Abstract
Ubiquitination plays a crucial role in regulating proteins post-translationally. The focus of this review is on NEDD4, the founding member of the NEDD4 family of ubiquitin ligases that is evolutionarily conserved in eukaryotes. Many potential substrates of NEDD4 have been identified and NEDD4 has been shown to play a critical role in the regulation of a number of membrane receptors, endocytic machinery components and the tumour suppressor PTEN. In this review we will discuss the diverse pathways in which NEDD4 is involved, and the patho-physiological significance of this important ubiquitin ligase.
Keywords: Ubiquitin, Ubiquitin protein ligases, NEDD4 family, IGF signaling, PTEN
1. Introduction
Ubiquitination is a post-translational protein modification that is critical for a number of cellular processes. Ubiquitination involves the covalent attachment of the 8kDa protein ubiquitin to one or more lysine residues in the substrate protein to signal proteins for degradation, altered localisation, trafficking or function. Substrate proteins can be mono-ubiquitinated, multi-monoubiquitinated or poly-ubiquitinated, with the type of ubiquitination determining the fate of the protein. Ubiquitin itself has seven lysine residues, allowing for different ubiquitin linkage types; for example the well-studied K48-linkage typically targets proteins for proteasomal degradation (Hershko and Ciechanover, 1998) whereas K63 linkages are associated with protein trafficking and lysosomal degradation (Hicke and Dunn, 2003).
Ubiquitin is covalently attached to a protein substrate via an energy dependent three step process, involving an E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme and an E3 ubiquitin protein ligase. The E3 ubiquitin ligase largely determines the substrate specificity of the system and in mammals there are several hundred ubiquitin protein ligases (Hershko and Ciechanover, 1998). These can be grouped into two main classes; the RING (Really Interesting New Gene) E3s which mediate the direct transfer of ubiquitin to the substrate (Deshaies and Joazeiro, 2009), and the HECT (Homologous to E6-AP C-Terminus) E3s which are involved in the transfer of activated ubiquitin from the E2 to the substrate by forming an intermediate complex with the C-terminus of the E3 (Rotin and Kumar, 2009). This review will focus on the HECT type ubiquitin ligase NEDD4, one of the first HECT E3 ligases discovered, and the founding member of the NEDD4 family of HECT ubiquitin ligases.
2. History of NEDD4 discovery
The NEDD4 gene was cloned in 1992 as one of a number of murine Nedd (Neural precursor cell expressed developmentally down-regulated) genes differentially expressed in the central nervous system (Kumar et al., 1992). At the time of its cloning, the predicted protein had only one known domain – an N-terminal calcium/lipid-binding domain (C2 domain). The presence of three partial repeats of approximately 40 amino acids containing two conserved tryptophan residues in the middle part of the protein was also noted. These repeats, now known to occur in numerous proteins, are widely known as WW domains (Bork and Sudol, 1994). Subsequently, in the following year the C-terminal region of NEDD4 was found to be similar to human E6-AP, the papilloma virus oncoprotein E6-associated protein. E6-AP was the first discovered ubiquitin-protein ligase and it was shown to be involved in the E6-mediated ubiquitination of p53 (Scheffner et al., 1993). The C-terminus of E6-AP comprising the catalytic domain was named HECT (homologous to the E6-AP C-terminus) (Huibregtse et al., 1995). E6-AP became the founding member of the HECT type of E3 ubiquitin ligases, of which now there are 29 human members. In yeast the first HECT ligase was Rsp5p/Npi1p from Saccharomyces cerevisiae, which was originally discovered as a suppressor of mutations in the SPT3 gene (Huibregtse et al., 1995). NEDD4 and similar proteins discovered subsequently became a family of HECT ligases, comprising 9 human proteins including NEDD4, NEDD4–2 (NEDD4L), ITCH, SMURF1, SMURF2, WWP1, WWP2, NEDL1 AND NEDL2 (Scheffner and Kumar, 2014). Rsp5 is also a member of the NEDD4 family, suggesting that these ligases are highly conserved.
3. NEDD4 orthologues and structure
As mentioned above, NEDD4 is a highly evolutionarily conserved protein from yeast to man, and was initially cloned as a highly expressed gene in the early embryonic brain (Kumar et al., 1992; Kumar et al., 1997). There are 94 orthologues of NEDD4 in the NCBI database, all sharing the same modular structure consisting of an N-terminal C2 domain, 3–4 WW domains and a C-terminal catalytic HECT domain for ubiquitin protein ligation (Harvey and Kumar, 1999) (Figure 1A). The C2 domain is a calcium-dependent lipid-binding domain around 116 amino acids in length that targets proteins to phospholipid membranes (Dunn et al., 2004), and can also be involved in protein-protein interactions (Morrione et al., 1999; Plant et al., 2000). The C2 domain-mediated membrane translocation is required for some cellular functions of NEDD4 (Dunn et al., 2004). The WW domains are protein-protein interactions domains, usually around 40 amino acids in length, containing two conserved tryptophans (W) residues that are 21 amino acids apart (Bork and Sudol, 1994). The WW domains interact with proline rich PPxY (PY) motifs and can also interact with phospho-serine/threonine residues in substrates (Sudol et al., 1995). The number of WW domains can vary between NEDD4 family members, and also between species i.e. the NEDD4 protein in human, chicken and Xenopus contains four WW domains, whereas mouse, zebrafish, Drosophila and yeast contains three WW domains (Yang and Kumar, 2010) (Figure 1B). The HECT domain is a highly conserved domain that comprises around 350 amino acids, and contains a conserved cysteine residue that forms an intermediate thioester bond with the activated ubiquitin accepted from an E2, before catalysing the ubiquitination of a lysine in the substrate protein (Rotin and Kumar, 2009). There are a number of E2s that are able to transfer ubiquitin to NEDD4, including Ubc4, UbcH5B, UbcH5C, UbcH6 and UbcH7 (Anan et al., 1998; Fotia et al., 2006).
Figure 1. (A) Schematic structure of the NEDD4 protein.
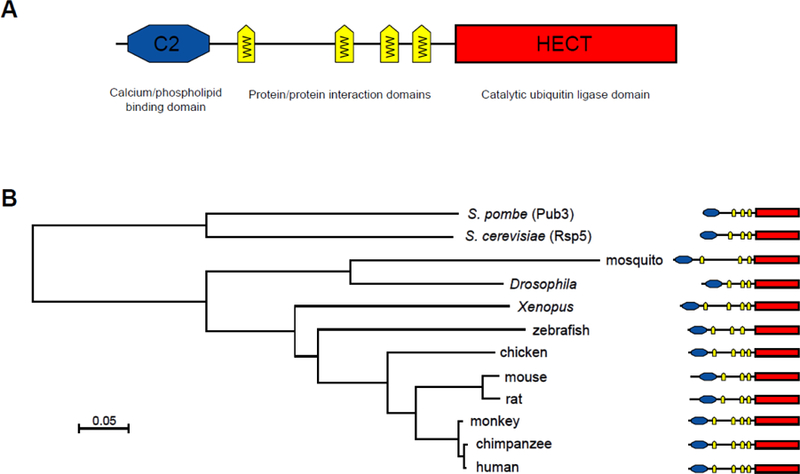
Schematic of the modular structure of the human NEDD4 protein. The C2 calcium/phospholipid binding domain mediates NEDD4 binding to membranes, and is also involved in substrate recognition. The WW domains are protein-protein interaction domains that bind to conserved PY motifs in substrates and regulatory proteins. The catalytic ubiquitin ligase domain binds the E2 conjugation enzyme and forms a thioester bond with ubiquitin before transferring ubiquitin to the substrate.
(B) Phylogenetic relationship of NEDD4 proteins from various species. NEDD4 sequences were obtained from the NCBI protein database as follows; S. pombe (Schizosaccharomyces pombe Pub3; NP_595793.1), S. cerevisiae (Saccharomyces cerevisiae Rsp5p; AAC03223.1), mosquito (Anopheles gambiae; XP_003436401.1), Drosophila [Drosophila melanogaster; NP_996116.1), Xenopus (Xenopus laevis; NP_001084258.1), zebrafish (Danio rerio; NP_001029358.1), chicken (Gallus gallus; XP_413791.3), mouse (Mus musculus; NP_035020.2), rat (Rattus norvegicus; NP_037118.1), monkey (Macaca fascicularis; XP_005559683.1), chimpanzee (Pan troglodytes; XP_523083.3),and human (Homo sapiens; NP_006145.2 ). Sequences were aligned using NCBI COBALT (Constraint Based Multiple Protein Alignment Tool) http://www.stva.ncbi.nlm.nih.gov/tools/cobalt/re_cobalt.cgi (Papadopoulos and Agarwala, 2007) and the minimum-evolution Phylogenetic Tree output displayed. The individual domains on the NEDD4 schematics were identified using NCBI Conserved Domain Database search http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi (Marchler-Bauer et al., 2011) and are drawn roughly to scale. The scale bar indicates evolutionary distance (Grishin, 1995).
