SUMMARY
Several ethylene‐response factor (ERF) transcription factors are believed to play a crucial role in the activation of plant defence responses, but little is known about the relationships between the diversity of this family and the functions of groups or individual ERFs in this process. In this study, 200 ERF genes from the unigene cotton database were identified. Conserved amino acid residues and phylogeny reconstruction using the AP2 conserved domain suggest that the classification into 10 major groups used for Arabidopsis and rice is applicable to the cotton ERF family. Based on in silico studies, we predict that group IX ERF genes in cotton are involved in jasmonate (JA), ethylene (ET) and pathogen responses. To test this hypothesis, we analysed the transcript profiles of the group IXa subfamily in the regulation of specific resistance to Xanthomonas campestris pathovar malvacearum. The expression of four members of group IXa was induced on challenge with X. campestris pv. malvacearum. Furthermore, the expression of several ERF genes of group IXa was induced synergistically by JA in combination with ET, suggesting that the encoded ERF proteins may play key roles in the integration of both signals to activate JA‐ and ET‐dependent responses.
INTRODUCTION
Plants undergo continuous exposure to various biotic and abiotic stresses in their natural environment. To survive under such conditions, plants have evolved intricate mechanisms to perceive external signals, allowing optimal responses to environmental stresses including attack by herbivores or microbial pathogens (Fujita et al., 2006). The perception of stress signals leads to the production of secondary signalling molecules, such as jasmonic acid, ethylene (ET) and salicylic acid (SA). Jasmonic acid belongs to a family of signalling molecules collectively known as jasmonates (JAs). In addition to their role in plant growth and development, JAs are major intermediates involved in response to wounding, herbivore attack and pathogen infection (Wasternack, 2007). An essential step in the JA‐dependent defence response is the rapid transcription of genes encoding antimicrobial proteins (Penninckx et al., 1998) or enzymes involved in the biosynthesis of secondary metabolites (Memelink et al., 2001). The study of mechanisms in which the expression of these defence‐related genes is regulated is of crucial importance to an understanding of signal transduction pathways and plant responses to environmental stress.
Transcription factors belonging to the AP2/ERF domain protein family, known as the ethylene‐response factors (ERFs), have emerged as important elements in plant defence responses (Gutterson and Reuber, 2004). These transcription factors are characterized by a single AP2/ERF‐type DNA‐binding domain with a highly conserved amino acid sequence, which consists of about 60 amino acids. The three‐dimensional structure of the AP2 DNA‐binding domain of AtERF1 from Arabidopsis thaliana complexed with its target DNA fragment has been determined by nuclear magnetic resonance (NMR) (Allen et al., 1998). The domain consists of a three‐stranded, anti‐parallel β‐sheet and an α‐helix packed approximately parallel to the β‐sheet. The AP2 domain of ERF proteins specifically interacts with a conserved AGCCGCC sequence called the GCC‐box (Menke et al., 1999).
The ERF transcription factor encoding genes have been identified exclusively in plants (Gutterson and Reuber, 2004) with a total of 122 and 139 in the Arabidopsis and rice genomes, respectively. The ERF family has been classified into 10 major groups according to the type of AP2 domain (Nakano et al., 2006). Several AP2/ERF genes have been shown to be regulated by various stress‐related stimuli, such as wounding, JA, ET, SA and pathogens (Chen et al., 2002). McGrath et al. (2005) have demonstrated that the ERF gene family is the predominant transcription factor family responsive to both JA and the incompatible necrotrophic pathogen Alternaria brassicicola. The importance of ERF factors in regulating the plant defence transcriptome has been further underlined by studies using overexpression and gene‐silencing approaches. The transcription factor ERF1 of group IX has been suggested to act as an integrator of JA and ET signalling pathways in Arabidopsis (Lorenzo et al., 2003). Constitutive overexpression of ERF1 activates the expression of several defence‐related genes, including plant defensin1. 2 (PDF1. 2) and basic‐chitinase (ChiB; Solano et al., 1998), and has been shown to confer resistance to several fungi (Berrocal‐Lobo and Molina, 2004).
Cotton (Gossypium hirsutum) is an important crop because the fibre is an excellent natural textile material. Cotton has been shown to be a convenient experimental tropical plant for the elucidation of resistance mechanisms to pathogens, including bacterial blight caused by Xanthomonas campestris pathovar malvacearum (Delannoy et al., 2005). Resistance of cotton to bacterial blight caused by X. campestris pv. malvacearum is mediated by gene for gene interactions (De Feyter et al., 1993). The resistance phenotype is characterized by rapid localized tissue collapse, resulting in necrotization (hypersensitive response, HR) and immobilization of the intruding pathogen at the sites of attack. At least 18 major resistance genes, so‐called B genes, have been reported to be dominant resistance factors in G. hirsutum. So far, defence responses to X. campestris pv. malvacearum have been investigated, such as the oxidative burst (Delannoy et al., 2003), accumulation of SA (Martinez et al., 2000), production of antimicrobial molecules (Daïet al., 1996) and triggering of the oxilipin pathway (Jalloul et al., 2002). The identification and expression profiles of a few ERF transcription factors have been reported recently from cotton. Among these, an ERF transcription factor, so‐called ERF1, is induced during fibre cell initiation, but repressed in the naked seed mutant (N1N1) that is impaired in fibre formation (Samuel Yang et al., 2006). Furthermore, GhERF1 has been found to be responsive to abiotic stresses (Qiao et al., 2008), and GhERF4 is possibly involved in the regulation of transcription by ET‐mediated signalling (Jin and Liu, 2008).
Based on the highly conserved sequences in the AP2 domain of known ERFs (Nakano et al., 2006), a computational analysis to identify novel ERF genes in cotton appears to be relevant. In this study, we report the identification and characterization of 200 ERF transcription factors based on the current availability of a large public cotton expressed sequence tag (EST) database. The phylogeny reconstruction based on the AP2 domains suggests that the classification into the 10 major groups used for Arabidopsis and rice is applicable to the cotton ERF family. From an analysis of the molecular diversity, conserved specific motifs of the AP2 domains of each group have been identified, and allow a rapid and easy classification of any plant ERF transcription factor. Based on our phylogenetic studies, we predict that cotton group IX ERF genes are involved in JA and ET responses. An accurate analysis of the temporal expression pattern of five ERF transcription factors of group IXa, in response to hormone defences, including JA, ET and SA, has been performed. Consistently, the expression of several ERF genes was induced by JA and ET, but not by SA, suggesting that the encoded ERF proteins may play roles in the integration of signals to activate both JA‐ and ET‐dependent responses. Moreover, it has been observed that the expression of several factors GhERF of group IXa is differentially regulated by virulent and avirulent strains of X. campestris pv. malvacearum.
RESULTS
Identification of the ERF family genes in Gossypium
To identify the members of the cotton AP2 domain family, Blast searches of the cotton databases were performed using the AP2/ERF domain (about 59 amino acids) of the 122 ERF proteins from Arabidopsis as a query sequence. Because G. hirsutum is an allotetraploid formed from progenitors represented by diploid A genome (G. arboreum) and D genome (G. raimondii) lineages, Blast searches were performed in databases from these three species. Two hundred and eighteen unique genes were identified as possibly encoding AP2/ERF domain(s) (Table 1). Eleven genes were predicted to encode proteins containing two AP2/ERF domains and were assigned to the AP2 family. Seven genes were predicted to encode one AP2/ERF domain together with one B3 domain and were assigned to the RAV family. Two hundred genes were predicted to encode proteins containing a single AP2/ERF domain and were assigned to the ERF family, as indicated by Sakuma et al. (2002). As shown in Table 1, 148 ERF genes were identified from G. hirsutum. For G. raimondii and G. arboreum species, 34 and 18 ERF genes were identified, respectively. The individual genes of the ERF family from the three species are listed in Table S1 (see Supporting Information). The 148 unigenes of the ERF family containing a complete AP2/ERF domain were analysed in detail.
