretory pathway is activated during the plant's resistance response (Wang et al., 2005). Membrane trafficking appears to be particularly important in the interaction of plants with cell wall‐penetrating fungi, such as powdery mildew fungi. The interaction of plants with powdery mildew fungi is considered as a model for the study of the cell biology of plant–pathogen interactions (Hückelhoven and Panstruga, 2011). This is because of the microscopically accessible, highly polarized immune response of plant epidermal cells at sites of attempted penetration. Polarized defence includes localized formation of plant cell wall appositions (Zeyen et al., 2002), as well as localized secretion of defence‐related compounds (Bednarek et al., 2011; von Röpenack et al., 1998). Focal secretion of plant defence compounds in Arabidopsis attacked by the nonadapted barley powdery mildew fungus Blumeria graminis f.sp. hordei depends on a ternary SNARE (soluble N‐ethylmaleimide‐sensitive‐factor attachment receptor) complex consisting of SYP121 (PEN1), SNAP33 and VAMP721/722 (Collins et al., 2003; Kwon et al., 2008). Some of these SNARE proteins may have functions specific for defence, whereas others may also participate in plant development (Assaad et al., 2004; Collins et al., 2003; Kwon et al., 2008). The SNARE complex appears to be conserved in structure and function in the monocot model barley (Hordeum vulgare) (Collins et al., 2003; Douchkov et al., 2005). Cell wall‐associated defence against B. graminis further involves polarization of the cytoskeleton, and can be inhibited by drugs that affect the polymerization of actin or tubulin (Hoefle et al., 2011; Kobayashi et al., 1997; Miklis et al., 2007; Opalski et al., 2005). In addition, the endoplasmic reticulum and the Golgi apparatus accumulate close to the site of attack from B. graminis f.sp. hordei (Eichmann and Hückelhoven, 2008; Hückelhoven and Panstruga, 2011). Together, these observations suggest that polar transport of defence‐related compounds is required for penetration defence.
The nature of vesicles that carry defence compounds to the site of attempted penetration is not entirely understood. Transmission electron microscopy in barley suggested that, in addition to the Golgi apparatus and endoplasmic reticulum, multivesicular bodies (MVBs) accumulate at the site of local cell wall defence (An et al., 2006b). Live cell imaging further showed the incorporation of plasma membrane‐resident proteins into cell wall appositions in Arabidopsis. These observations led to the hypothesis that exosome‐like vesicles are secreted during fungal attack (An et al., 2007; Meyer et al., 2009). MVBs also occur in cells successfully penetrated by powdery mildew fungi both inside the plant and the fungus (An et al., 2006a; Micali et al., 2011). In addition, a defence‐associated ADP ribosylation factor (ARF) GTPase, ARFA1b/c, which co‐localizes with the MVB/trans‐Golgi network RAB GTPase ARA7, is required for callose deposition and penetration defence of barley to B. graminis f.sp. hordei (Böhlenius et al., 2010).
The compatible interaction with powdery mildew fungi may also require proper membrane trafficking for building the haustorial complex, which consists of the fungal haustorium as the assumed feeding organ, the extrahaustorial matrix and the extrahaustorial membrane. The so‐called neckband seals the complex from the plant's apolast. The plant likely partially or entirely provides neckband, extrahaustorial matrix and extrahaustorial membrane to the complex (Green et al., 2002; Koh et al., 2005; Micali et al., 2011). Hence, host membrane transport is postulated to be key to both defence and pathogenesis of haustoria‐forming pathogens (Feechan et al., 2011; Hoefle et al., 2011; Hückelhoven and Panstruga, 2011; Koh et al., 2005; Lu et al., 2012). However, the precise components of the membrane trafficking complexes participating in defence mechanisms are largely unknown. Here, we report the application of a candidate gene approach used for identification of barley genes that code for potential membrane trafficking proteins required for penetration defence to B. graminis f.sp. hordei.
Results
Transient‐induced gene silencing (TIGS) screening identifies membrane trafficking proteins with function in barley penetration defence
The technique of TIGS can be used to identify genes involved in immunity or resistance to grass powdery mildew (e.g. Collins et al., 2003; Douchkov et al., 2005; Hoefle et al., 2011; Rayapuram et al., 2012). To identify candidates, we used a keyword‐based approach to search databases for proteins that had been described to be involved in membrane trafficking in yeast, metazoans or other plants. Keyword‐based searches were not comprehensive, but involved RAB and ARF GTPases, proteins regulating GTPase activity, vesicle coat proteins, proteins of vesicle tethering complexes, yeast secretion (SEC) proteins and proteins containing C2 or VHS (Vps‐27, Hrs and STAM) membrane‐targeting domains. Keyword‐based searches were carried out in the barley HarvEST 35 annotation (http://harvest.ucr.edu/) and led to 135 hits. Plasmids containing expressed sequence tags (ESTs) of corresponding contigs were provided by Patrick Schweizer (IPK Gatersleben, Germany) and, after polymerase chain reaction (PCR) amplification, cDNA fragments were cloned into the Gateway‐compatible vector pIPKTA30N for TIGS according to Douchkov et al. (2005). This was successful for 78% of the ESTs, which we then further tested for gene function in TIGS experiments. We challenged barley epidermal cells after microprojectile‐mediated transformation with spores of B. graminis f.sp. hordei. Subsequently, we microscopically scored the susceptibility of transformed cells to fungal penetration and compared it in all individual experiments with that of empty vector controls and of a positive control for TIGS‐mediated reduction of susceptibility [TIGS of the MILDEW LOCUS O (MLO) susceptibility gene] using the green fluorescence protein (GFP) as a transformation marker. The average susceptibility index (percentage of penetrated cells relative to all transformed cells) was 14.2% over all control experiments in the initial TIGS screening with empty vectors. When TIGS of a candidate halved or doubled the susceptibility index in individual experiments, we selected the candidate for experimental repetition. Statistical analyses are based on five or more biological repetitions of the TIGS effects. Three TIGS constructs caused statistically significant effects. All appeared to affect basal penetration resistance, because knockdown supported the establishment of haustoria, which was documented by an increase in the relative susceptibility index when compared with the controls (set to 100%, Table 1). These TIGS constructs contained cDNA fragments of an HvARFA1b/c gene (EST HO30E03S, susceptibility index of 169% of the control), recently published to be involved in basal penetration resistance to B. graminis f.sp. hordei (Böhlenius et al., 2010), an HvExo70F‐like gene (EST HD14N02r, susceptibility index of 173% of the control), coding for a potential subunit of an exocyst complex, and HvCOG3 (EST HA14A08r), also known as SEC34 in yeast, coding for subunit 3 of the conserved oligomeric Golgi (COG) complex. The average susceptibility index after TIGS of HvCOG3 was 174% of the control.
Table 1.
Membrane trafficking genes significantly affect basal resistance on transient‐induced gene silencing (TIGS)
| Target gene (TIGR or GenBank accession of the full‐length gene) | Relative susceptibility index (number of independent experiments)† |
|---|---|
| Empty TIGS vector | 100% |
| HvMLO (Z83834) | 26% (n = 29)*** , † |
| HvARFA1b/c (TA29770_4513) | 169% (n = 9)* |
| HvCOG3 (AK249208) | 174% (n = 20)* |
| HvEXO70F‐like (AK362856) | 173% (n = 7)** |
TIGR, Institute for Genomic Research.
*P < 0.05, **P < 0.01, ***P < 0.001 (one‐sided Student's t‐test against 100%).
†Each experiment contained an empty vector control and an HvMLO‐TIGS positive control. The number of MLO‐TIGS experiments differs from the sum of all experiments in the table (n = 36) because MLO was used as a common positive control for more than one test construct in some of the experiments.
