SUMMARY
GBB1, a heterotrimeric G‐protein β‐subunit gene, was shown to be a key regulator of fumonisin B1 (FB1) biosynthesis in the maize pathogen Fusarium verticillioides. In this study, we performed functional analyses of genes that encode putative RGS (regulators of G‐protein signalling) proteins and PhLPs (phosducin‐like proteins) in F. verticillioides. These proteins are known to regulate heterotrimeric G‐protein activity by altering the intrinsic guanosine triphosphatase (GTPase) activity, which, in turn, influences the signalling mechanisms that control fungal growth, virulence and secondary metabolism. Our aim was to isolate and characterize gene(s) that are under the transcriptional control of GBB1, and to test the hypothesis that these genes are directly associated with FB1 regulation and fungal development in F. verticillioides on maize kernels. We first identified eight genes (two PhLPs and six RGSs) in the F. verticillioides genome, and a subsequent transcriptional expression study revealed that three RGS genes were up‐regulated in the gbb1 deletion (Δgbb1) mutant and one RGS gene was up‐regulated in the wild‐type. To characterize their function, we generated knockout mutants using a homologous recombination strategy. When grown on autoclaved nonviable kernels, two mutants (ΔflbA2 and ΔrgsB) produced significantly higher levels of FB1 compared with the wild‐type progenitor, suggesting that the two mutated genes are negative regulators of FB1 biosynthesis. ΔflbA2 also showed a severe curly conidia germination pattern, which was contradictory to that observed in the Δgbb1 strain. Strikingly, when these mutants were grown on live maize kernels, we observed contrasting FB1 and conidiation phenotypes in fungal mutants, which strongly suggests that these G‐protein regulators have an impact on how F. verticillioides responds to host/environmental factors. Our data also provide evidence that fungal G‐protein signalling is important for modulating the ethylene biosynthetic pathway in maize kernels.
INTRODUCTION
Fusarium verticillioides (teleomorph Gibberella moniliformis) is a maize pathogen that causes seedling blight, root rot, stalk rot and kernel (or ear) rot worldwide (Kommedahl et al., 1979; Munkvold and Desjardins, 1997). In particular, kernel rot by F. verticillioides not only results in yield loss, but also lower grain quality, because of contamination by mycotoxins, namely fumonisins. Fumonisins are a group of polyketide‐derived mycotoxins, which are structurally similar to sphingolipid intermediates, and can lead to the inhibition of ceramide synthase and a disruption of sphingolipid metabolism (Marasas et al., 2004; Merrill et al., 1996). Fumonisin‐contaminated foods and feeds have been linked to a variety of illnesses in animals and humans (Gelderblom et al., 1991; Marasas, 2001; Marasas et al., 1988; Rheeder et al., 1992; Voss et al., 2002).
Over 15 structurally related fumonisins are known to date, and they all share a 19‐ or 20‐carbon polyketide backbone, but with variations in functional groups (1999, 2006; Seo et al., 1996). Among these, fumonisin B1 (FB1) is the predominant form in nature (Gelderblom et al., 1992; Sydenham et al., 1990). Fumonisins are synthesized by a cluster of co‐regulated genes, designated as the FUM cluster (Proctor et al., 2003), which spans a region of approximately 45 kb. The cluster harbours over 20 genes, including FUM1, a polyketide synthase gene, and FUM8, an aminotransferase gene (Proctor et al., 1999; Seo et al., 2001), which are transcriptionally activated when the fungus encounters environmental conditions conducive to fumonisin production (2005, 2007; Proctor et al., 2003). Significantly, one of the key questions that remains is how the fungus perceives these external environmental cues, transduces these signals and ultimately activates the FUM cluster. We now have a certain level of understanding of the nature of these fumonisin‐conducive conditions and of a select number of regulatory genes associated with toxin production (Bluhm and Woloshuk, 2005; Choi and Shim, 2008b; Flaherty and Woloshuk, 2004; Flaherty et al., 2003; Sagaram and Shim, 2007; Shim and Woloshuk, 2001).
G‐protein‐mediated signalling is one of the most important mechanisms by which eukaryotic cells sense extracellular signals and integrate them into intrinsic signal transduction pathways (McCudden et al., 2005; Neves et al., 2002). In fungi, the heterotrimeric G‐protein system, with key components such as G‐protein‐coupled receptors (GPCRs), Gα, Gβ and Gγ subunits, and regulatory proteins, is well recognized as playing a critical role in a variety of cellular functions in response to external signals. Excellent review articles are available discussing the role of these components in cell signalling in fungi (Lengeler et al., 2000; Li et al., 2007; Yu, 2006; Yu and Keller, 2005). In plant‐pathogenic fungi, G‐protein signalling components, in particular Gα, Gβ and Gγ subunits, have been functionally characterized in a number of species, and have been shown to play critical roles in virulence, morphogenesis and secondary metabolism (Li et al., 2007; Nishimura et al., 2003; Sagaram and Shim, 2007; Yu et al., 2008). However, we have a limited understanding of how regulatory proteins, namely RGS (regulators of G‐protein signalling) and PhLP (phosducin‐like proteins), function in pathogenic fungi. RGS proteins regulate cell signalling by acting as negative regulators of heterotrimeric G‐protein cascades that enable eukaryotic cells to perceive and respond to external stimuli. These proteins contain a conserved ∼130‐amino‐acid RGS box that interacts with an activated guanosine triphosphate (GTP)‐Gα subunit and increases its intrinsic guanosine triphosphatase (GTPase) activity, thereby rapidly turning off the GPCR‐mediated signalling pathways (Chidiac and Roy, 2003; McCudden et al., 2005). In addition, RGS proteins can enhance G‐protein pathway activation, serving as effector antagonists, and can also act as scaffolding proteins to congregate receptors, G proteins, effectors and other regulatory molecules (Zhong and Neubig, 2001). Phosducin and PhLPs are a group of evolutionarily conserved proteins that were initially recognized as negative regulators of Gβγ activity by binding and sequestering Gβγ heterodimer from its interaction with Gα or downstream effectors (Bauer et al., 1992; Blüml et al., 1997; Flanary et al., 2000). However, previous genetic studies with the chestnut blight fungus Cryphonectria parasitica (Kasahara et al., 2000) and the social amoeba Dictyostelium discoideum (Blaauw et al., 2003) have shown that PhLPs are positive regulators of Gβγ signalling. Furthermore, biochemical studies of PhLP in humans (Lukov et al., 2005) and D. discoideum (Knol et al., 2005) have clearly demonstrated that PhLP is essential for Gβγ dimer assembly and for normal levels of Gβ and Gγ subunits.
The genome of Aspergillus nidulans harbours three PhLPs (PhnA, PhnB and PhnC) and four RGSs (FlbA, RgsA, RgsB and RgsC) (Yu, 2006). One of the earliest known RGSs is A. nidulans FlbA, and research has shown that it is required for the control of mycelial proliferation and the activation of asexual sporulation (Yu et al., 1996). FlbA transcriptionally and post‐transcriptionally regulates the sterigmatocystin transcription factor aflR in a PkaA‐dependent manner (Shimizu et al., 2003). In Aspergillus fumigatus, FlbA has been shown to regulate conidiation and hyphal proliferation, indicating a conserved function in Aspergillus sp. (Mah and Yu, 2006). Another RGS protein, A. nidulans RgsA, downregulates pigment production and conidial germination, but stimulates asexual sporulation (Han et al., 2004). In the rice blast fungus Magnaporthe grisea, Rgs1 serves as a negative regulator of all three Gα subunits, thus regulating pathogenesis, asexual reproduction and thigmotropism (Liu et al., 2007). The deletion of Cryphonectria parasitica RGS‐1 resulted in reduced growth, sparse aerial mycelium and loss of pigmentation, sporulation and virulence (Segers et al., 2004). Furthermore, recently, a PhLP PhnA has been reported to be involved in Gβγ‐mediated signalling required for vegetative growth, developmental control and toxin biosynthesis in A. nidulans (Seo and Yu, 2006).
In an earlier study, we investigated the role of the heterotrimeric G‐protein complex β‐subunit gene, GBB1, in F. verticillioides (Sagaram and Shim, 2007). Our data showed that, although GBB1 was not involved directly in fungal virulence, it served as a key positive regulator of FB1 biosynthesis and asexual spore production. Our subsequent question was whether regulatory components of the heterotrimeric G‐protein signalling complex are involved directly in FB1 biosynthesis and conidiation in F. verticillioides. In the present study, we identified the regulators of the heterotrimeric G‐protein complex, and investigated their roles in the F. verticillioides–maize interaction, particularly on nonviable (autoclaved) versus viable (live) maize kernels. In addition to conidiation and FB1 production, we analysed ethylene emission by live maize kernels when inoculated with fungal mutants.
