Summary
The fungal nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (Nox) complex, which has been implicated in the production of low‐level reactive oxygen species (ROS), contains mainly NoxA, NoxB (gp91phox homologues) and NoxR (p67phox homologue). Here, we report the developmental and pathological functions of NoxB and NoxR in the tangerine pathotype of Alternaria alternata. Loss‐of‐function genetics revealed that all three Nox components are required for the accumulation of cellular hydrogen peroxide (H2O2). Alternaria alternata strains lacking NoxA, NoxB or NoxR also displayed an increased sensitivity to H2O2 and many ROS‐generating oxidants. These phenotypes are highly similar to those previously seen for the Δyap1 mutant lacking a YAP1 transcriptional regulator and for the Δhog1 mutant lacking a HOG1 mitogen‐activated protein (MAP) kinase, implicating a possible link among them. A fungal strain carrying a NoxA NoxB or NoxA NoxR double mutation was more sensitive to the test compounds than the strain mutated at a single gene, implicating a synergistic function among Nox components. The ΔnoxB mutant strain failed to produce any conidia; both ΔnoxA and ΔnoxR mutant strains showed a severe reduction in sporulation. Mutant strains carrying defective NoxB had higher chitin content than the wild‐type and were insensitive to calcofluor white, Congo red and the fungicides vinclozolin and fludioxonil. Virulence assays revealed that all three Nox components are required for the elaboration of the penetration process. The inability to penetrate the citrus host, observed for Δnox mutants, could be overcome by wounding and by reacquiring a dominant Nox gene. The A. alternata NoxR did not influence the expression of NoxB, but negatively regulated NoxA. Importantly, the expression of both YAP 1 and HOG 1 genes, whose products are involved in resistance to ROS, was down‐regulated in fungi carrying defective NoxA, NoxB or NoxR. Our results highlight the requirement of Nox in ROS resistance and provide insights into its critical role in regulating both YAP1 and HOG1 in A. alternata.
Introduction
The tangerine pathotype of Alternaria alternata is a necrotrophic fungal pathogen that causes brown spots on citrus leaves and fruit. This pathogen produces a host‐selective toxin, called ACT (A. citri toxin), which kills host cells prior to invasion, and acquires nutrients exclusively from dead tissues (Akimitsu et al., 2003). The tangerine pathotype of A. alternata rarely produces an appressorium in axenic culture or in planta and does not rely on it for penetration. The colonization of A. alternata in the leaves of citrus induces rapid lipid peroxidation, increased accumulation of hydrogen peroxide (H2O2) and cell death (Lin et al., 2011). It appears that effective scavenging or detoxification of H2O2 and other reactive oxygen species (ROS) is required to ensure fungal survival in the host plant.
Previously, we have demonstrated an essential role of the A. alternata YAP1 gene, encoding a redox‐responsive YAP1‐like transcription factor, for ROS detoxification and fungal pathogenicity (Lin et al., 2009; Yang et al., 2009). The A. alternata YAP1 is responsible for the detoxification of H2O2 and, perhaps, other oxidants by the regulation of a number of enzymatic activities, including catalase, superoxide dismutase (SOD), glutathione‐S‐transferase, glutathione peroxidase, glutathione reductase and ligninolytic peroxidase (Lin et al., 2011). Further studies have revealed that a HOG1 mitogen‐activated protein (MAP) kinase and a SKN7 response regulator are also required for cellular resistance to oxidative stress and pathogenicity in A. alternata (Chen et al., 2012; Lin and Chung, 2010), confirming further that A. alternata is able to detoxify or obviate the ROS‐mediated plant defence barriers. Both A. alternata YAP1 and HOG1 are localized in the cytoplasm under normal conditions; on exposure to oxidative stresses, they are transported into the nucleus (Lin et al., 2009, 2010). In contrast, A. alternata SKN7 resides constitutively in the nucleus. In Saccharomyces cerevisiae, SKN7 has been shown to interact physically with YAP1 in response to oxidative stress (He et al., 2009). The A. alternata YAP1 and HOG1 are required for cellular resistance to oxidative stress induced by H2O2 and several superoxide‐generating compounds [KO2, menadione (MND) and diamide]. The A. alternata SKN7, perhaps interacting with YAP1, is primarily required for cellular resistance to H2O2‐induced oxidative stress. In A. alternata, SKN7 and HOG1 probably function independently in resistance to oxidative stress (Chen et al., 2012). HOG1 is responsible for resistance to salt‐induced osmotic stress, whereas the ‘two‐component’ histidine kinase (HSK1) is the primary regulator for cellular resistance to sugar osmotic stress (Lin and Chung, 2010). However, SKN7, probably regulated by the HSK1 signalling pathway, is involved in resistance to sugar‐induced osmotic stress (Chen et al., 2012). Hence, A. alternata is able to differentiate environmental stimuli using distinct or shared signalling pathways.
In mammalian cells, membrane‐bound nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (Nox) transfer electrons from NADPH to oxygen molecules via flavin adenine dinucleotides (FAD) and iron, leading to the production of superoxide, which is rapidly converted to H2O2 by SOD (Chen et al., 2009; Cheng et al., 2006; Vignais, 2002). Nox homologues, which are widely found in animals, plants and many multicellular microorganisms, yet completely absent in prokaryotes, have a vital role in both cellular differentiation and the defence response. The mammalian phagocytic Nox system, composed of a catalytic subunit (gp91phox), an adaptor protein (p22phox), a small guanosine triphosphatase (GTPase) (Rac2) and several cytosolic regulatory subunits (p67phox, p47phox and p40phox), is responsible for the oxidative burst on encountering a pathogen (Diebold and Bokoch, 2001; Lambeth, 2004). The Nox complex also plays a role in cell proliferation, apoptosis and hormone responses in animals (Lambeth et al., 2000; Lardy et al., 2005; Suh et al., 1999). In a cell‐free system, p67phox and Rac2 (a small GTP‐binding protein) are sufficient to activate gp91phox (Diebold and Bokoch, 2001). Plants also have Nox enzymes, designated respiratory burst oxidase homologues (Rboh), that are involved in a wide range of physiological processes (Simon‐Plas et al., 2002; Torres et al., 2002; Yoshioka et al., 2003). The generation of H2O2 mediated by specific plasma membrane Nox enzymes of plants often leads to programmed cell death and cellular defence against pathogen attack (Nanda et al., 2010; Torres et al., 2002).
Fungi possess three Nox isoforms: NoxA (Nox1), NoxB (Nox2) and NoxC (Aguirre et al., 2005; Kawahara et al., 2007; Lalucque and Silar, 2003; Lara‐Ortiz et al., 2003). NoxA and NoxB are homologous to the mammalian gp91phox, whereas NoxC, containing a calcium‐binding EF‐hand motif, is similar to the mammalian Nox5 and the plant Rboh enzymes. The function of NoxA and NoxB is thought to be coordinately regulated by the regulatory subunit NoxR (p67phox homologue) and Rac2 (Scott and Eaton, 2008). Functional inactivation of fungal Nox homologues often leads to the reduced production of H2O2 in many fungi (Cano‐Domínguez et al., 2008; Kim et al., 2011; Lara‐Ortiz et al., 2003; Rolke and Tudzynski, 2008; Segmüller et al., 2008; Semighini and Harris, 2008; Takemoto et al., 2007; Yang and Chung, 2012). Nox enzymes are conserved, but their actual roles in the regulation of multicellular development and pathogenicity vary considerably among fungal species that possess them (Heller and Tudzynski, 2011). For example, NoxA has been shown to be required for the development of the sexual fruiting body in Neurospora crassa, Aspergillus nidulans and Podospora anserina (Cano‐Domínguez et al., 2008; Lara‐Ortiz et al., 2003; Malagnac et al., 2004). NoxB is required for ascospore germination in N. crassa and P. anserina, but either NoxA or NoxB is required for ascospore germination in Botrytis cinerea. Both NoxA and NoxB are required for sclerotia formation in B. cinerea and Sclerotinia sclerotiorum (Kim et al., 2011; Segmüller et al., 2008). NoxA (Nox1), NoxB (Nox2) and NoxR are involved in cellulose degradation in P. anserina (Brun et al., 2009). In Magnaporthe grisea, NoxA and NoxB are required for the formation of a penetration peg under the appressorium, thus playing an important role in pathogenesis (Egan et al., 2007). In B. cinerea, only NoxB is required for the formation of the penetration structure, even though both Nox isoforms have a role in pathogenicity. NoxA, but not NoxB, plays a vital role in the mutualistic symbiotic association between the fungal endophyte Epichloë festucae and perennial ryegrass (Takemoto et al., 2007). NoxA‐catalysed production of ROS limits hyphal branching and E. festucae is capable of maintaining a symbiotic lifestyle within its host. Further studies in the grass–endophyte interaction have revealed that NoxA is activated by the small GTPase Rac, and NoxR, which interacts with homologues of the yeast polarity proteins, Bem1 and Cdc24 (Takemoto et al., 2011; Tanaka et al., 2008). The E. festucae strain lacking NoxA shows increased branching and causes severe stunting and premature senescence when inoculated into its grass host (Scott and Eaton, 2008).
The tangerine pathotype of A. alternata has NoxA, NoxB and NoxR homologues. Previously, we have shown that the A. alternata NoxA is required for the production of ROS, cellular resistance to oxidative stress and virulence (Yang and Chung, 2012), whereas the biological roles of NoxB and NoxR remain unknown. The objective of this work was to determine whether NoxB and NoxR are also involved in the production of H2O2, and to further understand the function of Nox enzymes in oxidative stress resistance and their role in pathogenesis. The A. alternata NoxB and NoxR genes were independently inactivated by targeted gene disruption, and the resulting phenotypes were compared with those of the ΔnoxA mutant. We report shared and independent roles of NoxA, NoxB and NoxR in developmental and physiological processes, and their regulatory functions in the expression of the YAP1 and HOG1 genes in A. alternata. We also provide genetic evidence which demonstrates the importance of fungal Nox in ROS detoxification and during plant infection.