Function of HvCOG3 in protein secretion and Golgi stack assembly
We decided to investigate HvCOG3 and its function in more detail, because little is known about the function of the COG complex in plants. For this purpose, we isolated the full‐length cDNA sequence of HvCOG3 prepared from barley leaf RNA and generated an over‐expression construct driven by the cauliflower mosaic virus (CaMV) 35S promoter. Subsequent transient over‐expression of HvCOG3 in barley epidermal cells resulted in a significant reduction in the penetration rate of B. graminis f.sp. hordei (percentage of penetrated cells relative to all attacked cells) to 71% of the penetration observed in empty vector control experiments (Fig. 1). Hence, over‐expression and TIGS of HvCOG3 had opposing effects on the susceptibility of barley epidermal cells.
Figure 1.
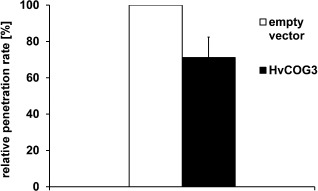
Transient over‐expression of HvCOG3 supports basal penetration resistance. Barley epidermal cells of detached barley cv. ‘Golden Promise’ leaves were transiently transformed with an HvCOG3 over‐expression construct, together with green fluorescence protein (GFP), by particle bombardment. As control, the empty pGY‐1 vector was used. Inoculation with spores of Blumeria graminis f.sp. hordei took place 2 days after transformation and microscopic analysis followed 2 days after inoculation. The columns represent the mean values of the relative penetration rate. The respective control was set to 100%. The mean values are based on seven independent over‐expression experiments. The bar represents the standard error; the difference of the means is significant at P < 0.05 according to Student's t‐test.
In yeast and mammals, COG3 is one of eight subunits of the COG tethering complex, which is involved in retrograde transport processes at the Golgi apparatus (Ungar et al., 2006). In addition to its function in retrograde trafficking, the COG complex is also required to maintain Golgi integrity and the functionality of the secretory pathway (Ungar et al., 2002; Zolov and Lupashin, 2005). Therefore, we examined the possibility that the interference with the HvCOG3 expression level in TIGS experiments may lead to defects in secretory processes, which may result in a defective plant defence against B. graminis f.sp. hordei. Therefore, we transiently expressed a secreted GFP (sGFP) and red‐fluorescing mCherry, together with the HvCOG3 RNA interference (RNAi) construct or the empty vector pIPKTA30N, in barley epidermal cells. Transient expression of sGFP leads to very low fluorescence intensities inside cells when compared with cells expressing cytosolic GFP (data not shown), because the majority of the protein is secreted into the apoplast where GFP does not fluoresce because of its sensitivity to low pH values (see Bartetzko et al., 2009). We examined the transformed cells for GFP fluorescence 2 days after bombardment using a confocal laser scanning microscope. To eliminate differences in the general expression potential of the analysed cells, the GFP fluorescence of each cell was normalized to the fluorescence intensity of co‐expressed mCherry. As expected, the fluorescence intensity in empty vector control cells expressing sGFP was low. By comparison, cells transformed with the HvCOG3 RNAi construct showed significantly enhanced sGFP fluorescence inside the cell (Fig. 2 ). This suggests that the secretion of sGFP is hampered when HvCOG3 is silenced. Fluorescence of sGFP was often strong at the nucleus or tubular structures in HvCOG3 RNAi experiments. This may reflect GFP restricted in the endoplasmic reticulum or GFP leaking out into the cytoplasm when Golgi transport is hampered.
Figure 2.
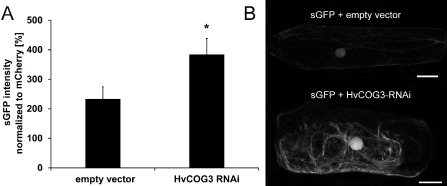
Analysis of green fluorescence protein (GFP) secretion in cells expressing the HvCOG3 RNA interference (RNAi) construct. GFP was N‐terminally fused with a signal peptide. The resulting construct (sGFP in pGY‐1) was transiently co‐transformed, together with either the empty vector pIPKTA30N as a control or the HvCOG3 RNAi construct and the transformation marker mCherry, into epidermal cells of detached barley leaves (cv. ‘Golden Promise’) and analysed by confocal laser scanning microscopy 2 days after transformation. (A) The intensity of sGFP in the cytoplasm of transformed cells was normalized against the fluorescence intensity of mCherry. In each of three independent repetitions, 50 cells were examined. All cells were imaged with the same excitation and detection settings. To warrant detection within the dynamic range of the system, the brightly fluorescing mCherry was recorded at low detector gain, whereas weakly fluorescing sGFP was recorded at higher detector gain. Average GFP fluorescence in HvCOG3 deficient cells is significantly higher than in control cells (Student's t‐test, P < 0.05). (B) Confocal laser scanning micrographs of barley epidermal cells expressing sGFP together with either the empty vector (top) or the HvCOG3 RNAi construct (bottom) 2 days after transformation. Photographs were taken using the same detection settings. Size bars, 20 μm.
The depletion of mammalian COG3 transcript leads to an extensive fragmentation of the Golgi complex into Golgi ministacks (Zolov and Lupashin, 2005). To test whether TIGS of HvCOG3 expression had a similar effect, we transiently co‐expressed the HvCOG3 RNAi construct together with a green fluorescing Golgi marker protein, sGFPHDEL (Hückelhoven and Panstruga, 2011). sGFPHDEL represents an in‐frame substitution of a part of barley CALRETICULIN 3 by GFP. It co‐localizes with the known Golgi marker GmMAN1‐RFP (Yang et al., 2005), but provides a much brighter fluorescence signal in barley. We analysed transformed cells 3 days after bombardment using fixed settings for confocal laser scanning microscopy. The microscope detector gain was adjusted such that it was able to distinguish small potentially fragmented Golgi bodies, which fell below the detection limit, from larger detectable Golgi bodies. Detectable Golgi bodies were counted in whole‐cell maximum projections and cells were assigned into one of the following categories: 0–10, 11–20, 21–50 and more than 50 Golgi bodies per cell. The results indicated a significantly different distribution of brightly fluorescing Golgi bodies in HvCOG3 RNAi cells 3(Fig. 3). Cells with only 0–10 Golgi bodies per cell were ten times more abundant in HvCOG3 RNAi cells, whereas the number of cells exhibiting 50 or more Golgi bodies was clearly reduced in these cells when compared with the controls. This suggests that TIGS of HvCOG3 affects the formation or induces fragmentation of Golgi bodies/stacks.
Figure 3.
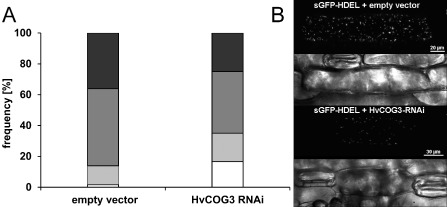
Analysis of Golgi body frequency in cells expressing the HvCOG3 RNA interference (RNAi) construct. Epidermal cells of detached barley leaves (cv. ‘Golden Promise’) were simultaneously transformed with a green fluorescing Golgi marker (sGFPHDEL) and the empty vector pIPKTA30N or the HvCOG3 RNAi construct. Transformed cells were analysed 3 days after particle bombardment using confocal laser scanning microscopy, and the number of brightly fluorescing Golgi bodies per cell was counted. (A) The cells were classified into four categories with 0–10 (white), 11–20 (light grey), 21–50 (dark grey) and >50 (black) Golgi bodies per cell. Statistical analysis using a χ 2 test showed that the expression of the two constructs led to a significantly different distribution concerning the number of Golgi bodies per cell, with a probability of 99.5%. Seventy cells were examined per variant in three independent repetitions. All cells of the same experiment were imaged with the same excitation and detection settings. (B) Confocal laser scanning whole‐cell projections of barley epidermal cells expressing sGFPHDEL together with either the empty pIPKTA30N vector (top two photographs) or the HvCOG3 RNAi construct (bottom two photographs) 3 days after bombardment. The transmission channels show the cell borders of scanned cells.
