Summary
Plant pathogenic bacteria utilize complex signalling systems to control the expression of virulence genes at the cellular level and within populations. Quorum sensing (QS), an important intercellular communication mechanism, is mediated by different types of small molecules, including N‐acyl homoserine lactones (AHLs), fatty acids and small proteins. AHL‐mediated signalling systems dependent on the LuxI and LuxR family proteins play critical roles in the virulence of a wide range of Gram‐negative plant pathogenic bacteria belonging to the Alphaproteobacteria, Betaproteobacteria and Gammaproteobacteria. Xanthomonas spp. and Xylella fastidiosa, members of the Gammaproteobacteria, however, possess QS systems that are mediated by fatty acid‐type diffusible signal factors (DSFs). Recent studies have demonstrated that Ax21, a 194‐amino‐acid protein in Xanthomonas oryzae pv. oryzae, plays dual functions in activating a rice innate immune pathway through binding to the rice XA21 pattern recognition receptor and in regulating bacterial virulence and biofilm formation as a QS signal molecule. In xanthomonads, DSF‐mediated QS systems are connected with the signalling pathways mediated by cyclic diguanosine monophosphate (c‐di‐GMP), which functions as a second messenger for the control of virulence gene expression in these bacterial pathogens.
Introduction
A diverse range of virulence factors enable plant pathogenic bacteria to overcome host defence systems, including both pre‐existing and induced barriers, during the process of infection and colonization in host plants. For successful infection and multiplication, as well as survival during various environmental conditions throughout a life cycle, plant pathogenic bacteria must be able to control the production of virulence factors in response to dynamic changes in the surrounding environment and their interactions with other members of the microbial community and host plants. Failure to regulate precisely the production of virulence factors to avoid excessive production at inappropriate times would compromise the ecological fitness of the bacterial pathogen through the wastage of the cellular resources necessary for survival, as well as by early detection by the host and consequent activation of host defence systems. In nature, bacteria operate complex systems for signal perception, transduction and exchange at the cellular level and within populations. These signalling systems also play pivotal roles in the fine tuning of the production of virulence factors in pathogenic bacteria.
In this review, bacterial signalling and communication systems utilized by plant pathogenic bacteria are discussed, with a focus on their regulatory functions in bacterial pathogenesis in plants. Intercellular and intracellular signalling systems mediated by different types of signal molecule are described in terms of their functional mechanisms and roles in bacterial virulence. In particular, quorum‐sensing systems based on N‐acyl homoserine lactones (AHLs), diffusible signal factors and Ax21 family small proteins are highlighted, as well as recent advancements in cyclic diguanosine monophosphate (c‐di‐GMP)‐based signalling systems.
Intercellular Signalling (Quorum‐Sensing) Systems
Quorum sensing (QS) is an important cell–cell communication mechanism that enables a bacterial population to control the expression of genes primarily based on its density or growth stage. In pathogenic bacteria, QS increases the propensity for successful infection in various hosts as a result of the coordinated expression of virulence genes in accordance with bacterial population levels. Different types of signal molecule are utilized by different bacterial pathogens for QS. The major groups of known QS signal molecules include AHLs, oligopeptides and autoinducer‐2 (AI‐2) molecules (Roy et al., 2011). AHLs are major QS signal molecules in many Gram‐negative bacteria, whereas oligopeptides are more commonly used in Gram‐positive bacteria. AI‐2, a furanosyl borate diester (Cao and Meighen, 1989), is considered to be ‘a universal language’ in the bacterial world because it is found in both Gram‐negative and Gram‐positive bacteria (Bassler and Losick, 2006). In plant pathogenic Xanthomonas spp. and Xylella fastidiosa, the primary QS signal molecules are unsaturated fatty acids, called diffusible signal factors (DSFs) (Deng et al., 2011). A new type of QS signal, the small protein Ax21, has been discovered recently in these bacteria (Han et al., 2011a, 2011b; Ronald, 2011).
QS systems mediated by AHLs
Bacterial QS systems mediated by AHL‐type signal compounds are typically composed of two major genetic elements that are homologous to luxI and luxR of Vibrio fischeri, the first QS genes identified and characterized (Fuqua et al., 1994). The primary function of the LuxI/LuxR QS system encoded by luxI and luxR is the autoinduction of luminescence in the marine bacterium V. fischeri, in which genes for luminescence are expressed in a cell density‐dependent manner (Fuqua et al., 1994). LuxI family proteins encoded by luxI homologues are AHL synthases, which are responsible for the biosynthesis of AHL molecules, whereas LuxR family proteins encoded by luxR homologues are AHL receptor proteins, which are activated by binding to AHL molecules (Fuqua et al., 1994; Tsai and Winans, 2010). QS mediated by AHL signal compounds is present in a wide range of Gram‐negative bacteria, including many plant and animal pathogens. Here, three representative AHL‐mediated QS systems are described in detail. These and other similar QS systems in plant pathogenic bacteria are summarized in Table 1.
Table 1.
Representative N‐acyl homoserine lactone (AHL)‐mediated quorum‐sensing (QS) systems present in plant pathogenic bacteria
| Bacterium | Gene names for luxI/luxR homologues | Primary AHL compounds for QS | Functions | References |
|---|---|---|---|---|
| Agrobacterium tumefaciens | traI/traR | N‐3‐oxooctanoyl‐l‐homoserine lactone | Conjugal transfer of Ti plasmid | (Fuqua and Winans, 1994; Hwang et al., 1994; More et al., 1996; Zhang et al., 1993) |
| Burkholderia cepacia | cepI/cepR | N‐octanoyl‐l‐homoserine lactone | Production of protease and polygalacturonase | (Aguilar et al., 2003a) |
| Burkholderia glumae | tofI/tofR | N‐octanoyl‐l‐homoserine lactone | Production of toxoflavin and lipase; flagellar biogenesis/motility | (Devescovi et al., 2007; Kim et al., 2004, 2007) |
| Pantoea ananatis | eanI/eanR | N‐3‐oxohexanoyl‐l‐homoserine lactone | Production of extracellular polysaccharide; biofilm formation; infection of onion leaves | (Morohoshi et al., 2007) |
| Pantoea stewartii | esaI/esaR | N‐3‐oxohexanoyl‐l‐homoserine lactone | Production of extracellular polysaccharide; biofilm formation; dissemination and specific localization of bacterial cells at the xylem vessel | (von Bodman et al., 1998; Koutsoudis et al., 2006) |
| Pectobacterium carotovorum ssp. atrosepticum | expI/virR | N‐3‐oxohexanoyl‐l‐homoserine lactone | Production of virulence factors, including extracellular enzymes | (Burr et al., 2006; Liu et al., 2008) |
| Pectobacterium carotovorum ssp. carotovorum | carI/carR | N‐3‐oxohexanoyl‐l‐homoserine lactone | Production of carbapenem antibiotic | (McGowan et al., 2005) |
| expI/expR | N‐3‐oxohexanoyl‐l‐homoserine lactone | Production of virulence factors, including extracellular enzymes | (Jones et al., 1993; Pirhonen et al., 1993) | |
| Pectobacterium chrysanthemi | expI/expR | N‐3‐oxohexanoyl‐l‐homoserine lactone | Biosynthesis of pectinases | (Reverchon et al., 1998) |
| Pseudomonas syringae pv. syringae | ahlI/ahlR | N‐3‐oxohexanoyl‐l‐homoserine lactone | Production of extracellular polysaccharide; motility | (Quinones et al., 2004, 2005) |
TraI/TraR of Agrobacterium tumefaciens
QS systems of A. tumefaciens mediated by the traI AHL synthase genes and the traR AHL receptor genes have been relatively well studied compared with other QS systems of plant pathogenic bacteria. The LuxI family protein, TraI, synthesizes N‐3‐oxooctanoyl‐l‐homoserine lactone (OOHL), and the LuxR family protein, TraR, is activated by binding to OOHL (Hwang et al., 1994; More et al., 1996; Zhang et al., 1993). OOHL molecules are accumulated at a high cell density and, at a threshold level, TraR‐OOHL activates target genes, including the tra genes required for Ti plasmid conjugation (Fuqua and Winans, 1994, 1996). TraR is unstable in the absence of the ligand (OOHL) and functions as a homodimer by binding directly to specific DNA sequences, called tra boxes (Vannini et al., 2002; Zhang R. G. et al., 2002; Zhu and Winans, 1999).
TraI/TraR QS systems are under the control of sophisticated fine‐tuning systems involving traM, trlR, attM and attJ. traM is located downstream of traR and TraM acts as a negative regulator for TraR activity on the conjugal transfer of Ti plasmids (Fuqua et al., 1995; Hwang et al., 1995). The inhibitory activity of TraM on TraR may be through direct protein–protein interaction with TraR (Hwang et al., 1999). trlR encodes a TraR homologue that lacks the C‐terminal DNA‐binding domain; this TrlR protein is thought to inhibit the activity of TraR on octopine‐type Ti plasmids through the formation of inactive heterodimers with TraR (Chai et al., 2001; Oger et al., 1998; Zhu and Winans, 1998). Because TrlR requires OOHL for solubility, TrlR may function as a TrlR–OOHL complex, like TraR (2001) (Fig. 1).
Figure 1.
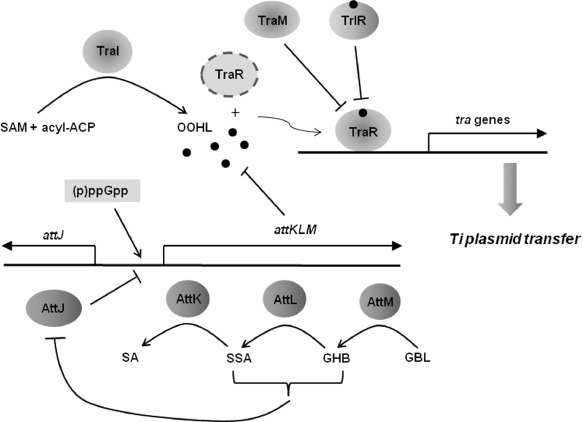
Schematic view of the TraI/TraR‐mediated quorum‐sensing system in Agrobacterium tumefaciens. TraR alone (depicted by broken line) is unstable. acyl‐ACP, acyl‐acyl carrier protein; GBL, γ‐butyrolactone; GHB, γ‐hydroxybutylic acid; OOHL, N‐3‐oxooctanoyl‐l‐homoserine lactone; SA, succinic acid; SAM, S‐adenosylmethionine; SSA, succinic acid semialdehyde.
