SUMMARY
Most Bursaphelenchus species are fungal feeding nematodes that colonize dead or dying trees. However, Bursaphelenchus xylophilus, the pine wood nematode, is also a pathogen of trees and is the causal agent of pine wilt disease. B. xylophilus is native to North America and here it causes little damage to trees. Where it is introduced to new regions it causes huge damage. The most severely affected areas are found in the Far East but more recently B. xylophilus has been introduced into Portugal and the potential for damage here is also high. As incidence and severity of pine wilt disease are linked to temperature we suggest that climate change is likely to exacerbate the problems caused by B. xylophilus and, in addition, will extend (northwards in Europe) the range in which pine wilt disease can occur. Here we review what is currently known about the interactions of B. xylophilus with its hosts, including recent developments in our understanding of the molecular biology of pathogenicity in the nematode. We also examine the potential developments that could be made by more widespread use of genomics tools to understand interactions between B. xylophilus, bacterial pathogens that have been implicated in disease and host trees.
INTRODUCTION
Plant‐parasitic nematodes attack their hosts by using a wide range of strategies. Plant‐parasitic nematodes can be sedentary or migratory (within a host) and can be ectoparasites or endoparasites. For some nematodes, including most migratory ectoparasites, the interaction between the nematode and the plant is limited whereas for other nematodes the interactions are far more complex and long‐lasting. This is particularly true for endoparasitic nematodes, which must overcome plant defences in order to survive and feed. The most complex interactions are perhaps those of the sedentary endoparasitic nematodes (including cyst and root‐knot nematodes), which are biotrophic and induce profound changes in the roots of their hosts as they feed. Migratory endoparasitic nematodes, including Bursaphelenchus xylophilus, the subject of the present study, do not feed from a single site but move through the plant, causing extensive damage as they move and feed (Duncan and Moens, 2006).
The variety of feeding strategies used by plant‐parasitic nematodes is reflected in the diversity of their phylogenetic origins. Classifying nematodes is a notoriously difficult task as they often have a very conserved morphology. Fortunately, modern molecular techniques, coupled to developmental and morphological studies, have now allowed the detailed phylogeny of the phylum to be unravelled (e.g. Baldwin et al., 2004; Blaxter et al., 1998). One of the intriguing findings of these studies is that parasitism of plants, and even the most complex biotrophic interactions, has arisen on several independent occasions within the Nematoda (Baldwin et al., 2004, Holterman et al., 2006).
Bursaphelenchus spp. are part of the family Aphelenchoididae and 70 species are described for the genus (Ryss et al., 2005; Ye et al., 2007). Most nematodes within this clade are fungal feeding, and the same is also true for most Bursaphelenchus species, which are usually found in dead or dying trees feeding on fungi colonizing the tree. The life cycle of Bursaphelenchus is summarized in Fig. 1. The nematodes develop through a series of moults to the adult stage via four juvenile stages. All stages of the nematode feed. As food becomes scarce or nematode density increases, a specialized third juvenile stage (J3) forms; the pre‐dauer juvenile. Formation of the pre‐dauer J3 occurs at the same time as late instar larvae or pupae of Cerambycid beetles that the nematode uses as a vector are formed. Cerambycid beetles (Monochamus spp.) are longhorn beetles that exist in the same area of the tree occupied by the nematodes but not all species in this genus are used as vectors by Bursaphelenchus. Monochamus alternatus and M. galloprovincalis are the most important vector species (Mamiya and Enda, 1972; Sousa et al., 2001). The pre‐dauer J3 aggregate around the pupal chambers of the beetles and moult to the dauer stage larva, a stage that is adapted for long‐term survival in the absence of food. This moult to the dauer stage requires the presence of the beetle (Maehara et al., 1996). The dauer larvae are picked up by the adult beetles as they emerge and they settle beneath the elytra and inside the trachea of the beetles. All Bursaphelenchus species can be transmitted to a dying tree or newly cut log by the beetle during oviposition and the cycle begins again. B. xylophilus is unusual in that the nematodes have an additional transmission route from the beetles. This transmission route results in the nematodes infecting live trees during the maturity feeding period by the beetle that occurs between May and September. It is this infection route that leads to disease (Li et al., 2007; Shibata and Okuda, 1989).
Figure 1.
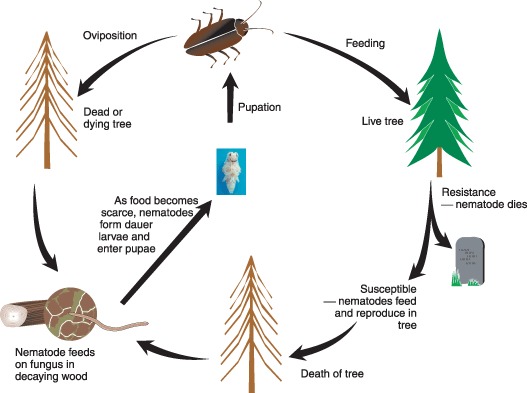
Life cycle of B. xylophilus. Image of Monochamus pupa provided by P. Naves, EFN‐INRB, Portugal.
B. xylophilus is thought to have originated in North America and was described in 1934 from Louisiana, as Aphelenchoides xylophilus (Steiner and Buhrer, 1934) although no connection was made at the time between the nematode and disease. Native American conifers are mostly resistant or tolerant to B. xylophilus and consequently here it causes little damage, other than on exotic species of trees (Mamiya, 1983). However, B. xylophilus was introduced into Japan at the start of the 20th century, with the first report of pine wilt disease attributable to the nematode in 1905 (Mamiya, 1983), although it was not until 1971 that the nematode was confirmed as the causal agent of pine wilt disease (Kiyohara and Tokushige, 1971). From here it has subsequently spread to other Asian countries including China, Korea and Taiwan. Many native tree species in this region, such as Pinus thunbergii (Japanese black pine) and P. densiflora (Japanese red pine), are highly susceptible to B. xylophilus and the damage caused is extensive. Infected trees can die less than a year after infection given appropriate environmental conditions (see below). Damage in Japan peaked in 1978–1979 due to abnormally warm weather with over 2 million cubic metres of timber lost to the disease (Nose and Shiraishi, 2007). Over 600 thousand cubic metres of forest are destroyed and damage estimated at US$10 million is caused each year in this region.
Long‐range spread of B. xylophilus occurs as a result of human activity. Most often the nematodes are thought to be transported in timber, frequently that used for production of packaging materials. Nematodes and their vectors may be transported separately or the nematode may be transported within the beetle in the wood. These factors drive the development of quarantine regulations that aim to prevent spread and introduction of the pathogen. These regulations may include a requirement for inspections of wood, insistence that all imported wood material is heat treated or a complete ban on importation of timber products from infected regions (Dwinell, 1997). The fallibility of these methods is demonstrated by the discovery of B. xylophilus in Portugal in 1999 (Mota et al., 1999), the first report of this nematode in native conifer stands within the EU. Achieving control of B. xylophilus once introduced is extremely difficult and clear cutting and replanting of infested areas is widely practised. In Portugal, a national programme for the control of the pine wood nematode, ‘PROLUNP’, was implemented immediately after the discovery of the nematode. This action was backed with EU financial support and includes yearly inspections of infected regions. A national survey was also undertaken in order to determine the area in which the nematode was located. Where the nematode was detected (a 30‐km‐radius area south‐east of Lisbon) all symptomatic trees were felled, approximately 50 000 trees per year (Rodrigues, 2006). More recently, in 2007, a 5‐km‐wide phytosanitary strip surrounding the affected zone was been established in which all P. pinaster trees have been felled. These actions have been complemented by additional inspections by forest authorities and police and are similar to those used in other regions of the world in which B. xylophilus is detected for the first time. Treatment of clear‐cut trees in infected areas is also used and may include methyl bromide fumigation (where legally permissible), high‐temperature treatment of wood before use, or chipping and burial. Vector beetles can be targeted using terpene‐baited traps or sprays of insecticides and this treatment can reduce death rates of trees in infested areas. Although one report exists describing eradication of B. xylophilus from an island in Japan (Muramoto, 1999), where large areas are infested these methods do not usually give effective control and for most regions improved quarantine measures to prevent introduction or spread of the pathogen are the only effective tool available.
