Summary
The mechanisms determining the host range of Xanthomonas are still undeciphered, despite much interest in their potential roles in the evolution and emergence of plant pathogenic bacteria. Xanthomonas citri pv. citri (Xci) is an interesting model of host specialization because of its pathogenic variants: pathotype A strains infect a wide range of Rutaceous species, whereas pathotype A*/AW strains have a host range restricted to Mexican lime (Citrus aurantifolia) and alemow (Citrus macrophylla). Based on a collection of 55 strains representative of Xci worldwide diversity assessed by amplified fragment length polymorphism (AFLP), we investigated the distribution of type III effectors (T3Es) in relation to host range. We examined the presence of 66 T3Es from xanthomonads in Xci and identified a repertoire of 28 effectors, 26 of which were shared by all Xci strains, whereas two (xopAG and xopC1) were present only in some A*/AW strains. We found that xopAG (=avrGf1) was present in all AW strains, but also in three A* strains genetically distant from AW, and that all xopAG‐containing strains induced the hypersensitive response (HR) on grapefruit and sweet orange. The analysis of xopAD and xopAG suggested horizontal transfer between X. citri pv. bilvae, another citrus pathogen, and some Xci strains. A strains were genetically less diverse, induced identical phenotypic responses and possessed indistinguishable T3E repertoires. Conversely, A*/AW strains exhibited a wider genetic diversity in which clades correlated with geographical origin and T3E repertoire, but not with pathogenicity, according to T3E deletion experiments. Our data outline the importance of taking into account the heterogeneity of Xci A*/AW strains when analysing the mechanisms of host specialization.
Introduction
Several factors may contribute to the emergence of new diseases: the introduction of an existing pathogen into new geographical areas, environmental changes that favour the spread of a pathogen or the broadening of the pathogen host range through rapid evolution to overcome host defences and an increase in fitness (Anderson et al., 2004). The investigation of the mechanisms underlying the host range broadening or restriction of a pathogen might aid in the understanding of emergences. Asiatic canker caused by Xanthomonas citri pv. citri (Xci) is the most common and most aggressive form of citrus canker. This invasive disease occurs in more than 30 countries throughout the world, and is currently emerging in Africa (Balestra et al., 2008; Derso et al., 2009; Leduc et al., 2011; Traore et al., 2008). It affects most commercial varieties of citrus, limiting citrus production worldwide (Gottwald et al., 2002). In South America, it has been reported to supplant the milder, geographically and host‐restricted South American canker (Xanthomonas citri pv. aurantifolii), referred to as canker B and C, which has a much lower incidence on citrus (Schubert et al., 2001). Xci enters host plant tissues through stomata or wounds, and multiplies in the mesophyll. This pathogen induces erumpent, callus‐like lesions with a water‐soaked margin on leaves, fruit and stem tissue. Severe attacks result in extensive defoliation, premature fruit drop and twig dieback. Strains of Xci have been classified into three pathogenic variants (i.e. pathotypes) as they differ in host range but not in symptomatology (Vernière et al., 1998). Strains of pathotype A are present worldwide and have the widest host range (nearly all citrus species and some related genera), although with differences in susceptibility among species or cultivars. In contrast, strains of pathotypes A* and AW have a narrow host range, mainly restricted to Mexican lime (Citrus aurantifolia) and alemow (Citrus macrophylla) in natural conditions (Sun et al., 2004; Vernière et al., 1998). Strains of pathotype AW have been distinguished from strains of pathotype A* on the basis of hypersensitive response (HR)‐like symptoms on grapefruit (Citrus paradisi), sweet orange (Citrus sinensis), sour orange (Citrus aurantium), citron (Citrus medica), Orlando tangelo (Citrus reticulate × Citrus paradisi) and trifoliate orange (Poncirus trifoliata) (Sun et al., 2004). A previous description of the diversity of a collection of Xci by amplified fragment length polymorphism (AFLP) analysis, insertion sequence ligation‐mediated polymerase chain reaction (IS‐LM‐PCR) typing and multi‐locus variable‐number tandem‐repeat analysis (MLVA) showed a congruent structure of Xci separating pathotypes A and A*/AW (Bui‐Thi‐Ngoc et al., 2009). Furthermore, this study emphasized that the Xci population probably had a predominant clonal structure and that A* strains showed a higher genetic diversity than A strains. This result raised interesting questions about the phylogenetic relationship and evolutionary histories of Xci pathotypes A, A* and AW.
Today, much remains to be learned about the molecular determinants involved in pathogenicity and host range variations reported among Xci pathotypes. The pathogenicity of most Xanthomonas depends on the conserved syringe‐like type III secretion system (T3SS, encoded by the chromosomal hrp—HR and pathogenicity—gene cluster) capable of traversing the host cell wall and membrane to inject effector proteins, called type III effectors (T3Es), into the host cell cytoplasm (Alfano and Collmer, 1997). In compatible interactions, T3Es promote bacterial growth and virulence. In incompatible interactions, they trigger specific defences in resistant plants, and induce HR (Gurlebeck et al., 2006; White et al., 2009). They constitute prime targets for mutation and selection to evade the host immune system.
Recent work has speculated the likely role of T3Es as a group of factors or ‘repertoire’ shaping the host ranges of individual Pseudomonas syringae strains (Sarkar et al., 2006) and many xanthomonads (Hajri et al., 2009, 2012a, 2012b). Documented examples have shown the contribution of one or a few effectors in the virulence of a specific strain on a specific host or cultivar (Arnold and Jackson, 2011). In the Xanthomonas genus, effectors named Xanthomonas outer proteins (Xops) have been described according to studies based on functional assays, combined with sequence and structural similarity searches of genomic data. They are classified into 66 families (White et al., 2009) (Xanthomonas resource website: http://www.xanthomonas.org/; R. Koebnik, IRD, Montpellier, France, personal communication). In Xci, a functional T3SS is necessary for infection, the formation of canker symptoms on host plants and HR induction in nonhost plants (Dunger et al., 2005). Twenty‐six effector families are represented in the genome of Xci IAPAR 306 (Da Silva et al., 2002). Furthermore, the effector avrGf1 (=xopAG) was subsequently identified in AW strains (Rybak et al., 2009), enlarging the Xci repertoire to 27 T3Es. XopAG has been shown to be responsible for the induction of HR‐like reactions on grapefruit, putatively contributing to the specific interaction of the pathogen. The deletion of xopAG partially restored canker development on host species differentiating A and AW strains, yet canker symptoms were less severe. This effector is also found in Xanthomonas campestris pv. campestris sequenced genomes (strains ATCC33913, B100, 8004) (Da Silva et al., 2002; Qian et al., 2005; Vorhoelter et al., 2008), in Xanthomonas vesicatoria ATCC35937 (Potnis et al., 2011), but not in the Xanthomonas euvesicatoria sequenced strain (85‐10) (Thieme et al., 2005), which is phylogenetically closer to Xci IAPAR 306. The involvement in pathogenicity of 13 of the 26 Xops found in the genome of the model strain Xci IAPAR 306 was assessed by site‐directed mutagenesis on each individual effector (Figueiredo et al., 2011). This study also included other candidate coding sequences revealed by the presence of a conserved promoter element, known in Xanthomonas as plant‐inducible promoter (PIP) boxes (Koebnik et al., 2006; Wengelnik and Bonas, 1996). Apart from hrp genes and the mutation of the key canker‐inducing protein PthA4 belonging to the transcription activator‐like (TAL) effector family (Swarup et al., 1991, 1992), none of the tested mutants reduced the ability to cause citrus canker disease on grapefruit and Mexican lime leaves. An understanding of the role of these effectors in host range variations among Xci pathotypes requires further investigation.
The aim of this work was to investigate the role of T3Es in defining the host specialization of Xci pathotypes. For this purpose, we worked on a comprehensive worldwide collection of Xci strains chosen to represent the known genetic diversity of the pathovar. We characterized the pathogenicity of the strains based on several tests to better assess their aggressiveness and host range, and we identified their T3E repertoires. We based this screen on the potential Xop candidates described in the Xanthomonas genus, and conducted PCR and Southern blot hybridization. In the case of pathotype‐specific candidate effectors, site‐directed mutagenesis was conducted to evaluate the role of single T3Es in the plant–bacterium interaction. We also investigated how the different T3E repertoires correlate with the genetic diversity of Xci.
Results
AFLP analysis reveals the Xci global diversity and geographical structure of A*/AW strain clusters
A collection of 55 Xci strains was built to represent Xci genetic diversity based on a previous analysis and including recently isolated strains (Bangladesh) and strains corresponding to recent outbreaks (Mali and Senegal). A total of 169 AFLP fragments was recorded, 105 of which were polymorphic. A total of 49 haplotypes was identified within the strain collection. Graphical representations, a neighbour‐joining (NJ) tree (Fig. 1) and metric multidimensional scaling (MDS) plots (Fig. S1, see Supporting Information) suggested a larger polymorphism for A*/AW strains. This higher diversity was supported by Nei's diversity indices: He = 0.10 for A*/AW strains vs. He = 0.06 for A strains. A majority of A*/AW clusters were supported by high bootstrap values (≥80%). Some of these clusters contained strains originating from a single country (e.g. Ethiopia), whereas others contained strains from several countries (e.g. Oman and India). Most A strains clustered irrespective of their geographical origin, except in rare cases (e.g. Bangladesh). A cluster of the three A* strains JF90‐8*, LG100* and LG116* from Oman and India (bootstrap of 71%) was found close to a cluster of A strains containing LE116‐1, LE117‐1, LH1‐3, LH37‐1 and NCPPB 3562 from Mali, Senegal and India (supported by a bootstrap of 98%). Strains LG115* and NCPPB 3608* from India and X2002‐1035W and X2003‐3218W from Florida formed a subclade supported by a maximal bootstrap value (97%), and were closer to A strains than to the other A* strains, based on the MDS plot and NJ tree (Figs S1 and 1).