Meanwhile, TraI/TraR QS systems are also modulated via sophisticated pathways for QS signal turnover. Zhang H. B. et al. (2002) found that an AHL‐lactonase encoded by attM degrades OOHL molecules and that the expression of attM was repressed by an IclR family protein encoded by attJ. Mutation of attJ resulted in the constitutive expression of attM, which led to the failure of AHL accumulation and conjugal transfer of Ti plasmids (Zhang H. B. et al., 2002). attM and two adjacent genes, attK and attL, comprise the attKLM operon, and attJ is divergently transcribed from this operon (Zhang H. B. et al., 2002). attK and attL encode a succinic acid semialdehyde dehydrogenase and an alcohol dehydrogenase, respectively (Zhang H. B. et al., 2002). The attKLM operon participates in an assimilative pathway of γ‐butyrolactone (GBL) that produces the intermediate metabolites γ‐hydroxybutylic acid (GHB), succinic acid semialdehyde (SSA) and succinic acid (SA) through the actions of AttM, AttL and AttK, respectively (Carlier et al., 2004; Chai et al., 2007). An accepted model for the control of OOHL turnover is that GHB and SSA induce the expression of the attKLM operon through the inactivation of the AttJ repressor protein, which, in turn, increases the production of the AttM AHL‐lactonase, resulting in the depletion of OOHL and the cessation of Ti plasmid transfer (Carlier et al., 2004; Chai et al., 2007) (Fig. 1). Degradation of OOHL at the stationary phase, resulting from the expression of attK, attL and attM, is also promoted by either carbon or nitrogen starvation, and dependent on a stress alarmone (p)ppGpp synthase encoded by relA, indicating that the degradation of the QS signal is regulated by the stress response machinery involving alarmone (p)ppGpp (Zhang et al., 2004) (Fig. 1). It has also been demonstrated that both (p)ppGpp and SSA modulate separately the two succinic acid semialdehyde dehydrogenases, AldH and AttK, which presumably control the intracellular levels of SSA and, consequently, determine the QS signal turnover (Wang et al., 2006).
TofI/TofR of Burkholderia glumae
QS mediated by LuxI/LuxR homologues and AHL‐type signal molecules is a major regulatory mechanism that controls the production of various virulence factors in Burkholderia spp. pathogenic to plants and animals (Aguilar et al., 2003b; Devescovi et al., 2007; Eberl, 2006; Goo et al., 2010; Huber et al., 2004; Malott et al., 2009; Ulrich et al., 2004). It also controls antifungal activities (Schmidt et al., 2009), antibiotic production (Duerkop et al., 2009; Seyedsayamdost et al., 2010) and biofilm formation (Aguilar et al., 2003b) in beneficial Burkholderia spp. The LuxI homologues of Burkholderia spp., represented by the AHL synthase CepI of the Burkholderia cepacia complex, mostly catalyse the synthesis of N‐octanoyl‐l‐homoserine lactone (OHL) and N‐hexanoyl‐l‐homoserine lactone (HHL); however, only OHL has shown signalling functions for known LuxI/LuxR‐type QS systems of Burkholderia spp. (Aguilar et al., 2003b; Duerkop et al., 2007; Kim et al., 2007). Recently, other types of signalling molecules for cell–cell communication, 2‐heptyl‐4(1H)‐quinolone (HHQ) and cis‐2‐dodecenoic acid (BDSF), have been found in B. pseudomallei and B. cenocepacia, respectively (Deng et al., 2009; Diggle et al., 2006).
In B. glumae, which causes bacterial panicle blight and grain rot of rice, the QS system composed of the luxI and luxR homologues, tofI and tofR, respectively, globally controls the production of all known major virulence factors: toxoflavin (Kim et al., 2004), lipase (Devescovi et al., 2007) and flagella (Kim et al., 2007) (Fig. 2). tofI encodes an AHL synthase that produces HHL and OHL, whereas tofR encodes a QS signal receptor that binds to OHL (Kim et al., 2004, 2007) (Fig. 2). In the known regulatory cascade for the production and transport of toxoflavin, the main phytotoxin utilized by B. glumae as a major virulence factor, the TofR–OHL complex activates the expression of toxJ which encodes the transcriptional activator ToxJ; ToxJ induces the expression of toxR which encodes the LysR family regulatory protein ToxR (Kim et al., 2004) (Fig. 2). Finally, in the presence of the co‐inducer toxoflavin, ToxR activates the expression of the toxABCDE and toxFGHI operons, which are responsible for the biosynthesis and transport of toxoflavin, respectively (Kim et al., 2004, 2009) (Fig. 2). ToxJ is also required for the expression of toxABCDE and toxFGHI operons, and TofR–OHL positively controls the expression of tofI, forming a positive autoregulatory feedback loop (Kim et al., 2004, 2009). The TofI/TofR QS system is also a central regulatory element of the known regulatory system for flagellar biogenesis and, consequently, flagellum‐mediated swimming and swarming motilities of B. glumae (Kim et al., 2007) (Fig. 2). In this regulatory system, the TofR–OHL complex renders the expression of qsmR which encodes the IclR family regulatory protein QsmR; QsmR expresses the flhDC genes, encoding the regulatory protein complex FlhDC. FlhDC is, in turn, essential for the expression of flagellar biogenesis genes (Kim et al., 2007) (Fig. 2). Although it has been demonstrated that another major virulence gene, lipA, which encodes a lipase, is also dependent on the TofI/TofR QS system for its expression (Devescovi et al., 2007), intermediate regulatory factors that connect the QS system and the expression of lipA have not yet been reported (Fig. 2).
Figure 2.
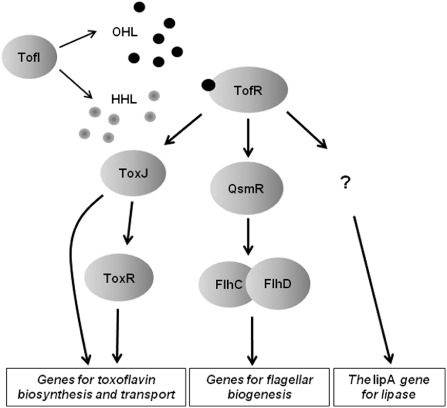
Schematic view of the global regulatory system mediated by the TofI/TofR quorum‐sensing system for the virulence of Burkholderia glumae. HHL, N‐hexanoyl‐l‐homoserine lactone; OHL, N‐octanoyl‐l‐homoserine lactone.
Meanwhile, an open reading frame (ORF) divergently transcribed from tofR and present in the intergenic region of tofI and tofR has been found to encode an RsaM homologue and to be highly conserved among Burkholderia spp. (Chen et al., 2012). Recently, rsaM has been discovered as a novel negative regulator of AHL‐mediated QS systems in another rice pathogenic bacterium, Pseudomonas fuscovaginae (Mattiuzzo et al., 2011). rsaM of Pseudomonas fuscovaginae is also present in the intergenic region of the luxI and luxR homologues, pfsI and pfsR, respectively, and is divergently transcribed from pfsR, suggesting that the ORF encoding a RsaM homologue in B. glumae may also have a regulatory function for the TofI/TofR QS system. Indeed, modulation of QS‐dependent toxoflavin production by this ORF has been observed recently in preliminary experiments (Chen et al., 2012). The elucidation of the function of this new regulatory factor would substantially expand our knowledge of the QS systems of Burkholderia spp.
EsaI/EsaR of Pantoea stewartii ssp. stewartii
The QS system present in the corn pathogen P. stewartii ssp. stewartii is unusual when compared with the majority of AHL‐mediated QS systems in at least two aspects. First, the EsaR AHL receptor negatively regulates the production of extracellular polysaccharides (EPS), important virulence factors of P. stewartii ssp. stewartii, in the absence of the cognate AHL compound N‐3‐oxohexanoyl‐l‐homoserine lactone (OHHL) (von Bodman et al., 1998) (Fig. 3). EsaR exerts this negative regulatory function through direct repression of the transcription of rcsA, which encodes an essential transcriptional activator for the expression of cps genes involved in the production of EPS (Minogue et al., 2005) (Fig. 3). Second, the cognate AHL produced by the EsaI AHL synthase, OHHL, antagonizes this repressive function of EsaR, resulting in the derepression of EPS production (von Bodman et al., 1998) (Fig. 3). esaR mutants producing excessive amounts of EPS show reduced virulence, poor adhesion to the surface and less compact biofilm structures, suggesting that the repression of EPS production by EsaR at the early stages of infection is critical for normal disease development caused by P. stewartii ssp. stewartii (Koutsoudis et al., 2006; von Bodman et al., 1998).
Figure 3.
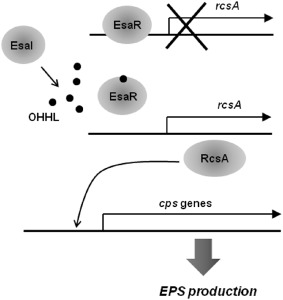
Schematic view of the regulatory mechanism for the production of extracellular polysaccharide (EPS) in Pantoea stewartii ssp. stewartii. OHHL, N‐3‐oxohexanoyl‐l‐homoserine lactone.
Similar to EsaR, several AHL receptors, including ExpR proteins of Pectobacterium carotovorum and Pectobacterium chrysanthemi, have also been shown to have repressive activities on the expression of virulence genes or virulence‐related phenotypes in the absence of cognate AHL molecules, and their regulatory activities are neutralized by the addition of cognate AHL molecules (Andersson et al., 2000; Cui et al., 2005, 2006; Sjoblom et al., 2006).
QS systems mediated by DSFs
A DSF‐mediated QS system was first discovered in the plant pathogenic bacterium Xanthomonas campestris pv. campestris (Tang et al., 1991), and later found to be widely distributed among plant pathogenic Xanthomonas spp. and Xy. fastidiosa, as well as other bacteria, including the B. cepacia complex, Pseudomonas aeruginosa and Stenotrophomonas maltophilia (Deng et al., 2011). The DSF of X. campestris pv. campestris was identified as an unsaturated fatty acid, cis‐11‐methyl‐2‐dodecenoic acid (Wang et al., 2004), whereas that of Xy. fastidiosa was identified as a saturated fatty acid, 12‐methyl‐tetradecanoic acid (Colnaghi Simionato et al., 2007). Additional structural variants of DSF molecules have also been identified from various Gram‐negative bacteria (Deng et al., 2011). The biosynthesis of DSF is dependent on the rpfF gene, which encodes a putative enoyl‐CoA hydratase in Xanthomonas spp. and Xy. fastidiosa (Barber et al., 1997; Newman et al., 2004). rpfB, which encodes a putative long‐chain fatty acyl CoA ligase, is also involved in DSF biosynthesis in both X. campestris pv. campestris and Xy. fastidiosa (Almeida et al., 2012; Barber et al., 1997). A recent study by Bi et al. (2012) on Bcam0581, an RpfF homologue in the opportunistic human pathogen B. cenocepacia, provided an important clue for the functional mechanism of RpfF in the biosynthesis of DSF molecules. In that study, Bcam0581 was shown to have dual enzymatic activity: (i) dehydratase activity for the dehydration of 3‐hydroxydodecanoyl‐acyl carrier protein (ACP) to cis‐2‐dodecenoyl‐ACP; and (ii) thioesterase activity for the cleavage of a thiolester bond to liberate the free fatty acid and produce BDSF, the DSF molecule of B. cenocepacia (Bi et al., 2012). DSF signal perception and transduction into target molecules in the cell are conferred by rpfC and rpfG genes, which encode a hybrid sensor kinase and a response regulator, respectively, comprising the RpfC/RpfG two‐component regulatory system (Fig. 4). These rpf genes are clustered and conserved in a wide range of bacteria that possess a DSF‐mediated QS system (He and Zhang, 2008).
Figure 4.