Results
Characterization of NoxB and NoxR homologues
The A. alternata NoxB gene contains an 1840‐bp open reading frame (ORF) interrupted by three introns (48, 47 and 59 bp). The A. alternata NoxB polypeptide contains 561 amino acids, showing strong similarity to many Nox of fungi. NoxB is most similar (94% identity and 97% similarity) to Nox 5 of Pyrenophora tritici‐repentis (accession number XP_001936616). NoxB has a FAD‐binding domain, a NADPH‐binding domain and six transmembrane domains, resembling those found in the A. alternata NoxA (Fig. S1, see Supporting Information).
The A. alternata NoxR gene has an 1811‐bp ORF interrupted by three introns (62, 54 and 53 bp), encoding 546 amino acids. Analysis of the predicted NoxR polypeptide identified an N‐terminal tetratricopeptide repeat (TPR) domain and a carboxyl‐terminal PB1 domain, commonly found in the mammalian p67phox and fungal NoxR‐like family (Fig. S2, see Supporting Information).
Targeted disruption of NoxB and NoxR in A. alternata
The Nox genes were disrupted with a split marker approach, using two DNA fragments overlapping within the selection marker. This approach has been shown to increase the frequency of targeted gene disruption and homologous integration to as high as 100% in the tangerine pathotype of A. alternata (Lin and Chung, 2010; Lin et al., 2009, 2010; Wang et al., 2010). The A. alternata NoxB gene was disrupted by transforming two DNA fragments (5′NoxB::5′HYG and 3′NoxB::3′HYG), overlapping within the hygromycin phosphotransferase gene (HYG) cassette (Fig. S3, see Supporting Information), into protoplasts of the wild‐type strain (EV‐MIL31) of A. alternata. More than 30 fungal transformants were selected from medium containing hygromycin and tested for H2O2 sensitivity. Two strains (DB5 and DB6) displayed an increased sensitivity to H2O2 and were examined further by polymerase chain reaction (PCR) with different primer sets (Fig. S3). A 1.8‐kb fragment was amplified from genomic DNA of the wild‐type using the primers NoxB‐1F and NoxB‐tag. In contrast, an expected 3.4‐kb fragment was amplified from DNA purified from both DB5 and DB6 strains. When the primer pro2F, whose sequence is not present in split marker fragments, was paired with the hyg3 primer, an expected 2.9‐kb fragment was amplified from genomic DNA of DB5 and DB6, but not the wild‐type, confirming further that the 2.5‐kb HYG gene cassette was integrated at the NoxB locus. Similarly, a single expected 2.5‐kb fragment was amplified from DNA of DB5 and DB6, but not the wild‐type, using the 3′‐end primer paired with hyg4. Northern blot analysis indicated that hybridization of total RNA from the wild‐type strain with a NoxB‐specific probe identified an expected 1.8‐kb transcript that was absent in both DB5 and DB6 strains (Fig. S3). In contrast, truncated transcripts (<1 kb) were detected in RNA from the putative mutant strains, confirming the successful disruption of NoxB in A. alternata. Because both DB5 and DB6 strains displayed phenotypes that were different from that of the wild‐type (see below for details), it is unlikely that the truncated transcript encodes a polypeptide with normal functionality.
The introduction of 5′NoxR::5′HYG and 3′NoxR::3′HYG DNA fragments (Fig. S4, see Supporting Information) into the EV‐MIL31 strain recovered 39 transformants. Fungal strains were tested for H2O2 sensitivity and screened by PCR using different primer sets, revealing that two strains (DR2 and DR5) had an integrated fragment specifically at the NoxR site in the genome (Fig. S4). Northern blot analysis revealed that the 1.7‐kb transcript of NoxR with a NoxR‐specific probe was detected in total RNA purified from the wild‐type. In contrast, a truncated transcript of approximately 1.2 kb in size was detected in RNA purified from the DR2 and DR5 strains, confirming the successful disruption of NoxR in A. alternata.
The NoxA NoxB (ΔnoxAB1 and AB6) and NoxA NoxR (ΔnoxAR2 and AR3) double‐mutant strains were created by transforming split sulphonylurea‐resistance gene (Sur) fragments fused with truncated NoxA (Fig. S5, see Supporting Information) into protoplasts prepared from a ΔnoxB mutant (DB5) and a ΔnoxR mutant (DR2), respectively. Fungal transformants were recovered from medium containing sulphonylurea and screened for an elevated sensitivity to H2O2 compared with DB5 or DR2. PCR diagnosis validated the targeted gene disruption of NoxA in the DB5 and DR2 strains (Fig. S5).
NoxB and NoxR are involved in conidia formation
ΔnoxB (DB5 and DB6) and ΔnoxR (DR2 and DR5) mutants accumulated little or no pigmentation compared with the wild‐type strain grown on potato dextrose agar (PDA) in the light (Fig. 1). Quantitative analysis of conidia formation revealed that ΔnoxR mutants showed conidiation reduced by 90%; neither the DB5 nor DB6 mutant produced conidia. Conidia produced by the ΔnoxR mutants germinated at the same rate as those of the wild‐type (data not shown). Pigmentation and conidiation were restored in a CpB24 strain by transforming the DB5 mutant protoplasts with a wild‐type copy of NoxB. Similarly, the expression of a NoxR gene cassette in the DR2 mutant identified the CpR40 strain that produced wild‐type levels of conidia. The NoxA NoxB double‐mutant strain did not produce any conidia; the NoxA NoxR double‐mutant strain produced conidia at levels similar to the strain mutated at either NoxA or NoxR alone (data not shown). The ΔnoxA mutant was also impaired in pigmentation and conidiation (Yang and Chung, 2012).
Figure 1.
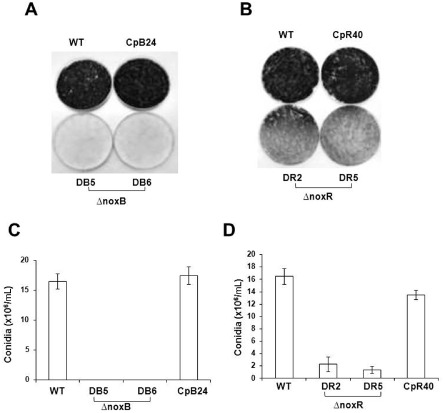
Conidia produced by the EV‐MIL31 strain (wild‐type, WT), the strains lacking NoxB (DB5 and DB6), the strains lacking NoxR (DR2 and DR5) and the rescued strains (CpB24 and CpR40) of Alternaria alternata. (A,B) Conidiation was evaluated by growing fungal strains on potato dextrose agar plates in the light for 3 days. (C,D) Quantification of conidia. Each column represents the mean number of conidia ± the standard error from two independent experiments, with at least three replicates.
NoxB and NoxR are required for the accumulation of H2O2
Mutational inactivation of the A. alternata NoxA gene reduced the accumulation of both extra‐ and intracellular H2O2 (Yang and Chung, 2012). The content of H2O2 within fungal hyphae was examined by staining with 3,3′‐diaminobenzidine (DAB). The presence of H2O2 induced DAB polymerization, resulting in a dark‐brown pigmentation. The H2O2 levels, based on the intensity of pigmentation, were much higher within incipient hyphae of the wild‐type strain than those seen for ΔnoxB (DB5 and DB6) or ΔnoxR (DR2 and DR5) mutants (Fig. 2). Hyphae of the genetically complemented strains CpB24 and CpR40 were stained dark brown, indicating restoration of H2O2 accumulation within hyphae (Fig. 2).
Figure 2.

NoxB and NoxR are required for the accumulation of cellular hydrogen peroxide (H2O2). The EV‐MIL31 strain (wild‐type, WT), the strains lacking NoxB (DB5 and DB6), the strains lacking NoxR (DR2 and DR5) and the rescued strains (CpB24 and CpR40) of Alternaria alternata were grown on potato dextrose agar for 5 days, stained with 5 mm 3,3′‐diaminobenzidine (DAB) solution (>8 h) and examined microscopically. DAB reacts with H2O2 to form a brownish polymer. Only one representative replicate of the treated hyphae is shown. Bar, 10 μm.
Nox contributes to oxidative stress resistance
Compared with the wild‐type, ΔnoxA, ΔnoxB and ΔnoxR mutants showed reduced growth on PDA by 12%, 11% and 5%, respectively. The double‐mutant strains (ΔnoxAB and ΔnoxAR) showed reduced growth by c. 20%. Growth reduction of the mutant greater than the respective threshold was considered to be indicative of hypersensitivity to the compounds tested. The A. alternata strain impaired for NoxA displayed a growth reduction on PDA amended with H2O2, cumyl H2O2, rose bengal (RB), diamide, MND or KO2, consistent with previous findings with this mutant (Yang and Chung, 2012). Likewise, mutational inactivation of NoxB or NoxR apparently resulted in an increased sensitivity to these ROS‐inducing compounds to varying degrees (Fig. 3A,B), suggesting distinct functions and regulations among the Nox components. For example, ΔnoxA and ΔnoxB single‐mutant strains were more sensitive than the ΔnoxR mutant to H2O2. ΔnoxB and ΔnoxR mutant strains displayed greater sensitivity than the ΔnoxA mutant to cumyl H2O2. ΔnoxA and ΔnoxR single‐mutant strains were more sensitive than ΔnoxB to diamide. ΔnoxB and ΔnoxR mutant strains were less sensitive than ΔnoxA to KO2 (Fig. 3B). Mutant strains carrying impaired NoxA, NoxB or NoxR displayed an increased sensitivity to tert‐butyl‐hydroxyperoxide (t‐BHP) and, to a lesser extent, haematoporphyrin (HP) (Fig. S6, see Supporting Information).
