Abstract
Background
OXA-2 is a class D β-lactamase that confers resistance to penicillins, as well as narrow-spectrum cephalosporins. OXA-2 was recently reported to also possess carbapenem-hydrolysing activity. Here, we describe a KPC-2-encoding Klebsiella pneumoniae isolate that demonstrated reduced susceptibility to ceftazidime and ertapenem due to production of OXA-2.
Objectives
To elucidate the role of OXA-2 production in reduced ceftazidime and ertapenem susceptibility in a K. pneumoniae ST258 clinical isolate.
Methods
MICs were determined by the agar dilution method. WGS was conducted to identify and compare resistance genes between isolates. Expression of KPC-2 was quantified by quantitative RT–PCR and immunoblotting. OXA-2 was expressed in Escherichia coli TOP10, as well as in K. pneumoniae ATCC 13883, to define the relative contribution of OXA-2 in β-lactam resistance. Kinetic studies were conducted using purified OXA-2 enzyme.
Results
K. pneumoniae 1761 belonged to ST258 and carried both blaKPC-2 and blaOXA-2. However, expression of blaKPC-2 was substantially reduced due to an IS1294 insertion in the promoter region. K. pneumoniae 1761, K. pneumoniae ATCC 13883 and E. coli TOP10 carrying blaOXA-2-harbouring plasmids showed reduced susceptibility to ertapenem and ceftazidime, but meropenem, imipenem and cefepime were unaffected. blaOXA-2 was carried on a 2910 bp partial class 1 integron containing aacA4-blaOXA-2-qacEΔ1-sul1 on an IncA/C2 plasmid, which was not present in the earlier ST258 isolates possessing blaKPC-2 with intact promoters. Hydrolysis of ertapenem by OXA-2 was confirmed using purified enzyme.
Conclusions
Production of OXA-2 was associated with reduced ceftazidime and ertapenem susceptibility in a K. pneumoniae ST258 isolate.
Introduction
β-Lactams are the most widely used antimicrobial agents, with carbapenems often being last-resort agents in the treatment of infections among critically ill patients. However, multiple carbapenem resistance mechanisms have emerged among Enterobacteriaceae in recent years, putting at jeopardy our antimicrobial armamentarium. While production of KPC, NDM and OXA-48-group carbapenemases is considered to be the major carbapenem resistance mechanism among Enterobacteriaceae, the roles of other less common carbapenemases are yet to be elucidated. OXA-2 is a class D β-lactamase primarily found in Pseudomonas aeruginosa.1 It was the first class D β-lactamase to be described in 1965 and is considered to be a narrow-spectrum β-lactamase.2 Recently, OXA-2 was shown to also hydrolyse carbapenems and when expressed in Acinetobacter baumannii resulted in resistance to meropenem and doripenem. Thus, it was proposed that OXA-2 could be considered a carbapenemase when produced in A. baumannii.3 Here, we report a blaKPC-2-positive clinical Klebsiella pneumoniae ST258 isolate whose reduced susceptibility to ceftazidime and ertapenem was attributed to production of OXA-2 rather than KPC-2.
Methods
Bacterial isolates
K. pneumoniae isolates 1081, 1701 and 1761 were identified in blood (1081 and 1701) and bronchoalveolar lavage (1761) cultures from a patient admitted to a hospital in Western Pennsylvania in 2016.
WGS
K. pneumoniae isolates 1081, 1701 and 1761 were subjected to WGS by NextSeq (Illumina, San Diego, CA, USA) using the paired-end method. Additionally, isolates 1701 and 1761 were sequenced using the MinION platform (Oxford Nanopore Technologies, Oxford Science Park, UK). Illumina reads were assembled de novo using CLC Genomics Workbench v11 (CLC Bio, Aarhus, Denmark). Hybrid assembly of Illumina and MinION reads was performed using Unicycler v0.4.6 with default parameters.4 Draft genome assemblies of isolates 1081, 1701 and 1761 are deposited in the NCBI database under accession numbers QUWO00000000, CP039968-CP039973 and CP039974-CP039987, respectively. Sequence Read Archive (SRA) accession numbers are SRR7784156, SRR7784155 and SRR7784157.