Table 1.
Summary of the diversity of the AP2/ethylene‐response factor (ERF) superfamily in cotton and Arabidopsis.
| Classification | Arabidopsis genes | Gossypium hirsutum genes | G. raimondii genes | G. arboreum genes | Total cotton genes |
|---|---|---|---|---|---|
| ERF family | 122 | 148 | 34 | 18 | 200 |
| AP2 family | 18 | 4 | 5 | 2 | 11 |
| RAV family | 6 | 4 | 2 | 1 | 7 |
| Total | 146 | 156 | 41 | 21 | 218 |
Phylogeny reconstruction of Arabidopsis and Gossypium ERF genes based on the sequence similarity of the AP2/ERF domains
To study the phylogenetic relationships between the genes in the Gossypium ERF family, a multiple alignment analysis was performed using amino acid sequences in the AP2/ERF domain. With the identification of 122 ERF genes described previously (Sakuma et al., 2002) in the model species Arabidopsis, it was possible to perform a phylogeny reconstruction (Fig. 1). The tree was constructed with the neighbour‐joining method using sequences of 58–59 amino acids of the AP2 domain from cotton and Arabidopsis. This tree is globally conserved and distinguished the 10 major groups, namely groups I–X, as described by Nakano et al. (2006). The phylogram shows low resolution and no bootstrap support for the lower branches. By contrast, there is good bootstrap support for a number of groups in the higher branches of the tree (data not shown). However, several modifications of the groups are observed (Fig. 1). Group III was reported to be divided into five subgroups, IIIa–IIIe. In this study, the phylogenetic tree shows that subgroup IIIab is closer to group II than to subgroup IIIcde. Group II was reported to be composed of three subgroups. Subgroups IIc and IIb are closer to subgroup IIIab than to subgroup IIa. Otherwise, we observed that six sequences of Arabidopsis from group VIIIb branched together with group V.
Figure 1.
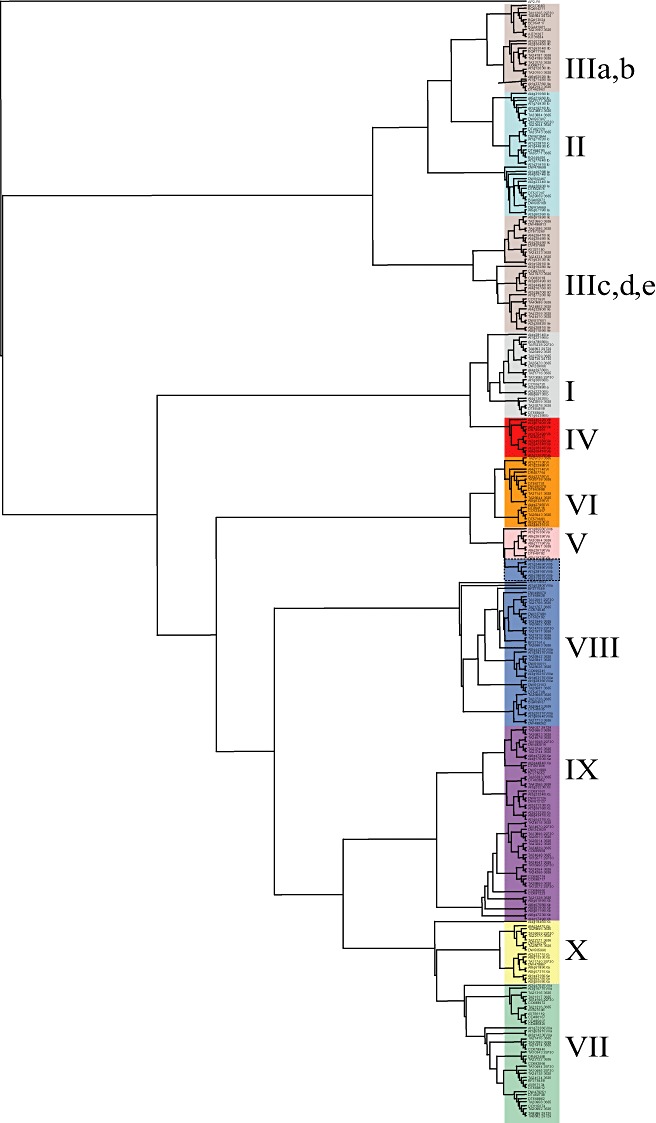
Neighbour‐joining phylogenetic tree of the ethylene‐response factor (ERF) protein family of Arabidopsis and cotton. The tree was created by the bootstrap option of the ClustalX multiple alignment package and the neighbour‐joining method using the AP2 domain sequences, except members of group VI‐L and Xb‐L of Arabidopsis. The tree, rooted to APETALA2 protein At4g36920, contains 115 Arabidopsis AP2 domains and 200 cotton AP2 domains. The classification by Nakano et al. (2006) is indicated by coloured rectangles.
The distributions of ERF genes were compared in Arabidopsis, cotton and rice (Table 2). One of the characteristic features of the G. hirsutum ERF family is that the number of genes in group VIII is twofold larger than that in Arabidopsis and rice. The number of genes in group VII is fourfold larger than that in Arabidopsis. Another feature is the absence of candidates for groups VI‐L and Xb‐L (Table 2). These two groups are characterized in Arabidopsis by a very low homology in the C‐terminal regions of the AP2/ERF domains. As in Arabidopsis and in contrast with rice, no cotton sequences were assigned to groups XI, XII, XIII and XIV. We therefore conclude that the classification into the 10 major groups used for Arabidopsis is applicable to the cotton ERF family.
Table 2.
Comparison of group size between Gossypium, Arabidopsis and rice ethylene‐response factor (ERF) families. Classification by Nakano et al. (2006).
| Group | AtERF genes | OsERF genes | GhERF genes | GrERF genes | GaERF genes | Total cotton genes |
|---|---|---|---|---|---|---|
| I | 10 | 9 | 9 | 4 | 1 | 14 |
| II | 15 | 15 | 17 | 1 | 2 | 20 |
| III | 23 | 26 | 25 | 3 | 7 | 35 |
| IV | 9 | 6 | 2 | 0 | 0 | 2 |
| V | 5 | 8 | 3 | 0 | 0 | 3 |
| VI | 8 | 6 | 11 | 1 | 0 | 12 |
| VI‐L | 4 | 3 | 0 | 0 | 0 | 0 |
| VII | 5 | 15 | 22 | 9 | 2 | 33 |
| VIII | 15 | 13 | 26 | 4 | 3 | 33 |
| IX | 17 | 18 | 26 | 11 | 2 | 39 |
| X | 8 | 13 | 7 | 2 | 0 | 9 |
| Xb‐L | 3 | 0 | 0 | 0 | 0 | 0 |
| XI | 0 | 4 | 0 | 0 | 0 | 0 |
| XII | 0 | 1 | 0 | 0 | 0 | 0 |
| XIII | 0 | 1 | 0 | 0 | 0 | 0 |
| XIV | 0 | 1 | 0 | 0 | 0 | 0 |
| Total | 122 | 139 | 148 | 35 | 17 | 200 |
Identification of group‐specific amino acids in the AP2/ERF domain
Detailed analysis of alignments of the Arabidopsis and cotton AP2 domain sequences allowed the identification of group‐specific amino acids, i.e. conserved in all sequences belonging to the same group and distinct in other groups. These data are summarized in Fig. 2B. The amino acid positions of the AP2 domain are numbered according to the three‐dimensional structure of AtERF1 (Allen et al., 1998). For example, group VIII was characterized by both P153 and K168 and at least one of the two residues R156 and Y157. By contrast, group IX was characterized by the presence of one supplementary residue in turn 2. Only five of the 315 sequences appeared as false negatives, no sequence appeared as a false positive and 10 sequences were positive for two groups including the right one (Table 3).
