Abstract
The molybdenum cofactor (Moco) is a redox-active prosthetic group found in the active site of Mocodependent enzymes, which are vitally important for life. Moco biosynthesis involves several enzymes that catalyze the subsequent conversion of GTP to cyclic pyranopterin monophoshpate (cPMP), molybdopterin (MPT), adenylated MPT (MPT-AMP) and finally Moco. While the underlying principles of cPMP, MPT, and MPT-AMP formation are well-understood, the molybdenum insertase (Mo-insertase) catalyzed final Moco maturation step is not. In this study we analyzed high resolution X-ray data sets of the plant Mo-insertase Cnx1E that revealed two molybdate binding sites within the active site, hence improving the current view on Cnx1E functionality. The presence of molybdate anions in either of these sites is tied to a distinctive backbone conformation, which we suggest to be essential for Mo-insertase molybdate selectivity and insertion efficiency.
Keywords: Molybdenum cofactor synthesis, Mo-insertase, molybdenum cofactor maturation
INTRODUCTION
The molybdenum cofactor (Moco) is a pyranopterin derivative found in the active site of molybdenum enzymes (Mo-enzymes). Here, at least in the enzymes characterized structurally, a complex hydrogen bond network is required for its proper positioning within the active site [1] allowing for a diverse set of two electron transfer reactions to be catalyzed. The versatile redox chemistry of Moco makes it the irreplaceable key component of the global nitrogen, carbon, and sulfur cycles. The most prominent Mo-enzyme in autotrophs is nitrate reductase, which catalyzes the key reaction of nitrate assimilation [2]. In mammals, sulfite oxidase is a vitally important enzyme that inactivates neurotoxic sulfite [3]. Next to these, Moco was also found being essential for the reduction of N-hydroxylated compounds (mARC, [4] [5]) and the detoxification of N-hydroxylated base analogous (Ycbx and Yiim, [6]), respectively. Notably although Ycbx and Yiim are known for a decade as yet the underlying reaction mechanism is still open. For plants, aldehyde oxidase is an essential Moco dependent enzyme as it is involved in phytohormone biosynthesis [7]. Moco biosynthesis (Fig. 1) is carried out by an evolutionarily old and highly conserved multistep pathway that employs a diverse set of enzymes and reaction mechanisms [8]. The Moco biosynthesis pathway was found to be widespread amongst prokaryotes, archaea and eukaryotes [9]. As a notable exception amongst eukaryotes the yeasts Saccharomyces cerivisiae and Schizosaccharomyces pombe were found to lack Moco [9]. In the first step of Moco biosynthesis, GTP is circularized to cyclic pyranopterin monophosphate (cPMP) [10]. The catalyzing enzyme is the iron sulfur cluster dependent cPMP synthase, which in eukaryotes is localized in the mitochondrial matrix [11]. As all subsequent Moco biosynthesis reaction steps are localized in the cytoplasm of the cell [12], mitochondrial cPMP export is handled by an ABC type transporter [11, 13]. In the cytosol, cPMP is converted into molybdopterin (MPT), a reaction characterized by the sequential introduction of two sulfur atoms forming the MPT dithiolene motif [14]. Subsequently, molybdate is inserted herein, which involves two partial reaction steps attributed to the two functional domains of molybdate insertases (Mo-insertases [15–17]). Historically, these domains are termed E and G domain [18] and are either found fused together (in eukaryotes, except the lower alga Chlamydomonas reinhardtii [19]) or as separate entities (in prokaryotes). The G domain adenylates MPT, yielding MPT-AMP [15, 16], which is subsequently used as substrate by the E-domain [17]. The latter catalyzes both molybdate insertion into the MPT dithiolene motif and MPT-AMP hydrolysis [17, 18], whereby it is assumed that MPT-AMP hydrolyzation is the prerequisite of the molybdate insertion reaction [17, 20]. Recently, progress has been made in identifying the AMP and molybdate binding sites, both in the mammalian Mo-insertase gephyrin [21] and the plant Mo-insertase Cnx1 [20]. Work with recombinant Cnx1E attributed an anchoring function to the AMP moiety of MPT-AMP and suggests that MPT-AMP binds to Cnx1E in hitherto unknown conformation [20]. In the latest model for Mo-insertase functionality, the AMP anchoring function was suggested to be required for proper positioning of the MPT moiety within the Cnx1E active site. Thus placed, the MPT dithiolene motif will be positioned directly opposite to the enzyme bound molybdate, and hydrolysis of the MPT-AMP phosphor-anhydride bond was suggested to trigger molybdate insertion into the MPT-dithiolene motif [20]. In the present work, structure based analysis revealed the basic principles behind the Cnx1 catalyzed molybdate insertion reaction which depends on a highly conserved set of functional relevant residues in eukaryotic Mo-insertases.
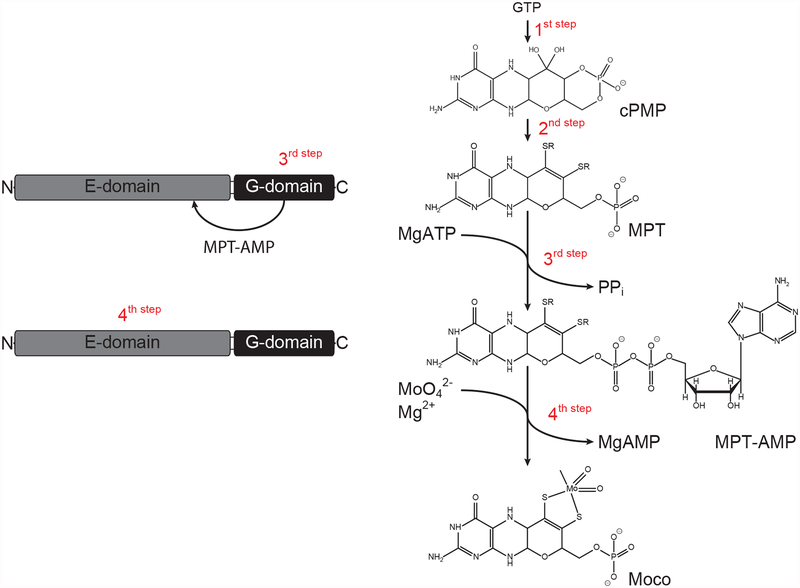
Figure 1:
Shown is a general scheme of the molybdenum cofactor (Moco) biosynthesis pathway. With the exception of 3,8 ˙-cH2 GTP (a precursor of cyclic pyranopterin monophosphate, cPMP), known and characterized intermediates of the Moco biosynthesis pathway are shown sequentially. GTP = guanosine triphosphate, MPT = molybdopterin, MPT-AMP = adenylated molybdopterin. The domain structure of Arabidopsis molybdenum insertase Cnx1 is shown and catalyzed reactions are indicated. This figure and the corresponding caption were originally published in [http://www.biochemj.org/content/474/1/163, Joern Krausze, Corinna Probst, Ute Curth, Joachim Reichelt, Sayantan Saha, David Schafflick, Dirk W. Heinz, Ralf R. Mendel, Tobias Kruse, Biochemical Journal, 2016] [20].
MATERIALS AND METHODS
Recombinant expression and purification of Cnx1E –
Cnx1E was recombinant expressed in Escherichia coli strains RK5204 [22, 23] and RK5206 [22] and purified as described previously [20]. In order to obtain protein preparations suitable for high-resolution structural biology, the purification protocol was modified by adding a washing step with high-salt buffer (0.2 M Tris-HCl, 0.5 M NaCl, 5 % (v/v) glycerol, pH 8.0) while the protein was bound to the Strep-Tactin resin. The protein was then eluted in buffer suitable for crystallization (0.02 M Tris-HCl, 0.15 M NaCl, 0.015 M MgCl2, 5 % (v/v) glycerol, pH 8.0) containing 5 mM desthiobiotin. Without further purification, the protein was concentrated to about 30 g/L using Vivaspin concentrator columns with a molecular weight cut-off of 30 kDa and then subjected to crystallization experiments.
