Summary
Auxin plays a fundamental role in plant growth and development, and also influences plant defence against various pathogens. Previous studies have examined the different roles of the auxin pathway during infection by biotrophic bacteria and necrotrophic fungi. We now show that the auxin signalling pathway was markedly down‐regulated following infection of rice by Rice black streaked dwarf virus (RBSDV), a dsRNA virus. Repression of the auxin receptor TIR1 by a mutant overexpressing miR393 increased rice susceptibility to RBSDV. Mutants overexpressing the auxin signalling repressors OsIAA20 and OsIAA31 were also more susceptible to RBSDV. The induction of jasmonic acid (JA) pathway genes in response to RBSDV was supressed in auxin signalling mutants, suggesting that activation of the JA pathway may be part of the auxin signalling‐mediated rice defence against RBSDV. More importantly, our results also revealed that OsRboh‐mediated reactive oxygen species levels played important roles in this defence. The results offer novel insights into the regulatory mechanisms of auxin signalling in the rice–RBSDV interaction.
Keywords: auxin signalling, jasmonic acid, OsRboh, reactive oxygen species, rice black streaked dwarf virus
Introduction
Auxins play key roles in almost every aspect of plant growth and development, regulating many processes by changes in their concentration. Indole‐3‐acetic acid (IAA) is the major form of natural auxin in plants (Zhao, 2010) and three components are known to be involved in auxin signal transduction: IAA receptors TIR1/AFB, transcriptional repressors Aux/IAA and auxin response factors (ARF) transcription factors (Mockaitis and Estelle, 2008; Weijers and Wagner, 2016). In the absence of auxin, Aux/IAA proteins bind to the transcription factor ARFs, repressing the expression of auxin response genes. The F‐box protein TIR1/AFB is a component of the SCFTIR1/AFB ubiquitination E3 complex. When the IAA concentration increases, TIR1/AFB receptors bind Aux/IAA repressor proteins and form a SCFTIR1/AFB–auxin–Aux/IAA complex leading to the polyubiquitination and degradation of Aux/IAA proteins. Therefore, the degradation of Aux/IAAs allows a set of ARFs to activate their target genes. This process is essential for most physiological processes regulated by auxin (Weijers and Wagner, 2016).
Besides its role in plant development, recent studies have demonstrated that auxin is also involved in plant defence (Fu and Wang, 2011; Vidhyasekaran, 2015). For example, repressing auxin signalling by the overexpression of miR393 increased resistance of Arabidopsis to the bacterial pathogen Pseudomonas syringae (Navarro et al., 2006). Auxin signalling was believed to antagonize the salicylic acid (SA)‐mediated disease resistance pathway in response to bacterial invasion (Wang et al., 2007). Auxin signalling plays multiple roles in plant defence against necrotrophic fungal pathogens (Llorente et al., 2008; Qi et al., 2012). Disruption of the auxin signalling pathway increased susceptibility to Alternaria brassicicola, possibly because the induction of the jasmonic acid (JA) pathway related genes was suppressed (Qi et al., 2012). However, auxin signalling and transport increased the susceptibility of Arabidopsis to Fusarium oxysporum (Kidd et al., 2011). Hence, the effect of the auxin pathway on plant defence is complicated, depending on the pathogen and host.
Rice black streaked dwarf virus (RBSDV), a member of the genus Fijivirus, family Reoviridae, is transmitted to rice, maize, barley and wheat by the small brown planthopper (Laodelphax striatellus Fallén, SBPH) in a persistent, circulative manner (Wei and Li, 2016). RBSDV has an icosahedral, double‐layered particle with a diameter of about 75–80 nm and a genome that consists of ten segments of double‐stranded RNA (dsRNA) (Wang et al., 2003). RBSDV infection causes severe plant growth abnormalities, such as dwarfism, and darkening of leaves, symptoms that are associated with a change of hormone homeostasis (Huang et al., 2018). Indeed, our previous research revealed that the jasmonic acid (JA) and brassinosteroid (BR) pathways were changed in contrasting ways in response to RBSDV infection (He et al., 2017). While the JA pathway played an important role in defence against RBSDV, the BR pathway facilitated infection (Zhang et al., 2019b). In addition, the abscicic acid (ABA) pathway was also shown to negatively regulate plant defence against RBSDV (Xie et al., 2018).
Here, the role of auxin signalling in RBSDV infection was investigated by using various auxin signalling mutants. The results showed that auxin signalling played a positive role in rice defence against RBSDV infection. Further results demonstrated that the JA pathway and OsRboh‐mediated reactive oxygen species (ROS) levels contributed to this defence.
Results
The effect of RBSDV infection on auxin homeostasis
Rice plants were severely dwarfed after RBSDV infection (Fig. 1A) and therefore the effect of RBSDV infection on the auxin pathway was investigated in rice. First, the effects of RBSDV on the expression of genes involved in auxin transport (OsPINs), biosynthesis (OsYUCCAs and OsTAAs), signalling (OsIAAs) and metabolism (OsGH3s) was assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) 30 days post‐infection (dpi). As shown in Fig. 1B, the auxin transport genes OsPIN1, OsPINL2 and OsPIN6 (but not OsPINL1) and the auxin biosynthesis genes OsYUCCA1 and OsYUCCA6 were significantly down‐regulated in RBSDV‐infected plants compared to the controls. The OsGH3 family genes are reported to reduce auxin content by catalysing the conjugation of IAA to amino acids and we found that the expression of OsGH3.2 and OsGH3.8 was markedly increased in our RBSDV‐infected plants. Moreover, the expression of genes involved in auxin signalling (OsIAA20, OsIAA31 and OsIAA7) was also obviously changed in response to virus infection. Interestingly, the expression pattern of OsIAA20 and OsIAA31 genes was converse. This might be due to the different cis‐element in the promoters of OsIAA20 and OsIAA31 (Fig. S1).
Figure 1.
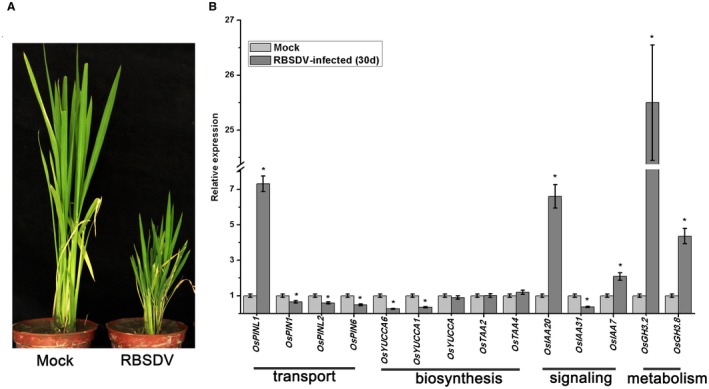
The effects of RBSDV infection on gene expression in the auxin pathway. (A) Typical symptoms of RBSDV‐infected rice compared with mock‐inoculated plant. The symptoms were observed 30 days post‐RBSDV inoculation (dpi). (B) Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) verification of the expression of auxin pathway genes (transport, biosynthesis, signalling and metabolism) in response to RBSDV infection at 30 dpi in rice. Data are shown as relative expression levels of virus‐infected plants in comparison to the control plants. UBQ5 was used as the internal reference gene. Values are means ± SD of three biological replicates. Statistically significant differences from the control are indicated: *P ≤ 0.05.