Figure 1.
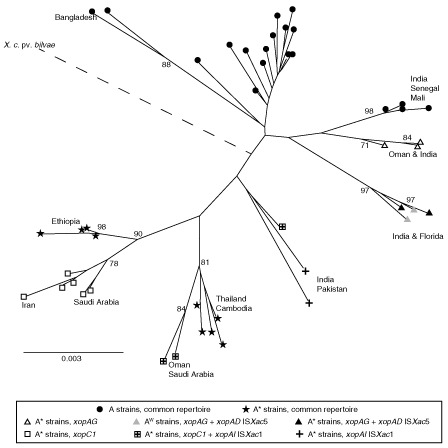
Amplified fragment length polymorphism (AFLP) neighbour‐joining tree. Tree constructed using evolutionary genome divergences among 55 Xanthomonas citri pv. citri strains based on 169 AFLP markers. Bootstrap values under 70% are not indicated. The branch containing X. citri pv. bilvae should be twofold longer and is represented by a broken line. Symbols indicate the type III effector (T3E) contents of each strain, either as the common repertoire, the common repertoire with additional genes or the common repertoire with genes containing an insertion. Geographical origins are indicated when they are consistent with branching.
All Xci share a common repertoire of 26 T3Es and only the host‐restricted A* and AW strains contain a variable repertoire
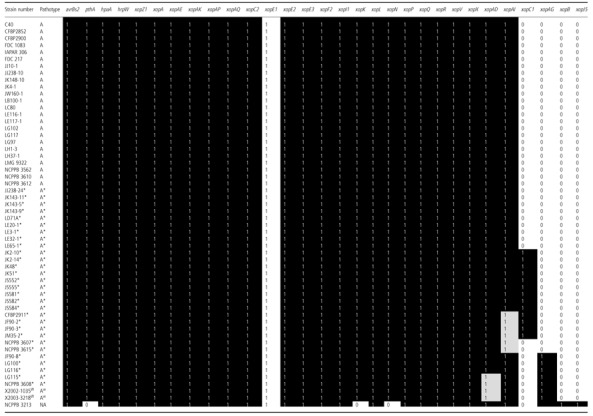
Repertoires of T3Es were defined in terms of the absence or presence of portions of the corresponding sequence after PCR amplification with two pairs of specific primers (Table S4, see Supporting Information). We were able to define a common repertoire of 26 candidate T3Es, present in all tested Xci strains, and a variable set of two candidate T3Es: xopC1 and xopAG (Table 1). xopC1 was present in only 13 A* strains. xopAG was present in the two AW tested strains and in five A* strains (Fig. 1 and Table 1). Among the 26 common T3Es, the two T3Es xopAD and xopAI were detected by PCR with larger sizes than expected (2 kb and 1.2 kb, respectively) in some A* and AW strains, suggesting the presence of ISs (see below). The pathogen X. citri pv. bilvae strain NCPPB 3213, responsible for spot lesions on Rutaceae, shared 23 effectors with Xci and contained two additional effectors: xopJ5 and xopB. Southern blots were performed to validate the presence of T3Es that were detected by PCR in only a few Xci strains or that were not detected by PCR and previously reported in canker‐forming strains. We looked for the presence of two T3Es detected in a few Xci strains by PCR: xopC1 and xopAG (false‐negative results putatively induced by primer mismatches). We also looked for the presence of four T3Es [xopB, xopE4, xopAF and xopJ5 (=avrXccB)] reported in South American citrus canker‐forming strains Xanthomonas citri pv. aurantifolii type B ICBP 11122 (XauB) and type C ICPB 10535 (XauC) (Moreira et al., 2010), and xopF1 previously reported in other repertoire studies, including strains of Xci (Hajri et al., 2009). The two effectors xopC1 and xopAG were only detected from strains with positive results in PCR, and no hybridization was detected in Xci strains for the five markers xopB, xopE4, xopAF, xopJ5 and xopF1. Overall, strains sharing the same variable repertoire cluster in the same AFLP clades were supported by high bootstrap values (>70%) (Fig. 1).
Table 1.
Distribution of 66 type III effector (T3E) genes among 55 strains of Xanthomonas citri pv. citri and Xanthomonas citri pv. bilvae. Presence or absence of an orthologue of each selected T3E was determined by polymerase chain reaction (PCR) and Southern blot when necessary. Black, grey and white squares represent the presence, interruption by an insertion sequence and absence, respectively
Hierarchical clustering based on T3E content
Hierarchical clustering by Ward's method was performed using a matrix of T3E presence/absence built after homology searches of the T3E candidate list on 27 Xci, X. citri pv. bilvae strain NCPPB 3213 draft genome sequences and genomes of XauB and XauC. This clustering showed that the T3E repertoire does not distinguish pathotypes, but rather reflects strain phylogenetic divergence, the X. citri pv. bilvae strain NCPPB 3213 being closer to the Xci strains than the two tested X. citri pv. aurantifolii strains (Fig. 2).
Figure 2.
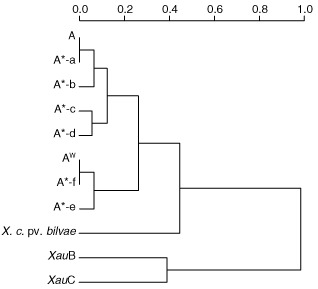
Relationships between strains according to their variable repertoire. Dendrogram built by Ward's method (Maechler et al., 2012) based on the variable repertoires of Xanthomonas citri pv. citri (Xci), X. citri pv. bilvae strain NCPPB 3213, and X. citri pv. aurantifolii strains ICPB 11122 (XauB) and ICPB 10535 (XauC). All strains contain the common repertoire with the following variations for A* strains: (a) common repertoire; (b) xopAIΔISXac1; (c) xopC1; (d) xopAIΔISXac1 and xopC1; (e) xopAG; (f) xopAG and xopADΔISXac5. AW strains contain the common repertoire with xopADΔISXac5 and xopAG.
Sequencing analysis of two variable T3Es of Xci reveals potential horizontal gene transfer (HGT) events with other Xanthomonas strains
Sequencing analyses of the two variable T3Es and their flanking regions were performed to better understand their distribution and to assess whether differences in sequence could explain some phenotypic variability among strains sharing the same variable repertoire.
Sequence analysis of xopC1
The xopC1 gene was sequenced from four A* strains belonging to three different AFLP clusters: CFBP 2911* from Pakistan, JF90‐2* from Oman and both JS584* from Iran and JK2‐10* from Saudi Arabia belonging to the same cluster as they displayed distinct phenotypes on sweet orange and grapefruit (see below). Our analysis showed that xopC1 DNA coding sequences were completely identical between Xci A* strains and had 98.8% homology with xopC1 from X. euvesicatoria strains 85‐10 and 75‐3. Analysis of the xopC1 upstream flanking region (1460 bp) and downstream region (750 bp) for all four strains revealed that they were highly homologous (98.8%–99.0%) to the corresponding regions bordering xopC1 in X. euvesicatoria 85‐10. xopC1 had a G + C content of 48%, contrasting with the genomic average of 65.2%, and was flanked by the insertion sequences IS1595 and IS1478. The presence of IS1595 had not been documented in Xci previously.
Sequence analysis of xopAG
The xopAG gene was sequenced from four Xci strains found in two AFLP clades and of various geographical origins: X2003‐3218W from Florida and NCPPB 3608* from India, which were found in the same subclade, and strains JF90‐8* and LG116* from Oman and India, respectively. The gene xopAG was also sequenced from X. citri pv. bilvae strain NCPPB 3213. xopAG from AW strains differed by one nonsynonymous single nucleotide polymorphism (SNP) from xopAG found in A* strains NCPPB 3608*, JF90‐8* and LG116*, and had a G + C content of 56.9%, which was again lower than the average value of the rest of the genome (65.2%). Orthologues of xopAG were found in X. campestris pv. campestris B100 (80% homology), XauC (55% homology) and interrupted by IS1479 in XauB (55% homology) and X. citri pv. bilvae NCPPB 3213 (99% homology), which induced a strong HR on grapefruit and sweet orange. Based on unpublished draft genome sequence data, blast analyses revealed the presence of the right‐end inverted repeat of ISXac4, 1240 bp downstream of xopAG (data not shown). Both G + C content and the presence of IS elements suggested that both xopC1 and xopAG might have been acquired through HGT.