The human NEDD4 gene is located on chromosome 15q21.3 and comprises 30 exons (HGNC:7727) shown to encode a ~120 KDa protein. There are five NEDD4 protein variants in the NCBI database, all of which share 100% identity from the first WW domain through to the end of the protein, only varying in the N-terminal region which includes the C2 domain. Recently it was reported that there is a 75kDa NEDD4 isoform found exclusively in myotonic dystrophy type 2 muscle in addition to full length NEDD4 (Screen et al., 2014). NEDD4 protein is localised to the cytoplasm, mainly in the perinuclear region and cytoplasmic periphery of human cultured cells (Anan et al., 1998). NEDD4 is also found in exosomes when recruited by the NEDD4 family interacting protein Ndfip1 (Putz et al., 2008).
4. NEDD4 binding partners and targets
A number of in vitro binding studies and proteomic approaches have been used to identify potential NEDD4 substrates (see below for a summary and Table 1 for a list of interacting proteins).
Table 1.
Potential binding partners of NEDD4
| Interactor | Function | References |
|---|---|---|
| α-synuclein | Pre-synaptic neural protein | (Tofaris et al., 2011) |
| Ack | Activated cdc42-associated tyrosine kinase | (Lin et al., 2010) |
| Alix | Vacuolar protein sorting factor adaptor protein | (Sette et al., 2010) |
| AMPAR (GluR1) | Alpha-amino-3-hydroxy-5 methyl isoxazole-4 propianic acid receptor ion channel | (Lin et al., 2011) |
| ARRDC3 | Arrestin domain containing 3 adaptor protein | (Nabhan et al., 2010) |
| β2-AR | Beta-2 androgen receptor | (Nabhan et al., 2010) |
| β‒arrestin | Adaptor protein | (Shenoy et al., 2008) |
| Β‒TRCP | F box component of the SCF complex | (Liu et al., 2014) |
| Beclin1 | Tumor suppressor protein | (Platta et al., 2012) |
| Calpain 2 | Calcium-dependent cysteine protease | (Cui et al., 2014) |
| CKIδ | Casein kinase I δ | (Liu et al., 2014) |
| Caspase 9 | Cysteinyl aspartate protease | (Fombonne et al., 2012) |
| Cav1.2 | Voltage-gated calcium channel | (Rougier et al., 2011) |
| Cbl-b | E3 RING finger ubiquitin protein ligase | (Magnifico et al., 2003; Yang et al., 2008) |
| c-Myc | Transcription factor | (Liu et al., 2013) |
| Cnx43 | Connexin43 gap junction protein | (Leykauf et al., 2006; Girao et al., 2009) |
| Cullin | Scaffolding protein for E3s | (Liu et al., 2014) |
| Deltex | Regulator of Notch signalling | (Sakata et al., 2004) |
| Dvl1 | Dishevelled-1 adaptor protein | (Nethe et al., 2012) |
| EGFR | Epidermal growth factor receptor | (Katz et al., 2002) |
| ENaC | Epithelial sodium channel | (Staub et al., 1996) |
| Eps15 | Epidermal growth factor receptor substrate 15 | (Woelk et al., 2006) |
| FGFR1 | Fibroblast growth factor receptor 1 | (Persaud et al., 2011) |
| FoxM1B | Forkhead box protein M1B, transcription factor | (Kwak et al., 2012) |
| Gag | Core viral protein | (Ingham et al., 2004) |
| GGA3 | Golgi-localised γ-ear-containing ADP ribosylation factor binding protein | (Pak et al., 2006) |
| Grb10 | Growth factor receptor bound protein 10, adaptor protein | (Morrione et al., 1999) |
| HER3, HER4 | Epidermal growth factor receptor family | (Zeng et al., 2009; Huang et al., 2014) |
| Hgs | Ubiquitin-interacting adaptor | (Katz et al., 2002) |
| IGF-1R | Insulin-like growth factor 1 receptor | (Vecchione et al., 2003; Cao et al., 2008; Monami et al., 2008) |
| ISG15 | Small ubiquitin-like molecule | (Malakhova and Zhang, 2008) |
| γ2-adaptin | Ubiquitin-interacting adaptor protein | (Rost et al., 2008) |
| LAPTM5 | Lysosomal associated protein transmembrane 5 | (Pak et al., 2006) |
| LATS1 | Large tumor suppressor kinase 1 | (Salah et al., 2013) |
| LMP2A | Latent membrane protein 2A | (Ikeda et al., 2000) |
| MDM2 | Mouse double minute 2, E3 ubiquitin ligase | (Xu et al., 2014) |
| MTMR4 | Inositol phosphatase | (Plant et al., 2009) |
| Nav1.2, Na v1.7 | Voltage-gated sodium channels | (Fotia et al., 2004) |
| Ndifp1, Ndfip2 | NEDD4 family interacting proteins, adaptor proteins | (Shearwin-Whyatt et al., 2006) |
| NHE1 | Sodium/hydrogen exchange ion transporter | (Simonin and Fuster, 2010) |
| n-Myc | Myc family transcription factor | (Liu et al., 2013) |
| Notch | Transmembrane signalling protein | (Sakata et al., 2004) |
| p34 | Cyclin dependent kinase binding protein | (Hong et al., 2014) |
| p63 | p53 family member | (Li and Xiao, 2014) |
| pAkt | Phospo-AKT, serine/threonine kinase | (Fan et al., 2013) |
| pol II | RNA polymerase II | (Anindya et al., 2007) |
| PPARγ | Peroxisome proliferator-activated receptor γ | (Han et al., 2013) |
| Ptc | Patched receptor | (Fombonne et al., 2012) |
| PTEN | Phosphatase and tensin homologue depleted on chromosome ten | (Wang et al., 2007; Christie et al., 2012; Aronchik et al., 2014) |
| Rac | Rho GTPase | (Nethe et al., 2012) |
| Rap2A | Ras-related protein 2A | (Kawabe et al., 2010) |
| SAG | Sensitive to apoptosis gene, zinc RING finger protein | (Zhou et al., 2014) |
| SIRT2 | Binds to the promoter of NEDD4 | (Liu et al., 2013) |
| Smo | Smoothened receptor | (Luo et al., 2012) |
| Spy1A | Cyclin-dependent kinase activator | (Al Sorkhy et al., 2009) |
| TINK | Traf2 and NCK interacting kinase, | (Kawabe et al., 2010) |
| Traf3 | TNFR associated factor 3 | (Fang et al., 2014) |
| VEGF-R2 | Vascular endothelial growth factor receptor 2 | (Murdaca et al., 2004) |
4.1. Ion channels and membrane transporters
The epithelial sodium channel (ENaC) is a transmembrane ion channel that contains a PY motif in its cytoplasmic tail. Using yeast two hybrid studies, rat NEDD4 was shown to bind to the PY motif in ENaC via its WW domains (Staub et al., 1996). Furthermore, in response to increased intracellular sodium, NEDD4 binds and ubiquitinates ENaC to mediate the down-regulation of sodium channel activity (Dinudom et al., 1998). However, in vivo studies indicate that NEDD4–2 is the main NEDD4 family member that is responsible for ENaC regulation (Kamynina et al., 2001; Fotia et al., 2003; Boase et al., 2011; Goel et al., 2015).
A number of ion channels found in the brain are subject to NEDD4 regulation. Alpha-Amino-3-hydroxy-5-methyl-isoxazole-4-propionic acid receptors (AMPARs) are ion channels that mediate excitatory synaptic transmission in the brain. In rat neurons, NEDD4 was shown to ubiquitinate the AMPAR GLUR1 subunit, leading to lower levels of AMPAR at the cell surface, and a decrease in synaptic transmission (Lin et al., 2011). The voltage-gated calcium ion channel Cav1.2 is regulated by NEDD4, inducing down-regulation of cell surface expression at the plasma membrane to affect whole cell Cav currents, and also directing Cav1.2 sorting from the trans-golgi network to endosomes (Rougier et al., 2011). Voltage gated sodium channels 1.2 and 1.7 (Nav1.2 and 1.7) contain PY motifs that interact with the WW domains of NEDD4 to result in down-regulation of the channel and reduced sodium channel activity in Xenopus oocytes (Fotia et al., 2004).
4.2. Growth factor signalling
In the epidermal growth factor signalling pathway, there are a number of components that are regulated by NEDD4. The epidermal growth factor receptor (EGFR) family of tyrosine kinase members, HER3 and HER4, both undergo NEDD4-mediated ubiquitination and degradation to down-regulate signalling (Zeng et al., 2009; Huang et al., 2014). Activated Cdc42-associated tyrosine kinase (ACK) is an ubiquitin-binding protein involved in EGFR regulation. The PY motif of ACK binds the WW3 domain of NEDD4, resulting in ubiquitination and lysosomal degradation of ACK and EGFR in response to EGF stimulation (Lin et al., 2010). Epidermal growth factor receptor substrate 15 (EPS15) is a protein in the EGFR pathway that contains a ubiquitin interacting motif (UIM), and has been shown to interact with ubiquitinated NEDD4 to promote mono-ubiquitination of itself in a process termed coupled monoubiquitination (Woelk et al., 2006).
