SUMMARY
Multilocus sequence analysis (MLSA) and type III effector (T3E) repertoire mining were performed to gain new insights into the genetic relatedness of Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc), two major bacterial pathogens of rice. Based on a collection of 45 African and Asian strains, we first sequenced and analysed three housekeeping genes by MLSA, Bayesian clustering and a median‐joining network approach. Second, we investigated the distribution of 32 T3E genes, which are known to be major virulence factors of plant pathogenic bacteria, in all selected strains, by polymerase chain reaction and dot‐blot hybridization methods. The diversity observed within housekeeping genes, as well as within T3E repertoires, clearly showed that both pathogens belong to closely related, but distinct, phylogenetic groups. Interestingly, these evolutionary groups are differentiated according to the geographical origin of the strains, suggesting that populations of Xoo and Xoc might be endemic in Africa and Asia, and thus have evolved separately. We further revealed that T3E gene repertoires of both pathogens comprise core and variable gene suites that probably have distinct roles in pathogenicity and different evolutionary histories. In this study, we carried out a functional analysis of xopO, a differential T3E gene between Xoo and Xoc, to determine the involvement of this gene in tissue specificity. Altogether, our data contribute to a better understanding of the evolutionary history of Xoo and Xoc in Africa and Asia, and provide clues for functional studies aiming to understand the virulence, host and tissue specificity of both rice pathogens.
INTRODUCTION
Bacterial leaf blight, caused by Xanthomonas oryzae pv. oryzae (Xoo), and bacterial leaf streak, caused by Xanthomonas oryzae pv. oryzicola (Xoc), are two major diseases of rice (Mew, 1987; Opina and Exconde, 1971; Ou, 1985). Interestingly, both pathogens present distinct tissue specificities: Xoo enters the rice leaf through hydathodes and invades the plant through the xylem, whereas Xoc penetrates the leaf through stomata and colonizes the intercellular spaces of the parenchyma tissue (Niño‐Liu et al., 2006). Xoo and Xoc are not easily differentiated by phenotypic tests and are genetically closely related, as they exhibit more than 90% similarity by DNA–DNA hybridizations (Rademaker et al., 2000; Swings et al., 1990; Vauterin et al., 1995). Despite their close relatedness, both pathogens show a high diversity among isolates, as revealed by pathogenicity assays and DNA polymorphism analyses (1995, 1999; Leach et al., 1992; Nelson et al., 1994; Raymundo et al., 1999; 1996, 2000). Genetic variations among isolates have been confirmed by the whole‐genome sequence of the Korean Xoo strain KACC10331 (Lee et al., 2005), the Japanese Xoo strain MAFF311018 (Ochiai et al., 2005), the Philippine Xoo strain PXO99A (Salzberg et al., 2008) and the Philippine Xoc strain BLS256 (Bogdanove et al., 2011).
The presence of Xoo and Xoc has long been reported in most rice‐growing areas in Asia, Latin America and Australia (Mew et al., 1993). In Africa, both pathogens were first described in the 1980s (Basso et al., 2011; Buddenhagen, 1985; Gonzalez et al., 2007; Ryba‐White et al., 1995; Sere et al., 2005; Wonni et al., 2011). New emergences in Africa are probably a result of the intensification of rice cultivation, the lack of breeding programmes for resistance and the absence of efficient prophylactic measures (Buddenhagen, 1985; Ryba‐White et al., 1995). The first comprehensive molecular and pathotypic characterization of African Xoo and Xoc strains was performed only recently (Gonzalez et al., 2007). Although both Asian and African Xoo strains induce identical symptoms, they appear to be genetically distinct (Gonzalez et al., 2007; 2010a, 2010b). Nonetheless, the genetic relatedness of these strains is not clear, as African Xoo strains are close to either Asian Xoo strains or Asian Xoc strains, depending on the method employed to measure diversity (Gonzalez et al., 2007). To clarify the relationships between African and Asian strains of Xoo and Xoc, a multilocus sequence analysis (MLSA) on housekeeping genes can be performed. Indeed, MLSA has the following advantages: (i) it analyses phylogenetic relationships of large sets of strains with a better portability than genotyping techniques, such as amplified fragment length polymorphism (AFLP) or repetitive extragenic palindromic sequence‐polymerase chain reaction (Rep‐PCR); (ii) it detects and measures recombination. Mutations within housekeeping genes are largely assumed to be selectively neutral, and therefore are more likely to correctly reflect the phylogeny of the strains (Gevers et al., 2005; Maiden et al., 1998). In addition, using multiple loci provides a buffer against the distorting effect of recombination at a single locus (Gevers et al., 2005; Maiden et al., 1998). A recent phylogenetic study based on the analysis of nine housekeeping genes from eight sequenced genomes confirmed that African and Asian Xoo strains are closely related, and revealed that the US strains of X. oryzae form a group substantially divergent from a clade formed by Xoo and Xoc (Triplett et al., 2011). However, this study was conducted on a few strains. In order to obtain a deeper knowledge on the origin and evolution of Xoo and Xoc, MLSA should be conducted on a larger collection of strains.
Today, much remains to be learned about the genes involved in the pathogenicity of Xoo and Xoc and, more particularly, for African strains. Until now, studies have shown that Xoo and Xoc pathogenicity is highly dependent on the type III secretion system (T3SS) that delivers effector (T3E) proteins into the eukaryotic host cell (Makino et al., 2006; White and Yang, 2009). In Xoo, most knowledge on T3SS is based on studies of TAL effector genes (Boch et al., 2009; Moscou and Bogdanove, 2009; Schornack et al., 2006; White and Yang, 2009; Yang and White, 2004; Yu et al., 2011). Interestingly, Gonzalez et al. (2007) showed that African Xoo strains exhibit a smaller number of TAL genes in their genomes relative to Asian Xoo strains. However, the whole‐genome sequencing of Xoo and Xoc strains has revealed the presence of many other non‐TAL‐related T3Es in both pathogens (Bogdanove et al., 2011; Lee et al., 2005; Ochiai et al., 2005; Salzberg et al., 2008). Some T3Es, such as XopX and XopZ, have been shown to be involved in the Xoo–rice interaction (Song and Yang, 2010; Soto‐Suarez et al., 2010b). Nevertheless, despite the availability of complete genome sequences, the diversity of T3Es that are present within Xoo and Xoc populations in the field and, particularly, in African strains is not known. Moreover, nothing is currently known about the role played by T3Es in host and tissue specificity for Xoo and Xoc. Numerous T3Es have been demonstrated to trigger and subvert host defences (Alfano and Collmer, 2004; Jones and Dangl, 2006). Thus, T3E repertoires represent candidate determinants of the pathological adaptation of plant pathogenic bacteria on their hosts (Hajri et al., 2009).The determination of the presence or absence of T3E genes in a large collection of strains will document the involvement of T3E repertoires in the host and tissue specialization of Xoo and Xoc.
Sustainable control measures for bacterial leaf blight and bacterial leaf streak will therefore be dependent on improvements in the understanding of Xoo and Xoc evolution, as well as on the analysis of the bacterial genes involved in the interaction with rice. To provide data addressing these questions, we selected 45 X. oryzae strains from the collection previously used by Gonzalez et al. (2007). From all strains, we sequenced three housekeeping genes and performed PCR and dot‐blot hybridizations to test for the presence of 32 T3E genes. These experiments allowed us to complete MLSA studies, to compare the diversity within T3E repertoires of both pathogens, and to provide clues for evolutionary and functional studies aiming to understand the virulence, host and tissue specificity of both pathogens.
RESULTS AND DISCUSSION
African and Asian strains of Xoo and Xoc belong to closely related, but distinct, phylogenetic groups
To assess the phylogenetic relationships of 45 X. oryzae strains, we sequenced and analysed three housekeeping genes (glnA, gyrB and rpoD). We selected these genes because they appeared among the most polymorphic loci in previous MLSA studies on Xanthomonas species (Ah‐You et al., 2009; Bui Thi Ngoc et al., 2010; Fargier et al., 2011; Parkinson et al., 2009; Triplett et al., 2011; Young et al., 2008). The sizes of the three gene fragments are 737 (rpoD), 801 (gyrB) and 887 bp (glnA), respectively, leading to a total of 2425 bp for the concatenated data (Table 1). The number of haplotypes at each locus ranges from four (glnA) to 10 (gyrB) (Table 1 and Fig. 1), and the number of polymorphic nucleotide sites varies from 12 for the least polymorphic locus (rpoD) to 24 for the most polymorphic locus (gyrB). The nucleotide transitions exceed transversions, with ratios ranging from 1.005 to 2.552. The nucleotide diversity (π) varies from 0.5% (glnA) to 1.1% (gyrB), and the percentage of variable sites ranges from 1.5% (glnA) to 3% (gyrB). These percentages confirm that Xoo and Xoc are highly homogeneous and genetically closely related. Indeed, these percentages are slightly lower than those observed in X. campestris (1.8%–10.2%) (Fargier et al., 2011) and Salmonella (3.3%–5.6%) (Kotetishvili et al., 2002) and considerably lower than those observed in Helicobacter pylori (19.8%–23.7%) (Solcàet al., 2001). The low K a/K s ratios, from 0.027 (glnA) to 0.150 (gyrB), are similar to those observed for housekeeping genes in X. campestris (Fargier et al., 2011) and X. citri (Ah‐You et al., 2009). Taken together, these data indicate that gyrB is the most diverse gene among the three housekeeping genes analysed in this study, and confirm that this gene is well suited to the determination of the phylogenetic structure of Xanthomonas species and pathovars (Parkinson et al., 2009). However, it is worth noting that glnA and rpoD, in some cases, are more informative than gyrB. Indeed, for instance, strains 12898 and 12896 appear to be identical on the basis of gyrB, but are discriminated on the basis of rpoD. Another example concerns strains CFBP2287 and CFBP7109, which are identical on the basis of gyrB, but are differentiated on the basis of glnA. Therefore, all three genes are informative, even for this bacterial species, X. oryzae, which is highly homogeneous. Finally, neutrality tests revealed positive values of Tajima estimators of 1.98 (gyrB) to 2.5 (rpoD). Although these values are not significant, they might reveal certain demographic effects, such as a structured population.
Table 1.