Figure 3.
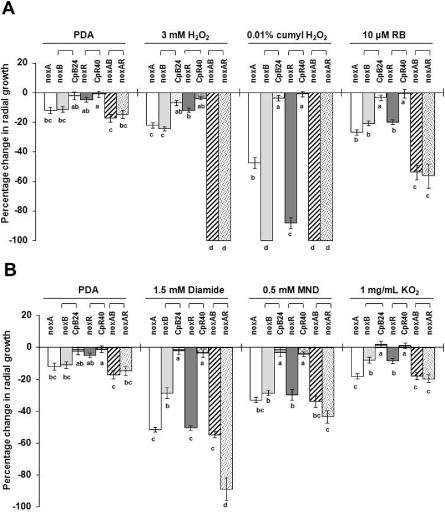
(A,B) Assays for the sensitivity of the EV‐MIL31 strain (wild‐type, WT), the strains carrying a single deletion of NoxA, NoxB or NoxR, the rescued strains (CpB24 and CpR40) and the NoxA NoxB (ΔnoxAB) and NoxA NoxR (ΔnoxAR) double‐mutant strains of Alternaria alternata. Two independent mutant strains of each type were used for the assays. Fungi were grown on potato dextrose agar (PDA) amended with the test compounds for 4–6 days. Radial growth was measured. The percentage change in radial growth was calculated as the percentage of growth of the deletion mutants in relation to the wild‐type grown on the same plate. A negative percentage change indicates growth reduction in relation to the wild‐type. The mutant was considered to be hypersensitive to the test compounds when the percentage change in growth reduction was greater than that measured in untreated PDA. The data presented are the mean and standard error of two independent mutants with at least two replicates of two experiments (n = 8). For each test compound, means indicated by the same letter within a test compound are not significantly different from one another, P < 0.05. RB, rose bengal; MND, menadione.
Re‐introduction of a functional copy of NoxB under the control of its own promoter in the DB5 mutant strain resulted in a fungal strain (CpB24) that exhibited wild‐type resistance to all test compounds, confirming that the NoxB disruption was indeed a contributor to all phenotypes. Similarly, all phenotypes seen in the ΔnoxR mutant (DR2) strain were fully or nearly restored, as seen in the CpR40 strain expressing a wild‐type copy of the NoxR gene (Fig. 3). The NoxA NoxB and NoxA NoxR double‐mutant strains displayed severe hypersensitivity to H2O2 and RB compared with the sensitivity observed in the strains mutated at NoxA, NoxB and NoxR alone. The NoxA NoxR, but not NoxA NoxB, double‐mutant strain displayed greater sensitivity than ΔnoxA or ΔnoxR to cumyl H2O2 and diamide. The NoxA NoxB and NoxA NoxR double mutants displayed MND and KO2 sensitivity similar to the strains carrying a single gene mutation.
Expression of the Nox genes is responsive to oxidative stress
Previous studies have revealed that the 1.8‐kb NoxA gene transcript was barely detectable when the wild‐type strain of A. alternata was grown on PDA (Yang and Chung, 2012). Expression of NoxA was up‐regulated by H2O2, KO2, MND, t‐BHP and HP (Yang and Chung, 2012). Northern blotting indicated that the 1.8‐kb NoxB transcript was detected abundantly when the fungus was grown on PDA, and was elevated to varying degrees when the fungus was shifted to medium supplemented with KO2, t‐BHP or HP (Fig. 4). NoxR was weakly expressed when the fungus was grown on PDA. The accumulation of the 1.7‐kb NoxR gene transcript was apparently elevated in the wild‐type strain responding to KO2, HP or t‐BHP. In contrast, H2O2 and MND had a moderate effect on the expression of NoxB and NoxR.
Figure 4.
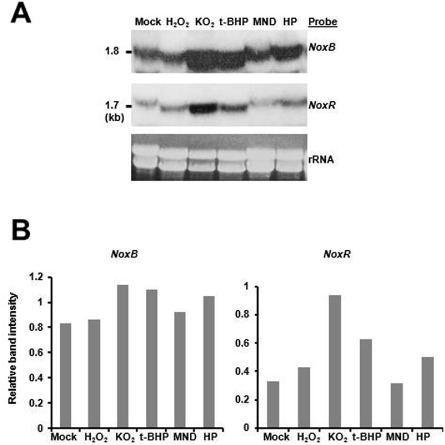
(A) Northern blot analyses for the expression of the NoxB and NoxR genes in response to reactive oxygen species (ROS). The wild‐type strain of Alternaria alternata grown on cellophane overlaid onto potato dextrose agar (PDA) for 2 days was shifted to PDA containing H2O2 (3 mm), KO2 (0.5 mg/mL), tert‐butyl‐hydroxyperoxide (t‐BHP, 0.05%), menadione (MND, 1 mm) or haematoporphyrin (HP, 50 μm) and incubated for an additional 24 h. The mock treatment contains RNA obtained from fungal mycelia shifted to the nonamended PDA. Fungal RNA was purified, electrophoresed in a formaldehyde‐containing, denaturing gel, blotted, and washed at high stringency after hybridization with a NoxB or NoxR probe. Gels staining with ethidium bromide indicate relative loading of the RNA samples. The sizes of the hybridization bands are indicated in kilobase pairs (kb). (B) The intensity of hybridizing bands normalized to that of rRNA was quantified using an Image J program available at http://rsb.info.nih.gov/ij/.
ΔnoxB and ΔnoxR mutants appear to be unable to penetrate citrus leaves
The A. alternata strain impaired for NoxA induced smaller and fewer necrotic lesions than the wild‐type on detached Minneola or calamondin leaves (Yang and Chung, 2012). Because the ΔnoxB single‐ and ΔnoxAB double‐mutant strains did not produce any conidia, fungal pathogenicity was assayed by placing a mycelial mass (with agar removed) on calamondin leaves. Necrotic lesions induced by the wild‐type strain appeared at 2–4 days post‐inoculation (dpi). In contrast, the ΔnoxB single‐ and ΔnoxAB double‐mutant strains did not produce visible lesions at 4 dpi (Table 1). The rescued strain (CpB24) produced necrotic lesions comparable with those induced by the wild‐type (Fig. S7, see Supporting Information). When citrus leaves were wounded prior to inoculation, the ΔnoxB and ΔnoxAB mutant strains induced necrotic lesions similar to those produced by the wild‐type and CpB24 strains.
Table 1.
Production of necrotic lesions on detached calamondin leaves inoculated with the wild‐type (EV‐MIL31) and the genetically modified strains of Alternaria alternata
| Fungal strain | Genotype | Inoculum | Disease incidence (%)a | |
|---|---|---|---|---|
| Unwounded | Wounded | |||
| Mock | H2O | 0/26 (0) | 0/15 | |
| EV‐MIL31 | Wild‐type | Mycelial mass | 32/32 (100) | 17/17 |
| DB5 | NoxB disruption | 0/22 (0) | 11/11 | |
| DB6 | 0/22 (0) | 11/11 | ||
| AB1 | NoxA NoxB double disruption | 0/11 (0) | 10/10 | |
| AB6 | 0/11 (0) | 10/10 | ||
| CpB24 | NoxB rescued | 21/22 (95.5) | 11/11 | |
| Mock | H2O | 0/24 (0) | 0/12 | |
| EV‐MIL31 | Wild‐type | Conidia | 63/63 (100) | 20/20 |
| DN2 | NoxA disruption | 32/42 (76.2)b | 20/20 | |
| DN6 | 39/48 (81.3)b | 20/20 | ||
| NCp16 | NoxA rescued | 40/45 (88.9) | 16/16 | |
| DR2 | NoxR disruption | 5/27 (18.5) | 24/24 | |
| DR5 | 6/27 (22.2) | 24/24 | ||
| AR2 | NoxA NoxR double disruption | 3/44 (6.8) | 10/10 | |
| AR3 | 3/44 (6.8) | 10/10 | ||
| CpR40 | NoxR rescued | 27/27 (100) | 20/20 | |
Pathogenicity was assayed on detached calamondin leaves by placing 5 μL of conidial suspension (104 conidia/mL) in each of the spots. The inoculated leaves were incubated in a mist chamber for lesion development. The number of necrotic lesions and the total number of leaves inoculated are to the right and left of the slash (/), respectively. Except for double‐mutant strains, all strains were tested at least twice. The mock control was treated with water only.
Lesions induced by NoxA disruption were significantly smaller (by ∼40%) than those induced by the wild‐type.
Pathogenicity assays using a point inoculation method indicated that the ΔnoxA mutant strains (DN2 and DN6) produced necrotic lesions on detached calamondin leaves at frequencies ranging from 76% to 81% (Table 1). The lesions induced by the ΔnoxA mutant were significantly smaller than those induced by the wild‐type and NCp16 strain re‐carrying a functional copy of NoxA, consistent with previous findings with this mutant (Yang and Chung, 2012). In contrast, the ΔnoxR single‐ and ΔnoxAR double‐mutant strains induced pinpoint or no visible lesions on detached leaves inoculated with conidial suspension (Fig. S8, see Supporting Information). The wild‐type and genetically reverted CpR40 strains induced visible lesions at 3 dpi. Pathogenicity assays performed on pre‐wounded calamondin leaves revealed that strains lacking NoxA, NoxR or both produced necrotic lesions at frequencies and magnitudes similar to those of the wild‐type, NoxA and NoxR complementation strains at 2 dpi (Table 1).
