SUMMARY
Microarray technology was used to identify the genes associated with disease defence responses in the model legume Medicago truncatula. Transcript profiles from M. truncatula cv. Jemalong genotype A17 leaves inoculated with Colletotrichum trifolii and Erysiphe pisi and roots infected with Phytophthora medicaginis were compared to identify the genes expressed in response to all three pathogens and genes unique to an interaction. The A17 genotype is resistant to C. trifolii and E. pisi, exhibiting a hypersensitive response after inoculation, and is moderately susceptible to P. medicaginis. Among the most strongly up‐regulated genes in all three interactions were those encoding a hevein‐like protein, thaumatin‐like protein (TLP) and members of the pathogenesis response (PR)10 family. Transcripts of genes for enzymes in the phenylpropanoid pathway leading to the production of isoflavonoid phytoalexins increased dramatically in response to inoculation with the foliar pathogens. In P. medicaginis‐inoculated roots, transcripts of genes in the phenylpropanoid pathway peaked at 5 days post‐inoculation, when symptoms became visible. Transcript accumulation of three PR10 family members, a TLP and chalcone synthase (CHS) was assessed in M. truncatula genotype R108 plants. The R108 plants are resistant to C. trifolii and moderately susceptible to E. pisi and P. medicaginis. Transcript accumulation paralleled the stages of pathogen development. To evaluate the role of a TLP, a PR10 family member and CHS in disease resistance, transgenic R108 plants containing interfering RNA (RNAi) constructs were produced. Reduced expression of PR10 and TLP had no effect on the disease phenotype, whereas reduced expression of CHS resulted in increased susceptibility to necrotrophic pathogens.
INTRODUCTION
The infection of plants with virulent pathogens sets in motion a cascade of defence responses. Sequencing of expressed sequence tag (EST) libraries from pathogen‐inoculated or elicitor‐treated plants and microarray transcript analyses have enabled the elucidation of genome‐wide gene expression changes associated with defence (Ameline‐Torregrosa et al., 2006). Among the genes that are strongly up‐regulated in many plant species after infection with viruses, bacteria, fungi and oomycetes are the pathogenesis response (PR) protein genes (van Loon et al., 2006). Although these genes and the proteins they encode have been the subject of many investigations, an understanding of their regulation and role in plant–microbe interactions is still rudimentary.
The PR proteins are currently divided into 17 classes (van Loon et al., 2006). Originally defined as proteins that were expressed only on pathogen infection, in‐depth investigations of genes in several classes have found that some family members are expressed in one or more plant parts during the development and growth of healthy plants, and their expression is enhanced on pathogen infection. Several classes of PR protein genes are expressed in most plant–pathogen interactions, whereas others are more specific. In certain pathogen interactions with Arabidopsis, the expression of several classes of PR protein genes is associated with elaborate and complex defence response networks (Thatcher et al., 2005). Salicylic acid (SA)‐dependent pathways are triggered in response to biotrophic pathogens, leading to the induction of the PR protein genes PR1, PR2 and PR5. PR1 has an unknown biochemical property, whereas PR2 has β‐1,3‐glucanase activity and PR5 proteins have similarity to a sweet‐tasting seed protein called thaumatin (van Loon et al., 2006). In contrast, infection by necrotrophs elicits jasmonic acid (JA)‐ and ethylene‐dependent pathways and the up‐regulation of defensin, PR8 (CHI‐B, type III chitinase) and PR13 (thionin‐like protein) (Thatcher et al., 2005). However, the relationship between the type of pathogen, defence pathway and defence responses defined in Arabidopsis may not be totally representative of defence responses in other plant families. In particular, defence responses appear to be divergent in legumes, which establish mutualistic relationships with nitrogen‐fixing bacteria and mycorrhizal fungi.
Legumes are a rich source of several subclasses of PR10 protein genes. In general, PR10 proteins are acidic, 15–18 kDa, intracellular and cytoplasmic (Liu and Ekramoddoullah, 2006). PR10 proteins are constitutively expressed in roots and accumulate in roots and other organs in response to biotic and abiotic stresses and wounding. Their expression is regulated by SA, JA and abscisic acid (ABA) and they are able to bind to cytokinins, brassinosteroids, fatty acids and flavonoids. Proteins from the PR10, PR10.1, monocot and solanaceous groups have been shown to have ribonuclease activity (Bantignies et al., 2000; Chadha and Das, 2006; Kim et al., 2008; Park et al., 2004; Srivastava et al., 2006a). The peanut PR10 protein, which clusters with PR10.1 proteins, has been demonstrated to have antifungal activity in vitro (Chadha and Das, 2006). Interestingly, Arabidopsis does not appear to express any PR10‐like proteins (Liu and Ekramoddoullah, 2006).
Transcript and protein profiling experiments have provided evidence for the up‐regulation of several PR10 genes in Medicago truncatula in response to diverse pathogens. Inoculation with the powdery mildew fungus Erysiphe pisi resulted in the strong up‐regulation of PR10 and PR10.1 protein genes in both susceptible and resistant plants (Foster‐Hartnett et al., 2007). The up‐regulation of specific PR10 genes was also observed in the resistance response of M. truncatula to Colletotrichum trifolii (Torregrosa et al., 2004). Genes from PR10, PR10.1 and PR10.2 families were strongly up‐regulated in both compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae (Bozsóet al., 2009). PR10‐like proteins have been observed to accumulate in M. truncatula roots after inoculation with the oomycete pathogen Aphanomyces euteiches, but the increased abundance of several PR10 proteins was specifically associated with a susceptible reaction (Colditz et al., 2005). Silencing of a PR10 protein resulted in the accumulation of PR5b and reduced susceptibility to A. euteiches (Colditz et al., 2007), suggesting that PR10 proteins facilitate disease, and that PR5b plays an important role in reducing pathogen damage.
The PR5 proteins are members of the thaumatin‐like proteins (TLPs), which occur in a wide range of organisms, including fungi, nematodes and insects, as well as plants (Liu et al., 2010). In some plant species, TLPs are abundant; there are at least 28 family members in Arabidopsis and 31 in rice. However, in legumes, the TLP families are smaller and little is known about their expression patterns or roles. In general, TLPs accumulate to high concentrations in plants after inoculation with pathogens, treatment with SA, JA or ethylene, and after wounding or cold stress. Purified TLPs have growth inhibitory activity against fungal and oomycete plant pathogens in vitro and over‐expression of certain TLPs enhances disease resistance, cold tolerance and/or salt tolerance in transgenic plants (Liu et al., 2010; Velazhahan et al., 1999). The mechanism of antifungal activity is thought to be related to the disruption of cell walls, because several TLPs have β‐1,3‐glucanase and glucan‐binding activity (Liu et al., 2010).
Legumes utilize flavonoid compounds, notably isoflavones and isoflavanones, which are not found in Arabidopsis, in defence against pathogens and as signal molecules (Dixon et al., 2002). A number of phenylpropanoid compounds have antimicrobial activity and are important in restricting pathogen growth and disease symptoms (He and Dixon, 2000; O'Neill and Saunders, 1994). Transcript profiling studies in soybean found rapid and strong up‐regulation of the genes encoding enzymes in the phenylpropanoid pathway in response to bacterial (Zabala et al., 2006; Zou et al., 2005), fungal (Iqbal et al., 2005; van de Mortel et al., 2007) and oomycete (Moy et al., 2004) pathogens. In general, transcript accumulation of phenylpropanoid pathway genes in soybean is higher and more rapid in incompatible interactions than compatible interactions. Similar patterns of gene expression were observed in M. truncatula in response to pathogen infection (Foster‐Hartnett et al., 2007; Torregrosa et al., 2004). Flavonoids have multiple roles in the initiation of effective symbiotic interactions with rhizobia (Cooper, 2004). In M. truncatula, transcript profiling following inoculation with Sinorhizobium meliloti indicated that the expression of phenylpropanoid pathway genes is highly regulated during nodule initiation (Lohar et al., 2006). Silencing of chalcone synthase (CHS), the first committed step in flavonoid biosynthesis, resulted in almost complete loss of flavonoid compounds from roots and prevented nodule formation (Wasson et al., 2006).