Crystallization, data collection, and model building –
Prior to crystallization, the protein solution was supplemented with 0.015 M AMP. For co-crystallization experiments, 0.015 M Na2MoO4 or Na2WO4was added. Despite the knowledge of previously published crystallization conditions [20], we subjected the protein to a new round of initial screening, assuming that the improved purification protocol would allow crystallization in hitherto unknown conditions. Best crystals of Cnx1E with and without molybdate or tungstate were found in a wide range of conditions from the Morpheus screen (Molecular dimensions). In average, these crystals diffracted significantly better (deff~2.0 Å) than the ones published before (deff~2.7 Å). As an advantage over previous conditions, the new crystals now grew from conditions not containing any sulfate or other oxo-anions that could compete with molybdate or tungstate for the binding to Cnx1E. Prior to crystallographic experiments, Cnx1E crystals were flash cooled in liquid nitrogen. X-ray diffraction data of Cnx1E wildtype crystals were collected on beamline BL14.1 operated by the Helmholtz-Zentrum Berlin (HZB) at the BESSY II electron storage ring (Berlin-Adlershof, Germany) [24]. Datasets for Cnx1E not containing molybdate or tungstate were collected at a wavelength of 0.9184 Å, whereas datasets for Cnx1E with molybdate and tungstate were recorded at a wavelength of 1.7700 Å and 1.2094Å, respectively, to give rise to an anomalous signal from the metal atoms. Data were indexed and integrated with XDS [25]. Since some degree of anisotropic diffraction was already apparent from visual inspection of the diffraction images, the datasets were subjected to further analysis and processing by the StarAniso server [26]. StarAniso confirmed considerable anisotropy of both the diffraction limits and the attenuation of the diffracted intensity, which was corrected by defining an anisotropic resolution cut-off surface and by applying an anisotropic correction for Debye-Waller factor to the dataset. Afterwards, 5% of the diffraction data were flagged for later exclusion from structural refinement and use in the calculation of Rfree. The structures were then solved by transplanting the phase information from previously published structures [20] using Phaser [27]. Attempts to solve the structure of Cnx1E in complex with tungstate through experimental phasing with ShelxC/D/E [28] by using the anomalous signal arising from the tungsten atoms were also successful. ShelxC found two tungsten sites per Cnx1E molecule and ShelxE was able to build a poly-alanine model that was 80 % complete. In contrast, the anomalous signal arising from the molybdenum atoms in the Cnx1E molybdate dataset was too weak for ShelxC to reliably identify the heavy metal positions let alone for experimental phasing. Hence, the molybdate binding sites were identified with ANODE [29] using the phase information from the final model ANODE found two adjacent peaks in the anomalous density that were at close distance to each other and that could not be explained by sulfur atoms of methionine or cysteine residues. The two peaks were of different strength with the stronger being thrice as intense as the weaker, and twice as intense as the strongest sulfur peak. These anomalous peaks were located in proximity to the location of the molybdate anion that we reported before [20]. Because of their close distance, each peak was interpreted as a single molybdate anion that has only partial occupancy. X-ray diffraction data of Cnx1E variant crystals were collected at a wavelength of 1.0 Å on beamlines X06SA and X06DA. The beamlines were equipped with an EIGER 16M X and a PILATUS 2M-F detector, respectively, and operated by the Paul Scherrer Institute at the Swiss Light Source (Villigen, Switzerland). Data of variant crystals were processed using AutoProc and StarAniso [30]. The initial structures were improved by alternating steps of refinement with Buster 2.10.3 [31] and manual rebuilding in Coot [32]. During the refinement the atomic displacement factors were treated as being isotropic and domain displacement was accounted for by modeling the domains as rigid bodies undergoing translation/libration/screw vibrational motion. The refinement was stopped after Rwork and Rfree converged. The files containing the structure factors and the structural models were deposited with the Protein Data Bank with accession numbers 6ETD (Cnx1E+AMP), 6ETF (Cnx1E+Mo), 6ETH (Cnx1E+W), 6GB0 (K294A), 6GAX (K294A+Mo), 6GB9 (S328A), 6GB4 (S328A+Mo), 6GBF (R369A) and 6GBC (R369A+Mo). The complete data collection and refinement statistics are shown in Tables 1 and S2 for structures of Cnx1E wildtype and variants, respectively. Figures depicting 3D images were created with PyMOL [33]. Two-dimensional schematics representing protein-ligand interactions were created with LIGPLOT [34] using the LigPlot+ frontend [35]. The calculation of electrostatic surface potentials was carried out with APBS [36]. The analysis of protein surface conservation was done using the ConSurf server [37].
Table 1: Data collection and refinement statistics.
Numbers in parentheses account for the shell of highest resolution.
| Cnx1E+AMP | Cnx1E+Mo | Cnx1E+W | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.9184 | 1.7700 | 1.2094 |
| Space group | I222 | ||
| Unit cell parameters | |||
| a (Å) | 66.13 ± 0.04 | 67.11 ± 0.04 | 65.50 ± 0.05 |
| b (Å) | 123.11 ± 0.05 | 123.59 ± 0.04 | 123.47 ± 0.08 |
| c (Å) | 132.11 ± 0.05 | 131.78 ± 0.03 | 133.60 ± 0.09 |
| α≡β≡γ (°) | := 90 | := 90 | := 90 |
| Resolution (Å) | |||
| dhkl,max – dhkl,min | 45.14–1.72 (1.95–1.72) | 45.07–1.78 (1.96–1.78) | 42.58–1.64 (1.77–1.64) |
| dh00,min | 2.05 | 2.29 | 1.78 |
| d0k0,min | 2.17 | 2.20 | 2.01 |
| d00l,min | 1.72 | 1.78 | 1.64 |
| deffa [dopt] | 1.95 [~1.7] | 2.02 [~1.8] | 1.78 [~1.7] |
| No. of reflections | |||
| total | 170,016 (11,741) | 531,596 (16,809) | 753,889 (43,169) |
| unique | 38,415 (2,744) | 34,559 (1,728) | 50,634 (2,532) |
| Completeness | |||
| spherical | 0.668 (0.151) | 0.656 (0.132) | 0.757 (0.184) |
| ellipsoidalb | 0.954 (0.800) | 0.947 (0.778) | 0.960 (0.758) |
| Multiplicity | 4.4 (4.3) | 15.4 (9.7) | 14.9 (17.0) |
| Mean I/σ(I) | 8.1 (1.7) | 26.6 (1.5) | 21.7 (1.6) |
| Wilson B (Å2) | 22.9 | 29.1 | 31.7 |
| Rmerge | 0.090 (0.659) | 0.053 (1.268) | 0.057 (1.864) |
| Rmeas | 0.117 (0.865) | 0.056 (1.413) | 0.061(1.980) |
| Rpim | 0.073 (0.555) | 0.020 (0.610) | 0.022 (0.665) |
| CC1/2 | 0.995 (0.674) | 1.000 (0.719) | 0.999 (0.818) |
| Refinement | |||
| No. of reflections used | 38,404 (2,744) | 34,543 (1,691) | 50,624 (2,532) |
| Rwork / Rfree | 0.1774 / 0.2088 | 0.1726 / 0.2160 | 0.1862 (0.2000) |
| No. of non-hydrogen atoms | |||
| total | 3,548 | 3,647 | 3,493 |
| in protein | 3,156 | 3,193 | 3,139 |
| in ligands | 26 | 27 | 29 |
| in ordered solvent | 366 | 427 | 325 |
| Atomic B-factors (Å2) | |||
| Average | 31.7 | 39.2 | 42.1 |
| Protein/Ligands/Solvent | 30.9 / 46.3 / 38.0 | 37.9 / 48.9 / 48.1 | 41.5 / 53.3 / 47.8 |
| No. of amino acid residues | |||
| total / ordered | 470 / 419 | 470 / 422 | 470 / 422 |
| RMSD from ideal | |||
| bonds (Å) | 0.014 | 0.014 | 0.014 |
| angles (°) | 1.65 | 1.95 | 1.65 |
| Ramachandran (%) | |||
| favored | 98.54 | 98.56 | 95.56 |
| allowed | 1.22 | 1.20 | 1.20 |
| outliers | 0.24 | 0.24 | 0.24 |
Effective (deff) and corresponding optical (dopt) resolution of the dataset determined with EFRESOL [43].
