Summary
Autophagy is a major intracellular process for the degradation of cytosolic macromolecules and organelles in the lysosomes or vacuoles for the purposes of regulating cellular homeostasis and protein and organelle quality control. In complex metazoan organisms, autophagy is highly engaged during the immune responses through interfaces either directly with intracellular pathogens or indirectly with immune signalling molecules. Studies over the last decade or so have also revealed a number of important ways in which autophagy shapes plant innate immune responses. First, autophagy promotes defence‐associated hypersensitive cell death induced by avirulent or related pathogens, but restricts unnecessary or disease‐associated spread of cell death. This elaborate regulation of plant host cell death by autophagy is critical during plant immune responses to the types of plant pathogens that induce cell death, which include avirulent biotrophic pathogens and necrotrophic pathogens. Second, autophagy modulates defence responses regulated by salicylic acid and jasmonic acid, thereby influencing plant basal resistance to both biotrophic and necrotrophic pathogens. Third, there is an emerging role of autophagy in virus‐induced RNA silencing, either as an antiviral collaborator for targeted degradation of viral RNA silencing suppressors or an accomplice of viral RNA silencing suppressors for targeted degradation of key components of plant cellular RNA silencing machinery. In this review, we summarize this important progress and discuss the potential significance of the perplexing role of autophagy in plant innate immunity.
Keywords: autophagy, cell death, innate immunity, jasmonic acid, RNA silencing, salicylic acid
Introduction
Autophagy is an evolutionarily conserved process by which cytoplasmic constituents, including proteins and organelles, are delivered to lysosomes or vacuoles for degradation (Boya et al., 2013). Autophagy can occur through at least three pathways, known as macroautophagy, chaperone‐mediated autophagy and microautophagy. Among these autophagic pathways, macroautophagy has been the most extensively analysed and is usually referred to simply as autophagy (Boya et al., 2013). Basal‐level autophagy is usually very low under normal conditions, but can be induced in response to a variety of stimuli, and induced autophagy plays a critical role in achieving cell homeostasis under starvation, pathogen infection and other environmental stresses (Boya et al., 2013). In the classical induction pathway, autophagy is initiated by the formation of an isolation membrane or ‘phagophore’, which can grow to engulf cytoplasmic components and close around the sequestered cargo, resulting in the formation of a double‐membrane autophagosome (Boya et al., 2013; He and Klionsky, 2009). The autophagosome formed fuses with lysosomes or vacuoles so that its inner cargo can be degraded by a series of lysosomal/vacuolar hydrolases (Boya et al., 2013; He and Klionsky, 2009). In yeast, more than 30 autophagy‐related (ATG) genes have been identified, and the products of these ATG genes often form functional groups involved in several physiologically continuous, but mechanistically distinct, steps in the autophagy process. Well‐characterized functional groups of ATG proteins include the ATG1–ATG13–ATG17 scaffold, formed on activation of the ATG1 kinase activity during the induction of autophagy, the class III phosphatidylinositol 3‐kinase (PtdIns3K)–ATG14–ATG6 (Beclin 1) complex, required for the nucleation and assembly of the initial phagophore membrane, and two interrelated ubiquitin‐like conjugation systems, ATG12–ATG5–ATG16 and ATG8–PE (phosphatidylethanolamine), for the regulation of membrane elongation and expansion of the forming autophagosomes (Boya et al., 2013; He and Klionsky, 2009).
Recent developments in metazoan organisms have revealed a crucial role for the autophagy process in host innate immune mechanisms. In these organisms, the recognition of pathogens, such as viruses, is mediated by membrane‐bound or cytosolic pattern recognition receptors (PRRs), which detect pathogen‐associated molecular patterns (PAMPs) and transduce the signals to activate innate immune responses (Deretic, 2012). Autophagy is induced by different families of PRRs, including Toll‐like receptors (TLRs), NOD‐like receptors and the double‐stranded (ds)RNA‐binding protein PKR (Deretic, 2012). Activated autophagy can target directly intracellular pathogens, such as bacteria and viruses, for degradation in lysosomes/vacuoles in a selective form, termed xenophagy (Deretic, 2012; Levine et al., 2011; Shoji‐Kawata and Levine, 2009). Autophagy can also control innate immunity indirectly. On activation by TLRs on recognition of PAMPs, for example, autophagy can play a role in the production and delivery of cytosolic PAMPs to endosomic TLRs, thereby facilitating the interaction between PAMPs and TLRs (Deretic, 2012; Levine et al., 2011; Shoji‐Kawata and Levine, 2009). In addition, autophagy affects the production and secretion of cytokines, such as interferon and interleukin molecules, important effector components in immune responses (Deretic, 2012; Levine et al., 2011; Shoji‐Kawata and Levine, 2009). In many of these analysed organisms, including Drosophila, Dictyostelium, Caenorhabditis and mice, mutations of autophagy genes increase host susceptibility to bacterial, viral and protozoan pathogens (Deretic, 2012; Levine et al., 2011; Shoji‐Kawata and Levine, 2009). The critical role of autophagy in host innate immunity is also underscored by the fact that pathogens have evolved mechanisms to avoid death by autophagy pathways through the evasion of autophagic recognition, inhibition of autophagy, modulation of autophagosomal maturation and modification of the overall state of autophagy (Deretic, 2012; Levine et al., 2011; Shoji‐Kawata and Levine, 2009).