Sequence variations for three housekeeping genes of Xanthomonas oryzae pv. oryzae and Xanthomonas oryzae pv. oryzicola strains used in this study.
| Gene | gyrB | rpoD | glnA |
|---|---|---|---|
| Fragment size (bp) | 801 | 737 | 887 |
| G + C content (mol.%) | 65.4 | 65.1 | 62.4 |
| Haplotype (H) | 10 | 5 | 4 |
| Polymorphic sites (N) | 24 | 12 | 13 |
| Percentage of variables sites | 3 | 1.6 | 1.5 |
| Nucleotide diversity per site (π) | 0.01097 | 0.00688 | 0.00577 |
| Ts/Tv* | 1.005 | 1.932 | 2.552 |
| K a † | 0.05199 | 0.02654 | 0.00820 |
| K s † | 0.28605 | 0.23444 | 0.24899 |
| K a/K s † | 0.150 | 0.096 | 0.027 |
| Tajima's D ‡ | 1.98808 | 2.55462 | 2.18633 |
Transition/transversion ratios (Ts/Tv) were determined using Kimura's two‐parameter method (Kimura, 1980).
Synonymous (K s) and nonsynonymous (K a) substitution rates were determined using the method of Nei and Gojobori (1986).
The values of Tajima's D are not significant (P > 0.05).
Figure 1.
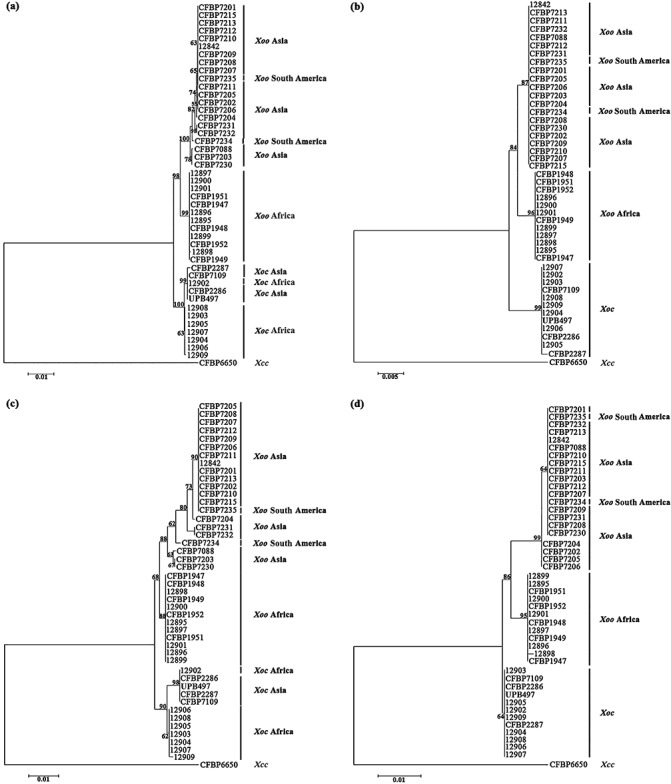
Phylogenetic trees of Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc) strains based on partial housekeeping gene sequences. Phylogenetic trees resulting from the concatenated dataset (a), glnA (b), gyrB (c) and rpoD (d) sequence analyses. These trees were constructed using the neighbour‐joining method and rooted with the strain CFBP6650 of Xanthomonas campestris pv. campestris (Xcc). Confidence on the nodes was tested with 1000 bootstrap replicates. The scale bars indicate the number of nucleotide substitutions per site.
Intragenic recombination events were estimated on each locus, as well as on the concatenated dataset, using dnasp and rdp (Recombination Detection Program). No recombination event was detected whatever the software and data analysed. Furthermore, using network 4.6, which generated an evolutionary network reflecting the mutational relationships among Xoo and Xoc haplotypes, no complex reticulation was found, supporting the absence of recombination events (Fig. 2). The same result was obtained with X. citri (Bui Thi Ngoc et al., 2010), but not with X. campestris, for which recombination was shown to play an important role in the genetic diversity of this plant pathogen (Fargier et al., 2011).
Figure 2.
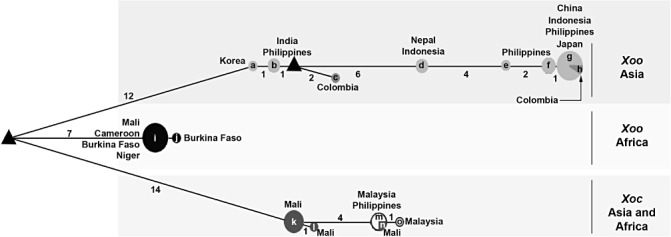
Median‐joining network of haplotypes inferred on the basis of polymorphisms observed in housekeeping genes (glnA, gyrB and rpoD) of Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc) strains. Haplotypes are depicted as circles, the radius of which is proportional to their allele frequency. The geographical origins of the haplotypes are shown next to the circles as well as the number of strains per haplotype. The branch length is proportional to the number of nucleotide changes, these numbers being mentioned next to the branches. Predicted intermediate (median) vectors are shown as triangles. Letters correspond to the following strains: a, CFBP7088; b, CFBP7203 and CFBP7230; c, CFBP7234; d, CFBP7231and CFBP7232; e, CFBP7204; f, CFBP7202, CFBP7205 and CFBP7206; g, CFBP7201, CFBP7202, CFBP7206, CFBP7207, CFBP7208, CFBP7209, CFBP7210, CFBP7211, CFBP7213, CFBP7215 and 12842; h, CFBP7235; i, 12895, 12896, 12897, 12899, 12900, 12901, CFBP1947, CFBP1948, CFBP1949, CFBP1951 and CFBP1952; j, 12898; k, 12903, 12904, 12905, 12906, 12907 and 12908; l, 12909; m, 497, CFBP2286 and CFBP7109; n, 12902; o, CFBP2287.
On the basis of the concatenated dataset, we evaluated the nucleotide diversity of Xoo and Xoc strains by the two estimators π and θ w. Our data indicate that Xoo strains (π= 0.00553, θ w= 0.00305) are more diverse than Xoc strains (π= 0.00101, θ w= 0.00082). This result is confirmed by median‐joining network analysis, as Xoo comprises nine haplotypes, whereas Xoc is composed of only four haplotypes (Fig. 2). Moreover, when we consider the geographical origin of the strains, Asian Xoo strains (π= 0.00174, θ w= 0.00149) appear to be more diverse than African Xoo strains (π= 0.00007, θ w= 0.00014), African Xoc strains (π= 0.00052, θ w= 0.00080) and Asian Xoc strains (π= 0.00021, θ w= 0.00022). The highest genetic diversity of Asian Xoo strains compared with African Xoo strains or African and Asian Xoc strains is also confirmed with the haplotype network (Fig. 2).
Phylogenetic trees were constructed by the neighbour‐joining method using individual gene sequences and concatenated sequence data (Fig. 1). Altogether, phylogenetic trees based on each locus are congruent with the concatenated tree, confirming the absence of intergenic and intragenic recombination. Indeed, all phylogenetic trees clearly show two major branches that are supported by high bootstrap values (98% and 100% based on the concatenated tree). These two major clusters comprise the Xoo and Xoc strains, respectively. This result confirms that pathovars, which have been defined in plant pathogenic bacteria at the infraspecific level by reference to host range or ability to cause distinctive symptoms (Dye et al., 1980), are distinct genetic and evolutionary lineages, as already revealed for other pathovars in the genus Xanthomonas (Bui Thi Ngoc et al., 2010; Fargier et al., 2011).
With regard to Xoo strains, all of the phylogenetic trees (Fig. 1) reveal that these strains can be separated into two groups: the first group comprises African Xoo strains, and the second Asian Xoo strains. Thus, this study shows that African Xoo strains form a group phylogenetically distant from Asian Xoo strains, and thus confirms the previous molecular and phylogenetic characterizations of African X. oryzae strains (Gonzalez et al., 2007; Triplett et al., 2011). This genetic differentiation associated with the geographical origin of the Xoo strains is supported by the Bayesian clustering and median‐joining network methods. Indeed, three major clusters (Fig. S1, see Supporting Information) and three major branches (Fig. 2), corresponding to Asian Xoo strains, African Xoo strains and African Xoc strains, respectively, are identified. Concerning Xoc strains, differentiation is also observed according to the geographical origin of the strains, but not with all of the methods used in this study. Indeed, the separation of African and Asian Xoc strains is only observed on the phylogenetic trees generated with gyrB (the most diverse housekeeping gene in this study; Table 1, Fig. 1) and the concatenated sequence data (Fig. 1). The genetic differentiation between African and Asian Xoc strains is also revealed by the median‐joining network analysis (Fig. 2), but not by the Bayesian clustering approach, as all Xoc strains are gathered into only one cluster (Fig. S1). Interestingly, based on the median‐joining network (Fig. 2), Asian Xoc strains might have been derived from African Xoc strains during the course of evolution. To support this hypothesis, further analysis of a larger collection of Xoc strains is required, as we used only a few Xoc strains in this study. Moreover, it will be useful to include more housekeeping genes, such as atpD or dnaK, which have been used previously in MLSA studies of Xanthomonas strains (Ah‐You et al., 2009; Bui Thi Ngoc et al., 2010; Fargier et al., 2011; Parkinson et al., 2009; Triplett et al., 2011; Young et al., 2008), as they might reveal more differences between African and Asian Xoc strains.
With regard to the genetic relatedness of X. oryzae strains, the respective positions of African and Asian Xoo strains remain unclear and seem to depend on the method used to measure diversity, as well as on the gene studied. Indeed, our phylogenetic trees, showing African Xoo strains to be more closely related to Asian Xoo strains than to Xoc strains (Fig. 1), are in complete agreement with the fluorescent AFLP results obtained by Gonzalez et al. (2007). Conversely, our results do not support the phylogenetic trees generated by Triplett et al. (2011) or the restriction fragment length polymorphism (RFLP) and Rep‐PCR analyses performed by Gonzalez et al. (2007), who reported African Xoo strains to be more closely related to Xoc strains than to Asian Xoo strains.
