SUMMARY
Pantoea stewartii subsp. stewartii is a Gram‐negative enteric bacterium that primarily infects sweet corn. Studies of this bacterium have provided useful insight into how xylem‐dwelling bacteria establish themselves and incite disease in their hosts. Pantoea stewartii subsp. stewartii is a remarkable bacterial system for laboratory studies because of its relative ease of propagation and genetic manipulation, and the fact that it appears to employ a minimal number of pathogenicity mechanisms. In addition, P. stewartii subsp. stewartii produces copious amounts of its quorum sensing (QS) signal, acyl‐homoserine lactone (AHL), making it an excellent organism for studying QS‐controlled gene regulation in a plant‐pathogenic bacterium. In fact, P. stewartii subsp. stewartii has become the microbial paradigm for QS control of gene expression by both repression and activation via a QS regulator that binds DNA in the absence and dissociates in the presence of the signal ligand. Moreover, P. stewartii subsp. stewartii is a member of the Enterobacteriaceae, and lessons learned from its interaction with plants may be extrapolated to other plant‐associated enterics, such as Erwinia, Dickeya and Pectobacterium spp., or enteric human pathogens associated with plants, such as Escherichia coli and Salmonella spp.
Taxonomy: Bacteria; Gammaproteobacteria; family Enterobacteriaceae; genus Pantoea; species stewartii (Mergaert et al., 1993).
Microbiological properties: Gram‐negative, motile, yellow pigmented, mucoid, facultative anaerobe.
Host range: Pantoea stewartii subsp. stewartii (Smith, 1898) Dye causes Stewart's wilt of corn (Zea mays). Early‐maturing sweet corn varieties and some elite inbred maize lines are particularly susceptible.
Disease symptoms: There are two major phases of Stewart's wilt disease: (i) wilt and (ii) leaf blight. The wilt phase occurs when young seedlings are infected with P. stewartii subsp. stewartii (Fig. 1A). Water‐soaked lesions first appear on the young expanding leaves and, later, seedlings may become severely wilted (Fig. 1B). The plants usually die when infected at the seedling stage. The leaf blight phase occurs when mature plants are infected (Fig. 1C). The bacteria enter the xylem and cause long linear yellow–grey lesions with a wavy margin that run parallel to the leaf veins. These lesions later turn necrotic and dark in colour. The leaf blight phase is most apparent after tasselling and does not generally cause death of the plant. In addition, the bacteria can sometimes break out of the xylem and cause pith rot in mature sweet corn plants. In resistant varieties, lesions are usually limited to only a few centimetres depending on the level of resistance of the particular hybrid (Claflin, 2000; Pataky, 2003).
Figure 1.

Disease symptoms associated with Stewart's wilt of sweet corn. (A) The seedling wilt phase of the disease which occurs when young plants are systemically infected. (B) Leaf lesions run parallel to the leaf veins. They begin as water‐soaked areas that turn into long, pale‐green to yellow gray streaks with wavy margins. (C) The leaf blight phase of the disease. This phase occurs when plants are infected after the seedling stage. Images used with permission from 2003, 2004).
Useful websites: http://www.apsnet.org/publications/apsnetfeatures/Pages/StewartsWilt.aspx
INTRODUCTION
Pantoea stewartii subsp. stewartii (syn. Erwinia stewartii) is a Gram‐negative bacterium that is the causal agent of Stewart's wilt of sweet corn. Stewart's wilt of corn has been reported worldwide from rare seed transmission, but it only occurs naturally in North America. The disease is endemic to the USA and was first reported on Long Island, New York in the late 1890s by F. C. Stewart (Claflin, 2000; Stewart, 1897). Indeed, E. F. Smith characterized the causal agent of the disease and named it Pseudomonas stewartii in honour of Stewart. Stewart's wilt is now known to occur from the Mid‐Atlantic States to the Ohio River Valley, and in Missouri and Iowa (Rand and Cash, 1921; Robert, 1955). Stewart's wilt caused severe losses for the corn industry in the 1930s, but the importance of the disease has diminished in the USA because of the use of resistant corn hybrids (Claflin, 2000; Freeman and Pataky, 2001; Pataky et al., 1988). However, Stewart's wilt remains a problem where susceptible sweet corn varieties are grown.
Pantoea stewartii subsp. stewartii is vectored by the corn flea beetle (Chaetocnema pulicaria) (Fig. 2) and, on initial introduction, the bacterium colonizes both the intercellular spaces of the leaf tissue, where it causes a characteristic water‐soaked lesion, and the xylem, which leads to systemic spread and wilting. The bacterium preferentially colonizes the xylem tissue, where it proliferates. Precisely what nutrients it extracts from the xylem parenchyma, tissue, cell walls and fluid remain unknown. Once established in the xylem, the bacteria grow to high cell densities and establish dense biofilms encased in an exopolysaccharide (EPS) capsule and slime, called ‘stewartan’. These dense biofilms, including the associated EPS, block the water flow in the xylem and lead to the wilting and death of the plant. As the xylem pit membranes become plugged, EPS expands and ruptures the pit membrane, allowing the bacteria to move to the next vessel, or the vessel wall, releasing them into the pith or intercellular spaces (Leigh and Coplin, 1992). EPS is thus required for systemic movement throughout the plant and for full virulence of the pathogen (Beck von Bodman and Farrand, 1995; Dolph et al., 1988). Infection of susceptible varieties at the young seedling stage leads to rapid wilting and death of the plant, whereas infections that occur in more mature plants later in the season result primarily in vascular chlorosis and necrosis with little wilting, but pronounced stunting of growth.
Figure 2.

The corn flea beetle (Chaetocnema pulicaria). This insect is the primary vector of the Stewart's wilt pathogen, Pnss. The bacteria are introduced into the plant via scratching wounds created by the beetle as it feeds on the corn plant. Image used with permission from Pataky (2003).
EPIDEMIOLOGY AND CONTROL OF STEWART'S WILT
The bacteria are present in the gut (Correa et al., 2008) and overwinter in the adult corn flea beetle. After it emerges in the spring, the insect deposits the bacterial cells into the feeding wounds via its faeces (Claflin, 2000). The vector–bacterium association of P. stewartii subsp. stewartii with the corn flea beetle is in large part unexplored because of the difficulty of rearing the insect in the laboratory, but recent evidence suggests that a second type III secretion system (T3SS), with homology to those found in mammalian pathogens, is involved in insect colonization (Correa et al., 2008). However, much is known about the importance of the corn flea beetle in the context of Stewart's wilt epidemiology, as discussed below.
