SUMMARY
Phytophthora ramorum is an oomycete plant pathogen classified in the kingdom Stramenopila. P. ramorum is the causal agent of sudden oak death on coast live oak and tanoak as well as ramorum blight on woody ornamental and forest understorey plants. It causes stem cankers on trees, and leaf blight or stem dieback on ornamentals and understorey forest species. This pathogen is managed in the USA and Europe by eradication where feasible, by containment elsewhere and by quarantine in many parts of the world. Genomic resources provide information on genes of interest to disease management and have improved tremendously since sequencing the genome in 2004. This review provides a current overview of the pathogenicity, population genetics, evolution and genomics of P. ramorum.
Taxonomy: Phytophthora ramorum (Werres, De Cock & Man in't Veld): kingdom Stramenopila; phylum Oomycota; class Peronosporomycetidae; order Pythiales; family Pythiaceae; genus Phytophthora.
Host range: The host range is very large and the list of known hosts continues to expand at the time of writing. Coast live oak and tanoak are ecologically, economically and culturally important forest hosts in the USA. Rhododendron, Viburnum, Pieris, Syringa and Camellia are key ornamental hosts on which P. ramorum has been found repeatedly, some of which have been involved in moving the pathogen via nursery shipments.
Disease symptoms: P. ramorum causes two different diseases with differing symptoms: sudden oak death (bleeding lesions, stem cankers) on oaks and ramorum blight (twig dieback and/or foliar lesions) on tree and woody ornamental hosts.
Useful websites: http://nature.berkeley.edu/comtf/, http://rapra.csl.gov.uk/, http://www.aphis.usda.gov/plant_health/plant_pest_info/pram/index.shtml, http://genome.jgi‐psf.org/Phyra1_1/Phyra1_1.home.html, http://pamgo.vbi.vt.edu/, http://pmgn.vbi.vt.edu/, http://vmd.vbi.vt.edu./, http://web.science.oregonstate.edu/bpp/labs/grunwald/resources.htm, http://www.defra.gov.uk/planth/pramorum.htm, http://www.invasive.org/browse/subject.cfm?sub=4603, http://www.forestry.gov.uk/forestry/WCAS‐4Z5JLL
INTRODUCTION
Phytophthora ramorum (Werres, De Cock & Man in't Veld) is an exotic pathogen of tanoak (Lithocarpus densiflorus), coast live oak (Quercus agrifolia) and other tree species as well as a number of woody and herbaceous perennials (Frankel, 2008; 2005, 2002; Werres et al., 2001). Broadly speaking, this pathogen causes two kinds of disease although distinctions are not absolute: sudden oak death on oaks in native forests in the USA and ramorum blight on trees and woody ornamentals in forest, nursery and garden environments in Europe and North America (Table 1). The pathogen causes sudden mortality in tanoak and coast live oak in the USA and has been responsible for the rapid decline of forest populations of these species in the California coastal forests and parts of coastal southern Oregon (Rizzo et al., 2002, 2005) (Fig. 1A). This pathogen is also of concern with respect to native and ornamental trees such as southern red oak (Q. falcata), American red oak (Q. rubra) and European beech (Fagus sylvatica) in urban gardens in Europe and heathland plants in the UK (Brasier et al., 2004a,b). In the USA, P. ramorum causes bleeding cankers and mortality of coast live oak, Shreve oak (Q. parvula) and tanoak and tip blighting and die‐back on woody and herbaceous perennials including Rhododendron and Viburnum (Davidson et al., 2003).
Table 1.
Broadly speaking, two distinct disease symptoms can be observed for infections by P. ramorum. Sudden oak death is mainly observed in oak and tanoak forests in coastal forests of California and Oregon, but can also be observed on specimen trees in European or American gardens. Ramorum blight is observed in forest, nursery and garden environments in both North America and Europe on trees, ornamentals and woody shrubs. Sudden oak death is the ecologically and economically important disease in North America.
| Disease | Symptoms | Host categories | Typical hosts* | Geography and environment |
|---|---|---|---|---|
| Sudden oak death | Stem cankers; bleeding cankers | Forest trees; garden trees | Coast live oak, tanoak, European beech | North American forests; European gardens |
| Ramorum blight | Foliar and twig blight; tip and shoot dieback; leaf blight | Ornamental trees and woody shrubs; forest understorey plants | Viburnum, rhododendron, pieris, lilac; coast redwood, Douglas fir, huckleberry, madrone, yew, tanoak, California bay laurel | European nurseries and gardens; North American nurseries and forest |
Only a small selection of typical hosts is presented. For a complete list of hosts refer to section on host range for appropriate references.
Figure 1.

(A) Tanoak mortality near Bolinas Ridge, Mt. Tamalpais, Marin County, California. Photo courtesy: Janet Klein, Marin Municipal Open Space District. (B) Eradication of sudden oak death in Curry County, Oregon, through slash and burn. Photo courtesy: Oregon Department of Forestry.
The rapid onset and death of infected tanoaks in Marin County, California, in the early 1990s raised alarm among residents and researchers and led to the term sudden oak death. The cause of the outbreak was not known until 2000 when the pathogen, a then unknown Phytophthora sp., was isolated from cankers on dying trees (Frankel, 2008; Rizzo et al., 2002). Simultaneously, it was recognized to be the same pathogen causing tip dieback, leaf blight and stem cankers on Rhododendron and Viburnum in European greenhouse settings (Frankel, 2008; Werres et al., 2001). This discovery prompted an examination of the nursery stock in the infested areas in California and soon thereafter infected Rhododendron were found (Frankel, 2008). This pathogen is currently managed by containment in woodlands in California, the UK and the Netherlands, by eradication in US nurseries and Oregon forests (Fig. 1B), and by quarantine in many other parts of the world. In the USA, the economic impact of losses due to P. ramorum is estimated to be in the tens of millions of dollars due to the direct loss of nursery and ornamental crops, the decrease of property values due to dead/dying trees, the cost of monitoring, tracking and eradicating the disease, the societal impact through loss of recreational value and cultural value, and the ecological impact through loss of food resources for native fauna (Cave et al., 2005; Dart and Chastagner, 2007; Frankel, 2008).
TAXONOMY
Morphology
P. ramorum can be readily identified based on morphology. It is different from other Phytophthora species in that it forms large and abundant chlamydospores (Werres and Kaminsky, 2005; Werres et al., 2001). Sporangia are elongate, ellipsoid, semi‐papillate and deciduous with a short pedicel, thus resembling P. palmivora. However, P. palmivora has smaller chlamydospores, papillate sporangia and higher temperature requirements for growth (Werres et al., 2001). Optimal growth for P. ramorum occurs at 20 °C over a range of 2–26 °C. P. ramorum is heterothallic requiring a compatible response between opposite mating types to form oospores (Brasier and Kirk, 2004; Werres et al., 2001), but oospores are not readily produced in culture and there has been no evidence of oospore formation reported in nursery settings where both mating types of the pathogen have been found (Grünwald et al., 2008a).