NoxB plays a negative role in cell wall integrity
To determine whether fungal strains lacking NoxA, NoxB or NorR were impaired in cell wall integrity, fungi were tested for sensitivity to cell wall‐targeting compounds, including SDS, calcofluor white (CFW) and Congo red (CR). The wild‐type EV‐MIL31 strain showed reduced radial growth by 35%–40% when grown on medium amended with 0.01% SDS. Fungal strains impaired for each of the Nox genes displayed an increased sensitivity to SDS (Fig. 5A). The CpB24 and CpR40 complementation strains displayed wild‐type sensitivity to SDS, whereas the ΔnoxAB and ΔnoxAR double‐mutant strains displayed severe hypersensitivity to SDS.
Figure 5.
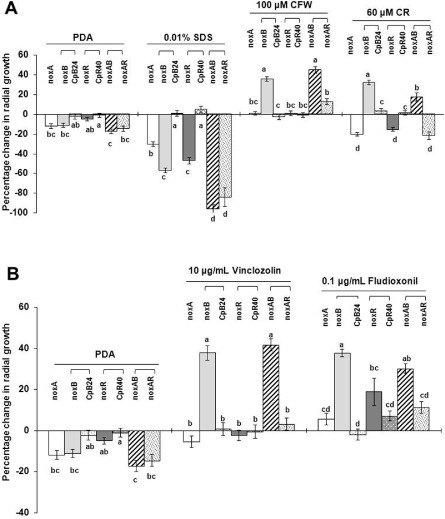
(A,B) Assays for cellular sensitivity of the strains carrying a single Nox gene mutation (ΔnoxA, ΔnoxB and ΔnoxR), the rescued strains (CpB24 and CpR40) and the NoxA NoxB (ΔnoxAB) and NoxA NoxR (ΔnoxAR) double‐mutant strains of Alternaria alternata to the cell wall‐targeting compounds sodium dodecylsulphate (SDS), calcofluor white (CFW) and Congo red (CR), as well as vinclozolin and fludioxonil fungicides. Fungal strains were grown on potato dextrose agar (PDA) amended with or without chemicals for 5 days. Two independent mutant strains of each type were used for the assays. The percentage change in radial growth was calculated as the percentage of growth of the deletion mutants in relation to the wild‐type grown on the same plate. A negative percentage change indicates growth reduction in relation to the wild‐type. A positive percentage change indicates that the mutants grew more rapidly than the wild‐type. The data presented are the means and standard errors of two independent mutants with at least two replicates of two experiments (n = 8). For each test compound, means indicated by the same letter within a test compound are not significantly different from one another, P < 0.05.
The wild‐type strain of A. alternata showed reduced radial growth by 34%–40% when grown on medium amended with CFW or CR. Both ΔnoxB and ΔnoxAB mutant strains grew significantly more rapidly than the wild‐type in the presence of CFW or CR. In contrast, mutant strains lacking NoxA, NoxR or both displayed a slightly increased growth in the presence of CFW, but an elevated sensitivity to CR, compared with the strains grown on PDA alone. The CpB24 and CpR40 complementation strains displayed a sensitivity to CFW and CR at levels not significantly different from that of the wild‐type (Fig. 5A).
The wild‐type strain and the genetically complemented strain CpB24 of A. alternata were sensitive to vinclozolin and fludioxonil fungicides, whereas ΔnoxB and ΔnoxAB mutant strains were insensitive to these fungicides (Fig. 5B). Disruption of NoxA or NoxR had a lesser effect on fungicide resistance. Fungal strains lacking NoxA, NoxR or both displayed slightly increased resistance to vinclozolin compared with the same strains grown on untreated PDA. On PDA amended with fludioxonil, all nox mutant strains, particularly ΔnoxB and ΔnoxAB, displayed an increased growth (a positive percentage change).
Quantification assays revealed that fungal strains carrying defective NoxB or NoxAB had significantly higher chitin content in relation to the levels measured in the wild‐type and other Δnox disruption strains (Fig. 6). The chitin level detected in the CpB24 strain was not significantly different from that of the wild‐type. The fungal strains lacking NoxA, NoxR or both had lower chitin contents than the wild‐type and the CpR40 strains.
Figure 6.
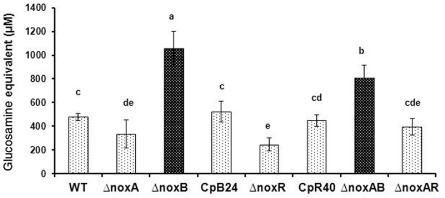
Quantification of chitin in the cell wall obtained from the wild‐type EV‐MIL31 (WT), the strains carrying a single Nox gene mutation (ΔnoxA, ΔnoxB and ΔnoxR), the rescued strains (CpB24 and CpR40) and the NoxA NoxB (ΔnoxAB) and NoxA NoxR (ΔnoxAR) double‐mutant strains of Alternaria alternata. Chitin was extracted with HCl and determined by measuring the acid‐released glucosamine from chitin using p‐dimethylaminobenzaldehyde as a chromogen. Except for CpB24 and CpR40 strains, all strains were tested at least twice. Means indicated by the same letter are not significantly different from one another, P < 0.05.
NoxB and NoxR regulate the YAP1‐like transcription factor and the HOG1 MAP kinase implicated in ROS resistance
Northern blot analyses revealed that the expression of both YAP1 and HOG1 genes was down‐regulated in fungal strains lacking NoxB or NoxR. The introduction of a wild‐type copy of NoxB into the DB5 mutant restored the expression of the YAP1 and HOG1 genes, as seen in the CpB24 strain (Fig. 7A). Similarly, the introduction of a wild‐type NoxR gene cassette into the respective mutant restored the expression of the YAP1 and HOG1 genes (Fig. 7B). Inactivation of NoxB reduced slightly the accumulation of the NoxA and NoxR gene transcripts, compared with the levels seen for the wild‐type and the CpB24 strains. In contrast, disruption of NoxR resulted in an elevated expression of NoxA, but not NoxB. Conversely, deletion of YAP1 or HOG1 apparently promoted the expression of both NoxB and NoxR genes (Fig. 7C,D).
Figure 7.
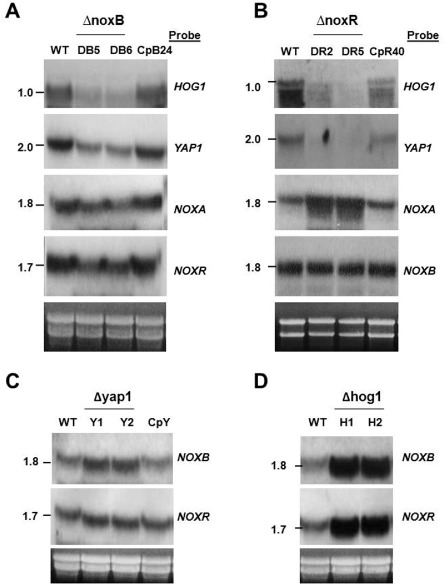
Images of RNA blotting. Fungal RNA, purified from the wild‐type (WT), the ΔnoxB mutants and the NoxB rescued strain (CpB24) (A), from the ΔnoxR mutants and the NoxR rescued strain (CpR40) (B), from the Δyap1 mutants and the YAP1 rescued strain (CpY) (C), and from the Δhog1 mutants of A. alternata (D), was electrophoresed in a formaldehyde‐containing gel, blotted and hybridized to DNA probes as indicated. Gels staining with ethidium bromide indicate relative loading of the RNA samples. The sizes of the hybridization bands are indicated in kilobase pairs (kb).
Discussion
We report the cloning and functional characterization of the A. alternata NoxB gene, homologous to the human phagocytic oxidase gp91phox catalytic subunit, and the A. alternata NoxR gene, homologous to the p67phox regulatory subunit. The work highlights the shared roles of Nox components in the regulation of conidia formation, H2O2 accumulation, oxidative stress resistance and fungal virulence in the tangerine pathotype of A. alternata. Similar to NoxA, disruption of the NoxB or NoxR gene with a hygromycin‐resistance cassette yielded mutants that displayed pleiotropic phenotypes. The phenotypes associated with mutational inactivation of the Nox gene were fully restored in the DB5 and DR2 mutants expressing the wild‐type NoxB and NoxR, respectively. Hence, the NoxB or NoxR disruption was indeed a contributor to the deformed phenotypes. Similar to ΔnoxA mutants (Yang and Chung, 2012), the A. alternata strain lacking NoxB or NoxR accumulated low quantities of H2O2 within hyphae relative to the wild‐type. H2O2 may serve as a secondary messenger to regulate various physiological, developmental and pathological processes. In striking contrast, deletion of a NoxA homologue facilitates ROS production in P. anserina and M. grisea (Egan et al., 2007; Malagnac et al., 2004).
We also report previously unidentified phenotypes that are associated with fungal Nox. The elevated sensitivity of ΔnoxB and ΔnoxR mutants to ROS was accompanied by a decreased expression of two redox‐responsive genes YAP1 and HOG1. In A. alternata, both YAP1 and HOG1 have been demonstrated previously by genetic analysis to be required for ROS detoxification (Lin and Chung, 2010; Lin et al., 2009; Yang et al., 2009). Although the expression of YAP1 and HOG1 was apparently activated by Nox, both YAP1 and HOG1 repressed the expression of NoxB and NoxR. This transcriptional feedback loop could avoid the excessive production of toxic ROS. Expression of the NoxB and NoxR genes in A. alternata was up‐regulated in response to a number of oxidants, confirming further the involvement of Nox enzymes in cellular resistance to ROS. However, the changes in gene expression induced by different oxidants are not always in agreement with their toxicities to the nox mutants. For example, although KO2 shows the strongest induction of the expression of NoxB and NoxR, this compound has little or no inhibitory effect on the growth of both noxB and noxR mutants.