Altogether, our results show that Xoo and Xoc in Africa and Asia are closely related, but phylogenetically distinct, and that Xoo from Asia exhibits the highest genetic diversity compared with Xoc from Asia or Xoo and Xoc from Africa. These data strongly suggest that African and Asian Xoo and Xoc strains have different evolutionary histories. Xoo and Xoc strains might be endemic populations in Africa and Asia, and the genetic differences observed in this study might reflect the impact of different control measures of both diseases on both continents. With regard to Xoo, the presence of this pathogen has long been reported in Asia, and control of the disease is mainly based on the intensive use of major resistance (R) genes (Niño‐Liu et al., 2006; Vera Cruz et al., 2000). In Africa, intensive cultivation of rice started only in the 1960s, Xoo was reported in the 1980s, but no breeding programmes have been conducted against this pathogen (Gonzalez et al., 2007). Thus, selection pressures imposed by the host defence system are clearly different in Asia and Africa, and this might explain the highest genetic diversity of Asian Xoo strains. However, we can also hypothesize that this highest genetic diversity might be explained by the fact that Asia might be the centre of origin of Xoo strains. With regard to Xoc, no major resistance gene has been found and resistance to the disease is believed to be quantitative (Niño‐Liu et al., 2006; Zhao et al., 2004). Selection pressures on Xoc populations are certainly fairly weak in Asia and Africa, and might explain the low genetic diversity of this pathogen. Thus, it is tempting to speculate that Xoo and Xoc in Africa and Asia are endemic populations that have evolved separately. Nonetheless, we cannot completely rule out the possibility of transfer between continents through contaminated germplasm or seeds, even though seed transmission of Xoo and Xoc remains controversial (Niño‐Liu et al., 2006). Indeed, in our study, the Xoc 12902 strain from Mali constantly clusters with Asian Xoc strains (Fig. 1). This suggests exchanges between Africa and Asia. Another example concerns two strains (CFBP7234 and CFBP7235) originating from South America (i.e. Colombia) that fall into the Asian Xoo strains group (Fig. 1). The same result was obtained by Triplett et al. (2011). The most likely explanation would be that both strains have been introduced into Colombia from Asia.
Determination of T3E repertoires confirms that African and Asian strains of Xoo and Xoc are closely related, but distinct, evolutionary groups
By PCR and dot‐blot hybridization methods, we investigated the distribution of 32 T3E genes in our collection of African and Asian strains of Xoo and Xoc. Our study does not reveal many differences in the composition of T3E repertoires (Fig. 3), even though these strains were collected from various locations and at different times (Table 2). Notably, we were not able to discriminate between African Xoo strains isolated in 1979 (from strains CFBP1947–CFBP1952) and those isolated in 2003 and 2004 (from strains 12895–12901). During this period, intensive rice cultivation was conducted in Africa, which has probably had an impact on the population structure of Xoo. Genetic variations in Xoo and Xoc T3E repertoires might reside in the T3E genes that have not been included in this study (http://www.xanthomonas.org/) or T3E genes of unidentified effector families. It is worth noting that genetic variations allowing the differentiation of African and Asian Xoo and Xoc strains were revealed by studying TAL effector genes. Indeed, there are multiple copies of TAL effectors in Xoo and Xoc, and the copy number varies among the strains. For instance, the Asian Xoo PXO99A strain has 19 TAL effector genes in its genome (Salzberg et al., 2008), whereas the Asian Xoc BLS256 strain has 26 copies in its genome (Bogdanove et al., 2011). The repertoire of TALs in African Xoo strains is reduced compared with that in Asian strains, with only eight TAL effector genes in their genomes (Gonzalez et al., 2007). Mutation in one of the eight TALs (TALc) severely affects the capacity of the bacteria to produce disease symptoms (Yu et al., 2011). African Xoc strains have as many TAL copy numbers as Asian Xoc strains (Gonzalez et al., 2007).
Figure 3.
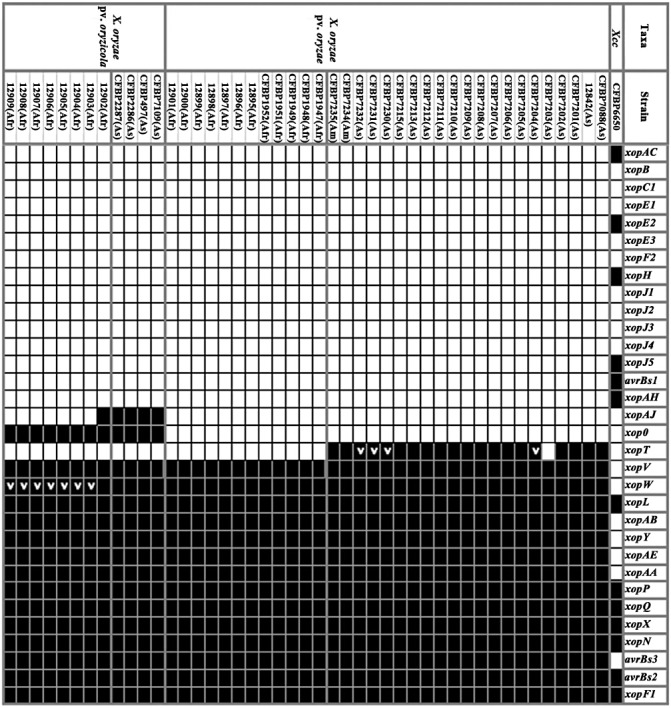
Distribution of type III effector (T3E) genes among African and Asian strains of Xanthomonas oryzae pv. oryzae and Xanthomonas oryzae pv. oryzicola. The presence or absence of an orthologue of each selected T3E gene was determined by polymerase chain reaction (PCR) and dot‐blot hybridization methods. Black squares and white squares indicate the presence and absence of the corresponding gene, respectively. A gene may be absent when its sequence is too divergent to be detected through PCR and dot‐blot. The T3E repertoire of Xanthomonas campestris pv. campestris (Xcc) strain CFBP6650 is also mentioned based on its genome sequence (http://www.xanthomonas.org). ‘>’ indicates that a DNA rearrangement was identified within the T3E gene [insertion sequence (IS) element in xopW and tandem duplication in xopT].
Table 2.
Bacterial strains used in this study.
| Xanthomonas species and pathovars | Strain number* | Other collections* | Host of isolation | Geographical origin | Year of isolation |
|---|---|---|---|---|---|
| X. oryzae pv. oryzae | CFBP7088 | KACC10331 | Oryza sativa | Korea | NA |
| X. oryzae pv. oryzae | 12842 | MAFF311018 | Oryza sativa | Japan | NA |
| X. oryzae pv. oryzae | CFBP7201 | PXO61 | Oryza sativa | Philippines | 1973 |
| X. oryzae pv. oryzae | CFBP7202 | PXO86 | Oryza sativa | Philippines | 1977 |
| X. oryzae pv. oryzae | CFBP7203 | PXO99A | Oryza sativa | Philippines | 1980 |
| X. oryzae pv. oryzae | CFBP7204 | PXO112 | Oryza sativa | Philippines | NA |
| X. oryzae pv. oryzae | CFBP7205 | PXO145 | Oryza sativa | Philippines | 1982 |
| X. oryzae pv. oryzae | CFBP7206 | PXO280 | Oryza sativa | Philippines | NA |
| X. oryzae pv. oryzae | CFBP7207 | PXO339 | Oryza sativa | Philippines | NA |
| X. oryzae pv. oryzae | CFBP7208 | PXO340 | Oryza sativa | Philippines | NA |
| X. oryzae pv. oryzae | CFBP7209 | PXO341 | Oryza sativa | Philippines | NA |
| X. oryzae pv. oryzae | CFBP7210 | PXO345 | Oryza sativa | Philippines | NA |
| X. oryzae pv. oryzae | CFBP7211 | PXO448 | Oryza sativa | Philippines | NA |
| X. oryzae pv. oryzae | CFBP7212 | HN35 | Oryza sativa | China | NA |
| X. oryzae pv. oryzae | CFBP7213 | IXO60 | Oryza sativa | Indonesia | NA |
| X. oryzae pv. oryzae | CFBP7215 | PXO71 | Oryza sativa | Philippines | 1979 |
| X. oryzae pv. oryzae | CFBP7230 | A3857 | Oryza sativa | India | NA |
| X. oryzae pv. oryzae | CFBP7231 | IXO57 | Oryza sativa | Indonesia | NA |
| X. oryzae pv. oryzae | CFBP7232 | NXO622 | Oryza sativa | Nepal | NA |
| X. oryzae pv. oryzae | CFBP7234 | LMG634 | Oryza sativa | Colombia | NA |
| X. oryzae pv. oryzae | CFBP7235 | LMG635 | Oryza sativa | Colombia | NA |
| X. oryzae pv. oryzae | CFBP1947 | LMG12464 | Oryza sativa | Cameroon | 1979 |
| X. oryzae pv. oryzae | CFBP1948 | LMG12465 | Oryza sativa | Cameroon | 1979 |
| X. oryzae pv. oryzae | CFBP1949 | LMG12466 | Oryza sativa | Mali | 1979 |
| X. oryzae pv. oryzae | CFBP1951 | LMG12467 | Oryza sativa | Mali | 1979 |
| X. oryzae pv. oryzae | CFBP1952 | LMG12468 | Oryza sativa | Mali | 1979 |
| X. oryzae pv. oryzae | 12895 | BAI1 | Oryza sativa | Burkina Faso | 2004 |
| X. oryzae pv. oryzae | 12896 | BAI2 | Oryza sativa | Burkina Faso | 2004 |
| X. oryzae pv. oryzae | 12897 | BAI3 | Oryza sativa | Burkina Faso | 2004 |
| X. oryzae pv. oryzae | 12898 | BAI4 | Oryza glaberrima | Burkina Faso | 2004 |
| X. oryzae pv. oryzae | 12899 | NAI5 | Oryza sativa | Niger | 2004 |
| X. oryzae pv. oryzae | 12900 | NAI9 | Oryza sativa | Niger | 2004 |
| X. oryzae pv. oryzae | 12901 | MAI1 | Oryza sativa | Mali | 2003 |
| X. oryzae pv. oryzicola | 12902 | MAI3 | Oryza sativa | Mali | 2003 |
| X. oryzae pv. oryzicola | 12903 | MAI4 | Oryza sativa | Mali | 2003 |
| X. oryzae pv. oryzicola | 12904 | MAI5 | Oryza sativa | Mali | 2003 |
| X. oryzae pv. oryzicola | 12905 | MAI6 | Oryza sativa | Mali | 2003 |
| X. oryzae pv. oryzicola | 12906 | MAI7 | Oryza sativa | Mali | 2003 |
| X. oryzae pv. oryzicola | 12907 | MAI8 | Oryza sativa | Mali | 2004 |
| X. oryzae pv. oryzicola | 12908 | MAI10 | Oryza sativa | Mali | 2004 |
| X. oryzae pv. oryzicola | 12909 | MAI11 | Oryza sativa | Mali | 2004 |
| X. oryzae pv. oryzicola | CFBP7109 | BLS256 | Oryza sativa | Philippines | 1985 |
| X. oryzae pv. oryzicola | UPB497 | UPB497 | Oryza sativa | Malaysia | NA |
| X. oryzae pv. oryzicola | CFBP2286 | LMG797 | Oryza sativa | Malaysia | 1964 |
| X. oryzae pv. oryzicola | CFBP2287 | NCPPB2921 | Oryza sativa | Malaysia | 1973 |
| X. campestris pv. campestris | CFBP6650 | LMG8004 | Brassica oleracea | UK | 1958 |
CFBP, Collection Française de Bactéries Phytopathogènes, Institut National de la Recherche Agronomique (INRA), Angers, France; 12842, 12895, our own local collection; PXO, Philippines Xanthomonas oryzae pv. oryzae strains collection, The International Rice Research Institute (IRRI), Los Baños, Philippines; KACC, Korean Agricultural Culture Collection, National Institute of Agricultural Biotechnology, Suwon, South Korea; MAFF, Ministry of Agriculture, Forestry and Fisheries, National Institute of Agrobiological Sciences, Tsukuba, Japan; LMG/BCCM, Belgian Coordinated Collection of Microorganisms, University of Gent, Belgium; NCPPB, National Collection of Plant Pathogenic Bacteria, UK; BLS, strains of bacterial leaf streak of cereals; MAI, NAI and BAI, Xanthomonas oryzae WARDA‐IRD (Africa Rice Centre—Institut de la Recherche pour le Développement) Collection from Mali, Niger and Burkina Faso, respectively; UPB, Unité de Phytopathologie Bacterial Collection, Louvain La Neuve, Belgium. DNA of A, HN, IX, MXO and NXO strains was provided by J. Leach (Colorado State University, Fort Collins, CO).
