Summary
The recognition of pathogen effectors by plant immune receptors leads to the activation of immune responses that often include a hypersensitive response (HR): rapid and localized host cell death surrounding the site of attempted pathogen ingress. We have demonstrated previously that the recognition of the Verticillium dahliae effector protein Ave1 by the tomato immune receptor Ve1 triggers an HR in tomato and tobacco. Furthermore, we have demonstrated that tomato Ve1 provides Verticillium resistance in Arabidopsis upon Ave1 recognition. In this study, we investigated whether the co‐expression of Ve1 and Ave1 in Arabidopsis results in an HR, which could facilitate a forward genetics screen. Surprisingly, we found that the co‐expression of Ve1 and Ave1 does not induce an HR in Arabidopsis. These results suggest that an HR may occur as a consequence of Ve1/Ave1‐induced immune signalling in tomato and tobacco, but is not absolutely required for Verticillium resistance.
Introduction
Immunity in plants against pathogens is generally governed by immune receptors that detect pathogen (‐induced) ligands of various nature (Boller and Felix, 2009; Thomma et al., 2011). The recognition of such ligands by immune receptors results in the activation of defence responses, which are often accompanied by a hypersensitive response (HR), in which necrosis of plant tissue surrounding the site of attempted penetration is activated to stop further pathogen colonization.
Verticillium spp. are economically important pathogens that cause vascular wilt diseases in a wide range of plant species worldwide, with V. dahliae and V. albo‐atrum as the main pathogenic species (Fradin and Thomma, 2006; Klosterman et al., 2009). The interaction between tomato and V. dahliae has been established as a model to study the interaction between plants and vascular pathogens (Fradin and Thomma, 2006; Fradin et al., 2009). In this model, immunity against V. dahliae is governed by the interaction between the tomato gene Ve1 and the V. dahliae gene Ave1 (de Jonge et al., 2012; Fradin et al., 2009). Ve1 encodes a receptor‐like protein (RLP)‐type immune receptor that carries extracellular leucine‐rich repeats (eLRRs), a single‐pass transmembrane (TM) domain and a short cytoplasmic tail that lacks obvious motifs for intracellular signalling (Fradin and Thomma, 2006; Fradin et al., 2009; Kawchuk et al., 2001). Ave1 encodes an effector protein that has a high degree of homology to plant natriuretic peptides and is secreted by Verticillium during host colonization (de Jonge et al., 2012).
Various RLPs have been shown to play roles in plant development or in pathogen resistance in several plant species (Wang et al., 2010). However, the genetics of RLP‐mediated disease resistance signalling has been most extensively studied in solanaceous plants, making use of the tomato Cf, Ve and LeEix proteins, and also exploiting tobacco as a heterologous model species (Bar et al., 2010; Gabriels et al., 2006, 2007; van der Hoorn et al., 2000; Ron and Avni, 2004; Vossen et al., 2010). One of the tools that has been exploited is the progeny of a cross of Cf‐4 tomato with transgenic tomato lines expressing the cognate Cladosporium fulvum effector gene Avr4, which results in Cf‐4/Avr4 offspring that display lethality at the seedling stage, but can be rescued on incubation at 33 °C (Cai et al., 2001; de Jong et al., 2002; Thomas et al., 1997). On transfer to 20 °C, a synchronous systemic HR is activated, which has been employed successfully to study Cf‐4 signalling (de Jong et al., 2002; Gabriels et al., 2006; Stulemeijer et al., 2007).
As a result of the lack of RLPs that have been implicated in the immune signalling of Arabidopsis thaliana, the many resources that are available for this model species have only been exploited to a limited extent thus far (Wang et al., 2008, 2010). Interestingly, it has been demonstrated recently that interfamily transfer of tomato Ve1 into Arabidopsis results in resistance against race 1 strains of V. dahliae and V. albo‐atrum (Fradin et al., 2011). Moreover, based on mutant analysis, the requirement of SERK (somatic embryogenesis receptor‐like kinase) family members for Ve1 resistance has been investigated in Arabidopsis, demonstrating a critical role for SERK1 in addition to SERK3/BAK1 (brassinosteroid‐associated kinase 1). With virus‐induced gene silencing, the requirement of SERK1 for Ve1‐mediated resistance was subsequently confirmed in tomato, demonstrating that Arabidopsis can be used to characterize Ve1 signalling (Fradin et al., 2011).
We have demonstrated recently that the Potato virus X (PVX)‐mediated transient expression of Ave1 specifically triggers HR on tomato carrying Ve1 (de Jonge et al., 2012). In addition, transient co‐expression of Ave1 and Ve1 through Agrobacterium tumefaciens‐mediated transient transformation (agroinfiltration) in Nicotiana tabacum and N. glutinosa similarly induces HR (de Jonge et al., 2012; Zhang et al., 2012). In this study, we investigated whether the co‐expression of Ve1 and Ave1 in Arabidopsis results in an HR that can be used as read‐out to investigate RLP signalling mediated by Ve1. Surprisingly, we found that, although Ave1 is able to trigger an HR in resistant tomato and Nicotiana plants, such an HR does not occur in Arabidopsis. However, our results show that the HR is not required for Ve1‐mediated resistance in this species.