Data completeness for a volume in reciprocal space bounded by an ellipsoid centered on {000} and with the dimensions a= 1/dh00,min, b= 1/d0k0,min, c= 1/d00l,min.
Generation of Cnx1E variants –
The Cnx1E variants K294A, S328A, and R369A were generated following the QuikChange (Agilent Technologies) protocol modified for the use of Phusion® High-Fidelity DNA Polymerase (NEB). The Cnx1E wildtype expression vector pGPlus [20] served as template for the polymerase reaction. The primer pairs were designed for the modified protocol according to Xia et al. [38]. Their sequences are 5’-tttcagcaaggtattgatggcacccgggaaacctttgaccttcg-3’ and 5’-gtcaaaggtttcccgggtgccatcaataccttgctgaaatatac-3’ for K294A, 5’-tacctggaaatcctgtggcctgtttggtttgtttcaatatctttgt-3’ and 5’-gatattgaaacaaaccaaacaggccacaggatttccaggtaatcca-3’ for S328A, and 5’-ctgatcccatcgcccccgagtttcatcgggccat-3’ and 5’-ccgatgaaactcgggggcgatgggatcagacttg-3’ for R369A. The identity of the generated constructs was confirmed by sequencing.
Phylogenetic analysis –,
The protein sequence of either the gephyrin E-domain (GephE, Q03555.3, [39] metazoa and fungi or the Cnx1 E-domain (Cnx1E, Q39054.2 [18]), plants, were used as queries in pBLAST [40] searches. For phylogentic analysis – if possible – two sequences from each phylum possessing highest / lowest sequence identity to the respective input sequence were used. For plants the analysis was expanded to the classes Chlorophyceae and Trebouxiophyceae (phylum Chlorophyta) and Klebsormidiophyceae and the taxon Embryophyta (phylum Streptophyta), respectively.
Inductively coupled Plasma Mass Spectrometry (ICP-MS) –
Molybdenum content was quantified with Agilent 7700 Series ICP-MS (Agilent Technologies) using a standard calibration curve of inorganic molybdenum (Fluka). The calibration range of the ICP-MS for molybdenum ranged between 1 – 20 μg/L. Protein solutions and standards were mixed automatically with rhodium as an internal standard. All values were corrected for the molybdenum content of control samples consisting solely of buffer. All data were collected and processed using MassHunter work station software.
Quantification of Cnx1E bound Moco/MPT –
Quantification was carried out via FormA based HPLC analysis, essentially as described in [41] [42]. MPT-AMP amounts were calculated by taking into account FormA-dephospho amounts in a sample treated with phosphodiesterase I (Biomol) prior to dephosphorylation. Other than described earlier [41], upon dephosphorylation FormA was not further purified via an anion exchange step but analyzed directly on a reversed phase C-18 column (250 × 4.6 mm, 5 μm, ReproSil-Pur Basic). Data acquisition and processessing was performed using Open LAB CDS ChemStation software.
RESULTS
The Cnx1E structure complexed with Mg2+-AMP, molybdate, and tungstate –
In order to shed more light on the basic principles behind the Cnx1E catalyzed molybdate insertion into MPT, we optimized already published protein purification and crystallization protocols [20]. Doing so allowed us to determine the structures of Cnx1E complexed with Mg2+-AMP alone (at 1.95 Å) and in the presence of either molybdate (at 2.02 Å) or tungstate (at 1.78 Å), respectively. These structures are henceforth referred to as Cnx1E+AMP, Cnx1E+Mo, and Cnx1E+W.
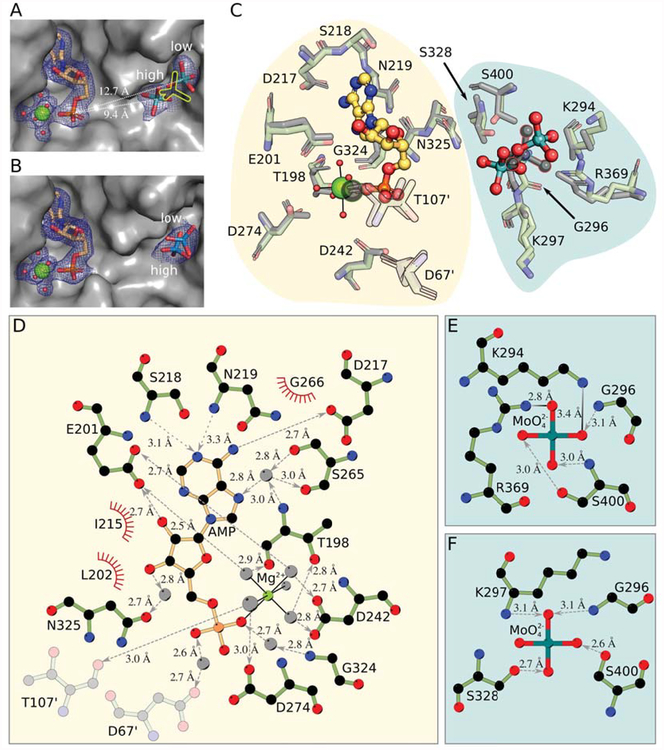
Careful inspection of the Cnx1E+Mo structure led to the identification of two molybdate anions bound 4.1 Å apart within the oxo-anion binding pocket (Fig. 2A, supplementary Fig. S1A). The single Cnx1E molybdate anion reported before [20] lies halfway between these newly identified two anions. This discrepancy between the recently published and the current structure is probably due to their different optical resolutions [43] of 2.5 Å and 1.8 Å, respectively, with the low-resolution structure [20] failing to resolve the two molybdate anions. Consistent with this new finding for Cnx1E+Mo, two tungstate anions are also found in the oxo-anion binding pocket of Cnx1E+W (Fig. 2B, supplementary Fig. S1B). In contrast to Cnx1E+Mo, the tungstate anions in Cnx1E+W are located in positions slightly different from the molybdate anions and found to be only 1.8 Å apart from one another (Fig. 2B). For Cnx1E+Mo as well as for Cnx1E+W the close proximity of the two anions in the binding pocket rules out the simultaneous presence of both due to a mutual steric hindrance and hints at a partial occupancy of the sites. B-factor analysis of both structures suggests that each of the corresponding oxo-anion pairs has a conjoint occupancy of 1.0 with their individual oxo-anions sharing an occupancy ratio of 2 to 1. Accordingly, the binding sites will henceforth be referred to as the high-occupancy site and the low-occupancy site. The molybdate anion in the high-occupancy site is well defined in the electron density map with its tetrahedral geometry clearly visible (Fig 2A). In contrast, the electron density of the molybdate anion bound in the low-occupancy site is much less well-defined. This is due to the lower occupancy and residual freedom of motion and is reflected by the high observed B-factors (Fig 2A).
Figure 2: The Cnx1E active site comprising the oxo-anion and AMP binding pockets.
