SUMMARY
To gain a better understanding of the molecular changes taking place in citrus fruit tissue following the application of the yeast biocontrol agent Metschnikowia fructicola, microarray analysis was performed on grapefruit surface wounds using an Affymetrix Citrus GeneChip. Using a cut‐off of P < 0.05 and a 1.5‐fold change difference as biologically significant, the data indicated that 1007 putative unigenes showed significant expression changes following wounding and yeast application relative to wounded controls. Microarray results of selected genes were validated by reverse transcription‐quantitative real‐time polymerase chain reaction (RT‐qPCR). The data indicated that yeast application induced the expression of the genes encoding Respiratory burst oxidase (Rbo), mitogen‐activated protein kinase (MAPK) and mitogen‐activated protein kinase kinase (MAPKK), G‐proteins, chitinase (CHI), phenylalanine ammonia‐lyase (PAL), chalcone synthase (CHS) and 4‐coumarate‐CoA ligase (4CL). In contrast, three genes, peroxidase (POD), superoxide dismutase (SOD) and catalase (CAT), were down‐regulated in grapefruit peel tissue treated with yeast cells. Moreover, suppression was correlated with significantly higher levels of hydrogen peroxide, superoxide anion and hydroxyl radical production in yeast‐treated surface wounds. Interestingly, large amounts of hydrogen peroxide were detected inside yeast cells recovered from wounded fruit tissue, indicating the ability of the yeast to activate reactive oxygen species when it is in contact with plant tissue. This study provides the first global picture of gene expression changes in grapefruit in response to the yeast antagonist M. fructicola.
INTRODUCTION
Yeasts used to control postharvest pathogens have been shown to induce several biochemical defence responses in surface wounds of fruit (El‐Ghaouth et al., 1998). The capability to elicit these responses in fruit tissue is regarded as one of the possible mechanisms of action by which yeast biocontrol agents inhibit the development of decay (Droby et al., 2009). Detailed information on the mechanisms of action for most of the antagonists investigated, however, is still rudimentary because of the difficulties associated with the investigation of interactions between a host, pathogen and an antagonist, as well as other resident microorganisms (Droby et al., 2009). Although competition for nutrient and space appears to play a significant role in the biocontrol activity of yeasts, other mechanisms, including the exudation of lytic enzymes, direct parasitism and the elicitation of defence responses, have been suggested to contribute.
Antagonistic yeasts have been reported to induce several biochemical defence responses in host tissues of surface‐wounded fruit. The treatment of wounds in lemons with Pichia guilliermondii, or of wounds in oranges with Candida famata, enhanced the production of the phytoalexins scoparone and scopoletin relative to untreated wounds (Arras, 1999; Rodov et al., 1994). Candida saitoana induced chitinase (CHI) activity in apple surface wounds and caused the deposition of papillae in host cell walls (El‐Ghaouth et al., 1998), whereas Aureobasidium pullulans caused a transient increases in β‐1,3‐glucanase (GLU), CHI and peroxidase (POD) activities (Ippolito et al., 2000). These increases started 24 h after yeast treatment was administered and reached maximum levels at 48–96 h later, depending on the specific enzyme. The induction of resistance mechanisms in citrus fruit following the application of the yeast Candida oleophila has also been reported by Droby et al. (2009).
Castoria et al. (2003) indicated that the ability to tolerate high levels of reactive oxygen species (ROS) produced in fruit tissue in response to wounding is an essential characteristic of effective yeast antagonists. This hypothesis raised many new questions about the role of ROS in biocontrol activity. Macarisin et al. (2010) reported the stimulation of ROS production in wounded citrus fruit tissue by the biocontrol agent Metschnikowia fructicola, and suggested that ROS played a key role in biocontrol activity. They demonstrated that the yeast antagonists, C. oleophila and M. fructicola, have the ability to produce significant levels of superoxide anions in vitro and even larger amounts in and around surface wounds of fruit. They concluded that strong O2− production by these biocontrol yeasts may act as a signal to stimulate rapid multiplication in fruit wounds (abundant in nutrients) to ensure maximal colonization within a favourable biological niche. Droby et al. (2009) suggested that ROS production by yeast antagonists, together with host ROS, may serve as an oxidative burst in wounded fruit tissue, triggering the induction of an array of host resistance responses.
ROS has been reported to serve a signalling function, mediating defence gene activation by redox control of transcription factors or by interaction with other signalling components, such as mitogen‐activated protein kinase (MAPK) cascades (Zhang and Klessig, 2001). The activation of MAPK leads to the phosphorylation of transcription factors, which, in turn, activate the gene expression involved in the defence response against pathogen invasion (Yang et al., 2001). ROS could also mediate the generation of phytoalexins and secondary metabolites that arrest pathogen growth (Torres, 2010), or contribute to the establishment of physical barriers via oxidative cross‐linking of precursors during the localized biosynthesis of lignin and suberin polymers (Huckelhoven, 2007).
The down‐regulation of scavenging/antioxidant systems can also contribute to increased ROS accumulation (Mittler et al., 2004). Changes in the level of detoxifying enzymes, such as superoxide dismutases (SODs), catalases (CATs) and POD, have been reported in response to many pathogens or biological agents and linked to an increase in ROS and the activation of host defences. Chan and Tian (2006) showed that the application of the yeast antagonist Pichia membranefaciens enhanced POD activity, but inhibited SOD and CAT activities, in sweet cherry fruit. When combined with the pathogen Penicillium expansum, yeast treatment increased SOD activity, but decreased CAT activity, compared with the pathogen alone, indicating that the yeast was directly involved in ROS metabolism in fruit tissue. In contrast, Monilinia fructicola stimulated the activities and expression levels of CAT and POD in peach (Xu et al., 2008).
In comparison with our understanding of plant responses to other stresses, such as drought, salinity, heat, chilling and pathogen attack, little is known about the molecular basis underlying the induction of host defences in response to antagonistic yeasts, or about the signalling networks involved in this response. Although several studies have been reported on global changes in fruit gene expression in response to cold stress (Maul et al., 2008) and pathogen infection (González‐Candelas et al., 2010), only a limited amount of information is available on fruit transcriptome changes associated with the application of yeast biocontrol agents (Jiang et al., 2009). This study was undertaken to determine the effect of the yeast M. fructicola on the following: (i) global gene expression in wounded grapefruit peel tissue using a citrus microarray; and (ii) regulation of host‐related oxidative stress responses in wounded fruit tissue. We specifically examined defence‐related oxidative genes [Respiratory burst oxidase homologues (Rbo), SOD, POD, CAT], signalling genes (G‐protein, MAPK and MAPKK), genes associated with secondary metabolism [phenylalanine ammonia‐lyase (PAL), 4‐coumarate‐CoA ligase (4CL), chalcone synthase (CHS)] and CHI, all of which are involved in the enhancement of the fruit response to and defence against pathogen attack.
RESULTS
Analysis of changes in grapefruit peel gene expression after application of the yeast biocontrol agent M. fructicola
Microarray analysis gene changes in grapefruit peel tissue after 24 h of incubation revealed 1007 probe sets, corresponding to 830 putatively differentially expressed unigenes that showed a significant (P < 0.05) change (up or down) in expression (≥1.5‐fold). The probes obtained were hierarchically clustered (Euclidean dissimilarity, by average linkage method) according to their expression patterns (Fig. 1A). Sixty‐five per cent (650 probes) of the probes were down‐regulated in the treated samples compared with controls, and 35% (357 probes) were up‐regulated. Principal component analysis (PCA) was performed to validate the repeatability of microarray data across the three replications. PCA revealed marked differences in gene expression patterns among the yeast‐treated and control surface wounds collected after 24 h of incubation (Fig. 1B).
Figure 1.