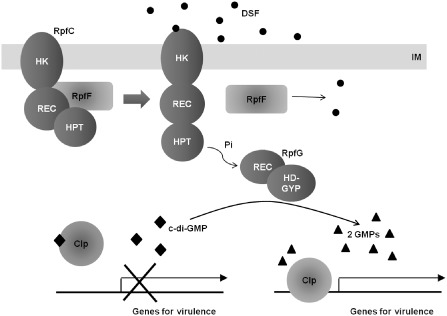
Schematic model of the regulatory mechanism involving DSF and c‐di‐GMP for the virulence of Xanthomonas campestris pv. campestris. c‐di‐GMP, cyclic diguanosine monophosphate; DSF, diffusible signal factor; GMP, guanosine monophosphate; HK, histidine kinase domain; HPT, histidine transfer domain; IM, inner membrane; Pi, phosphate group; REC, receiver domain.
The structural features of individual rpf gene products have been elucidated by several recent studies. Structural analyses of the crystal structure and sequence alignment of RpfF revealed a predicted substrate‐binding pocket composed of hydrophobic residues (G85, L136, G137, G138, M170, W258 and L276) and two putative catalytic residues (E141 and E161) within the RpfF molecule (Cheng et al., 2010). Point mutations of these core residues resulted in the abrogation of enzymatic activity for DSF biosynthesis, indicating the importance of these structural elements for RpfF function (Cheng et al., 2010). RpfC is a histidine sensor kinase containing several functionally discrete domains, including a histidine kinase (HK) domain, five transmembrane (TM) domains, a receiver (REC) domain and a histidine transfer (HPT) domain (Slater et al., 2000) (Fig. 4). The phosphorelay is seemingly an essential part of RpfC function, because substitution of the conserved histidine residues in the HK and HPT domains or the conserved aspartate residue in the REC domain, which are predicted to be involved in the phosphorelay, abrogated the RpfC function in DSF‐mediated QS (He et al., 2006). Intriguingly, RpfC also plays a negative regulatory role in DSF biosynthesis without much change in the transcription of rpf genes (Slater et al., 2000; Wang et al., 2004). He et al. (2006) found that RpfC controls DSF production at a post‐translational level via direct physical interaction with RpfF, suggesting the following working model: in the absence (or at a lower than threshold level) of DSF, both RpfC and RpfF are in an inactive state because of their physical binding to each other; in the presence (or at a higher than threshold level) of DSF, RpfC changes its conformation on reception of the DSF signal and consequently releases RpfF, which activates both proteins for their signal production and transduction functions (Deng et al., 2011) (Fig. 4). This mode of action allows a positive feedback of RpfF activity without a substantial increase in rpfF transcription (He et al., 2006).
RpfG is a response regulator that contains a typical REC domain as well as an HD‐GYP domain with phosphodiesterase (PDE) activity (Slater et al., 2000) (Fig. 4). Functional analyses of the RpfG protein indicated that both the REC and HD‐GYP domains are essential for DSF signal transduction in X. campestris pv. campestris (Deng et al., 2011). In particular, it has been shown that RpfG functions as a juncture that connects the QS signalling mediated by DSF and the intracellular signalling mediated by c‐di‐GMP (Deng et al., 2011). The function of RpfG in the c‐di‐GMP signalling system is addressed in detail in the section on Intracellular signalling systems mediated by second messenger molecules.
rpf/DSF QS systems play substantially different roles in Xanthomonas spp. and Xy. fastidiosa even though the rpf gene cluster, which contains the rpfG, rpfC and rpfF genes, is conserved in both genera. Basically, the rpf/DSF QS system plays a positive role for the virulence of Xanthomonas spp., whereas it exerts a negative role in the virulence of Xy. fastidiosa. In X. campestris pv. campestris, the disruption of any gene in the rpf gene cluster caused reduced biosynthesis of extracellular enzymes and EPS, and, consequently, reduced virulence (Barber et al., 1997; Slater et al., 2000; Tang et al., 1991). In Xy. fastidiosa, however, rpfF mutants, which were unable to synthesize DSF signal molecules, were more virulent than the wild‐type strain even though they were defective in vector transmission and biofilm formation in the insect vector (Newman et al., 2004). rpfF mutants also showed enhanced movement and multiplication in the xylem vessels relative to the wild‐type, suggesting that the DSF‐mediated QS system of Xy. fastidiosa was adapted to restrain bacterial virulence activities within host plants for the endophytic lifestyle of this bacterium (Chatterjee et al., 2008a). In contrast, rpfC mutants of Xy. fastidiosa, which overproduce DSFs and hyperexpress rpfF, showed reduced virulence, an inability to migrate in xylem vessels and increased expression of genes for the adhesion proteins FimA, HxfA and HxfB (Chatterjee et al., 2008b). Nevertheless, rpfC mutants of Xy. fastidiosa also showed impaired vector transmission similar to rpfF mutants (Chatterjee et al., 2008b). These differential regulatory functions of the rpf/DSF QS system in virulence between Xanthomonas spp. and Xy. fastidiosa suggest that this signalling system functions differently in different pathogens depending on the pathogenic behaviour and lifestyle of the pathogen.
rpf/DSF QS systems were also studied in other plant pathogenic Xanthomonas spp., including the bacterial citrus canker pathogen, X. axonopodis pv. citri (Andrade et al., 2006), the bacterial rice leaf blight pathogen, X. oryzae pv. oryzae (Chatterjee and Sonti, 2002; He et al., 2010; Tang et al., 1996), the bacterial rice leaf streak pathogen, X. oryzae pv. oryzicola (Zhao et al., 2011), and the bacterial soybean pustule pathogen, X. axonopodis pv. glycines (Thowthampitak et al., 2008). Even though positive roles of the rpf/DSF QS system in virulence were well conserved among all of the Xanthomonas spp. studied, species‐ and strain‐specific functional variations of the dpf/DSF QS system were observed within the genus. For example, extracellular enzyme production patterns of rpfF mutants were contradictory between two X. oryzae pv. oryzae strains from different geographical locations (Chatterjee and Sonti, 2002; He et al., 2010; Rai et al., 2012). rpfF was shown to positively regulate extracellular enzyme production in a strain of X. oryzae pv. oryzae from Korea (He et al., 2010), but to negatively regulate extracellular enzyme production in a strain from India (Chatterjee and Sonti, 2002; Rai et al., 2012). In addition, extracellular enzyme production was positively regulated by rpfC in X. campestris pv. campestris, but was not affected by rpfC in X. oryzae pv. oryzae (Slater et al., 2000; Tang et al., 1996).
QS systems mediated by peptide‐type signal molecules (Ax21 family small proteins)
Recent seminal works by Pamela Ronald's research group have elucidated the function of a small bacterial protein, Ax21, as a QS signal in the rice pathogenic bacterium X. oryzae pv. oryzae (Han et al., 2011b, 2011a). This is the first example of signalling by peptide molecules in Gram‐negative bacteria and provides great insight into the signalling networks of bacterial pathogens involving multiple signal molecules and pathways.
Ax21 was first identified as the ligand of XA21, a receptor kinase (or pattern recognition receptor, PRR) of rice responsible for the rice disease resistance derived from the gene‐for‐gene interaction between the resistance gene Xa21 and the avirulence gene avrXa21 (Fig. 5) (Lee et al., 2009). The rice resistance gene Xa21 was first identified via fine genetic mapping (Song et al., 1995). The product of this resistance gene, XA21, is a membrane‐spanning protein composed of the extracellular sensor domain and the cytoplasmic serine/threonine kinase domain (Song et al., 1995). The avirulence protein Ax21, which corresponds to XA21, was identified thorough serial purifications and accompanying tests for the avirulence activity of the extracellular fraction of the bacterial cell culture using reverse‐phase high‐performance liquid chromatography and nuclear magnetic resonance (Lee et al., 2009). Ax21 is a 194‐amino‐acid protein secreted through a type I secretion system, and its orthologues are present in plant and animal pathogens, including Xanthomonas spp. (90%–98% identity), Xy. fastidiosa (48% identity) and Stenotrophomonas maltophilia (61% identity).
Figure 5.
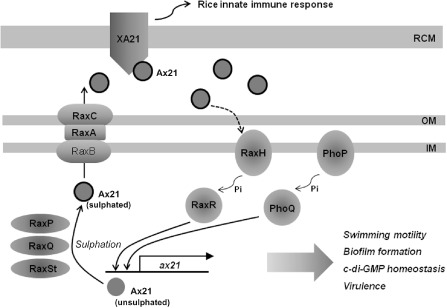
Schematic model of Ax21‐mediated quorum sensing and rice defence induction by Xanthomonas oryzae pv. oryzae. c‐di‐GMP, cyclic diguanosine monophosphate; IM, inner membrane; OM, outer membrane; Pi, phosphate group; RCM, rice cell membrane.
Intriguingly, Ax21 contains two predicted tyrosine (Tyr) sulphation sites and can be reduced to the 17‐amino‐acid functional peptide that includes sulphated Tyr‐22 (AxYS22) (Lee et al., 2009). Sulphation of Tyr‐22 is essential for the function of AxYS22 for Ax21 activity, and the sulphated form of this peptide binds to XA21 with high affinity (Lee et al., 2009). It is also noteworthy that the amino acid sequence of AxYS22 is highly conserved in the Ax21 family proteins of all the sequenced Xanthomonas spp. (100% identity), Xy. fastidiosa (77% identity) and S. maltophilia (65% identity) (Lee et al., 2009). Probably as a result of this high level of similarity, Ax21 orthologues may be functionally interchangeable among Xanthomonas spp. (Lee et al., 2009; Ronald, 2011) and other related genera (McCarthy et al., 2011). Genes encoding Ax21 are conserved in all of the sequenced species of the genus Xanthomonas and other related genera, suggesting that this protein exerts an important function for the ecological or parasitic fitness of the bacteria in nature, including hosts without XA21 or equivalent host defence components (Ronald, 2011). In this regard, Ax21 can be considered as a pathogen‐associated molecular pattern (PAMP) (Bent and Mackey, 2007), and the XA21‐mediated rice innate immune pathway may be a promising target to promote stable rice disease resistance against X. oryzae pv. oryzae.
Genetic studies of X. oryzae pv. oryzae have revealed that three groups of bacterial genes are required for the function of Ax21 in the activation of the XA21‐dependent rice disease resistance: (i) three genes for a type I secretion system (raxA, raxB and raxC); (ii) three genes for the sulphation of Ax21 (raxP, raxQ and raxSt); and (iii) four genes for two two‐component regulatory systems (raxR, raxH, phoP and phoQ) (Fig. 5) (Ronald, 2011). Recently, Han et al. (2011b) found that the expression of these rax (required for Ax21 activity) genes is dependent on bacterial density, and that Ax21 functions as the signal molecule for this QS. In the same study, a Δax21 derivative of the wild‐type strain, PXO99, showed reduced biofilm formation, swimming motility and bacterial growth in rice leaves compared with the wild‐type, suggesting that Ax21‐mediated QS also controls these phenotypic traits of X. oryzae pv. oryzae (Fig. 5) (Han et al., 2011b). In particular, the Δax21 strain, PXO99Δax21, showed reduced ability to cause symptoms and grow in rice leaves at a low cell density, which may mimic natural inoculation conditions, suggesting that Ax21‐mediated QS is critical to the initiation of infection at a low level of inoculum in nature. Microarray data presented by the same study supported the observed phenotypes of PXO99Δax21, in which genes involved in bacterial motility (including fliC, fliD and pilG), biofilm formation (including gumJ, gumE and gumK) and c‐di‐GMP metabolism (nine genes containing putative HD‐GYP, EAL and GGDEF domains) were up‐regulated in the wild‐type relative to the Δax21 strain (Han et al., 2011b; Ronald, 2011). The expression of the rpf operon was not affected significantly by Ax21, suggesting that the Ax21‐mediated QS system has little overlap with the DSF‐mediated QS system, and that each system plays an independent role in density‐dependent gene expression in X. oryzae pv. oryzae (Ronald, 2011).
