SUMMARY
The pioneering research of Harold Flor on flax and the flax rust fungus culminated in his gene‐for‐gene hypothesis. It took nearly 50 years before the first fungal avirulence (Avr) gene in support of his hypothesis was cloned. Initially, fungal Avr genes were identified by reverse genetics and map‐based cloning from model organisms, but, currently, the availability of many sequenced fungal genomes allows their cloning from additional fungi by a combination of comparative and functional genomics. It is believed that most Avr genes encode effectors that facilitate virulence by suppressing pathogen‐associated molecular pattern‐triggered immunity and induce effector‐triggered immunity in plants containing cognate resistance proteins. In resistant plants, effectors are directly or indirectly recognized by cognate resistance proteins that reside either on the plasma membrane or inside the plant cell. Indirect recognition of an effector (also known as the guard model) implies that the virulence target of an effector in the host (the guardee) is guarded by the resistance protein (the guard) that senses manipulation of the guardee, leading to activation of effector‐triggered immunity. In this article, we review the literature on fungal effectors and some pathogen‐associated molecular patterns, including those of some fungi for which no gene‐for‐gene relationship has been established.
INTRODUCTION
The gene‐for‐gene hypothesis proposed by Flor states that, for every dominant avirulence (Avr) gene in the pathogen, there is a cognate resistance (R) gene in the host, and the interaction between their gene products leads to the activation of host defence responses, such as the hypersensitive response (HR), that arrests the growth of biotrophic fungi (Flor, 1942). Since the pioneering genetic research of Flor, many plant pathologists have searched for molecular and biochemical evidence of the gene‐for‐gene concept. The molecular cloning of the first bacterial Avr gene was reported in 1984 (Staskawicz et al., 1984), the first fungal Avr gene in 1991 (van Kan et al., 1991) and the first oomycete Avr gene in 2004 (Shan et al., 2004). Over the last two decades, numerous novel Avr genes and cognate R genes have been identified, and have increased our molecular understanding of plant–microbe interactions considerably. In response to pathogen attacks, plants have evolved at least two lines of active defence. The first line provides basal defence against all potential pathogens and is based on the recognition of conserved pathogen‐associated molecular patterns (PAMPs), by so‐called PAMP recognition receptors (PRRs) that activate PAMP‐triggered immunity (PTI) and prevent further colonization of the host (De Wit, 2007; Jones and Dangl, 2006). One of the best‐known fungal PAMPs is chitin, a major structural component of fungal cell walls, for which two LysM‐type receptors have been characterized in rice and Arabidopsis (Kaku et al., 2006; Miya et al., 2007). There is now evidence that some Avr genes encode effectors that suppress PTI, enabling a pathogen to infect a plant and cause disease. Once the basal defence system of plants had been overcome by pathogens, during evolution plants responded by the development of a second recognition system based on effector perception by R proteins and subsequent activation of effector‐triggered immunity (ETI), which leads to rapid and enhanced defence responses in plants, including HR. This, in turn, triggered a second wave of co‐evolutionary arms race between pathogens and plants, during which pathogens responded by mutating effectors or developing novel effectors that could avoid or suppress ETI, whereas plants developed novel R proteins mediating the recognition of these novel effectors (De Wit, 2007; Jones and Dangl, 2006). One intriguing question relates to the molecular interaction between necrotrophic fungal pathogens and their host plants. It is assumed that necrotrophic pathogens do not comply with the gene‐for‐gene model. However, recently, several necrotrophic fungal pathogens have been reported to produce ribosomal proteins that might function as effectors that induce necrosis in plants with responsive genes to facilitate disease (Friesen et al., 2008a). The same could be true for the necrosis and ethylene‐inducing peptide 1 (Nep1)‐like proteins (NLPs) that are produced by several organisms, including fungi (Ottmann et al., 2009).
Many reviews on bacterial, fungal and oomycete Avr genes have been written in recent years (Bent and Mackey, 2007; Gohre and Robatzek, 2008; Kamoun, 2007; Stergiopoulos and de Wit, 2009). In this review, we also briefly discuss some fungal PAMPs and provide an update on fungal effectors, also including those of fungi for which no gene‐for‐gene type of interaction has been established with their hosts.
PAMPS OF FUNGAL PLANT AND HUMAN PATHOGENS
The first layer of plant defence is triggered after recognition of PAMPs by PRRs, leading to the activation of basal defence. Chitin and β‐glucan represent major fungal and oomycete PAMPs for which PRRs have been identified (Kaku et al., 2006; Miya et al., 2007). Several other cell wall components have been reported previously to be nonspecific elicitors, such as the galactoglucomannans of Cladosporium fulvum (De Wit and Kodde, 1981).
Likewise, several PAMPs have been described for the fungal human pathogen Candida albicans (Jouault et al., 2009). In general, the core structure of the fungal cell wall is composed of a skeleton of polysaccharide fibrils containing β‐(1,3)‐glucan, which is covalently linked to β‐(1,6)‐glucan and chitin. The outer layer also consists of proteins that are mainly glycosylated through N‐linked (Cutler, 2001) or O‐linked (Ernst and Prill, 2001) mannosylation. Although this basic model of the fungal cell wall is shared by many fungi, at the molecular level these structures differ between fungal species. In Aspergillus species, an important component of the cell wall is galactomannan, whereas the outermost cell wall layer is composed of hydrophobic proteins (hydrophobins) that contribute to the shielding properties of the cell wall (Bernard and Latge, 2001). In Cryptococcus neoformans, a capsule of mannoproteins, galactoxylomannan and glucoronoxylomannan masks the recognition and activation of host defence mechanisms (McFadden et al., 2006). This diversity in cell wall components will result in different qualities of PRR–ligand interaction and the activation of different sets of PRRs, leading to specific host responses. In addition to the recognition of PAMPs (infectious nonself molecules), plants have evolved the ability to sense their own degraded polymer molecules (infectious self molecules). These self‐released molecules, termed danger‐associated molecular patterns (DAMPs), often result from the degradation of plant cell walls by secreted fungal hydrolytic enzymes, such as xylanases, during host invasion (Matzinger, 2007). For example, plants can recognize oligo‐α‐galacturonides released from their damaged cell walls by fungal degrading enzymes, and subsequently activate the defence response (Denoux et al., 2008). It is anticipated that additional fungal PAMPs and DAMPs will be discovered in the near future that are recognized by novel plant and human PRRs.
EFFECTORS OF FUNGAL PLANT PATHOGENS
Cladosporium fulvum
To date, four Avr genes have been cloned from C. fulvum that all encode small cysteine‐rich proteins that are secreted during infection, including Avr2, Avr4, Avr4E and Avr9, whose recognition in tomato is mediated by the cognate Cf (for C. fulvum) proteins Cf‐2, Cf‐4, Cf‐4E and Cf‐9, respectively (De Wit et al., 1997; Joosten and De Wit, 1999; Thomma et al., 2005). Also, additional extracellular proteins (Ecps), namely Ecp1, Ecp2, Ecp4 and Ecp5, have been characterized from this fungus that invoke an HR in tomato accessions that carry a cognate Cf‐Ecp gene (De Kock et al., 2005; Laugéet al., 2000). Recently, Ecp6 and Ecp7 have been identified but, for these two effectors, no HR‐responding tomato accessions have been detected to date (Bolton et al., 2008). All Avrs and Ecps are assumed to be virulence factors (Bolton et al., 2008; 2007, 2008; Thomma et al., 2005).
The Avr2 effector inhibits tomato cysteine proteases that are presumed to be important in basal host defence, including Rcr3, Pip1, aleurain and TDI‐65 (Doehlemann et al., 2009; van Esse et al., 2008; Kruger et al., 2002; Muller et al., 2008; Rooney et al., 2005; R. Kahmann, unpublished; Shabab et al., 2008; Yoshida et al., 2009). Avr2 also facilitates the virulence of other fungal tomato pathogens, such as Botrytis cinerea and Verticillium dahliae, which are more virulent on Arabidopsis thaliana transgenic for Avr2 (van Esse et al., 2008). In the presence of Cf‐2, Avr2 behaves as an avirulence factor and its recognition is mediated by Rcr3pimp (required for C. fulvum resistance), a cysteine protease originating from Lycopersicon pimpinellifolium (Kruger et al., 2002; Rooney et al., 2005). Structural modification of Rcr3 by Avr2, rather than Rcr3 inhibition, is the most likely cause of the triggering of Cf‐2‐mediated defence signalling, as a natural variant of Rcr3 occurs in Lycopersicon esculentum (Rcr3esc) that is still an active enzyme but causes spontaneous HR in the presence of Cf‐2 in an Avr2‐independent manner. The Rcr3esc protein probably has a modified tertiary structure when compared with the Rcr3pimp protein (Kruger et al., 2002). Circumvention of Avr2‐triggered Cf‐2‐mediated HR can be achieved by point mutations, deletions or transposon insertions in the Avr2 gene (Luderer et al., 2002b; Stergiopoulos et al., 2007).
The Avr4 effector contains a functional chitin‐binding domain that protects chitinous fungi against plant chitinases, including C. fulvum (2006, 2004, 2003; van Esse et al., 2007). In the presence of Cf‐4, Avr4 induces an HR, but natural isoforms of this effector occur that no longer trigger Cf‐4‐mediated HR but can still bind chitin (van den Burg et al., 2003; Joosten et al., 1997; Stergiopoulos et al., 2007).