Functional characterization of putative barley COG complex subunits
Because COG3 belongs to a complex that contains seven additional subunits, we used human and Arabidopsis COG sequence information (Koumandou et al., 2007; Quental et al., 2010) for blast research in the National Center for Biotechnology Information (NCBI) and Institute for Genomic Research (TIGR) sequence databases to identify additional putative homologues of COG complex components in barley. The barley genome seemed to harbour a single copy gene of each COG complex subunit, and the same is true for rice (data not shown). According to ClustalW2.0 protein alignment, the identities between the human and plant COG complex subunits range between 18% and 35%, whereas the identities between the plant subunits of the barley and Arabidopsis COG complex range between 47% and 71%. COG3, COG4 and COG6 represent the most conserved subunits of the COG complex. The ClustalW2.0 protein alignment scores, derived from the comparison of the human, Arabidopsis and barley COG subunits, are summarized in Table S1 (see Supporting Information). Accession numbers are given in Table S2 (see Supporting Information).
For further investigations on the potential involvement of the COG complex in the interaction of barley with B. graminis f.sp. hordei, we isolated cDNA fragments of the remaining seven barley COG complex subunits for TIGS. In addition to HvCOG3 (Table 1), knockdown of HvCOG1 led to a significant increase (to 167% of the control) in the susceptibility index. Interestingly, COG1 is the central subunit of the complex and is supposed to be a direct interaction partner of COG3 within the COG complex (Loh and Hong, 2004; Ungar et al., 2006). The abundance of both COG subunits seems to be important for basal penetration resistance to B. graminis f.sp. hordei. TIGS of the other HvCOG complex subunits did not reveal significant effects. Nevertheless, there was a tendency to enhanced susceptibility of barley to B. graminis f.sp. hordei after TIGS of HvCOG2, HvCOG4 and HvCOG5 (Fig. 4).
Figure 4.
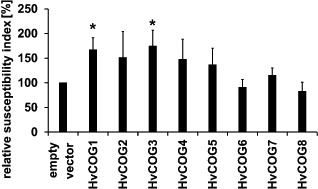
Functional analysis of the barley conserved oligomeric Golgi (COG) complex subunits in barley interaction with Blumeria graminis f.sp. hordei. Epidermal cells of detached barley leaves (cv. ‘Golden Promise’) were transiently transformed with the marker gene green fluorescence protein (GFP) and the transient‐induced gene silencing (TIGS) construct of one of the eight HvCOG subunit genes or empty pIPKTA30N vector via particle bombardment. Leaves were inoculated 2 days after transformation and the microscopic evaluation took place 2 days after inoculation. The susceptibility index of the empty vector control was set to 100%, and the TIGS results of HvCOG complex subunits are given as the mean values of at least five independent experiments. The bars represent standard errors. In addition to HvCOG3 (please note data for HvCOG3 are identical to those in Table 1, given here for better comparability of the results), TIGS of HvCOG1 significantly enhanced the susceptibility according to Student's t‐test (P < 0.05).
Functional characterization of potential HvCOG3 interaction partners
Suvorova et al. (2002) identified protein interaction partners of yeast COG3/SEC34 by tandem affinity purification tagging. A nucleotide blast search against the TIGR database identified barley homologues of the proteins co‐purified with COG3 from yeast. We successfully isolated cDNAs of a small RAB GTPase (HvYPT1‐like), a subunit of the COPI vesicle coat (HvCOPIγ‐like) and three vesicle‐SNAREs (HvSEC22‐like, HvYKT6‐like and HvVTI1‐like) and introduced them into TIGS and over‐expression vectors [for accession numbers, see Table S3 (Supporting Information)]. Database search identified only a potential C‐terminal fragment of the vesicle‐SNARE HvGOS1‐like sequence. Isolation of the homologous sequence of a t‐SNARE protein, SED5, failed in several attempts. Transient transformation experiments revealed that knockdown of HvYPT1‐like (susceptibility index of 148% of the control) and HvCOPIγ‐like (susceptibility index of 129% of the control) and over‐expression of HvVTI1‐like (susceptibility index of 197% of the control) significantly enhanced the susceptibility to B. graminis f.sp. hordei (Fig. 5). Taken together, the functional analysis of HvCOG3 and further proteins that might be involved in Golgi transport suggests that this transport is important for barley defence against B. graminis f.sp. hordei, as several of these proteins enhanced susceptibility when they were knocked down or over‐expressed.
Figure 5.
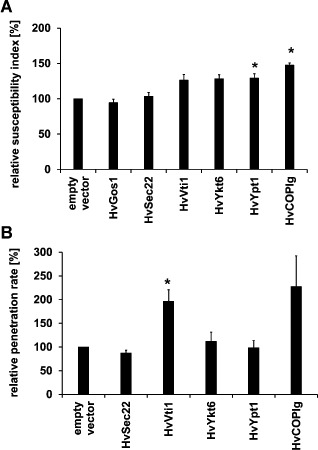
Functional characterization of potential HvCOG3 interaction partners in transient‐induced gene silencing (TIGS) and over‐expression experiments. Detached leaf segments of the barley cv. ‘Golden Promise’ were transiently transformed via particle bombardment. Potential interaction partners were either knocked down by TIGS (A) or over‐expressed (B), together with the transformation marker green fluorescence protein (GFP). The empty vectors pIPKTA30N (in A) and pGY‐1 (in B) served as controls. The leaf segments were inoculated 2 days after transformation and the microscopic evaluation was conducted 2 days after infection. The susceptibility index (frequency of penetrated cells relative to all cells transformed, A) and penetration rate (frequency of penetrated cells relative to all cells attacked, B) of the empty vector control were set to 100%. Columns for the potential HvCOG3 interaction partners represent mean values relative to the control of five to six independent experiments in (A) and three to six over‐expression experiments in (B). Knockdown of HvYPT1‐like, HvCOPIγ‐like and over‐expression of HvVTI1‐like significantly enhanced the susceptibility at P < 0.05 according to Student's t‐test.
Subcellular localization of the RAB GTPase HvYPT1‐like
We generated N‐terminal fusions of GFP with HvCOG3 and HvYPT1‐like and a C‐terminal fusion with HvCOPIγ‐like. However, only GFP fusions of HvYPT1‐like gave fluorescence signals sufficiently bright and specific to study subcellular distribution by confocal laser scanning microscopy in vivo. To determine the localization in nonattacked cells, the construct was transformed transiently into barley epidermal cells by particle bombardment, together with mCherry as a cytosolic and nuclear marker or with the Golgi marker GmMAN1‐RFP (Yang et al., 2005). The GFP fusion constructs of HvYPT1‐like produced bright fluorescence signals and partly co‐localized with GmMAN1‐RFP (see Fig. 6B,C). In addition, GFP‐YPT1‐like labelled the cytoplasm. Moreover, GFP‐HvYPT1‐like labelled Golgi bodies that accumulated, together with cytoplasmic aggregations, at the site of interaction with B. graminis f.sp. hordei (Fig. 6D).
Figure 6.
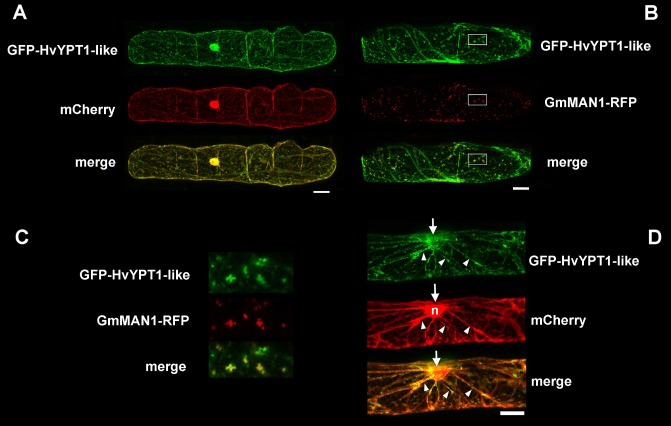
Subcellular localization of GFP‐HvYPT1‐like. GFP‐HvYPT1‐like was transiently expressed in barley epidermal cells (cv. ‘Golden Promise’), together with either the cytoplasmic and nuclear marker protein (mCherry) (A, D) or the Golgi marker protein GmMAN1‐RFP (B, C). (C) Enlargements of the boxed areas in (B) showing the overlay of GFP‐HvYPT1‐like with the Golgi marker (yellow pixel in the merge image). (D) Polarization of the cytoplasm (mCherry) and GFP‐HvYPT1‐like‐labelled Golgi bodies in cytoplasmic strands (arrow heads) to the site of nonsuccessful attack by Blumeria graminis f.sp. hordei (arrows). The nucleus (n in the mCherry channel) is also attracted to the site of interaction by 24 h post‐inoculation. Photographs are whole‐cell projections of single optical sections at 2‐μm increments recorded by confocal laser scanning microscopy. Size bars, 20 μm.