The complex ecology of B. xylophilus means that in order for the disease to spread, all three factors—a susceptible host, the nematode and the vector insect—need to be present and combined with permissive temperatures. Unfortunately, this appears to be the case in Portugal and, as the most important Pinus species grown in Europe are susceptible to B. xylophilus (Braasch, 1997), the presence of the nematode in Europe poses a serious problem to the forestry industry. Furthermore, the problems posed by B. xylophilus are likely to be exacerbated by climate change. Serious pine wilt disease is associated with higher temperatures, occurring only where average summer temperatures exceed 20 °C (e.g. Melakeberhan et al., 1992). If summer temperatures in Europe rise as predicted, the frequency of pine wilt disease events will potentially increase and the area of Europe in which it becomes possible for pine wilt disease to occur will enlarge further towards the north. B. xylophilus therefore represents a problem that will increase in Europe in coming years in spite of the measures taken to control its spread, thus causing both economic damage to the forest and forest products industries, as well as threatening forest ecosystems (Vieira and Mota, 2007).
PATHOGENICITY
Development of disease
The pathogenic process and development of disease have been summarized in a seminal review by Mamiya (1983). This article was of particular significance because it made much important early work on pine wilt disease originally published in Japanese accessible to a wider audience for the first time. Nematode infection of trees occurs via wounds made by beetles feeding on twigs (Shibata and Okuda, 1989). Nematodes can infect trees up to 79 days after emergence of adult beetles (Li et al., 2007). After invasion the nematodes begin feeding on parenchymal cells. Numerous experiments have shown that nematodes move rapidly from the inoculation point and enter woody tissues via resin canals of the xylem and cortex, feeding on their epithelial cells (Ichihara et al., 2000). Within 2 days the majority of the nematodes have moulted to the adult stage (Kiyohara et al., 1975) and it has been shown that woody tissues promote moulting of dauer‐stage larvae to adults in vitro (see Mamiya, 1983). The nematodes then begin to migrate, feed and reproduce within the resin canals, with the cambium cells seemingly used by the nematodes as a favoured route of migration. Vascular function is compromised due to two distinct phenomena. The vascular system may become blocked by secondary resin originating from radial parenchyma cells damaged as a result of nematode infection. In addition, cavitation, possibly caused as a result of increased production of volatile defence compounds, may disrupt water transport; volatile terpenes can be detected by gas chromatography as soon as 3 days after infection (Kuroda, 2007). Further evidence for the important role of cavitation in the disease process is provided by the observation that xylem dysfunction begins 2–3 weeks after inoculation before other visible symptoms and before the physical occlusion of water conduits by resin (Kuroda, 2007). Nematode levels in the infested tissues rise to between 1000 and 10 000 nematodes per gram of wood tissue, depending on the age of the host and on the extent of colonization by blue‐stain fungi (Kiyohara et al., 1975; Mamiya, 1972). As the host becomes increasingly diseased the nematode population decreases and the proportion of pre‐dauer larvae increases (Mamiya, 1972).
The pathological response of trees to nematode invasion can be seen as soon as 24 h after infection with death of the parenchymal cells near the axial resin canals and these symptoms continue to develop as the nematode spreads and multiplies. Tissues are destroyed mechanically as the nematode migrates through the host (Mamiya, 1980) but it is notable that the first cell death occurs before any major increase in nematode numbers. The severity of the observed symptoms and of the incidence of pine wilt are related to host species and also to temperature and hence to the time of year that inoculation or infection occurs. Inoculation of trees in summer when temperatures are high results in rapid death of trees (as little as 40–60 days) whereas inoculations in spring take longer to develop symptoms and inoculations in autumn or winter may result in no development of symptoms (Kiyohara and Tokushige, 1971). Pine seedlings inoculated experimentally and kept at 25–30 °C develop symptoms of disease whereas those maintained at lower temperatures (15–20 °C) do not (Kiyohara, 1973). Some of the most severe symptoms of disease are caused by failure of water transport and consequently water stress can exacerbate the symptoms of B. xylophilus infection (Suzuki and Kiyohara, 1978). Although discoloration of the pine needles and death of the tree are the most obvious symptoms of disease, these are not visible until some considerable time after infection. One of the earliest detectable symptoms of pine wilt disease is a reduction or cessation of oleoresin flow from artificial wounds made on trunks of trees. This encouraged development of a technique for measurement of oleoresin flow as a means of identifying diseased trees and which was particularly useful before the nematode was identified as the causal agent of pine wilt disease (Oda, 1967).
The observation that cell death in the host seems to occur in advance of any major increase in nematode population led to speculation that the nematode may produce phytotoxins responsible for the cell death in the host. Phytotoxins, including 8‐hydroxycarbotanacetone and 10‐hydroxyverbenone, were subsequently identified and characterized from nematode‐infested plants (Bolla et al., 1982; Oku et al., 1979). More recently a potential role for bacterial symbionts in the disease process has been proposed. A series of studies have shown that bacteria from various genera including Pseudomonas, Pantoea (Han et al., 2003) and Xanthomonas (Higgins et al., 1999) are associated with B. xylophilus. Structural studies have not shown bacteria within B. xylophilus, as has been described for entomopathogenic nematodes that use bacteria as symbionts to parasitize insects (Goodrich‐Blair, 2007), but the presence of bacteria on the nematode surface has been described (Kusonoki, 1987). Large numbers of bacteria have been described as being associated with damaged areas of plants infected with B. xylophilus (Kusonoki, 1987). In addition, it has been shown that Pseudomonas and Pantoea were present in trees infected with B. xylophilus but were absent from uninfected trees (Han et al., 2003). This study also provided perhaps the most convincing evidence available for a role for bacteria in the disease process as it demonstrated that trees infected with any of the strains of bacteria in the absence of nematodes or with asceptic nematodes alone did not develop disease symptoms but that a combination of nematode and bacteria led to disease. Previous studies had also suggested that axenic nematodes were not as pathogenic as B. xylophilus extracted from natural infections (Cao et al., 2001; Kawazu and Kaneko, 1997). It is possible that the bacteria associated with nematodes are responsible for production of phytotoxins associated with cell death in the host as bacterial culture filtrate causes death of callus material derived from trees (Han et al., 2003) and nematodes cultured axenically do not produce phytotoxin (Cao et al., 2001). It has also been shown that certain Pseudomonas species enhanced reproduction rates of B. xylophilus on trees and that the presence of the nematode also enhanced the reproduction rate of these bacteria, indicating a mutualistic symbiotic relationship (Zhao and Lin, 2005). Bacteria associated with the nematode that produces phytoxins also increased egg production and accelerated growth and development of B. xylophilus in callus cultures (Zhao et al., 2007).
Although the studies outlined above apparently provide convincing evidence for the role of bacteria we feel that a note of caution needs to be applied. Bacteria alone cannot cause disease (Han et al., 2003). In addition, the tests used to assess pathogenicity used callus or very young seedlings as host material and allowed B. mucronatus to be scored as highly pathogenic. This is a situation that is never observed in the field. The issue of testing for pathogenicity is discussed in more detail below. Our own studies using nematode infections applied in a more natural context (see below) suggest that the presence of bacteria may bring about earlier death of a host but that axenic nematodes will eventually cause death of a host (Li and Moens, in preparation). Further studies in this area are merited.