Fibroblast growth factor receptor 1 (FGFR1) has important roles in regulating cellular proliferation and differentiation. FGFR1 contains a novel site (VL***PSR) that binds the C2 and WW3 domain of NEDD4, leading to the ubiquitination and down-regulation of FGFR1 at the cell surface, and attenuation of downstream signalling (Persaud et al., 2011). The vascular endothelial growth factor receptor-2 (VEGF-R2) interacts with NEDD4 leading to the degradation of VEGF-R2, although this degradation is not due to NEDD4-mediated ubiquitination. Grb10 acts as a positive regulator of VEGF-R2 signalling by interacting with NEDD4 to inhibit the NEDD4-mediated degradation of VEGF-R2 (Murdaca et al., 2004). The beta 2 androgen receptor (β2-AR) interacts with NEDD4 via the adaptor protein beta-arrestin 2 in vitro, leading to the ubiquitination and degradation of activated β2-AR in the lysosome (Shenoy et al., 2008).
4.3. Tumor suppressors
Beclin 1 is a tumor suppressor involved in the PI3 kinase pathway. The PY motif of Beclin1 interacts with NEDD4 WW domains, leading to polyubiquitination and subsequent down-regulation of Beclin1 (Platta et al., 2012). Interestingly, the polyubiquitination involved ubiquitin-K11 linkages to promote Beclin1 down-regulation, rather than the typical ubiquitin-K48 linkages. Large tumor suppressor kinase 1 (LATS1) is a serine/threonine kinase that is a negative regulator of YAP1 in the Hippo signalling pathway, and directly binds NEDD4 leading to its ubiquitination and subsequent degradation, implicating NEDD4 as an additional regulator of the Hippo pathway (Salah et al., 2013). NEDD4 has also been shown to interact with the tumor suppressor PTEN (phosphatase and tensin homologue depleted on chromosome ten) (Wang et al., 2007), as discussed further in 5.2.
4.4. Endocytic regulation proteins
γ2-adaptin is a member of the heterotetrameric clathrin adaptor complex family that contains a UIM, which binds to the ubiquitinated C2 domain of NEDD4 to facilitate its own ubiquitination (Rost et al., 2008). Furthermore, the high affinity of γ2-adaptin for the C2 domain of NEDD4 allows not only mono-ubiquitination of γ2-adaptin, but also multi and poly-ubiquitination chains. Another UIM-containing protein, Hgs (a mammalian homologue of a yeast vacuolar sorting adaptor), interacts with NEDD4 to target substrate proteins such as the EGFR receptor for ubiquitination to induce endocytosis (Katz et al., 2002). This UIM is often found in endocytic adaptors, indicating that ubiquitination, and NEDD4, could play a general role in vesicular trafficking network.
Lysosomal associated protein transmembrane 5 (LAPTM5) is a lysosomal transmembrane protein that is expressed in hematopoietic cells, and resides in the late endosome/lysosome. The PY motifs of LAPTM5 bind the WW domains of NEDD4 to control the sorting of Laptm5 from the golgi to the lysosome, but this does not require the ubiquitination of LAPTM5 (Pak et al., 2006). In addition, The NEDD4-LAPTM5 complex recruits GGA3 (Golgi-localised, γ-ear-containing, ADP ribosylation factor binding protein), a protein involved in regulating cargo trafficking, and NEDD4 ubiquitinates GGA3 (Pak et al., 2006).
4.5. NEDD4 and viral budding
A number of viruses contain the PY late budding domain expressed within their matrix proteins, mediating interactions with the WW domains of NEDD4 to facilitate viral budding. These include the matrix proteins of Ebola virus (Harty et al., 2000), Rous sarcoma virus (Kikonyogo et al., 2001), Mason-Pfizer monkey virus (Gottwein et al., 2003), the murine leukemia virus (Segura-Morales et al., 2005), the Marburg virus (Urata and Yasuda, 2010) and Human Immunodeficiency virus (HIV) (Sette et al., 2010). NEDD4 also interacts with components of the ESCRT machinery required for viral budding. For example, the vacuolar protein sorting protein Alix recruits NEDD4 to HIV-1 Gag protein to facilitate HIV-1 release via a mechanism that involves Alix ubiquitination (Sette et al., 2010). Finally, NEDD4 binds and ubiquitinates the latent membrane protein 2A (LMP2A) of the Epstein-Bar virus to modulate B-cell signal transduction (Ikeda et al., 2000).
4.6. Other NEDD4 targets
Alpha-synuclein is an abundant protein in the human brain that leads to the neurodegenerative disorder Parkinson’s disease when it is misfolded and accumulates in the brain. NEDD4 recognises the c-terminus of α-synuclein and attaches K63-type ubiquitin chains, resulting in lysosomal degradation (Tofaris et al., 2011).
The WW domains of NEDD4 bind to the PY motif of the gap junction protein connexin43 (Cx43) (Leykauf et al., 2006). This results in the mono-ubiquitination of Cx43 and its interaction with Eps15, an adaptor protein involved in endocytosis, leading to the internalisation of Cx43 from the plasma membrane (Girao et al., 2009). Recently it was shown that the interaction between NEDD4 and Cx43 is enhanced in rat astrocytes after lipopolysaccharide stimulation (Liao et al., 2013). NEDD4 also mediates the ubiquitination of the adaptor protein dishevelled-1, and associates with Rho-GTPase Rac1 to stimulate the maturation of epithelial cell-cell contacts (Nethe et al., 2012).
Notch is an importing regulator of cell differentiation and proliferation, and with Deltex has been shown to interact with NEDD4 resulting in the ubiquitination of Notch and Deltex, promoting their endocytosis and turnover (Sakata et al., 2004). In Drosophila, Ndfip1 acts as an adaptor protein to promote NEDD4 interaction with Notch (Dalton et al., 2011).
Spy1A is a constitutively expressed cyclin-like protein required for progression through the G1/Sphase of the cell cycle that is ubiquitinated by NEDD4 to be degraded in a cell cycle manner (Al Sorkhy et al., 2009). RNA polymerase II was shown to be ubiquitinated by NEDD4 in response to UV-induced DNA damage in human cells (Anindya et al., 2007).
The calcium-dependent cysteine protease calpain 2 undergoes NEDD4-mediated ubiquitination and degradation in response to Brucella bacterial infection in macrophages (Cui et al., 2014). The cysteinyl aspartate protease 9 (caspase-9) is directly ubiquitinated by NEDD4 in the Patched complex, which is required for Patched-mediated apoptosis (Fombonne et al., 2012). Sensitive to apoptosis gene (SAG) is an anti-apoptotic cellular survival protein that has been shown to promote cell proliferation and protect cancer cells from apoptosis. SAG was initially identified as a potential NEDD4 substrate in a proteomics study (Persaud et al., 2009), and was validated as a target by Zhou et al (2014). The RING domain of SAG interacts with NEDD4 in an atypical fashion via the HECT domain, leading to the ubiquitination and subsequent degradation of SAG (Zhou et al., 2014). NEDD4 levels are inversely correlated with protein levels of SAG in human lung cancer cell lines (Zhou et al., 2014).
5. Physiological functions of NEDD4
NEDD4 is widely expressed in mammalian tissues. To investigate the physiological functions of NEDD4, a number of studies have focussed on NEDD4−/− mice that lack NEDD4 expression. The first described NEDD4−/− mice were neonatal lethal, with delayed embryonic development and reduced growth and body weight due (Cao et al., 2008). Consistent with this, NEDD4−/− murine embryonic fibroblasts (MEFs) display reduced mitogenic activity (Cao et al., 2008). NEDD4−/− embryos have reduced skeletal muscle size, decreased motor neuron and axon numbers and abnormal neuromuscular junction structure and function (Liu et al., 2009). NEDD4−/− embryos also have severe craniofacial defects, and are deficient in cranial neural crest cells (Wiszniak et al., 2013). Another independently generated NEDD4−/− mouse line also showed embryonic lethality, and NEDD4−/− embryos have pronounced heart defects, vasculature abnormalities and impaired dendrite development (Fouladkou et al., 2010; Kawabe et al., 2010). Recently, a conditional skeletal muscle-specific NEDD4−/− mouse demonstrated an important role for NEDD4 in mediating denervation-induced skeletal muscle atrophy (Nagpal et al., 2012). Due to the lethality of NEDD4−/− mice, studies have been performed in NEDD4 heterozygous mice, which are viable and live to maturity. NEDD4 heterozygous mice have normal motor function, but show significant age-dependent gait abnormalities (Camera et al., 2014).
5.1. NEDD4 and IGF signalling
One of the most pronounced phenotypes of the NEDD4−/− mice is reduced growth, attributed to decreased insulin like growth factor-1 (IGF-1) and insulin signalling, highlighting the important role of NEDD4 in the regulation of cell proliferation, survival, differentiation and cell motility (Cao et al., 2008). Previous in vitro work showed that NEDD4 bound to the adaptor protein Grb10 that is known to negatively regulate IGF1 signalling, to form a bridge between NEDD4 and the Insulin-like growth factor 1 receptor (IGF-1R) (Morrione et al., 1999). This NEDD4/Grb10 complex regulates the ubiquitination and stability of the IGR-1R (Vecchione et al., 2003) by mediating the multi-ubiquitination of IGF-1R, leading to receptor internalisation (Monami et al., 2008). These results suggest that NEDD4 acts as a negative regulator of IGF-1R signalling, and are inconsistent with the growth deficiency seen in the NEDD4−/− mice, as described below.