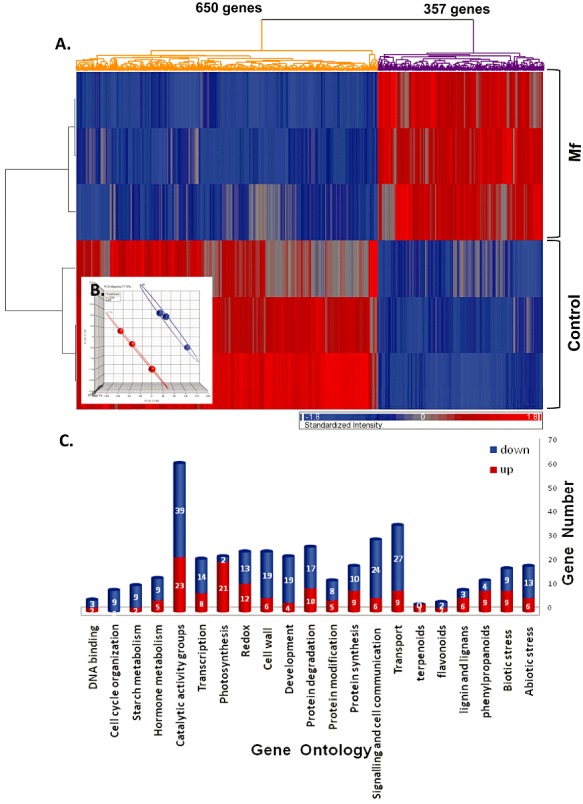
(A) Hierarchical clustering of genes. Mf, Metschnikowia fructicola. (B) Principal component analysis (PCA) of the three yeast‐treated and three control arrays used in the experiment. (C) Functional analysis of microarray citrus probes. Genes for major functional categories (gene ontology) are shown; up‐regulated genes are coloured blue and down‐regulated genes are coloured red.
Functional analysis was applied to the identified microarray probes of citrus genes using Arabidopsis orthologues (Forment et al., 2005) and citrus ontologies (http://bioinfo.cau.edu.cn/agriGO/index.php) (Du et al., 2010). Five hundred and seventy of the 1007 probes had an assigned Arabidopsis orthologue. The distribution of the genes among the MapMan and AgriGO functional categories indicated that application of the biological agent, M. fructicola, to grapefruit peel wounds involved genes from several major functional categories (Fig. 1C). Most genes (65% of annotated genes) were down‐regulated by M. fructicola treatment, and only 35% were up‐regulated. Only down‐regulated genes fell into the category of cell cycle activities. Photosynthesis and categories associated with secondary metabolism showed more up‐regulated than down‐regulated genes. Catalytic activity, signalling and transport were the functional categories with the most differentially expressed genes (total of 31% of all annotated genes) (Fig. 1B).
Transcriptional profile of grapefruit peel wounds following M. fructicola application
To further analyse the changes in gene expression in grapefruit peel tissue in response to M. fructicola application, microarray data for the probes of the significant dataset were mapped to the Arabidopsis biotic stress pathway using MapMan software. The data obtained from this analysis are presented in Fig. S1 and Table S1 (see Supporting Information), and 1, 2, 3 summarize the changes in defence‐related oxidative genes (Table 1), the activation of genes encoding proteins with potential functions in signalling mechanisms (Table 2) and the changes in genes encoding enzymes involved in secondary metabolic processes (Table 3). Based on our previous studies (Liu et al., 2011; Macarisin et al., 2010), the relationship between gene expression and ROS accumulation was of particular interest.
Table 1.
Description of the genes involved in oxidative and abiotic stresses in response to yeast antagonist (Metschnikowia fructicola) treatment of grapefruit, based on microarray analyses in MapMan software (http://gabi.rzpd./de/projects/MapMan/) (fold change, P≤ 0.05).
| ID No. | Probeset ID | TAIR | Symbol | Description | Fold change |
|---|---|---|---|---|---|
| Oxidative stress | |||||
| 264647_at | Cit.25134.1.S1_at | at1g09090 | Rbo | Respiratory burst oxidase homologue B | 4.27 |
| 267544_at | Cit.585.1.S1_s_at | at2g32720 | CB5‐B | Cytochrome B5 isoform B | 2.74 |
| 263096_at | Cit.8683.1.S1_s_at | at2g16060 | GLB1 | Oxygen transporter | 2.06 |
| 266941_at | Cit.26030.1.S1_at | at2g18980 | Peroxidase | 1.81 | |
| 256892_at | Cit.15260.1.S1_at | at3g19000 | 2OG‐Fe(II) | Oxidoreductase | 1.70 |
| 261412_at | Cit.8235.1.S1_at | at1g07890 | APX1 | l‐Ascorbate peroxidase 1 | 1.59 |
| 261751_at | Cit.2633.1.S1_at | at1g76080 | CDSP32 | Chloroplastic drought‐induced stress protein | −1.52 |
| 250133_at | Cit.1832.1.S1_s_at | at5g16400 | ATF2 | Enzyme activator | −1.71 |
| 264001_at | Cit.15540.1.S1_at | at2g22420 | POD | Peroxidase 17 | −2.02 |
| 264809_at | Cit.28102.1.S1_s_at | at1g08830 | SOD | Cu/Zn superoxide dismutase | −2.02 |
| Abiotic stress | |||||
| 255807_at | Cit.5237.1.S1_s_at | at4g10270 | – | Wound‐responsive family protein | 5.55 |
| 259478_at | Cit.21686.1.S1_at | at1g18980 | GLP1 | Germin‐like protein, putative | 2.92 |
| 254889_at | Cit.22710.1.S1_at | at4g11650 | ATOSM34 | Osmotin 34 | 1.60 |
| 258751_at | Cit.21797.1.S1_at | at3g05890 | RCI2B | Rare‐cold‐inducible 2b | 1.52 |
| 260248_at | Cit.280.1.S1_at | at1g74310 | HSP101 | Heat shock protein 101 | −1.57 |
| 262958_at | Cit.17622.1.S1_at | at1g54410 | – | Dehydrin family protein | −1.59 |
| 253884_at | Cit.9638.1.S1_s_at | at4g27670 | HSP21 | Heat shock protein 21 | −1.64 |
| 254701_at | Cit.4893.1.S1_s_at | at4g18030 | – | Dehydration‐responsive protein | −1.69 |
| 248352_at | Cit.11985.1.S1_at | at5g52300 | RD29B | Low‐temperature‐induced 65 | −1.71 |
| 256310_at | Cit.28436.1.S1_s_at | at1g30360 | ERD4 | Early‐responsive to dehydration 4 | −1.91 |
| 255088_at | Cit.6382.1.S1_at | at4g09350 | – | DNAJ heat shock protein | −2.18 |
Table 2.
Description of the genes involved in signalling and defence processes in response to yeast antagonist (Metschnikowia fructicola) treatment of grapefruit, based on microarray analyses in MapMan software (http://gabi.rzpd./de/projects/MapMan/) (fold change, P≤ 0.05).
| ID No. | Probeset ID | TAIR | Symbol | Description | Fold change |
|---|---|---|---|---|---|
| Signalling processes | |||||
| 267366_at | Cit.5257.1.S1_s_at | at2g44310 | – | Calcium‐binding EF hand family protein | 4.42 |
| 245731_at | Cit.25223.1.S1_at | at1g75840 | MAPKK | Mitogen‐activated protein kinase kinase | 1.98 |
| 266394_at | Cit.12225.1.S1_at | at2g43130 | ARA4 | GTP binding | 1.82 |
| 251513_at | Cit.35804.1.S1_at | at3g59220 | PRN | G protein | 1.75 |
| 258184_at | Cit.15243.1.S1_at | at3g21510 | AHP1 | Histidine phosphotransfer kinase | −1.56 |
| 256516_at | Cit.14463.1.S1_at | at1g66150 | TMK1 | Transmembrane receptor protein | −1.58 |
| 258321_at | Cit.26060.1.S1_at | at3g22840 | ELIP1 | Chlorophyll binding | −1.63 |
| 266447_at | Cit.19859.1.S1_at | at2g43290 | MSS3 | Calcium ion binding | −1.71 |
| 260587_at | Cit.19786.1.S1_s_at | at1g53210 | Calcium‐binding EF hand family protein | −1.76 | |
| 261378_at | Cit.22339.1.S1_s_at | at1g18890 | ATCDPK1 | Calmodulin‐dependent protein kinase | −1.79 |
| 262360_at | Cit.9672.1.S1_at | at1g73080 | PEP1 | ATP binding/threonine kinase | −1.87 |
| 261498_at | Cit.5665.1.S1_s_at | at1g28440 | HSL1 | ATP binding/threonine kinase | −2.18 |
| 259531_at | Cit.40040.1.S1_at | at1g12460 | – | Leucine‐rich repeat transmembrane protein kinase | −2.19 |
| Pathogenesis‐related protein | |||||
| 251895_at | Cit.15242.1.S1_at | at3g54420 | CHI | Chitinase 1 | 4.89 |
| 255904_at | Cit.17136.1.S1_s_at | at1g17860 | – | Trypsin and protease inhibitor family protein | 2.06 |
| 249767_at | Cit.15331.1.S1_s_at | at5g24090 | CHIB1 | Acidic endochitinase | 1.92 |
| 251673_at | Cit.21778.1.S1_at | at3g57240 | BG3 | β‐1,3‐Glucanase 3 | 1.69 |
| 249639_at | Cit.18657.1.S1_at | at5g36930 | TIR‐NBS‐LRR class | Disease resistance protein | −1.63 |
| 267411_at | Cit.6332.1.S1_at | at2g34930 | – | Disease resistance family protein | −1.86 |
Table 3.
