SUMMARY
Understanding the factors driving pathogen emergence and re‐emergence is a major challenge, particularly in agriculture, where the use of resistant plant cultivars imposes strong selective pressures on plant pathogen populations and leads frequently to ‘resistance breakdown’. Presently, durable resistances are only identified after a long period of large‐scale cultivation of resistant cultivars. We propose a new predictor of the durability of plant resistance. Because resistance breakdown involves modifications in the avirulence factors of pathogens, we tested for correlations between the evolutionary constraints acting on avirulence factors or their diversity and the durability of the corresponding resistance genes in the case of plant–virus interactions. An analysis performed on 20 virus species–resistance gene combinations revealed that the selective constraints applied on amino acid substitutions in virus avirulence factors correlate with the observed durability of the corresponding resistance genes. On the basis of this result, a model predicting the potential durability of resistance genes as a function of the selective constraints applied on the corresponding avirulence factors is proposed to help breeders to select the most durable resistance genes.
INTRODUCTION
Understanding the mechanisms linked to the emergence of pathogenic microorganisms is a major challenge because of their impact on human health and activities. Most attention has been paid to the role of ecological factors (including migration, climate, agricultural practices and spread of biological disease vectors) in the emergence of plant diseases (Anderson et al., 2004). However, the role of ecological factors is often difficult to disentangle from that of the genetic changes operating within the pathogen populations (Desprez‐Loustau et al., 2007; Fargette et al., 2006; Holmes and Drummond, 2007). The emergence of pathogens requires both their genetic adaptation to new hosts and their subsequent spread and maintenance within host populations. Diverse strategies have been developed to control plant pathogens, the use of resistant varieties by growers being the most efficient and simplest. However, the widespread use of genetic resistances imposes strong selective pressures on the targeted pathogens which are able to adapt, resulting in the breakdown of the resistance. Resistance breakdown can be defined as a case of pathogen emergence which involves the jump of a host barrier at the intraspecific level and whose genetic bases have been best documented (Flor, 1971; Gabriel, 1999; Skamnioti and Ridout, 2005). Combining surveys of resistance breakdown in the long term with genetic analyses of pathogen populations can therefore help in understanding the respective roles of intrinsic evolutionary processes and external ecological factors in pathogen emergence.
As defined by Johnson (1979), a resistance is durable if it remains effective in a cultivar for a long period of time during its widespread cultivation in environments favourable to disease development. According to this definition, the durability of resistances can be measured only a posteriori after their large‐scale deployment. The ability to predict the durability of resistance represents a major economic issue in agriculture. However, the criteria currently retained by breeders to evaluate resistance sources do not include their potential durability because of the lack of predictors. According to several authors, the mechanism of action and the genetic determinism of plant resistance to pathogens explain only a small part of their durability (Fraser, 1990; García‐Arenal and McDonald, 2003; McDonald and Linde, 2002). The first predictor of resistance durability to pathogens was proposed by McDonald and Linde (2002), and further applied to virus resistances by García‐Arenal and McDonald (2003). This predictor, called ‘evolutionary potential’ of pathogens, focused on processes that govern pathogen population evolution (mainly effective population size, migration and reproduction system). However, this approach ignores the molecular events involved in resistance breakdown or the fitness cost associated with virulence, i.e. the ability of a pathogen genotype to overcome a resistance in the case of gene‐for‐gene interactions (Sacristán and García‐Arenal, 2008), and therefore cannot explain why different resistance genes directed towards the same pathogen can display different durabilities.
1955, 1971) was the first to show that resistance or susceptibility of plants to pathogens results from an intimate molecular relationship governed by a gene‐for‐gene interaction model. In this model, the interaction between two partners, the resistance gene of the plant (with at least two allelic forms: ‘resistant’ and ‘susceptible’) and the avirulence gene of the pathogen (with at least two allelic forms: ‘avirulent’ and ‘virulent’), determine the resistance or susceptibility of a plant to a pathogen (Flor, 1971). Breakdown of resistance occurs when the following two steps can be fulfilled by the pathogen: (i) mutation and/or recombination events should appear in the avirulence gene of a pathogen to generate virulent variants; and (ii) the fitness of these virulent variants should permit them to increase in frequency in the plant population and spread in the agroecosystem (Leach et al., 2001). As resistance durability is linked to the mechanisms and dynamics that control these two steps, we propose a new predictor of plant resistance durability based on the properties of pathogen avirulence factors. We focused our study on plant viruses for which a large number of avirulence factors corresponding to diverse plant resistance genes have been determined (Kang et al., 2005b), and for which estimates of resistance durability are available from field data. We checked whether the evolutionary constraints acting on amino acid substitutions in avirulence factors determine the durability of the corresponding resistance. Indeed, it is likely that the greater the constraints on the avirulence factor, the greater the fitness penalty conferred by mutations and/or recombination in this avirulence factor, and consequently the greater the resistance durability. In addition, we tested the hypotheses that the nucleotide or amino acid diversity of avirulence factors determines resistance durability. Indeed, the higher the diversity in avirulence factors, the higher the probability for the virus to evolve a virulent form of the avirulence factor with a minimum number of nucleotide or amino acid changes, and hence the lower the resistance durability. It should be noted that, in theory, there is no expected correlation between the selective constraints applied on genes and their diversity (Kimura, 1985; Wyckoff et al., 2005), justifying the consideration of these hypotheses separately.
RESULTS
Dataset construction and evaluation
The first step to test the correlation between the resistance durability and its potential predictors (evolutionary constraints and diversities of avirulence factors) was to build a dataset allowing a precise estimation of all of these variables. Altogether, 20 virus species–plant resistance gene combinations satisfying this condition were retained (see Experimental procedures). These combinations corresponded to 17 different avirulence factors, 13 different virus species and seven virus genera (Table 1). All viruses were RNA viruses, with a majority of potyviruses and tobamoviruses, and they infected a large diversity of host species. Seven viruses were transmitted by insects, two by soil‐borne fungi and four by contacts between plants. Six recessive and 14 dominant resistance genes were included.
Table 1.
Dataset used for the analysis of resistance durability and diversity and evolutionary constraints of avirulence factors.