On the basis of the T3E presence/absence matrix, we constructed a dendrogram showing the genetic relationships of Xoo and Xoc strains (Fig. 4). Despite the close relatedness of these strains, the dendrogram allows the differentiation of four major groups that are well supported by high bootstrap values (Fig. 4). Interestingly, these groups gather strains according to their pathovar affiliation and to their geographical origin. The first two groups include Xoo strains from Asia and Africa, respectively, and the last two groups comprise Xoc strains from Asia and Africa, respectively. However, three exceptions can be noticed: the African Xoc strain 12902 that clusters with Asian Xoc strains, and the two South American Xoo strains (CFBP7234 and CFBP7235) that group with the Asian Xoo strains. The unexpected positions of these three strains on the T3E dendrogram (Fig. 4) are actually in agreement with the atypical phylogenetic positions of these strains revealed by MLSA (Fig. 1). This study based on T3E genes shows that African Xoo strains are genetically different from Asian Xoo strains, and thus confirms the phylogenetic structure of X. oryzae (Fig. 1; Triplett et al., 2011).
Figure 4.
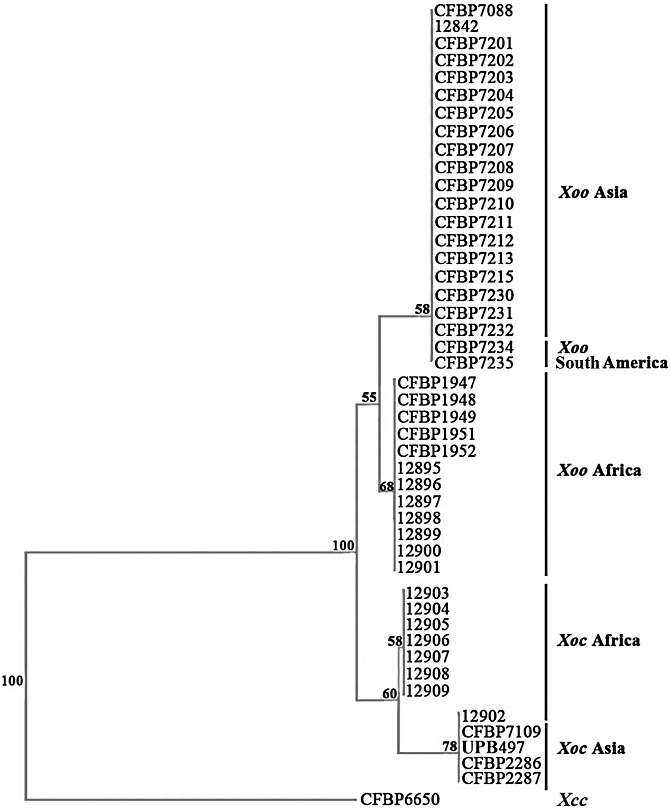
Dendrogram constructed on the basis of the results of the presence/absence of type III effectors (T3Es) in African and Asian strains of Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc). The dendrogram was built with the neighbour‐joining method using Jaccard distances and rooted with the strain CFBP6650 of Xanthomonas campestris pv. campestris (Xcc). Confidence on the nodes was tested with 1000 bootstrap replicates.
Altogether, our results clearly show a correspondence between the composition of T3E repertoires and the pathovars of X. oryzae, and thus confirm our findings obtained with a collection of X. axonopodis strains (Hajri et al., 2009). This is in accordance with the concept that T3E repertoires represent candidate determinants of host and tissue specificity in plant pathogenic bacteria. However, it should be kept in mind that other candidate determinants may be involved in host and tissue specificity, such as genes involved in chemotaxis, adhesion, sensing, biofilm formation, quorum sensing, motility or lipopolysaccharide synthesis (Büttner and Bonas, 2010; Mhedbi‐Hajri et al., 2011).
The mining of T3E repertoires of African and Asian strains of Xoo and Xoc provides insight into the evolutionary history of T3E genes in xanthomonads
Our work confirms that Xoo and Xoc are genetically closely related, as we showed that T3E repertoires are highly conserved within both pathogens. However, we have uncovered some differences that allow the distinction of three categories of genes within T3E repertoires (Fig. 3). The first group can be considered as the ubiquitous (= core) suite of T3Es for Xoo and Xoc, as it comprises 14 T3E genes that are present in all tested strains whatever their geographical origin. The second can be considered as the variable suite of T3Es for Xoo and Xoc, as it contains three T3E genes that are present in only some of the tested strains. The last group is composed of the remaining 15 T3E genes that we consider to be absent or sufficiently divergent in Xoo and Xoc. Indeed, our approach cannot completely exclude the possibility that some T3E genes may have been subjected to diversifying selection that may have resulted in a sufficient divergence sequence to avoid detection by PCR and dot‐blot hybridization methods.
Within the first group of genes that comprise the core suite of T3E genes for Xoo and Xoc (Fig. 3), it is interesting to note that some T3E genes, such as avrBs2, avrBs3/pthA, xopF1, xopN, xopQ and xopX, are also found to be ubiquitous in X. axonopodis pathovars (Hajri et al., 2009). Furthermore, when one looks at the Xanthomonas resource website (http://www.xanthomonas.org/), which gathers strains whose genomes have been sequenced, it can be seen that these T3E genes are also widely present, except avrBs3/pthA, within the strains of the species X. campestris, X. axonopodis, X. vasicola and X. fuscans. The fact that phylogenetically distant Xanthomonas strains harbour the same T3E gene suite suggests that these genes might have been acquired by the ancestor of Xanthomonas species before the diversification of pathovars, and thus before host specialization occurred. This observation strongly suggests that these T3E genes may have evolved from this ancestor by vertical descent among xanthomonads. To further support this hypothesis, it would be interesting to search for the presence of these T3E genes in large collections of Xanthomonas strains.
The second group of genes includes three T3E genes (xopAJ, xopO and xopT) that are present in only some of the Xoo and Xoc tested strains (Fig. 3). The variable presence of these T3E genes may reflect specific evolutionary histories. Indeed, these three genes exhibit a G + C content (51.1%, 53.1% and 61.3%, respectively) lower than the average value (∼64%) of Xoo and Xoc genomes, and are all flanked by insertion sequence (IS) elements (Bogdanove et al., 2011; Ochiai et al., 2005). These features, which are not observed for the majority of the core T3E genes, strongly suggest that these three T3E genes have been acquired by Xoo or Xoc through horizontal gene transfer events. This hypothesis is supported by previous analyses of Xanthomonas genomes which have clearly shown that these bacteria have been subjected to numerous horizontal gene transfers during evolution (Comas et al., 2006; Lu et al., 2008). For xopT and xopAJ, their presence can be correlated with the geographical origin of the strains, as xopT is detected only in South American and Asian Xoo strains (except for the CFBP7203 strain), and xopAJ is detected only in Asian Xoc strains (except for the 12902 strain). With regard to xopO, its presence can be correlated with a pathovar, as it is detected only in Xoc. These three T3E genes may thus reflect specific horizontal gene transfer events, as they are not widely distributed in xanthomonads (Hajri et al., 2009; http://www.xanthomonas.org/). This finding supports the observations made on Xanthomonas and other bacteria that, once acquired, genes are rarely transferred among lineages (Gillings et al., 2005; Lerat et al., 2005).
The mining of T3E repertoires provides clues for functional studies aiming to understand the virulence, as well as host and tissue specificity, of African and Asian strains of Xoo and Xoc
The characterization of T3E repertoires of Xoo and Xoc provides clues for functional studies on virulence, host and tissue specificity of both pathogens. With regard to the core T3E genes (Fig. 3), it is tempting to speculate, as has been proposed earlier (Grant et al., 2006; Rohmer et al., 2004; Stavrinides et al., 2008), that this gene set widely distributed in xanthomonads might provide virulence functions of broad utility and target defence components broadly conserved among a wide range of hosts. Thus, the loss of these genes would lead to a loss of fitness for the pathogens. Such an hypothesis is supported by functional studies carried out notably on avrBs2, xopN and xopX (Kearney and Staskawicz, 1990; Metz et al., 2005; Roden et al., 2004). It would be interesting to set up functional studies on T3E genes of this suite in Xoo and Xoc, for instance, on xopX, as it has been shown recently that this gene is highly expressed in an African Xoo strain during the early steps of host infection (Soto‐Suarez et al., 2010b). Otherwise, with regard to the other T3E genes (xopAA, xopAB, xopAE, xopL, xopP, xopQ, xopU, xopV and xopW) that are included in the ubiquitous T3E gene suite in Xoo and Xoc, it appears that their presence is not widely conserved in X. axonopodis pathovars (Hajri et al., 2009) or in other Xanthomonas strains (http://www.xanthomonas.org/). This suggests that these T3E genes might be critically important for Xoo and Xoc to target specific defence components of rice. These T3E genes may represent candidate determinants of Xoo and Xoc virulence, host and tissue specificity. Interestingly, a recent mutagenesis of T3E genes in the Philippine Xoo strain PXO99A has revealed that XopZ contributes to the virulence of this strain (Song and Yang, 2010).