Phylogenetic context
Although Phytophthora species resemble fungi morphologically, based on their hyphal growth patterns and nutritional requirements, they differ from true fungi in being diploid through much of their life cycle and in producing swimming zoospores. Rather, they are taxonomically placed within the kingdom Stramenopila (Baldauf, 2003; Förster et al., 1990), a distinct eukaryotic branch that includes golden‐brown algae, diatoms and brown algae. Futhermore, analyses of the P. ramorum and P. sojae genome sequences revealed genes of apparent plant origin, which suggests that the ancestor of oomycetes and other Stramenopiles was photosynthetic (Tyler et al., 2006).
Within the genus Phytophthora, P. ramorum falls into clade 8c (Blair et al., 2008; Cooke et al., 2000; Martin and Tooley, 2003). Clade 8 is the second most basal Phytophthora group (Fig. 2) (Blair et al., 2008). P. ramorum shares clade 8c with P. lateralis, P. hibernalis and P. foliorum; all except P. hibernalis are thought to be exotic in their known ranges (Donahoo et al., 2006; Erwin and Ribeiro, 1996; Ivors et al., 2004). Given that these four species are fairly genetically distant from each other, that their centres of origin are unknown and that new Phytophthora species are continually being discovered, it is likely that the closest relative of P. ramorum is unknown to science. P. lateralis is fairly well supported by molecular phylogenetics as the closest known relative to P. ramorum, yet it remains unclear whether P. hibernalis or P. foliorum is the sister taxon of the P. ramorum/P. lateralis lineage (Blair et al., 2008; Ivors et al., 2004; Kroon et al., 2004; Martin and Tooley, 2003).
Figure 2.
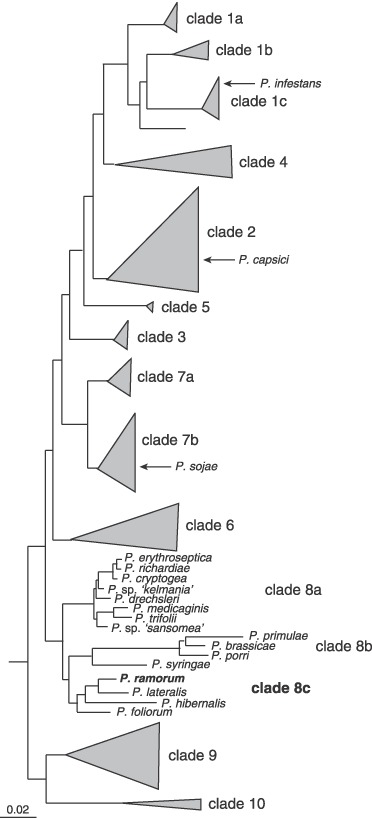
Phylogeny of the genus Phytophthora highlighting clade 8. Clade locations of other Phytophthora species with published or forthcoming whole genome sequences are also indicated (modified from Blair et al., 2008).
LIFE HISTORY AND DISEASE BIOLOGY
Natural history
The life cycle of P. ramorum is similar to that of other Phytophthora species and is broadly outlined in Fig. 3. The P. ramorum life and disease cycles are not well understood and are thus a major focus of research, both in natural and in nursery settings. P. ramorum produces sporangia on the surfaces of infected leaves and twigs of foliar hosts (Davidson et al., 2005). These sporangia can be splash‐dispersed to neighbouring hosts or driven longer distances by wind and rain (Davidson et al., 2005). P. ramorum has also been baited from rivers and streams downstream of infested areas (Davidson et al., 2005; Frankel, 2008). Inoculum can additionally be carried on soil or debris attached to boots of walkers, tyres, etc. (Davidson et al., 2005). At the same time, the distribution of P. ramorum within infested areas is patchy both in California (Rizzo et al., 2005) and in the UK (Denman et al., 2006a), suggesting some limitations in its ability to colonize new areas. Upon contact with a suitable host environment, it is thought that the sporangia germinate to produce zoospores that encyst, penetrate the host and initiate a new infection (Fig. 3). Direct germination of sporangia has not been documented in P. ramorum, but occurs in other Phytophthora species (Erwin and Ribeiro, 1996). Although P. ramorum is primarily a foliar pathogen, it appears to be able to survive in soil (Fichtner et al., 2007; Shishkoff, 2007), and has been shown to infect the roots of Rhododendron, colonize the vascular tissue and spread to the stem (Parke and Lewis, 2007). Chlamydospores are readily produced in infected plant material and can serve as resting structures, allowing the pathogen to survive adverse conditions, and may be particularly important for survival in soil (Shishkoff, 2007; Tooley et al., 2008). Oospores are currently missing in the life cycle but have been produced under controlled conditions in the laboratory, although very few and largely unviable oospores are formed in these pairings (Brasier and Kirk, 2004; Werres et al., 2001). Nevertheless, the presence of both mating types in US (Grünwald et al., 2008a) and Belgian nurseries (Werres and De Merlier, 2003) presents a potential for sexual recombination: establishment of sexual reproduction after migration of individuals of opposite mating type from a centre of sexual reproduction has been well documented for P. infestans (Drenth et al., 1995; Fry et al., 1993; Grünwald and Flier, 2005).
Figure 3.
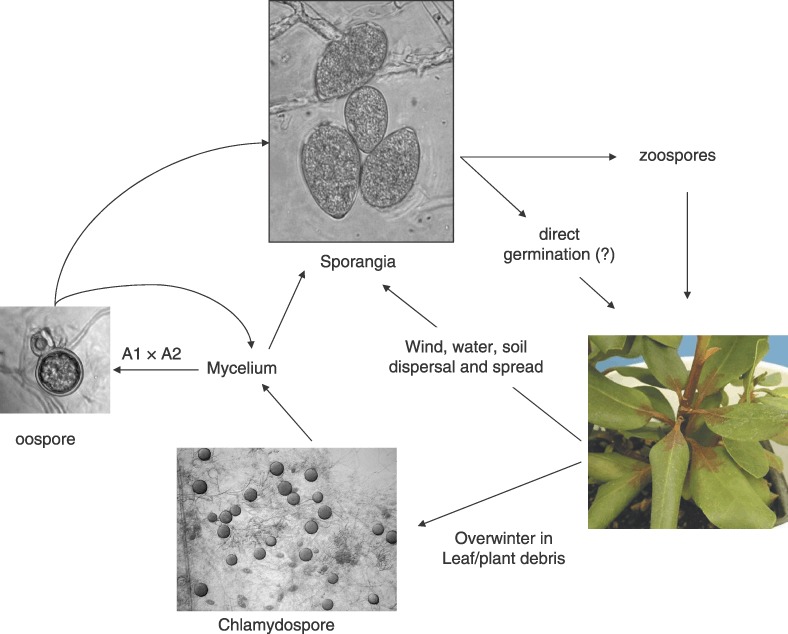
Generic life cycle of Phytophthora ramorum, broadly outlining the major life stages. Note that the sexual cycle to date has only been described under controlled conditions in the laboratory and has not been documented in nature (Brasier and Kirk, 2004; Werres and Kaminsky, 2005).