Figure 2.
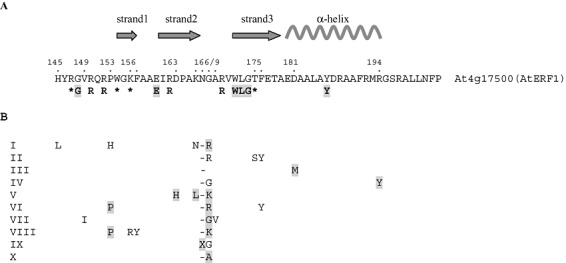
(A) Location of amino acids involved in DNA binding in AP2/ethylene‐response factor (ERF) domains. The sequence of AtERF1, used to resolve the AP2 structure, is shown as a reference number from 145 to 203. The arrangement of the secondary structural elements and the numbering of the Protein Data Bank (PDB) structure are shown. Strictly conserved residues are highlighted in grey; variable positions involved in DNA binding are indicated by asterisks. (B) Location of group‐specific amino acids in AP2/ERF domains. For each ERF group, specific amino acids are shown with the strictly conserved residues highlighted in grey.
Table 3.
Validation of the group‐specific residues on the collections of ethylene‐response factor (ERF) sequences. Bold residues are strictly conserved. The ratios represent, for each group, the number of sequences containing the group‐specific residues out of the total number of sequences. The error rates are indicated.
| Group | Specific residues | Arabidopsis | Gossypium | Oryza |
|---|---|---|---|---|
| I | L145 H153 N166 R168 | 10/10 | 14/14 | 8/8 |
| II | R168 S175 Y176 | 15/15 | 20/20 | 12/12 |
| III | M181 | 23/23 | 35/35 | 17/24 |
| IV | G168 Y194 | 9/9 | 2/2 | 5/6 |
| V | H163, L166 K168 | 4/6 | 2/3 | 5/8 |
| VI | P153 R168 Y176 | 7/8 | 12/12 | 2/6 |
| VII | I149 G168 V169 | 5/5 | 33/33 | 12/15 |
| VIII | P153 R156 Y157 K168 | 15/15 | 31/33 | 12/13 |
| IX | +X167 | 17/17 | 39/39 | 17/17 |
| X | A168 | 8/8 | 9/9 | 12/12 |
| Total | 113/115 | 198/200 | 103/121 | |
| False negative | 1.7% | 1.5% | 15% | |
| False positive | 0% | 0% | 5.8% | |
| Double assignment | 3.5% | 3% | 6.6% |
Among the 14 positions identified as markers of groups, only two positions, 156 and 175, are involved in DNA binding (Fig. 2A). Position 156, occupied by K in AtERF1, binds the A4 phosphate with ionic interaction. Position 175, occupied by T, forms hydrogen bonds with the G5 phosphate and A4 sugar of DNA. The group‐specific residues were tested on the corpus of rice sequences (Table 3). The presence of the group‐specific residues was validated for 103 of the 121 sequences.
The group‐specific residues were localized on the crystal structure of AtERF1 (Fig. 3). They were observed to cluster mostly at the vicinity of the loops. Most were oriented at the surface of the AP2 domain and would probably not influence the global structure of the AP2 domain. Only three group‐specific residues were buried in the core; however, the similarity of the biochemical properties of the new residues (I149V, F157Y, F176Y) would probably not modify the global AP2 domain structure. Turn 2 and its extremities were the most variable area, with a wide range of substitutions at five of the seven positions and with some indels.
Figure 3.
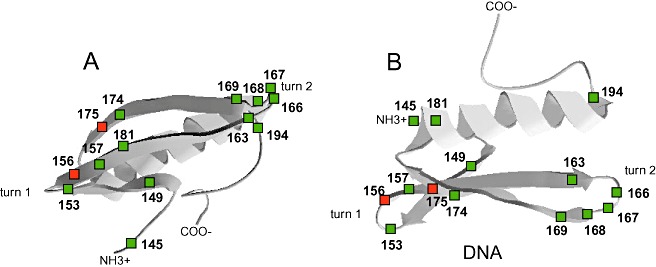
Three‐dimensional location and diversity of group‐specific residues of AP2/ethylene‐response factor (ERF) domain. The residues are labelled with their position numbers on the structure of the core of AtERF1 (PDB2GCC). The group‐specific residues are indicated by green squares. Red squares indicate group‐specific residues involved in DNA binding of the GCC‐box. N‐ and C‐termini are labelled NH3+ and COO−, respectively. The DNA‐binding interface is viewed face on (A), and looking along the double helix axis (B). The figure was drawn using PDBViewer.
The position of the conserved residues for a group was similar for Arabidopsis, cotton and rice, suggesting that this group‐specific AP2 domain structure preceded the divergence of monocots and dicots.
ERF and JA‐responsive transcription factors belonging to group IX
Previous reports have indicated that several ERF transcription factors are JA responsive. In Table 4, we list the entire group of previously published early JA‐responsive ERF transcription factors in Arabidopsis. McGrath et al. (2005) performed screening by quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR) of the 1534 members of the transcription factors of Arabidopsis to identify JA‐regulated signalling proteins. A total of 14 AtERF genes showed up‐regulation 6 h following JA treatment. Recently, Pauwels et al. (2008) have analysed transcript profiling of early JA responses in Arabidopsis cell suspension culture using microarray experiments. Seven ERF genes were up‐ or down‐regulated by elicitation with 50 µm methyl jasmonate (MeJA). Three of the JA‐responsive ERF genes (AtERF1, AtERF2 and AtERF13) have been identified previously by McGrath et al. (2005). Interestingly, 40% of the identified JA‐regulated ERF genes are located within group IX in Arabidopsis. This group includes ORA59, ERF1, AtERF1 and AtERF2, which have been shown to act as positive regulators of defence‐related transcription (Brown et al., 2003; Préet al., 2008; Solano et al., 1998). Furthermore, three more genes within this group, AtERF1, AtERF13 and AtERF14, have been reported to be responsive to virulent and avirulent Pseudomonas syringae pv. tomato (Oñate‐Sánchez and Singh, 2002).
Table 4.
Jasmonate (JA)‐responsive AP2/ethylene‐response factor (ERF) domain transcription factors in Arabidopsis.
| Generic name | Gene name | Group name | JA response | Method | References | Function |
|---|---|---|---|---|---|---|
| At3g50260 | IIa | Up‐regulated | Q‐PCR | * | ||
| At1g74930 | IIb | Up‐regulated | Microarray | † | ||
| At1g33760 | Tiny‐like | IIIa | Up‐regulated | Q‐PCR | * | |
| At1g03800 | AtERF10 | VIIIa | Up‐regulated | Q‐PCR | * | |
| At1g28360 | VIIIa | Down‐regulated | Microarray | † | ||
| At1g28370 | AtERF11 | VIIIa | Up‐regulated | Q‐PCR | * | |
| At3g15210 | AtERF4 | VIIIa | Up‐regulated | Q‐PCR | * ‡ | |
| At2g44840 | AtERF13 | IXa | Up‐regulated | Q‐PCR/microarray | * † | |
| At4g17500 | AtERF1 | IXa | Up‐regulated | Q‐PCR/microarray | * † | |
| At5g47220 | AtERF2 | IXa | Up‐regulated | Q‐PCR/microarray | * † | |
| At1g04370 | AtERF14 | IXc | Up‐regulated | Q‐PCR | * | Disease resistance |
| At1g06160 | ORA59 | IXc | Up‐regulated | Q‐PCR | * § | Disease resistance |
| At3g23230 | TDR1 | IXc | Up‐regulated | Q‐PCR | * | |
| At3g23240 | ERF1 | IXc | Up‐regulated | Q‐PCR | * | Disease resistance |
| At1g43160 | RAP2.6 | Xa | Up‐regulated | Microarray | † | |
| At5g50080 | Xa | Up‐regulated | Q‐PCR | * | ||
| At5g67010 | XbL | Up‐regulated | Q‐PCR | * |
Q‐PCR, quantitative polymerase chain reaction.