NEDD4−/− MEFs show reduced cell surface expression of both the Insulin Receptor (IR) and the Insulin-like growth factor 1 receptor (IGF-1R) (Cao et al., 2008). Upon ligand binding, IGF-1R undergoes auto-phosphorylation, leading to phosphorylation of substrates such as insulin receptor substrate (IRS) and the activation of the phosphoinositide 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signalling pathways. NEDD4−/− MEFS showed reduced IGF-1 induced tyrosine phosphorylation of IGF-1R and IRS-1, as well as reduced activation of Akt (PI3K pathway) and ERK (MAPK Pathway), although the total abundance of these proteins was not altered (Cao et al., 2008). Recently it was shown that defective IGF signalling in NEDD4−/− MEFs, including the phosphorylation of IRS1 and Akt, could be rescued by ablation of the phosphatase PTEN (Shi et al., 2014). The role of NEDD4 in this growth hormone signalling was specific for insulin and IGF-1, and not observed with epidermal growth factor (EGF) or serum signalling (Fan et al., 2013; Shi et al., 2014). Moreover, IGF-1 signalling stimulates NEDD4 K63-type poly-ubiquitination of pAKT at the plasma membrane, without altering total AKT ubiquitination, to promote pAKT nuclear trafficking (Fan et al., 2013). The in vivo data above indicate that IGF-1 and insulin signalling require NEDD4 function (see Figure 2).
Figure 2. The proposed role of NEDD4 in IGF-1R signaling.
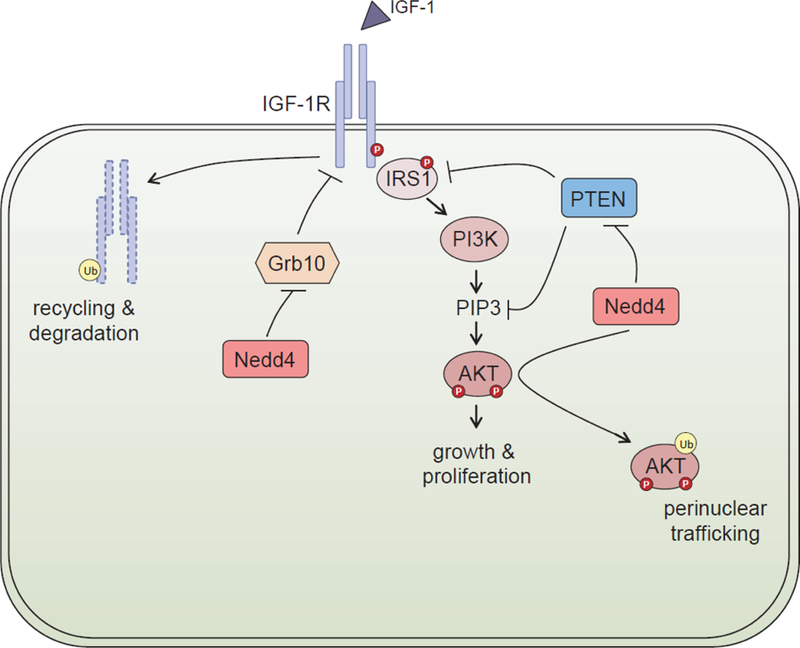
Stimulation of the IGF-1R with IGF-1 ligand results in auto-phosphorylation of the receptor, leading to phosphorylation of insulin receptor substrate 1 (IRS1) and the activation of the PI3K pathway that ultimately results in cellular growth and proliferation. PTEN has recently been shown to be a protein phosphatase for IRS-1, and NEDD4 antagonizes this phosphatase activity (Shi et al., 2014). IGF-1 signalling stimulates the ubiquitination of pAKT by NEDD4 to promote the peri-nuclear trafficking of pAKT (Fan et al., 2013). The adaptor protein Grb10 negatively regulates IGF-1 signalling by binding the IGF-1R, and in vivo data indicates that NEDD4 down-regulates Grb10 expression (Cao et al., 2008). IGF-1R is also targeted by ubiquitin ligases such as MDM2 and c-Cbl for endocytosis and degradation.
In vitro NEDD4 has been shown to bind to Grb10, a negative regulator of IGF-1 signalling (Morrione et al., 1999). NEDD4−/− MEFs showed increased levels of the Grb10, and simultaneous loss of NEDD4 and Grb10 restores the cell-surface levels and signalling of IGF-1R, indicating that NEDD4 acts to oppose the function of the negative regulator Grb10 (Cao et al., 2008). Furthermore NEDD4−/− lethality is partially rescued by the maternal inheritance of a disrupted Grb10 allele (Cao et al., 2008). The role of NEDD4 in IGF-R1 signalling may be context specific. It was recently reported that during neurodegeneration in mice, NEDD4 was upregulated in cultured neurons and this led to the ubiquitination and proteasomal degradation of IGF-IRβ receptor (Kwak et al., 2012). Other ubiquitin ligases such as Mdm2 and c-Cbl have been shown to ubiquitinate IGF-1R leading to receptor endocytosis (Sehat et al., 2008) and Mdm2 has recently been identified as substrate of NEDD4 (Xu et al., 2014). Clearly, the exact role of NEDD4 on the regulation of IGR-1R is yet to be fully elucidated.
5.2. NEDD4 and PTEN
PTEN is a tumour suppressor protein that functions as a phosphatase for PIP3 (phosphatidylinositol-3,4,5-tri-phosphate), which is an important secondary messenger of the PI3K/AKT/mTOR pathway that regulates cell survival, proliferation and differentiation. PTEN was identified as a direct target of NEDD4 in vitro, resulting in PTEN ubiquitination and subsequent proteasomal degradation (Wang et al., 2007). Mono-ubiquitination of PTEN by NEDD4 was shown to lead to PTEN nuclear import in both murine and human cells, thereby protecting PTEN from cytoplasmic degradation (Trotman et al., 2007). Thus NEDD4 can act as a proto-oncogene to degrade PTEN, or in a tumor-suppressive role to shuttle PTEN into the nucleus to protect PTEN, as even from the nucleus PTEN is still able to antagonize the AKT pathway and cause apoptosis (Trotman et al., 2007).
The role of NEDD4 in the regulation of PTEN in vivo however remains somewhat controversial. Increased NEDD4 expression leads to increased PTEN degradation in various human cancer cell lines overexpressing K-ras or treated with EGF (Zeng et al., 2014). In human melanoma cells, in the presence of indole-3-carbinol (I3C), stabilization of PTEN results from disrupting NEDD4 binding, thereby preventing the ubiquitination and proteasomal degradation of PTEN (Aronchik et al., 2014). Similarly, Rak functions as a tumour suppressor by phosphorylating PTEN to block NEDD4 binding, therefore stabilizing PTEN (Yim et al., 2009). NEDD4 co-localises with PTEN within sensory neurons in vivo and in vitro, and also within regenerating axons at an injury site (Christie et al., 2012). NEDD4 RNAi knockdown supports the role of NEDD4 mediated PTEN degradation in axon growth in Xenopus retinal ganglion cells (Drinjakovic et al., 2010) and in rat dorsal root ganglion cells (Christie et al., 2012). However, PTEN protein levels are equivalent in wild-type and NEDD4−/− MEFs (Cao et al., 2008; Fouladkou et al., 2008), and PTEN stability and nuclear import are not affected in NEDD4−/− MEFs (Fouladkou et al., 2008). In conditional neuronal-specific NEDD4−/− mice axon growth was affected by the lack of NEDD4, without changes in the activity, ubiquitination levels, or localisation of PTEN (Hsia et al., 2014). PTEN has been shown to interact with other E3 ligases (XIAP, WWP2, CHIP), so perhaps the interaction between NEDD4 and PTEN is context specific. Indeed, NEDD4 function in PTEN regulation may primarily occur in pathological contexts such as cancer or cellular stress.
5.3. NEDD4 in the neuron
Given that NEDD4 was originally identified as a neuronally expressed, developmentally downregulated gene, it is not surprising that NEDD4 is highly expressed in the central nervous system and plays an important physiological role in neuronal development (recently reviewed in (Donovan and Poronnik, 2013)). Using both conventional and neuron-specific conditional NEDD4−/− mouse models, NEDD4 was identified as an important regulator of dendrite formation and arborisation in both hippocampal and cortical neurons (Kawabe et al., 2010). NEDD4 forms a complex with the Traf2 and NCK Interacting Kinase (TINK), and Ras-related protein 2A (Rap2A), a negative regulator of dendritogenesis, and this trimeric complex formation results in the NEDD4 dependent mono- and di-ubiqutination of Rap2A to reduce downstream signalling and promote dendrite growth and arborisation (Kawabe et al., 2010). NEDD4-mediated ubiquitination and subsequent down-regulation of PTEN in Xenopus root ganglion cell axons also promotes axon branching in vivo (Drinjakovic et al., 2010). Upon zinc-mediated damage of the central nervous system, NEDD4 ubiquitinates PTEN to mediate the neuronal response (Kwak et al., 2010). Similarly during the regenerative response to axonal injury, NEDD4 associates with PTEN in dorsal root ganglion neurons to promote nerve regeneration (Christie et al., 2012). Recently NEDD4-binding protein 3 (N4BP3) has also been implicated in axonal and dendritic branching in developing neurons (Schmeisser et al., 2013). NEDD4 is also required for the normal formation and functioning of neuromuscular junctions, normal motor neuron and axon numbers and cranial neural crest (Liu et al., 2009; Wiszniak et al., 2013).