Enormous progress has been made over the last two decades or so in our understanding of the molecular basis of plant innate immune responses, particularly to biotrophic pathogens, which parasitize on plant living tissue. Plants respond to biotrophic pathogens using two innate immune systems: PAMP‐triggered immunity (PTI) and effector‐triggered immunity (ETI) (Jones and Dangl, 2006). PTI is activated by PAMPs such as bacterial flagellin. To overcome PTI, pathogens deliver virulence factors or effectors to plant cells to inhibit immune responses. Some of the effectors can be recognized by plant resistance (R) proteins and activate ETI (Jones and Dangl, 2006). ETI is often manifested as a hypersensitive response (HR) associated with rapid cell death, the production of reactive oxygen species (ROS) and salicylic acid (SA), and the expression of defence‐related genes (Jones and Dangl, 2006). Relatively less is known about plant innate immunity against necrotrophic pathogens, which actively kill host cells before colonizing them (van Kan, 2006). Unlike gene‐for‐gene resistance to biotrophic pathogens, plant resistance to necrotrophic pathogens is often polygenic (Brodersen et al., 2006; Mengiste et al., 2003; Nandi et al., 2005; Veronese et al., 2006), and HR is promoted by necrotrophic pathogens as it facilitates infection (Govrin and Levine, 2000). In Arabidopsis, resistance to necrotrophic pathogens depends on jasmonate (JA) and ethylene signalling, and the synthesis of the phytoalexin camalexin (Ferrari et al., 2003; Penninckx et al., 1996, 1998; Thomma et al., 1998, 1999).
Over the past two decades, many ATG genes involved in the core process of autophagy have been identified and functionally analysed in plants (Chung et al., 2009; Liu et al., 2005; Shin et al., 2009; Su et al., 2006). These studies have shown that autophagy plays an important role in the plant response to nutrient starvation and a spectrum of abiotic stresses, including heat, salt, drought and ROS (Bassham, 2007; Liu et al., 2009, 2012; Xiong et al., 2007a, b; Zhou et al., 2013). Using well‐established molecular and genetic approaches, including mutants and transgenic silenced plants, a number of groups have also examined the roles of autophagy in plant innate immunity. Through the examination of autophagy‐deficient mutants for responses to virulent and avirulent biotrophic pathogens, several studies have discovered that autophagy shapes several important facets of plant innate immunity, including pathogen‐induced programmed cell death (PCD), in a complex pattern influenced by a multitude of other factors, such as plant age (Hof et al., 2013; Hofius et al., 2009; Kabbage et al., 2013; Liu et al., 2005; Patel and Dinesh‐Kumar, 2008; Yoshimoto et al., 2009). However, there is strong evidence for a critical and positive role of autophagy in plant resistance to necrotrophic pathogens (Kabbage et al., 2013; Katsiarimpa et al., 2013; Lai et al., 2011; Lenz et al., 2011a, b). More recent studies have also uncovered the involvement of autophagy in plant antiviral RNA silencing, either deployed as a defence system against viral RNA silencing suppressors or subverted as a counter‐defence mechanism by invading viruses for the inactivation of components of the plant RNA silencing machinery (Derrien et al., 2012; Nakahara et al., 2012; Tadamura et al., 2012). In this review, we focus on the role of autophagy in plant interactions with various types of microbial pathogen, and discuss its complex crosstalk with other factors and pathways in controlling the outcome of plant–pathogen interactions.
Autophagy in Pathogen‐Induced Hypersensitive Cell Death
A number of studies have analysed the role of autophagy in plant HR triggered by the infection of avirulent strains of pathogens with seemingly conflicting results. Liu et al. (2005) first analysed this process using the tobacco–Tobacco mosaic virus (TMV) system. TMV infection of tobacco plants containing the N resistance gene induces rapid tobacco immune responses, including hypersensitive cell death, increased production of SA and the synthesis of PATHOGENESIS‐RELATED (PR) proteins at the infection sites, which limit viral replication, restrict viral spread and prevent disease development in uninfected parts of the plant (Erickson et al., 1999). From a high‐throughput, virus‐induced gene silencing (VIGS) screen in Nicotiana benthamiana, Liu et al. (2005) discovered that the silencing of the tobacco BECLIN1/ATG6 gene resulted in the spread of cell death to uninfected tissues and leaves distal to the TMV‐inoculated leaf. To confirm that TMV‐induced cell death in BECLIN1‐silenced plants spread beyond infected areas, they used Agrobacterium‐mediated transient expression of a TMV replicase protein fragment (TMV‐p50), the N gene elicitor, and found that elicitor‐induced cell death initiated in the agro‐infiltrated area and spread to the entire leaf (Liu et al., 2005). A number of other non‐spreading inducers, including Agrobacterium‐mediated transient expression of the INF1 elicitor gene from Phytophthora infestans, or the co‐expression of the bacterial resistance gene Pto with its cognate Avr gene AvrPto and the fungal resistance gene Cf9 with its cognate Avr gene Avr9, all led to spread of cell death from the infiltrated area to the entire leaf or even to upper uninfected leaves (Liu et al., 2005). Silencing of other autophagy genes, including PI3K/Vps34, ATG3 and ATG7, in N‐containing tobacco plants produced a similar phenotype of uncontrolled cell death in response to TMV infection (Liu et al., 2005). Further analysis revealed that TMV infection of resistant tobacco plants induced autophagy not only in infected cells, but also in distal uninfected areas (Liu et al., 2005). These results indicated that, during pathogen‐induced HR, autophagy is induced to limit the spread of hypersensitive cell death beyond infected cells. This anti‐death role of BECLIN1/ATG6 was also investigated in Arabidopsis using ATG6 antisense plants (in the Col‐0 ecotype). When infected with the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (PstDC3000) containing the AvrRpm1 effector protein, pathogen‐induced cell death was restricted to the infiltrated areas in wild‐type plants, but expanded beyond the infiltrated areas to the entire leaf in the ATG6 antisense plants (Patel and Dinesh‐Kumar, 2008). Thus, ATG6 is also required for the restriction of pathogen‐induced cell death in Arabidopsis.