Description of the genes involved in secondary and hormone metabolism in response to yeast antagonist (Metschnikowia fructicola) treatment of grapefruit, based on microarray analyses in MapMan software (http://gabi.rzpd./de/projects/MapMan/) (fold change, P≤ 0.05).
| ID No. | Citrus Probeset ID | TAIR | Symbol | Description | Fold change |
|---|---|---|---|---|---|
| Secondary metabolism | |||||
| 248639_at | Cit.14337.1.S1_at | at5g48930 | HCT | Quinate O‐hydroxycinnamoyl transferase 5 | 5.88 |
| 267470_at | Cit.21686.1.S1_at | at2g30490 | C4H | trans‐cinnamate 4‐monooxygenase | 2.95 |
| 261907_at | Cit.11987.1.S1_s_at | at1g65060 | 4CL | 4‐Coumarate‐CoA ligase | 2.22 |
| 250794_at | Cit.25997.1.S1_x_at | at5g05270 | CFI | Chalcone–flavanone isomerase family protein | 1.90 |
| 248200_at | Cit.24459.1.S1_at | at5g54160 | ATOMT | Quercetin 3‐O‐methyltransferase | 1.84 |
| 251984_at | Cit.13407.1.S1_s_at | at3g53260 | PAL | Phenylalanine ammonia‐lyase | 1.81 |
| 253088_at | Cit.14813.1.S1_s_at | at4g36220 | FAH1 | Ferulate 5‐hydroxylase | 1.69 |
| 253276_at | Cit.17602.1.S1_s_at | at4g34050 | – | Caffeoyl‐CoA 3‐O‐methyltransferase | −1.63 |
| 250207_at | Cit.19520.1.S1_s_at | at5g13930 | CHS | Naringenin–chalcone synthase | −2.15 |
| 252983_at | Cit.9171.1.S1_at | at4g37980 | ELI3‐1 | Oxidoreductase/zinc ion binding | −2.18 |
| 258116_at | Cit.18744.1.S1_at | at3g14520 | – | Terpene synthase | −2.53 |
| 252611_at | Cit.11683.1.S1_at | at3g45130 | LAS1 | Lanosterol synthase | −2.73 |
| Hormone metabolism | |||||
| 265948_at | Cit.1718.1.S1_s_at | at2g19590 | ACO | 1‐Aminocyclopropane‐1‐carboxylic acid oxidase | 10.45 |
| 262981_at | Cit.12252.1.S1_at | at1g75590 | – | Auxin‐responsive family protein | 6.75 |
| 260427_at | Cit.15712.1.S1_at | at1g72430 | – | Auxin‐responsive protein‐related | 2.45 |
| 252184_at | Cit.33172.1.S1_at | at3g50660 | SNP2 | Steroid 22‐α hydroxylase | 2.11 |
| 253579_at | Cit.16081.1.S1_at | at4g30610 | BRS1 | BRI1 suppressor 1 | 2.11 |
| 261228_at | Cit.10986.1.S1_s_at | at1g20050 | HYD1 | C‐8 sterol isomerase | 1.52 |
| 246864_at | Cit.26661.1.S1_at | at5g25900 | GA3 | GA requiring 3 | −1.73 |
Oxidative stress
The expression profile of the genes involved in the response to stress is presented in Table 1. Yeast application induced the expression of the oxygen transporter, cytochrome B, oxidoreductase and l‐ascorbate peroxidase genes. The highest induction, about four‐fold, was found in the Rbo transcript level. The level of genes corresponding to detoxifying enzymes, such as SOD, POD and CAT, however, was lower in yeast‐treated wounds (Table 1). Genes corresponding to abiotic stress were also altered by yeast treatment: wound‐responsive family, germin, osmotin and cold‐inducible genes were up‐regulated in yeast‐treated relative to control wounds, whereas two groups of heat shock protein (HSP) and dehydration‐responsive family member genes were down‐regulated (Table 1).
Signalling pathways and pathogenesis‐related (PR) proteins
The resulting dataset included 14 probes corresponding to various signalling genes (Table 2). Five of these encoded genes for calcium binding, G‐proteins and MAPKs, all of which were up‐regulated by M. fructicola application. The other nine were down‐regulated and corresponded to genes encoding for ATP binding, chlorophyll binding, transmembrane receptors and two additional calcium‐binding proteins (Table 2). In addition, genes encoding for CHI, β‐1,3‐glucanase (GLU), endochitinase and trypsin protease inhibitor were stimulated by M. fructicola application (Table 2).
Secondary metabolic processes
The data shown in Table 3 indicate that many genes involved in secondary metabolism, such as those corresponding to the phenylpropanoid pathway, were activated following M. fructicola application. A significant increase in the expression of the genes encoding for PAL, 4CL, trans‐cinnamate 4‐monooxygenase, chalcone–flavanone isomerase, quinate O‐hydroxycinnamoyltransferase 5 and quercetin 3‐O‐methyltransferase (enzymes involved in lignin and flavanol biosynthesis) was observed (Table 3). However, genes involved in the biosynthesis of other secondary metabolites, such as terpenes and anthocyanins, were down‐regulated (Table 3), as evidenced by a strong decrease in the expression of caffeoyl‐CoA 3‐O‐methyltransferase, naringenin–chalcone synthase and terpene synthase genes. In addition, there was an increase in the expression of the gene encoding for 1‐aminocyclopropane‐1‐carboxylic acid oxidase (ACO), a gene involved in the biosynthesis of ethylene, two auxin response‐related genes and genes involved in the brassinosteroid (BR) synthesis pathway (steroid 22‐α hydroxylase, C‐8 sterol isomerase and BRI1 suppressor 1). All were up‐regulated following yeast application. In contrast, a gene related to gibberellic acid (GA) hormone metabolism (GA requiring 3) was down‐regulated (Table 3).
Reverse transcription‐quantitative real‐time polymerase chain reaction (RT‐qPCR) analyses of gene expression changes
To confirm the expression profiles obtained from the microarray gene expression data, RT‐qPCR analysis was carried out for 10 genes selected on the basis of their biological significance. The time points included in the analysis were 2, 8, 24 and 48 h after yeast treatment (Fig. 2). Transcript levels of Rbo were significantly higher after 24 and 48 h following yeast application relative to the controls (Fig. 2A). The expression level of G‐protein was higher in yeast‐treated wounds at all time points relative to control wounds (Fig. 2B). CAT and POD expression levels, however, were lower at all time points in yeast‐treated samples relative to controls (Fig. 2C,E). The MAPKK expression profile was similar to that found for Rbo, with a significant increase in transcript levels after 24 and 48 h in yeast‐treated wounds relative to the non‐treated control (Fig. 2A,D). Metschnikowia fructicola application also resulted in higher MAPK expression levels at 8, 24 and 48 h relative to controls (Fig. 2F). The expression profiles of two genes involved in secondary metabolism, PAL and CHI, and a gene encoding 4CL were up‐regulated to higher levels by M. fructicola application after 24 and 48 h relative to controls (Fig. 2G, I and J). In contrast, relative to the controls, the expression level of CHS was reduced by yeast treatment at all time points and exhibited a gradual decrease in expression between 24 and 48 h in both treatments (Fig. 2H). Overall, the RT‐qPCR data supported the microarray gene expression data.