The virulence function of Avr4E is not yet known but, in the presence of Cf‐4E, Avr4E triggers Cf‐4E‐mediated resistance (Westerink et al., 2004). Evasion of Cf‐4E‐mediated recognition is achieved by point mutations in the Avr4E gene or jettison of the Avr4E gene, suggesting that the fitness penalty associated with the loss of this gene is not very high (Stergiopoulos et al., 2007). However, Avr4E‐expressing tomato plants are more susceptible than control plants to natural C. fulvum strains that lack Avr4E, suggesting that Avr4E is a virulence factor (H. P. van Esse and B. P. H. J. Thomma, Wageningen, Laboratory of Phytopathology, Wageningen University).
The Avr9 effector contains a cystine knot with structural but, so far, no functional homology to carboxypeptidase inhibitors (van den Ackerveken et al., 1993; van den Hooven et al., 2001; van Kan et al., 1991; Vervoort et al., 1997). Disruption of Avr9 in C. fulvum did not affect the virulence on tomato plants, suggesting that Avr9 is not required for full virulence (Marmeisse et al., 1993). However, Avr9‐expressing tomato plants appear to be more susceptible than control plants to natural C. fulvum strains that lack Avr9, suggesting that Avr9 is a virulence factor with redundant activity (H. P. van Esse and B. P. H. J. Thomma, personal communication). Expression of Avr9 in vitro is induced under nitrogen‐limiting conditions (van den Ackerveken et al., 1994; Perez‐Garcia et al., 2001; Thomma et al., 2006) and an Nrf1 gene (for nitrogen‐responsive factor) has been identified in C. fulvum. Nrf1 deletion mutants no longer express Avr9 under nitrogen‐limiting conditions in vitro and are compromised in their virulence on Cf‐0 tomato plants. However, these strains are still avirulent on Cf‐9 tomato plants, suggesting that Nrf1 is a major, but not only, positive regulator of Avr9 expression (Perez‐Garcia et al., 2001). The expression of all other Avr and Ecp effector genes of C. fulvum is not induced under nitrogen‐limiting conditions (Bolton and Thomma, 2008; Thomma et al., 2006), indicating that nitrogen starvation is not a general environmental condition that induces effectors of C. fulvum.
In addition, effector genes coding for Ecps have been cloned from C. fulvum, including Ecp1, Ecp2, Ecp4, Ecp5, Ecp6 and Ecp7 (van den Ackerveken et al., 1993; Bolton et al., 2008; Laugéet al., 2000). Ecps are abundantly secreted by all strains of C. fulvum during infection, and possess an even number of cysteine residues that are most probably involved in intramolecular disulphide bridges (Luderer et al., 2002a). Ecp6, Ecp1 and Ecp2 are all virulence genes, as silencing or disruption of these genes compromises the virulence of C. fulvum on tomato (Bolton et al., 2008; Laugéet al., 1997). For Ecp1, Ecp2, Ecp4 and Ecp5, tomato accessions have been identified that carry single cognate dominant Cf‐Ecp genes mediating Ecp‐triggered HR, which, however, have not yet been cloned (2000, 1998; Soumpourou et al., 2007).
Until recently, no homologues of Avr and Ecp effectors could be found in public databases because of the small numbers of fungal plant pathogens sequenced, except for Avr4 and Ecp6 which contain orthologues in many other fungal species. This is mainly a result of the presence of CBM14 and LysM domains in these proteins, which are implicated in carbohydrate binding, including chitin, suggesting that Ecp6 might be a functional homologue of Avr4 (Bolton et al., 2008). However, Ecp6 most probably does not protect chitinous fungi against plant chitinases, but is most likely to be involved in the scavenging of chitin fragments that are released from fungal cell walls during infection, thus preventing them from acting as PAMPs that trigger PTI (Bolton et al., 2008).
Rhynchosporium secalis
Rhynchosporium secalis secretes three low‐molecular‐weight peptides, designated Nip1–Nip3. Nip1 triggers specific (non‐HR) defence responses in barley cultivars carrying the as yet uncloned Rrs1 resistance gene (Hahn et al., 1993). Mature Nip1 (also known as AvrRrs1) contains 10 cysteine residues (Rohe et al., 1995) that are all involved in intramolecular disulphide bonds (Van't Slot et al., 2003). Strains of R. secalis virulent on Rrs1 plants either lack Nip1 or contain alleles with point mutations that translate into single amino acid substitutions (Rohe et al., 1995). It has been found that Nip1 interacts with a single plasma membrane receptor that is involved in both the mediation of virulence and the triggering of defence, but the exact receptor has not yet been characterized (Van't Slot et al., 2007). A study of field populations of the pathogen showed clear evidence of positive diversifying selection operating on the Nip1 locus (Schurch et al., 2004). In total, 14 Nip1 isoforms were identified, at least three of which were correlated with gain of virulence on Rrs1 plants, whereas a high deletion frequency of Nip1 was also observed that was much higher than that seen for Nip2 and Nip3. As single amino acid substitutions in Nip1 that correlated with gain of virulence on Rrs1 plants were observed at much lower frequencies than gene deletions, the fitness cost associated with the loss of this gene is probably not high.
Recently, the Nip2 and Nip3 genes have also been cloned (W. Knogge, Halle, Leibniz Institute for Plant Biochemistry). Nip2 encodes a 109‐amino‐acid protein with a predicted signal peptide of 16 amino acids, whereas Nip3 encodes a 115‐amino‐acid protein with a predicted signal peptide of 17 amino acids. Mature Nip2 and Nip3 presumably carry six and eight cysteines, respectively (W. Knogge, Halle, Leibniz Institute for Plant Biochemistry).
Fusarium oxysporum f. sp. lycopersici
The so‐called Six effectors (for secreted in xylem) are produced by F. oxysporum f. sp. lycopersici during infection of tomato. Six1 (renamed Avr3) is required for full virulence on tomato (2004, 2005), but also triggers ETI in the presence of the cognate I‐3 resistance gene (Huang and Lindhout, 1997; Rep et al., 2004). In addition, Six3 (renamed Avr2) contributes to virulence, but also triggers ETI in the presence of the cognate I‐2 resistance gene (Houterman et al., 2009). Avr3 resides on a small chromosome that contains additional effector genes, including Six2 and Six3. This chromosome is not found outside the F. oxysporum f. sp. lycopersici lineage, nor in nonpathogenic Fusarium species, suggesting that it confers virulence specifically on tomato (van der Does et al., 2008). This suggests that the ability to cause disease has probably arisen once during the evolution of F. oxysporum f. sp. lycopersici by the acquisition or emergence of the genomic region harbouring the effector genes necessary for the infection of tomato. Subsequently, this region might have spread to other clonal F. oxysporum f. sp. lycopersici strains by horizontal gene transfer (van der Does et al., 2008). Six4 (renamed Avr1) confers avirulence to F. oxysporum f. sp. lycopersici strains on tomato lines carrying the I or I‐1 resistance gene. However, Avr1 is not required for full virulence of F. oxysporum f. sp. lycopersici strains on tomato plants that lack the cognate I or I‐1 gene. This suggests that Avr1 has evolved as an effector that suppresses ETI rather than PTI. Indeed, Avr1 functions as a suppressor of I‐2‐ and I‐3‐mediated resistance (Houterman et al., 2008) and is always present in F. oxysporum f. sp. lycopersici strains virulent on these lines. When F. oxysporum f. sp. lycopersici strains avirulent on I‐2 and/or I‐3 lines were transformed with Avr1, they gained virulence on these lines, indicating that Avr1 suppresses both I‐2‐ and I‐3‐mediated resistance. Avr1 might have been acquired by F. oxysporum f. sp. lycopersici in order to avoid the fitness penalty associated with the loss of Avr3 and probably also Avr2 in overcoming I‐2‐ and I‐3‐mediated resistance. This could explain why all F. oxysporum f. sp. lycopersici strains analysed to date contain Avr3, whereas Avr1 is only present in strains that are virulent on I‐2 and/or I‐3 lines. Possibly, the I‐3 protein fits the guard model, where not the Avr3 protein itself, but the modulation of its virulence target, is recognized by I‐3, whereas F. oxysporum f. sp. lycopersici strains can regain virulence towards I‐3‐containing lines by the acquisition of Avr1. During evolution, tomato responded to this adaptation with the development of the I or unlinked I‐1 resistance gene that specifically mediates the recognition of Avr1 (Houterman et al., 2008). Interestingly, the Avr2 protein is secreted in the xylem when F. oxysporum f. sp. lycopersici colonizes tomato and is recognized intracellularly by I‐2, implying uptake by the host (Houterman et al., 2009). Point mutations resulting in single amino acid changes are the mechanism for gaining virulence on I‐2 genotypes.
Leptosphaeria maculans
At least nine distinct Avr genes, designated AvrLm1–AvrLm9, have been genetically mapped in L. maculans on unlinked genomic regions (Balesdent et al., 2002). AvrLm1 and AvrLm6 (Fudal et al., 2007; Gout et al., 2006) are in relatively close proximity at a locus that also harbours AvrLm2 (Balesdent et al., 2002). AvrLm1 and AvrLm6 reside in a gene‐poor heterochromatin‐like region surrounded by GC‐rich isochors that comprises long‐terminal repeat (LTR) retrotransposons, whereas both genes show a low GC content. AvrLm1 and AvrLm6 are single‐copy genes, encoding small secreted proteins (SSPs) that lack any characteristic signatures. In contrast with AvrLm1, AvrLm6 contains six cysteine residues and might be secreted in the apoplast, resembling the cysteine‐rich effectors of C. fulvum and F. oxysporum f. sp. lycopersici (Gout et al., 2006). Races virulent on Rlm1 cultivars mostly lack the AvrLm1 gene (2005, 2006; Rouxel et al., 2003).
