Summary
Xanthomonas translucens pv. graminis (Xtg) is a gammaproteobacterium that causes bacterial wilt on a wide range of forage grasses. To gain insight into the host–pathogen interaction and to identify the virulence factors of Xtg, we compared a draft genome sequence of one isolate (Xtg29) with other Xanthomonas spp. with sequenced genomes. The type III secretion system (T3SS) encoding a protein transport system for type III effector (T3E) proteins represents one of the most important virulence factors of Xanthomonas spp. In contrast with other Xanthomonas spp. assigned to clade 1 on the basis of phylogenetic analyses, we identified an hrp (hypersensitive response and pathogenicity) gene cluster encoding T3SS components and a representative set of 35 genes encoding putative T3Es in the genome of Xtg29. The T3SS was shown to be divergent from the hrp gene clusters of other sequenced Xanthomonas spp. Xtg mutants deficient in T3SS regulating and structural genes were constructed to clarify the role of the T3SS in forage grass colonization. Italian ryegrass infection with these mutants led to significantly reduced symptoms (P < 0.05) relative to plants infected with the wild‐type strain. This showed that the T3SS is required for symptom evocation. In planta multiplication of the T3SS mutants was not impaired significantly relative to the wild‐type, indicating that the T3SS is not required for survival until 14 days post‐infection. This study represents the first major step to understanding the bacterial colonization strategies deployed by Xtg and may assist in the identification of resistance (R) genes in forage grasses.
Introduction
The pathogen Xanthomonas translucens pv. graminis (Xtg) is a gammaproteobacterium that causes bacterial wilt of forage grasses, a disease leading to considerable forage yield losses depending on host susceptibility (Egli et al., 1975). Its host range comprises a broad variety of forage grasses, including Lolium and Festuca spp. (Egli and Schmidt, 1982). Xtg invades the plant through wounded tissue and initially colonizes the protoxylem lacuna, from where it migrates to the vascular tissue, resulting in symptoms such as wilting of leaves and necrosis of the entire plant (Masuch et al., 1989). Breeding for resistant cultivars based on recurrent phenotypic selection has led to cultivars with improved partial resistance to bacterial wilt. Complete resistance has not been achieved and hypersensitive response (HR) symptoms have never been observed on resistant plants. In addition, highly susceptible plants still occur after continuous recurrent selection (Michel, 2001). Breeding for resistance is also complicated by the population‐based breeding schemes and the obligate cross‐pollination mode of reproduction of many species, which result in highly heterogeneous populations (Brummer, 1999). A thorough understanding of this complex host–pathogen interaction will enable the development of molecular genetics tools for improved resistance breeding and the development of superior cultivars.
In plants, resistance to diseases caused by Xanthomonas spp. is most often based on the specific recognition of effector proteins secreted through the type III secretion system (T3SS). These type III effectors (T3Es) are recognized by plant resistance (R) genes, which promote an HR, limiting pathogen spread and leading to effector‐triggered immunity (ETI). The T3SS is encoded by a large gene cluster on either the chromosome or a plasmid (Arnold et al., 2003; Zou et al., 2006). Expression of the hrp (hypersensitive response and pathogenicity) gene cluster results in the formation of a membrane‐spanning secretion apparatus (Hrp‐pilus), which mediates T3E translocation into the host cell (Furutani et al., 2009; Roden et al., 2004; Thieme et al., 2005; White et al., 2009). The first key component in the hrp regulatory cascade is HrpG, together with HrpX, part of a two‐component regulatory system (Noël et al., 2001). Point mutations in the hrpG gene of Xanthomonas campestris pv. campestris (Xcc) and X. campestris pv. vesicatoria (Xcv) result in the constitutive expression of the hrp gene cluster (Jiang et al., 2006; Wengelnik et al., 1999). Secreted T3Es can modulate the physiology of the plant by suppressing pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) of the host (Jones and Dangl, 2006) or by facilitating nutritional and virulence processes of the pathogen (Büttner and He, 2009). Mutants deficient in T3SS genes of Xanthomonas spp. are no longer able to trigger ETI on resistant plants, and cannot cause symptoms on susceptible plants (Cho et al., 2008; Darsonval et al., 2008; Wengelnik et al., 1996; Zou et al., 2006).
Although the genome sequences of several Xanthomonas spp. are available and have revealed many shared virulence factors (Bogdanove et al., 2011; Da Silva et al., 2002; Lee et al., 2005; Moreira et al., 2005, 2010; Ochiai et al., 2005; Pieretti et al., 2009; Salzberg et al., 2008; Studholme et al., 2010; Thieme et al., 2005; Vorhölter et al., 2008; reviewed in Ryan et al., 2011; Studholme et al., 2011), Xtg is only distantly related to the other Xanthomonas spp. with sequenced genomes (Hauben et al., 1997; Parkinson et al., 2009). This has hindered the identification and analysis of homologous virulence factors of Xtg by comparative analyses. Based on gyrB and 16S rRNA gene sequencing, Xtg seems to be most closely related to two recently sequenced Xanthomonas spp., which have been shown not to have an hrp gene cluster and, in the case of X. albilineans, cannot produce xanthan (Pieretti et al., 2009).
The objectives of this study were to use the draft genome sequence of one Xtg isolate and to compare the hrp gene cluster of Xtg with the hrp gene clusters of other Xanthomonas spp. Our aim was also to elucidate the importance of the T3SS for infection and in planta multiplication using mutants deficient in genes encoding T3SS structural components and a regulatory gene. Further, homologous genes encoding T3Es were identified in Xtg. An understanding of the host colonization strategies deployed by Xtg may provide the necessary information to predict the T3Es that potentially induce ETI on forage grasses, and therewith may assist R gene identification in the future.
Results and Discussion
The T3SS of Xtg strain 29 (Xtg29) is distinctly different from the T3SS of other Xanthomonas spp
A shotgun approach with the Genome Sequencer FLX (GS FLX) was used to sequence the Xtg29 genome (DDBJ/EMBL/GenBank accession numbers ANGG01000001:ANGG01000788). Using this draft sequence, we constructed a phylogenetic tree based on the core genes of selected fully sequenced genomes (Fig. 1). Although most of the strains sequenced to date fall into clade 2 of the Xanthomonas genus, Xtg29 was assigned to clade 1, together with X. sacchari and X. albilineans. Therefore, this analysis confirmed the previous phylogenetic positions assessed using gyrB and 16S rRNA gene sequencing (Hauben et al., 1997; Parkinson et al., 2009). As a next step, we assessed the genome sequence for the presence of genes encoding the T3SS, as the two other Xanthomonas spp. of clade 1, X. albilineans and X. sacchari, do not have an hrp‐type T3SS (Pieretti et al., 2009; Studholme et al., 2011). Interestingly, we managed to identify a region covering 17 751 bp putatively encoding an hrp gene cluster in Xtg29. This region consisted of 11 hrc genes, eight hrp genes and three hpa genes (Fig. 2a). Sanger sequencing across a gap observed in the original draft sequence between the genes encoding hrpX and hrcT revealed an 815‐bp sequence with high nucleotide sequence identity (88.3%) to two insertion sequence (IS) elements of X. oryzae pv. oryzae (Xoo) (ISXoo11 and ISXoo12). Another gap affected the 5′ region of the hrpB4 gene between hrcJ and hrcL. Again Sanger sequencing was performed and resulted in a spanning sequence of 145 bp in length with a G + C content of almost 80%. Analysis by the EMBOSS tool palindrome (http://emboss.bioinformatics.nl/cgi‐bin/emboss/palindrome) revealed two palindromic sequences within this fragment, which might have caused difficulties during amplification and sequencing of this region in the first instance.
Figure 1.
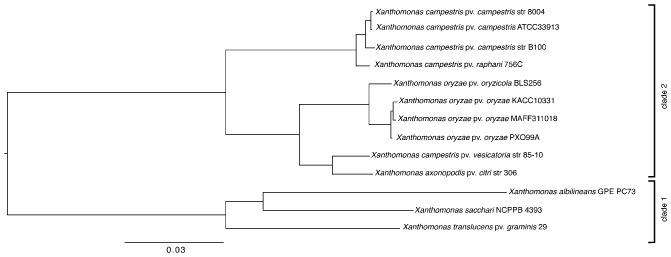
Neighbour‐joining tree of concatenated nucleotide sequences of all genes included in the core genome of completely sequenced X anthomonas strains compared with X . translucens pv. graminis strain 29 (X tg29). The strains include: X . campestris pv. campestris 8004 (CP000050; Qian et al., 2005), ATCC 33913 (AE008922; Da Silva et al., 2002) and B100 (AM920689; Vorhölter et al., 2008), X . campestris pv. raphani 756 (can be downloaded from: http://cmr.jcvi.org; Bogdanove et al., 2011), X . oryzae pv. oryzae KACC10331 (AE013598; Ochiai et al., 2005), MAFF311018 (AP008229; Lee et al., 2005) and PXO99A (CP000967; Salzberg et al., 2008), X . oryzae pv. oryzicola BLS256 (AAQN01000001; Bogdanove et al., 2011), X . campestris pv. vesicatoria strain 85‐10 (AM039952; Thieme et al., 2005), X . axonopodis pv. citri strain 306 (AE008924; Da Silva et al., 2002), X anthomonas albilineans str. GPE PC73 (FP565176; Pieretti et al., 2009) and X . sacchari NCPPB 4393 (AGDB01000024; Studholme et al., 2011).
Figure 2.