RESULTS
RGSs and PhLPs in the F. verticillioides genome
In silico analysis of the F. verticillioides genome revealed the presence of six RGS genes and two PhLP genes: the RGS genes were designated as RGSA, RGSB, RGSC1, RGSC2, FLBA1 and FLBA2, and the PhLP genes were designated as PHNA and PHNB, based on shared homology with the respective proteins in other fungal species (Fig. S1, see Supporting Information). Namely, when we initially compared the similarity of these proteins to the respective proteins in A. nidulans, employing SMART (Letunic et al., 2009), we recognized that F. verticillioides did not contain a PhnC homologue, but had two copies of RgsC and FlbA (Fig. 1A). Fusarium verticillioides contains two paralogues of FlbA and, although both conceptually translated proteins are smaller in size than the A. nidulans counterpart, FlbA1 and FlbA2 share greater than 40% identity with A. nidulans FlbA at the amino acid level and harbour an RGS domain and DEP (dishevelled, Egl‐10 and pleckstrin) domain (McCudden et al., 2005; Yu, 2006). For RgsC, however, on further analysis, we concluded that F. verticillioides RgsC2 cannot be considered as a functional RGS as it lacks a conserved RGS motif. A comparison of A. nidulans and F. verticillioides RGSs and PhLPs (percentage amino acid identity) and their putative functions are listed in Table 1.
Figure 1.
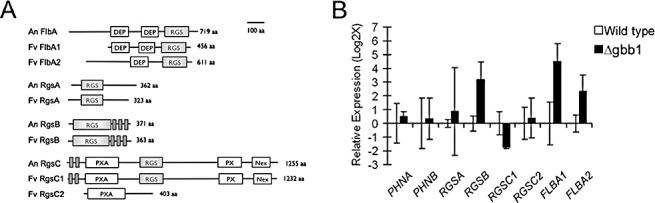
(A) Comparison of RGS proteins in Aspergillus nidulans and Fusarium verticillioides. Schematic representation of protein orthologues shows the location and alignment of conserved domains. DEP (dishevelled, Egl‐10 and pleckstrin); Nex (nexin family protein C‐terminus domain); PX (phox homology domain); PXA (PX association domain); RGS (regulator of G‐protein‐signalling domain); TM (transmembrane domain). Grey boxes depict transmembrane domains. (B) Expression analysis of G‐protein regulator genes in the wild‐type and Δgbb1 mutant. Total RNA samples were prepared from fungal strains grown in cracked‐corn medium for 10 days, and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis was performed with SYBR‐Green as the fluorescent reporter. The levels of transcription were evaluated using the 2−ΔΔ Ct method, with TUB2 as the endogenous control. The graph represents the logarithmic fold differences in gene expression. The range of expression was calibrated using 2−ΔΔ Ct − S− 2−ΔΔ Ct + S, where S is the standard deviation of the ΔCt value. Ct, threshold cycle. Three biological replications and two technical replications were performed.
Table 1.
The regulators of heterotrimeric G‐protein signalling in Fusarium verticillioides—PhLPs (phosducin‐like proteins) and RGSs (regulators of G‐protein signalling).
| Aspergillus nidulans protein* | Fusarium verticillioides protein | Identity (%)† | Putative function in fungi‡ |
|---|---|---|---|
| PhnA: An0082 | PhnA: FVEG_10292.3 | 36.3 | Regulation of vegetative growth, development and secondary metabolite biosynthesis |
| PhnB: An4561 | PhnB: FVEG_06475.3 | 61.1 | Apoptosis |
| PhnC: An8847 | — | — | Unknown |
| RgsA: An5755 | RgsA: FVEG_11363.3 | 63.2 | Stress response, conidiation, germination and pigmentation |
| RgsB: An3622 | RgsB: FVEG_09572.3 | 69.3 | Pheromone and cyclic‐AMP signalling |
| RgsC: An1377 | RgsC1: FVEG_03826.3 | 69.9 | Unknown |
| RgsC: An1377 | RgsC2: FVEG_05340.3 | 36.6 | Unknown |
| FlbA: An5893 | FlbA1: FVEG_08855.3 | 44.7 | Regulation of conidiation, autolysis and pathogenesis |
| FlbA: An5893 | FlbA2: FVEG_06192.3 | 71.4 | Regulation of conidiation and autolysis |
A. nidulans and F. verticillioides protein sequences can be found at the Broad Institute (http://www.broadinstitute.org/scientific‐community/data) using the locus number listed in the table.
References for A. nidulans genes are: PhnA (Seo and Yu, 2006), PhnB (Seo and Yu, 2006), PhnC (Seo and Yu, 2006), RgsA (Han et al., 2004), RgsB (Han et al., 2004), RgsC (Yu, 2006) and FlbA (Yu et al., 1996).
Identity (%) is based on amino acid comparison.
Putative functions were deduced from recent research and review articles (Li et al., 2007; Liu et al., 2007; Xue et al., 2008; Yu, 2006).
Our next aim was to identify RGSs and PhLPs associated with Gbb1 that were involved in the regulation of FB1 biosynthesis and conidiation. Published reports have indicated that RGS and PhLP genes are transcriptionally regulated in filamentous fungi (Han et al., 2004; Lee and Adams, 1994; Seo and Yu, 2006). Therefore, we studied the relative expression level of these genes in the wild‐type and Δgbb1 (GBB1 deletion) strains grown in cracked‐corn medium via quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis. Statistical analyses of qRT‐PCR revealed that four genes showed significant differential gene expression between the wild‐type and Δgbb1; three genes, RGSB, FLBA1 and FLBA2, were up‐regulated in the Δgbb1 background, whereas RGSC1 was up‐regulated in the wild‐type (Fig. 1B). This outcome suggested that RgsB, FlbA1 or FlbA2 serves as a negative regulator of FB1 biosynthesis and conidiation. Conversely, RgsC1 serves as a positive regulator.
Generation of RGS gene knockout mutants
To further study gene function, we generated F. verticillioides mutants by a double homologous recombination strategy (Sagaram and Shim, 2007). We also used a F. verticillioides SF41 strain, a Ku70‐deletion strain, to improve homologous recombination efficiency (Choi and Shim, 2008a). We made gene disruption constructs for flbA1, flbA2, rgsB and RGSC1 by a double‐joint PCR strategy with the hygromycin phosphotransferase (HPH) gene as the selectable marker (Fig. 2) (Sagaram and Shim, 2007). Hygromycin‐resistance transformants were isolated and tested for targeted gene replacement using PCR (data not shown) and Southern analysis (Fig. 2), and we obtained at least three independent knockout mutants for FLBA1 (ΔflbA1), FLBA2 (ΔflbA2), RGSB (ΔrgsB) and RGSC1 (ΔrgsC1) (Fig. 2). We selected these mutant strains and the wild‐type for phenotypic characterization, focusing on FB1 biosynthesis and conidiation.
Figure 2.
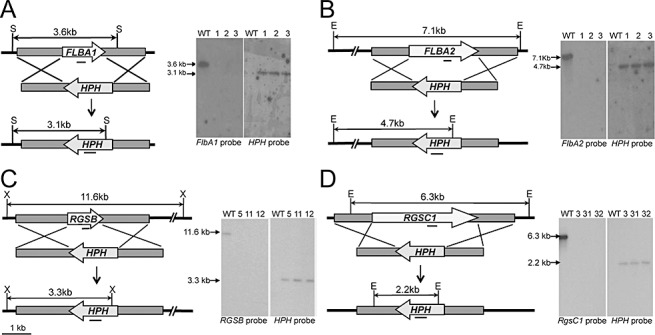
Targeted gene knockout of FLBA1 (A), FLBA2 (B), RGSB (C) and RGSC1 (D) from the genome of Fusarium verticillioides. Left of panel: schematic representation of homologous gene recombination strategy resulting in knockout mutant strains. E, EcoRI; HPH, hygromycin phosphotransferase gene; S, SalI; X, XbaI. Solid bar indicates DNA fragment used as the probe for Southern hybridization. Right of panel: Southern analyses of wild‐type and three mutants with targeted gene probe and HPH probe labelled with 32P. Genomic DNA samples were digested with the restriction enzyme shown on the left. Anticipated band sizes before and after recombination are indicated on the left.
Prior to testing these mutants for toxin and conidia production, we investigated whether these strains had been altered in radial growth (on solid agar) and mycelial mass (in liquid broth). No significant differences were observed between the wild‐type, ΔflbA1 and ΔrgsC1 when grown on defined agar medium. However, ΔflbA2 and ΔrgsB showed 10% and 25% reduction in radial growth, respectively, when compared with the wild‐type progenitor (Figs 3A,B and S2A, see Supporting Information). In defined liquid culture, only ΔrgsB showed a statistically significant difference (80% reduction) in mycelial mass (dry weight) after 1 week of incubation (Fig. 3C).
Figure 3.