Disease symptoms
P. ramorum causes two symptom classes consisting of the foliar phase (ramorum blight) or the bole canker phase (sudden oak death) that are considered to be separate and distinct (Table 1). On oaks, tanoaks and European beech, as well as poison oak, P. ramorum causes bleeding cankers on the stem. The cankers themselves may only be visible under the cambium after removal of the bark (Fig. 4A). It is these cankers that cause ‘sudden oak death’ through killing of the inner bark (phloem) and girdling of the tree, resulting in the canopy often rapidly and completely wilting and trees dying even though the plant might have been infected for several years before the apparent ‘sudden death’.
Figure 4.

Typical symptoms observed on different hosts of Phytophthora ramorum. (A) Bleeding canker of tanoak in the exposed inner bark (phloem) after removal of the outer bark: note typical lesions with clearly differentiated margins; (B) leaf tip necrosis or leaf spots on leaf margins of California bay laurel; (C) rhododendron—arrow marks necrosis of leaf petiole; (D) leaf necrosis on nursery ornamental rhododendron.
P. ramorum also causes ramorum blight on foliar and twig hosts. This form of the disease can be evident as a shoot tip dieback or leaf spots (Fig. 4B–D). In forests in the USA, hosts that experience dieback include California bay laurel (Umbellularia californica), coast redwood (Sequoia sempervirens), Douglas fir (Pseudotsuga menziesii), evergreen huckleberry (Vaccinium ovatum), Pacific madrone (Arbutus menziesii), Pacific yew (Taxus brevifolia), Pacific rhododendron (Rhododendron macrophyllum) and tanoak. Affected shoots may develop discoloration or wilting. Many species have been documented as exhibiting leaf blight in response to P. ramorum infection, the most prominent being California bay laurel, tanoak, rhododendron and other Ericaceae species. Lesions on California bay laurel are distinctive as they are often found on the downward pointing tips of leaves where water puddles (Fig. 4B). Leaf infections may also become evident as water‐soaked lesions on leaf margins, petioles or tips, rapidly become necrotic, and spread to cover the entire leaf and the shoot. At least two research groups have observed sporulation of P. ramorum on asymptomatic leaves (Denman et al., 2006b; Frankel, 2008; Vettraino et al., 2007) and rhododendron roots can be infected from infested potting media while remaining asymptomatic (Parke and Lewis, 2007).
Host range and susceptibility
The known host range for P. ramorum continues to expand and currently includes over 109 plant species (Denman et al., 2005; Hansen et al., 2005; Tooley and Kyde, 2007; Tooley et al., 2004). An up‐to‐date list of hosts is maintained by the US Department of Agriculture (USDA) Animal and Plant Health Inspection Service (APHIS) (http://www.aphis.usda.gov/plant_health/plant_pest_info/pram/index.shtml). Although many nursery hosts are regulated at the genus level for interstate shipping, as is the case for Rhododendron or Viburnum, it has now become clear that plants can exhibit a considerable range of susceptibility to P. ramorum by species and even by cultivar within species. For example, most Viburnum cultivars evaluated for resistance to P. ramorum were not susceptible or developed only small lesions (Grünwald et al., 2008b).
Oaks in the red oak group (section Lobatae) are taking the brunt of the coastal California epidemic, yet Quercus species appear to be a dead‐end host for P. ramorum. They do not support foliar infections and sporulation has not been observed on stem cankers (Davidson et al., 2005; Garbelotto et al., 2003). Coast live oak (Quercus agrifolia) exhibits up to an eight‐fold difference in susceptibility among individuals within populations, which may spare some of these trees in coastal populations from P. ramorum‐induced mortality (Dodd et al., 2005). Tanoaks (Lithocarpus densiflorus), by contrast, develop lethal stem cankers and also support sporulation on leaves and twigs. Tanoaks appear to be more susceptible to P. ramorum than true oaks (Davidson et al., 2003). P. ramorum colonizes the xylem of tanoaks and reduces stem water transport (Parke et al., 2007). In fact, P. ramorum, P. kernoviae and other Phytophthora species have been repeatedly isolated from discoloured xylem of oaks, beech and maples in the UK (Brown and Brasier, 2007), so colonization of the xylem may be more common than previously thought. In California forests, P. ramorum epidemics are driven by California bay laurel, on which P. ramorum sporulates prolifically (Davidson et al., 2005). These trees exhibit genetic variation in susceptibility, but this variation is overwhelmed in the field by local environmental conditions, such as temperature and humidity, which ultimately determine disease levels (Anacker et al., 2008). In south‐west Oregon, bay laurel is not the key foliar host and producer of inoculum, and instead tanoak itself is a major source of sporangia (Rizzo et al., 2005). In Europe, heavily infected wild Rhododendron ponticum in the understorey appears to be responsible for producing inoculum that infects neighbouring tree trunks (Brasier et al., 2004a).
Management
P. ramorum is managed differently in nursery versus forest systems. In Europe, nurseries are under EU directives to prevent the spread of P. ramorum within the European Community by eradicating outbreaks, using good phytosanitary practices and employing plant passports for trade in host species. Currently, a passport is required both for movements within and between member states, and additional requirements apply for movements into and within EC Protected Zones. A passport is issued by the authorized and registered grower of the plant material that is also subject to official, annual inspections of this material. Due to market demands it is highly unlikely that the nursery industry will limit its trade to resistant cultivars or species, and therefore the emphasis in managing the pathogen in nurseries is on best management practices and vigilance. In the USA, nurseries in Oregon, Washington, and California that ship host plants out of state are subject to a federal quarantine. When P. ramorum is found during routine inspections, a government‐regulated protocol must be followed to attempt to eradicate the pathogen.
Sudden oak death in California is managed largely through containment, with a focus on limiting the spread of the pathogen to the north. Individual high‐value oaks can be treated with a phosphite fungicide preventively or very early in the course of infection to slow the progress of the disease (Garbelotto et al., 2007). In Oregon, the infested area is geographically limited and a policy of eradication has been pursued, which includes cutting and burning all hosts within an infected area and surrounding buffer zone and has thus far succeeded in limiting the extent of the epidemic (Frankel, 2008) (Fig. 1B).