| Plant | Virus | Resistance durability | Avirulence factor variables | Selected references** | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crop | Resistance gene* | Species | Genus | Avirulence factor† | Number of sequence | EP‡ | Timescale of protection efficiency | Status of the resistance | Virulent isolates | Resistance durability class | Nucleotide diversity§ | Amino acid diversity§ | ω ratio¶ | |
| Pepper | Tsw | TSWV | Tospovirus | NSs | 28 | 7 | 1 year | Broken down | Common | 1 | 0.034 | 0.036 | 0.291 | Margaria et al. (2007) |
| Tomato | Tm‐1 | ToMV | Tobamovirus | 184‐kDa protein | 8 | 6 | 2 years | Broken down | Common | 1 | 0.007 | 0.005 | 0.169 | Meshi et al. (1988); Strasser and Pfitzner (2007) |
| Tomato | Tm‐2 | ToMV | Tobamovirus | MP | 11 | 6 | 2–3 years | Broken down | Common | 1 | 0.005 | 0.006 | 0.314 | Meshi et al. (1989); Strasser and Pfitzner (2007) |
| Pepper | L3 | PMMoV | Tobamovirus | CP | 18 | 6 | 5 years | Broken down | Common | 1 | 0.029 | 0.020 | 0.204 | Berzal‐Herranz et al. (1995); Hamada et al. (2007) |
| Sugar beet | Rz‐1 | BNYVV | Benyvirus | P25 | 33 | 4 | 6–11 years | Broken down | Common | 1 | 0.022 | 0.037 | 0.969 | Acosta‐Leal and Rush (2007); Chiba et al. (2008) |
| Soybean | Rsv‐1 | SMV | Potyvirus | P3‐HcPro | 8 | 7 | 15 years | Broken down | Common | 2 | 0.034 | 0.022 | 0.132 | Eggenberger and Hill (1997); Eggenberger et al. (2008); Hajimorad et al. (2006) |
| Barley | rym‐4 | BaYMV | Bymovirus | VPg | 15 | 4 | >20 years | Broken down | Common | 2 | 0.053 | 0.050 | 0.258 | Kühne et al. (2003) |
| Pepper | pvr21 | PVY | Potyvirus | VPg | 11 | 6 | >20 years | Broken down | Common | 2 | 0.090 | 0.065 | 0.137 | Moury et al. (2004) |
| Potato | Nb | PVX | Potyvirus | 25‐kDa protein | 9 | 4 | >20 years | Broken down | Common | 2 | 0.035 | 0.021 | 0.096 | Malcuit et al. (1999) |
| Rapeseed | TuRB01 | TuMV | Potyvirus | CI | 29 | 9 | >20 years | Broken down | Common | 2 | 0.138 | 0.030 | 0.029 | Jenner et al. (2000) |
| Pepper | pvr22 | TEV | Potyvirus | VPg | 15 | 7 | >20 years | Broken down | Common | 2 | 0.068 | 0.050 | 0.194 | Kang et al. (2005a) |
| Rice | rymv‐1 | RYMV | Sobemovirus | VPg | 18 | 4 | >20 years | Broken down | Common | 2 | 0.076 | 0.031 | 0.075 | Hébrard et al. (2006) |
| Tomato | Tm‐22 | ToMV | Tobamovirus | MP | 11 | 6 | >20 years | Not broken down | Rare | 3 | 0.005 | 0.006 | 0.314 | Weber et al. (1993); Strasser and Pfitzner (2007) |
| Potato | Nx | PVX | Potexvirus | CP | 26 | 4 | >20 years | Not broken down | Rare | 3 | 0.086 | 0.040 | 0.058 | Santa Cruz and Baulcombe (1995) |
| Potato | Rx | PVX | Potexvirus | CP | 26 | 4 | >20 years | Not broken down | Rare | 3 | 0.086 | 0.040 | 0.058 | Bendahmane et al. (1995) |
| Pepper | pvr22 | PVY | Potyvirus | VPg | 11 | 6 | >20 years | Not broken down | Rare | 3 | 0.090 | 0.065 | 0.137 | Moury et al. (2004) |
| Bean | wlv | BYMV | Potyvirus | VPg | 10 | 8 | >20 years | Not broken down | Rare | 3 | 0.160 | 0.109 | 0.100 | Bruun‐Rasmussen et al. (2007) |
| Potato | Ry | PVY | Potyvirus | Nia‐Pro | 23 | 7 | >20 years | Not broken down | Not reported | 4 | 0.128 | 0.053 | 0.068 | Mestre et al. (2000) |
| Tobacco | N | TMV | Tobamovirus | 183 kDa protein | 10 | 6 | >20 years | Not broken down | Not reported | 4 | 0.025 | 0.011 | 0.087 | Padgett et al. (1997) |
| Pepper | L1 | ToMV | Tobamovirus | CP | 17 | 6 | >20 years | Not broken down | Not reported | 4 | 0.014 | 0.017 | 0.186 | Dardick et al. (1999); Hamada et al. (2007) |
Upper‐ and lower‐case initials indicate dominant and recessive resistances, respectively.
CI, cylindrical inclusion protein; CP, coat protein; MP, movement protein; Nia‐Pro, nuclear inclusion a protein; NSs, nonstructural S protein; P3‐HcPro, protein 3 and helper component protease; P25, 25‐kDa protein; VPg, genome‐linked viral protein.
Evolutionary potential overall risk defined by García‐Arenal and McDonald (2003) or from bibliographical data (see Supporting Information).
Average pairwise p distance estimated with mega 3.1 software.
Nonsynonymous/synonymous substitution rate ratio estimated by the single‐likelihood ancestor counting (SLAC) method implemented in HyPhy software.
Selected references for avirulence gene identification and amino acid substitutions involved in the resistance breakdown (when these are known).
BaYMV, Barley yellow mosaic virus; BNYVV, Beet necrotic yellow vein virus; BYMV, Bean yellow mosaic virus; PMMoV, Pepper mild mottle virus; PVX, Potato virus X; PVY, Potato virus Y; RYMV, Rice yellow mottle virus; SMV, Soybean mosaic virus; TEV, Tobacco etch virus; TMV, Tobacco mosaic virus; ToMV, Tomato mosaic virus; TSWV, Tomato spotted wilt virus; TuMV, Turnip mosaic virus.
The 20 virus species–plant resistance gene combinations composing the dataset were classified among four durability classes of increasing durability according to the timescale of protection efficiency and the proportion of virulent variants described in the agroecosystem (Table 1 and see Experimental procedures). The diversity of avirulence factors and the evolutionary constraints acting on these factors were estimated from sequences available in GenBank (table S2, see Supporting Information). The p distance was used to estimate the diversity of avirulence factors at both the nucleotide and amino acid levels, and the ω ratio, which is the ratio between the nonsynonymous and synonymous substitution rates (Kimura, 1983), was used to estimate the evolutionary constraints acting on amino acid substitutions in the avirulence factors (Yang and Swanson, 2002; Yang et al., 2000) (see Experimental procedures). We observed that the nucleotide diversity of the avirulence factors retained in the dataset was usually low, and varied from 0.005 [movement protein of Tomato mosaic virus (ToMV)] to 0.160 [VPg (genome‐linked viral protein) of Bean yellow mosaic virus (BYMV)]. Similarly, the amino acid diversity varied from 0.005 to 0.109 (Table 1). The ω ratio was generally low (≤0.3), indicating that avirulence factors were globally submitted to negative selection and that most amino acid changes were deleterious in these proteins (Kimura, 1983). However, for the P25 protein (25‐kDa protein) of Beet necrotic yellow vein virus (BNYVV), a higher value was estimated (ω = 0.97). For all the avirulence factors analysed, no amino acid sites were found to be under significant positive selection (ω > 1) with the single‐likelihood ancestor counting (SLAC) method.
To remove possible biases in the estimation of the putative predictors of resistance durability, we investigated the minimum number of sequences required for the correct estimation of the diversities and ω ratio. We noticed that eight to 10 sequences were usually sufficient to obtain satisfactory estimates (see Experimental procedures and Fig. 1), and that the lower the variable, the smaller the number of sequences required. Consequently, for the analyses, we retained only datasets with a minimum of 10 sequences of the avirulence factor, except in three datasets [P3‐HcPro (protein 3 and helper component protease) of Soybean mosaic virus (SMV), 183‐kDa protein of ToMV and 25‐kDa protein of Potato virus X (PVX)], for which only eight or nine sequences of the avirulence factor were available, but which presented relatively low ω ratios and diversities. We also validated that the number of geographical origins of isolates or the presence of virulent isolates did not introduce biases in our estimates of the diversity or ω ratio of the avirulence factors (see Experimental procedures for details).
