SUMMARY
Penicillium spp. are the major postharvest pathogens of citrus fruit in Mediterranean climatic regions. The induction of natural resistance constitutes one of the most promising alternatives to avoid the environmental contamination and health problems caused by chemical fungicides. To understand the bases of the induction of resistance in citrus fruit against Penicillium digitatum, we have used a 12k citrus cDNA microarray to study transcriptional changes in the outer and inner parts of the peel (flavedo and albedo, respectively) of elicited fruits. The elicitor treatment led to an over‐representation of biological processes associated with secondary metabolism, mainly phenylpropanoids and cellular amino acid biosynthesis and methionine metabolism, and the down‐regulation of genes related to biotic and abiotic stresses. Among phenylpropanoids, we detected the over‐expression of a large subset of genes important for the synthesis of flavonoids, coumarins and lignin, especially in the internal tissue. Furthermore, these genes and those of ethylene biosynthesis showed the highest induction. The involvement of both phenylpropanoid and ethylene pathways was confirmed by examining changes in gene expression and ethylene production in elicited citrus fruit. Therefore, global results indicate that secondary metabolism, mainly phenylpropanoids, and ethylene play important roles in the induction of resistance in citrus fruit.
INTRODUCTION
In nature, plants are permanently in contact with a broad range of pathogens. However, disease is not widespread in plants and only a limited number of pathogens are capable of successfully invading a plant and causing disease. Plants have evolved an intricate and elaborate set of defensive barriers in order to protect themselves against pathogens (Mysore and Ryu, 2004). Preformed physical or chemical barriers constitutively present on the plant surface may initially stop the establishment of infection structures. Later, the recognition of the pathogen may lead to the activation of defence mechanisms, such as the hypersensitive response, increased expression of defence‐related genes, such as pathogenesis‐related (PR) genes, and the oxidative burst (Ferreira et al., 2006; Glazebrook, 2005; Jones and Dangl, 2006; Király et al., 2007). Moreover, induced resistance activates the plant's defence mechanisms, thereby enabling the plant to better restrict the growth of a pathogen on subsequent attack. Although the molecular bases of induced resistance have been studied extensively in the vegetative parts of plants (Bostock, 2005; Conrath, 2009; Durrant and Dong, 2004), our knowledge of the processes underlying the establishment of induced resistance in fruits is still very poor, in most cases being limited to single metabolites, enzymes or genes. Moreover, we cannot assume that the mechanisms operating in mature fruits are equal to those found in vegetative parts of model plants. These factors strengthen the relevance of studying the mechanisms of induced resistance in crop fruits.
Penicillium digitatum (Pers.:Fr.) Sacc. is the causal agent of green mould rot, and represents the major postharvest pathogen of citrus fruit in Mediterranean regions, accounting for up to 60%–80% of total losses as a result of fungal decay during fruit storage at ambient temperature. For many years, the control of this postharvest pathogen has mainly relied on the use of chemicals. With the current concerns about the harmful effects of synthetic fungicides on human health and the environment, there is a trend to adopt new and safer control alternatives. In citrus fruit, the induction of natural resistance constitutes one of these alternatives.
Various treatments are known to trigger induced resistance in citrus fruit against fungal infections (Ben‐Yehoshua et al., 2000), including the application of physical (Arcas et al., 2000; Droby et al., 1993; Rodov et al., 1992), chemical (2001, 2002; Venditti et al., 2005) and microbial antagonist treatments (Arras, 1996; Droby et al., 2002; Fajardo et al., 1998). Among these treatments, the highest induction of the antimicrobial phytoalexin scoparone was achieved in fruits subjected to pathogen inoculation followed by a heat treatment at 37 °C for 3 days (curing) (Ben‐Yehoshua et al., 1992; Kim et al., 1991). Infected–cured citrus fruits showed a significant reduction in the incidence of green mould (Ballester et al., 2010a). This treatment induced a higher level of resistance than a wounding–curing treatment, whereas curing alone increased the susceptibility to pathogen infection.
So far, the analysis of the molecular and physiological bases of induced resistance in citrus fruit has only been addressed at individual gene or enzyme activity levels. The application of elicitors, such as UV light, jasmonic acid (JA), β‐aminobutyric acid (BABA), wounding or brushing and hot water treatment, led to an induction of the genes coding for chitinase, β‐1,3‐glucanase, phenylalanine ammonia‐lyase (PAL) and heat‐shock proteins (Lers et al., 1998; Pavoncello et al., 2001; 2001, 2002). The induction of chitinase, β‐1,3‐glucanase, PAL and peroxidase activities has also been described in oranges subjected to different biotic or abiotic treatments that elicit induced resistance (Ballester et al., 2010a; Fajardo et al., 1998). We have shown recently that induced resistance in oranges against P. digitatum, elicited by the combination of an inoculation with the fungus followed, 1 day later, by a curing treatment (37 °C for 3 days with high relative humidity), coincided with the induction of PAL, soluble peroxidase, basic β‐1,3‐glucanase and chitinase at both gene expression and enzyme activity levels (Ballester et al., 2010a).
Ethylene may stimulate senescence, but also plays a protective role against stress conditions causing postharvest losses in citrus fruit (Lafuente and Sala, 2002; Lafuente et al., 2001; Marcos et al., 2005; Porat et al., 1999b). Its production increases in citrus fruit infected with P. digitatum, being synthesized by both the fruit and the pathogen (1985a, 1985b). Application of the ethylene inhibitor 1‐methylcyclopropene (1‐MCP) increases the susceptibility of oranges to infection by P. digitatum (Marcos et al., 2005; Porat et al., 1999b). Moreover, the fact that many genes induced in citrus fruit upon P. digitatum attack are also up‐regulated by ethylene highlights the role of this hormone in the defence response of citrus to this pathogen (González‐Candelas et al., 2010). However, it is still unknown whether this hormone is also able to induce pathogen resistance in citrus fruit.
As citrus is one of the most important and widely grown fruit crops, several genetic, genomic and proteomic tools have been quickly adopted in recent years by the citrus research community to address major challenges of this fruit crop (Talon and Gmitter, 2008). The genome sequences of both Citrus sinensis (Sweet Orange Genome Project 2010, http://www.phytozome.net/citrus.php) and Citrus clementina (Haploid Clementine Genome, International Citrus Genome Consortium, 2011, http://www.phytozome.net/clementine) have just been released. One of the strategies developed for transcriptomic analysis has been the construction of microarrays. In citrus, the first global transcriptomic study was reported by Shimada et al. (2005), who developed a citrus cDNA microarray containing 2213 independent genes, which was used for gene expression profiling during fruit development. The Spanish Citrus Functional Genomic Project (CFGP, http://bioinfo.ibmcp.upv.es/genomics/cfgpDB/) has developed different generations of cDNA microarrays containing up to 20 000 probes obtained from 53 cDNA libraries covering different tissues, developmental stages and stress conditions (Forment et al., 2005; Martinez‐Godoy et al., 2008). Two other microarrays, namely a citrus GeneChip from Affymetrix containing 30 171 probe sets representing up to 33 879 citrus transcripts based on expressed sequence tags (ESTs) obtained from several citrus species and citrus hybrids, and a citrus 22k oligoarray containing 21 495 independent ESTs from citrus species (Fujii et al., 2007), have been developed recently. Currently, additional work is being performed within various research groups using large cDNA citrus microarrays or smaller custom arrays based on subtractive cDNA libraries (Bernardi et al., 2010). These citrus microarrays have been used to study gene expression in relation to fruit development and quality (Bernardi et al., 2010; Mayuoni et al., 2011; Shimada et al., 2005) or under various biotic (Albrecht and Bowman, 2008; Gandía et al., 2007; Kim et al., 2009) and abiotic (Gimeno et al., 2009) stresses. A first transcriptomic approach with cDNA macroarrays has recently been used to unravel the molecular processes underlying the response of citrus fruit to P. digitatum infection (González‐Candelas et al., 2010). However, large‐scale gene expression studies addressing the process of induced resistance in citrus fruit against pathogen attack have not been conducted so far.
In this work, with the aim of better understanding the mechanisms underlying induced resistance in citrus fruit, we have carried out a large‐scale gene expression analysis in elicited fruits, using the 12k citrus cDNA microarray developed by the CFGP. In addition, we have examined the involvement of ethylene in the induction of resistance in citrus fruits.
RESULTS
Efficacy of elicitation of induced resistance in citrus fruits against P. digitatum
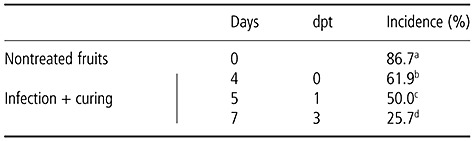
The combination of pathogen inoculation (denoted as I) followed by a curing treatment (3 days at 37 °C; denoted as C) reduced the incidence of a subsequent P. digitatum infection in oranges, with the efficacy of the treatment being dependent on the elapsed time between the curing treatment and subsequent infection (Table 1). Thus, the highest effectiveness of the infection plus curing treatment (IC) was observed when P. digitatum inoculation was conducted 3 days post‐treatment (dpt) (7 days after the start of the experiment), although, at 0 and 1 dpt (4 and 5 days after the start, respectively) the incidence of the infection was also reduced when compared with nontreated fruits.
Table 1.