Discussion
Vesicular traffic is a major mechanism of cytoplasmic transport in eukaryotes. It includes the budding and release of vesicles from a donor membrane, as well as its tethering to and fusion with the target membrane. One can distinguish the transport of cargo in two general directions. Anterograde transport represents the transport of material towards its destination, e.g. the transport of newly synthesized proteins from the endoplasmic reticulum via the Golgi to the plasma membrane or the vacuole. The retrograde transport route is primarily responsible for the recycling of escaped proteins and lipids back to the home compartments, and also comprises endocytosis. A balance of anterograde and retrograde trafficking may be pivotal for membrane homeostasis and function of vesicle transport. Plant vesicle transport is comparably weakly understood. However, many proteins of the well‐studied transport and secretory machinery of yeast are conserved in other eukaryotes, including plants (Koumandou et al., 2007). Their function in plants is often unknown and their possible involvement in plant–pathogen interactions is an emerging field of research. Recently, an increasing number of publications have suggested an important function of membrane trafficking in interactions of plants with mycorrhizal fungi and pathogens, including powdery mildew fungi (e.g. Böhlenius et al., 2010; Eschen‐Lippold et al., 2012; Genre et al., 2012; Hückelhoven and Panstruga, 2011; Koh et al., 2005; Lu et al., 2012; Nielsen et al., 2012; Uemura et al., 2012). We have shown here that proteins, which are homologues of well‐characterized mammalian or yeast membrane trafficking components, are required for basal penetration resistance to B. graminis f.sp. hordei. Basal resistance is expressed as a quantitative trait in barley, and vesicle trafficking genes are over‐represented in loci for quantitative resistance to fungal leaf pathogens (Schweizer and Stein, 2011).
In a first round of screening, three candidates changed significantly basal resistance on knockdown by TIGS. HvARFA1b/c was one of the candidate genes apparently required for basal penetration resistance to B. graminis f.sp. hordei. HvARFA1b/c has been identified previously by Böhlenius et al. (2010) to be required for penetration resistance, papillary callose deposition and subcellular accumulation of HvROR2, a barley subunit of a ternary SNARE complex at the plasma membrane involved in basal penetration resistance to powdery mildew. HvARFA1b/c was independently included into the screening because of its annotation as an ARF GTPase potentially involved in membrane budding and was pulled out because of its strong and statistically significant effect. The recovery of HvARFA1b/c supports the results of Böhlenius et al. (2010) and validates the methodology adapted from Douchkov et al. (2005) for the identification of components of basal resistance to powdery mildew.
HvEXO70F‐like was also found to be involved in basal resistance to fungal entry. The protein contains an exocyst70 domain and may thus be involved in vesicle tethering via an exocyst complex. The exocyst is an oligomeric vesicle tethering complex possibly involved in exocytosis. Recently, an Arabidopsis EXO70B2 protein has been described to modulate the cellular defence response to attack from nonadapted B. graminis f.sp. hordei. Interestingly EXO70B2 can interact in a yeast two‐hybrid assay with SNAP33, a part of the ternary SNARE complex involved in basal penetration resistance (Collins et al., 2003; Pecenková et al., 2011). In plants, the EXO70 family has diversified when compared with animals or yeast. This is particularly true for the EXO70F subfamily in grasses. Hence, one may speculate about a specific function of HvEXO70F‐like in pathogen defence (Cvrčková et al., 2012).
The third TIGS construct that gave significant results during screening targeted HvCOG3, a subunit of the COG complex involved in vesicle tethering in retrograde trafficking at the Golgi apparatus (Miller and Ungar, 2012). Little is known about the function of the COG complex in plants (Ishikawa et al., 2008), but its structural conservation suggests similar functions to those described for yeast and mammals. An intact COG complex is also a prerequisite for protein N‐glycosylation in the Golgi apparatus (Pokrovskaya et al., 2011).
Over‐expression of HvCOG3 enhanced basal resistance to B. graminis f.sp. hordei. This was consistent with the result that TIGS of HvCOG3 had the opposite effect. However, as COG3 is supposedly a subunit of an oligomeric complex, over‐expression of HvCOG3 alone was not necessarily expected to have a defence‐supporting effect. This could be explained if HvCOG3 has an as yet unknown regulatory function on Golgi membrane trafficking. An alternative explanation could be that HvCOG3 is a possible target of fungal virulence, and over‐expression compensates for the inhibitory effects of fungal effectors. Interestingly, the COG complex is hijacked for virulence by Chlamydia trachomatis, an obligate intracellular bacterium, that survives and multiplies in nonlytic membrane‐surrounded inclusions in human host cells (Pokrovskaya et al., 2012).
In a more targeted approach, focusing on the other seven subunits of the COG complex and six potential interaction partners of HvCOG3, we identified three of 13 TIGS constructs that significantly weakened basal penetration resistance to fungal entry. In addition, over‐expression of VTI1‐like weakened basal resistance. This, together with the finding that GFP‐fused YPT1‐like proteins co‐localized with Golgi stacks in barley epidermal cells, indicates that Golgi membrane trafficking is important for basal penetration resistance to B. graminis f.sp. hordei.
In yeast, homologues of the HvCOG complex components investigated here, as well as potential COG3 interaction partners (HvSEC22‐like, HvYKT6‐like, HvCOPIγ‐like, HvVTI1‐like and HvYPT1‐like), participate in retrograde Golgi transport (Suvorova et al., 2001, 2002). The yeast COG complex, which resides mainly at the cis‐Golgi compartment (Kim et al., 2001), is thought to be responsible for the tethering of COPI vesicles to the cis‐Golgi membrane (Ungar et al., 2006). During this work, HvCOPIγ‐like, HvVTI1‐like and HvYPT1‐like, like HvCOG3, were found to alter the susceptibility of barley to the barley powdery mildew fungus. COPIγ is one subunit of the coat of COPI vesicles, which are responsible for retrograde transport and recycling of proteins in between the Golgi and from the Golgi to the endoplasmic reticulum (e.g. McMahon and Mills, 2004). Interestingly, in an independent screening, HvCOPIγ has been identified as being required for nonhost resistance of barley to penetration by nonadapted B. graminis f.sp. tritici (D. Douchkov and P. Schweizer, personal observation).
Over‐expression of VTI1‐like and HvCOPIγ‐like weakened basal resistance, although this was not significant for HvCOPIγ‐like. The over‐expression of single components of protein complexes might sequester interaction partners from functional complexes and therefore exert a negative effect on protein function. VTI1 is a SNARE protein responsible for the fusion of vesicles during several transport steps. In yeast, this SNARE protein is involved in the transport from the trans‐Golgi network to the prevacuolar compartment, in transport to the vacuole and in retrograde transport to the cis‐Golgi (Fischer von Mollard et al., 1997; Fischer von Mollard and Stevens, 1999; Lupashin et al., 1997). The third, HvYPT1‐like, is a small RAB GTPase similar to yeast YPT1, rice RAB1C1 (locus Os01g0179700) and Arabidopsis ARA5/RABD2A (locus At1g02130), which is involved in protein secretion (Pinheiro et al., 2009). Yeast YPT1 is involved in different tethering events occurring during anterograde and retrograde transport (Hutagalung and Novick, 2011; Jones et al., 2000; Suvorova et al., 2002).