The unravelling of the roles of individual PR protein and phenylpropanoid pathway genes is confounded by the presence of multiple family members in a single species. We utilized a custom microarray to identify genes up‐regulated in M. truncatula in response to three pathogens, C. trifolii (anthracnose), E. pisi (powdery mildew) and Phytophthora medicaginis (Phytophthora root rot), to identify pathogen‐specific or organ‐specific responses, as well as responses common to all pathogenic interactions. Following these experiments, we focused on the measurement of gene expression of three PR10 family members, a TLP and CHS in M. truncatula cv. Jemalong genotype A17, a well‐characterized genotype, and in M. truncatula genotype R108, a genotype that is easily transformed. Finally, we evaluated disease resistance in transgenic M. truncatula R108 plants expressing interfering RNA (RNAi) for a PR10.1, TLP and CHS gene.
RESULTS
Microarray analysis of pathogen‐infected M. truncatula A17 plants
The cDNA set used to prepare microarrays contained a total of 1152 clones (GEO accession GLP2929) selected by sequence analysis of M. truncatula ESTs to represent genes that play roles in plant–microbe interactions, symbiosis, disease response and resistance, signal transduction pathways and primary metabolism. Gene expression profiles were obtained from three plant–pathogen interactions using RNA from M. truncatula cv. Jemalong genotype A17. The A17 genotype is resistant to E. pisi and C. trifolii and exhibits a rapid hypersensitive response after inoculation that halts pathogen development after spore germination. No visible symptoms occur on leaves after E. pisi inoculation, but small necrotic flecks are seen on cotyledons several days after inoculation with C. trifolii. The roots of A17 plants are moderately susceptible to P. medicaginis. The pathogen completes its life cycle on A17 roots, but symptoms are limited to small necrotic lesions and rarely does infection result in plant death. RNA samples from mock‐inoculated and infected plants were extracted and labelled in parallel from E. pisi‐inoculated leaves harvested 36 h after inoculation, at the onset of cell death associated with a hypersensitive response, from C. trifolii‐inoculated cotyledons at 10 days after inoculation (dai), which showed necrotic damage from the pathogen, and from roots of P. medicaginis‐inoculated plants at 10 dai. Significance analysis of microarrays (SAM) (Tusher et al., 2001) was used to identify genes that showed a statistically significant change in expression on infection. Genes with expression ratios above 1.2 or below 0.8 were considered to be up‐ or down‐regulated, respectively (Tables S1 and S2, see Supporting Information).
A striking pattern from the analysis of these data was the coordinated up‐regulation of several groups of genes in response to all three pathogens. This expression pattern was remarkable in that they occurred in different organs (leaves, cotyledons and roots), in early and late stages of interactions with the pathogens, and in both compatible and incompatible host–pathogen interactions. Among the most strongly up‐regulated genes in interactions with all three pathogens were a TLP (TC113538), PR4A (a hevein‐like protein; TC125088) and several members of the PR10 family (TC131880, TC117333, TC122461). In both the C. trifolii and P. medicaginis interactions, the up‐regulation of CHS genes (TC117768, TC116581) and other genes involved in isoflavonoid production was observed. In addition, strong up‐regulation of genes involved in flavonoid production was observed at 12 h post‐inoculation (hpi) in interactions of M. truncatula lines with E. pisi (Foster‐Hartnett et al., 2007).
A time course study of the P. medicaginis interaction with M. truncatula A17 was performed to further our understanding of the genes expressed with increasing symptom development. The pathogen completes its life cycle on roots of A17 plants, but symptoms are limited to small lesions. We identified 179 genes that changed in expression significantly throughout the time course (Table S2). At 1 dai, when no symptoms were visible, 37 genes were up‐regulated, most encoding enzymes involved in secondary metabolism, particularly enzymes in the phenylpropanoid and isoflavonoid pathways, defence, cell wall modification, and ethylene synthesis and response. At 2 dai, in addition to many of the genes up‐regulated at 1 dai, a TLP and PR4A were strongly expressed, and genes encoding a berberine bridge enzyme, peroxidases and a cysteine protease inhibitor were up‐regulated. The 5‐dai sampling point marked a distinct change in gene expression accompanied by the presence of necrotic symptoms. The highest level of expression of defence response‐related genes occurred at this time point and there was a marked up‐regulation of genes involved in primary metabolism. The maximum number of up‐regulated and down‐regulated genes occurred at 10 dai, when severe necrotic symptoms were observed. The prominent co‐expression of TLP, PR10 family members and CHS genes in all three pathogen interactions led us to conduct a deeper investigation of their expression patterns and roles in defence reactions.
PR10, TLP and CHS expression in uninfected M. truncatula
Alignment and phylogenetic analysis of the predicted PR10 proteins from legumes identified three related clusters (Fig. 1): a canonical PR10 group, a PR10.1 subclass represented by the Pisum sativum ABR17 gene (Z15128) and a PR10.2 subclass represented by the Pisum sativum ABR18 gene (Z15127). No legume sequences clustered with the large pollen allergen group of PR10‐like sequences represented by the birch Betv1 gene (CAA96547), the Solanaceae PR10 subclass or parsley PR10 subclass. The three legume groups are well represented in the DFCI Medicago Gene Index version 9 (http://compbio.dfci.harvard.edu/tgi/cgi‐bin/tgi/gimain.pl?gudb=medicago). The PR10 and PR10.1 family members are expressed in roots and in response to pathogens, cell wall elicitors, mycorrhizal fungi, S. meliloti and abiotic stress. PR10.2 family members are expressed in response to pathogens, symbionts and treatment with JA. Based on their expression patterns after inoculation with pathogens, we selected several family members for further investigation: TC131880, representing the canonical PR10 group; TC122461, representing the PR10.1 group; and TC123318, representing the PR10.2 group.
Figure 1.
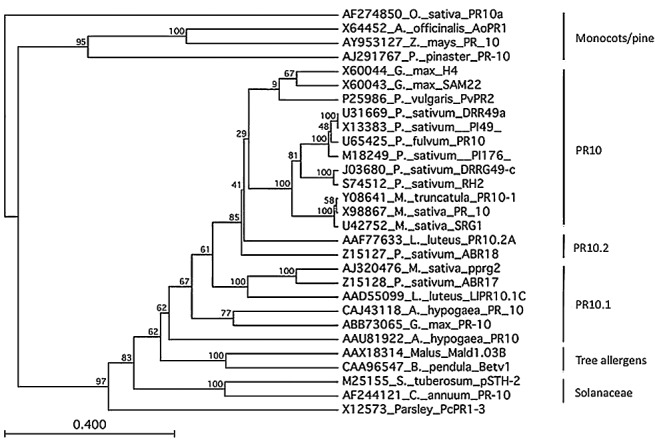
Phylogenetic tree based on the alignment of the deduced amino acid sequences of pathogenesis response (PR)10 proteins from legumes and representative members of the monocot/pine, Solanaceae and tree allergen groups. Protein sequences were aligned with Clustal W. Based on the alignment, a phylogenetic tree was constructed by the neighbour‐joining method. The parsley PR1‐3 protein (X12573; PcPR1‐3) is included as the outgroup. Bootstrap values are given at branch points. The scale indicates genetic distance proportional to the amino acid substitutions per site. Sequence abbreviations of PR10 proteins are given with GenBank accession numbers in parentheses as follows: Arachis hypogaea PR10 (AAU81922, CAJ43118), Asparagus officinalis AoPR1 (X64452), Betula pendula Betv1 (CAA96547), Capsicum annuum PR10 (AF244121), Glycine max H4 (X60044), G. max PR10 (ABB73065), G. max SAM22 (X60043), Lupinus luteus LlPR10.1C (AAD55099), L. luteus PR10.2 (AAF77633), Malus×domestica Mald1.03B (AAX18314), Medicago sativa pprg2 (AJ320476), M. sativa PR10 (X98867), M. sativa SRG1 (U42752), M. truncatula PR10‐1 (Y08641), Oryza sativa PR10a (AF274850), Petroselinum cripsum PcRR1‐3 (X12573), Phaseolus vulgaris PvPR2 (P25986), Pinus pinaster PR‐10 (AJ291767), Pisum fulvum PR10 (U65425), P. sativum ABR17 (Z15128), P. sativum ABR18 (Z15127), P. sativum DRR49a (U31669), P. sativum DRRG49‐c (J03680), P. sativum PI49 (X13383), P. sativum PI176 (M18249), P. sativum RH2 (S74512), Solanum tuberosum pSTH‐2 (M25155), Zea mays PR‐10 (AY953127).