A unique feature of Ax21 is that this protein exerts dual functions: as a PAMP triggering a rice innate immune system through direct binding to a PRR, and as a QS signal molecule controlling bacterial genes (Han et al., 2011b; Lee et al., 2009). Even though the Ax21‐mediated QS system has been studied mainly with X. oryzae pv. oryzae and rarely with other bacteria, the presence of ax21 and rax gene orthologues in other Xanthomonas spp. and closely related genera strongly suggests that this novel QS system is a common signalling system in at least certain groups of bacterial pathogens, especially those belonging to Xanthomonas spp. and Xy. fastidiosa.
QS systems mediated by other types of signalling molecule
AI‐2 molecules are synthesized by S‐ribosylhomocysteinase (LuxS) encoded by luxS genes (Pereira et al., 2012; Roy et al., 2011). Specifically, LuxS catalyses the conversion of S‐ribosylhomocysteine (SRH) to 4,5‐dihydroxy‐2,3‐pentanedione (DPD). In turn, DPD undergoes intramolecular cyclization, forming different isoforms, which are collectively referred to as AI‐2 (Pereira et al., 2012; Roy et al., 2011). In V. harveyi, where AI‐2 was first found, the AI‐2 molecule is a borated form of S‐2‐methyl‐2,3,3,4‐tetrahydroxytetrahydrofuran (S‐THMF), S‐THMF‐borate (Chen et al., 2002). A markedly different isomer of DPD without boron, R‐2‐methyl‐2,3,3,4‐tetrahydroxytetrahydrofuran (R‐THMF), was found to be the AI‐2 molecule in Salmonella enterica ssp. enterica serovar Typhimurium (Miller et al., 2004). The production of AI‐2 molecules is widespread among various species of Gram‐positive and Gram‐negative bacteria, and these signal molecules are considered to be a ‘universal language’ for interspecies' communication in the bacterial community (Bassler and Losick, 2006; Lowery et al., 2008; Pereira et al., 2012; Sun et al., 2004). Since AI‐2 molecules were first reported to function as a second QS signal for bioluminescence in V. harveyi (Bassler et al., 1994) and for virulence and biofilm formation in Vibrio cholerae (Miller et al., 2002), numerous studies have demonstrated diverse functions of AI‐2 molecules, including biofilm formation, motility and virulence factor production, in various bacterial species (Pereira et al., 2012).
In spite of the recent advancement in AI‐2 studies, the signalling function of luxS/AI‐2 in plant pathogenic bacteria has not been widely studied and is somewhat ambiguous. In Pe. carotovorum ssp. carotovorum and Pe. carotovorum ssp. atrosepticum, the production of extracellular enzymes was reduced in luxS mutants when compared with the wild‐type parental strains (Coulthurst et al., 2006). In Erwinia amylovora, QS mediated by LuxS was not observed and inactivation of luxS did not cause any significant change in virulence‐related phenotypes, other than reduced expression of the virulence genes, hrpL and dspA/E (Rezzonico and Duffy, 2007). However, another study by Gao et al. (2009) with the same strain of E. amylovora showed that mutation of luxS caused impaired motility, tolerance to reactive oxygen species and virulence. Despite the different conclusions made by these research groups on the role of LuxS in the virulence of E. amylovora, it should be noted that genes encoding known AI‐2 receptors required for AI‐2 QS, LuxP or LsrB, could not be found in E. amylovora (Rezzonico and Duffy, 2007) or in other plant pathogenic bacteria (Rezzonico and Duffy, 2008). This suggests that any effect caused by a luxS mutation may be a result of impaired metabolic function rather than signalling function (Rezzonico and Duffy, 2008). Nevertheless, it cannot be ruled out that unknown, alternative AI receptors might exist for AI‐2 QS signalling in some plant pathogenic bacteria.
QS systems mediated by oligopeptides have been investigated in several Gram‐positive bacteria, including Staphylococcus aureus, Bacillus subtilis, Enteroccocus faecalis and the Bacillus cereus group (Antunes et al., 2010; Rocha‐Estrada et al., 2010; Shank and Kolter, 2011). However, not much is known about this type of QS signal molecule in Gram‐positive plant pathogenic bacteria. In addition, 2‐heptyl‐3‐hydroxy‐4‐quinolone (Pseudomonas quinolone signal, PQS) and its immediate precursor, 2‐heptyl‐4‐quinolone (HHQ), are known to act as QS signal molecules to control global gene expression for many virulence factors and quinolone biosynthesis in the opportunistic human pathogen Ps. aeruginosa (Antunes et al., 2010). However, as with the oligopeptides, the signalling functions of PQS and HHQ in plant pathogenic bacteria remain unknown.
Intracellular Signalling Systems Mediated by Second Messenger Molecules
Signalling systems mediated by c‐di‐GMP
c‐di‐GMP is a small diffusible signal molecule that influences a wide range of cellular processes, including flagellum‐mediated motility, cell cycle and EPS biosynthesis, as well as bacterial virulence (Romling, 2012). c‐di‐GMP signalling systems are generally composed of three major components: diguanylate cyclases (DGCs), PDEs and c‐di‐GMP‐binding effectors (Mills et al., 2011; Sondermann et al., 2012). DGCs synthesize c‐di‐GMP from two guanosine triphosphate (GTP) molecules, whereas PDEs degrade c‐di‐GMP, generating a single pGpG molecule or two GMP molecules (Sondermann et al., 2012). In general, the GGDEF domain of DGCs and the EAL or HD‐GYP domains of PDEs are responsible for DGC and PDE activities, respectively, and balanced control of these opposite activities of DGCs and PDEs determines c‐di‐GMP homeostasis within the cell (Sondermann et al., 2012). DGC and PDE activities are modulated through various mechanisms, including protein–protein interaction and phosphorylation, in accordance with environmental cues and cell–cell communication. Thus, cellular levels of c‐di‐GMP are an integral output of bacterial sensory systems that perceive various biotic and abiotic conditions. Finally, c‐di‐GMP translates input signals into the modulation of cellular behaviour by binding to diverse downstream effector molecules, including specific c‐di‐GMP receptor proteins with a characteristic ‘PilZ’ domain, degenerate GGDEF/EAL domains and c‐di‐GMP‐specific transcriptional factors (i.e. Clp) and RNA motifs (riboswitches) (Mills et al., 2011; Sondermann et al., 2012).
Although an early study revealed the regulatory function of c‐di‐GMP on cellulose production by the plant pathogen A. tumefaciens (Amikam and Benziman, 1989), not much was known about the c‐di‐GMP signalling system in plant pathogenic bacteria until recently. Recent seminal studies on X. campestris pv. campestris and X. axonopodis pv. citri have provided great insights into the function of c‐di‐GMP in virulence, in conjunction with the QS mediated by DSF molecules and the RpfC/RpfG two‐component regulatory system (Ryan and Dow, 2010; Zhang, 2010). Ryan et al. (2006) demonstrated with X. campestris pv. campestris that RpfG containing the HD‐GYP domain degraded c‐di‐GMP, and that this PDE activity was dependent on the conserved H and D residues of the HD‐GYP domain. In the same study, the conserved H and D residues were also required for the regulatory activity of RpfG in the biosynthesis of virulence factors (Ryan et al., 2006). This regulatory function associated with the key residues for PDE activity is probably exerted through the c‐di‐GMP‐binding protein, Clp (Tao et al., 2010). Clp, a Crp [cyclic adenosine monophosphate (cAMP) receptor protein] family protein, is an essential component of the rpf/DSF‐mediated QS system that regulates the expression of virulence genes of X. campestris pv. campestris, including the genes for flagellar biogenesis, the Hrp type III secretion system, extracellular enzymes and EPS synthesis (He et al., 2007). Unlike most Crp family proteins, Clp has a high level of binding affinity to its target DNA in the absence of any ligand, and c‐di‐GMP allosterically inhibits the DNA‐binding activity of Clp (Chin et al., 2010; Leduc and Roberts, 2009). This inhibitory action of c‐di‐GMP on Clp is probably a result of the direct binding of c‐di‐GMP to the Clp protein, because mutations of the Clp residues predicted to be involved in c‐di‐GMP binding through structural modelling processes caused substantial reductions in the binding affinity of Clp to c‐di‐GMP, the inhibitory effect of c‐di‐GMP on DNA binding of Clp and virulence (Chin et al., 2010; Tao et al., 2010).
However, it was also revealed that RpfG exerted a subset of its functions, including motility, through physical interactions with DDGEF domain proteins in X. campestris pv. campestris (Ryan et al., 2010). Similarly, the RpfG protein of X. axonopodis pv. citri was observed to interact with GGDEF domain proteins in yeast two‐hybrid (YTH) experiments (Andrade et al., 2006). The interaction of RpfG with DDGEF domain proteins and the RpfG functions associated with the protein–protein interaction were dependent on the G, Y and P residues of the HD‐GYP domain, but not on the H and D residues and their associated PDE activity (Ryan et al., 2010). In the same study, two GGDEF domain proteins (XC0249 and XC0420) were confirmed through YTH and fluorescence resonance energy transfer (FRET) assays to be RpfG‐interacting proteins; in addition, simultaneous mutation of the two genes encoding the RpfG interactors caused the loss of pilus‐dependent motility and reduced virulence, as observed with alanine substitution of the G, Y or P residue in the HD‐GYP domain of the RpfG protein (Ryan et al., 2010).
Meanwhile, McCarthy et al. (2008) reported that X. campestris pv. campestris contains at least four genes that produce proteins with a PilZ or c‐di‐GMP‐binding domain. In animal pathogens, PilZ domain proteins play important roles in c‐di‐GMP signalling for the control of specific cellular processes, including virulence (Cotter and Stibitz, 2007; Schirmer and Jenal, 2009). Among the four genes that encode proteins with a PilZ domain (XC0965, XC2249, XC2317 and XC3221) in X. campestris pv. campestris, mutation of XC0965 and XC2249 caused reduced production of extracellular enzymes, whereas mutation of XC2249 and XC3221 impaired motility (McCarthy et al., 2008). It was also demonstrated that XC6012, a PilZ domain protein that is identical to XC2249, but from another strain of X. campestris pv. campestris, was important for virulence and formed a unique tetrameric structure (Li et al., 2011).
A recent study by Ryan et al. (2012) has illustrated how the physical interaction of RpfG with GGEDF domain proteins controls pilus‐dependent motility through the PilZ domain protein XC2249. In this study, RpfG‐GGDEF domain protein complexes recruit the PilZ domain protein XC2249 as an adaptor for further interaction with the pilus motor proteins PilU and PilT (Ryan et al., 2012).