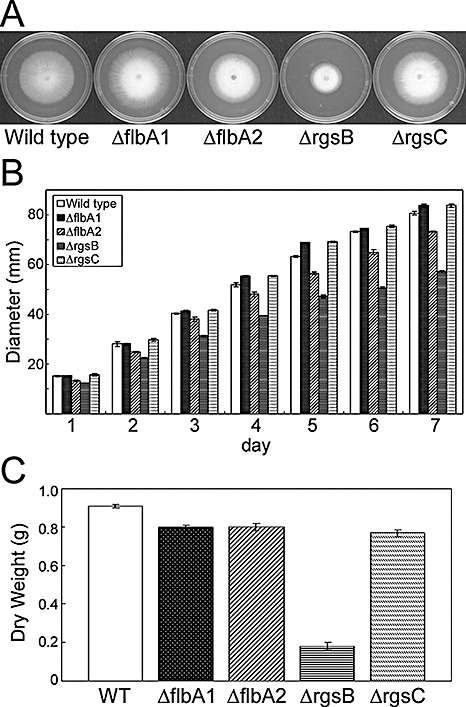
Growth rate of Fusarium verticillioides strains. (A) Hyphal growth rate of wild‐type (WT), ΔflbA1, ΔflbA2, ΔrgsB and ΔrgsC1 strains 4 days after inoculation on defined liquid (DL) agar medium. (B) Time‐course growth rate analysis of wild‐type (WT), ΔflbA1, ΔflbA2, ΔrgsB and ΔrgsC strains. Fungal strains were point inoculated with an agar block (0.5 cm in diameter). Results are the means of three biological replications. (C) Dry weight of wild‐type, ΔflbA1, ΔflbA2, ΔrgsB and ΔrgsC1 strains. The mycelia of each strain were harvested after 5 days of incubation in DL medium and dried at 70 °C for 12 h.
Impact of gene deletion on FB1 biosynthesis, conidiation, stalk rot and sexual mating
Wild‐type and mutant strains were grown on autoclaved cracked‐corn medium (nonviable kernels) to determine the impact of gene mutation on FB1 biosynthesis. All mutants grew similarly to the wild‐type progenitor on B73‐line nonviable kernels and, after a 7‐day incubation, FB1 was extracted from each sample and analysed with a high‐performance liquid chromatography (HPLC) system. The result showed that two mutants, ΔflbA2 and ΔrgsB, produced higher levels of FB1 than did the wild‐type (Fig. 4A). In particular, the ΔflbA2 strain produced a drastically higher level of FB1, approximately six times higher than that of the wild‐type, on nonviable kernels. However, two other mutants, ΔflbA1 and ΔrgsC1, showed no significant difference in FB1 level when compared with the wild‐type (data not shown). When these strains were grown in liquid medium [bovine serum albumin liquid (BSAL) and 0.2 × potato dextrose broth (PDB)], we observed aberrant pigment production in all four mutant strains (Fig. S2B). The most intense pigmentation was observed in the ΔflbA1 mutant grown in 0.2 × PDB medium.
Figure 4.
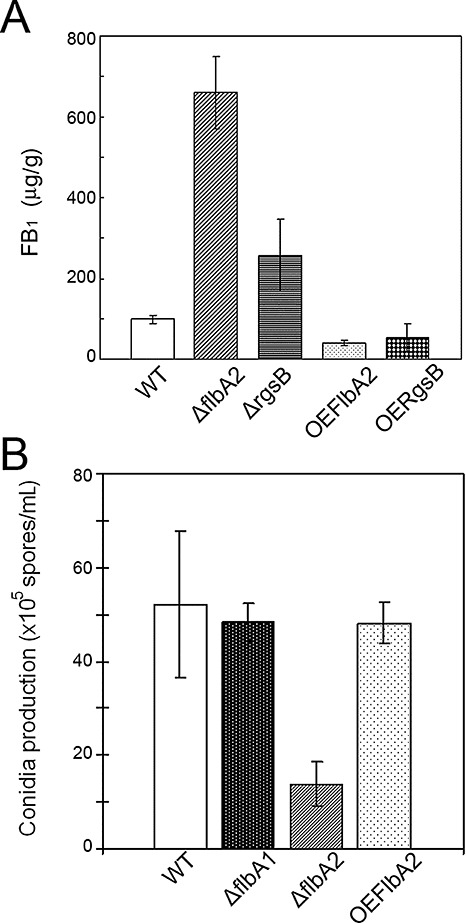
Quantification of fumonisin B1 (FB1) production and conidiation in Fusarium verticillioides strains when grown in nonviable autoclaved B73 maize kernels. (A) FB1 production in wild‐type (WT), ΔflbA2, ΔrgsB, OEFlbA2 and OERgsB strains was quantified by high‐performance liquid chromatography (HPLC) analysis. Autoclaved cracked corn (2 g) was inoculated with an agar block (0.5 cm in diameter) of WT and mutant strains. After 10 days of incubation at 25 °C, FB1 was extracted with 10 mL of 50% acetonitrile in water, purified through SPE C18 columns and eluted in 2 mL of 70% acetonitrile in water and quantified by HPLC. All values represent the means of three biological replications with standard deviations shown as error bars. (B) Wild‐type (WT), ΔflbA1, ΔflbA2 and OEFlbA2 strains were spot inoculated with an agar block (0.5 cm in diameter) on KCl agar plates and incubated for 7 days at 25 °C under a 14‐h light/10‐h dark cycle. Conidia were harvested and quantified with a haemocytometer. Three biological replications were performed to obtain standard deviations.
We also studied conidia production in the mutant strains. A fungal agar block (0.5 cm in diameter) was inoculated on KCl agar plates, and the plates were incubated for 7 days. We observed a substantial decrease (>75%) in conidia production in the ΔflbA2 isolate (Fig. 4B), but no statistically significant difference in the three other mutants (data not shown). Germination efficiency was not affected in the mutants when compared with the wild‐type progenitor. When incubated in defined liquid medium, the conidia of three mutants, ΔflbA1, ΔrgsB and ΔrgsC, produced standard ‘meandering’ germ tubes typically observed in the wild‐type strain (Fig. 5) (Sagaram and Shim, 2007). Meanwhile, ΔflbA2 conidia germinated in a severe curly, wavy fashion not commonly observed in the wild‐type strain (Fig. 5) and quite contrary to the ‘undeviating straight’ hyphal germination seen in the Δgbb1 mutant (Sagaram and Shim, 2007). However, the overexpression of FLBA2 did not result in Δgbb1‐like conidia germination, but rather was similar to that of the wild‐type strain (data not shown).
Figure 5.
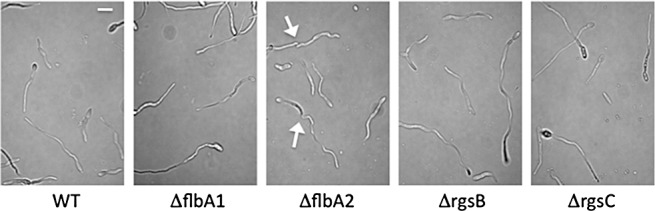
Effect of RGS (regulators of G‐protein signalling) gene mutation on colony hyphal development. Microconidia of the designated strains were allowed to germinate in 0.5 × potato dextrose broth for 24 h at 23 °C. Scale bar, 20 µm. It should be noted that the ΔflbA2 strain produces hyphae that germinate in a severe curly, wavy fashion (as indicated with arrows), not commonly observed in the wild‐type and other mutants.
When we investigated whether the mutants were affected with regard to maize stalk rot virulence, we found that the mutants were all capable of colonizing and rotting maize (Fig. S3, see Supporting Information). We measured the vertical length of the rot in every stalk sample tested, and the difference was not statistically significant (P < 0.05). We also determined that all mutants could carry out sexual development. The mutants were crossed to the opposite mating type wild‐type strain A102 on carrot agar and, after 21 days of incubation under a 14‐h light/10‐h dark cycle, they produced perithecia with viable ascopores (data not shown).
Effects of FLBA2 and RGSB overexpression in F. verticillioides
Deletion of FLBA2 and RGSB resulted in increased FB1 production (Fig. 4A), but a drastic reduction in conidiation (Fig. 4B) and reduced mycelial growth on solid medium (Fig. 3A,B). This led us to question the impact of the overexpression of these genes. We placed these genes under the control of the GpdA promoter, and transformed them into wild‐type protoplasts. The integration and overexpression of these genes were confirmed by PCR and northern analyses (data not shown). The overexpression of FLBA2 and RGSB resulted in suppression of FB1 production (Fig. 4A), suggesting that these genes are associated with the regulation of FB1 biosynthesis in F. verticillioides. However, the overexpression of FLBA2 or RGSB did not result in hyperconidiation (Fig. 4B) or hypermycelial growth (data not shown), respectively.