Interestingly, a subsequent study using Arabidopsis atg7 and atg9 mutants (in the Ws‐0 ecotype) showed no spread of pathogen‐induced cell death in the mutant plants after inoculation with PstDC3000(AvrRps4) or PstDC3000(AvrRpm1) (Hofius et al., 2009). Indeed, the onset of cell death in the autophagy‐defective mutants caused by infection of PstDC3000(AvrRps4) was delayed by 4–5 h, based on electrolyte leakage, when compared with that in wild‐type plants (Hofius et al., 2009). Suppression of cell death was also observed in the atg7 and atg9 mutants after inoculation with an avirulent isolate (Noco2) of the oomycete Hyaloperonospora arabidopsidis (Hofius et al., 2009). This suppression of cell death by autophagy appears to be specific to plants containing Toll/Interleukin‐1 (TIR)‐type immune receptors, which activate ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1)‐dependent signalling and are correlated with induced autophagy in an EDS1‐dependent manner. By contrast, little induction of autophagy was observed in Arabidopsis plants on infection with PstDC3000 containing AvrRpt2, the cognate effector for the coiled‐coil (CC)‐type immune receptor RPS2, which activates NONSPECIFIC DISEASE RESISTANCE 1 (NDR1)‐dependent signalling. In addition, HR cell death after inoculation with PstDC3000(AvrRpt2) strain was not suppressed in the atg7 and atg9 mutants (Hofius et al., 2009). Based on these results, the authors proposed that pathogen‐induced autophagy contributes positively to HR cell death initiated through TIR‐type immune receptors in Arabidopsis (Hofius et al., 2009).
The reasons for the seemingly conflicting results are not fully understood, but may be related to the ages of the plants and the timescales of cell death monitoring. Yoshimoto et al. (2009) showed that, when the older leaves of 7–8‐week‐old Arabidopsis atg5 mutant plants (in the Col‐0 ecotype) were inoculated with PstDC3000(avrRpm1), there was spread of chlorotic cell death. However, no clear spread of cell death was observed if the younger leaves of 7–8‐week‐old plants or leaves of 4–5‐week‐old atg5 mutant plants were inoculated with the same avirulent pathogen (Yoshimoto et al., 2009). Further analysis revealed that SA is hyperaccumulated in the autophagy‐defective mutants and this increase in SA levels was higher in the older (7‐week‐old) plants than in the younger (4‐week‐old) plants (Yoshimoto et al., 2009). Genetic analysis using Arabidopsis sid2 and npr1 mutants defective in SA biosynthesis and signalling, respectively, showed that SA hyperaccumulation and signalling are required for both pathogen‐induced chlorotic cell death and early senescence in the autophagy‐defective mutants (Yoshimoto et al., 2009). In addition, autophagy‐defective mutants accumulated increased levels of ROS in a partly SA‐dependent manner. Importantly, SA or its agonist benzothiadiazole (BTH) and ROS induce pro‐survival autophagy in wild‐type plants, but induce runaway cell death in autophagy‐defective plants (Yoshimoto et al., 2009). Apparently pathogen and age‐induced production and signalling of SA and ROS can be accelerated through a positive feedback amplification loop, and culminate in cell death in the autophagy‐defective mutants (Fig. 1). The highly diffusible and autopropagating nature of ROS would also make them probable candidates as diffusible pro‐death signals for the promotion of runaway cell death in pathogen‐infected plants. In wild‐type plants, however, increased SA and ROS levels in pathogen‐infected tissues induce autophagy, which can apparently suppress SA and ROS production and signalling, thereby restricting pathogen‐induced cell death to the site of infection (Yoshimoto et al., 2009) (Fig. 1). How autophagy suppresses SA and ROS production and signalling to promote plant cell survival is currently unclear. Studies in both mammalian cells and yeast have shown that, under stress conditions, excess mitochondria, the most important ROS in these organisms, are degraded by autophagy (mitophagy), and ROS production from mitochondria is suppressed (Ashrafi and Schwarz, 2013). In autophagy‐defective cells, however, excess mitochondria are not degraded, leading to the production of surplus ROS. The surplus ROS can damage mitochondria, and the damaged mitochondria produce more ROS in a vicious feedback cycle (Ashrafi and Schwarz, 2013). A similar feedback cycle of ROS production from damaged organelles, such as endoplasmic reticulum, mitochondria and chloroplasts, could also operate in plant cells with elevated levels of SA and ROS during pathogen infection (Fig. 1). By removing excess and damaged organelles, pathogen‐induced autophagy may contribute to cell survival by maintaining quality organelles to a basal level to fulfil cellular requirements and preventing excess ROS production.
Figure 1.
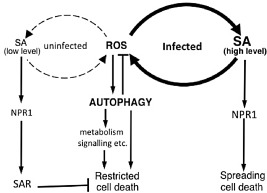
Autophagy in hypersensitive cell death induced by avirulent and related pathogens. Autophagy functions both as a promoter of restricted hypersensitive response (HR) cell death at infected sites and as a suppressor of cell death spreading to uninfected areas. Autophagy promotes restricted HR cell death either directly through the removal of specific negative regulators of HR cell death or pro‐survival signals of host plant cells, or indirectly through effects on cellular metabolism and signalling. Autophagy could also promote HR cell death by reducing basal salicylic acid (SA) levels, thereby preventing the establishment of pre‐existing systemic acquired resistance (SAR), which is known to suppress HR cell death. Autophagy suppresses the spread of cell death by the inhibition of pathogen‐induced production and signalling of SA and reactive oxygen species (ROS), which could be accelerated through a positive feedback amplification loop. NPR1, NON‐EXPRESSOR OF PATHOGENESIS‐RELATED 1.