There are 15 M. truncatula tentative consensus sequences (TCs) with sequence similarity to thaumatin; however, only two, with limited sequence similarity to each other, appear to be induced by pathogen infection. We selected TC113538 for additional characterization, as it is expressed in response to diverse pathogens, insect herbivory and abiotic stress, over TC116203, which does not exhibit strong up‐regulation by pathogen infection.
There are 31 M. truncatula TCs with sequence similarity to CHS, which cluster into six groups based on their similarity to the M. sativa proteins MsCHS2, MsCHS4, MsCHS8 and MsCHS9 (Dixon et al., 2002) and Pisum sativum proteins, all of which contain specific members (TCs) that are up‐regulated with pathogen infection. We designed real‐time polymerase chain reaction (PCR) primers (Table S3, see Supporting Information) based on an MsCHS2 family member (AW776018; TC118157) that will also amplify transcripts from other MsCHS2 family members and some members from MsCHS4 and MsCHS8.
Quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) assays were performed using RNA extracted from healthy 3‐week‐old plants of M. truncatula A17 and R108. As shown in Table 1, the assays detected transcripts of the selected PR10 family members, TLP and CHS in foliar and root tissues of M. truncatula R108. Root‐enhanced expression was observed for PR10, PR10.1, PR10.2 and CHS, whereas foliage‐enhanced expression was observed for TLP. Similar patterns were observed for each gene in foliar and root tissues of M. truncatula A17 (data not shown).
Table 1.
Transcript accumulation of defence response genes in healthy tissues of 3‐week‐old Medicago truncatula R108 relative to transcripts in leaves.
| Average relative expression (SD) | |||||
|---|---|---|---|---|---|
| PR10 | PR10.1 | PR10.2 | TLP | CHS | |
| Root | 4.2 (1.0) | 2.2 (0.1) | 4.2 (1.0) | 0.4 (0.6) | 5.3 (2.2) |
| Leaf | 1 | 1 | 1 | 1 | 1 |
| Stem | 0.4 (0.06) | 1.2 (0.03) | 0.5 (0.09) | 0.4 (0.2) | 0.4 (0.1) |
| Petiole | 0.8 (0.3) | 0.9 (0.07) | 0.6 (0.2) | 1.7 (1.0) | 0.5 (0.1) |
Relative expression (fold difference) of five genes was determined by quantitative reverse transcriptase‐polymerase chain reaction assays using actin as the endogenous reference. Values are the average and standard deviation (SD) as determined by the ΔΔC t method (n= 3). Primers used are listed in Table S3.
CHS, chalcone synthase; PR, pathogenesis response; TLP, thaumatin‐like protein.
Comparison of pathogen development on M. truncatula R108 and A17
Three‐week‐old M. truncatula R108 and A17 plants were inoculated with C. trifolii race 1. Lactophenol trypan blue staining of R108 leaves at 24, 36, 48 and 72 hpi revealed that 91% of the spores had germinated by 24 hpi and an appressorium had formed. At 36 hpi, approximately 20% of the spores had produced a mycelium greater than the length of the spore; at 48 hpi, 39% of spores had developed mycelial growth. However, at 72 hpi, only 7% of the spores observed had formed mycelia. The spores were highly vacuolated, often with multiple appressoria, and the mycelium was often highly vacuolated, indicating that the germination of spores was arrested and that few, if any, germinated spores continued to develop. The apparent reduction in number of germinated spores from 48 to 72 hpi is probably a result of a lack of staining of dead spores or dead mycelia or the loss of dead spores from leaves during staining. We did not observe anthracnose symptoms, formation of acervuli or sporulation of the fungus on M. truncatula R108 plants. Thus, although some fungal development occurs after germination, R108 is resistant to the pathogen. A17 also demonstrated a resistant reaction, but the response was more rapid. By 24 hpi, 90% of spores had germinated and formed an appressorium; however, no further development was observed at later time points.
In response to inoculation with E. pisi, genotypes A17 and R108 also showed distinct phenotypes. R108 is moderately susceptible to E. pisi, whereas A17 is highly resistant. Staining the leaves of R108 inoculated with E. pisi at 24 hpi revealed that 71% of spores had germinated and a mycelium greater than the length of the spore had developed. At 48 hpi, some host epidermal cells demonstrated a hypersensitive response characterized by cell browning and collapse; however, approximately 78% of the spores continued to develop a branched mycelium. By 6 dai, mycelium had colonized leaflets and the fungus had begun to sporulate. However, the amount of mycelium on leaf surfaces of R108 plants was less than that observed on accession DZA315.16, which is highly susceptible to powdery mildew (Ameline‐Torregrosa et al., 2008; Foster‐Hartnett et al., 2007). In contrast, only approximately 3% of spores inoculated onto A17 leaves had formed a mycelium greater than the length of the spore at 48 hpi, and many epidermal cells demonstrated a hypersensitive response. We did not observe sporulation of E. pisi on A17.
We compared the responses of A17 and R108 to inoculation with P. medicaginis using a quantitative real‐time PCR assay to measure the amount of pathogen DNA in individual root systems. Both genotypes are moderately susceptible and supported the growth of P. medicaginis; however, larger amounts of pathogen DNA were extracted from R108 than from A17 at 4 and 6 dai (Fig. 2). The pathogen can complete its life cycle on both A17 and R108 plants. Symptoms were generally more severe on R108.
Figure 2.
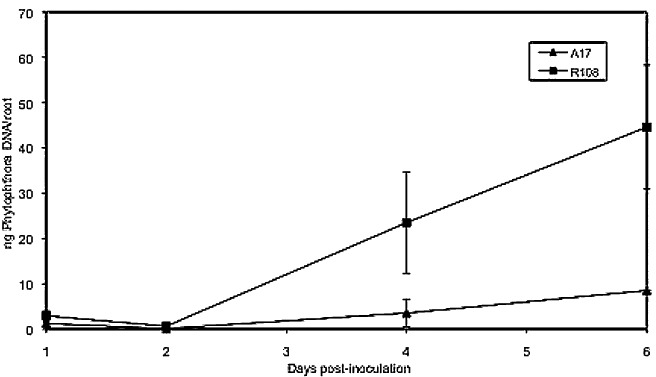
Phytophthora medicaginis DNA in roots of Medicago truncatula A17 and R108 as measured by quantitative real‐time polymerase chain reaction. Bars indicate standard deviation (n= 6).
PR10, TLP and CHS expression in pathogen‐infected M. truncatula R108
Transcript accumulation in leaves of R108 plants was measured using quantitative RT‐PCR assays after inoculation with C. trifolii. A biphasic pattern of gene expression was observed that paralleled fungal development. This pattern was observed in three independent experiments; representative results of one experiment are shown in Fig. 3a. Up‐regulation of all assayed genes compared with the mock‐inoculated control was observed at 12 hpi, corresponding to fungal germination and appressorium development. Expression of PR10 increased over 50‐fold. Transcript accumulation of all genes, except PR10.2, declined by 24 hpi, but was elevated at 48 hpi, with CHS showing the largest increase. This pattern of expression is probably a response to the development of some fungal mycelia by this time point. Transcripts of PR10 remained elevated to 72 hpi, when all fungal development had arrested, whereas the transcripts of the other genes were close to the levels in mock‐inoculated controls.
Figure 3.