The three variable T3Es (xopAJ, xopO and xopT) (Fig. 3) in Xoo and Xoc may reflect specific strategies during the interaction with the host, or may have an impact on the host range of the strains. For instance, the presence of xopAJ (=avrRxo1) in Asian Xoc strains is known to restrict the host range of these strains, this T3E gene being recognized by plants carrying the corresponding Rxo1 resistance gene (Zhao et al., 2004). Interestingly, the presence of xopO is detected in Xoc, but not in Xoo strains. This gene is not broadly distributed among xanthomonads, as its presence was detected only in X. axonopodis pv. vesicatoria strains (Hajri et al., 2009; http://www.xanthomonas.org/). xopO might represent a determinant of tissue specificity, as it is carried by two nonvascular Xanthomonas species. We carried out a functional study to examine the role of xopO in tissue specificity. Using a knockout and complementation approach, we inactivated xopO in the Xoc CFBP7109 (= BLS256) strain and introduced xopO into Xoo CFBP7203 (= PXO99A). Both experiments failed to show any phenotypic alterations on inoculation into rice compared with the wild‐type strains (Fig. S2, see Supporting Information). No change in tissue specificity was observed on expression of xopO in Xoo (evaluated by leaf‐clipping and infiltration pathogenicity assays), or on disruption of xopO in Xoc (tested by infiltration) (Fig. S2). Strikingly, the disruption of xopO did not alter the pathogenicity of X. axonopodis pv. vesicatoria (Roden et al., 2004). Such results may be explained by functional redundancy among T3Es as, in plant pathogenic bacteria, a mutation of a single T3E gene often has no detectable effect on pathogenicity (Kvitko et al., 2009).
Further investigation of T3E repertoires in a large collection of Xoo and Xoc strains is still required to identify new candidate determinants of host and tissue specificity. Indeed, in this study, we considered only 32 T3E genes and did not include other T3E genes that have been revealed by genome sequencing of Xanthomonas strains (http://www.xanthomonas.org). For instance, xopAF and xopAK, which we did not analyse in our study, might represent tissue specificity determinants, as both genes seem to be present in Xoc and not in Xoo (http://www.xanthomonas.org).
Pathoadaptation of X. oryzae strains is suggested by sequence variations revealed in two T3E genes
Sequence variation analysis of T3E genes is a complementary approach for the identification of candidate determinants of host and tissue specificity. Indeed, it has been suggested that subtle changes in some genes may be responsible for host and tissue specificity in Xanthomonas (Lu et al., 2008). DNA polymorphisms can modify the expression of the genes, the function of the gene products, the pathogenicity of the strains and thus the outcome of the plant–pathogen interaction, as has been demonstrated for Pseudomonas syringae (Ma et al., 2006; Zhou et al., 2009). Furthermore, it has been shown that the host defence may accelerate the generation of genetic rearrangements to provide a selective advantage to the pathogens (Arnold et al., 2007). In our study, we identified two types of DNA rearrangement within T3E genes that may represent examples of pathoadaptation (Sokurenko et al., 1999) for Xoo and Xoc strains. The first is a tandem duplication of 120 bp in size and affects xopT in four Asian Xoo strains (CFBP7204, CFBP7230, CFBP7231 and CFBP7232) (Fig. 3). Another tandem duplication has already been found in the xopD gene of an X. axonopodis pv. vesicatoria strain (Hajri et al., 2009). Interestingly, both tandem duplications do not shift the reading frames of both T3E genes, suggesting that these strains used these strategies to generate modified T3E forms to avoid recognition by the plant. Therefore, the evolutionary history of xopT, which is present in Asian and South American Xoo strains, but not in African strains (Fig. 3), might have been driven by exposure to a specific host defence system, as this gene is altered by a DNA rearrangement in four strains, and one can speculate that this gene has been deleted in strain CFBP7203, as it is not detected in this Asian strain (Fig. 3; Salzberg et al., 2008). Further sequencing of xopT in other Asian and South American Xoo strains is required to examine small DNA polymorphisms that were not detected by our approach. Further functional studies are necessary to analyse the role of allelic variation in the avoidance of host recognition. The second type of DNA rearrangement was identified in xopW, which is disrupted by the IS1112 element at position 432 in all African Xoc strains except one (strain 12902) (Fig. 3). This IS element of 1050 bp in size, belonging to the IS30 family, is widely distributed in Xoo and Xoc genomes (for instance, 20 complete copies and 12 truncated copies in Xoo strain MAFF311018; Ochiai et al., 2005). As many T3E genes have also been shown to be disrupted by ISs in X. axonopodis strains (Hajri et al., 2009), in P. syringae (Ma et al., 2006) and in Ralstonia solanacearum (Lavie et al., 2004), it can be speculated that the inactivation of T3E genes by ISs is a frequent strategy used by plant pathogenic bacteria to control the expression of virulence genes and to avoid detection by the plant defence system. Therefore, it can be speculated that African Xoc strains, in response to specific selection pressure imposed by host plants in Africa, may have driven the inactivation of xopW by insertion of IS1112. To further support this hypothesis, it would be interesting to complement African Xoc strains with a functional xopW. Altogether, our data reinforce our interest in mobile genetic elements, as numerous ISs in one Xoo African strain have been shown to be differentially expressed in planta during infection, suggesting that these mobile elements may play a significant role in bacterial pathogenicity (Soto‐Suarez et al., 2010b).
Finally, the discovery of DNA rearrangements in xopT and xopW reinforces the need to examine the allelic diversity of T3E genes in our collection of Xoo and Xoc strains. Thus, we now plan to sequence and analyse T3E gene product polymorphisms for comparative analyses with the phylogeny of the X. oryzae strains determined during the course of this study. By analysing amino acid residues, hpaA and xpsD have been revealed as candidate determinants of tissue specificity in Xanthomonas (Lu et al., 2008). Such an approach will provide resources for functional and evolutionary studies aiming to understand host and tissue specificity, functional redundancy between T3Es and the driving forces shaping T3E repertoires in African and Asian Xoo and Xoc strains.
EXPERIMENTAL PROCEDURES
Bacterial strains, growth conditions and DNA extraction
The bacterial strains used in this study are listed in Table 2, including those whose genomes have been sequenced: Xoc strain CFBP7109 (BLS256; http://cmr.jcvi.org/tigr‐scripts/CMR/cmrHomePage.cgi) and Xoo strains CFBP7088 (KACC10331; Lee et al., 2005) 12842 (MAFF311018; Ochiai et al., 2005) and CFBP7203 (PXO99A; Salzberg et al., 2008). All of these sequenced strains were used as positive or negative controls for PCR and dot‐blot hybridizations. All strains were routinely cultured on YPGA (yeast extract, 7 g/L; peptone, 7 g/L; glucose, 7 g/L; agar, 15 g/L) at 28 °C. Genomic DNA was extracted from all bacterial strains grown overnight at 28 °C in YP medium (yeast extract, 7 g/L; peptone, 7 g/L) using the standard hexadecyltrimethylammonium bromide method (Ausubel et al., 1991). The quality and quantity of DNA were evaluated spectrophotometrically (Nanodrop ND‐1000; Nanodrop Technologies, Wilmington, DE).
Phylogenetic and molecular evolutionary analyses
Three housekeeping genes, glnA (glutamine synthetase I), gyrB (DNA gyrase subunit B) and rpoD (RNA polymerase sigma‐70 factor), were sequenced with the primers shown in Table S1 (see Supporting Information). All amplifications were carried out in a final volume of 20 µL containing 1 × ColorlessGoTaq® Flexi Buffer (Promega, Verrières, France), 200 µm of each deoxynucleoside triphosphate (dNTP) (Promega), 400 nm of each primer, 1.5 mm MgCl2, 1 unit of GoTaq® Flexi DNA polymerase (Promega) and 50 ng of genomic DNA. The reactions were run for 35 cycles, each consisting of 50 s at 94 °C, 50 s at 60 °C and 1 min at 72 °C, with initial denaturation of 3 min at 94 °C and final extension of 7 min at 72 °C. Amplification products were separated on a 1.5% agarose gel in Tris/Borate/EDTA buffer, stained with ethidium bromide and visualized under UV light. PCR amplicons were sequenced at the Biogenouest platform (http://www.ifr26.univ‐nantes.fr), using the primers shown in Table S1. A subset of genes was replicated twice to assess the reproducibility of the PCR and sequencing results. The glnA, gyrB and rpoD sequences from X. campestris pv. campestris CFBP6650 were used as the outgroup for phylogenetic analyses. The sequences of all alleles were deposited in GenBank under accession numbers JF815233–JF815366.
All sequences were edited, assembled, aligned and concatenated using the geneious v 4.7.6 software package. If a polymorphic nucleotide was present in only one sequence, the sequence was re‐examined to ensure that the base call was accurate. The network 4.6 software package was used to construct the minimum‐mutation network, which reflects the mutational relationships among the inferred haplotypes by means of the median‐joining algorithm (Bandelt et al., 1999). Tajima's D test of selective neutrality (Tajima, 1989), the evaluation of synonymous/nonsynonymous substitution ratios (K a/K s) by the method of Nei and Gojobori (1986), the nucleotide diversity π (probability that two randomly selected sequences possess different nucleotides at a site) and θ w (estimate of the population mutation rate calculated from the total number of segregating sites with correction for sample size) were calculated using dnasp version 5.10 (Librado and Rozas, 2009). Transition/transversion ratios were determined using Kimura's two‐parameter model (Kimura, 1980) with the mega version 4.1 program (Tamura et al., 2007). rdp software v3.44 (Martin et al., 2003) was used to infer recombination events with the MaxChi matrix and the chimaera, geneconv, rdp, maxchi, bootscan, 3seq, lard, phylpro and siscan methods. The analyses were performed with default settings and a Bonferroni corrected P value cut‐off of 0.05. The concatenated X. oryzae sequences were assigned to bacterial clusters using the ‘no admixture’ model of the program structure 2.1 (Pritchard et al., 2000). Xmfa2 software was used to convert the sequence data into the input file format of structure (http://www.Xavierdidelot.xtreemhost.com/clonalframe.htm). In each run, a Markov Chain Monte Carlo (MCMC) simulation of 500 000 iterations approximated the posterior probability of K, following a burn‐in of 50 000 iterations. The optimal number of bacterial clusters K was determined by comparing the posterior probability from 10 runs assuming 1 ≤K≤ 8 according to the method developed by Evanno et al. (2005). K= 3 yielded the highest probability and consistent strain assignment to clusters.