As an important pro‐survival mechanism that limits the spread of pathogen‐infected plant cell death, it is intriguing that autophagy displays a pro‐death activity that contributes to the onset of restricted HR cell death in Arabidopsis plants after infection by certain avirulent pathogens (Hofius et al., 2009). The pro‐death activity of autophagy during plant–pathogen interactions has also been demonstrated by more recent studies. One of these reported studies focused on the necrotrophic fungal pathogen Sclerotinia sclerotiorum, which induces cell death in host plant tissue through the non‐selective phytotoxin, oxalic acid (OA) (Kabbage et al., 2013). OA‐deficient S. sclerotiorum mutants are non‐pathogenic and induce a pronounced oxidative burst and restricted cell death in host plant tissue, similar to HR induced by an avirulent pathogen (Kabbage et al., 2013). Interestingly, the inhibition of autophagy in Arabidopsis autophagy‐defective mutants, or by treatment with autophagy inhibitors, led to enhanced susceptibility to OA‐deficient S. sclerotiorum mutants (Kabbage et al., 2013). Furthermore, the pronounced oxidative burst associated with the restrictive cell death of wild‐type plant tissues was significantly reduced in the autophagy‐deficient mutants in response to OA‐deficient S. sclerotiorum mutants (Kabbage et al., 2013). Thus, autophagy plays a positive role in mounting a rapid and strong oxidative burst during the early stage of infection of the host plant tissue by the necrotrophic pathogen. In another recent study, Arabidopsis plants over‐expressing a small GTP‐binding protein, RabG3b, displayed accelerated, unrestricted HR cell death and accumulation of autophagic structures in response to avirulent bacterial pathogens (Kwon et al., 2013). In an Arabidopsis autophagy‐defective mutant over‐expressing RabG3b, HR cell death was still observed, but its progression was significantly suppressed, demonstrating that autophagy positively influences pathogen‐induced HR cell death (Kwon et al., 2013). In barley plants, the maize smut Ustilago maydis triggers a non‐host response of epidermal cell death, which can be partially suppressed by the over‐expression of Bax Inhibitor‐1 (BI‐1), a PCD suppressor (Hof et al., 2013). By contrast, cell death induced by Ustilago mutants for pep1, which encodes a secreted effector, was not suppressed by BI‐1 over‐expression and involved hallmarks of autophagy (Hof et al., 2013). These observations suggest that the cell death response triggered by the Ustilago pep1 mutants differs from that triggered by the wild‐type pathogen, and may be activated or promoted by autophagy.
As a catabolic process usually for quality control and nutrient generation, autophagy is a common and critical mechanism for the promotion of cell survival under a wide spectrum of biotic and abiotic stress conditions. The promotion of cell death by autophagy is relatively uncommon and is often associated with development, particularly in insects (Denton et al., 2012). In many cases, the association of cell death with the accumulation of autophagosomes has been interpreted as cell death by autophagy, when often it is actually really cell death with autophagy (Kroemer and Levine, 2008). Even for the cases in which the promotion of cell death by autophagy has been clearly demonstrated, the precise molecular mechanisms for the pro‐death activity of autophagy often remain unclear (Denton et al., 2012; Kroemer and Levine, 2008). As pathogen‐induced HR cell death is subjected to regulation by a spectrum of regulatory and metabolic pathways, it is of interest to distinguish whether autophagy contributes to HR cell death through a direct role or a secondary effect. Autophagy could constitute a direct death effector mechanism through the removal of specific negative regulators of HR cell death or pro‐survival signals of host plant cells (Fig. 1). Alternatively, delayed HR cell death after infection of a pathogen in the autophagy‐defective mutants could be caused by indirect effects of altered physiological/metabolic state or basal defence signalling, which can prime plant cell death (Fig. 1). SA, which is elevated in autophagy‐defective mutants (Yoshimoto et al., 2009), is a signalling molecule with paradoxical roles as both a promoter and suppressor of pathogen‐induced cell death. Although it is widely known that excessive SA promotes cell death, pre‐existing SA‐regulated systemic acquired resistance (SAR) or pretreatment of Arabidopsis plants with SA actually suppresses HR cell death (Devadas and Raina, 2002; Yu et al., 1998). In autophagy‐defective mutants, there are increased basal SA levels and expression of SA‐regulated SAR genes (e.g. PR1) prior to pathogen infection, even at the young stages (4 weeks) (Yoshimoto et al., 2009). These pre‐existing low levels of SA in autophagy‐defective mutants could serve as a signal for the induction of anti‐death SAR that renders plants less sensitive to the onset of pathogen‐triggered HR cell death (Fig. 1). Similar paradoxical roles, both as a promoter and suppressor of cell death, have also been known and extensively analysed for ROS, which, at high levels, induces cell death, but, at low levels, can promote mitogenic signalling in animal cells through the enhancement of protein tyrosine phosphorylation and the activation of transcription factors, including NF‐kB, antioxidant enzymes and Bcl‐2 (Das and Maulik, 2004). Regardless of the underlying molecular mechanisms, the paradoxical roles of autophagy, as a promoter of cell death at the onset of pathogen‐triggered HR cell death and as an inhibitor of cell death following containment of invading pathogens, illustrate an elaborate programme by plant host cells for the rapid activation of HR and prevention of uncontrolled cell death (Fig. 1).
Autophagy in Plant Basal Resistance to Biotrophic Pathogens
Plant basal resistance results from complex pre‐formed and inducible defence mechanisms that protect plants against a broad spectrum of pathogens. Plant basal resistance can be induced by virulent pathogens and is often associated with the induced production of ROS, SA, cell wall‐fortifying compounds (e.g. callose) and defence‐related proteins (e.g. PR proteins). The role of autophagy in plant basal resistance to virulent biotrophic and hemibiotrophic pathogens has been analysed by several reported studies. For example, when infected with the virulent PstDC3000, there was a slight increase in bacterial growth in ATG6‐silenced plants over wild‐type plants at early stages after infection, but this difference disappeared at later times (Patel and Dinesh‐Kumar, 2008). Enhanced growth of the virulent bacterial pathogen was also observed in an atg7 mutant over the wild‐type plants (both in the WS ecotype) (Hofius et al., 2009). No difference in basal resistance to PstDC3000 was observed between Arabidopsis and a mutant for ATG2 (Wang et al., 2011a). However, a more comprehensive analysis using mutants for ATG5, ATG10 and ATG18a indicated that autophagy negatively regulated plant basal immunity to the virulent bacterial pathogens (Lenz et al., 2011a). In all these autophagy‐defective mutants, bacterial growth was strongly reduced when compared with that in wild‐type plants. Enhanced basal resistance in the autophagy‐defective mutants was not associated with altered pathogen‐induced cell death, leaf sugar and amino acid contents or PTI‐, ethylene‐ or JA‐related defence responses (Lenz et al., 2011a). However, there was a significant difference in the basal SA levels between the wild‐type and the autophagy‐defective mutants. A more significant difference in SA levels was observed between the wild‐type and the autophagy‐defective mutants after PstDC3000 infection (Lenz et al., 2011a). In addition, both the basal and pathogen‐induced levels of SA‐regulated PR1 expression were higher in the autophagy‐defective mutants than in wild‐type plants. Given the well‐established role of SA‐regulated defence in plant basal resistance to this hemibiotrophic bacterial pathogen, it has been suggested that the elevated SA levels are responsible for the enhanced immunity of autophagy‐defective mutants to PstDC3000 (Lenz et al., 2011a) (Fig. 2). As SA levels in the autophagy‐defective mutants are sensitive to developmental and environmental conditions, the discrepancy in the assessments of basal immunity to the bacterial pathogen could be explained by the differences in the autophagy‐defective mutants used in the studies, as well as their ages and growth conditions.