(A) Active site in Cnx1E+Mo with the oxo-anion pocket occupied by two molybdate anions with partial occupancy. The protein is shown as Connolly surface and colored gray. AMP and molybdate are shown as sticks; magnesium ions are shown as big, water molecules are shown as small spheres. Mg2+-AMP and the oxo-anions are surrounded by their corresponding 2Fo-Fc electron density contoured to ı UHSUHVHQWHG E\ D EOXH PHVK 7KH SRVLWLRQ RI WKH PRO\EGDWH DQLRQ IRXQG LQ WKH SUHYLRXVO\ published structure [20] is shown as yellow silhouette. The high- and low-occupancy sites and their distance to the phosphate group of the AMP molecule are indicated. (B) Active site in Cnx1E+W structure with the oxo-anion pocket occupied by two tungstate anions with partial occupancy. The presentation is the same is in panel A. (C) Superposition of the binding pockets of the Cnx1E molybdate co-structure published previously ([20], PDB 5G2S) with the higher resolution Cnx1E+Mo structure. Amino acid residues interacting with Mg2+-AMP or molybdate are shown in simple stick representation whereas the AMP molecule and the molybdate anion are shown in ball-and-stick representation. Magnesium ions are shown as spheres, coordinated water molecules as smaller spheres. The atoms of the previously published structure (2.8 Å, [20]) are colored gray. Residues shown in semi-transparent fashion are located in the other monomer of the physiological dimer. The single previously identified molybdate ion ([20], PDB 5G2S) and the corresponding magnesium ion are highlighted by colored outlines. The background shades are meant to distinguish the two binding pockets of the Cnx1E active site. (D) Schematic representation of the interactions between Cnx1E and Mg2+-AMP. (E) Interactions in the molybdate low-occupancy site, (F) Interactions in the molybdate high-occupancy site. Involved residues and molecules are shown in ball-and-stick representation with disregard for double bonds. The magnesium ion and water molecules are shown as light gray spheres. Interactions within the octahedral Mg2+-water complex are shown as solid black lines. Hydrogen bonds are shown as broken lines. If discernible, arrowheads point to the hydrogen bond acceptor. Salt bridges are represented as solid lines with a color gradient ranging from black to light gray with black indicating the origin of the negative charge in the ionic interaction. Amino acids involved in hydrophobic interactions are represented by red coronas. If not stated otherwise, atoms are colored throughout Fig. 2 as follows: carbon in Cnx1E, green, other carbon, yellow-orange; oxygen, red; nitrogen, blue; phosphorous, orange; magnesium, bright green; molybdenum, cyan; tungsten, teal. Hydrogen atoms are omitted. All distances shown are given in Ångströms.
Mg2+-AMP is bound in all three crystal structures in a rigid conformation with little flexibility, regardless of the presence or absence of oxo-anions. The full occupancy and low B-factors for the Mg2+-AMP molecule present in the structures are evidence for a strong interaction between the adenosine moiety and Cnx1E. This is noteworthy as AMP is a product of the reaction catalyzed by Cnx1E and is meant to leave the active site [17]. This observation supports the potential anchoring function of the adenosine moiety, which we postulated earlier [20]. The AMP molecule is found to be at a large distance to either of the molybdate anions (~9.4 Å and ~12.7 Å) in the Cnx1E+Mo structure. Thus, activation of molybdate via the formation of an anhydride bond with the alpha-phosphate of AMP [17] appears to be implausible.
The binding of Mg2+-AMP to Cnx1E –
The binding position of Mg2+-AMP found in the structures presented in this work is in agreement with the position reported for Rattus norvegicus (R. norvegicus) gephyrin [21], and certainly represents the pocket that accommodates the nucleotide moiety of MPT-AMP. Mg2+-AMP binding involves both directed and undirected interactions with amino acids (Fig. 2D)[20]. The amino acids Leu202, Ile215, and Gly266 form the core of the pocket and ensure the binding through hydrophobic interaction with the aromatic π-system of the purine ring. A high specificity for the adenosine moiety is achieved by an intricate network of hydrogen bonds, four of which are realized through bridging water molecules (Fig. 2C, D). Of the amino acids involved in a direct interaction with the adenosine moiety, Ser218 and Asn219 act as hydrogen bond donors, whereas Glu201 and Asp217 assume the role of hydrogen bond acceptors. Hydrogen bonds realized through bridging water molecules link the adenosine with donors Ser265 and Thr198 and acceptors Ser265 and Asn325. Further stabilization of Mg2+-AMP is achieved through interactions of Thr198, Asp242, Gly324, Asp274, Glu201, Asp67’ and Thr107’ with the phosphate group and the associated octahedral Mg2+-water complex, all of which involve linking water molecules. Despite the fact that the AMP binding pocket of Cnx1E is located at the dimer interface, Asp67’ and Thr107’ constitute the only cross-monomer contribution to AMP binding
Structural characterization of the Cnx1E oxo-anion binding pocket –
The coordination of the molybdate anion by amino acid residues differs between the high- and low-occupancy sites (Fig. 2F, E). In the high-occupancy site, molybdate is coordinated through hydrogen bonds to four amino acid residues: the side chains of Ser328 and Ser400, and the amide bond nitrogen atoms of Gly296 and Lys297. In contrast, the low-occupancy site provides the molybdate anion with salt bridges to the side chains of the amino acids Lys294 and Arg369, in addition to hydrogen bonds with the side chain of Ser400 and to the backbone amide of Gly296. Comparison of the Cnx1E+Mo structure with R. norvegicus GephE (PDB: 5ERU) and E. coli MoeA (PDB: 1FC5) shows that aforementioned residues are conserved both in sequence and structure (Fig. 3B, supplementary Fig. S2, Fig. 4A). Exceptions include GephE Leu605 (equivalent to Cnx1E Lys297) and Ser698 (equivalent to Cnx1E Ser400), which deviate from their counterparts in Cnx1E and MoeA in terms of side-chain characteristics and position, respectively. Superposition of the Cnx1E and GephE structures [20, 21] reveals Lys297 and Leu605 to be positional homologs. As a result of this and the fact that the interaction of Lys297 with molybdate is realized through its amide bond rather than through the amino function of its side chain, we conclude that this residue can be replaced by Leu605 in GephE. In contrast to the good spatial agreement of these two residues, the positional mismatch of GephE Ser698 with its Cnx1E Ser400 equivalent amounts to a distance larger than 23 Å (Fig. 4A). This makes Ser698 incapable of molybdate binding despite the side chain conservation. However, close examination of the GephE structure ([21], 5ERU) revealed that Ser698 is located at the edge of a disordered and probably nicked region, which could come into contact with the molybdate anion upon reorientation of this flexible stretch of amino acids. The Ser400 and Arg369 in Cnx1E were previously reported by us to be molybdate interacting residues [20]. This was also reported for residues Ser630 and Arg670 in mammalian gephyrin [21], which are equivalent to Cnx1E Ser328 and Arg369 (Fig. 4A). For the first time, Lys294 is identified as a molybdate interacting residue in the higher resolution Cnx1E structure that we report here, hence providing a complete view of Cnx1E molybdate interacting residues. Similar to molybdate, tungstate binds to Cnx1E in mutually exclusive low- and high-occupancy sites (Fig. 2B, supplementary Fig. S1B). Whereas the tungstate low-occupancy site is identical to the one found for molybdate, the high-occupancy site differs in position by ~2.3 Å. As consequence, the hydrogen bond with Lys297 cannot be formed, whereas the interactions with Ser328, Ser400 and the amide bond nitrogen of Gly296 are preserved. Additionally, a salt bridge with Arg369, which is not observed for the high-occupancy molybdate site, is formed. The tungstate oxo-anion identified in the low-occupancy site forms the same interactions with amino acid residues as its molybdate counterpart in Cnx1E+Mo, with the exception of the hydrogen bond to the backbone amide of Gly296.
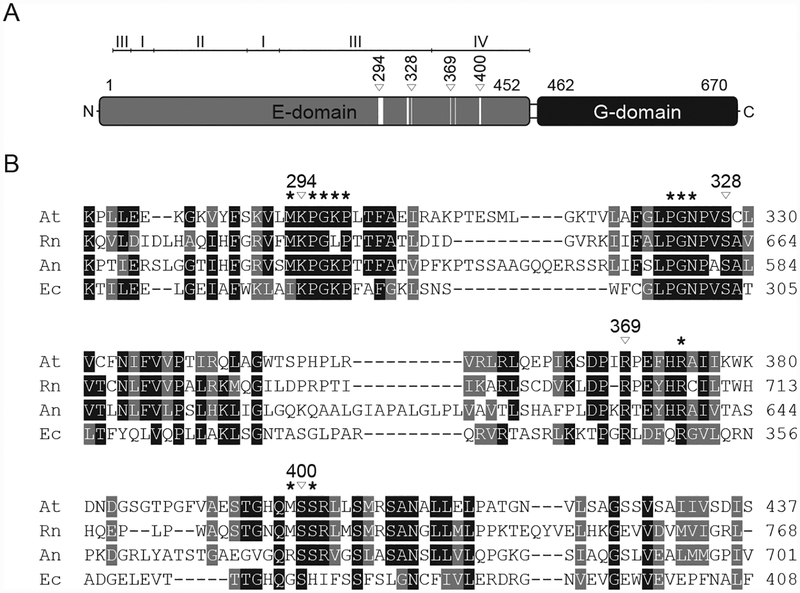
Figure 3: Cnx1 domain organization and sequence comparison of Mo-insertases from various species.