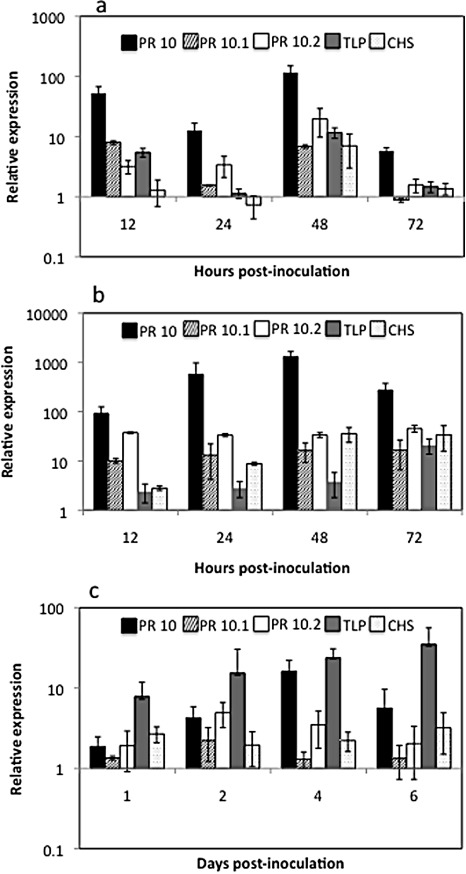
Relative expression of pathogenesis response (PR)10, PR10.1, PR10.2, thaumatin‐like protein (TLP) and chalcone synthase (CHS) in Medicago truncatula R108 after pathogen inoculation by quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR). Values are the average fold difference of inoculated plants relative to mock‐inoculated plants as determined by the ΔΔC t method. Bars indicate standard deviation (n= 3). (a) Relative gene expression in leaves inoculated with Colletotrichum trifolii race 1. (b) Relative gene expression in leaves inoculated with Erysiphe pisi. (c) Relative gene expression in roots of plants inoculated with Phytophthora medicaginis.
A different pattern of gene expression was observed in the moderately susceptible interaction of R108 plants inoculated with E. pisi. All genes assayed were up‐regulated at 12 hpi and transcript accumulation remained stable or increased for all genes to 48 hpi, with PR10 transcripts accumulating more than 1000‐fold over mock‐inoculated controls (Fig. 3b). Transcripts of TLP and CHS increased over time and remained elevated to 72 hpi, with CHS transcripts 34‐fold greater and TLP transcripts 21‐fold greater than in mock‐inoculated controls. The sustained transcript accumulation paralleled sustained pathogen growth on inoculated leaves. This pattern of transcript accumulation was consistent in repeated experiments, although fold differences were higher in experiments in which higher inoculum density occurred. The results from one experiment are shown in Fig. 3b.
The inoculation of roots of R108, which is moderately susceptible to P. medicaginis, resulted in the accumulation of all defence response genes assayed (Fig. 3c). However, the pattern of expression was distinct from the response to foliar pathogens. Although PR10 family members predominated in infected foliar tissues, TLP predominated in infected roots. PR10 and PR10.1 reached a maximum of 16‐fold and 10‐fold greater than the control, respectively, at 4 dpi. Accumulation of TLP increased with time, reaching a maximum of 35‐fold greater than the control at 6 dpi. The accumulation of CHS was constant over the course of infection at two‐ to three‐fold greater than the control. Similar patterns of transcript accumulation were observed in repeated experiments, with results from one experiment shown in Fig. 3c. In all experiments, sustained gene expression occurred in R108 with sustained pathogen development.
Construction and identification of PR10, TLP and CHS RNAi lines
Transgenic R108 plants were produced by Agrobacterium tumefaciens‐mediated transformation with constructs to produce RNAi for PR10.1 (AW560467), TLP (AW560163) and CHS (AW207939). Amplification of the nptII selectable marker gene and the insert encoding the hairpin RNA (hpRNA) in each vector identified eight T0 plants with the PR10.1 construct, seven with the TLP construct and two with the CHS construct. Leaves of T1 and selected T2 plants were assayed for the expression of the target genes using qRT‐PCR. Of the plants with the PR10.1 construct, four T1 lines had plants with a reduction in accumulation of PR10.1 transcripts compared with vector control plants. The accumulation of PR10.1 transcripts ranged from 2% to 34% of the PR10.1 transcripts in vector control plants. Interestingly, these plants showed enhanced amounts of TLP transcripts two‐ to four‐fold greater than vector control plants. Of the plants with the TLP construct, four T1 lines had plants with a reduction in the accumulation of TLP transcripts, with 0.1%–39% of the TLP transcripts measured in vector control plants. The down‐regulation of TLP did not result in the altered expression of PR10 genes. The production of seed from plants with the CHS construct was difficult as the majority of plants died before the seed matured, even when grown in controlled growth chamber conditions. From the original two T0 plants, only 18 seeds were produced from one plant and three from the other. Of these, only a single plant survived to produce viable seed. As a result of high sequence conservation among CHS genes, it was not possible to design gene‐specific primers to specifically quantify transcripts corresponding to the CHS gene from which the RNAi vector was constructed. Thus, the expression of multiple CHS family members was measured. The reduced fitness observed for plants with the CHS RNAi construct suggested that plants were affected by the hpRNA derived from the construct. However, CHS transcript accumulation in leaves from the RNAi line was only slightly reduced to 75% of the CHS transcripts found in leaves of vector control plants.
Transcript and phenotypic analysis of RNAi lines
T2 plants from lines showing the greatest down‐regulation in the target gene in foliar tissues were inoculated with C. trifolii to measure transcript accumulation after pathogen infection. As shown in Table 2, plants from three lines with the PR10.1 hpRNA construct (ABR20, ABR21 and ABR42) exhibited greatly reduced levels of PR10.1 compared with vector control plants before inoculation (Day 0). At 2 dai, transcripts in inoculated RNAi plants were compared with those in mock‐inoculated vector control plants. The average relative expression of PR10.1 increased in RNAi lines after C. trifolii inoculation, but was lower than in vector control plants. Thus, the PR10.1 hpRNA was effective in down‐regulating PR10.1 gene expression in noninoculated plants and, to a lesser degree, in inoculated plants.
Table 2.
Relative expression of PR10.1, TLP and CHS in transgenic Medicago truncatula R108 expressing hairpin RNAs (hpRNAs) after inoculation with Colletotrichum trifolii.
| Line | Day 0* | Day 2† |
|---|---|---|
| Average relative expression of PR10.1 (SD) | ||
| ABR20 | 0.1 (0.05) | 0.5 (0.2) |
| ABR21 | 0.09 (0.01) | 0.6 (0.3) |
| ABR42 | 0.03 (0.06) | 0.7 (0.3) |
| Average relative expression of TLP (SD) | ||
| TLP55 | 0.1 (0.1) | 0.03 (0.02) |
| TLP56 | 0.4 (0.3) | 1.8 (0.6) |
| TLP71 | 0.03 (0.02) | 0.01 (0.005) |
| Average relative expression of CHS (SD) | ||
| CHS11 | 0.7 (0.1) | 1.4 (0.8) |
Relative expression (fold difference) was determined by quantitative reverse transcriptase‐polymerase chain reaction assays in leaves of noninoculated 3‐week‐old plants and 2 days after inoculation with a conidial suspension of Colletotrichum trifolii. Values are the average and standard deviation (SD) as determined by the ΔΔC t method (n= 3).
Fold difference compared with vector control plants before inoculation.
Fold difference compared with inoculated vector control plants, 2 days post‐inoculation.
CHS, chalcone synthase; PR, pathogenesis response; TLP, thaumatin‐like protein.
In contrast, the TLP hpRNA appeared to be effective in down‐regulating gene expression in both mock‐inoculated and inoculated plants (Table 2). At Day 0, plants from three RNAi lines (TLP55, TLP56 and TLP71) showed reduced accumulation of TLP transcripts compared with the vector control. At 2 dai, little to no increase in TLP transcripts occurred with inoculation in lines TLP55 and TLP71.
Plants from RNAi line CHS11 with the CHS hpRNA showed a 30% reduction in CHS transcripts compared with the vector control at Day 0 (Table 2). After inoculation, the accumulation of CHS transcripts was similar in RNAi and vector control lines.