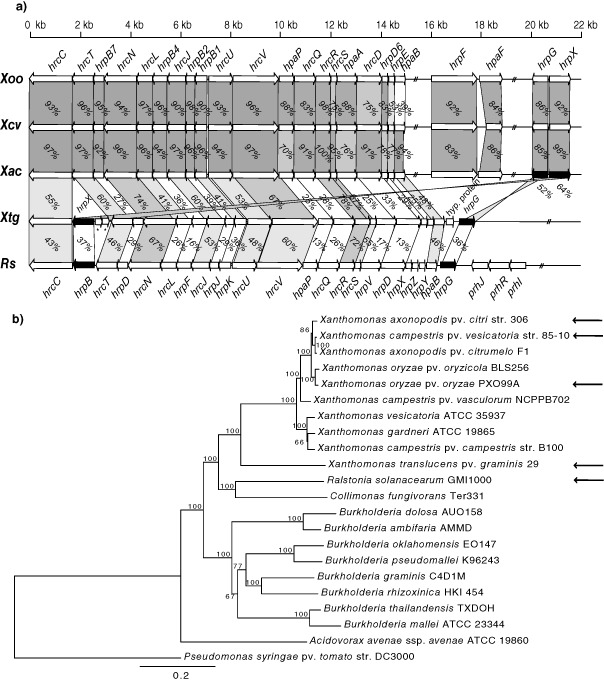
(a) Genetic organization and protein sequence identity of Hrp, Hrc and Hpa proteins from different sequenced X anthomonas spp. and R alstonia solanacearum GMI1000 compared with X . translucens pv. graminis strain 29 (X tg29) hrp genes. Genes encoding the type III secretion system (T3SS) of the following strains are shown: X oo, X . oryzae pv. oryzae PXO99A (CP000967; Salzberg et al., 2008); X cv, X . campestris pv. vesicatoria strain 85‐10 (AM039952; Thieme et al., 2005); X ac, X . axonopodis pv. citri strain 306 (AE008922; Da Silva et al., 2002); R s, R . solanacearum GMI1000 megaplasmid pGM1000MP (AL646053; Salanoubat et al., 2002). Arrows indicate the sizes, positions and orientations of the hrp, hrc and hpa genes. The two genes indicated with asterisks were shown to be similar to ISXoo11 and ISXoo12 transposases. The identity of each protein sequence with its homologue in X . translucens pv. graminis is indicated by the percentage identity and the use of a gradual black/grey colour scale. (b) Phylogenetic tree inferred with maximum likelihood for the alignment of concatenated sequences of eight different Hrc proteins (i.e. HrcC, HrcJ, HrcN, HrcR, HrcS, HrcT, HrcU and HrcV) from different X anthomonas spp., R . solanacearum (AL646053), C ollimonas fungivorans (NC_015856), B urkholderia spp. and A cidovorax avenae ssp. avenae (NC_015138). P seudomonas syringae pv. tomato strain DC3000 (AE016853) was used as an outgroup. Numbers represent bootstrap values for 100 replications. Arrows indicate that these sequences were used in (a) for the comparison of T3SS synteny.
The hrpF and hpaF genes encoding a substantial part of the type III secretion translocon present in some Xanthomonas spp. (Sugio et al., 2005) were not found in the genome of Xtg29 (Fig. 2a). HrpF from Xcv has been shown to be dispensable for type III secretion, but is presumably required for effector translocation into the host cell, as it binds to lipid bilayers and induces channel formation from the intracellular space (Büttner et al., 2002; Rossier et al., 2000). As Xtg makes use of breaches to gain direct access to the xylem cells in which it resides, and as these cells do not contain a cell membrane, the hrpF gene may be dispensable for effector translocation in Xtg29 and, consequently, may have been lost during evolution. However, as Xtg29 is so distantly related to the other Xanthomonas spp. with a sequenced T3SS, the hrpF and hpaF genes might nevertheless be present, but cannot be recognized on the basis of sequence identity.
Interestingly, the two genes hrpG and hrpX, encoding the response regulators of the T3SS, were found to be localized within the hrp gene cluster in Xtg29, whereas, in other Xanthomonas spp., they are encoded outside of the hrp gene cluster (Fig. 2a). A similar genetic organization with the genes encoding hrpG and hrpB (which is homologous to hrpX of Xanthomonas spp.) within the T3SS has been reported for Ralstonia solanacearum (Rs; Salanoubat et al., 2002). Furthermore, pairwise protein sequence identity of homologous genes revealed a lower sequence identity to Rs. Genealogy analysis of T3SSs based on the concatenated sequences of eight different Hrc proteins (i.e. HrcC, HrcJ, HrcN, HrcR, HrcS, HrcT, HrcU and HrcV) showed that the T3SS of Xtg29 clustered together with the T3SSs of other sequenced Xanthomonas spp. (Fig. 2b). The same topology was observed for separate phylogenetic trees of each of these proteins (Fig. S1, see Supporting Information). This analysis demonstrated that the T3SS of Xtg29 is clearly divergent from other T3SSs of Xanthomonas spp. Based on a comprehensive phylogenetic analysis of the HrcN, HrcS and HrcR proteins, the T3SS of Xanthomonas spp. has been hypothesized previously to have been horizontally acquired from Rs during evolution (Gophna et al., 2003). The higher degree of synteny within the T3SS, observed between Xtg29 and Rs compared with other Xanthomonas and Rs, bolsters such an evolutionary scenario and also suggests that the hrp gene cluster of Xtg29 possibly represents a more ancestral state of the T3SSs within the Xanthomonas genus. The T3SS of Xtg29 has very distinct characteristics concerning genetic organization and sequence composition, indicating potential differences in functionality during infection.
Other major virulence factors of Xanthomonas present in the genome of Xtg29
In addition to the T3SS, the type II secretion system (T2SS) is highly important for some Xanthomonas spp., as it facilitates host colonization by the secretion of toxins and extracellular enzymes, such as proteases, lipases and cell wall‐degrading enzymes (reviewed in Büttner and Bonas, 2010). In Xtg29, the xps gene cluster encoding components of a T2SS covered a region of 11 732 bp and consisted of the genes xpsEFGHIJKLMD. A gum gene cluster was identified that included 11 genes for xanthan biosynthesis. Surprisingly, the gene gumF, which encodes an acetyltransferase (Becker et al., 1998) and is well conserved in other Xanthomonas genomes (Vorhölter et al., 2008), was missing in the otherwise complete gum gene cluster of Xtg29. Although the gum gene cluster is absent in X. albilineans (Pieretti et al., 2009), the other sequenced member of clade 1, i.e. X. sacchari, has also been shown to be capable of xanthan biosynthesis (Studholme et al., 2011). Flanked on one side by precursor genes for xanthan and lipopolysaccharide (LPS) biosynthesis (Vorhölter et al., 2008) and by the electron transfer genes etfAB in a generally conserved Xanthomonas genomic organization, and, on the other side, by metBC genes involved in amino acid biosynthesis (Schatschneider et al., 2011), an LPS biosynthesis gene cluster was identified that was made up of 19 genes (XTG29_00287 to XTG29_00305). Apart from two wzm and wzt genes coding for a polysaccharide‐specific ABC transporter, also found in other Xanthomonas genomes (Patil et al., 2007; Vorhölter et al., 2003), other genes differed from the Xanthomonas LPS gene clusters characterized to date, thereby indicating a distinct LPS structure for Xtg29. Furthermore, an rpf gene cluster that regulates the synthesis of pathogenicity factors was also found in the genome of Xtg29 (Ryan and Dow, 2011).
A homologue to the type IV secretion system (T4SS), present in many Xanthomonas spp. (Alegria et al., 2005; Qian et al., 2005; Thieme et al., 2005), was not found in Xtg29. However, the function of the T4SS has not been clarified and experimental data on the extent of its contribution to disease development are still missing. In addition to the type I secretion system (T1SS), which secretes the AvrXa21 molecule in Xoo and is encoded by raxABC, all rax gene orthologues were identified in Xtg29 (Table S1, see Supporting Information). In addition, a large number of other genes with functions related to type I secretion of toxins (bacteriocins), lipases and proteins or encoding components of T1SSs were found in Xtg29 (data not shown).
Predicted genes encoding T3Es of Xtg29
An extensive analysis of all known Xanthomonas T3E protein sequences (summarized at http://www.xanthomonas.org) was performed using blastp and tblastn (Altschul et al., 1990) against all coding DNA sequences (CDSs) and the nucleotide sequence of Xtg29. In addition, the genome was searched for plant‐inducible promoter (PIP) box sequences. All approaches combined revealed 35 genes homologous to genes encoding T3Es (Table 1). This number reflects the abundance and variety of T3Es typically found in other Xanthomonas spp. (Kay and Bonas, 2009).
Table 1.