Results
Agroinfiltration in Arabidopsis leaves
Previously, it has been demonstrated that agroinfiltration can be employed to study immune receptor‐mediated HR in Arabidopsis (Lee and Yang, 2006). To investigate whether agroinfiltration can similarly be exploited in Arabidopsis, A. tumefaciens carrying a construct for the constitutive expression of VdNLP2 was infiltrated into Arabidopsis leaves. It has been shown recently that the VdNLP2 protein from V. dahliae exhibits cytotoxic activity on infiltration into Arabidopsis leaves (Santhanam et al., 2013; Zhou et al., 2012). Indeed, agroinfiltration of VdNLP2 resulted in clear necrosis within 7 days (Fig. 1A). Next, A. tumefaciens carrying a construct for the constitutive expression of C‐terminally green fluorescent protein (GFP)‐tagged Ve1 was infiltrated into the leaves of 3‐week‐old Arabidopsis plants. As a control, A. tumefaciens carrying a construct for the constitutive expression of untagged Ve1 was infiltrated. At 2 days post‐infiltration (dpi), GFP fluorescence was detected in leaves infiltrated with A. tumefaciens carrying a construct for the expression of GFP‐tagged Ve1, which was not observed on expression of untagged Ve1 (Fig. 1B). Closer inspection of the localization of GFP‐tagged Ve1 in N. benthamiana revealed that Ve1 localizes to the plasma membrane (Fig. S1, see Supporting Information), as predicted previously (Fradin et al., 2009; Kawchuk et al., 2001). Collectively, these results confirm that agroinfiltration can be used for the transgenic expression of Ve1 in Arabidopsis.
Figure 1.
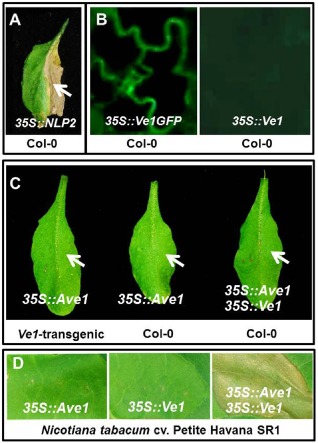
Co‐expression of Ave1 and Ve1 in Arabidopsis leaves does not induce a hypersensitive response (HR). (A) Infiltration of Agrobacterium tumefaciens carrying a construct for constitutive VdNLP2 expression into Arabidopsis leaves results in clear necrosis at 7 days post‐infiltration (dpi). (B) Leaves of wild‐type Arabidopsis Col‐0 plants were infiltrated with A. tumefaciens cultures carrying 35S::Ve1GFP or 35S::Ve1. Green fluorescent protein (GFP) fluorescence was only detected in the leaf infiltrated with A. tumefaciens carrying a construct for the expression of GFP‐tagged Ve1 at 2 dpi. (C) Leaves of wild‐type or Ve1‐expressing Arabidopsis plants were infiltrated with A. tumefaciens carrying 35S::Ave1. In addition, leaves of wild‐type Arabidopsis were co‐infiltrated with A. tumefaciens carrying 35S::Ave1 and 35S::Ve1. No necrosis was observed in infiltrated Arabidopsis leaves. (D) Co‐expression of Ve1 and Ave1 in Nicotiana tabacum cv. Petite Havana SR1 results in HR.
Co‐expression of Ave1 and Ve1 in Arabidopsis does not induce HR
To test whether Ve1‐mediated recognition of Ave1 results in HR in Arabidopsis, transient expression of Ave1 by agroinfiltration was pursued in Ve1‐transgenic Arabidopsis. However, up to 7 dpi, no signs of necrosis could be observed in agroinfiltrated Arabidopsis plants (Fig. 1C). Similarly, the co‐expression of Ave1 and Ve1 in wild‐type Arabidopsis did not lead to HR (Fig. 1C). In contrast, the co‐expression of Ave1 and Ve1 in Nicotiana tabacum cv. Petite Havana SR1 resulted in clear HR within 5 dpi (Fig. 1D; Zhang et al., 2012).
To further investigate whether Ve1‐mediated recognition of Ave1 induces HR in Arabidopsis, stable co‐expression of Ave1 and Ve1 was pursued. To this end, the Ave1 coding sequence was cloned into the binary vector pFAST‐R02 (Shimada et al., 2010) to generate the expression construct pFAST::Ave1, in which Ave1 expression was driven by the cauliflower mosaic virus (CaMV) 35S promoter. This construct contains a nondestructive red fluorescent protein (RFP) marker to identify transgenic seeds by UV microscopy (Shimada et al., 2010). The pFAST::Ave1 construct was subsequently transformed into Ve1‐transgenic and wild‐type Arabidopsis (Clough and Bent, 1998), and Ave1‐transgenic seeds were selected. Unexpectedly, the seeds germinated, and seedlings developed into mature plants that successfully set seeds. To evaluate the growth in more detail, three independent transgenic lines (named Ve1/Ave1‐1, Ve1/Ave1‐2 and Ve1/Ave1‐3), which carry both the Ve1 and Ave1 genes, were grown on Murashige and Skoog (MS) medium in a growth chamber at 22 °C, or on soil in the glasshouse, alongside Ve1‐transgenic, Ave1‐transgenic and nontransgenic control plants. No phenotypic alterations were observed in plants that co‐expressed Ve1 and Ave1 when compared with plants that expressed either of the transgenes alone or nontransgenic controls (Fig. 2A). As it has been demonstrated for tomato that the HR can be suppressed by elevated temperature (de Jong et al., 2002), we also grew the plants at 16 °C. However, also under these conditions, no necrosis or growth inhibition was observed (Fig. 2A). Reverse transcription‐polymerase chain reaction (RT‐PCR) was performed to confirm the simultaneous expression of Ve1 and Ave1 in these lines (Fig. 2B).
Figure 2.
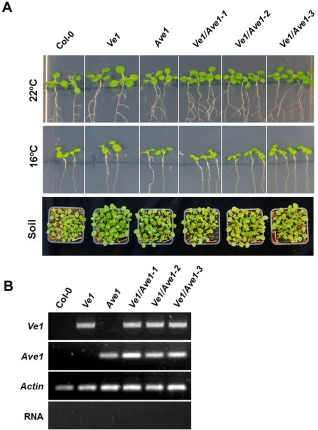
Stable co‐expression of Ve1 and Ave1 does not affect Arabidopsis viability. (A) No phenotypical alterations were observed in plants that co‐expressed Ve1 and Ave1 when compared with plants that expressed either of the transgenes alone or nontransgenic control plants. Arabidopsis plants were grown on Murashige and Skoog (MS) medium in a growth chamber at 22 °C or 16 °C, or on soil in the glasshouse. (B) Reverse transcription‐polymerase chain reaction (RT‐PCR) was performed to confirm the expression of Ve1 and Ave1 in the transgenic lines.