Figure 2.
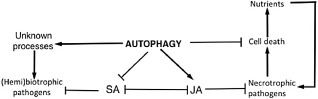
Autophagy in plant basal resistance to biotrophic and necrotrophic pathogens. Autophagy generally has a negative role in plant basal resistance to biotrophic pathogens by suppressing salicylic acid (SA) signalling or through effects on other unknown processes. Autophagy plays a critical role in plant resistance to necrotrophic pathogens by inhibiting pathogen‐induced cell death and promoting jasmonate (JA) signalling.
Arabidopsis plants are susceptible to several obligately biotrophic powdery mildew fungal species. Yoshimoto et al. (2009) compared Arabidopsis atg2 and atg5 mutants with wild‐type plants in terms of the response to Golovinomyces orontii, and found mildew‐induced cell death but no difference in powdery mildew resistance. However, when using another powdery mildew species, G. cichoracearum, Wang et al. (2011a) discovered both mildew‐induced cell death and enhanced resistance to powdery mildew in an Arabidopsis atg2 mutant. Using a number of Arabidopsis mutants defective in pathogen‐induced SA biosynthesis or signalling, it was further demonstrated that enhanced powdery mildew resistance of the atg2 mutant required the SA pathway (Wang et al., 2011a). Interestingly, although age‐related cell death and senescence in the atg2 mutant also required the SA pathway, mildew‐induced cell death was only partially suppressed by defective SA signalling, indicating that de‐regulated SA signalling cannot fully explain the associated cell death (Wang et al., 2011a). Mutations of other ATG genes, including ATG5, ATG7, ATG10 and ATG18A, also led to enhanced powdery mildew resistance and mildew‐induced cell death (Wang et al., 2011a, b). In addition, Arabidopsis mutant plants for AMSH1, encoding a deubiquitinating enzyme required for autophagic degradation, were more resistant to the powdery mildew pathogen Erysiphe cruciferarum, and this enhanced resistance was also associated with increased spontaneous cell death (Katsiarimpa et al., 2013). Thus, autophagy plays a negative role in basal immunity against some powdery mildew fungal species, at least, in part, by suppression of the SA signalling pathway (Fig. 2).
Downy mildew of Arabidopsis is caused by an obligate biotrophic oomycete Hyaloperonospora arabidopsidis and has also been used to assess the role of autophagy in plant basal resistance. Using Ws‐0 wild‐type and the atg7 mutant, Hofius et al. (2009) reported that mutation of the ATG gene led to increased growth of the virulent Emwa isolate of H. arabidopsidis. However, when the Col‐0 wild‐type and autophagy‐defective mutant plants were infected with the virulent Noco2 isolate of H. arabidopsidis, no differences in conidiophore formation or host cell death were found (Lenz et al., 2011a). It remains to be determined whether the different isolates of the pathogen or other factors were responsible for the variable results.
Autophagy in Plant Resistance to Necrotrophic Pathogens
Unlike the variable results with biotrophic pathogens, a number of studies have provided consistent and compelling lines of evidence indicating that autophagy plays a critical and positive role in plant resistance to necrotrophic pathogens. The expression of many ATG genes was induced by infection of the necrotrophic fungal pathogen Botrytis cinerea (Lai et al., 2011). With both green fluorescent protein (GFP)‐tagged ATG8a, as a marker of autophagosome, and LysoTracker Green dye, which stains autophagosomes, it was shown that Botrytis infection led to increased accumulation of autophagosomes, not only in the lesions and the areas surrounding the lesions, but also in more distal areas in which no fungal mycelium was observed (Lai et al., 2011). Furthermore, when Arabidopsis mutants for ATG5, ATG7 and ATG18a were inoculated with Botrytis, they developed more extensive symptoms of chlorosis and supported more fungal growth than did Col‐0 wild‐type plants (Lai et al., 2011). In a detached leaf assay, these autophagy‐defective mutants also developed more severe disease symptoms and supported more fungal spore formation than did Col‐0 wild‐type plants after infection with another necrotrophic fungal pathogen, Alternaria brassicicola (Lai et al., 2011). Compromised resistance to A. brassicicola was also observed in other studies using Arabidopsis atg5, atg10, atg18a and amsh1 mutants (Katsiarimpa et al., 2013; Lenz et al., 2011a, b). In addition, the Arabidopsis autophagy‐defective mutants become more susceptible to infection by the necrotrophic ascomycete Plectosphaerella cucumerina (Lenz et al., 2011a).
The critical role of autophagy in plant resistance to necrotrophic pathogens is further supported by the recent finding that S. sclerotiorum, a necrotrophic pathogen, has evolved mechanisms to suppress autophagy in plant host cells to promote pathogenesis (Kabbage et al., 2013). Sclerotinia sclerotiorum secretes the non‐selective phytotoxin OA, a critical pathogenesis factor that promotes fungal colonization of host plants through a complex set of mechanisms, including acidification, chelation of Ca2+, low pH and activation of degradative enzymes (Kabbage et al., 2013). Recent studies, however, have revealed that OA contributes to pathogenesis through additional mechanisms of suppression of the oxidative burst and induction of apoptotic cell death of plant host cells (Kabbage et al., 2013). Unlike other necrotrophic pathogens, such as Botrytis, wild‐type S. sclerotiorum did not induce the production of autophagosomes, even in severely diseased plant tissue, whereas the OA‐deficient mutant S. sclerotiorum induced the production of autophagosomes in plant host tissue despite very restricted mycelial growth. Furthermore, pretreatment of OA prior to challenge with the OA‐deficient mutant S. sclerotiorum significantly reduced the number of autophagosomes. Thus, the phytotoxin is capable of suppressing autophagy, either directly or indirectly, to promote the pathogenicity of S. sclerotiorum.