NoxR is homologous to the p67phox regulatory subunit and is presumably required for the activation of NoxA and NoxB. NoxA and NoxB have been shown to be regulated by NoxR in As. nidulans, E. festucae, P. anserina, N. crassa and B. cinerea (Brun et al., 2009; Cano‐Domínguez et al., 2008; Segmüller et al., 2008; Semighini and Harris, 2008; Tanaka et al., 2008). Although Nox components are well conserved in fungi that possess them, the regulatory mechanisms of each component may vary considerably in different fungal species. In E. festucae, NoxR physically interacts with homologues of the yeast polarity proteins, Bem1 and Cdc24, and a small GTPase RacA; both NoxR and RacA are essential for the activation of NoxA (Takemoto et al., 2011; Tanaka et al., 2008). In N. crassa, NoxR is required for the function of both NoxA and NoxB in cell differentiation and growth (Cano‐Domínguez et al., 2008). NoxR is required for the function of both NoxA and NoxB in cellulose degradation in P. anserina (Brun et al., 2009). In mammalian cells, the Nox complex has been demonstrated to be regulated by a p21‐activated kinase (Pak) (Martyn et al., 2005). In addition, both p47 phox and p67 phox (NoxR homologue) are known to be phosphorylated by the p38 MAP kinase (HOG1 homologue) (Brown et al., 2009). In the ergot fungus Claviceps purpurea, mutational inactivation of a Pak homologue decreased the Nox1 gene transcript (Rolke and Tudzynski, 2008). In contrast with mammalian cells, the expression of a NoxA homologue in As. nidulans was down‐regulated by SakA, a HOG1 MAP kinase homologue (Lara‐Ortiz et al., 2003). However, disruption of a SakA homologue did not influence NoxA or NoxR in the endophytic fungus E. festucae (Eaton et al., 2008). Expression of Nox‐coding genes has also been implicated to be regulated by SLT2 (cell wall integrity) and FUS3/KSS1 (pheromone signalling) MAP kinases (Cano‐Domínguez et al., 2008; Segmüller et al., 2008). Moreover, NoxA facilitates nuclear localization of the PaMpk1 MAP kinase in P. anserina (Kicka et al., 2006; Malagnac et al., 2004). In the present study, we found that NoxR transcriptionally activated NoxA, because disruption of NoxR resulted in an elevated expression of NoxA, but not NoxB. Unlike the findings with N. crassa, B. cinerea and P. anserina (Cano‐Domínguez et al., 2008; Lara‐Ortiz et al., 2003; Malagnac et al., 2004), the phenotypes seen for the ΔnoxR and ΔnoxAB mutant strains of A. alternata were also very different, indicating further that NoxR may not regulate NoxA and NoxB directly. That the A. alternata NoxR has no impact on the expression of NoxB, but negatively regulates the expression of NoxA, is surprising. In this context, regulation of the Nox system in response to environmental stimuli in A. alternata might be somewhat unique.
Conidia formation by A. alternata has been shown to be positively regulated by the FUS3 MAP kinase‐ and the G protein/protein kinase A (PKA)‐mediated signalling pathways (Lin et al., 2010; Wang et al., 2010). In contrast, a cAMP‐dependent PKA suppresses conidia formation in the tangerine pathotype of A. alternata (Tsai et al., 2012), indicating the complexity of conidia formation. An A. alternata strain lacking NoxA is also defective for conidia production (Yang and Chung, 2012). Nox enzymes and their ability to produce ROS apparently play a crucial role in conidia formation. NoxB is probably the primary determinant for the initiation of conidiation because inactivation of NoxB completely blocked their formation. Impairment of NoxA or NoxR also resulted in a severe reduction in conidia formation, confirming the important role of Nox enzymes in fungal development. The A. alternata Nox system has no impact on hyphal branching and conidia germination.
In addition to conidia formation, the A. alternata Nox system is required for vegetative growth and cellular resistance to ROS in axenic culture. Although NoxA, NoxB and NoxR are core components of the Nox system, each may have a unique function or regulation in response to different environmental stimuli, because the degree of impairment varied considerably among individual Δnox mutants. For example, ΔnoxA and ΔnoxB mutants displayed much greater growth reduction and cellular sensitivity than the ΔnoxR mutant to H2O2. In contrast, ΔnoxB and ΔnoxR mutants were more sensitive than the ΔnoxA mutant to cumyl H2O2 and SDS. However, a fungal strain impaired for NoxA NoxB or NoxA NoxR was more sensitive to H2O2, MND, diamide, RB and SDS than the strain mutated at each of the genes, indicating an additive effect between Nox components. These results also suggest that Nox isoforms have alternative regulatory mechanisms under different physiological conditions. Nevertheless, the ROS sensitivity observed for Δnox mutants may be caused by the decreased expression of YAP1 and HOG1. Both YAP1 and HOG1 have been demonstrated to be essential for cellular resistance to oxidative stress induced by H2O2 and several superoxide‐generating compounds in A. alternata (Lin et al., 2009, 2010).
Alternaria alternata secretes a host‐selective toxin to rapidly kill host cells prior to invasion and obtains nutrients exclusively from dead tissues. A considerable amount of H2O2, as a result of cell death, has been shown to be accumulated in the vicinity of the infection area (Lin et al., 2011). In order to colonize within the oxidative environment of necrotic tissues, A. alternata must have evolved effective ROS detoxification machinery. The detoxification of ROS produced by the host plants is achieved by both the fungal YAP1 redox‐responsive transcriptional regulator and the HOG1 MAP kinase‐mediated signalling pathway, and is absolutely required for A. alternata pathogenicity in citrus (Lin and Chung, 2010; Lin et al., 2009; Yang et al., 2009). Although we have shown here that the expression of both YAP1 and HOG1 is regulated by the Nox system‐mediated network, this virulence deficiency observed for Δnox mutants is not completely caused by the decreased expression of YAP1 and HOG1. Fungal strains carrying defective Nox enzymes apparently are arrested in the penetration stage. Although ΔnoxB and ΔnoxR mutants induced little or no necrotic lesion on unwounded calamondin leaves, both mutants induced wild‐type lesions on citrus leaves that were wounded before inoculation. In contrast, YAP1 and HOG1 mutants are impaired in both penetration and colonization stages (Lin et al., 2009, 2010).
NoxA, NoxB and NoxR may independently and cooperatively interact with other yet unidentified components under different environmental conditions and during different developmental stages. As shown in this study, the A. alternata NoxB, but not NoxA, has a unique role in cell wall integrity and fungicide sensitivity. Fungal strains lacking NoxA, NoxR or both were sensitive to CFW and CR to the same level as the wild‐type strain. Our data reveals that NoxB down‐regulated chitin biosynthesis, because ΔnoxB and ΔnoxAB mutants accumulated more chitin than the wild‐type and the complementation strains, indicating a negative regulatory role of NoxB in cell wall integrity. This novel phenotype has not been identified previously in any fungus. Previous studies have revealed that mutational inactivation of the A. alternata SLT2 MAP kinase causes a reduced accumulation of chitin (Yago et al., 2011). Disruption of the HOG1, but not class III histidine kinase (HSK1), coding gene resulted in a fungal mutant that was highly resistant to cell wall‐degrading enzymes (Lin et al., 2010). Fungal strains carrying the mutated NoxB gene, HSK1 gene or both Hog1 and SKN7 genes are highly resistant to dicarboximide (vinclozolin) and phenylpyrrole (fludioxonil) fungicides (Chen et al., 2012; Lin et al., 2010). The results imply a possible interplay between these different signalling elements in the context of cell wall integrity and fungicide sensitivity. Because the expression of HOG1 is regulated by NoxB, it will be of great interest to determine whether HSK1, SKN7 and SLT2 are regulated by NoxB at the transcriptional and/or post‐translational level in the future. Overall, we have demonstrated here that Nox enzymes are a key ROS producer, and that the maintenance of ROS homeostasis via a close interaction among the Nox system, the YAP1 redox responsive regulator and the HOG1 MAP kinase‐mediated signalling pathway is critical for fungal survival, development and pathogenesis. Our study further underlines an important regulatory role of Nox enzymes in fungi.
Experimental Procedures
Fungal strains and sensitivity test
The wild‐type EV‐MIL31 strain of A. alternata (Fr.) Keissler was used as both a recipient host for transformation and in the mutagenesis experiments. This strain was single spore isolated from diseased leaves of Minneola tangelo, a hybrid between Duncan grapefruit (Citrus paradisi Macfad.) and Dancy tangerine (C. reticulata Blanco) in Florida, and has been characterized elsewhere (Lin et al., 2009, 2010, 2011). Fungal strains lacking and regaining an oxidative stress‐responsive transcription activator‐coding gene (YAP1) and strains lacking a HOG1 MAP kinase‐coding gene (HOG1) were generated in separate studies (Lin and Chung, 2010; Lin et al., 2009). Fungi were cultured on PDA (Difco, Sparks, MD, USA) at 28 °C. Conidia were collected by flooding with sterile water and centrifugation (5000 × g) from fungal cultures grown on PDA in the light for 3–4 days. The concentration of conidia was determined with the aid of a haemocytometer. Chemical sensitivity was assessed by transferring hyphae/conidia as a toothpick point inoculation onto medium containing the test chemical. The diameter of the colonies was measured from 4 to 7 days. The difference in the growth of the disruption mutant relative to that of the wild‐type grown on the same plate was calculated. The percentage change, which could be positive or negative, was determined by dividing the relative difference in growth by that of the wild‐type, followed by multiplication by 100. The mutant was considered to be hypersensitive to the test compounds when the percentage change in growth reduction was greater than that measured in untreated PDA. The significance of the treatments was determined by analysis of variance and the treatment means were separated by Tukey's test or Student's t‐test (P < 0.05).