Determination and analysis of T3E repertoires
To characterize the T3E repertoires of our collection of strains, we carried out PCR and dot‐blot hybridization methods, as described previously (Hajri et al., 2009). A subset of T3E genes was replicated twice to assess the reproducibility of the PCR and dot‐blot results. Table S1 presents a list of the 32 selected T3E genes, as well as the primer pairs used for PCR amplifications and for the preparation of probes. We used the T3E gene nomenclature available on the Xanthomonas resource website (http://www.xanthomonas.org/). We selected 30 T3E genes identified from the sequenced genomes of Xanthomonas strains (Xoo strain 12842, X. axonopodis pv. vesicatoria strain CFBP5618, X. campestris pv. campestris strain CFBP5241, X. axonopodis pv. citri strain 306) (Da Silva et al., 2002; Ochiai et al., 2005; Thieme et al., 2005). We also selected two T3E genes from X. axonopodis pv. vesicatoria strains, whose genome has not been sequenced: xopJ2 from strain 75‐3 and xopJ4 from strain 91‐118 (http://www.xanthomonas.org/).
A few PCR products of an unexpected size were obtained on agarose gels, purified with the Nucleospin Extract II Kit (Macherey‐Nagel EURL, Hoerdt, France) and sequenced at the Genoscreen genomic analysis platform (Lille, France). We sequenced xopT from the Xoo CFBP7204, CFBP7230, CFBP7231 and CFBP7232 strains, as well as xopW from the Xoc 12903, 12904, 12905, 12906, 12907, 12908 and 12909 strains. Sequence data were examined using the Blast search programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the Mobyle portal (http://mobyle.pasteur.fr/cgi‐bin/portal.py) and the IS Finder database (http://www‐is.biotoul.fr/is.html).
Based on the presence/absence matrix of T3E genes for each of the strains, we constructed a dendrogram using Jaccard distances and the neighbour‐joining method. Bootstrapping was performed with 1000 replicates to assess the robustness of the dendrogram. The resulting dendrogram was visualized using past 2.01 software (http://folk.uio.no/ohammer/past/).
Functional analysis of the T3E gene xopO in Xanthomonas oryzae
The nonpolar mutant of xopO in the Xoc strain CFBP7109 (= BLS256) was constructed by homologous integration with a suicide plasmid using pK18mob as a vector (Schäfer et al., 1994), as described previously (Windgassen et al., 2000). A 303‐bp internal fragment of the xopO gene (from 61 bp downstream of the start codon to 363 bp) was amplified using total DNA of Xoc CFBP7109 as the template and the primer set xopOMF/xopOMR (Table S2, see Supporting Information). The amplified DNA fragment was digested with restriction enzymes EcoRI and BamHI, recovered and ligated to the same digested vector pKMob18 to create the recombinant plasmid pKxopO, and confirmed by sequencing. The plasmid pKxopO was transferred into Xoc strain CFBP7109 by triparental conjugation, as described by Turner et al. (1985), using pRK2073 as the helper plasmid (Leong et al., 1982). With a mutation in the xopO gene, transconjugants were confirmed by PCR using total DNA as the template and the primer set pKMob18F/xopOYR (Table S2). Total DNA from the wild‐type strain as a template was used as a negative control. The expected 1005‐bp PCR products were further confirmed by sequencing. One of the confirmed mutants, designated as BLSMxopO, was used for further study.
To complement the Xoc xopO mutant or to express the xopO gene in Xoo, a 1199‐bp DNA fragment containing the entire xopO gene (from 246 bp upstream of the start codon to 300 bp downstream of the stop codon) was amplified by PCR using total DNA from Xoc strain CFBP7109 as the template and the primer set xopOCF/xopOCR (Table S2). After the amplified DNA fragment had been cut by enzymes XbaI and SacI, the recovered DNA fragment was ligated to the same digested plasmid pBBR1MCS‐5 (Kovach et al., 1995) to create the recombinant plasmid pBxopO, which was confirmed by sequencing. The plasmid pBxopO was transferred into Xoc xopO mutant BLSMxopO or Xoo strain CFBP7203 (= PXO99A), respectively, by triparental mating as described above. The transconjugants were confirmed by PCR using total DNA as the template and the primer set T7Promotor/M13‐26 (Table S2). Total DNA from the recipient strain as a template was used as a negative control. The expected 1367‐bp PCR products were further confirmed by sequencing. One confirmed complemented strain of Xoc xopO mutant BLSMxopO was named BLSCxopO, and one verified Xoo transconjugant was designated as PXO99A/xopO. Similarly, the empty plasmid vector pBBR1MCS‐5 was introduced into the Xoo strain CFBP7203 to generate the strain named PXO99A/pBBR1.
For plant assays, rice cultivar Nipponbare plants were grown in a glasshouse with 12 h of light alternating with 12 h of darkness at temperatures of 28 °C during the day and 25 °C at night. Bacterial cells were grown on an oryzae broth medium agar plate (Tang et al., 1996) at 28 °C for 48 h. The bacterial cells from the agar plate were suspended in sterilized water and the cell concentration was adjusted to an optical density at 600 nm (OD600) of 0.2 [2 × 108 colony‐forming units (cfu)/mL]. The virulence of Xoc strains was tested on 14‐day‐old seedlings of rice Nipponbare by leaf infiltration inoculation. The virulence of Xoo strains was assayed on 30‐day‐old Nipponbare plants by leaf clipping (Kauffman et al., 1973) and on 14‐day‐old rice seedlings by leaf infiltration inoculation. Thirty leaves were inoculated with each strain in each treatment and maintained under the growth conditions described above. The lesion length was measured 7 days after inoculation with Xoc strains, 14 days after inoculation with Xoo strains by leaf clipping and 7 days after inoculation with Xoo strains by leaf infiltration. Each treatment was repeated three times in each experiment. The same experiment was repeated three times.
Supporting information
Fig. S1 Plot of the Bayesian assignment of Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc) strains to clusters based on the analysis of the concatenated dataset (glnA, gyrB and rpoD sequences) using the no admixture model of structure 2.1 software. Each haplotype is represented by a line and the geographical origin of the strains is given.
Fig. S2 The type III effector (T3E) XopO is not essential for the virulence and tissue specificity of Xanthomonas oryzae in rice Nipponbare. The bacterial cells were suspended in sterilized water and the cell concentration was adjusted to an optical density at 600 nm (OD600) of 0.2 '2 × 108 colony‐forming units (cfu)/mL'. Data shown are the means ± standard deviation from three replications, each with 30 leaves, from one representative experiment; similar results were obtained in the other two independent experiments. (a) Average length of lesions caused by Xanthomonas oryzae pv. oryzicola 7 days post‐inoculation. Approximately 20 µL of bacterial culture suspension were infiltrated into leaf mesophyll tissue with a 1‐mL blunt‐end plastic syringe. (b) Average length of lesions caused by Xanthomonas oryzae pv. oryzae inoculated by the leaf‐clipping method, 14 days after inoculation. (c) Average length of lesions caused by Xanthomonas oryzae pv. oryzae inoculated by the leaf infiltration method, 7 days after inoculation.
Table S1 The type III effector (T3E) genes analysed in this study: gene functions and primer sequences used for polymerase chain reaction (PCR) amplifications and for the preparation of probes for dot‐blot hybridizations.
Table S2 Polymerase chain reaction (PCR) primers used for the functional analysis of the type III effector (T3E) gene xopO in Xanthomonas oryzae.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This study was funded by the Region Pays de la Loire (Xanthost program) and by the Plant Health and Environment Department of Institut National de la Recherche Agronomique (INRA). Ahmed Hajri was supported by a grant from the Tunisian Government, from the Conseil Général du Maine‐et‐Loire and from INRA. Shuai Zhao was supported by ‘Bourses Doctorales en Alternance’ of the French Embassy in China and programs for the joint training of doctoral students and academic visiting of postgraduates of Guangxi University.