STs were determined in silico using the online MLST database (https://pubmlst.org/kpneumoniae/). ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) was used to identify antimicrobial resistance genes.5 PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/) was used to identify plasmid incompatibility types. Pairwise SNP differences between the genomes were identified using snippy (https://github.com/tseemann/snippy) and processed using custom python and Stata (StataCorp LP, College Station, TX, USA) scripts with isolate 1081 used as an internal reference. Potentially recombinant SNP clusters (more than five SNPs per kb) and SNPs located on poorly assembled contigs were excluded.
Generation of Escherichia coli TOP10 transformants
Native plasmids were extracted from K. pneumoniae 1761 by standard phenol–chloroform extraction and used to transform E. coli TOP10 by electroporation. The transformants were selected on lysogenic broth agar plates containing 50 mg/L ampicillin. Additionally, the native plasmids were partially digested with Sau3AI and cloned into the BamHI restriction site of cloning vector pBC-SK(−). Transformants with recombinant plasmids were selected on lysogenic broth agar plates with 30 mg/L chloramphenicol and 50 mg/L ampicillin. Nucleotide sequences were determined by primer walking with Sanger sequencing. blaOXA-1 and blaOXA-10 were amplified by PCR with the primers listed in the Supplementary data available at JAC Online. PCR products were digested with XbaI and HindIII, and EcoRI and HindIII, respectively, and ligated to pBC-SK(−). E. coli TOP10 was transformed with the ligated product by electroporation and sequences were confirmed with Sanger sequencing.
Generation of K. pneumoniae ATCC 13883 transformants
pOXA-2, consisting of pBC-SK(−) and a 2.9 kb insert containing blaOXA-2, was transferred to K. pneumoniae ATCC 13883 by electroporation. The transformants were selected on lysogenic broth agar plates containing 30 mg/L chloramphenicol. The presence of blaOXA-2 was confirmed by PCR. pBC-SK(−) plasmids containing blaOXA-1 and blaOXA-10 were transferred to K. pneumoniae ATCC 13883 by electroporation. Transformants were identified by growth on lysogenic broth agar plates containing chloramphenicol and the presence of blaOXA-1 and blaOXA-10 was confirmed by PCR. Empty vector controls were also generated.
Susceptibility testing
The MICs of cephalosporins (cefotaxime, ceftazidime and cefepime) and carbapenems (ertapenem and meropenem) were determined by the standard agar dilution method using Mueller–Hinton agar plates. E. coli ATCC 25922 was used as the quality control strain. MICs for constructs generated in K. pneumoniae ATCC 13883 and E. coli TOP10 were also determined by Etest using strips for ertapenem, meropenem, imipenem, cefepime and ceftazidime. Strains carrying empty vector were used as controls.
Quantitative RT–PCR (qRT–PCR)
qRT–PCR was performed using the Power SYBR green RNA-to-CT 1-step kit (Thermo Fisher Scientific, Pittsburgh, PA, USA). Twenty microlitre reactions were performed in a 96-well format and reaction mixtures contained 200 nM final concentrations of rplB, blaOXA-2 or blaKPC-2 primers (primer sequences are available as Supplementary data at JAC Online) and the reaction mix diluted to 1× RNA was then added to a final amount of 50 ng per reaction. The cycle conditions were as follows: 1 cycle at 48°C for 30 min, 1 cycle at 95°C for 10 min and 40 cycles of 95°C for 15 s and 56°C for 1 min. Melting curves were generated by 80 cycles of 60°C for 15 s with 0.5°C increments. The results were analysed using the comparative CT method (2−ΔΔCT), where relative amounts of RNA transcript were normalized to that of the rplB gene.6 Changes in fluorescence were monitored using an Applied Biosystems 7500 real-time system (Thermo). No template and no reverse transcriptase controls were included in each run. qRT–PCRs were performed in triplicate at least twice with comparable results.