A tree was generated using 17 Arabidopsis, 18 rice and 39 cotton members of group IX ERF proteins (Fig. 4). As reported by Nakano et al. (2006), group IX is subdivided into three subgroups, namely a, b and c. However, the tree generated in this study shows a fourth group (IXd) composed of At3g23230, Os01g54890, TA43265_3635, TA35383_3635 and DT463852. The reliability of the major group clustering was supported by the presence of common motifs outside of the AP2/ERF domain. A Multiple Em for Motif Elicitation (MEME) search using amino acid sequences of Arabidopsis, rice and cotton ERFs from group IX confirmed the presence of these conserved motifs. As illustrated in Fig. 4, nine cotton members of subgroup IXa possess either the CMIX‐2 or CMIX‐3 motifs, or both, as in rice and Arabidopsis (this study; Nakano et al., 2006). CMIX‐2 and CMIX‐3 motifs are acidic regions that could act as transcriptional activation domains (Fujimoto et al., 2000).
Figure 4.
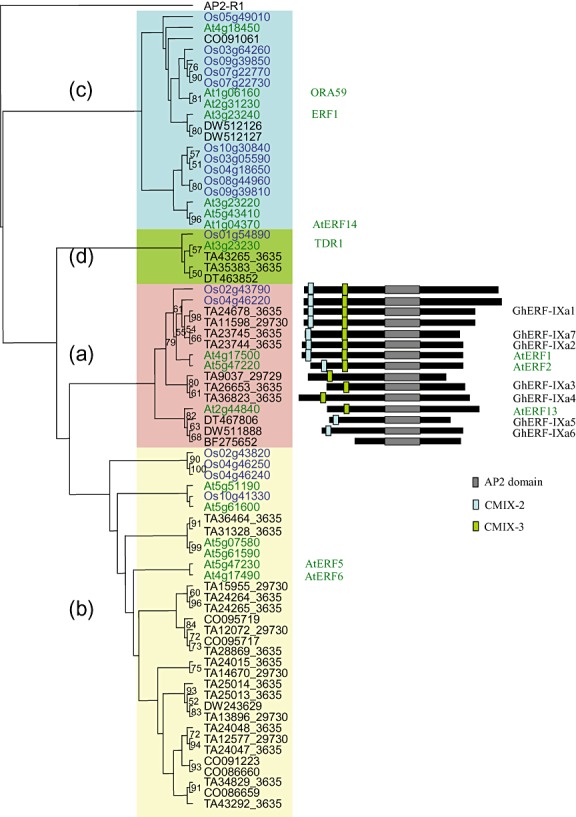
Domain localization and phylogenetic tree showing the predicted relationships between group IX ethylene‐response factor (ERF) genes from rice, Arabidopsis and cotton. The tree was created by the bootstrap option of the ClustalX multiple alignment package and the neighbour‐joining method using the AP2 domain sequences. The tree, rooted to APETALA2 protein At4g36920, contains 17 Arabidopsis AP2 domains (green), 18 rice AP2 domains (blue) and 39 cotton AP2 domains (black). Bootstrap values from 100 replicates were used to assess the robustness of the tree. Bootstrap values over 50 are shown. The classification by Nakano et al. (2006) is indicated by coloured rectangles. On the right, comparisons of the diagrams with conserved domains of cotton group IXa proteins with the homologous Arabidopsis and rice proteins are shown. Coloured boxes represent the AP2/ERF domain and the two conserved motifs CMIX‐2 and CMIX‐3.
Identification of JA‐ and ET‐responsive members of the cotton AP2 domain transcription factor gene family
ERF genes of group IX have been shown to play crucial roles in biotic stress responses and have been linked to JA and ET signalling pathways. Accordingly, we first investigated whether JA induces genes of subgroup GhERF‐IXa (Fig. 5) using quantitative RT‐PCR. Gene expression in seedlings treated with SA and the ET‐releasing agent ethephon was also analysed to determine the specificity of the gene expression response. The gene GhLOX1, which is responsive to JA and SA, was used as a control to verify that the hormone treatments were effective (Marmey et al., 2007).
Figure 5.
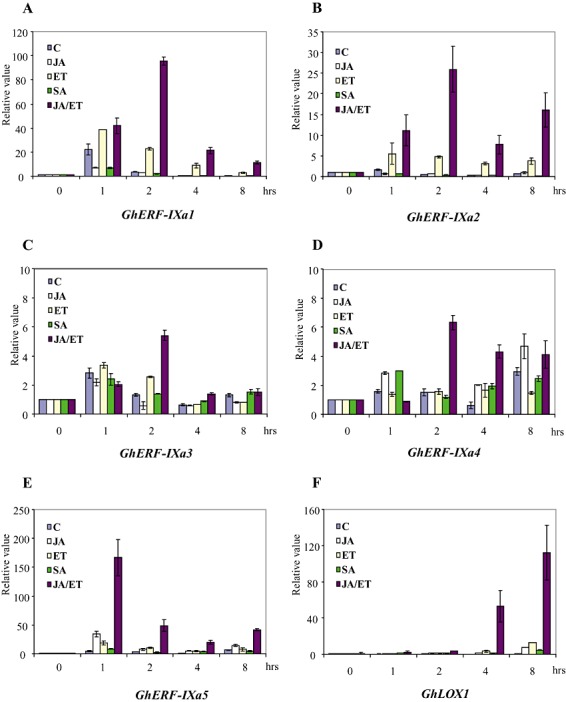
Relative expression profiles of five GhERF‐IXa genes in response to jasmonate (JA), ethephon, salicylic acid (SA) and a combined treatment with JA and ethephon. Real‐time quantitative polymerase chain reaction (PCR) analysis of GhERF‐IXa gene expression was performed in 10‐day‐old cotton plants treated with 50 µm JA, 1 mm ethephon (ethylene, ET), 50 µm SA, both JA/ET or solvents (C) for the indicated times. (A) GhERF‐IXa1 gene; (B) GhERF‐IXa2 gene; (C) GhERF‐IXa3 gene; (D) GhERF‐IXa4 gene; (E) GhERF‐IXa5 gene; and (F) GhLOX1 gene. The data presented correspond to one representative of three experiments with similar results (12 cotyledons per treatment). The data are mean values and standard errors (bar) of three independent real‐time quantitative PCR experiments. The transcript level is represented as the ratio of the Ct value of the studied gene calibrated to the T0 time point and normalized to the Ct value of the GhACT2 gene.
One ERF gene GhERF‐IXa5 was found to be responsive to JA (Fig. 5), showing a rapid transient expression (Fig. 5E). The expression of this gene was induced by JA with a peak at 1 h of treatment (34‐fold), and returned to a low level within 4 h (fivefold) of exposure to JA. A second peak of induction (15‐fold) was observed after 8 h of treatment.