To determine the effect of hpRNAs on resistance to C. trifolii infection, a detached leaf assay was used. Spot inoculation facilitated microscopic observations of spore development after inoculation. At 48 hpi, leaflets were stained and the number of spores arrested in development after the formation of an appressorium and those forming a mycelium were counted. The proportions of spores arrested in development were similar for the resistant lines R108, A17 and the vector control line, with 74%, 69% and 63% spores arrested, respectively (Table 3). The PR10.1 and TLP hpRNAs did not affect the proportion of arrested spores. However, only 22% of the spores inoculated on line CHS11 with the CHS hpRNA were arrested in development and there was extensive mycelium development (Fig. 4). Thus, CHS hpRNA appeared to affect defence responses such that the plants were much more susceptible to C. trifolii infection.
Table 3.
Pathogen development in Medicago truncatula R108 plants expressing hairpin RNA (hpRNA) for PR10.1, TLP or CHS.
| Colletotrichum trifolii | Erysiphe pisi | Phytophthora medicaginis | |
|---|---|---|---|
| Line | Percentage spores arrested | ng DNA/root (SD) | |
| ABR20 | 68 | 37 | 2.2 (2.8) |
| ABR21 | 75 | 19 | 7.9 (7.1) |
| TLP55 | 69 | 38 | 4.3 (9.8) |
| TLP56 | 73 | 16 | 4.1 (4.9) |
| CHS11 | 22* | 23 | 63.3 (22.2)† |
| R108 | 76 | 22 | 10.8 (10.9) |
| A17 | 69 | 97 | 2.9 (3.4) |
| Vector control | 63 | 34 | 8.1 (7.0) |
The percentage of Colletotrichum trifolii and Erysiphe pisi spores arrested in development (not forming a mycelium) after germination on leaves and the amount of Phytophthora medicaginis DNA in seedling roots of transgenic and control lines were measured.
Plants from each line were inoculated to test the effect of gene down‐regulation on disease resistance. Lines ABR20 and ABR21 contain an RNAi construct for down‐regulation of PR10.1, lines TLP55 and TLP56 contain an RNAi construct for down‐regulation of TLP, and line CHS11 contains an RNAi construct for down‐regulation of CHS. Line R108 and the vector control are resistant to C. trifolii and moderately susceptible to E. pisi and P. medicaginis. Line A17 is resistant to C. trifolii and E. pisi, and moderately susceptible to P. medicaginis. Detached leaflets (n= 24) were spot inoculated with C. trifolii spores, fixed and stained to visualize mycelia by light microscopy at 2 days after inoculation. Whole plants were inoculated with E. pisi conidia and leaflets (n= 36) were stained to visualize mycelia at 2 days after inoculation. The numbers of spores that formed a mycelium or that were arrested in development after germination were counted. Seedlings (n= 10) were inoculated with P. medicaginis mycelium and the amount of pathogen DNA in each seedling root was measured 4 days after inoculation by a quantitative real‐time polymerase chain reaction assay.
Significantly different from the vector control at P= 0.0116. The proportions of arrested spores from each leaflet were square root arcsin transformed and analysed by Student's t‐test. All other transgenic lines were not significantly different from the vector control. No significant differences were observed between vector control and transgenic lines when inoculated with E. pisi.
Significantly different from the vector control at P= 0.0021 by Student's t‐test. All other transgenic lines were not significantly different from the vector control.
CHS, chalcone synthase; SD, standard deviation; TLP, thaumatin‐like protein.
Figure 4.
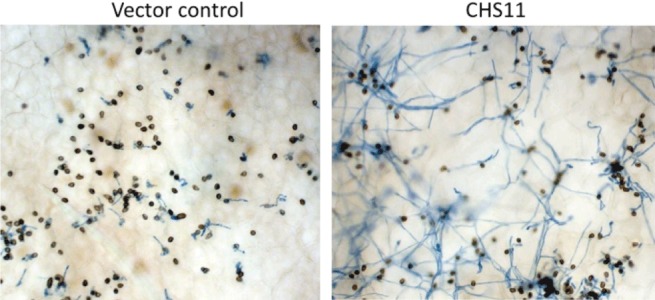
Colletotrichum trifolii development on detached leaves of vector control Medicago truncatula and line CHS11 RNAi plants. Leaflets were placed in moist chambers, spot inoculated with a 6‐µL drop of spores at 7 × 104 conidia/mL and held on the laboratory bench at 23 °C under ambient light conditions. At 48 h post‐inoculation, leaflets were fixed and stained with lactophenol trypan blue for light microscopy. Appressoria appear as brown melanized structures and mycelia stain blue. The vector control (R108 background) is resistant to C. trifolli.
To further investigate the effect of hpRNAs on disease resistance responses, entire plants from the same lines were inoculated with E. pisi and the proportion of spores arrested in development and those in which mycelial development continued were counted. As expected, plants from both R108 and the vector control were susceptible to powdery mildew, with only 22% and 34% of the spores arrested in development. The proportion of arrested spores was similar in all RNAi lines, including the line with CHS hpRNA (Table 3), indicating that the presence of hpRNAs did not increase disease susceptibility.
Ten seedlings from each line were tested for response to P. medicaginis infection, and the amount of pathogen DNA in the roots of each seedling was quantified by a Taqman assay at 4 dai. As shown in Table 3, the average amount of P. medicaginis DNA was similar in the roots of all lines, except for the line with the CHS hpRNA. In that line (CHS11), pathogen DNA per root system was significantly greater than in the root systems of vector control plants, and the roots showed increased necrosis and were less developed than vector control roots. The CHS hpRNA appears to increase susceptibility to P. medicaginis above the susceptibility observed in R108 and vector control plants.
DISCUSSION
We identified several genes coordinately up‐regulated in defence responses in diverse host–pathogen interactions in M. truncatula by microarray transcript profiling, and evaluated the expression patterns and roles in defence of selected genes by knocking down expression using RNAi. For transcript profiling, we used genotype A17, which is resistant to C. trifolii and E. pisi and moderately susceptible to P. medicaginis. A TLP (TC113538) similar to apple TLP1 was the most highly up‐regulated gene in all three interactions. Little is known about the role of TLPs in legumes. Previously, a TLP with similarity to osmotin, PR5b, was associated with resistance to A. euteiches in M. truncatula (Colditz et al., 2007). In Arabidopsis, PR5, a TLP, is induced in response to biotrophic, but not necrotrophic, pathogens. We found that TC113538 was expressed at low levels in healthy plants, with greater expression in foliar tissues, and expression was elevated in response to biotrophic (E. pisi), hemibiotrophic (C. trifolii) and necrotrophic (P. medicaginis) pathogens.
For functional analyses, we used R108, an easily transformable genotype (Trinh et al., 1998) that is resistant to C. trifolii, but is moderately susceptible to E. pisi and P. medicaginis. Silencing a gene critical to a successful pathogen defence response in R108 should result in increased susceptibility to one or more pathogens. Alternatively, if a gene facilitates disease, silencing should increase resistance. Expression of hpRNA derived from 475 bp from the 5′ region of TC113538 resulted in the suppression of TLP transcripts in healthy and C. trifolii‐inoculated leaves. The sequence used to construct hpRNA has little sequence similarity to other TLPs in M. truncatula. Thus, we expect the construct used to specifically knock down the expression of TLP corresponding to TC113538. We found that RNAi lines with small amounts of TLP transcripts showed disease phenotypes similar to vector control plants, indicating that the gene is not required for resistance, nor is it a facilitator of disease. However, it is possible that sufficient protein and sufficient activity were produced from the TLP mRNA available, as has been observed for other PR proteins (Samac and Shah, 1994), or that other proteins compensated for the loss of TLP. It has also been documented that pathogens have developed mechanisms to tolerate some PR proteins (Misas‐Villamil and van der Hoorn, 2008). Although a tolerance mechanism has not been described for TLPs, over‐expression studies of a variety of TLP genes found that enhanced disease resistance occurred for some, but not all, genes (Liu et al., 2010), suggesting differential sensitivity of pathogens to TLPs.