Immunoblotting for KPC carbapenemase
For one-dimensional electrophoresis, 10 μg of membrane proteins was separated by SDS–PAGE with a Proteon II xi Cell gel apparatus (Bio-Rad, Hercules, CA, USA). Gels were either stained with Coomassie or transferred to nitrocellulose with a Bio-Rad Trans Blot Cell (60 V for 1.5 h at 4°C).7 After transfer, proteins were visualized by amido blue (0.1% amido blue dye in 1.0% acetic acid) and standards were marked. Membranes were blocked with blocking buffer [5% (w/v) non-fat dry milk in 1× Tris-buffered saline with 0.1% Tween 20 (TBS-T20)] overnight at 4°C and probed with a polyclonal KPC rabbit antibody (1:5000) in blocking buffer for 1 h at 25°C. The membrane was washed with 1× TBS-T20 three times for 5 min. After washing, the membrane was probed with a goat anti-rabbit horseradish peroxidase (‘HRP’)-conjugated IgG antibody (1:10000) (Biorbyt) for 1 h at 25°C. Membranes were incubated with Clarity Western ECL Substrate (Bio-Rad) in accordance with the manufacturer's specifications and visualized on a ChemiDoc XRS+ System (Bio-Rad).7
Kinetic measurements of OXA-2
E. coli BL21 (DE33) cells harbouring pET SUMO carrying blaOXA-2 were grown in lysogenic broth supplemented with 50 mg/L kanamycin until an OD600 of 0.8 was achieved. Expression was induced with 1 mM IPTG for 2 h at 37°C. The bacterial cells were harvested by centrifugation, resuspended in 1× PBS and disrupted by sonication. The cell extract was then centrifuged for 10 min at 8000 g and 4°C. The recovered supernatant was mixed with HisPur Cobalt Resin (Thermo Fisher) and incubated for 1 h at room temperature. The cell lysate/resin mixture was then loaded onto a Poly-Prep chromatography column (Bio-Rad), which was equilibrated with 500 mM sodium chloride buffer supplemented with 20 mM sodium phosphate and 30 mM imidazole. After washing, the protein was eluted with a linear gradient of 500 mM imidazole. Fractions containing pure enzyme were identified with nitrocefin and SDS–PAGE. The enzyme was concentrated by buffer exchange using Amicon® Ultra centrifugal filters 10MWCO (Millipore). The protein was >90% pure as determined by SDS–PAGE.
Kinetics were measured on a V-750 UV-Visible Spectrophotometer (JASCO) at room temperature in 100 mM sodium phosphate buffer (pH 7.0) supplemented with 50 mM sodium bicarbonate. Substrate hydrolysis was measured at 260 nm for oxacillin and 295 nm for ertapenem. Enzyme kinetic data were analysed with GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA) and fitted to the Michaelis–Menten equation.
Results
Resistance phenotype of K. pneumoniae 1761 and the prior isolates
K. pneumoniae isolate 1761 was identified in a bronchoalveolar lavage culture from a hospitalized long-term care facility resident. The patient had a history of multiple hospitalizations for recurrent infections due to carbapenem-resistant, KPC-producing K. pneumoniae and was treated with multiple antimicrobial agents. Over the course of 2 years, a total of 20 K. pneumoniae strains were collected from the patient.
Unlike the preceding K. pneumoniae isolates, isolate 1761 was susceptible to cefepime and ertapenem. MICs by agar dilution were 0.125 and 0.5 mg/L, respectively, yet PCR for blaKPC was positive (Table 1). Of the 20 K. pneumoniae isolates available from the patient, two additional isolates representing unique infection episodes were selected for further analysis: isolate 1701, isolated 80 days prior to isolate 1761; and isolate 1081, the original infecting isolate that was isolated 2 years prior to isolate 1761. Isolates 1081 and 1701 were isolated from blood, while isolate 1761 was obtained from bronchoalveolar lavage as described above.
Table 1.