To determine whether the change of gene expression affected auxin homeostasis, IAA concentrations were measured in RBSDV‐infected leaves at 40 dpi by UPLC‐MS/MS (Fu et al., 2012). As shown in Fig. 2A, IAA concentrations were significantly less in the infected plants (15.8 pg mg−1 FW) than in the mock‐inoculated controls (23.7 pg mg−1 FW). We next measured the IAA concentration in DR5:GUS transgenic plants inoculated with RBSDV. DR5:GUS constructs use highly active auxin‐responsive promoter elements to drive β‐glucuronidase (GUS) expression (Ulmasov et al., 1997) and can therefore be used as an indicator to monitor auxin homeostasis. Consistent with the direct IAA measurements, the GUS activity in RBSDV‐infected transgenic plants was obviously reduced compared to the mock‐inoculated controls (Fig. 2B). The results all demonstrate that the expression of auxin pathway genes and auxin homeostasis was significantly affected by RBSDV infection.
Figure 2.
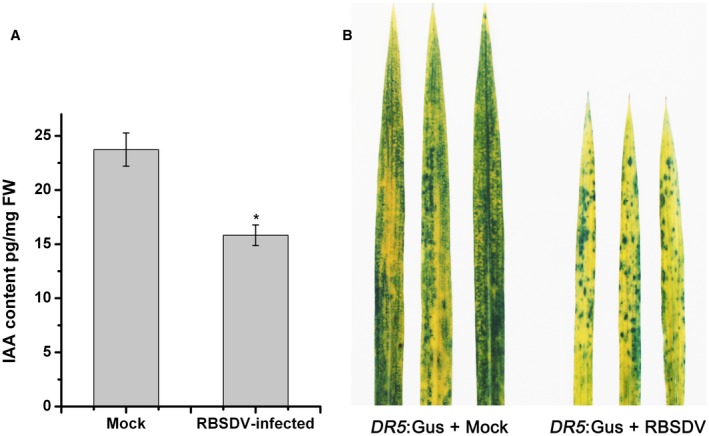
The effects of RBSDV infection on the homeostasis of the auxin pathway. (A) The IAA contents of mock‐inoculated and RBSDV‐infected rice plants at 40 days post‐inoculation (dpi). (B) Activity of the DR5 promoter in the leaves of mock‐inoculated and RBSDV‐infected DR5:GUS rice transgenic plants at 40 dpi.
The effect of the auxin signalling pathway on RBSDV infection
To explore the relationship between the auxin pathway and viral infection, we examined the susceptibility of auxin signalling mutants to RBSDV. First, we used a mutant overexpressing miR393 (35S:miR393) in which the expression of the auxin receptor TIR1 was significantly decreased (Bian et al., 2012). 30 days after inoculating 35S:miR393 transgenic and non‐transgenic (Nip) plants with RBSDV using viruliferous small brown planthopper (SBPH) (or virus‐free insects for the controls), viral symptoms were examined and the concentration of viral RNA transcripts (S6, S7 and S10) was determined. Dwarfing symptoms were more severe in the 35S:miR393 mutant than in the Nip controls (Fig. 3A) and the mutant plants had a significant two‐ to three‐fold increase in expression of the viral RNAs compared with Nip (P ≤ 0.05) (Fig. 3B). The viral P10 outer capsid protein levels also showed a similar increase (Fig. 3C) and there was an increase in viral incidence (percentage of plants infected as determined by RT‐PCR) in the mutant compared to Nip (Fig. 3D). Thus disruption of auxin receptor TIR1 increased rice susceptibility to RBSDV infection.
Figure 3.
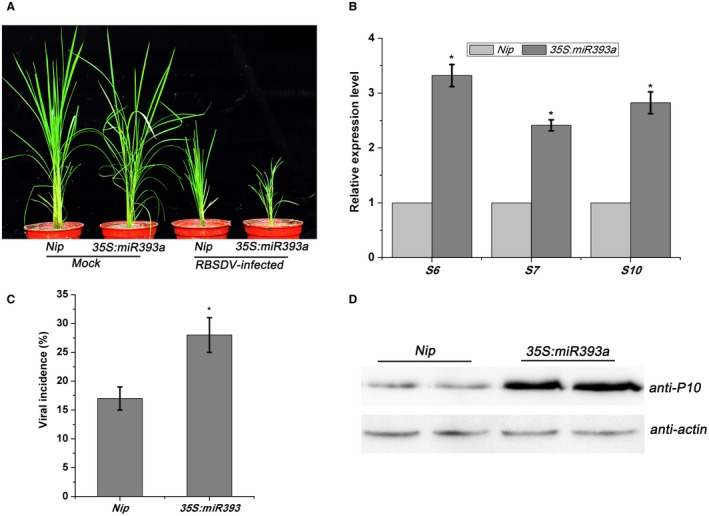
The effect of overexpression of miR393 on RBSDV infection. (A) The symptoms on mock‐inoculated or RBSDV‐infected Nip and 35S:miR393a mutant plants, respectively. The symptoms were observed at 30 days post‐inoculation (dpi). (B) The relative expression levels of RBSDV S6, S7 and S10 in RBSDV‐infected Nip and 35S:miR393a mutant plants assessed by RT‐qPCR. Three biological replicates were performed. The average values from three biological replicates are shown. Error bars represent ±SD. *P ≤ 0.05. (C) Viral incidence: the numbers of healthy and diseased plants in each treatment were determined by RT‐PCR at 30 dpi. Each treatment used at least 40 seedlings and at least three biological replicates were performed. * at the top of columns indicates significant difference at P ≤ 0.05 (n ≥ 3). (D) Western blot of leaf extracts showing detection of the RBSDV P10 outer capsid protein using an anti‐P10 antibody. Actin antibody was used as an internal reference.
Aux/IAA proteins play important roles in repressing auxin signalling. In normal conditions, Aux/IAA proteins bind to the transcription factor ARFs, repressing the expression of auxin response genes. When the IAA concentration increases, the TIR1 receptor can bind Aux/IAA proteins to form a SCFTIR1/AFB–auxin–Aux/IAA complex leading to polyubiquitination and degradation of Aux/IAA proteins. Recently, Aux/IAA proteins have been shown to be involved in plant–virus interactions (Jin et al., 2016; Padmanabhan et al., 2005). Since RBSDV infection altered the expression of OsIAA20 and OsIAA31 (Fig. 1B), we then produced transgenic rice overexpressing these two genes driven by the 35S promoter, with a 4XHA epitope tag at the N‐terminus (OE‐IAA20 and OE‐IAA31). Western blot assays confirmed the expression of OsIAA20 and OsIAA31 in the corresponding transgenic rice plants (Fig. S2). The transgenic plants overexpressing OE‐IAA20 and OE‐IAA31 and wildtype Nip were then inoculated with RBSDV. Thirty days after inoculation, both transgenic plants had more severe dwarfing than the Nip controls (Fig. 4A). There were corresponding increases in RBSDV RNA levels as measured by RT‐qPCR (Fig. 4B) and in the expression levels of viral P10 as shown by western blot (Fig. 4C). These results indicate that repressing auxin signalling by overexpression of Aux/IAA proteins promotes rice susceptibility to RBSDV infection.