PR10 proteins have been found in many legumes and may play diverse roles in development and stress responses. The PR10.1 proteins were initially identified as ABA‐responsive proteins, named ABR17 and ABR18, in pea seeds, but were also found to be up‐regulated in response to diverse abiotic stresses (Barratt and Clark, 1993). Over‐expression of ABR17 increases salt, drought and cold tolerance (Dunfield et al., 2007; Srivastava et al., 2006b). Recombinant protein produced in Escherichia coli was shown to have in vitro RNase activity (Srivastava et al., 2006a). A PR10.1 protein from peanut has in vitro antifungal activity that is dependent on RNase activity (Chadha and Das, 2006). Expression of RNAi for a PR10 (TC109466) in M. truncatula hairy roots resulted in the down‐regulation of the target gene, as well as additional PR10 (TC94217) and PR10.1 (TC106355) family members (Colditz et al., 2007). Similarly, we found that the down‐regulation of a PR10.1 resulted in reduced transcript accumulation of PR10 transcripts, and confirmed that reduced expression of PR10 results in enhanced expression of TLP, as observed by Colditz et al. (2007). However, although a reduction in PR10 was associated with increased resistance to A. euteiches in hairy roots, we did not observe alteration of the disease phenotypes of PR10.1 RNAi plants inoculated with P. medicaginis, an oomycete root‐invading pathogen with a similar mechanism of pathogenesis to A. euteiches, or after inoculation with two foliar pathogens. Nonetheless, we cannot rule out that sufficient PR10 protein accumulated for activity or that the function of PR10.1 is unrelated to defence against the pathogens tested.
In contrast with the lack of phenotype change observed in TLP and PR10.1 RNAi plants, the line expressing CHS RNAi was clearly altered in disease phenotype, exhibiting increased susceptibility to C. trifolii and P. medicaginis compared with vector control plants. The difficulty in obtaining transgenic plants with CHS hpRNA and the low vigour of the few plants obtained attest to the importance of phenylpropanoid pathway products in stress tolerance and plant development in M. truncatula. The product of the CHS reaction, naringenin chalcone, is a substrate for the production of a wide array of secondary metabolites, including isoflavonoid phytoalexins, flavones, proanthocyanidins (tannins) and anthocyanins. Detailed transcript profiling experiments of phenylpropanoid pathway genes in soybean found a strong bias for the production of isoflavonoid phytoalexins and coordinate down‐regulation of anthocyanins and proanthocyanidins during a resistance response (Zabala et al., 2006). In M. truncatula leaves inoculated with Phoma medicaginis, flavonoids accumulate rapidly from undetectable amounts to high concentrations (Jasinski et al., 2009). Surprisingly, we found that RNAi for CHS increased susceptibility to necrotrophic, but not biotrophic, pathogens. Phenylpropanoid products are produced as part of the hypersensitive reaction in M. truncatula to E. pisi (Foster‐Hartnett et al., 2007), but the fungus may not come into contact with these compounds during the major biotrophic phase of the disease. Taken together, our results indicate that phenylpropanoid products, probably isoflavonoid phytoalexins, are important for limiting damage from P. medicaginis and C. trifolii.
This work demonstrates that RNAi‐expressing transgenic plants can be used to identify genes critical to defence responses. More extensive transcript profiling identified genes of unknown function that are strongly up‐regulated in incompatible interactions (Foster‐Hartnett et al., 2007). We are presently assessing the role of a set of these genes using the RNAi approach.
EXPERIMENTAL PROCEDURES
Microarray analysis of gene expression
Medicago truncatula cv. Jemalong genotype A17 seedlings were grown and maintained in growth chambers under a 12‐h photoperiod (300 µmol/m2/s) with a 21 °C/19 °C day/night regimen and 50% humidity. Seeds were sown directly in a vermiculite–sand–perlite mixture (1:2:1, v/v) in bedding plant tray inserts with 3.8‐cm × 6‐cm cells. Phytophthora medicaginis strain M2019 was grown on V8 agar medium for 10 days at room temperature. The inoculum was prepared by homogenizing mycelium and oospores from six 100‐mm plates of the pathogen in 1 L of water. Ten millilitres of inoculum were dispensed onto the growing medium in each cell with 10, 1‐week‐old seedlings. The flats were flooded for 2 days and then watered as needed thereafter. The control plants were subjected to mock inoculation with water and grown in parallel under identical environmental conditions. Roots were harvested for RNA extraction at 10 dai when approximately one‐half of the root system showed necrotic symptoms. Inoculations with C. trifolii race 1 strain 2sp2 and E. pisi were performed as described previously (Foster‐Hartnett et al., 2007). Colletotrichum trifolii‐infected and control cotyledonary leaves were harvested at 7 dai, whereas E. pisi‐infected and control mature leaves were collected at 36 hpi. Plant materials were frozen in liquid nitrogen and held at −80 °C until use. Three biological replicates were used for transcript profiling.
Total RNA extractions were performed using RNeasy Plant Mini Kits (Qiagen, Valencia, CA, USA), according to the manufacturer's procedure, starting from approximately 100 mg of frozen tissues. cDNA synthesis and fluorescent labelling were performed using the 3DNA SubmicroEX Expression Array Detection Kit from Genisphere (Hatfield, PA, USA) with a modified bulk synthesis protocol. Glass slide cDNA microarrays (GEO accession GLP2929) containing 1152 clones in triplicate were fabricated in the same manner as a larger array (Lohar et al., 2006). The 1152 clones were selected to represent genes that play roles in plant–microbe interactions, genes involved in symbiosis, disease response and resistance, signal transduction pathways and primary metabolism. The set also included organ‐specific controls, positive controls (genes known to be expressed) for microbial responses, and genes with putative functions as cell cycle/cell death regulators, regulators of cell organization and morphogenesis, transcription factors, metabolic enzymes, as well as several genes with unknown function or no hits in GenBank. Hybridizations were performed using a dye‐swap design, and washing, slide scanning and normalization were performed as described previously (Lohar et al., 2006). At least two separate slides containing three replicated spots of each probe sequence were used for the assay. A stringent threshold for acceptance was set so that the false discovery rate ranged from <1% to 1.4% (Tusher et al., 2001).
Phylogenetic analysis of legume PR10 proteins
A tblastx search was performed of the GenBank nr database with AW561042 (M. truncatula PR10) to identify PR10 family members (E < 10−20). ClustalW was used to align amino acid sequences of representative species. A phylogenetic tree was constructed by the neighbour‐joining method with 1000 bootstrap replications using CLC Sequence Viewer 6 with the parsley PcPR1‐3 (X12573) sequence used as the outgroup.
Inoculation of M. truncatula R108 and pathogen detection by microscopy and real‐time PCR assays
Seeds were mechanically scarified, imbibed at 23 °C overnight on moist filter paper and then transferred to pots containing Metro‐Mix 300 (Sungrow USA, Bellevue, WA, USA), and grown in a growth chamber with a 16‐h photoperiod at 21 °C and 8 h of darkness at 19 °C. Three‐week‐old plants were sprayed to runoff with a suspension of 1 × 106/mL C. trifolii race 1 conidiospores with 0.01% Tween20. Plants were covered with a clear plastic dome for 48 h to maintain the high relative humidity needed for spore germination. Leaves were harvested at 12, 24, 48 and 72 hpi, fixed and stained as described by Torregrosa et al. (2004) with lactophenol trypan blue to visualize germinated spores by light microscopy. Leaf samples from the same time points were also used for RNA extraction and qRT‐PCR assays as described below.
The E. pisi inoculum was produced on pea plants as described previously (Foster‐Hartnett et al., 2007). Three‐week‐old plants were inoculated with fresh conidia in a settling tower to achieve approximately 40 condia/mm2 leaf area. Leaves were harvested at 12, 24, 48 and 72 hpi and processed in the same way as for the C. trifolii‐inoculated plants for microscopy and RNA extraction.