5.4. NEDD4 and T-cell function
The NEDD4 family E3s NEDD4 and Itch are both expressed in T-cells (Heissmeyer et al., 2004) and although in vitro data shows these ligases share a number of substrates involved in T-cell regulation, Itch and NEDD4 knockout mouse models display unique phenotypes, suggesting discrete functions in vivo (Fang et al., 2002; Yang et al., 2008). Fetal liver chimeras that lack NEDD4 expression in cells of haemopoietic origin show that NEDD4 is not required for T-cell development, or for initial T-cell antigen receptor mediated activation events (Yang et al., 2008). However NEDD4−/− mice have fewer effector T-cells, and in response to antigen immunization T-cells lacking NEDD4 proliferate poorly, and produce less interleukin 2, suggesting the role of NEDD4 is to convert naïve T-cells into activated T-cells. The hypo-responsiveness of NEDD4−/− T-cells can be explained by the impaired ubiquitination and degradation of Cbl-b, a ubiquitin ligase that plays a critical role in T-cell activation and tolerance induction, as NEDD4 is required for the poly-ubiquitination of Cbl-b (Magnifico et al., 2003; Yang et al., 2008). Recently it was shown that Cbl-b inhibits T-cell activation by impeding the association of NEDD4 with PTEN in T-cells to suppress PTEN inactivation (Guo et al., 2012). In addition NEDD4 is not required for B-cells to become activated, but NEDD4−/− T-cells are unable to provide adequate help for B-cells to undergo immunoglobulin class switching (Yang et al., 2008).
5.5. NEDD4 and cancer
NEDD4 is frequently overexpressed in many different types of cancer (for reviews see (Chen and Matesic, 2007; Ye et al., 2014)). NEDD4 was first described as a proto-oncogene for its role in negatively regulating the tumor suppressor PTEN via ubiquitination in vitro (Wang et al., 2007). An inverse correlation between (increased) NEDD4 and (decreased) PTEN has been observed in many human cancer cell lines, including breast cancer MDA-MB-231 and prostate cancer DU145 cell lines (Liu et al., 2014). After overexpressing K-ras or EGF treatment, increased NEDD4 levels and PTEN degradation are observed in various type of human cancer cell lines including cervical adenocarcinoma HeLa, colorectal adenocarcinoma HT-29, gastric adenocarcinoma BGC-823 and hepatocellular carcinoma HepG2 (Zeng et al., 2014). Given the lack of NEDD4 regulation of PTEN in NEDD4−/− mice, this led to the hypothesis that perhaps NEDD4-mediated PTEN degradation primarily occurs in cancer cells under certain oncogenic circumstances (Zeng et al., 2014). Furthermore, the CDK-4 binding partner p34 has been identified as an interactor of NEDD4 in cancer cells lines, and co-expression of p34 and NEDD4 is correlated with lowered PTEN levels in colon cancer tissues, suggesting that NEDD4 positively regulates tumorigenesis via the p34-dependent PTEN proteasomal degradation (Hong et al., 2014). Contrary to this, there is also evidence of NEDD4 overexpression in cancer that promotes cell growth independent of PTEN signalling, such as in human colorectal cancer lines HCT-15 and LoVo (Eide et al., 2013).
Decreased levels of NEDD4 can also be associated with cancer. NEDD4 directly ubiquitinates oncoproteins N-Myc in neuroblastoma and c-Myc in pancreatic cancer cells to target these Myc proteins for proteasomal degradation (Liu et al., 2013). The histone deacetylase SIRT2 enhances expression of N-Myc and c-Myc by directly binding to the NEDD4 promoter and repressing NEDD4 gene expression by deacetylating histone H4 lysine 16 (Liu et al., 2013). Importantly, NEDD4 gene expression could be reactivated by the addition of SIRT2 inhibitors, resulting in reduced N-Myc and c-Myc protein expression, and suppressing neuroblastoma and pancreatic cancer cell proliferation (Liu et al., 2013). An inverse relationship between protein levels of (low) NEDD4 and (high) HER3 (an EGFR tyrosine kinase member) is observed in ductal cells of prostate cancer tumors compared to surrounding tissues, and knockdown of NEDD4 in human prostate and breast cancer cell lines leads to increased HER3-mediated cell migration and proliferation in vitro, and xenoplant tumor growth in vivo (Huang et al., 2014).
6. Regulation of NEDD4
6.1. NEDD4 auto-inhibition
NEDD4 activity is in part regulated by auto-inhibition. In the absence of calcium, an auto-inhibitory conformation of NEDD4 is formed by the C2 domain of NEDD4 binding to the HECT domain via intramolecular interactions, thereby inhibiting the enzymatic activity of NEDD4 (Wang et al., 2010). In the presence of calcium, the binding of the C2 domain to the HECT domain is disrupted and the C2 domain recruits NEDD4 to the lipid membrane promoting its ubiquitin ligase activity (Wang et al., 2010). Proteins or compounds that prohibit this auto-inhibitory conformation of NEDD4 serve as activators. The adaptor proteins Ndfip1 and Ndfip2 can stimulate NEDD4 activity by binding to the WW domains of NEDD4 via their PY motifs to release the auto-inhibitory conformation between the C2 and HECT domains of NEDD4 (Mund and Pelham, 2009). Recently, the c-Src kinase was demonstrated to phosphorylate NEDD4 at specific tyrosine residues Y43 (in the C2 domain) and Y585 (in the HECT domain), resulting in the disruption of the auto-inhibitory conformation of NEDD4 and consequently activating NEDD4 ubiquitin ligase activity (Persaud et al., 2014).
Conversely, stabilizing the C2 and HECT domain auto-inhibitory conformation would lead to an inhibition of NEDD4 activity, as has been proposed by in silico molecular modelling for the chemical indole-3-carbinol that binds to the HECT domain of NEDD4 (Aronchik et al., 2014). Increasing intracellular calcium can also act to release the auto-inhibitory conformation of NEDD4, for example gram-negative Brucella infection induces increases in NEDD4 activity in an intracellular calcium-dependent manner (Cui et al., 2014).
6.2. Ndfip adaptors
In addition to the activating roles of Ndfips in abrogating the auto-inhibitory conformation of NEDD4 (see above), these adaptors can facilitate substrate binding to target proteins that lack PY motifs (Foot et al., 2008; Foot et al., 2011). The Ndfip proteins contain 3 PY motifs with which they interact with the WW domains of NEDD4 (Shearwin-Whyatt et al., 2006). Ndfip1 is also involved in exosomal secretion, and overexpression of Ndfip1 results in NEDD4 being recruited to the exosome (Howitt et al., 2009).
6.3. Oxidative stress
In the presence of neurotoxins that elicit oxidative stress in neurons, such as camptothecin, hydrogen peroxide and zinc, NEDD4 protein expression was up-regulated (Kwak et al., 2012). Reactive oxygen species (ROS) induces up-regulation of NEDD4 in primary rat cortical neurons after zinc treatment, and pre-treatment of neurons with antioxidants prevents the zinc-induced NEDD4 up-regulation. The oxidative stress induces NEDD4 transcriptional activation, with FOXM1B (Forkhead box protein M1B) identified as the key transcription factor mediating this ROS response (Kwak et al., 2012). The transcription factor FoxM1 is a member of the Forkhead box (Fox) family, and has been shown to bind directly to the endogenous human Nedd4 promoter at two FoxM1 binding sites to result in up-regulation of Nedd4 expression (Dai et al., 2010).
6.4. Phosphorylation
NEDD4 protein stability is governed by the SCFβ-TRCP (Skp, Cullin, F-box containing complex, a multi-protein E3 ligase complex) (Liu et al., 2014). NEDD4 bound both Cullin1 and the F-box β-TRCP component of the SCF directly, and depletion of endogenous Cullin1 or β-TRCP led to the up-regulation of NEDD4 protein in human cells. Furthermore, Casein Kinase Iδ was identified as the kinase that phosphorylated NEDD4 at both S347 and S348 to trigger its interaction with β-TRCP and lead to the ubiquitination and degradation of NEDD4 (Liu et al., 2014). As mentioned above, NEDD4 phosphorylation by c-Src enhances its catalytic activity (Persaud et al., 2014).
6.5. Other NEDD4 regulators
The interferon inducible ISG15 is a small ubiquitin-like protein that negatively regulates NEDD4 by binding directly to NEDD4 to block its interaction with the conjugating E2 enzyme, thus preventing the ubiquitin transfer from the E2 to NEDD4 (Malakhova and Zhang, 2008). Another inhibitor of NEDD4, heclin (HECT ligase inhibitor) was recently identified that broadly inhibits HECT ligases in cells by binding to the HECT domain to cause a conformational change that allows the oxidation of the active Cys site, blocking formation of a Ub-E3 thioester bond (Mund et al., 2014) The p34 protein was recently identified as interacting with the WW1 domain of NEDD4 to enhance stability of NEDD4 by inhibiting NEDD4 auto-ubiquitination and subsequent proteasomal degradation (Hong et al., 2014). Similarly, both wild-type and catalytically inactive Cbl-b ligase inhibited NEDD4 auto-ubiquitination to unexpectedly enhances NEDD4 ubiquitination activity (Guo et al., 2012). These data suggest that Cbl-b regulates NEDD4 ubiquitin ligase activity independently of its RING finger domain.