Figure 4.
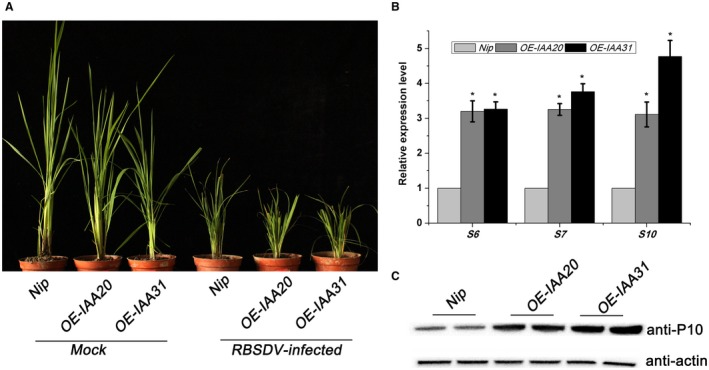
The effect of overexpression OE‐IAA20 and OE‐IAA31 on RBSDV infection. (A) The symptoms on mock‐inoculated or RBSDV‐infected Nip and mutant plants, respectively. The symptoms were observed at 30 days post‐inoculation. (B) The relative expression levels of RBSDV S6, S7 and S10 in RBSDV‐infected Nip, OE‐IAA20 and OE‐IAA31 transgenic plants assessed by RT‐qPCR. The average values from three biological replicates are shown. Error bars represent ±SD. *, P ≤ 0.05. (C) Western blot of leaf extracts to detect the levels of RBSDV in inoculated control (Nip) and OE‐IAA20 and OE‐IAA31 plants. The RBSDV P10 outer capsid protein was detected using an anti‐P10 antibody. Actin antibody was used as an internal reference.
To exclude the possibility that the auxin signalling mutants had altered resistance to the insect vector, we estimated SBPH resistance as described previously (He et al., 2017). Ten‐day‐old seedlings were infested with SBPH at seven insects per seedling, and insect survival was evaluated 3 and 5 days later. As shown in Fig. S3, there was no significant difference in SBPH mortality between the wildtype plants and the auxin signalling mutants. Together, the results indicate that auxin signalling plays an important role in plant defence against RBSDV.
The involvement of the JA pathway in auxin‐mediated defence against RBSDV infection
Previous research has shown that auxin signalling interacts either positively or antagonistically with the SA or JA pathways in response to pathogen invasion (Qi et al., 2012; Wang et al., 2007). We therefore examined the expression of JA and SA pathway genes in our auxin signalling mutants following viral infection. As shown in Fig. 5A, the expression of JA biosynthesis‐related genes (OsOPR7, OsLOXs) and JA signalling‐related genes (OsMYC2, OsJAZ12) was markedly increased in RBSDV‐infected Nip plants compared with mock‐inoculated Nip plants (P ≤ 0.05) (Fig. 5A, upper panel), but there was little obvious difference in expression between RBSDV‐infected and mock‐inoculated 35S:miR393 mutant plants (Fig. 5A, bottom panel). In our previous study, the defence‐related OsPR genes (such as OsPR1b, OsJiPR10) were activated upon RBSDV infection (He et al., 2017; Zhang et al., 2019b). The expression of OsPR1b and OsJiPR10 was also tested in RBSDV‐infected 35S:miR393 mutant and Nip plants. Consistent with our previous results, the expression of OsPR1b and OsJiPR10 was remarkably enhanced in RBSDV‐infected Nip compared to non‐infected control plants (P ≤ 0.05) (Fig. 5A, upper panel). Interestingly, the expression of these genes in RBSDV‐infected 35S:miR393 mutant was slightly increased compared to the mock‐inoculated 35S:miR393 plants (Fig. 5A, bottom panel). Similar results were obtained using the OE‐IAA20 and OE‐IAA31 transgenic plants (Fig. S4). These results indicate that the JA pathway and defence‐related OsPR genes normally activated by RBSDV infection were repressed when auxin signalling was disrupted.
Figure 5.
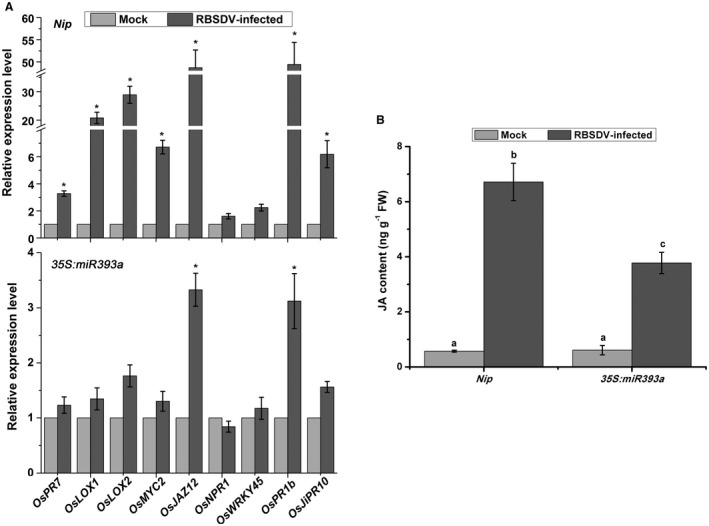
The effect of repressing auxin signalling on jasmonic acid (JA) or salicylic acid (SA) pathway genes in response to RBSDV infection. (A) The expression of JA pathway or SA pathway genes in the Nip controls (upper panel) or 35S:miR393a mutant (bottom panel) in response to RBSDV infection. Data are shown as relative expression levels in virus‐infected plants in comparison to the control plants. UBQ5 was used as the internal reference gene. Values are means ± SD of three biological replicates. Statistically significant differences from the control are indicated: *P ≤ 0.05. (B) The content of JA in Nip controls or 35S:miR393a mutant in response to RBSDV infection. Values are means ± SD of three biological replicates. Different letters at the top of columns indicate significant differences at P ≤ 0.05 (n = 3).
To confirm whether the JA content was affected by repressing auxin signalling, the JA concentration was measured in RBSDV‐infected rice plants at 40 dpi (Fig. 5B). Consistent with our previous results, the JA concentration was more than six‐fold greater in RBSDV‐infected Nip than in mock‐inoculated Nip plants, while its concentration in RBSDV‐infected 35S:miR393 mutants was only about three‐fold greater than that in the corresponding controls. Thus the induction of JA in response to RBSDV was impaired in the auxin signalling mutant.