Pathogenic status of other Bursaphelenchus species
Although it is widely recognized that the only truly pathogenic species of Bursaphelenchus in a natural (field) context is B. xylophilus, there are many reports in the literature of other species exerting a pathogenic effect. For example, several of the studies on the role of bacteria in the pathogenic process discussed above describe experiments in which B. mucronatus exhibits a profound pathogenic effect, particularly when mixed with the most virulent bacterial strains (Han et al., 2003). B. mucronatus has also been described as being associated with trees exhibiting symptoms of pine wilt in France (Baujard et al., 1979). Other species, including B. sexdentati and B. leoni, have been described as pathogenic (e.g. Skarmoutsos and Michalopoulos‐Skarmoutsos, 2000). However, there is no evidence that any of these species are truly pathogenic under field conditions and phylogenetic analysis does not suggest that the allegedly pathogenic species are clustered (Ye et al., 2007). A potential problem with the work described above relates to the methods used to assess pathogenicity. In particular, the methods used to inoculate trees with nematodes (injection of thousands of juveniles) may not relate to the situation in nature where limited numbers (no more than 300 —Li et al., 2007) of nematodes infect wounds made by vector beetles during feeding. The host material used in these studies may also give rise to false‐positive results—many studies are based on artificial inoculation of very young pine seedlings or even pine callus material. Such tests have been criticised due to their tendency to produce variable results (McNamara, 2004). Methods for delivering nematodes to larger trees from insects under field conditions have been developed (Li and Moens, in preparation). Under these conditions B. mucronatus is not pathogenic and wider application of these methods will help shed light on the pathogenic status of other Bursaphelenchus species.
THE PLANT RESPONSE
Resistance
Natural resistance is the most effective means of controlling almost all pests and diseases and different pine species vary in their susceptibility to B. xylophilus. For the purposes of this article we consider ‘resistance’ in the sense that it is best applied to plant–nematode interactions; a resistant plant prevents multiplication of a nematode population so that the nematode numbers after infection (Pf) are lower than the number of nematodes that infect a plant (Pi) (Trudgill, 1991). Such a definition implies the action of a resistance gene (or genes) and, possibly, a ‘classical’ hypersensitive response. Although a hypersensitive response against infection sites of B. xylophilus has been reported in resistant trees (Myers, 1988), it is clear that the ‘resistance’ described in many studies of B. xylophilus—pine interactions is in fact tolerance; this is defined in plant nematology as an ability to withstand or recover from damage caused by a nematode (Trudgill, 1991).
Two main strategies have been used, particularly in China and Japan, in resistance breeding programmes: selection and cross breeding. Selection programmes exploit the rare, naturally occurring disease ‘resistant’ individual plants in forests of normally susceptible species that occur in Japan and China. It has been calculated that such plants exist at a frequency of between 0.1 and 1 in 1 million (Ohoba, 1976). These plants are readily identified in areas of forests devastated by B. xylophilus. One advantage of using this strategy is that the starting point is mature trees, enabling selections on the basis of other desirable traits (e.g. growth, straightness) to be made before the resistance breeding programme has started. Several different breeding programmes using this strategy are currently underway and numerous potentially useful lines have been identified (Peng and Moens, 2003). Given that indigenous tree species have had no prior exposure to B. xylophilus it seems unlikely that genuine resistance will be identified using this approach and that tolerant lines are in fact being identified. Indeed, analysis of one such selected line suggested that the failure of nematode development was due to differences in structure of the resin canals that impeded normal nematode migration, rather than being due to a defence response of the plant to the nematode (Kuroda, 2004). Various marker development programmes are underway; these suggest that identified ‘resistance’ may be additive when combined and that several independent quantitative trait loci (QTLs) can be identified that are linked to resistance. Marker programmes are being developed in order to allow cloning of QTLs controlling resistance to nematodes (Isoda et al., 2007; Kuramoto et al., 2007).
Cross breeding exploits natural resistance found in tree species that have co‐evolved with B. xylophilus. Although this approach is more likely to identify genuine resistance genes, a complication is that material that progresses through such breeding programmes needs to be further analysed for required traits other than resistance. Japanese red and black pine (Pinus densiflora and P. thunbergii) have been used in crossing programmes against loblolly pine (P. banksiana), eastern white pine (P. strobus) and table mountain pine (P. pungens), each of which is highly resistant to B. xylophilus (Nose and Shiraishi, 2007). Masson pine (P. massoniana), which is common in China, has also been used in these programmes and genetic analysis of the resulting progeny suggested the presence of a single dominant resistance gene (Sasaki et al., 1983). The progeny resulting from interspecific crosses have been used in reforestation programmes in some areas (Peng and Moens, 2003).
Breeding for natural resistance (or indeed almost any other trait) in trees presents a number of challenges. Most notable is the time taken for the ‘crop’ to reach a sufficient level of maturity for the desired trait to be assessed and it is this reason that drives development of marker‐assisted selection programmes. In addition, grafting and tissue culture can be used to speed the multiplication of important lines (Peng and Moens, 2003). For most crop breeding programmes desirable characteristics are introgressed into cultivars possessing desirable agronomic characteristics by repeated crossing and backcrossing, a process that is not practical in forest tree breeding. In these programmes F1 lines are put to use directly, meaning that care needs to be taken in selection of parental lines (Nose and Shiraishi, 2007). Further complexity is added by the fact that much of the ‘resistance’ that has been identified in the selection strategy described below is controlled by QTLs rather than single dominant R genes. An additional challenge when breeding for resistance against B. xylophilus is the requirement for robust tests of pathogenicity; the problems in assessing nematode pathogenicity discussed above apply equally to experiments that aim to assess resistance against a defined population. Time limitations mean that seedling tests are most commonly used in breeding programmes for resistance. It would seem likely that false negatives (i.e. potentially useful resistance being discarded as susceptible material) are common, given the tendency of seedling tests to score non‐pathogenic nematodes as pathogens. A further concern is that seedling tests will not expose trees to the full extent of virulence that is present in the field, meaning that it is difficult to assess the sustainability of the resistance sources that are being developed. Nematodes in the field vary widely in their virulence (or aggressiveness) (Kiyohara and Bolla, 1990). It is also known that a great deal of movement of B. xylophilus has taken place since its initial introduction; for example B. xylophilus is thought to have migrated to the Kyushu region of Japan four times since the initial introduction (Nose and Shiraishi, 2007). In spite of the potential concerns outlined above field testing of resistant pine lines selected in artificial inoculation tests on seedlings has shown promising levels of durable resistance (Toda, 2004).
It has been suggested that resistance can be induced in susceptible trees by prior inoculation with avirulent nematodes (reviewed by Kosaka et al., 1991). Little is known about the mechanisms underlying this phenomenon and, in the absence of a detailed understanding of the molecular basis of pathogenicity and virulence in the nematodes, it seems unlikely that this will be a widely used method of controlling pine wilt disease.
The molecular biology of the host response to nematode infection
Many Pinus species are economically important for the forestry industry throughout the world and consequently several, including Pinus species, are the focus of genomics projects (see http://www.pinegenome.org for details). Significant publicly accessible expressed sequence tag (EST) data sets exist for P. taeda (loblolly pine) and P. pinaster (maritime pine) and these have given rise to the development of tools and techniques for functional genomics in many tree species (e.g. Gion et al., 2005; Plomion et al., 2003). In spite of this, little is known about the molecular basis of the response of Pinus to infection with B. xylophilus and indeed about the nature of tree defences against pathogens more widely. Herbaceous plants respond dramatically to challenge with pathogens; changes in expression patterns of 20% of transcribed genes in response to the presence of a pathogen have been described (Nimchuk et al., 2003). Plant responses may include localized cell death, activation of signalling pathways, synthesis of PR proteins that may directly affect pathogens, cell‐wall strengthening and synthesis of toxins. One recent large‐scale study identified Populus genes whose expression profiles changes in response to infection with poplar leaf rust (Melampsora medusae) (Miranda et al., 2007). Genes encoding components of the flavonoid pathway were among the most strongly induced genes and biochemical analysis confirmed an accumulation of proanthocyanidins in response to infection. The flavonoid pathway clearly forms an important aspect of the response of trees to pathogens and one set of compounds produced from one branch of the flavonoid pathway, stilbenoids, are known to be toxic to B. xylophilus (Suga et al., 1993). Enzymes that control production of stilbenoids have been identified from P. densiflora and also regulate other enzymes in the flavonoid synthetic pathway (Kodan et al., 2002). In studies specifically analysing the effects of B. xylophilus on the host, ESTs whose expression patterns change in response to infection have been identified from P. densiflora. In this study a subtracted library was generated enriched for genes differentially expressed in resistant and susceptible tree lines 19 days after infection. A thaumatin‐like protein, chitinase, metallothionein and antimicrobial peptides were among the most abundantly represented genes in this library and qRT‐PCR was used to confirm the differential expression of some of the genes after inoculation (Watanabe et al., 2007). Several of the genes identified have previously been identified as PR proteins, suggesting a systemic response to nematode infection.