Figure 1.
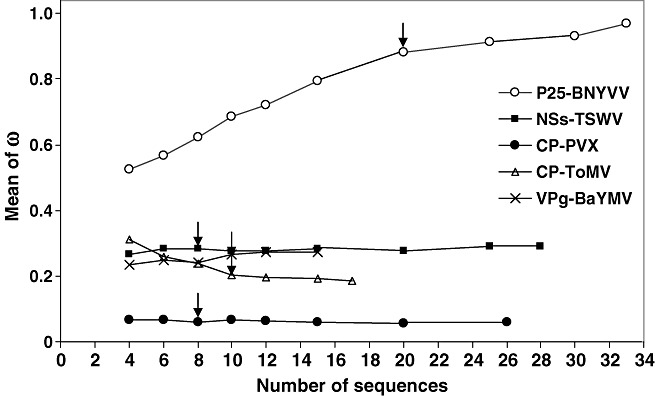
Estimates of the ω ratio of virus avirulence factors according to the size of the sequence set. Five large sequence sets were used to analyse random samples of sequences of increasing size (10 random samples for each size). Arrows indicate the smallest sequence set size for which the estimate of the ω ratio is not significantly different from the estimates obtained with larger datasets. BaYMV, Barley yellow mosaic virus; BNYVV, Beet necrotic yellow vein virus; PVX, Potato virus X; ToMV, Tomato mosaic virus; TSWV, Tomato spotted wilt virus.
Statistical analyses of the correlation between resistance durability and its potential predictors
The distribution of the virus species–resistance gene combinations in the durability classes was slightly unbalanced, with class 4 of ‘very high resistance durability’ under‐represented compared with the other classes (Table 1). Consequently, classes 3 and 4 were grouped into a single durability class named 3 + 4. Correlations between the three variables related to the avirulence factors or the evolutionary potential of viruses, as defined by García‐Arenal and McDonald (2003), and the durability of the corresponding resistances were tested using two statistical methods: an ordinal multinomial model and a Kruskal–Wallis test (see Experimental procedures and Table 2). In agreement with our hypothesis, a clear relationship was observed between the ω ratio of the avirulence factors and the durability of the corresponding resistance: the lower the ω ratio, the higher the resistance durability (Fig. 2A). This relationship was highly significant with the ordinal multinomial model (P = 0.008) and not determined solely by the particularly high ω value of the P25 protein of BNYVV. Indeed, a nonparametric Kruskal–Wallis test, which has the advantage of giving the same weight to each datum whatever its ω value, was also significant (P = 0.031) (Table 2). Because no noticeable differences were observed between the ω ratio of classes 2 and 3 + 4 (Fig. 2A), an analysis of the correlation between resistance durability and the ω ratio was carried out using only two durability classes: class 1 and class 2 + 3 + 4. Considering only two durability classes, the relationship observed between the resistance durability and the ω ratio was slightly better (P = 0.003; ordinal multinomial model) than with three durability classes (P = 0.008) (Table 2). The relationship between the diversity of avirulence factors and resistance durability was, on the whole, opposite to that expected: the higher the diversity, the higher the resistance durability (Fig. 2B,C). For the nucleotide diversity, a marginally significant relationship was detected using the Kruskal–Wallis test (P = 0.052, Table 2). It should be noted that the results obtained with the ordinal multinomial model cannot be exploited because the proportional odds assumption was not satisfied (score test = 0.039). No significant relationship was found between the resistance durability and the amino acid diversity, the evolutionary potential of viruses defined by García‐Arenal and McDonald (2003) or the type of resistance (dominant or recessive) (Fig. 2C,D, Table 2). Similarly, the separate analysis of the relationships between resistance durability and the three components of the evolutionary potential of viruses did not reveal any significance (data not shown). The smaller size of the dataset used here, as well as differences in the analytical methods, could explain this difference from the analyses of García‐Arenal and McDonald (2003). A likelihood ratio test indicates that a model including the ω ratio estimated on avirulence factors and the evolutionary potential of viruses or the type of resistance does not explain the resistance durability better than a model involving only the ω ratio (Table 2).
Table 2.
Statistical analysis of the relationships between the variable ‘durability of resistance’ and the four potential predictor variables computed with the ordinal multinomial model and with the nonparametric Kruskal–Wallis test (the under‐represented durability classes 3 and 4 were grouped together for the analyses). Values in bold indicate significant tests (P < 0.05) which satisfied test assumptions.
| Variable | Ordinal multinomial model | Kruskal–Wallis test | |||
|---|---|---|---|---|---|
| Odds assumption | Likelihood ratio test | P value | |||
| Score test† | Chi‐square | d.f.‡ | P value§ | ||
| ω ratio | 0.120 | 7.008 | 1 | 0.008 ** | 0.031 * |
| ω ratio (avirulent isolates only) | 0.372 | 5.488 | 1 | 0.019 * | 0.110 ns |
| ω ratio (two durability classes: 1 and 2 + 3 + 4) | Not applicable | 8.582 | 1 | 0.003 ** | 0.008 ** |
| Nucleotide diversity | 0.039 | 4.330 | 1 | 0.037* | 0.052 ns |
| Amino acid diversity | 0.157 | 2.454 | 1 | 0.118 ns | 0.216 ns |
| Evolutionary potential (EP) | 0.976 | 0.006 | 1 | 0.935 ns | 0.995 ns |
| Resistance type | 0.051 | 0.264 | 1 | 0.607 ns | 0.107 ns |
| ω ratio + EP | 0.238 | 7.014 | 2 | 0.028 * | Not applicable |
| ω ratio + resistance type | 0.015 | 7.277 | 2 | 0.026* | Not applicable |
A nonsignificant test (P > 0.05) indicates that the proportional odds assumption is valid.
d.f., degree(s) of freedom.
P value of the global test (β = 0): a significant test supports the evidence that at least one of the covariates is correlated with the resistance durability.
P > 0.05.
0.01 < P < 0.05.
P < 0.01.
Figure 2.
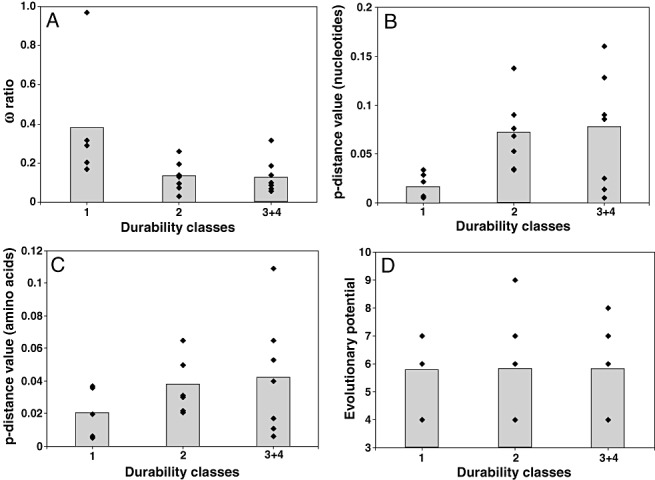
Distribution into the three resistance durability classes of the putative predictor variables related to virus avirulence factors or to the biology of viruses (durability classes 3 and 4 are grouped together): (A) ω ratio of avirulence factors; (B) nucleotide diversity of avirulence factors; (C) amino acid diversity of avirulence factors; (D) evolutionary potential of viruses (García‐Arenal and McDonald, 2003). Each virus species–resistance gene combination is represented by a diamond, and grey bars indicate average values.