The expression levels of SA signalling genes (OsNPR1, OsWRKY45) were also measured by RT‐qPCR in RBSDV‐infected plants. As shown in Fig. 5A, the expression of these genes did not appear to respond significantly to RBSDV in either the 35S:miR393 mutant or Nip plants. Similar results were obtained using the OE‐IAA20 and OE‐IAA31 mutants (Fig. S4). These results suggest that the SA pathway was not involved in the interaction between RSBDV and auxin signalling. Therefore, disruption of auxin signalling affected the activation of the JA pathway, but not the SA pathway, in response to RBSDV infection.
The involvement of OsRboh‐mediated ROS levels in the interaction between auxin signalling and RBSDV infection
It has recently been reported that accumulation of basal ROS plays an important role in antiviral immunity (Wu et al., 2017; Xie et al., 2018). In plants, NADPH oxidases (respiratory burst oxidase homologs, Rboh) play key roles in generating ROS by catalysing the conversion of O2 to superoxide (O2 ‐) (Suzuki et al., 2011). The rice genome encodes nine Rboh proteins (A–I) (Wang et al., 2013) of which OsRbohA, OsRbohB and OsRbohE are reported to be involved in ROS production during pathogen invasion (Wong et al., 2007; Yoshie et al., 2005). To verify the impact of auxin signalling on OsRboh‐mediated ROS levels, the expression of OsRboh transcripts was monitored in auxin signalling mutants. As shown in Fig. 6A, the expression of OsRbohB and OsRbohD was significantly decreased in all auxin signalling mutants compared to the wildtype Nip (P ≤ 0.05). In addition, the expression of OsRbohA in the OE‐IAA20 and OE‐IAA31 transgenic plants and of OsRbohE in the 35S:miR393 mutant were all significantly decreased (P ≤ 0.05). The ROS levels as measured by nitroblue tetrazolium (NBT) staining were obviously less in the auxin signalling mutants than in the Nip controls (Fig. 6B). Thus, the expression of OsRboh and basal ROS levels was affected by disrupting auxin signalling.
Figure 6.
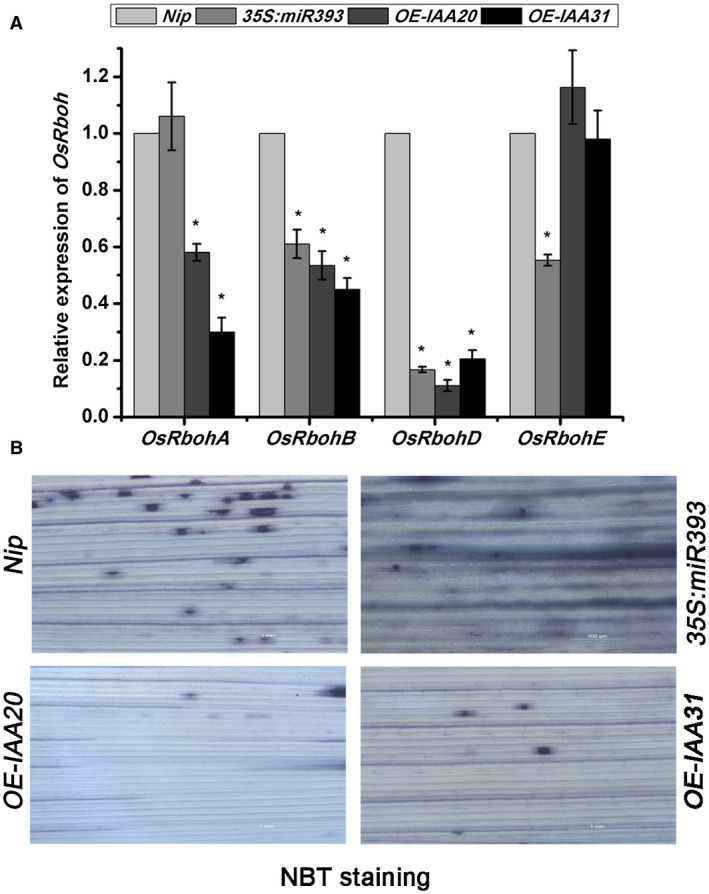
The effect of auxin signalling mutants on reactive oxygen species (ROS) homeostasis. (A) RT‐qPCR analysis of the expression of OsRboh genes in the seedlings of auxin signalling mutants compared with that in Nip. Data are shown as relative expression levels in the mutants in comparison to the wildtype Nip. UBQ5 was used as the internal reference gene. Values are means ± SD of three biological replicates. Statistically significant differences from the control are indicated: *P ≤ 0.05. (B) In situ detection of leaf ROS levels in Nip or auxin signalling mutants using nitroblue tetrazolium staining.
To further elucidate the interaction between the auxin pathway and the OsRboh‐mediated ROS levels, 10‐day‐old rice seedlings were treated with the synthetic auxins 2,4‐dichlorophenoxyacetic (2,4‐D) or 1‐naphthylacetic acid (NAA). The expression of OsRbohB and OsRbohD was significantly increased after treatment with 1 μM 2,4‐D or 1 μM NAA (P ≤ 0.05) (Fig. 7A,B), and ROS levels, as shown by NBT staining, were also greater than in the controls treated with 0.1% Triton X‐100 (Fig. 7C). These results show that the auxin pathway positively modulated the OsRboh‐mediated ROS levels. Since increased ROS levels enhance resistance to RBSDV, we conclude that OsRboh‐mediated accumulation of ROS is at least partly responsible for the auxin signalling‐regulated plant defence against RBSDV.
Figure 7.
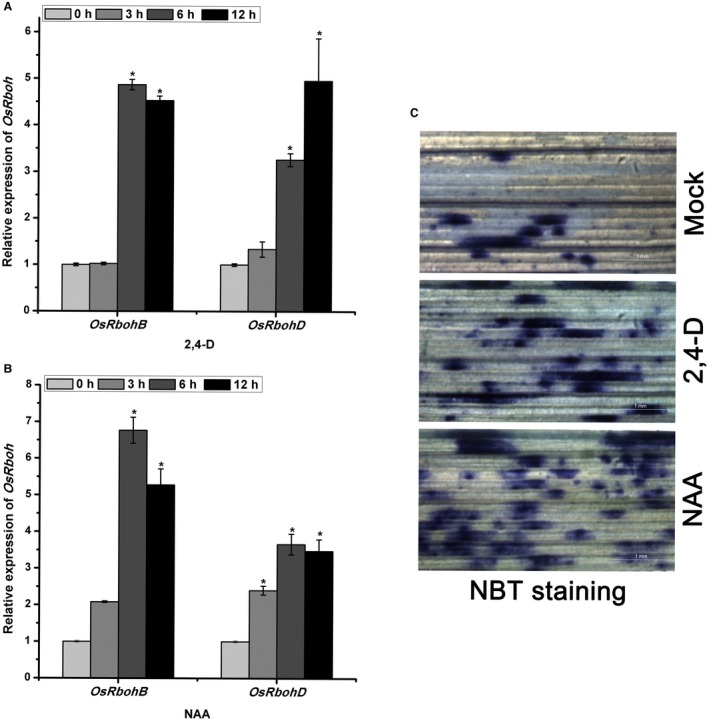
The effect of synthetic auxin treatments on OsRboh‐mediated ROS levels. (A) and (B) RT‐qPCR verification of relative expression of OsRbohB and OsRbohD genes in rice seedlings treated with 1 μM 2,4‐D (A) or 1 μM NAA (B) compared with 0.1% Triton X‐100 treated rice controls. Treatments used 10‐day‐old rice seedlings. Each sample contained a pool of eight to ten plants and was collected 0, 3, 6 or 12 h after spraying. The average values from three biological replicates are shown. * at the top of columns indicates significant difference at P ≤ 0.05. (C) Nitroblue tetrazolium (NBT) staining of rice leaves treated with 0.1% Triton X‐100 (mock), 1 μM 2,4‐D or 1 μM NAA.