The transient nature of the TIGS assay may allow for the identification of genes which would have pleiotropic effects when permanently silenced or knocked out during plant development. Hence, the question arises as to whether the TIGS effects observed here indicate a specific function in defence of COG complex proteins and associated proteins, or simply reflect a general weakening of the plant cells. We favour the interpretation that these proteins are probably generally involved in the maintenance of Golgi integrity and function. However, the COG complex and associated proteins may be co‐opted for defence‐associated functions in cells attacked by B. graminis f.sp. hordei (see also Assaad et al., 2004; Kwon et al., 2008). This view is supported by the following indications. COG complex genes are single copy genes and probably essential for general Golgi membrane transport. An essential function of COG3 in plants is supported because attempts to produce stable transgenic knockdown barley plants of HvCOG3 failed as a result of lethality of the RNAi construct during regeneration in tissue culture (G. Hensel, IPK Gatersleben, Germany, personal communication). In addition, TIGS of HvCOG3 inhibited general secretion of sGFP, such that the protein accumulated intracellularly. TIGS of HvCOG3 also reduced the number of Golgi bodies which reached a size sufficient to be visualized under the fixed settings of confocal laser scanning microscopy. Together, these observations indicate that HvCOG3 is a general factor of Golgi membrane homeostasis, and hence is generally required for protein secretion, which is similar to its function in yeast and mammals. Protein secretion may be pivotal to polarized plant defence of fungal penetration (Lipka et al., 2010). Up‐regulation of the protein secretory pathway at the transcriptional level may reflect a higher demand for flux through the pathway during defence (Wang et al., 2005). For the interaction of barley with B. graminis, this has already been shown for proteins of a ternary SNARE complex at the plasma membrane (Zierold et al., 2005) and for chaperones from the endoplasmic reticulum (Eichmann et al., 2006). We analysed the expression of the eight COG genes of barley and six potential interaction partners of COG3 in pathogen response in public databases. This supported the pathogen‐responsive expression of most of these 14 genes in barley. A cluster analysis of all barley challenge experiments in Genevestigator (Hruz et al., 2008) further revealed a large cluster of independent experiments, in which most of the genes were up‐regulated in response to either B. graminis f.sp. hordei or the rust fungus Puccinia graminis (Fig. S1, see Supporting Information). This indicates that these genes are responsive to pathogenesis‐related signalling. Together, our data suggest that the barley COG complex and, potentially, COG3‐associated proteins are general Golgi trafficking components that are involved in plant defence against penetration by B. graminis f.sp. hordei.
Experimental Procedures
Plant material, growth conditions and pathogen
Transient transformation experiments were carried out with the powdery mildew‐susceptible barley (Hordeum vulgare L.) cv. ‘Golden Promise’. Total RNA from inoculated and noninoculated leaves of the susceptible barley cv. ‘Ingrid’ served as template for the isolation of cDNAs encoding COG complex subunits or their interactors. Plants were grown in a growth chamber (Conviron, Winnipeg, MB, Canada) at 18 °C, with a relative humidity of 65% and a photoperiod of 16 h light at 150 μmol/s/m2 photon flux density. Barley leaves were infected with the powdery mildew fungus B. graminis f.sp. hordei (Bgh) race A6 at a density of about 150 conidia/mm2, which was maintained on the barley cultivar ‘Golden Promise’ at the conditions described above.
Cloning of membrane transport‐associated ESTs into the TIGS vector pIPKTA30N
The candidate genes for RNAi screening, which were supposedly involved in membrane trafficking and secretion, were selected via a keyword‐based search in the EST database (http://harvest.ucr.edu/) of the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK, Gatersleben, Germany; Zhang et al., 2004). ESTs were provided in one of the following vectors: pCRblunt, pBluescript SK+, pSORT or pLambdaZAP. cDNAs of these ESTs were amplified by PCR using vector‐specific primer pairs (Table S4, see Supporting Information), and subcloned via the Gateway entry vector pIPKTA38 into the pIPKTA30N Gateway destination vector, as described by Douchkov et al. (2005).
Isolation and cloning of COG complex‐associated cDNA fragments
Further COG3 interaction candidate cDNAs were isolated from a cDNA pool comprising inoculated and noninoculated leaves of the barley cultivar ‘Ingrid’. Primers used for the amplification of cDNA fragments for TIGS or full‐length cDNAs for over‐expression are listed in Tables S5 and S6 (see Supporting Information). For TIGS experiments, the respective cDNA fragments were subcloned into the Gateway‐based vector system pIPKTA38 and pIPKTA30N (Douchkov et al., 2005, see above), whereas the plant expression vector pGY1 was used for over‐expression experiments (Schweizer et al., 1999). Subcloning for over‐expression was carried out via PCR‐mediated introduction of unique restriction sites XbaI and PstI at the end of full‐length cDNAs.
Transient transformation of barley epidermal cells for gene function analysis
The first leaves of 7‐day‐old barley plants were transiently transformed via gold particle delivery. Per shot, 1.65 mg of 1.0‐μm gold particles were coated with 7 mg of pGY1‐GFP (GFP under the control of the CaMV 35S promoter; Schweizer et al., 1999) as transformation marker and 7 mg of pGY1/pIPKTA30N vector containing the gene of interest or the empty vector, respectively. The ballistic transformation of barley epidermal cells was performed using the biolistic PDS‐1000/He system (Biorad, Munich, Germany) according to Douchkov et al. (2005). Detached leaf segments were inoculated for TIGS experiments 2 days after transformation. Microscopic analysis took place 2 days after inoculation. For TIGS, we recorded the relative susceptibility index (percentage of penetrated cells relative to all cells transformed) and, for over‐expression, we recorded the relative penetration rate (percentage of penetrated cells relative to all cells attacked by the fungus). Both parameters gave similar results (Douchkov et al., 2005). For statistics, we calculated P values based on Student's t‐test for relative values against the empty vector control.
Analysis of the impact of HvCOG3 depletion on protein secretion
To examine secretory defects in HvCOG3 deficient cells, an sGFP protein was used. To create this, we amplified GFP with primers 5′‐GGATCCATGGTGAGCAAGGGCGAG‐3′ (creating a BamHI restriction site) and 5′‐TCATTTGTACAGCTCGTCCAT‐3′ from pGY‐1‐GFP (Schweizer et al., 1999), and cloned it into the BamHI restriction site of empty pGY‐1. The coding sequence of the signal peptide of an Arabidopsis basic chitinase protein was amplified by PCR from mGFP5 endoplasmic reticulum (Haseloff et al., 1997) using primers 5′‐CCCGGGGATCGATCCAAGGA‐3′ and 5′‐CCCGGGGGAATTCGGCCGAGG‐3′ (both creating a SmaI restriction site) and inserted in frame in front of GFP using the SmaI site. We co‐transformed barley epidermal cells with 1 μg of pGY‐1‐sGFP, together with 1 μg of pGY‐1‐mCherry and 1 μg of either empty pIPKTA30N vector or pIPKTA30N‐HvCOG3. sGFP‐expressing cells were analysed using a Leica TSC SP5 confocal laser scanning microscope (Leica Microsystems CMS GmbH, Mannheim, Germany). GFP fluorescence was excited with a 488‐nm laser line and detected between 496 and 530 nm. mCherry was excited at 561 nm and detected between 570 and 625 nm. To show disturbed secretion, we measured the sGFP fluorescence intensities in HvCOG3 RNAi and empty vector control cells, and normalized them to the fluorescence intensities of mCherry in each cell. In three independent experiments, we measured GFP fluorescence in 50 cells per variant. Statistical analysis was performed using a two‐sided Student's t test.