For inoculation with P. medicaginis, germinated seeds were placed in CYG seed germination pouches (Mega International, West St. Paul, MN, USA), seven seeds per pouch, and placed in a growth chamber at 25 °C with a 16‐h photoperiod and 8 h of darkness at 21 °C. After 6 days, plants were inoculated with 7 mL of inoculum placed in the pouch trough. The inoculum consisted of comminuted mycelium from a 7‐day‐old culture of P. medicaginis M2019 grown on V8 agar (1 g culture/10 mL water). Two days later, plants were fertilized with 2 mL of 1% (w/v) Peters All Purpose Fertilizer 20‐20‐20 (J. R. Peters, Allentown, PA, USA). Water was added as needed to keep the pouches moist. Entire root systems from six plants were harvested at 1, 2, 4 and 6 dai and used for RNA extraction. To quantify the amount of P. medicaginis DNA in inoculated roots, total DNA of entire root systems was extracted using the Fast DNA Kit and FastPrep Instrument (MP Biomedicals, Solon, OH, USA) and used in a TaqMan assay as described previously (Vandemark and Barker, 2003).
qRT‐PCR assays
To measure gene expression in healthy plant tissues, leaves, stems, petioles and roots were harvested from healthy 3‐week‐old plants; leaves and roots were also harvested from plants inoculated with pathogens as described above. All plant materials were frozen immediately in liquid nitrogen and held at −80 °C. Total RNA was extracted from three replicate samples using the RNAeasy Plant Mini Kit (Qiagen) with on‐column DNase treatment. cDNA was made with 1 µg total RNA using the iScript cDNA Synthesis Kit (Bio‐Rad Laboratories, Hercules, CA, USA). PCRs were conducted in triplicate in an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA, USA) using iTaq SYBR Green Supermix with ROX (Bio‐Rad Laboratories) with the following parameters: 2 min at 95 °C, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Primer Express (Applied Biosystems) was used to design primers (Table S3). The specificity of amplification was confirmed by a single disassociation curve peak generated after the last PCR cycle. Data analysis and calculations to compare transcript accumulation data were performed using the ΔΔC t (threshold cycle) method with actin as the endogenous reference. Each gene–actin primer combination was tested for equal amplification efficiency.
Production of transgenic plants expressing RNAi
Plant transformation vectors were synthesized using DNA sequences from the 5′ proximal region of the following clones: CHS (AW207939; 279 bp), ABR17 (AW560467; 314 bp) and TLP (AW560163; 475 bp). Sequences were PCR amplified and cloned into the RNAi‐inducing pHellsgate8 (Helliwell et al., 2002). Clones were introduced into Agrobacterium tumefaciens LBA4404 by electroporation and used to transform M. truncatula R108 by the method described by Trinh et al. (1998). Regenerated plants (T0) were selected using kanamycin and screened for the presence of the transgenes by PCR. Seeds were collected from PCR‐positive plants and subsequent plants (T1) were analysed by qRT‐PCR for the expression of target genes. Seeds were collected from plants showing reduced gene expression compared with vector control plants. T2 plants homozygous for the RNAi transgene were analysed for gene expression and disease phenotype as detailed below.
Inoculation of transgenic RNAi plants and assays for pathogen development
Three‐week‐old transgenic R108 plants expressing hpRNA for PR10.1 (ABR20, ABR21, ABR42), TLP (TLP55, TLP56, TLP71) or CHS (CHS11), and vector control plants, were sprayed to runoff with a suspension of 1 × 106/mL C. trifolii conidiospores with 0.01% Tween20. Leaves were harvested immediately before inoculation and at 48 hpi. RNA was extracted from replicate samples and used in qRT‐PCR assays for the expression of PR10.1, TLP and CHS as described above.
Transgenic and control plants were inoculated with each pathogen to determine whether hpRNA expression affected pathogen development. For C. trifolii inoculation, detached leaflets of 3‐week‐old plants were placed on moist filter paper in Petri dishes and the adaxial surface spot was inoculated with a 6‐µL droplet of spores at 7 × 104 conidia/mL with 0.01% Tween20. Plates were sealed with parafilm and held on the laboratory bench at 23 °C under ambient light conditions. Six plants were assayed from each line using four leaflets from each plant. At 48 hpi, leaflets were fixed, stained with lactophenol trypan blue for light microscopy (Torregrosa et al., 2004) and the number of spores arrested in development after the formation of an appressorium or those forming a mycelium were counted. Approximately 100 spores were evaluated from each leaflet.
For E. pisi inoculation, 3‐week‐old plants were inoculated in a settling tower to achieve approximately four conidia/mm2. Six plants were assayed from each line and six leaflets from each plant were fixed and stained at 2 dai for light microscopy. The observed proportion of arrested spores on each leaflet was square root arcsin converted and analysed by Student's t‐test.
Ten seedlings from each line were inoculated with P. medicaginis as described above using CYG pouches. Roots were harvested at 4 dai and frozen immediately in liquid nitrogen. The amount of P. medicaginis per root was quantified as described above and the data were analysed by Student's t‐test.
Supporting information
Table S1 Differentially expressed genes significant in more than one interaction.
Table S2 Differentially expressed genes significant at more than one time point of the time course study with Phytophthora medicaginis.
Table S3 Primers used in real‐time polymerase chain reaction assays.
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This research was supported by the NSF‐Plant Genome Research awards #0196179 and #0110206. We thank Melinda Dornbusch for excellent technical assistance and Natasha Sharopova for microarray analysis. We acknowledge the Computational Genetics Laboratory of the University of Minnesota Supercomputing Institute for computational support. This article is a joint contribution from the Plant Science Research Unit, United States Department of Agriculture. Agricultural Research Service (USDA‐ARS) and the Minnesota Agricultural Experiment Station. Mention of a trademark, proprietary product or vendor does not constitute a guarantee or warranty of the product by the USDA, and does not imply its approval to the exclusion of other products and vendors that might also be suitable.
REFERENCES
- Ameline‐Torregrosa, C. , Dumas, B. , Krajinksi, F. , Esquerre‐Tugayé, M.‐T. and Jacquet, C. (2006) Transcriptomic approaches to unravel plant–pathogen interactions in legumes. Euphytica, 147, 25–36. [Google Scholar]
- Ameline‐Torregrosa, C. , Cazaux, M. , Danesh, D. , Chardon, F. , Cannon, S.B. , Esquerré‐Tugayé, M.‐T. , Dumas, B. , Young, N.D. , Samac, D.A. , Huguet, T. and Jacquet, C. (2008) Genetic dissection of resistance to anthracnose and powdery mildew in Medicago truncatula . Mol. Plant–Microbe Interact. 21, 61–69. [DOI] [PubMed] [Google Scholar]
- Bantignies, B. , Séguin, J. , Muzac, I. , Dédaldéchamp, F. , Gulick, P. and Ibrahim, R. (2000) Direct evidence for ribonucleolytic activity of a PR‐10‐like protein from white lupin roots. Plant Mol. Biol. 42, 871–881. [DOI] [PubMed] [Google Scholar]
- Barratt, D.H.P. and Clark, J.A. (1993) A stress‐induced, developmentally regulated, highly polymorphic protein family in Pisum sativum L. Planta, 191, 7–17. [Google Scholar]
- Bozsó, Z. , Maunoury, N. , Szatmari, A. , Mergaert, P. , Ott, P.G. , Zsíros, L. , Szabó, E. , Kondorosi, E. and Klement, Z. (2009) Transcriptome analysis of a bacterially induced basal and hypersensitive response of Medicago truncatula . Plant Mol. Biol. 70, 627–646. [DOI] [PubMed] [Google Scholar]