Plant‐to‐plant spread does not occur without the presence of the corn flea beetle and disease incidence is correlated directly with the numbers of corn flea beetles present in corn fields (Claflin, 2000; Menclas et al., 2006). There are two major cycles of Stewart's wilt infection throughout the growing season. The first cycle occurs when the emerged overwintering adults transmit the bacteria to young corn seedlings. This first cycle of Stewart's wilt is the most damaging, because corn is infected at the seedling stage, the most vulnerable developmental stage of the plant. The second cycle of infection occurs when the bacteria are transmitted by the first summer generation of the insect that have acquired the bacterium from the infected plants. The timing of the emergence of this population of flea beetles coincides with a decrease in the susceptibility of the corn plant that occurs after tasselling. Therefore, plants infected at this stage usually only display leaf blight symptoms of Stewart's wilt and do not succumb to wilting as do seedlings. There is also a second generation of corn flea beetles that occurs 4–6 weeks later than the first generation. In addition, there can be several more subsequent generations of flea beetles that lead to later season infections. These infections that occur late in the season in mature plants also only progress to the leaf blight stage of infection. The last beetle generation of the season acquires the bacterium from infected plants and becomes the overwintering population (Pataky, 2003).
Control for Stewart's wilt is largely based on resistant hybrid cultivars. Much of the commercial corn grown today has been bred for resistance to Stewart's wilt as part of the criteria for selection. These varieties show adequate levels of resistance that prevent systemic infection (Freeman and Pataky, 2001; Pataky, 2004; Pataky et al., 2008). However, there are still some desirable sweet corn hybrids and some elite inbred lines used for hybrid corn seed production that remain highly susceptible to Stewart's wilt. The early maturing sweet corn varieties are the most susceptible, and some growers risk planting these to get to market early. Most of these varieties are grown in areas in which P. stewartii subsp. stewartii and the corn flea beetle do not occur.
Stewart's wilt disease is relatively easy to forecast and is based on the overwintering potential of the corn flea beetle. The severity of the disease is related directly to the numbers of corn flea beetles that survive the winter. Predictions of vector survival are based on the sum of the mean temperatures (F) for December, January and February (Agrios, 2005; Claflin, 2000; Stevens, 1934). Basically, during mild winters, growers can expect higher Stewart's wilt incidence because of the greater survival rates of the corn flea beetle over the winter. Cold winters greatly decrease the corn flea beetle populations, decreasing the risk for Stewart's wilt in the following growing season. In addition to the index, growers also take into account the previous incidence of disease in the field, because it takes several years to build up. Insecticides used at planting are effective at decreasing the initial wilt phase of the disease, and the disease forecasting index plays a large role in determining the need for an early insecticide spray. Recent studies have indicated that the corn flea beetle is a very efficient vector of P. stewartii subsp. stewartii, confirming that appropriately timed insecticides (i.e. at the seedling stage) are beneficial in years with high corn flea beetle incidence (Menclas et al., 2006). Insecticides are generally not economical when used later in the season.
In addition to corn, several other monocot species have been reported to serve as hosts of the species P. stewartii, such as sudan grass, oat and triticale, sorghum, millet and sugarcane, to name a few (Azad et al., 2000; Claflin, 2000). Notably, the P. stewartii isolate recovered from infected sudan grass was pathogenic on corn, although the disease did not progress beyond the lesion stage and never induced wilting in corn plants (Azad et al., 2000). However, it should be emphasized that the isolate was not characterized to the subspecies' level.
ECONOMIC IMPACT
Stewart's wilt is the most serious bacterial disease of sweet corn and maize in the north‐central and eastern USA. Significant economic impact can occur in both sweet and dent corn. Economic impact caused by Stewart's wilt is minimal in fields planted with resistant cultivars. Costly measures, such as seed outgrow tests and enzyme‐linked immunosorbent assay (ELISA), must be undertaken to certify that the corn seed destined for export markets is free of P. stewartii subsp. stewartii (Lamka et al., 1991; Michener et al., 2002). For this reason, most seed corn is produced in locations in which Stewart's wilt does not occur. In addition, there are quarantine restrictions placed on P. stewartii subsp. stewartii to avoid introduction of the bacterium into areas in which it does not occur.
PATHOGENICITY FACTORS
There are two main pathogenicity systems that contribute to Stewart's wilt disease caused by P. stewartii subsp. stewartii: (i) the Hrp T3SS; and (ii) stewartan EPS production. The visible symptomatology associated with the Hrp T3SS gene system involves the water‐soaked lesions observed in the early phase of infection on juvenile leaves of seedlings. The Hrp T3SS is also required for the systemic phase of the infection, where biofilm formation and EPS production also play a major role (Koutsoudis et al., 2006). It is this phase that leads to the systemic wilting of seedlings and the characteristic long wilted lesions on leaves, stunting and pith rot in mature plants. Both T3SS and the EPS pathogenicity system are critical for Stewart's wilt disease development (Beck von Bodman and Farrand, 1995; 1992a, 1992b; Dolph et al., 1988).
T3SS
Many Gram‐negative plant‐pathogenic bacteria possess a T3SS, which acts as a conduit through which bacteria can send their own proteins, called effectors, directly into eukaryotic cells. The delivery of effector proteins is essential for successful disease development in host plants, as they are often involved in the inhibition of host defences. However, in nonhost or resistant plants, the recognition of effector proteins or their subversive activities can instead elicit a second wave of defences, including the hypersensitive response (HR). In plant‐pathogenic bacteria, the hrp (HR and pathogenicity) gene cluster encodes a T3SS and effectors. For P. stewartii subsp. stewartii, this interaction seems to be important after the initial deposition of inoculum by the corn flea beetle into the intercellular spaces, where the bacteria are in close proximity and in contact with living host cells, or during xylem colonization, where the bacteria reside in later stages of the disease. Xylem is a nonliving tissue, but living parenchymal cells often form extrusions into the xylem lumen. These are subject to interactions with bacterial T3SS effectors, so that they may be an important source of added nutrients, enabling the pathogen to grow to high levels.
The P. stewartii subsp. stewartii hrp/hrc genes (previously named wts) are found in a 29‐kb gene cluster in the following map order: hrpN, hrpV, hrpT, hrcC, hrpG, hrpF, hrpE, hrpD, hrcJ, hrpB, hrpA, hrpS, hrpY, hrpX and hrpL. The genetic organization is very similar to that of the cognate gene clusters in P. agglomerans pv. gypsophilae and Erwinia amylovora (Frederick et al., 2001). The P. stewartii subsp. stewartii hrp locus is required to elicit HR in tobacco and Nicotiana benthamiana and to cause both water‐soaking and wilting in corn (1992a, 1992b; Ham et al., 2008).