GENETICS AND GENOMICS
Population genetics
Currently P. ramorum is known only in Europe and North America. Populations on both continents are clonal and belong to three distinct lineages (Table 2). Population genetic analyses have shown them to be genetically distinct and asexually reproducing yet clearly conspecific (Ivors et al., 2004, 2006; Prospero et al., 2007). In 2006 there was an informal agreement within the P. ramorum research community on designated names for these clonal lineages: EU1, NA1 and NA2. The EU1 clonal lineage was first identified on Rhododendron and Viburnum in European nurseries (Werres et al., 2001). It is the only lineage found in Europe to date, but it is now regularly found in nurseries on the west coast of the USA (Grünwald et al., 2008a; Hansen et al., 2003; Ivors et al., 2006). Clonal lineage NA1 is responsible for the natural infestations in California and Oregon and many of the nursery infections in North America (Ivors et al., 2006; Prospero et al., 2007). The third clonal lineage, NA2, has a limited distribution. It has only been isolated in a few instances from nurseries in North America (Ivors et al., 2006). All NA1 and NA2 isolates that have been tested are of mating type A2 (Table 2). Interestingly, EU1 is predominately mating type A1 (Brasier and Kirk, 2004; Werres and Kaminsky, 2005), yet rare A2 EU1 isolates have been identified in Belgium (Werres and De Merlier, 2003). It is unclear how the mating type switch occurred in the EU1 lineage as the genetics of mating type are complex and poorly understood. In the case of P. infestans, the A1 mating type behaves as a heterozygote and the A2 as a homozygote (Judelson and Blanco, 2005). It is thus possible that the A2 genotypes observed in the EU1 lineage are the product of a mutation to the homozygous state. As already discussed, there is currently no evidence for sexual reproduction in the nurseries in which both mating types are present, yet the levels of heterozygosity observed in P. ramorum are consistent with an outcrossing species (Tyler et al., 2006). In addition, sequencing both alleles of several nuclear genes in each clonal lineage reveals that the alleles for each lineage do not cluster together and, in fact, the genealogical relationships among the lineages change with each gene (E. M. Goss and N. J. Grünwald, unpublished data). This suggests that the clonal lineages are descendants of sexually recombining populations.
Table 2.
Currently recognized nomenclature and behavioural characteristics of Phytophthora ramorum clonal lineages (adapted from Ivors et al., 2006).
| Clonal lineage | Current distribution | Habitat | Mating type | Colony growth | Colony stability | Aggressiveness |
|---|---|---|---|---|---|---|
| EU1 | EU, North America | Nurseries | A1 (A2)† | fast | Stable | Higher |
| NA1 | North America | Forests, nurseries | A2 | slow | Unstable | Lower |
| NA2 | USA | Nurseries | A2 | fast | Stable | ND* |
ND = not determined.
EU1 is predominately mating type A1 (Brasier and Kirk, 2004; Werres and Kaminsky, 2005), yet several A2 EU1 isolates have been identified in Belgium (Werres and De Merlier, 2003).
The three clonal lineages also show phenotypic differences (Table 2). The most notable difference is relative homogeneity among EU1 isolates as compared with phenotypic variation and intrinsic instability in NA1. Instability in NA1 has been suggested by variation in colony morphology and vegetative growth rates among subcultures (Brasier et al., 2006) as well as the observation of abnormally shaped sporangia in single colony segments and between subcultures (Werres and Kaminsky, 2005). On average, EU1 isolates have shown faster growth rates in culture and larger chlamydospore size, although the growth rate of NA1 isolates can equal that of EU1 isolates (Brasier, 2003; Brasier et al., 2006; Werres and Kaminsky, 2005). NA2 exhibited a relatively rapid growth rate in culture, similar to EU1 isolates (Ivors et al., 2006). EU1 isolates have been found to produce larger lesions than NA1 isolates on cut stems of Quercus rubra (Brasier et al., 2006) and detached Rhododendron leaves (Fig. 5), but differences in other measures of fitness were not consistent across experiments. No differences in aggressiveness between the two major lineages were observed on a variety of other host species (Denman et al., 2005; Tooley et al., 2004). Variation in aggressiveness among isolates within both NA1 and EU1 lineages has been documented on coast live oak seedlings, detached bay laurel leaves and Q. rubra cut stems (Brasier et al., 2006; Huberli et al., 2006). None of these studies has observed genotype‐specific (gene‐for‐gene‐like) variation in virulence.
Figure 5.
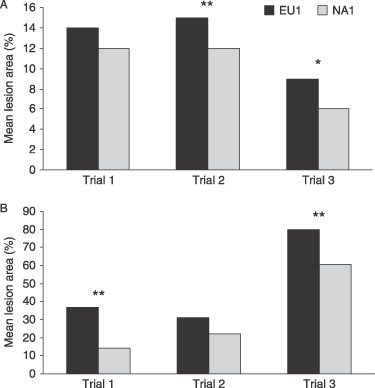
Difference in aggressiveness of P. ramorum clonal lineages EU1 and NA1. (A) Leaf lesion area 10 days post inoculation on two Rhododendron cultivars as a function of clonal lineage. The difference between EU1 and NA1 was significant in trials 2 and 3 (P < 0.005 and P < 0.05, respectively) (V. T. McDonald and N. J. Grünwald, unpublished data). (B) Mean lesion area in wound inoculated lower stems of Quercus rubra (data from Brasier et al., 2006). The differences between EU1 and NA1 were significant in trials 1 and 3 (P < 0.001 and P < 0.0012, respectively).
Amplified fragment length polymorphism (AFLP) and fast‐evolving microsatellites (simple sequence repeats) have revealed genetic variation within the lineages, which has allowed for studies of population structure and change. Given the asexual reproduction of these populations, this variation is hypothesized to be the result of mutation or mitotic recombination (Goodwin, 1997). The first AFLP and microsatellite studies found the European population to be more variable than the NA1 population (Ivors et al., 2004, 2006). Conversely, Prospero et al. (2007) found microsatellites that are variable within NA1 but not EU1. Yet these loci may be evolving so quickly in NA1 as to be considered unstable, providing an interesting parallel to the phenotypic data. These markers significantly differentiated the nursery and forest populations in Oregon and showed that the genotypic composition of nursery populations had changed from 2003 to 2004 (Prospero et al., 2007). Microsatellite variation has also proven valuable for rapid and accurate diagnosis of clonal lineages and is being used to monitor nursery finds in the USA. There are numerous simple sequence repeats identified in the P. ramorum genome sequence that have not been screened for variation and may yet provide useful markers (Garnica et al., 2006; Tyler et al., 2006) (http://web.science.oregonstate.edu/bpp/labs/grunwald/resources.htm).
Genome sequence
A 7X draft whole‐genome sequence for P. ramorum (an NA1 isolate from California) was published in 2006 along with that of the soybean pathogen P. sojae (Tyler et al., 2006), and the genome sequences of P. infestans and P. capsici are forthcoming (Broad Institute and Joint Genome Institute, respectively). The complete mitochondrial genome of P. ramorum has also been sequenced (Martin et al., 2007) and there are several other oomycete mitochondrial genomes to which it can be compared (Avila‐Adame et al., 2006; Grayburn et al., 2004; Martin et al., 2007; Paquin et al., 1997). The mitochondrial genomes are largely conserved between Phytophthora species, although there are a number of more rapidly evolving genes, some of which may be specific to Phytophthora, as well as apparent hotspots for gene rearrangement. P. ramorum, in particular, contains a short inverted repeat, which among Phytophthora has previously been observed in only one other species, as inversions in other Stramenopile mitochondrial genomes typically represent much larger proportions of the genome (Martin et al., 2007).