Incidence (percentage of infection) of green mould disease caused by Penicillium digitatum in elicited ‘Navelate’ oranges. Wounded fruits were inoculated with 10 µL of a P. digitatum spore suspension containing 105 conidia/mL and incubated for 24 h at 20 °C and then for 3 days at 37 °C and 90%–95% relative humidity. Nontreated oranges were included as a control. At 0, 1 or 3 days post‐treatment (dpt) (4, 5 or 7 days after the beginning of the experiment, respectively) fruits were inoculated with 10 µL of a P. digitatum spore suspension containing 104 conidia/mL. Inoculation sites were 0.5 cm apart from the previous wound in the infected–cured fruits. Incidence was determined for up to 6 days of incubation at 20 °C following inoculation. The data shown correspond to 6 days post‐inoculation. Different letters indicate significant differences in the treatments according to Tukey's test with P < 0.05.
Comparative analysis of transcriptional profiles in elicited citrus fruits
A functional genomics approach employing cDNA microarrays was used to investigate the molecular responses associated with induced resistance in citrus fruits elicited by the combination of pathogen infection followed by a curing treatment. For this purpose, five different samples were analysed: nontreated ‘Navelate’ oranges (NT), fruits infected with P. digitatum during 1 day (I1) and infected–cured samples taken 4 days (IC4), 5 days (IC5) and 7 days (IC7) after the beginning of the experiment, thus corresponding to 0, 1 and 3 dpt, respectively. In all cases, three biological replicates of each sample, consisting of 10 individual fruits each, divided into two tissues, i.e. flavedo (F, outer coloured part of the peel) and albedo (A, inner white part of the peel), were used for transcriptomic analysis. Each sample was hybridized against a reference sample composed of a mix of equal amounts of RNA of all the samples included in the analysis. We used the CFGP 12k microarray, which contains 24 288 clones, corresponding to 11 241 unigenes, 77.1% of which have an Arabidopsis thaliana homologue candidate. After discarding low‐quality spot signals and normalization, 10 769 genes were analysed.
Figure 1 shows a summary of the genes differentially expressed [significant analysis of microarrays (SAM), P < 0.01] in the flavedo and albedo of fruits subjected to the elicitor treatment compared with nontreated fruits. In the flavedo, no single gene was differentially expressed 1 day after inoculation (FI1), whereas 299, 127 and 229 genes showed altered expression levels at 4, 5 and 7 days after the beginning of the experiment (FIC4, FIC5 and FIC7), respectively, when compared with nontreated fruits (FNT). 2, 3 show the genes with the highest induction (log2 > 2, Table 2) or repression (log2 < −2, Table 3) levels in the flavedo and/or albedo of elicited oranges, 5 and/or 7 days after the beginning of the experiment, which are the time points with the largest reduction in disease incidence, when compared with control samples. The most highly induced genes revealed the relevance of the phenylpropanoid pathway in elicited fruits: among the 29 most up‐regulated genes in the flavedo, 13 were involved in either phenylpropanoid metabolism or coumarin biosynthesis, including seven different O‐methyltransferases, an isoflavone reductase, a hydroxycinnamoyl transferase, two leucoanthocyanidin dioxygenases and two SRG1 proteins. Genes related to methionine and ethylene biosynthetic processes, such as 1‐aminocyclopropane‐1‐carboxylic acid oxidase (ACO), and proteins related to defence and response to stress were also induced in the flavedo (Table 2). Among the 18 most down‐regulated genes in FIC5 and/or FIC7, half were involved in responses to biotic and abiotic stresses, whereas the other half had unknown functions (Table 3). In the albedo, the expression of 521, 245 and 176 genes changed significantly in AIC4, AIC5 and AIC7, respectively, when compared with ANT (Fig. 1). Similarly to the flavedo, genes related to the biosynthesis of phenylpropanoids, coumarins and methionine were highly up‐regulated in AIC5 and/or AIC7 relative to ANT (Table 2). In addition to these genes, the expression of different PR genes was also induced in the albedo of elicited fruits. Among the 18 genes with the highest down‐regulation in the albedo of elicited fruits were cold‐regulated genes (COR15) and genes with unknown function (Table 3).
Figure 1.
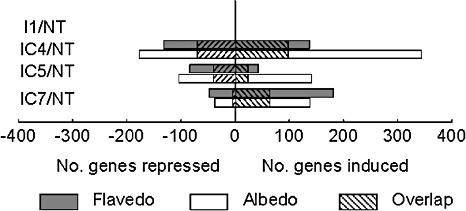
Summary of differentially expressed genes [significant analysis of microarrays (SAM), P < 0.01] in the flavedo (grey bars) and albedo (white bars) of fruits infected with Penicillium digitatum during 1 day (I1) and infected and cured fruits at 4 days (IC4), 5 days (IC5) and 7 days (IC7) after the beginning of the experiment, compared with nontreated (NT) fruits. Genes differentially expressed in both tissues are represented as striped bars.
Table 2.
Citrus genes with the highest induction level (log2 > 2) in the flavedo and/or albedo of elicited oranges 5 and/or 7 days after the beginning of the experiment. Values represent the log2 ratio of: a, I1/NT; b, IC4/NT; c, IC5/NT; d, IC7/NT. Numbers in bold indicate differential expression in the compared conditions according to significant analysis of microarrays (SAM) (P < 0.01) and log2 > 2. The symbol ‘+’ indicates no expression in NT and the symbol ‘–’ indicates expression in NT fruits but not in the compared treatment. NT, nontreated fruits; I1, infected fruits 1 day after pathogen inoculation; IC4, infected and cured fruits 4 days after the beginning of the experiment; IC5, infected and cured fruits 5 days after the beginning of the experiment; IC7, infected and cured fruits 7 days after the beginning of the experiment.
| Citrus unigene | Description | Arabidopsis homologue | Flavedo | Albedo | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | a | b | c | d | |||
| Phenylpropanoid and flavonoid biosynthetic process | ||||||||||
| aCL3343Contig1 | Caffeic acid 3‐O‐methyltransferase | AT5G54160 | 0.23 | 2.38 | 4.22 | 3.18 | 0.28 | 2.47 | 4.18 | 3.41 |
| aCL38Contig7 | Catechol O‐methyltransferase | AT5G54160 | 0.37 | 1.17 | 3.56 | 1.78 | 0.09 | 1.66 | 3.86 | 2.49 |
| aC06052D07T7 | Caffeic acid 3‐O‐methyltransferase 1 | AT5G54160 | –0.82 | 1.45 | 3.58 | 1.92 | 0.56 | 2.14 | 3.83 | 3.08 |
| aCL38Contig8 | Eugenol O‐methyltransferase | AT5G54160 | –0.41 | 2.29 | 3.43 | 3.28 | 0.74 | 2.39 | 3.79 | 4.04 |
| aC08010D03SK | Caffeic acid O‐methyltransferase | AT5G54160 | –0.53 | 1.53 | 3.41 | 2.08 | 0.03 | 1.36 | 3.36 | 2.10 |
| aC31502H09EF | SRG1 protein | AT1G17020 | 0.37 | 0.49 | 2.36 | 2.85 | 0.55 | 1.33 | 2.84 | 3.40 |