MOLECULAR BASIS OF PARASITISM
Studies on the molecular biology of the interactions between B. xylophilus and its host and on the molecular basis of pathogenicity of the nematode were, until recently, few and far between. Molecular studies on B. xylophilus tended instead to be focused on understanding the phylogenetic relationships of various Bursaphelenchus species and, in particular, on development of diagnostic tools that distinguish between B. xylophilus and other species (e.g. Kang et al., 2004; Kanzaki and Futai, 2002; Leal et al., 2005; Matsunaga and Togashi, 2004; Ye et al., 2007). Such studies continue and have been used, for example, to demonstrate that the B. xylophilus present in Portugal probably arose from a single introduction from East Asia (Vieira et al., 2007). This contrasts markedly with the situation for sedentary endoparasitic nematodes, including the root‐knot and cyst nematodes. For these nematodes progress has been made in understanding the nature of the proteins used by the nematodes to successfully invade plants (e.g. Smant et al., 1998), avoid host defences (e.g. Robertson et al., 2000) and establish feeding sites (Huang et al., 2006; Wang et al., 2005; Weerasinghe et al., 2005). Much of this progress has been underpinned by EST projects that have allowed identification of genes important in host–parasite interactions. In addition, a variety of approaches, including microarrays, have been used to identify host genes whose expression is modified as a result of nematode infection (Gheysen and Fenoll, 2002; Ithal et al., 2007). Resistance genes targeting root‐knot and cyst‐forming nematodes have been identified (Williamson and Kumar, 2006). However, this imbalance has now begun to be rectified as a result of a large‐scale EST project undertaken for B. xylophilus and B. mucronatus (Kikuchi et al., 2007) and subsequent functional studies on candidate pathogenicity genes.
One of the key findings from the analysis of B. xylophilus ESTs is that, like sedentary endoparasitic nematodes, the nematode has a variety of endogenous plant cell‐wall‐degrading enzymes that allow it to invade and migrate within its host. A family of secreted cellulases (β‐1,4 endoglucanases) are present (Kikuchi et al., 2004) as well as pectate lyases (Kikuchi et al., 2006) and these enzymes are produced in the oesophageal gland cells and secreted via the stylet. Immunological studies have shown that cellulases are detectable in tracheid cells and in resin canals in infected tissues and it has been suggested that the activity of these cell‐wall‐degrading enzymes is the cause of early disease symptoms (Kojima et al., 1994; Zhang et al., 2006). It has also been suggested that differences in cellulase activities and isoforms may be correlated with differences in virulence (Kojima et al., 1994) but further work in this area is needed to confirm this now that sequences of the genes are available. A preliminary analysis of variation in cellulase‐ and pectate lyase‐encoding cDNA sequences from B. xylophilus populations of varying virulence did not reveal any polymorphisms associated with virulence (H. Li, M. Moens and J. Jones, unpublished observations). Although these cell‐wall‐degrading enzymes clearly play an important role in the interaction of B. xylophilus with its host it is clear that they are not the sole determinants of pathogenicity, as they are also present in ESTs derived from the non‐pathogenic B. mucronatus. It is likely therefore that they are present simply to aid the nematode's migration through plant tissues, whether the plant is alive or dead. Further analysis of these sequences in unequivocally non‐parasitic species would confirm this.
Bursaphelenchus is not related to cyst and root‐knot nematodes and it is thought that plant parasitism in these groups has arisen independently (Baldwin et al., 2004). The availability of comprehensive EST data sets from Bursaphelenchus and from cyst/root‐knot nematodes therefore raises the possibility that comparative genomics could be used to help understand the basic requirements for plant parasitism by nematodes or genes associated with traits specific to each group. A comparison of the ‘secreteomes’ of B. xylophilus with those of root‐knot and cyst nematodes may be particularly informative. For example, Bursaphelenchus contains genes encoding secreted endo‐β‐1,3 glucanases (Kikuchi et al., 2005) while similar genes are not present in cyst and root‐knot nematodes. It is thought that this protein may help Bursaphelenchus spp. metabolize the cell wall of the fungi on which they feed. Comparisons of EST data sets may also be useful in helping to identify root‐knot/cyst nematode proteins important in the biology of the feeding sites induced by these species.
One intriguing finding that has emerged from the analysis of B. xylophilus and B. mucronatus ESTs and comparative studies with cyst and root‐knot nematodes is that multiple independent horizontal gene transfer events have driven evolution of plant parasitism in both groups. The cellulases present in the two groups are not related to each other and it is thought that those present in Bursaphelenchus were originally obtained from fungi while those present in cyst and root‐knot nematodes are likely to have been transferred from bacteria (Jones et al., 2005). The endo‐β‐1,3 glucanases present in Bursaphelenchus are thought to have been obtained via horizontal gene transfer from bacteria but are not present in root‐knot or cyst nematodes. Similarly, root‐knot nematodes contain a gene encoding a nod‐L‐like protein and this is not present in cyst nematodes or in Bursaphelenchus (Scholl et al., 2003). A case for at least five independent horizontal gene transfer events can be made on the basis of the ESTs available to date (Jones et al., 2005) and analysis of genome sequences (see below) will allow the full extent of horizontal gene transfer within these groups to be determined.
Although it has proved relatively simple to identify candidate pathogenicity genes in EST data sets, analysis of their function, particularly in vivo, is likely to prove more challenging. Biochemical tests have been used to test predicted biochemical functions of cellulases and endo‐β‐1,3 glucanases from B. xylophilus and in situ hybridization has been used to confirm spatial expression patterns of the genes (Kikuchi et al., 2004, 2005). RNA interference (RNAi) offers the prospect of analysing gene function in vitro on a routine basis and methods have been developed for use of RNAi with both cyst and root‐knot nematodes that have been used for studies of pathogenicity genes (Chen et al., 2005; Rosso et al., 2005; Urwin et al., 2002). However, no reports of similar studies exist for B. xylophilus. Our own work has shown that B. xylophilus and B. mucronatus take up fluorescently labelled double‐stranded RNA from solution (Fig. 2) without the need for the presence of neurotransmitters or other stimulants as used for root‐knot and cyst nematodes (Rosso et al., 2005; Urwin et al., 2002). Efforts to develop a system for RNAi in Bursaphelenchus are continuing in two of our laboratories (J. Jones, T. Kikuchi, H. Li and M. Moens, unpublished data). An alternative approach to functional analysis may be the use of forward genetic analysis. The life cycle of B. xylophilus suggests that such analysis should be feasible. The nematode reproduces sexually and studies investigating inheritance of virulence/pathogencity that use crossing of closely related species have been reported (Bolla and Boschert, 1993; Hajaukiewicz and Myers, 1988; Riga et al., 1992). The nematode can be maintained on fungal cultures for its entire life cycle and then used to infect plants. This offers the possibility of maintaining crossed, or even mutant, lines of the nematode in the laboratory that can subsequently be used in biological experiments investigating pathogenicity. Although establishing genetics as a routine tool for B. xylophilus would require a major investment of time and resources, there appear to be no practical obstacles to the use of this powerful technology.
Figure 2.

Uptake of fluorescently labelled dsRNA by B. xylophilus adult. (A) Nematode viewed under light microscope; (B) same nematode viewed under fluorescence optics. Fluorescently labelled dsRNA was generated using the Megascript RNAi kit (Ambion) following the manufacturer's instructions but substituting Cy3‐labelled UTP (GE Healthcare) for unlabelled UTP in the synthesis reaction. Nematodes were incubated in the labelled dsRNA for 24 h in the dark on a rotator in the presence of 3 mm spermidine and 0.05% gelatin.