We further examined the potential occurrence of micro‐HR‐like lesions microscopically on trypan blue staining of Ve1/Ave1‐1, Ve1/Ave1‐2 and Ve1/Ave1‐3 plants. No micro‐HR‐like lesions were observed in the three independent transgenic lines that co‐expressed Ve1 and Ave1 when compared with plants that expressed either of the transgenes alone or nontransgenic controls (Fig. 3). These results further confirm that HR is not induced in Arabidopsis on co‐expression of Ve1 and Ave1.
Figure 3.
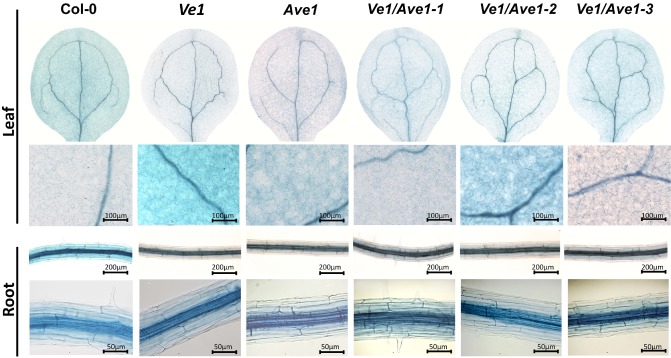
Microscopic examination of trypan blue‐stained Arabidopsis seedlings co‐expressing Ve1 and Ave1. No micro‐hypersensitive response (HR)‐like lesions were observed in plants that co‐expressed Ve1 and Ave1 or in plants that expressed either of the transgenes alone or in nontransgenic control plants.
In planta‐expressed Ave1 activates Ve1‐mediated HR in N. tabacum
Previously, we have shown that transient expression of Ave1 by PVX specifically induces HR in resistant tomato carrying Ve1 (de Jonge et al., 2012). In addition, HR can also be induced in tobacco on co‐expression of Ave1 and Ve1 by agroinfiltration (Zhang et al., 2012). These experiments demonstrate that in planta‐expressed Ave1 is able to activate Ve1‐mediated HR. However, as both transient and stable expression of Ave1 did not induce HR in Ve1‐transgenic Arabidopsis, we investigated whether the Ave1 protein produced in Arabidopsis can be recognized by Ve1. To this end, apoplastic fluid (AF) was extracted from leaf tissue of Ave1‐transgenic and wild‐type Arabidopsis by the vacuum infiltration–centrifugation technique (Joosten, 2012). The AF obtained was subsequently infiltrated into the leaves of N. tabacum transiently expressing Ve1 or its nonfunctional homologue Ve2 (Zhang et al., 2012). By 3 dpi, Ve1‐expressing leaves developed clear necrosis when AF of Ave1‐transgenic Arabidopsis was infiltrated (Fig. 4). In contrast, AF of wild‐type Arabidopsis did not induce necrosis in Ve1‐expressing N. tabacum. Furthermore, AF of neither Ave1‐transgenic nor wild‐type Arabidopsis induced HR in N. tabacum leaves expressing Ve2 (Fig. 4). These data demonstrate that the Ave1 protein expressed in transgenic Arabidopsis potentially can activate Ve1‐mediated HR.
Figure 4.
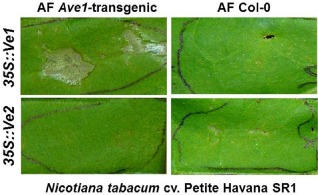
In planta‐expressed Ave1 triggers a Ve1‐mediated hypersensitive response (HR) in Nicotiana tabacum. Apoplastic fluid (AF) extracted from Ave1‐transgenic Arabidopsis induced HR in Ve1‐expressing, but not in Ve2‐expressing, N. tabacum leaves. AF from wild‐type Arabidopsis (Col‐0) did not induce HR.
Inoculation of Ve1/Ave1‐transgenic Arabidopsis
We hypothesized that the co‐expression of Ve1 with Ave1 may result in the constitutive activation of plant immunity in Arabidopsis in the absence of HR. However, considering the absence of a visible phenotype, such as dwarfing, which is typically observed in Arabidopsis constitutive defence mutants, such as cpr, cim and other constitutive PR expression mutants (Bowling et al., 1994; Cheng et al., 2011; Gou et al., 2009; Maleck et al., 2002), strongly elevated defence is not expected in the lines that co‐express Ve1 and Ave1. We challenged all transgenic lines and wild‐type plants with the V. dahliae race 1 strain JR2. As expected, nontransgenic and Ave1‐transgenic plants displayed typical Verticillium wilt symptoms on V. dahliae inoculation, including wilting, stunting, chlorosis and necrosis (Fig. 5A). In contrast, Ve1‐expressing plants, as well as plants that co‐expressed Ve1 and Ave1, showed clear resistance against V. dahliae (Fig. 5). Next, we inoculated Ave1 deletion mutants of V. dahliae strain JR2 (de Jonge et al., 2012) on the various genotypes. Nontransgenic and Ave1‐transgenic plants displayed similar symptoms when compared with inoculation with the wild‐type fungal strain and, as shown previously, Ve1‐expressing plants were not able to provide resistance against Ave1 deletion mutants (Fig. 5; de Jonge et al., 2012). Surprisingly, however, also plants that co‐expressed Ve1 and Ave1 were susceptible to the Ave1 deletion mutants (Fig. 5). These data suggest that the co‐expression of Ve1 and Ave1 does not activate basal defence against fungal infection.
Figure 5.