On the basis of these studies, the c‐di‐GMP signalling function mediated by RpfG may consist of two levels. The first level of RpfG function is dependent on the PDE activity of the HD‐GYP domain (H and D are key residues) and controls bacterial motility and extracellular enzyme biosynthesis (Ryan et al., 2006, 2010). This signalling pathway may function through the following mechanism: the HD‐GYP domain of RpfG degrades c‐di‐GMP, resulting in decreased c‐di‐GMP levels in bacterial cells; Clp is then liberated from its inhibitory ligand, c‐di‐GMP, to promote the expression of virulence genes (Fig. 4). The second level of RpfG function is dependent on the physical interaction of the HD‐GYP domain with GGDEF domain proteins (G, Y and P are key residues) and is responsible for only a subset of the functions of RpfG, such as bacterial motility (Ryan et al., 2010, 2012). This signalling function of RpfG for motility involves direct physical interactions with the motor proteins PilU and PilT, using the PilZ domain protein XC2249 as a juncture (Ryan et al., 2012).
Several studies have also revealed additional c‐di‐GMP signalling systems in plant pathogenic bacteria. In X. campestris pv. campestris, another two‐component regulatory system, RavS/RavR, has been shown to regulate bacterial virulence factors through Clp, like the Rpf/DSF QS system (He et al., 2009). In this system, RavR, like RpfG, contains an EAL domain that shows PDE activity, whereas RavS includes two PAS domains implicated in the sensing of low‐oxygen conditions; these domains have been shown to be essential for the regulatory functions of RavS and RavR in the production of virulence factors (EPS and extracellular enzymes), transcription of clp and bacterial virulence in host plants (He et al., 2009). Additive roles of the RavS/RavR and rpf/DSF systems in the production of virulence factors, clp expression and virulence have also been observed from single and double mutants of X. campestris pv. campestris for these systems, suggesting that Clp, a global regulator for X. campestris pv. campestris virulence, is modulated by the two separate signalling pathways that sense population density and hypoxia (He et al., 2009). In Pe. carotovorum ssp. atrocepticum, it has been found recently that the intracellular level of c‐di‐GMP affects the secretion of a multi‐repeat adhesion protein (MRP), motility, biofilm formation and virulence (Perez‐Mendoza et al., 2011a, 2011b), and that two novel genes encoding a putative PDE and a putative DGC modulate the intracellular c‐di‐GMP level in this plant pathogen (Perez‐Mendoza et al., 2011a). A genetic study by Perez‐Mendoza et al. (2011a) revealed that the putative PDE gene, ECA3271, suppressed the accumulation of secreted MRP and increased swimming motility via depletion of intracellular c‐di‐GMP, whereas the putative DGC gene, ECA3270, accounted for opposite phenotypes via elevation of the c‐di‐GMP level in the cell. Nevertheless, mutation of either ECA3270 or ECA3271 caused a significant reduction in bacterial virulence, suggesting that fine tuning of the cellular c‐di‐GMP level via balanced actions of DGC and PDE is crucial for host infection (Perez‐Mendoza et al., 2011a). In another soft rot‐causing plant pathogen, Dickeya dadantii (syn. Pe. chrysanthemi), two proteins containing an EAL domain (putative PDEs) were found to control multiple cellular behaviours, including biofilm formation and motility, and virulence gene expression (Yi et al., 2010). Both ecpB and ecpC were revealed to play a positive role in swarming/swimming motility, pectate lyase production and gene expression for the type III secretion system and type III effectors, but a negative role in biofilm formation (Yi et al., 2010). In the same study, EcpC was shown to have PDE activity that hydrolyses c‐di‐GMP into linear pGpG, suggesting that these EAL domain proteins exert their regulatory functions through the modulation of c‐di‐GMP levels (Yi et al., 2010).
Signalling pathways mediated by other cyclic nucleotide molecules
cAMP has long been known to play an important role in various cellular processes from the catabolism of alternative sugars to motility and virulence (Gomelsky, 2011). In soft rot‐causing plant pathogenic bacteria, the cAMP–CRP (cAMP receptor protein) complex has been reported to positively control the genes for pectolysis (Nasser et al., 1997; Reverchon et al., 1997; Thomson et al., 1999).
In recent studies, cyclic di‐AMP (c‐di‐AMP) and cGMP have also been found to function as signalling molecules in some bacteria. In B. subtilis, c‐di‐AMP was reported to be an important messenger controlling bacterial sporulation in a DNA integrity‐dependent manner (Oppenheimer‐Shaanan et al., 2011; Witte et al., 2008), and to play an essential role in peptidoglycan (cell wall) homeostasis (Luo and Helmann, 2012). In Staphylococcus aureus, c‐di‐AMP was found to play a role in controlling cell size and in coping with stresses on cell membranes and walls (Corrigan et al., 2011). c‐di‐AMP may also be essential for viability in many bacteria because genes encoding di‐adenylate cyclase domains, which are responsible for the biosynthesis of c‐di‐AMP, are conserved in a wide range of bacterial genomes, and mutations of these genes were lethal in tested bacteria belonging to Listeria, Streptococcus and Mycoplasma (French et al., 2008; Glass et al., 2006; Song et al., 2005; Woodward et al., 2010). Little is known about the signalling functions of cGMP in prokaryotes, but it has been demonstrated recently that cyst formation of Rhodospirillum centenum is dependent on cGMP and that the guanylyl cyclase gene cluster involved in cGMP production is conserved in a number of cyst‐forming bacteria, including Sinorhizobium meliloti (Marden et al., 2011). Even though little is known about the signalling functions of these additional cyclic nucleotide signal molecules in plant pathogenic bacteria, this is an interesting research subject to pursue to expand our knowledge of signalling systems in these bacteria.
Concluding Remarks and Future Perspectives
Signalling systems involved in the regulation of virulence genes in plant pathogenic bacteria have been discussed in this review. Even though individual signalling systems have been discussed separately, a single bacterial pathogen often possesses multiple signalling systems that are functionally interconnected with each other and comprise a global signalling network. For example, the signalling systems for the expression of virulence genes in Xanthomonas spp. are integrated forms of the DSF‐mediated QS and c‐di‐GMP signalling systems. Recent studies on RpfG functions involving interactions with GGDEF and PilZ domain proteins in X. campestris pv. campestris (Ryan et al., 2010, 2012) and on the involvement of DSF in strong interactions of these c‐di‐GMP signalling components indicate that the QS and c‐di‐GMP signalling systems constitute an integrated sensory/regulatory pathway in this pathogen (Ryan et al., 2010). Similarly, it has been observed that QS is linked to c‐di‐GMP for the control of swarming activity in the animal pathogen Vibrio parahaemolyticus (Trimble and McCarter, 2011).
AHL‐mediated QS systems in diverse bacterial species, including numerous plant pathogenic bacteria, have been studied extensively in recent decades. Nevertheless, signalling and regulatory mechanisms associated with AHL molecules have yet to be fully understood. In particular, it is extremely challenging to characterize precisely the QS systems of some bacteria that possess multiple luxI and luxR homologues because of the complexity derived from interactions of more than one set of LuxI/LuxR family proteins. Unknown accessory components that function in QS systems may also add to the complexity of QS systems. Recently, negative regulatory genes of QS systems have been discovered in the intergenic regions of various sets of luxI and luxR homologues (Venturi et al., 2011). It is very probable that additional accessory components may play crucial roles in AHL‐mediated QS systems, even though they still remain to be identified.
DSF‐mediated QS systems connected with c‐di‐GMP signalling systems are essential for the cell–cell communication of important plant pathogenic bacteria belonging to Xanthomonas and Xy. fastidiosa. More information is needed to comprehensively understand this type of QS. The Ax21 system is a newly identified QS system, especially for Xanthomonas spp. and Xy. fastidiosa. The functional relationship of this QS system with the known signalling and regulatory systems of Xanthomonas spp. and Xy. fastidiosa is an emerging research subject, and studies elucidating these relationships will provide a better understanding of the integrated signalling networks mediated by DSF, c‐di‐GMP and Ax21.
The control of plant diseases caused by bacterial pathogens is difficult because only a few antibiotics are permissible for use for agricultural purposes and because antibiotic‐resistant strains are easily generated from the repeated use of antibiotics. The intercellular and intracellular signalling systems of bacterial pathogens are promising targets for new disease control strategies because they tend to be fundamentally different from those of eukaryotic organisms and they globally control multiple virulence factors and biofilm formation. A better understanding of bacterial signalling systems would provide important clues for the development of methods to inhibit signalling, which would result in the suppression of bacterial infection. Several approaches can be proposed for the inhibition of, or interference with, bacterial signalling systems including: (i) the development of synthetic or natural compounds that perturb the bacterial signalling systems; (ii) the employment of enzymes that degrade bacterial signal molecules; and (iii) the development of host plants that constitutively produce bacterial signal molecules to provide false signals to bacterial cells.
Recent exciting findings from the study of mammalian cells include the stimulation of innate immune systems on recognition of the bacterial signal molecules, c‐di‐GMP (Burdette et al., 2011) and c‐di‐AMP (Woodward et al., 2010). A TM protein of mammalian cells, STING (stimulator of interferon genes), was found to activate an innate immune pathway via the direct recognition of cyclic dinucleotides secreted by bacterial pathogens (Burdette et al., 2011). The interaction between XA21 in rice and Ax21 in X. oryzae pv. oryzae is an excellent example which illustrates that plants also possess similar innate immune systems that induce defence reactions through the recognition of bacterial signal molecules (Lee et al., 2009). The identification and characterization of the plant defence components that recognize other bacterial signal molecules and subsequently activate defence responses are important research subjects for the study of plant–bacterial interactions, as well as for the development of new control strategies for bacterial plant diseases.
Acknowledgements
This work was supported by the Louisiana State University Agricultural Center and the Research and Development Subprogram of the Louisiana Board of Regents Support Fund (Grant number: LEQSF(2008‐11)‐RD‐A‐02). I thank Rebecca Melanson, Bishnu Shrestha, Hari Karki and Ruoxi Chen for critical review of the manuscript.