(A) Schematic representation of the Arabidopsis thaliana Cnx1 enzyme domain structure [20]. The first and last residues of the domains are indicated. For Cnx1E subdomains are indicated. Within the schematic representation of the Cnx1E domain, white boxes indicate residues forming the molybdate binding sites. From these, four are involved in directed side-chain molybdate interactions, indicated by white triangles with the corresponding Cnx1 amino acid positions given above. (B) Partial sequence comparison of Arabidopsis thaliana (At), Rattus norvegicus (Rn), Aspergillus nidulans (An) and Escherichia coli (Ec) Cnx1E homologs. Residues forming the molybdate binding site are indicated by asterisks. Cnx1 residues involved in directed molybdate interactions through their side-chains are indicated by white triangles with the corresponding Cnx1 amino acid positions given above. Strictly conserved residues are highlighted in black, conserved residues are highlighted in grey. The alignment was generated with Clustal Omega.
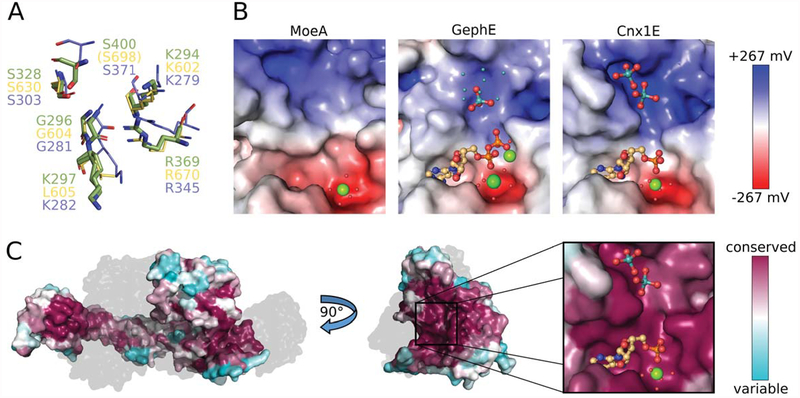
Figure 4: Properties of the oxo-anion binding pocket.
(A) Superposition of Cnx1E molybdate interacting residues with equivalent residues from GephE (Rattus norvegicus, [21]) and MoeA (Escherichia coli, [50]). Residues from Cnx1E are shown as sticks with the carbon atoms colored green. GephE and MoeA residues are shown in line representation with their carbon atoms yellow and light-blue, respectively. The residue labels are in the corresponding colors. In GephE, the amino acid residue Ser698 (corresponding to Cnx1E Ser400) is located too far away from the binding site to be displayed (see text). (B) Electrostatic properties of the molybdate binding pocket from MoeA (left, PDB 1FC5), GephE (middle, PDB 5ERU) and Cnx1E (right). The Connolly surfaces are colored according to their electrostatic potentials ranging from −267 mV(red) to +267 mV (blue). Potentials were calculated with APBS [36] for 310 K. AMP/ADP molecules and molybdate anions are shown as sticks. Mg2+ ions and water molecules are shown as big and small spheres, respectively. The small cyan-colored spheres in the GephE structure correspond to the partially occupied molybdate sites, identified by the authors only by the anomalous signal arising from molybdenum atoms [21]. The subpanels of Fig. 4C present the same view of the active site as the close-up in Fig. 4B. (C) Connolly surface of Cnx1E colored according to the degree of amino acid conservation with magenta corresponding to a high, cyan to a low degree of conservation. The conservation was calculated from a sequence alignment of Cnx1E with 27 homologous proteins from eukaryotes (see also supplementary Fig. S2). On the left, the physiological dimer of Cnx1E is depicted with the second monomer shown gray and transparent for better visibility of the dimer interface. The highest degree of conservation is found in the Cnx1E active site, which is revealed by a rotation by 90°. The active site comprises the oxo-anion and AMP binding pockets, which are shown in close-up. Molybdate and AMP are shown in ball-and-stick representation the single magnesium ion is shown as sphere. The color scheme for hetero-atoms is the same as in Fig. 2.
Analysis of the electrostatic potential of the oxo-anion binding pocket from plant, mammalian, and bacterial Mo-insertases shows a predominantly positively charged surface, which is essential for compensating the negative charge of the oxo-anion (Fig. 4B). The positive electrostatic potential arises from the presence of Lys294 and Lys297 (located in subdomain III, Fig. 3A, B), Arg347 and Arg369 (located in subdomain IV, Fig. 3A, B) and from the absence of any negatively charged amino acid residues. The uncharged amino acids that line the oxo-anion binding pocket are Met293, Pro295, Gly296, Pro297, Pro323, Gly324, Asn325, and Met399. Consistent with the fact that Moco biosynthesis is an evolutionarily old process [9, 11], residues forming the oxo-anion binding pocket are found in large part to be conserved among Mo-insertases from various species (Fig. 3B, supplementary Fig. S2). Accordingly the electrostatic properties are highly comparable among bacterial (E. coli), mammalian (R. norvegicus) and plant (A. thaliana) Mo-insertases (Fig. 4B).
Biochemical characterization of the Cnx1E oxo-anion binding pocket –
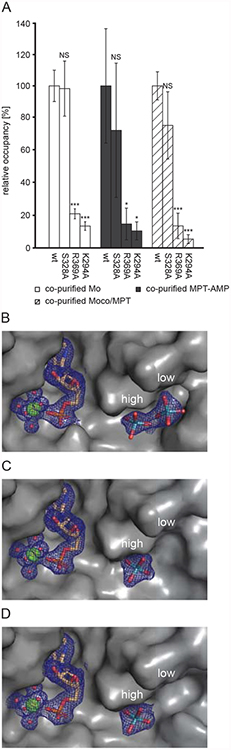
To reveal the impact of residues Ser328, Arg369 and Lys294 on molybdate, MPT-AMP and Moco/MPT binding, we next went on and characterized the respective alanine exchange variants. As compared to wildtype Cnx1E, for variants R369A and K294A we found the amount of co-purified MPT-AMP and molybdenum (molybdate) but also Moco/MPT being significantly reduced (Fig. 5A). We conclude that (i) both residues are required for molybdate binding to the Cnx1E low-occupancy site and that (ii) reduced molybdate binding here goes hand in hand with reduced MPT-AMP binding and hence reduced Moco formation. For the Cnx1E variant S328A ICP-MS-based Mo quantification combined with significance evaluations revealed wildtype like molybdenum, MPT-AMP and Moco/MPT binding properties (Fig. 5A). Since the majority of molybdate is bound to the low-occupancy site as apparent from Cnx1E R369A and K294A characterization, a hidden effect of Ser328 on Cnx1E molybdate binding to the high-occupancy site cannot be excluded with the biochemical methods applied. Therefore structures of Cnx1E variants R396A, K294A and S328A with and without complexed molybdate were solved and analyzed.
Figure 5: Biochemical and structural characterization of Cnx1E variants.
(A) Recombinant Cnx1E wildtype protein and variants S328A, R369A and K294A were analyzed for molybdenum cofactor (Moco) / molybdopterin (MPT), Mo and adenylated MPT (MPT-AMP) content after expression and purification in E. coli strain RK5206. Analysis of the recombinant proteins was carried out as described in the Materials and Methods section. For wildtype Cnx1E the molybdate binding stoichiometrie determined was 0.074 +/− 0.0072, the MPT-AMP binding stoichiometry was found to be 0.0136 +/− 0.0048 and the Moco/MPT binding stoichiometrie was 0.133 +/− 0.012. Bars represent the standard deviation, resulting from three full replicas. A Student’s t-test was carried out to evaluate the significance of differences; with very high significance (p≤0.001): *** high significance (p≤0.01):**, significance (p≤0.05):*, no significance: NS. Comparison of the oxo-anion binding pocket of Cnx1E variants S328A (B), R369A (C) and K294A (D). View, representation and the electron density contour level are identical to Fig. 2A. Occupied low- and high-occupancy sites are indicated.