REFERENCES
- Adhikari, T.B. , Vera Cruz, C.M. , Zhang, Q. , Nelson, R.J. , Skinner, D.Z. , Mew, T.W. and Leach, J.E. (1995) Genetic diversity of Xanthomonas oryzae pv. oryzae in Asia. Appl. Environ. Microbiol. 61, 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari, T.B. , Mew, T.W. and Leach, J.E. (1999) Genotypic and pathotypic diversity in Xanthomonas oryzae pv. oryzae in Nepal. Phytopathology, 89, 687–694. [DOI] [PubMed] [Google Scholar]
- Ah‐You, N. , Gagnevin, L. , Grimont, P.A. , Brisse, S. , Nesme, X. , Chiroleu, F. , Bui Thi Ngoc, L. , Jouen, E. , Lefeuvre, P. , Vernière, C. and Pruvost, O. (2009) Polyphasic characterization of xanthomonads pathogenic to members of the Anacardiaceae and their relatedness to species of Xanthomonas . Int. J. Syst. Evol. Microbiol. 59, 306–318. [DOI] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Physiol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Arnold, D.L. , Jackson, R.W. , Waterfield, N.R. and Mansfield, J.W. (2007) Evolution of microbial virulence: the benefits of stress. Trends Genet. 23, 293–300. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. , Brent, R. , Kingston, R.E. , Moore, D.D. , Seidman, J.G. , Smith, J.A. and Struhl, K. (1991) Current Protocols in Molecular Biology. New York: Greene Publishing Associates‐Wiley Interscience. [Google Scholar]
- Bandelt, H.J. , Forster, P. and Rohl., A. (1999) Median‐joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. [DOI] [PubMed] [Google Scholar]
- Basso, A. , Onasanya, A. , Issaka, S. , Sido, A.Y. , Haougui, A. , Adam, T. , Séré, Y. and Saadou, M. (2011) Le flétrissement bactérien du riz au Niger: diversité pathologique d'isolats collectés sur les périmètres irrigués. J. Appl. Biosci. 38, 2551–2563. [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye, T. , Nickstadt, A. and Bonas, U. (2009) Breaking the code of DNA‐binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A. , Koebnik, R. , Lu, H. , Furutani, A. , Angiuoli, S. , Patil, B. , Van Sluys, M.A. , Ryan, R. , Meyer D., Han, S.W. , Aparna, G. , Rajaram, M. , Delcher, A. , Phillippy, A. , Puiu, D. , Schatz, M. , Shumway, M. , Sommer, D. , Trapnell, C. , Benahmed, F. , Dimitrov, G. , Madupu, R. , Radune, D. , Sullivan, S. , Jha, G. , Ishihara H., Lee, SW. , Pandey, A. , Sharma, V. , Sriariyanun, M. , Szurek, B. , Vera‐Cruz, C. , Dorman, K. , Ronald, P. , Verdier, V. , Dow, M. , Sonti, R. , Tsuge, S. , Brendel, V. , Rabinowicz, D. , Leach, J. , White, F. and Salzberg, S. (2011) Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp . J. Bacteriol. DOI: 10.1128/JB.05262‐11 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddenhagen, I.W. (1985) Rice disease evaluation in Madagascar. Int. Rice Comm. Newsl. 34, 74–78. [Google Scholar]
- Bui Thi Ngoc, L. , Vernière, C. , Jouen, E. , Ah‐You, N. , Lefeuvre, P. , Chiroleu, F. , Gagnevin, L. and Pruvost, O. (2010) Amplified fragment length polymorphism and multilocus sequence analysis‐based genotypic relatedness among pathogenic variants of Xanthomonas citri pv. citri and Xanthomonas campestris pv. bilvae . Int. J. Syst. Evol. Microbiol. 60, 515–525. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and Bonas. U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Comas, I. , Moya, A. , Azad, R.K. , Lawrence J.G. and Gonzalez‐Candelas, F. (2006) The evolutionary origin of Xanthomonadales genomes and the nature of the horizontal gene transfer process. Mol. Biol. Evol. 23, 2049–2057. [DOI] [PubMed] [Google Scholar]
- Da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vittorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐ Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Dye, D.W. , Bradbury, J.F. , Goto, M. , Hayward, A.C. , Lelliot, R.A. and Schroth, M.N. (1980) International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev. Plant. Pathol. 59, 153–168. [Google Scholar]
- Evanno, G. , Regnaut, S. and Goudet, J. (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Fargier, E. , Fischer‐Le Saux, M. and Manceau, C. (2011) A multilocus sequence analysis of Xanthomonas campestris reveals a complex structure within crucifer‐attacking pathovars of this species. Syst. Appl. Microbiol. 34, 156–165. [DOI] [PubMed] [Google Scholar]
- Gevers, D. , Cohan, F.M. , Lawrence, J.G. , Spratt, B.G. , Coenye, T. , Feil, E.J. , Stackebrandt, E. , Van de Peer, Y. , Vandamme, P. , Thompson, F.L. and Swings, J. (2005) Opinion: reevaluating prokaryotic species. Nat. Rev. Microbiol. 3, 733–739. [DOI] [PubMed] [Google Scholar]
- Gillings, M.R. , Holley, M.P. , Stokes, H.W. and Holmes, A.J. (2005) Integrons in Xanthomonas: a source of species genome diversity. Proc. Natl. Acad. Sci. USA, 102, 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, C. , Szurek, B. , Manceau, C. , Mathieu, T. , Sere, Y. and Verdier, V. (2007) Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol. Plant–Microbe Interact. 20, 534–546. [DOI] [PubMed] [Google Scholar]
- Grant, S.R. , Fisher, E.J. , Chang, J.H. , Mole, B.M. and Dangl, J.L. (2006) Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60, 425–449. [DOI] [PubMed] [Google Scholar]
- Hajri, A. , Brin, C. , Hunault, G. , Lardeux, F. , Lemaire, C. , Manceau, C. , Boureau, T. and Poussier, S. (2009) A ‘repertoire for repertoire’ hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas . Plos ONE, 4, e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kauffman, H.E. , Reddy, A.P.K. , Hsieh, S.P.Y. and Merca, S.D. (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae . Plant Dis. Rep. 57, 537–541. [Google Scholar]
- Kearney, B. and Staskawicz, B.J. (1990) Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2 . Nature, 346, 385–386. [DOI] [PubMed] [Google Scholar]
- Kimura, M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. [DOI] [PubMed] [Google Scholar]
- Kotetishvili, M. , Stine, O.C. , Kreger, A. , Morris, J.G. and Sulakvelidze, A. (2002) Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40, 1626–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M. II and Peterson, K.M. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Kvitko, B.H. , Park, D.H. , Velasquez, A.C. , Wei, C.‐F. , Russell, A.B. , Martin, G.B. , Schneider, D.J. and Collmer, A. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. Plos Pathog. 5, e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie, M. , Seunes, B. , Prior, P. and Boucher, C. (2004) Distribution and sequence analysis of a family of type III‐dependent effectors correlate with the phylogeny of Ralstonia solanacearum strains. Mol. Plant–Microbe Interact. 17, 931–940. [DOI] [PubMed] [Google Scholar]
- Leach, J.E. , Rhoads, M.L. , Vera Cruz, C.M. , White, F.F. , Mew, T.W. and Leung, H. (1992) Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae with a repetitive DNA element. Appl. Environ. Microbiol. 58, 2188–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.M. , Park, Y.J. , Park, D.S. , Kang, H.W. , Kim, J.G. , Song, E.S. , Park, I.C. , Yoon, U.H. , Hahn, J.H. , Koo, B.S. , Lee, G.B. , Kim, H. , Park, H.S. , Yoon, K.O. , Kim, J.H. , Jung, C.H. , Koh, N.H. , Seo, J.S. and Go, S.J. (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331 the bacterial blight pathogen of rice. Nucleic Acids Res. 33, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, S.A. , Ditta, G.S. and Helinski, D.R. (1982) Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta‐aminolevulinic acid synthetase from Rhizobium meliloti . J. Biol. Chem. 257, 8724–8730. [PubMed] [Google Scholar]
- Lerat, E. , Daubin, V. , Ochman, H. and Moran, N.A. (2005) Evolutionary origins of genomic repertoires in bacteria. Plos Biol. 3, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado, P. and Rozas, P. (2009) DNASP v5.1: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Patil, P. , Van Sluys, M.‐A. , White, F.F. , Ryan, R.P. , Dow, J.M. , Rabinowicz, P. , Salzberg, S.L. , Leach, J.E. , Sonti, R. , Brendel, V. and Bogdanove, A.J. (2008) Acquisition and evolution of plant pathogenesis‐associated gene clusters and candidate determinants of tissue‐specificity in Xanthomonas . Plos ONE. 3, e3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W. , Dong, F. , Stavrinides, J. and Guttman, D.S. (2006) Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. Plos Genet. 2, e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden, M.C. , Bygraves, J.A. , Feil, E. , Morelli, G. , Russell, J.E. , Urwin, R. , Zhang, Q. , Zhou, J. , Zurth, K. , Caugant, D.A. , Feavers, I.M. , Achtman, M. and Spratt, B.G. (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA, 95, 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino, S. , Sugio, A. , White, F.F. and Bogdanove, A.J. (2006) Inhibition of resistance gene‐mediated defense in rice by Xanthomonas oryzae pv. oryzicola . Mol. Plant–Microbe Interact. 19, 240–249. [DOI] [PubMed] [Google Scholar]
- Martin, D.P. , Lemey, P. , Lott, M. , Moulton, V. , Posada, D. and Lefeuvre, P. (2003) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics, 26, 2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz, M. , Dahlbeck, D. , Morales, C.Q. , Al Sady, B. , Clark, E.T. and Staskawicz, B.J. (2005) The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana . Plant J. 41, 801–814. [DOI] [PubMed] [Google Scholar]
- Mew, T. (1987) Current status and future prospects of research on bacterial blight of rice. Annu. Rev. Physiol. 25, 359–382. [Google Scholar]
- Mew, T.W. , Alvarez, A.M. , Leach, J.E. and Swings, J. (1993) Focus on bacterial blight of rice. Plant Dis. 77, 5–12. [Google Scholar]
- Mhedbi‐Hajri, N. , Darrasse, A. , Pigne, S. , Durand, K. , Fouteau, S. , Barbe, V. , Manceau, C. , Lemaire, C. and Jacques, M.‐A. (2011) Sensing and adhesion are adaptive functions in the plant pathogenic xanthomonads. BMC Evol. Biol. 11, 11–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou, M.J. and Bogdanove, A.J. (2009) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. [DOI] [PubMed] [Google Scholar]
- Nei, M. and Gojobori, T. (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426. [DOI] [PubMed] [Google Scholar]
- Nelson, R.J. , Baraoidan, M.R. , Vera‐Cruz, C.M. , Yap, I.V. , Leach, J.E. , Mew, T.W. and Leung, H. (1994) Relationships between phylogeny and pathotype for the bacterial blight pathogen of rice. Appl. Environ. Microbiol. 60, 3275–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño‐Liu, D. , Ronald, P. and Bogdanove, A. (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. [DOI] [PubMed] [Google Scholar]