- Chadha, P. and Das, R.H. (2006) A pathogenesis related protein, AhPR10, from peanut: an insight of its mode of antifungal activity. Planta, 225, 213–222. [DOI] [PubMed] [Google Scholar]
- Colditz, F. , Braun, H.‐P. , Jacquet, C. , Niehaus, K. and Krajinski, F. (2005) Proteomic profiling unravels insights into the molecular background underlying increased Aphanomyces euteiches‐tolerance of Medicago truncatula . Plant Mol. Biol. 59, 387–406. [DOI] [PubMed] [Google Scholar]
- Colditz, F. , Niehaus, K. and Krajinski, F. (2007) Silencing of PR‐10‐like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches . Planta, 226, 57–71. [DOI] [PubMed] [Google Scholar]
- Cooper, J.E. (2004) Multiple responses of rhizobia to flavonoids during legume root infection. Adv. Bot. Res. 41, 1–62. [Google Scholar]
- Dixon, R.A. , Achnine, L. , Kota, P. , Liu, C.‐J. , Reddy, M.S.S. and Wang, L. (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol. Plant Pathol. 3, 371–390. [DOI] [PubMed] [Google Scholar]
- Dunfield, K. , Srivastava, S. , Shah, S. and Kav, N.N.V. (2007) Constitutive expression of ABR17 cDNA enhances germination and promotes early flowering in Brassica napus . Plant Sci. 173, 521–532. [Google Scholar]
- Foster‐Hartnett, D. , Danesh, D. , Peñuela, S. , Sharopova, N. , Endre, G. , VandenBosch, K.A. , Young, N.D. and Samac, D.A. (2007) Molecular and cytological responses of Medicago truncatula to Erysiphe pisi . Mol. Plant Pathol. 8, 307–319. [DOI] [PubMed] [Google Scholar]
- He, X.Z. and Dixon, R.A. (2000) Genetic manipulation of isoflavones 7‐O‐methyltransferase enhances biosynthesis of 4′‐O‐methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell, 12, 1689–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A. , Wesley, S.V. , Wielopolska, A.J. and Waterhouse, P.M. (2002) High‐throughput vectors for efficient gene silencing in plants. Funct. Plant Biol. 29, 1217–1225. [DOI] [PubMed] [Google Scholar]
- Iqbal, M.J. , Yaegashi, S. , Ahsan, R. , Shopinski, K.L. and Lightfoot, D.A. (2005) Root response to Fusarium solani f. sp. glycines: temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor. Appl. Genet. 110, 1429–1438. [DOI] [PubMed] [Google Scholar]
- Jasinski, M. , Kachlicki, P. , Rodziewicz, P. , Figlerowicz, M. and Stobiecki, M. (2009) Changes in the profile of flavonoid accumulation in Medicago truncatula leaves during infection with fungal pathogen Phoma medicaginis . Plant Physiol. Biochem. 47, 847–853. [DOI] [PubMed] [Google Scholar]
- Kim, S.T. , Yu, S. , Kang, Y.H. , Kim, S.G. , Kim, J.‐Y. , Kim, S.‐H. and Kang, K.Y. (2008) The rice pathogen‐related protein 10 (JIOsPR10) is induced by abiotic and biotic stresses and exhibits ribonuclease activity. Plant Cell Rep. 27, 593–603. [DOI] [PubMed] [Google Scholar]
- Liu, J.‐J. and Ekramoddoullah, A.K.M. (2006) The family 10 of plant pathogenesis‐related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. 68, 3–13. [Google Scholar]
- Liu, J.‐J. , Sturrock, R. and Ekramoddoullah, A.K.M. (2010) The superfamily of thaumatin‐like proteins: its origin, evolution, and expression towards biological function. Plant Cell Rep. 29, 419–436. [DOI] [PubMed] [Google Scholar]
- Lohar, D.P. , Sharapova, N. , Endre, G. , Peñuela, S. , Samac, D.A. , Town, C. , Silverstein, K.A. and VandenBosch, K.A. (2006) Transcript analysis of early nodulation events in Medicago truncatula . Plant Physiol. 140, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Misas‐Villamil, J.C. and van der Hoorn, R.A.L. (2008) Enzyme–inhibitor interactions at the plant–pathogen interface. Curr. Opin. Plant Biol. 11, 380–388. [DOI] [PubMed] [Google Scholar]
- van de Mortel, M. , Recknor, J.C. , Graham, M.A. , Nettleton, D. , Dittman, J.D. , Nelson, R.T. , Godoy, C.V. , Abdelnoor, R.V. , Almeida, A.M.R. , Baum, T.J. and Whitham, S.A. (2007) Distinct biphasic mRNA changes in response to Asian soybean rust infection. Mol. Plant–Microbe Interact. 20, 887–899. [DOI] [PubMed] [Google Scholar]
- Moy, P. , Qutob, D. , Chapman, B.P. , Atkinson, I. and Gijzen, M. (2004) Patterns of gene expression upon infection of soybean plants by Phytophthora sojae . Mol. Plant–Microbe Interact. 17, 1051–1062. [DOI] [PubMed] [Google Scholar]
- O'Neill, N.R. and Saunders, J.A. (1994) Compatible and incompatible responses in alfalfa cotyledons to races 1 and 2 of Colletotrichum trifolii . Phytopathology, 84, 283–287. [Google Scholar]
- Park, C.‐J. , Kim, K.‐J. , Shin, R. , Park, J.M. , Shin, Y.‐C. and Paek, K.‐H. (2004) Pathogenesis‐related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 37, 186–198. [DOI] [PubMed] [Google Scholar]
- Samac, D.A. and Shah, D.M. (1994) Effect of chitinase antisense RNA expression on disease susceptibility of Arabidopsis plants. Plant Mol. Biol. 25, 587–596. [DOI] [PubMed] [Google Scholar]
- Srivastava, S. , Emery, R.J.N. , Kurepin, L.V. , Reid, D.M. , Fristensky, B. and Kav, N.N.V. (2006a) Pea PR 10.1 is a ribonuclease and its transgenic expression elevates cytokinin levels. Plant Growth Regul. 49, 17–25. [Google Scholar]
- Srivastava, A. , Rahman, M.H. , Shah, S. and Kav, N.N.V. (2006b) Constitutive expression of the pea ABA‐responsive 17 (ABR17) cDNA confers multiple stress tolerance in Arabidopsis thaliana . Plant Biotechnol. J. 4, 529–549. [DOI] [PubMed] [Google Scholar]
- Thatcher, L.F. , Anderson, J.P. and Singh, K.B. (2005) Plant defence responses: what have we learnt from Arabidopsis? Funct. Plant Biol. 32, 1–19. [DOI] [PubMed] [Google Scholar]
- Torregrosa, C. , Cluzet, S. , Fournier, J. , Huguet, T. , Gamas, P. , Prospéri, J.M. , Esquerre‐Tugayé, M.T. , Dumas, B. and Jacquet, C. (2004) Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii . Mol. Plant–Microbe Interact. 17, 909–920. [DOI] [PubMed] [Google Scholar]
- Trinh, T.H. , Ratet, P. , Kondorosi, E. , Durand, P. , Kamaté, K. , Bauer, P. and Kondorosi, A. (1998) Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp. falcata lines improved in somatic embryogenesis. Plant Cell Rep. 17, 345–355. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G. , Tibshirani, R. and Chu, G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandemark, G.J. and Barker, B.M. (2003) Quantifying Phytophthora medicaginis in susceptible and resistant alfalfa with a real‐time fluorescent PCR assay. J. Phytopathol. 151, 577–583. [Google Scholar]
- Velazhahan, R. , Datta, S.K. and Muthukrishnan, S. (1999) The PR‐5 family: thaumatin‐like proteins In: Pathogenesis‐Related Proteins in Plants (Datta S.K. and Muthukrishnan S., eds), pp. 107–129. Boca Raton, FL: CRC Press. [Google Scholar]
- Wasson, A.P. , Pellerone, F.I. and Mathesius, U. (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell, 18, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala, G. , Zou, J. , Tuteja, J. , Gonzalez, D.O. , Clough, S.J. and Vodkin, L.O. (2006) Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol. 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Rodriguez‐Zas, S. , Aldea, N. , Li, M. , Zhu, J. , Gonzalez, D.O. , Vodkin, L.O. , DeLucia, E. and Clough, S.J. (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense‐related genes and rapid HR‐specific downregulation of photosynthesis. Mol. Plant–Microbe Interact. 18, 1161–1174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Differentially expressed genes significant in more than one interaction.
Table S2 Differentially expressed genes significant at more than one time point of the time course study with Phytophthora medicaginis.
Table S3 Primers used in real‐time polymerase chain reaction assays.
Supporting info item
Supporting info item
Supporting info item