Characteristics of K. pneumoniae clinical isolates used in the study
| Isolate | Source | Date | MIC (mg/L) |
ST | β-Lactamases | ||||
|---|---|---|---|---|---|---|---|---|---|
| ETP | MEM | FEP | CTX | CAZ | |||||
| 1081 | blood | September 2014 | 16 | 8 | 16 | 16 | 32 | ST258 | KPC-2, OXA-9, SHV-11, TEM-1 |
| 1701 | blood | October 2016 | 16 | 8 | 16 | 16 | 32 | ST258 | KPC-2, SHV-11 |
| 1761 | BAL | December 2016 | 0.5 | 0.125 | 0.125 | 0.125 | 4 | ST258 | KPC-2, OXA-2, SHV-11 |
ETP, ertapenem; MEM, meropenem; FEP, cefepime; CTX, cefotaxime; CAZ, ceftazidime.
Detection of SNPs and β-lactamase genes by WGS
All isolates belonged to K. pneumoniae ST258, a high-risk epidemic clone with worldwide distribution and close association with blaKPC carriage.8 There were >200 non-recombinant core genome SNPs between the initial isolate 1081, identified in 2014, and isolates 1701 and 1761, both isolated in 2016. Isolate 1761 had 17 non-recombinant SNPs compared with isolate 1701 (Supplementary data available at JAC Online). Isolates 1081, 1701 and 1761 all possessed blaKPC-2 encoding carbapenemase and blaSHV-11 encoding a non-ESBL. Additionally, isolate 1081 possessed blaTEM-1 and blaOXA-9, while isolate 1761 possessed blaOXA-2, all encoding non-ESBLs. These findings suggest that isolate 1081, the original infecting strain, persisted and evolved over 2 years to give rise to isolates 1701 and 1761.
Overall, the broad β-lactam resistance phenotype of isolates 1081 and 1701, which included carbapenems, was consistent with production of the KPC-2 carbapenemase. In contrast, despite possessing blaKPC-2, the MICs of ertapenem and ceftazidime for isolate 1761 were in the susceptible ranges, though higher than the WT ranges, at 0.5 and 4 mg/L, respectively.
KPC-2 expression in K. pneumoniae 1761
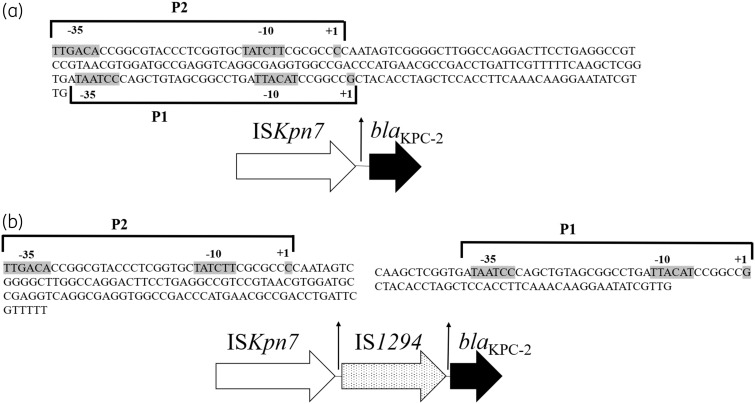
The expression of blaKPC-2 as determined by qRT–PCR was 56210 ± 1811 and 26646 ± 1273 copies/genome for isolates 1701 and 1761, respectively. Western blot showed robust expression of the KPC-2 protein by isolate 1701, but not by isolate 1761 (Figure 1). To investigate the mechanism underlying decreased KPC-2 expression in isolate 1761, MinION sequencing of isolates 1701 and 1761 was conducted to examine the blaKPC-2 genetic environment, which could not be resolved by Illumina sequencing alone. In isolate 1701, blaKPC was found to be part of a Tn4401 element and was located on a plasmid (p1701_01) that was 168369 bp in size and contained two distinct replicons, IncFIB(K) and IncFII(K). The overall structure of the plasmid was highly similar (97% query coverage and 99.9% maximum nucleotide identity by Blast) to that of the plasmid pBK32179 (GenBank accession number JX430448.1), harboured by carbapenem-resistant ST258 K. pneumoniae strain BK32179, which was isolated from a urine culture in 2010 from a patient in New York.9 In contrast, the KPC-2-encoding plasmid in isolate 1761 appeared to be fused with another plasmid originally detected in isolate 1701, which possessed a class 1 integron containing dfrA12-aadA2-qacEΔ1-sul1. This resulted in a chimeric plasmid (p1761_01) measuring 282961 bp in size, which contained four distinct predicted rep genes [IncFIB(K), IncFII(K), IncFII(Yp) and IncR]. Importantly, insertion of an IS1294 element was identified between the P2 and P1 promoter sites upstream of blaKPC-2 (Figure 2a and b). P2 has been reported as the primary promoter driving expression of blaKPC.10
Figure 1.