Since the first release of the P. ramorum and P. sojae whole genome sequences, there has been rapid progress in the area of Phytophthora genomics, especially as pertains to genes involved in pathogenicity (Kamoun, 2006; Lamour et al., 2007; Morgan and Kamoun, 2007). The follow‐up functional studies are mostly being conducted in the established model systems P. infestans and P. sojae, which are both fairly host‐specific pathogens and may rely on different strategies for infection than the generalist P. ramorum. As such, it is interesting to note how the P. ramorum genome differs from these other Phytophthora genomes.
The P. ramorum genome size is 65 Mb, smaller than P. sojae (95 Mb) and P. infestans (240 Mb), but approximately the same size as P. capsici (Lamour et al., 2007). Proteins that are secreted from the pathogen are thought to have functions specific to the host–pathogen interaction, and thus the predicted secretomes are of particular interest to the plant pathology community. Comparison of the P. ramorum and P. sojae genomes showed these genes to be evolving faster than the rest of the proteome (Tyler et al., 2006). In addition, there has been family‐specific expansion of pathogenicity‐related genes. Figure 6 shows the families that are expanded in P. ramorum as compared with P. sojae and the autotrophic diatom Thalassiosira pseudonana. P. ramorum has more protease genes than P. sojae, but fewer of the other groups of hydrolytic enzymes, proteins that may be important to the necrotrophic stage of infection. Among effectors, the necrosis and ethylene‐inducing protein (NPP) family shows an expansion in P. ramorum, while P. sojae contains relatively more of the necrosis‐inducing PcF and Crn families, which are also large and diverse families in P. infestans (Kamoun, 2006).
Figure 6.
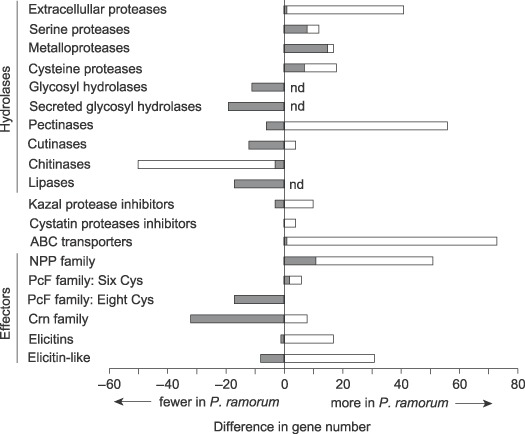
Relative number of genes potentially involved in infection in P. ramorum compared with P. sojae (grey bars) and the diatom Thalassiosira pseudonana (white bars). For three gene families, the number of genes in T. pseudonana was not determined (nd). Data are from Tyler et al. (2006: table 1).
The avirulence genes cloned from oomycetes to date have all contained the conserved amino acid motif RXLR (Arg, any amino acid, Leu, Arg), which has recently been shown to be required for translocation of the protein into the host cell (Dou et al., 2008b; Whisson et al., 2007). Following the RXLR is an additional motif, dEER, which takes on more variable functional forms (Allen et al., 2004; Jiang et al., 2008; Shan et al., 2004) but is also required for translocation (Dou et al., 2008b; Whisson et al., 2007). Fascinatingly, the RXLR motif shares similarity with the motif in the malaria parasite Plasmodium falciparum (RXLX followed by E, D or Q) that serves the equivalent function of exporting pathogen proteins to the host erythrocyte (Bhattacharjee et al., 2006).
When homologues of these genes were examined in the P. ramorum and P. sojae draft genome sequences, a large and diverse family of potential effectors was found (Jiang et al., 2008; Tyler et al., 2006). The number of ‘true’ RXLR‐class effectors (i.e. avirulence gene homologues) in the Phytophthora genomes is under debate, as the vast majority of them have not been functionally validated; certainly none has been validated in P. ramorum. As a result, different numbers exist based on the bioinformatic method used to mine them (Table 3). These genes are so variable between Phytophthora genomes that orthology is difficult to determine. Of the nearly 400 genes identified in the P. ramorum and P. sojae genomes by Jiang et al. (2008), only 34 syntenic orthologues could be definitively assigned as such. Most of these genes have only distant homologues based on weak similarity between inferred amino acid sequences.
Table 3.
Estimates of numbers of RXLR‐class effectors in three Phytophthora genomes.
| Method | P. ramorum | P. sojae | P. infestans |
|---|---|---|---|
| Jiang et al. (2008) | 374 | 396 | − |
| Win et al. (2007) * | 531 | 672 | 716‡ |
| Whisson et al. (2007) † | 314 | 400 | 425 |
Upper limit established by combining results of two methods.
Upper limit (signal peptide plus RXLR between amino acids 30 and 60).
From Lamour et al. (2007).
RXLR‐type effectors have a modular structure: the RXLR and dEER motifs are localized to the N‐terminus, while the C‐terminal domain is thought to be responsible for activity within the host plant cell (Dou et al., 2008a; Jiang et al., 2008; Kamoun, 2007; Whisson et al., 2007). The majority of likely RXLR‐class effectors in both P. ramorum and P. sojae contain a conserved C‐terminal motif, called the W motif, which has recently been shown in P. sojae to suppress programmed cell death and thus increase virulence in the initial biotrophic phase of infection (Dou et al., 2008a; Jiang et al., 2008). The W motif is often followed by two other conserved motifs (Y and L) forming a W–Y–L module that may be repeated up to eight times (Jiang et al., 2008). As the function of these and other motifs are characterized in model organisms, they may elucidate the function of effectors with these motifs in P. ramorum.
Effector evolution
Within genomes, recently duplicated genes (paralogues) can be identified based on sequence similarity. Analysis of paralogous RXLR‐class effectors in P. ramorum showed many to be under detectable positive selection, often localized to the C‐terminal region and within the W and Y motifs (Jiang et al., 2008; Win et al., 2007). Of 59 groups of closely related paralogues examined by Win et al. (2007), 41 showed evidence of positive selection. Of those 41 groups, 21 had two to four members clustered in the genome (within 100 kb). The P. sojae genome contained a comparable number of putative RXLR‐type effectors, but only 28 groups of closely related paralogues were identified, of which 18 had experienced positive selection and only five of these were clustered. These data indicate that P. ramorum may have experienced a more recent expansion of RXLR‐class effectors than P. sojae. We examined a group of four paralogues in P. ramorum more closely and found only a single homologue in P. lateralis and P. hibernalis (Fig. 7; E. M. Goss and N. J. Grünwald, unpublished), suggesting that this group is a result of gene duplication since the divergence of P. ramorum and P. lateralis. Win et al. (2007) found that for those P. ramorum RXLR‐type effectors that have paralogues, an estimated 67–75% had between 70 and 99% amino acid identity to their closest paralogue. The most divergent of the four genes in Fig. 7 have 72% amino acid identity (21% divergence at synonymous sites in the whole genome sequence isolate Pr102) and thus fall in the lower end of the above range. Therefore, many of the expansions may indeed have occurred since divergence from P. lateralis, although one would expect rates of evolution to vary widely among effector genes.
Figure 7.