Genes encoding predicted type III secreted effectors found in the genome of Xanthomonas translucens pv. graminis strain 29 (Xtg29) and putative functions or homologues from other pathogens containing a type III secretion system (e.g. Pseudomonas syringae, Ralstonia solanacearum or Yersinia spp.)
| Gene name | Sequence name | PIP box | Function/family/homology to Pseudomonas effectors | Best blastx hit | E‐value | % protein identity | Length (bp) | Reference |
|---|---|---|---|---|---|---|---|---|
| avrBs2 | XTG29_03687 | − | Putative glycerophosphoryl‐diester phosphodiesterase | Avirulence protein (X. campestris pv. musacearum NCPPB 4381) | 0.0 | 62 | 2148 | Swords et al. (1996) |
| avrRxv | XTG29_02581 | + | YopJ/AvrRxv family, putative cysteine protease | Type III effector HopZ2 (P. syringae pv. coronafaciens) | 3 E‐14 | 44 | 246 | Whalen et al. (1993) |
| avrRxv | XTG29_02457 | + | YopJ/AvrRxv family, putative cysteine protease | YopP/AvrRxv family protein (R. solanacearum GMI1000) | 8 E‐90 | 49 | 1698 | Whalen et al. (1993) |
| xopB | XTG29_02183 | + | Homology to HopD1 (P. syringae pv. tomato) | Type III effector HopD1 (P. syringae pv. oryzae str. 1_6) | 0.0 | 80 | 1887 | Noël et al. (2001) |
| xopC2 | XTG29_01319 | − | Putative Xanthomonas outer protein C2 (X. perforans 91‐118) | 0.0 | 62 | 1416 | Noël et al. (2003) | |
| xopE1 | XTG29_03655 | + | Homology to avrPphE/HopX, predicted transglutaminase | Type III secretion system effector protein (X. fuscans ssp. aurantifolii str. ICPB 10535) | 2 E‐170 | 70 | 1089 | Thieme et al. (2007) |
| xopF1 | XTG29_00871 | + | Unknown function | Outer protein F1 (X. campestris pv. vasculorum NCPPB 702) | 1 E‐70 | 33 | 2058 | Roden et al. (2004) |
| xopF2‐1 | XTG29_02713 | − | Unknown function | Xanthomonas outer protein F2 (X. perforans 91‐118) | 1 E‐109 | 73 | 813 | Roden et al. (2004) |
| xopF2‐2 | XTG29_02714 | − | Unknown function | Outer protein F2 (X. campestris pv. vesicatoria str. 85‐10) | 2 E‐148 | 83 | 855 | Roden et al. (2004) |
| xopI | XTG29_02986 | − | F‐box protein | Type III secretion system effector protein (X. fuscans ssp. aurantifolii str. ICPB 11122) | 3 E‐86 | 83 | 618 | Thieme et al. (2007) |
| xopK | XTG29_03500 | + | Unknown function | Putative Xanthomonas outer protein K (X. gardneri ATCC 19865) | 0.0 | 67 | 2610 | Furutani et al. (2009) |
| xopN | XTG29_00130 | − | Homology to HopAU1 (P. syringae), α‐helical ARM/HEATS repeats | XopN effector (X. oryzae pv. oryzae PXO99A) | 0.0 | 61 | 1881 | Kim et al. (2009) |
| xopP | XTG29_02884 | − | Unknown function | Type III effector protein XopP (X. campestris pv. raphani 756C) | 0.0 | 56 | 1635 | Roden et al. (2004) |
| xopP | XTG29_00851 | − | Unknown function | Type III secretion system effector protein (X. fuscans ssp. aurantifolii str. ICPB 11122) | 8 E‐99 | 82 | 555 | Roden et al. (2004) |
| xopQ | XTG29_00093 | + | HopQ1‐1 family protein, inosine‐uridine nucleoside N‐ribohydrolase | Type III effector RipB protein (R. solanacearum PSI07) | 1 E‐138 | 54 | 1407 | Roden et al. (2004) |
| xopR | XTG29_00207 | − | Unknown function | Putative Xanthomonas outer protein R (X. gardneri ATCC 198650 | 2 E‐58 | 54 | 831 | Furutani et al. (2009) |
| xopV | XTG29_00797 | + | Unknown function | Type III effector protein (X. arboricola pv. pruni) | 2 E‐90 | 53 | 930 | Furutani et al. (2009) |
| xopX1 | XTG29_01318 | − | Homology to HopAE1 (P. syringae) | Xanthomonas outer protein X (X. vesicatoria ATCC 35937) | 0.0 | 74 | 1893 | Metz et al. (2005) |
| xopX2‐1 | XTG29_01078 | − | Unknown function | Xanthomonas outer protein X (X. gardneri ATCC 19865) | 0.0 | 54 | 2124 | Noël et al. (2002) |
| xopX2‐2 | XTG29_01080 | − | Unknown function | Xanthomonas outer protein X2 (X. perforans 91‐118) | 0.0 | 55 | 2124 | Noël et al. (2002) |
| xopY | XTG29_00726 | + | Unknown function | Type III effector protein XopY (X. oryzae pv. oryzicola BLS256) | 6 E‐05 | 39 | 408 | |
| xopZ1‐1 | XTG29_02419 | − | Unknown function | Type III effector protein XopZ1 (X. oryzae pv. oryzicola BLS256) | 0.0 | 64 | 3000 | Song and Yang (2010) |
| xopZ1‐2 | XTG29_02420 | − | Unknown function | Type III effector protein XopZ1 (X. oryzae pv. oryzicola BLS256) | 2 E‐60 | 54 | 681 | Song and Yang (2010) |
| xopAD | XTG29_02463 | − | Unknown function | Type III effector protein XopAD (X. oryzae pv. oryzicola BLS256) | 0.0 | 64 | 7896 | |
| xopAK | XTG29_03476 | − | Homology to HopK1 (P. syringae) | Type III effector protein XopAK (X. oryzae pv. oryzicola BLS256) | 2 E‐89 | 56 | 771 | Wei et al. (2007) |
| XTG29_01559 | − | Homology to HopH 1 (P. syringae) and xopG | Type III effector protein (R. solanacearum IPO1609) | 1 E‐34 | 50 | 531 | Wei et al. (2007) | |
| XTG29_01817 | − | Glycerophosphodiester phosphodiesterase | Hypothetical protein XsacN4_02427 (X. sacchari NCPPB 4393) | 6 E‐39 | 32 | 1017 | ||
| XTG29_03294 | − | Glycerophosphodiester phosphodiesterase | Glycerophosphodiester phosphodiesterase (X. albilineans GPE PC73) | 0.0 | 86 | 945 | ||
| XTG29_01857 | + | Homology to avrPphE/HopX | Hypothethical protein, partial )P. syringae pv. actinidiae str. M302091) | 1 E‐178 | 90 | 987 | ||
| XTG29_01339 | − | Homology to HopX1 | Type III effector protein (X. arboricola pv. pruni) | 5 E‐30 | 34 | 1131 | Wei et al. (2007) | |
| XTG29_00200 | + | Homology to HopR | Xanthomonas outer protein AM (X. vesicatoria ATCC 35937) | 0.0 | 55 | 5166 | Wei et al. (2007) | |
| XTG29_02140 | + | Homology to HopAJ1 (P. syringae) | Outer membrane antigen precursor (X. sacchari NCPPB 4393) | 0.0 | 91 | 2457 | ||
| XTG29_02185 | + | Homology to putative type III effector protein (R. solanacearum) | Hypothetical protein XGA_1070 (X. gardneri ATCC 19865) | 3 E‐62 | 56 | 798 | ||
| XTG29_01881 | − | Putative type III effector protein (R. solanacearum) | Type III effector protein (R. solanacearum) | 5 E‐30 | 38 | 846 | ||
| XTG29_00627 | + | Putative cysteine protease, yopT‐like | HopAY1 (P. syringae pv. mori str. 301020) | 2 E‐68 | 57 | 1071 |
In Xtg, the genes putatively encoding T3Es included at least three different effectors of the xopX family. Furthermore, two genes with predicted protein sequence similarity to AvrRxv effectors were identified in Xtg29. The genes that encode these two candidate effectors showed rather low G + C contents (53% and 48%), which may be indicative of acquisition by horizontal gene transfer (Dobrindt et al., 2004). AvrRxv has been described in Xcv and belongs to the AvrRxv/XopJ effector group, including members with different enzymatic activities which target multiple host physiological pathways (Mukherjee et al., 2007). Therefore, it is difficult to predict a function for these two effectors. Both avrRxv‐like genes harboured an upstream PIP box, and therefore we hypothesize that these two effectors are important for the Xtg–forage grass interaction.
A homologous gene encoding the AvrBs2 effector has been identified in Xtg29. AvrBs2 encodes a putative glycerophosphoryl‐diester phosphodiesterase and has been shown to be involved in both osmotic adaptation and plant host signalling (Swords et al., 1996). The avrBs2 gene is highly conserved among a very broad range of Xanthomonas spp. (Hajri et al., 2009). However, experimental evidence that translocation of AvrBs2 contributes to virulence and in planta multiplication has only been provided for Xcv (Gurlebeck et al., 2006; Mudgett et al., 2000).
A number of genes presumably encoding T3Es found in Xtg29 are members of a ‘core set’ of T3Es typically found in Xanthomonas genomes (e.g. xopB, xopF1, xopN, xopP, xopQ and xopX), and some are less conserved. Effector composition can vary significantly between strains and pathovars, as has been demonstrated for X. axonopodis (Hajri et al., 2009). We therefore expect effector composition to vary among Xtg isolates. As differences in the virulence of Xtg isolates have consistently been observed on Lolium multiflorum genotypes (Kölliker et al., 2006), we hypothesize that this could be a result of differential effector composition.