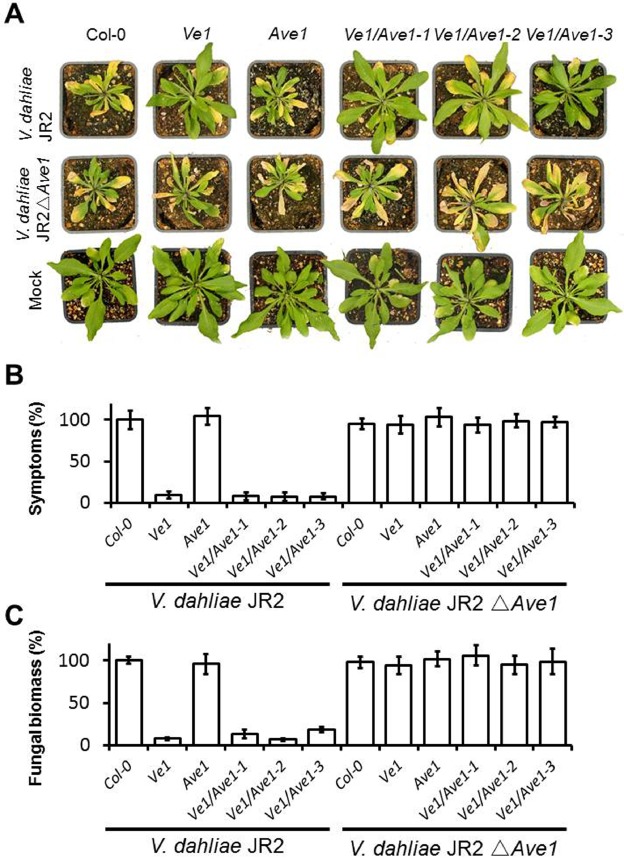
Inoculation of Ve1/Ave1‐transgenic Arabidopsis with Verticillium dahliae. (A) Typical appearance of nontransgenic and transgenic Arabidopsis lines on mock inoculation or inoculation with V. dahliae race 1 isolate JR2 or the Ave1 deletion strain (JR2ΔAve1). (B) Quantification of Verticillium wilt symptoms in wild‐type and transgenic Arabidopsis. Bars represent quantification of symptom development shown as the percentage of diseased rosette leaves. Symptoms on Col‐0 are set to 100%. (C) Fungal biomass determined by real‐time polymerase chain reaction (PCR) in wild‐type Arabidopsis and transgenic lines. Bars represent Verticillium internal transcribed spacer (ITS) transcript levels relative to Arabidopsis RuBisCo transcript levels (for equilibration). The fungal biomass in Col‐0 is set to 100%. Data from a representative experiment are shown.
To corroborate that the co‐expression of Ve1 and Ave1 does not activate basal defence, inoculation of the transgenic lines with the bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000 was performed. Also, in this case, no increased resistance was observed in plants that co‐expressed Ve1 and Ave1 (Fig. 6), confirming that basal defence is not activated through the co‐expression of Ve1 and Ave1.
Figure 6.
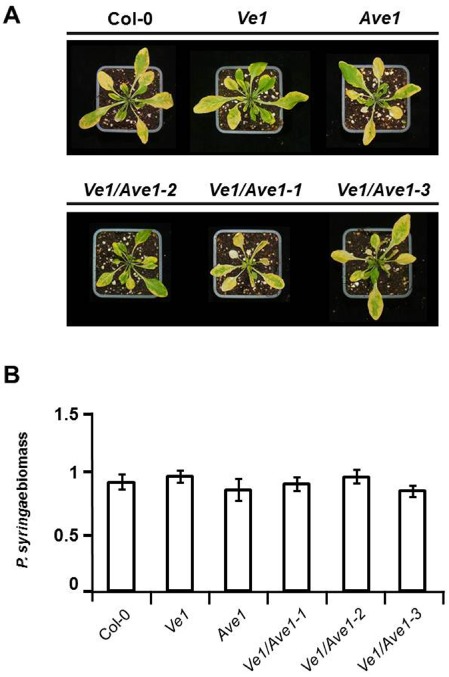
Inoculation of Ve1/Ave1‐transgenic Arabidopsis with Pseudomonas syringae pv. tomato strain DC3000. (A) Typical appearance of nontransgenic and transgenic Arabidopsis lines on mock inoculation or inoculation with P. syringae. (B) Bacterial biomass determined by real‐time polymerase chain reaction (PCR) in wild‐type Arabidopsis and transgenic lines. Bars represent levels of the P. syringae Oprf gene relative to Arabidopsis RuBisCo transcript levels (for equilibration).
Discussion
Recently, the V. dahliae effector that activates immunity in Ve1 tomato plants has been identified through population genomics as Ave1, a protein that has homology to plant natriuretic peptides (de Jonge et al., 2012). When expressed through PVX in Ve1‐carrying tomato, and when co‐expressed with Ve1 through agroinfiltration in N. tabacum and N. glutinosa, it has been demonstrated that the combination of Ve1 and Ave1 induces HR (de Jonge et al., 2012). Furthermore, it has been shown recently that Ve1‐transgenic Arabidopsis is resistant to race 1 strains of V. dahliae and V. albo‐atrum, demonstrating that Ve1 remains fully functional after interfamily transfer into Arabidopsis (Fradin et al., 2011). Tomato crosses, in which the cross involves plant lines that express a pathogen effector and corresponding RLP immune receptor, set seeds normally, but develop seedling lethality on germination of the seeds (Cai et al., 2001; de Jong et al., 2002; Thomas et al., 1997). With this knowledge, we aimed to develop a similar ‘dying seedling’ in Arabidopsis. The mutagenesis of such seeds would allow for a simple forward genetics screen, as seeds that survive after germination are probably affected in signalling components downstream of the immune receptor if they are not mutagenized in either the Ave1 or Ve1 transgene. Unexpectedly, however, we failed to identify the HR on co‐expression of Ve1 and Ave1 in transient assays and on stable transformation in Arabidopsis. Moreover, the progreny of a cross of Arabidopsis plants constitutively expressing Ve1 and Ave1 resulted in progeny that did not show any obvious phenotypical differences when compared with nontransgenic wild‐type plants.