| aCL38Contig2 | Phloroglucinol O‐methyltransferase | AT5G54160 | –0.18 | 1.12 | 3.81 | – | – | 0.37 | 2.85 | – |
| aCL3152Contig1 | Hydroxycinnamoyl transferase | AT5G48930 | –0.03 | 1.14 | 2.23 | 1.51 | 0.79 | 1.67 | 2.60 | 2.27 |
| aC08017C07SK | Isoflavone reductase‐like protein | AT1G19540 | –0.79 | 1.20 | 2.37 | 0.94 | 0.36 | 1.17 | 2.34 | 1.26 |
| aC31207A03EF | Eugenol O‐methyltransferase | AT5G54160 | 0.20 | 1.11 | 2.26 | 2.02 | 0.43 | 0.95 | 1.79 | 0.85 |
| aCL5465Contig1 | SRG1 protein | AT1G17020 | 0.45 | 0.51 | 1.26 | 2.21 | 0.63 | 1.56 | 1.63 | 2.24 |
| aCL1474Contig1 | Cinnamyl alcohol dehydrogenase | AT5G19440 | 0.66 | 1.01 | 1.35 | 1.60 | 0.64 | 1.80 | 1.82 | 2.20 |
| aC31705B10EF | Eugenol O‐methyltransferase | AT5G54160 | –0.13 | 1.03 | 2.14 | 1.55 | 0.43 | 0.24 | 1.18 | 0.98 |
| Coumarin biosynthetic process | ||||||||||
| aC31108D04EF | Leucoanthocyanidin dioxygenase‐like protein | AT3G13610 | –1.21 | 3.08 | 3.77 | 3.96 | 0.49 | 3.46 | 4.36 | 4.26 |
| aCL18Contig10 | Caffeoyl‐CoA O‐methyltransferase 2 | AT4G34050 | 0.13 | 1.61 | 2.24 | 1.79 | 0.69 | 2.99 | 4.16 | 3.33 |
| acl7037Contig1 | Caffeoyl‐CoA O‐methyltransferase | AT4G34050 | 0.22 | 0.48 | 1.30 | 0.59 | 0.02 | 1.33 | 2.32 | 1.68 |
| aCL139Contig2 | Caffeoyl‐CoA O‐methyltransferase 2 | AT4G34050 | 0.14 | –0.26 | 1.03 | –0.36 | 0.56 | 0.93 | 2.24 | 0.94 |
| aCL8378Contig1 | Leucoanthocyanidin dioxygenase‐like protein | AT3G13610 | – | 1.53 | 2.22 | – | + | + | + | |
| Methionine and ethylene biosynthetic processes | ||||||||||
| aC31605B08EF | ACC oxidase | AT1G05010 | 0.24 | 2.24 | 4.22 | 1.47 | 1.61 | 2.76 | 4.70 | 2.68 |
| aCL3488Contig1 | ACCO2 | AT2G19590 | – | 0.85 | 2.48 | – | 0.03 | 0.66 | 1.03 | 0.35 |
| aCL90Contig4 | 5‐Methyltetrahydropteroyltriglutamate‐homocysteine methyltransferase | AT5G17920 | –0.07 | 0.41 | 2.13 | 0.16 | –0.22 | 0.73 | 2.09 | 0.36 |
| Defence‐related proteins and response to stress | ||||||||||
| aCL46Contig3 | PR10A | AT1G24020 | 0.87 | 1.56 | 1.09 | 1.31 | 1.27 | 3.61 | 3.45 | 3.03 |
| aCL1Contig14 | PR‐4A precursor | AT3G04720 | –0.50 | 1.47 | 1.34 | 2.04 | –0.21 | 2.06 | 2.42 | 3.09 |
| aCL7008Contig1 | Germin‐like protein subfamily 1 member 13 precursor | AT5G39120 | 1.51 | 3.26 | 2.39 | 2.69 | 1.31 | 3.72 | 3.06 | 3.05 |
| aCL311Contig2 | Germin‐like protein subfamily 1 member 13 precursor | AT5G39150 | 1.70 | 2.24 | 2.31 | 2.68 | 1.03 | 3.67 | 3.02 | 2.96 |
| aCL3449Contig1 | Dicyanin | AT5G20230 | 0.51 | 1.22 | 2.12 | 1.51 | 1.27 | 1.89 | 3.01 | 2.47 |
| aCL2358Contig2 | Putative embryo‐abundant protein | AT2G41380 | –0.98 | 0.45 | 1.69 | 0.23 | 1.06 | 1.66 | 2.87 | 2.04 |
| aCL3319Contig1 | Chitinase CHI1 | AT3G54420 | –1.03 | 1.86 | 1.61 | 1.40 | 0.68 | 2.38 | 2.06 | 2.82 |
| aCL20Contig7 | β‐1,3‐Glucanase precursor | AT3G57270 | –0.33 | –0.40 | 1.61 | 0.35 | –0.61 | 0.68 | 2.81 | 1.18 |
| aCL9Contig16 | Lea5 | AT4G02380 | 0.07 | 1.99 | 1.25 | 2.31 | –0.41 | 0.95 | 0.50 | 0.90 |
| aCL9353Contig1 | Homogentisate phytylprenyltransferase (HPT1) | AT2G18950 | 1.03 | 1.59 | 1.95 | 1.63 | 1.20 | 2.13 | 2.28 | 2.05 |
| aC34104H09EF | Receptor kinase Lecrk | AT2G37710 | 0.10 | 1.10 | 1.69 | 1.01 | 0.17 | 1.25 | 2.13 | 0.98 |
| aC18021E12Rv | Isoleucine‐tRNA ligase‐like protein | AT4G10320 | 0.51 | 1.20 | 0.86 | 1.05 | 1.03 | 2.07 | 2.10 | 1.89 |
| aC31304H09EF | Putative glutathione S‐transferase T3 | AT3G09270 | –0.15 | 0.95 | 2.07 | 1.55 | 0.36 | 1.15 | 1.99 | 2.13 |
| aCL7071Contig1 | PR4b | AT3G04720 | –0.56 | 1.24 | 1.25 | 1.57 | 0.41 | 1.54 | 1.62 | 2.24 |
| aCL5030Contig1 | PR5‐1 | AT4G11650 | –0.20 | 1.58 | 0.96 | 1.49 | –0.27 | 1.39 | 1.53 | 2.04 |
| aCL9Contig11 | Lea5 | –0.02 | 1.84 | 1.19 | 2.02 | –0.31 | 0.95 | 0.55 | 0.93 | |
| Others | ||||||||||
| aC34005H07EF | Expressed protein | AT3G48770 | –0.09 | 1.58 | 2.28 | 1.39 | 0.66 | 3.02 | 4.06 | 3.32 |
| aCL2Contig10 | No annotation available | 0.05 | 1.65 | 1.97 | 3.69 | 0.16 | 0.85 | 1.49 | 2.75 | |
| aC05803G03SK | No annotation available | 0.20 | 0.67 | 2.00 | –0.40 | 1.83 | 1.81 | 3.69 | 1.72 | |
| aC31305B02EF | No annotation available | 0.11 | 1.40 | 1.54 | 3.34 | 0.68 | 1.51 | 2.07 | 3.50 | |
| aC08012E03SK | No annotation available | 0.06 | 1.45 | 1.29 | 3.04 | –0.05 | 1.03 | 1.94 | 3.48 | |
| aCL257Contig1 | Putative DNA‐binding protein | AT4G27000 | 0.06 | 2.16 | 2.28 | 1.68 | 0.53 | 2.59 | 3.29 | 2.48 |
| aCL8302Contig1 | No annotation available | –1.00 | 1.07 | 1.03 | 1.52 | 0.62 | 2.17 | 2.37 | 3.20 | |
| aCL5505Contig1 | β‐Cyanoalanine synthase | AT3G61440 | –0.51 | –0.30 | 2.72 | 1.70 | 0.14 | 0.74 | 3.16 | 2.23 |
| aCL363Contig1 | Expressed protein | AT4G32480 | –0.26 | 0.90 | 2.49 | 0.47 | 0.98 | 1.63 | 3.08 | 1.76 |
| aCL5472Contig1 | No annotation available | 0.06 | 0.13 | 1.51 | –0.30 | 1.39 | 1.23 | 2.93 | 1.24 | |
| aCL7118Contig1 | FAD‐binding domain‐containing protein | AT4G20820 | 0.29 | 1.12 | 1.99 | 1.06 | 1.25 | 2.18 | 2.77 | 2.06 |
| aC08027C08SK | Agglutinin‐2 precursor | AT5G65600 | –0.62 | 1.53 | 2.42 | 0.93 | –0.07 | 1.88 | 2.74 | 1.74 |
| aCL8719Contig1 | Globulin‐like protein | AT1G07750 | –0.27 | – | –0.20 | 0.64 | 0.92 | – | 0.79 | 2.73 |
| aC05811H08SK | Transferase family protein | AT1G28680 | 0.03 | 0.71 | 1.82 | 0.59 | 0.62 | 1.46 | 2.45 | 1.16 |
| aCL3612Contig1 | Cytochrome P450 79A2 | AT5G05260 | –0.53 | 0.41 | 1.31 | 0.73 | 0.59 | 1.49 | 2.44 | 1.73 |
| aC08016G11SK | No annotation available | 0.27 | 1.00 | 0.97 | 2.37 | 0.13 | 0.24 | 0.66 | 1.92 | |
| aCL214Contig1 | 3‐Deoxy‐d‐arabino‐heptulosonate 7‐phosphate synthase 3 | AT1G22410 | –0.09 | 0.27 | 1.25 | –0.10 | 0.86 | 1.53 | 2.36 | 0.94 |
| aC01008H04SK | Phosphatidate cytidylyltransferase family protein | AT3G45040 | 0.37 | 1.23 | 1.94 | 0.53 | 0.19 | 1.45 | 2.30 | 0.79 |
| aCL1084Contig1 | FAD‐binding domain‐containing protein | AT4G20820 | 0.62 | 1.39 | 2.26 | 1.48 | 0.56 | 1.65 | 2.17 | 1.51 |
| aCL944Contig2 | EIG‐I24 protein | AT1G28680 | 0.80 | 1.07 | 2.16 | 0.85 | + | + | ||
| aC08019H09SK | No annotation available | 0.30 | 1.39 | 0.93 | 2.14 | 0.29 | 1.02 | 0.90 | 2.05 | |
| aCL866Contig1 | CYP81E8 | AT4G37370 | 1.09 | 1.46 | 1.59 | 2.05 | 0.22 | 0.69 | 1.01 | 1.08 |
| aC18010B08Rv | No annotation available | 0.11 | 0.72 | 2.30 | 0.40 | 0.42 | 0.74 | 2.11 | 0.78 | |
| aCL632Contig3 | Proline‐rich protein | AT4G38770 | 0.12 | 1.14 | 1.48 | 0.20 | 0.44 | 1.41 | 2.08 | 1.00 |
| aCL1591Contig2 | Expressed protein | AT5G65520 | – | – | 1.90 | 1.37 | 0.30 | 0.67 | 2.07 | 1.49 |
| aCL3641Contig1 | Ripening regulated protein DDTFR18 | AT5G17700 | –0.11 | 1.98 | 1.14 | 2.03 | –0.43 | 1.28 | 0.73 | 0.92 |
| aCL8293Contig1 | CYP82C1p | AT4G31940 | 0.75 | 1.69 | 1.45 | 2.03 | 0.34 | 1.16 | 0.81 | 1.16 |
| aC19007C04T7 | No annotation available | –0.11 | 1.84 | 1.12 | 2.03 | –0.49 | 0.94 | 0.46 | 0.84 | |
Table 3.