Genes encoding effectors belonging to the highly conserved avrBs3/pth family, also called transcription activator‐like effectors (TALEs), present in many Xanthomonas spp. and Rs, were not found in the genome of Xtg29. TALEs have been demonstrated to have certain amino acid residues that correspond to one nucleotide in the promoter sequence of the corresponding R genes (Moscou and Bogdanove, 2009; Römer et al., 2009a, b). Although other T3Es and resistance mechanisms that can trigger ETI are known in plant–Xanthomonas interactions, the absence of TALEs could be an indication for the absence of an HR, even on forage grasses with considerable resistance to Xtg. So far, HR symptoms have never been described in studies on bacterial wilt in forage grasses (Egli and Schmidt, 1982; Leyns, 1993; Michel, 2001). In a needle inoculation experiment using resistant L. multiflorum genotypes and the wild‐type Xtg29 isolate, we failed to observe HR symptoms (data not shown). The lack of HR could be a further explanation for the absence of race specificity reported previously for the interaction of different Xtg isolates with Italian ryegrass genotypes (Wichmann et al., 2011). However, this needs to be clarified in further studies targeting directly the translocation of effector proteins into the host cell using immunochemical approaches.
The T3SS of Xtg29 is required for symptom evocation
In order to test whether the T3SS of Xtg29 is required for disease development, deletion mutants of different genes encoding T3SS components were constructed. These included two mutants of structural T3SS genes (i.e. ΔhrpE and ΔhrcR), one mutant of a response regulator gene (i.e. ΔhrpG) and a double mutant (i.e. ΔhrpG/hrcR). In addition, the ΔhrpG mutant was complemented with the hrpG gene and its promoter sequence (322 bp upstream of the start codon) in a plasmid with a broad host range. Infection of a highly susceptible Italian ryegrass (Lolium multiflorum Lam.) genotype with all T3SS mutants led to significantly reduced symptoms (P < 0.05) when compared with the wild‐type Xtg29 isolate (Fig. 3a; Fig. S2, see Supporting Information). However, when compared with the negative control treatment, although not significant, all T3SS mutants still caused weak but detectable symptoms, particularly the ΔhrpG mutant. Although drastically reduced symptom development has also been observed for the T3SS mutants of other Xanthomonas spp. (Cho et al., 2008; Darsonval et al., 2008; Zou et al., 2006), it cannot be excluded that other virulence factors, such as extracellular polysaccharides (EPSs) or cell wall‐degrading enzymes, secreted by the T2SS, contribute to the remaining observed symptoms.
Figure 3.
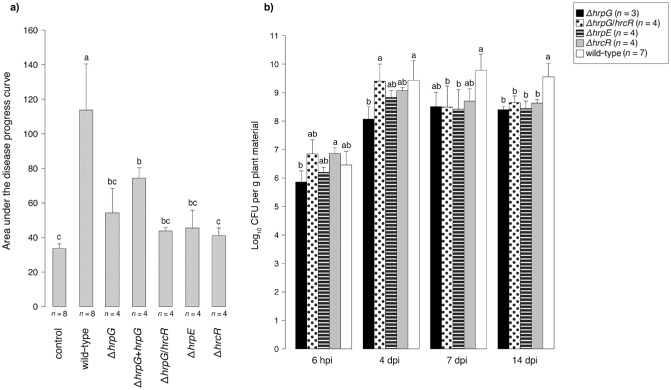
(a) Mean areas under the disease progress curve (AUDPCs) from four replications for L olium multiflorum genotype LmK‐01 infected with Xanthomonas translucens pv. graminis strain 29 (X tg29) deficient in genes encoding the type III secretion system (T3SS) compared with the wild‐type strain. Means with different letters are significantly different (P < 0.05) on the basis of a two‐sided t‐test using the Holm P value adjustment. The negative control treatment consisted of cutting the plants with scissors dipped in sterile physiological sodium chloride solution. (b) Colonization of L . multiflorum by X tg29 and the X tg29 strains deficient in T3SS genes. Bacterial population densities were determined for leaves of three or four different plants of LmK‐01 sampled at 6 h post‐infection (hpi) and 4, 7 and 14 days post‐infection (dpi), counting the colony‐forming units (CFU) of serial dilutions. For each time point, mean population densities and standard deviations were calculated. Means indicated with different letters are significantly (P < 0.05) different on the basis of a two‐sided t‐test using the Holm P value adjustment.
Infection with the complemented ΔhrpG mutant caused 33.83% higher areas under the disease progress curve (AUDPCs) when compared with the ΔhrpG mutant and the wild‐type strain. Significant differences (P < 0.05) between the complemented ΔhrpG mutant and other T3SS mutants were only observed for the ΔhrcR mutant. Previously, functional complementation of the hrpG gene resulted in a complete restoration of the wild‐type phenotype (i.e. induction of HR on resistant plants) in Xcv and X. oryzae pv. oryzicola (Wengelnik et al., 1996; Zou et al., 2006). However, an HR was not observed on forage grasses after Xtg infection. Therefore, it was possible that the complete function of the hrpG gene had been restored in Xtg29, but the levels of expression were not as fine tuned as necessary, which resulted in less pronounced symptoms.
Mutation of T3SS components does not prevent in planta survival of Xtg29
In order to test whether the in planta survival of mutants deficient in T3SS genes is affected, the dynamics of bacterial population densities for the T3SS mutants and the wild‐type strain (Xtg29) were determined. Bacteria were isolated from harvested plant material at different time points after infection [i.e. 6 h post‐infection (hpi), and 4, 7 and 14 days post‐infection (dpi)], and serial dilutions were plated. The population densities of all T3SS mutants quantified in leaves were not significantly different from those of the wild‐type, except at 14 dpi (Fig. 3b), but also at this time point the mutants were still quantified in substantial numbers. This indicated that the T3SS of Xtg29 is not crucial for plant colonization. This is in contrast with most other mutant studies with Xanthomonas spp. deficient in T3SS genes (Büttner et al., 2007; Cho et al., 2008; Darsonval et al., 2008). One isolate of Xcc deficient in the hrpE gene (i.e. str. B305) could also establish populations comparable with those of the wild‐type strain (Sun et al., 2011), but, in the other studies, in planta multiplication was drastically impaired. The in planta survival of ΔhrpG or ΔhrpX mutants of X. fuscans pv. fuscans was even more impaired relative to the T3SS mutants deficient in structural components (Darsonval et al., 2008). These results therefore implied that HrpG and HrpX regulate processes during early infection and the epiphytic phase (reviewed in Büttner and Bonas, 2010). Surprisingly, total in planta populations of the Xtg29 ΔhrpG mutant were not significantly lower than those of the other T3SS mutants or the wild‐type, which demonstrates that the hrpG gene, despite initial significant differences at 4 dpi, establishes comparable bacterial populations later during infection.
Therefore, to characterize in more detail their role in type III secretion, gene expression of the hrpE and hrcR genes was quantified in plant material infected with the wild‐type Xtg29 and the ΔhrpG mutant. Four biological replicates and three technical replicates of each sample were used to quantify hrpE and hrcR expression relative to the genomic DNA quantity obtained from the same nucleic acid extraction. We observed that, in infected plant material, the hrpE gene was up‐regulated significantly in the wild‐type isolate relative to the ΔhrpG mutant at all sampled time points after infection (Fig. 4). For the hrcR gene, significant up‐regulation according to the permutation test was only observed at 4 and 14 dpi. That the hrpG gene activates the expression of T3SS structural genes is consistent with findings in other Xanthomonas spp. and Rs (Noël et al., 2001; Wengelnik et al., 1996; Yoshimochi et al., 2009). However, up‐regulation was particularly pronounced at 14 dpi, where a 321‐fold up‐regulation of the hrpE gene and a 121‐fold up‐regulation of the hrcR gene were observed. The hrpE and hrcR genes remained up‐regulated until 28 dpi in wild‐type Xtg, but gene expression differences were less pronounced. Therefore, we conclude from our gene expression data and in planta multiplication assays that major virulence processes of Xtg29 occur at 14 dpi, which is the time point at which the presence of T3SS genes matters.
Figure 4.
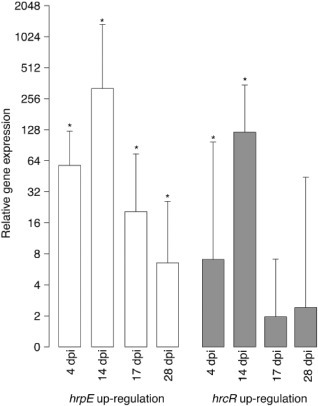
Average log2 fold change of the hrpE and hrcR genes monitored in plant material infected with the X anthomonas translucens pv. graminis strain 29 (X tg29) wild‐type compared with the X tg29 ΔhrpG mutant. The average and standard errors were obtained from four biological replicates (each with three technical replicates). Genes significantly up‐regulated in the wild‐type (P < 0.05) were determined according to the pairwise fixed reallocation randomization test (using 2000 randomizations) and are indicated with asterisks. dpi, days post‐infection.
Conclusions
Using the whole genome sequence of Xtg29, we showed that the T3SS of Xtg29 is present and required for symptom development, but is divergent from that of other sequenced Xanthomonas spp. In addition, our genealogy analyses propose that Xtg29 harbours a more ancestral state of the T3SS, and is important to infect a broad host range. Overall, our results show that the hrpG gene activates the expression of the T3SS, and that the T3SS is not primarily responsible for in planta survival. From our results and data published by others, we hypothesize that the T3SS may not be required for xylem‐colonizing pathogens, as some vascular pathogens, such as Xylella fastidiosa, Pectobacterium carotovorum and Xanthomonas albilineans (Kim et al., 2009; Pieretti et al., 2009; Van Sluys et al., 2002), do not even have an hrp gene cluster. Furthermore, Xtg gains direct access to the protoxylem lacuna, which is the area of primary multiplication (Masuch et al., 1989), and from where it infects the xylem. Therefore, it is thought that, initially, the T2SS, which secretes necessary cell wall‐degrading enzymes to gain access to the xylem vessels, is more important for initial in planta growth. Nonetheless, the T3SS of Xtg must provide some advantage to bacterial fitness, especially during later infection stages, otherwise we would not have observed significant differences at any time point after infection.