Three genes were found to be responsive to ET, GhERF‐IXa1, GhERF‐IXa2 and GhERF‐IXa5, characterized by a more prolonged expression after 1 h up to the longest time point of 8 h (Fig. 5A, B, E). One hour after all treatments, strong transient induction of GhERF‐IXa1 was observed (22‐fold), presumably caused by hypo‐osmotic or wounding stimuli following infiltration. Although GhERF‐IXa1 expression was shut off 2 h after solvent treatment, its expression continued in ET‐treated leaves up to 8 h after treatment, confirming that GhERF‐IXa1 expression is induced by ET treatment. Interestingly, mock induction of GhERF‐IXa1 was repressed by SA or JA treatment, but not by ET. Except for the repressive effect of SA on GhERF‐IXa1 gene expression, SA alone did not have rapid or strong effects on the other genes tested.
Weak induction (two‐ to threefold) of the expression of GhERF‐IXa3 and GhERF‐IXa4 was detected during the 8 h of JA, ET, SA or control treatment (Fig. 5C, D). Expression analysis of the GhERF‐IXa6 and GhERF‐IXa7 genes was not carried out, as no PCR products were amplified using the three sets of primer pairs attempted. GhERF‐IXa6 is an EST singleton and may be a less reliable representation of the actual sequence. The mRNA level of GhLOX1 was induced after 8 h of JA treatment (Fig. 5F). GhLOX1 was induced after 4 h in response to ethephon, up to 8 h, and after 8 h following SA infiltration (Fig. 5F).
Cotton seedlings were also treated with a combination of JA and ET in order to test the effect on GhERF‐IXa gene expression. Quantitative RT‐PCR analysis showed that the combined JA/ET treatment led to a super‐induction of GhERF‐IXa1, GhERF‐IXa2 and GhERF‐IXa5 gene expression (Fig. 5A, B, E). The combined treatment triggered this gene expression at the same early time point as JA treatment, but the maximum expression level was higher and the expression was more prolonged. The GhERF‐IXa3 and GhERF‐IXa4 genes were rapidly induced by combined JA/ET treatment with a peak at 2 h of treatment, and returned to control levels within 4 and 8 h of exposure to JA and ET, respectively (Fig. 5C, D). We therefore conclude that JA enhances the stress‐induced expression of specific members of group IXa ERF transcription factors and not the group as a whole, by contrast with the combined treatment with ET.
Identification of cotton ERF genes induced by X. campestris pv. malvacearum infection
As members of subgroup GhERF‐IXa were induced by ET or JA, it was tested whether GhERF‐IXa gene expression could also be induced by X. campestris pv. malvacearum attack. During the cotton HR to X. campestris pv. malvacearum race 18, both JA and SA induced cotton GhLOX1 gene expression and activity (Marmey et al., 2007). During the incompatible interaction, quantitative RT‐PCR analysis (Fig. 6D) revealed that GhERF‐IXa4 mRNA did not accumulate at any of the time points analysed relative to the control treatment. In contrast, mRNA levels of GhLOX1 started to accumulate 8 h after inoculation, and continued to increase at the 10 h time point (Fig. 6F). GhERF‐IXa1 was induced by the avirulent race 8 h after infection, and by the virulent pathogen (X. campestris pv. malvacearum race 20) and water earlier (2 h, Fig. 6A). Three of the ERF genes were specifically induced by avirulent strain X. campestris pv. malvacearum race 18 infection. GhERF‐IXa3 showed a rapid response at 1 h and returned to basal levels within 4 h (Fig. 6C). GhERF‐IXa2 and GhERF‐IXa5 showed quite similar induction patterns, with a large increase in mRNA levels within 8 h after infection by X. campestris pv. malvacearum race 18 (Fig. 6B, E).
Figure 6.
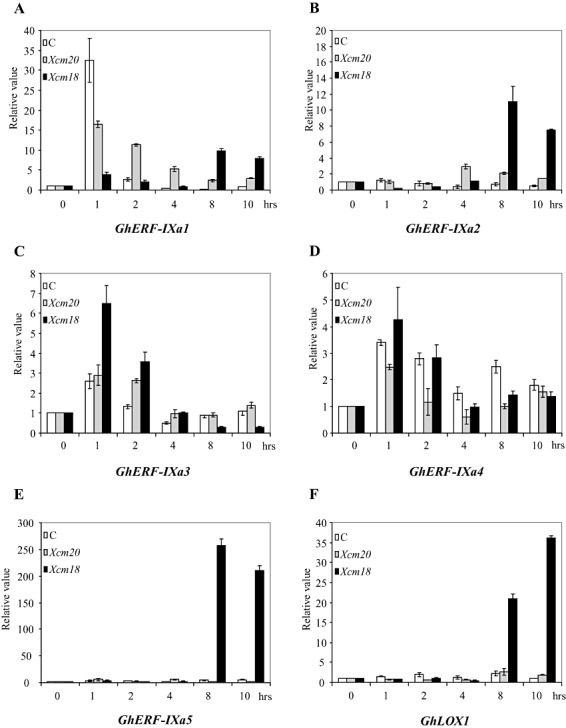
Analysis of GhERF‐IXa gene expression in water‐, Xanthomonas campestris pv. malvacearum race 20‐ and X. campestris pv. malvacearum race 18‐infiltrated Reba B50 cotyledons. Real‐time quantitative polymerase chain reaction (PCR) analysis of GhERF‐IXa gene expression was investigated during the incompatible (X. campestris pv. malvacearum race 18) and compatible (X. campestris pv. malvacearum race 20) interaction and in water‐infiltrated cotyledons (C). Cotyledons were harvested at the indicated times (0–10 h). The transcript level is represented as the ratio of the Ct value of the studied gene calibrated to the T0 time point and normalized to the Ct value of the GhACT2 gene. The data presented correspond to one representative of three experiments with similar results (10 cotyledons per treatment).
DISCUSSION
JA‐responsive gene expression is mediated via transcription factors, such as ORCA2, ORCA3 and ERF1 (van der Fits and Memelink, 2000; Lorenzo et al., 2003), which belong to the class of AP2 domain transcription factors. The expression of the ORCA and ERF1 genes themselves is JA responsive. Based on these observations, we hypothesized that JA‐responsive gene expression in cotton could also be mediated by members of the AP2 domain transcription factor family, and that the corresponding genes would also be expressed in a JA‐responsive manner. The objectives of this study were (i) to perform an annotation of the ERF family genes in cotton, (ii) to provide a new tool (i.e. identification of group‐specific amino acids in the AP2 domain) to predict the group‐based classification in plants, and (iii)to identify JA‐, ET‐ and X. campestris pv. malvacearum‐responsive members of the group IXa genes in cotton.
The ERF family of transcription factors experienced significant expansion during the evolution of plants, revealing that many groups and subgroups have evolved, resulting in a high level of functional divergence. This family comprises 122 and 139 genes in Arabidopsis and rice, divided into 12 and 15 groups, respectively (Nakano et al., 2006). Based on the fact that 11 of these groups are present in both Arabidopsis and rice, the authors concluded that functional diversification within the ERF family predated the monocot/dicot divergence. The actual number of ERF genes in our cotton dataset is 148 unigenes in G. hirsutum. Gossypium hirsutum is an allotetraploid (2n= 4x= 52) resulting from interspecific hybridization between an A2 genome‐like ancestral African species and a D genome‐like American species (Chen et al., 2007). As a consequence of its polyploid nature and the fact that we excluded the ERF sequences that contain partial AP2 domains, it is presently difficult to predict the total number of G. hirsutum ERF genes, but this number is likely to be higher. Furthermore, 34 ERF genes from the D genome species G. raimondii and 18 ERF genes from the A2 genome species G. arboreum have been annotated. Gossypium hirsutum was found to have more genes than the combined total of the two progenitor diploid species. This difference could be associated with the number of ESTs available for G. hirsutum, which is higher than those for the two diploids. These data allowed the first analysis of ERF genes from three species of the Malvales using a large dataset, and may be useful to analyse specific gene expression among paralogous, orthologous and homologous sequences in diploid and allotetraploid species of cotton.