Sequencing analysis of the larger amplicons of xopAD and xopAI reveals the presence of IS elements
The larger amplicon obtained for xopAI was sequenced in strain JF90‐2*, and sequence analysis revealed that it was interrupted at position 714 after the start codon (total length, 906 bp) by ISXac1, a member of the IS4 family. Similarly, the larger amplicons obtained by PCR for xopAD were sequenced from strains NCPPB 3608* and X2003‐3218w, representative of the two pathotypes, and sequence analysis revealed that xopAD was interrupted at position 7989 (total length, 8652 bp) by ISXac5 related to ISRso19, which belongs to the IS21 family. ISXac5 and ISRso19 had a similar size and shared 72.5% DNA homology, similar inverted repeats and displayed two putative peptides which were 67% and 74% identical, respectively. Both ISs were bordered by direct repeats of 6 bp. Comparison of the xopAD sequence (without considering the IS element) showed that xopAD from strains NCPPB 3608* and X2003‐3218W shared 99.2% DNA homology with the corresponding xopAD allele from X. citri pv. bilvae strain NCPPB 3213. Draft genome sequence data indicated that, in strains possessing xopAG (including JF90‐8*, NCPPB 3608* and X2003‐3218W), the sequence of xopAD showed less homology with xopAD orthologues in the other A* strains (95.9%) and in A strains (97.6%) (data not shown) than with X. citri pv. bilvae strain NCPPB 3213.
Phenotypic characterization on detached leaf assay reveals that all A strains produce an identical phenotypic response, whereas A*/AW strains show heterogeneous responses depending on the host and the strains
Phenotypic characterization was conducted to assess the role of T3Es in the observed responses during the interaction with various hosts. A disease scale was used to assess strain pathogenicity each day post‐inoculation (dpi) (Fig. S2, see Supporting Information), and a disease development index (DDi) was calculated to assess the global response of each tested strain, as values of the area under the disease progression curve. As expected, X. citri pv. bilvae strain NCPPB 3213 did not cause canker‐like symptoms, but produced extensive water‐soaked lesions on Mexican and Tahiti limes and alemow, and HR‐like responses on Pineapple sweet orange and Marsh grapefruit.
All tested Xci strains showed similar typical erumpent, callus‐like lesions at 14 dpi on the nondiscriminant host species (Mexican lime, Tahiti lime and alemow) with minor differences in disease development and time of appearance of the epidermis rupture (data not shown). As expected, inoculation of two pathotype‐discriminating host species, Pineapple sweet orange and Marsh grapefruit, showed heterogeneous responses depending on the strains. On both host species, A strains produced typical canker‐like symptoms at 14 dpi, whereas A* strains elicited a range of reactions without any correlation with the T3E content, from no symptoms to erumpent symptoms for the three A* strains from Iran (JS581*, JS582* and JS584*). The majority of A* strains induced defence‐like responses, which consisted of necrotic tissues on grapefruit and/or sweet orange with variability in days of appearance from 3 to 21 dpi. Representation of the disease progression curves combined with the appearance of HR for a subset of strains representative of the various pathogenic profiles observed in this assay showed no HR‐like responses for A strains on these host species (Fig. S3, see Supporting Information). Strains JS581*, JS582* and JS584*, which are further described below, showed the same disease progression curve, and only strain JS584* is represented. Based on DDi on sweet orange and grapefruit, three clusters of strains were identified. Cluster I contained all 17 A strains which showed a homogeneous response to inoculations, cluster II contained the three Iranian A* strains with intermediate phenotypes (JS581* and JS582* showed exactly the same phenotypic response and thus are not distinguishable on Fig. 3), and cluster III contained all other A* and AW strains with no typical canker lesions on discriminant host species. Overall, sweet orange provided a better discriminative ability than grapefruit, with Iranian A* strains being distinguished from A strains by a delayed development of canker‐like lesions. Strains of cluster II were further inoculated, together with three A strains, on Temple mandarin, Fairchild mandarin SRA30, Meyer lemon and Ortanique tangor (mandarin C. reticulata × sweet orange). Inoculation on Meyer lemon and the two mandarin cultivars did not allow the cluster II strains to be distinguished more clearly than on sweet orange (data not shown). Interestingly, all cluster II strains induced defence‐like responses on Ortanique tangor without canker‐like lesions. A strains produced canker‐like lesions, but the observed reactions suggested a moderate aggressiveness on this host species (Figs 4 and S2). Overall, the Xci phenotypic response was variable for A* strains, but did not correlate with their T3E content.
Figure 3.
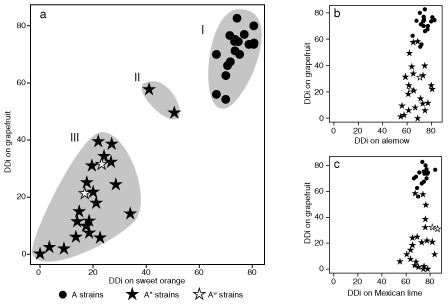
Disease development indices (DDi) on four host species. Forty‐seven strains were wound inoculated on detached leaves of grapefruit and sweet orange (a), grapefruit and alemow (b), and grapefruit and Mexican lime (c). Clusters I, II and III represent subsamples according to Tukey's tests.
Figure 4.
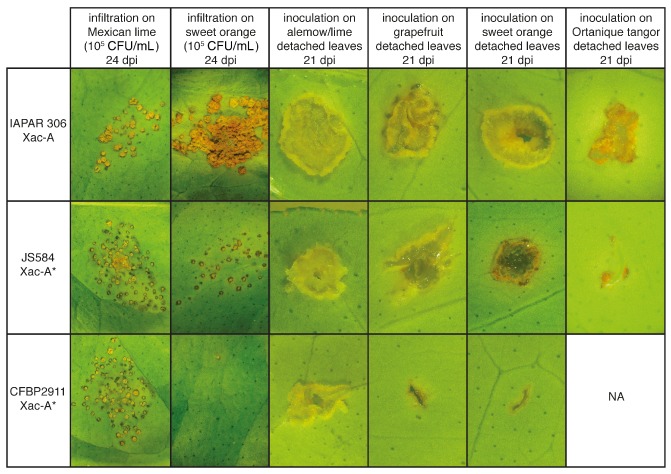
Symptomatology of A and A* strains. Symptoms observed after infiltration of attached leaves (first two columns) and after wound inoculation of detached leaves (last four columns) with A strain IAPAR 306, and A* strains JS584* and CFBP 2911*. The width of each photograph is approximately 15 mm for the first two columns and 1.5 mm for the last four columns. CFU, colony‐forming units.
Comparison of bacterial population in planta confirms A* heterogeneous response on sweet orange
In order to better characterize the atypical interaction on detached leaf assay of A* strains from Iran, which correspond to cluster II (Fig. 3), we compared in planta bacterial multiplication of strain JS584* (selected as a reference for cluster II) with A strain IAPAR 306 and A* strain CFBP 2911* (selected as references for clusters I and III, respectively). As shown in Fig. 5, on Mexican lime at 24 dpi, all three strains reached population densities of 2.5 × 107 to 1.3 × 108 colony‐forming units (CFU)/cm2, which is typical of canker symptoms. On sweet orange at 8 dpi, strains JS584* and IAPAR 306 showed nonsignificantly different mean population sizes, whereas strain CFBP 2911* showed a much smaller population size (Fig. 5). At 24 dpi, only strain IAPAR 306 reached large population sizes (6.3 × 108 CFU/cm2), whereas strains CFBP 2911* and JS584* showed significantly smaller population sizes (according to Tukey's tests), thus confirming previous results (Fig. 5) (Bui‐Thi‐Ngoc et al., 2010). Visual comparison at 24 dpi of the symptoms caused by all three strains showed overall differences in symptomatology that illustrated the differences observed in terms of bacterial population sizes (Fig. 4), and confirmed that strain JS584* is much less aggressive than A strains on sweet orange. Strain JS584* induced small canker‐like symptoms on sweet orange, which is unexpected for A* strains, compared with the symptomless strain CFBP 2911*. However, JS584* symptoms were attenuated compared with the typical canker symptoms induced by the A strain IAPAR 306, and this is consistent with population sizes recovered in planta.
Figure 5.
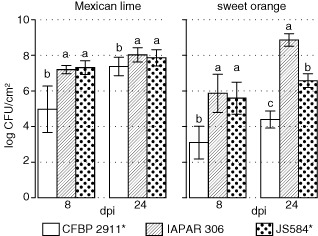
Bacterial multiplication in Mexican lime and sweet orange. Means of log colony‐forming units (CFU) from 20 leaf discs (two experiments) after inoculation at 105 CFU/mL. Error bars indicate ± standard deviation (SD). Distinct letters indicate significant differences between strains at a given time on a given host according to Tukey's test.