Figure 2.
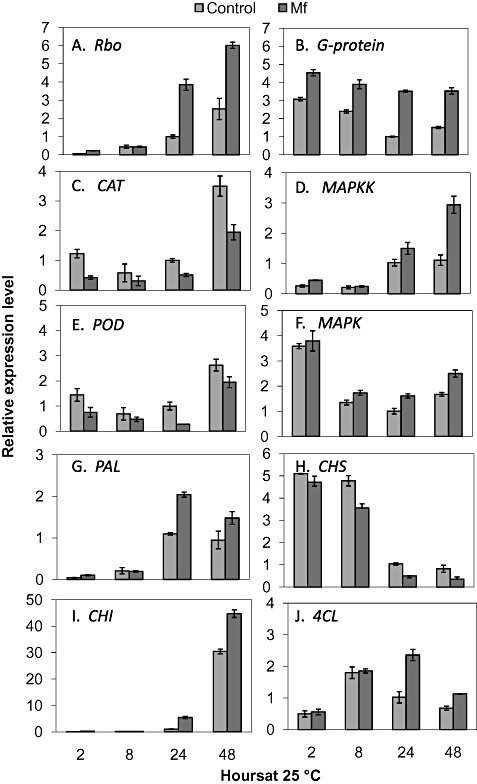
Relative expression levels (transcript accumulation) determined by reverse transcription‐quantitative real‐time polymerase chain reaction (RT‐qPCR) of respiratory burst oxidase (Rbo, Cit.25134.1.S1_at) (A), G‐protein (Cit.35804.1.S1_at) (B), catalase (CAT, ABG49115) (C), mitogen‐activated protein kinase kinase (MAPKK, Cit.25223.1.S1_at) (D), peroxidase (POD, ABG49115) (E), mitogen‐activated protein kinase (MAPK, EF185418) (F), phenylalanine ammonia‐lyase 2 (PAL, AJ238753) (G), chalcone synthase 1 (CHS, AB009351) (H), chitinase I (CHI, AF090336.1) (I) and 4‐coumarate‐CoA ligase (4CL, Cit.11987.1.S1_s_at) (J) in Metschnikowia fructicola (Mf)‐ and water (control)‐treated grapefruit wounds. Values were normalized to the control at 24 h, arbitrarily set to unity. Vertical lines represent the standard error for an average of three biological replicates.
Hydrogen peroxide (H2O2) level in yeast cells
Metschnikowia fructicola applied to grapefruit wounds exhibited an intense fluorescence, indicating a high level of intracellular H2O2, 5 h following the application of yeast to grapefruit wounds (Fig. 3A); however, this intensity decreased dramatically after 24 h and remained low at 48 h (Fig. 3B,C). Quantification of the fluorescence signal showed that yeast cells reached their highest level of fluorescence 5 h after application to the fruit and decreased after 24 and 48 h [relative pixel intensities (rpi) of 1200 and 250, respectively] (Fig. 3D). In contrast, yeast cells grown in nutrient yeast dextrose broth (NYDB) displayed elevated accumulation of H2O2 only after 48 h (Fig. 4).
Figure 3.
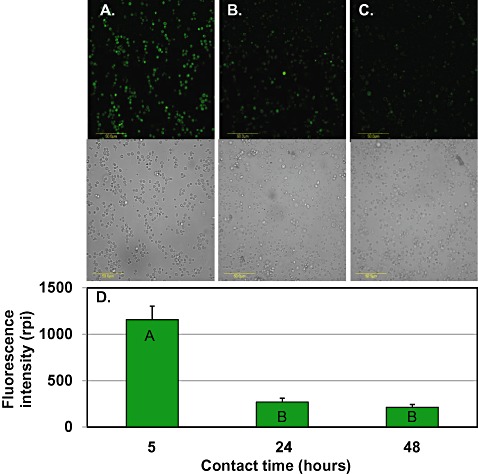
Laser scanning confocal fluorescence images (top panel) of H2O2 accumulation in Metschnikowia fructicola cells washed from grapefruit wounds, 5 h (A), 24 h (B) and 48 h (C) after application, combined with bright field images (bottom panel). Scale bar, 50 µm. Quantification of reactive oxygen species (ROS) level in yeast cells, expressed as the relative pixel intensity (rpi) (D). Histograms represent the mean ± standard error (n= 6). Histograms with different letters indicate significant differences according to Tukey's test (P < 0.05).
Figure 4.
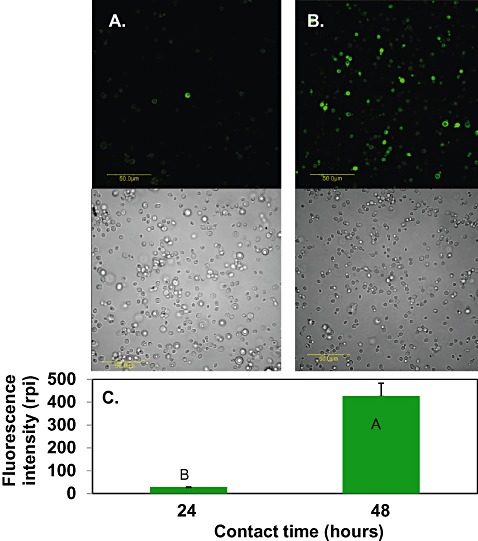
Laser scanning confocal fluorescence images (top panel) of H2O2 accumulation in Metschnikowia fructicola cells, combined with bright field images (bottom panel). Yeast was grown in nutrient yeast dextrose broth (NYDB) for 24 h (A) or 48 h (B) at 25 °C. Scale bar, 50 µm. Quantification of H2O2 level in yeast cells, expressed as the relative pixel intensity (rpi) (C). Histograms represent the mean ± standard error (n= 6). Histograms with different letters indicate significant differences according to Tukey's test (P < 0.05).
H2O2, superoxide anion and hydroxyl radical production in fruit
As illustrated in Fig. 5, the application of M. fructicola to discs of grapefruit peel resulted in increased levels of H2O2 in fruit discs relative to water control peel discs, with a maximum value at 5 h after yeast application (Fig. 5A). Superoxide anion and hydroxyl radical, however, displayed different trends compared with H2O2. Significantly higher levels of superoxide anion and hydroxyl radical in response to yeast antagonist were found after 5 h of application relative to water‐treated discs. These levels continued to increase during 24 and 48 h following application relative to the basal levels in control discs (Fig. 5B,C).
Figure 5.

Time course of levels of H2O2 (A), superoxide anion (B) and hydroxyl radical (C) in grapefruit peel discs following Metschnikowia fructicola (Mf) treatment and control. The level of H2O2 is expressed as the fluorescence intensity (530 nm/590 nm) × 1000, the superoxide anion level is expressed as the increased absorbance at 580 nm/h/g/fresh weight (FW) and the hydroxyl radical level is expressed as the increased absorbance at 540 nm/g/FW × 1000. Values represent the mean ± standard error (n= 8–12). Value with different letters indicate significant differences according to Tukey's test (P < 0.05).
DISCUSSION
Interactions between postharvest yeast biocontrol agents and host tissue are poorly understood (Droby et al., 2009). In this study, we used a microarray to identify global changes in gene expression occurring in grapefruit tissue following the application of the yeast biological control agent M. fructicola to peel wounds. Special emphasis was placed on the metabolic pathways involved in oxidative‐related defence mechanisms, particularly those associated with early host resistance signalling.
The results of this study demonstrate that significant changes in gene expression occur in grapefruit wounded tissue in response to the application of M. fructicola. These changes include a variety of genes involved in response to biotic and abiotic stresses, signalling, defence and secondary metabolism (Fig. 1C). These findings imply that complex biochemical and molecular processes are involved in the reaction of fruit host tissue to the introduction of yeast cells, which have the potential to influence the efficacy of the biocontrol agent.