Traditionally, the HR was considered as a defence mechanism that prevents pathogen growth directly (Spoel and Dong, 2012). However, a growing number of examples have reported on immunity in the absence of HR mediated by nucleotide‐binding site leucine‐rich repeat (NBS‐LRR)‐type immune receptors, suggesting that HR and immunity to infection are genetically separable. These examples include Rx1‐ and Rx2‐mediated resistance to PVX in potato (Bendahmane et al., 1999, 2000), Mla1‐mediated resistance against the powdery mildew pathogen Blumeria graminis f. sp. hordei in barley (Bieri et al., 2004), Rrs1‐mediated resistance against the scald pathogen Rhynchosporium secalis (Lehnackers and Knogge, 1990; Rohe et al., 1995), Rdg2a‐mediated resistance to the leaf stripe pathogen Pyrenophora graminea in barley (Bulgarelli et al., 2010) and RPS4‐mediated resistance to Pseudomonas syringae in Arabidopsis (Gassmann, 2005). In Arabidopsis, a genetic separation of disease resistance and the HR was first described for the dnd1 mutant (defence, no death 1; Clough et al., 2000), which shows resistance to Pseudomonas syringae bacteria expressing the avirulence genes avrRpt2, avrB, avrRpm1 and avrRps4 in the absence of an HR. Similarly, the Arabidopsis mutant hlm1 (HR‐like lesion mimic; Balagué et al., 2003) and its allelic mutant dnd2 (Jurkowski et al., 2004) were found to display resistance in the absence of HR. In addition to NBS‐LRR‐type immune receptors, also for Cf‐4, an LRR‐RLP‐type receptor, HR and resistance to C. fulvum on Avr4 recognition could be separated (Stulemeijer et al., 2007). Our data suggest that Ve1‐mediated Verticillium resistance in Arabidopsis also does not involve an HR. Presently, we cannot rule out the possibility that Ave1 needs to be processed in order to be recognized by Ve1, and that the lack of HR in Arabidopsis is caused by a lack of an enzyme in the apoplast of Arabidopsis that is required for the maturation of Ave1. However, the observation that Ve1‐expressing Arabidopsis plants are resistant to Verticillium infection, which is based on the recognition of Ave1 (de Jonge et al., 2012), suggests that Ave1 perception by Ve1 is functional in Arabidopsis, even if this requires Ave1 processing by host enzymes.
To date, several types of immune receptor have been identified, which can be divided into extracellular receptors and cytoplasmic receptors. Both perceive pathogen‐derived ligands or ligands that are released as a consequence of pathogen colonization to activate immune responses (Boller and Felix, 2009). Although many of these receptors activate an HR on ligand perception, others generally do not (Thomma et al., 2011). Here, we demonstrate that the occurrence of the HR may be determined by the plant species in which the receptor is expressed, as treatment with Ave1 leads to HR in tomato and tobacco plants that express Ve1, but not in N. benthamiana or in Arabidopsis (de Jonge et al., 2012; Zhang et al., 2012; this study). Nevertheless, Ve1‐expressing Arabidopsis is resistant to race 1 Verticillium strains (Fradin et al., 2011). These data suggest that the HR is not absolutely required for Verticillium wilt resistance, and may occur as a consequence of escalated signalling on Ave1 recognition in tomato and tobacco. The mechanism by which plants actually stop V. dahliae infection still requires further investigation.
Experimental Procedures
Plant materials
Arabidopsis plants were grown in the glasshouse or climate chamber with the following settings: 22/19 °C (unless mentioned otherwise) with 70% relative humidity and 16‐h/8‐h day/night periods. Supplemental light (100 W/m2) was supplied in the glasshouse when the light intensity dropped below 150 W/m2.
Generation of the constructs
To generate Ve1 fused at the 3′ end to GFP, the Ve1 coding sequence lacking the stop codon was PCR amplified using the primers attB‐Ve1‐F (5′‐GGGGACAAGTTTGTACAAAAAAGCAGGCTATGAAAATGATGGCAACTCT‐3′) and attB‐Ve1‐R‐SC (5′‐GGGGACCACTTTGTACAAGAAAGCTGGGTACTTTCTTGAAAACCAAAG‐3′). The PCR fragment was cloned into pDONR207 (Invitrogen, Carlsbad, CA, USA) through a Gateway BP reaction to generate entry vector pDONR207::Ve1‐SC. Subsequently, pDONR207::Ve1‐SC was recombined with the Gateway‐compatible destination vector pSol2095 (Zhang et al., 2012) to generate an expression construct for GFP‐tagged Ve1 driven by the constitutive CaMV 35S promoter. The fusion construct was transformed into A. tumefaciens strain GV3101 by electroporation. For the agroinfiltration of untagged Ve1, construct pMOG800::Ve1 was used (Fradin et al., 2009). pFAST::Ave1 has been described by Zhang et al. (2012).
Agrobacterium tumefaciens‐mediated transient expression
Agrobacterium tumefaciens‐containing expression constructs were infiltrated into Arabidopsis plants as described previously (van der Hoorn et al., 2000). Briefly, an overnight culture of A. tumefaciens cells was harvested at an optical density at 600 nm (OD600) of 0.8–1 by centrifugation and resuspended to a final OD of 2. Agrobacterium tumefaciens cultures containing constructs to express Ave1 or Ve1 were infiltrated into leaves of 3‐/4‐week‐old Arabidopsis plants.