The AvrLm4‐7 gene shows the same characteristics as the AvrLm1‐2‐6 locus, including the presence of multiple LTR retrotransposons (Parlange et al., 2009). AvrLm7 confers avirulence on both Rlm7 and Rlm4 genotypes, and has been renamed AvrLm4‐7. AvrLm4‐7 encodes a putatively secreted protein with eight cysteine residues with no homology to proteins currently present in public databases. Complete or partial deletion of the AvrmL4‐7 gene is the main mechanism for gaining virulence on both Rlm4 and Rlm7 genotypes, whereas most isolates virulent on Rlm4 genotypes alone show a single point mutation in the AvrLm4‐7 gene. Strains with the wild‐type AvrLm4‐7 allele appear to be more virulent than those lacking it, suggesting that AvrLm4‐7 is a virulence factor (Parlange et al., 2009).
Magnaporthe oryzae
Several Avr genes have been cloned and characterized from M. oryzae, including Avr–Pita (Orbach et al., 2000; Valent et al., 1991), Avr1–CO39 (Farman and Leong, 1998), Ace1 (Bohnert et al., 2004; Collemare et al., 2008) and the Pwl effectors (Kang et al., 1995; Sweigard et al., 1995). The Avr‐Pita effector shows homology to fungal zinc‐dependent metalloproteases and is dispensable for virulence on rice (Jia et al., 2000; Orbach et al., 2000). Avr‐Pita interacts with the cognate Pi‐ta resistance protein (Jia et al., 2000). Recognition specificity for Avr‐Pita is determined by a difference in one amino acid residue (Ala‐918) of the Pi‐ta protein present in resistant vs. susceptible rice varieties. An array of mutations has been described at the Avr‐Pita locus causing virulence of the fungus, including various size deletions, point mutations and a transposon insertion (Bryan et al., 2000; Jia et al., 2003; Kang et al., 2001; Orbach et al., 2000; Zhou et al., 2007). Recently, it has been shown that Avr‐Pita (currently renamed Avr‐Pita1) belongs to a gene family with at least two additional members, Avr‐Pita2 and Avr‐Pita3 (Khang et al., 2008). Avr‐Pita2 acts as an elicitor of defence responses mediated by Pi‐ta, but Avr‐Pita3 does not. Members of the Avr‐Pita family are widely distributed among strains of M. grisea isolated from diverse hosts, including isolates that are not pathogenic on rice. However, although Avr‐Pita1 and Avr‐Pita2 are present in both M. oryzae and M. grisea, Avr‐Pita3 is only present in M. oryzae isolates, suggesting that Avr‐Pita1 and Avr‐Pita3 are derived from a gene duplication event that must have occurred after the separation of M. oryzae from M. grisea (Khang et al., 2008).
The Ace1 effector is a putative cytoplasmic fusion polypeptide containing a polyketide synthase (PKS) and a nonribosomal peptide synthetase (NRPS), two distinct classes of enzymes that are involved in the production of microbial secondary metabolites (Bohnert et al., 2004; Collemare et al., 2008). Ace1 seems to mediate avirulence indirectly by producing a secondary metabolite that activates Pi33. Ace1 is exclusively expressed in appressoria, suggesting that the secondary metabolite produced might have a role in virulence, although mutants in which Ace1 was deleted were not compromised in virulence (Bohnert et al., 2004; Fudal et al., 2005).
The Pwl effectors, encoded by the Pwl (pathogenicity towards weeping lovegrass) gene family, are rapidly evolving, small, glycine‐rich secreted proteins that are generally found in rice pathogens. At least four members of this family, designated Pwl1–Pwl4, are present in M. oryza, confer species‐specific avirulence on weeping lovegrass and finger millet, but have no effect on rice (Kang et al., 1995; Sweigard et al., 1995). Pwl2 is located on a highly unstable locus, where frequent genetic rearrangements associated with large deletions have led to the emergence of spontaneous mutants virulent on weeping lovegrass. Pwl1, the allelic Pwl3 and Pwl4 were identified on the basis of homology to Pwl2, but only Pwl1 is a functional homologue of Pwl2, conferring avirulence on weeping lovegrass. However, Pwl4 could become functional when expressed under the control of the Pwl2 promoter, which was not the case for Pwl3.
To isolate additional Avr genes from M. grisea, Yoshida et al. (2009) recently retrieved 1032 putative secreted protein genes from the genomic sequence of isolate 70‐15 (an experimental isolate obtained from a sexual cross between a rice and grass isolate), examined their DNA polymorphisms among 46 isolates and looked for association with Avr function on a set of differential rice cultivars harbouring different R genes. However, no association was found with Avr activity, indicating that isolate 70‐15 may have lost several functional Avr genes through sexual recombination. Indeed, sequencing of the genome of another natural isolate, Ina 168, revealed that isolate 70‐15 lacked a total of 1.68 Mb of regions comprising 316 candidate effector genes. Association analyses of these 316 genes revealed three novel Avr genes, AVR‐Pia, AVR‐Pii and AVR‐Pik/km/kp. AVR‐Pia and AVR‐Pii have probably evolved by gene gain/loss processes, whereas AVR‐Pik/km/kp has evolved by a combination of gene gain/loss processes and nucleotide substitutions (Yoshida et al., 2009).
Blumeria graminis f. sp. hordei
Powdery mildews are a large group of ascomycete obligate biotrophic fungi that produce haustoria in the epidermis of their host plants (Glawe, 2008). Blumeria graminis f. sp. hordei causes powdery mildew on barley and interacts with its host in a gene‐for‐gene manner (Zhang et al., 2005). At least 85 dominant or semi‐dominant mildew R genes (Ml) have been characterized in barley, including Mlk genes and 28 highly homologous genes that all map to the Mla (for mildew A) locus of barley chromosome 5 (Jensen et al., 1980; Jorgensen, 1994). Six of the genes present at this locus, including Mla1, Mla6, Mla7, Mla10, Mla12 and Mla13, have been cloned, and they all encode highly related intracellular coiled‐coil nucleotide‐binding site leucine‐rich repeat (CC‐NBS‐LRR)‐type R proteins that all recognize isolate‐specific effectors of Blumeria graminis f. sp. hordei (Halterman and Wise, 2004; Halterman et al., 2001; Shen et al., 2003). Two Avr genes, designated Avrk1 and Avra10, have been cloned that induce defence responses in barley varieties containing the cognate Mlk1 and Mla10 proteins, respectively. Both genes belong to a large multigene family of more than 30 paralogues in Blumeria graminis f. sp. hordei, whereas homologues are present in formae speciales that are pathogenic on other grasses. The predicted proteins encoded by Avra10 and Avrk1 both lack an N‐terminal signal sequence or a signature for uptake by host cells. However, recently, it has been shown by fluorescence microscopy that the majority of the Mla10 protein is localized in the cytoplasm and approximately 5% in the nucleus (Bieri et al., 2004; Shen et al., 2007). Perturbation of nucleocytoplasmic Mla10 partitioning by the expression of an Mla10 fusion protein containing a nuclear export signal (NES), that enhances nuclear export over import, decreased Mla10‐specified disease resistance (Shen et al., 2007). In the nucleus, Mla10 showed an Avr10‐dependent physical association with two WRKY transcription factors (HvWRKY1 and HvWRKY2 transcription factors), suggesting that these transcription factors serve as immediate downstream targets of the activated receptor.
Melampsora lini
At least 30 Avr genes corresponding to cognate flax R genes have been identified in genetic analyses of Melampsora lini (Ellis et al., 1997). They have been cloned from four different loci, including AvrL567, AvrM, AvrP123 and AvrP4, that code for haustorially expressed secreted proteins (HESPs) and elicit HR in flax plants that carry the cognate R genes (Catanzariti et al., 2006).
The AvrL567A, AvrL567B and AvrL567C genes cluster at the AvrL567 locus and trigger HR in flax lines that carry the L5, L6 and L7 resistance genes, respectively. Virulent variants that no longer trigger HR have been identified (Dodds et al., 2006), which exhibit substitutions in amino acid residues that are exposed to the surface of the protein and interact directly with the cognate R proteins (Ellis et al., 2007; Wang et al., 2007).
AvrM is recognized by the M resistance protein, AvrP4 by P4 and the complex AvrP123 proteins are variously recognized by P, P1, P2 and/or P3 resistance proteins (Catanzariti et al., 2006). At least five different paralogues (AvrMA–AvrME) have been detected at the AvrM locus of an avirulent strain, whereas one paralogue encodes an effector that is not recognized by any known flax R protein. The six AvrM proteins have no known homologues in the public databases and show significant sequence and size variations caused by DNA insertions, deletions or polymorphisms in the location of stop codons. AvrP123 proteins contain 10 cysteine residues, including the characteristic CX7CX6YX3CX2‐3C signature present in the Kazal family of serine protease inhibitors, suggesting that host proteases might be a target of these effectors. AvrP4 encodes a protein with six cysteine residues at the C‐terminal part of the mature protein which show a spacing (CX3–7CX4–6CX0–5CX1–4CX4–10C) typical for cystine‐knotted peptides (Pallaghy et al., 1994). Both AvrM and AvrP4 are expressed in planta. Transient intracellular expression of AvrM and AvrP4 in flax plants carrying the cognate R genes triggers an HR, suggesting that effector translocation into the host cells occurs during infection, which is consistent with the predicted cytoplasmic location of M and P resistance proteins (Anderson et al., 1997; Lawrence et al., 1995). However, both effectors also induce an HR in flax when targeted into the apoplast, suggesting re‐entry from the apoplast into host cells after secretion (Catanzariti et al., 2006; Dodds et al., 2004). A role in virulence for the Avr genes of M. lini has not been shown to date.