DISCUSSION
McDonald and Linde (2002) first proposed a predictor of the durability of resistances based on the biological properties of the targeted pathogen. Later, García‐Arenal and McDonald (2003) applied the same approach to virus resistances and defined a risk index for virus species which correlates with resistance durability. With the increasing knowledge of viral avirulence factors, it has been postulated that certain relationships might exist between the resistance durability and the following: (i) the number of mutations involved in virulence (Ayme et al., 2007; Harrison, 2002); and (ii) the fitness cost associated with these mutations (Jenner et al., 2002; Leach et al., 2001). The fitness cost associated with mutations involved in virulence has been reported to be important in predicting the resistance durability in several plant–virus pathosystems (Boyd, 2006; Lecoq et al., 2004). Consequently, we explored the possible relationships between the observed resistance durability and properties of virus avirulence factors.
Causality relationships between properties of viral avirulence factors and resistance durability
We observed a marginal relationship between resistance durability and the nucleotide diversity of the avirulence factor (Table 2). This relationship was opposite to that expected, as the higher the diversity, the higher the resistance durability (Fig. 2B). This results mainly from the lower diversity observed in durability class 1. A possible explanation is that three [Tomato spotted wilt virus (TSWV), BNYVV and Pepper mild mottle virus (PMMoV)] of the four viruses in durability class 1 emerged relatively recently on a large geographical scale (independent of the resistance breakdown phenomenon), whereas the emergence of viruses in other classes was less recent (at least 20 years ago given our definition of durability classes 2, 3 and 4). As a consequence, viruses in durability class 1 may have had less time to diversify than viruses corresponding to classes 2, 3 and 4. The marginal correlation between the resistance durability and nucleotide diversity of avirulence factors would then be both indirect and caused by bias in our dataset. More precise data on resistance breakdowns are necessary to validate this assumption. It is important to note that this putative bias has, in theory, no consequences on our analyses involving the ω ratio (Kimura, 1985; Wyckoff et al., 2005).
A significant relationship was detected between resistance durability and the constraints exerted on amino acid changes (ω ratio) in avirulence factors (Table 2). Predicting resistance durability from the value of the ω ratio implies a causality relationship. The observed association between resistance durability and the ω ratio could be a result of the following relationships: (i) the ω ratio determines the resistance durability; (ii) the resistance durability determines the ω ratio; (iii) both the resistance durability and the ω ratio are determined by at least one additional factor. As we could not find any evident additional factor that would determine a negative correlation between resistance durability and the ω ratio of avirulence factors, only the first two scenarios were examined. Could resistance durability determine variations in the ω ratio of avirulence factors? Datasets corresponding to durability classes 1 and, to a lesser extent, 2 contain a proportion of sequences of virulent isolates which have undergone amino acid substitutions in their avirulence factors. As these particular amino acid positions could be subjected to positive selection (Moury, 2004; Schirmer et al., 2005), this could increase the ω ratio for durability classes 1 and 2, whilst having a lesser effect (or not at all) on the ω ratio for durability classes 3 and 4. However, the following observations suggest that the ω ratio in viral avirulence factors indeed determines the level of resistance durability: (i) in durability classes 1 and 2, a limited proportion of virulent isolates (0%–22%) are characterized [except for BNYVV–Rz1, Barley yellow mosaic virus (BaYMV)–rym4, BYMV–wlv, PMMoV–L3, SMV–Rsv1 and Potato virus Y (PVY)–pvr21] (Table S2, see Supporting Information); (ii) the change in virulence properties generally depends only on a small number of amino acid substitutions, usually one or two (Harrison, 2002); and (iii) the presence of virulent isolates in datasets does not introduce significant biases into the ω ratio estimates for the BNYVV–Rz1, BaYMV–rym4 and PMMoV–L3 combinations (the BYMV–wlv, SMV–Rsv1 and PVY–pvr21 datasets did not include sufficient virulent and avirulent isolates for such analyses). In addition, and most convincingly, when all virulent isolates were withdrawn from the sequence dataset, the negative correlation between the ω ratio and the resistance durability was still significant (P = 0.019; ordinal multinomial model) (Table 2). It should be noted, however, that this relationship was not significant (P = 0.11) using the Kruskal–Wallis test, probably because four pathosystems were excluded from the analysis as a result of an insufficient number of sequences, and because the Kruskal–Wallis test has a low statistical power (see Experimental procedures). The mechanism behind this causal link resides in the fact that the ω ratio measures the propensity of viruses to change amino acids in their avirulence factors without high fitness costs under the assumption that synonymous nucleotide changes are neutral.
For viruses and other plant pathogens, gains of virulence towards resistant genotypes are frequently associated with fitness costs in susceptible genotypes (Ayme et al., 2007; Desbiez et al., 2003; Jenner et al., 2002; Leach et al., 2001; Meshi et al., 1989). Although limited numbers of amino acid positions have been shown to be involved in virulence properties (Harrison, 2002), measurement of the evolutionary constraints on the whole avirulence factor seemed to be appropriate to our study for two reasons. First, it is plausible that many more virulence mutations than presently known exist for a given virus species–resistance gene interaction because reverse genetic analyses of virulence mutations have frequently been performed with a small number of virulent and avirulent isolates. Second, when virulence mutations in the virus impose a fitness cost, compensatory mutations may appear somewhere else in the avirulence factor, making it worthwhile to measure the ω ratio in these parts of the protein as well.
Predicting resistance durability as a function of the ω ratio
As the evolutionary constraints acting on amino acid substitutions (ω ratio) in avirulence factors are correlated with the durability of resistance, it is possible to assess, for a given resistance gene, the probability of being in a given durability class as a function of a measured ω ratio using Eq. (1) (see Experimental procedures) (Fig. 3). According to this model, resistance genes corresponding to avirulence factors with ω values higher than 0.50 are associated with the class of lower durability (class 1) with a fairly high probability (P > 0.80). In contrast, resistance genes for which avirulence factors have ω values lower than 0.15 are assigned to classes of either medium or high durability (classes 2 or 3 + 4) with a probability P ≥ 0.80. On the whole, cross validation tests (see Experimental procedures) allowed assignments of resistance genes to the correct durability class in 44% of cases (33% of correct assignations would be expected by chance). Even though our model still lacks power, 65% of resistance genes were correctly assigned in durability class 3 + 4, corresponding to the class of genes of highest interest for resistance durability. It should also be noted that our model cannot account for the fact that different resistance alleles or genes interacting with the same viral avirulence gene show different durabilities, such as, for example, pvr21 and pvr22 for PVY (Table 1). Additional virus species–resistance gene combinations or additional predictive variables would be needed to improve the precision of these predictors. In the same manner, using ω ratio estimates on the specific domain of avirulence proteins which interact with resistance factors could improve the power of this model.
Figure 3.