Experimental Procedures
Bacterial isolates and cultivation conditions
The bacterial isolates and plasmids used in this study are listed in Table S2 (see Supporting Information). Xtg29 has been characterized previously for virulence on different genotypes and cultivars (Kölliker et al., 2006; Wichmann et al., 2011). Xtg was grown either in Circle Grow (CG) broth (Molecular Probes, Eugene, OR, USA) or on CG and GYC (glucose, yeast extract, CaCO3) plates containing 1.5% agar at 28 °C. Escherichia coli cells were cultivated in Luria–Bertani (LB) broth or on LB plates at 37 °C. Antibiotics were used in E. coli cultures at the following concentrations: 50 μg/mL ampicillin, 50 μg/mL kanamycin and 25 μg/mL streptomycin. We prepared electrocompetent Xtg29 cells and conducted electroporation using the protocol described in Oshiro et al. (2006). Sucrose (5%) was added to the medium when selecting for the second crossing‐over event.
Sequencing, genome assembly and gap closure
Genomic DNA of isolate Xtg29 (Kölliker et al., 2006) was extracted from 30 mL of bacterial culture grown at 28 °C in CG broth (Molecular Probes) using the cetyltrimethylammonium bromide (CTAB) method (Ausubel et al., 1987). The DNA was nebulized to obtain random fragments with an average size of 3 kb. The GS Titanium Library Paired End Adaptors Kit (Roche, Mannheim, Germany) was employed to generate a 3‐K Paired End fragment library. After titration with a GS Titanium SV emPCR Kit (Lib‐L) v2 (Roche), an emulsion polymerase chain reaction (PCR) was carried out with the GS Titanium LV emPCR Kit (Lib‐L) v2 (Roche). Samples were analysed using a GS Titanium Sequencing Kit XLR70t and the GS Titanium PicoTiterPlate Kit 70x75 (both from Roche) by means of a 454 Genome Sequencer FLX System (Roche). Sequencing reads were assembled using the GS de novo Assembler software (Newbler; release 2.5.3, 454 Life Sciences, Roche Corporation). Relative coverages of individual contigs were calculated as described previously for another bacterium with high G + C content (Schwientek et al., 2012).
Gap closure was performed using the BigDye® Terminator v3.1 kit (Applied Biosystems, Foster City, CA, USA). A sequencing premix of 12 μL containing 3 μg genomic DNA, 833 mm betaine and 1 pmol of primer was prepared and denatured at 98 °C for 5 min (Kieleczawa, 2006); 8 μL of BigDye® Terminator v3.1 ready reaction mix (Applied Biosystems) was added and cycle sequencing was performed using an initial step of 95 °C for 5 min, followed by 99 cycles of 95 °C for 5 min, 50–55 °C for 20 s and 60 °C for 4 min. The samples were purified using ethanol/ethylenediaminetetraacetic acid (EDTA) precipitation, and analysed with an ABI PRISM® 3130xl Genetic Analyzer (Applied Biosystems).
Genome analysis and annotation
Functional annotation of the genome was achieved comparing several software tools employed for previous Xanthomonas genome projects. The gene predictor Prodigal (Hyatt et al., 2010) outperformed Glimmer 3.02 (Delcher et al., 2007), Gismo (Krause et al., 2007) and Reganor (Linke et al., 2006), the latter implementing combined activity of Glimmer and Critica in terms of identifying translational start sites (data not shown). Hence, Prodigal was used to predict CDSs that received locus tags with the prefix XTG29. Functional information was obtained for the CDSs by means of the GenDB Metanor pipeline (Meyer et al., 2003) for Gram‐negative bacteria. Functional annotation was copied from orthologous genes of the strain Xcc B100 for which the annotation of several CDSs with metabolic functions has been updated recently (Schatschneider et al., 2011). Genome comparisons were performed using edgar (Blom et al., 2009) and are made available by means of the public edgar project ‘Xanthomonas translucens graminis 29’ at http://edgar.cebitec.uni‐bielefeld.de/. DNA and protein sequences were compared using blastn, blastx or blastp (Altschul et al., 1990). Predicted Xtg29 amino acid sequences of T3SS genes were compared after ClustalW alignment using the resulting identity matrices (Hall, 1999). T3Es were found using edgar software, blastp and tblastn programs against all proteins and the nucleotide sequence of Xtg29, and searching the whole genome sequence for PIP box sequences, i.e. TTCGB‐N15‐TTCGB, where B is any base other than adenine (Fenselau and Bonas, 1995).
Cloning of plasmids, plasmid isolations and PCR conditions
Plasmid isolations were performed with the PureYield™ Plasmid Miniprep System (Promega, Madison, WI, USA). Restriction enzymes were used according to the manufacturer's recommendations (New England Biolabs, Ipswich, MA, USA). Cloning reactions were performed using the Dephos and Ligation kit (Roche, Penzberg, Germany). PCRs were conducted in 20‐μL volumes using Hotstar DNA Polymerase (Qiagen, Hilden, Germany) or Phusion (Qiagen), depending on the required proofreading activity. The PCR conditions were as follows: initial denaturation at 94 °C for 15 min, followed by 35 cycles of 94 °C for 30 s, 50–60 °C annealing for 40 s, 72 °C extension for 1 min/kb and a final extension of 7 min at 72 °C. Constructs were verified by sequencing plasmid DNA using gene‐specific or M13 primers. Sequencing reactions were performed using 20 ng of DNA.
The ΔhrpG, ΔhrcR and ΔhrpE fragments used for site‐directed mutation were generated using Soeing PCR (Horton, 1995) by connecting the PCR fragments amplified from the corresponding flanking regions with primers containing the sequence of a SalI or BamHI restriction site at the 5′ end of each primer. PCR fragments were subcloned into the pCR®4Blunt‐TOPO® vector (Invitrogen, Carlsbad, CA, USA). The ΔhrpG, ΔhrcR and ΔhrpE fragments were cut from the pCR®4Blunt‐TOPO® vector and ligated into the suicide vector pKNG101 (Kaniga et al., 1991). Control and selection of single and double crossing‐over events were performed using PCR on single colonies and the primers listed in Table S3 (see Supporting Information). For the complementation of the ΔhrpG mutant, the GFPuv gene of the plasmid pDSK‐GFPuv (Wang et al., 2007) was excised using EcoRI and PstI, and replaced with the open reading frame (ORF) and the promoter region (321 bp upstream) of hrpG.
Generation of rifampicin‐resistant Xtg strains
To selectively re‐isolate Xtg29 and Xtg29 ΔhrpG, Xtg29 ΔhrpE and Xtg29 ΔhrcR mutants from plant material, rifampicin resistance was induced. The rifampicin concentration was increased continuously in CG broth from an initial value of 10 μg/mL over intervals of 30, 50, 100 and 150 μg/mL, to a final concentration of 300 μg/mL, in a rotary shaker at 28 °C and 150 rpm. One millilitre of bacterial culture was used to inoculate 4 mL of CG medium supplemented with the required amount of rifampicin. Cells were grown for 24 h until the rifampicin concentration had increased.
Screening for Xtg symptoms and in planta growth
To monitor Xtg symptoms and in planta multiplication, the highly susceptible genotype LmK‐01 (Wichmann et al., 2011) was used. Inoculation was performed by cutting plants with scissors dipped into a bacterial suspension [optical density at 600 nm (OD600) = 0.6]. Plants were kept at 16 h light per day at 19 °C/23 °C (average day/night temperature). The assessment of bacterial wilt symptoms was performed using four replications per genotype × treatment combination. Scoring for bacterial wilt symptoms was performed at 7, 14, 21 and 28 dpi according to a scale ranging from completely healthy (1) to dead (9), with intervals as described in Wichmann et al. (2011). A negative control treatment consisted of cutting the plants without inoculum.
In planta growth was determined by counting the colony‐forming units (CFU) per gram of fresh plant material, including leaves and tillers (2 cm above the soil). To ensure exhaustive sampling, two preliminary experiments using the ΔhrpG mutant and the Xtg29 wild‐type were carried out (data not shown). As highly similar results were obtained in these two preliminary experiments, the four most meaningful time points after infection were monitored for all resulting mutants: 6 hpi and 4, 7 and 14 dpi. Experiments were performed using three or four biological replicates. Plant surface sterilization was performed using a 1% Chloramine‐T solution (Honeywell Riedel de‐Haën, Seelze, Germany). Serial dilutions were prepared on CG (Molecular Probes) plates supplemented with 300 μg/mL of rifampicin (AppliChem, Darmstadt, Germany). Bacterial cell counts per gram of fresh weight plant material were determined after incubation of the plates at 28 °C for 7 days. In order to verify the identity of the isolated bacteria, PCR using primers targeting the flanking regions of the corresponding mutation (Table S3) was conducted on 10 different colonies per treatment and time point.