References
- Aguilar, C. , Bertani, I. and Venturi, V. (2003a) Quorum‐sensing system and stationary‐phase sigma factor (RpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69, 1739–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar, C. , Friscina, A. , Devescovi, G. , Kojic, M. and Venturi, V. (2003b) Identification of quorum‐sensing‐regulated genes of Burkholderia cepacia . J. Bacteriol. 185, 6456–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, R.P. , Killiny, N. , Newman, K.L. , Chatterjee, S. , Ionescu, M. and Lindow, S. (2012) Contribution of rpfB to cell–cell signal synthesis, virulence, and vector transmission of Xylella fastidiosa . Mol. Plant–Microbe Interact. 25, 453–462. [DOI] [PubMed] [Google Scholar]
- Amikam, D. and Benziman, M. (1989) Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens . J. Bacteriol. 171, 6649–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, R.A. , Eriksson, A.R. , Heikinheimo, R. , Mae, A. , Pirhonen, M. , Koiv, V. , Hyytiainen, H. , Tuikkala, A. and Palva, E.T. (2000) Quorum sensing in the plant pathogen Erwinia carotovora subsp. carotovora: the role of expREcc . Mol. Plant–Microbe Interact. 13, 384–393. [DOI] [PubMed] [Google Scholar]
- Andrade, M.O. , Alegria, M.C. , Guzzo, C.R. , Docena, C. , Rosa, M.C. , Ramos, C.H. and Farah, C.S. (2006) The HD‐GYP domain of RpfG mediates a direct linkage between the Rpf quorum‐sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri . Mol. Microbiol. 62, 537–551. [DOI] [PubMed] [Google Scholar]
- Antunes, L.C. , Ferreira, R.B. , Buckner, M.M. and Finlay, B.B. (2010) Quorum sensing in bacterial virulence. Microbiology, 156, 2271–2282. [DOI] [PubMed] [Google Scholar]
- Barber, C.E. , Tang, J.L. , Feng, J.X. , Pan, M.Q. , Wilson, T.J. , Slater, H. , Dow, J.M. , Williams, P. and Daniels, M.J. (1997) A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. [DOI] [PubMed] [Google Scholar]
- Bassler, B.L. and Losick, R. (2006) Bacterially speaking. Cell, 125, 237–246. [DOI] [PubMed] [Google Scholar]
- Bassler, B.L. , Wright, M. and Silverman, M.R. (1994) Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13, 273–286. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Bi, H. , Christensen, Q.H. , Feng, Y. , Wang, H. and Cronan, J.E. (2012) The Burkholderia cenocepacia BDSF quorum sensing fatty acid is synthesized by a bifunctional homologue having both dehydratase and thioesterase activities. Mol. Microbiol. 83, 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bodman, S.B. , Majerczak, D.R. and Coplin, D.L. (1998) A negative regulator mediates quorum‐sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii . Proc. Natl. Acad. Sci. USA, 95, 7687–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette, D.L. , Monroe, K.M. , Sotelo‐Troha, K. , Iwig, J.S. , Eckert, B. , Hyodo, M. , Hayakawa, Y. and Vance, R. (2011) STING is a direct innate immune sensor of cyclic di‐GMP. Nature, 478, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, T. , Barnard, A.M. , Corbett, M.J. , Pemberton, C.L. , Simpson, N.J. and Salmond, G.P. (2006) Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol. Microbiol. 59, 113–125. [DOI] [PubMed] [Google Scholar]
- Cao, J.G. and Meighen, E.A. (1989) Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi . J. Biol. Chem. 264, 21 670–21 676. [PubMed] [Google Scholar]
- Carlier, A. , Chevrot, R. , Dessaux, Y. and Faure, D. (2004) The assimilation of gamma‐butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N‐acyl‐homoserine lactone signal. Mol. Plant–Microbe Interact. 17, 951–957. [DOI] [PubMed] [Google Scholar]
- Chai, Y. , Zhu, J. and Winans, S.C. (2001) TrlR, a defective TraR‐like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40, 414–421. [DOI] [PubMed] [Google Scholar]
- Chai, Y. , Tsai, C.S. , Cho, H. and Winans, S.C. (2007) Reconstitution of the biochemical activities of the AttJ repressor and the AttK, AttL, and AttM catabolic enzymes of Agrobacterium tumefaciens . J. Bacteriol. 189, 3674–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. and Sonti, R.V. (2002) rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol. Plant–Microbe Interact. 15, 463–471. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. , Newman, K.L. and Lindow, S.E. (2008a) Cell‐to‐cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol. Plant–Microbe Interact. 21, 1309–1315. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. , Wistrom, C. and Lindow, S.E. (2008b) A cell–cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa . Proc. Natl. Acad. Sci. USA, 105, 2670–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R. , Barphagha, I.K. , Karki, H.S. and Ham, J.H. (2012) Dissection of quorum‐sensing genes in Burkholderia glumae reveals non‐canonical regulation and the new regulatory gene tofM for toxoflavin production. PLoS ONE. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Schauder, S. , Potier, N. , Van Dorsselaer, A. , Pelczer, I. , Bassler, B.L. and Hughson, F.M. (2002) Structural identification of a bacterial quorum‐sensing signal containing boron. Nature, 415, 545–549. [DOI] [PubMed] [Google Scholar]
- Cheng, Z. , He, Y.W. , Lim, S.C. , Qamra, R. , Walsh, M.A. , Zhang, L.H. and Song, H. (2010) Structural basis of the sensor‐synthase interaction in autoinduction of the quorum sensing signal DSF biosynthesis. Structure, 18, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Chin, K.H. , Lee, Y.C. , Tu, Z.L. , Chen, C.H. , Tseng, Y.H. , Yang, J.M. , Ryan, R.P. , McCarthy, Y. , Dow, J.M. , Wang, A.H. and Chou, S.H. (2010) The cAMP receptor‐like protein CLP is a novel c‐di‐GMP receptor linking cell–cell signaling to virulence gene expression in Xanthomonas campestris . J. Mol. Biol. 396, 646–662. [DOI] [PubMed] [Google Scholar]
- Colnaghi Simionato, A.V. , da Silva, D.S. , Lambais, M.R. and Carrilho, E. (2007) Characterization of a putative Xylella fastidiosa diffusible signal factor by HRGC‐EI‐MS. J. Mass Spectrom. 42, 490–496. [DOI] [PubMed] [Google Scholar]
- Corrigan, R.M. , Abbott, J.C. , Burhenne, H. , Kaever, V. and Grundling, A. (2011) c‐di‐AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7, e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, P.A. and Stibitz, S. (2007) c‐di‐GMP‐mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10, 17–23. [DOI] [PubMed] [Google Scholar]
- Coulthurst, S.J. , Lilley, K.S. and Salmond, G.P. (2006) Genetic and proteomic analysis of the role of luxS in the enteric phytopathogen, Erwinia carotovora . Mol. Plant Pathol. 7, 31–45. [DOI] [PubMed] [Google Scholar]
- Cui, Y. , Chatterjee, A. , Hasegawa, H. , Dixit, V. , Leigh, N. and Chatterjee, A.K. (2005) ExpR, a LuxR homolog of Erwinia carotovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA‐binding protein. J. Bacteriol. 187, 4792–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Chatterjee, A. , Hasegawa, H. and Chatterjee, A.K. (2006) Erwinia carotovora subspecies produce duplicate variants of ExpR, LuxR homologs that activate rsmA transcription but differ in their interactions with N‐acylhomoserine lactone signals. J. Bacteriol. 188, 4715–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Boon, C. , Eberl, L. and Zhang, L.H. (2009) Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum‐sensing signal BDSF and its synthase. J. Bacteriol. 191, 7270–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Wu, J. , Tao, F. and Zhang, L.H. (2011) Listening to a new language: DSF‐based quorum sensing in Gram‐negative bacteria. Chem. Rev. 111, 160–173. [DOI] [PubMed] [Google Scholar]
- Devescovi, G. , Bigirimana, J. , Degrassi, G. , Cabrio, L. , LiPuma, J.J. , Kim, J. , Hwang, I. and Venturi, V. (2007) Involvement of a quorum‐sensing‐regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl. Environ. Microbiol. 73, 4950–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle, S.P. , Lumjiaktase, P. , Dipilato, F. , Winzer, K. , Kunakorn, M. , Barrett, D.A. , Chhabra, S.R. , Camara, M. and Williams, P. (2006) Functional genetic analysis reveals a 2‐alkyl‐4‐quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem. Biol. 13, 701–710. [DOI] [PubMed] [Google Scholar]
- Duerkop, B.A. , Ulrich, R.L. and Greenberg, E.P. (2007) Octanoyl‐homoserine lactone is the cognate signal for Burkholderia mallei BmaR1‐BmaI1 quorum sensing. J. Bacteriol. 189, 5034–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop, B.A. , Varga, J. , Chandler, J.R. , Peterson, S.B. , Herman, J.P. , Churchill, M.E. , Parsek, M.R. , Nierman, W.C. and Greenberg, E.P. (2009) Quorum‐sensing control of antibiotic synthesis in Burkholderia thailandensis . J. Bacteriol. 191, 3909–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl, L. (2006) Quorum sensing in the genus Burkholderia . Int. J. Med. Microbiol. 296, 103–110. [DOI] [PubMed] [Google Scholar]
- French, C.T. , Lao, P. , Loraine, A.E. , Matthews, B.T. , Yu, H. and Dybvig, K. (2008) Large‐scale transposon mutagenesis of Mycoplasma pulmonis . Mol. Microbiol. 69, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua, C. and Winans, S.C. (1996) Conserved cis‐acting promoter elements are required for density‐dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua, C. , Burbea, M. and Winans, S.C. (1995) Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J. Bacteriol. 177, 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua, W.C. and Winans, S.C. (1994) A LuxR‐LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176, 2796–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua, W.C. , Winans, S.C. and Greenberg, E.P. (1994) Quorum sensing in bacteria: the LuxR‐LuxI family of cell density‐responsive transcriptional regulators. J. Bacteriol. 176, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Song, J. , Hu, B. , Zhang, L. , Liu, Q. and Liu, F. (2009) The luxS gene is involved in AI‐2 production, pathogenicity, and some phenotypes in Erwinia amylovora . Curr. Microbiol. 58, 1–10. [DOI] [PubMed] [Google Scholar]
- Glass, J.I. , Assad‐Garcia, N. , Alperovich, N. , Yooseph, S. , Lewis, M.R. , Maruf, M. , Hutchison, C.A. , Smith, H.O. and Venter, J.C. (2006) Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA, 103, 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelsky, M. (2011) cAMP, c‐di‐GMP, c‐di‐AMP and now cGMP: bacteria use them all! Mol. Microbiol. 79, 562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo, E. , Kang, Y. , Kim, H. and Hwang, I. (2010) Proteomic analysis of quorum sensing‐dependent proteins in Burkholderia glumae . J. Proteome Res. 9, 3184–3199. [DOI] [PubMed] [Google Scholar]
- Han, S.W. , Lee, S.W. and Ronald, P.C. (2011a) Secretion, modification, and regulation of Ax21. Curr. Opin. Microbiol. 14, 62–67. [DOI] [PubMed] [Google Scholar]