Citrus genes with the highest repression level (log2 < –2) in the flavedo and/or albedo of elicited oranges 5 and/or 7 days after the beginning of the experiment. Values represent the log2 ratio of: a, I1/NT; b, IC4/NT; c, IC5/NT; d, IC7/NT. Numbers in bold indicate differential expression in the compared conditions according to significant analysis of microarrays (SAM) (P < 0.01) and log2 < –2. The symbol ‘–’ indicates expression in NT fruits but not in the compared treatment. See nomenclature in Table 2.
| Citrus unigene | Description | Arabidopsis homologue | Flavedo | Albedo | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | a | b | c | d | |||
| Defence‐related proteins and response to stress | ||||||||||
| aCL2916Contig1 | Probable polygalacturonase noncatalytic subunit JP650 precursor | AT1G70370 | 0.55 | –2.01 | –2.72 | –3.38 | 0.92 | –1.77 | – | –2.91 |
| aCL6368Contig1 | Probable polygalacturonase noncatalytic subunit JP650 precursor | AT1G70370 | 0.84 | –1.65 | –2.64 | –3.14 | 0.98 | –1.54 | – | – |
| aCL5Contig15 | Putative early light‐induced protein | AT3G22840 | –0.85 | –2.77 | –2.36 | –3.10 | –0.43 | –0.70 | –0.84 | –1.53 |
| aCL80Contig2 | Chloroplast small heat‐shock protein class I | AT3G46230 | –0.07 | 0.97 | –0.65 | –1.87 | –0.38 | 0.01 | –1.60 | –3.08 |
| aCL2349Contig1 | Putative β‐1,3‐glucanase | AT2G16230 | 0.23 | –1.46 | –1.87 | –2.83 | 0.30 | 0.21 | –0.54 | –0.98 |
| aC31403B04EF | Dehydrin family protein | AT1G54410 | –0.18 | –2.88 | –2.07 | –1.27 | –0.43 | –3.13 | –2.80 | –1.60 |
| aCL6Contig15 | Dehydrin family protein | AT1G54410 | –0.97 | –3.60 | –2.71 | –1.98 | –0.44 | –3.04 | –2.57 | –1.56 |
| aC31502B11EF | Plasma membrane intrinsic protein | AT4G00430 | 0.35 | –1.36 | –2.33 | –1.96 | 1.20 | 0.41 | –1.25 | –1.03 |
| aC20006B02SK | MYB91 | AT2G37630 | –0.28 | –2.29 | –2.07 | –1.32 | –0.44 | –2.29 | – | –1.27 |
| Others | ||||||||||
| aCL2649Contig1 | No annotation available | –0.77 | – | – | – | –0.64 | –1.26 | –2.42 | –3.36 | |
| aCL1642Contig3 | No annotation available | –0.51 | –2.14 | –1.82 | –3.23 | –0.76 | –1.29 | –2.06 | –2.66 | |
| aCL3246Contig1 | Steroid sulphotransferase‐like protein | AT5G07010 | 0.37 | –1.73 | – | –3.26 | 0.92 | 0.32 | –0.44 | –1.48 |
| aC20005G09SK | Importin β‐2 | AT5G53480 | 0.32 | –1.52 | –1.84 | –3.17 | 0.11 | 0.35 | 0.03 | –0.83 |
| aCL4849Contig1 | Expressed protein | AT5G01750 | –0.20 | 0.92 | –0.76 | –1.84 | –0.42 | –0.44 | –1.46 | –3.10 |
| aCL6348Contig1 | T2J13.20 protein | AT3G59300 | –0.36 | –2.71 | –2.01 | –1.21 | –0.32 | –2.97 | –2.70 | –1.75 |
| aC06003B11SK | Expressed protein | AT5G14790 | –0.26 | –2.69 | –2.01 | –1.24 | –0.23 | –2.75 | –2.54 | –1.53 |
| aCL6Contig6 | No annotation available | AT1G54410 | –0.33 | –2.56 | –2.04 | –1.30 | –0.34 | –2.56 | –2.49 | –1.63 |
| aC08006G03SK | Proline‐rich extension‐like family protein | AT1G21310 | 0.05 | –0.88 | –1.83 | –2.44 | 0.18 | –0.61 | –1.97 | –2.46 |
| aCL1Contig17 | No annotation available | 0.02 | –1.16 | –2.39 | –2.03 | 0.49 | 0.46 | –0.38 | –1.36 | |
| aC31504C04EF | Pollen Ole e 1 allergen and extension family protein | AT4G08685 | –0.56 | –0.81 | –2.28 | –2.01 | –0.45 | –0.95 | –1.96 | –1.90 |
| aCL939Contig3 | No annotation available | –0.29 | –0.04 | –0.82 | –0.50 | –0.78 | –1.11 | –1.75 | –2.27 | |
| aC04006E11SK | Flavin reductase‐related | AT2G34460 | –0.28 | –2.10 | –1.63 | –1.28 | –0.55 | –2.21 | –2.24 | –1.62 |
| aCL533Contig3 | CP12 precursor | AT3G62410 | –0.29 | –1.64 | –1.79 | –1.65 | –0.77 | –1.37 | –1.78 | –2.21 |
| aCL5404Contig1 | Hypothetical protein | AT5G38050 | –0.15 | –2.21 | –1.35 | –1.39 | –0.12 | –2.32 | –2.15 | –1.42 |
| aCL3235Contig1 | Expressed protein | AT3G62370 | 0.09 | –1.88 | –1.40 | –0.98 | –0.24 | –2.18 | –2.12 | –1.34 |
| aCL381Contig1 | Galactinol synthase | AT1G56600 | –0.96 | –0.09 | – | –0.55 | –1.14 | –0.52 | –0.98 | –2.11 |
| aCL3Contig33 | No annotation available | 0.22 | –0.84 | –1.22 | –0.74 | –0.25 | –0.94 | –1.41 | –2.10 | |
| aC16015G06SK | Expressed protein | AT3G01370 | –0.12 | –2.03 | –1.57 | –1.01 | –0.36 | –2.05 | –2.05 | –1.32 |
| aCL310Contig2 | Expressed protein | AT4G26850 | –0.07 | –1.60 | –2.04 | –2.03 | –0.14 | –1.26 | –1.86 | –1.93 |
| aC04023D07SK | No annotation available | –0.22 | –1.92 | –1.55 | –1.07 | –0.29 | –1.97 | –2.04 | –1.43 | |
Functional analysis based on the transcriptome profiles
Hierarchical cluster analysis of all differentially expressed genes (SAM, P < 0.01) revealed the presence of a few major clusters containing genes that shared a common expression pattern, as shown in the gene‐to‐gene correlation matrix (Fig. 2a). Genes within these clusters and having an A. thaliana homologue were subjected to a singular enrichment analysis (SEA) using AgriGO to determine which biological processes were significantly over‐represented (Table 4). Genes included in clusters f and g, whose expression increased in response to elicitor treatment (Fig. 2b), belonged to three major biological processes: phenylpropanoid biosynthesis, methionine metabolism and lipid biosynthesis. Most of the other pathways that appeared in these two clusters were ancestors of these three processes.
Figure 2.
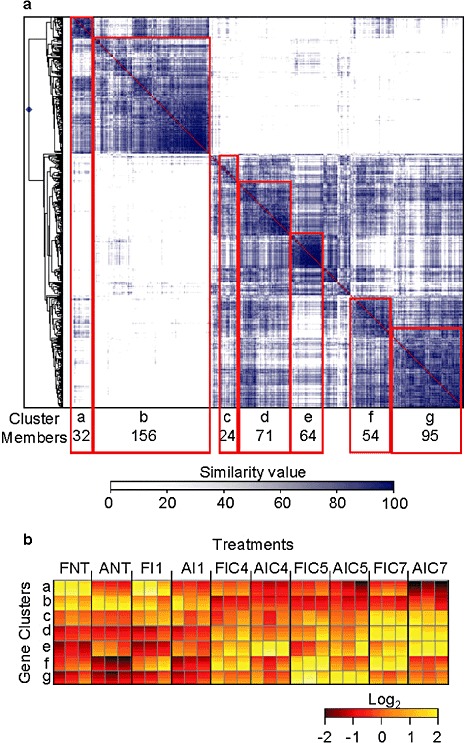
Gene‐to‐gene correlation matrix of differentially expressed genes. (a) Main gene clusters are situated along the diagonal line (groups a–g). Correlations between genes are shown by the blue scale: the darker the blue colour, the higher the percentage of similarity between gene expression patterns. (b) Patterns of expression of flavedo (F) and albedo (A) genes included in each cluster. NT, nontreated fruits; I1, fruits infected with Penicillium digitatum during 1 day; IC, infected and cured fruits at 4, 5 and 7 days after the beginning of the experiment.
Table 4.
Gene ontology (GO) biological process categories over‐represented in the clusters based on the gene‐to‐gene correlation matrix (Fig. 2). Only three clusters (b, f and g) showed significant biological processes with Bonferroni multi‐test adjustment (P < 0.05).