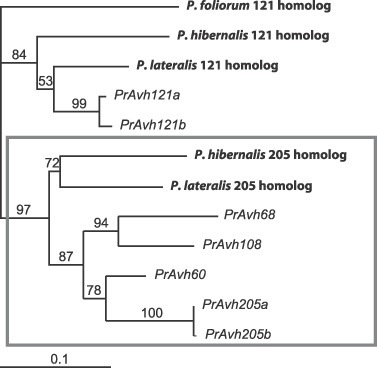
Maximum likelihood genealogy of a subfamily of RXLR‐class effector genes (avr gene homologues, Avh) in Phytophthora clade 8c. PrAvh gene sequences are from P. ramorum isolate Pr102. For heterozygous genes, the two alleles are distinguished by the suffixes ‘a’ and ‘b’. Homologous sequences are in bold typeface. The P. lateralis and P. hibernalis homologues to PrAvh60, PrAvh68, PrAvh108 and PrAvh205 (genes in box) appear to be syntenic to PrAvh205 based on flanking sequence similarity. Bootstrap support is shown for all branches (E. M. Goss and N. J. Grünwald, unpublished data).
PROSPECTS
Several features make P. ramorum a particularly interesting candidate for further genomic and genetic analysis. P. ramorum is unique among the sequenced oomycete pathogens in that it has a very wide host range. Thus, genes involved in host–pathogen interactions must have been subject to very different evolutionary dynamics. Similarly, in contrast to P. infestans, P. sojae and P. capsici, P. ramorum can infect mature trees and is able to penetrate bark and colonize the xylem (Brown and Brasier, 2007; Parke et al., 2007). It is thus expected that a unique set of biochemical pathways and novel chemical functions will have evolved to enable these distinct infection pathways. Yet, these characteristics of P. ramorum also make it a challenging organism for molecular genetics because its host plants are primarily woody perennials that exhibit poorly characterized multigene resistance.
ACKNOWLEDGEMENTS
This work was supported in part by funds from USDA ARS CRIS Project 5358‐22000‐034‐00, the Northwest Center for Nursery Crop Research, the US Forest Service, the USDA ARS Floriculture Nursery Initiative and Oregon Department of Agriculture/Oregon Association of Nurseries. We thank Clive Brasier and Brett Tyler for suggested improvement to the manuscript. Mention of trade names or commercial products in this manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
REFERENCES
- Allen, R.L. , Bittner‐Eddy, P.D. , Grenville‐Briggs, L.J. , Meitz, J.C. , Rehmany, A.P. , Rose, L.E. and Beynon, J.L. (2004) Host–parasite coevolutionary conflict between arabidopsis and downy mildew. Science, 306, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Anacker, B.L. , Rank, N.E. , Huberli, D. , Garbelotto, M. , Gordon, S. , Harnik, T. , Whitkus, R. and Meentemeyer, R. (2008) Susceptibility to Phytophthora ramorum in a key infectious host: landscape variation in host genotype, host phenotype, and environmental factors. New Phytol. 177, 756–766. [DOI] [PubMed] [Google Scholar]
- Avila‐Adame, C. , Gomez‐Alpizar, L. , Zismann, V. , Jones, K.M. , Buell, C.R. and Ristaino, J.B. (2006) Mitochondrial genome sequences and molecular evolution of the Irish potato famine pathogen, Phytophthora infestans . Curr. Genet. 49, 39–46. [DOI] [PubMed] [Google Scholar]
- Baldauf, S.L. (2003) The deep roots of eukaryotes. Science, 300, 1703–1706. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. , Hiller, N.L. , Liolios, K. , Win, J. , Kanneganti, T.‐D. , Young, C. , Kamoun, S. and Haldar, K. (2006) The malarial host‐targeting signal is conserved in the Irish potato famine pathogen. PLOS Pathogens 2, e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair, J.E. , Coffey, M.D. , Park, S.‐Y. , Geiser, D.M. and Kang, S. (2008) A multi‐locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet. Biol. doi: 10.1016/j.fgb.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Brasier, C. (2003) Sudden oak death: Phytophthora ramorum exhibits transatlantic differences. Mycol. Res. 107, 258–259. [DOI] [PubMed] [Google Scholar]
- Brasier, C. and Kirk, S. (2004) Production of gametangia by Phytophthora ramorum in vitro. Mycol. Res. 108, 823–827. [DOI] [PubMed] [Google Scholar]
- Brasier, C. , Denman, S. , Brown, A. and Webber, J. (2004a) Sudden Oak Death (Phytophthora ramorum) discovered on trees in Europe. Mycol. Res. 108, 1108–1110. [Google Scholar]
- Brasier, C. , Kirk, S. and Rose, J. (2006) Differences in phenotypic stability and adaptive variation between the main European and American lineages of Phytophthora ramorum In: Progress in Research on Phytophthora Diseases of Forest Trees (Brasier C. Jung T. and Oßwald W., eds), pp. 166–173. Farnham: Forest Research, [Google Scholar]
- Brasier, C.M. , Denman, S. , Rose, J. , Kirk, S.A. , Hughes, K.J.D. , Griffin, R.L. , Lane, C.R. , Inman, A.J. and Webber, J.F. (2004b) First report of ramorum bleeding canker on Quercus falcata, caused by Phytophthora ramorum . Plant Pathol. 53, 804. [Google Scholar]
- Brown, A.V. and Brasier, C.M. (2007) Colonization of tree xylem by Phytophthora ramorum, P. kernoviae and other Phytophthora species. Plant Pathol. 56, 227–241. [Google Scholar]
- Cave, G.L. , Randall‐Schadel, B. and Redlin, S.C. (2005) Risk analysis for Phytophthora ramorum Werres, de Cock & In't Veld, causal agent of Phytophthora Canker (Sudden Oak Death), Ramorum Leaf blight, and Ramorum Dieback . USDA APHIS report.