We have proposed previously that the production of ROS by yeast antagonists may serve as a signal to trigger an oxidative burst in host tissue, leading to the activation of host defence mechanisms (Droby et al., 2009; Macarisin et al., 2010). This hypothesis was supported by other reports demonstrating a significant increase in POD activity in harvested commodities, such as apple (Yu et al., 2008), peach (Xu et al., 2008), sweet cherry (Chan and Tian, 2006) and pear (Yu and Zheng, 2007), in response to the application of yeast antagonists used to control postharvest diseases. In the current study, a significant accumulation of ROS, including H2O2, superoxide anions and hydroxyl radicals, was observed in host tissue (Fig. 5A) as early as 6 h following the application of M. fructicola cells (Fig. 3). In addition, we have also shown the ability of this yeast to produce and accumulate intracellular H2O2 in cells dislodged from grapefruit peel discs as early as 5 h after coming into contact with fruit peel tissue (Fig. 3). These findings are consistent with an earlier report by Macarisin et al. (2010) which demonstrated the intense production of superoxide anions by yeast cells shortly after application to intact or wounded apple peel tissue, with a concomitant accumulation of H2O2 in the host fruit tissue itself. Together, these results support the assumption that ROS may play a major role in the early stages of the interaction between yeast antagonist cells and host tissue. However, other effectors, such as pathogen‐associated molecular patterns (PAMPs), are likely to be involved in such interactions, and the relative role of each component is still not understood. The involvement of PAMPs in the induction of host defence mechanisms has been the subject of several studies, indicating their role in the modulation of host defences in response to pathogen attack through ROS production (Boller and Felix, 2009; Boller and He, 2009; Felix et al., 1999; Zhao and Qi, 2008).
Plant NADPH (nicotinamide adenine dinucleotide phosphate) oxidases, called ‘Respiratory burst oxidase homologues (Rbo)’, have been reported as a source of the oxidative burst in most plant–pathogen interactions (Torres et al., 2006). In our microarray analysis data, we detected a significant increase in gene expression in yeast‐treated wounds for a probe corresponding to Rbo (Table 1). This was further supported by RT‐qPCR of Rbo gene expression (Fig. 2). The activation of Rbo in fruit host tissue by M. fructicola is likely to be responsible for the early increase in superoxide anion and H2O2 levels, which serve as a precursor of hydroxyl radicals shown to be increased in peel surface wounds at 24 and 48 h after the application of yeast cells. ROS accumulation in yeast‐treated wounds is accompanied by a decrease in expression levels of genes encoding for ROS‐detoxifying enzymes, such as SOD, CAT and POD (Table 1, 2, 5). Changes in antioxidant gene expression, leading to an increase in ROS levels, and the activation of defence mechanisms have been reported to be a response to cold stress (Maul et al., 2008), pathogen attack (2006, 2010) and biological agents (Xu et al., 2008). The up‐regulation of additional genes related to the production of an oxidative burst (Table 1) suggests that the presence of the yeast actively contributes to the ROS level in host tissue. The balance between ROS levels and the different scavenging enzymes is considered to be crucial in determining the steady‐state levels of H2O2 and O2 – (Mittler et al., 2004). Additional research is needed to determine whether specific plant/yeast genes contribute to the ROS scavenging capacity and should be included in the ROS gene network of plant–yeast interactions.
ROS contribute to the activation of plant defences by inducing changes in gene expression, either directly through the redox regulation of transcription factors or indirectly by interaction with other signalling components, such as MAPK cascades, a specific class of serine/threonine protein kinases. MAPK cascades transfer signals from upstream receptors to downstream cellular effectors, and rapid MAPK activation allows the modification of downstream signalling proteins (Zhang and Klessig, 2001). In plants, these cascades have been implicated in typical defence responses, such as the production of PR proteins, ethylene synthesis and cell death (Yang et al., 2001). In this regard, our study revealed the up‐regulation of two genes, encoding MAPK and MAPKK, following treatment with the biocontrol yeast (Table 2; Fig. 2D,F). A similar effect on the expression of MAPK and MAPKK was reported by Jiang et al. (2009) when the yeast, Cryptococcus laurentii, was applied to tomato fruit.
In addition to the increased expression of key genes involved in the MAPK cascade, the plant defence‐related genes CHI and GLU were up‐regulated in grapefruit peel tissue following yeast treatment. An oxidative burst is known to induce a broad range of PR proteins. Plant PR proteins are represented by 17 protein families, including GLUs, CHIs and PODs (Van Loon et al., 2006). PR proteins have been shown to be directly involved in plant immunity and associated with protective mechanisms (Pieterse and Van Loon, 1999). The expression levels of PR proteins can be regulated by various stress‐related situations, including wounding, salinity, chemical elicitors, hormones and UV light (Van Loon et al., 2006). In postharvest systems, several reports have demonstrated the induction of CHI or GLU, including apple (El‐Ghaouth et al., 1998), citrus (Ballester et al., 2010; Droby et al., 2002), peach (Xu et al., 2008) and cherry tomato fruit (Jiang et al., 2009), in response to the application of a biocontrol agent.
The HSPs are one of the major classes of chaperone molecules which may act as a primary defence during oxidative stress caused by pathogens by ameliorating ROS damage to proteins. In our study, however, the microarray data indicated a decrease in the expression level of genes encoding for HSPs (Table 1). The reason for this reduction is not known, but may be related to the maturation stage of the grapefruit. HSP has been reported to play a role in the resistance response of sweet cherry fruit at the later stages of maturity (Chan et al., 2008). Therefore, additional experiments are needed to better understand the role of HSPs in the biocontrol of systems and resistance responses in harvested fruits.
Dehydrins are a subfamily of late‐embryogenesis‐abundant (LEA) proteins produced by plants in response to water, freezing and chilling stress (Borovskii et al., 2002; Hara et al., 2004), as well as other dehydrating stresses. In our microarray data, which characterized the changes in gene expression 24 h following yeast treatment, we detected a decrease in dehydrin gene expression (Table 1); however, immediately after treatment and during the first 8 h, dehydrin gene expression was significantly elevated by biocontrol agent application (data not shown), suggesting a possible radical scavenging activity. Hara et al. (2004) reported antioxidative activity for a citrus dehydrin and suggested that this activity played an important role in adaptation to abiotic stress. Our data suggest that it may also play a role in the response to biotic stress.
The present study revealed that M. fructicola application enhanced the expression levels of PAL, trans‐cinnamate 4‐monooxygenase (C4H) and 4CL (Table 3, Fig. 2G,J), all of which are products of the phenylpropanoid metabolism pathway, which is associated with the synthesis of a wide range of plant defence compounds (Dixon and Paiva, 1995). PAL is the first enzyme in this pathway responsible for the biosynthesis of secondary products, such as the phytoalexin scoparone, p‐coumaric acid derivatives and lignins. The involvement of PAL gene expression and the synthesis of phenylpropanoid‐derived compounds have been reported previously in the citrus fruit response to biocontrol agents and to the citrus postharvest pathogen, Penicillium digitatum (Ballester et al., 2006; González‐Candelas et al., 2010; Mittler et al., 1999).
The microarray data indicated an increase in expression of a gene encoding for ACO in M. fructicola‐treated grapefruit peel wounds (Table 3). The increase in ACO, which converts 1‐aminocyclopropane‐1‐carboxylic acid to ethylene, could explain the marked increase in ethylene production level previously observed in yeast‐treated grapefruit discs (Droby et al., 2002). In Droby et al. (2002), ethylene production in citrus peel tissue following application of the yeast antagonist Candida oleophila was noted among several fruit responses to yeast cells, and was suggested to play a role in biocontrol activity.
Plant hormones are known to play essential roles in the plant response to stresses, and the roles of ethylene, salicylic acid (SA) and jasmonic acid (JA) in plant defence are well established. Ethylene is believed to be directly involved in pathogen resistance mechanisms (Mehta et al., 2007). Other plant hormones, such as gibberellins, auxins and BRs, have also been associated with plant defence and microbial pathogenesis (Koornneef and Pieterse, 2008). Our results identified an increase in the expression of two auxin response‐related genes and three genes related to the BR hormone (Table 3). It is known that the auxin signalling pathway modulates plant disease resistance both directly and indirectly (Kazan and Manners, 2009). The direct effects of auxin might involve interference with plant defence pathways (SA signalling), whereas indirect effects might involve changes in the progression of host–pathogen interaction caused by the effect of auxin on plant development (Kazan and Manners, 2009).