It is likely that the critical role of plant autophagy in resistance to necrotrophic pathogens involves complex mechanisms. Necrotrophic pathogens actively kill plant host cells from the very early stages of infection to extract nutrients for their growth and reproduction. These pathogens rely on a variety of pathogenesis/virulence factors, including toxins and degradative enzymes, to promote plant cell death. As a pro‐survival mechanism, autophagy may promote plant resistance to necrotrophic pathogens simply by suppressing pathogen‐induced cell death, thereby depriving the pathogens of the necessary nutrients from dead plant tissues (Fig. 2). After infection with Botrytis, autophagy‐defective mutants developed extensive and rapidly expanding chlorosis, which is associated with the accelerated breakdown of cellular proteins (Lai et al., 2011). Thus, during the early stage of infection, rapidly induced autophagy may be responsible for the removal of damaged and potentially toxic cellular constituents to promote cell survival. In autophagy‐defective mutants, however, these damaged and potentially toxic cellular constituents will accumulate at increased rates to trigger PCD, during which an alternative protein degradation route is apparently activated for the massive breakdown of cellular constituents, which would provide necessary nutrients for the growth and reproduction of the invading pathogens (Lai et al., 2011) (Fig. 2).
Autophagy may also promote plant resistance to necrotrophic pathogens through the modulation of SA‐ and JA‐regulated defence signalling pathways (Fig. 2). In the mutants for a number of ATG genes examined, there were higher basal levels of JA‐regulated PDF1.2 and SA‐regulated PR1 gene transcripts (Lai et al., 2011), suggesting the activation of JA and SA signalling mutants before the onset of visible senescence. After Botrytis infection, the transcript levels of SA‐regulated PR1 were substantially higher in the autophagy‐defective mutants than in the wild‐type plants (Lai et al., 2011). By contrast, despite its increased basal expression, Botrytis‐induced expression of JA‐regulated PDF1.2 was reduced in the autophagy‐defective mutants when compared with that in wild‐type plants (Lai et al., 2011). Reduced induction of PDF1.2 was also observed in the SA‐deficient atg5/sid2 double mutant, indicating that reduced expression of the JA‐regulated defence genes in the autophagy‐defective mutants did not result from increased SA signalling. Thus, in Botrytis‐infected plants, autophagy positively modulates JA signalling, which is known to be required for plant resistance to necrotrophic pathogens (Fig. 2).
Autophagy in Plant Antiviral Immune Response
As a recognized player in the innate immune response, autophagy has been extensively analysed in animal systems for its role in protecting cells from diverse intracellular pathogens, such as viruses. As an endolysosomal delivery system, autophagy can promote antiviral immunity through the transfer of viruses from the cytoplasm to the lysosome for degradation or the transfer of viral components to specific subcellular compartments for the activation of innate or adaptive antiviral immunity (Shoji‐Kawata and Levine, 2009). Likewise, autophagy plays a significant role in plant antiviral immunity. In TMV‐infected BECLIN 1‐, VPS34‐, ATG3‐ and ATG7‐silenced resistant tobacco plants, there was increased accumulation of TMV, in addition to increased spread of HR cell death (Liu et al., 2005). The increase in TMV accumulation is not a result of increased spread because the virus and viral RNA are both confined only to the infection site in the plants. This result indicates that plant autophagy functions to limit virus replication and/or movement either directly through degradation of viruses in vacuoles or indirectly through effects on other antiviral defence (Fig. 3).
Figure 3.
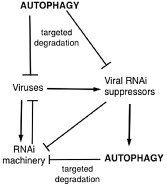
Autophagy in plant responses to viral pathogens. Autophagy can play a positive role in plant antiviral defence by targeting viruses and viral RNA interference (RNAi) suppressors for degradation. Autophagy can also be hijacked by viral RNAi suppressors for the degradation of critical components of the host cell RNAi machinery.
Recent studies have shown that autophagy is involved in the degradation of plant and viral proteins associated with dsRNA‐induced RNA silencing, which plays a critical role in plant antiviral defence (Agius et al., 2012). Plant antiviral RNA silencing involves the processing of viral dsRNA by the enzyme Dicer into small viral RNAs. One of the two viral‐derived small RNA strands is incorporated into a protein complex called the RNA‐induced silencing complex (RISC) to guide the degradation of the corresponding viral RNA. As a counter‐defence mechanism, viruses have evolved viral suppressors that inhibit the antiviral RNA silencing process (Agius et al., 2012). A recent study has shown that P0, a viral suppressor of RNA silencing from polerovirus, targets the autophagic degradation of ARGONAUTE 1 (AGO1), a key component of RISC that binds to small interfering RNA (siRNA) and carries the RNA slicer activity (Derrien et al., 2012). The viral P0 protein is an F‐box protein that promotes the degradation of AGO1 in cooperation with the host S‐phase kinase‐associated protein1 (SKP1)–Cullin 1(CUL1)–F‐box protein (SCF) ubiquitin‐protein ligase (E3). Although P0‐targeted AGO1 degradation requires ubiquitination by an SCF‐type E3 ligase, it is insensitive to inhibition of the proteasome (Baumberger et al., 2007), arguing against the degradation by the ubiquitin–proteasome system (UPS). However, P0‐targeted degradation of AGO1 is inhibited by both 3‐methyladenine (3‐MA), an inhibitor of autophagosome formation, and E64d, a cysteine protease inhibitor of the degradation of autophagic cargo inside autolysosomes (Derrien et al., 2012). When its degradation was inhibited, AGO1 co‐localized with ATG‐8a in autophagic vesicles (Derrien et al., 2012). Thus, the viral suppressor hijacks a host cellular SCF‐type ubiquitin E3 ligase to ubiquitinate AGO1, and the ubiquitinated AGO1 is targeted for degradation before the RISC assembly by selective autophagy (Fig. 3).