The regulation of the hrp regulon relies on a complex network of HrpX, HrpY, HrpS and HrpL. The hrpX/hrpY operon encodes a two‐component signal transduction system, which constitutes the first step in the regulatory cascade controlling the hrp regulon (Merighi et al., 2003). HrpX is the sensor kinase and HrpY is the corresponding response regulator. It is not known what plant signals trigger the Hrp regulatory system, although it is induced in vitro in an apoplast‐mimicking minimal medium that is limiting in nitrogen and inorganic phosphate (Pi) and low in pH. The HrpX sensor has PAS domains that may enable it to sense the redox state of the bacterial cells. It is likely that some host‐specific signals are needed because the hrp genes are not fully induced in tobacco. In order to obtain an HR in this nonhost, the bacteria must be pre‐induced in inducing medium or hrp gene expression must be constitutively activated by the overexpression of HrpS on a plasmid.
The hrpX/hrpY operon is constitutively expressed at a low level. Phosphorylated HrpY then activates the expression of the hrpS gene (previously wtsA), which encodes a transcriptional enhancer. The HrpS transcriptional enhancer then activates hrpL, which encodes an alternative sigma factor that controls the expression of all the hrp and wtsE structural genes (Merighi et al., 2003). The hrpL–hrpX–hrpY–hrpS region is also autoregulated by a novel indirect mechanism that involves read‐through transcription from the hrpL promoter into hrpXY and hrpS, rather than directly autoregulating itself, which may be a mechanism that provides P. stewartii subsp. stewartii with the ability to rapidly turn on the expression of hrp genes when it initially enters the plant host (2005, 2006). Moreover, HrpS appears to be the point at which several specific and global post‐transcriptional and post‐translational regulators (RmsA and Lon protease; D. Coplin, Ohio State University, Columbus, Ohio, personal communication) converge to control hrp gene regulation, either directly or indirectly (Merighi et al., 2006), making HrpS a key step in the regulatory cascade that controls hrp gene expression (Fig. 3). hrpY mutants are totally nonpathogenic. Interestingly, peptone, alkaline pH and high osmolarity, hallmarks of extensively damaged plant tissue and insect guts, repress hrp gene expression in vitro. These conditions may represent environmental stimuli that turn off hrp gene expression in planta during the late phase of infection, or in the vector when T3SS is no longer needed (Merighi et al., 2006).
Figure 3.
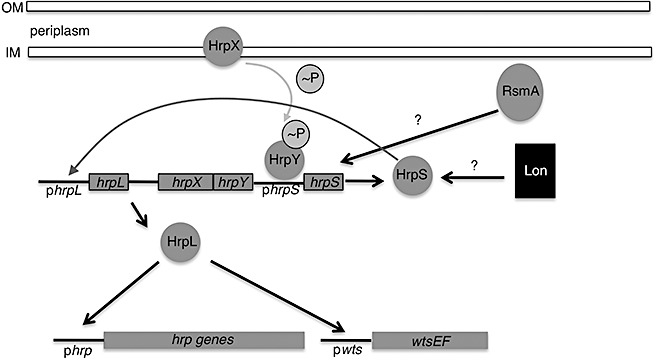
Model depicting the regulatory cascade of the Hrp system in Pnss. HrpX and HrpY constitute a two‐component regulatory system where HrpX is the sensor and HrpY is the corresponding response regulator. hrpXY expression is both constitutive and induced. HrpX transfers phosphate to HrpY and phosphorylated HrpY activates transcription of hrpS. HrpS is an enhancer binding protein that is needed for expression of hrpL. HrpL is an alternate sigma factor that activates transcription of both the entire hrp gene cluster and the wts gene cluster. The hrpS promoter appears to be a point in the cascade where several specific and global regulators (RsmA and Lon protease) converge to control hrp gene expression. In addition, there is a novel transcriptional autoregulatory mechanism of the hrpL/hrpX/hrpX/hrpS gene cluster that is triggered by HrpS (dark grey arrow) and may allow Pnss to rapidly turn on central hrp genes when it initially enters the plant. Figure adapted from Merighi et al. (2005). Note: Figure not drawn to scale.
Immediately adjacent to the hrp locus is a disease‐specific effector locus containing two genes, designated wtsE and wtsF (Ham et al., 2006). WtsE encodes a type III effector protein that belongs to the AvrE family of effector proteins that are conserved in the enteric plant pathogens and Pseudomonas syringae pathovars (E. amylovora, P. agglomerans pv. gypsophilae DspE and Pseudomonas syringae AvrE1 are members of this group). In all cases, the AvrE effectors are or can be essential for pathogenicity.
WtsE is secreted into the culture medium in an Hrp‐dependent manner. A mutation in wtsE completely abolishes pathogenicity (Ham et al., 2006). This species does not appear to have many other candidate effector proteins, indicating that it may have acquired its Hrp T3SS relatively recently. Moreover, no effectors, other than WtsE, have been found to contribute to virulence. WtsE is necessary for the development of early water‐soaked symptoms during the early phase of infection when P. stewartii subsp. stewartii still resides in the intercellular spaces of the plant host. Recombinant WtsE causes water‐soaking in sweet corn leaves and HR in tobacco when secreted through the heterologous Dickeya dadantii Hrp secretion system expressed in an E. coli background. It can also elicit HR in tobacco and N. benthamiana when delivered by Agrobacterium transient expression (D. Coplin and D. Mackey, Ohio State University, Columbus, Ohio, personal communication) (Ham et al., 2008). In addition, expression of WtsE is also lethal to yeast cells, indicating that it may be generally toxic to eukaryotic cells (Ham et al., 2008). WtsE is an extremely large protein (1830 amino acids) and is likely to be multifunctional. Ham et al. (2009) demonstrated that, when delivered by Pseudomonas syringae pv. phaseolicola into Arabidopsis leaves, WtsE is able to suppress basal defence responses, such as callose formation.