Virulence tests
Fungal virulence was evaluated on detached calamondin (Citrus mitis Blanco) leaves inoculated with conidial suspension (1 × 104 conidia/mL) or mycelial mass. Conidial suspension was applied (5 μL) to detached calamondin leaves and the inoculated leaves were incubated in a mist chamber for 3–4 days for lesion development.
Manipulation of nucleic acids
DNA was isolated using a DNeasy Plant kit (Qiagen, Valencia, CA, USA); RNA was purified with Trizol reagent (Molecular Research Center, Cincinnati, OH, USA). A chromosome library of A. alternata was constructed from genomic DNA cleaved with four different restriction enzymes (DraI, EcoRI, PvuI and StuI), using a Universal GenomeWalker kit (BD Biosciences, San Jose, CA, USA). The sequences of oligonucleotide primers used in this study are listed in Table S1 (see Supporting Information). A 2.8‐kb DNA fragment containing the entire NoxB ORF and its 5′ and 3′ untranslated regions was amplified by PCR with the primers NoxB‐pro1F and NoxB‐TAG from the genome of A. alternata. The A. alternata NoxR gene fragment (430 bp) was amplified by PCR with two degenerate primers, p67f1 and p67r1, as reported by Takemoto et al. (2006). The full‐length NoxR gene was obtained by PCR using a chromosome walking strategy, as described previously (Chen et al., 2005; Choquer et al., 2005; You et al., 2007). PCR amplicons were sequenced at Eton Bioscience (Research Triangle Park, NC, USA). ORF and exon/intron positions were predicted using Softberry gene‐finding software (http://www.softberry.com). RNA was blotted onto a nylon membrane and hybridized to a digoxigenin (DIG)‐11‐dUTP (Roche Applied Science, Indianapolis, IN, USA)‐labelled DNA probe. The probe was amplified and labelled by PCR with gene‐specific primers. The probe was detected by an immunological assay using CSPD disodium (3‐[4‐methoxyspiro{1,2‐dioxetane‐3,2′‐(5′‐chloro)tricyclo [3.3.1.1]decan}‐4‐yl]phenyl phosphate; Roche Applied Science) as a chemofluorescent substrate for alkaline phosphatase.
Creation of genetically modified fungi
The Nox genes were disrupted using a split marker approach employing two hybrid DNA fragments, overlapping within a selection marker gene, as described previously (Yang and Chung, 2012). The bacterial phosphotransferase B gene (HYG) cassette under the control of the As. nidulans trpC gene promoter, conferring resistance to hygromycin, was used as a selection marker. This approach has been shown to decrease ectopic integration and to increase the frequency of targeted gene disruption and homologous integration to as high as 100% in A. alternata (Lin and Chung, 2010; Lin et al., 2009, 2010). PCR diagnosis using different primer sets was performed to identify the putative mutants. For each of the Nox genes, a primer whose sequence is located outside the split marker fragments was included for amplification to confirm the successful integration of the marker gene cassette within the gene of interest and to rule out ectopic integration or tandem insertion at the integration site.
To disrupt the NoxB gene, a 5′NoxB::5′HYg fusion fragment (0.9 + 1.2 kb) was amplified by two‐round PCR with the primers NoxB‐3F, NoxB::M13R, M13R and hyg3 (Fig. S3). A 3′NoxB::3′hYG fusion fragment (0.7 + 1.8 kb) was amplified with primers hyg4, M13F, M13F:NoxB and NoxB‐tag. The underlined sequence in the primers NoxB::M13R and M13F::NoxB represents the oligonucleotides completely complementary to the sequence of primers M13R and M13F, respectively.
Similar approaches were performed to disrupt the NoxR gene. A 5′NoxR::5′HYg fusion fragment (0.7 + 1.2 kb) was amplified by two‐round PCR with the primers NoxR‐pro2F, M13R::NoxR, M13R and hyg3 (Fig. S4). A 3′NoxR::3′hYG fragment (1.0 + 1.8 kb) was amplified with the primers hyg4, M13F, M13F:NoxR and NoxR‐tail. PCR fragments were transformed directly into protoplasts prepared from the EV‐MIL31 strain, using CaCl2 and polyethylene glycol (Chung et al., 2002). Fungal transformants were recovered from medium containing 250 μg/mL hygromycin (Calbiochem, La Jolla, CA, USA) and tested for sensitivity to H2O2. Successful integration of HYG within the targeted gene was examined by PCR with multiple sets of primers and by Northern blot hybridization to a gene‐specific probe.
A Sur gene cassette, conferring resistance to sulphonylurea, was used as a dominant selectable marker to construct double mutants from a ΔnoxB mutant (DB5) or ΔnoxR mutant (DR2) (Fig. S5). The NoxA NoxB and NoxA NoxR double‐mutant strains were created by transforming split Sur marker fragments fused with truncated NoxA into protoplasts prepared from DB5 and DR2, respectively. Transformants were recovered from medium supplemented with 5 μg/mL sulphonylurea (Chem Service, West Chester, PA, USA) and tested for H2O2 sensitivity.
Genetic complementation was achieved by co‐transformation of a functional Nox gene under the control of its own promoter with the pCB1532 plasmid, as described previously (Lin et al., 2009). A functional NoxB gene cassette was amplified by PCR with the primers NoxB‐2F and NoxB‐tag. A NoxR cassette was amplified with the primers NoxR‐pro2F and NoxR‐tail.
Detection of H2O2
The H2O2 content within A. alternata was assessed by staining fungal hyphae with DAB dissolved in water. The fungal isolates were grown on PDA for 5 days and colonies were flooded with a DAB solution (5 mm) for 12–16 h. Hyphae were cut from the edge of the colony and examined for the formation of a brown pigmentation, indicating the presence of H2O2 (Torres et al., 2002), with a Leitz Laborlux phase contrast microscope (Leica Microsystems, Exton, PA, USA).
Preparation and quantification of fungal chitin
Fungal mycelium was ground in liquid nitrogen, dissolved in a buffer containing 50 mm Tris‐HCl (pH 7.8), 2% SDS, 0.3 m β‐mercaptoethanol and 1 mm ethylenediaminetetraacetic acid (EDTA), and boiled at 100 °C for 15 min. After centrifugation at 8000 × g, the pellet was washed three times with water and dried completely. The fungal cell wall was dissolved in water to make a solution of 25 mg/mL. Cell wall (5 mg) was acidified in 6 M HCl, boiled at 100 °C for at least 4 h and used for chitin quantification. The chitin content was quantified by measuring the acid‐released glucosamine from chitin using p‐dimethylaminobenzaldehyde as a chromogen and measuring spectrophotometrically at A 520 (Selvaggini et al., 2004). The quantity of glucosamine was determined from a regression line established using pure glucosamine (Sigma, St. Louis, MO, USA).
Nucleotide sequence
Sequence data reported in this article have been deposited in the GenBank/EMBL Data Libraries under Accession Nos. JX136700 (AaNoxB), JX207117 (AaNoxR) and JN900389 (AaNoxA).
Supporting information
Fig. S1 Alignment and comparison of the deduced amino acid sequences of NoxA and NoxB of Alternaria alternata. Conserved amino acids are shaded. Both proteins share 34.9% identity and 52.4% similarity. Conserved histidine residues potentially for haem binding are indicated by asterisks. Six putative transmembrane domains are underlined. Putative flavin adenine dinucleotide (FAD)‐ and nicotinamide adenine dinucleotide phosphate (NADPH)‐binding domains are also indicated. Alignment was obtained by BioEdit using clustalw.
Fig. S2 Alignment of the deduced amino acid sequence of the Alternaria alternata NoxR with the Epichloe festucae EfNoxR, Botrytis cinerea BcNoxR and human Hspp67phox. Conserved amino acids are shaded. Proline‐rich (P‐rich), Src homology 3 (SH3) and Phox and Bem1 (PB1) domains of human p67phox are boxed. Four tetratricopeptide repeats (TPRs) and a putative Nox activation domain are also indicated. Alignment was obtained by BioEdit using clustalw with default settings.
Fig. S3 Targeted disruption of AaNoxB in Alternaria alternata using a split marker approach. (A) Schematic depiction of the generation of truncated, but overlapping, hygromycin phosphotransferase gene (HYG) under the control of the Aspergillus nidulans trpC promoter (P) and terminator (T), and gene disruption within AaNoxB. Oligonucleotide primers used to amplify each fragment are also indicated. (B) Image of DNA fragments amplified from genomic DNA of fungi with the primers indicated. The primer pro2F sequence is not present in the split marker fragment. (C) RNA gel blotting.
Fig. S4 Targeted disruption of AaNoxR in Alternaria alternata using a split marker approach. (A) Schematic depiction of the generation of truncated, but overlapping, hygromycin phosphotransferase gene (HYG) under the control of the Aspergillus nidulans trpC promoter (P) and terminator (T), and gene disruption within AaNoxR. Oligonucleotide primers used to amplify each fragment are also indicated. (B) Image of DNA fragments amplified from genomic DNA of fungi with the primers indicated. (C) RNA gel blotting.