All A* and AW strains containing xopAG induce HR‐like responses on sweet orange and grapefruit
Considering that AW strains were first described as causing HR‐like reaction on various hosts, and that such responses depend on the T3E xopAG (Rybak et al., 2009), all xopAG‐containing strains were submitted to infiltration to look for HR or symptom development, and compared with a subset of 17 strains representative of Xci genetic diversity and repertoire variability. After inoculation with bacterial suspensions (1 × 108 CFU/mL), typical water‐soaked lesions that evolved as canker symptoms at 4–6 dpi were observed on Mexican lime, irrespective of the pathotype. On sweet orange, mandarin and grapefruit, HR‐like symptoms appeared at 3–6 dpi for strains X2003‐3218W and X2002‐1035W, and for strains NCBBP 3608*, LG100*, LG115* and LG116* (from India) and JF90‐8* (from Oman), which all carry xopAG. Interestingly, none of the strains that did not contain xopAG produced HR‐like symptoms on sweet orange, mandarin and grapefruit (Fig. S4, see Supporting Information, and data not shown). On these hosts, A strains induced typical canker symptoms, whereas A* strains exhibited variable responses: chlorosis, blister‐like lesions with water soaking in the infiltrated area or canker‐like symptoms, depending on the strain and host species. Xanthomonas citri pv. bilvae strain NCPPB 3213 induced water‐soaking lesions on Mexican lime and HR‐like symptoms at 3 dpi on mandarin, sweet orange and grapefruit. This strain contains xopAG, but also possesses other T3Es absent from Xci strains, and we cannot conclude that xopAG is responsible for the HR‐like symptoms observed for this pathovar.
Deletion of xopC1 or xopAD does not alter Xci pathogenicity
Xci is characterized by a variable repertoire of two T3Es: xopAG, whose role in HR‐like symptoms has already been shown (Rybak et al., 2009), and xopC1. We also showed that both xopAD and xopAI are interrupted by IS elements in several strains. The role of xopAI has already been assessed by Figueiredo et al. (2011). They showed that the pathogenicity of A strain IAPAR 306 was not altered when xopAI was deleted. Therefore, we focused our study on xopC1 and xopAD. xopC1 was deleted in strains JS584*R and CFBP 2911*R, as these two strains showed different phenotypic responses on sweet orange and pomelo. xopAD is interrupted in several A* and AW strains by ISXac5, and so may not be functional. To assess the role of the xopAD full‐length protein in A strains, strain IAPAR 306 was deleted for xopAD. Mutants JS584*RΔxopC1, CFBP 2911*RΔxopC1 and IAPAR 306RΔxopAD showed similar symptoms to the rifampicin‐derivative strains when inoculated on detached leaves, as well as when infiltrated at a concentration of 1 × 105 CFU/mL (Fig. S5, see Supporting Information). In planta population sizes between a given mutant and its wild‐type‐resistant derivative were not significantly different at 8 and 24 dpi, according to Tukey's tests. Population size differences did not exceed 0.48 log CFU/cm2 at 8 dpi and 0.30 log CFU/cm2 at 24 dpi (Table S2, see Supporting Information).
Discussion
Xci pathotype A* shows a wider genetic diversity than pathotype A and a broad extent of reaction on several Citrus species
Most studies aimed at understanding Xci virulence mechanisms and host range specialization have focused on one strain (Dunger et al., 2012; Figueiredo et al., 2011; Rybak et al., 2009; Yan and Wang, 2012) or a set of strains (Al‐Saadi et al., 2007). The choice of a particular strain often relies on the availability of genomic data, or of well‐characterized phenotypes obtained by random mutagenesis. Therefore, only one of few pathological profiles can be tested simultaneously. In both cases, the representativeness of a particular strain or set of strains remains arbitrary and the genetic diversity of Xci was not taken into account when studying determinants of its host range. In this work, we wanted to characterize the role of T3Es in Xci host range specialization, and we rationalized our choice of strains to represent the worldwide genetic and phenotypic diversity of the bacterium, based on previous studies (Bui‐Thi‐Ngoc et al., 2009) and including strains from recent outbreaks (Balestra et al., 2008; Derso et al., 2009; Leduc et al., 2011; Traore et al., 2008). We first analysed the genetic relatedness between Xci pathotypes using AFLP markers, which have the advantage of covering the genome and being theoretically neutral. This analysis indicated that strains from Xci pathotypes are differentiated into several clusters which do not correlate with Xci pathotypes: all A* and AW strains which contain xopAG cluster closer to A strains than to A* strains (Figs 1 and S1). Pathotype A* strains are genetically more diverse than pathotype A strains. In addition, pathogenicity tests revealed that this difference in terms of homogeneity/heterogeneity is correlated with their aggressiveness and virulence. Our results clearly revealed that A strains are homogeneous in terms of host, types of symptom and aggressiveness, whereas the pathogenicity of A* strains is highly diverse on sweet orange, grapefruit and mandarin (Figs 3, 4 and S3). The two tested AW strains exhibited one of the several reactions observed for A* strains and, consistent with previous data (Bui‐Thi‐Ngoc et al., 2010), AW and most A* strains were not distinguished on the basis of their host range. Within pathotype A*, no correlation was found between pathogenicity profiles and genetic structure. We observed that A* Iranian strains of our collection induced canker‐like symptoms when inoculated on detached or attached leaves of grapefruit and sweet orange, although the size of the canker symptoms was often more moderate than those induced by A strains. Our results highlighted the difficulty of assigning strains to a given pathotype on the sole basis of detached leaf assay on grapefruit and sweet orange, and suggested that Ortanique tangor may be a useful species for pathotype identification based on pathogenicity test data. The genetic relatedness between A* strains did not correlate with differences in symptomatology, as the latter A* strains from Iran (corresponding to cluster II in Fig. 3), which had a high virulence on sweet orange and grapefruit, belonged to the same clade as strains from Saudi Arabia, which induced no symptoms on detached leaf assay or when infiltrated (Figs 1 and 4). The classification of Iranian strains within pathotype A* was confirmed by the cumulative information obtained by in planta population measurements on sweet orange (two log units lower than those obtained for strain IAPAR 306 at 24 dpi), response to inoculations on several host species and genotyping data (this study and data not shown).
The Xci variable repertoire does not explain host specialization between pathotypes
Our T3E gene distribution analysis showed that Xci possesses a common repertoire of 26 known T3Es which corresponds to the average size of Xanthomonas repertoires described so far (Hajri et al., 2012a, b; White et al., 2009). Xci repertoire variations do not explain host specialization between Xci pathotypes. Several A* strains shared the same repertoire as A strains, but belonged to distant genetic clusters and induced different symptoms (Fig. 1). Only A* strains exhibited variability in known T3Es (xopC1 and xopAG), which showed a restricted distribution among Xanthomonas sequenced strains (Xanthomonas resource website: http://www.xanthomonas.org/; R. Koebnik, personal communication). However, the distribution of T3Es among A* strains was not correlated with symptomatology variation within this pathotype. Deletion of xopC1 did not seem to alter pathogenicity, regardless of the inoculation technique used (Fig. S5). The secretion of xopC1 in planta was shown for X. euvesicatoria, but no function modification could be evidenced after deletion (Noel et al., 2003). The close relatedness between the xopC1 orthologues and the presence of identical IS elements in the flanking regions suggests that this T3E belongs to a ‘mobile module’ that has been inserted in genomes by HGT mediated by mobile elements. The region in which it is inserted in X. euvesicatoria contains multiple coding sequences associated with mobile elements (Noel et al., 2003) and is homologous with two nonadjacent regions of the genome of Xci strain IAPAR 306 (one on the chromosome, one on plasmid pXac64), in which the mobile module containing xopC1 is absent (Fig. S6, see Supporting Information). We can therefore speculate that this region is favourable for the insertion of mobile elements, which could explain why xopAI (if it is located in the same area) is interrupted by an IS in several A* strains. However, deletion of xopC1 (this study, Fig. S5) and xopAI (Figueiredo et al., 2011) did not show any selective advantage related to the plasticity of this region for colonization on the tested citrus species. Similarly, the effector xopAD is interrupted by the ISXac5 related to ISRso19 in several AW strains, but the deletion of this effector (this study, Fig. S5) did not modify its host range.
The Xci A*/AW T3E repertoire is closer to that of Xci A than to that of South American canker‐forming strains despite homologies in host range
The variability of the response of A* strains on two pathotype‐discriminating host species, Pineapple sweet orange and Marsh grapefruit; can be compared with the symptomatology and host ranges characterizing South American canker‐causing strains XauB and XauC. A* strains from Iran (corresponding to cluster II in Fig. 3) share many similarities in symptomatology with strains causing canker B, which was described as primarily restricted to lemon (C. limon), but also mildly pathogenic to sweet orange and grapefruit, and was characterized by an in planta multiplication ability lower than that of A strains (Moreira et al., 2010). Similarly, A*/AW strains that contain xopAG exhibit the same pattern of response as strains causing canker C, which is restricted to Mexican lime and induces HR‐like reactions on most other species (Moreira et al., 2010). All strains of our collection shared a common T3E repertoire identical to the repertoire of the sequenced Xci A strain IAPAR 306. Although some T3Es could be missed in our PCR screen, because of sequence divergence between Xci and strains from which primers were designed, homology searches of the effector candidate list on 28 Xci draft genome sequences confirmed that T3E repertoires determined in this study correspond to the T3E deduced from the genomic sequences (data not shown). Despite the fact that host range‐restricted Xci A* strains and X. citri pv. aurantifolii display similar host ranges, searches against the whole genome shotgun database revealed several differences between citrus canker‐forming strains in their non‐TAL effector repertoires. In addition, the 17.5‐repeat TAL‐effector PthA4 from Xci IAPAR 306 and its homologues PthB in XauB and PthC in XauC have been reported to be functionally interchangeable to induce canker symptoms when transformed in strains A, A*, AW, B and C, without incidence on the host range (Al‐Saadi et al., 2007). Therefore, our results suggest that the T3E repertoire may reflect phylogenetic divergence rather than pathogenicity features, such as host range (Fig. 2).