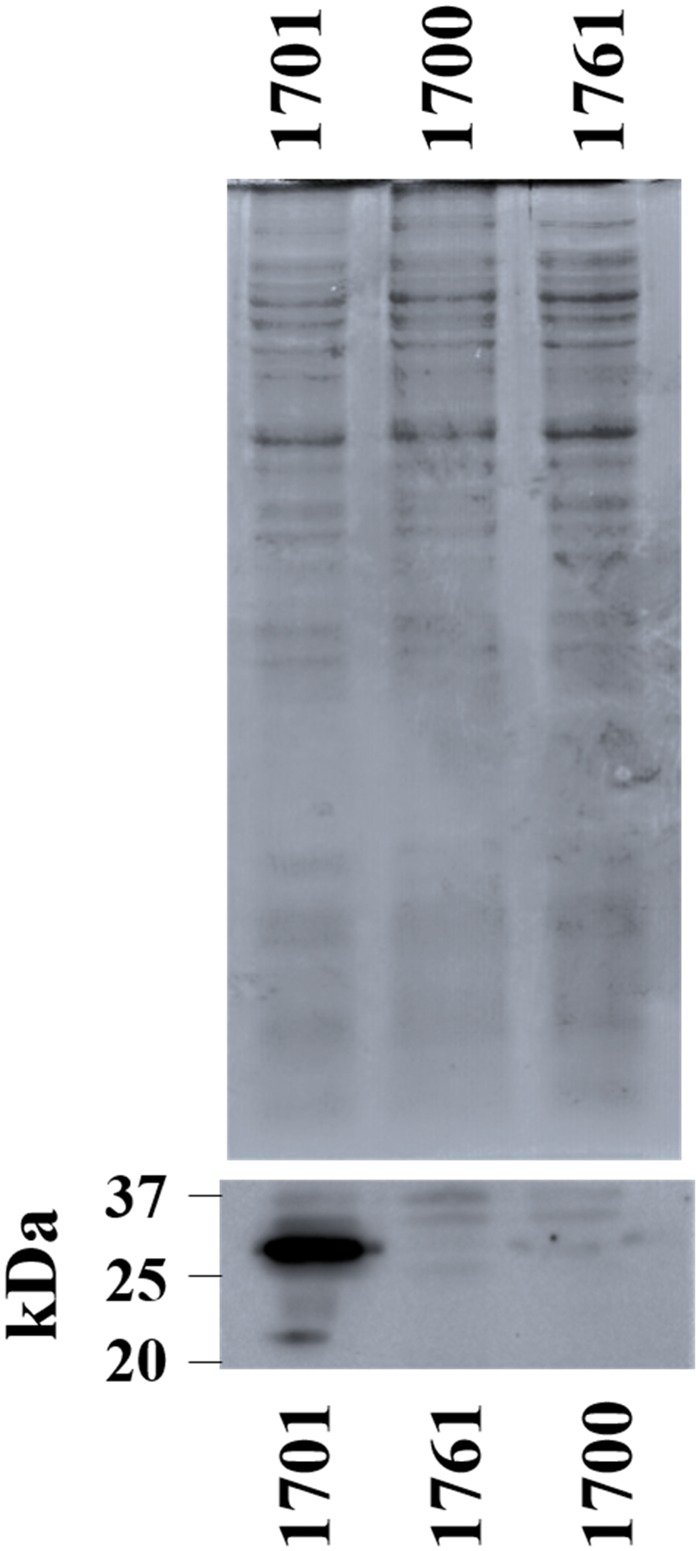
Western blot of KPC in clinical isolates 1701, 1700 and 1761. Total cell lysate (10 μg) was separated by SDS–PAGE and gels were either stained with Coomassie (top) or immunoblotted with a KPC-specific antibody (bottom). Isolate 1700 is a CTX-M-producing K. pneumoniae isolate identified from the same patient and was included as a negative control.