To confirm the effect of OsRboh‐mediated ROS accumulation on RBSDV infection, diphenyliodide (DPI)‐treated rice plants were inoculated with RBSDV. DPI is a well‐known inhibitor of RBOH activity (Hyodo et al., 2017; Lv et al., 2018). As shown in Fig. 8A, the expression of OsRbohB and OsRbohD in DPI‐treated rice seedlings was significantly reduced compared with that in untreated controls (P ≤ 0.05). The ROS levels as measured by NBT staining were much lower in the DPI‐treated rice seedlings than in mock‐treated plants (Fig. 8B). When these DPI‐treated rice plants were inoculated with RBSDV, viral symptoms at 30 dpi were more severe in DPI‐treated plants than in mock‐treated plants (Fig. 8C). An RT‐qPCR assay indicated that the RBSDV RNA levels (S6, S7 and S10) were about two‐fold greater in DPI‐treated plants than in the untreated controls (P ≤ 0.05) (Fig. 8D). These results demonstrate that OsRboh‐mediated ROS accumulation plays an important role in rice defence against RBSDV infection.
Figure 8.
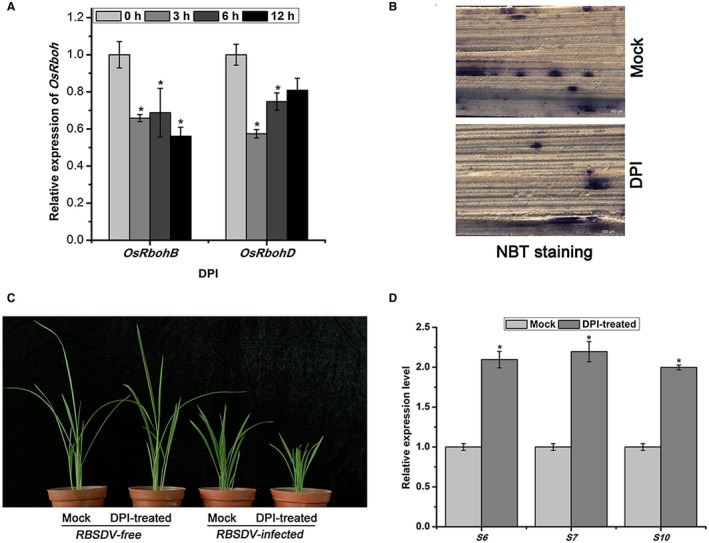
The effect of OsRboh‐mediated reactive oxygen species on RBSDV infection. (A) The effect of treatment with 100 μM diphenyliodide (DPI) on the expression of OsRboh. Each result represents not less than three biological repeats. * at the top of columns indicates significant difference from the mock‐treated control at P < 0.05. (B) Nitroblue tetrazolium (NBT) staining of rice seedlings treated with 100 μM DPI. (C) The effect of treatment with 100 μM DPI on the symptoms of RBSDV at 30 days post‐inoculation. (D) The effect of treatment with 100 μM DPI on the relative expression levels of RBSDV S6, S7 and S10 in RBSDV‐infected plants assessed by RT‐qPCR. Each treatment consisted of three biological replicates of at least 30 seedlings each. Different letters at the top of columns indicate significant differences at P < 0.05 (n ≥ 3) by Fisher's least significant difference tests.
Discussion
Previous studies have mainly focused on the influence of viral infection on plant auxin homeostasis and signalling. For example, Tobacco mosaic virus (TMV) p126 protein was shown to interact with several Aux/IAA proteins to affect the expression of a large number of auxin‐responsive genes (Padmanabhan et al., 2005, 2008). Rice dwarf virus (RDV) P2 protein interfered with auxin signalling by directly interacting with and stabilizing the OsIAA10 protein to enhance viral infection and disease development (Jin et al., 2016). However, the role of auxin signalling in the antiviral mechanism remains unclear. Here, we have shown that auxin signalling plays an important role against RBSDV. The induction of the JA pathway in response to RBSDV was undermined in auxin signalling mutants. These results suggest that auxin signalling is required to activate the JA pathway upon viral infection. Recent studies showed that auxin defective mutants of Arabidopsis were more susceptible than wild‐type plants to a necrotrophic pathogen (Qi et al., 2012). Application of IAA and methyl jasmonate together synergistically induced the expression of defence‐related genes PDF1.2 and HEL. Those results indicated that auxin positively interacted with the JA pathway against necrotrophic pathogens. Moreover, JA was shown to induce the auxin biosynthesis genes ASA and YUCCA. However, the influence of auxin signalling on the JA pathway during pathogen invasion has been little reported. Here, our results suggest that auxin plays important roles in the activation of the JA pathway, and that induction of the JA pathway may be part of the auxin signalling‐mediated defence against RBSDV.
ROS were initially considered to be harmful to cells, but are now known to act as signalling molecules to regulate cell growth and defence (Mittler et al., 2011). The crosstalk between auxin and ROS has mainly been reported from studies of plant growth and development (Joo et al., 2001, 2005). A recent report showed that auxin‐induced ROS production was important for polar root hair growth by inducing the expression of ROS‐generating enzymes (NADPH oxidases) (Mangano et al., 2017). Auxin signalling could activate RSL4, which directly targets the NADPH oxidases through auxin response factors to modulate ROS levels. However, the interplay between auxin and ROS has been little reported in the context of plant defence. Recently, ROS were demonstrated to be involved in plant–virus interactions (Hyodo et al., 2017; Wu et al., 2017; Xie et al., 2018). Two positive‐strand RNA viruses, Red clover necrotic mosaic virus (RCNMV) and Brome mosaic virus, were shown to hijack the host's ROS‐generating machinery and trigger intracellular ROS bursts (Hyodo et al., 2017). These bursts are required for viral RNA replication. However, ROS were shown to play vital roles in plant defence against rice viruses (Wu et al., 2017). Our previous results also showed that high basal ROS levels can enhance antiviral defence against RBSDV infection (Xie et al., 2018). Here, our results indicate that auxin can induce the expression of OsRboh and that disruption of auxin signalling decreased the expression of OsRboh. Repressing the expression of OsRboh increased rice susceptibility to RBSDV infection. These results indicate that ROS reduction by repression of auxin signalling contributes in part to rice susceptibility to RBSDV.