It has been reported recently that members of a family of type III effectors from animal pathogens (including E. coli, Shigella spp. and Salmonella spp.) can perturb functions in the host by mimicking constitutively active Ras‐like G‐proteins (Alto et al., 2006). These effectors have the same effects as activated G‐proteins, but do not bind guanine nucleotides or possess apparent GTPase domains, suggesting that they are functional and not structural mimics. They all contain one or more WxxxE motifs that are essential for their function as activated G‐protein mimics, as well as various C‐terminal subcellular localization signals. Pantoea stewartii subsp. stewartii WtsE (and other AvrE family effectors) all contain one or more WxxxE motifs and a putative C‐terminal endoplasmic reticulum membrane retention/retrieval signal (ERMRS) motif. Therefore, WtsE probably uses a similar, but not identical, mechanism to disrupt host cell functions as other type III effectors found in several animal pathosystems. Either of the two WtsE WxxxE motifs and the ERMRS motif are required for virulence in corn, HR elicitation in nonhosts and suppression of basal defences in Arabidopsis (Ham et al., 2009). Recently, the interaction of WtsE with several maize leucine‐rich repeat‐receptor‐like kinases (LRR‐RLKs) and two regulatory subunits of protein phosphatase 2A (PP2A) has been shown by yeast two‐hybrid assays and confirmed by a screen for host mutations that suppress WtsE lethality in yeast (Ham et al., 2008) (J. Ham, D. Mackey and D. Coplin, Ohio State University, Columbus, Ohio, personal communication). Thus, WtsE may be a broad host range cytotoxin that interferes with normal endoplasmic reticulum function and may manipulate host signal transduction pathways by mimicking activated plant G‐protein activity (Ham et al., 2009).
In addition, WtsF has been identified as a molecular chaperone for WtsE (Ham et al., 2006). WtsF is required for WtsE accumulation within the bacterial cell, possibly for protection against degradation by Lon protease, rather than being directly involved in the secretion of WtsE (Ham et al., 2006).
Pantoea stewartii subsp. stewartii produces harpin, encoded by the hrpN gene. The harpin protein is secreted into the medium in an Hrp‐dependent manner and shares biochemical similarities with other well‐characterized harpins produced by Erwinia spp. (Ahmad et al., 2001). HrpN can also cause HR in tobacco. However, a mutation in hrpN has no effect on pathogenicity, symptom severity or endophytic growth in corn, even when tested at different inoculum doses (Ahmad et al., 2001).
Stewartan EPS production
Stewartan EPS production is the major pathogenicity factor responsible for the vascular streaking, bacterial oozing and wilting characteristic of P. stewartii subsp. stewartii infection of sweet corn (Beck von Bodman and Farrand, 1995; Dolph et al., 1988). Stewartan is associated with the cells as both a bound capsule and a loose slime. Pantoea stewartii subsp. stewartii also produces large amounts of stewartan under in vitro conditions, but only if the culture medium contains fermentable sugars, such as glucose. The anionic stewartan polysaccharide is composed of galactose, glucose and glucuronic acid in a 3 : 3 : 1 ratio (Nimtz et al., 1996a; Yang et al., 1996). On the basis of predictions derived from the genome sequence, the mechanism of assembly of stewartan appears to be very closely related to colanic acid synthesis in E. coli (Carlier et al., 2009). Stewartan EPS is similar in structure and composition to amylovoran EPS produced by the closely related species, E. amylovora (1996a, 1996b). Moreover, the E. amylovora ams (amylovoran biosynthetic operon) and the P. stewartii subsp. stewartii wceI genes (formerly cps) are similar in nucleotide sequence and gene organization (Coplin et al., 1996) to the extent that some, but not all, of these functions are cross‐complementary (Bernhard et al., 1996); the genes that differ are primarily glycosyltransferases involved in the determination of the EPS structure.
Until recently, it was thought that the well‐characterized wceI gene cluster was solely responsible for stewartan production (Dolph et al., 1988). However, recent discoveries have identified two novel loci, denoted wceII and wceIII, that contribute to EPS production. wceII and wceIII lie outside of the original cps or wceI gene cluster. Interestingly, this entire tripartite biosynthetic system is controlled by the EsaI/EsaR quorum sensing (QS) system and the downstream multicomponent Rcs signal transduction system (Carlier et al., 2009).
QS and the Rcs environmental sensing phosphorelay
Pantoea stewartii subsp. stewartii possesses only one canonical QS system. esaI (a luxI homologue) encodes the signal (N‐3‐oxo‐hexanoyl homoserine lactone; AHL) synthase and esaR (a luxR homologue) encodes the response regulator. In the absence of the AHL signal, EsaR dimerizes and binds DNA. Therefore, under signal‐limiting or noninducing conditions, EsaR acts as a repressor and prevents transcription of its target genes. This is essentially the reverse of the luxI/luxR paradigm for the QS system described for Vibrio spp., where LuxR binds DNA and acts as a transcriptional activator only in the presence of the AHL ligand (von Bodman et al., 1998; Carlier and von Bodman, 2006; Minogue et al., 2002; Waters and Bassler, 2005).
Stewartan EPS production is a cell density‐dependent phenotype, in which the onset of stewartan production coincides with c. 3 × 108 cells/mL culture. Mutants that lack the EsaI AHL signal synthase remain repressed for EPS synthesis and are nonvirulent. The esaI mutant is locked into a low cell density mode. In contrast, mutant strains lacking a functional EsaR protein synthesize stewartan EPS constitutively, rendering the mutants hypermucoid. These mutants appear to be locked into a high cell density mode. Surprisingly, these EPS‐overproducing strains are significantly attenuated in virulence, even though they produce large amounts of the EPS pathogenicity factor. In both cases, the mutants cannot systemically colonize the xylem or form the spatially defined three‐dimensional biofilms typical of wild‐type P. stewartii subsp. stewartii (Koutsoudis et al., 2006). One can deduce from these observations that deregulated or developmentally premature EPS synthesis interferes with or blocks critical low cell density functions, such as surface adhesion (Koutsoudis et al., 2006). These observations demonstrate a very important point, namely that bacterial infection processes rely on a highly structured developmental program which, when perturbed, diminishes the colonization potential of the bacterium in question.
Although EsaR normally governs gene expression by repression and AHL‐dependent derepression, it also retains the ability to interact with RNA polymerase and functions as an activator of transcription similar to other LuxR homologues. Importantly, however, unlike LuxR‐type proteins, which bind DNA in an AHL ligand‐bound state, EsaR binds to the DNA in the nonligand‐bound state; regardless of whether it functions as a repressor or activator (von Bodman et al., 2003). It was first demonstrated that EsaR could activate the transcription of a reporter gene in an artificial E. coli system provided that the cognate DNA‐binding motif is positioned at the −42 site within a regulated promoter region, rather than at the −10 site position required for repressor activity (von Bodman et al., 2003; Schu et al., 2009). Recent work has identified EsaR activation of a native promoter upstream of a potential small regulatory RNA, confirming the biological relevance of EsaR activator activity in P. stewartii subsp. stewartii (Schu et al., 2009). The function of this small RNA, tentatively named EsaS, has yet to be determined, but it may play a role in gene regulation and/or contribute to the robustness of the QS regulatory scheme (Schu et al., 2009). The dual functionality of EsaR distinguishes itself from the AHL‐dependent activation paradigm of QS, in that it controls target genes at both low and high cell density, and hence serves as a central developmental switch in the developmental cycle of P. stewartii subsp. stewartii.