The proto-oncogene ΔNp63α negatively regulates NEDD4. p63−/− MEFs display increased NEDD4 protein, and silencing of ΔNp63α in adult human keratinocytes led to increased transcript and protein levels of NEDD4 (Leonard et al., 2013)
Recently it was demonstrated that PTEN negatively regulates NEDD4 expression at the translational level in mouse hippocampal neurons by antagonising the PI3K pathway (Hsia et al., 2014). Another recent study showed Ras signalling up-regulates NEDD4 transcription (Zeng et al., 2014). It was observed in HEK293T cells that over-expressing K-Ras or treatment with EGF significantly increased endogenous protein levels of NEDD4 by enhancing transcription of NEDD4, indicating a negative feedback loop between Ras signalling and NEDD4 (Zeng et al., 2014).
7. Conclusions
NEDD4 is the founding member of the NEDD4 family of ubiquitin protein ligases that function in the ubiquitin proteasome system. NEDD4 has many important cellular functions, as indicated by the high degree of evolutionary conservation observed across many species. NEDD4 has roles in regulating viral budding, IGF-1 signalling, in T-cell function and in PTEN signalling, although this is yet to be fully understood as there are currently a number of conflicting models. The role of NEDD4 in cancer suggests that NEDD4 could be a potential therapeutic target for the treatment of human cancer.
Acknowledgements
The ubiquitin work in our laboratory is supported by the National Health and Medical Research Council (NHMRC) Project Grants 1020755 & 1059393, and a NHMRC Senior Principal Research Fellowship 1002863 to SK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al Sorkhy M, Craig R, Market B, Ard R and Porter LA, 2009. The cyclin-dependent kinase activator, Spy1A, is targeted for degradation by the ubiquitin ligase NEDD4. J Biol Chem 284, 2617–27. [DOI] [PubMed] [Google Scholar]
- Anan T, Nagata Y, Koga H, Honda Y, Yabuki N, Miyamoto C, Kuwano A, Matsuda I, Endo F, Saya H and Nakao M, 1998. Human ubiquitin-protein ligase Nedd4: expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes Cells 3, 751–63. [DOI] [PubMed] [Google Scholar]
- Anindya R, Aygun O and Svejstrup JQ, 2007. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell 28, 386–97. [DOI] [PubMed] [Google Scholar]
- Aronchik I, Kundu A, Quirit JG and Firestone GL, 2014. The Anti-proliferative Response of Indole-3-carbinol in human melanoma cells is Triggered by an Interaction with NEDD4–1 and Disruption of Wild-type PTEN Degradation. Mol Cancer Res, Epub 9 July 2014. [DOI] [PMC free article] [PubMed]
- Boase NA, Rychkov GY, Townley SL, Dinudom A, Candi E, Voss AK, Tsoutsman T, Semsarian C, Melino G, Koentgen F, Cook DI and Kumar S, 2011. Respiratory distress and perinatal lethality in Nedd4–2-deficient mice. Nat Commun 2, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P and Sudol M, 1994. The WW domain: a signalling site in dystrophin? Trends Biochem Sci 19, 531–3. [DOI] [PubMed] [Google Scholar]
- Camera D, Boase NA, Kumar S, Pow DV and Poronnik P, 2014. Subtle gait abnormalities in Nedd4 heterozygous mice. Behav Brain Res 260, 15–24. [DOI] [PubMed] [Google Scholar]
- Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, Daly RJ, Kumar S and Yang B, 2008. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal 1, ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C and Matesic LE, 2007. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev 26, 587–604. [DOI] [PubMed] [Google Scholar]
- Christie KJ, Martinez JA and Zochodne DW, 2012. Disruption of E3 ligase NEDD4 in peripheral neurons interrupts axon outgrowth: Linkage to PTEN. Mol Cell Neurosci 50, 179–92. [DOI] [PubMed] [Google Scholar]
- Cui G, Wei P, Zhao Y, Guan Z, Yang L, Sun W, Wang S and Peng Q, 2014. Brucella infection inhibits macrophages apoptosis via Nedd4-dependent degradation of calpain2. Vet Microbiol, Epub 11 Sept 2014. [DOI] [PubMed]
- Dai B, Pieper RO, Li D, Wei P, Liu M, Woo SY, Aldape KD, Sawaya R, Xie K and Huang S, 2010. FoxM1B regulates NEDD4–1 expression, leading to cellular transformation and full malignant phenotype in immortalized human astrocytes. Cancer Res 70, 2951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton HE, Denton D, Foot NJ, Ho K, Mills K, Brou C and Kumar S, 2011. Drosophila Ndfip is a novel regulator of Notch signaling. Cell Death Differ 18, 1150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ and Joazeiro CA, 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78, 399–434. [DOI] [PubMed] [Google Scholar]
- Dinudom A, Harvey KF, Komwatana P, Young JA, Kumar S and Cook DI, 1998. Nedd4 mediates control of an epithelial Na+ channel in salivary duct cells by cytosolic Na+. Proc Natl Acad Sci U S A 95, 7169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan P and Poronnik P, 2013. Nedd4 and Nedd4–2: ubiquitin ligases at work in the neuron. Int J Biochem Cell Biol 45, 706–10. [DOI] [PubMed] [Google Scholar]
- Drinjakovic J, Jung H, Campbell DS, Strochlic L, Dwivedy A and Holt CE, 2010. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron 65, 341–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R, Klos DA, Adler AS and Hicke L, 2004. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J Cell Biol 165, 135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide PW, Cekaite L, Danielsen SA, Eilertsen IA, Kjenseth A, Fykerud TA, Agesen TH, Bruun J, Rivedal E, Lothe RA and Leithe E, 2013. NEDD4 is overexpressed in colorectal cancer and promotes colonic cell growth independently of the PI3K/PTEN/AKT pathway. Cell Signal 25, 12–8. [DOI] [PubMed] [Google Scholar]
- Fan CD, Lum MA, Xu C, Black JD and Wang X, 2013. Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4–1, in the insulin-like growth factor-1 response. J Biol Chem 288, 1674–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Elly C, Gao B, Fang N, Altman Y, Joazeiro C, Hunter T, Copeland N, Jenkins N and Liu YC, 2002. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol 3, 281–7. [DOI] [PubMed] [Google Scholar]
- Fang DF, He K, Wang N, Sang ZH, Qiu X, Xu G, Jian Z, Liang B, Li T, Li HY, Li AL, Zhou T, Gong WL, Yang B, Karin M, Zhang XM and Li WH, 2014. NEDD4 ubiquitinates TRAF3 to promote CD40-mediated AKT activation. Nat Commun 5, 4513. [DOI] [PubMed] [Google Scholar]
- Fombonne J, Bissey PA, Guix C, Sadoul R, Thibert C and Mehlen P, 2012. Patched dependence receptor triggers apoptosis through ubiquitination of caspase-9. Proc Natl Acad Sci U S A 109, 10510–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foot NJ, Dalton HE, Shearwin-Whyatt LM, Dorstyn L, Tan SS, Yang B and Kumar S, 2008. Regulation of the divalent metal ion transporter DMT1 and iron homeostasis by a ubiquitin-dependent mechanism involving Ndfips and WWP2. Blood 112, 4268–75. [DOI] [PubMed] [Google Scholar]
- Foot NJ, Leong YA, Dorstyn LE, Dalton HE, Ho K, Zhao L, Garrick MD, Yang B, Hiwase D and Kumar S, 2011. Ndfip1-deficient mice have impaired DMT1 regulation and iron homeostasis. Blood 117, 638–46. [DOI] [PubMed] [Google Scholar]
- Fotia AB, Cook DI and Kumar S, 2006. The ubiquitin-protein ligases Nedd4 and Nedd4–2 show similar ubiquitin-conjugating enzyme specificities. Int J Biochem Cell Biol 38, 472–9. [DOI] [PubMed] [Google Scholar]
- Fotia AB, Dinudom A, Shearwin KE, Koch JP, Korbmacher C, Cook DI and Kumar S, 2003. The role of individual Nedd4–2 (KIAA0439) WW domains in binding and regulating epithelial sodium channels. FASEB J 17, 70–2. [DOI] [PubMed] [Google Scholar]
- Fotia AB, Ekberg J, Adams DJ, Cook DI, Poronnik P and Kumar S, 2004. Regulation of neuronal voltage-gated sodium channels by the ubiquitin-protein ligases Nedd4 and Nedd4–2. J Biol Chem 279, 28930–5. [DOI] [PubMed] [Google Scholar]
- Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, Brose N, Stambolic V and Rotin D, 2008. The ubiquitin ligase Nedd4–1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci U S A 105, 8585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouladkou F, Lu C, Jiang C, Zhou L, She Y, Walls JR, Kawabe H, Brose N, Henkelman RM, Huang A, Bruneau BG and Rotin D, 2010. The ubiquitin ligase Nedd4–1 is required for heart development and is a suppressor of thrombospondin-1. J Biol Chem 285, 6770–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girao H, Catarino S and Pereira P, 2009. Eps15 interacts with ubiquitinated Cx43 and mediates its internalization. Exp Cell Res 315, 3587–97. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Bodem J, Muller B, Schmechel A, Zentgraf H and Krausslich HG, 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J Virol 77, 9474–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV, 1995. Estimation of the number of amino acid substitutions per site when the substitution rate varies among sites. J Mol Evol 41, 675–9. [DOI] [PubMed] [Google Scholar]