Analysis of the impact of HvCOG3 depletion on Golgi stacks
sGFPHDEL was used to investigate the number of brightly fluorescing Golgi bodies in HvCOG3 deficient cells. For the cloning of sGFPHDEL, the barley CALRETICULIN 3 (TIGR ID TA32081_4513) full‐length sequence was amplified from barley cDNA using primers 5′‐GTCGACGCCACCACCTACTCTTCGTC‐3′ (creating a SalI restriction site) and 5′‐CTGCAGTGTCAAATCCCAGCTTCTCC‐3′ (creating a PstI restriction site), and cloned into the SalI/PstI restriction sites of the pGY‐1 expression vector (Schweizer et al., 1999). GFP was amplified by PCR using primers 5′‐GGATCCCATGGTGAGCAAGGGCGAG‐3′ and 5′‐GGATCCTTGTACAGCTCGTCCAT‐3′ (both creating a BamHI restriction site) from pGY‐1‐GFP (Schweizer et al., 1999). GFP was inserted in frame into the HvCRT3 coding sequence using two internal BamHI restriction sites, thereby replacing a 501‐bp fragment in between the predicted signal peptide and the HDEL endoplasmic reticulum retention signal. Barley epidermal cells were co‐bombarded with 1 μg of sGFPHDEL plasmid, together with 1 μg of either empty pIPKTA30N vector or pIPKTA30N‐HvCOG3. sGFPHDEL‐expressing cells were analysed using confocal laser scanning microscopy. GFP fluorescence was excited with a 488‐nm laser line and detected between 496 and 530 nm. To quantify the differences in the assembly of Golgi bodies in HvCOG3 RNAi and empty vector control cells, brightly fluorescing Golgi bodies per cell were counted, and classified into four categories: 0–10, 10–20, 20–50 or more than 50 Golgi bodies per cell. The detection settings were chosen at an intentionally low level to obtain GFP signals only from brightly fluorescing Golgi bodies. All cells of the same experiment were imaged with the same excitation and detection settings. We counted Golgi bodies in whole‐cell maximum projections of 2‐μm increments for 20–30 cells per variant. The results were averaged over three independent experiments. To test for a significantly different distribution of the different categories in empty vector and HvCOG3 RNAi cells, we used the χ 2 test.
Subcelllular localization of GFP‐tagged HvYPT1‐like
For the generation of GFP‐HvYPT1‐like, GFP (ΔSTOP) cDNA was excised from pGY1‐GFP (Schultheiss et al., 2003) with BamHI and inserted in frame into the BamHI restriction site of pGY1‐YPT1‐like, creating an N‐terminal fusion. The subcellular localization of GFP‐HvYPT1‐like was examined in planta by confocal laser scanning microscopy. Barley cv. ‘Golden Promise’ epidermal cells were transformed by particle bombardment with 0.24 mg of 1.0‐μm gold particles coated with 1 μg of GFP‐HvYPT1‐like together with 1 μg of mCherry or GmMAN1‐RFP (Yang et al., 2005) expression plasmids, and analysed 2 days after bombardment. GFP was excited with a 488‐nm laser line and detected between 496 and 530 nm; red fluorescing GmMAN1‐RFP and mCherry were excited with a 561‐nm laser line and detected between 570 and 625 nm. In order to avoid channel cross‐talk, cells were scanned sequentially, if required.
Accession numbers: HvMLO (Z83834), HvARFA1b/c (TA29770_4513), HvEXO70F‐like
(AK362856), HvCOG1 (AK370785), HvCOG2 (AK370312), HvCOG3 (AK249208), HvCOG4
(AK356539), HvCOG5 (AK252011), HvCOG6 (AK354922), HvCOG7 (AK363189), HvCOG8
(AK248490), HvYPT1‐like (TA34665_4513), HvGOS1‐like (TA36193_4513), HvYKT6‐like
(TA38338_4513), HvSEC22‐like (TA49055_4513), HvVTI1‐like (TA38030_4513), HvCOPIglike
(BAJ99125.1).
Supporting information
Fig. S1 Hierarchical clustering of barley gene expression experiments at Genevestigator 4. Analysis was carried out for conserved oligomeric Golgi (COG) subunit genes and genes of putative interaction partners of COG3. Gene IDs on barley 1 chips are: HvCOG1, Contig13332_at; HvCOG2, Contig11948_at; HvCOG3, Contig7930_at; HvCOG4, Contig11310_at; HvCOG5, Contig13262_at; HvCOG6, Contig10684_at; HvCOG7, Contig14245_at; HvCOG8, Contig9113_at; HvGOS1, Contig12134_at; HvYKT6, Contig8337_at; HvSEC22, Contig18727_at; HvVTI1, Contig9598_at; HvYPT1, Contig4865_s_at; HvCOPIg, Contig7764_at and/or Contig7765_at. Colour code shows log2 transformed changes in expression (from −2.5 to +2.5, i.e. from 18% to 566% of the expression level of the respective control).
Table S1 Protein sequence similarity scores for subunits of the human, Arabidopsis and barley conserved oligomeric Golgi (COG) complex.
Table S2 Accession/locus numbers of human and plant conserved oligomeric Golgi (COG) subunits.
Table S3 Interaction partners of conserved oligomeric Golgi 3 (COG3) identified in yeast by Suvorova et al. (2002) and their barley homologues.
Table S4 Oligo DNA primers for the isolation of vector‐provided sequences for transient‐induced gene silencing (TIGS) construct generation.
Table S5 Oligo DNA primers for isolation of cDNA fragments for transient‐induced gene silencing (TIGS) construct generation.
Table S6 Oligo DNA primers for isolation of full‐length cDNA sequences for over‐expression constructs.
Acknowledgements
We are grateful to David Robinson (University of Heidelberg, Germany) for providing the GmMAN1‐RFP construct and to Gregor Langen (University of Giessen, Germany) for providing pGY‐1‐mCherry. We are grateful to Angela Alkofer and Ernst Bernges for excellent technical assistance. This project was supported by a grant from the Federal Ministry of Research and Education (GABI‐phenome‐0315056D). The authors declare no conflict of interest.
Present address: The School of Life Sciences, University of Warwick, Gibbet Hill Campus, Coventry, CV4 7AL, UK.