FlbA1 and FlbA2 are necessary for mediating the host response during fungal infection in viable maize kernels
To elucidate the potential roles of the RGS gene family in F. verticillioides–maize interactions, particularly with viable maize kernels, we inoculated surface‐sterilized B73 maize kernels with F. verticillioides mutants and analysed conidiation, FB1 production and endogenous ethylene levels. The disruption of FLBA1 and FLBA2 resulted in lower conidiation compared with that of the wild‐type strain, whereas ΔrgsB and ΔrgsC1 showed no statistically significant difference in conidiation from the wild‐type (Fig. 6A). Quantification of FB1 in maize kernels for these four strains showed that FB1 levels in mutant strains were suppressed in comparison with the level of the wild‐type strain (Fig. 6B). The role of ethylene as a plant defence hormone is well characterized (for reviews, see Bari and Jones, 2009; Wang et al., 2002). A recent study demonstrating that F. graminearum exploits host ethylene signalling in wheat and Arabidopsis seeds (Chen et al., 2009) raised a question regarding the role of ethylene in the F. verticillioides–maize kernel interaction. Here, we tested whether F. verticillioides mutants generated in this study could alter the emission of ethylene in infected kernels. For the assay, we quantified ethylene levels in maize kernels in response to fungal strains at designated time points. As shown in Fig. 6C, significantly lower ethylene levels were detected in kernels inoculated with ΔflbA1 and ΔflbA2 mutants at 7 and 10 days post‐inoculation when compared with the wild‐type and other mutant strains. Taken together, our data indicate a positive correlation between ethylene production in the host and conidiation and mycotoxin production in select fungal mutants (ΔflbA1 and ΔflbA2) when grown on viable maize kernels.
Figure 6.
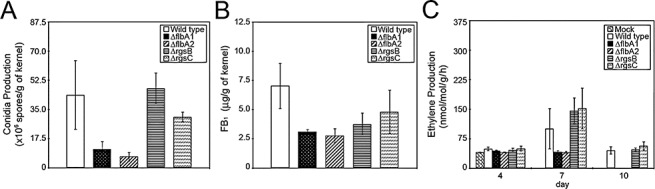
Fungal infection assay on viable B73 maize kernels by Fusarium verticillioides wild‐type and mutant strains. (A) Conidiation of wild‐type and mutant strains of F. verticillioides grown on seeds at 7 days post‐inoculation. (B) Fumonisin B1 (FB1) was quantified at 7 days after inoculation with F. verticillioides wild‐type and mutant strains using high‐performance liquid chromatography (HPLC). (C) Ethylene was measured at 4, 7 and 10 days post‐inoculation with F. verticillioides strains. The vials containing infected kernels were tightly sealed for 3 h prior to the withdrawal of headspace gases to allow the detection of ethylene accumulation by gas chromatography. The values are the mean ± standard deviation of four or five replicates per strain.
DISCUSSION
Heterotrimeric G proteins in filamentous fungi were first cloned by Turner and Borkovich (1993) and, since then, researchers have identified and characterized these G proteins in a number of fungal species (Li et al., 2007). In the maize pathogen F. verticillioides, the Gβ subunit has been shown to be associated directly with mycotoxin FB1 biosynthesis and hyphal development (Sagaram and Shim, 2007). Gene deletion of GBB1 did not impact fungal radial growth on solid medium and mycelial mass production in liquid medium. However, HPLC analysis showed that FB1 production in the mutant (10 µg/g) was drastically suppressed when compared with that of the wild‐type progenitor (140 µg/g). In contrast with other plant‐pathogenic ascomycetes (Mehrabi et al., 2009; Nishimura et al., 2003; Yu et al., 2008), the mutation of the Gβ subunit did not have an impact on the ability of the fungus to infect and colonize maize kernels and stalks (Sagaram and Shim, 2007). This observation was similar to that in Ustilago maydis, where the Gβ subunit regulates cyclic‐AMP signalling that ultimately induces pheromone gene expression, but not pathogenicity (Muller et al., 2004). However, we know from studies in other fungi that G‐protein activities are regulated by RGS proteins and PhLPs (Li et al., 2007; Ross and Wilkie, 2000).
If Gβ is associated with the regulation of FB1 biosynthesis and fungal development in F. verticillioides, are these regulated through the same signalling pathway or do they diverge to independent signalling mechanisms? To identify regulatory proteins that mediate G‐protein signalling associated with FB1 production and hyphal development, we searched for RGS and PhLP genes that were transcriptionally impacted by the Gβ mutation (Fig. 1B). As RGS proteins transcriptionally regulate downstream gene activation/suppression (Han et al., 2004; Lee and Adams, 1994; Shimizu et al., 2003), we reasoned that RGS genes differentially expressed in the Gβ mutant when compared with the wild‐type strain could be associated directly with mutant phenotypes, positively or negatively. Based on our results, we hypothesized that RgsB, FlbA1 and FlbA2 are negative regulators of FB1 production and hyphal germination phenotypes in Δgbb1 (Sagaram and Shim, 2007), as these genes were up‐regulated in the mutant strain. RgsC1 would be considered as a positive regulator. Phenotypic analyses of four knockout mutants showed that RGSB deletion resulted in FB1 overproduction but hyphal growth suppression, and that FLBA2 deletion led to FB1 overproduction but suppression of conidiation. In contrast with our anticipation, deletion of FLBA1 and RGSC1 resulted in no observable phenotypes on synthetic medium and nonviable maize kernels. RGS proteins are GTPase‐activating proteins, which are known to interact with specific Gα subunits and promote GTP hydrolysis, thus negatively regulating Gα‐mediated cell signalling (Li et al., 2007; McCudden et al., 2005; Ross and Wilkie, 2000). In addition to the RGS domain, a conserved domain required for Gα interaction, these proteins carry other motifs that may facilitate Gβ binding, Ras binding, GPCR phosphorylation and membrane localization.
In A. nidulans, FlbA is known to interact genetically with FadA, a group I Gα subunit, and to promote asexual development and sterigmatocystin biosynthesis (Hicks et al., 1997; Tag et al., 2000; Yu et al., 1996). Hicks et al. (1997) showed that a loss‐of‐function mutation in FlbA blocks sterigmatocystin production and conidiation, and the converse is true when the FLBA gene is overexpressed (Hicks et al., 1997; Lee and Adams, 1994). However, when we investigated FlbA function in F. verticillioides, we discovered that the regulatory circuitry could be quite different from that of A. nidulans. First, F. verticillioides harbours two FlbA paralogues, both carrying RGS motifs, which suggests that these FlbA proteins bind to Gα subunits. We hypothesized that the two FlbA proteins bind to group I Gα protein, but physical interaction has yet to be tested. Second, mutation in FlbA1 did not result in any recognizable phenotype on nonviable maize kernels, whereas FlbA2 null mutants resulted in the overproduction of FB1 (Fig. 4A), but suppressed conidiation (Fig. 4B). Overexpression of FLBA2 suppressed FB1 production but, with regard to asexual reproduction, only maintained wild‐type level conidiation. The FlbA orthologue in M. grisea, Rgs1, physically interacts with group I and II Gα subunits to facilitate intrinsic GTPase activity, and serves as a negative regulator of Gα proteins impacting on asexual development as well as appressoria formation (Liu et al., 2007). Mutation in Rgs1 resulted in precocious appressoria formation on noninductive surfaces, and this was suggested to be the result of the unrestrained intracellular cyclic‐AMP level. Xue et al. (2008) also demonstrated this in the Cryptococcus neoformans system. However, our result showed that mutation in FlbA1 and FlbA2 did not have an impact on fungal virulence, positively or negatively. We can postulate that FlbA is associated with fungal development, particularly infection structures, in select pathogenic fungi. Therefore, it is reasonable to predict that FlbA is not associated with pathogenicity in F. verticillioides, as Fusarium species do not utilize specialized infection structures to penetrate the host.
RgsB proteins can be easily identified in silico in filamentous fungi, with a distinct RGS motif and multiple transmembrane domains, but the biological understanding is very limited. RgsB, together with RgsA, is known to be a fungal‐specific RGS protein, and participates in the regulation of bipolar budding in Saccharomyces cerevisiae. However, in filamentous fungi, the role of RgsB is unclear. In our study, we discovered that the RGSB gene is transcriptionally regulated by the Gβ subunit in the GBB1 deletion, but whether RgsB physically interacts with Gbb1 at the protein level remains to be tested. We also cannot rule out the possibility of Gβ having an indirect impact on RGSB gene expression through a dissociated Gα protein that is the target of RgsB. Loss‐of‐function mutation in RgsB resulted in slower hyphal growth on solid medium and the overproduction of FB1 (approximately 150% higher level) when compared with the wild‐type progenitor (3, 4). Our results suggest that FlbA2 and RgsB work together to regulate the downstream FB1 biosynthesis machinery; however, they also independently control proper conidiation and hyphal proliferation, respectively, in F. verticillioides.
As a result of our qRT‐PCR analyses, we identified four RGS genes that were transcriptionally impacted by the deletion of the Gβ subunit. Does this imply then that the other four genes, PHNA, PHNB, RGSA and RGSC2, are not involved in F. verticillioides development and secondary metabolism via a heterotrimeric G‐protein complex? Based on the literature, we can presume that these regulators interact and control downstream signalling pathways at post‐translational levels (Liu et al., 2007; Xue et al., 2008), or that these other genes may be under the transcriptional control of Gα subunits (Han et al., 2004; Lafon et al., 2005). PhLPs have been shown to serve as molecular chaperones for Gβ and Gγ assembly (Knol et al., 2005; Lukov et al., 2005), and it is perhaps through these physical interactions that PhLPs perform regulatory roles. For instance, in fungi, Seo and Yu (2006) have shown that PhnA in A. nidulans is required for wild‐type level fungal biomass and conidiation, phenotypes that are known to be regulated by the Gβ subunit SfaD. With regard to RGS, A. nidulans RgsA downregulates pigment production, but stimulates conidiation (Han et al., 2004). These cellular responses are triggered when RgsA elicits GTPase activity of GanB, a Gα subunit, which is known to repress brlA, a primary regulator of conidiophore development in A. nidulans, and, ultimately, conidiation (Chang et al., 2004).