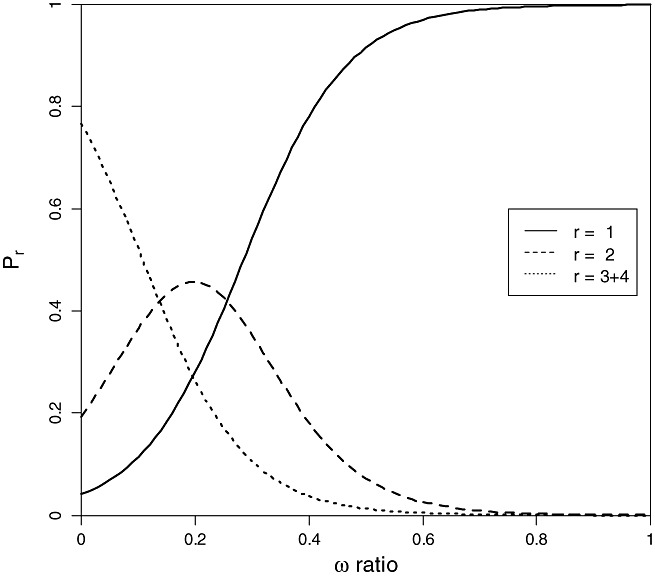
Probability (P r) that a resistance gene belongs to the durability class r ∈ {1, 2, 3 + 4} as a function of the ω ratio of the targeted avirulence factor based on Eq. (1) (see Experimental procedures) and our dataset (Table 1).
A novel strategy for resistance screening: targeted search for durable resistance
Resistance durability is presently evaluated downstream of breeding programmes, when the resistance genes have been deployed in commercial varieties at a large scale and over several years. The incorporation of resistance durability earlier in the breeding scheme would be highly desirable (Boyd, 2006). Beyond their predictive value for the durability of resistances already incorporated in plant cultivars, ω ratios estimated from viral avirulence genes could also be integrated upstream into resistance breeding programmes in two ways. First, the comparison of estimates of the ω ratios from avirulence factors corresponding to several candidate resistance sources could allow breeders to evaluate and select the potentially more durable resistances. Second, new resistance sources could be identified via the directed search for resistance factors elicited by candidate avirulence factors. Thus, it has been shown that transient expression of avirulence proteins in plant tissues enables the detection of resistance genes in plants through the elicitation of a necrotic response characteristic of dominant resistances (Bendahmane et al., 2000; Laugéet al., 1998; Mestre et al., 2000). Even if the efficiency of the detected resistance should be further confirmed by inoculation tests, as necrosis can also be induced independently of the specific resistance phenomenon, using this method with the most constrained viral proteins could allow the a priori identification of the most durable resistances targeting them.
CONCLUSION
As proposed by McDonald and Linde (2002) and García‐Arenal and McDonald (2003), we analysed the ability of some pathogen traits to predict the capacity of emergence of adapted pathogens in crop plants carrying specific resistance genes. We focused on plant–virus interactions, but our analytical approach could have broader interest. The validity of the method depends on two functional assumptions: (i) the control mechanism (here plant resistance genes) interacts either directly or indirectly with a specific protein target; and (ii) adaptation of the targeted organism to circumvent the control mechanism occurs via mutations in this protein. The observed correlation between the ω ratio of viral avirulence factors and the durability of corresponding plant resistance genes strongly suggests that these assumptions are widely satisfied in the case of plant–virus interactions. Notably, the fact that molecular interactions between both partners operate at the protein but not the RNA level, from the virus side, is not trivial, as it has been shown that RNA segments can act as viral avirulence factors independent of their possible coding capacity (Diaz et al., 2004; Szittya and Burgyan, 2001). As the above two assumptions are commonplace among plant–pathogen interactions, it would be worthwhile evaluating the generalization of our approach to other kinds of plant pathogen.
EXPERIMENTAL PROCEDURES
Dataset
To analyse the durability of plant resistance to viruses, we retained virus species–resistance gene (or allele) combinations that satisfied the following three conditions: (i) the virus avirulence factor corresponding to the resistance gene had been identified; (ii) nucleotide sequences encoding the avirulence factor could be obtained from a sufficient number of virus isolates to estimate their diversity and evolutionary constraints; and (iii) plant cultivars carrying the resistance gene had been largely exploited and field data on the durability of this resistance were available. All resistance genes composing the dataset controlled qualitative resistance (Table 1). The nucleotide sequences of avirulence factors were retrieved from GenBank, aligned with the ClustalW program (Thompson et al., 1994) and analysed with RDP version 3 software (Martin et al., 2005) to remove putative recombinant sequences which could create biases in the estimation of the evolutionary constraints (Schierup and Hein, 2000). For Potato virus Y (PVY) and Tobacco etch virus (TEV), five and 12 additional sequences of the VPg coding region, respectively, were obtained (accession numbers EU334778 to EU334794). For the pvr21 and pvr22 resistance alleles in pepper, only VPg sequences from pepper isolates of PVY belonging to the C phylogenetic group were analysed. In the case of overlapping open reading frames in the cistron coding for the avirulence factor (183‐ or 184‐kDa proteins and movement protein of tobamoviruses and 25‐kDa protein of PVX), the overlapping region was removed to avoid artefacts in the estimation of their diversity and evolutionary constraints. The GenBank accession numbers of the sequences retained are available in Table S2 (see Supporting Information). To avoid artefactual polymorphism, we discarded the sequences of virus isolates that had been maintained in the long term in the laboratory through repeated inoculations and sequences of virulent isolates selected artificially.
Estimation of avirulence factor diversity, constraints exerted on amino acid substitutions and evolutionary potential of viruses
For each virus species–resistance gene combination, the following four putative predictors of resistance durability were determined: (i) the diversity of avirulence factors at the nucleotide level; (ii) the diversity of avirulence factors at the amino acid level; (iii) the evolutionary constraints on amino acid substitutions in avirulence factors; and (iv) the evolutionary potential of viruses as defined by García‐Arenal and McDonald (2003). Sequence alignments of avirulence factors were used to estimate the first three predictors. The average pairwise p distance was used to estimate the diversity of the avirulence factors, at both the nucleotide and amino acid levels, using mega version 3.1 (Kumar et al., 1994). The ω ratio, which is the ratio between the nonsynonymous (amino acid altering) and synonymous (silent at the amino acid level) substitutions, was used to estimate the evolutionary constraints acting on amino acid substitutions in the avirulence factors (Kimura, 1983; Yang and Swanson, 2002; Yang et al., 2000). This ω ratio was estimated on whole avirulence factor sequences using the SLAC method in HyPhy software (Kosakovsky Pond and Frost, 2005), a modification of the site‐by‐site analytical method of Suzuki and Gojobori (1999). The probability threshold to detect particular amino acid sites undergoing positive selection (ω > 1) was set to P = 0.10.
The evolutionary potential of viruses (García‐Arenal and McDonald, 2003) corresponds to a compound risk index based on the biology of the viruses, and takes into account: (i) the population size of the virus; (ii) the amount of gene and genotype flow in the virus population; and (iii) its frequency of recombination. The evolutionary potential of virus species was obtained from García‐Arenal and McDonald (2003), or estimated according to the criteria defined by these authors (Table 1 and Table S1 in Supporting Information).