Gene expression analyses of T3SS genes in Xtg29 and Xtg29 ΔhrpG
To quantify hrpE and hrcR expression in planta, plant material was sampled at 4, 14, 17 and 28 dpi with Xtg29 and the ΔhrpG mutant, and four biological and three technical replicates were performed per treatment and time point. The harvested plant leaves were ground in liquid nitrogen and 200 mg of plant material were used for total nucleic acid extraction. This was performed using a modified hot phenol extraction: 1 mL of neutral phenol and 500 μL of RNase‐free water were added to the plant material and incubated at 65 °C for 6 min. After cooling and centrifugation, the aqueous phase was transferred to a new tube and an equal volume of phenol–chloroform–isoamylalcohol (25 : 24 : 1) mix was added. After two extractions using an equal volume of chloroform, the nucleic acids were precipitated at −70 °C overnight using 100% ethanol. After washing once with 70% ethanol, pellets were resuspended in 200 μL of RNase‐free water. After this step, RNA and DNA were treated separately. RNA purification was performed using the RNeasy MinElute Cleanup Kit (Qiagen) including DNase treatment. For DNA purification, RNase A was added to the samples and incubated for 30 min at 37 °C. After RNase inactivation at 70 °C for 10 min, the sample was purified using the NucleoSpin Extract II kit (Macherey‐Nagel, Düren, Germany).
Reverse transcription was performed with 5 μL of purified RNA (1 μg/μL) using gene‐specific primers of the hrpE and hrcR genes (Table S3) and Superscript II (Invitrogen), according to the manufacturer's recommendations. PCR amplifications of DNA and cDNA were performed using hrpE and hrcR primers in 20‐μL volumes using 2 μL of cDNA or DNA. Quantitative real‐time PCR was performed using SsoFast EvaGreen® Supermix (Bio‐Rad, Hercules, CA, USA). The thermal cycling conditions were as follows: initial denaturation step at 98 °C for 3 min, 45 cycles of 5 s at 98 °C and 10 s at 60 °C. The specificity of the primer pairs was verified by melting curve analysis. The analysis of relative gene expression was performed using REST® software (Qiagen).
Data analyses and statistics
Geneoius software version 5.5.3 was used for protein sequence comparison and genealogy analyses. We used ClustalW (Larkin et al., 2007) for sequence alignments, and the phylogenetic trees were inferred using maximum likelihood. Bootstrap values were calculated on the basis of 100 replications. Disease symptoms were described using the AUDPC values (Wichmann et al., 2011). In order to compare AUDPC values resulting from infection with the T3SS mutants or the wild‐type Xtg strain, multiple t‐tests using Bonferroni family wise error rate correction were performed. The same procedure was used when testing for significant differences in CFU values in planta after infection with the T3SS mutants or the wild‐type Xtg strain. P < 0.05 was considered to be significant. These statistical analyses were all performed in R (The R Development Core Team, 2008) using the packages: stats, graphics and coin (Hothorn et al., 2006).
Nucleotide sequences are accessible through DDBJ/EMBL/GenBank under the accession numbers ANGG01000001:ANGG01000788
Supporting information
Fig. S1 Phylogenetic trees inferred with maximum likelihood for the separate alignment of eight different Hrc proteins (HrcC, HrcJ, HrcN, HrcR, HrcS, HrcT, HrcU and HrcV) from different Xanthomonas spp., Ralstonia solanacearum, Collimonas fungivorans, Burkholderia spp. and Acidovorax avenae ssp. avenae. Pseudomonas syringae pv. tomato strain DC3000 was used as an outgroup. Numbers represent bootstrap values for 100 replications.
Fig. S2 Symptoms on Lolium multiflorum seedlings at 7 days post‐infection with different Xanthomonas translucens pv. graminis type III secretion system mutants (i.e. ΔhrcR, ΔhrpE, ΔhrpG, ΔhrpG/ΔhrcR, ΔhrpG + hrpG) and the wild‐type or negative control treatment, i.e. cutting the plants with scissors dipped in sterile physiological sodium chloride solution. Symptoms are primarily characterized by increased wilting of the entire seedling when compared with the control treatment.
Table S1 rax gene homologues from Xanthomonas oryzae pv. oryzae found in the genome of Xanthomonas translucens pv. graminis 29.
Table S2 Strains and plasmids used in this study.
Table S3 Primer sequences and their application used in this study. Underlined sequences indicate recognition sites for restriction enzymes used for cloning.
Acknowledgements
This research was funded by the Swiss National Science Foundation (SNF) Project (3100A0‐112582). We would like to thank Sabrina Kuhnen, Philipp Streckeisen, Marius Liesch, Bernhard Mueller‐Hug and Luisa Last (Agroscope Reckenholz‐Tänikon, Ettenhausen, Switzerland), as well as Anika Hegemann (Bielefeld University, Bielefeld, Germany), for technical assistance. The authors also thank Philipp Engel (Yale University, New Haven, CT, USA) for assistance with the phylogenetic analyses and critical review of the manuscript.
References
- Alegria, M.C. , Souza, D.P. , Andrade, M.O. , Docena, C. , Khater, L. , Ramos, C.H.I. , da Silva, A.C.R. and Farah, C.S. (2005) Identification of new protein–protein interactions involving the products of the chromosome‐ and plasmid‐encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri . J. Bacteriol. 187, 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Arnold, D.L. , Pitman, A. and Jackson, R.W. (2003) Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 4, 407–420. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. , Brent, R. , Kinston, R. , Moore, D. , Seidman, J. , Smith, J. and Struhl, K. (1987) Current Protocols in Molecular Biology. New York: J. Wiley & Sons. [Google Scholar]
- Becker, A. , Katzen, F. , Pühler, A. and Ielpi, L. (1998) Xanthan gum biosynthesis and application: a biochemical/genetic perspective. Appl. Microbiol. Biotechnol. 50, 145–152. [DOI] [PubMed] [Google Scholar]
- Blom, J. , Albaum, S.P. , Doppmeier, D. , Pühler, A. , Vorhölter, F.J. , Zakrzewski, M. and Goesmann, A. (2009) EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics, 10, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Koebnik, R. , Lu, H. , Furutani, A. , Angiuoli, S.V. , Patil, P.B. , Van Sluys, M.‐A. , Ryan, R.P. , Meyer, D.F. , Han, S.‐W. , Aparna, G. , Rajaram, M. , Delcher, A.L. , Phillippy, A.M. , Puiu, D. , Schatz, M.C. , Shumway, M. , Sommer, D.D. , Trapnell, C. , Benahmed, F. , Dimitrov, G. , Madupu, R. , Radune, D. , Sullivan, S. , Jha, G. , Ishihara, H. , Lee, S.‐W. , Pandey, A. , Sharma, V. , Sriariyanun, M. , Szurek, B. , Vera‐Cruz, C.M. , Dorman, K.S. , Ronald, P.C. , Verdier, V. , Dow, J.M. , Sonti, R.V. , Tsuge, S. , Brendel, V.P. , Rabinowicz, P.D. , Leach, J.E. , White, F.F. and Salzberg, S.L. (2011) Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193, 5450–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer, E.C. (1999) Capturing heterosis in forage crop cultivar development. Crop Sci. 39, 943–954. [Google Scholar]
- Büttner, D. and Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and He, S.Y. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. , Nennstiel, D. , Klusener, B. and Bonas, U. (2002) Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 184, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. , Noël, L. , Stuttmann, J. and Bonas, U. (2007) Characterization of the nonconserved hpaB‐hrpF region in the hrp pathogenicity island from Xanthomonas campestris pv. vesicatoria . Mol. Plant–Microbe Interact. 20, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Cho, H.J. , Park, Y.J. , Noh, T.H. , Kim, Y.T. , Kim, J.G. , Song, E.S. , Lee, D.H. and Lee, B.M. (2008) Molecular analysis of the hrp gene cluster in Xanthomonas oryzae pathovar oryzae KACC10859. Microb. Pathog. 44, 473–483. [DOI] [PubMed] [Google Scholar]
- Da Silva, A.C.R. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M.C. , Do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E.A. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Clapina, L.P. , Cicarelli, R.M.B. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J.S. , Ferreira, R.C.C. , Ferro, M.I.T. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G.M. , Lemos, M.V.F. , Locali, E.C. , Machado, M.A. , Madeira, A. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F.M. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T.M. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A.D. , Silva, C. , De Souza, R.F. , Spinola, L.A.F. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I.D. , Dos Santos, M.T. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Darsonval, A. , Darrasse, A. , Meyer, D. , Demarty, M. , Durand, K. , Bureau, C. , Manceau, C. and Jacques, M.A. (2008) The type III secretion system of Xanthomonas fuscans subsp fuscans is involved in the phyllosphere colonization process and in transmission to seeds of susceptible beans. Appl. Environ. Microbiol. 74, 2669–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher, A.L. , Bratke, K.A. , Powers, E.C. and Salzberg, S.L. (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics, 23, 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt, U. , Hochhut, B. , Hentschel, U. and Hacker, J. (2004) Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. [DOI] [PubMed] [Google Scholar]