One of the intriguing and conceptually important findings of this study is the striking differential response of the fungal mutant strains when grown on either nonviable (autoclaved cracked kernels) or viable (live) maize kernels of the same host genotype. Although the ΔflbA2 mutant grown on cracked kernels produced approximately six times greater FB1 levels and approximately 4.5 times fewer conidia, this same strain, when grown on live kernels, produced about three‐ to four‐fold less FB1 and conidia. Similarly, although the ΔflbA1 strain showed no detectable phenotypes associated with FB1 and conidiation when grown on nonviable kernels, both processes were inhibited when the mutant strain was grown on live maize kernels. These results provide significant evidence for the impact of the fungal G‐protein signalling pathway on pathogen‐triggered metabolism and/or signalling in the host, which is in contrast with the hypothesis that preformed metabolites in maize seeds regulate pathogen development and secondary metabolism. The importance of signalling cross‐talk between hosts and pathogens in determining the outcomes of plant–pathogen interactions has been emphasized recently in several reviews (Christensen and Kolomiets, 2011; Gao and Kolomiets, 2009; Tsitsigiannis and Keller, 2007). Although the requirement for host‐derived chemical signals in the regulation of FB1 biosynthesis and conidia production has been recognized previously (Gao et al., 2007), the precise nature of most of these signalling molecules remains obscure. One such potent molecular signal is the plant hormone ethylene. Seed‐derived ethylene is implicated in the regulation of F. verticillioides growth and conidiation, as fungal growth was reduced on ethylene‐deficient maize mutants (M. V. Kolomiets, unpublished data). Interestingly, a recent screen of diverse Botrytis cinerea signalling mutants revealed that a Gα null mutant Δbcg1 was insensitive to exogenous ethylene, and showed considerable changes in expression of the fungal genes when compared with the wild‐type strain (Chague et al., 2006). This result suggests that the fungal pathways mediated by GPCRs may be involved in the sensing and/or response to exogenous ethylene, resulting in transcriptional reprogramming of B. cinerea. In our current study, we showed that ΔflbA1 and ΔflbA2 mutants are incapable of triggering the production of normal levels of ethylene by the host, suggesting that the GPCR signalling cascade is important in the modulation of host ethylene biosynthesis.
EXPERIMENTAL PROCEDURES
Fungal strains and media
Wild‐type F. verticillioides strain 7600 (Fungal Genetics Stock Center, University of Missouri‐Kansas City, Kansas City, MO, USA) was stored in 30% glycerol at −80 °C. The fungus was routinely maintained on V8 juice agar (200 mL of V8 juice, 3 g of CaCO3 and 20 g agar per litre) and potato dextrose agar (PDA; Difco, Detroit, MI, USA). For genomic DNA extraction, the fungus was grown in YEPD medium (3 g of yeast extract, 10 g of peptone and 20 g of dextrose per litre) on a rotary shaker (150 r.p.m.) at 25 °C. For RNA isolation, the fungus was grown in defined liquid (DL) medium, pH 4.5–6.0 (1 g NH4H2PO4, 40 g sucrose, 3 g KH2PO4, 2 g MgSO4.7H2O and 5 g NaCl per litre) or BSAL, pH 6.0 (1 g BSA, 40 g sucrose, 3 g KH2PO4, 0.5 g MgSO4.7H2O and 5 g NaCl per litre) (Shim and Woloshuk, 2001) with shaking (150 r.p.m.) at 25 °C. For microconidia counts, 104 microconidia were inoculated at the centre of V8 agar or KCl agar (Shim et al., 2006) and allowed to grow for 9 days. The spores were harvested in sterile water, passed through sterile Miracloth (Calbiochem, La Jolla, CA, USA) to eliminate mycelia and counted using a haemocytometer.
Nucleic acid manipulation and PCR
Fungal genomic DNA was extracted as described previously (Shim and Woloshuk, 2001). Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. Southern and northern analyses were performed following standard procedures described previously (Sagaram and Shim, 2007). The probes used in all hybridization experiments were 32P‐labelled with the Prime‐It Random Primer Labelling Kit (Stratagene, La Jolla, CA, USA). The primers used in this study are listed in Table S1 (see Supporting Information).
PCR was performed in a GeneAmp PCR system 9700 thermocycler (PE Applied Biosystems, Norwalk, CT, USA). DNA amplification was performed in a 25‐ or 50‐µL volume with either Taq DNA polymerase (New England Biolabs, Ipswich, MA, USA) or Expand Long Polymerase (Roche, Indianapolis, IN, USA). The PCR conditions included 2 min of initial denaturation at 94 °C, followed by 30 cycles of 30 s of denaturation at 94 °C, 30 s of annealing at 55–57 °C (based on primer pair) and 1–5 min (based on length of the anticipated amplicon) of elongation at 72 °C (68 °C for Expand Long Polymerase amplifications). A 10‐min final extension was carried out after 30 cycles.
Identification of F. verticillioides RGS and phosducin genes impacted by Δgbb1 mutation
qRT‐PCR experiments were carried out as described previously (Sagaram et al., 2006) with some modifications. All primers used in qRT‐PCR for the eight genes (PHNA, PHNB, RGSA, RGSB, RGSC1, RGSC2, FLBA1 and FLBA2) are given in Table S1. For this experiment, the wild‐type and the Δgbb1 mutant were grown on cracked‐corn medium for 7 days before total RNA samples were extracted using Trizol (Invitrogen). All qRT‐PCR experiments were performed in the ABI 7500FAST system (Applied Biosystems, Carlsbad, CA, USA) with the QuantiTech SYBR Green RT‐PCR Kit (Qiagen, Valencia, CA, USA). Reactions were carried out with 30 min of RT at 50 °C, followed by 15 min of predenaturation at 95 °C and 35 cycles of 15 s of denaturation at 95 °C, 30 s of annealing at 56 °C and 30 s of extension at 72 °C. The β‐tubulin gene (TUB2) (GenBank U27303) expression was determined with TUB2 F1 and TUB2 R31 primers, and was subsequently used as a reference. The levels of transcription were evaluated using the 2−ΔΔCt method (Schmittgen and Livak, 2008), with TUB2 as the endogenous control. Three biological replications and two technical replications were performed.
Gene deletion and overexpression constructs
The individual gene disruption cassettes were constructed by a double‐joint PCR strategy (Yu et al., 2004). DNA fragments corresponding to the 5′ (1.5 kb) and 3′ (1.5 kb) ends of each gene were amplified from the F. verticillioides genomic DNA with the primer pairs listed in Table S1. An HPH gene cloned into pBluescript II (Stratagene) was amplified with the primers M13‐F and M13‐R. Nested primer pairs were used to amplify the amplicon carrying the HPH marker fused to the flanking regions of the corresponding individual genes.
FlbA2 and RgsB overexpression strains were generated by transforming the wild‐type strain with constructs that harboured the gene of interest under the control of a constitutive GpdA promoter. For FlbA2 overexpression strain generation, the GpdA promoter amplified from the gGFP vector (Maor et al., 1998) with the primer pair tTrpF and OEFlbA2revt was fused to FlbA2 (amplified from the F. verticillioides genome by the primer pair OEFlbA2for and OEFlbA2rev) by single‐joint PCR. The joint‐PCR product was amplified with primers tTrpF and OEFlbA2rev. The geneticin‐resistant gene (gen) cloned into pBluescript II (Stratagene) was amplified with primers M13F and M13R. Two linear PCR products were co‐transformed in the wild‐type strain and the geneticin‐resistant colonies were screened for integration of the overexpression cassette. For RgsB overexpression strain generation, we followed the same protocol, except that the RGSB gene was amplified with the primer pair OERgsBfor and OERgsBrev and fused to the GpdA promoter.
Fungal transformation
For the generation of high‐frequency knockout strains, protoplasts were prepared from F. verticillioides KU70 deletion strain SF41 (Choi and Shim, 2008a) and transformed as described previously (Shim and Woloshuk, 2001). Transformants were selected on regeneration medium (Shim and Woloshuk, 2001) containing 150 µg/mL hygromycin B (Calbiochem). Hygromycin‐resistant colonies were screened for appropriate knockout construct integration by PCR and further verified by Southern analysis. To obtain overexpression strains, the wild‐type F. verticillioides protoplasts were used for transformation and the transformants were selected on either 150 µg/mL geneticin G‐418 (Cellgro, Herndon, VA, USA) or hygromycin B, depending on the marker used in the experiment. The respective drug‐resistant colonies were screened as described above.