RT‐PCR
Arabidopsis seedlings were collected and total RNA was extracted using the QIAGEN RNeasy extraction kit (Qiagen, Valencia, CA, USA). First‐strand cDNA was synthesized from 1 μg of total RNA, using the SuperScript™ III cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. PCRs were performed in a total volume of 25 μL with 17.9 μL water, 5 μL 5 × PCR buffer, 0.5 μL deoxynucleoside triphosphates (dNTPs), 0.5 μL of each primer, 0.1 μL GoTaq polymerase (Promega, Madison, WI, USA) and 1 μL of first‐strand cDNA. PCR was performed for 30 cycles, with denaturation at 95 °C for 15 s, annealing at 55 °C for 45 s and elongation at 72 °C for 60 s. The generated PCR products were evaluated by agarose gel electrophoresis. RT‐PCR was conducted with Ve1‐specific primers Ve1F3 (5′‐GGAACAATTTACTCAGCGGGAGC‐3′) and Ve1R4 (5′‐CCATGACTGATTCTTGAGATCGG‐3′), or Ave1‐specific primers Ave1F (5′‐CACTGGTCACTGCCGATCTA‐3′) and Ave1R (5′‐CTTGCAGGACCCTCTAGCAC‐3′). As an endogenous control, AtRub‐F3 (5′‐GCAAGTGTTGGGTTCAAAGCTGGTG‐3′) and AtRub‐R3 (5′‐CCAGGTTGAGGAGTTACTCGGAATGCTG‐3′) were used to amplify a fragment of the Arabidopsis RuBisCo gene from cDNA and from 1 μL of total RNA as control for DNA contamination.
Trypan blue staining of Arabidopsis seedlings
Two‐week‐old Arabidopsis seedlings were stained with trypan blue. To this end, whole seedlings were collected in a 1.5‐mL centrifuge tube. An adequate volume of lactophenol (1:1:1:1 volume of lactic acid : glycerol : phenol : water) with trypan blue (1 mg/mL) was added. The tubes were placed in a boiling water bath for 1–2 min. Seedlings were de‐stained in chloral hydrate and placed in 50% glycerol. The tubes were placed in a speed‐vacuum infiltrator for 1 min to remove air bubbles from the seedlings, and seedlings were mounted on microscope slides in 50% glycerol. Cell death was monitored by differential interference contrast microscopy.
Verticillium inoculations
Verticillium dahliae race 1 strain JR2 and the corresponding Ave1 deletion strain (ΔAve1) were grown on potato dextrose agar (PDA) at 22 °C. Verticillium dahliae conidia were harvested from 7–14‐day‐old fungal plates and washed with tap water. The resuspended conidia were adjusted to a final concentration of 106 conidia/mL. For inoculation, the plants were gently uprooted and rinsed in tap water. Subsequently, the roots were dipped in the conidial suspension for 3 min. As a control, plants were mock inoculated in tap water. After inoculation, plants were immediately transplanted to new pots. The inoculated plants were evaluated by observing the wilting of leaves at 21 dpi. The quantification of Verticillium biomass was performed as described previously (Ellendorff et al., 2009).
Pseudomonas syringae inoculations
Pseudomonas syringae pv. tomato strain DC3000 was cultured on King's B medium containing 200 μg/mL rifampicin. Inoculation was performed as described previously (van Esse et al., 2008). Briefly, a bacterial suspension of 5 × 108 colony‐forming units/mL in 10 mm MgCl2 and 0.05% Silwet L‐77 (Lehle Seeds) was sprayed onto the leaves until droplet runoff. Plants were incubated at 100% relative humidity for 1 h, followed by incubation at 24 °C, 60% relative humidity and a 16‐h/8‐h light/dark regime. Disease progression was scored at 4 days after inoculation.
Bacterial quantification in infected Arabidopsis plants was performed with real‐time PCR, as described previously (Brouwer et al., 2003). Briefly, real‐time PCR was conducted on DNA isolated from P. syringae‐infected Arabidopsis with primers amplifying the Arabidopsis RuBisCo gene as endogenous loading control (AtRub‐F3, GCAAGTGTTGGGTTCAAAGCTGGTG; AtRub‐R3, CCAGGTTGAGGAGTTACTCGGAATGCTG) and primers amplifying the P. syringae Oprf gene (OWB575, AACTGAAAAACACCTTGGGC; OWB576, CCTGGGTTGTTGAAGTGGTA). Real‐time PCR was conducted using an ABI7300 PCR machine (Applied Biosystems, Foster City, CA, USA) in combination with the qPCR SensiMix kit (BioLine, Taunton, MA, USA). Real‐time PCR conditions were as follows: an initial 95 °C hot start activation step for 10 min was followed by denaturation for 15 s at 95 °C, annealing for 30 s at 60 °C and extension for 30 s at 72 °C for 40 cycles.
Supporting information
Fig. S1 Ve1 localizes to the plasma membrane. (A) Localization of untagged Ve1 in Nicotiana benthamiana leaf epidermis with fluorescence microscopy at 30 h after agroinfiltration. (B) Localization of green fluorescent protein (GFP)‐tagged Ve1 in Nicotiana benthamiana leaf epidermis with fluorescence microscopy at 30 h after agroinfiltration. (C) Localization of GFP‐tagged Ve1 in Nicotiana benthamiana leaf epidermis on plasmolysis by incubation in 750 mm mannitol using confocal microscopy. (D) Bright field image of (C). (E) Overlay of (C) and (D). The arrows indicate plasma membrane detached from the cell wall. (F) Localization of GFP‐tagged Ve1 in protoplasts of Nicotiana benthamiana leaf epidermis.
Acknowledgements
Z.Z. was supported by a sandwich fellowship from Wageningen University; Z.Z and C.M.L. are partially supported by the National Natural Science Foundation of China (30625018); H.P.V.E and B.P.H.J.T. were supported by Veni and Vidi grants, respectively, from the Netherlands Organization for Scientific Research (NWO‐ALW). This research was further supported by the Centre for BioSystems Genomics (CBSG). We thank Bert Essenstam and Henk Smid for excellent plant care.