References
- An, Q. , Ehlers, K. , van Kogel, K.‐H., Bel, A.J.E. and Hückelhoven, R. (2006a) Multivesicular compartments proliferate in susceptible and resistant MLA12‐barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 172, 563–576. [DOI] [PubMed] [Google Scholar]
- An, Q. , Hückelhoven, R. , van Kogel, K.‐H. and Bel, A.J.E. (2006b) Multivesicular bodies participate in a cell wall‐associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell. Microbiol. 8, 1009–1019. [DOI] [PubMed] [Google Scholar]
- An, Q. , van Bel, A.J.E. and Hückelhoven, R. (2007) Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2, 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad, F.F. , Qiu, J.L. , Youngs, H. , Ehrhardt, D. , Zimmerli, L. , Kalde, M. , Wanner, G. , Peck, S.C. , Edwards, H. , Ramonell, K. , Somerville, C.R. and Thordal‐Christensen, H. (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15, 5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartetzko, V. , Sonnewald, S. , Vogel, F. , Hartner, K. , Stadler, R. , Hammes, U.Z. and Börnke, F. (2009) The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall‐associated defense responses. Mol. Plant–Microbe Interact. 22, 655–664. [DOI] [PubMed] [Google Scholar]
- Bednarek, P. , Piślewska‐Bednarek, M. , Ver Loren van Themaat, E. , Maddula, R.K. , Svatoš, A. and Schulze‐Lefert, P. (2011) Conservation and clade‐specific diversification of pathogen‐inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 192, 713–726. [DOI] [PubMed] [Google Scholar]
- Böhlenius, H. , Mørch, S.M. , Godfrey, D. , Nielsen, M.E. and Thordal‐Christensen, H. (2010) The multivesicular body‐localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin‐dependent preinvasive basal defense in barley. Plant Cell, 22, 3831–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Collins, N.C. , Thordal‐Christensen, H. , Lipka, V. , Bau, S. , Kombrink, E. , Qiu, J.‐L. , Hückelhoven, R. , Stein, M. , Freialdenhoven, A. , Somerville, S.C. and Schulze‐Lefert, P. (2003) SNARE‐protein‐mediated disease resistance at the plant cell wall. Nature, 425, 973–977. [DOI] [PubMed] [Google Scholar]
- Cvrčková, F. , Grunt, M. , Bezvoda, R. , Hála, M. , Kulich, I. , Rawat, A. and Zárský, V. (2012) Evolution of the land plant exocyst complexes. Front. Plant Sci. 3, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov, D. , Nowara, D. , Zierold, U. and Schweizer, P. (2005) A high‐throughput gene‐silencing system for the functional assessment of defense‐related genes in barley epidermal cells. Mol. Plant–Microbe Interact. 18, 755–761. [DOI] [PubMed] [Google Scholar]
- Eichmann, R. and Hückelhoven, R. (2008) Accommodation of powdery mildew fungi in intact plant cells. J. Plant Physiol. 165, 5–18. [DOI] [PubMed] [Google Scholar]
- Eichmann, R. , Biemelt, S. , Schäfer, P. , Scholz, U. , Jansen, C. , Felk, A. , Schafer, W. , Langen, G. , Sonnewald, U. , Kogel, K.H. and Hückelhoven, R. (2006) Macroarray expression analysis of barley susceptibility and nonhost resistance to Blumeria graminis . J. Plant Physiol. 163, 657–670. [DOI] [PubMed] [Google Scholar]
- Eschen‐Lippold, L. , Landgraf, R. , Smolka, U. , Schulze, S. , Heilmann, M. , Heilmann, I. , Hause, G. and Rosahl, S. (2012) Activation of defense against Phytophthora infestans in potato by down‐regulation of syntaxin gene expression. New Phytol. 193, 985–996. [DOI] [PubMed] [Google Scholar]
- Feechan, A. , Kabbara, S. and Dry, I.B. (2011) Mechanisms of powdery mildew resistance in the Vitaceae family. Mol. Plant Pathol. 12, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard, G. and Stevens, T.H. (1999) The Saccharomyces cerevisiae v‐SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell 10, 1719–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard, G. , Nothwehr, S.F. and Stevens, T.H. (1997) The yeast v‐SNARE Vti1p mediates two vesicle transport pathways through interactions with the t‐SNAREs Sed5p and Pep12p. J. Cell Biol. 137, 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei dit Frey, N. and Robatzek, S. (2009) Trafficking vesicles: pro or contra pathogens? Curr. Opin. Plant Biol. 12, 437–443. [DOI] [PubMed] [Google Scholar]
- Genre, A. , Ivanov, S. , Fendrych, M. , Faccio, A. , Zársky, V. , Bisseling, T. and Bonfante, P. (2012) Multiple exocytotic markers accumulate at the sites of perifungal membrane biogenesis in arbuscular mycorrhizas. Plant Cell Physiol. 53, 244–255. [DOI] [PubMed] [Google Scholar]
- Green, J.R. , Carver, T.L.W. and Gurr, S.J. (2002) The formation and function of infection structures In: The Powdery Mildews: A Comprehensive Treatise (Bélanger R.R., Bushnell W.R., Dik A.J. and Carver T.L.W., eds), pp. 66–82. St. Paul, MN: APS Press, American Phytopatholgical Society. [Google Scholar]
- Haseloff, J. , Siemering, K.R. , Prasher, D.C. and Hodge, S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA, 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy, K. , Cherk, C. and Somerville, S. (2009) Host–pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 12, 406–413. [DOI] [PubMed] [Google Scholar]
- Hoefle, C. , Huesmann, C. , Schultheiss, H. , Börnke, F. , Hensel, G. , Kumlehn, J. and Hückelhoven, R. (2011) A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell, 23, 2422–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz, T. , Laule, O. , Szabo, G. , Wessendorp, F. , Bleuler, S. , Oertle, L. , Widmayer, P. , Gruissem, W. and Zimmermann, P. (2008) Genevestigator v3, a reference expression database for the meta‐analysis of transcriptomes. Adv. Bioinformatics, 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven, R. and Panstruga, R. (2011) Cell biology of the plant–powdery mildew interaction. Curr. Opin. Plant Biol. 14, 738–746. [DOI] [PubMed] [Google Scholar]
- Hutagalung, A.H. and Novick, P.J. (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, T. , Machida, C. , Yoshioka, Y. , Ueda, T. , Nakano, A. and Machida, Y. (2008) EMBRYO YELLOW gene, encoding a subunit of the conserved oligomeric Golgi complex, is required for appropriate cell expansion and meristem organization in Arabidopsis thaliana . Genes Cells, 13, 521–535. [DOI] [PubMed] [Google Scholar]
- Jones, S. , Newman, C. , Liu, F. and Segev, N. (2000) The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell, 11, 4403–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.‐W. , Massey, T. , Sacher, M. , Pypaert, M. and Ferro‐Novick, S. (2001) Sgf1, a new component of the Sec34/Sec35 complex. Traffic, 2, 820–830. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y. , Kobayashi, I. , Funaki, Y. , Fujimoto, S. , Takemoto, T. and Kunoh, H. (1997) Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non‐host resistance in barley coleoptile cells. Plant J. 11, 525–537. [Google Scholar]
- Koh, S. , Andre, A. , Edwards, H. , Ehrhardt, D. and Somerville, S. (2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 44, 516–529. [DOI] [PubMed] [Google Scholar]
- Koumandou, V.L. , Dacks, J.B. , Coulson, R.M. and Field, M.C. (2007) Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol. Biol. 7, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C. , Neu, C. , Pajonk, S. , Yun, H.S. , Lipka, U. , Humphry, M. , Bau, S. , Straus, M. , Kwaaitaal, M. , Rampelt, H. , El Kasmi, F. , Jürgens, G. , Parker, J. , Panstruga, R. , Lipka, V. and Schulze‐Lefert, P. (2008) Co‐option of a default secretory pathway for plant immune responses. Nature, 451, 835–840. [DOI] [PubMed] [Google Scholar]
- Lipka, U. , Fuchs, R. , Kuhns, C. , Petutschnig, E. and Lipka, V. (2010) Live and let die—Arabidopsis nonhost resistance to powdery mildews. Eur. J. Cell Biol. 89, 194–199. [DOI] [PubMed] [Google Scholar]
- Loh, E. and Hong, W. (2004) The binary interacting network of the conserved oligomeric Golgi tethering complex. J. Biol. Chem. 279, 24 640–24 648. [DOI] [PubMed] [Google Scholar]
- Lu, Y.J. , Schornack, S. , Spallek, T. , Geldner, N. , Chory, J. , Schellmann, S. , Schumacher, K. , Kamoun, S. and Robatzek, S. (2012) Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell. Microbiol. 14, 682–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin, V.V. , Pokrovskaya, I.D. , McNew, J.A. and Waters, M.G. (1997) Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol. Biol. Cell, 8, 2659–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, H.T. and Mills, I.G. (2004) COP and clathrin‐coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 16, 379–391. [DOI] [PubMed] [Google Scholar]