| Cluster | GO term | Description | GO level | FDR |
|---|---|---|---|---|
| b | GO:0006950 | Response to stress | 3 | 0.021 |
| GO:0009628 | Response to abiotic stimulus | 3 | 0.034 | |
| f | GO:0019438 | Aromatic compound biosynthetic process | 5 | 0.0041 |
| GO:0044249 | Cellular biosynthetic process | 4 | 0.0055 | |
| GO:0006519 | Cellular amino acid and derivative metabolic process | 4 | 0.0087 | |
| GO:0009058 | Biosynthetic process | 3 | 0.0094 | |
| GO:0044283 | Small molecule biosynthetic process | 4 | 0.017 | |
| GO:0006725 | Cellular aromatic compound metabolic process | 4 | 0.026 | |
| GO:0008610 | Lipid biosynthetic process | 4 | 0.026 | |
| GO:0044281 | Small molecule metabolic process | 3 | 0.027 | |
| GO:0019748 | Secondary metabolic process | 3 | 0.030 | |
| g | GO:0044283 | Small molecule biosynthetic process | 4 | 3.10E‐11 |
| GO:0006519 | Cellular amino acid and derivative metabolic process | 4 | 3.70E‐09 | |
| GO:0044281 | Small molecule metabolic process | 3 | 2.10E‐07 | |
| GO:0006520 | Cellular amino acid metabolic process | 5 | 7.40E‐07 | |
| GO:0044106 | Cellular amine metabolic process | 5 | 1.70E‐06 | |
| GO:0009308 | Amine metabolic process | 4 | 3.70E‐06 | |
| GO:0019752 | Carboxylic acid metabolic process | 6 | 1.00E‐05 | |
| GO:0043436 | Oxoacid metabolic process | 5 | 1.00E‐05 | |
| GO:0006082 | Organic acid metabolic process | 4 | 1.10E‐05 | |
| GO:0008652 | Cellular amino acid biosynthetic process | 6 | 1.10E‐05 | |
| GO:0006790 | Sulphur metabolic process | 4 | 1.40E‐05 | |
| GO:0042180 | Cellular ketone metabolic process | 4 | 1.50E‐05 | |
| GO:0042398 | Cellular amino acid derivative biosynthetic process | 5 | 1.90E‐05 | |
| GO:0009309 | Amine biosynthetic process | 5 | 2.70E‐05 | |
| GO:0034641 | Cellular nitrogen compound metabolic process | 4 | 3.20E‐05 | |
| GO:0019438 | Aromatic compound biosynthetic process | 5 | 3.60E‐05 | |
| GO:0046394 | Carboxylic acid biosynthetic process | 6 | 3.80E‐05 | |
| GO:0016053 | Organic acid biosynthetic process | 5 | 3.80E‐05 | |
| GO:0000096 | Sulphur amino acid metabolic process | 6 | 6.50E‐05 | |
| GO:0006575 | Cellular amino acid derivative metabolic process | 5 | 0.00015 | |
| GO:0044271 | Cellular nitrogen compound biosynthetic process | 5 | 0.00025 | |
| GO:0006555 | Methionine metabolic process | 7 | 0.00033 | |
| GO:0009058 | Biosynthetic process | 3 | 0.00034 | |
| GO:0046483 | Heterocycle metabolic process | 4 | 0.00042 | |
| GO:0044249 | Cellular biosynthetic process | 4 | 0.00044 | |
| GO:0006725 | Cellular aromatic compound metabolic process | 4 | 0.00089 | |
| GO:0009066 | Aspartate family amino acid metabolic process | 6 | 0.0028 | |
| GO:0019748 | Secondary metabolic process | 3 | 0.0073 | |
| GO:0009699 | Phenylpropanoid biosynthetic process | 7 | 0.015 | |
| GO:0044237 | Cellular metabolic process | 3 | 0.029 | |
| GO:0009698 | Phenylpropanoid metabolic process | 6 | 0.036 | |
| GO:0008152 | Metabolic process | 2 | 0.04 | |
| GO:0006807 | Nitrogen compound metabolic process | 3 | 0.043 |
A principal component analysis (PCA) was conducted in order to identify those genes that mostly accounted for the differences among tissues and treatments. The result of this analysis (Fig. 3) showed a clear difference between elicited and nonelicited tissues in the first principal component (X‐axis, explaining 45.7% of the total variation). In addition, the PCA revealed a variation between the two fruit tissues, flavedo and albedo, in the second component (variation Y= 19.7%) (Fig. 3a). The genes determining both tissue and treatment variations can be found by the projection of the sample differentiation vectors onto the PCA plot showing the distribution of genes (SAM, P < 0.01) (Fig. 3b).
Figure 3.
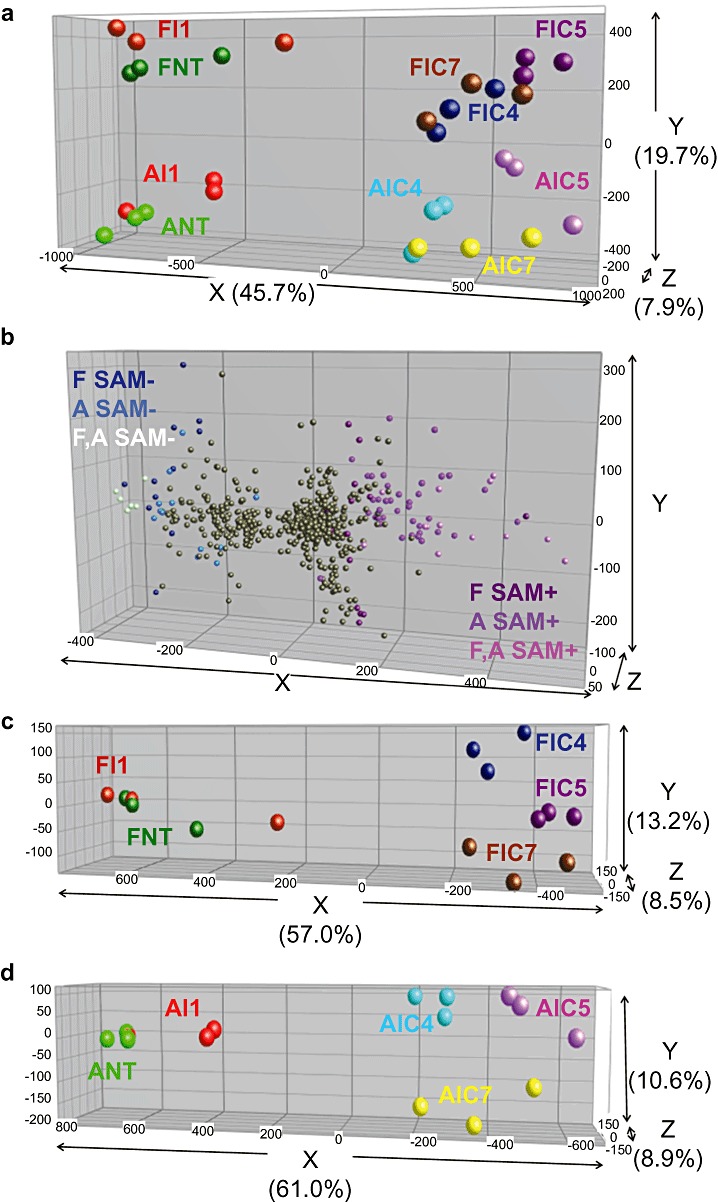
Multivariate analysis of differentially expressed genes in elicited citrus fruits. (a) Principal component analysis (PCA) showing the variation between fruits subjected or not to elicitor treatment in the first component, and the variation between tissues, flavedo (F) and albedo (A), in the second component. (b) PCA showing the distribution of genes. Genes with log2 > 2 for induced and log2 < –2 for repressed expression are indicated in different colours. Independent PCAs for the flavedo (c) and albedo (d) samples. See nomenclature in Fig. 1.
When the two fruit tissues were subjected to PCA independently, a clear separation of samples based on treatments was observed (Fig. 3c,d for flavedo and albedo, respectively). Independent of treatment, six genes showed higher expression in flavedo than in albedo, including a putative ABC transporter, two lipoxygenases, a cinnamoyl‐CoA reductase, a glucosyltransferase and a senescence‐related (SRG1) protein (Table S1, see Supporting Information). Four genes were more expressed in albedo samples than in flavedo samples: two proteins without any A. thaliana homologue and two different Citrus tristeza virus (CTV) proteins (Table S2, see Supporting Information). Based on gene ontology (GO) terms, the biological processes over‐represented in the nontreated flavedo compared with nontreated albedo were associated with flavonoid and fatty acid biosynthesis (Fig. S1 and Table S3, see Supporting Information).
To elucidate the key processes that were altered in elicited citrus fruits, we searched for functional enrichment categories in the set of differentially expressed genes. This analysis showed that genes induced in the flavedo of elicited fruits (FIC5), when compared with nontreated fruits (FNT), were mainly involved in biological processes associated with secondary metabolism, in particular phenylpropanoids (Fig. 4 and Table S4, see Supporting Information). The same trend was observed in the flavedo of elicited fruits at 7 days after the beginning of the experiment (FIC7; Table S5 and Fig. S2, see Supporting Information). In the albedo, processes related to flavonoid and cellular amino acid biosynthesis and methionine metabolism were over‐represented in AI5 (Table S6 and Fig. S3, see Supporting Information), whereas, 2 days later (AIC7), the response to cadmium ion was the most significant over‐represented biological process (Table S7 and Fig. S4, see Supporting Information).
Figure 4.
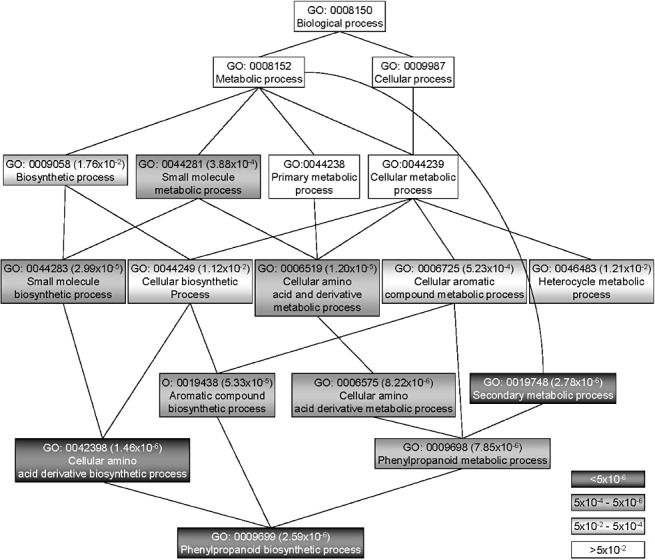
Hierarchical view of gene ontology (GO) biological categories significantly over‐represented in the elicited flavedo 5 days after the beginning of the experiment (FIC5) compared with nontreated flavedo (FNT), obtained with AgriGO. Significant categories (adjusted P < 0.05) are shown using grey scaling according to their significance level. Other categories required to complete the hierarchy are shown in white.
To better visualize the results of gene expression profiling experiments in a metabolic pathway context, the ‘OMICS Viewer’ was used. Based on the results described above, two biosynthetic pathways over‐represented in elicited flavedo and albedo tissues were selected for further analysis: the methionine–ethylene biosynthesis pathway (Fig. S5, see Supporting Information) and the phenylpropanoid–phenolic acid–suberin pathway (Fig. 5).