References
- Balagué, C. , Lin, B. , Alcon, C. , Flottes, G. , Malmström, S. , Köhler, C. , Neuhaus, G. , Pelletier, G. , Gaymard, F. and Roby, D. (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide‐gated channel ion channel family. Plant Cell, 15, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar, M. , Sharfman, M. , Ron, M. and Avni, A. (2010) BAK1 is required for the attenuation of ethylene‐inducing xylanase (Eix)‐induced defense responses by the decoy receptor LeEix1. Plant J. 63, 791–800. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A. , Querci, M. , Kanyuka, K. and Baulcombe, D.C. (2000) Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21, 73–81. [DOI] [PubMed] [Google Scholar]
- Bieri, S. , Mauch, S. , Shen, Q.H. , Peart, J. , Devoto, A. , Casais, C. , Ceron, F. , Schulze, S. , Steinbiss, H.H. , Shirasu, K. and Schulze‐Lefert, P. (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell, 16, 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A. , Guo, A. , Cao, H. , Gordon, A.S. , Klessig, D.F. and Dong, X.I. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell, 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer, M. , Lievens, B. , Van Hemelrijck, W. , van den Ackerveken, G. , Cammue, B.P.A. and Thomma, B.P.H.J. (2003) Quantifcation of disease progression of several microbial pathogens on Arabidopsis thaliana using real‐time fluorescence PCR. FEMS Microbiol. Lett. 228, 241–248. [DOI] [PubMed] [Google Scholar]
- Bulgarelli, D. , Biselli, C. , Collins, N.C. , Consonni, G. , Stanca, A.M. , Schulze‐Lefert, P. and Vale, G. (2010) The CC‐NB‐LRR‐type Rdg2a resistance gene confers immunity to the seed‐borne b leaf stripe pathogen in the absence of hypersensitive cell death. Plos ONE, 5, e12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, X. , Takken, F.L. , Joosten, M.H.A.J. and De Wit, P.J.G.M. (2001) Specific recognition of AVR4 and AVR9 results in distinct patterns of hypersensitive cell death in tomato, but similar patterns of defence‐related gene expression. Mol. Plant Pathol. 2, 77–86. [DOI] [PubMed] [Google Scholar]
- Cheng, Y.T. , Li, Y.Z. , Huang, S.A. , Huang, Y. , Dong, X.N. , Zhang, Y.L. and Li, X. (2011) Stability of plant immune‐receptor resistance proteins is controlled by SKP1‐CULLIN1‐F‐box (SCF)‐mediated protein degradation. Proc. Natl. Acad. Sci. USA, 108, 14 694–14 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. , Fengler, K.A. , Yu, I.C. , Lippok, B. , Smith, R.K. and Bent, A.F. (2000) The Arabidopsis dnd1 ‘defense, no death’ gene encodes a mutated cyclic nucleotide‐gated ion channel. Proc. Natl. Acad. Sci. USA, 97, 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellendorff, U. , Fradin, E.F. , de Jonge, R. and Thomma, B.P.H.J. (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse, H. , van't Klooster, J.W. , Bolton, M.D. , Yadeta, K.A. , van Baarlen, P. , Boeren, S. , Vervoort, J. , de Wit, P.J.G.M. and Thomma, B.P.H.J. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell, 20, 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. and Thomma, B.P.H.J. (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo‐atrum . Mol. Plant Pathol. 7, 71–86. [DOI] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Juarez Ayala, J.C. , Castroverde, C.D. , Nazar, R.N. , Robb, J. , Liu, C.M. and Thomma, B.P.H.J. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. , Abd‐El‐Haliem, A. , Masini, L. , van den Berg, G.C. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriels, S.H.E.J. , Takken, F.L.W. , Vossen, J.H. , de Jong, C.F. , Liu, Q. , Turk, S.C.H.J. , Wachowski, L.K. , Peters, J. , Witsenboer, H.M.A. , de Wit, P.J.G.M. and Joosten, M.H.A.J. (2006) cDNA‐AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol. Plant–Microbe Interact. 19, 567–576. [DOI] [PubMed] [Google Scholar]
- Gabriels, S.H.E.J. , Vossen, J.H. , Ekengren, S.K. , van Ooijen, G. , Abd‐El‐Haliem, A.M. , van den Berg, G.C.M. , Rainey, D.Y. , Martin, G.B. , Takken, F.L.W. , de Wit, P.J.G.M. and Joosten, M.H.A.J. (2007) An NB‐LRR protein required for HR signalling mediated by both extra‐ and intracellular resistance proteins. Plant J. 50, 14–28. [DOI] [PubMed] [Google Scholar]
- Gassmann, W. (2005) Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol. Plant–Microbe Interact. 18, 1054–1060. [DOI] [PubMed] [Google Scholar]
- Gou, M. , Su, N. , Zheng, J. , Huai, J. , Wu, G. , Zhao, J. , He, J. , Tang, D. , Yang, S. and Wang, G. (2009) An F‐box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60, 757–770. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. , Laurent, F. , Roth, R. and De Wit, P.J.G.M. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf‐9‐induced and Avr4/Cf‐4‐induced necrosis. Mol. Plant–Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- de Jong, C.F. , Takken, F.L.W. , Cai, X.H. , de Wit, P.J.G.M. and Joosten, M.H.A.J. (2002) Attenuation of Cf‐mediated defense responses at elevated temperatures correlates with a decrease in elicitor‐binding sites. Mol. Plant–Microbe Interact. 15, 1040–1049. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M.K. , Zhang, Z. , Usami, T. , Lievens, B. , Subbarao, K.V. and Thomma, B.P.H.J. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, M.H.A.J. (2012) Isolation of apoplastic fluid from leaf tissue by the vacuum infiltration–centrifugation technique. Methods Mol. Biol. 835, 603–610. [DOI] [PubMed] [Google Scholar]