Figure 2.
Schematic of the blaKPC-2 promoter region of isolates 1701 (a) and 1761 (b).
OXA-2 as a cause of reduced ceftazidime and ertapenem susceptibility in K. pneumoniae 1761
We suspected that the IS1294 insertion between the P2 promoter and the blaKPC-2 coding sequence likely accounted for the discrepancy between the presence of an intact blaKPC-2 gene and susceptibility to cephalosporins and carbapenems in isolate 1761. To identify an alternative β-lactamase gene that might be causing reduced ceftazidime and ertapenem susceptibility, functional genomic cloning was conducted, which yielded a pBC-SK(−) construct containing a 2910 bp insert carrying blaOXA-2 (pOXA-2). Expression of OXA-2 was confirmed by qRT–PCR, showing an expression level of 1.8 × 108 copies/genome. E. coli TOP10 (pOXA-2) had an ertapenem MIC of 0.016 mg/L (2-fold increase compared with vector-only control) and a ceftazidime MIC of 4 mg/L (16-fold increase), indicating that OXA-2 production was associated with reduced susceptibility to ceftazidime and ertapenem. However, production of OXA-2 did not impact the MICs of cefepime, imipenem or meropenem (Table 2). For comparison, we also evaluated the impact of OXA-1 and OXA-10, two representatives of other OXA subgroups. E. coli TOP10 producing OXA-1 and OXA-10 did not show changes in the MICs of carbapenems or ceftazidime when compared with empty vector controls (Table 3). Through MinION sequencing we determined that blaOXA-2 was part of a class 1 integron consisting of aacA4-blaOXA-2-qacEΔ1-sul1, which was located on an IncA/C2-like plasmid (p1761_02) and shared 100% identity over 150 kb with pCR14-2 (CP015394.1), which was identified in an MDR, ST258 K. pneumoniae clinical isolate identified in New York in 2012.11 This blaOXA-2-carrying plasmid was only present in isolate 1761 and not in the other two isolates subjected to WGS, including isolate 1701, which shared only 17 non-recombinant SNPs with isolate 1761, suggesting that the p1761_02 plasmid was acquired by the ST258 K. pneumoniae strain during the course of the patient’s illness.
Table 2.
Etest MICs for the OXA-2-producing K. pneumoniae isolate 1761 and corresponding E. coli TOP10 and K. pneumoniae ATCC 13883 transformants
| Isolate | bla OXA-2 | MIC (mg/L) |
||||
|---|---|---|---|---|---|---|
| ETP | MEM | IPM | FEP | CAZ | ||
| K. pneumoniae 1761 | + | 0.38 | 0.094 | 0.38 | 0.25 | 4 |
| E. coli TOP10 [pBC-SK(−)] | − | 0.008 | 0.023 | 0.38 | 0.064 | 0.25 |
| E. coli TOP10 (p1761_02) | + | 0.023 | 0.032 | 0.38 | 0.064 | 4 |
| E. coli TOP10 (pOXA-2) | + | 0.016 | 0.032 | 0.38 | 0.064 | 4 |
| K. pneumoniae ATCC 13883 [pBC-SK(−)] | − | 0.008 | 0.023 | 0.75 | 0.64 | 0.38 |
| K. pneumoniae ATCC 13883 (pOXA-2) | + | 0.125 | 0.094 | 1 | 0.94 | 6 |
ETP, ertapenem; MEM, meropenem; IPM, imipenem; FEP, cefepime; CAZ, ceftazidime.
Table 3.