- Meyer, D. , Pajonk, S. , Micali, C. , O'Connell, R. and Schulze‐Lefert, P. (2009) Extracellular transport and integration of plant secretory proteins into pathogen‐induced cell wall compartments. Plant J. 57, 986–999. [DOI] [PubMed] [Google Scholar]
- Micali, C.O. , Neumann, U. , Grunewald, D. , Panstruga, R. and O'Connell, R. (2011) Biogenesis of a specialized plant–fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell. Microbiol. 13, 210–226. [DOI] [PubMed] [Google Scholar]
- Miklis, M. , Consonni, C. , Bhat, R.A. , Lipka, V. , Schulze‐Lefert, P. and Panstruga, R. (2007) Barley MLO modulates actin‐dependent and actin‐independent antifungal defense pathways at the cell periphery. Plant Physiol. 144, 1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, V.J. and Ungar, D. (2012) Re'COG'nition at the Golgi. Traffic, 13, 891–897. [DOI] [PubMed] [Google Scholar]
- Nielsen, M.E. , Feechan, A. , Böhlenius, H. , Ueda, T. and Thordal‐Christensen, H. (2012) Arabidopsis ARF‐GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. USA, 109, 11 443–11 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalski, K.S. , Schultheiss, H. , Kogel, K.‐H. and Hückelhoven, R. (2005) The receptor‐like MLO protein and the RAC/ROP family G‐protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei . Plant J. 41, 291–303. [DOI] [PubMed] [Google Scholar]
- Pecenková, T. , Hála, M. , Kulich, I. , Kocourková, D. , Drdová, E. , Fendrych, M. , Toupalová, H. and Zársky, V. (2011) The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant–pathogen interaction. J. Exp. Bot. 62, 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro, H. , Samalova, M. , Geldner, N. , Chory, J. , Martinez, A. and Moore, I. (2009) Genetic evidence that the higher plant Rab‐D1 and Rab‐D2 GTPases exhibit distinct but overlapping interactions in the early secretory pathway. J. Cell Sci. 122, 3749–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaya, I.D. , Willett, R. , Smith, R.D. , Morelle, W. , Kudlyk, T. and Lupashin, V.V. (2011) Conserved oligomeric Golgi complex specifically regulates the maintenance of Golgi glycosylation machinery. Glycobiology, 21, 1554–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaya, I.D. , Szwedo, J.W. , Goodwin, A. , Lupashina, T.V. , Nagarajan, U.M. and Lupashin, V.V. (2012) Chlamydia trachomatis hijacks intra‐Golgi COG complex‐dependent vesicle trafficking pathway. Cell. Microbiol. 14, 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quental, R. , Azevedo, L. , Matthiesen, R. and Amorim, A. (2010) Comparative analyses of the conserved oligomeric Golgi (COG) complex in vertebrates. BMC Evol. Biol. 10, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram, C. , Jensen, M.K. , Maiser, F. , Shanir, J.V. , Hornshøj, H. , Rung, J.H. , Gregersen, P.L. , Schweizer, P. , Collinge, D.B. and Lyngkjær, M.F. (2012) Regulation of basal resistance by a powdery mildew‐induced cysteine‐rich receptor‐like protein kinase in barley. Mol. Plant Pathol. 13, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Röpenack, E. , Parr, A. and Schulze‐Lefert, P. (1998) Structural analyses and dynamics of soluble and cell wall‐bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J. Biol. Chem. 272, 9013–9022. [DOI] [PubMed] [Google Scholar]
- Schultheiss, H. , Dechert, C. , Kogel, K.‐H. and Hückelhoven, R. (2003) Functional analysis of barley RAC/ROP G‐protein family members in susceptibility to the powdery mildew fungus. Plant J. 36, 589–601. [DOI] [PubMed] [Google Scholar]
- Schweizer, P. and Stein, N. (2011) Large‐scale data integration reveals colocalization of gene functional groups with meta‐QTL for multiple disease resistance in barley. Mol. Plant–Microbe Interact. 24, 1492–1501. [DOI] [PubMed] [Google Scholar]
- Schweizer, P. , Pokorny, J. , Abderhalden, O. and Dudler, R. (1999) A transient assay system for the functional assessment of defense‐related genes in wheat. Mol. Plant–Microbe Interact. 12, 647–654. [Google Scholar]
- Spoel, S.H. and Dong, X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. [DOI] [PubMed] [Google Scholar]
- Suvorova, E.S. , Kurten, R.C. and Lupashin, V.V. (2001) Identification of a human orthologue of Sec34p as a component of the cis‐Golgi vesicle tethering machinery. J. Biol. Chem. 276, 22 810–22 818. [DOI] [PubMed] [Google Scholar]
- Suvorova, E.S. , Duden, R. and Lupashin, V.V. (2002) The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra‐Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T. , Kim, H. , Saito, C. , Ebine, K. , Ueda, T. , Schulze‐Lefert, P. and Nakano, A. (2012) Qa‐SNAREs localized to the trans‐Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc. Natl. Acad. Sci. USA, 109, 1784–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar, D. , Oka, T. , Brittle, E.E. , Vasile, E. , Lupashin, V.V. , Chatterton, J.E. , Heuser, J.E. , Krieger, M. and Waters, M.G. (2002) Characterization of a mammalian Golgi‐localized protein complex, COG, that is required for normal Golgi morphology and function. J. Cell Biol. 157, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar, D. , Oka, T. , Krieger, M. and Hughson, F.M. (2006) Retrograde transport on the COG railway. Trends Cell Biol. 16, 113–120. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Weaver, N.D. , Kesarwani, M. and Dong, X. (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science, 308, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Yang, Y.D. , Elamawi, R. , Bubeck, J. , Pepperkok, R. , Ritzenthaler, C. and Robinson, D.G. (2005) Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY‐2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell, 17, 1513–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyen, R.J. , Carver, T.L.W. and Lyngkjaer, M.F. (2002) Epidermal cell papillae In: The Powdery Mildews: A Comprehensive Treatise (Bélanger R.R., Bushnell W.R., Dik A.J. and Carver T.L.W. eds), pp. 107–124. St. Paul, MN: APS Press, American Phytopatholgical Society. [Google Scholar]
- Zhang, H. , Sreenivasulu, N. , Weschke, W. , Stein, N. , Rudd, S. , Radchuk, V. , Potokina, E. , Scholz, U. , Schweizer, P. , Zierold, U. , Langridge, P. , Varshney, R.K. , Wobus, U. and Graner, A. (2004) Large‐scale analysis of the barley transcriptome based on expressed sequence tags. Plant J. 40, 276–290. [DOI] [PubMed] [Google Scholar]
- Zierold, U. , Scholz, U. and Schweizer, P. (2005) Transcriptome analysis of mlo‐mediated resistance in the epidermis of barley. Mol. Plant Pathol. 6, 139–151. [DOI] [PubMed] [Google Scholar]
- Zolov, S.N. and Lupashin, V.V. (2005) Cog3p depletion blocks vesicle‐mediated Golgi retrograde trafficking in HeLa cells. J. Cell Biol. 168, 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Hierarchical clustering of barley gene expression experiments at Genevestigator 4. Analysis was carried out for conserved oligomeric Golgi (COG) subunit genes and genes of putative interaction partners of COG3. Gene IDs on barley 1 chips are: HvCOG1, Contig13332_at; HvCOG2, Contig11948_at; HvCOG3, Contig7930_at; HvCOG4, Contig11310_at; HvCOG5, Contig13262_at; HvCOG6, Contig10684_at; HvCOG7, Contig14245_at; HvCOG8, Contig9113_at; HvGOS1, Contig12134_at; HvYKT6, Contig8337_at; HvSEC22, Contig18727_at; HvVTI1, Contig9598_at; HvYPT1, Contig4865_s_at; HvCOPIg, Contig7764_at and/or Contig7765_at. Colour code shows log2 transformed changes in expression (from −2.5 to +2.5, i.e. from 18% to 566% of the expression level of the respective control).
Table S1 Protein sequence similarity scores for subunits of the human, Arabidopsis and barley conserved oligomeric Golgi (COG) complex.
Table S2 Accession/locus numbers of human and plant conserved oligomeric Golgi (COG) subunits.
Table S3 Interaction partners of conserved oligomeric Golgi 3 (COG3) identified in yeast by Suvorova et al. (2002) and their barley homologues.
Table S4 Oligo DNA primers for the isolation of vector‐provided sequences for transient‐induced gene silencing (TIGS) construct generation.
Table S5 Oligo DNA primers for isolation of cDNA fragments for transient‐induced gene silencing (TIGS) construct generation.
Table S6 Oligo DNA primers for isolation of full‐length cDNA sequences for over‐expression constructs.