Fig. S5 Targeted disruption of AaNoxA in a fungal strain lacking NoxB (DB5) or NoxR (DR2) of Alternaria alternata using a split marker approach. (A) Schematic depiction of the generation of truncated, but overlapping, sulphonylurea‐resistance gene (SUR), and gene disruption within AaNoxA. Oligonucleotide primers used to amplify each fragment are also indicated. (B) Image of DNA fragments amplified from genomic DNA of fungi with the primers indicated. The primer pro1F sequence is not located within the split marker fragment. Fragment patterns indicate successful disruption at the AaNoxA locus. Fungal strains AB1 and AB6 carry impaired NoxA and NoxB genes; strains AR2 and AR3 carry impaired NoxA and NoxR genes.
Fig. S6 Assays for the sensitivity of the EV‐MIL31 strain (wild‐type, WT), the strains carrying a single deletion of NoxA, NoxB or NoxR, and the NoxA NoxB (ΔnoxAB) and NoxA NoxR (ΔnoxAR) double‐mutant strains of Alternaria alternata. Two independent mutant strains of each type were used for the assays. Fungi were grown on potato dextrose agar (PDA) amended with haematoporphyrin (50 μm) or tert‐butyl‐hydroxyperoxide (0.05%) for 4–6 days. Radial growth was measured. The percentage change in radial growth was calculated as the percentage of growth of the deletion mutants in relation to the wild‐type grown on the same plate. A negative percentage change indicates growth reduction in relation to the wild‐type. The data presented are the means and standard errors of two independent mutants with at least two replicates.
Fig. S7 Virulence assays. Detached calamondin leaves inoculated with mycelial mass obtained from the wild‐type (WT) and the rescued (CpB24) strains of Alternaria alternata resulted in necrotic lesions at 4 days post‐inoculation (dpi). ΔnoxB mutants (DB5 and DB6) failed to induce necrotic lesions on unwounded leaves. ΔnoxB mutants induced necrotic lesions on calamondin leaves with wounding prior to inoculation. The mock control was treated with water only. The inoculated leaves were incubated in a moist chamber for lesion development.
Fig. S8 Virulence assays. ΔnoxR mutants (DR2 and DR5) induced little or no necrotic lesions on detached calamondin leaves inoculated with conidial suspension (5 μL, 1 × 104 conidia/mL). The rescued strain (CpR40) carrying a functional NoxR induced wild‐type (WT)‐like necrotic lesions. Wounding prior to inoculation rendered ΔnoxR mutants able to produce necrotic lesions. The mock control was treated with water only.
Table S1 Oligonucleotide primers used in this study.
References
- Aguirre, J. , Ríos‐Momberg, M. , Hewitt, D. and Hansberg, W. (2005) Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13, 111–118. [DOI] [PubMed] [Google Scholar]
- Akimitsu, K. , Peever, T.L. and Timmer, L.W. (2003) Molecular, ecological and evolutionary approaches to understanding Alternaria diseases of citrus. Mol. Plant Pathol. 4, 435–446. [DOI] [PubMed] [Google Scholar]
- Brown, A.J.P. , Haynes, K. and Quinn, J. (2009) Nitrosative and oxidative stress responses in fungal pathogenicity. Curr. Opin. Microbiol. 12, 384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun, S. , Malagnac, F. , Bidard, F. , Lalucque, H. and Silar, P. (2009) Functions and regulation of the Nox family in the filamentous fungus Podospora anserina: a new role in cellulose degradation. Mol. Microbiol. 74, 480–496. [DOI] [PubMed] [Google Scholar]
- Cano‐Domínguez, N. , Álvarez‐Delfín, K. , Hansberg, W. and Aguirre, J. (2008) NADPH oxidases NOX‐1 and NOX‐2 require the regulatory subunit NOR‐1 to control cell differentiation and growth in Neurospora crassa . Eukaryot. Cell, 7, 1352–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.Q. , Dekkers, K.L. , Rollins, J.A. , Ko, N.J. , Timmer, L.W. and Chung, K.‐R. (2005) A gene with domains related to transcription regulation is required for pathogenicity in Colletotrichum acutatum causing Key lime anthracnose. Mol. Plant Pathol. 6, 513–525. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Craige, S.E. and Keaney, J.F. Jr (2009) Downstream targets and intracellular compartmentalization in Nox signaling. Antioxid. Redox Signal. 11, 2467–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐H. , Lin, C.‐H. and Chung, K.‐R. (2012) Roles for SKN7 response regulator in stress resistance, conidiation and virulence in the citrus pathogen Alternaria alternata . Fungal Genet. Biol. 49, 802–813. [DOI] [PubMed] [Google Scholar]
- Cheng, G. , Diebold, B.A. , Hughes, Y. and Lambeth, J.D. (2006) Nox1‐dependent reactive oxygen generation is regulated by Rac1. J. Biol. Chem. 281, 17 718–17 726. [DOI] [PubMed] [Google Scholar]
- Choquer, M. , Dekkers, K.A. , Chen, H.Q. , Ueng, P.P. , Daub, M.E. and Chung, K.‐R. (2005) The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae . Mol. Plant–Microbe Interact. 18, 468–476. [DOI] [PubMed] [Google Scholar]
- Chung, K.‐R. , Shilts, T. , Li, W. and Timmer, L.W. (2002) Engineering a genetic transformation system for Colletotrichum acutatum, the causal fungus of lime anthracnose and postbloom fruit drop. FEMS Microbiol. Lett. 213, 33–39. [DOI] [PubMed] [Google Scholar]
- Diebold, B.A. and Bokoch, G.M. (2001) Molecular basis for Rac2 regulation of phagocyte NADPH oxidase. Nat. Immunol. 2, 211–215. [DOI] [PubMed] [Google Scholar]
- Eaton, C.J. , Jourdain, I. , Foster, S.J. , Hyams, J.S. and Scott, B. (2008) Functional analysis of a fungal endophyte stress‐activated MAP kinase. Curr. Genet. 53, 163–174. [DOI] [PubMed] [Google Scholar]
- Egan, M.J. , Wang, Z.‐Y. , Jones, M.A. , Smirnoff, N. and Talbot, N.J. (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA, 104, 11 772–11 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X.‐J. , Mulford, K.E. and Fassler, J.S. (2009) Oxidative stress function of the Saccharomyces cerevisiae Skn7 receiver domain. Eukaryot. Cell, 8, 768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller, J. and Tudzynski, P. (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390. [DOI] [PubMed] [Google Scholar]
- Kawahara, T. , Quinn, M.T. and Lambeth, J.D. (2007) Molecular evolution of the reactive oxygen‐generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol. Biol. 7, 109–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kicka, S. , Bonnet, C. , Sobering, A.K. , Ganesan, L.P. and Silar, P. (2006) A mitotically inheritable unit containing a MAP kinase module. Proc. Natl. Acad. Sci. USA, 103, 13445–13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.‐J. , Chen, C. , Kabbage, M. and Dickman, M.B. (2011) Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases. Appl. Environ. Microbiol. 77, 7721–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalucque, H. and Silar, P. (2003) NADPH oxidase: an enzyme for multicellularity? Trends Microbiol. 11, 9–12. [DOI] [PubMed] [Google Scholar]
- Lambeth, J.D. (2004) NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4, 181–189. [DOI] [PubMed] [Google Scholar]
- Lambeth, J.D. , Cheng, G. , Arnold, R.S. and Edens, W.A. (2000) Novel homologs of gp91phox . Trends Biochem. Sci. 25, 459–461. [DOI] [PubMed] [Google Scholar]
- Lara‐Ortiz, T. , Riverose‐Rosas, H. and Aguirre, J. (2003) Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans . Mol. Microbiol. 50, 1241–1256. [DOI] [PubMed] [Google Scholar]
- Lardy, B. , Bof, M. , Aubry, L. , Paclet, M.H. , Morel, F. , Satre, M. and Klein, G. (2005) NADPH oxidase homologs are required for normal cell differentiation and morphogenesis in Dictyostelium discoideum . Biochim. Biophys. Acta, 1744, 199–212. [DOI] [PubMed] [Google Scholar]
- Lin, C.‐H. and Chung, K.‐R. (2010) Specialized and shared functions of the histidine kinase‐ and HOG1 MAP kinase‐mediated signaling pathways in Alternaria alternata, the filamentous fungal pathogen of citrus. Fungal Genet. Biol. 47, 818–827. [DOI] [PubMed] [Google Scholar]
- Lin, C.‐H. , Yang, S.L. and Chung, K.‐R. (2009) The YAP1 homolog‐mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant–Microbe Interact. 22, 942–952. [DOI] [PubMed] [Google Scholar]
- Lin, C.‐H. , Yang, S.L. , Wang, N.‐Y. and Chung, K.‐R. (2010) The FUS3 MAPK signaling pathway of the citrus pathogen Alternaria alternata functions independently or cooperatively with the fungal redox‐responsive AP1 regulator for diverse developmental, physiological and pathogenic processes. Fungal Genet. Biol. 47, 381–391. [DOI] [PubMed] [Google Scholar]
- Lin, C.‐H. , Yang, S.L. and Chung, K.‐R. (2011) Cellular responses required for oxidative stress tolerance, colonization and lesion formation by the necrotrophic fungus Alternaria alternata in citrus. Curr. Microbiol. 62, 807–815. [DOI] [PubMed] [Google Scholar]
- Malagnac, F. , Lalucque, H. , Lepère, G. and Silar, P. (2004) Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina . Fungal Genet. Biol. 41, 982–997. [DOI] [PubMed] [Google Scholar]
- Martyn, K.D. , Kim, M.‐J. , Quinn, M.T. , Dinauer, M.C. and Knaus, U.G. (2005) p21‐activated kinase (Pak) regulates NADPH oxidase activation in human neutrophils. Blood, 106, 3962–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, A.K. , Andrio, E. , Marino, D. , Pauly, N. and Dunand, C. (2010) Reactive oxygen species during plant–microorganism early interactions. J. Integr. Plant Biol. 52, 195–204. [DOI] [PubMed] [Google Scholar]