EFFECTORS OF FUNGAL PATHOGENS FOR WHICH NO GENE‐FOR‐GENE RELATIONSHIP HAS BEEN ESTABLISHED
Ustilago maydis
For the corn smut fungus Ustilago maydis, no gene‐for‐gene interaction has been established, but several effector genes from this fungus have been analysed for a function in virulence (Kahmann and Kamper, 2004). Several hydrophobins or repellent genes that encode secreted proteins were examined for their roles in virulence. Single knock‐outs of these genes did not affect virulence, but a double knock‐out of Hum3 (a gene encoding a protein containing both hydrophobin repellent domains) and the repellent‐encoding gene Rsp1 were arrested at an early stage of penetration. This indicates that Hum3 and Rsp1 are effectors with a partly redundant virulence function during the early stages of infection (Muller et al., 2008).
Pep1 is a novel effector protein from U. maydis that is also essential during penetration. Pep1 is secreted into the apoplast and accumulated at sites of cell‐to‐cell passages. Disruption mutants of pep1 are not affected in saprophytic growth and develop normal infection structures, but are arrested during the penetration of epidermal cells and elicit a strong plant defence response. In addition, two of the four cysteine residues in Pep1 are essential for its function. Ustilago hordei contains an orthologue of Pep1 which is also required for the penetration of barley and is able to complement the U. maydis ΔPep1 mutant (Doehlemann et al., 2009).
Stp1 encodes an effector that is secreted by U. maydis into the apoplast (R. Kahmann, unpublished), and stp1 deletion mutants are avirulent as a result of growth arrest directly after the penetration of plant cells. This coincides with induction of strong plant defence responses. However, transient expression of Stp1 lacking the N‐terminal signal peptide in Nicotiana benthamiana and Zea mays revealed that the protein localizes specifically to subcompartments of the nucleus. This suggests that Stp1 is transferred from the apoplast to the plant cell after infection (R. Kahmann, unpublished).
It would be interesting to determine whether some of the effectors of U. maydis could give differential responses on accessions of wild cereal and corn species, providing evidence that they could also act as Avr factors.
Necrotrophic fungi
Obligate and biotrophic fungi retrieve their nutrients from living host cells, whereas necrotrophic pathogens thrive on killed host cells. As necrotrophic pathogens produce proteinaceous effectors (some also known as host‐selective toxins) that promote disease, and the host produces receptors that are required for susceptibility, these systems are often seen as a mirror image of the classical gene‐for‐gene systems found in biotrophic pathosystems, where matching of dominant host and pathogen gene products triggers resistance (Friesen et al., 2008a).
Stagonospora nodorum and Pyrenophora tritici‐repentis are two necrotrophic fungal pathogens that produce several necrogenic host‐specific peptide effectors which can be recognized by host susceptibility genes to cause disease. Stagonospora nodorum is a fungal pathogen of wheat, causing Stagonospora nodorum blotch (SNB) disease.
SnTox1 was the first toxic peptide produced by S. nodorum that interacts with a corresponding host susceptibility gene Snn1 (Friesen et al., 2007; Liu et al., 2006). SnToxA is encoded by a gene with a high degree of similarity to the Ptr ToxA gene from P. tritici‐repentis, the causal agent of tan spot of wheat with the matching susceptibility gene Tsn1. Tsn1‐disrupted mutants were insensitive to both Ptr ToxA and SnToxA, suggesting that both toxins are functionally similar as they are recognized by the same locus in the host. Therefore, the Tsn1–ToxA interaction in the wheat–S. nodorum pathosystem parallels that of the wheat–tan spot system, and the wheat Tsn1 gene serves as the major determinant for susceptibility to both SNB and tan spot (Liu et al., 2006).
The SnTox2–Snn2 interaction was the third gene pair identified in this system. SnTox2 is a small secreted peptide of about 7 kDa. Sensitivity to SnTox2 is conferred by the single dominant gene Snn2. In contrast with the classical gene‐for‐gene model, the Tsn1–SnToxA and Snn2–SnTox2 interactions are additive in their contribution to susceptibility (Friesen et al., 2007).
SnTox3 is the fourth necrosis‐inducing peptide that gives Snn3‐dependent necrosis. However, unlike the SnToxA–Tsn1 and SnTox2–Snn2 interactions, which both have largely additive effects relative to each other, both the SnToxA–Tsn1 and SnTox2–Snn2 interactions are epistatic to the SnTox3–Snn3 interaction (Friesen et al., 2008b; Zhang et al., 2009).
ToxA was the first peptide toxin produced by the most common races of P. tritici‐repentis. Ptr ToxA is a 13.2‐kDa protein that causes necrosis in particular genotypes of wheat (Ciuffetti et al., 1997). ToxA interacts with a high‐affinity binding site present on wheat mesophyll cells through the Arg‐Gly‐Asp (RGD)‐containing, solvent‐exposed loop, resulting in toxin internalization causing cell death (Manning et al., 2008). ToxA interacts with a chloroplast ToxA binding protein 1 (ToxABP1). ToxABP1 contains a lysine‐rich region within a coiled‐coil domain that is similar to the phosphatidylinositol binding sites present in animal proteins involved in endocytosis. ToxABP1 protein is present in both chloroplast membranes and chloroplast stroma. Surprisingly, ToxABP1 is expressed at similar levels and encodes an identical protein in both ToxA‐sensitive and ToxA‐insensitive cultivars, indicating that ToxA should have other targets apart from ToxABP1 (Manning et al., 2007).
Just like the effectors of biotrophic fungal pathogens, toxic peptides from necrotrophic pathogens represent effectors that interact with host targets. However, the effectors of necrotrophic pathogens cause necrosis, facilitating disease, and might therefore be considered as basic effectors for which no true gene‐for‐gene‐based resistance has been developed to date as a result of a low level of co‐evolution. It could be that host targets of effectors of necrotrophic and biotrophic pathogens are intrinsically similar, but that those of biotrophic pathogens have evolved further as a result of co‐evolution and no longer induce necrosis, but manipulate host targets in a more gentle manner.
EFFECTORS OF ECTOMYCORRHIZAL FUNGI
Little is known about effectors from ectomycorrhizal fungi that show both a saprophytic and biotrophic lifestyle. Recently, the genome of Laccaria bicolor was sequenced, which revealed that this basidiomycete shows highest sequence similarity with the sequenced basidiomycete plant pathogens U. maydis and M. lini (Martin and Selosse, 2008; Martin et al., 2008). The 65‐Mb genome is expected to code for approximately 20 000 proteins, approximately 3000 of which are predicted to be secreted, including approximately 300 effector‐type SSPs. A number of these SSPs show homology to HESPs of rust fungi. Some of the genes encoding SSPs are strongly up‐regulated during infection, whereas others are down‐regulated, suggesting a complex regulation of SSPs during colonization of the host. Many SSPs are also cysteine‐rich and probably belong to the cystine‐knotted peptides, such as Avr9 of C. fulvum (van den Hooven et al., 2001). One of those mycorrhiza‐induced cysteine‐rich SSPs of 7 kDa (MISSP7) was highly up‐regulated in the mycorrhizal tips of mycelium present in the Hartig net. However, additional studies are required to learn more about its function and that of other MISSPs. Further functional analysis of the L. bicolor genome will provide more insight into the differences between symbiotic, saprophytic and pathogenic fungi.
DISCOVERY OF NOVEL EFFECTORS
Recently, whole‐genome sequencing of fungal pathogens has provided an enormous amount of data that can be analysed for the presence of putative (secreted) effectors. Enrichment for effector candidates can be achieved by the integration of genome, transcriptome, proteome and metabolome data, when available. Comparative secretome analysis and blast sequence similarity searches can also be used to identify putative effectors in sequenced genomes, but will only prove to be successful when sufficient homology exists among effector genes, such as seen for Avr4, Ecp6 and the functional homologue of ToxA peptide from S. nodorum in P. tritici‐repentis (2008a, 2006). In addition, the genome of L. bicolor (Martin et al., 2008) has identified some MISSPs homologous to effectors in rust and other fungi. However, despite the great potential of genome‐wide searches for the identification of candidate effector genes, their function still needs to be confirmed experimentally by overexpression, gene disruption or silencing in fungal isolates and subsequent (a)virulence assays on host plants.
TRANSLOCATION OF FUNGAL EFFECTORS
Fungal effector proteins can be roughly grouped into extracellular effectors that are secreted into the apoplast or xylem of their host plants, and cytoplasmic effectors that are translocated into host cells. Despite the low degree of sequence conservation amongst fungal effectors, most code for small secreted proteins, some of which are translocated into host cells by an as yet unknown mechanism. Extracellular effectors are often N‐ and sometimes C‐terminally processed, as has been shown for the Avr and Ecp effectors of C. fulvum (Stergiopoulos and de Wit, 2009) and the Six effectors of F. oxysporum f. sp. lycopersici (Houterman et al., 2007). Another common feature of many fungal effectors is the presence of multiple cysteine residues that might be involved in disulphide bridge formation, providing protein stability. Some effectors active inside the host cell, such as those of rusts, possibly require appropriate folding and disulphide bridge formation outside the host before being taken up (Dodds et al., 2004; Kemen et al., 2005). Magnaporthe oryzae induces a biotrophic interfacial complex (BIC) in the host which contains secreted effectors, such as Avr‐Pita, Pwl1 and Pwl2. It has been shown by live‐cell fluorescence imaging that these effectors that accumulate in BICs are translocated into invaded host cells (Valent et al., 2009).