- Cooke, D.E.L. , Drenth, A. , Duncan, J.M. , Wagels, G. and Brasier, C.M. (2000) A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 30, 17–32. [DOI] [PubMed] [Google Scholar]
- Dart, N.L. and Chastagner, G.A. (2007) Estimated economic losses associated with the destruction of plants due to Phytophthora ramorum quarantine efforts in Washington State. Plant Health Progress doi: 10.1094/PHP-2007-0508-02-RS. [DOI] [Google Scholar]
- Davidson, J.M. , Werres, S. , Garbelotto, M. , Hansen, E.M. and Rizzo, D.M. (2003) Sudden oak death and associated diseases caused by Phytophthora ramorum . Plant Health Prog. doi: 10.1094/PHP-2003-0707-01-DG. [DOI] [Google Scholar]
- Davidson, J.M. , Wickland, A.C. , Patterson, H.A. , Falk, K.R. and Rizzo, D.M. (2005) Transmission of Phytophthora ramorum in mixed‐evergreen forest in California. Phytopathology, 95, 587–596. [DOI] [PubMed] [Google Scholar]
- Denman, S. , Kirk, S. , Whybrow, A. , Orton, E. and Webber, J.F. (2006a) Phytophthora kernoviae and P. ramorum: host susceptibility and sporulation potential on foliage of susceptible trees 1. EPPO Bull. 36, 373–376. [Google Scholar]
- Denman, S. , Kirk, S.A. , Brasier, C.M. and Webber, J.F. (2005) In vitro leaf inoculation studies as an indication of tree foliage susceptibility to Phytophthora ramorum in the UK. Plant Pathol. 54, 512–521. [Google Scholar]
- Denman, S. , Moralejo, E. , Kirk, S. , Orton, E. and Webber, J. (2006b) Sporulation of Phytophthora ramorum and P. kernoviae on asymptomatic foliage. Phytopathology, 96, S28. [Google Scholar]
- Dodd, R.S. , Huberli, D. , Douhovnikoff, V. , Harnik, T.Y. , Afzal‐Rafii, Z. and Garbelotto, M. (2005) Is variation in susceptibility to Phytophthora ramorum correlated with population genetic structure in coast live oak (Quercus agrifolia)? New Phytol. 165, 203–214. [DOI] [PubMed] [Google Scholar]
- Donahoo, R. , Blomquist, C.L. , Thomas, S.L. , Moulton, J.K. , Cooke, D.E.L. and Lamour, K.H. (2006) Phytophthora foliorum sp nov., a new species causing leaf blight of azalea. Mycol. Res. 110, 1309–1322. [DOI] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Chen, Y. , Wang, Q. , Jiang, R.H.Y. , Arredondo, F.D. , Anderson, R.G. , Thakur, P.B. , McDowell, J.M. , Wang, Y. and Tyler, B.M. (2008a) Conserved C‐terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell, 20, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H.Y. , Bruce, N.A. , Arredondo, F.D. , Zhang, X. and Tyler, B.M. (2008b) RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen encoded machinery. Plant Cell, doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenth, A. , Janssen, E.M. and Govers, F. (1995) Formation and survival of oospores of Phytophthora infestans under natural conditions. Plant Pathol. 44, 86–94. [Google Scholar]
- Erwin, D.C. and Ribeiro, O.K. (1996) Phytophthora Diseases Worldwide. St. Paul, MN: APS Press. [Google Scholar]
- Fichtner, E.J. , Lynch, S.C. and Rizzo, D.M. (2007) Detection, distribution, sporulation, and survival of Phytophthora ramorum in a california redwood‐tanoak forest soil. Phytopathology, 97, 1366–1375. [DOI] [PubMed] [Google Scholar]
- Förster, H. , Coffey, M.D. , Elwood, H. and Sogin, M.L. (1990) Sequence analysis of the small subunit ribosomal RNAs of three zoosporic fungi and implications for fungal evolution. Mycologia, 82, 306–312. [Google Scholar]
- Frankel, S.J. (2008) Sudden oak death and Phytophthora ramorum in the USA: a management challenge. Australas. Plant Pathol. 37, 19–25. [Google Scholar]
- Fry, W.E. , Goodwin, S.B. , Dyer, A.T. , Matuszak, J.M. , Drenth, A. , Tooley, P.W. , Sujkowski, L.S. , Koh, Y.J. , Cohen, B.A. , Spielman, L.J. Deahl, K.L. , Inglis, D.A. and Sandlan, K.P. (1993) Historical and recent migrations of Phytophthora infestans: chronology, pathways, and implications. Plant Dis. 77, 653–661. [Google Scholar]
- Garbelotto, M. , Davidson, J.M. , Ivors, K. , Maloney, P.E. , Hüberli, D. , Koike, S.T. and Rizzo, D.M. (2003) Non‐oak native plants are the main hosts for the sudden oak death pathogen in California. Calif. Agri. 57, 18–23. [Google Scholar]
- Garbelotto, M. , Schmidt, D.J. and Harnik, T.Y. (2007) Phosphite injections and bark application of phosphite + Pentrabark™ control sudden oak death in coast live oak. Arboriculture Urban Forestry, 33, 309–317. [Google Scholar]
- Garnica, D.P. , Pinzon, A.M. , Quesada‐Ocampo, L.M. , Bernal, A.J. , Barreto, E. , Grünwald, N.J. and Restrepo, S. (2006) Survey and analysis of microsatellites from transcript sequences in Phytophthora species: frequency, distribution, and potential as markers for the phylum Oomycota. BMC Genomics, 7, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, S.B. (1997) The population genetics of Phytophthora . Phytopathology, 87, 462–473. [DOI] [PubMed] [Google Scholar]
- Grayburn, W.S. , Hudspeth, D.S.S. , Gane, M.K. and Hudspeth, M.E.S. (2004) The mitochondrial genome of Saprolegnia ferax: organization, gene content and nucleotide sequence. Mycologia, 96, 981–989. [PubMed] [Google Scholar]
- Grünwald, N.J. and Flier, W.G. (2005) The biology of Phytophthora infestans at its center of origin. Ann. Rev. Phytopathol. 43, 171–190. [DOI] [PubMed] [Google Scholar]
- Grünwald, N.J. , Goss, E.M. , Larsen, M.M. , Press, C.M. , McDonald, V.T. , Blomquist, C. and Thomas, S.L. (2008a) First report of the European lineage of Phytophthora ramorum in a California nursery. Plant Dis. 2, 314–314. [DOI] [PubMed] [Google Scholar]
- Grünwald, N.J. , Kitner, K. , McDonald, V. and Goss, E.M. (2008b) Susceptibility in Viburnum to Phytophthora ramorum . Plant Dis. 92, 210–214. [DOI] [PubMed] [Google Scholar]
- Hansen, E.M. , Parke, J.L. and Sutton, W. (2005) Susceptibility of Oregon forest trees and shrubs to Phytophthora ramorum: a comparison of artificial inoculation and natural infection. Plant Dis. 89, 63–70. [DOI] [PubMed] [Google Scholar]
- Hansen, E.M. , Reeser, P.W. , Sutton, W. and Winton, L.M. (2003) First report of A1 mating type of Phytophthora ramorum in North America. Plant Dis. 87, 1267–1267. [DOI] [PubMed] [Google Scholar]
- Huberli, D. , Harnik, T.Y. , Meshriy, M. , Miles, L. and Garbelotto, M. (2006) Phenotypic variation among Phytophthora ramorum isolates from California and Oregon In: Proceedings of the Sudden Oak Death Second Science Symposium: The State Of Our Knowledge. January 18–21, 2005, Monterey, CA. Gen. Tech. Rep. PSW‐GTR‐196 (Frankel S.J., Shea P.J. and Haverty M.I., eds), pp. 131–134. Albany, CA: US Department of Agriculture, Forest Service, Pacific Southwest Research Station. [Google Scholar]