- Rolke, Y. and Tudzynski, P. (2008) The small GTPase Rac and the p21‐activated kinase Cla4 in Claviceps purpurea: interaction and impact on polarity, development and pathogenicity. Mol. Microbiol. 68, 405–423. [DOI] [PubMed] [Google Scholar]
- Scott, B. and Eaton, C.J. (2008) Role of reactive oxygen species in fungal cellular differentiations. Curr. Opin. Microbiol. 11, 488–493. [DOI] [PubMed] [Google Scholar]
- Segmüller, N. , Kokkelink, L. , Giesbert, S. , van Odinius, D., Kan, J. and Tudzynski, P. (2008) NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea . Mol. Plant–Microbe Interact. 21, 808–819. [DOI] [PubMed] [Google Scholar]
- Selvaggini, S. , Munro, C.A. , Paschoud, S. , Sanglard, D. and Gow, N.A.R. (2004) Independent regulation of chitin synthase and chitinase activity in Candida albicans and Saccharomyces cerevisiae . Microbiology, 150, 921–928. [DOI] [PubMed] [Google Scholar]
- Semighini, C.P. and Harris, S.D. (2008) Regulation of apical dominance in Aspergillus nidulans hyphae by reactive oxygen species. Genetics, 179, 1919–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon‐Plas, F. , Elmayan, T. and Blein, J. (2002) The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J. 31, 137–147. [DOI] [PubMed] [Google Scholar]
- Suh, Y.A. , Arnold, R.S. , Lassehue, B. , Shi, J. , Xu, X. , Sorescu, D. , Chung, A.B. , Griendling, K.K. and Lambeth, J.D. (1999) Cell transformation by the superoxide‐generating oxidase Mox1. Nature, 401, 79–82. [DOI] [PubMed] [Google Scholar]
- Takemoto, D. , Tanaka, A. and Scott, B. (2006) A p67Phox‐like regulator is recruited to control hyphal branching in a fungal–grass mutualistic symbiosis. Plant Cell, 18, 2807–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, D. , Tanaka, A. and Scott, B. (2007) NADPH oxidase in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet. Biol. 44, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Takemoto, D. , Kamakura, S. , Saikia, S. , Becker, Y. , Wrenn, R. , Tanaka, A. , Sumimoto, H. and Scott, B. (2011) Polarity proteins Bem1 and Cdc24 are components of the filamentous fungal NADPH oxidase complex. Proc. Natl. Acad. Sci. USA, 108, 2861–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A. , Takemoto, D. , Hyon, G.‐S. , Park, P. and Scott, B. (2008) NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Mol. Microbiol. 68, 1165–1178. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Dangl, J.L. and Jones, J.D. (2002) Arabidopsis gp91phox homologs AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA, 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H.‐C. , Yang, S.L. and Chung, K.‐R. (2012) Cyclic AMP‐dependent protein kinase A negatively regulates conidia formation by the tangerine pathotype of Alternaria alternata . World J. Microbiol. Biotechnol. 29, 289–300. [DOI] [PubMed] [Google Scholar]
- Vignais, P.V. (2002) The superoxide‐generating NADPH oxidase: structural aspects and activation mechanism. Cell. Mol. Life Sci. 59, 1428–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N.‐Y. , Lin, C.‐H. and Chung, K.‐R. (2010) A Gα subunit gene is essential for conidiation and potassium efflux but dispensable for pathogenicity of Alternaria alternata in citrus. Curr. Genet. 56, 43–51. [DOI] [PubMed] [Google Scholar]
- Yago, J.I. , Lin, C.‐H. and Chung, K.‐R. (2011) The SLT2 mitogen‐activated protein kinase‐mediated signalling pathway governs conidiation, morphogenesis, fungal virulence and production of toxin and melanin in the tangerine pathotype of Alternaria alternata . Mol. Plant Pathol. 12, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.L. and Chung, K.‐R. (2012) The NADPH oxidase‐mediated production of H2O2 and resistance to oxidative stress in the necrotrophic pathogen Alternaria alternata of citrus. Mol. Plant Pathol. 13, 900–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.L. , Lin, C.‐H. and Chung, K.‐R. (2009) Coordinate control of oxidative stress, vegetative growth and fungal pathogenicity via the AP1‐mediated pathway in the rough lemon pathotype of Alternaria alternata . Physiol. Mol. Plant Pathol. 74, 100–110. [Google Scholar]
- Yoshioka, H. , Numata, N. , Nakajima, K. , Katou, S. , Kawakita, K. , Rowland, O. , Jones, J.D.G. and Doke, N. (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans . Plant Cell, 15, 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, B.‐J. , Choquer, M. and Chung, K.‐R. (2007) The Colletotrichum acutatum gene encoding a putative pH‐responsive transcription regulator is a key virulence determinant during fungal pathogenesis on citrus. Mol. Plant–Microbe Interact. 20, 1149–1160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Alignment and comparison of the deduced amino acid sequences of NoxA and NoxB of Alternaria alternata. Conserved amino acids are shaded. Both proteins share 34.9% identity and 52.4% similarity. Conserved histidine residues potentially for haem binding are indicated by asterisks. Six putative transmembrane domains are underlined. Putative flavin adenine dinucleotide (FAD)‐ and nicotinamide adenine dinucleotide phosphate (NADPH)‐binding domains are also indicated. Alignment was obtained by BioEdit using clustalw.
Fig. S2 Alignment of the deduced amino acid sequence of the Alternaria alternata NoxR with the Epichloe festucae EfNoxR, Botrytis cinerea BcNoxR and human Hspp67phox. Conserved amino acids are shaded. Proline‐rich (P‐rich), Src homology 3 (SH3) and Phox and Bem1 (PB1) domains of human p67phox are boxed. Four tetratricopeptide repeats (TPRs) and a putative Nox activation domain are also indicated. Alignment was obtained by BioEdit using clustalw with default settings.
Fig. S3 Targeted disruption of AaNoxB in Alternaria alternata using a split marker approach. (A) Schematic depiction of the generation of truncated, but overlapping, hygromycin phosphotransferase gene (HYG) under the control of the Aspergillus nidulans trpC promoter (P) and terminator (T), and gene disruption within AaNoxB. Oligonucleotide primers used to amplify each fragment are also indicated. (B) Image of DNA fragments amplified from genomic DNA of fungi with the primers indicated. The primer pro2F sequence is not present in the split marker fragment. (C) RNA gel blotting.
Fig. S4 Targeted disruption of AaNoxR in Alternaria alternata using a split marker approach. (A) Schematic depiction of the generation of truncated, but overlapping, hygromycin phosphotransferase gene (HYG) under the control of the Aspergillus nidulans trpC promoter (P) and terminator (T), and gene disruption within AaNoxR. Oligonucleotide primers used to amplify each fragment are also indicated. (B) Image of DNA fragments amplified from genomic DNA of fungi with the primers indicated. (C) RNA gel blotting.
Fig. S5 Targeted disruption of AaNoxA in a fungal strain lacking NoxB (DB5) or NoxR (DR2) of Alternaria alternata using a split marker approach. (A) Schematic depiction of the generation of truncated, but overlapping, sulphonylurea‐resistance gene (SUR), and gene disruption within AaNoxA. Oligonucleotide primers used to amplify each fragment are also indicated. (B) Image of DNA fragments amplified from genomic DNA of fungi with the primers indicated. The primer pro1F sequence is not located within the split marker fragment. Fragment patterns indicate successful disruption at the AaNoxA locus. Fungal strains AB1 and AB6 carry impaired NoxA and NoxB genes; strains AR2 and AR3 carry impaired NoxA and NoxR genes.
Fig. S6 Assays for the sensitivity of the EV‐MIL31 strain (wild‐type, WT), the strains carrying a single deletion of NoxA, NoxB or NoxR, and the NoxA NoxB (ΔnoxAB) and NoxA NoxR (ΔnoxAR) double‐mutant strains of Alternaria alternata. Two independent mutant strains of each type were used for the assays. Fungi were grown on potato dextrose agar (PDA) amended with haematoporphyrin (50 μm) or tert‐butyl‐hydroxyperoxide (0.05%) for 4–6 days. Radial growth was measured. The percentage change in radial growth was calculated as the percentage of growth of the deletion mutants in relation to the wild‐type grown on the same plate. A negative percentage change indicates growth reduction in relation to the wild‐type. The data presented are the means and standard errors of two independent mutants with at least two replicates.
Fig. S7 Virulence assays. Detached calamondin leaves inoculated with mycelial mass obtained from the wild‐type (WT) and the rescued (CpB24) strains of Alternaria alternata resulted in necrotic lesions at 4 days post‐inoculation (dpi). ΔnoxB mutants (DB5 and DB6) failed to induce necrotic lesions on unwounded leaves. ΔnoxB mutants induced necrotic lesions on calamondin leaves with wounding prior to inoculation. The mock control was treated with water only. The inoculated leaves were incubated in a moist chamber for lesion development.
Fig. S8 Virulence assays. ΔnoxR mutants (DR2 and DR5) induced little or no necrotic lesions on detached calamondin leaves inoculated with conidial suspension (5 μL, 1 × 104 conidia/mL). The rescued strain (CpR40) carrying a functional NoxR induced wild‐type (WT)‐like necrotic lesions. Wounding prior to inoculation rendered ΔnoxR mutants able to produce necrotic lesions. The mock control was treated with water only.
Table S1 Oligonucleotide primers used in this study.