Figure 5.
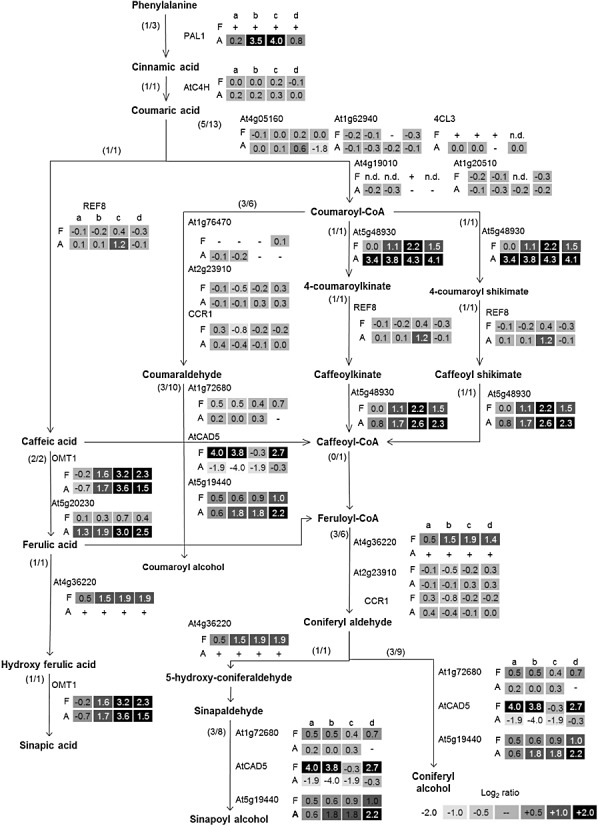
Graphical representation of the phenylpropanoid, free phenolic acids and suberin pathways showing gene expression values in the flavedo (F) and albedo (A) of elicited oranges. The first number in parentheses indicates the number of genes spotted onto the microarray that have an Arabidopsis thaliana homologue, and the second number indicates the total number of A. thaliana genes for each step in the pathway. Numbers in squares indicate the log2 ratio of: a, I1/NT; b, IC4/NT; c, IC5/NT; d, IC7/NT. n.d., not detected. The symbol ‘+’ indicates no expression in NT and the symbol ‘–’ indicates expression in NT fruits but not in the compared treatment. NT, nontreated fruits. I1, fruits infected with Penicillium digitatum during 1 day; IC, infected and cured fruits at 4, 5 or 7 days after the beginning of the experiment.
Involvement of ethylene in the induction of resistance in citrus fruits
Several genes involved in the methionine and ethylene biosynthesis pathway were up‐regulated upon pathogen challenge followed by curing treatment (Table 2 and Fig. S5). As the highest induction in the elicited flavedo (FIC5) was observed for a homologue of the A. thaliana EFE gene, which encodes an ACO that is 4.7 times more strongly expressed than in FNT, we decided to study the involvement of ethylene in the fruit's response in more detail by analysing both the expression of the CsACO gene and the production of ethylene in elicited fruits (Fig. 6). In nontreated fruits, CsACO expression was not detectable in either flavedo or albedo. However, in infected–cured fruits, high expression was observed in both tissues, especially 5 days after the beginning of the experiment (IC5). The level of CsACO mRNA was 16.9‐ and 30.0‐fold higher in the flavedo (FIC5) and albedo (AIC5) of infected–cured fruits, respectively, compared with that of flavedo infected with P. digitatum during 1 day (FI1). In both tissues, the expression of CsACO decreased 7 days after the beginning of the experiment. This trend was also observed in the unigene aC31605B08 spotted onto the 12k microarray (Table 2), which putatively also encoded another ACO. CsACO expression was induced in response to the wounding–curing treatment (WC7), but to a lesser extent than in infected–cured fruits (IC7). However, no expression was detected in response to the curing treatment alone (C7), in either the flavedo or albedo. It is interesting to note the lack of hybridization signal with the P. digitatum ribosomal probe (Fig. 6a), a fact that indicates that the pathogen did not progress in the tissue that had been inoculated previously with the pathogen (treatment IC).
Figure 6.
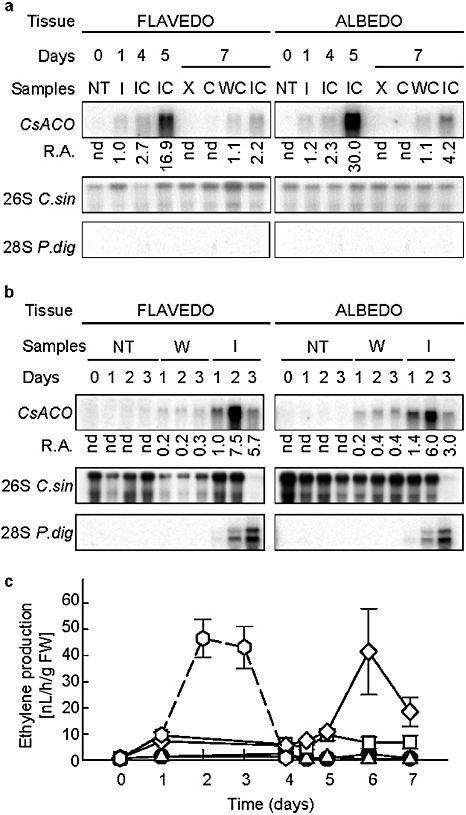
Involvement of ethylene in the response of citrus fruits to elicitor treatment and to Penicillium digitatum infection. (a) Northern blot analysis of CsACO in the flavedo and albedo of oranges: NT, nontreated; I, infected; IC, infected and cured; X, nonwounded; C, cured; WC, wounded and cured. (b) CsACO mRNA accumulation in oranges infected with P. digitatum during 1, 2 and 3 days. Fruits were nontreated (NT), wounded and water inoculated (W) or wounded and inoculated with a suspension of 106 conidia/mL of P. digitatum (I). In (a) and (b), relative accumulation (R.A.) values of CsACO mRNA in arbitrary units are shown at the bottom. Normalization was carried out with respect to the hybridization signal of the Citrus sinensis 26S rRNA using the flavedo infected with P. digitatum for 1 day (I1) as a reference. Hybridization with the P. digitatum 28S rDNA probe is shown at the bottom. (c) Ethylene production in discs of nontreated oranges (●), infected fruits ( ), cured fruits (
), cured fruits ( ), wounded and cured fruits (□) and infected and cured fruits (◊). Values represent the average of three replicates, including 10 discs in each replicate, ± SD.
), wounded and cured fruits (□) and infected and cured fruits (◊). Values represent the average of three replicates, including 10 discs in each replicate, ± SD.
It is known that wounding and also P. digitatum infection induce the expression of ethylene biosynthetic genes in the peel of C. sinensis fruits (Marcos et al., 2005). Therefore, we aimed to compare the effect of wounding and pathogen infection alone with that observed in response to elicitor treatment, which consisted of an infection with P. digitatum before the curing treatment. In this experiment, ‘Navelina’ oranges were inoculated with 106 conidia/mL of P. digitatum, and flavedo and albedo tissues were analysed 1, 2 and 3 days after inoculation. The development of P. digitatum throughout both tissues was rapid, as determined by the accumulation of the P. digitatum 28S rRNA (Fig. 6b). Complementarily, the accumulation of the C. sinensis 26S rRNA was hardly detectable at 3 days post‐inoculation, reflecting the degradation of fruit tissue as a result of fungal invasion. The pattern of CsACO expression was similar in flavedo and albedo. Wounded fruits showed only a small increase in CsACO levels 3 days after mock‐inoculation in both tissues, compared with nontreated fruits. In contrast, a marked increase in CsACO expression was detected in both tissues during the development of infection by P. digitatum. The highest levels of CsACO mRNA were detected at 2 days after inoculation. Nevertheless, the increase in CsACO expression in response to P. digitatum infection was lower than that observed in infected–cured fruits.
Ethylene production (Fig. 6c) was determined in discs centred on the point of inoculation. No change in ethylene production was detected in either nontreated or cured fruits, whereas a slight increase was detected in wounded–cured fruits 5 days after the beginning of the experiment. In contrast, high ethylene production (42 nL/h/g FW) was detected in infected–cured fruits at day 6 of the experiment. Similar high levels were observed in infected fruits 2–3 days after inoculation with P. digitatum.
Involvement of the phenylpropanoid pathway in the induction of resistance in citrus fruits
It is known that genes involved in the biosynthesis of phenylpropanoids and compounds derived from this pathway are also involved in the resistance of citrus fruit against biotic and abiotic stresses (Ballester et al., 2006; Kim et al., 1991; Sánchez‐Ballesta et al., 2000). Taking into account this previous result and the current transcriptomic observations, we decided to conduct a deeper study into the expression of genes specifically related to the phenylpropanoid pathway. Northern blot hybridization was used to both validate the microarray results and to better define the expression patterns of selected genes putatively involved in the phenylpropanoid pathway. In total, 17 genes were analysed (Fig. 7 and Table S8, see Supporting Information) and the expression profiles for 12 of them were compared with their expression levels on the 12k microarray. Overall patterns were similar in both approaches, thus confirming our microarray results.
Figure 7.