- Egli, T. and Schmidt, D. (1982) Pathogenic variation among the causal agents of bacterial wilt of forage grasses. J. Phytopathol. 104, 138–150. [Google Scholar]
- Egli, T. , Goto, M. and Schmidt, D. (1975) Bacterial wilt, a new forage grass disease. J. Phytopathol. 82, 111–121. [Google Scholar]
- Fenselau, S. and Bonas, U. (1995) Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol. Plant–Microbe Interact. 8, 845–854. [DOI] [PubMed] [Google Scholar]
- Furutani, A. , Takaoka, M. , Sanada, H. , Noguchi, Y. , Oku, T. , Tsuno, K. , Ochiai, H. and Tsuge, S. (2009) Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 22, 96–106. [DOI] [PubMed] [Google Scholar]
- Gophna, U. , Ron, E.Z. and Graur, D. (2003) Bacterial type III secretion systems are ancient and evolved by multiple horizontal‐transfer events. Gene, 312, 151–163. [DOI] [PubMed] [Google Scholar]
- Gurlebeck, D. , Thieme, F. and Bonas, U. (2006) Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163, 233–255. [DOI] [PubMed] [Google Scholar]
- Hajri, A. , Brin, C. , Hunault, G. , Lardeux, F. , Lemaire, C. , Manceau, C. , Boureau, T. and Poussier, S. (2009) A ‘repertoire for repertoire’ hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas . PLoS ONE, 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.A. (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hauben, L. , Vauterin, L. , Swings, J. and Moore, E.R.B. (1997) Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int. J. Syst. Bacteriol. 47, 328–335. [DOI] [PubMed] [Google Scholar]
- Horton, R.M. (1995) PCR‐mediated recombination and mutagenesis. Mol. Biotechnol. 3, 93–99. [DOI] [PubMed] [Google Scholar]
- Hothorn, T. , Hornik, K. , Van de Wiel, M.A. and Zeileis, A. (2006) A lego system for conditional inference. Am. Stat. 60, 257–263. [Google Scholar]
- Hyatt, D. , Chen, G.‐L. , LoCascio, P.F. , Land, M.L. , Larimer, F.W. and Hauser, L.J. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, B. , Xu, R.Q. , Li, X.Z. , Wei, H.Y. , Bai, F. , Hu, X. , He, Y.Q. and Tang, J.L. (2006) Construction and characterization of a hrpG mutant rendering constitutive expression of hrp genes in Xanthomonas campestris pv. campestris . Prog. Nat. Sci. 16, 480–485. [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaniga, K. , Delor, I. and Cornelis, G.R. (1991) A wide‐host‐range suicide vector for improving reverse genetics in Gram‐negative bacteria—inactivation of the blaA gene of Yersinia enterocolitica . Gene, 109, 137–141. [DOI] [PubMed] [Google Scholar]
- Kay, S. and Bonas, U. (2009) How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12, 37–43. [DOI] [PubMed] [Google Scholar]
- Kieleczawa, J. (2006) Fundamentals of sequencing of difficult templates—an overview. J. Biomol. Tech., 17, 207–217. [PMC free article] [PubMed] [Google Scholar]
- Kim, J.G. , Li, X.Y. , Roden, J.A. , Taylor, K.W. , Aakre, C.D. , Su, B. , Lalonde, S. , Kirik, A. , Chen, Y.H. , Baranage, G. , McLane, H. , Martin, G.B. and Mudgett, M.B. (2009) Xanthomonas T3S effector XopN suppresses PAMP‐triggered immunity and interacts with a tomato atypical receptor‐like kinase and TFT1. Plant Cell, 21, 1305–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölliker, R. , Kraehenbuehl, R. , Boller, B. and Widmer, F. (2006) Genetic diversity and pathogenicity of the grass pathogen Xanthomonas translucens pv. graminis . Syst. Appl. Microbiol. 29, 109–119. [DOI] [PubMed] [Google Scholar]
- Krause, L. , McHardy, A.C. , Nattkemper, T.W. , Pühler, A. , Stoye, J. and Meyer, F. (2007) GISMO—gene identification using a support vector machine for ORF classification. Nucleic Acids Res. 35, 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. , Wallace, I.M. , Wilm, A. , Lopez, R. , Thompson, J.D. , Gibson, T.J. and Higgins, D.G. (2007) ClustalW and ClustalX version 2. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee, B.M. , Park, Y.J. , Park, D.S. , Kang, H.W. , Kim, J.G. , Song, E.S. , Park, I.C. , Yoon, U.H. , Hahn, J.H. , Koo, B.S. , Lee, G.B. , Kim, H. , Park, H.S. , Yoon, K.O. , Kim, J.H. , Jung, C. , Koh, N.H. , Seo, J.S. and Go, S.J. (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyns, F. (1993) Xanthomonas campestris pv. graminis: cause of bacterial wilt of forage grasses In: Xanthomonas (Swings J.G. and Civerolo E.L., eds), pp. 55–57. London: Chapman & Hall. [Google Scholar]
- Linke, B. , McHardy, A.C. , Neuweger, H. , Krause, L. and Meyer, F. (2006) REGANOR: a gene prediction server for prokaryotic genomes and a database of high quality gene predictions for prokaryotes. Appl. Bioinformatics, 5, 193–198. [DOI] [PubMed] [Google Scholar]
- Masuch, G. , Schoene, K. and Paul, V.H. (1989) Histological investigations on the pathogenesis of Xanthomonas campestris pv. graminis to Lolium multiflorum . EPPO Bull. 19, 73–80. [Google Scholar]
- Metz, M. , Dahlbeck, D. , Morales, C.Q. , Al Sady, B. , Clark, E.T. and Staskawicz, B.J. (2005) The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana . Plant J. 41, 801–814. [DOI] [PubMed] [Google Scholar]
- Meyer, F. , Goesmann, A. , McHardy, A.C. , Bartels, D. , Bekel, T. , Clausen, J. , Kalinowski, J. , Linke, B. , Rupp, O. , Giegerich, R. and Pühler, A. (2003) GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31, 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, V.V. (2001) Interactions between Xanthomonas campestris pv. graminis strains and meadow fescue and Italian rye grass cultivars. Plant Dis. 85, 538–542. [DOI] [PubMed] [Google Scholar]
- Moreira, L.M. , De Souza, R.F. , Digiampietri, L.A. , Da Silva, A.C.R. and Setubal, J.C. (2005) Comparative analyses of Xanthomonas and Xylella complete genomes. Omics, 9, 43–76. [DOI] [PubMed] [Google Scholar]
- Moreira, L.M. , Almeida, N.F. , Potnis, N. , Digiampietri, L.A. , Adi, S.S. , Bortolossi, J.C. , da Silva, A.C. , da Silva, A.M. , de Moraes, F.E. , de Oliveira, J.C. , de Souza, R.F. , Facincani, A.P. , Ferraz, A.L. , Ferro, M.I. , Furlan, L.R. , Gimenez, D.F. , Jones, J.B. , Kitajima, E.W. , Laia, M.L. , Leite, R.P. , Nishiyama, M.Y. , Neto, J.R. , Nociti, L.A. , Norman, D.J. , Ostroski, E.H. , Pereira, H.A. , Staskawicz, B.J. , Tezza, R.I. , Ferro, J.A. , Vinatzer, B.A. and Setubal, J.C. (2010) Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii . BMC Genomics, 11, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou, M.J. and Bogdanove, A.J. (2009) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B. , Chesnokova, O. , Dahlbeck, D. , Clark, E.T. , Rossier, O. , Bonas, U. and Staskawicz, B.J. (2000) Molecular signals required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl. Acad. Sci. USA, 97, 13 324–13 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S. , Hao, Y.H. and Orth, K. (2007) A newly discovered post‐translational modification—the acetylation of serine and threonine residues. Trends Biochem. Sci. 32, 210–216. [DOI] [PubMed] [Google Scholar]
- Noël, L. , Thieme, F. , Nennstiel, D. and Bonas, U. (2001) cDNA analysis unravels a genome‐wide HrpG‐regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria . Mol. Microbiol. 41, 1271–1281. [DOI] [PubMed] [Google Scholar]
- Noël, L. , Thieme, F. , Nennstiel, D. and Bonas, U. (2002) Two novel type III‐secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184, 1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël, L. , Thieme, F. , Gabler, J. , Büttner, D. and Bonas, U. (2003) XopC and XopJ, two novel type III effector proteins from Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 185, 7092–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai, H. , Inoue, V. , Takeya, M. , Sasaki, A. and Kaku, H. (2005) Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Japan Agric. Research Quart. 39, 275–287. [Google Scholar]
- Oshiro, E.E. , Nepomuceno, R.S.L. , Faria, J.B. , Ferreira, L.C.S. and Ferreira, R.C.C. (2006) Site‐directed gene replacement of the phytopathogen Xanthomonas axonopodis pv. citri . J. Microbiol. Methods, 65, 171–179. [DOI] [PubMed] [Google Scholar]
- Parkinson, N. , Cowie, C. , Heeney, J. and Stead, D. (2009) Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int. J. Syst. Evol. Microbiol. 59, 264–274. [DOI] [PubMed] [Google Scholar]