In summary, our results showed that RBSDV infection affected auxin homeostasis and down‐regulated the expression of auxin pathway genes. Repressing auxin signalling uncovered the positive roles of auxin signalling in RBSDV infection. Further research revealed that activation of JA‐mediated defence and accumulation of OsRboh‐mediated ROS played vital roles in the interaction between auxin signalling and RBSDV infection. Our results confirm that auxin signalling plays a positive role in rice defence against RBSDV, but more research is needed to determine the mechanism by which RBSDV represses the auxin pathway.
Experimental Procedures
Plant materials and growth conditions
Rice seeds of Wuyujing No.3 and Huaidao No. 5 were used for this study. The cultivar rice Nipponbare (O. sativa, NIP) was used as the wild‐type control. The auxin signalling mutant 35S:miR393 was obtained from Dr Hongwu Bian (Zhejiang University) (Bian et al., 2012). The DR5:GUS transgenic plants were gifts from Dr Yanhua Qi (Zhejiang University). RBSDV‐infected plants collected from fields (Shandong Province, China) were used as a virus source. Rice plants were grown in the greenhouse with14/10 h light/dark cycle at 28–30 °C.
Construction and rice transformation of overexpression vector
To prepare constructs to overexpress the OsIAA20 and OsIAA31 genes, the respective open reading frames were first amplified from cDNA of NIP using primers incorporating BamHI and SacI sites (Table S1). The pCV1300 vector was digested with BamHI and SacI, which contains the cauliflower mosaic virus (CaMV) doubled 35S promoter to drive transcription. The OsIAA20 and OsIAA31 PCR products were then combined into the pCV1300 vector (Sun et al., 2013a,b) to create the respective recombinant vectors OE‐IAA20 and OE‐IAA31. These vectors were introduced into Agrobacterium (strain GV3101) using electroporation and transformed into NIP rice as described previously (Zhang et al., 2019a).
RBSDV inoculation assay
The detailed assays were conducted as described previously (Zhang et al., 2019a). Non‐viruliferous SBPH were reared on Wuyujing No. 3 seedlings at 26 °C in a room under artificial light. Briefly, to acquire virus, large numbers of first‐ or second‐instar SBPH were prepared and then released onto RBSDV‐infected rice plants. After feeding for 3–5 days, instar nymphs were transferred to healthy Wuyujing No. 3 seedlings for 10–12 days (an incubation period). For RBSDV inoculation, planthoppers were transferred to 10‐day‐old rice seedlings (two to three leaf stage) to feed for 3 days (about three SBPH on each of 35–40 seedlings per treatment). The rice seedlings were then grown in the greenhouse to develop symptoms. RBSDV‐infected plants were stunted and had darkened leaves. Plant height was measured and RT‐PCR using virus‐specific primers (Table S1) was used to confirm infection and determine the percentage of plants infected (viral incidence). Simultaneously, to detect any effect of mutant rice plants on insect mortality, the rice seedlings were infested with SBPH at seven insects per seedling and their survival was measured 3 and 5 days later. The inoculation experiments had at least three biological repeats.
Foliar application of chemicals
Ten‐day‐old Huaidao No. 5 seedlings were grown in glass beakers (35–40 seedlings per beaker) and sprayed with 25 mL 1 μM 2,4‐dichlorophenoxyacetic acid (2,4‐D, Sigma‐Aldrich), 1 μM 1‐naphthylacetic acid (NAA, Sigma‐Aldrich, St Louis, MO, USA) or 100 μM DPI (MedChemExpress, Monmouth Junction, NJ, USA). Solutions also contained 0.1% Triton X‐100 and control plants were sprayed with 25 ml 0.1% Triton X‐100. For RT‐qPCR, samples of eight to ten plants per beaker were collected and pooled 0, 3, 6 or 12 h after spraying. Each experiment used not less than three biological repeats.
NBT staining assay
Superoxide was detected with nitroblue tetrazolium (NBT). The detailed procedures were based on those described previously (Xia et al., 2009; Zhang et al., 2019a). Thirty‐day‐old rice leaves were cut into 1–2 cm pieces and infiltrated with 2 mL NBT solution. The solution contained 6 mM NBT (Sigma) and 10 mM sodium citrate (pH = 6.0). The leaves were vacuum infiltrated (10 min, 60 kPa pressure) and incubated at 37 °C for 2 h. Then the rice leaves were rinsed in 75% ethanol twice for 30 min each and finally rinsed with absolute ethanol to bleach out the chlorophyll. The samples were then stored in 20% glycerol and photographed under a stereomicroscope.
Analysis of IAA and JA concentrations
RBSDV‐infected and mock‐inoculated rice leaves were collected at 40 dpi and powdered in liquid nitrogen. The phytohormones were extracted and analysed as described previously with slight modifications (Fu et al., 2012; He et al., 2017). To measure IAA concentrations, an internal standard 2H2‐IAA (100 pmol) was added to a 20 mg sample. Samples were purified through a column purification and examined by ultra‐high‐performance liquid chromatography–triple quadrupole mass spectrometry (UPLC‐MS/MS) with negative electrospray ionization. To quantify JA, 2 mL of methanol containing 2H5‐JA (95 pmol) was added to 200 mg of leaf powder, and then purified by an Oasis mode anion exchange (MAX) solid phase extraction (SPE) column. The supernatants were collected and then analysed with UPLC‐MS/MS. Each sample consisted of at least four to five pooled plants and was replicated at least three times.
RNA extraction and RT‐qPCR
To analyse gene expression levels, total RNA was isolated from rice leaves of 30‐day‐old plants using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. cDNA was synthesised using the Tiangen fast quant RT kit (Tiangen, Beijing, China). The RT‐qPCR assay was performed using the ABI7900HT Sequence Detection System (Applied Biosystems, CA, USA) with ChamQTM SYBR qPCR Master Mix (Low ROX Premixed). OsUBQ5 (AK061988) was used as an internal control (Sun et al., 2015). The relative expression levels of genes were determined using the 2‐ΔΔC (t) method. Each treatment had at least three biological replicates of three to five plants each and there were three technical replicates per sample. The primers used for RT‐qPCR are listed in Table S1.