Unique to P. stewartii subsp. stewartii, the EsaI/EsaR QS system feeds into the Rcs phosphorelay regulatory network, which is an environmental monitoring system common to the Enterobacteriaceae (Fig. 4).This system was originally identified as a regulator of capsular polysaccharide synthesis in E. coli, hence the name ‘Rcs’ (Gottesman et al., 1985; Majdalani and Gottesman, 2005). The Rcs phosphorelay is a multicomponent signal transduction system comprising two sensor kinases, RcsC and RcsD, two co‐activators of transcription, RcsA and RcsB, and one membrane‐bound lipoprotein, RcsF, whose proposed function is to sense membrane perturbations (Majdalani and Gottesman, 2005). The conserved Rcs phosphorelay controls stewartan EPS production in P. stewartii subsp. stewartii (Torres‐Cabassa et al., 1987). However, unique to P. stewartii subsp. stewartii, the EsaI/EsaR QS system feeds into the Rcs phosphorelay by negatively regulating the expression of the rcsA gene, which encodes an essential transcriptional activation component of the well‐characterized two‐partite RcsA/RcsB response regulatory complex of the Rcs phosphorelay (Minogue et al., 2005). This finding represents the first demonstration that the QS cell–cell communication regulatory pathway converges with the environmental signal sensing Rcs regulatory network for the appropriate control of stewartan EPS synthesis. This dual control system ensures that EPS production occurs only when the bacteria sense conducive environmental conditions and the population has reached a threshold cell density. Indeed, poorly timed EPS production disrupts normal surface adhesion, biofilm formation and virulence (Koutsoudis et al., 2006). The specific environmental signals that stimulate the Rcs phosphorelay system remain undefined in P. stewartii subsp. stewartii and other bacteria, although conditions such as desiccation, which perturb the bacterial membrane structure, are proposed to be key factors. Interestingly, growth on a solid surface also activates the Rcs system, reiterating the link between the Rcs phosphorelay and its role in biofilm formation (Ferrieres and Clarke, 2003). The identification of the specific environmental signal(s) that triggers the Rcs phosphorelay remains an interesting area of research.
Figure 4.
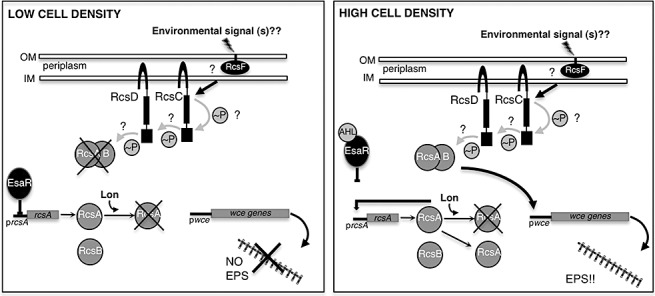
The EsaI/R quorum sensing pathway and the Rcs environmental sensing pathway converge to control stewartan EPS production. At low cell density, AHL ligand‐free EsaR binds to the promoter region of rcsA and represses transcription. There is also low, basal level expression of RcsA. However, degradation by Lon protease prevents significant RcsA accumulation (Gottesman and Stout, 1991) leaving insufficient RcsA to form the RcsA/B activation complex. At high cell density (3‐oxo‐C6‐HSL inducing conditions), the AHL ligand binds to EsaR which relieves EsaR repression of rcsA. RcsA production now exceeds the degradation capability of Lon protease. RcsA recruits RcsB and the RcsA/B heterodimer activates transcription of the wce gene cluster leading to production of stewartan EPS. The role of the remaining components of the Rcs phosphorelay are less clear in Pnss but are likely similar to what occurs in E. coli where rcsC and rcsD encode 2 sensor kinases that participate in a phosphorelay that transfers phosphate to the RcsA/B complex. In E.coli, RcsF is a membrane bound protein that is likely the receiver of an environmental signal (s) that serves as the trigger of the Rcs phosphorelay. The precise environmental signal (s) that stimulate the Rcs phoshorelay are unknown. Figure adapted from Minogue et al. (2005) and Madjalani and Gottesman (2005). Note: Figure not drawn to scale.
SURFACE‐BASED MOTILITY
Historically and taxonomically, P. stewartii subsp. stewartii has been classified as aflagellate and nonmotile (Pepper, 1967). However, recent discoveries have indicated that, in fact, P. stewartii subsp. stewartii exhibits robust flagellar‐mediated surface motility on semi‐solid agar medium (Herrera et al., 2008). Pantoea stewartii subsp. stewartii flagella were observed by electron microscopy and are relatively thin and fragile in comparison with flagella produced by other bacterial species (Fig. 5). Interestingly, although P. stewartii subsp. stewartii is motile on semi‐solid surfaces, it is unable to swim under planktonic conditions or in typical swimming assays. The fragility of the flagella and the lack of swimming motility may explain why this bacterium was previously classified as nonmotile and aflagellate. The P. stewartii subsp. stewartii genome sequence revealed several orthologues of flagellar biosynthetic and assembly genes (2003, 2006). The fliC gene, encoding the flagellin subunit protein, is required for flagellar production. Not surprisingly, fliC mutants are nonmotile (Herrera et al., 2008).
Figure 5.
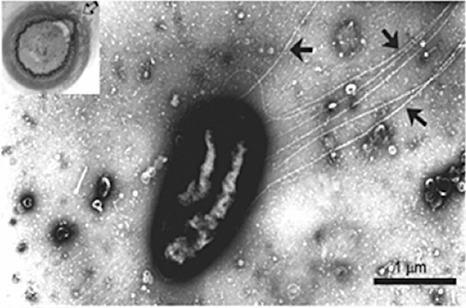
An individual Pnss cell displaying flagella. Cells were harvested from the swarming ring (arrow in left corner inset) where cells have transitioned into a vigorous motility mode. Flagella are indicated by the black arrows. Image used with permission from Herrera et al. (2008).