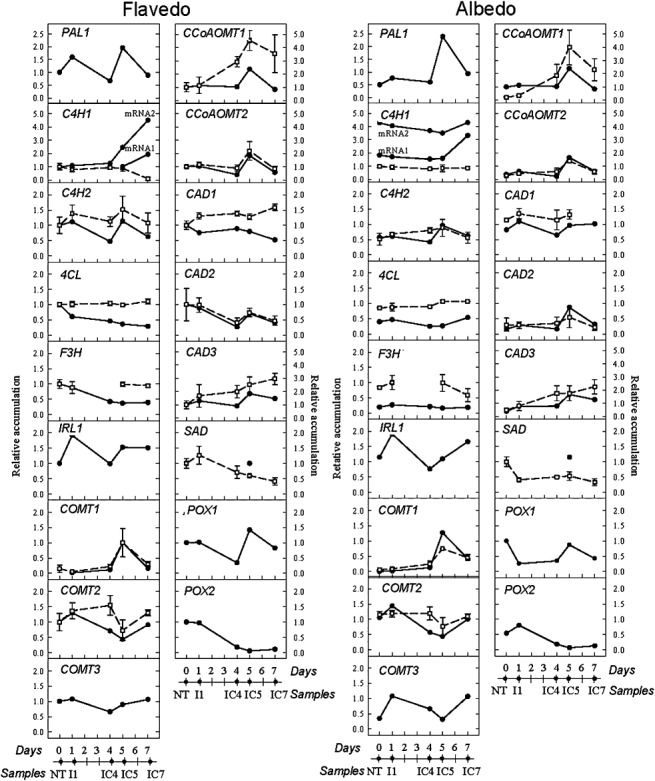
mRNA relative accumulation of genes putatively involved in phenylpropanoid metabolism in elicited citrus fruits. The expression level was determined by Northern blot (●) or microarray (□) hybridizations. In general, a reference value of 1.0 was assigned to the nontreated flavedo, except for COMT1 and SAD genes as a result of a lack of expression in nontreated flavedo. Values from microarray hybridizations represent the average of three biological replicates ± SD. NT, nontreated fruits; I1, fruits infected with Penicillium digitatum during 1 day; IC, infected and cured fruits at 4, 5 and 7 days after the beginning of the experiment.
The expression of the 17 genes putatively involved in the phenylpropanoid pathway increased in the elicited fruits, generally reaching the highest induction levels 5 days after the beginning of the experiment. In this regard, it is noteworthy that the expression of PAL, the first gene in the phenylpropanoid pathway, and also the expression of most of the O‐methyltransferases analysed increased in elicited oranges, suggesting the implication of these genes in the induction of resistance in citrus fruit. In general, the highest absolute expression levels were detected in the flavedo, whereas induction ratios compared with nontreated fruits were higher in the albedo.
DISCUSSION
The search for and optimization of new pathogen control systems that could help to reduce the amount of chemicals needed to control postharvest pathogens in citrus fruit will benefit from a better understanding of the biological basis of induced resistance against major pathogens. Among the different elicitor treatments to induce resistance in citrus fruits, we have chosen the combination of pathogen inoculation followed by curing treatment as a tool to study the biological processes underlying the induction of resistance in citrus fruit in more detail. This combined treatment is highly reproducible, triggers the highest induction of the phytoalexin scoparone and leads to a greater reduction in disease incidence than wounding–curing or curing alone (Ballester et al., 2010a). The build up of endogenous pathogen resistance in citrus fruit by postharvest treatments is usually achieved between 1 and 3 days after elicitor treatment (Ballester et al., 2010a; Droby et al., 2002; Pavoncello et al., 2001). Accordingly, we have found that the combination of pathogen infection followed by curing treatment achieves maximum efficacy at 3 dpt, when a reduction of up to 70% in subsequent disease incidence is observed relative to control fruits (Table 1).
Functional genomics to elucidate induced resistance in citrus fruits
High‐throughput technologies have allowed rapid progress in the understanding of the resistance of model plants against pathogens (De Vos et al., 2005; van Loon et al., 2006). Despite the progress made in the characterization of defence‐related responses against P. digitatum infection (Angioni et al., 1998; Ballester et al., 2010a; Kim et al., 1991; Ortuño et al., 2006; 1999a, 2001), remarkably little is known about the complex network regulating induced resistance in citrus fruit against P. digitatum.
Penicillium digitatum penetrates citrus fruits through wounds that affect the albedo, the inner tissue. In more superficial wounds, the chance for this pathogen to cause disease is much lower (Kavanagh and Wood, 1967). In a comparison between both tissues, we found that the expression of genes involved in the phenylpropanoid pathway is higher in the flavedo (Fig. 7). However, the largest elicitation was in the albedo. Thus, the transcriptomic approach supports the concept that the flavedo is more resistant than the albedo to P. digitatum infection (Afek et al., 1999; Ballester et al., 2006, 2010a), although the internal tissue responds more intensively to elicitor treatment. This greater response of the albedo is also reflected in the greater number of induced/repressed genes in elicited fruits when compared with that of the flavedo (Fig. 1).
The application of different elicitors to citrus fruit has been related to the induction of genes coding for PR proteins, such as chitinases and β‐1,3‐glucanases (Ballester et al., 2010a; 2001, 2002). Indeed, a chitinase and a β‐1,3‐glucanase gene were induced more than two‐fold in the albedo of elicited fruits (Table 2). However, only three of nine chitinase and three of 11 β‐1,3‐glucanase genes present on the 12k microarray were induced in the albedo, whereas the number of induced genes was even lower in the flavedo. Other PR encoding genes were also induced, such as a ribonuclease‐like protein (PR‐10), two hevein‐like PR4 proteins and two germin‐like PR16 proteins (Table 2). Notably, most of the induced PR encoding genes showed a higher induction in the albedo.
Hierarchical cluster analysis of the differentially expressed genes revealed the presence of several clusters of co‐expressed genes. Functional analysis of the genes included in these clusters showed that clusters f and g, which contained up‐regulated genes in both tissues at 5 days after the beginning of the experiment, were enriched in processes related to methionine metabolism and phenylpropanoid biosynthesis, and, to a lesser extent, lipid biosynthesis, most specifically to isopentenyl diphosphate biosynthesis. Thus, although different factors/signals are probably involved in eliciting resistance in citrus fruit against P. digitatum, phenylpropanoids and ethylene seem to be the most important players in the complex network that regulates induced resistance. It is interesting to note that the metabolism of phenylpropanoids is also induced in response to P. digitatum infection in citrus fruits (González‐Candelas et al., 2010).
Implication of ethylene and related hormones in induced resistance in citrus fruit
The transcriptomic analysis showed an implication of genes related to methionine and ethylene biosynthetic processes in the defence response of citrus fruit. The expression of CsACO was induced by elicitor treatment (Table 2 and Fig. 6). Three additional putative ACO genes were present on the 12k microarray. However, only the expression of one of them (aCL3488contig1) increased in infected–cured samples. A comparative analysis of the nucleotide sequences of these four ACO genes showed a 29%–60% identity among them (data not shown), indicating that they are different genes and not alleles of the same gene. It is well known that ethylene regulates the induction of genes related to plant defence against pathogens, including several β‐1,3‐glucanases and basic chitinases, PR‐1 and hydroxyproline‐rich proteins (Ecker, 1995). We have shown previously that the expression of genes coding for two basic β‐1,3‐glucanase and chitinase isoforms is induced in elicited citrus fruits (Ballester et al., 2010a). These previous results have been extended in the present study, where we found the induction of several ethylene responsive genes, including additional β‐1,3‐glucanases and chitinases, as well as other PR‐encoding genes mentioned above. However, many other genes encoding PR proteins were not induced by elicitor treatment.
The infection of citrus fruit by P. digitatum induces ethylene production and the expression of genes involved in the synthesis of ethylene and phenylpropanoids (Achilea et al., 1985a; Marcos et al., 2005). Elicitor treatment also leads to an increase in ethylene production of the same magnitude as observed in P. digitatum‐infected fruits (Fig. 6). In infected fruits, ethylene is produced by both the fruit and the pathogen (1985a, 1985b). However, in elicited fruits, ethylene only originates from the fruit as the fungus does not grow within the peel, as confirmed by the lack of hybridization with the P. digitatum rDNA 28S probe (Fig. 6).
Although ethylene plays a role in the resistance of citrus fruit (Marcos et al., 2005; Porat et al., 1999b), we found only a limited overlap between genes induced by elicitor treatment and those induced by ethylene (González‐Candelas et al., 2010). Moreover, the reduction in disease incidence achieved by ethylene treatment (Marcos et al., 2005) was much lower than that observed with the infection–curing treatment, suggesting that other factors are involved in triggering induced resistance in citrus fruits.
Ethylene and JA usually show a synergistic relationship in regulating the expression of defence genes effective against necrotrophic pathogens (Glazebrook, 2005; Lorenzo et al., 2003; Schenk et al., 2000). The elicitor treatment altered the expression of marker genes for SA, such as PR‐1, PR‐2, PR5, PR‐10 or class II β‐1,3‐glucanase, but there was no clear trend in the observed changes, and functional analysis of the differentially expressed genes did not reveal any biological process related to SA or JA in elicited citrus fruits. However, we cannot rule out the implication of these hormones in the induction of resistance of citrus.
Involvement of the phenylpropanoid pathway in the induced resistance of citrus fruits
Transcriptomic analysis revealed that the metabolism and biosynthesis of phenolic compounds were involved in the induced resistance of citrus fruits against P. digitatum. PAL is the first enzyme in the phenylpropanoid pathway leading to the synthesis of coumarins and flavonoids (Dixon et al., 2002). Studies in citrus fruit have focused on changes in PAL expression in response to pathogen attack (Ballester et al., 2006; McCollum, 2000) or elicitor treatment (Ballester et al., 2010a; Droby et al., 1993; Fajardo et al., 1998). We have shown previously that PAL expression increases in both flavedo and albedo of infected–cured fruits. In the present work, we observed that the expression of not only PAL, but also a large subset of genes important for the synthesis of phenylpropanoids and flavonoids, such as C4H, 4CL, COMT, CCoAOMT, CAD, SAD and POX, was increased in elicited fruits (Table 2 and 5, 7). Transcript levels of selected phenylpropanoid genes were further investigated by Northern blot hybridization, validating the results obtained with the 12k microarray (Fig. 7). Although the highest expression of these genes was detected in the flavedo of infected–cured fruits, the highest inductions were observed in the internal tissue (Fig. 7).