- Ochiai, H. , Inoue, Y. , Takeya, M. , Sasaki, A. and Kaku, H. (2005) Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39, 275–287. [Google Scholar]
- Opina, O. and Exconde, O. (1971) Assessment of yield loss due to bacterial leaf streak of rice. Philipp. Phytopathol. 7, 35–39. [Google Scholar]
- Ou, S.H. (1985) Rice Diseases. Kew, Surrey: Commonwealth Agricultural Bureau. [Google Scholar]
- Parkinson, N. , Cowie, C. , Heeney, J. and Stead, D. (2009) Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int. J. Syst. Evol. Microbiol. 59, 264–274. [DOI] [PubMed] [Google Scholar]
- Pritchard, J.K. , Stephens, M. and Donnelly, P. (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker, J.L.W. , Hoste, B. , Louws, F.J. , Kersters, K. , Swings, J. , Vauterin, L. , Vauterin, P. and de Bruijn, F.J. (2000) Comparison of AFLP and rep‐PCR genomic fingerprinting with DNA–DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 50, 665–677. [DOI] [PubMed] [Google Scholar]
- Raymundo, A.K. , Briones, A.M. Jr , Ardales, E.Y. , Perez, M.T. , Fernandez, L.C. , Leach, J.F. , Mew, T.W. , Ynalvez, M.A. , McLaren, C.G. and Nelson, R.J. (1999) Analysis of DNA polymorphism and virulence in Philippine strains of Xanthomonas oryzae pv. oryzicola . Plant Dis. 83, 434–440. [DOI] [PubMed] [Google Scholar]
- Roden, J.A. , Belt, B. , Ross, J.B. , Tachibana, T. , Vargas, J. and Mudgett, M.B. (2004) A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. USA, 101, 16624–16629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer, L. , Guttman, D.S. and Dangl, J.L. (2004) Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in the plant pathogen Pseudomonas syringae . Genetics, 167, 1341–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba‐White, M. , Notteghem, J.L. and Leach, J.E. (1995) Comparison of Xanthomonas oryzae pv. oryzae strains from Africa, North America, and Asia by restriction fragment length polymorphism analysis. Int. Rice Res. Notes, 20, 25–26. [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Philippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Futurani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Won Lee, S. , Seo, Y.S. , Sriariyanum, M. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A . BMC Genomics, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, A. , Tauch, A. , Jäger, W. , Kalinowski, J. , Thierbach, G. and Pühler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- Schornack, S. , Meyer, A. , Romer, P. , Jordan, T. and Lahaye, T. (2006) Gene‐for‐gene‐mediated recognition of nuclear‐targeted AvrBs3‐like bacterial effector proteins. J. Plant Physiol. 163, 256–272. [DOI] [PubMed] [Google Scholar]
- Sere, Y. , Onasanya, A. , Verdier, V. , Akator, K. , Ouedraogo, L.S. , Segda, Z. , Mbare, M.M. , Sido, A.Y. and Baso, A. (2005) Rice bacterial leaf blight in West Africa: preliminary studies on disease in farmer's field and screening. Asian J. Plant Sci. 4, 577–579. [Google Scholar]
- Sokurenko, E.V. , Hasty, D.L. and Dykhuizen, D.E. (1999) Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7, 191–195. [DOI] [PubMed] [Google Scholar]
- Solcà, N.M. , Bernasconi, M.V. , Valsangiacomo, C. , VanDoorn, L.J. and Piffaretti, J.C. (2001) Population genetics of Helicobacter pylori in the southern part of Switzerland analysed by sequencing of four housekeeping genes (atpD, glnA, scoB and recA), and by vacA, cagA, iceA and IS605 genotyping. Microbiology, 147, 1693–1707. [DOI] [PubMed] [Google Scholar]
- Song, C. and Yang, B. (2010) Mutagenesis of 18 type III effectors reveals virulence function of XopZPXO99 in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 23, 893–902. [DOI] [PubMed] [Google Scholar]
- Soto‐Suarez, M. , Gonzalez, C. , Piegu, B. , Tohme, J. and Verdier, V. (2010a) Genomic comparison between Xanthomonas oryzae pv. oryzae and Xanthomonas oryzae pv. oryzicola, using suppression subtractive hybridization. FEMS Microbiol. Lett. 308, 16–23. [DOI] [PubMed] [Google Scholar]
- Soto‐Suarez, M. , Bernal, D. , Gonzalez, C. , Szurek, B. , Guyot, R. , Tohme, J. and Verdier, V. (2010b) In planta gene expression analysis of Xanthomonas oryzae pv. oryzae, African strain MAI1. BMC Microbiol. 10, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides, J. , McCann, H.C. and Guttman, D.S. (2008) Host–pathogen interplay and the evolution of bacterial effectors. Cell. Microbiol. 10, 285–292. [DOI] [PubMed] [Google Scholar]
- Swings, J. , Van Den Mooter, M. , Vauterin, L. , Hoste, B. , Gillis, M. , Mew, T.W. and Kersters, K. (1990) Reclassification of the causal agents of bacterial blight Xanthomonas campestris pathovar oryzae and bacterial leaf streak Xanthomonas campestris pathovar oryzicola of rice as pathovars of Xanthomonas oryzae new. Int. J. Syst. Bacteriol. 40, 309–311. [Google Scholar]
- Tajima, F. (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. and Kumar, S. (2007) mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tang, J.L. , Feng, J.X. , Li, Q.Q. , Wen, H.X. , Zhou, D.L. , Wilson, T.J. , Dow, J.M. , Ma, Q.S. and Daniels, M.J. (1996) Cloning and characterization of the rpfC gene of Xanthomonas oryzae pv. oryzae: involvement in exopolysaccharide production and virulence to rice. Mol. Plant–Microbe Interact. 9, 664–666. [DOI] [PubMed] [Google Scholar]
- Thieme, F. , Koebnik, R. , Bekel, T. , Berger, C. , Boch, J. , Büttner, D. , Caldana, C. , Gaigalat, L. , Goesmann, A. , Kay, S. , Kirchner, O. , Lanz, C. , Linke, B. , McHardy, A.C. , Meyer, F. , Mittenhuber, G. , Nies, D.H. , Niesbach‐ Klosgen, U. , Patschkowski, T. , Ruckert, C. , Rupp, O. , Schneiker, S. , Schuster, S.C. , Vorholter, F.J. , Weber, E. , Puhler, A. , Bonas, U. , Bartels, D. and Kaiser, O. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett, L.R. , Hamilton, J.P. , Buell, C.R. , Tisserat, N.A. , Verdier, V. , Zink, F. and Leach, J.E. (2011) Genomic analysis of Xanthomonas oryzae from US rice reveals substantial divergence from known X. oryzae pathovars. Appl. Environ. Microbiol. 77, 3930–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, P. , Barber, C.E. and Daniels, M.J. (1985) Evidence for clustered pathogenicity genes in Xanthomonas campestris pv. campestris . Mol. Gen. Genet. 199, 338–343. [Google Scholar]
- Vauterin, L. , Hoste, B. , Kersters, K. and Swings, J. (1995) Reclassification of Xanthomonas . Int. J. Syst. Bacteriol. 45, 472–489. [Google Scholar]
- Vera Cruz, C.M. , Ardales, E.Y. , Skinner, D.Z. , Talag, J. , Nelson, R.J. , Louws, F.J. , Leung, H. , Mew, T.W. and Leach, J.E. (1996) Measurement of haplotypic variation in Xanthomonas oryzae pv. oryzae within a single field by rep‐PCR and RFLP analyses. Phytopathology, 86, 1352–1359. [Google Scholar]
- Vera Cruz, C.M. , Bai, J. , Ona, I. , Leung, H. , Nelson, R.J. , Mew, T.W. and Leach, J.E. (2000) Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. USA, 97, 13500–13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, F.F. and Yang, B. (2009) Host and pathogen factors controlling the rice–Xanthomonas oryzae interaction. Plant Physiol. 150, 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen, M. , Urban, A. and Jaeger, K.E. (2000) Rapid gene inactivation in Pseudomonas aeruginosa . FEMS Microbiol. Lett. 193, 201–205. [DOI] [PubMed] [Google Scholar]
- Wonni, I. , Ouedraogo, L. and Verdier, V. (2011) First report of bacterial leaf streak caused by Xanthomonas oryzae pv. oryzicola on rice in Burkina Faso. Plant Dis. 1, 72. [DOI] [PubMed] [Google Scholar]
- Yang, B. and White, F. (2004) Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant–Microbe Interact. 17, 1192–1200. [DOI] [PubMed] [Google Scholar]
- Young, J.M. , Park, D.C. , Shearman, H.M. and Fargier, E. (2008) A multilocus sequence analysis of the genus Xanthomonas . Syst. Appl. Microbiol. 31, 366–377. [DOI] [PubMed] [Google Scholar]
- Yu, Y. , Streubel, J. , Balzergue, S. , Champion, A. , Boch, J. , Koebnik, R. , Feng, J.‐X. , Verdier, V. and Szurek, B. (2011) Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector which induces the rice nodulin‐3 Os11N3 gene. Mol. Plant–Microbe Interact. (in press). [DOI] [PubMed] [Google Scholar]
- Zhao, B. , Ardales, E.Y. , Raymundo, A. , Bai, J. , Trick, H.N. , Leach, J.E. and Hulbert, S.H. (2004) The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1 . Mol. Plant–Microbe Interact. 17, 771–779. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Morgan, R.L. , Guttman, D.S. and Ma, W. (2009) Allelic variants of the Pseudomonas syringae type III effector HopZ1 are differentially recognized by plant resistance systems. Mol. Plant–Microbe Interact. 22, 176–189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Plot of the Bayesian assignment of Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc) strains to clusters based on the analysis of the concatenated dataset (glnA, gyrB and rpoD sequences) using the no admixture model of structure 2.1 software. Each haplotype is represented by a line and the geographical origin of the strains is given.
Fig. S2 The type III effector (T3E) XopO is not essential for the virulence and tissue specificity of Xanthomonas oryzae in rice Nipponbare. The bacterial cells were suspended in sterilized water and the cell concentration was adjusted to an optical density at 600 nm (OD600) of 0.2 '2 × 108 colony‐forming units (cfu)/mL'. Data shown are the means ± standard deviation from three replications, each with 30 leaves, from one representative experiment; similar results were obtained in the other two independent experiments. (a) Average length of lesions caused by Xanthomonas oryzae pv. oryzicola 7 days post‐inoculation. Approximately 20 µL of bacterial culture suspension were infiltrated into leaf mesophyll tissue with a 1‐mL blunt‐end plastic syringe. (b) Average length of lesions caused by Xanthomonas oryzae pv. oryzae inoculated by the leaf‐clipping method, 14 days after inoculation. (c) Average length of lesions caused by Xanthomonas oryzae pv. oryzae inoculated by the leaf infiltration method, 7 days after inoculation.
Table S1 The type III effector (T3E) genes analysed in this study: gene functions and primer sequences used for polymerase chain reaction (PCR) amplifications and for the preparation of probes for dot‐blot hybridizations.
Table S2 Polymerase chain reaction (PCR) primers used for the functional analysis of the type III effector (T3E) gene xopO in Xanthomonas oryzae.
Supporting info item
Supporting info item
Supporting info item
Supporting info item