Protein Analysis
To detect the OsIAA20 and OsIAA31 proteins in the transgenic OE‐IAA20 and OE‐IAA31 rice plants, western blot analysis was performed using HA‐tag antibody (TransGen Biotech, Beijing, China). Western blots to test viral accumulation in RBSDV‐infected plants used an anti‐P10 antibody to detect the RBSDV P10 outer capsid protein (Lu et al., 2019). Fresh leaf samples were ground in liquid nitrogen and extracted as described previously (Zhang et al., 2019b). Briefly, 0.1 g samples were extracted in 300 μL SDS lysis buffer (100 mM Tris‐HCl, pH = 6.8, 10% SDS) and boiled with 5 × protein loading buffer for 10 min. The proteins were separated by 10–12% sodium dodecyl sulphate polyacrylamide gel electrophoresis, and then transferred to a polyvinylidene fluoride membrane (previously activated with methanol for 15 s). The membrane was blocked with 10% non‐fat dry milk diluted in tris buffered saline tween for 2 h at 25 °C, and then the primary antibody (1:5000) in blocking buffer was added for 2 h. The secondary antibody in blocking buffer was then added (1:10 000) and incubated for 1.5 h. Actin (Abbkine, A01050‐3) antibody was used as a reference protein. The proteins were detected using an ECL kit (Pierce, Rockford, USA) in the BIO‐RAD ChemiDoc MP Imaging System (Bio‐Rad, CA, USA).
GUS staining assays
GUS staining assays were done as described previously (Shen et al., 2013). Briefly, the rice leaves were cut into 10 cm pieces and infiltrated with 15 mL GUS staining solution using a brief vacuum infiltration. The solution included 0.5 mM X‐gluc (Sigma), 100 mM phosphate buffer (NaH2PO4·2H2O, Na2HPO4·12H2O, pH = 7.0), 1 mM K3[Fe(CN)6], 1 mM K4[Fe(CN)6], 10 mM Na2EDTA, 20%(v/v) methanol and 0.5% (v/v) Triton X‐100. After incubation at 37 °C for 1–2 h, the leaf pieces were washed with 75% ethanol twice for 30 min each time and finally rinsed with absolute ethanol before microscopic examination to determine GUS activity.
Statistical analysis
Data were analysed using one‐way or two‐way ANOVA with Fisher's least significant difference tests. A P‐value ≤ 0.05 was considered statistically significant. All analyses were performed using ORIGIN 8.0 software.
Competing Interests
The authors declare that they have no competing interests.
Supporting information
Fig. S1 The cis‐element in the promoters of OsIAA20 and OsIAA31.
Fig. S2 Western blot of leaf extracts to confirm the overexpression of OE‐IAA20 and OE‐IAA31 in the respective transgenic plants compared to the Nip control.
Fig. S3 The mortality of small brown planthoppers on control (Nip) and auxin signalling mutant plants.
Fig. S4 The expression levels of jasmonic acid (JA) or salicylic acid (SA) pathway genes in OE‐IAA20 (upper panel) and OE‐IAA31 (lower panel) mutants in response to RSBDV.
Table S1 The primers used in this study.
Acknowledgements
The authors are indebted to Dr Hongwu Bian (Zhejiang University) for providing the 35S:miR393 mutant, to Dr Yanhua Qi (Zhejiang University) for the DR5:GUS transgenic plants (Shen et al., 2013), and to Professor Jianxiang Wu (Zhejiang University) for providing P10 antibody. We are grateful for the expertise of Shuang Fang and Dr Jinfang Chu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China) in determining the JA and IAA concentrations. We thank Mike Adams for critically reading and improving the manuscript. This work was supported by grants from the National Key Research and Development Plan (2016YFD0200804), the International Science & Technology Cooperation Program of China (2015DFA30700), State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (2010DS700124‐ZZ1801). This work was sponsored by the K.C. Wong Magna Fund in Ningbo University.
References
- Bian, H. , Xie, Y. , Guo, F. , Han, N. , Ma, S. , Zeng, Z. , Wang, J. , Yang, Y. and Zhu, M. ( 2012) Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 196, 149 – 161. [DOI] [PubMed] [Google Scholar]
- Fu, J. and Wang, S. (2011) Insights into auxin signaling in plant–pathogen interactions. Front. Plant Sci. 2, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J. , Chu, J. , Sun, X. , Wang, J. and Yan, C. (2012) Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC‐MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28, 1081–1087. [DOI] [PubMed] [Google Scholar]
- He, Y. , Zhang, H. , Sun, Z. , Li, J. , Hong, G. , Zhu, Q. , Zhou, X. , MacFarlane, S. , Yan, F. and Chen, J. (2017) Jasmonic acid‐mediated defense suppresses brassinosteroid‐mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 214, 388–399. [DOI] [PubMed] [Google Scholar]
- Huang, R. , Li, Y. , Tang, G. , Hui, S. , Yang, Z. , Zhao, J. , Liu, H. , Cao, J. and Yuan, M. (2018) Dynamic phytohormone profiling of rice upon rice black‐streaked dwarf virus invasion. J. Plant Physiol. 228, 92–100. [DOI] [PubMed] [Google Scholar]
- Hyodo, K. , Hashimoto, K. , Kuchitsu, K. , Suzuki, N. and Okuno, T. ( 2017) Harnessing host ROS‐generating machinery for the robust genome replication of a plant RNA virus. Proc. Natl. Acad. Sci. USA. 114, E1282 – E1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L. , Qin, Q. , Wang, Y. , Pu, Y. , Liu, L. , Wen, X. , Ji, S. , Wu, J. , Wei, C. , Ding, B. and Li, Y. (2016) Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 12, e1005847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, J.H. , Bae, Y.S. and Lee, J.S. (2001) Role of auxin‐induced reactive oxygen species in root gravitropism. Plant Physiol. 126, 1055–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, J.H. , Yoo, H.J. , Hwang, I. , Lee, J.S. , Nam, K.H. and Bae, Y.S. (2005) Auxin‐induced reactive oxygen species production requires the activation of phosphatidylinositol 3‐kinase. FEBS Lett. 579, 1243–1248. [DOI] [PubMed] [Google Scholar]
- Kidd, B.N. , Kadoo, N.Y. , Dombrecht, B. , Tekeoglu, M. , Gardiner, D.M. , Thatcher, L.F. , Aitken, E.A. , Schenk, P.M. , Manners, J.M. and Kazan, K. ( 2011) Auxin signaling and transport promote susceptibility to the root‐infecting fungal pathogen Fusarium oxysporum in Arabidopsis . Mol. Plant Microbe Interact. 24, 733 – 748. [DOI] [PubMed] [Google Scholar]
- Llorente, F. , Muskett, P. , Sánchez‐Vallet, A. , López, G. , Ramos, B. , Sánchez‐Rodríguez, C. , Jordá, L. , Parker, J. and Molina, A. (2008) Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Molecular Plant, 1, 496–509. [DOI] [PubMed] [Google Scholar]