Etest MICs for the OXA-1 and OXA-10 E. coli TOP10 and K. pneumoniae ATCC 13883 transformants
| Isolate | MIC (mg/L) |
|||
|---|---|---|---|---|
| ETP | MEM | IPM | CAZ | |
| E. coli TOP10 [pBC-SK(−)] | 0.008 | 0.023 | 0.38 | 0.25 |
| E. coli TOP10 (pOXA-1) | 0.008 | 0.023 | 0.38 | 0.125 |
| E. coli TOP10 (pOXA-10) | 0.006 | 0.023 | 0.38 | 0.125 |
| K. pneumoniae ATCC 13883 [pBC-SK(−)] | 0.008 | 0.023 | 0.75 | 0.25 |
| K. pneumoniae ATCC 13883 (pOXA-1) | 0.016 | 0.023 | 0.75 | 0.38 |
| K. pneumoniae ATCC 13883 (pOXA-10) | 0.016 | 0.032 | 0.75 | 0.38 |
ETP, ertapenem; MEM, meropenem; IPM, imipenem; CAZ, ceftazidime.
Expression of OXA-2 in K. pneumoniae
Transfer of blaOXA-2 (pOXA-2) into K. pneumoniae ATCC 13883 resulted in a 16-fold increase in ertapenem MIC from 0.008 to 0.125 mg/L, as well as a 16-fold increase in ceftazidime MIC from 0.38 to 6 mg/L, indicating that production of OXA-2 has a greater impact on reduced susceptibility of ertapenem and ceftazidime in K. pneumoniae than in E. coli. The MICs of meropenem, imipenem and cefepime were still not substantially affected by OXA-2 expression in this strain. OXA-1 and OXA-10 production in K. pneumoniae ATCC 13883 was also tested and neither resulted in substantial MIC changes of any of the agents compared with empty vector controls.
Outer membrane proteins, in particular OmpK35 and OmpK36, have been widely implicated in augmenting reduced carbapenem susceptibility in K. pneumoniae.12,13 The ompK35 and ompK36 sequences of K. pneumoniae 1761 were identical to those of K. pneumoniae ATCC 13883, suggesting that these porins did not contribute to the reduced ertapenem susceptibility observed in isolate 1761.
Kinetic parameters of OXA-2
The Km values for oxacillin and ertapenem were 692 ± 77.79 and 0.898 ± 0.23 μM, respectively. kcat for oxacillin was 111.5 ± 5 s−1 and that for ertapenem was 0.0093 ± 0.002 s−1. Thus, the catalytic efficiency (kcat/Km) against oxacillin and ertapenem was 160 M−1 s−1 and 9.5 M−1 s−1, respectively. The catalytic efficiency for oxacillin was lower than previously reported, possibly due to the presence of the Sumo-His tag.3 Nonetheless, the findings confirmed the ability of OXA-2 to hydrolyse ertapenem and also corroborated the resistance phenotype observed among the OXA-2-producing strains.
Discussion
Carbapenem resistance in Enterobacteriaceae is widely recognized as one of the most problematic issues in antimicrobial resistance.14 Globally, production of three groups of β-lactamases—KPC, NDM and OXA-48-group carbapenemases—are the predominant carbapenem resistance mechanisms in Enterobacteriaceae, with regional differences in epidemiology. In the USA, KPC is by far the most commonly encountered carbapenemase, produced primarily by K. pneumoniae strains belonging to the ST258 clonal lineage, whereas strains producing NDM and OXA-48-group carbapenemases are increasingly reported.15
In the case presented here, the multiple carbapenem-resistant K. pneumoniae isolates identified prior to the identification of K. pneumoniae isolate 1761 had typical features of KPC-producing strains, represented by broad-spectrum β-lactam resistance that included oxyiminocephalosporins and carbapenems. The unusual phenotype of isolate 1761, with higher than expected MICs of ertapenem and ceftazidime but unaltered susceptibility to other carbapenems and cefepime despite the presence of blaKPC, therefore drew our attention and triggered the investigation. We found that expression of KPC-2 in this isolate was disrupted due to insertion of IS1924 between the P2 and P1 promoter sequences. This likely occurred within 1 month upon exposure to a carbapenem and broad-spectrum cephalosporins. The reduced susceptibility to ertapenem and ceftazidime was largely due to acquisition of blaOXA-2. It appears that isolate 1761 evolved from isolate 1701 by acquiring IncA/C2 plasmid p1761_02 carrying blaOXA-2, possibly to compensate for the loss of KPC-2 expression caused by the unusual insertion event. We also speculate that differential exposure to β-lactam agents allowed the loss of KPC-2 expression and acquisition of blaOXA-2 to occur, since isolate 1761 was isolated from bronchoalveolar lavage, whereas isolates 1081 and 1701 were isolated from blood. Alternatively, these changes may have been random events.