Precision of the estimates of diversity and evolutionary constraints of avirulence factors
The precision of the estimates for the diversity and evolutionary constraints of avirulence genes depends on representative samples of sequences of these genes. A method to evaluate the representativeness of samples is to measure the variance of the studied variables with random subsamples of increasing size (Hughes et al., 2001). In this study, subsamples of sequences of increasing size were randomly chosen among the five largest sequence sets [P25 of BNYVV, CP (coat protein) of ToMV, VPg of BaYMV, CP of PVX and NSs (nonstructural S protein) of TSWV] presenting contrasted diversity and evolutionary constraints (Table 1). In each subsample, 10 random sets of sequences were analysed, allowing an estimation of the variance of the three variables (nucleotide and amino acid diversities and ω ratio) and the performance of mean comparisons. The minimal number of sequences allowing satisfactory estimates of the ω ratio or diversities of avirulence factors (i.e. minimum sample delivering estimates that are not significantly different from those obtained with the whole dataset) to be obtained was between eight (CP of PVX and NSs of TSWV) and 10 (CP of ToMV and VPg of BaYMV) (Fig. 1). The only exception was the P25 protein of BNYVV that had a higher ω ratio, and consequently required a minimum of 20 sequences to be estimated correctly (Fig. 1).
The same approach was used to determine the effect of an increase in the number of geographical localities (countries) on the precision of variable estimates. Subsamples of 10 sequences with increasing numbers of geographical origins, chosen among the four sequence sets showing the highest diversity of geographical origins [NSs of TSWV, CI (cylindrical inclusion protein) of Turnip mosaic virus (TuMV), CP of PMMoV and NiaPro (nuclear inclusion a protein) of PVY], were used to estimate the variance of the variables. No significant differences in predictor estimates were revealed by rank tests performed on subsamples of sequences showing increasing diversity of geographical origins. To evaluate the impact of virulent isolates on our results, we also compared the estimates of the ω ratio obtained with sequence samples, including or not the virulent isolates for BNYVV–Rz1, BaYMV–rym4 and PMMoV–L3, which included sufficient numbers of virulent and avirulent isolates (Table S2, see Supporting Information). We also conducted our analyses with a dataset excluding the sequences of virulent isolates to remove any bias on the correlation between the ω ratio of avirulence factors and resistance durability. This dataset was made up of 16 virus species–plant resistance gene combinations satisfying the condition of a minimum of eight sequences of the avirulence gene (BYMV–wlv, SMV–Rsv1, PVY–pvr21 and PVX–Nb were excluded).
Estimation of the durability of resistance
The durability of resistance genes or alleles was assessed according to the timescale of the protection efficiency before the appearance of crop damage as a result of resistance‐breaking isolates, and according to the abundance of resistance‐breaking isolates in the agroecosystem (Table 1). The majority of these data were collected from García‐Arenal and McDonald (2003) and Harrison (2002), and additional references are available in Table S1 (see Supporting Information). All the resistances in the dataset had been used in the field for more than 20 years or were broken down rapidly (Table 1). Resistances were considered as ‘broken‐down’ if the resistance was defeated by resistance‐breaking isolates and led to important economic losses, at least in some growing regions. Resistance durability was categorized into four discrete durability classes as follows.
-
1
Class 1: rapid resistance breakdown (less than 12 years after initial deployment). Frequent virulent isolates in the field prevent the efficiency of the resistance, at least in some growing regions.
-
2
Class 2: slow resistance breakdown (more than 12 years after initial deployment). Frequent virulent isolates in the field prevent the efficiency of the resistance, at least in some growing regions.
-
3
Class 3: resistances deployed for more than 20 years with rare virulent isolates in the field. Virulent isolates are restricted to small areas and do not economically compromise the use of the resistant cultivars.
-
4
Class 4: resistances deployed for more than 20 years and no virulent strains described in the field.
In our dataset (Table 1), the distinction between durability classes 1 and 2 was clear‐cut as the breakdown of resistance in class 1 occurred in 1–11 years, whereas the breakdown of resistance in class 2 occurred mostly in more than 20 years. The SMV–Rsv‐1 gene combination only showed an intermediate durability (breakdown in about 15 years). However, similar results were obtained when it was classified into class 1 or 2. Owing to the small number of resistances in class 4, classes 3 and 4 were grouped together for further analyses.
Statistical analysis
Statistical analyses were used to evaluate the correlation between the response variable ‘durability of resistance’ and four potential predictor variables (nucleotide and amino acid diversities of avirulence factors, ω ratios of avirulence factors and evolutionary potential of viruses). We first used a nonparametric Kruskal–Wallis test for the null hypothesis: ‘The average values of the considered explanatory variable are similar for all resistance durability classes’. We also used a parametric approach based on ordinal multinomial models. Multinomial models extend logistic regression to the case in which the response variable has three (or more) discrete outcomes (i.e. the response variable follows a multinomial distribution) (McCullagh and Nelder, 1989). In our case, the durability of resistances is an ordinal response variable, because resistance durability increases from class 1 to class 3 + 4. Ordinal multinomial models allow both: (i) testing for a correlation between predictor and response variables by taking into account the ordinal and multinomial nature of the response variable; and (ii) modelling the probability of belonging to a particular durability class as a function of a set of predictor variables. More precisely, if Y is the response variable ‘durability of resistance’, Pr = p(Y ≤ r) denotes the probability of Y being in the r or lower class of durability (r ∈ {1, 2, 3 + 4}). In the proportional odds model, Pr is modelled with a logit link as
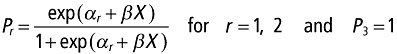 |
(1) |
where αr are intercept parameters that depend only on the categories, such that α 1 < α2, and β is a vector of the regression coefficients for the set of predictor variables X. Inherent in this model is the proportional odds assumption, which states that the odds [i.e. the quantity p(Y ≤ r)/p(Y > r)] of being in a category less than or equal to r is exp[β(x 1 − x 2)] times higher at X = x 1 than at X = x 2, with x 1 and x 2 being two particular values of X. PROC LOGISTIC of SAS software (version 8, SAS Inc., Cary, NC, USA) was used to estimate αr and β by maximum likelihood for five models differing by the set X of predictor variables included (amino acid diversity, nucleotide diversity, evolutionary potential, ω ratio and the combination of ω ratio and evolutionary potential). Before fitting these models, a score test for the proportional odds assumption was performed, a nonsignificant test supporting the evidence that the assumption is valid.
The predictive performance of the ordinal multinomial model was assessed using cross‐validation tests. Cross‐validation tests were performed with the R software environment (http://cran.r‐project.org/) by: (i) randomly leaving out three pathosystems (i.e. 15% of the dataset); (ii) fitting the model to the remaining dataset; and (iii) checking whether the true class of durability for each left‐out resistance gene–avirulence factor combination corresponded to the highest probability class predicted by the model (if so, the prediction is correct). The predictive performance of the model was assessed as the percentage of correct assignments and estimated by iterating the previous three steps 1500 times.
Supporting information
Table S1 Estimated risk categories for the three factors included in the evolutionary potential compound index of viruses and durability of the corresponding resistance gene for virus species–resistance gene combinations not described by García‐Arenal and McDonald (2003).