The relationship of hormones to induced resistance processes in plants against diseases is quite complex (Nakashita et al., 2003; Yu et al., 2008). Although changes in the expression levels of genes corresponding to ethylene, auxin and BR hormonal signalling pathways in M. fructicola‐treated grapefruit peel were observed in this study, their direct involvement in resistance mechanisms is still speculative.
In conclusion, our data provide further evidence that ROS produced by M. fructicola shortly after application to fruit tissue stimulates an oxidative burst in host tissue, leading to the activation of downstream complex responses associated with host pathogen resistance. We suggest that ROS acts as a signal to activate a MAPK cascade signalling pathway that ultimately triggers different physiological responses in fruit tissue. Overall, the main molecular mechanisms related to the host response following biocontrol agent application are summarized in Fig. 6. The significant changes in the various functional categories of genes detected following the application of yeast cells support our previous assumption on the complexity of the interactions taking place between yeast antagonists and the harvested commodity. The present study provides a detailed identification and characterization of genes and their expression in fruit tissue following the application of a yeast biocontrol agent. It would also be intriguing to study the effect of the host on the transcriptome of the biocontrol yeast cells. Having both sets of information would provide further insight into the mechanisms/processes involved in the biocontrol performance of yeast antagonists and how biocontrol efficacy can be increased.
Figure 6.
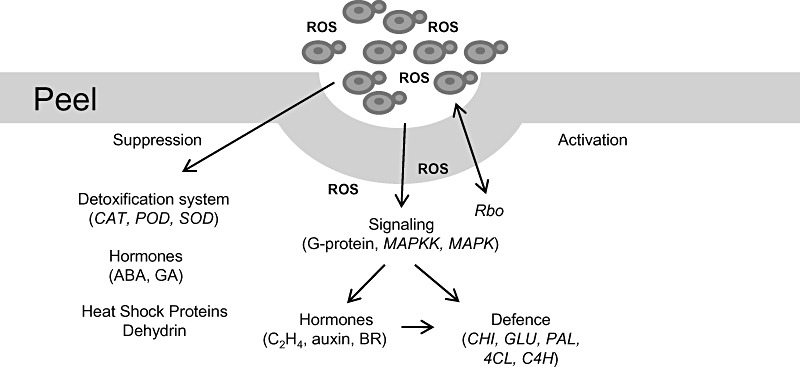
Model describing possible molecular mechanisms related to host response following biocontrol agent (Metschnikowia fructicola) application. ABA, abscisic acid; BR, brassinosteroid; CAT, catalase; C4H, trans‐cinnamate 4‐monooxygenase; CHI, chitinase; 4CL, 4‐coumarate‐CoA ligase; GA, gibberellic acid; GLU, β‐1,3‐glucanase; MAPK, mitogen‐activated protein kinase; MAPKK, mitogen‐activated protein kinase kinase; PAL, phenylalanine ammonia‐lyase; POD, peroxidase; Rbo, Respiratory burst oxidase; ROS, reactive oxygen species; SOD, superoxide dismutase.
EXPERIMENTAL PROCEDURES
Plant material and biocontrol agent
‘Ruby Star’ grapefruits were obtained from a local orchard and used within 24 h after harvest. Prior to treatment with the yeast, the fruits were washed with tap water and surface sterilized by dipping in 0.05% hypochloride for 2 min. Fruits used to prepare peel discs were wiped with 70% ethanol.
The yeast antagonist M. fructicola (Strain 277) was grown in a 250‐mL Ehrlenmeyer flask placed on an orbital shaker for 24 h at 26 °C. The growth medium was NYDB containing nutrient broth (0.8%), yeast extract (0.5%), d‐glucose (1%) and chloramphenicol (250 mg/L). Yeast cells were pelleted by centrifugation at 6000 g, washed once with sterile distilled water and resuspended in sterile water to its initial volume. The cell suspension concentration was adjusted to 1 × 108 cells/mL.
Peel disc preparation and treatment with M. fructicola
Surface‐sterilized fruits were peeled and discs were cut using a 9‐mm‐diameter cork borer under aseptic conditions. Discs were immersed for 1 min in a water suspension of M. fructicola (1 × 108 cells/mL) or in sterile water (control), and then transferred into Petri dishes lined with moist sterile filter paper. Treated and control discs were sampled following 2, 4, 24 and 48 h of incubation at 25 °C for the analysis of ROS production, as described below.
Application of M. fructicola to surface wounds
Fruits were wounded at four sites around the blossom end using a dissecting needle. Wounds were 3 × 3 mm2 in size. Thirty microlitres of either a yeast cell suspension or sterile water (control) were pipetted into each wound and allowed to dry at room temperature. Wound sites were removed using a 9‐mm cork borer under aseptic conditions after 2, 8, 24 and 48 h of incubation at 25 °C and immediately frozen in liquid nitrogen and stored at −80 °C for subsequent RNA preparation. Each sample consisted of fruit tissue pooled from five wounds collected from three fruits. Each treatment and sampling period within a treatment had three biological replicates.
RNA extraction and cDNA synthesis
For each biological replicate, five frozen peel discs were ground in liquid nitrogen with a mortar and pestle, and total RNA was extracted using an SV Total RNA Isolation System (Promega, Madison, WI, USA), according to the manufacturer's instructions. The quality and concentration of total RNA were analysed by gel electrophoresis and an ND‐1000 spectrophotometer, respectively (NanoDrop, Wilmington, DE, USA). First‐strand cDNA was synthesized with a Reverse‐iTM 1st Strand Synthesis Kit (ABgene, Epsom, Surrey, UK) from 1 µg of total RNA that had been pretreated with 1.5 units of RQ1 (Promega).
Microarray of grapefruit peel transcriptome and data analysis
RNA samples isolated from yeast‐treated and control peel discs after 24 h of incubation were further prepared for hybridization according to the protocols outlined in the GeneChip Expression Analysis Technical Manual, and hybridized to the Affymetrix Citrus GeneChip microarray (Affymetrix, Santa Clara, CA, USA). Hybridizations were performed at The Center for Genomic Technologies, Faculty of Natural Sciences, The Alexander Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem, Israel. Data analysis was performed on CEL files using Partek® Genomics Suite™ (Partek Incorporated, St. Louis, MO, USA; http://partek.com). For comparison of global gene expression in treated versus untreated samples (three biological replicates each), the array data were normalized to the mean gene expression levels with the robust multi‐average method (Irizarry et al., 2003). The t‐test was used to identify probe sets that exhibited significant changes in signal levels between treatment and control after 24 h of incubation at P≤ 0.05 and a cut‐off factor of at least 1.5. Differentially expressed genes were further analysed for gene ontology functional annotations by AgriGO, a gene ontology (GO) analysis toolkit and database for the agricultural community http://bioinfo.cau.edu.cn/agriGO/index.php (Du et al., 2010), to perform functional categorization. To analyse changes in gene expression associated with metabolic pathways and functional classification, expression data for the probes of significant datasets were mapped to the Arabidopsis metabolic pathways using MapMan software (http://gabi.rzpd./de/projects/MapMan/) (Thimm et al., 2004). The microarray data were deposited at GEO (Gene Expression Omnibus) at the National Center for Biotechnology Information (NCBI) http://www.ncbi.nlm.nih.gov/geo/. The accession number is GSE31669.
RT‐qPCR
Transcript accumulation of citrus genes encoding Rbo 2 (Rbo, Cit.25134.1.S1_at), POD (POD, ABG49115), CAT (CAT, ABG49115), G‐protein (Cit.35804.1.S1_at), MAPKK (MAPKK, Cit.25223.1.S1_at), MAPK (MAPK, EF185418), PAL 2 (PAL, AJ238753), CHI I (CHI, AF090336.1), CHS 1 (CHS, AB009351) and 4CL (4CL, Cit.11987.1.S1_s_at) were determined using a GENE 6000 instrument (Corbett Life Science, Sydney, Australia). Gene‐specific primers reported in Table S2 (see Supporting Information) were used at an optimal concentration of 200 nm. Reactions were carried out with 5 µL of Absolute QPCR SYBR Green ROX Mix (ABgene), according to the manufacturer's instructions. The RT‐qPCR conditions were as follows: incubation for 15 min at 95 °C, followed by 40 cycles of 95 °C for 15 s, 60 °C for 20 s and 72 °C for 20 s. Control, wounded, water‐treated peel discs at 24 h were designated as the calibration point, set as unity, for the relative expression levels, which were calculated according to the comparative C t method (Livak and Schmittgen, 2001), with actin (ADE05307) used as an internal standard.