A*/AW strains that contain xopAG are closely related to A strains, and therefore xopAG may have contributed to their host range restriction
The difficulty in demonstrating virulence roles for individual effectors has been documented previously and is generally imputed to functional redundancy (Kvitko et al., 2009). The role of single T3Es has often been characterized in nonhost interactions, where they are recognized by a corresponding resistance gene, resulting in local cell death (HR). Here, we investigated the role of the Xci variable T3E repertoire, but were unable to identify a single or combination of effectors to explain Xci host range specialization. In agreement with previous work showing that XopAG (AvrGf1) is responsible for HR‐like symptoms caused by Xci AW strains (Rybak et al., 2009), we found that all A* and AW strains carrying xopAG induced necrotic symptoms (HR‐like) on grapefruit and sweet orange within a few days. Mutants of Aw strains lacking xopAG produced small pustules, typical of reactions induced by some A* strains, but to a less severe extent than the disease reactions induced by A strains in grapefruit (Rybak et al., 2009), thus indicating a possible role of another determinant of pathogenicity in host range specialization of AW strains. All strains inducing similar phenotypic responses contained xopAG orthologues that were 99.9% homologous at the protein level between strains. Both groups of strains carrying xopAG were more closely related to A strains than to other A* strains (Fig. 1). Strains carrying xopAG might have a common ancestor in the Indian region and might have participated in the evolution of Xci local host range specializations driving to the current state: two clades of A*/AW strains with a narrow host range share homologies in symptomatology because of the effector xopAG, and both are closely related to A strains.
The variable T3E repertoire of Xci is related to phylogenetic placement and the geographical origins of Xci pathotypes
Interestingly, xopC1 was found only in strains originating from western Asia, but genetically distant. This finding is consistent with the hypothesis of HGT between Xci strains. xopAG was first described in Aw strains from Florida, but an Indian origin of the Florida Aw strains is very likely (Schubert et al., 2001). The Xci variable T3E repertoire, consisting of xopC1 and xopAG, but also DNA rearrangements within xopAI and xopAD, can therefore be linked to the geographical origin of the strains and their intrapathovar phylogenetic position, rather than to differences in host range. Such results suggest that these Xci AFLP clades might have distinct evolutionary histories.
Comparison of xopAG sequences among xanthomonads pathogenic to citrus showed that xopAG Xci was more closely related to xopAG from X. citri pv. bilvae, which also induces HR on grapefruit and sweet orange, than to xopAG from XauB and XauC (99% vs. 55% DNA homology). xopAD was present in all tested Xci strains but, in strains that also possessed xopAG (which were the only ones inducing HR on grapefruit and sweet orange), xopAD was more homologous to the corresponding orthologue found in X. citri pv. bilvae than to the orthologue present in Xci strains that did not contain xopAG. Xanthomonas citri pv. bilvae has been reported solely from India, where it shares Mexican lime (Chakravarti and Chaudhary, 1988) and, to a lesser extent, Aegle marmelos and Feronia elephantum (Khan and Hingorani, 1970) as host species with Xci. On the basis of these data, we hypothesize that several T3Es may have been horizontally transferred between X. citri pv. bilvae and some Xci strains in the Indian subcontinent and that xopAG‐containing strains may have subsequently spread to Oman. This result is consistent with the very close genetic relatedness between the xopAG‐containing strains JF90‐8* from Oman and LG116* from India.
The monophyletic pathovar Xci is an interesting model for the study of the contribution of the T3E repertoire to the intrapathovar structure and host range variations reported among pathotypes. We showed that, at the pathotype level, the T3E repertoire did not explain Xci host range specialization. Previous studies have shown that, in other Xanthomonas species, including X. axonopodis, X. arboricola and X. oryzae, the composition of the T3E repertoires and the host specificity of pathovars are correlated (Hajri et al., 2009). A possible limitation is that we worked on Xops previously identified in the genus Xanthomonas. Although these effectors were identified on the basis of avirulence activity, co‐regulation with the T3SS or in silico prediction based on complete genome sequences (White et al., 2009), there are probably other unsuspected T3Es to be discovered. Determinants of pathogenicity, other than those associated with the T3SS, could be involved in the specialization of the host range in Xci. However, the fact that Xci pathotypes A and A* induce canker symptoms on both limes and alemow suggests that common mechanisms allow them to escape both innate and specific host immunity, and to colonize citrus species to produce canker symptoms. The presence and/or functionality of a few or even single T3Es can be a founder event to colonize a new plant host (Schulze‐Lefert and Panstruga, 2011). Therefore, the hypothesis of the role of T3Es in the definition of the host range of Xci pathotypes remains strong. Screening was made for the presence of T3Es, without considering whether or not a gene was functional. Alternatively, sequence allelic variation of a given T3E may also be involved in host specificity. Minute variations in these genes can have important implications for host range and pathogenicity, as illustrated in X. campestris pv. vesicatoria, in which point mutations in avrBs2 resulted in the breakdown of Bs2‐based field resistance in pepper (Gassmann et al., 2000), and for P. syringae, in which allelic variants of the T3E HopZ1 were shown to be differentially recognized by Nicotiana benthamiana and soybean (Zhou et al., 2009), and where a key amino‐acid polymorphism in AvrPto led the pathogen to overcome tomato resistance (Kunkeaw et al., 2010). Host range changes have also been observed via inactivation of the T3E avrBs2 by a 5‐bp insertion in X. euvesicatoria (Swords et al., 1996). We are now working on Xci draft genome sequences to assess the role of xop in the definition of the pathotypes by exploring their allelic variation. Access to Xci draft genome sequences might lead to the identification of new T3Es potentially involved in Xci pathogenicity. Together with the description of the phenotypic responses of Xci pathotypes presented here, the study of T3E sequences, combined with a better knowledge of housekeeping genes, will help us to better understand the mechanism underlying the interaction of the bacterium with its host, what determines the host range specialization and to provide clues about the evolution of the pathogenicity of Xci pathotypes. Our results also illustrated the dynamic nature of the host range of a pathogen (Schulze‐Lefert and Panstruga, 2011). Working on a large collection of strains revealed the complexity of Xci pathotype A*, for which no correlation was found between the reported genetic diversity and the observed phenotypic responses. Xci host specialization may involve several evolutionary mechanisms affecting the capacity of a strain to have a narrow or a wide host range. It is therefore tempting to speculate that the diversity of Xci A* phenotypes described here corresponds to several steps in this dynamic process. Under this hypothesis, a better characterization of the clades which encompass A* Iranian strains with unexpected aggressiveness on sweet orange and grapefruit and A* strains causing no symptoms on both hosts should be of great interest, and might help us to understand the contribution of mutations or loss/gain of virulence genes in Xci host range specialization. Similarly, the close relatedness of some A*/AW strains to A strains highlights that Xci variability should not be ignored when describing the mechanisms involved in pathogen evolution and host specialization.
Experimental Procedures
Bacterial strain culture and DNA extraction conditions
The bacterial strains used in this study are listed in Tables S1 and S3 (see Supporting Information). All 55 Xci strains were isolated from citrus canker lesions on various Citrus species and collected from 21 different countries. These strains are a subset of the collection used previously (Bui‐Thi‐Ngoc et al., 2009) and were selected in order to maximize the diversity in terms of host, year and country of isolation. Xanthomonas citri pv. bilvae strain NCPPB 3213, which is responsible for spot lesions on Rutaceae, was included in the study for comparative purposes. Positive and negative controls for T3E screening consisted of Xanthomonas published genomes or draft genome sequences. Xanthomonas strains were stored and cultivated on YPGA (yeast extract 7 g/L, glucose 7 g/L, peptone 7 g/L, agar 18 g/L, pH 7.2), as described previously (Bui‐Thi‐Ngoc et al., 2010). Suspensions were used for DNA extraction with the Wizard® Genomic DNA Purification Kit (Promega, Charbonnières, France). Rifampicin derivatives from strains IAPAR 306, JS584* and CFBP 2911* were selected on plating of YPGA supplemented with 1000 μg/mL rifampicin. For each strain, two isolates showing resistance to rifampicin were inoculated on detached leaves of Pineapple sweet orange, Mexican lime, Tahiti lime and Marsh grapefruit, and confirmed to be as pathogenic as the corresponding wild‐type strains. One of each was selected for further experiments and referred to as IAPAR 306R, JS584*R and CFBP 2911*R, respectively. Prior to triparental conjugation, Xanthomonas strains were grown at 28 °C on MOKA‐rich medium (yeast extract 4 g/L, casamino acids 8 g/L, K2HPO4 2 g/L, MgSO4·7H2O 0.3 g/L, agar 18 g/L, pH 7.2; Blanvillain et al., 2007). Escherichia coli cells were cultivated on Luria–Bertani medium at 37 °C. Antibiotics were used at the following concentrations: for Xci: rifampicin, 100 μg/mL; kanamycin, 50 μg/mL; tetracycline, 10 μg/mL; chloramphenicol, 10 μg/mL; for E. coli: ampicillin, 50 μg/mL; chloramphenicol, 100 μg/mL; kanamycin, 25 μg/mL; tetracycline, 10 μg/mL; spectinomycin, 40 μg/mL.