Structural characterization of Cnx1E variants R369A, K294A and S328A –
Cnx1E variants S328A, R369A and K294A crystallized in the absence of molybdate exhibit a structure identical to that of Cnx1E+AMP. The Cnx1E R369A+Mo and K294+Mo structures revealed that even when present in excess, molybdate is abolished from the low-occupancy site of both variants (Fig. 5C, D), indicating that the residual molybdenum (molybdate) amounts quantified by ICP-MS are either bound to the high-occupancy site of these variants or originate from bound Moco (Fig. 5A, [41, 44]). For Cnx1E variant S328A in complex with molybdate (Fig. 5B) our analysis revealed wildtype like crystal structure characteristics with respect to the molybdate occupancy ratio and backbone conformation. We conclude that there is no hidden effect of Ser328 on Cnx1E molybdate binding.
DISCUSSION
While the mechanistic principles of the initial two steps of Moco biosynthesis (the formation of cPMP and its subsequent conversion to MPT) are well understood, (summarized in [45]) the last step of Moco biosynthesis is not. Earlier fundamental work [16] identified that MPT is adenylated by the Mo-insertase G-domain, a step that precedes the E-domain catalyzed insertion reaction that comprises hydrolysis of the MPT-AMP phosphor-anhydride bond and the insertion of molybdate into the MPT dithiolene motif. These two reaction steps were found to be intrinsically connected to one another and, as a result, it was suggested that the AMP released through MPT-AMP hydrolysis is required for the formation of adenosine-5´-phosphomolybdate [17], which was supposed to be the activated molybdate species required for enzyme catalyzed molybdate insertion. This can now be ruled out on a structural basis (this work and [20]) and due to the fact that it is consistently observed that adenosine-5´-phosphomolybdate formation is not an energetically favoured reaction [46, 47]. As an alternative function, the AMP moiety of MPT-AMP was suggested to serve as an anchor guaranteeing the precise positioning of the MPT dithiolene motif with respect to enzyme bound molybdate [20]. Thus, the proper arrangement of molybdate relative to the dithiolene moiety of MPT will result in an enhanced rate for the molybdate insertion reaction in Cnx1E. The general molybdate insertion reaction can be simply described as a second order chemical reaction of the form
for which the reaction rate can be expressed by
with k being the rate constant for the forward reaction. In the context of Cnx1E catalyzed Moco formation, the rate constant may be described by the Arrhenius equation,
where EA is the activation energy, R is the gas constant, and T is the temperature. A is the pre-exponential frequency factor, which can be expressed as a product of collision frequency (z) and orientation (p) factors, and represents the rate of the reaction in the high-temperature limit. Thus, rate enhancement, outside of a simple increase in the molybdate [11] or MPT concentrations, is a function of a large pre-exponential factor and a reduced energy of activation [48]. Since Cnx1E binds both MPT-AMP and molybdate, the enzyme can enhance the rate of Moco formation by properly orienting molybdate and the dithiolene chelate of MPT for complexation [49]. This requires the dithiolene component of MPT-AMP to be oriented toward molybdate, thereby maximizing the orientation factor contribution to A. Understanding how Cnx1E lowers the activation energy for the reaction is still under investigation, but it likely involves the modulation of ionizable moieties (e.g. pKa changes) in the active site, bond polarizations, and how the active site controls the effective nuclear charge of the Mo ion during the catalytic sequence in order to accelerate the rate of molybdate insertion and hence Moco formation. Tungstate is known to mimick molybdate and trigger MPT-AMP transfer onto Cnx1E [17]. However, Cnx1E does not catalyze the formation of the tungsten cofactor [17], indicating that Cnx1E specificity is realized not through selective molybdate binding but through selective molybdate insertion. In this work, we show that Cnx1E possesses high-occupancy and low-occupancy binding sites for both molybdate and tungstate. The coordinating amino acids and the position of molybdate and tungstate within the respective low-occupancy sites are highly comparable to each other, whereas they differ in the high-occupancy sites. Assuming that for molecular mimicry tungstate is required to be bound in the identical position as molybdate, we conclude that initially either of these two oxo-anions must bind to the low-occupancy site to trigger cooperative MPT-AMP binding [17]. We deduce that for insertion to occur, the oxo-anion must move to the high-occupancy site, which provides the necessary hydrogen bonds that are required for its proper orientation relative to the MPT dithiolene moiety. Since Cnx1E fails to catalyze tungstate insertion into the MPT dithiolene moiety, we conclude that the coordination and position of tungstate in the high-occupancy site, which is significantly different from that of molybdate, are not adequate for insertion to occur. Moco biosynthesis is an evolutionary conserved pathway [9, 11] and consistently, extensive sequence analyses revealed that among eukaryotes, all molybdate side chain interacting residues (i.e. Lys294, Arg369, Ser328 and Ser400) are strictly conserved (supplementary Fig. S2). Biochemical and structure based characterization of the Cnx1E variants K294A and R369A revealed that both residues are crucial for molybdate binding to the Cnx1E low-occupancy site. Therefore (i) structure-based (ii) biochemical and (iii) phylogenetic evidence document the importance of Lys294 and Arg369 for Mo-insertase functionality of Cnx1E.
Next to the molybdate side chain interacting residues Cnx1E Lys294, Arg369, Ser328 and Ser400 also residue Cnx1E Gly296 was found to be conserved amongst eukaryotes. This residue interacts with molybdate through its backbone amide bond. Its conservation is best explained by the fact that only glycine allows the observed torsion angles in this part of the Cnx1E active site. However as sole exception, the fungus Linderina pennispora (supplementary Fig. S2) was found to possess a serine residue at the Cnx1E Gly296 homologous position, but it remains open whether or not the analyzed sequence from Linderina pennispora is a Mo-insertase.
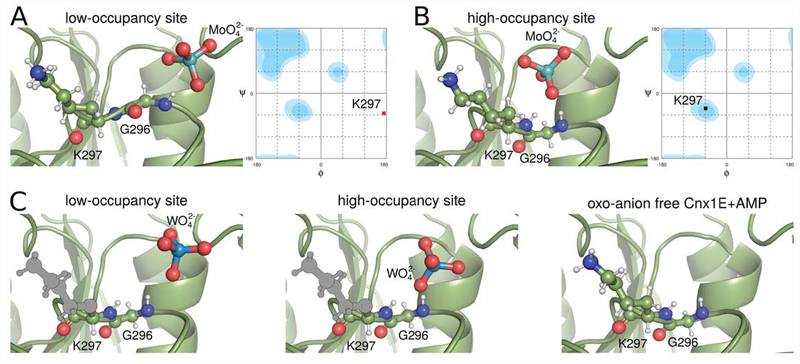
In the Cnx1E+Mo structure, the region comprising amino acids Gly296 and Lys297 was found to adopt a relaxed and tense backbone conformation, each of which is linked to the presence of a molybdate anion either in the high- or low-occupancy binding site, respectively (Fig. 6A, B). In contrast, in the Cnx1E+AMP structure the Gly296 Lys297 region only adopts the relaxed conformation as do the variants R369A+Mo and K294A+Mo for which molybdate was exclusively found in the high-occupancy site (Fig. 6C). Likewise, when co-crystallized with tungstate, the Cnx1E Gly296 Lys297 region was found to adopt exclusively the relaxed backbone conformation (Fig. 6C). Therefore, we conclude that the tense conformation is induced upon molybdate but not tungstate binding. Since Cnx1E fails to insert tungstate into the MPT dithiolene motif [17], we suggest this conformational transition to be of mechanistic relevance for the molybdate insertion process. Consistent with our biochemical and structural data, we propose that the Cnx1E low-occupancy site represents the entry site for oxo-anion binding (Fig. 6A, C) and that binding is facilitated mostly through ionic interactions with Arg369 and Lys294 here. The presence of a molybdate anion in the entry site leads to the reorientation of the amide bond Gly296-Lys297 leading to the tense conformation. The subsequent relaxation of the backbone then assists in the relocation of the molybdate anion from the low-occupancy to the high-occupancy site. We deduce this to be crucial for the insertion reaction to occur. In contrast, a tungstate anion bound to the entry site fails to induce a tense conformation as the amide of Gly296 does not provide an H-bond for interaction with tungstate (Fig. 6C). Accordingly, tungstate is not assisted to relocate to the high-occupancy site and therefore cannot be inserted into the MPT dithiolene motif. However it remains an open question as to whether the lack of Gly296 hydrogen bonding to tungstate is the cause or the effect of the loss of tungstate insertion capability. Subsequent work may answer the question whether or not the observed backbone flip contributes to lowering EA.