Biofilm formation usually requires some form of motility over solid surfaces, and P. stewartii subsp. stewartii has retained the ability to translocate over semi‐solid surfaces, indicating that this behaviour is important for the structured multicellular community development. Indeed, Herrera et al. (2008) demonstrated that this motility is critically important for bacterial aggregation, one of the early steps in the complex biofilm developmental cycle. Pantoea stewartii subsp. stewartii motility can be classified in general terms as swarming motility, which is defined as a surface‐based social migration. Curiously, P. stewartii subsp. stewartii swarms in a distinctly unidirectional pattern, which differs from classic swarming behaviour that usually progresses in a radial pattern. During colony morphogenesis, P. stewartii subsp. stewartii undergoes a distinct spatial organization around the periphery of the colony that eventually forms into a palisade ring‐like structure. This ring bursts open on one side, presumably from the force created by cells transitioning to the vigorous motility mode (Fig. 6). The position at which the ring breaks open dictates the direction in which the bacteria will migrate. The basis for the unidirectional motility in P. stewartii subsp. stewartii is unknown.
Figure 6.
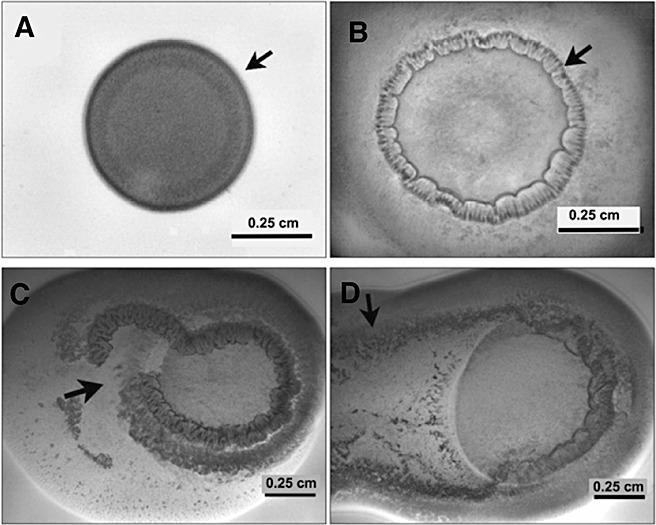
A time lapse series of images showing the different phases on Pnss motility following inoculation on a semi‐solid agar surface. A drop of a 107 cfu/mL suspension was placed on Nutrient agar containing 0.4% agar and 0.4% glucose. (A) A colony of Pnss after 5 h of incubation exhibiting the initiation of colony organization along the periphery (arrow). (B) Periphery of the colony organizing into a palisade ring‐like structure (arrow) after 10 h post‐inoculation. (C) Ring rupture (arrow) after 16 h post inoculation. (D) Rapid surface colonization (arrow) after 30 h. Images used with permission from Herrera et al. (2008).
In planta, fliC mutants are compromised in their ability to migrate from the point of inoculation; thus, flagellar‐based motility is required for the characteristic basipetal dissemination within the xylem tissue (Herrera et al., 2008).
PERSPECTIVES
To date, the only known pathogenicity factors for P. stewartii subsp. stewartii are the Hrp T3SS and stewartan EPS production. A cursory analysis of the recently partially sequenced P. stewartii subsp. stewartii (DC283) genome has provided insight into other potential virulence factors that may be employed by P. stewartii subsp. stewartii during plant infection, colonization and insect colonization (Glasner et al., 2006). Genes or gene systems that stand out as obvious candidates for virulence factors in the host include those encoding for host cell wall‐degrading enzymes and cytolytic toxins. Historically, P. stewartii subsp. stewartii is not known to produce plant cell wall‐degrading enzymes, but the presence of genes encoding several putative endoglucanases, xylanases and a β‐1,4–β‐1,3 mixed‐linkage glucan glucanohydrolase has prompted a more thorough biochemical study of the secretome. Preliminary biochemical data indicate that this bacterium has the potential to digest carbohydrates containing β‐1,4 and mixed‐linkage 1,3–1,4 β‐d‐glucan backbones associated with the plant cell wall (M. C. Roper, unpublished data). The latter is particularly relevant because corn cell walls contain a mixed‐linkage 1,3–1,4 β‐d‐glucan polymer unique to the Poaceae. These enzymes are probably critical for the bacteria to access the carbohydrates associated with the xylem cell wall structure and development. It has yet to be determined whether these enzymes play a role in virulence. There is also evidence that at least one cytolytic toxin is involved in the early stages of P. stewartii subsp. stewartii virulence, perhaps by contributing to the cell lysis observed during the water‐soaking phase of infection (M. C. Roper and S. B. von Bodman, unpublished data).
In conclusion, P. stewartii subsp. stewartii is an excellent bacterial pathosystem for studies involving host–pathogen interactions of xylem‐dwelling bacteria. Findings in the P. stewartii subsp. stewartii system may also be extrapolated to other xylem‐dwelling bacterial pathogens. Because of its interesting forms of gene regulation, P. stewartii subsp. stewartii is also fascinating from a microbiological perspective.
ACKNOWLEDGEMENTS
I would sincerely like to thank S. von Bodman, D. Coplin and J. Asselin for critical review of the manuscript and their helpful suggestions and perspectives.