- Jurkowski, G.I. , Smith, R.K. , Yu, I.C. , Ham, J.H. , Sharma, S.B. , Klessig, D.F. , Fengler, K.A. and Bent, A.F. (2004) Arabidopsis DND2, a second cyclic nucleotide‐gated ion channel gene for which mutation causes the ‘defense, no death’ phenotype. Mol. Plant–Microbe Interact. 17, 511–520. [DOI] [PubMed] [Google Scholar]
- Kawchuk, L.M. , Hachey, J. , Lynch, D.R. , Kulcsar, F. , van Rooijen, G. , Waterer, D.R. , Robertson, A. , Kokko, E. , Byers, R. , Howard, R.J. , Fischer, R. and Prufer, D. (2001) Tomato Ve disease resistance genes encode cell surface‐like receptors. Proc. Natl. Acad. Sci. USA, 98, 6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman, S.J. , Atallah, Z.K. , Vallad, G.E. and Subbarao, K.V. (2009) Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. [DOI] [PubMed] [Google Scholar]
- Lee, M.W. and Yang, Y. (2006) Transient expression assay by agroinfiltration of leaves. Methods Mol. Biol. 323, 225–229. [DOI] [PubMed] [Google Scholar]
- Lehnackers, H. and Knogge, W. (1990) Cytological studies on the infection of barley cultivars with known resistance genotypes by Rhynchosporium secalis . Can. J. Bot. 68, 1953–1961. [Google Scholar]
- Maleck, K. , Neuenschwander, U. , Cade, R.M. , Dietrich, R.A. , Dangl, J.L. and Ryals, J.A. (2002) Isolation and characterization of broad‐spectrum disease‐resistant Arabidopsis mutants. Genetics, 160, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohe, M. , Gierlich, A. , Hermann, H. , Hahn, M. , Schmidt, B. , Rosahl, S. and Knogge, W. (1995) The race‐specific elicitor, Nip1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 14, 4168–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, M. and Avni, A. (2004) The receptor for the fungal elicitor ethylene‐inducing xylanase is a member of a resistance‐like gene family in tomato. Plant Cell, 16, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam, P. , Esse, H.P. , Albert, I. , Faino, L. , Nürnberger, T. and Thomma, B.P.H.J. (2013) Evidence for functional diversification within a fungal NEP1‐like protein family. Mol. Plant–Microbe Interact. 26, 278–286. [DOI] [PubMed] [Google Scholar]
- Shimada, T.L. , Shimada, T. and Hara‐Nishimura, I. (2010) A rapid and non‐destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana . Plant J. 61, 519–528. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. and Dong, X.N. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. [DOI] [PubMed] [Google Scholar]
- Stulemeijer, I.J.E. , Stratmann, J.W. and Joosten, M.H.A.J. (2007) Tomato mitogen‐activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf‐4/Avr4‐induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol. 144, 1481–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M. , Jones, D.A. , Parniske, M. , Harrison, K. , Balint‐Kurti, P.J. , Hatzixanthis, K. and Jones, J.D.G. (1997) Characterization of the tomato Cf‐4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf‐4 and Cf‐9. Plant Cell, 9, 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. , Nürnberger, T. and Joosten, M.H.A.J. (2011) Of PAMPs and effectors: the blurred PTI–ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen, J.H. , Abd‐El‐Haliem, A. , Fradin, E.F. , van den Berg, G.C.M. , Ekengren, S.K. , Meijer, H.J.G. , Seifi, A. , Bai, Y.L. , ten Have, A. , Munnik, T. , Thomma, B.P.H.J. and Joosten, M.H.A.J. (2010) Identification of tomato phosphatidylinositol‐specific phospholipase‐C (PI‐PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J. 62, 224–239. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Ellendorff, U. , Kemp, B. , Mansfield, J.W. , Forsyth, A. , Mitchell, K. , Bastas, K. , Liu, C.M. , Woods‐Tor, A. , Zipfel, C. , de Wit, P.J. , Jones, J.D.G. , Tör, M. and Thomma, B. (2008) A genome‐wide functional investigation into the roles of receptor‐like proteins in Arabidopsis. Plant Physiol. 147, 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.D. , Fiers, M. , Ellendorff, U. , Wang, Z.Z. , de Wit, P.J.G.M. , Angenent, G.C. and Thomma, B.P.H.J. (2010) The diverse roles of extracellular leucine‐rich repeat‐containing receptor‐like proteins in plants. Crit. Rev. Plant Sci. 29, 285–299. [Google Scholar]
- Zhang, Z. , Fradin, E.F. , Jonge, R. , Esse, H.P. , Smit, P. , Liu, C.M. and Thomma, B.P.H.J. (2012) Optimized agroinfiltration and virus‐induced gene silencing to study Ve1‐mediated Verticillium resistance in tobacco. Mol. Plant–Microbe Interact. 26, 182–190. [DOI] [PubMed] [Google Scholar]
- Zhou, B.J. , Jia, P.S. , Gao, F. and Guo, H.S. (2012) Molecular characterization and functional analysis of a necrosis‐ and ethylene‐inducing, protein‐encoding gene family from Verticillium dahliae . Mol. Plant–Microbe Interact. 25, 964–975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Ve1 localizes to the plasma membrane. (A) Localization of untagged Ve1 in Nicotiana benthamiana leaf epidermis with fluorescence microscopy at 30 h after agroinfiltration. (B) Localization of green fluorescent protein (GFP)‐tagged Ve1 in Nicotiana benthamiana leaf epidermis with fluorescence microscopy at 30 h after agroinfiltration. (C) Localization of GFP‐tagged Ve1 in Nicotiana benthamiana leaf epidermis on plasmolysis by incubation in 750 mm mannitol using confocal microscopy. (D) Bright field image of (C). (E) Overlay of (C) and (D). The arrows indicate plasma membrane detached from the cell wall. (F) Localization of GFP‐tagged Ve1 in protoplasts of Nicotiana benthamiana leaf epidermis.