- Ivors, K. , Garbelotto, M. , Vries, I.D.E. , Ruyter‐Spira, C. , Hekkert, B.T. , Rosenzweig, N. and Bonants, P. (2006) Microsatellite markers identify three lineages of Phytophthora ramorum in US nurseries, yet single lineages in US forest and European nursery populations. Mol. Ecol. 15, 1493–1505. [DOI] [PubMed] [Google Scholar]
- Ivors, K.L. , Hayden, K.J. , Bonants, P.J.M. , Rizzo, D.M. and Garbelotto, M. (2004) AFLP and phylogenetic analyses of North American and European populations of Phytophthora ramorum . Mycol. Res. 108, 378–392. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tripathy, S. , Govers, F. and Tyler, B.M. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA, 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson, H.S. and Blanco, F.A. (2005) The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 3, 47–58. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Ann. Rev. Phytopathol. 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2007) Groovy times: filamentous pathogen effectors revealed. Curr. Opin. Plant Biol. 10, 358–365. [DOI] [PubMed] [Google Scholar]
- Kroon, L.P.N.M. , Bakker, F.T. , Van Den Bosch, G.B.M. , Bonants, P.J.M. and Flier, W.G. (2004) Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet. Biol. 41, 766–782. [DOI] [PubMed] [Google Scholar]
- Lamour, K.H. , Win, J. and Kamoun, S. (2007) Oomycete genomics: new insights and future directions. FEMS Microbiol. Lett. 274, 1–8. [DOI] [PubMed] [Google Scholar]
- Martin, F.N. and Tooley, P.W. (2003) Phylogenetic relationships of Phytophthora ramorum, P. nemorosa, and P. pseudosyringae, three species recovered from areas in California with sudden oak death. Mycol. Res. 107, 1379–1391. [DOI] [PubMed] [Google Scholar]
- Martin, F.N. , Bensasson, D. , Tyler, B.M. and Boore, J.L. (2007) Mitochondrial genome sequences and comparative genomics of Phytophthora ramorum and P. sojae . Curr. Genet. 51, 285–296. [DOI] [PubMed] [Google Scholar]
- Morgan, W. and Kamoun, S. (2007) RXLR effectors of plant pathogenic oomycetes. Curr. Opin. Microbiol. 10, 332–8. [DOI] [PubMed] [Google Scholar]
- Paquin, B. , Laforest, M.‐J. , Forget, L. , Roewer, I. , Wang, Z. , Longcore, J. and Lang, B.F. (1997) The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr. Genet. 31, 380–395. [DOI] [PubMed] [Google Scholar]
- Parke, J.L. and Lewis, C. (2007) Root and stem infection of Rhododendron from potting medium infested with Phytophthora ramorum . Plant Dis. 91, 1265–1270. [DOI] [PubMed] [Google Scholar]
- Parke, J.L. , Oh, E. , Voelker, S. , Hansen, E.M. , Buckles, G. and Lachenbruch, B. (2007) Phytophthora ramorum colonizes tanoak xylem and is associated with reduced stem water transport. Phytopathology, 97, 1558–1567. [DOI] [PubMed] [Google Scholar]
- Prospero, S. , Hansen, E.M. , Grünwald, N.J. and Winton, L.M. (2007) Population dynamics of the sudden oak death pathogen Phytophthora ramorum in Oregon from 2001 to 2004. Mol. Ecol. 16, 2958–2973. [DOI] [PubMed] [Google Scholar]
- Rizzo, D.M. , Garbelotto, M. and Hansen, E.M. (2005) Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Ann. Rev. Phytopathol. 43, 309–335. [DOI] [PubMed] [Google Scholar]
- Rizzo, D.M. , Garbelotto, M. , Davidson, J.M. , Slaughter, G.W. and Koike, S.T. (2002) Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis. 86, 205–214. [DOI] [PubMed] [Google Scholar]
- Shan, W. , Cao, M. , Leung, D. and Tyler, B.M. (2004) The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant–Microbe Interact. 17, 394–403. [DOI] [PubMed] [Google Scholar]
- Shishkoff, N. (2007) Persistence of Phytophthora ramorum in soil mix and roots of nursery ornamentals. Plant Dis. 91, 1245–1249. [DOI] [PubMed] [Google Scholar]
- Tooley, P.W. and Kyde, K.L. (2007) Susceptibility of some Eastern forest species to Phytophthora ramorum . Plant Dis. 91, 435–438. [DOI] [PubMed] [Google Scholar]
- Tooley, P.W. , Browning, M. and Berner, D. (2008) Recovery of Phytophthora ramorum following exposure to temperature extremes. Plant Dis. 92, 431–437. [DOI] [PubMed] [Google Scholar]
- Tooley, P.W. , Kyde, K.L. and Englander, L. (2004) Susceptibility of selected Ericaceous ornamental host species to Phytophthora ramorum . Plant Dis. 88, 993–999. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X. , Dehal, P. , Jiang, R.H.Y. , Aerts, A. , Arredondo, F.D. , Baxter, L. , Bensasson, D. , Beynon, J.L. , Chapman, J. , Damasceno, C.M.B. , Dorrance, A.E. , Dou, D. , Dickerman, A.W. , Dubchak, I.L. , Garbelotto, M. , Gijzen, M. , Gordon, S.G. , Govers, F. , Grünwald, N.J. , Huang, W. , Ivors, K.L. , Jones, R.W. , Kamoun, S. , Krampis, K. , Lamour, K.H. , Lee, M.‐K. , McDonald, W.H. , Medina, M. , Meijer, H.J.G. , Nordberg, E.K. , Maclean, D.J. , Ospina‐Giraldo, M.D. , Morris, P.F. , Phuntumart, V. , Putnam, N.H. , Rash, S. , Rose, J.K.C. , Sakihama, Y. , Salamov, A.A. , Savidor, A. , Scheuring, C.F. , Smith, B.M. , Sobral, B.W.S. , Terry, A. , Torto‐Alalibo, T.A. , Win, J. , Xu, Z. , Zhang, H. , Grigoriev, I.V. , Rokhsar, D.S. and Boore, J.L. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Vettraino, A.M. , Ceccarelli, B. and Vannini, A. (2007) Potential risk of spread of Phytophthora ramorum in Mediterranean wild environments and nurseries 4th IUFRO Workshop: Phytophthoras in Forests & Natural Ecosystems, Monterrey, CA. [Google Scholar]
- Werres, S. and De Merlier, D. (2003) First detection of Phytophthora ramorum mating type A2 in Europe. Plant Dis. 87, 1266–1266. [DOI] [PubMed] [Google Scholar]
- Werres, S. and Kaminsky, K. (2005) Characterization of European and North American Phytophthora ramorum isolates due to their morphology and mating behaviour in vitro with heterothallic Phytophthora species. Mycol. Res. 109, 860–871. [DOI] [PubMed] [Google Scholar]
- Werres, S. , Marwitz, R. , Veld, W. , De Cock, A. , Bonants, P.J.M. , De Weerdt, M. , Themann, K. , Ilieva, E. and Baayen, R.P. (2001) Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and Viburnum . Mycol. Res. 105, 1155–1165. [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , Van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R.J. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Win, J. , Morgan, W. , Bos, J. , Krasileva, K.V. , Cano, L.M. , Chaparro‐Garcia, A. , Ammar, R. , Staskawicz, B.J. and Kamoun, S. (2007) Adaptive evolution has targeted the C‐terminal domain of the RXLR Effectors of plant pathogenic oomycetes. Plant Cell, tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]