Despite the recent progress outlined above, our understanding of the mechanisms of pathogenicity by Bursaphelenchus remains sketchy. No obvious clues as to why B. xylophilus is such a formidable pathogen whereas B. mucronatus is not are immediately obvious from comparisons of ESTs from the two species. No genes encoding enzymes that may synthesize phytotoxins, for example, are present in B. xylophilus that are absent from B. mucronatus, although identification of such genes currently relies on BLAST hits against annotated matches in databases. More detailed bioinformatic analysis of the sequences present in each species coupled to functional studies may lead to identification of genes associated with pathogenicity in B. xylophilus. However, the absence of clear differences between the two species may support the idea that bacteria or other factors (such as behavioural differences) play an important role in pathogenicity.
OPPORTUNITIES FOR GENOMICS
From the above it should be clear that B. xylophilus represents a significant threat to forestry in the Far East and in the EU and that, as a result of climate change, the threat from this pathogen is likely to increase. It is also clear that many of the significant advances in our understanding of the biology of this organism have come from application of molecular techniques, for example through analysis of EST data sets. We believe that a strong case can be made for genome‐scale analysis of the interactions between B. xylophilus and its hosts and that such studies present enormous opportunities for scientific progress at several levels. Whereas in the past genome‐scale projects were prohibitively expensive, the costs associated with these projects are decreasing and the infrastructure is becoming more efficient. At the time of writing, 634 genome sequencing projects are complete, with a further 2136 genome projects in progress (http://www.genomesonline.org/index.htm). Even where genome sequences are not available, more accessible means of generating large‐scale data sets, such as ESTs, may be present. It is probably fair to say that obtaining raw DNA sequence no longer represents the major bottleneck for genome projects. The major challenge is for functional analysis and, in particular, high‐quality bioinformatic analysis of the sequences that are obtained.
Genomic analysis of bacteria carried by B. xylophilus
The relatively small genome size for most bacterial species means that genome sequencing projects for bacteria are becoming almost routine. At the time of writing, 523 bacterial genome sequences have been completed and a further 1305 sequencing projects are in progress. The large numbers of bacterial sequences that have been obtained mean that comparative genomics can be used to identify genome regions associated with specific biological traits (Galperin and Kolker, 2006). As described above, various species of bacteria seem to be associated with B. xylophilus and there is some evidence that suggests these bacteria play a role in the disease process. The bacteria most commonly described as being associated with B. xylophilus are from the genus Pseudomonas. There are currently 13 different published Pseudomonas genome sequences and a further 14 sequences in progress. These sequences include animal and plant pathogenic bacteria. Powerful tools have been developed that allow comparison of entire bacterial genome sequences and identification of regions of the genome that contain important pathogenicity‐associated genes (Pritchard et al., 2006). Using this and other bioinformatics tools would allow the scale of the involvement of bacteria in pine wilt disease and a better understanding of the pathogenic process to be determined. Sequencing of the genomes of other bacteria associated with nematodes, particularly bacteria associated with entomopathogenic nematodes, has led to the identification of genes encoding novel toxins (e.g. Marokhazi et al., 2003) and it is reasonable to assume that similar findings might emerge from the sequence of a bacterium associated with B. xylophilus. Given the relatively low costs of bacterial genome sequencing, a project in this area would seem to offer excellent value for money.
Exploitation of resources available from the tree genomics community
Wood is one of the most important natural resources exploited by humans and, as a consequence, much effort has been put into developing genomics tools for a range of tree species. Most work using these resources has focused on agronomically important traits such as wood formation and responses to abiotic stress. However, analysis of changes in the transcriptome of poplar in response to insect and viral pathogens has been undertaken and has yielded insights into responses of these trees to these pathogens (Major and Constabel, 2006; Ralph et al., 2006; Smith et al., 2004). Similar collections of ESTs and cDNA arrays are available for Pinus species. A large amount of information about the nature of the response of Pinus to infection with Bursaphelenchus could be collected by exploitation of these resources, particularly by comparisons of infections with virulent and avirulent species/lines.
Genomics of B. xylophilus
Clearly a full genome sequence would provide an enormous amount of information on the biology of B. xylophilus, which would allow the mechanisms underlying pathogenicity in this nematode to be far better analysed. Scientifically valuable information, including a full understanding of the extent of horizontal gene transfer in this species, would also be obtained. It should be noted, however, that well‐defined systems for functional analysis, including robust pathogenicity assays, will be required in order for a full understanding of the role of various nematode genes in pathogenicity to be obtained.
A genome sequence would also provide information that could lead to novel control avenues for this pathogen; knowledge of the full complement of genes present in B. xylophilus would provide information on targets such as enzyme pathways or essential nematode genes. Although the size of the B. xylophilus genome has not been published, the genome sizes of the closely related B. mucronatus and another aphelenchid nematode Aphelenchus avenae have both been calculated to be fairly small, at around 34 Mbp (Leroy et al., 2007). The enormous economic damage attributable to this nematode may mean that the practical benefits emerging would therefore outweigh the still considerable costs of sequencing and annotating the B. xylophilus genome. In addition to the direct benefits that could be obtained, genome sequencing projects are now underway for several plant parasitic nematodes including root‐knot (Meloidogyne hapla, M. incognita) and cyst nematodes (Globodera pallida). Genome sequences for several free‐living (Caenorhabditis) and animal‐parasitic (e.g. Brugia malayi, Haemonchus contortus) species are also available or in progress. Comparative genomics within the Nematoda is therefore a genuine prospect that offers exciting possibilities. Comparing all available plant‐parasitic nematode genomes with those of free‐living or animal‐parasitic nematodes will allow the essential gene complements required for parasitism of plants by nematodes to be identified as well as provide an evolutionary history of parasitism within the nematodes. Comparisons of a B. xylophilus genome sequence with those of root‐knot and/or cyst nematodes will allow the genes associated with the various forms of plant parasitism by nematodes (sedentary parasites and migratory parasites) to be identified. When linked to information on the responses of the various hosts, the development of this area clearly offers the potential for spectacular progress.
ACKNOWLEDGEMENTS
Some of the work described in this article was funded by an International Joint Project of the Royal Society awarded to J. Jones and M. Moens. T. Kikuchi has been funded by Japanese Ministry of Education, Science, Sports and Culture, Grant‐in‐Aid for Encouragement of Young Scientists (B) 18780032 and FFPRI Research grant no. 200302, M. Mota has been funded by ICAM (University of Évora) and two EU projects EU‐SSPE‐RTD‐502348 (PORTCHECK) and QLK5‐CT‐2002–00672 (PHRAME). H. Li received a scholarship from the VLIR for PhD studies. SCRI receives funding from the Rural and Environment Research and Analysis Directorate of the Scottish Government.