- Goel P, Manning JA and Kumar S, 2015. NEDD4–2 (NEDD4L): The ubiquitin ligase for multiple membrane proteins. Gene 10.1016/j.gene.2014.11.051. [DOI] [PMC free article] [PubMed]
- Guo H, Qiao G, Ying H, Li Z, Zhao Y, Liang Y, Yang L, Lipkowitz S, Penninger JM, Langdon WY and Zhang J, 2012. E3 ubiquitin ligase Cbl-b regulates Pten via Nedd4 in T cells independently of its ubiquitin ligase activity. Cell Rep 1, 472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Wang P, Zhao G, Wang H, Wang M, Chen J and Tong T, 2013. Upregulation of SIRT1 by 17beta-estradiol depends on ubiquitin-proteasome degradation of PPAR-gamma mediated by NEDD4–1. Protein Cell 4, 310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Brown ME, Wang G, Huibregtse J and Hayes FP, 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci U S A 97, 13871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Dinudom A, Cook DI and Kumar S, 2001. The Nedd4-like protein KIAA0439 is a potential regulator of the epithelial sodium channel. J Biol Chem 276, 8597–601. [DOI] [PubMed] [Google Scholar]
- Harvey KF and Kumar S, 1999. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol 9, 166–9. [DOI] [PubMed] [Google Scholar]
- Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML and Rao A, 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol 5, 255–65. [DOI] [PubMed] [Google Scholar]
- Hershko A and Ciechanover A, 1998. The ubiquitin system. Annu Rev Biochem 67, 425–79. [DOI] [PubMed] [Google Scholar]
- Hicke L and Dunn R, 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19, 141–72. [DOI] [PubMed] [Google Scholar]
- Hong SW, Moon JH, Kim JS, Shin JS, Jung KA, Lee WK, Jeong SY, Hwang JJ, Lee SJ, Suh YA, Kim I, Nam KY, Han S, Kim JE, Kim KP, Hong YS, Lee JL, Lee WJ, Choi EK, Lee JS, Jin DH and Kim TW, 2014. p34 is a novel regulator of the oncogenic behavior of NEDD4–1 and PTEN. Cell Death Differ 21, 146–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt J, Putz U, Lackovic J, Doan A, Dorstyn L, Cheng H, Yang B, Chan-Ling T, Silke J, Kumar S and Tan SS, 2009. Divalent metal transporter 1 (DMT1) regulation by Ndfip1 prevents metal toxicity in human neurons. Proc Natl Acad Sci U S A 106, 15489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia HE, Kumar R, Luca R, Takeda M, Courchet J, Nakashima J, Wu S, Goebbels S, An W, Eickholt BJ, Polleux F, Rotin D, Wu H, Rossner MJ, Bagni C, Rhee JS, Brose N and Kawabe H, 2014. Ubiquitin E3 ligase Nedd4–1 acts as a downstream target of PI3K/PTEN-mTORC1 signaling to promote neurite growth. Proc Natl Acad Sci U S A, Epub 25 Aug 2014. [DOI] [PMC free article] [PubMed]
- Huang Z, Choi BK, Mujoo K, Fan X, Fa M, Mukherjee S, Owiti N, Zhang N and An Z, 2014. The E3 ubiquitin ligase NEDD4 negatively regulates HER3/ErbB3 level and signaling. Oncogene, Epub 24 March 2014. [DOI] [PubMed]
- Huibregtse JM, Scheffner M, Beaudenon S and Howley PM, 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A 92, 2563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Ikeda A, Longan LC and Longnecker R, 2000. The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology 268, 178–91. [DOI] [PubMed] [Google Scholar]
- Ingham RJ, Gish G and Pawson T, 2004. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23, 1972–84. [DOI] [PubMed] [Google Scholar]
- Kamynina E, Tauxe C and Staub O, 2001. Distinct characteristics of two human Nedd4 proteins with respect to epithelial Na(+) channel regulation. Am J Physiol Renal Physiol 281, F469–77. [DOI] [PubMed] [Google Scholar]
- Katz M, Shtiegman K, Tal-Or P, Yakir L, Mosesson Y, Harari D, Machluf Y, Asao H, Jovin T, Sugamura K and Yarden Y, 2002. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic 3, 740–51. [DOI] [PubMed] [Google Scholar]
- Kawabe H, Neeb A, Dimova K, Young SM Jr., Takeda M, Katsurabayashi S, Mitkovski M, Malakhova OA, Zhang DE, Umikawa M, Kariya K, Goebbels S, Nave KA, Rosenmund C, Jahn O, Rhee J and Brose N, 2010. Regulation of Rap2A by the ubiquitin ligase Nedd4–1 controls neurite development. Neuron 65, 358–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C and Leis J, 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci U S A 98, 11199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Harvey KF, Kinoshita M, Copeland NG, Noda M and Jenkins NA, 1997. cDNA cloning, expression analysis, and mapping of the mouse Nedd4 gene. Genomics 40, 435–43. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tomooka Y and Noda M, 1992. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochemical and Biophysical Research Communications 185, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Kwak YD, Wang B, Li JJ, Wang R, Deng Q, Diao S, Chen Y, Xu R, Masliah E, Xu H, Sung JJ and Liao FF, 2012. Upregulation of the E3 ligase NEDD4–1 by oxidative stress degrades IGF-1 receptor protein in neurodegeneration. J Neurosci 32, 10971–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak YD, Wang B, Pan W, Xu H, Jiang X and Liao FF, 2010. Functional interaction of phosphatase and tensin homologue (PTEN) with the E3 ligase NEDD4–1 during neuronal response to zinc. J Biol Chem 285, 9847–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard MK, Hill NT, Grant ED and Kadakia MP, 2013. DeltaNp63alpha represses nuclear translocation of PTEN by inhibition of NEDD4–1 in keratinocytes. Arch Dermatol Res 305, 733–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leykauf K, Salek M, Bomke J, Frech M, Lehmann WD, Durst M and Alonso A, 2006. Ubiquitin protein ligase Nedd4 binds to connexin43 by a phosphorylation-modulated process. J Cell Sci 119, 3634–42. [DOI] [PubMed] [Google Scholar]
- Li C and Xiao ZX, 2014. Regulation of p63 protein stability via ubiquitin-proteasome pathway. Biomed Res Int 2014, 175721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CK, Jeng CJ, Wang HS, Wang SH and Wu JC, 2013. Lipopolysaccharide induces degradation of connexin43 in rat astrocytes via the ubiquitin-proteasome proteolytic pathway. PLoS One 8, e79350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Hou Q, Jarzylo L, Amato S, Gilbert J, Shang F and Man HY, 2011. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J Neurochem 119, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wang J, Childress C, Sudol M, Carey DJ and Yang W, 2010. HECT E3 ubiquitin ligase Nedd4–1 ubiquitinates ACK and regulates epidermal growth factor (EGF)-induced degradation of EGF receptor and ACK. Mol Cell Biol 30, 1541–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wan L, Liu P, Inuzuka H, Liu J, Wang Z and Wei W, 2014. SCF(beta-TRCP)-mediated degradation of NEDD4 inhibits tumorigenesis through modulating the PTEN/Akt signaling pathway. Oncotarget 5, 1026–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PY, Xu N, Malyukova A, Scarlett CJ, Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK, Koach J, Tee AE, Haber M, Norris MD, Toon C, Rooman I, Xue C, Cheung BB, Kumar S, Marshall GM, Biankin AV and Liu T, 2013. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ 20, 503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Oppenheim RW, Sugiura Y and Lin W, 2009. Abnormal development of the neuromuscular junction in Nedd4-deficient mice. Dev Biol 330, 153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo QF, Chen W and Zhang ST, 2012. Identification of Nedd4 as a novel regulator in Hedgehog signaling. Chin Med J (Engl) 125, 3851–5. [PubMed] [Google Scholar]
- Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, Lipkowitz S and Weissman AM, 2003. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J Biol Chem 278, 43169–77. [DOI] [PubMed] [Google Scholar]
- Malakhova OA and Zhang DE, 2008. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem 283, 8783–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C and Bryant SH, 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39, D225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monami G, Emiliozzi V and Morrione A, 2008. Grb10/Nedd4-mediated multiubiquitination of the insulin-like growth factor receptor regulates receptor internalization. J Cell Physiol 216, 426–37. [DOI] [PubMed] [Google Scholar]
- Morrione A, Plant P, Valentinis B, Staub O, Kumar S, Rotin D and Baserga R, 1999. mGrb10 interacts with Nedd4. J Biol Chem 274, 24094–9. [DOI] [PubMed] [Google Scholar]
- Mund T, Lewis MJ, Maslen S and Pelham HR, 2014. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc Natl Acad Sci U S A 111, 16736–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund T and Pelham HR, 2009. Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep 10, 501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdaca J, Treins C, Monthouel-Kartmann MN, Pontier-Bres R, Kumar S, Van Obberghen E and Giorgetti-Peraldi S, 2004. Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J Biol Chem 279, 26754–61. [DOI] [PubMed] [Google Scholar]