Figure 6: Low- and high-occupancy sites in the oxo-anion binding pocket.
Cnx1E is shown in cartoon representation with relevant amino acids and the oxo-anions shown in ball-and-stick representation. The use of colors is the same as in Fig. 2. Hydrogen atoms are indicated as smaller white balls and sticks. Hydrogen positions were derived from formal geometric considerations (“riding hydrogens”) and not from experiment. Amino acid side-chains not visible in the structure due to flexibility were modeled in their most likely conformation and are shown in gray. (A) Low-occupancy site with molybdate bound. The hydrogen bond between the amide of Gly296 with molybdate forces the protein backbone in a tense conformation with the φ and ψ angles of Lys297 in the disallowed regions of the Ramachandran plot on the right-hand site. (B) The high-occupancy site occupied with molybdate allows the adoption of a relaxed backbone conformation. The φ and ψ angles of Lys297 are in a favored region of the Ramachandran plot. (C) The backbone conformations associated with the tungstate-occupied low- and high-occupancy sites as well as the empty oxo-anion binding pocket are all relaxed and resemble the situation of the molybdate occupied high-occupancy site. Their Ramachandran plots look similar to the one in panel B.
Supplementary Material
Figure S1: Omit and anomalous maps of the Cnx1E active site. The omit maps were calculated in the absence of AMP, the magnesium water complex, and the oxo-anions. The 2Fo-Fc and Fo-Fc omit maps are represented by a magenta and green mesh, respectively. Anomalous maps are represented by a black mesh. The molecules bound to the active site are shown in the same fashion as in Fig. 2. (A) The situation in Cnx1E+Mo. The 2Fo-Fc map is contoured to 1.5σ the Fo-Fc to 3.5σ and the anomalous map to 4σ. (B) The situation in Cnx1E+W. The 2Fo-Fc map is contoured to 1.5σ, the Fo-Fc to 3.5σ, and the anomalous map to 12.0σ.
Figure S2: Partial amino acid sequence comparison of molybdate (Mo) insertase E-domains from various species. For species and Mo insertase annotation please see supplementary table S1. The conserved Cnx1E amino acid positions are highlighted in magenta (Lys294), green (Gly296) yellow (Ser328), teal (Arg369) and cyan (Ser400) respectively.
Table S1: Phylogentic analysis of eukaryotic molybdate insertases. Homologs to the protein sequences of either the gephyrin E-domain (Metazoa and Fungi) or the Cnx1 E-domain (Viridiplantae) are tabulated. According to the NCBI taxonomy database [51]: *no rank, **genus, ***subphylum.
Table S2: Data collection and refinement statistics of Cnx1E variants. Numbers in parentheses account for the shell of highest resolution. aEffective (deff) and corresponding optical (dopt) resolution of the dataset determined with EFRESOL [43].bData completeness for a volume in reciprocal space bounded by an ellipsoid centered on {000} and with the dimensions a= 1/dh00,min, b= 1/d0k0,min,c= 1/d00l,min.
ACKNOWLEDGMENTS
We thank Ute Nielaender, Luise Fricke, Franziska Holtkotte and Archna (Braunschweig University of Technology) for excellent technical assistance. M.L.K. acknowledges the National Institutes of Health for generous and continued financial support of our work on molybdoenzymes.
FUNDING INFORMATION
This work was financed by grants of the Deutsche Forschungsgemeinschaft to TK and RRM (GRK 2223/1) and the National Institutes of Health MLK (GM-057378).
Abbreviations list:
- Cnx1E+AMP
Cnx1E in complex with Mg2+-AMP alone
- Cnx1E+Mo
Cnx1E in complex with molybdate and Mg2+-AMP
- Cnx1E+W
Cnx1E in complex with tungstate and Mg2+-AMP
- cPMP
cyclic pyranopterin monophosphate
- Moco
molybdenum cofactor
- Mo-enzymes
molybdenum enzymes
- Mo-insertase
molybdenum insertase
- MPT
molybdopterin
- MPT-AMP
adenylated MPT
Footnotes
DECLARATIONS OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Rees Douglas C., H. Y, Kisker Caroline and Schindelin Hermann. (1997) A crystallographic view of the molybdenum cofactor. Journal of the Chemical Society, Dalton Transactions 21, 3909–3914 [Google Scholar]
- 2.Campbell WH (2001) Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell. Mol. Life Sci 58, 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwahn BC, Van Spronsen FJ, Belaidi AA, Bowhay S, Christodoulou J, Derks TG, Hennermann JB, Jameson E, Konig K, McGregor TL, Font-Montgomery E, Santamaria-Araujo JA, Santra S, Vaidya M, Vierzig A, Wassmer E, Weis I, Wong FY, Veldman A and Schwarz G (2015) Efficacy and safety of cyclic pyranopterin monophosphate substitution in severe molybdenum cofactor deficiency type A: a prospective cohort study. Lancet 386, 1955–1963 [DOI] [PubMed] [Google Scholar]
- 4.Gruenewald S, Wahl B, Bittner F, Hungeling H, Kanzow S, Kotthaus J, Schwering U, Mendel RR and Clement B (2008) The fourth molybdenum containing enzyme mARC: cloning and involvement in the activation of N-hydroxylated prodrugs. J Med Chem 51, 8173–8177 [DOI] [PubMed] [Google Scholar]
- 5.Ott G, Havemeyer A and Clement B (2015) The mammalian molybdenum enzymes of mARC. J Biol Inorg Chem 20, 265–275 [DOI] [PubMed] [Google Scholar]
- 6.Kozmin SG, Leroy P, Pavlov YI and Schaaper RM (2008) YcbX and yiiM, two novel determinants for resistance of Escherichia coli to N-hydroxylated base analogues. Mol Microbiol 68, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y and Koshiba T (2000) The arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci U S A 97, 12908–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendel RR and Kruse T (2012) Cell biology of molybdenum in plants and humans. Biochim Biophys Acta 1823, 1568–1579 [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y and Gladyshev VN (2008) Molybdoproteomes and evolution of molybdenum utilization. J Mol Biol 379, 881–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanzelmann P and Schindelin H (2006) Binding of 5’-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc Natl Acad Sci U S A 103, 6829–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendel RR (2013) The molybdenum cofactor. J Biol Chem 288, 13165–13172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufholdt D, Baillie CK, Bikker R, Burkart V, Dudek CA, von Pein L, Rothkegel M, Mendel RR and Hansch R (2016) The molybdenum cofactor biosynthesis complex interacts with actin filaments via molybdenum insertase Cnx1 as anchor protein in Arabidopsis thaliana. Plant Sci 244, 8–18 [DOI] [PubMed] [Google Scholar]
- 13.Teschner J, Lachmann N, Schulze J, Geisler M, Selbach K, Santamaria-Araujo J, Balk J, Mendel RR and Bittner F (2010) A novel role for Arabidopsis mitochondrial ABC transporter ATM3 in molybdenum cofactor biosynthesis. Plant Cell 22, 468–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuebbens MM and Rajagopalan KV (2003) Mechanistic and Mutational Studies of Escherichia coli Molybdopterin Synthase Clarify the Final Step of Molybdopterin Biosynthesis. J Biol Chem 278, 14523–14532. [DOI] [PubMed] [Google Scholar]
- 15.Llamas A, Mendel RR and Schwarz G (2004) Synthesis of adenylated molybdopterin: an essential step for molybdenum insertion. J Biol Chem 279, 55241–55246 [DOI] [PubMed] [Google Scholar]
- 16.Kuper J, Llamas A, Hecht HJ, Mendel RR and Schwarz G (2004) Structure of molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430, 806–806 [DOI] [PubMed] [Google Scholar]