Table S2 GENBANK accession numbers of sequences of avirulence factors and related biological characteristics.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
ACKNOWLEDGEMENTS
We acknowledge Dr D. Fargette (Institut de Recherche pour le Développement, Montpellier, France) for sharing information about the durability of the Rymv‐1 gene, C. Marchal (Clause‐Tézier, Saint‐Rémy‐de‐Provence, France) for providing TEV isolates, and K. Ezzaier (Institut National de la Recherche Agronomique de Tunisie, Ariana, Tunisia) and Dr G. Marchoux (INRA, Avignon, France) for providing PVY isolates. We also acknowledge the ANRT Foundation and the companies Gautier Semences, Clause Vegetable Seeds, Vilmorin SA, Sakata Seeds Europe and Rijk Zwaan for financial support.
REFERENCES
- Acosta‐Leal, R. and Rush, C.M. (2007) Mutations associated with resistance‐breaking isolates of beet necrotic yellow vein virus and their allelic discrimination using TaqMan technology. Phytopathology, 97, 325–330. [DOI] [PubMed] [Google Scholar]
- Anderson, P.K. , Cunningham, A.A. , Patel, N.G. , Morales, F.J. , Epstein, P.R. and Daszak, P. (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544. [DOI] [PubMed] [Google Scholar]
- Ayme, V. , Petit‐Pierre, J. , Souche, S. , Palloix, A. and Moury, B. (2007) Molecular dissection of the potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88, 1594–1601. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Kohm, B.A. , Dedi, C. and Baulcombe, D.C. (1995) The coat protein of potato virus X is a strain‐specific elicitor of Rx1‐mediated virus resistance in potato. Plant J. 8, 933–941. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Querci, M. , Kanyuka, K. and Baulcombe, D.C. (2000) Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21, 73–81. [DOI] [PubMed] [Google Scholar]
- Berzal‐Herranz, A. , De la Cruz, A. , Tenllado, F. , Díaz‐Ruiz, J.R. , López, L. , Sanz, A.I. , Vaquero, C. , Serra, M.T. and García‐Luque, I. (1995) The Capsicum L3 gene‐mediated resistance against the tobamoviruses is elicited by the coat protein. Virology, 209, 498–505. [DOI] [PubMed] [Google Scholar]
- Boyd, L.A. (2006) Can the durability of resistance be predicted? J. Sci. Food Agric. 86, 2523–2526. [Google Scholar]
- Bruun‐Rasmussen, M. , Møller, I.S. , Hensen, J.K.R. , Lund, O.S. and Johansen, I.E. (2007) The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum . Mol. Plant–Microbe Interact. 20, 1075–1082. [DOI] [PubMed] [Google Scholar]
- Chiba, S. , Miyanishi, M. , Andika, I.B. , Kondo, H. and Tamada, T. (2008) Identification of amino acids of the beet necrotic yellow vein virus p25 protein required for induction of the resistance response in leaves of Beta vulgaris plants. J. Gen. Virol. 89, 1314–1323. [DOI] [PubMed] [Google Scholar]
- Dardick, C.D. , Taraporewala, Z. , Lu, B. and Culver, J.N. (1999) Comparison of tobamovirus coat protein structural features that affect elicitor activity in pepper, eggplant, and tobacco. Mol. Plant–Microbe Interact. 12, 247–251. [Google Scholar]
- Desbiez, C. , Gal‐On, A. , Girard, M. , Wipf‐Scheibel, C. and Lecoq, H. (2003) Increase in Zucchini yellow mosaic virus symptom severity in tolerant zucchini cultivars is related to a point mutation in P3 protein and is associated with a loss of relative fitness on susceptible plants. Phytopathology, 93, 1478–1484. [DOI] [PubMed] [Google Scholar]
- Desprez‐Loustau, M.L. , Robin, C. , Buee, M. , Courtecuisse, R. , Garbaye, J. , Suffert, F. , Sache, I. and Rizz, D.M. (2007) The fungal dimension of biological invasions. Trends Ecol. Evol. 22, 472–480. [DOI] [PubMed] [Google Scholar]
- Diaz, J. , Nieto, C. , Moriones, E. , Truniger, V. and Aranda, M. (2004) Molecular characterization of a Melon necrotic spot virus strain that overcomes the resistance in melon and nonhost plants. Mol. Plant–Microbe Interact. 17, 668–675. [DOI] [PubMed] [Google Scholar]
- Eggenberger, A.L. and Hill, J.H. (1997) Analysis of resistance‐breaking determinants of soybean mosaic virus. Phytopathology, 87, S27. [Google Scholar]
- Eggenberger, A.L. , Hajimorad, M.R. and Hill, J.H. (2008) Gain of virulence on Rsv1‐genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC‐Pro. Mol. Plant–Microbe Interact. 21, 931–936. [DOI] [PubMed] [Google Scholar]
- Fargette, D. , Konate, G. , Fauquet, C. , Muller, E. , Peterschmitt, M. and Thresh, J.M. (2006) Molecular ecology and emergence of tropical plant viruses. Annu. Rev. Phytopathol. 44, 235–260. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1955) Host–parasite interaction in flax rust—its genetics and other implications. Phytopathology, 45, 680–685. [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Fraser, R.S.S. (1990) The genetics of resistance to plant viruses. Annu. Rev. Phytopathol. 28, 179–200. [Google Scholar]
- Gabriel, D.W. (1999) Why do pathogens carry avirulence genes? Physiol. Mol. Plant Pathol. 55, 205–214. [Google Scholar]
- García‐Arenal, F. and McDonald, B.A. (2003) An analysis of the durability of resistance to plant viruses. Phytopathology, 93, 941–952. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2006) Strain‐specific P3 of Soybean mosaic virus elicits Rsv1‐mediated extreme resistance, but absence of P3 elicitor function alone is insufficient for virulence on Rsv1‐genotype soybean. Virology, 345, 156–166. [DOI] [PubMed] [Google Scholar]
- Hamada, H. , Tomita, R. , Iwadate, Y. , Kobayashi, K. , Munemura, I. , Takeuchi, S. , Hikichi, Y. and Suzuki, K. (2007) Cooperative effect of two amino acid mutations in the coat protein of Pepper mild mottle virus overcomes L3‐mediated resistance in Capsicum plants. Virus Genes, 34, 205–214. [DOI] [PubMed] [Google Scholar]
- Harrison, B.D. (2002) Virus variation in relation to resistance breaking in plants. Euphytica, 124, 181–192. [Google Scholar]
- Hébrard, E. , Pinel‐Galzi, A. , Bersoult, A. , Sire, C. and Fargette, D. (2006) Emergence of a resistance‐breaking isolate of Rice yellow mottle virus during serial inoculations is due to a single substitution in the genome‐linked viral protein VPg. J. Gen. Virol. 87, 1369–1373. [DOI] [PubMed] [Google Scholar]
- Holmes, E.C. and Drummond, A. (2007) The evolutionary genetics of viral emergence. Curr. Top. Microbiol. Immunol. 315, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J.B. , Hellmann, J.J. , Ricketts, T.H. and Bohannan, B.J.M. (2001) Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67, 4399–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner, C.E. , Sanchez, F. , Nettleship, S.B. , Foster, G.D. , Ponz, F. and Walsh, J.A. (2000) The cylindrical inclusion gene of turnip mosaic virus encodes a pathogenic determinant to the Brassica resistance gene TuRB01. Mol. Plant–Microbe Interact. 13, 1102–1108. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E. , Wang, X. , Ponz, F. and Walsh, J.A. (2002) A fitness cost for Turnip mosaic virus to overcome host resistance. Virus Res. 86, 1–6. [DOI] [PubMed] [Google Scholar]
- Johnson, R. (1979) The concept of durable resistance. Phytopathology, 69, 198–199. [Google Scholar]