REFERENCES
- Baldwin, J.G. , Nadler, S.A. and Adams, B.J. (2004) Evolution of plant parasitism among nematodes. Annu. Rev. Phytopathol. 42, 83–105. [DOI] [PubMed] [Google Scholar]
- Baujard, P. , Boulbria, A. , Ham, R. , Laumond, C. and Scotto la Massese, C. (1979) First data on the nematode‐fauna related to decays of maritime‐pine in Western France. Ann. Sci. Forestieres, 36, 331–339. [Google Scholar]
- Blaxter, M.L. , De Ley, P. , Garey, J.R. , Liu, L.X. , Scheldeman, P. , Vierstraete, A. , Vanfleteren, J.R. , Mackey, L.Y. , Dorris, M. , Frisse, L.M. , Vida, J.T. and Thomas, W.K. (1998) A molecular evolutionary framework for the Phylum Nematoda. Nature, 392, 71–75. [DOI] [PubMed] [Google Scholar]
- Bolla, R. , Shaheen, F. and Winters, R.E.K. (1982) Phytotoxin produced in Bursaphelenchus xylophilus infected Pinus sylvestris In: Proceedings of the National Pine Wilt Disease Workshop, Rosemont, Illinois, 17–31. Campaign, IL ( Appleby J.E. and Marck R., eds): Springfield Illinois Department of Energy and Natural Resources. [Google Scholar]
- Bolla, R.I. and Boschert, M. (1993) Pinewood nematode species complex: interbreeding potential and chromosome number. J. Nematol. 25, 227–238. [PMC free article] [PubMed] [Google Scholar]
- Braasch, H. (1997) Host and pathogenicity tests with pine wood nematode (Bursaphelenchus xylophilus) from North America under Central European weather conditions. Nachrichtenblatt Deutschen Pflanzenschutzdienstes, 49, 209–214. [Google Scholar]
- Cao, Y. , Han, Z.M. and Li, C.D. (2001) Studies on wilting toxic substances produced in pines infected by pine wood nematodes. Scienta Silvae Sinicae, 37, 75–79. [Google Scholar]
- Chen, Q. Rehman, S. , Smant, G. and Jones, J.T. (2005) Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Mol. Plant–Microbe Interact. 18, 621–625. [DOI] [PubMed] [Google Scholar]
- Duncan, L.W. and Moens, M. (2006) Migratory endoparasitic nematodes In: Plant Nematology (Perry R.N. and Moens M., eds), pp. 123–152. Wallingford, UK: CABI. [Google Scholar]
- Dwinell, L.D. (1997) The pinewood nematode: Regulation and mitigation. Annu. Rev. Phytopathol. 35, 153–166. [DOI] [PubMed] [Google Scholar]
- Galperin, M.Y. and Kolker, E. (2006) New metrics for comparative genomics. Curr. Opin. Biotechnol. 17, 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen, G. and Fenoll, C. (2002) Gene expression in nematode feeding sites. Annu. Rev. Phytopathol. 40, 191–219. [DOI] [PubMed] [Google Scholar]
- Gion, J.M. , Lalanne, C. , Le Provost, G. , Ferry‐Dumazet, H. , Paiva, J. , Chaumeil, P. , Frigerio, J.M. , Brach, J. , Barre, A. , De Daruvar, A. , Claverol, S. , Bonneu, M. , Sommerer, N. , Negroni, L. and Plomion, C. (2005) The proteome of maritime pine word forming tissue. Proteomics, 5, 3731–3751. [DOI] [PubMed] [Google Scholar]
- Goodrich‐Blair, H. (2007) They've got a ticket to ride: Xenorhabdus nematophila–Steinernema carpocapsae symbiosis. Curr. Opin. Microbiol. 10, 225–230. [DOI] [PubMed] [Google Scholar]
- Hajaukiewicz, P.T. and Myers, R.T. (1988) Experimental crossing of selected isolates of Bursaphelenchus xylophilus and B. mucronatus . Phytopathology, 78, 1507–1509. [Google Scholar]
- Han, Z.M. , Hong, Y.D. and Zhao, B.G. (2003) A study on pathogenicity of bacteria carried by pine wood nematodes. J. Phytopathol. 151, 683–689. [Google Scholar]
- Higgins, D.F. , Harmey, M.A. and Jones, D.L. (1999) Pathogenicity related gene expression in Bursaphelenchus xylophilus In: Sustainability of Pine Forests in relation to Pine Wilt and Decline. Proceedings of an International Symposium (Kazuyoshi F., Togashi K. and Ikedu T., eds), pp. 23–28. Kyoto, Japan: Shokado Shote. [Google Scholar]
- Holterman, M. , Van Der Wurff, A. , Van Den Elsen, S. , Van Megen, H. , Bongers, T. , Holovachov, O. , Bakker, J. and Helder, J. (2006) Phylum‐wide analysis of SSU rDNA reveals deep phylogenetic relationships among Nematodes and accelarated evolution towards crown clades. Mol. Biol. Evol. 23, 1792–1800. [DOI] [PubMed] [Google Scholar]
- Huang, G.Z. , Dong, R.H. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2006) A root‐knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol. Plant–Microbe Interact. 19, 463–470. [DOI] [PubMed] [Google Scholar]
- Ichihara, Y. , Fukuda, K. and Suzuki, K. (2000) Early symptom development and histological changes associated with migration of Bursaphelenchus xylophilus in seedling tissues of Pinus thunbergii . Plant Dis. 84, 675–680. [DOI] [PubMed] [Google Scholar]
- Isoda, K. , Watanabe, A. , Kuramoto, N. and Kondo, T. (2007) Linkage mapping of Pinus densiflora and QTL analysis for resistance to the pine wilt disease Plant Anim. Genomes XV Conference, p. 513. San Diego (Abs). [Google Scholar]
- Ithal, N. , Recknor, J. , Nettleton, D. , Maier, T. , Baum, T.J. and Mitchum, M.G. (2007) Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant–Microbe Interact. 20, 510–525. [DOI] [PubMed] [Google Scholar]
- Jones, J.T. , Furlanetto, C. and Kikuchi, T. (2005) Horizontal gene transfer from bacteria and fungi as a driving force in the evolution of plant parasitism in nematodes. Nematology, 7, 641–646. [Google Scholar]
- Kang, J.S. , Choi, K.S. , Shin, S.H. , Moon, I.S. , Lee, S.G. and Lee, S.H. (2004) Development of an efficient PCR‐based diagnostic protocol for the identification of the pinewood nematode Bursaphelenchus xylophilus (Nematoda. Parasitaphelenchidae). Nematology, 6, 279–285. [Google Scholar]
- Kanzaki, N. and Futai, K. (2002) A PCR primer set for determination of phylogenetic relationships of Bursaphelenchus species within the xylophilus group. Nematology, 4, 35–41. [Google Scholar]
- Kawazu, K. and Kaneko, N. (1997) Asepsis of the pine wood nematode isolate OKD‐3 causes it to lose its pathogenicity. Jpn J. Nematol. 27, 76–80. [Google Scholar]
- Kikiuchi, T. , Shibuya, H. and Jones, J.T. (2005) Molecular and biochemical characterization of an endo‐β‐1,3‐glucanase from the pine wood nematode Bursaphelenchus xylophilus . Biochem. J. 389, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, T. , Aikawa, T. , Kosaka, H. , Pritchard, L. , Ogura, N. and Jones, J.T. (2007) EST analysis of the pine wood nematode Bursaphelenchus xylophilus and B. mucronatus . Mol. Biochem. Parasitol. 155, 9–17. [DOI] [PubMed] [Google Scholar]
- Kikuchi, T. , Jones, J.T. , Aikawa, T. , Kosaka, H. and Ogura, N. (2004) A family of GHF45 cellulases from the pine wood nematode Bursaphelenchus xylophilus . FEBS Lett. 572, 201–205 [DOI] [PubMed] [Google Scholar]
- Kikuchi, T. , Shibuya, H. , Aikawa, T. and Jones, J.T. (2006) Cloning and characterization of pectate lyases secreted by the pine wood nematode Bursaphelenchus xylophilus . Mol. Plant–Microbe Interact. 19, 280–287. [DOI] [PubMed] [Google Scholar]
- Kiyohara, T. and Bolla, R.I. (1990) Pathogenic variability among populations of the pinewood nematode, Bursaphelenchus xylophilus . Forest Sci. 36, 1061–1076. [Google Scholar]
- Kiyohara, T. (1973) Effect of temperature in the disease incidence of pine seedlings inoculated with Bursaphelenchus lignicolus . Trans. Meeting Jpn Forestry Society, 84, 334–335. [Google Scholar]
- Kiyohara, T. and Tokushige, Y. (1971) Inoculation experiments of a nematode Bursaphelenchus sp. onto pine trees. Jpn J. Forestry Society, 53, 210–218. [Google Scholar]