- 17.Llamas A, Otte T, Simons A, Multhaupt G, Mendel RR and Schwarz G (2006) The mechanism of nucleotide-assisted molybdenum insertion into molybdopterin: Novel routes towards metal cofactor assembly. J Biol Chem 281, 18343–18340 [DOI] [PubMed] [Google Scholar]
- 18.Stallmeyer B, Nerlich A, Schiemann J, Brinkmann H and Mendel RR (1995) Molybdenum co-factor biosynthesis: the Arabidopsis thaliana cDNA cnx1 encodes a multifunctional two-domain protein homologous to a mammalian neuroprotein, the insect protein Cinnamon and three Escherichia coli proteins. Plant J 8, 751–762 [DOI] [PubMed] [Google Scholar]
- 19.Llamas A, Tejada-Jimenez M, Gonzalez-Ballester D, Higuera JJ, Schwarz G, Galvan A and Fernandez E (2007) Chlamydomonas reinhardtii CNX1E reconstitutes molybdenum cofactor biosynthesis in Escherichia coli mutants. Eukaryot Cell 6, 1063–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krausze J, Probst C, Curth U, Reichelt J, Saha S, Schafflick D, Heinz DW, Mendel RR and Kruse T (2017) Dimerization of the plant molybdenum insertase Cnx1E is required for synthesis of the molybdenum cofactor. Biochem J 474, 163–178 [DOI] [PubMed] [Google Scholar]
- 21.Kasaragod VB and Schindelin H (2016) Structural Framework for Metal Incorporation during Molybdenum Cofactor Biosynthesis. Structure 24, 782–788 [DOI] [PubMed] [Google Scholar]
- 22.Stewart V and MacGregor CH (1982) Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J. Bacteriol 151, 788–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivers SL, McNairn E, Blasco F, Giordano G and Boxer DH (1993) Molecular genetic analysis of the moa operon of Escherichia coli K-12 required for molybdenum cofactor biosynthesis. Mol. Microbiol 8, 1071–1081 [DOI] [PubMed] [Google Scholar]
- 24.Mueller U, Förster R, Hellmig M,Huschmann FU, Kastner A, Malecki P, Pühringer S, Röwer M, Sparta K, Steffien M, Ühlein M, Wilk P, Weiss MS (2015) The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur. Phys. J. Plus 130, 141–150 [Google Scholar]
- 25.Kabsch W (2010) XDS. Acta Cryst, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tickle IJ, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. (2017) STARANISO Cambridge, United Kingdom: Global Phasing Ltd. [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC and Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 29.Thorn A and Sheldrick GM (2011) ANODE: anomalous and heavy-atom density calculation. J Appl Crystallogr 44, 1285–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T and Bricogne G (2011) Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr 67, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bricogne G, B. E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO (2017) BUSTER version 2.10.3 Cambridge, United Kingdom: Global Phasing Ltd. [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG and Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC. [Google Scholar]
- 34.Wallace AC, Laskowski RA and Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 35.Laskowski RA and Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51, 2778–2786 [DOI] [PubMed] [Google Scholar]
- 36.Baker NA, Sept D, Joseph S, Holst MJ and McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashkenazy H, Erez E, Martz E, Pupko T and Ben-Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38, W529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Y, Chu W, Qi Q and Xun L (2015) New insights into the QuikChange process guide the use of Phusion DNA polymerase for site-directed mutagenesis. Nucleic Acids Res 43, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stallmeyer B, Schwarz G, Schulze J, Nerlich A, Reiss J, Kirsch J and Mendel RR (1999) The neurotransmitter receptor-anchoring protein gephyrin reconstitutes molybdenum cofactor biosynthesis in bacteria, plants, and mammalian cells. Proc. Natl. Acad. Sci. U. S. A 96, 1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 41.Schwarz G, Boxer DH and Mendel RR (1997) Molybdenum cofactor biosynthesis. The plant protein Cnx1 binds molybdopterin with high affinity. J Biol Chem 272, 26811–26814 [DOI] [PubMed] [Google Scholar]
- 42.Klewe A, Kruse T and Lindel T (2017) Aminopyrazine Pathway to the Moco Metabolite Dephospho Form A. Chemistry 23, 11230–11233 [DOI] [PubMed] [Google Scholar]
- 43.Urzhumtsev L U. a. A. (2015) EFRESOL: effective resolution of a diffraction data set. Journal of Applied Crystallography 48, 589–597 [Google Scholar]
- 44.Schwarz G, Schulze J, Bittner F, Eilers T, Kuper J, Bollmann G, Nerlich A, Brinkmann H and Mendel RR (2000) The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell 12, 2455–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leimkuhler S (2017) Shared function and moonlighting proteins in molybdenum cofactor biosynthesis. Biol Chem 398, 1009–1026 [DOI] [PubMed] [Google Scholar]
- 46.Lipmann F (1941) Metabolic gneration and utilization of phosphate bond energy. Advances in Enzymology and Related Areas of Molecular Biology 1, 99–161 [Google Scholar]
- 47.Huxtable RJ (1986) Biochemistry of Sulfur Biochemistry of the Elements, Springer Science+Business media, LLC [Google Scholar]
- 48.Roduner E (2014) Understanding catalysis. Chem Soc Rev 43, 8226–8239 [DOI] [PubMed] [Google Scholar]
- 49.Dafforn A and Koshland DE Jr. (1971) Theoretical aspects of orbital steering. Proc Natl Acad Sci U S A 68, 2463–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrag JD, Huang W, Sivaraman J, Smith C, Plamondon J, Larocque R, Matte A and Cygler M (2001) The crystal structure of Escherichia coli MoeA, a protein from the molybdopterin synthesis pathway. J. Mol. Biol 310, 419–431. [DOI] [PubMed] [Google Scholar]
- 51.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E and Ye J (2009) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 37, D5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Omit and anomalous maps of the Cnx1E active site. The omit maps were calculated in the absence of AMP, the magnesium water complex, and the oxo-anions. The 2Fo-Fc and Fo-Fc omit maps are represented by a magenta and green mesh, respectively. Anomalous maps are represented by a black mesh. The molecules bound to the active site are shown in the same fashion as in Fig. 2. (A) The situation in Cnx1E+Mo. The 2Fo-Fc map is contoured to 1.5σ the Fo-Fc to 3.5σ and the anomalous map to 4σ. (B) The situation in Cnx1E+W. The 2Fo-Fc map is contoured to 1.5σ, the Fo-Fc to 3.5σ, and the anomalous map to 12.0σ.
Figure S2: Partial amino acid sequence comparison of molybdate (Mo) insertase E-domains from various species. For species and Mo insertase annotation please see supplementary table S1. The conserved Cnx1E amino acid positions are highlighted in magenta (Lys294), green (Gly296) yellow (Ser328), teal (Arg369) and cyan (Ser400) respectively.
Table S1: Phylogentic analysis of eukaryotic molybdate insertases. Homologs to the protein sequences of either the gephyrin E-domain (Metazoa and Fungi) or the Cnx1 E-domain (Viridiplantae) are tabulated. According to the NCBI taxonomy database [51]: *no rank, **genus, ***subphylum.
Table S2: Data collection and refinement statistics of Cnx1E variants. Numbers in parentheses account for the shell of highest resolution. aEffective (deff) and corresponding optical (dopt) resolution of the dataset determined with EFRESOL [43].bData completeness for a volume in reciprocal space bounded by an ellipsoid centered on {000} and with the dimensions a= 1/dh00,min, b= 1/d0k0,min,c= 1/d00l,min.