FB1 analysis
For FB1 analysis, fungal strains were grown on autoclaved cracked‐corn medium (B73 line; 2 g dry weight) in a 20‐mL glass vial (VWR, West Chester, PA, USA) for 14 days at room temperature (22–23 °C). FB1 was extracted with acetonitrile–water (1:1, v/v) for 24 h. The crude extracts were passed through equilibrated PrepSep SPE C18 columns (Fisher Scientific, Pittsburgh, PA, USA). The FB1 concentration of the samples was analysed on a Shimadzu LC‐20AT HPLC system (Shimadzu Scientific Instruments, Inc., Kyoto, Japan) equipped with an analytical Zorbax ODS column (4.6 × 150 mm2) (Agilent Technologies, Santa Clara, CA, USA) and a Shimadzu RF‐20A fluorescence detector. The HPLC system was operated following the protocol described previously (Shim and Woloshuk, 1999). FB1 was quantified by comparing HPLC peak areas with FB1 standards (Sigma, St. Louis, MO, USA). The experiment was repeated with three biological replicates.
Maize stalk rot assay
Stalk rot severity was assayed on 8‐week‐old B73 maize plants as described previously (Shim et al., 2006). Internodal regions of the stalk were punctured to approximately 2 mm depth with a sterile needle, and fungal spore suspension (108 spores/µL water) was inoculated into the wound with the help of sterile cotton applicators. Plants were incubated in a growth chamber at 25 °C, 70% humidity with a 14‐h light/10‐h dark cycle. After 14 days of inoculation, stalks were split longitudinally to examine the extent of rot. The experiment was performed thrice with three independent biological replications.
Conidiation and ethylene analyses on viable maize kernels
Maize genotype B73 was used to determine the conidiation ability. Seeds were surface sterilized with Clorox bleach (containing 6% sodium hypochlorite) for 15 min, and rinsed with sterilized, distilled water at least five times. In order to provide an infection site for fungal inoculation, embryos of each kernel were cut longitudinally (0.5 cm) using a razor blade to a depth of about 0.5 mm. Seeds were dried with paper towels and placed in a 20‐mL glass scintillation vial (Wheaton Science, Millville, NJ, USA). The fungal suspension (200 µL of 1 × 106 conidia) of the tested strains of F. verticillioides was applied to the glass vial. Mock‐inoculated kernels were treated with 200 µL of 0.01% Tween‐20 solution. The inoculated kernels were placed in a container with moisture and incubated with a 12‐h light/12‐h dark cycle at 27 °C for 7 days. Incubated kernels were vortexed with 2 mL of sterilized water and the spores were counted using a haemocytometer. Five replicates of maize kernels were used per fungal strain.
The quantification of ethylene production in live maize kernels was carried out as described previously (Gao et al., 2008) with some modifications. Conidial suspensions were applied to the kernels as above. Ethylene was quantified at 4, 7 and 10 days post‐inoculation (dpi). Vials were sealed with screw caps with septa. One millilitre of the headspace gas was withdrawn from vials by a syringe and analysed using digital gas chromatography (Photovac 10 plus; Perkin‐Elmer, Inc., Norwalk, CT, USA) with a photodetector and compressed air (ultra zero grade; Praxair, Inc., Danbury, CT, USA) as carrier gas.
Supporting information
Fig. S1 Phylogenetic analyses of Fusarium verticillioides RGS (regulators of G‐protein signalling) proteins (A) and PhLPs (phosducin‐like proteins) (B). Homologues of F. verticillioides RGSs and PhLPs in other fungal species were identified in GenBank by blastp search. Multiple alignments were performed using muscle software, curated with Gblocks, phylogeny derived using PhyML and tree drawn with TreeDyn on (http://www.phylogeny.fr). Abbreviated names of fungal species in the figure are as follows: Anidulans, Aspergillus nidulans; Cglobosum, Chaetomium globosum; Cparasitica, Cryphonectria parasitica; Fv, Fusarium verticillioides; Mgrisea, Magnaporthe grisea; Ncrassa, Neurospora crassa; Panserina, Podospora anserina.
Fig. S2 Fusarium verticillioides wild‐type and mutant strains grown on solid agar medium (A) to assess the hyphal growth rate and in liquid medium (B) to study pigmentation. (A) Fusarium verticillioides strains (wild‐type and mutants) were first grown on V8 agar medium for 5 days, and used as an inoculum source. Agar blocks were prepared using a cork borer (0.5 cm in diameter) and inoculated at the centre of Petri plates containing five different solid agar media [defined liquid medium (DL) with glucose, DL with sucrose, DL with sorbitol, 1 × potato dextrose agar (PDA) and V8] for 5 days at room temperature before being photographed. (B) Fusarium verticillioides strains (wild‐type and mutants, 107 conidia/mL) were inoculated into 50 mL of bovine serum albumin liquid (BSAL) (DL with BSA as nitrogen source) and 0.2 × potato dextrose broth (PDB) for 7 days with continuous shaking at room temperature. A 2‐mL sample of each culture was transferred to a 24‐well sterile culture plate and photographed. A noninoculated (blank) sample is shown as a negative control.
Fig. S3 Maize stalk rot assay with Fusarium verticillioides wild‐type and mutant strains. Eight‐week‐old maize stalks were inoculated with wild‐type (WT) and mutant strains (108 spores) at the internodal region, and incubated in a growth chamber for 14 days at 25 °C, 70% humidity with a 14‐h light/10‐h dark cycle. Subsequently, the stalks were split longitudinally and photographed. Three independent biological replications were used in three independent experiments. Maize stalks inoculated with sterile water are shown as a negative control.
Table S1 Primers used in this study.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This work was supported in part by the National Research Initiative Competitive Grants Program of the United States Department of Agriculture (USDA) National Institute of Food and Agriculture Grant (No. 2007‐35319‐18334) to W.‐B. Shim, and by the National Science Foundation Grant (IOB‐0925561) to M. V. Kolomiets.
REFERENCES
- Bari, R. and Jones, J.D.G. (2009) Role of plant hormones in plant defense responses. Plant Mol. Biol. 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Bauer, P.H. , Muller, S. , Puzicha, M. , Pippig, S. , Obermaier, B. , Helmreich, E.J.M. and Lohse, M.J. (1992) Phosducin is a protein kinase A‐regulated G‐protein regulator. Nature, 358, 73–76. [DOI] [PubMed] [Google Scholar]
- Blaauw, M. , Knol, J.C. , Kortholt, A. , Roelofs, J. , Ruchira, Postma, M. , Visser, A.J. and van Haastert, P.J. (2003) Phosducin‐like proteins in Dictyostelium discoideum: implications for the phosducin family of proteins. EMBO J. 22, 5047–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm, B. and Woloshuk, C. (2005) Amylopectin induces fumonisin B1 production by Fusarium verticillioides during colonization of maize kernels. Mol. Plant–Microbe Interact. 18, 1333–1339. [DOI] [PubMed] [Google Scholar]
- Blüml, K. , Schnepp, W. , Schröder, S. , Beyermann, M. , Macias, M. , Oschkinat, H. and Lohse, M.J. (1997) A small region in phosducin inhibits G‐protein bg‐subunit function. EMBO J. 16, 4908–4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.W. , Cheung, F. , Proctor, R.H. , Butchko, R.A.E. , Zheng, L. , Lee, Y. , Utterback, T. , Smith, S. , Feldblyum, T. , Glenn, A.E. , Plattner, R.D. , Kendra, D.F. , Town, C.D. and Whitelaw, C.A. (2005) Comparative analysis of 87,000 expressed sequence tags from the fumonisin‐producing fungus Fusarium verticillioides . Fungal Genet. Biol. 42, 848–861. [DOI] [PubMed] [Google Scholar]
- Brown, D.W. , Butchko, R.A.E. , Busman, M. and Proctor, R.H. (2007) The Fusarium verticillioides FUM gene cluster encodes a Zn(II)2Cys6 protein that affects FUM gene expression and fumonisin production. Eukaryot. Cell, 6, 1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chague, V. , Danit, L.V. , Siewers, V. , Schulze‐Gronover, C. , Tudzynski, P. , Tudzynski, B. and Sharon, A. (2006) Ethylene sensing and gene activation in Botrytis cinerea: a missing link in ethylene regulation of fungus–plant interactions? Mol. Plant–Microbe Interact. 19, 33–42. [DOI] [PubMed] [Google Scholar]
- Chang, M.‐H. , Chae, K.‐S. , Han, D.‐M. and Jahng, K.‐Y. (2004) The GanB Gα‐protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans . Genetics, 167, 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Steed, A. , Travella, S. , Keller, B. and Nicholson, P. (2009) Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 182, 975–983. [DOI] [PubMed] [Google Scholar]