Determination of ROS production in fruit peel tissue
H2O2 was measured in eight peel discs using the Amplex Red Hydrogen Peroxide assay kit (Molecular Probes, Eugene, OR, USA), according to the manufacturer's instructions. In the presence of POD, the Amplex Red reagent reacts with H2O2 and produces the red‐fluorescent oxidation product, resorufin. Extracellular H2O2 was determined by measuring fluorescence using an EnSpire 2300 Multiabel Reader (PerkinElmer™, Turku, Finland), equipped with a 530‐nm excitation and a 590‐nm reading filter. The results are given as three replications of fluorescence intensity for each time point.
Superoxide anion (O2 –) was measured using nitroblue tetrazolium (NBT) (Sigma‐Aldrich, St. Louis, MO, USA) reduction, as described by Doke (1983). Eight peel discs were immersed in 0.5 mL of 0.01 m potassium phosphate buffer, pH 7.8, containing 0.05% NBT and 10 mM NaN3 (Sigma‐Aldrich) for 1 h. After removing the discs, the mixture was heated at 85 °C for 15 min and cooled. The NBT‐reducing activity of the discs was expressed as the increase in absorbance at 580 nm/g/fresh weight (FW). The effect of SOD on the reduction of NBT by the discs was determined by adding SOD (100 mg/mL) to the reaction solution from which NaN3 was omitted.
Hydroxyl radical (OH·)was measured by the method described by Tiedemann (1997). Eight peel discs were immersed in 550 mL of 1 mm 2‐deoxyribose (DOR) as a scavenger for 45 min of incubation. Then, 0.5 mL of thiobarbituric acid (TBA) (Sigma‐Aldrich), 1% w/v in 0.05 m NaOH, and 0.5 mL of trichloroacetic acid (TCA) (Sigma‐Aldrich), 2.8% w/v, were added and immediately boiled for 10 min. Finally, samples were cooled on ice for 10 min and were measured at 540 nm using an EnSpire 2300 Multiabel Reader (PerkinElmer). The results are reported as the absorbance at 540 nm/g/FW × 1000.
Intracellular accumulation of H2O2 in M. fructicola
H2O2 accumulation in M. fructicola cells was examined using the fluorescent probe 2′,7′‐dichlorodihydrofluorescein diacetate (H2DCF‐DA) [dissolved in dimethylsulphoxide (DMSO); Molecular Probes Invitrogen, Eugene, OR, USA), as described by Deveau et al. (2010) with slight modifications. Yeast cells were collected from treated surface wounds after 5, 24 and 48 h of incubation at 25 °C under moist conditions. Cells were washed from 10 discs prepared from wounds, as described above, with 10 mL of 50 mm 2‐(N‐morpholino)ethanesulphonic acid (MES) buffer (pH 6.5) in 50‐mL sterile conical tubes shaken at 180 rpm for 30 min on an orbital shaker. Yeast cells were then pelleted by centrifugation at 6000 g for 10 min and resuspended in 500 µL of MES containing 10 µm H2DCF‐DA. The suspension was incubated in the dark for 30 min. After washing twice with MES, cells were examined by a laser scanning confocal microscope (FLUOVIEW 500, Olympus, Tokyo, Japan) as described previously (Macarisin et al., 2007). Yeast cells grown in NYDB for 24 or 48 h were used as control, following the removal of the growth medium by centrifugation and washing with 50 mm MES. Five microscopic fields containing at least 200 cells each were randomly chosen and viewed. The quantification of H2O2 accumulation in yeast cells was expressed in rpi. There were two replicates at each time point, and the experiment was repeated three times.
Statistical analysis
The H2O2, hydroxyl radical and superoxide anion level data were analysed statistically with JMP 5.0 software (SAS Institute, Cary, NC, USA) and expressed as the mean ± standard error.
Supporting information
Fig. S1 Description of the mode of action in the signalling and defence processes in response to yeast antagonist (Metschnikowia fructicola) treatment of grapefruit, based on microarray analyses in MapMan software (http://gabi.rzpd./de/projects/MapMan/) (fold change, P ≤ 0.05).
Table S1 Specific primers used for transcription analysis of mRNA by reverse transcription‐quantitative real‐time polymerase chain reaction.
Table S2 Description of the genes involved in biotic stress in response to biocontrol antagonist (Metschnikowia fructicola) treatment of grapefruit, based on microarray analyses in MapMan software (fold change, P ≤ 0.05).
Supporting info item
ACKNOWLEDGEMENTS
This research was supported in part by a grant (IS‐4268‐09) from the US–Israel Binational Agricultural Research and Development (BARD) Fund to Samir Droby and Michael Wisniewski.
REFERENCES
- Arras, G. (1999) Mode of action of an isolate of Candida famata in biological control of Penicillium digitatum in orange fruits. Postharvest Biol. Technol. 8, 191–198. [Google Scholar]
- Ballester, A. , Lafuente, M. and Gonzalez‐Candelas, L. (2006) Spatial study of antioxidant enzymes, peroxidase and phenylalanine ammonia‐lyase in the citrus fruit–Penicillium digitatum interaction. Postharvest Biol. Technol. 39, 115–124. [Google Scholar]
- Ballester, A. , Izquierdo, A. , Lafuente, M. and Gonzalez‐Candelas, L. (2010) Biochemical and molecular characterization of induced resistance against Penicillium digitatum in citrus fruit. Postharvest Biol. Technol. 56, 31–38. [Google Scholar]
- Boller, T. and Felix, J. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Boller, T. and He, S.Y. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 8, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovskii, G. , Stupnikova, I. , Antipina, A. , Vladimirova, S. and Voinikov, V. (2002) Accumulation of dehydrin‐like proteins in the mitochondria of cereals in response to cold, freezing, drought and ABA treatment. BMC Plant Biol. 2, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoria, R. , Caputo, L. , De Curtis, F. and De Cicco, V. (2003) Resistance of postharvest biocontrol yeasts to oxidative stress: a possible new mechanism of action. Phytopathology, 93, 564–572. [DOI] [PubMed] [Google Scholar]
- Chan, Z. and Tian, S. (2006) Induction of H2O2‐metabolizing enzymes and total protein synthesis by antagonistic yeast and salicylic acid in harvested sweet cherry fruit. Postharvest Biol. Technol. 39, 314–321. [Google Scholar]
- Chan, Z. , Wang, Q. , Xu, X. , Meng, X. , Qin, G. , Li, B. and Tian, S. (2008) Functions of defense‐related proteins and dehydrogenases in resistance response induced by salicylic acid in sweet cherry fruits at different maturity stages. Proteomics, 8, 4791–4807. [DOI] [PubMed] [Google Scholar]
- Deveau, A. , Pispanen, A. , Jackson, A. and Hogan, D. (2010) Farnesol induces hydrogen peroxide resistance in Candida albicans yeast by inhibiting the Ras‐cyclic AMP signaling pathway. Eukaryot. Cell, 9, 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R. and Paiva, N. (1995) Stress‐induced phenylpropanoid metabolism. Plant Cell, 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke, N. (1983) Involvement of superoxide generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophtora infestans and to hyphal wall contents. Physiol. Plant Pathol. 23, 345–357. [Google Scholar]
- Droby, S. , Vinokur, V. , Weiss, B. , Cohen, L. , Daus, A. , Goldschmidt, E. and Porat, R. (2002) Induction of resistance to Penicillium digitatum in grapefruit by the yeast biocontrol agent Candida oleophila . Biol. Control, 92, 393–399. [DOI] [PubMed] [Google Scholar]
- Droby, S. , Wisniewski, M. , Macarisin, D. and Wilson, C. (2009) Twenty years of postharvest biocontrol research: is it time for a new paradigm? Postharvest Biol. Technol. 52, 137–145. [Google Scholar]