In our study, we observed that the 10 major groups from Arabidopsis also occurred in the genus Gossypium. These data are in accordance with the fact that the Malvales (including G. hirsutum) are the closest relatives of Arabidopsis outside of the Brassicales (Bowers et al., 2003). We identified conserved group‐specific residues which allowed a rapid and easy classification of plant ERF transcription factors based on inter‐ and intragroup comparisons of AP2 domains. The role of these amino acids is still unknown. Most were not directly involved in the interaction with the GCC‐box. Part of such residues would participate in intramolecular interactions to stabilize the ERF protein. Several identified conserved motifs correlate with the major groups defined on the basis of the relationships among AP2 domains (Nakano et al., 2006). Such a correlation suggests the existence of coordinated amino acids inside and outside the AP2 domain. The group‐specific residues of the AP2 domain defined in this study are good candidates for future investigation to further understand the structure–function relationships of ERF transcription factors.
Molecular phylogenetics of environmental responses has been suggested to provide insights into the mechanism of the defence response (Gutterson and Reuber, 2004). Taking advantage of the fact that many ERF genes are inducible, our study focused on a transcriptional response to defence‐related stimuli. As a first step in a molecular phylogenetic analysis of plant defence regulation by ERF transcription factors, we mapped published data concerning JA responses of these genes onto the Arabidopsis ERF phylogeny (Table 3). This set of observations is surely incomplete, as (i) JA‐responsive AP2‐like genes were expressed only at earlier or later time points, or expressed only in specific tissues present at low abundance, and (ii) AP2 domain transcription factor genes that are responsive only to a combination of JA and another signalling molecule would not be characterized. Despite these limitations, patterns were observed, in particular within group IX, which contains 40% of the characterized JA‐responsive ERF genes in Arabidopsis. Overexpression of certain ERF transcription factors from group IX results in resistance to multiple pathogens. For example, constitutive overexpression of AtERF2 of subgroup IXa has been shown to induce PDF1.2 and ChiB gene expression (Brown et al., 2003; Préet al., 2008). Similarly, overexpression of the Arabidopsis AtERF1 gene, a close homologue of AtERF2, confers resistance to Botrytis cinerea, Sclerotinia sclerotiorum and Erysiphe orontii in Arabidopsis (Gutterson and Reuber, 2004). Recently, Rushton et al. (2008) have identified 13 tobacco genes of the group IX ERF genes that show increased mRNA levels following JA treatment. Together, these observations prompted us to focus our study on the cotton members of group IX.
In cotton, we demonstrated that JA induces the expression of the GhERF‐IXa5 gene. By contrast, ET, but not JA alone, induces GhERF‐IXa1 and GhERF‐IXa2 expression. The expression of GHERF‐IXa5, as well as GhERF‐IXa1 and GhERF‐IXa2 genes, was super‐induced by combined treatment with JA and ET. Super‐induction of gene expression by JA and ET has also been shown for some defence‐related genes, such as CHIB, PDF1.2 and ERF1 (Penninckx et al., 1998). During the time course experiments on cotton, we observed that the expression of each gene of subgroup GhERF‐IXa was altered more than fivefold by either JA, ET or combined treatment (JA/ET) for at least one of the time points analysed. Together, these results suggest that crosstalk between the JA and ET signalling pathways may occur at the level of multiple ERF transcription factors.
Resistance of cotton cv. Reba B50 to the bacterial pathogen X. campestris pv. malvacearum race 18 is characterized by an HR, during which a strong increase in the production of JA and SA signalling molecules was observed (Delannoy et al., 2005). To our knowledge, ET production has not been reported as yet during X. campestris pv. malvacearum and cotton interactions. To relate our findings on responses to defence hormones to responses to pathogens, we investigated GhERF‐IXa gene expression in response to X. campestris pv. malvacearum race 18. In cotyledon cells, induction of the JA‐ and ET‐responsive GhERF‐IXa transcription factors was correlated with resistance (5, 6), with variation in the kinetics of gene expression, as the GhERF‐IXa3 gene showed a faster induction than GhERF‐IXa2 and GhERF‐IXa5. A transcriptome study by Patil et al. (2005) on the incompatible cotton–X. campestris pv. malvacearum interaction showed that 121 transcripts in leaves over a period of 8–60 h after inoculation were up‐regulated. In particular, the EST CF93196 with high similarity to GhERF‐IXa2 was differentially expressed in X. campestris pv. malvacearum‐inoculated tissues compared with non‐inoculated control tissues. Only a few studies have reported the role of ERF transcription factors in triggering or limiting HR. In Arabidopsis, the AtEBP gene from group VII was identified as a suppressor of Bax‐induced cell death by functional screening in yeast (Pan et al., 2001). Overexpression of AtEBP confers resistance to Bax‐induced cell death and H2O2‐induced cell death in plant cells (Ogawa et al., 2005). In contrast, in N. benthamiana, NbCD1 from group VIII positively regulates cell death and contributes to non‐host resistance (Nasir et al., 2005).
Taken together, these results increase our knowledge about the involvement of the ERF transcription factors in plant resistance and indicate that one given member of the ERF gene family is involved in JA or ET perception. Further experiments are needed to understand the role of the cotton ERF transcription factor in the X. campestris pv. malvacearum‐induced HR.
EXPERIMENTAL PROCEDURES
Database search
Database searches were performed to collect all members of the cotton ERF family using the 122 conserved AP2 domains of A. thaliana. Arabidopsis and rice AP2 domain protein sequences were retrieved from the Arabidopsis Transcription Database (ArabTFDB) (version 1.0, http://arabtfdb.bio.uni‐potsdam.de/v1.0) and the rice TIGR website (http://www.tigr.org/tdb/e2k1/osa1/LocusNameSearch.shtml), respectively. We surveyed the database of coding sequences from genes in the current version (July 2007) of TIGR cotton (http://plantta.tigr.org/search.shtml) using a TBlastN search. These libraries were derived from allopolyploid cotton (G. hirsutum; AT and DT genomes; 70667 unique sequences) as well as its two diploid progenitors: G. arboreum (A2 genome; 28642 unique sequences) and G. raimondii (D genome, 26884 unique sequences).
Many sequences initially collected had incomplete AP2 domains or appeared to contain incorrect open reading frames (ORFs). These sequences were excluded from further analysis. As some sequence differences in ESTs are a result of sequencing errors or DNA polymorphism in cotton cultivars, nucleotide sequences with 98% or greater identities over their length were considered as the same gene (many homeologous genes in G. hirsutum are also likely to fall into this category), and a representative was chosen.
Tree building
The phylogenetic tree was constructed with AP2‐domains of around 60 amino acids derived from AP2‐domain proteins with a single DNA‐binding domain using ClustalX program with the default settings, including 100 bootstraps. The tree was displayed using Treeview (Page, 1996; http://taxonomy.zoology.gla.ac.uk/rod/treeview.html), with APETALA2 AP domain repeat 1 defined as an outgroup.
Identification and three‐dimensional localization of group‐specific amino acids
The phylogenetic tree allowed assignments of cotton sequences to groups. Comparison of the sequence alignments with the group assignments allowed the identification of group‐specific amino acids, i.e. conservation in all sequences belonging to the same group and distinct in other groups. The presence of the group‐specific amino acids was searched in the rice sequences and the predicted assignments were compared with those published.