- Nabhan JF, Pan H and Lu Q, 2010. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep 11, 605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Plant PJ, Correa J, Bain A, Takeda M, Kawabe H, Rotin D, Bain JR and Batt JA, 2012. The ubiquitin ligase Nedd4–1 participates in denervation-induced skeletal muscle atrophy in mice. PLoS One 7, e46427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nethe M, de Kreuk BJ, Tauriello DV, Anthony EC, Snoek B, Stumpel T, Salinas PC, Maurice MM, Geerts D, Deelder AM, Hensbergen PJ and Hordijk PL, 2012. Rac1 acts in conjunction with Nedd4 and dishevelled-1 to promote maturation of cell-cell contacts. J Cell Sci 125, 3430–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak Y, Glowacka WK, Bruce MC, Pham N and Rotin D, 2006. Transport of LAPTM5 to lysosomes requires association with the ubiquitin ligase Nedd4, but not LAPTM5 ubiquitination. J Cell Biol 175, 631–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos JS and Agarwala R, 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–9. [DOI] [PubMed] [Google Scholar]
- Persaud A, Alberts P, Amsen EM, Xiong X, Wasmuth J, Saadon Z, Fladd C, Parkinson J and Rotin D, 2009. Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4–2 using proteome arrays. Mol Syst Biol 5, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud A, Alberts P, Hayes M, Guettler S, Clarke I, Sicheri F, Dirks P, Ciruna B and Rotin D, 2011. Nedd4–1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J 30, 3259–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud A, Alberts P, Mari S, Tong J, Murchie R, Maspero E, Safi F, Moran MF, Polo S and Rotin D, 2014. Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci Signal 7, ra95. [DOI] [PubMed] [Google Scholar]
- Plant PJ, Correa J, Goldenberg N, Bain J and Batt J, 2009. The inositol phosphatase MTMR4 is a novel target of the ubiquitin ligase Nedd4. Biochem J 419, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant PJ, Lafont F, Lecat S, Verkade P, Simons K and Rotin D, 2000. Apical membrane targeting of Nedd4 is mediated by an association of its C2 domain with annexin XIIIb. J Cell Biol 149, 1473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, Abrahamsen H, Thoresen SB and Stenmark H, 2012. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem J 441, 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz U, Howitt J, Lackovic J, Foot N, Kumar S, Silke J and Tan SS, 2008. Nedd4 family-interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. J Biol Chem 283, 32621–7. [DOI] [PubMed] [Google Scholar]
- Rost M, Doring T and Prange R, 2008. gamma2-Adaptin, a ubiquitin-interacting adaptor, is a substrate to coupled ubiquitination by the ubiquitin ligase Nedd4 and functions in the endosomal pathway. J Biol Chem 283, 32119–30. [DOI] [PubMed] [Google Scholar]
- Rotin D and Kumar S, 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10, 398–409. [DOI] [PubMed] [Google Scholar]
- Rougier JS, Albesa M, Abriel H and Viard P, 2011. Neuronal precursor cell-expressed developmentally down-regulated 4–1 (NEDD4–1) controls the sorting of newly synthesized Ca(V)1.2 calcium channels. J Biol Chem 286, 8829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K and Hayashi S, 2004. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol 14, 2228–36. [DOI] [PubMed] [Google Scholar]
- Salah Z, Cohen S, Itzhaki E and Aqeilan RI, 2013. NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle 12, 3817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD and Howley PM, 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505. [DOI] [PubMed] [Google Scholar]
- Scheffner M and Kumar S, 2014. Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim Biophys Acta 1843, 61–74. [DOI] [PubMed] [Google Scholar]
- Schmeisser MJ, Kuhl SJ, Schoen M, Beth NH, Weis TM, Grabrucker AM, Kuhl M and Boeckers TM, 2013. The Nedd4-binding protein 3 (N4BP3) is crucial for axonal and dendritic branching in developing neurons. Neural Dev 8, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screen M, Jonson PH, Raheem O, Palmio J, Laaksonen R, Lehtimaki T, Sirito M, Krahe R, Hackman P and Udd B, 2014. Abnormal splicing of NEDD4 in myotonic dystrophy type 2: possible link to statin adverse reactions. Am J Pathol 184, 2322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Morales C, Pescia C, Chatellard-Causse C, Sadoul R, Bertrand E and Basyuk E, 2005. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J Biol Chem 280, 27004–12. [DOI] [PubMed] [Google Scholar]
- Sehat B, Andersson S, Girnita L and Larsson O, 2008. Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis. Cancer Res 68, 5669–77. [DOI] [PubMed] [Google Scholar]
- Sette P, Jadwin JA, Dussupt V, Bello NF and Bouamr F, 2010. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4–1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol 84, 8181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin-Whyatt L, Dalton HE, Foot N and Kumar S, 2006. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays 28, 617–28. [DOI] [PubMed] [Google Scholar]
- Shenoy SK, Xiao K, Venkataramanan V, Snyder PM, Freedman NJ and Weissman AM, 2008. Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. J Biol Chem 283, 22166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Wang J, Chandarlapaty S, Cross J, Thompson C, Rosen N and Jiang X, 2014. PTEN is a protein tyrosine phosphatase for IRS1. Nat Struct Mol Biol 21, 522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin A and Fuster D, 2010. Nedd4–1 and beta-arrestin-1 are key regulators of Na+/H+ exchanger 1 ubiquitylation, endocytosis, and function. J Biol Chem 285, 38293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J and Rotin D, 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J 15, 2371–80. [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A and Bork P, 1995. Characterization of a novel protein-binding module--the WW domain. FEBS Lett 369, 67–71. [DOI] [PubMed] [Google Scholar]
- Tofaris GK, Kim HT, Hourez R, Jung JW, Kim KP and Goldberg AL, 2011. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc Natl Acad Sci U S A 108, 17004–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X and Pandolfi PP, 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128, 141–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata S and Yasuda J, 2010. Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40. J Gen Virol 91, 228–34. [DOI] [PubMed] [Google Scholar]
- Vecchione A, Marchese A, Henry P, Rotin D and Morrione A, 2003. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol Cell Biol 23, 3363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Peng Q, Lin Q, Childress C, Carey D and Yang W, 2010. Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. J Biol Chem 285, 12279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, Pandolfi PP and Jiang X, 2007. NEDD4–1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniak S, Kabbara S, Lumb R, Scherer M, Secker G, Harvey N, Kumar S and Schwarz Q, 2013. The ubiquitin ligase Nedd4 regulates craniofacial development by promoting cranial neural crest cell survival and stem-cell like properties. Dev Biol 383, 186–200. [DOI] [PubMed] [Google Scholar]
- Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP and Polo S, 2006. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol 8, 1246–54. [DOI] [PubMed] [Google Scholar]
- Xu C, Fan CD and Wang X, 2014. Regulation of Mdm2 protein stability and the p53 response by NEDD4–1 E3 ligase. Oncogene, Epub 13 Jan 2014. [DOI] [PubMed]
- Yang B, Gay DL, MacLeod MK, Cao X, Hala T, Sweezer EM, Kappler J, Marrack P and Oliver PM, 2008. Nedd4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of Cbl-b in activated T cells. Nat Immunol 9, 1356–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B and Kumar S, 2010. Nedd4 and Nedd4–2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ 17, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Wang L, Shang B, Wang Z and Wei W, 2014. NEDD4: A Promising Target for Cancer Therapy. Curr Cancer Drug Targets 14, 549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim EK, Peng G, Dai H, Hu R, Li K, Lu Y, Mills GB, Meric-Bernstam F, Hennessy BT, Craven RJ and Lin SY, 2009. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell 15, 304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Xu J and Harris RC, 2009. Nedd4 mediates ErbB4 JM-a/CYT-1 ICD ubiquitination and degradation in MDCK II cells. FASEB J 23, 1935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T, Wang Q, Fu J, Lin Q, Bi J, Ding W, Qiao Y, Zhang S, Zhao W, Lin H, Wang M, Lu B, Deng X, Zhou D, Yin Z and Wang HR, 2014. Impeded Nedd4–1-mediated Ras degradation underlies Ras-driven tumorigenesis. Cell Rep 7, 871–82. [DOI] [PubMed] [Google Scholar]
- Zhou W, Xu J, Zhao Y and Sun Y, 2014. SAG/RBX2 is a novel substrate of NEDD4–1 E3 ubiquitin ligase and mediates NEDD4–1 induced chemosensitization. Oncotarget 5, 6746–55. [DOI] [PMC free article] [PubMed] [Google Scholar]