Pathogenicity tests
Detached leaf assays
Inoculations of the 47 strains (Table S1), submitted to pathogenicity tests on detached leaves, were performed as described previously (Vernière et al., 1998) on five citrus species: Pineapple sweet orange (C. sinensis), grapefruit (C. paradisi), Mexican lime (C. aurantifolia), alemow (C. macrophylla) and Tahiti lime (C. latifolia). Leaves were inoculated with bacterial suspension containing 1 × 106 CFU/mL, and were observed for the development of symptoms at 5, 7, 10, 14 and 21 dpi. Each strain was inoculated on three leaves. The experiment was repeated at least twice, which made a total of at least 48 replicates on six different leaves for each strain–host combination. A subset of six strains was subsequently inoculated onto Mandarin temple (C. temple), Mandarin Fairchild SRA30 (C. reticulata), Meyer lemon (C. meyeri) and Ortanique tangor (C. reticulata × C. sinensis) to further describe the atypical pathogenicity of Iranian strains and to compare them with the typical A strain responses and with another A* strain (Table S1).
Disease intensity was evaluated visually, and scored using a scale of 0–5 as follows: 0, no symptoms; 0.5, water‐soaked margin surrounding the wound sites; 1, more extensive water soaking (diameter ≥ 1 mm); 2, small pustules with no visible rupture of the epidermis or isolated pustules with rupture of the epidermis; 3, numerous pustules with visible rupture of the epidermis; 4, small canker (observation of callus‐like material); 5, wide canker formation (at least twice the diameter of the pinprick). Detailed illustrations of these symptoms are presented in Fig. S2. Although no typical HR could be visualized using the detached leaf assay, defence‐like reactions that appeared over time were also recorded (Fig. S1). To assess the global response of each tested strain, a DDi was calculated as the area under the disease progression curve (see the ‘Data analysis’ section below for further details). DDis on grapefruit, combined with the DDis of each of three other species (sweet orange, alemow or Mexican lime), were then plotted to give an overview of Xci pathogenicity using this mode of inoculation.
Attached leaf assays
All plants for leaf infiltration assays were pruned to produce uniformly aged shoots. Twenty‐four strains were submitted to infiltration to look for HR or symptom development (Table S1). Leaves of Mexican lime, Pineapple sweet orange, Marsh grapefruit and Page mandarin were inoculated by infiltration of bacterial suspensions containing 1 × 108 and 1 × 105 CFU/mL, as described previously (Ah‐You et al., 2007). Symptom development was evaluated at 4, 6, 8, 10 and 14 dpi, and HR was scored at 4 and 6 dpi.
In planta bacterial population sizes were determined from infiltrated leaves (1 × 105 CFU/mL) of Mexican lime and Washington Navel sweet orange, as described previously (Bui‐Thi‐Ngoc et al., 2010) in two experiments: (1) the first aimed to characterize the atypical interaction of cluster II A* strains from Iran and strain JS584* (selected as a reference for cluster II) in comparison with A strain IAPAR 306 and A* strain CFBP 2911*; (2) mutant strains and their corresponding rifampicin‐resistant derivatives were compared. Each experiment was performed twice. Inoculated leaf fragments were sampled at 8 and 24 dpi. Xanthomonas enumerations were performed on YPGA‐KC semi‐selective medium (YPGA complemented with 20 mg/L of kasugamycin and 40 mg/L of cephalexin; Pruvost et al., 2005) according to the method described earlier (Ah‐You et al., 2007; experiment 1), or on YPGA‐KC and YPGA supplemented with 100 mg/L rifampicin (YPGAR; experiment 2).
T3E screening
We screened for the presence of 66 Xanthomonas T3Es (Xop) described on the Xanthomonas resource website (http://www.xanthomonas.org/; R. Koebnik, personal communication). The screen was performed using two pairs of primers for each Xop (Table S4). Screening of the 26 Xops found in Xci strain IAPAR 306 was performed using pairs of primers targeting different regions of the T3E genes. One of the two pairs of primers was designed to amplify at least 80% of the open reading frame (ORF), except for xopAD, which is too long (8652 bp) to be amplified in a single reaction, for which we amplified two fragments of 1660 and 608 bp. Screening of xopAG (avrGf1) was performed using primers designed on the sequence of avrGf1 (accession number GI: 82571048, LOCUS DQ275469). In order to screen the 40 remaining effectors absent in Xci IAPAR 306, primers were designed in conserved regions based on the alignments of Xop orthologues from Xanthomonas available sequences to avoid false‐negative results caused by point mutations or length differences between orthologues. Positive and negative controls are listed in Table S1. PCR amplification was verified in silico (http://insilico.ehu.es/PCR/Amplify.php) on the available Xanthomonas sequenced genomes. All PCR runs were performed with a GeneAmp PCR system 9700 thermocycler (Applied Biosystems, Saint Aubin, France). PCRs were performed in 20‐μL reaction mixtures containing 1 × Gotaq® green buffer (Promega), 1.5 mm MgCl2, 200 μm of each deoxynucleoside triphosphate (dNTP), 0.3 μm of each primer, 2 ng of template genomic DNA and 0.8 U of GoTaq® Polymerase. The amplification programme consisted of 35 cycles of denaturation at 95 °C for 45 s, annealing at 55 °C for 45 s and extension at 72 °C for 0.5 to 2 min, depending on the length of the PCR product (1 kb/min). Positive and negative controls were systematically included in the PCR amplification. PCR amplicons of the expected size were considered as positive responses for the presence of each marker. Amplified PCR products showing an unexpected size were sequenced to confirm identity.
DNA sequencing
All PCR products to be sequenced were sent to Beckman Coulter Genomics (Stansted, Essex, UK). They consisted of Xop ORF and flanking regions for mutagenesis, and fragments from the screen with unexpected size. Sequence alignments and sequence analyses were performed using Geneious software (v5.5.6; Biomatters Ltd, Auckland, New Zealand). Sequences were submitted to the nucleotide blast program available online (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi; accessed July 2012) when necessary.
Southern blot hybridization
DNAs (10 μg) from the 56 strains of the collection were digested with BamHI (New England Biolabs, Evry, France), according to the supplier's instructions. DNA fragment separation and blotting were performed as described previously (Gagnevin et al., 1997). Hybridization probes were obtained by PCR with primers used for the screen, under the same PCR conditions (Table S4). Probe labelling, hybridization and detection were performed with the ECL kit (Amersham ECL Direct Nucleic Acid Labelling and Detection Systems, GE Healthcare Limited, Amersham, Buckinghamshire, UK), according to the manufacturer's instructions, under moderate stringency. Blots were then exposed to HyperfilmTM ECL for several minutes. Southern blot was preferred to dot blot to prevent false‐positive results.
Construction of Xci mutants
Deletion mutant strains were constructed using the sacB system (Schäfer et al., 1994). Briefly, 400–600‐bp upstream and downstream regions of the full‐length targeted gene were amplified by PCR on the DNA of the targeted strain as template. PCR for plasmid construction was performed with PHusion® High Fidelity DNA polymerase (Finnzymes, Waltham, MA, USA) following the manufacturer's instructions. Both amplicons were then cloned into GoldenGate‐compatible nonreplicative suicide plasmid pK18::sacB (L. Noël, CNRS, Castanet‐Tolosan, France, unpublished data; Schäfer et al., 1994). GoldenGate is a cloning method based on the use of type II restriction enzymes (BsaI in our study), which cuts outside of the enzyme recognition sites (Engler et al., 2008). On triparental conjugation using pRK2073 as a helper (Leong et al., 1982), primary transformants were selected on MOKA medium supplemented with kanamycin. Then, KanR colonies were plated onto MOKA medium containing 10% sucrose to select for second recombination. Colonies found to be sensitive to kanamycin and resistant to 10% sucrose were assayed by PCR for deletion of the targeted gene. Primers used for deletion are listed in Table S5.
AFLP analysis
The experiments were performed in 96‐well plates in a GeneAmp PCR system 9700 thermocycler (Applied Biosystems), as described previously (Ah‐You et al., 2007; Bui‐Thi‐Ngoc et al., 2010). Four AFLP conditions, each involving the two restriction enzymes MspI and SacI and two selective nucleotides, were used; two independent DNA extractions were used for all strains, and strain IAPAR 306 was used as a control in each experiment. The amplified fragments were scored as present (1) or absent (0) to create binary matrices. Only bands with an intensity above a preset level (500 relative fluorescence units) were scored.