Effectors of rusts, powdery mildews, M. oryzae and F. oxysporum f. sp. lycopersici are putatively translocated into the host cell, where they interact with cytoplasmic or nuclear R proteins (Catanzariti et al., 2007; 2004, 2006; Ellis et al., 2007; Houterman et al., 2009; Jia et al., 2000; Shen et al., 2007). However, so far, in all fungal effectors, no clear consensus signature, such as the RXLR motif present in oomycete effectors, has been identified. The cytoplasmic effectors Avra10 and Avrk1 of B. graminis f. sp. hordei even lack a signal sequence for secretion (Ridout et al., 2006). The host‐selective protein toxin ToxA of P. tritici‐repentis (Ciuffetti et al., 1997) contains a solvent‐exposed RGD motif that interacts with the host plasma membrane and is probably required for internalization (Manning et al., 2008). A similar RGD motif mediating interaction with the plasma membrane is also present in the IpiO (AvrBlb1) effector proteins of the oomycete Phytophthora infestans (Senchou et al., 2004; Vleeshouwers et al., 2008).
DIRECT AND INDIRECT INTERACTIONS BETWEEN EFFECTORS AND R PROTEINS
Direct interaction between the cognate R protein and effector is also referred to as the ‘receptor–ligand’ model. Direct interactions between a fungal effector and plant R protein have been described for Avr‐Pita (Avr‐Pita176) from M. oryzae and the cognate Pi‐ta resistance protein from rice (Jia et al., 2000), and all effectors of M. lini and their cognate R proteins in flax (Dodds et al., 2006; Ellis et al., 2007; Wang et al., 2007). One implication of the ‘receptor–ligand’ model is that plants must carry large numbers of R proteins to enable them to recognize all individual effectors. This problem seems to be partly overcome by plants that have developed R proteins that monitor modifications in plant targets of fungal effectors. In this so‐called ‘guard’ model, R proteins do not interact directly with an effector, but guard its host target and respond to alterations in this target caused by the effector. The ‘guard’ model also enables the detection of multiple unrelated effectors that interact with the same host target guarded by a single R protein. A classical example of the ‘guard’ model is represented by the indirect interaction between Avr2 and Cf‐2, mediated by Rcr3, in the C. fulvum–tomato pathosystem (Rooney et al., 2005). In many other (nonfungal) pathosystems, indirect interaction between R protein and cognate Avr has been reported, suggesting that the majority of interactions would fit into the ‘guard’ model (Jones and Dangl, 2006). Direct interaction between effector and R protein can be overcome more readily by mutations in effectors that abolish recognition, whereas jettison of effectors seems to mediate evasion of R‐mediated resistance in the case of indirect interactions (De Wit, 2007; Jones and Dangl, 2006).
CONCLUSIONS AND FUTURE PROSPECTS
In Fig. 1, a short overview of the putative roles of effectors from necrotrophic, ectomycorrhizal and biotrophic fungi, and different types of interaction with their hosts, is depicted. Much additional research is required to confirm or reject the proposed models as they are in part speculative.
Figure 1.
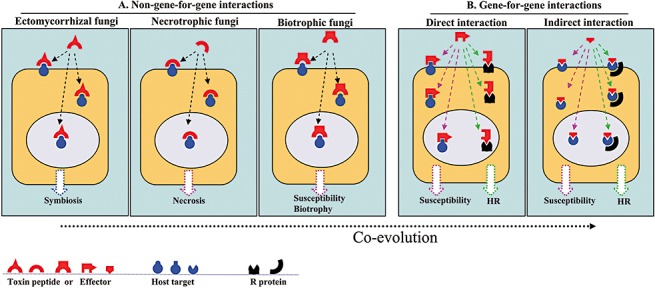
Diagram showing effectors from non‐gene‐for‐gene ectomycorrhizal, necrotrophic and biotrophic fungi, and from gene‐for‐gene biotrophic fungi, interacting with host targets and resistance proteins. (A) Non‐gene‐for‐gene interactions. Ectomycorrhizal fungi: putative effectors of (ecto)mycorrhizal fungi interact with host targets to facilitate symbiosis and suppress or avoid host defence responses. Necrotrophic fungi: effectors of necrotrophic fungi interact with host targets, eventually causing necrosis of host plant cells to facilitate disease. Biotrophic fungi: effectors of biotrophic fungi interact with host targets and facilitate disease without killing host plant cells. (B) Gene‐for‐gene interactions. It is assumed that, as a result of co‐evolution with their host plants, biotrophic fungi have evolved further. Direct interaction: it is assumed that effectors have two domains, one of which interacts with the virulence target, leading to host susceptibility, whereas the other is recognized by the R protein present in resistant plants, leading to the hypersensitive response (HR) and resistance. Indirect interaction: the effectors interact with the virulence target leading to susceptibility. Resistant plants have developed resistant proteins that do not interact directly with effectors, but guard the virulence target, and HR is triggered after sensing manipulation of the virulence target by the effector. Interaction between effectors and host targets (virulence targets and R proteins) can take place on the plasma membrane, in the cytoplasm or in the nucleus.
With the rapid advances in novel sophisticated and efficient sequencing platforms, many fungal genomes will be sequenced in the near future and, with parallel advances in bioinformatics tools, this will speed up the discovery of novel fungal effectors by comparative genomics. However, so far, homology among fungal effectors is limited. For most fungal effectors, a function in virulence can be investigated experimentally by gene disruption, gene silencing or overexpression, and it is expected that a number play a role in the suppression of PTI induced by fungal PAMPs. The discovery of more fungal PAMPs is anticipated and will enrich our understanding of the evolution of the plant defence system and the interplay between PTI and ETI. The finding that some fungal effector genes reside on pathogenicity islands is intriguing, and it will be interesting to determine how these islands have arisen during evolution. The elucidation of the mechanisms of translocation and the identification of interactors of fungal effectors in host cells remain major challenges for the future. Future research on effectors of mycorrhizal fungi, and of necrotrophic and biotrophic fungal pathogens, will reveal whether they represent similar biological functions, but only differ in their degree of co‐evolution.
REFERENCES
- Van Den Ackerveken, G.F.J.M. , Dunn, R.M. , Cozijnsen, A.J. , Vossen, J.P.M.J. , Van Den Broek, H.W.J. and De Wit, P.J.G.M. (1994) Nitrogen limitation induces expression of the avirulence gene avr9 in the tomato pathogen Cladosporium fulvum . Mol. Gen. Genet. 243, 277–285. [DOI] [PubMed] [Google Scholar]
- Van Den Ackerveken, G.F.J. , Van Kan, J.A. , Joosten, M.H.A.J. , Muisers, J.M. , Verbakel, H.M. and De Wit, P.J.G.M. (1993) Characterization of two putative pathogenicity genes of the fungal tomato pathogen Cladosporium fulvum . Mol. Plant–Microbe Interact. 6, 210–215. [DOI] [PubMed] [Google Scholar]
- Anderson, P.A. , Lawrence, G.J. , Morrish, B.C. , Ayliffe, M.A. , Jean Finnegan, E. and Ellis, J.G. (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine‐rich repeat coding region. Plant Cell, 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balesdent, M.H. , Attard, A. , Kuhn, M.L. and Rouxel, T. (2002) New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans . Phytopathology, 92, 1122–1133. [DOI] [PubMed] [Google Scholar]
- Balesdent, M.H. , Barbetti, M.J. , Li, H. , Sivasithamparam, K. , Gout, L. and Rouxel, T. (2005) Analysis of Leptosphaeria maculans race structure in a worldwide collection of isolates. Phytopathology, 95, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Balesdent, M.H. , Louvard, K. , Pinochet, X. and Rouxel, T. (2006) A large‐scale survey of races of Leptosphaeria maculans occurring on oilseed rape in France. Eur. J. Plant Pathol. 114, 53–65. [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Bernard, M. and Latge, J.P. (2001) Aspergillus fumigatus cell wall: composition and biosynthesis. Med. Mycol. 39, 9–17. [PubMed] [Google Scholar]
- Bieri, S. , Mauch, S. , Shen, Q.H. , Peart, J. , Devoto, A. , Casais, C. , Ceron, F. , Schulze, S. , Steinbiss, H.H. , Shirasu, K. and Schulze‐Lefert, P. (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell, 16, 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert, H.U. , Fudal, I. , Dioh, W. , Tharreau, D. , Notteghem, J.L. and Lebrun, M.H. (2004) A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell, 16, 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, M.D. and Thomma, B.P.H. (2008) The complexity of nitrogen metabolism and nitrogen‐regulated gene expression in plant pathogenic fungi. Physiol. Mol. Plant Pathol. 72, 104–110. [Google Scholar]
- Bolton, M.D. , Van Esse, H.P. , Vossen, J.H. , De Jonge, R. , Stergiopoulos, I. , Stulemeijer, I.J.E. , Van, G.C.M. , Borras‐Hidalgo, O. , Dekker, H.L. , De Koster, C.G. , De Wit, P.J.G.M. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69, 119–136. [DOI] [PubMed] [Google Scholar]