- Chidiac, P. and Roy, A.A. (2003) Activity, regulation, and intracellular localization of RGS proteins. Recept. Channels, 9, 135–147. [PubMed] [Google Scholar]
- Choi, Y.E. and Shim, W.B. (2008a) Enhanced homologous recombination in Fusarium verticillioides by disruption of FvKU70, a gene required for a non‐homologous end joining mechanism. Plant Pathol. J. 24, 1–7. [Google Scholar]
- Choi, Y.E. and Shim, W.B. (2008b) Identification of genes associated with fumonisin biosynthesis in Fusarium verticillioides via proteomics and quantitative real‐time PCR. J. Microbiol. Biotechnol. 18, 648–657. [PubMed] [Google Scholar]
- Christensen, S.A. and Kolomiets, M.V. (2011) The lipid language of plant–fungal interactions. Fungal Genet. Biol. 48, 4–14. [DOI] [PubMed] [Google Scholar]
- Flaherty, J.E. and Woloshuk, C.P. (2004) Regulation of fumonisin biosynthesis in Fusarium verticillioides by a zinc binuclear cluster‐type gene, ZFR1 . Appl. Environ. Microbiol. 70, 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty, J.E. , Pirttila, A.M. , Bluhm, B.H. and Woloshuk, C.P. (2003) PAC1, a pH‐regulatory gene from Fusarium verticillioides . Appl. Environ. Microbiol. 69, 5222–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanary, P.L. , DiBello, P.R. , Estrada, P. and Dohlman, H.G. (2000) Functional analysis of plp1 and plp2, two homologues of phosducin in yeast. J. Biol. Chem. 275, 18 462–18 469. [DOI] [PubMed] [Google Scholar]
- Gao, X. and Kolomiets, M.V. (2009) Host‐derived lipids and oxylipins are crucial signals in modulating mycotoxin production by fungi. Toxin Rev. 28, 79–88. [Google Scholar]
- Gao, X. , Shim, W.B. , Göbel, C. , Kunze, S. , Feussner, I. , Meeley, R. , Balint‐Kurti, P. and Kolomiets, M.V. (2007) Disruption of a maize 9‐lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol. Plant–Microbe Interact. 20, 922–933. [DOI] [PubMed] [Google Scholar]
- Gao, X.Q. , Starr, J. , Gobel, C. , Engelberth, J. , Feussner, I. , Tumlinson, J. and Kolomiets, M. (2008) Maize lipoxygenase ZmLOX3 controls development, root‐specific expression of defense genes, and resistance to root‐knot nematodes. Mol. Plant–Microbe Interact. 21, 98–109. [DOI] [PubMed] [Google Scholar]
- Gelderblom, W.C.A. , Kriek, N.P.J. , Marasas, W.F.O. and Thiel, P.G. (1991) Toxicity and carcinogenicity of the Fusarium moniliforme metabolite, Fumonisin B1, in rats. Carcinogenesis, 12, 1247–1251. [DOI] [PubMed] [Google Scholar]
- Gelderblom, W.C.A. , Semple, E. , Marasas, W.F.O. and Farber, E. (1992) The cancer‐initiating potential of the Fumonisin B mycotoxins. Carcinogenesis, 13, 433–437. [DOI] [PubMed] [Google Scholar]
- Han, K.H. , Seo, J.A. and Yu, J.H. (2004) Regulators of G‐protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Gα) signalling. Mol. Microbiol. 53, 529–540. [DOI] [PubMed] [Google Scholar]
- Hicks, J.K. , Yu, J.H. , Keller, N.P. and Adams, T.H. (1997) Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein‐dependent signaling pathway. EMBO J. 16, 4916–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, S. , Wang, P. and Nuss, D.L. (2000) Identification of bdm‐1, a gene involved in G protein β subunit function and α subunit accumulation. Proc. Natl. Acad. Sci. USA, 97, 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knol, J.C. , Engel, R. , Blaauw, M. , Visser, A.J.W.G. and van Haastert, P.J.M. (2005) The phosducin‐like protein PhLP1 is essential for G βγ dimer formation in Dictyostelium discoideum . Mol. Cell. Biol. 25, 8393–8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommedahl, T. , Windels, C.E. and Stucker, R.E. (1979) Occurrence of Fusarium species in roots and stalks of symptomless corn plants during the growing season. Phytopathology, 69, 961–966. [Google Scholar]
- Lafon, A. , Seo, J.A. , Han, K.H. , Yu, J.H. and d'Enfert, C. (2005) The heterotrimeric G‐protein GanB(α)‐SfaD(β)‐GpgA(γ) is a carbon source sensor involved in early cAMP‐dependent germination in Aspergillus nidulans . Genetics, 171, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.N. and Adams, T.H. (1994) Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol. Microbiol. 14, 323–334. [DOI] [PubMed] [Google Scholar]
- Lengeler, K.B. , Davidson, R.C. , D'Souza, C. , Harashima, T. , Shen, W.C. , Wang, P. , Pan, X.W. , Waugh, M. and Heitman, J. (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I. , Doerks, T. and Bork, P. (2009) SMART 6: recent updates and new developments. Nucleic Acids Res. 37, D229–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Wright, S.J. , Krystofova, S. , Park, G. and Borkovich, K.A. (2007) Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61, 423–452. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Suresh, A. , Willard, F.S. , Siderovski, D.P. , Lu, S. and Naqvi, N.I. (2007) Rgs1 regulates multiple Gα subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 26, 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukov, G.L. , Hu, T. , McLaughlin, J.N. , Hamm, H.E. and Willardson, B.M. (2005) Phosducin‐like protein acts as a molecular chaperone for G protein βγ dimer assembly. EMBO J. 24, 1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah, J.H. and Yu, J.H. (2006) Upstream and downstream regulation of asexual development in Aspergillus fumigatus . Eukaryot. Cell, 5, 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor, R. , Puyesky, M. , Horwitz, B.A. and Sharon, A. (1998) Use of green fluorescent protein (GFP) for studying development and fungal–plant interaction in Cochliobolus heterostrophus . Mycol. Res. 102, 491–496. [Google Scholar]
- Marasas, W.F.O. (2001) Discovery and occurrence of the fumonisins: a historical perspective. Environ. Health Persp. 109, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasas, W.F.O. , Kellerman, T.S. , Gelderblom, W.C.A. , Coetzer, J.A.W. , Thiel, P.G. and Vanderlugt, J.J. (1988) Leukoencephalomalacia in a horse induced by fumonisin B1 isolated from Fusarium moniliforme . Onderstepoort J. Vet. 55, 197–203. [PubMed] [Google Scholar]
- Marasas, W.F.O. , Riley, R.T. , Hendricks, K.A. , Stevens, V.L. , Sadler, T.W. , Gelineau‐van Waes, J. , Missmer, S.A. , Cabrera, J. , Torres, O. , Gelderblom, W.C.A. , Allegood, J. , Martinez, C. , Maddox, J. , Miller, J.D. , Starr, L. , Sullards, M.C. , Roman, A.V. , Voss, K.A. , Wang, E. and Merrill, A.H. (2004) Fumonisins disrupt sphingolipid metabolism, folate transport, and neural tube development in embryo culture and in vivo: a potential risk factor for human neural tube defects among populations consuming fumonisin‐contaminated maize. J. Nutr. 134, 711–716. [DOI] [PubMed] [Google Scholar]
- McCudden, C.R. , Hains, M.D. , Kimple, R.J. , Siderovski, D.P. and Willard, F.S. (2005) G‐protein signaling: back to the future. Cell. Mol. Life Sci. 62, 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi, R. , Ben M'Barek, S. , van der Lee, T.A.J. , Waalwijk, C. , de Wit, P.J.G.M. and Kema, G.H.J. (2009) Gα and Gβ proteins regulate the cyclic AMP pathway that is required for development and pathogenicity of the phytopathogen Mycosphaerella graminicola . Eukaryot. Cell, 8, 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill, A.H. , Liotta, D.C. and Riley, R.T. (1996) Fumonisins: fungal toxins that shed light on sphingolipid function. Trends Cell Biol. 6, 218–223. [DOI] [PubMed] [Google Scholar]
- Muller, P. , Leibbrandt, A. , Teunissen, H. , Cubasch, S. , Aichinger, C. and Kahmann, R. (2004) The Gβ‐subunit‐encoding gene bpp1 controls cyclic‐AMP signaling in Ustilago maydis . Eukaryot. Cell, 3, 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold, G.P. and Desjardins, A.E. (1997) Fumonisins in maize—can we reduce their occurrence? Plant Dis. 81, 556–565. [DOI] [PubMed] [Google Scholar]
- Neves, S.R. , Ram, P.T. and Iyengar, R. (2002) G protein pathways. Science, 296, 1636–1639. [DOI] [PubMed] [Google Scholar]
- Nishimura, M. , Park, G. and Xu, J.R. (2003) The Gβ subunit MGB1 is involved in regulating multiple steps of infection‐related morphogenesis in Magnaporthe grisea . Mol. Microbiol. 50, 231–243. [DOI] [PubMed] [Google Scholar]
- Proctor, R.H. , Desjardins, A.E. and Plattner, R.D. (1999) Biosynthetic and genetic relationships of B‐series fumonisins produced by Gibberella fujikuroi mating population A. Nat. Toxins, 7, 251–258. [DOI] [PubMed] [Google Scholar]