Data analysis
Data were analysed using R software (version R 2.15.0; R Development Core Team, Vienna, Austria). Evolutionary genome divergence (EGD) values were calculated on the basis of Dice similarity indices (Mougel et al., 2002) and were used as distances to construct a weighted NJ tree with the ‘ape’ package version 3.0–2 in R (Paradis, 2006). The robustness of the tree was assessed by bootstrap (1000 resamplings). MDS was used to represent distances between strains based on the Dice dissimilarity matrix. MDS transforms a distance matrix (which cannot be analysed by eigendecomposition) into a cross‐product matrix, and then solves the eigenvector problem to find the coordinates of individuals, so that distortions to the distance matrix are minimized. As in principal component analysis, individuals are projected into n dimensions (Abdi, 2007). MDS was performed using the cmd‐scale function. Populations in planta were analysed by analysis of variance (ANOVA). Whenever F values were significant (P = 0.05), mean population sizes were compared using Tukey's contrasts at level 0.05. The area under the disease progress curve (AUDPC) was calculated using the audpc function from the package agricolae. Hierarchical agglomerative clustering was performed on a simple matching dissimilarity matrix based on a matrix of T3E presence/absence built after homology searches of the T3E candidate list on 28 Xci draft genome sequences and genomes of XauB (accession number ACPX00000000) and XauC (accession number ACPY00000000). We used Ward's method implemented in ‘cluster’ package version 1.14.2 in R (Maechler et al., 2012).
Supporting information
Fig. S1 Amplified fragment length polymorphism (AFLP)‐based metric multidimensional scaling (MDS).
Fig. S2 Disease scale used in this study.
Fig. S3 Disease severity evolution on detached leaves.
Fig. S4 Hypersensitive response (HR) symptoms induced by Aw and A* strains containing xopAG.
Fig. S5 Symptomatology of A and A* mutants.
Fig. S6 xopC1 regions.
Table S1 Bacterial strains used in this study.
Table S2 In planta populations of mutants and corresponding rifampicin‐derivative strains.
Table S3 Bacterial strains and plasmids used for mutagenesis experiments.
Table S4 Primers for polymerase chain reaction (PCR) amplifications of Xanthomonas type III effector (T3E) genes.
Table S5 Polymerase chain reaction (PCR) primers for PCR amplification for deletion of Xanthomonas type III effector (T3E) genes.
Acknowledgements
We thank C. Boyer and F. Maillot for their technical expertise, and F. Chiroleu and P. Lefeuvre for advice on data analysis. The European Union (FEDER), Conseil Régional de La Réunion and Centre de Coopération Internationale en Recherche Agronomique pour le Développement provided financial support. Financial support for LDN was provided by the LABEX TULIP (ANR‐10‐LABX‐41) and an Agence Nationale de la Recherche–Jeunes Chercheurs grant (Xopaque ANR‐10‐JCJC‐1703‐01).
GenBank accession numbers for original sequences in this work are as follows: ISXac5 from Xci strain NCPPB 3608: JX566665; xopAG from Xci strain NCPPB 3608: JX566667; xopC1 from Xci strain JK2‐10: JX566666; xopAG from X. citri pv. bilvae strain NCPPB 3213: JX556152.
References
- Abdi, H. (2007) Metric multidimensional scaling (MDS): analyzing distance matrices In: Encyclopedia of Measurement and Statistics (Salkind N., ed.), pp. 1–13. Thousand Oaks, CA: Sage. [Google Scholar]
- Ah‐You, N. , Gagnevin, L. , Chiroleu, F. , Jouen, E. , Neto, J.R. and Pruvost, O. (2007) Pathological variations within Xanthomonas campestris pv. mangiferaeindicae support its separation into three distinct pathovars that can be distinguished by Amplified Fragment Length Polymorphism. Phytopathology, 97, 1568–1577. [DOI] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (1997) The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179, 5655–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Saadi, A. , Reddy, J.D. , Duan, Y.P. , Brunings, A.M. , Yuan, Q.P. and Gabriel, D.W. (2007) All five host‐range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host‐range variation. Mol. Plant–Microbe Interact. 20, 934–943. [DOI] [PubMed] [Google Scholar]
- Anderson, P.K. , Cunningham, A.A. , Patel, N.G. , Morales, F.J. , Epstein, P.R. and Daszak, P. (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544. [DOI] [PubMed] [Google Scholar]
- Arnold, D.L. and Jackson, R.W. (2011) Bacterial genomes: evolution of pathogenicity. Curr. Opin. Plant Biol. 14, 385–391. [DOI] [PubMed] [Google Scholar]
- Balestra, G.M. , Sechler, A. , Schuenzel, E. and Schaad, N.W. (2008) First report of citrus canker caused by Xanthomonas citri in Somalia. Plant Dis. 92, 981. [DOI] [PubMed] [Google Scholar]
- Blanvillain, S. , Meyer, D. , Boulanger, A. , Lautier, M. , Guynet, C. , Denance, N. , Vasse, J. , Lauber, E. and Arlat, M. (2007) Plant carbohydrate scavenging through TonB‐dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE, 2, e224. doi: 10.1371/journal.pone.0000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui‐Thi‐Ngoc, L. , Verniere, C. , Jarne, P. , Brisse, S. , Guerin, F. , Boutry, S. , Gagnevin, L. and Pruvost, O. (2009) From local surveys to global surveillance: three high‐throughput genotyping methods for epidemiological monitoring of Xanthomonas citri pv. citri pathotypes. Appl. Environ. Microbiol. 75, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui‐Thi‐Ngoc, L. , Verniere, C. , Jouen, E. , Ah‐You, N. , Lefeuvre, P. , Chiroleu, F. , Gagnevin, L. and Pruvost, O. (2010) Amplified fragment length polymorphism and multilocus sequence analysis‐based genotypic relatedness among pathogenic variants of Xanthomonas citri pv. citri and Xanthomonas campestris pv. bilvae . Int. J. Syst. Evol. Microbiol. 60, 515–525. [DOI] [PubMed] [Google Scholar]
- Chakravarti, B.P. and Chaudhary, S.L. (1988) Citrus canker in India with special reference to Rajasthan In: Proceedings of the International Symposium on Citrus Canker, Declinio/Blight and Similar Diseases (Rossetti V., ed.), pp. 26–35. Campinas, SP: Fundaçao Cargill Sao Paulo. [Google Scholar]
- Da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , Do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , De Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Derso, E. , Verniere, C. and Pruvost, O. (2009) First report of Xanthomonas citri pv. citri‐A* causing citrus canker on lime in Ethiopia. Plant Dis. 93, 203. [DOI] [PubMed] [Google Scholar]
- Dunger, G. , Arabolaza, A.L. , Gottig, N. , Orellano, E.G. and Ottado, J. (2005) Participation of Xanthomonas axonopodis pv. citri hrp cluster in citrus canker and nonhost plant responses. Plant Pathol. 54, 781–788. [Google Scholar]
- Dunger, G. , Garofalo, C.G. , Gottig, N. , Garavaglia, B.S. , Rosa, M.C.P. , Farah, C.S. , Orellano, E.G. and Ottado, J. (2012) Analysis of three Xanthomonas axonopodis pv. citri effector proteins in pathogenicity and their interactions with host plant proteins. Mol. Plant Pathol. 13, 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, C. , Kandzia, R. and Marillonnet, S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE, 3, e3647. doi: 10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, J. , Minsavage, G. , Graham, J. , White, F. and Jones, J. (2011) Mutational analysis of type III effector genes from Xanthomonas citri subsp. citri . Eur. J. Plant Pathol. 130, 339–347. [Google Scholar]
- Gagnevin, L. , Leach, J.E. and Pruvost, O. (1997) Genomic variability of the Xanthomonas pathovar mangiferaeindicae, agent of mango bacterial black spot. Appl. Environ. Microbiol. 63, 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann, W. , Dahlbeck, D. , Chesnokova, O. , Minsavage, G.V. , Jones, J.B. and Staskawicz, B.J. (2000) Molecular evolution of virulence in natural field strains of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 182, 7053–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald, T.R. , Graham, J.H. and Schubert, T.S. (2002) Citrus canker: the pathogen and its impact. Online. Plant Health Progress doi: 10.1094/PHP-2002-0812-01-RV. Available at http://www.apsnet.org/publications/apsnetfeatures/Pages/CitrusCanker.aspx [accessed on Feb 18, 2013]. [DOI] [Google Scholar]
- Gurlebeck, D. , Thieme, F. and Bonas, U. (2006) Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163, 233–255. [DOI] [PubMed] [Google Scholar]
- Hajri, A. , Brin, C. , Hunault, G. , Lardeux, F. , Lemaire, C. , Manceau, C. , Boureau, T. and Poussier, S. (2009) A ‘repertoire for repertoire’ hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas . PLoS ONE, 4, e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajri, A. , Brin, C. , Zhao, S. , David, P. , Feng, J.‐X. , Koebnik, R. , Szurek, B. , Verdier, V. , Boureau, T. and Poussier, S. (2012a) Multilocus sequence analysis and type III effector repertoire mining provide new insights into the evolutionary history and virulence of Xanthomonas oryzae . Mol. Plant Pathol. 13, 288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajri, A. , Pothier, J.F. , Saux, M.F.‐L. , Bonneau, S. , Poussier, S. , Boureau, T. , Duffy, B. and Manceau, C. (2012b) Type three effector gene distribution and sequence analysis provide new insights into the pathogenicity of plant‐pathogenic Xanthomonas arboricola . Appl. Environ. Microbiol. 78, 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, I.D. and Hingorani, M.K. (1970) Strain studies in Xanthomonas citri (Hasse) Dowson. J. Hortic. Sci. 45, 15–17. [Google Scholar]