- Kiyohara, T. , Suzuki, K. and Hashimoto, H. (1975) Migration of the pine wood nematode in a pine tree at an early stage of infection. Trans. Meeting Jpn Forestry Society, 86, 299–300. [Google Scholar]
- Kodan, A. , Kuroda, H. and Sakai, F. (2002) A stilbene synthase from Japanese red pine (Pinus densiflora): implications for phytoalexin accumulation and down‐regulation of flavonoid biosynthesis. Proc. Natl Acad Sci. USA, 99, 3335–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, K. , Kamuyo, A. , Masumori, M. and Sasaki, S. (1994) Cellulase activities of pine‐wood nematode isolates with different virulences. J. Jpn Forestry Society, 76, 258–262. [Google Scholar]
- Kosaka, H. , Aikawa, T. , Oggura, N. , Tabata, K. and Kiyohara, T. (2001) Pine wilt disease caused by the pine wood nematode: the induced resistance of pine trees by the avirulent isolates of nematode. Eur. J. Plant Pathol. 107, 667–675. [Google Scholar]
- Kuramoto, N. , Isoda, K. , Watanabe, A. , Fujisawa, Y. and Kondo, T. (2007) Genetic analysis of the resistance against the pine wilt disease in Japanese pines In: Plant Anim. Genomes XV Conference, p. 500. San Diego (Abs). [Google Scholar]
- Kuroda, K. (2004) Inhibiting factors of symptom development in several Japanese red pine (Pinus densiflora) families selected as resistant to pine wilt. J. Forestry Res. 9, 217–224. [Google Scholar]
- Kuroda, K. (2007) Physiological incidences related to symptom development and wilting mechanism. In: Pine Wilt Disease (Zhao B. and Futai K., eds), in press. Springer: Japan. [Google Scholar]
- Kusonoki, M. (1987) Symptom development of pine wilt disease—histopathological observations with electron microscopy. Ann. Phytopathol. Soc Jpn, 53, 622–629. [Google Scholar]
- Leal, I. , Green, M. , Allen, E. , Humble, L. and Rott, M. (2005) An effective PCR‐based diagnostic method for the detection of Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae) in wood samples from lodgepole pine. Nematology, 7, 833–842. [Google Scholar]
- Leroy, S. , Bouamer, S. , Morand, S. and Fargette, M. (2007) Genome size of plant‐parasitic nematodes. Nematology, 9, 449–450. [Google Scholar]
- Li, H. , Shen, P. , Fu, P. , Lin, M. and Moens, M. (2007) Characteristics of the emergence of Monochamus alternatus, the vector of Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae), from Pinus thunbergii logs in Nanjing, China, and of the transmission of the nematodes through feeding wounds. Nematology, 9, 807–816. [Google Scholar]
- Maehara, N. and Futai, K. (1996) Factors affecting both the numbers of the pinewood nematode Bursaphelenchus xylophilus (Nematodoa: Aphelenchoididae), carried by the Japanese pine sawyer Monochamus alternatus (Coleoptera: Cerambycidae), and the nematode's life history. Appl. Entomol. Zool. 31, 443–452. [Google Scholar]
- Major, I.T. and Constabel, C.P. (2006) Molecular analysis of poplar defense against herbivory. Comparison of wound‐ and insect elicitor‐induced gene expression. New Phytol. 172, 617–635. [DOI] [PubMed] [Google Scholar]
- Mamiya, Y. (1972) Reproduction of pine lethal wilting disease by the inoculation of young trees with Bursaphelenchus lignicolus . Jpn J. Nematol. 2, 40–44. [Google Scholar]
- Mamiya, Y. (1980) Inoculation of the first year pine (Pinus densiflora) seedlings with the pine wood nematode Bursaphelenchus lignicolus and histopathology of diseased seedlings. Jpn J. Forestry Soc. 62, 176–183. [Google Scholar]
- Mamiya, Y. (1983) Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus . Annu. Rev. Phytopathol. 21, 201–220. [DOI] [PubMed] [Google Scholar]
- Mamiya, Y. and Enda, N. (1972) Transmission of Bursaphelenchus lignicolus (Nematoda: Aphelenchoididae) by Monochamus alternatus (Coleoptera: Cerambycidae). Nematologica, 18, 159–166. [Google Scholar]
- Marokhazi, J. , Waterfield, N. , LeGeoff, G. , Feil, E. , Stabler, R. , Hinds, J. , Fodor, A. and Ffrench‐Constant, R.H. (2003) Using a DNA microarray to investigate the distribution of insect virulence factors in strains of Photorhabdus bacteria. J. Bacteriol. 185, 4648–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, K. and Togashi, K. (2004) A simple method for discriminating Bursaphelenchus xylophilus and B. mucronatus by species specific polymerase chain reaction primer pairs. Nematology, 6, 273–277. [Google Scholar]
- McNamara, D. (2004) Quarantine concerns about the methods used to demonstrate pathogenicity of Bursaphelenchus spp. In: The Pinewood Nematode, Bursaphelenchus xylophilus. Nematology Monographs and Perspectives volume 1 (Mota M. and Vieira P., eds), pp. 187–197. Brill Academic Publishers. [Google Scholar]
- Melakeberhan, H. , Rutherford, T.A. and Webster, J.M. (1992) Influence of temperature on reproduction of Bursaphelenchus xylophilus and Pinus sylvestris mortality. Nematologica, 38, 80–87. [Google Scholar]
- Miranda, M. , Ralph, S.G. , Mellway, R. , White, R. , Heath, M.C. , Bohlmann, J. and Constabel, C.P. (2007) The transcriptional response of hybrid Polar (Populus trichocarpa×P. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Mol. Plant–Microbe Interact. 20, 816–831. [DOI] [PubMed] [Google Scholar]
- Mota, M.M. , Braasch, H. , Bravo, M.A. , Penas, A.C. , Burgermeister, W. , Metge, K. and Sousa, E. (1999) First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology, 1, 727–734. [Google Scholar]
- Muramoto, M. (1999) Ending of pine wilt disease in Okinerabu island, Kgoshima prefecture In: Sustainability of Pine Forests in relation to Pine Wilt and Decline. Proceedings of an International Symposium (Kazuyoshi F., Togashi K. and Ikedu T., eds), pp. 193–195. Kyoto, Japan: Shokado Shote. [Google Scholar]
- Myers, R.F. (1988) Pathogenesis in pine wilt caused by pinewood nematode, Bursaphelenchus xylophilus . J. Nematol. 20, 236–244. [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z. , Eulgem, T. , Holt, B.F. and Dangl, J.L. (2003) Recognition and response in the plant immune system. Annu. Rev. Genet. 37, 579–609. [DOI] [PubMed] [Google Scholar]
- Nose, M. and Shiraishi, S. (2007) Breeding for resistance to pine wilt disease In: Pine Wilt Disease (Futai K. and Sutherland J., eds), in press. Tokyo: Springer Japan. [Google Scholar]
- Oda, K. (1967) The applicability of measurement of the oleoresin yield in determining the susceptibility of pine trees to beetle infestation. Forest Pest News (Tokyo), 16, 263–266. [Google Scholar]
- Ohoba, K. (1976) Breeding for resistance to pine wilt disease. Forest Tree Breeding, 99, 1–6. [Google Scholar]
- Oku, H. , Shiraishi, T. and Kurozomi, S. (1979) Participation of toxin in wilting of Japanese pines caused by a nematode. Naturwissenschaften, 67, 210. [Google Scholar]
- Peng, Y. and Moens, M. (2003) Host resistance and tolerance to migratory plant‐parasitic nematodes. Nematology, 5, 145–177. [Google Scholar]
- Plomion, C. , Cooke, J. , Richardson, T. , Mackay, J. and Tuscan, G. (2003) Report on the forest trees workshop at the plant and animal genome conference. Comparative, 4, 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, L. , White, L.A. , Birch, P.R.J. and Toth, I.K. (2006) GenomeDiagram: A Python package for the visualisation of large‐scale genomic data. Bioinformatics, 22, 616–617. [DOI] [PubMed] [Google Scholar]