REFERENCES
- Agrios, G.N. (2005) Plant Pathology. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Ahmad, M. , Majerczak, D.R. , Pike, S. , Hoyos, M.E. , Novacky, A. and Coplin, D.L. (2001) Biological activity of harpin produced by Pantoea stewartii subsp. stewartii . Mol. Plant–Microbe Interact. 14, 1223–1234. [DOI] [PubMed] [Google Scholar]
- Alto, N.M. , Shao, F. , Lazar, C.S. , Brost, R.L. , Chua, G. , Mattoo, S. , McMahon, S.A. , Ghosh, P. , Hughes, T.R. , Boone, C. and Dixon, J.E. (2006) Identification of a bacterial type III effector family with G protein mimicry functions. Cell, 124, 133–145. [DOI] [PubMed] [Google Scholar]
- Azad, H.R. , Holmes, G.J. and Cooksey, D.A. (2000) A new leaf blotch disease of sudangrass caused by Pantoea ananas and Pantoea stewartii . Plant Dis. 84, 973–979. [DOI] [PubMed] [Google Scholar]
- Beck von Bodman, S. and Farrand, S.K. (1995) Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N‐acylhomoserine lactone autoinducer. J. Bacteriol. 177, 5000–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard, F. , Schullerus, D. , Bellemann, P. , Nimtz, M. , Coplin, D.L. and Geider, K. (1996) Genetic transfer of amylovoran and stewartan synthesis between Erwinia amylovora and Erwinia stewartii . Microbiol. SGM, 142, 1087–1096. [DOI] [PubMed] [Google Scholar]
- von Bodman, S.B. , Majerczak, D.R. and Coplin, D.L. (1998) A negative regulator mediates quorum‐sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii . Proc. Natl. Acad. Sci. USA, 95, 7687–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bodman, S.B. , Ball, J.K. , Faini, M.A. , Herrera, C.M. , Minogue, T.D. , Urbanowski, M.L. and Stevens, A.M. (2003) The quorum sensing negative regulators EsaR and ExpR(Ecc), homologues within the LuxR family, retain the ability to function as activators of transcription. J. Bacteriol. 185, 7001–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier, A. , Burbank, L. and von Bodman, S.B. (2009) Identification and characterization of three novel EsaI/EsaR quorum‐sensing controlled stewartan exopolysaccharide biosynthetic genes in Pantoea stewartii ssp. stewartii . Mol. Microbiol. 74, 903–913. [DOI] [PubMed] [Google Scholar]
- Carlier, A.L. and von Bodman, S.B. (2006) The rcsA promoter of Pantoea stewartii subsp. stewartii features a low‐level constitutive promoter and an EsaR quorum‐sensing‐regulated promoter. J. Bacteriol. 188, 4581–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claflin, L.E. (2000) Stewart's bacterial wilt In: Compendium of Corn Diseases (White D.G., ed.), pp. 3–4. St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Coplin, D.L. , Frederick, R.D. and Majerczak, D.R. (1992a) New pathogenicity loci in Erwinia stewartii identified by random Tn5 mutagenesis and molecular‐cloning. Mol. Plant–Microbe Interact. 5, 266–268. [Google Scholar]
- Coplin, D.L. , Frederick, R.D. , Majerczak, D.R. and Tuttle, L.D. (1992b) Characterization of a gene cluster that specifies pathogenicity in Erwinia stewartii . Mol. Plant–Microbe Interact. 5, 81–88. [Google Scholar]
- Coplin, D.L. , Majerczak, D.R. , Bugert, P. and Geider, K. (1996) Nucleotide sequence analysis of the Erwinia stewartii cps gene cluster for synthesis of stewartan and comparison to the Erwinia amylovora ams cluster for synthesis of amylovoran. Acta Hortic. 411, 251–257. [Google Scholar]
- Correa, V.R. , Majerczak, D.R. , Ammar, E. , Merighi, M. , Coplin, D.L. , Pratt, R.C. , Redinbaugh, M.G. and Hogenhout, S.A. (2008) Characterization of a Pantoea stewartii TTSS gene required for persistence in its flea beetle vector. Phytopathology, 98, S41. [Google Scholar]
- Dolph, P.J. , Majerczak, D.R. and Coplin, D.L. (1988) Characterization of a gene cluster for exopolysaccharide biosynthesis and virulence in Erwinia stewartii . J. Bacteriol. 170, 865–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrieres, L. and Clarke, D.J. (2003) The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K‐12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50, 1665–1682. [DOI] [PubMed] [Google Scholar]
- Frederick, R.D. , Ahmad, M. , Majerczak, D.R. , Arroyo‐Rodriguez, A.S. , Manulis, S. and Coplin, D.L. (2001) Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol. Plant–Microbe Interact. 14, 1213–1222. [DOI] [PubMed] [Google Scholar]
- Freeman, N.D. and Pataky, J.K. (2001) Levels of Stewart's wilt resistance necessary to prevent reductions in yield of sweet corn hybrids. Plant Dis. 85, 1278–1284. [DOI] [PubMed] [Google Scholar]
- Glasner, J.D. , Liss, P. , Plunkett, G., 3rd , Darling, A. , Prasad, T. , Rusch, M. , Byrnes, A. , Gilson, M. , Biehl, B. , Blattner, F.R. and Perna, N.T. (2003) ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31, 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasner, J.D. , Rusch, M. , Liss, P. , Plunkett, G., 3rd , Cabot, E.L. , Darling, A. , Anderson, B.D. , Infield‐Harm, P. , Gilson, M.C. and Perna, N.T. (2006) ASAP: a resource for annotating, curating, comparing, and disseminating genomic data. Nucleic Acids Res. 34, D41–D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman, S. and Stout, V. (1991) Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5, 1599–1606. [DOI] [PubMed] [Google Scholar]
- Gottesman, S. , Trisler, P. and Torrescabassa, A. (1985) Regulation of capsular polysaccharide synthesis in Escherichia‐coli K‐12—characterization of 3 regulatory genes. J. Bacteriol. 162, 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham, J.H. , Majerczak, D.R. , Arroyo‐Rodriguez, A.S. , Mackey, D.M. and Coplin, D.L. (2006) WtsE, an AvrE‐family effector protein from Pantoea stewartii subsp. stewartii, causes disease‐associated cell death in corn and requires a chaperone protein for stability. Mol. Plant–Microbe Interact. 19, 1092–1102. [DOI] [PubMed] [Google Scholar]
- Ham, J.H. , Majerczak, D. , Ewert, S. , Sreerekha, M.V. , Mackey, D. and Coplin, D. (2008) WtsE, an AvrE‐family type III effector protein of Pantoea stewartii subsp stewartii, causes cell death in non‐host plants. Mol. Plant Pathol. 9, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham, J.H. , Majerczak, D.R. , Nomura, K. , Mecey, C. , Uribe, F. , He, S.Y. , Mackey, D. and Coplin, D.L. (2009) Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Mol. Plant–Microbe Interact. 22, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera, C.M. , Koutsoudis, M.D. , Wang, X. and von Bodman, S.B. (2008) Pantoea stewartii subsp. stewartii exhibits surface motility, which is a critical aspect of Stewart's wilt disease development on maize. Mol. Plant–Microbe Interact. 21, 1359–1370. [DOI] [PubMed] [Google Scholar]