The group‐specific residues were located on the three‐dimensional structure of the AP2 domain of AtERF1 GBD (PDB2GCC) corresponding to the sequence At4g17500 (Allen et al., 1998).
Determination of conserved motifs of group IX outside of the AP2/ERF domain
The identification of conserved motifs outside of the highly conserved AP2 domain was performed on 74 protein sequences of group IX with multiple sequence alignments and MEME version 3.5.3 (http://meme.sdsc.edu) (Bailey and Elkan, 1994). Options for MEME were adjusted to find 3–25 amino acid motifs. Input sequence data were modified to exclude the conserved AP2 domain for each.
Bacterial strains
Xanthomonas campestris pv. malvacearum races 18 and 20 were maintained at 30 °C on YPG agar (0.5% w/v yeast extract, 0.5% w/v bacteriological peptone, 0.5% w/v glucose as a carbon source, solidified with 1.5% w/v agar; Difco, Detroit, MI, USA) in distilled water. Bacteria for inoculation were grown in 150 mL YPG medium at 150 r.p.m., 30 °C. After 18 h of growth, cultures were washed once with sterile water by centrifugation at 10 000 g for 10 min to remove nutrients and exopolysaccharides, and then the bacterial pellet was resuspended in sterile water and adjusted to 108 colony‐forming units (cfu)/mL.
Plant growth, inoculation and treatments
The cultivar Reba B‐50 (Allen × stoneville 2B) from G. hirsutum, similar to the 101‐102B line, contains the B2B3 genes for resistance to race 18 of X. campestris pv. malvacearum, but not to race 20. Plants were grown in a glasshouse under natural light and a 28/24 °C light/dark cycle with a relative humidity averaging 80%. These conditions were shown to favour bacterial development. Bacterial suspension (108 cfu/mL), or sterile water as a control, was injected into intercellular areas of 10‐day‐old cotyledons using a needle‐less syringe. Two interactions were tested: the incompatible Reba B50/X. campestris pv. malvacearum race 18 and the compatible Reba B50/X. campestris pv. malvacearum race 20. Plantlets were treated for different time periods with 50 µm JA (Sigma‐Aldrich, St. Louis, MO, USA), 50 µm of SA (Sigma‐Aldrich), 1 mm of the ET‐releasing compound ethephon (Sigma‐Aldrich), or a combination of JA and ethephon. As controls, seedlings were treated with the combination of solvents dimethyl sulfoxide (DMSO; 0.1%) and sodium phosphate, pH 7 (0.5 mm).
Preparation of total RNA and cDNA synthesis
Total RNA was isolated from the cotyledons using Plant RNA Reagent from Qiagen, France, following the manufacturer's directions. RNA was treated with the RNAse‐Free DNAse (Qiagen) and purified using the RNeasy Plant Mini‐kit column (Qiagen), following the manufacturer's instructions. First‐strand cDNA was synthesized using an oligo(dT25) primer from purified total RNA using Omniscript Reverse Transcriptase (Qiagen), following the manufacturer's instructions.
Quantitative RT‐PCR analysis
Transcript levels for the GhERF‐IXa group were quantified by real‐time PCR with MX3005P (Stratagene, France) using MESA Green qPCR Master Mix Plus (Eurogentec, France), according to the manufacturer's instructions. The gene‐specific primer pairs used are listed in Table S2 (see Supporting Information). All reactions contained 12.5 µL of 2 × MESA Green qPCR Master Mix Plus (Eurogentec), 2.0 ng cDNA and 300 nm of each gene‐specific primer in a final volume of 25 µL. Thermal cycling was as follows: 50 °C for 2 min; 95 °C for 10 min; 45 cycles of 95 °C for 15 s, 58 °C for 20 s, 72 °C for 40 s. Relative expression levels of reporter and target genes were determined based on the 2−ΔΔCt method (Livak and Schmittgen, 2001) using cotton Actin2 (AY305724) as internal control (Li et al., 2005).
Supporting information
Table S1 Ethylene‐response factor (ERF) family genes in cotton.
Table S2 Gene‐specific primer pairs used in real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) experiments.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank J. Aribi for technical assistance in the glasshouse work. For critical reading of the manuscript, we would like to thank Johan Memelink and Raoul Ranjeva.
REFERENCES
- Allen, M.D. , Yamasaki, K. , Ohme‐Takagi, M. , Tateno, M. and Suzuki, M. (1998) A novel mode of DNA recognition by a β‐sheet revealed by the solution structure of the GCC‐box binding domain in complex with DNA. EMBO J. 17, 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. and Elkan, C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers In: Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology (Altman R., Brutlog D., Karp P., Lathrop R. and Searls D., eds), pp. 28–36. Menlo Park, CA: American Association for Artificial Intelligence Press. [PubMed] [Google Scholar]
- Berrocal‐Lobo, M. and Molina, A. (2004) Ethylene Response Factor1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum . Mol. Plant–Microbe Interact. 17, 763–770. [DOI] [PubMed] [Google Scholar]
- Bowers, J.E. , Chapman, B.A. , Rong, J. and Paterson, A.H. (2003) Unraveling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature, 422, 433–438. [DOI] [PubMed] [Google Scholar]
- Brown, R.L. , Kazan, K. , McGrath, K.C. , Maclean, D.J. and Manners, J.M. (2003) A role for the GCC‐box in jasmonate‐mediated activation of the PDF1.2 gene of Arabidopsis . Plant Physiol. 132, 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Provart, N.J. , Glazebrook, J. , Katagiri, F. , Chang, H.S. , Eulgem, T. , Mauch, F. , Luan, S. , Zou, G. , Whitham, S.A. , Budworth, P.R. , Tao, Y. , Xie, Z. , Chen, X. , Lam, S. , Kreps, J.A. , Harper, J.F. , Si‐Ammour, A. , Mauch‐Mani, B. , Heinlein, M. , Kabayashi, K. , Hohn, T. , Dangl, J.L. , Wang, X. and Zhu, T. (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell, 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J. , Scheffler, B.E. , Dennis, E. , Triplett, B.A. , Zhang, T. , Guo, W. , Chen, X. , Stelly, D.M. , Rabinowicz, P.D. , Town, C.D. , Arioli, T. , Brubaker, C. , Cantrell, R.G. , Lacape, J.M. , Ulloa, M. , Chee, P. , Gingle, A.R. , Haigler, C.H. , Percy, R. , Saha, S. , Wilkins, T. , Wright, R.J. , Van Deynze, A. , Zhu, Y. , Yu, S. , Abdurakhmonov, I. , Katageri, I. , Kumar, P.A. , Rahman, M. , Zafar, Y. , Yu, J.Z. , Kohel, R.J. , Wendel, J.F. and Paterson, A.H. (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 145, 1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daï, G.H. , Nicole, M. , Martinez, C. , Bresson, E. , Daniel, J.F. , Andary, C. and Geiger, J.P. (1996) Flavonoids accumulate in cell walls, middle lamellae and callose‐rich papillae during an incompatible interaction between Xanthomonas campestris pv. malvacearum (Race 18) and cotton. Physiol. Mol. Plant. Pathol. 49, 285–306. [Google Scholar]
- De Feyter, R. , Yang, Y. and Gabriel, D.W. (1993) Gene‐for‐genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant–Microbe Interact. 6, 225–237. [DOI] [PubMed] [Google Scholar]
- Delannoy, E. , Jalloul, A. , Assigbetsé, K. , Marmey, P. , Geiger, J.P. , Lherminier, J. , Daniel, J.F. , Martinez, C. and Nicole, M. (2003) Activity of class III peroxidases during defense of cotton to bacterial blight. Mol. Plant–Microbe Interact. 16, 1030–1038. [DOI] [PubMed] [Google Scholar]
- Delannoy, E. , Lyon, B. , Marmey, P. , Jalloul, A. , Montillet, J.L. , Daniel, J.F. , Essenberg, M. and Nicole, M. (2005) Resistance of cotton to Xanthomonas campestris pv. malvacearum . Annu. Rev. Phytopathol. 43, 63–82. [DOI] [PubMed] [Google Scholar]