- Koebnik, R. , Krueger, A. , Thieme, F. , Urban, A. and Bonas, U. (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC‐type transcriptional activator HrpX to plant‐inducible promoter boxes. J. Bacteriol. 188, 7652–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkeaw, S. , Tan, S. and Coaker, G. (2010) Molecular and evolutionary analyses of Pseudomonas syringae pv. tomato race 1. Mol. Plant–Microbe Interact. 23, 415–424. [DOI] [PubMed] [Google Scholar]
- Kvitko, B.H. , Park, D.H. , Velasquez, A.C. , Wei, C.‐F. , Russell, A.B. , Martin, G.B. , Schneider, D.J. and Collmer, A. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388. doi: 10.1371/journal.ppat.1000388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc, A. , Verniere, C. , Boyer, C. , Vital, K. , Pruvost, O. , Niang, Y. and Rey, J.Y. (2011) First report of Xanthomonas citri pv. citri pathotype A causing Asiatic citrus canker on grapefruit and Mexican lime in Senegal. Plant Dis. 95, 1311. [DOI] [PubMed] [Google Scholar]
- Leong, S.A. , Ditta, G.S. and Helinski, D.R. (1982) Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta‐aminolevulinic acid synthetase from Rhizobium meliloti . J. Biol. Chem. 257, 8724–8730. [PubMed] [Google Scholar]
- Maechler, M. , Rousseeuw, P. , Struyf, A. , Hubert, M. and Hornik, K. (2012) Cluster: Cluster Analysis Basics and Extensions. R package version 1.14.2. [Google Scholar]
- Moreira, L.M. , Almeida, N.F. , Potnis, N. , Digiampietri, L.A. , Adi, S.S. , Bortolossi, J.C. , da Silva, A.C. , da de Silva, A.M., de Moraes, F.E., de Oliveira, J.C., Souza, R.F. , Facincani, A.P. , Ferraz, A.L. , Ferro, M.I. , Furlan, L.R. , Gimenez, D.F. , Jones, J.B. , Kitajima, E.W. , Laia, M.L. , Leite, R.P. , Nishiyama, M.Y. , Neto, J.R. , Nociti, L.A. , Norman, D.J. , Ostroski, E.H. , Pereira, H.A. , Staskawicz, B.J. , Tezza, R.I. , Ferro, J.A. , Vinatzer, B.A. and Setubal, J.C. (2010) Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp aurantifolii . BMC Genomics, 11, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel, C. , Thioulouse, J. , Perrière, G. and Nesme, X. (2002) A mathematical method for determining genome divergence and species delineation using AFLP. Int. J. Syst. Evol. Microbiol. 52, 573–586. [DOI] [PubMed] [Google Scholar]
- Noel, L. , Thieme, F. , Gabler, J. , Buttner, D. and Bonas, U. (2003) XopC and XopJ, two novel type III effector proteins from Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 185, 7092–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis, E. (2006) Analysis of Phylogenetics and Evolution with R. New York: Springer. [Google Scholar]
- Potnis, N. , Krasileva, K. , Chow, V. , Almeida, N.F. , Patil, P.B. , Ryan, R.P. , Sharlach, M. , Behlau, F. , Dow, J.M. , Momol, M.T. , White, F.F. , Preston, J.F. , Vinatzer, B.A. , Koebnik, R. , Setubal, J.C. , Norman, D.J. , Staskawicz, B.J. and Jones, J.B. (2011) Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics, 12, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruvost, O. , Roumagnac, P. , Gaube, C. , Chiroleu, F. and Gagnevin, L. (2005) New media for the semi‐selective isolation and enumeration of Xanthomonas campestris pv. mangiferaeindicae, the causal agent of mango bacterial black spot. J. Appl. Microbiol. 99, 803–815. [DOI] [PubMed] [Google Scholar]
- Qian, W. , Jia, Y. , Ren, S.X. , He, Y.Q. , Feng, J.X. , Lu, L.F. , Sun, Q. , Ying, G. , Tang, D.J. , Tang, H. , Wu, W. , Hao, P. , Wang, L. , Jiang, B.L. , Zeng, S. , Gu, W.Y. , Lu, G. , Rong, L. , Tian, Y. , Yao, Z. , Fu, G. , Chen, B. , Fang, R. , Qiang, B. , Chen, Z. , Zhao, G.P. , Tang, J.L. and He, C. (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris . Genome Res. 15, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak, M. , Minsavage, G.V. , Stall, R.E. and Jones, J.B. (2009) Identification of Xanthomonas citri ssp citri host specificity genes in a heterologous expression host. Mol. Plant Pathol. 10, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, S.F. , Gordon, J.S. , Martin, G.B. and Guttman, D.S. (2006) Comparative genomics of host‐specific virulence in Pseudomonas syringae . Genetics, 174, 1041–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, A. , Tauch, A. , Jäger, W. , Kalinowski, J. , Thierbach, G. and Pühler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- Schubert, T.S. , Rizvi, S.A. , Sun, X.A. , Gottwald, T.R. , Graham, J.H. and Dixon, W.N. (2001) Meeting the challenge of eradicating citrus canker in Florida – again. Plant Dis. 85, 340–356. [DOI] [PubMed] [Google Scholar]
- Schulze‐Lefert, P. and Panstruga, R. (2011) A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16, 117–125. [DOI] [PubMed] [Google Scholar]
- Sun, X.A. , Stall, R.E. , Jones, J.B. , Cubero, J. , Gottwald, T.R. , Graham, J.H. , Dixon, W.N. , Schubert, T.S. , Chaloux, P.H. , Stromberg, V.K. , Lacy, G.H. and Sutton, B.D. (2004) Detection and characterization of a new strain of citrus canker bacteria from key Mexican lime and Alemow in South Florida. Plant Dis. 88, 1179–1188. [DOI] [PubMed] [Google Scholar]
- Swarup, S. , De Feyter, R. , Brlansky, R.H. and Gabriel, D.W. (1991) A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of X. campestris to elicit cankerlike lesions on citrus. Phytopathology, 81, 802–809. [Google Scholar]
- Swarup, S. , Yang, Y. , Kingsley, M.T. and Gabriel, D.W. (1992) A Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant–Microbe Interact. 5, 204–213. [DOI] [PubMed] [Google Scholar]
- Swords, K.M.M. , Dahlbeck, D. , Kearney, B. , Roy, M. and Staskawicz, B.J. (1996) Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2 . J. Bacteriol. 178, 4661–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme, F. , Koebnik, R. , Bekel, T. , Berger, C. , Boch, J. , Buttner, D. , Caldana, C. , Gaigalat, L. , Goesmann, A. , Kay, S. , Kirchner, O. , Lanz, C. , Linke, B. , McHardy, A.C. , Meyer, F. , Mittenhuber, G. , Nies, D.H. , Niesbach‐Klosgen, U. , Patschkowski, T. , Ruckert, C. , Rupp, O. , Schneiker, S. , Schuster, S.C. , Vorholter, F.J. , Weber, E. , Puhler, A. , Bonas, U. , Bartels, D. and Kaiser, O. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traore, Y.N. , Ngoc, L.B.T. , Verniere, C. and Pruvost, O. (2008) First report of Xanthomonas citri pv. citri causing citrus canker in Mali. Plant Dis. 92, 977. [DOI] [PubMed] [Google Scholar]
- Vernière, C. , Hartung, J.S. , Pruvost, O.P. , Civerolo, E.L. , Alvarez, A.M. , Maestri, P. and Luisetti, J. (1998) Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. Eur. J. Plant Pathol. 104, 477–487. [Google Scholar]
- Vorhoelter, F.‐J. , Schneiker, S. , Goesmann, A. , Krause, L. , Bekel, T. , Kaiser, O. , Linke, B. , Patschkowski, T. , Rueckert, C. , Schmid, J. , Sidhu, V.K. , Sieber, V. , Tauch, A. , Watt, S.A. , Weisshaar, B. , Becker, A. , Niehaus, K. and Puehler, A. (2008) The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 134, 33–45. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. and Bonas, U. (1996) HrpXv, an AraC‐type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 178, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Q. and Wang, N. (2012) High‐throughput screening and analysis of genes of Xanthomonas citri subsp citri involved in citrus canker symptom development. Mol. Plant–Microbe Interact. 25, 69–84. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Morgan, R.L. , Guttman, D.S. and Ma, W. (2009) Allelic variants of the Pseudomonas syringae type III effector HopZ1 are differentially recognized by plant resistance systems. Mol. Plant–Microbe Interact. 22, 176–189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Amplified fragment length polymorphism (AFLP)‐based metric multidimensional scaling (MDS).
Fig. S2 Disease scale used in this study.
Fig. S3 Disease severity evolution on detached leaves.
Fig. S4 Hypersensitive response (HR) symptoms induced by Aw and A* strains containing xopAG.
Fig. S5 Symptomatology of A and A* mutants.
Fig. S6 xopC1 regions.
Table S1 Bacterial strains used in this study.
Table S2 In planta populations of mutants and corresponding rifampicin‐derivative strains.
Table S3 Bacterial strains and plasmids used for mutagenesis experiments.
Table S4 Primers for polymerase chain reaction (PCR) amplifications of Xanthomonas type III effector (T3E) genes.
Table S5 Polymerase chain reaction (PCR) primers for PCR amplification for deletion of Xanthomonas type III effector (T3E) genes.