In another study, it was shown that a tobacco calmodulin‐like protein (rgs‐CaM) provides a secondary antiviral mechanism by binding to and directing the degradation of viral RNA silencing suppressors by plant autophagy (Nakahara et al., 2012). rgs‐CaM was first identified as an interacting protein of the HC‐Pro protein, an RNA silencing suppressor from Tobacco etch virus (TEV). The tobacco rgs‐CaM protein binds to the dsRNA‐binding domains of not only Hc‐Pro, but also structurally unrelated 2b RNA silencing suppressors, and sequesters them from inhibiting RNA interference (RNAi) (Nakahara et al., 2012). Interestingly, the protein levels of endogenous rgs‐CaM and interacting viral RNA silencing suppressors increased in plant cells after treatment with an inhibitor of autophagy or by silencing of tobacco BECLIN1/ATG6 (Nakahara et al., 2012). In addition, accumulated endogenous rgs‐CaM and interacting viral RNA silencing suppressors co‐localized with LysoTracker‐stained autophagosomes, suggesting that they are recruited into autophagosomes for degradation after complex formation (Nakahara et al., 2012). Over‐expression of rgs‐CaM in transgenic plants enhanced resistance, whereas silencing rendered plants more susceptible to viral infection (Nakahara et al., 2012). These results indicate that tobacco rgs‐CaM is an antiviral factor that binds viral RNA silencing suppressors to both reduce the suppressor activity and promote their degradation by autophagy (Nakahara et al., 2012). Thus, autophagy can be deployed as a defence system against viral RNA silencing suppressors (Fig. 3).
Conclusions
Studies over the last decade or so have uncovered an important role of autophagy in plant innate immune responses. From these studies, it is also clear that autophagy influences the outcomes of plant pathogens through interfaces with a number of important facets of plant innate immunity, including pathogen‐induced hypersensitive cell death (Fig. 1), SA‐ and JA‐regulated defence (Fig. 2) and virus‐induced RNAi (Fig. 3). However, much less is known about the mechanistic basis of the critical and perplexing role of autophagy in plant innate immune responses. It remains to be determined whether the observed pro‐death activity of autophagy in pathogen‐induced HR cell death is caused by a direct autophagic death effector mechanism or by secondary effects of the altered physiological/metabolic state of host cells. SA and JA signalling are altered in autophagy‐deficient mutants, even without pathogen infection, but the underlying mechanisms are unclear. Autophagy influences plant antiviral defence through a number of pathways, including VIGS, but it has yet to be demonstrated whether autophagy directly targets the degradation of viruses and, if so, whether it constitutes an important antiviral mechanism in plants. Functional analysis of autophagy in plants has so far mostly relied on genes for the core process of autophagy. However, efforts to elucidate the complex roles of autophagy in plant innate immune responses will require the identification and characterization of additional components, including autophagy receptors, adaptors and cargo proteins, which determine the functional specificity and dictate the action mechanisms of autophagy in plant innate immunity. In addition, as an important catabolic process for cell homeostasis under stress conditions, autophagy undoubtedly shapes or reshapes some of the important facets of plant innate immunity through the modulation or reprogramming of globally important metabolic and signalling pathways. A particularly important effort will therefore involve the systematic identification and integration of these affected pathways and processes through systems biology to develop a comprehensive understanding of the network of complex functional interactions between autophagy and plant innate immune responses.
Acknowledgements
Research from the authors' laboratories described in this review is in part supported by grants from the US National Science Foundation (IOS0958066) and the Natural Science Foundation of China (2013C150203).
References
- Agius, C. , Eamens, A.L. , Millar, A.A. , Watson, J.M. and Wang, M.B. (2012) RNA silencing and antiviral defense in plants. Methods Mol. Biol. 894, 17–38. [DOI] [PubMed] [Google Scholar]
- Ashrafi, G. and Schwarz, T.L. (2013) The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham, D.C. (2007) Plant autophagy—more than a starvation response. Curr. Opin. Plant Biol. 10, 587–593. [DOI] [PubMed] [Google Scholar]
- Baumberger, N. , Tsai, C.H. , Lie, M. , Havecker, E. and Baulcombe, D.C. (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614. [DOI] [PubMed] [Google Scholar]
- Boya, P. , Reggiori, F. and Codogno, P. (2013) Emerging regulation and functions of autophagy. Nat. Cell Biol. 15, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P. , Petersen, M. , Bjørn Nielsen, H. , Zhu, S. , Newman, M.A. , Shokat, K.M. , Rietz, S. , Parker, J. and Mundy, J. (2006) Arabidopsis MAP kinase 4 regulates salicylic acid‐ and jasmonic acid/ethylene‐dependent responses via EDS1 and PAD4. Plant J. 47, 532–546. [DOI] [PubMed] [Google Scholar]
- Chung, T. , Suttangkakul, A. and Vierstra, R.D. (2009) The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8‐lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149, 220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, D.K. and Maulik, N. (2004) Conversion of death signal into survival signal by redox signaling. Biochemistry (Moscow), 69, 10–17. [DOI] [PubMed] [Google Scholar]
- Denton, D. , Nicolson, S. and Kumar, S. (2012) Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 19, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic, V. (2012) Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr. Opin. Immunol. 24, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien, B. , Baumberger, N. , Schepetilnikov, M. , Viotti, C. , De Cillia, J. , Ziegler‐Graff, V. et al (2012) Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA, 109, 15 942–15 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devadas, S.K. and Raina, R. (2002) Preexisting systemic acquired resistance suppresses hypersensitive response‐associated cell death in Arabidopsis hrl1 mutant. Plant Physiol. 128, 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, F.L. , Dinesh‐Kumar, S.P. , Holzberg, S. , Ustach, C.V. , Dutton, M. , Handley, V. et al (1999) Interactions between tobacco mosaic virus and the tobacco N gene. Philos. Trans. R. Soc. London B: Biol Sci. 354, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari, S. , Plotnikova, J.M. , De Lorenzo, G. and Ausubel, F.M. (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35, 193–205. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- He, C. and Klionsky, D.J. (2009) Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof, A. , Zechmann, B. , Schwammbach, D. , Huckelhoven, R. and Doehlemann, G. (2013) Alternative cell death mechanisms determine epidermal resistance in incompatible barley–Ustilago interactions. Mol. Plant–Microbe Interact. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Hofius, D. , Schultz‐Larsen, T. , Joensen, J. , Tsitsigiannis, D.I. , Petersen, N.H. , Mattsson, O. et al (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell, 137, 773–783. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathog. 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan, J.A. (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11, 247–253. [DOI] [PubMed] [Google Scholar]