- Patil, P.B. , Bogdanove, A.J. and Sonti, R.V. (2007) The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC Evol. Biol. 7, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti, I. , Royer, M. , Barbe, V. , Carrere, S. , Koebnik, R. , Cociancich, S. , Couloux, A. , Darrasse, A. , Gouzy, J. , Jacques, M.A. , Lauber, E. , Manceau, C. , Mangenot, S. , Poussier, S. , Segurens, B. , Szurek, B. , Verdier, V. , Arlat, M. and Rott, P. (2009) The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem‐limited Xanthomonadaceae . BMC Genomics, 10, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, W. , Jia, Y.T. , Ren, S.X. , He, Y.Q. , Feng, J.X. , Lu, L.F. , Sun, Q.H. , Ying, G. , Tang, D.J. , Tang, H. , Wu, W. , Hao, P. , Wang, L.F. , Jiang, B.L. , Zeng, S.Y. , Gu, W.Y. , Lu, G. , Rong, L. , Tian, Y.C. , Yao, Z.J. , Fu, G. , Chen, B.S. , Fang, R.X. , Qiang, B.Q. , Chen, Z. , Zhao, G.P. , Tang, J.L. and He, C.Z. (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris . Genome Resi. 15, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden, J.A. , Belt, B. , Ross, J.B. , Tachibana, T. , Vargas, J. and Mudgett, M.B. (2004) A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. USA, 101, 16 624–16 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer, P. , Recht, S. and Lahaye, T. (2009a) A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl. Acad. Sci. USA, 106, 20 526–20 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer, P. , Strauss, T. , Hahn, S. , Scholze, H. , Morbitzer, R. , Grau, J. , Bonas, U. and Lahaye, T. (2009b) Recognition of AvrBs3‐like proteins is mediated by specific binding to promoters of matching pepper Bs3 Alleles. Plant Physiol. 150, 1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier, O. , Van den Ackerveken, G. and Bonas, U. (2000) HrpB2 and HrpF from Xanthomonas are type III‐secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38, 828–838. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. and Dow, J.M. (2011) Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbioli. 19, 145–152. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorhölter, F.‐J. , Potnis, N. , Jones, J.B. , Van Sluys, M.‐A. , Bogdanove, A.J. and Dow, J.M. (2011) Pathogenomics of Xanthomonas: understanding bacterium × plant interactions. Nat. Rev. Microbiol. 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thebault, P. , Whalen, M. , Wincker, P. , Levy, M. , Weissenbach, J. and Boucher, C. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Lee, S.W. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99(A). BMC Genomics, 9, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatschneider, S. , Vorhölter, F.‐J. , Rueckert, C. , Becker, A. , Eisenreich, W. , Pühler, A. and Niehaus, K. (2011) Genome‐enabled determination of amino acid biosynthesis in Xanthomonas campestris pv. campestris and identification of biosynthetic pathways for alanine, glycine, and isoleucine by (13)C‐isotopologue profiling. Mol. Genet. Genomics, 286, 247–259. [DOI] [PubMed] [Google Scholar]
- Schwientek, P. , Szczepanowski, R. , Ruckert, C. , Kalinowski, J. , Klein, A. , Selber, K. , Wehmeier, U.F. , Stoye, J. and Pühler, A. (2012) The complete genome sequence of the acarbose producer Actinoplanes sp SE50/110. BMC Genomics, 13, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C. and Yang, B. (2010) Mutagenesis of 18 type III effectors reveals virulence function of XopZ(PXO99) in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 23, 893–902. [DOI] [PubMed] [Google Scholar]
- Studholme, D.J. , Kemen, E. , MacLean, D. , Schornack, S. , Aritua, V. , Thwaites, R. , Grant, M. , Smith, J. and Jones, J.D.G. (2010) Genome‐wide sequencing data reveals virulence factors implicated in banana Xanthomonas wilt. FEMS Microbiol. Lett. 310, 182–192. [DOI] [PubMed] [Google Scholar]
- Studholme, D.J. , Wasukira, A. , Paszkiewicz, K. , Aritua, V. , Thwaites, R. , Smith, J. and Grant, M. (2011) Draft genome sequences of Xanthomonas sacchari and two banana‐associated xanthomonads reveal insights into the Xanthomonas Group 1 Clade. Gene, 2, 1050–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. , Yang, B. and White, F.F. (2005) Characterization of the hrpF pathogenicity peninsula of Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 18, 546–554. [DOI] [PubMed] [Google Scholar]
- Sun, W.X. , Liu, L.J. and Bent, A.F. (2011) Type III secretion‐dependent host defence elicitation and type III secretion‐independent growth within leaves by Xanthomonas campestris pv. campestris . Mol. Plant Pathol. 12, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords, K.M.M. , Dahlbeck, D. , Kearney, B. , Roy, M. and Staskawicz, B.J. (1996) Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2. J. Bacteriol. 178, 4661–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Development Core Team (2008) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Thieme, F. , Koebnik, R. , Bekel, T. , Berger, C. , Boch, J. , Büttner, D. , Caldana, C. , Gaigalat, L. , Goesmann, A. , Kay, S. , Kirchner, O. , Lanz, C. , Linke, B. , McHardy, A.C. , Meyer, F. , Mittenhuber, G. , Nies, D.H. , Niesbach‐Klosgen, U. , Patschkowski, T. , Ruckert, C. , Rupp, O. , Schneiker, S. , Schuster, S.C. , Vorhölter, F.‐J. , Weber, E. , Pühler, A. , Bonas, U. , Bartels, D. and Kaiser, O. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme, F. , Szczesny, R. , Urban, A. , Kirchner, O. , Hause, G. and Bonas, U. (2007) New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N‐myristoylation motif. Mol. Plant–Microbe Interact. 20, 1250–1261. [DOI] [PubMed] [Google Scholar]
- Van Sluys, M.A. , Monteiro‐Vitorello, C.B. , Camargo, L.E.A. , Menck, C.F.M. , da Silva, A.C.R. , Ferro, J.A. , Oliveira, M.C. , Setubal, J.C. , Kitajima, J.P. and Simpson, A.J. (2002) Comparative genomic analysis of plant‐associated bacteria. Annu. Rev. Phytopathol. 40, 169–189. [DOI] [PubMed] [Google Scholar]
- Vorhölter, F.J. , Thias, T. , Meyer, F. , Bekel, T. , Kaiser, O. , Pühler, A. and Niehaus, K. (2003) Comparison of two Xanthomonas campestris pathovar campestris genomes revealed differences in their gene composition. J. Biotechnol. 106, 193–202. [DOI] [PubMed] [Google Scholar]
- Vorhölter, F.J. , Schneiker, S. , Goesmann, A. , Krause, L. , Bekel, T. , Kaiser, O. , Linke, B. , Patschkowski, T. , Rueckert, C. , Schmid, J. , Sidhu, V.K. , Sieber, V. , Tauch, A. , Watt, S.A. , Weisshaar, B. , Becker, A. , Niehaus, K. and Puehler, A. (2008) The genome of Xanthomonas campestris pv. campestris B 100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 134, 33–45. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Kang, L. , Anand, A. , Lazarovits, G. and Mysore, K.S. (2007) Monitoring in planta bacterial infection at both cellular and whole‐plant levels using the green fluorescent protein variant GFPuv. New Phytol. 174, 212–223. [DOI] [PubMed] [Google Scholar]
- Wei, C.F. , Kvitko, B.H. , Shimizu, R. , Crabill, E. , Alfano, J.R. , Lin, N.C. , Martin, G.B. , Huang, H.C. and Collmer, A. (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1‐1 is able to cause disease in the model plant Nicotiana benthamiana . Plant J. 51, 32–46. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. , VandenAckerveken, G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. , Rossier, O. and Bonas, U. (1999) Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181, 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen, M.C. , Wang, J.F. , Carland, F.M. , Heiskell, M.E. , Dahlbeck, D. , Minsavage, G.V. , Jones, J.B. , Scott, J.W. , Stall, R.E. and Staskawicz, B.J. (1993) Avirulence gene avrRxv from Xanthomonas campestris pv. vesicatoria specifies resistance on tomato line Hawaii‐7998. Mol. Plant–Microbe Interact. 6, 616–627. [DOI] [PubMed] [Google Scholar]
- White, F. , Potnis, N. , Jones, J. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann, F. , Hug, B.M. , Widmer, F. , Boller, B. , Studer, B. and Kölliker, R. (2011) Phenotypic and molecular genetic characterization indicate no major race‐specific interactions between Xanthomonas translucens pv. graminis and Lolium multiflorum . Plant Pathol. 60, 314–324. [Google Scholar]
- Yoshimochi, T. , Zhang, Y. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2009) Expression of hrpG and activation of response regulator HrpG are controlled by distinct signal cascades in Ralstonia solanacearum . J. Gen. Plant Pathol. 75, 196–204. [Google Scholar]
- Zou, L.F. , Wang, X.P. , Xiang, Y. , Zhang, B. , Li, Y.R. , Xiao, Y.L. , Wang, J.S. , Walmsley, A.R. and Chen, G.Y. (2006) Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72, 6212–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic trees inferred with maximum likelihood for the separate alignment of eight different Hrc proteins (HrcC, HrcJ, HrcN, HrcR, HrcS, HrcT, HrcU and HrcV) from different Xanthomonas spp., Ralstonia solanacearum, Collimonas fungivorans, Burkholderia spp. and Acidovorax avenae ssp. avenae. Pseudomonas syringae pv. tomato strain DC3000 was used as an outgroup. Numbers represent bootstrap values for 100 replications.
Fig. S2 Symptoms on Lolium multiflorum seedlings at 7 days post‐infection with different Xanthomonas translucens pv. graminis type III secretion system mutants (i.e. ΔhrcR, ΔhrpE, ΔhrpG, ΔhrpG/ΔhrcR, ΔhrpG + hrpG) and the wild‐type or negative control treatment, i.e. cutting the plants with scissors dipped in sterile physiological sodium chloride solution. Symptoms are primarily characterized by increased wilting of the entire seedling when compared with the control treatment.
Table S1 rax gene homologues from Xanthomonas oryzae pv. oryzae found in the genome of Xanthomonas translucens pv. graminis 29.
Table S2 Strains and plasmids used in this study.
Table S3 Primer sequences and their application used in this study. Underlined sequences indicate recognition sites for restriction enzymes used for cloning.