- Proctor, R.H. , Brown, D.W. , Plattner, R.D. and Desjardins, A.E. (2003) Co‐expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis . Fungal Genet. Biol. 38, 237–249. [DOI] [PubMed] [Google Scholar]
- Proctor, R.H. , Plattner, R.D. , Desjardins, A.E. , Busman, M. and Butchko, R.A.E. (2006) Fumonisin production in the maize pathogen Fusarium verticillioides: genetic basis of naturally occurring chemical variation. J. Agric. Food Chem. 54, 2424–2430. [DOI] [PubMed] [Google Scholar]
- Rheeder, J.P. , Marasas, W.F.O. , Thiel, P.G. , Sydenham, E.W. , Shephard, G.S. and Vanschalkwyk, D.J. (1992) Fusarium moniliforme and fumonisins in corn in relation to human esophageal cancer in Transkei. Phytopathology, 82, 353–357. [Google Scholar]
- Ross, E.M. and Wilkie, T.M. (2000) GTPase‐activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS‐like proteins. Ann. Rev. Biochem. 69, 795–827. [DOI] [PubMed] [Google Scholar]
- Sagaram, U.S. and Shim, W.B. (2007) Fusarium verticillioides GBB1, a gene encoding heterotrimeric G protein β subunit, is associated with fumonisin B1 biosynthesis and hyphal development but not with fungal virulence. Mol. Plant Pathol. 8, 375–384. [DOI] [PubMed] [Google Scholar]
- Sagaram, U.S. , Butchko, R.A.E. and Shim, W.B. (2006) The putative monomeric G‐protein GBP1 is negatively associated with fumonisin B1 production in Fusarium verticillioides . Mol. Plant Pathol. 7, 381–389. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative C‐T method. Nat. Protocol. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Segers, G.C. , Regier, J.C. and Nuss, D.L. (2004) Evidence for a role of the regulator of G‐protein signaling protein CPRGS‐1 in Gα subunit CPG‐1‐mediated regulation of fungal virulence, conidiation, and hydrophobin synthesis in the chestnut blight fungus Cryphonectria parasitica . Eukaryot. Cell, 3, 1454–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.A. and Yu, J.H. (2006) The phosducin‐like protein PhnA is required for Gβγ‐mediated signaling for vegetative growth, developmental control, and toxin biosynthesis in Aspergillus nidulans . Eukaryot. Cell, 5, 400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.A. , Kim, J.C. and Lee, Y.W. (1996) Isolation and characterization of two new type C fumonisins produced by Fusarium oxysporum . J. Nat. Prod. 59, 1003–1005. [DOI] [PubMed] [Google Scholar]
- Seo, J.A. , Proctor, R.H. and Plattner, R.D. (2001) Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides . Fungal Genet. Biol. 34, 155–165. [DOI] [PubMed] [Google Scholar]
- Shim, W.B. and Woloshuk, C.P. (1999) Nitrogen repression of fumonisin B1 biosynthesis in Gibberella fujikuroi . FEMS Microbiol. Lett. 177, 109–116. [DOI] [PubMed] [Google Scholar]
- Shim, W.B. and Woloshuk, C.P. (2001) Regulation of fumonisin B1 biosynthesis and conidiation in Fusarium verticillioides by a cyclin‐like (C‐type) gene, FCC1 . Appl. Environ. Microbiol. 67, 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, W.B. , Sagaram, U.S. , Choi, Y.E. , So, J. , Wilkinson, H.H. and Lee, Y.W. (2006) FSR1 is essential for virulence and female fertility in Fusarium verticillioides and F. graminearum . Mol. Plant–Microbe Interact. 19, 725–733. [DOI] [PubMed] [Google Scholar]
- Shimizu, K. , Hicks, J.K. , Huang, T.P. and Keller, N.P. (2003) Pka, Ras and RGS protein interactions regulate activity of aflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans . Genetics, 165, 1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydenham, E.W. , Gelderblom, W.C.A. , Thiel, P.G. and Marasas, W.F.O. (1990) Evidence for the natural occurrence of fumonisin B1, a mycotoxin produced by Fusarium moniliforme in corn. J. Agric. Food Chem. 38, 285–290. [Google Scholar]
- Tag, A. , Hicks, J. , Garifullina, G. , Ake, C. , Phillips, T.D. , Beremand, M. and Keller, N. (2000) G‐protein signalling mediates differential production of toxic secondary metabolites. Mol. Microbiol. 38, 658–665. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis, D.I. and Keller, N.P. (2007) Oxylipins as developmental and host–fungal communication signals. Trends Microbiol. 15, 109–118. [DOI] [PubMed] [Google Scholar]
- Turner, G.E. and Borkovich, K.A. (1993) Identification of a G‐protein α‐subunit from Neurospora crassa that is a member of the G(I) family. J. Biol. Chem. 268, 14 805–14 811. [PubMed] [Google Scholar]
- Voss, K.A. , Howard, P.C. , Riley, R.T. , Sharma, R.P. , Bucci, T.J. and Lorentzen, R.J. (2002) Carcinogenicity and mechanism of action of fumonisin B‐1: a mycotoxin produced by Fusarium moniliforme (=F. verticillioides). Cancer Detect. Prev. 26, 1–9. [DOI] [PubMed] [Google Scholar]
- Wang, K.L.‐C. , Li, H. and Ecker, J.R. (2002) Ethylene biosynthesis and signaling networks. Plant Cell, 14, S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, C.Y. , Hsueh, Y.P. , Chen, L.D. and Heitman, J. (2008) The RGS protein Crg2 regulates both pheromone and cAMP signalling in Cryptococcus neoformans . Mol. Microbiol. 70, 379–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.H. (2006) Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans . J. Microbiol. 44, 145–154. [PubMed] [Google Scholar]
- Yu, J.H. and Keller, N. (2005) Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43, 437–458. [DOI] [PubMed] [Google Scholar]
- Yu, J.H. , Wieser, J. and Adams, T.H. (1996) The Aspergillus flbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15, 5184–5190. [PMC free article] [PubMed] [Google Scholar]
- Yu, J.H. , Hamari, Z. , Han, K.H. , Seo, J.A. , Reyes‐Dominguez, Y. and Scazzocchio, C. (2004) Double‐joint PCR: a PCR‐based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 41, 973–981. [DOI] [PubMed] [Google Scholar]
- Yu, H.Y. , Seo, J.A. , Kim, J.E. , Han, K.H. , Shim, W.B. , Yun, S.H. and Lee, Y.W. (2008) Functional analyses of heterotrimeric G protein Gα and Gβ subunits in Gibberella zeae . Microbiology, 154, 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, H.L. and Neubig, R.R. (2001) Regulator of G protein signaling proteins: novel multifunctional drug targets. J. Pharmacol. Exp. Ther. 297, 837–845. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic analyses of Fusarium verticillioides RGS (regulators of G‐protein signalling) proteins (A) and PhLPs (phosducin‐like proteins) (B). Homologues of F. verticillioides RGSs and PhLPs in other fungal species were identified in GenBank by blastp search. Multiple alignments were performed using muscle software, curated with Gblocks, phylogeny derived using PhyML and tree drawn with TreeDyn on (http://www.phylogeny.fr). Abbreviated names of fungal species in the figure are as follows: Anidulans, Aspergillus nidulans; Cglobosum, Chaetomium globosum; Cparasitica, Cryphonectria parasitica; Fv, Fusarium verticillioides; Mgrisea, Magnaporthe grisea; Ncrassa, Neurospora crassa; Panserina, Podospora anserina.
Fig. S2 Fusarium verticillioides wild‐type and mutant strains grown on solid agar medium (A) to assess the hyphal growth rate and in liquid medium (B) to study pigmentation. (A) Fusarium verticillioides strains (wild‐type and mutants) were first grown on V8 agar medium for 5 days, and used as an inoculum source. Agar blocks were prepared using a cork borer (0.5 cm in diameter) and inoculated at the centre of Petri plates containing five different solid agar media [defined liquid medium (DL) with glucose, DL with sucrose, DL with sorbitol, 1 × potato dextrose agar (PDA) and V8] for 5 days at room temperature before being photographed. (B) Fusarium verticillioides strains (wild‐type and mutants, 107 conidia/mL) were inoculated into 50 mL of bovine serum albumin liquid (BSAL) (DL with BSA as nitrogen source) and 0.2 × potato dextrose broth (PDB) for 7 days with continuous shaking at room temperature. A 2‐mL sample of each culture was transferred to a 24‐well sterile culture plate and photographed. A noninoculated (blank) sample is shown as a negative control.
Fig. S3 Maize stalk rot assay with Fusarium verticillioides wild‐type and mutant strains. Eight‐week‐old maize stalks were inoculated with wild‐type (WT) and mutant strains (108 spores) at the internodal region, and incubated in a growth chamber for 14 days at 25 °C, 70% humidity with a 14‐h light/10‐h dark cycle. Subsequently, the stalks were split longitudinally and photographed. Three independent biological replications were used in three independent experiments. Maize stalks inoculated with sterile water are shown as a negative control.
Table S1 Primers used in this study.
Supporting info item
Supporting info item