- Kang, B.‐C. , Yeam, I. , Frantz, J.D. , Murphy, J.F. and Jahn, M.M. (2005a) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42, 392–405. [DOI] [PubMed] [Google Scholar]
- Kang, B.‐C. , Yeam, I. and Jahn, M.M. (2005b) Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Kimura, M. (1983) The Neutral Theory of Molecular Evolution. Cambridge University Press. [Google Scholar]
- Kimura, M. (1985) The role of compensatory neutral mutations in molecular evolution. J. Genet. 64, 7–19. [Google Scholar]
- Kosakovsky Pond, S.L. and Frost, S.D.W. (2005) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics, 21, 2531–2533. [DOI] [PubMed] [Google Scholar]
- Kühne, T. , Shi, N. , Proeseler, G. , Adams, M.J. and Kanyuka, K. (2003) The ability of a bymovirus to overcome the rym4‐mediated resistance in barley correlates with a codon change in the VPg coding region on RNA1. J. Gen. Virol. 84, 2853–2859. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Tamura, K. and Nei, M. (1994) MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10, 189–191. [DOI] [PubMed] [Google Scholar]
- Laugé, R. , Joosten, M.H.A.J. , Haanstra, J.P.W. , Goodwin, P.H. , Lindhout, P. and De Wit, P.J.G.M. (1998) Successful search for a resistance gene in tomato targeted against a virulence factor of a fungal pathogen. Proc. Natl. Acad. Sci. USA, 95, 9014–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, J.E. , Vera Cruz, C.M. , Bai, J. and Leung, H. (2001) Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu. Rev. Phytopathol. 39, 187–224. [DOI] [PubMed] [Google Scholar]
- Lecoq, H. , Moury, B. , Desbiez, C. , Palloix, A. and Pitrat, M. (2004) Durable virus resistance in plants through conventional approaches: a challenge. Virus Res. 100, 31–39. [DOI] [PubMed] [Google Scholar]
- Malcuit, I. , Marano, M.R. , Kavanagh, T.A. , De Jong, W. , Forsyth, A. and Baulcombe, D.C. (1999) The 25‐kDa movement protein of PVX elicits Nb‐mediated hypersensitive cell death in potato. Mol. Plant–Microbe Interact. 12, 536–543. [Google Scholar]
- Margaria, P. , Ciuffo, M. , Pacifico, D. and Turina, M. (2007) Evidence that the nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol. Plant–Microbe Interact. 20, 547–558. [DOI] [PubMed] [Google Scholar]
- Martin, D.P. , Williamson, C. and Posada, D. (2005) RDP2: recombination detection and analysis from sequence alignments. Bioinformatics, 21, 260–262. [DOI] [PubMed] [Google Scholar]
- McCullagh, P. and Nelder, J.A. (1989) Generalized linear models Vol. 37 of Monographs on Statistics and Applied Probability, 2 edn. London: Chapman & Hall/CRC. [Google Scholar]
- McDonald, B.A. and Linde, C. (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. [DOI] [PubMed] [Google Scholar]
- Meshi, T. , Motoyoshi, F. , Adachi, A. , Watanabe, Y. , Takamatsu, N. and Okada, Y. (1988) Two concomitant base substitutions in the putative replicase genes of Tobacco mosaic virus confer the ability to overcome the effects of a tomato resistance gene, Tm‐1. EMBO J. 7, 1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi, T. , Motoyoshi, F. , Maeda, T. , Yoshiwoka, S. , Watanabe, H. and Okada, Y. (1989) Mutations in the tobacco mosaic virus 30‐kD protein gene overcome Tm‐2 resistance in tomato. Plant Cell, 1, 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre, P. , Brigneti, G. and Baulcombe, D.C. (2000) An Ry‐mediated resistance response in potato requires the intact active site of the NIa proteinase from Potato virus Y . Plant J. 23, 653–661. [DOI] [PubMed] [Google Scholar]
- Moury, B. (2004) Differential selection of genes of Cucumber mosaic virus subgroups. Mol. Biol. Evol. 21, 1602–1611. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. , Guilbaud, L. , Souche, S. , Ayme, V. , Caranta, C. , Palloix, A. and Jacquemond, M. (2004) Mutations in Potato virus Y genome‐linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum . Mol. Plant–Microbe Interact. 17, 322–329. [DOI] [PubMed] [Google Scholar]
- Padgett, H.S. , Watanabe, Y. and Beachy, R.N. (1997) Identification of the TMV replicase sequence that activates the N gene‐mediated hypersensitive response. Mol. Plant–Microbe Interact. 10, 709–715. [Google Scholar]
- Sacristán, S. and García‐Arenal, F. (2008) The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Cruz, S. and Baulcombe, D.C. (1995) Analysis of potato virus X coat protein genes in relation to resistance conferred by the genes Nx, Nb and Rxl of potato. J. Gen. Virol. 76, 2057–2061. [DOI] [PubMed] [Google Scholar]
- Schierup, M.H. and Hein, J. (2000) Consequences of recombination on traditional phylogenetic analysis. Genetics, 156, 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, A. , Link, D. , Cognat, V. , Moury, B. , Beuve, M. , Meunier, A. , Bragard, C. , Gilmer, D. and Lemaire, O. (2005) Phylogenetic analysis of isolates of beet necrotic yellow vein virus collected worldwide. J. Gen. Virol. 86, 2897–2911. [DOI] [PubMed] [Google Scholar]
- Skamnioti, P. and Ridout, C.J. (2005) Microbial avirulence determinants: guided missiles or antigenic flak? Mol. Plant Pathol. 6, 551–559. [DOI] [PubMed] [Google Scholar]
- Strasser, M. and Pfitzner, A.J.P. (2007) The double‐resistance‐breaking Tomato mosaic virus strain ToMV1‐2 contains two independent single resistance‐breaking domains. Arch. Virol. 152, 903–914. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y. and Gojobori, T. (1999) A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 16, 1315–1328. [DOI] [PubMed] [Google Scholar]
- Szittya, G. and Burgyan, J. (2001) Cymbidium ringspot tombusvirus coat protein coding sequence acts as an avirulent RNA. J. Virol. 75, 2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, H. , Schultze, S. and Pfitznern, A.J.P. (1993) Two amino acid substitutions in the tomato mosaic virus 30‐kilodalton movement protein confer the ability to overcome the Tm‐22 resistance gene in the tomato. J. Virol. 67, 6432–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff, G.J. , Malcom, C.M. , Vallender, E.J. and Lahn, B.T. (2005) A highly unexpected strong correlation between fixation probability of nonsynonymous mutations and mutation rate. Trends Genet. 21, 381–385. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Swanson, W.J. (2002) Codon‐substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol. Biol. Evol. 19, 49–57. [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Nielsen, R. , Goldman, N. and Pedersen, A.M.K. (2000) Codon‐substitution models for heterogeneous selection pressure at amino acid sites. Genetics, 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Estimated risk categories for the three factors included in the evolutionary potential compound index of viruses and durability of the corresponding resistance gene for virus species–resistance gene combinations not described by García‐Arenal and McDonald (2003).
Table S2 GENBANK accession numbers of sequences of avirulence factors and related biological characteristics.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