- Bryan, G.T. , Wu, K.S. , Farrall, L. , Jia, Y. , Hershey, H.P. , McAdams, S.A. , Faulk, K.N. , Donaldson, G.K. , Tarchini, R. and Valent, B. (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi‐ta . Plant Cell, 12, 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Burg, H.A. , Harrison, S.J. , Joosten, M.H.A.J. , Vervoort, J. and De Wit, P.J.G.M. (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant–Microbe Interact. 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Van Den Burg, H.A. , Spronk, C.A.E. , Boeren, S. , Kennedy, M.A. , Vissers, J.P.C. , Vuister, G.W. , De Wit, P.J.G.M. and Vervoort, J. (2004) Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein–protein interactions: the chitin‐binding site of Avr4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin‐binding domain. J. Biol. Chem. 279, 16786–16796. [DOI] [PubMed] [Google Scholar]
- Van Den Burg, H.A. , Westerink, N. , Francoijs, K.J. , Roth, R. , Woestenenk, E. , Boeren, S. , De Wit, P.J.G.M. , Joosten, M.H.A.J. and Vervoort, J. (2003) Natural disulfide bond‐disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf‐4‐mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278, 27340–27346. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. and Ellis, J.G. (2007) Avirulence proteins from haustoria‐forming pathogens. FEMS Microbiol. Lett. 269, 181–188. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffetti, L.M. , Tuori, R.P. and Gaventa, J.M. (1997) A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell, 9, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collemare, J. , Pianfetti, M. , Houlle, A.E. , Morin, D. , Camborde, L. , Gagey, M.J. , Barbisan, C. , Fudal, I. , Lebrun, M.H. and Bo?hnert, H.U. (2008) Magnaporthe grisea avirulence gene ACE1 belongs to an infection‐specific gene cluster involved in secondary metabolism. New Phytol. 179, 196–208. [DOI] [PubMed] [Google Scholar]
- Cutler, J.E. (2001) N‐glycosylation of yeast, with emphasis on Candida albicans . Med. Mycol. 39, 75–86. [PubMed] [Google Scholar]
- De Kock, M.J.D. , Brandwagt, B.F. , Bonnema, G. , De Wit, P.J.G.M. and Lindhout, P. (2005) The tomato Orion locus comprises a unique class of Hcr9 genes. Mol. Breed. 15, 409–422. [Google Scholar]
- Denoux, C. , Galletti, R. , Mammarella, N. , Gopalana, S. , Werck, D. , De Lorenzo, G. , Ferrari, S. , Ausubel, F.M. and Dewdney, J. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant, 1, 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit, P.J.G. (2007) How plants recognize pathogens and defend themselves. Cell. Mol. Life Sci. 64, 2726–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit, P.J.G. and Kodde, E. (1981) Further characterization and cultivar‐specificity of glycoprotein elicitors from culture filtrates and cell walls of Cladosporium fulvum (syn Fulvia fulva). Physiol. Plant Pathol. 18, 297–314. [Google Scholar]
- De Wit, P.J.G.M. , Lauge, R. , Honee, G. , Joosten, M.H.A.J. , Vossen, P. , Kooman‐Gersmann, M. , Vogelsang, R. and Vervoort, J.J.M. (1997) Molecular and biochemical basis of the interaction between tomato and its fungal pathogen Cladosporium fulvum . Antonie Van Leeuwenhoek, 71, 137–141. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Ayliffe, M.A. and Ellis, J.G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Teh, T. , Wang, C.I.A. , Ayliffe, M.A. , Kobe, B. and Ellis, J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann, G. , Van Der Linde, K. , Amann, D. , Schwammbach, D. , Hof, A. , Mohanty, A. , Jackson, D. and Kahmann, R. (2009) Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5 (2), e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Does, C. , Lievens, B. , Claes, L. , Houterman, P.M. , Cornelissen, B.J.C. and Rep, M. (2008) The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 10, 1475–1485. [DOI] [PubMed] [Google Scholar]
- Ellis, J. , Lawrence, G. , Ayliffe, M. , Anderson, P. , Collins, N. , Finnegan, J. , Frost, D. , Luck, J. and Pryor, T. (1997) Advances in the molecular genetic analysis of the flax–flax rust interaction. Annu. Rev. Phytopathol. 35, 271–291. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G. , Lawrence, G.J. and Dodds, P.N. (2007) Further analysis of gene‐for‐gene disease resistance specificity in flax. Mol. Plant Pathol. 8, 103–109. [DOI] [PubMed] [Google Scholar]
- Ernst, J.F. and Prill, S.K.H. (2001) O‐glycosylation. Med. Mycol. 39, 67–74. [PubMed] [Google Scholar]
- Van Esse, H.P. , Bolton, M.D. , Stergiopoulos, I. , De Wit, P.J.G.M. and Thomma, B.P.H.J. (2007) The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant–Microbe Interact. 20, 1092–1101. [DOI] [PubMed] [Google Scholar]
- Van Esse, H.P. , Van't Klooster, J.W. , Bolton, M.D. , Yadeta, K. , VanBaarlen, P. , Boeren, S. , Vervoort, J. , De Wit, P.J.G. and Thomma, B.P.H.J. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell, 20, 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farman, M.L. and Leong, S.A. (1998) Chromosome walking to the AVR1‐CO39 avirulence gene of Magnaporthe grisea: discrepancy between the physical and genetic maps. Genetics, 150, 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1942) Inheritance of pathogenicity in Melampsora lini . Phytopathology, 32, 653–669. [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Solomon, P.S. and Oliver, R.P. (2008a) Host‐specific toxins: effectors of necrotrophic pathogenicity. Cell. Microbiol. 10, 1421–1428. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Meinhardt, S.W. and Faris, J.D. (2007) The Stagonospora nodorum–wheat pathosystem involves multiple proteinaceous host‐selective toxins and corresponding host sensitivity genes that interact in an inverse gene‐for‐gene manner. Plant J. 51, 681–692. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Stukenbrock, E.H. , Liu, Z. , Meinhardt, S. , Ling, H. , Faris, J.D. , Rasmussen, J.B. , Solomon, P.S. , McDonald, B.A. and Oliver, R.P. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Zhang, Z.C. , Solomon, P.S. , Oliver, R.P. and Faris, J.D. (2008b) Characterization of the interaction of a novel Stagonospora nodorum host‐selective toxin with a wheat susceptibility gene. Plant Physiol. 146, 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudal, I. , Bohnert, H.U. , Tharreau, D. and Lebrun, M.H. (2005) Transposition of MINE, a composite retrotransposon, in the avirulence gene ACE1 of the rice blast fungus Magnaporthe grisea . Fungal. Genet. Biol. 42, 761–772. [DOI] [PubMed] [Google Scholar]
- Fudal, I. , Ross, S. , Gout, L. , Blaise, F. , Kuhn, M.L. , Eckert, M.R. , Cattolico, L. , Bernard‐Samain, S. , Balesdent, M.H. and Rouxel, T. (2007) Heterochromatin‐like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map‐based cloning of AvrLm6 . Mol. Plant–Microbe Interact. 20, 459–470. [DOI] [PubMed] [Google Scholar]
- Glawe, D.A. (2008) The powdery mildews: a review of the world's most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 46, 27–51. [DOI] [PubMed] [Google Scholar]
- Gohre, V. and Robatzek, S. (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46, 189–215. [DOI] [PubMed] [Google Scholar]
- Gout, L. , Fudal, I. , Kuhn, M.L. , Blaise, F. , Eckert, M. , Cattolico, L. , Balesdent, M.H. and Rouxel, T. (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans . Mol. Microbiol. 60, 67–80. [DOI] [PubMed] [Google Scholar]
- Hahn, M. , Jungling, S. and Knogge, W. (1993) Cultivar‐specific elicitation of barley defense reactions by the phytotoxic peptide NIP1 from Rhynchosporium secalis . Mol. Plant–Microbe Interact. 6, 745–754. [DOI] [PubMed] [Google Scholar]
- Halterman, D. , Zhou, F. , Wei, F. , Wise, R.P. and Schulze‐Lefert, P. (2001) The MLA6 coiled‐coil, NBS‐LRR protein confers AvrMla6‐dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 25, 335–348. [DOI] [PubMed] [Google Scholar]
- Halterman, D.A. and Wise, R.P. (2004) A single‐amino acid substitution in the sixth leucine‐rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J. 38, 215–226. [DOI] [PubMed] [Google Scholar]
- Van Den Hooven, H.W. , Burg, H.A. , Vossen, P. , Boeren, S. , De Wit, P.J.G.M. and Vervoort, J. (2001) Disulfide bond structure of the AVR9 elicitor of the fungal tomato pathogen Cladosporium fulvum: evidence for a cystine knot. Biochemistry, 40, 3458–3466. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Cornelissen, B.J.C. and Rep, M. (2008) Suppression of plant resistance gene‐based immunity by a fungal effector. PLoS Pathog. 4, e1000061. doi:1000010.1001371/journal.ppat.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman, P.M. , Ma, L. , Van Ooijen, G. , De Vroomen, M.J. , Cornelissen, B.J.C. , Takken, F.L.W. and Rep, M. (2009) The effector protein Avr2 of the xylem‐colonizing fungus Fusarium oxysporum activates the tomato resistance protein I‐2 intracellularly. Plant J. 58, 970–978. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Speijer, D. , Dekker, H.L. , De Koster, C.G. , Cornelissen, B.J.C. and Rep, M. (2007) The mixed xylem sap proteome of Fusarium oxysporum‐infected tomato plants. Mol. Plant Pathol. 8, 215–221. [DOI] [PubMed] [Google Scholar]