- Du, Z. , Zhou, X. , Ling, Y. , Zhang, Z. and Su, Z. (2010) AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Ghaouth, A. , Wilson, C. and Wishniewski, M. (1998) Ultrastructural and cytochemical aspects of the biological control of Botrytis cinerea by Candida saitoana in apple fruit. Phytopathology, 88, 282–291. [DOI] [PubMed] [Google Scholar]
- Felix, J. , Duran, J.D. , Volko, S. and Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Forment, J. , Gadea, J. , Huerta, L. , Abizanda, L. , Agusti, J. , Alamar, S. , Alos, E. , Andres, F. , Arribas, R. , Beltran, J. , Berbel, A. , Blazquez, M. , Brumos, J. , Canas, L. , Cercos, M. , Colmenero‐Flores, J. , Conesa, A. , Estables, B. , Gandia, M. , Garcia‐Martinez, J. , Gimeno, J. , Gisbert, A. , Gomez, G. , Gonzalez‐Candelas, L. , Granell, A. , Guerri, J. , Lafuente, M. , Madueno, F. , Marcos, J. , Marques, M. , Martinez, F. , Martinez‐Godoy, M. , Miralles, S. , Moreno, P. , Navarro, L. , Pallas, V. , Perez‐Amador, M. , Perez‐Valle, J. , Pons, C. , Rodrigo, I. , Rodriguez, P. , Royo, C. , Serrano, R. , Soler, G. , Tadeo, F. , Talon, M. , Terol, J. , Trenor, M. , Vaello, L. , Vicente, O. , Vidal, C. , Zacarias, L. and Conejero, V. (2005) Development of a citrus genome‐wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol. Biol. 57, 375–391. [DOI] [PubMed] [Google Scholar]
- González‐Candelas, L. , Alamar, S. , Sánchez‐Torres, P. , Zacarías, L. and Marcos, J.F. (2010) A transcriptomic approach highlights induction of secondary metabolism in citrus fruit in response to Penicillium digitatum infection. BMC Plant Biol. 10, 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, M. , Fujinaga, M. and Kuboi, T. (2004) Radical scavenging activity and oxidative modification of citrus dehydrin. Plant Physiol. Biochem. 42, 657–662. [DOI] [PubMed] [Google Scholar]
- Huckelhoven, R. (2007) Cell wall‐associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. [DOI] [PubMed] [Google Scholar]
- Ippolito, A. , El Ghaouth, A. , Wilson, C.L. and Wisniewski, M. (2000) Control of postharvest decay of apple fruit by Aureobasidium pullulans and induction of defense responses. Postharvest Biol. Technol. 19, 265–272. [Google Scholar]
- Irizarry, R. , Hobbs, B. , Collin, F. , Beazer‐Barclay, Y. , Antonellis, K. and Scherf, U. (2003) Speed T: exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Jiang, F. , Chen, J. , Miao, Y. , Krupinska, K. and Zheng, X. (2009) Identification of differentially expressed genes from cherry tomato fruit (Lycopersicon esculentum) after application of the biological control yeast Cryptococcus laurentii . Postharvest Biol. Technol. 53, 131–137. [Google Scholar]
- Kazan, K. and Manners, J. (2009) Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci. 14, 373–382. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. and Pieterse, C. (2008) Cross talk in defense signaling. Plant Physiol. 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Wisniewski, M. , Droby, S. , Tian, S. , Hershkovitz, V. and Tworkoski, T. (2011) Effect of heat shock treatment on stress tolerance and biocontrol efficacy of Metschnikowia fructicola . FEMS Microbiol. Ecol. 76, 145–155. [DOI] [PubMed] [Google Scholar]
- Livak, K. and Schmittgen, T. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2–ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Macarisin, D. , Cohen, L. , Eick, A. , Rafael, G. , Belausov, E. , Wisniewski, M. and Droby, S. (2007) Penicillium digitatum suppresses production of hydrogen peroxide in host tissue during infection of citrus fruit. Phytopathology, 97, 1491–1500. [DOI] [PubMed] [Google Scholar]
- Macarisin, D. , Droby, S. , Bauchan, G. and Wisniewski, M. (2010) Superoxide anion and hydrogen peroxide in the yeast antagonist–fruit interaction: a new role for reactive oxygen species in postharvest biocontrol? Postharvest Biol. Technol. 58, 194–202. [Google Scholar]
- Maul, P. , McCollum, G. , Popp, M. , Guy, C. and Porat, R. (2008) Transcriptome profiling of grapefruit flavedo following exposure to low temperature and conditioning treatments uncovers principal molecular components involved in chilling tolerance and susceptibility. Plant Cell Environ. 31, 752–768. [DOI] [PubMed] [Google Scholar]
- Mehta, A. , Silva, M. , Guidetti‐Gonzalez, S. , Carrer, H. , Takita, M. and Martins, N. (2007) Signaling pathways in a Citrus EST database. Gen. Mol. Biol. 30, 734–751. [Google Scholar]
- Mittler, R. , Hallak Herr, E. , Orvar, B. , van Camp, W. , Willekens, H. , Inze, D. and Ellis, B. (1999) Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. 96, 14 165–14 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. , Vanderauwera, S. , Gollery, M. and Van Breusegem, F. (2004) Reactive oxygen network of plants. Trends Plant Sci. 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Nakashita, H. , Yasuda, M. , Nitta, T. , Asami, T. , Fujioka, S. , Arai, Y. , Sekimata, K. , Takatsuto, S. , Yamaguchi, I. and Yoshida, S. (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898. [DOI] [PubMed] [Google Scholar]
- Pieterse, C. and Van Loon, L. (1999) Salicylic acid‐independent plant defense pathways. Trends Plant Sci. 4, 52–58. [DOI] [PubMed] [Google Scholar]
- Rodov, V. , Ben‐Yehoshua, S. , D'hallewin, S. and Castia, T. (1994) Accumulation of phytoalexins scoparone and scolopetin in citrus fruits subjected to various postharvest treatments. Acta Hortic. 381, 517–523. [Google Scholar]
- Thimm, O. , Blasing, O. , Gibon, Y. , Nagel, A. , Meyer, S. , Kruger, P. , Selbig, J. , Muller, L. , Rhee, S. and Stitt, M. (2004) Mapman: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Tiedemann, A. (1997) Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea . Physiol. Mol. Plant Pathol. 50, 151–156. [Google Scholar]
- Torres, M. (2010) ROS in biotic interactions. Physiol. Plant. 138, 414–429. [DOI] [PubMed] [Google Scholar]
- Torres, M. , Jones, J. and Dangl, J. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon, L. , Rep, M. and Pieterse, C. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Xu, X. , Qin, G. and Tian, S. (2008) Effect of microbial biocontrol agents on alleviating oxidative damage of peach fruit subjected to fungal pathogen. Int. J. Food Microbiol. 126, 153–158. [DOI] [PubMed] [Google Scholar]
- Yang, K. , Liu, Y. and Zhang, S. (2001) Activation of a mitogen‐activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. 98, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T. and Zheng, X. (2007) Indole‐3‐acetic acid enhances the biocontrol of Penicillium expansum and Botrytis cinerea on pear fruit by Cryptococcus laurentii . Biol. Control, 7, 459–464. [DOI] [PubMed] [Google Scholar]
- Yu, T. , Zhang, H. , Li, X. and Zheng, X. (2008) Biocontrol of Botrytis cinerea in apple fruit by Cryptococcus laurentii and indole‐3‐acetic acid. Biol. Control, 46, 171–177. [Google Scholar]
- Zhang, S. and Klessig, D. (2001) MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]
- Zhao, S. and Qi, X. (2008) Signaling in plant disease resistance and symbiosis. J. Integr. Plant Biol. 50, 799–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Description of the mode of action in the signalling and defence processes in response to yeast antagonist (Metschnikowia fructicola) treatment of grapefruit, based on microarray analyses in MapMan software (http://gabi.rzpd./de/projects/MapMan/) (fold change, P ≤ 0.05).
Table S1 Specific primers used for transcription analysis of mRNA by reverse transcription‐quantitative real‐time polymerase chain reaction.
Table S2 Description of the genes involved in biotic stress in response to biocontrol antagonist (Metschnikowia fructicola) treatment of grapefruit, based on microarray analyses in MapMan software (fold change, P ≤ 0.05).
Supporting info item