- Lu, L. , Wang, Q. , Huang, D. , Xu, Q. , Zhou, X. and Wu, J. (2019) Rice black‐streaked dwarf virus P10 suppresses protein kinase C in insect vector through changing the subcellular localization of LsRACK1. Phil. Trans. R. Soc. B: Biol. Sci., 374, 20180315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, B. , Tian, H. , Zhang, F. , Liu, J. , Lu, S. , Bai, M. , Li, C. and Ding, Z. (2018) Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 14, e1007144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano, S. , Denita‐Juarez, S.P. , Choi, H.‐S. , Marzol, E. , Hwang, Y. , Ranocha, P. , Velasquez, S.M. , Borassi, C. , Barberini, M.L. , Aptekmann, A.A. and Muschietti, J.P. ( 2017) Molecular link between auxin and ROS‐mediated polar growth. Proc. Natl. Acad. Sci. USA. 114, 5289 – 5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. , Vanderauwera, S. , Suzuki, N. , Miller, G. , Tognetti, V.B. , Vandepoele, K. , Gollery, M. , Shulaev, V. and Van Breusegem, F. (2011) ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. [DOI] [PubMed] [Google Scholar]
- Mockaitis, K. and Estelle, M. (2008) Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24, 55–80. [DOI] [PubMed] [Google Scholar]
- Navarro, L. , Dunoyer, P. , Jay, F. , Arnold, B. , Dharmasiri, N. , Estelle, M. , Voinnet, O. and Jones, J.D. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, M.S. , Goregaoker, S.P. , Golem, S. , Shiferaw, H. and Culver, J.N. (2005) Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 Is associated with disease development. J. Virol. 79, 2549–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, M.S. , Kramer, S.R. , Wang, X. and Culver, J.N. (2008) Tobacco mosaic virus replicase‐auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. J. Virol. 82, 2477–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L. , Yan, J. , Li, Y. , Jiang, H. , Sun, J. , Chen, Q. , Li, H. , Chu, J. , Yan, C. , Sun, X. and Yu, Y. (2012) Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola . New Phytol. 195, 872–882. [DOI] [PubMed] [Google Scholar]
- Shen, C. , Wang, S. , Zhang, S. , Xu, Y. , Qian, Q. , Qi, Y. and Jiang, D.A. (2013) OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 36, 607–620. [DOI] [PubMed] [Google Scholar]
- Sun, Z. , Yang, D. , Xie, L. , Sun, L. , Zhang, S. , Zhu, Q. , Li, J. , Wang, X. and Chen, J. (2013a) Rice black‐streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana . Virology, 447, 131–139. [DOI] [PubMed] [Google Scholar]
- Sun, Z. , Zhang, S. , Xie, L. , Zhu, Q. , Tan, Z. , Bian, J. , Sun, L. and Chen, J. (2013b) The secretory pathway and the actomyosin motility system are required for plasmodesmatal localization of the P7–1 of rice black‐streaked dwarf virus. Adv. Virol. 158, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Sun, Z. , He, Y. , Li, J. , Wang, X. and Chen, J. (2015) Genome‐wide characterization of rice black streaked dwarf virus‐responsive microRNAs in rice leaves and roots by small RNA and degradome sequencing. Plant Cell Physiol. 56, 688–699. [DOI] [PubMed] [Google Scholar]
- Suzuki, N. , Miller, G. , Morales, J. , Shulaev, V. , Torres, M.A. and Mittler, R. (2011) Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14, 691–699. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T. , Murfett, J. , Hagen, G. and Guilfoyle, T.J. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell, 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidhyasekaran, P. (2015) Auxin signaling system in plant innate immunity In: Plant Hormone Signaling Systems in Plant Innate Immunity (Vidhyasekaran P.), pp. 311–357. Dordrecht: Springer Netherlands. [Google Scholar]
- Wang, Z.‐H. , Fang, S.‐G. , Xu, J.‐L. , Sun, L.‐Y. , Li, D.‐W. and Yu, J.‐L. (2003) Sequence analysis of the complete genome of rice black‐streaked dwarf virus isolated from maize with rough dwarf disease. Virus Genes, 27, 163–168. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Pajerowska‐Mukhtar, K. , Culler, A.H. and Dong, X. (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wang, G.‐F. , Li, W.‐Q. , Li, W.‐Y. , Wu, G.‐L. , Zhou, C.‐Y. and Chen, K.‐M. (2013) Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int. J. Mol. Sci. 14, 9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. and Li, Y. (2016) Rice reoviruses in insect vectors. Annu. Rev. Phytopathol. 54, 99–120. [DOI] [PubMed] [Google Scholar]
- Weijers, D. and Wagner, D. (2016) Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67, 539–574. [DOI] [PubMed] [Google Scholar]
- Wong, H.L. , Pinontoan, R. , Hayashi, K. , Tabata, R. , Yaeno, T. , Hasegawa, K. , Kojima, C. , Yoshioka, H. , Iba, K. , Kawasaki, T. and Shimamoto, K. (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N‐terminal extension. Plant Cell, 19, 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Yang, R. , Yang, Z. , Yao, S. , Zhao, S. , Wang, Y. , Li, P., Song, X., Jin, L., Zhou, T. and Lan, Y. (2017) ROS accumulation and antiviral defence control by microRNA528 in rice. Nature Plants, 3, 16203. [DOI] [PubMed] [Google Scholar]
- Xia, X.‐J. , Wang, Y.‐J. , Zhou, Y.‐H. , Tao, Y. , Mao, W.‐H. , Shi, K. , Asami, T. , Chen, Z. and Yu, J.Q. (2009) Reactive oxygen species are involved in brassinosteroid‐induced stress tolerance in cucumber. Plant Physiol. 150, 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Li, L. , Zhang, H. , Wang, R. , Tan, X. , He, Y. , Hong, G. , Li, J. , Ming, F. , Yao, X. and Yan, F. (2018) Abscisic acid negatively modulates plant defence against rice black‐streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 41, 2504–2514. [DOI] [PubMed] [Google Scholar]
- Yoshie, Y. , Goto, K. , Takai, R. , Iwano, M. , Takayama, S. , Isogai, A. and Che, F.S. (2005) Function of the rice gp91phox homologs OsrbohA and OsrbohE genes in ROS‐dependent plant immune responses. Plant Biotechnol. 22, 127–135. [Google Scholar]
- Zhang, H. , Tan, X. , He, Y. , Xie, K. , Li, L. , Wang, R. , Hong, G. , Li, J. , Li, J. , Taliansky, M. and MacFarlane, S. ( 2019a) Rice black‐streaked dwarf virus P10 acts as either a synergistic or an antagonistic determinant during superinfection with related or unrelated virus. Mol. Plant Pathol. 20, 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , He, Y. , Tan, X. , Xie, K. , Li, L. , Hong, G. , Li, J. , Cheng, Y. , Yan, F. , Chen, J. and Sun, Z. ( 2019b) The dual effect of the brassinosteroid pathway on rice black streaked dwarf virus infection by modulating the peroxidase‐mediated oxidative burst and plant defense. Mol. Plant Microbe Interact. 10.1094/mpmi-1010-1018-0285-R. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. (2010) Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The cis‐element in the promoters of OsIAA20 and OsIAA31.
Fig. S2 Western blot of leaf extracts to confirm the overexpression of OE‐IAA20 and OE‐IAA31 in the respective transgenic plants compared to the Nip control.
Fig. S3 The mortality of small brown planthoppers on control (Nip) and auxin signalling mutant plants.
Fig. S4 The expression levels of jasmonic acid (JA) or salicylic acid (SA) pathway genes in OE‐IAA20 (upper panel) and OE‐IAA31 (lower panel) mutants in response to RSBDV.
Table S1 The primers used in this study.