- Ralph, S. , Oddy, C. , Cooper, D. , Yueh, H. , Jancsik, S. , Kolosova, N. , Philippe, R.N. , Aeschliman, D. , White, R. , Huber, D. , Ritland, C.E. , Benoit, F. , Rigby, T. , Nantel, A. , Butterfield, Y.S.N. , Kirkpatrick, R. , Chun, E. , Liu, J. , Palmquist, D. , Wynhoven, B. , Stott, J. , Yang, G. , Barber, S. , Holt, R A. , Siddiqui, A. , Jones, S.J.M. , Marra, M.A. , Ellis, B.E. , Douglas, C.J. , Ritland, K. and Bohlmann, J. (2006) Genomics of hybrid poplar (Populus trichocarpa × deltoides) interacting with forest tent caterpillars (Malacosoma disstria): Normalized and full‐length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect‐induced defences in poplar. Mol. Ecol. 15, 1275–1297. [DOI] [PubMed] [Google Scholar]
- Riga, E. , Beckenbach, K. and Webster, J.M. (1992) Taxonomic relationships of Bursaphelenchus xylophilus and B. mucronatus based on intraspecific cross hybridization and DNA analysis. Fundam. Appl. Nematol. 15, 391–395. [Google Scholar]
- Robertson, L. , Robertson, W.M. , Sobczak, M. , Bakker, J. , Tetaud, E. , Arinagayayam, M.R. , Ferguson, M.A.J. , Fairlamb, A.H. and Jones, J.T. (2000) Cloning, expression and functional characterisation of a thioredoxin peroxidase from the potato cyst nematode Globodera rostochiensis . Mol. Biochem. Parasitol. 111, 41–49. [DOI] [PubMed] [Google Scholar]
- Rodrigues, J. (2006) Eradication program for the pinewood nematode in Portugal In: Pine Wilt Disease: a Worldwide Threat to Forest Ecosystems, http://www.nemalab.uevora.pt/ . Lisbon: Fundação Calouste Gulbenkian. [Google Scholar]
- Rosso, M.N. , Dubrana, M.P. , Cimbolini, N. , Jaubert, S. and Abad, P. (2005) Application of RNA interference to root‐knot nematode genes encoding esophageal gland proteins. Mol. Plant–Microbe Interact. 18, 615–620. [DOI] [PubMed] [Google Scholar]
- Ryss, A. , Vieira, P. , Mota, M. and Kulinich, O. (2005) A synopsis of the genus Bursaphelenchus Fuchs, 1937 (Aphelenchida: Parasitaphelenchidae) with keys to species. Nematology, 7, 393–458. [Google Scholar]
- Sasaki, M. , Tashima, M. , Kawamura, K. , Okada, S. , Furukoshi, T. , Tsuda, T. , Kobayashi, S. and Katayama, S. (1983) Crossing between the F1 hybrids from Japanese black pine and Masson pine (II) Resistance to pine wood nematode. In: Transaction of the 94th Annual Meeting of the Japanese Forestry Society, pp. 249–250. [Google Scholar]
- Scholl, E.H. , Thorne, J.L. , McCarter, J.P. and Bird, D.M. (2003) Horizontally transferred genes in plant‐parasitic nematodes: a high‐throughput genomic approach. Genome Biol. 4, art. no. R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, E. and Okuda, K. (1989) Transmission of the pine wood nematode, Bursaphelenchus xylophilus (Steiner et Buhrer) Nickle (Nematoda: Aphelenchiodidae), by the Japanese pine sawyer, Monochamus alternatus Hope (Coleoptera: Cerambycidae) to pine twigs under laboratory conditions. Jpn J. Nematol. 18, 6–14. [Google Scholar]
- Skarmoutsos, G. and Michalopoulos‐Skarmoutsos, H. (2000) Pathogenicity of Bursaphelenchus sexdentati, Bursaphelenchus leoni and Bursaphelenchus hellenicus on European pine seedlings. Forest Pathol. 30, 149–156. [Google Scholar]
- Smant, G. , Stokkermans, J.P. , Yan, Y. , De Boer, J.M. , Baum, T.J. , Wang, X. , Hussey, R.S. , Gommers, F.J. , Henrissat, B. , Davis, E.L. , Helder, J. , Schots, A. and Bakker, J. (1998) Endogenous cellulases in animals: isolation of beta‐1, 4‐endoglucanase genes from two species of plant‐parasitic cyst nematodes. Proc. Natl Acad. Sci. USA, 95, 4906–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.M. , Rodriguez‐Buey, M. , Karlsson, J. and Campbell, M.M. (2004) The response of the poplar transcriptome to wounding and subsequent infection by a viral pathogen. New Phytologist, 164, 123–136. [DOI] [PubMed] [Google Scholar]
- Sousa, E. , Bravo, M.A. , Pires, J. , Naves, P. , Penas, A.C. , Bonifacio, L. and Mota, M.M. (2001) Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae) associates with Monochamus galloproincialis (Coleoptera: Cerambycidae) in Portugal. Nematology, 3, 89–91. [Google Scholar]
- Steiner, S.G. and Buhrer, E.M. (1934) Aphelenchoides xylophilus, n. sp., a nematode associated with blue‐stain and other fungi in timber. J. Agr. Res. 48, 949–951 [Google Scholar]
- Suga, T. , Ohta, S. , Munesada, K. , Ide, N. , Kurokawa, M. , Shimizu, M. and Ohta, E. (1993) Endogenous pine wood nematicidal substances in pines, Pinus massoniana, P. strobus and P. palustris. Phytochemistry, 33, 1395–1401. [Google Scholar]
- Suzuki, K. and Kiyohara, T. (1978) Incidence of water stress on development of pine wilting disease caused by Bursaphelenchus lignicolus . Eur. J. Forest Pathol. 8, 97–107. [Google Scholar]
- Toda, T. (2004) Studies on the breeding for resistance to the pine wilt disease in Pinus densiflora and P. thunbergii . Bull. Forest Tree Breeding Center, 20, 83–217. [Google Scholar]
- Trudgill, D.L. (1991) Resistance to and tolerance of plant parasitic nematodes in plants. Annu. Rev. Phytopathol. 29, 167–192. [Google Scholar]
- Urwin, P.E. , Lilley, C.J. , and Atkinson, H.J. (2002) Ingestion of double‐stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant–Microbe Interact. 15, 747–752. [DOI] [PubMed] [Google Scholar]
- Vieira P. and Mota M. (eds) (2007) Pine wilt disease: a worldwide threat to forest ecosystems. Lisbon: Fundação Calouste Gulbenkian, http://www.nemalab.uevora.pt/ . [Google Scholar]
- Vieira, P. , Burgermeister, W. , Mota, M. , Metge, K and Silva, G. (2007) Lack of genetic variation of Bursaphelenchus xylophilus in Portugal revealed by RAPD‐PCR analyses. J. Nematol. 39, 118–126. [PMC free article] [PubMed] [Google Scholar]
- Wang, X.H. , Mitchum, M.G. , Gao, B.L. , Li, C.Y. , Diab, H. , Baum, T.J. , Hussey, R.S. and Davis, E.L. (2005) A parasitism gene from a plant‐parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana . Mol. Plant Pathol. 6, 187–191. [DOI] [PubMed] [Google Scholar]
- Watanabe, A. , Isoda, K. and Ozawa, H. (2007) Isolation of genes expressed in Japanese Black Pine infected with pine wilt disease In: Plant Anim. Genomes XV Conference, p. 501. San Diego (Abs). [Google Scholar]
- Weerasinghe, R.R. , Bird, D.M. and Allen, N.S. (2005) Root knot nematodes and bacterial Nod factors elicit common signal transduction pathways in Lotus japonicus. Proc. Natl Acad. Sci. USA, 102, 3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, V. and Kumar, A. (2006) Nematode resistance in plants: the battle underground. Trends Genet. 22, 396–403. [DOI] [PubMed] [Google Scholar]
- Ye, W. , Giblin‐Davies, R.M. , Braasch, H. , Morris, K. and Thomas, W.K. (2007) Phylogenetic relationships among Bursaphelenchus species (Nematoda: Parasitaphelenchidae) inferred from nuclear ribosomal and mitochondrial DNA sequence data. Mol. Phylogenet. Evol. 43, 1185–1197. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Bai, G. , Yang, W. , Li, H. and Xiong, H. (2006) Pathogenic cellulase assay of pine wilt disease and immunological localisation. Biosci. Biotechnol. Biochem. 70, 2727–2732. [DOI] [PubMed] [Google Scholar]
- Zhao, B.G. and Lin, F. (2005) Mutualistic symbiosis between Bursaphelenchus xylophilus and bacteria of the genus Pseudomonas . Forest Pathol. 35, 339–345. [Google Scholar]
- Zhao, B.G. , Liu Y. and Lin, F. (2007) Effects of bacteria associated with pine wood nematode (Bursaphelenchus xylophilus) on development and egg production of the nematode. J. Phytopathol. 155, 26–30. [Google Scholar]