- Huang, C.C. and Lindhout, P. (1997) Screening for resistance in wild Lycopersicon species to Fusarium oxysporum f. sp. lycopersici race 1 and race 2. Euphytica, 93, 145–153. [Google Scholar]
- Jensen, J. , Jorgensen, J.H. , Jensen, H.P. , Giese, H. and Doll, H. (1980) Linkage of the hordein loci Hor1 and Hor2 with the powdery mildew resistance loci Ml‐k and Ml‐a on barley chromosome 5. Theor. Appl. Genet. 58, 27–31. [DOI] [PubMed] [Google Scholar]
- Jia, Y. , Bryan, G.T. , Farrall, L. and Valent, B. (2003) Natural variation at the Pi‐ta rice blast resistance locus. Phytopathology, 93, 1452–1459. [DOI] [PubMed] [Google Scholar]
- Jia, Y. , McAdams, S.A. , Bryan, G.T. , Hershey, H.P. and Valent, B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H.A. , Vogelsang, R. , Cozijnsen, T.J. , Verberne, M.C. and De Wit, P.J.G.M. (1997) The biotrophic fungus Cladosporium fulvum circumvents Cf‐4‐mediated resistance by producing unstable AVR4 elicitors. Plant Cell, 9, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, M.H.A.J. and De Wit, P.J.G.M. (1999) The tomato–Cladosporium fulvum interaction: a versatile experimental system to study plant–pathogen interactions. Annu. Rev. Phytopathol. 37, 335–367. [DOI] [PubMed] [Google Scholar]
- Jorgensen, J.H. (1994) Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119. [Google Scholar]
- Jouault, T. , Sarazin, A. , Martinez‐Esparza, M. , Fradin, C. , Sendid, B. and Poulain, D. (2009) Host responses to a versatile commensal: PAMPs and PRRs interplay leading to tolerance or infection by Candida albicans . Cell. Microbiol. 11, 1007–1015. [DOI] [PubMed] [Google Scholar]
- Kahmann, R. and Kamper, J. (2004) Ustilago maydis: how its biology relates to pathogenic development. New Phytol. 164, 31–42. [DOI] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. and Shibuya, N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA, 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. (2007) Groovy times: filamentous pathogen effectors revealed. Curr. Opin. Plant Biol. 10, 358–365. [DOI] [PubMed] [Google Scholar]
- Van Kan, J.A.L. , Van Den Ackerveken, G.F.J.M. and De Wit, P.J.G.M. (1991) Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant–Microbe Interact. 4, 52–59. [DOI] [PubMed] [Google Scholar]
- Kang, S. , Lebrun, M.H. , Farrall, L. and Valent, B. (2001) Gain of virulence caused by insertion of a Pot3 transposon in a Magnaporthe grisea avirulence gene. Mol. Plant–Microbe Interact. 14, 671–674. [DOI] [PubMed] [Google Scholar]
- Kang, S. , Sweigard, J.A. and Valent, B. (1995) The PWL host specificity gene family in the blast fungus Magnaporthe grisea . Mol. Plant–Microbe Interact. 8, 939–948. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Kemen, A.C. , Rafiqi, M. , Hempel, U. , Mendgen, K. , Hahn, M. and Voegele, R.T. (2005) Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant–Microbe Interact. 18, 1130–1139. [DOI] [PubMed] [Google Scholar]
- Khang, C.H. , Park, S.Y. , Lee, Y.H. , Valent, B. and Kang, S. (2008) Genome organization and evolution of the AVR‐pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant–Microbe Interact. 21, 658–670. [DOI] [PubMed] [Google Scholar]
- Kruger, J. , Thomas, C.M. , Golstein, C. , Dixon, M.S. , Smoker, M. , Tang, S. , Mulder, L. and Jones, J.D.G. (2002) A tomato cysteine protease required for Cf‐2‐dependent disease resistance and suppression of autonecrosis. Science, 296, 744–747. [DOI] [PubMed] [Google Scholar]
- Laugé, R. , Goodwin, P.H. , De Wit, P.J.G.M. and Joosten, M.H.A.J. (2000) Specific HR‐associated recognition of secreted proteins from Cladosporium fulvum occurs in both host and non‐host plants. Plant J. 23, 735–745. [DOI] [PubMed] [Google Scholar]
- Laugé, R. , Joosten, M.H.A. , Haanstra, J.P.W. , Goodwin, P.H. , Lindhout, P. and De Wit, P.J.G.M. (1998) Successful search for a resistance gene in tomato targeted against a virulence factor of a fungal pathogen. Proc. Natl. Acad. Sci. USA, 95, 9014–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugé, R. , Joosten, M.H.A.J. , Van Den Ackerveken, G.F.J.M. , Van Den Broek, H.W.J. and De Wit, P.J.G.M. (1997) The in planta‐produced extracellular proteins ECP1 and ECP2 of Cladosporium fulvum are virulence factors. Mol. Plant–Microbe Interact. 10, 725–734. [Google Scholar]
- Lawrence, G.J. , Finnegan, E.J. , Ayliffe, M.A. and Ellis, J.G. (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N . Plant Cell, 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.H. , Friesen, T.L. , Ling, H. , Meinhardt, S.W. , Oliver, R.P. , Rasmussen, J.B. and Faris, J.D. (2006) The Tsn1–ToxA interaction in the wheat–Stagonospora nodorum pathosystem parallels that of the wheat–tan spot system. Genome, 49, 1265–1273. [DOI] [PubMed] [Google Scholar]
- Luderer, R. , De Kock, M.J.D. , Dees, R.H.L. , De Wit, P.J.G.M. and Joosten, M.H.A.J. (2002a) Functional analysis of cysteine residues of ECP elicitor proteins of the fungal tomato pathogen Cladosporium fulvum . Mol. Plant Pathol. 3, 91–95. [DOI] [PubMed] [Google Scholar]
- Luderer, R. , Takken, F.L.W. , De Wit, P.J.G.M. and Joosten, M.H.A.J. (2002b) Cladosporium fulvum overcomes Cf‐2‐mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45, 875–884. [DOI] [PubMed] [Google Scholar]
- Manning, V.A. , Hamilton, S.M. , Karplus, P.A. and Ciuffetti, L.M. (2008) The Arg‐Gly‐Asp‐containing, solvent‐exposed loop of Ptr ToxA is required for internalization. Mol. Plant–Microbe Interact. 21, 315–325. [DOI] [PubMed] [Google Scholar]
- Manning, V.A. , Hardison, L.K. and Ciuffetti, L.M. (2007) Ptr ToxA interacts with a chloroplast‐localized protein. Mol. Plant–Microbe. Interact. 20, 168–177. [DOI] [PubMed] [Google Scholar]
- Marmeisse, R. , Van Den Ackerveken, G.F.J.M. , Goosen, T. , De Wit, P.J.G.M. and Van Den Broek, H.W.J. (1993) Disruption of the avirulence gene avr9 in two races of the tomato pathogen Cladosporium fulvum causes virulence on tomato genotypes with the complementary resistance gene Cf9 . Mol. Plant–Microbe. Interact. 6, 412–417. [Google Scholar]
- Martin, F. , Aerts, A. , Ahren, D. , Brun, A. , Danchin, E.G.J. , Duchaussoy, F. , Gibon, J. , Kohler, A. , Lindquist, E. , Pereda, V. , Salamov, A. , Shapiro, H.J. , Wuyts, J. , Blaudez, D. , Buee, M. , Brokstein, P. , Canback, B. , Cohen, D. , Courty, P.E. , Coutinho, P.M. , Delaruelle, C. , Detter, J.C. , Deveau, A. , DiFazio, S. , Duplessis, S. , Fraissinet‐Tachet, L. , Lucic, E. , Frey‐Klett, P. , Fourrey, C. , Feussner, I. , Gay, G. , Grimwood, J. , Hoegger, P.J. , Jain, P. , Kilaru, S. , Labbe, J. , Lin, Y.C. , Legue, V. , Le Tacon, F. , Marmeisse, R. , Melayah, D. , Montanini, B. , Muratet, M. , Nehls, U. , Niculita‐Hirzel, H. , Oudot‐Le Secq, M.P. , Peter, M. , Quesneville, H. , Rajashekar, B. , Reich, M. , Rouhier, N. , Schmutz, J. , Yin, T. , Chalot, M. , Henrissat, B. , Kues, U. , Lucas, S. , Van de Peer, Y. , Podila, G.K. , Polle, A. , Pukkila, P.J. , Richardson, P.M. , Rouze, P. , Sanders, I.R. , Stajich, J.E. , Tunlid, A. , Tuskan, G. and Grigoriev, I.V. (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature, 452, 88–U87. [DOI] [PubMed] [Google Scholar]
- Martin, F. and Selosse, M.A. (2008) The Laccaria genome: a symbiont blueprint decoded. New Phytol. 180, 296–310. [DOI] [PubMed] [Google Scholar]
- Matzinger, P. (2007) Friendly and dangerous signals: is the tissue in control? Nat. Immunol. 8, 11–13. [DOI] [PubMed] [Google Scholar]
- McFadden, D. , Zaragoza, O. and Casadevall, A. (2006) The capsular dynamics of Cryptococcus neoformans . Trends Microbiol. 14, 497–505. [DOI] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. , Narusaka, Y. , Kawakami, N. , Kaku, H. and Shibuya, N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA, 104, 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, O. , Schreier, P.H. and Uhrig, J.F. (2008) Identification and characterization of secreted and pathogenesis‐related proteins in Ustilago maydis . Mol. Genet. Genomics, 279, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach, M.J. , Farrall, L. , Sweigard, J.A. , Chumley, F.G. and Valent, B. (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi‐ta . Plant Cell, 12, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann, C. , Luberacki, B. , Kufner, I. , Koch, W. , Brunner, F. , Weyand, M. , Mattinen, L. , Pirhonen, M. , Anderluh, G. , Seitz, H.U. , Nurnberger, T. and Oecking, C. (2009) A common toxin fold mediates microbial attack and plant defense. Proc. Natl. Acad. Sci. USA, 106, 10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallaghy, P.K. , Nielsen, K.J. , Craik, D.J. and Norton, R.S. (1994) A common structural motif incorporating a cystine knot and a triple‐stranded β‐sheet in toxic and inhibitory polypeptides. Protein Sci. 3, 1833–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlange, F. , Daverdin, G. , Fudal, I. , Kuhn, M.L. , Balesdent, M.H. , Blaise, F. , Grezes‐Besset, B. and Rouxel, T. (2009) Leptosphaeria maculans avirulence gene AvrLm4‐7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4‐mediated recognition through a single amino acid change. Mol. Microbiol. 71, 851–863. [DOI] [PubMed] [Google Scholar]