- Katsiarimpa, A. , Kalinowska, K. , Anzenberger, F. , Weis, C. , Ostertag, M. , Tsutsumi, C. et al (2013) The deubiquitinating enzyme AMSH1 and the ESCRT‐III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell, 25, 2236–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer, G. and Levine, B. (2008) Autophagic cell death: the story of a misnomer. Nat. Rev. Mol. Cell Biol. 9, 1004–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S.I. , Cho, H.J. , Kim, S.R. and Park, O.K. (2013) The Rab GTPase RabG3b positively regulates autophagy and immunity‐associated hypersensitive cell death in Arabidopsis. Plant Physiol. 161, 1722–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z. , Wang, F. , Zheng, Z. , Fan, B. and Chen, Z. (2011) A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66, 953–968. [DOI] [PubMed] [Google Scholar]
- Lenz, H.D. , Haller, E. , Melzer, E. , Gust, A.A. and Nurnberger, T. (2011a) Autophagy controls plant basal immunity in a pathogenic lifestyle‐dependent manner. Autophagy, 7, 773–774. [DOI] [PubMed] [Google Scholar]
- Lenz, H.D. , Haller, E. , Melzer, E. , Kober, K. , Wurster, K. , Stahl, M. et al (2011b) Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66, 818–830. [DOI] [PubMed] [Google Scholar]
- Levine, B. , Mizushima, N. and Virgin, H.W. (2011) Autophagy in immunity and inflammation. Nature, 469, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Czymmek, K. , Talloczy, Z. , Levine, B. and Dinesh‐Kumar, S.P. (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell, 121, 567–577. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Xiong, Y. and Bassham, D.C. (2009) Autophagy is required for tolerance of drought and salt stress in plants. Autophagy, 5, 954–963. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Burgos, J.S. , Deng, Y. , Srivastava, R. , Howell, S.H. and Bassham, D.C. (2012) Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell, 24, 4635–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste, T. , Chen, X. , Salmeron, J. and Dietrich, R. (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell, 15, 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K.S. , Masuta, C. , Yamada, S. , Shimura, H. , Kashihara, Y. , Wada, T.S. et al (2012) Tobacco calmodulin‐like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA, 109, 10 113–10 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, A. , Moeder, W. , Kachroo, P. , Klessig, D.F. and Shah, J. (2005) Arabidopsis ssi2‐conferred susceptibility to Botrytis cinerea is dependent on EDS5 and PAD4. Mol. Plant–Microbe Interact. 18, 363–370. [DOI] [PubMed] [Google Scholar]
- Patel, S. and Dinesh‐Kumar, S.P. (2008) Arabidopsis ATG6 is required to limit the pathogen‐associated cell death response. Autophagy, 4, 20–27. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A. , Eggermont, K. , Terras, F.R. , Thomma, B.P. , De Samblanx, G.W. , Buchala, A. et al (1996) Pathogen‐induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid‐independent pathway. Plant Cell, 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A. , Thomma, B.P. , Buchala, A. , Metraux, J.P. and Broekaert, W.F. (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell, 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J.H. , Yoshimoto, K. , Ohsumi, Y. , Jeon, J.S. and An, G. (2009) OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells, 27, 67–74. [DOI] [PubMed] [Google Scholar]
- Shoji‐Kawata, S. and Levine, B. (2009) Autophagy, antiviral immunity, and viral countermeasures. Biochim. Biophys. Acta, 1793, 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, W. , Ma, H. , Liu, C. , Wu, J. and Yang, J. (2006) Identification and characterization of two rice autophagy associated genes, OsAtg8 and OsAtg4. Mol. Biol. Rep. 33, 273–278. [DOI] [PubMed] [Google Scholar]
- Tadamura, K. , Nakahara, K.S. , Masuta, C. and Uyeda, I. (2012) Wound‐induced rgs‐CaM gets ready for counterresponse to an early stage of viral infection. Plant Signal. Behav. 7, 1548–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B. , Eggermont, K. , Penninckx, I. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P.A. et al (1998) Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA, 95, 15 107–15 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P. , Nelissen, I. , Eggermont, K. and Broekaert, W.F. (1999) Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola . Plant J. 19, 163–171. [DOI] [PubMed] [Google Scholar]
- Veronese, P. , Nakagami, H. , Bluhm, B. , Abuqamar, S. , Chen, X. , Salmeron, J. et al (2006) The membrane‐anchored BOTRYTIS‐INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell, 18, 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Nishimura, M.T. , Zhao, T. and Tang, D. (2011a) ATG2, an autophagy‐related protein, negatively affects powdery mildew resistance and mildew‐induced cell death in Arabidopsis. Plant J. 68, 74–87. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wu, Y. and Tang, D. (2011b) The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew‐induced cell death in Arabidopsis. Plant Signal. Behav. 6, 1408–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y. , Contento, A.L. and Bassham, D.C. (2007a) Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy, 3, 257–258. [DOI] [PubMed] [Google Scholar]
- Xiong, Y. , Contento, A.L. , Nguyen, P.Q. and Bassham, D.C. (2007b) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, K. , Jikumaru, Y. , Kamiya, Y. , Kusano, M. , Consonni, C. , Panstruga, R. et al (2009) Autophagy negatively regulates cell death by controlling NPR1‐dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell, 21, 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, I.C. , Parker, J. and Bent, A.F. (1998) Gene‐for‐gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA, 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Wang, J. , Cheng, Y. , Chi, Y.J. , Fan, B. , Yu, J.Q. et al (2013) NBR1‐mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9, e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]