Several nuclear acid amplification tests have been developed for rapid identification of major carbapenemase genes, including blaKPC. However, these techniques do not quantify gene expression and they do not detect uncommon carbapenemase genes like blaOXA-2 reported here. This case might have been dismissed as yet another KPC-producing K. pneumoniae if close attention had not been paid to the unusual resistance phenotype. The level of carbapenem resistance conferred by OXA-2 production is modest and its clinical significance is not yet defined. The patient presented here was treated with a 2 week course of cefepime and inhaled tobramycin, resulting in good clinical response. No further cultures were obtained from the patient and therefore it is not known whether the isolate would have gone on to accumulate additional mutations, such as in ompK35 and ompK36, to become fully resistant to carbapenems including meropenem and imipenem, as OXA-2 has been reported as being capable of hydrolysing these anti-pseudomonal carbapenems as well. This unusual case serves as a cautionary tale as more nucleic acid amplification-based diagnostic tests are adopted in the clinic for detection of carbapenemase genes. The findings also highlight the importance of continued surveillance of resistance mechanisms in the age of molecular diagnostics.
In conclusion, we report the contribution of OXA-2 production in reduced susceptibility to ertapenem and ceftazidime in a K. pneumoniae ST258 isolate in which KPC-2 production was compromised due to promoter disruption. Whether OXA-2 emerges as a clinically significant mechanism underlying carbapenem resistance in Enterobacteriaceae remains to be seen.
Supplementary Material
Acknowledgements
We thank Dr Robert Bonomo, for the kind gift of anti-KPC antibody, and Dan Evans, for his assistance with MinION sequencing.
Funding
This work was supported by internal funding from the University of Pittsburgh. The effort of R. K. S. was supported by National Institutes of Health (NIH) grant K08AI114883. The effort of V. S. C. was supported by NIH grants R01GM110444 and U01AI124302. The effort of Y. D. was supported by NIH grants R01AI104895, R21AI123747 and R21AI135522.
Transparency declarations
None to declare.
References
- 1. Antunes NT, Fisher JF.. Acquired class D β-lactamases. Antibiotics (Basel) 2014; 3: 398–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Danel F, Page M, Livermore D Class D β-lactamases. In: Bonomo R, Tolmasky ME, eds. Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition. Washington, DC, USA: ASM Press, 2007; 163–94.
- 3. Antunes NT, Lamoureaux TL, Toth M. et al. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 2014; 58: 2119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wick RR, Judd LM, Gorrie CL. et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zankari E, Hasman H, Cosentino S. et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- 7. Towbin H, Staehelin T, Gordon J.. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 1979; 76: 4350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pitout JD, Nordmann P, Poirel L.. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 2015; 59: 5873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L, Chavda KD, Melano RG. et al. Complete sequence of a blaKPC-2-harboring IncFIIK1 plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob Agents Chemother 2013; 57: 1542–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheruvanky A, Stoesser N, Sheppard AE. et al. Enhanced Klebsiella pneumoniae carbapenemase expression from a novel Tn4401 deletion. Antimicrob Agents Chemother 2017; 61: e00025‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang W, Wang G, Sebra R. et al. Emergence and evolution of multidrug-resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M integrated in the chromosome. Antimicrob Agents Chemother 2017; 61: e00076‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsai YK, Fung CP, Lin JC. et al. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 2011; 55: 1485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Landman D, Bratu S, Quale J.. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol 2009; 58: 1303–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tacconelli E, Magrini N.. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva, Switzerland: WHO, 2017. [Google Scholar]
- 15.CDC. Healthcare-Associated Infections: Tracking CRE https://www.cdc.gov/hai/organisms/cre/TrackingCRE.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.