- Koutsoudis, M.D. , Tsaltas, D. , Minogue, T.D. and von Bodman, S.B. (2006) Quorum‐sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii . Proc. Natl. Acad. Sci. USA, 103, 5983–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamka, G.L. , Hill, J.H. , Mcgee, D.C. and Braun, E.J. (1991) Development of an immunosorbent‐assay for seed‐borne Erwinia stewartii in corn seeds. Phytopathology, 81, 839–846. [Google Scholar]
- Leigh, J.A. and Coplin, D.L. (1992) Exopolysaccharides in plant–bacterial interactions. Annu. Rev. Microbiol. 46, 307–346. [DOI] [PubMed] [Google Scholar]
- Majdalani, N. and Gottesman, S. (2005) The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59, 379–405. [DOI] [PubMed] [Google Scholar]
- Menclas, B. , Block, C.C. , Esker, P.D. and Nutter, F.W. (2006) Quantifying the feeding periods required by corn flea beetles to acquire and transmit Pantoea stewartii . Plant Dis. 90, 319–324. [DOI] [PubMed] [Google Scholar]
- Mergaert, J. , Verdonck, L. and Kersters, K. (1993) Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartii to the Genus Pantoea emend. as Pantoea ananas (Serrano 1928) comb. nov. and Pantoea stewartii (Smith 1898) comb, nov., respectively, and description of Pantoea stewartii subsp. indologenes subsp. nov. Int. J. Syst. Microbiol. 43, 162–173. [Google Scholar]
- Merighi, M. , Majerczak, D.R. , Stover, E.H. and Coplin, D.L. (2003) The HrpX/HrpY two‐component system activates hrpS expression, the first step in the regulatory cascade controlling the Hrp regulon in Pantoea stewartii subsp. stewartii . Mol. Plant–Microbe Interact. 16, 238–248. [DOI] [PubMed] [Google Scholar]
- Merighi, M. , Majerczak, D.R. and Coplin, D.L. (2005) A novel transcriptional autoregulatory loop enhances expression of the Pantoea stewartii subsp. stewartii Hrp type III secretion system. FEMS Microbiol. Lett. 243, 479–487. [DOI] [PubMed] [Google Scholar]
- Merighi, M. , Majerczak, D.R. , Zianni, M. , Tessanne, K. and Coplin, D.L. (2006) Molecular characterization of Pantoea stewartii subsp. stewartii HrpY, a conserved response regulator of the Hrp type III secretion system, and its interaction with the hrpS promoter. J. Bacteriol. 188, 5089–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener, P.M. , Pataky, J.K. and White, D.G. (2002) Rates of transmitting Erwinia stewartii from seed to seedlings of a sweet corn hybrid susceptible to Stewart's wilt. Plant Dis. 86, 1031–1035. [DOI] [PubMed] [Google Scholar]
- Minogue, T.D. , Wehland‐von Trebra, M. , Bernhard, F. and von Bodman, S.B. (2002) The autoregulatory role of EsaR, a quorum‐sensing regulator in Pantoea stewartii ssp stewartii: evidence for a repressor function. Mol. Microbiol. 44, 1625–1635. [DOI] [PubMed] [Google Scholar]
- Minogue, T.D. , Carlier, A.L. , Koutsoudis, M.D. and von Bodman, S.B. (2005) The cell density‐dependent expression of stewartan exopolysaccharide in Pantoea stewartii ssp. stewartii is a function of EsaR‐mediated repression of the rcsA gene. Mol. Microbiol. 56, 189–203. [DOI] [PubMed] [Google Scholar]
- Nimtz, M. , Mort, A. , Domke, T. , Wray, V. , Zhang, Y. , Qiu, F. , Coplin, D. and Geider, K. (1996a) Structure of amylovoran, the capsular exopolysaccharide from the fire blight pathogen Erwinia amylovora . Carbohydr. Res. 287, 59–76. [DOI] [PubMed] [Google Scholar]
- Nimtz, M. , Mort, A. , Wray, V. , Domke, T. , Zhang, Y. , Coplin, D.L. and Geider, K. (1996b) Structure of stewartan, the capsular exopolysaccharide from the corn pathogen Erwinia stewartii . Carbohydr. Res. 288, 189–201. [DOI] [PubMed] [Google Scholar]
- Pataky, J.K. (2003) Stewart's wilt of corn. Available at: http://www.apsnet.org/publications/apsnetfeatures/Pages/StewartsWilt.aspx
- Pataky, J.K. (2004) Stewart's wilt of corn. The Plant Health Instructor. DOI:10.1094/PHI‐I‐2004‐0113‐01. Available at: http://www.apsnet.org/edcenter/intropp/lessons/prokaryotes/Pages/StewartWilt.aspx
- Pataky, J.K. and Headrick, J.M. (1988) Classification of sweet corn hybrid reactions to common rust, northern leaf‐blight, stewart wilt, and goss wilt and associated yield reductions. Phytopathology, 78, 172–178. [Google Scholar]
- Pataky, J.K. , Bohn, M.O. , Lutz, J.D. and Richter, P.M. (2008) Selection for quantitative trait loci associated with resistance to Stewart's wilt in sweet corn. Phytopathology, 98, 469–474. [DOI] [PubMed] [Google Scholar]
- Pepper, E.H. (1967) Stewart's bacterial wilt of corn. Monograph 4. American Phytopathological Society, St. Paul, MN, USA.
- Rand, F.V. and Cash, L. (1921) Stewarts disease of corn. J. Agric. Res. 21, A0262–A0263. [Google Scholar]
- Robert, A.L. (1955) Bacterial wilt and Stewart's leaf blight of corn. USDA Farmer's Bull. No. 2092. [Google Scholar]
- Schu, D.J. , Carlier, A.L. , Jamison, K.P. , von Bodman, S. and Stevens, A.M. (2009) Structure/function analysis of the Pantoea stewartii quorum‐sensing regulator EsaR as an activator of transcription. J. Bacteriol. 191, 7402–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, N.E. (1934) Stewart's disease in relation to winter temperatures. Plant Dis. Rep. 18, 141–149. [Google Scholar]
- Stewart, F.C. (1897) A bacterial disease of sweet corn. N. Y. Agric. Exp. Sta. Bull. 130, 422–439. [Google Scholar]
- Torres‐Cabassa, A. , Gottesman, S. , Frederick, R.D. , Dolph, P.J. and Coplin, D.L. (1987) Control of extracellular polysaccharide synthesis in Erwinia stewartii and Escherichia coli K‐12—a common regulatory function. J. Bacteriol. 169, 4525–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, C.M. and Bassler, B.L. (2005) Quorum sensing: cell‐to‐cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. [DOI] [PubMed] [Google Scholar]
- Yang, B.Y. , Gray, J.S. and Montgomery, R. (1996) The structure of stewartan, a capsular polysaccharide produced by Erwinia stewartii strain DC283. Carbohydr. Res. 296, 183–201. [DOI] [PubMed] [Google Scholar]