- Perez‐Garcia, A. , Snoeijers, S.S. , Joosten, M.H.A.J. , Goosen, T. and De Wit, P.J.G.M. (2001) Expression of the avirulence gene Avr9 of the fungal tomato pathogen Cladosporium fulvum is regulated by the global nitrogen response factor NRF1. Mol. Plant–Microbe Interact. 14, 316–325. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Van Der Does, H.C. , Meijer, M. , Van Wijk, R. , Houterman, P.M. , Dekker, H.L. , De Koster, C.G. and Cornelissen, B.J.C. (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Meijer, M. , Houterman, P.M. , Van Der Does, H.C. and Cornelissen, B.J.C. (2005) Fusarium oxysporum evades I‐3‐mediated resistance without altering the matching avirulence gene. Mol. Plant–Microbe Interact. 18, 15–23. [DOI] [PubMed] [Google Scholar]
- Ridout, C.J. , Skamnioti, P. , Porritt, O. , Sacristan, S. , Jones, J.D.G. and Brown, J.K.M. (2006) Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell, 18, 2402–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohe, M. , Gierlich, A. , Hermann, H. , Hahn, M. , Schmidt, B. , Rosahl, S. and Knogge, W. (1995) The race‐specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 14, 4168–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, H.C.E. , Van't Klooster, J.W. , Van Der Hoorn, R.A.L. , Joosten, M.H.A.J. , Jones, J.D.G. and De Wit, P.J.G.M. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf‐2‐dependent disease resistance. Science, 308, 1783–1786. [DOI] [PubMed] [Google Scholar]
- Rouxel, T. , Penaud, A. , Pinochet, X. , Brun, H. , Gout, L. , Delourme, R. , Schmit, J. and Balesdent, M.H. (2003) A 10‐year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene of oilseed rape. Eur. J. Plant Pathol. 109, 871–881. [Google Scholar]
- Schurch, S. , Linde, C.C. , Knogge, W. , Jackson, L.F. and McDonald, B.A. (2004) Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1 . Mol. Plant–Microbe. Interact. 17, 1114–1125. [DOI] [PubMed] [Google Scholar]
- Senchou, V. , Weide, R. , Carrasco, A. , Bouyssou, H. , Pont‐Lezica, R. , Govers, F. and Canut, H. (2004) High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell. Mol. Life Sci. 61, 502–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabab, M. , Shindo, T. , Gu, C. , Kaschani, F. , Pansuriya, T. , Chintha, R. , Harzen, A. , Colby, T. , Kamoun, S. and Van Der Hoorn, R.A. (2008) Fungal effector protein AVR2 targets diversifying defense‐related cys proteases of tomato. Plant Cell, 20, 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, W.X. , Cao, M. , Dan, L.U. and Tyler, B.M. (2004) The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant–Microbe Interact. 17, 394–403. [DOI] [PubMed] [Google Scholar]
- Shen, Q.H. , Saijo, Y. , Mauch, S. , Biskup, C. , Bieri, S. , Keller, B. , Seki, H. , Ulker, B. , Somssich, I.E. and Schulze‐Lefert, P. (2007) Nuclear activity of MLA immune receptors links isolate‐specific and basal disease‐resistance responses. Science, 315, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Shen, Q.H. , Zhou, F. , Bieri, S. , Haizel, T. , Shirasu, K. and Schulze‐Lefert, P. (2003) Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell, 15, 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumpourou, E. , Lakovidis, M. , Chartrain, L. , Lyall, V. and Thomas, C.M. (2007) The Solanum pimpinellifolium Cf‐ECP1 and Cf‐ECP4 genes for resistance to Cladosporium fulvum are located at the Milky Way locus on the short arm of chromosome 1. Theor. Appl. Genet. 115, 1127–1136. [DOI] [PubMed] [Google Scholar]
- Staskawicz, B.J. , Dahlbeck, D. and Keen, N.T. (1984) Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race‐specific incompatibility on Glycine max (L.) Merr. Proc. Natl. Acad. Sci. USA, 81, 6024–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. , De Kock, M.J.D. , Lindhout, P. and De Wit, P.J.G.M. (2007) Allelic variation in the effector genes of the tomato pathogen Cladosporium fulvum reveals different modes of adaptive evolution. Mol. Plant–Microbe Interact. 20, 1271–1283. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. and De Wit, P.J.G.M. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Sweigard, J.A. , Carroll, A.M. , Kang, S. , Farrall, L. , Chumley, F.G. and Valent, B. (1995) Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell, 7, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. , Bolton, M.D. , Clergeot, P.H. and De Wit, P.J.G.M. (2006) Nitrogen controls in planta expression of Cladosporium fulvum Avr9 but no other effector genes. Mol. Plant Pathol. 7, 125–130. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H. , Van Esse, H.P. , Crous, P.W. and De Wit, P.J.G.M. (2005) Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6, 379–393. [DOI] [PubMed] [Google Scholar]
- Valent, B. , Farrall, L. and Chumley, F.G. (1991) Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics, 127, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent, B. , Khang, C.H. , Giraldo, M.C. , Mosquera, G. , Berruyer, R. , Kankanala, P. , Yi, M. , Czymmek, K. , Park, S.Y. and Kang, S. (2009) The biotrophic interfacial complex and effector translocation during rice blast disease. Phytopathology, 99, S167–S167. [Google Scholar]
- Van't Slot, K.A.E. , Van Den Burg, H.A. , Kloks, C.P.A.M. , Hilbers, C.W. , Knogge, W. and Papavoine, C.H.M. (2003) Solution structure of the plant disease resistance‐triggering protein NIP1 from the fungus Rhynchosporium secalis shows a novel β‐sheet fold. J. Biol. Chem. 278, 45730–45736. [DOI] [PubMed] [Google Scholar]
- Van't Slot, K.A.E. , Gierlich, A. and Knogge, W. (2007) A single binding site mediates resistance‐ and disease‐associated activities of the effector protein NIP1 from the barley pathogen Rhynchosporium secalis . Plant Physiol. 144, 1654–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort, J. , Van Den Hooven, H.W. , Berg, A. , Vossen, P. , Vogelsang, R. , Joosten, M.H.A. and De Wit, P.J.G.M. (1997) The race‐specific elicitor AVR9 of the tomato pathogen Cladosporium fulvum: a cystine knot protein: sequence‐specific 1H NMR assignments, secondary structure and global fold of the protein. FEBS Lett. 404, 153–158. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S.K. , Wang, M. , Bouwmeester, K. , Vosman, B. , Visser, R.G.F. , Jacobsen, E. and Govers, F. , Kamoun, S. and Van der Vossen, E.A.G. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE, 3, published 6 Aug 2008, doi:10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.I.A. , Guncar, G. , Forwood, J.K. , Teh, T. , Catanzariti, A.M. , Lawrence, G.J. , Loughlin, F.E. , Mackay, J.P. , Schirra, H.J. , Anderson, P.A. , Ellis, J.G. , Dodds, P.N. and Kobe, B. (2007) Crystal structures of flax rust avirulence proteins AvrL567‐A and ‐D reveal details of the structural basis for flax disease resistance specificity. Plant Cell, 19, 2898–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerink, N. , Brandwagt, B.F. , De Wit, P.J.G. and Joosten, M.H.A.J. (2004) Cladosporium fulvum circumvents the second functional resistance gene homologue at the Cf‐4 locus (Hcr9‐4E) by secretion of a stable avr4E isoform. Mol. Microbiol. 54, 533–545. [DOI] [PubMed] [Google Scholar]
- Yoshida, K. , Saitoh, H. , Fujisawa, S. , Kanzaki, H. , Matsumura, H. , Yoshida, K. , Tosa, Y. , Chuma, I. , Takano, Y. , Win, J. , Kamoun, S. and Terauchi, R. (2009) Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae . Plant Cell, 21, 1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Henderson, C. , Perfect, E. , Carver, T.L.W. , Thomas, B.J. , Skamnioti, P. and Gurr, S.J. (2005) Of genes and genomes, needles and haystacks: Blumeria graminis and functionality. Mol. Plant Pathol. 6, 561–575. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.C. , Friesen, T.L. , Simons, K.J. , Xu, S.S. and Faris, J.D. (2009) Development, identification, and validation of markers for marker‐assisted selection against the Stagonospora nodorum toxin sensitivity genes Tsn1 and Snn2 in wheat. Mol. Breed. 23, 35–49. [Google Scholar]
- Zhou, E. , Jia, Y. , Singh, P. , Correll, J.C. and Lee, F.N. (2007) Instability of the Magnaporthe oryzae avirulence gene AVR‐Pita alters virulence. Fungal. Genet. Biol. 44, 1024–1034. [DOI] [PubMed] [Google Scholar]