- Han, S.W. , Sriariyanun, M. , Lee, S.W. , Sharma, M. , Bahar, O. , Bower, Z. and Ronald, P.C. (2011b) Small protein‐mediated quorum sensing in a Gram‐negative bacterium. PLoS ONE, 6, e29192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- He, Y.W. and Zhang, L.H. (2008) Quorum sensing and virulence regulation in Xanthomonas campestris . FEMS Microbiol. Rev. 32, 842–857. [DOI] [PubMed] [Google Scholar]
- He, Y.W. , Wang, C. , Zhou, L. , Song, H. , Dow, J.M. and Zhang, L.H. (2006) Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain–protein interaction. J. Biol. Chem. 281, 33 414–33 421. [DOI] [PubMed] [Google Scholar]
- He, Y.W. , Ng, A.Y. , Xu, M. , Lin, K. , Wang, L.H. , Dong, Y.H. and Zhang, L.H. (2007) Xanthomonas campestris cell–cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64, 281–292. [DOI] [PubMed] [Google Scholar]
- He, Y.W. , Boon, C. , Zhou, L. and Zhang, L.H. (2009) Co‐regulation of Xanthomonas campestris virulence by quorum sensing and a novel two‐component regulatory system RavS/RavR. Mol. Microbiol. 71, 1464–1476. [DOI] [PubMed] [Google Scholar]
- He, Y.W. , Wu, J. , Cha, J.S. and Zhang, L.H. (2010) Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF‐family signals in regulation of virulence factor production. BMC Microbiol. 10, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, B. , Feldmann, F. , Kothe, M. , Vandamme, P. , Wopperer, J. , Riedel, K. and Eberl, L. (2004) Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans . Infect. Immun. 72, 7220–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I. , Li, P.L. , Zhang, L. , Piper, K.R. , Cook, D.M. , Tate, M.E. and Farrand, S.K. (1994) TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N‐acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. USA, 91, 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I. , Cook, D.M. and Farrand, S.K. (1995) A new regulatory element modulates homoserine lactone‐mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177, 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I. , Smyth, A.J. , Luo, Z.Q. and Farrand, S.K. (1999) Modulating quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol. Microbiol. 34, 282–294. [DOI] [PubMed] [Google Scholar]
- Jones, S. , Yu, B. , Baiton, N.J. , Birdsall, M. , Bycoft, B.W. , Chhabra, S.R. , Cox, A.J.R. , Reeves, P.J. , Stephens, S. , Winson, M.K. , Salmond, G.P.C. , Stewart, G.S.A.B. and Williams, P. (1993) The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa . EMBO J. 12, 2477–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Kim, J.G. , Kang, Y. , Jang, J.Y. , Jog, G.J. , Lim, J.Y. , Kim, S. , Suga, H. , Nagamatsu, T. and Hwang, I. (2004) Quorum sensing and the LysR‐type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae . Mol. Microbiol. 54, 921–934. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Kang, Y. , Choi, O. , Jeong, Y. , Jeong, J.E. , Lim, J.Y. , Kim, M. , Moon, J.S. , Suga, H. and Hwang, I. (2007) Regulation of polar flagellum genes is mediated by quorum sensing and FlhDC in Burkholderia glumae . Mol. Microbiol. 64, 165–179. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Oh, J. , Choi, O. , Kang, Y. , Kim, H. , Goo, E. , Ma, J. , Nagamatsu, T. , Moon, J.S. and Hwang, I. (2009) Biochemical evidence for ToxR and ToxJ binding to the tox operons of Burkholderia glumae and mutational analysis of ToxR. J. Bacteriol. 191, 4870–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoudis, M.D. , Tsaltas, D. , von Minogue, T.D. and Bodman, S.B. (2006) Quorum‐sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii . Proc. Natl. Acad. Sci. USA, 103, 5983–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc, J.L. and Roberts, G.P. (2009) Cyclic di‐GMP allosterically inhibits the CRP‐like protein (Clp) of Xanthomonas axonopodis pv. citri . J. Bacteriol. 191, 7121–7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.W. , Han, S.W. , Sririyanum, M. , Park, C.J. , Seo, Y.S. and Ronald, P.C. (2009) A type I‐secreted, sulfated peptide triggers XA21‐mediated innate immunity. Science, 326, 850–853. [DOI] [PubMed] [Google Scholar]
- Li, T.N. , Chin, K.H. , Fung, K.M. , Yang, M.T. , Wang, A.H. and Chou, S.H. (2011) A novel tetrameric PilZ domain structure from xanthomonads. PLoS ONE, 6, e22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Coulthurst, S.J. , Pritchard, L. , Hedley, P.E. , Ravensdale, M. , Humphris, S. , Burr, T. , Takle, G. , Brurgerg, M.‐B. , Birch, P.R.J. , Salmond, G.P.C. and Toth, I.K. (2008) Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogenic Pectobacterium atrosepticum . PLoS Pathog. 4, e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery, C.A. , Dickerson, T.J. and Janda, K.D. (2008) Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem. Soc. Rev. 37, 1337–1346. [DOI] [PubMed] [Google Scholar]
- Luo, Y. and Helmann, J.D. (2012) Analysis of the role of Bacillus subtilis sigma(M) in beta‐lactam resistance reveals an essential role for c‐di‐AMP in peptidoglycan homeostasis. Mol. Microbiol. 83, 623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malott, R.J. , O'Grady, E.P. , Toller, J. , Inhulsen, S. , Eberl, L. and Sokol, P.A. (2009) A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum‐sensing regulation. J. Bacteriol. 191, 2447–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden, J.N. , Dong, Q. , Roychowdhury, S. , Berleman, J.E. and Bauer, C.E. (2011) Cyclic GMP controls Rhodospirillum centenum cyst development. Mol. Microbiol. 79, 600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiuzzo, M. , Bertani, I. , Ferluga, S. , Cabrio, L. , Bigirimana, J. , Guarnaccia, C. , Pongor, S. , Maraite, H. and Venturi, V. (2011) The plant pathogen Pseudomonas fuscovaginae contains two conserved quorum sensing systems involved in virulence and negatively regulated by RsaL and the novel regulator RsaM. Environ. Microbiol. 13, 145–162. [DOI] [PubMed] [Google Scholar]
- McCarthy, Y. , Ryan, R.P. , O'Donovan, K. , He, Y.Q. , Jiang, B.L. , Feng, J.X. , Tang, J.L. and Dow, J.M. (2008) The role of PilZ domain proteins in the virulence of Xanthomonas campestris pv. campestris . Mol. Plant Pathol. 9, 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, Y. , Dow, J.M. and Ryan, R.P. (2011) The Ax21 protein is a cell–cell signal that regulates virulence in the nosocomial pathogen Stenotrophomonas maltophilia . J. Bacteriol. 193, 6375–6378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- McGowan, S.J. , Barnard, A.M. , Bosgelmez, G. , Sebaihia, M. , Simpson, N.J. , Thomson, N.R. , Todd, D.E. , Welch, M. , Whitehead, N.A. and Salmond, G.P. (2005) Carbapenem antibiotic biosynthesis in Erwinia carotovora is regulated by physiological and genetic factors modulating the quorum sensing‐dependent control pathway. Mol. Microbiol. 55, 526–545. [DOI] [PubMed] [Google Scholar]
- Miller, M.B. , Skorupski, K. , Lenz, D.H. , Taylor, R.K. and Bassler, B.L. (2002) Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae . Cell, 110, 303–314. [DOI] [PubMed] [Google Scholar]
- Miller, S.T. , Xavier, K.B. , Campagna, S.R. , Taga, M.E. , Semmelhack, M.F. , Bassler, B.L. and Hughson, F.M. (2004) Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum‐sensing signal AI‐2. Mol. Cell, 15, 677–687. [DOI] [PubMed] [Google Scholar]
- Mills, E. , Pultz, I.S. , Kulasekara, H.D. and Miller, S.I. (2011) The bacterial second messenger c‐di‐GMP: mechanisms of signalling. Cell. Microbiol. 13, 1122–1129. [DOI] [PubMed] [Google Scholar]
- Minogue, T.D. , Carlier, A.L. , von Koutsoudis, M.D. and Bodman, S.B. (2005) The cell density‐dependent expression of stewartan exopolysaccharide in Pantoea stewartii ssp. stewartii is a function of EsaR‐mediated repression of the rcsA gene. Mol. Microbiol. 56, 189–203. [DOI] [PubMed] [Google Scholar]
- More, M.I. , Finger, L.D. , Stryker, J.L. , Fuqua, C. , Eberhard, A. and Winans, S.C. (1996) Enzymatic synthesis of a quorum‐sensing autoinducer through use of defined substrates. Science, 272, 1655–1658. [DOI] [PubMed] [Google Scholar]
- Morohoshi, T. , Nakamura, Y. , Yamazaki, G. , Ishida, A. , Kato, N. and Ikeda, T. (2007) The plant pathogen Pantoea ananatis produces N‐acylhomoserine lactone and causes center rot disease of onion by quorum sensing. J. Bacteriol. 189, 8333–8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser, W. , Robert‐Baudouy, J. and Reverchon, S. (1997) Antagonistic effect of CRP and KdgR in the transcription control of the Erwinia chrysanthemi pectinolysis genes. Mol. Microbiol. 26, 1071–1082. [DOI] [PubMed] [Google Scholar]
- Newman, K.L. , Almeida, R.P. , Purcell, A.H. and Lindow, S.E. (2004) Cell–cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. USA, 101, 1737–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oger, P. , Kim, K.S. , Sackett, R.L. , Piper, K.R. and Farrand, S.K. (1998) Octopine‐type Ti plasmids code for a mannopine‐inducible dominant‐negative allele of traR, the quorum‐sensing activator that regulates Ti plasmid conjugal transfer. Mol. Microbiol. 27, 277–288. [DOI] [PubMed] [Google Scholar]
- Oppenheimer‐Shaanan, Y. , Wexselblatt, E. , Katzhendler, J. , Yavin, E. and Ben‐Yehuda, S. (2011) c‐di‐AMP reports DNA integrity during sporulation in Bacillus subtilis . EMBO Rep. 12, 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, C.S. , Thompson, J.A. and Xavier, K.B. (2012) AI‐2‐mediated signaling in bacteria. FEMS Microbiol. Rev. doi: 10.1111/j.1574-6976.2012.00345.x. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Perez‐Mendoza, D. , Coulthurst, S.J. , Humphris, S. , Campbell, E. , Welch, M. , Toth, I.K. and Salmond, G.P. (2011a) A multi‐repeat adhesin of the phytopathogen, Pectobacterium atrosepticum, is secreted by a Type I pathway and is subject to complex regulation involving a non‐canonical diguanylate cyclase. Mol. Microbiol. 82, 719–733. [DOI] [PubMed] [Google Scholar]
- Perez‐Mendoza, D. , Coulthurst, S.J. , Sanjuan, J. and Salmond, G.P. (2011b) N‐Acetylglucosamine‐dependent biofilm formation in Pectobacterium atrosepticum is cryptic and activated by elevated c‐di‐GMP levels. Microbiology, 157, 3340–3348. [DOI] [PubMed] [Google Scholar]
- Pirhonen, M. , Flego, D. , Heikinheimo, R. and Palva, E.T. (1993) A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora . EMBO J. 12, 2467–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones, B. , Pujol, C.J. and Lindow, S.E. (2004) Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae . Mol. Plant–Microbe Interact. 17, 521–531. [DOI] [PubMed] [Google Scholar]
- Quinones, B. , Dulla, G. and Lindow, S.E. (2005) Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae . Mol. Plant–Microbe Interact. 18, 682–693. [DOI] [PubMed] [Google Scholar]