- Van Der Fits, L. and Memelink, J. (2000) ORCA3, a jasmonate‐responsive transcriptional regulator of plant primary and secondary metabolism. Science, 289, 295–297. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y. , Ohta, M. , Usui, A. , Shinshi, H. and Ohme‐Takagi, M. (2000) Arabidopsis ethylene‐responsive element binding factors act as transcriptional activators or repressors of GCC box‐mediated gene expression. Plant Cell, 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, M. , Fujita, Y. , Noutoshi, Y. , Takahashi, F. , Narusaka, Y. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. [DOI] [PubMed] [Google Scholar]
- Gutterson, N. and Reuber, T.L. (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7, 465–471. [DOI] [PubMed] [Google Scholar]
- Jalloul, A. , Montillet, J.L. , Assigbetsé, K. , Agnel, J.P. , Delannoy, E. , Daniel, J.F. , Marmey, P. , Geiger, J.P. and Nicole, M. (2002) Lipid peroxidation in cotton–Xanthomonas interactions. Role of lipoxygenases during the hypersensitive reaction and leaf blight. Plant J. 32, 1–12. [DOI] [PubMed] [Google Scholar]
- Jin, L.G. and Liu, J.Y. (2008) Molecular cloning, expression profile and promoter analysis of a novel ethylene responsive transcription factor gene GhERF4 from cotton (Gossypium hirsutum). Plant Physiol. Biochem. 46, 46–53. [DOI] [PubMed] [Google Scholar]
- Li, X.B. , Fan, X.P. , Wang, X.L. , Cai, L. and Yang, W.C. (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell, 17, 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O. , Piqueras, R. , Sánchez‐Serrano, J.J. and Solano, R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell, 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmey, P. , Jalloul, A. , Alhamdia, M. , Assigbetse, K. , Cacas, J.L. , Voloudakis, A.E. , Champion, A. , Clerivet, A. , Montillet, J.L. and Nicole, M. (2007) The 9‐lipoxygenase GhLOX1 gene is associated with the hypersensitive reaction of cotton Gossypium hirsutum to Xanthomonas campestris pv. malvacearum . Plant Physiol. Biochem. 45, 596–606. [DOI] [PubMed] [Google Scholar]
- Martinez, M. , Baccou, J.C. , Bresson, E. , Baissac, Y. , Daniel, J. , Jalloul, A. , Montillet, J.L. , Geiger, J.P. , Assigbetse, K. and Nicole, M. (2000) Salicylic acid mediated by the oxidative burst is a key molecule in the local and systemic resistance of cotton challenged by an avirulent race of Xanthomonas campestris pv. malvacearum race 18. Plant Physiol. 122, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, K.C. , Dombrecht, B. , Manners, J.M. , Schenk, P.M. , Edgar, C.I. , Maclean, D.J. , Scheible, W.R. , Udvardi, M.K. and Kazan, K. (2005) Repressor‐ and activator‐type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome‐wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink, J. , Verpoorte, R. and Kijne, J.W. (2001) ORCAnization of jasmonate‐responsive gene expression in alkaloid metabolism. Trends. Plant Sci. 6, 212–219. [DOI] [PubMed] [Google Scholar]
- Menke, F.L.H. , Champion, A. , Kijne, J.W. and Memelink, J. (1999) A novel jasmonate‐ and elicitor‐responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate‐ and elicitor‐inducible AP2‐domain transcription factor, ORCA2. EMBO J. 18, 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, T. , Suzuki, K. , Fujimura, T. and Shinshi, H. (2006) Genome‐wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir, K.H. , Takahashi, Y. , Ito, A. , Saitoh, H. , Matsumura, H. , Kanzaki, H. , Shimizu, T. , Ito, M. , Fujisawa, S. , Sharma, P.C. , Ohme‐Takagi, M. , Kamoun, S. and Terauchi, R. (2005) High‐throughput in planta expression screening identifies a class II ethylene‐responsive element binding factor‐like protein that regulates plant cell death and non‐host resistance. Plant J. 43, 491–505. [DOI] [PubMed] [Google Scholar]
- Ogawa, T. , Pan, L. , Kawai‐Yamada, M. , Yu, L.H. , Yamamura, S. , Koyama, T. , Kitajima, S. , Ohme‐Takagi, M. , Sato, F. and Uchimiya, H. (2005) Functional analysis of Arabidopsis ethylene‐responsive element binding protein conferring resistance to Bax and abiotic stress‐induced plant cell death. Plant Physiol. 138, 1436–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate‐Sánchez, L. and Singh, K.B. (2002) Identification of Arabidopsis ethylene‐responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128, 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Pan, L. , Kawai, M. , Yu, L.H. , Kim, K.M. , Hirata, A. , Umeda, M. and Uchimiya, H. (2001) The Arabidopsis thaliana ethylene‐responsive element binding protein (AtEBP) can function as a dominant suppressor of Bax‐induced cell death of yeast. FEBS Lett. 508, 375–378. [DOI] [PubMed] [Google Scholar]
- Patil, M.A. , Pierce, M.L. , Phillips, A.L. , Venters, B.J. and Essenberg, M. (2005) Identification of genes up‐regulated in bacterial‐blight‐resistant upland cotton in response to inoculation with Xanthomonas campestris pv malvacearum . Physiol. Mol. Plant Pathol. 67, 319–335. [Google Scholar]
- Pauwels, L. , Morreel, K. , De Witte, E. , Lammertyn, F. , Van Montagu, M. , Boerjan, W. , Inzé, D. and Goossens, A. (2008) Mapping methyl jasmonate‐mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA, 105, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A. , Thomma, B.P.H.J. , Buchala, A. , Metraux, J.P. and Broekaert, W.F. (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis . Plant Cell, 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré, M. , Atallah, M. , Champion, A. , De Vos, M. , Pieterse, C.J. and Memelink, J. (2008) The AP2/ERF‐domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Z.X. , Huang, B. and Liu, J.Y. (2008) Molecular cloning and functional analysis of an ERF gene from cotton (Gossypium hirsutum). Biochim. Biophys. Acta 1779, 122–127. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J. , Bokowiec, M.T. , Han, S. , Zhang, H. , Brannock, J.F. , Chen, X. , Laudeman, T.W. and Timko, M.P. (2008) Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae . Plant Physiol. 147, 280–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, Y. , Liu, Q. , Dubouzet, J.G. , Abe, H. , Shinozaki, K. and Yamaguchi‐ Shinozaki, K. (2002) DNA‐binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration and cold‐inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. [DOI] [PubMed] [Google Scholar]
- Samuel Yang, S. , Cheung, F. , Lee, J.J. , Ha, M. , Wei, N.E. , Sze, S.H. , Stelly, D.M. , Thaxton, P. , Triplett, B. , Town, C.D. and Jeffrey Chen, Z. (2006) Accumulation of genome‐specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 47, 761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, R. , Stepanova, A. , Chao, Q. and Ecker, J.R. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE‐INSENSITIVE3 and ETHYLENE‐RESPONSE‐FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Tian, L. , Latoszek‐Green, M. , Brown, D. and Wu, K. (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58, 585–596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Ethylene‐response factor (ERF) family genes in cotton.
Table S2 Gene‐specific primer pairs used in real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) experiments.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