- Rai, R. , Ranjan, M. , Pradhan, B.B. and Chatterjee, S. (2012) Atypical regulation of virulence‐associated functions by a diffusible signal factor in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 25, 789–801. [DOI] [PubMed] [Google Scholar]
- Reverchon, S. , Expert, D. , Robert‐Baudouy, J. and Nasser, W. (1997) The cyclic AMP receptor protein is the main activator of pectinolysis genes in Erwinia chrysanthemi . J. Bacteriol. 179, 3500–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon, S. , Bouillant, M.L. , Salmond, G. and Nasser, W. (1998) Integration of the quorum‐sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi . Mol. Microbiol. 29, 1407–1418. [DOI] [PubMed] [Google Scholar]
- Rezzonico, F. and Duffy, B. (2007) The role of luxS in the fire blight pathogen Erwinia amylovora is limited to metabolism and does not involve quorum sensing. Mol. Plant–Microbe Interact. 20, 1284–1297. [DOI] [PubMed] [Google Scholar]
- Rezzonico, F. and Duffy, B. (2008) Lack of genomic evidence of AI‐2 receptors suggests a non‐quorum sensing role for luxS in most bacteria. BMC Microbiol. 8, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha‐Estrada, J. , Aceves‐Diez, A.E. , Guarneros, G. and Torre, M. (2010) The RNPP family of quorum‐sensing proteins in Gram‐positive bacteria. Appl. Microbiol. Biotechnol. 87, 913–923. [DOI] [PubMed] [Google Scholar]
- Romling, U. (2012) Cyclic di‐GMP, an established secondary messenger still speeding up. Environ. Microbiol. 14, 1817–1829. [DOI] [PubMed] [Google Scholar]
- Ronald, P.C. (2011) Small protein‐mediated quorum sensing in a gram‐negative bacterium: novel targets for control of infectious disease. Discov. Med. 12, 461–470. [PubMed] [Google Scholar]
- Roy, V. , Adams, B.L. and Bentley, W.E. (2011) Developing next generation antimicrobials by intercepting AI‐2 mediated quorum sensing. Enzyme Microb. Technol. 49, 113–123. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. and Dow, J.M. (2010) Intermolecular interactions between HD‐GYP and GGDEF domain proteins mediate virulence‐related signal transduction in Xanthomonas campestris . Virulence, 1, 404–408. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , Fouhy, Y. , Lucey, J.F. , Crossman, L.C. , Spiro, S. , He, Y.W. , Zhang, L.H. , Heeb, S. , Camara, M. , Williams, P. and Dow, J.M. (2006) Cell–cell signaling in Xanthomonas campestris involves an HD‐GYP domain protein that functions in cyclic di‐GMP turnover. Proc. Natl. Acad. Sci. USA, 103, 6712–6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ryan, R.P. , McCarthy, Y. , Andrade, M. , Farah, C.S. , Armitage, J.P. and Dow, J.M. (2010) Cell–cell signal‐dependent dynamic interactions between HD‐GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris . Proc. Natl. Acad. Sci. USA, 107, 5989–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. , McCarthy, Y. , Kiely, P.A. , O'Connor, R. , Farah, C.S. , Armitage, J.P. and Dow, J.M. (2012) Dynamic complex formation between HD‐GYP, GGDEF and PilZ domain proteins regulates motility in Xanthomonas campestris . Mol. Microbiol. 86, 557–567. [DOI] [PubMed] [Google Scholar]
- Schirmer, T. and Jenal, U. (2009) Structural and mechanistic determinants of c‐di‐GMP signalling. Nat. Rev. Microbiol. 7, 724–735. [DOI] [PubMed] [Google Scholar]
- Schmidt, S. , Blom, J.F. , Pernthaler, J. , Berg, G. , Baldwin, A. , Mahenthiralingam, E. and Eberl, L. (2009) Production of the antifungal compound pyrrolnitrin is quorum sensing‐regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 11, 1422–1437. [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost, M.R. , Chandler, J.R. , Blodgett, J.A. , Lima, P.S. , Duerkop, B.A. , Oinuma, K. , Greenberg, E.P. and Clardy, J. (2010) Quorum‐sensing‐regulated bactobolin production by Burkholderia thailandensis E264. Org. Lett. 12, 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank, E.A. and Kolter, R. (2011) Extracellular signaling and multicellularity in Bacillus subtilis . Curr. Opin. Microbiol. 14, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom, S. , Brader, G. , Koch, G. and Palva, E.T. (2006) Cooperation of two distinct ExpR regulators controls quorum sensing specificity and virulence in the plant pathogen Erwinia carotovora . Mol. Microbiol. 60, 1474–1489. [DOI] [PubMed] [Google Scholar]
- Slater, H. , Alvarez‐Morales, A. , Barber, C.E. , Daniels, M.J. and Dow, J.M. (2000) A two‐component system involving an HD‐GYP domain protein links cell–cell signalling to pathogenicity gene expression in Xanthomonas campestris . Mol. Microbiol. 38, 986–1003. [DOI] [PubMed] [Google Scholar]
- Sondermann, H. , Shikuma, N.J. and Yildiz, F.H. (2012) You've come a long way: c‐di‐GMP signaling. Curr. Opin. Microbiol. 15, 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.H. , Ko, K.S. , Lee, J.Y. , Baek, J.Y. , Oh, W.S. , Yoon, H.S. , Jeong, J.Y. and Chun, J. (2005) Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells, 19, 365–374. [PubMed] [Google Scholar]
- Song, W.Y. , Wang, G.L. , Chen, L.L. , Kim, H.S. , Pi, L.Y. , Holsten, T. , Gardner, J. , Wang, B. , Zhai, W.X. , Zhu, L.H. , Fauquet, C. and Ronald, P. (1995) A receptor kinase‐like protein encoded by the rice disease resistance gene, Xa21 . Science, 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Daniel, R. , Wagner‐Dobler, I. and Zeng, A.P. (2004) Is autoinducer‐2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, J.L. , Liu, Y.N. , Barber, C.E. , Dow, J.M. , Wootton, J.C. and Daniels, M.J. (1991) Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol. Gen. Genet. 226, 409–417. [DOI] [PubMed] [Google Scholar]
- Tang, J.L. , Feng, J.X. , Li, Q.Q. , Wen, H.X. , Zhou, D.L. , Wilson, T.J. , Dow, J.M. , Ma, Q.S. and Daniels, M.J. (1996) Cloning and characterization of the rpfC gene of Xanthomonas oryzae pv. oryzae: involvement in exopolysaccharide production and virulence to rice. Mol. Plant–Microbe Interact. 9, 664–666. [DOI] [PubMed] [Google Scholar]
- Tao, F. , He, Y.W. , Wu, D.H. , Swarup, S. and Zhang, L.H. (2010) The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di‐GMP effectors. J. Bacteriol. 192, 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, N.R. , Nasser, W. , McGowan, S. , Sebaihia, M. and Salmond, G.P. (1999) Erwinia carotovora has two KdgR‐like proteins belonging to the IciR family of transcriptional regulators: identification and characterization of the RexZ activator and the KdgR repressor of pathogenesis. Microbiology, 145 (Pt 7), 1531–1545. [DOI] [PubMed] [Google Scholar]
- Thowthampitak, J. , Shaffer, B.T. , Prathuangwong, S. and Loper, J.E. (2008) Role of rpfF in virulence and exoenzyme production of Xanthomonas axonopodis pv. glycines, the causal agent of bacterial pustule of soybean. Phytopathology, 98, 1252–1260. [DOI] [PubMed] [Google Scholar]
- Trimble, M.J. and McCarter, L.L. (2011) Bis‐(3′‐5′)‐cyclic dimeric GMP‐linked quorum sensing controls swarming in Vibrio parahaemolyticus . Proc. Natl. Acad. Sci. USA, 108, 18 079–18 084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.S. and Winans, S.C. (2010) LuxR‐type quorum‐sensing regulators that are detached from common scents. Mol. Microbiol. 77, 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, R.L. , Deshazer, D. , Hines, H.B. and Jeddeloh, J.A. (2004) Quorum sensing: a transcriptional regulatory system involved in the pathogenicity of Burkholderia mallei . Infect. Immun. 72, 6589–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini, A. , Volpari, C. , Gargioli, C. , Muraglia, E. , Cortese, R. , De Francesco, R. , Neddermann, P. and Marco, S.D. (2002) The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21, 4393–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi, V. , Rampioni, G. , Pongor, S. and Leoni, L. (2011) The virtue of temperance: built‐in negative regulators of quorum sensing in Pseudomonas . Mol. Microbiol. 82, 1060–1070. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Zhang, H.B. , Wang, L.H. and Zhang, L.H. (2006) Succinic semialdehyde couples stress response to quorum‐sensing signal decay in Agrobacterium tumefaciens . Mol. Microbiol. 62, 45–56. [DOI] [PubMed] [Google Scholar]
- Wang, L.H. , He, Y. , Gao, Y. , Wu, J.E. , Dong, Y.H. , He, C. , Wang, S.X. , Weng, L.X. , Xu, J.L. , Tay, L. , Fang, R.X. and Zhang, L.H. (2004) A bacterial cell–cell communication signal with cross‐kingdom structural analogues. Mol. Microbiol. 51, 903–912. [DOI] [PubMed] [Google Scholar]
- Witte, G. , Hartung, S. , Buttner, K. and Hopfner, K.P. (2008) Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell, 30, 167–178. [DOI] [PubMed] [Google Scholar]
- Woodward, J.J. , Iavarone, A.T. and Portnoy, D.A. (2010) c‐di‐AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science, 328, 1703–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, X. , Yamazaki, A. , Biddle, E. , Zeng, Q. and Yang, C.H. (2010) Genetic analysis of two phosphodiesterases reveals cyclic diguanylate regulation of virulence factors in Dickeya dadantii . Mol. Microbiol. 77, 787–800. [DOI] [PubMed] [Google Scholar]
- Zhang, H.B. , Wang, L.H. and Zhang, L.H. (2002) Genetic control of quorum‐sensing signal turnover in Agrobacterium tumefaciens . Proc. Natl. Acad. Sci. USA, 99, 4638–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H.B. , Wang, C. and Zhang, L.H. (2004) The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p)ppGpp. Mol. Microbiol. 52, 1389–1401. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Murphy, P.J. , Kerr, A. and Tate, M.E. (1993) Agrobacterium conjugation and gene regulation by N‐acyl‐L‐homoserine lactones. Nature, 362, 446–448. [DOI] [PubMed] [Google Scholar]
- Zhang, L.H. (2010) A novel C‐di‐GMP effector linking intracellular virulence regulon to quorum sensing and hypoxia sensing. Virulence, 1, 391–394. [DOI] [PubMed] [Google Scholar]
- Zhang, R.G. , Pappas, K.M. , Brace, J.L. , Miller, P.C. , Oulmassov, T. , Molyneaux, J.M. , Anderson, J.C. , Bashkin, J.K. , Winans, S.C. and Joachimiak, A. (2002) Structure of a bacterial quorum‐sensing transcription factor complexed with pheromone and DNA. Nature, 417, 971–974. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Qian, G. , Yin, F. , Fan, J. , Zhai, Z. , Liu, C. , Hu, B. and Liu, F. (2011) Proteomic analysis of the regulatory function of DSF‐dependent quorum sensing in Xanthomonas oryzae pv. oryzicola . Microb. Pathog. 50, 48–55. [DOI] [PubMed] [Google Scholar]
- Zhu, J. and Winans, S.C. (1998) Activity of the quorum‐sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR‐like protein. Mol. Microbiol. 27, 289–297. [DOI] [PubMed] [Google Scholar]
- Zhu, J. and Winans, S.C. (1999) Autoinducer binding by the quorum‐sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA, 96, 4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]