Biosynthetic genes are not the only genes related to the accumulation of phenylpropanoids and flavonoids. It is well known that R2R3‐type MYB genes control many aspects of plant secondary metabolism (Stracke et al., 2001). Recently, it has been described that the lack of expression of the MYB12 transcription factor, which controls the expression of the biosynthetic flavonoid genes, is related to the lack of accumulation of naringenin chalcone in tomato (Adato et al., 2009; Ballester et al., 2010b). The A. thaliana MYB12 homologue was included in the 12k microarray, but was not affected by elicitor treatment. However, two MYB, two WRKY and one bHLH transcription factors clustered within the e, f and g groups in the gene‐to‐gene correlation analysis. Additional analysis will be performed in order to clarify the possible role of these transcription factors in the induced response.
In conclusion, the transcriptomic analysis of elicited oranges showed important up‐regulation in the expression of genes involved in the metabolism of phenylpropanoids and synthesis of ethylene and down‐regulation of genes related to diverse biotic and abiotic stresses. Our results indicate that the highest inductions were found in the albedo, whereas the highest expression values were detected in the external tissue. These results reinforce the concept that the internal tissue is more susceptible to P. digitatum infection, and it is this tissue that should increase its defensive barriers to a greater extent in order to avoid the progression of the fungus. To the best of our knowledge, this is the first study in any harvested fruit that has addressed the analysis of global changes in gene expression in the process of induced resistance.
EXPERIMENTAL PROCEDURES
Fruit and fungal material
Oranges (Citrus sinensis L. Osbeck) from a commercial orchard in Lliria (Valencia, Spain) were selected and used in the experiments before any commercial postharvest treatment was applied. ‘Navelate’ fruits were taken in three independent samplings and used for the induction of resistance treatment, and ‘Navelina’ oranges were employed to study the expression of CsACO during the infection of P. digitatum. Fruits were immediately surface sterilized with 5% commercial bleach solution for 5 min, extensively washed with tap water and allowed to dry at room temperature until the next day.
Petri dishes containing potato dextrose agar were inoculated with Penicillium digitatum (Pers.:Fr.) Sacc. isolate PHI‐26 and incubated at 24 °C for 7 days (López‐García et al., 2000). Conidia were rubbed from the agar surface by scraping them with a sterile spatula and transferred to sterile water. The conidial suspension was then filtered and the concentration was determined with a haemocytometer and adjusted to the desired value.
Induction of resistance treatment and P. digitatum infection
The treatment for eliciting resistance has been described previously by Ballester et al. (2010a). Briefly, the following treatments were applied to the three biological replicates of ‘Navelate’ oranges: (i) fruits were wounded by making punctures (3 mm in depth) with a sterilized nail and inoculated with 10 µL of a P. digitatum conidial suspension adjusted to 105 conidia/mL; the treated fruits were placed in plastic boxes and maintained at 90%–95% relative humidity at 20 °C for 1 day to allow pathogen development; fruits were then heat treated at 37 °C for 3 days under water‐saturated conditions (curing) in order to stop the progress of the pathogen (sample IC); (ii) control inoculations were carried out by injecting 10 µL of sterile water and holding the fruits under the same conditions (sample WC); (iii) additional controls consisted of intact nonwounded fruits held at 20 °C for 1 day and then at 37 °C for 3 days (sample C) and intact nonwounded fruits held at 20 °C for 1 day and then at 4 °C for 3 days (sample X). A sample from intact nonwounded fruits was obtained on the first day of the experiment (sample NT). Peel tissue discs of 13 mm around the inoculation point were sampled using a cork borer. Flavedo and albedo tissues were separated with a scalpel. Tissue discs obtained from 15 oranges with eight discs per fruit were immediately frozen in liquid nitrogen, mixed, ground to a fine powder with a coffee mill and stored at −80 °C until further analysis.
To check the influence of P. digitatum infection in the expression of CsACO, ‘Navelina’ oranges were wounded by making punctures (5 mm in depth) with a sterilized nail and inoculated with 10 µL of a suspension of P. digitatum conidia adjusted to 106 conidia/mL. This high inoculum level was used in order to synchronize fungal development in all wounds. Oranges were kept at 20 °C for up to 3 days (samples I). As controls, wounded fruits inoculated with sterile water (samples W) and nonwounded fruits (samples NT) were also taken. At 1, 2 and 3 days post‐inoculation, flavedo and albedo discs of 7 mm around the point of inoculation were sampled using a cork borer. Flavedo and albedo tissues were processed as described above and stored at −80 °C until RNA isolation.
Infections
To determine the effectiveness of the elicitor treatment in reducing pathogen infection and the importance of the elapsed time between the treatment and the infection, disease susceptibility was analysed at the beginning of the experiment in nontreated ‘Navelate’ fruits and at 4, 5 and 7 days (0, 1 and 3 dpt) for infected–cured fruits. Each infected–cured fruit was punched at a distance of 0.5 cm from the previous wound or in the equatorial axis in fruits that had not been inoculated previously. Then, 10 µL of a suspension (104 conidia/mL) of P. digitatum spores were applied to each wound. After inoculation, fruits were kept at 20 °C and 90%–95% relative humidity. The incidence of infection, as a percentage, was determined for up to 6 days of incubation at 20 °C. The experimental design consisted of three replicates of five fruits, with four wounds per fruit, for each treatment. To test the effect of the treatments, a one‐way analysis of variance (anova) was performed. Means were separated using Tukey's honestly significant difference test at P < 0.05. The analysis was performed with Statgraphics Plus 5.1 Software (Manugistics Inc., Rockville, MD, USA).
RNA isolation and preparation of labelled cDNA probes
Total RNA was isolated from frozen tissue as described by Ballester et al. (2006). RNA concentration was measured spectrophotometrically and the integrity was verified by agarose gel electrophoresis and ethidium bromide staining.
RNA samples for microarray hybridizations were labelled using the indirect method by the incorporation of 5‐(3‐aminoallyl)‐2‐deoxy‐UTP (aa‐dUTP) into single‐stranded cDNA during reverse transcription, followed by the conjugation of fluorescent cyanine 3 (Cy3) and cyanine 5 (Cy5) as reactive N‐hydroxyl succinimide dyes (NHS dyes). Reverse transcription, cDNA purification, dye coupling and fluorescent cDNA purification were performed as described by Forment et al. (2005), except that total RNA (30 µg) was used instead of poly(A)+ RNA. Sample RNA was labelled with Cy5, and reference RNA (pooled RNA consisting of an equal amount of RNA from each sample) was labelled with Cy3.
Microarray hybridization, data acquisition and data analysis
The 12k cDNA microarray developed by CFGP (http://bioinfo.ibmcp.upv.es/genomics/cfgpDB/) was used. This microarray contains probes corresponding to 11 241 putative unigenes from citrus. Microarray hybridization, washing and scanning were performed as described by Forment et al. (2005), with some modifications. Labelled cDNA from experimental and control samples was dried separately and resuspended in fresh hybridization solution [containing 50% (v/v) formamide, 5 × standard saline citrate (SSC), 0.1% (w/v) sodium dodecylsulphate (SDS) and 0.1 mg/mL salmon sperm DNA]. Samples were heated for 1 min at 95 °C before hybridization, which was conducted at 42 °C. Microarray slides were scanned with a GenePix 4000B (Axon Instruments, Sunnyvale, CA, USA) using ‘GenePix Pro 6.0’ image acquisition software (Axon Instruments) at 10‐µm resolution, 100% laser power and different photomultiplier tube (PMT) values to adjust the channels' intensity ratio to 1.0. Nonhomogeneous and aberrant spots were discarded. Only spots with background‐subtracted intensity greater than two‐fold the mean background intensity in at least one channel were selected and used for normalization and further analysis. Data were log transformed and analysed using GEPAS (Gene Expression Pattern Analysis Suite) software v3.1 (Montaner et al., 2006). Firstly, the DNMAD (Diagnosis and Normalization for MicroArray Data) module (Vaquerizas et al., 2004) was employed to normalize the data using background‐subtracted median values and an intensity‐based Lowess function within and among microarrays. The preprocessing application included in GEPAS (Herrero et al., 2003) was used to merge gene replicate values. Finally, genes detected in only one of the three biological replicates were discarded.
The identification of differentially expressed genes was performed using SAM (Tusher et al., 2001) included in the TM4 Microarray Software Suite (Saeed et al., 2003). Genes that satisfied the statistical threshold (false discovery rate, adjusted P < 0.01) were considered to be differentially expressed. Multivariate analysis, including hierarchical cluster analysis and PCA, was performed using the GeneMaths XT software package (http://www.applied‐maths.com/). Pearson's product‐moment correlation coefficient was used as a measure for gene‐to‐gene correlation.
AgriGO (Zhou and Su, 2007) was used to extract GO terms that were significantly over‐ or under‐represented in a particular set of genes relative to a reference group composed of all genes present in the microarray which have an Arabidopsis thaliana homologue. To overlay the gene expression data derived from the microarray hybridizations onto a metabolic map, the OMICS Viewer tool from AraCyc 3.5 was used (Mueller et al., 2003).
Northern blot analysis
Northern blot analysis was carried out by electrophoresis of denatured total RNA (10 µg) in 1.2% (w/v) agarose–formaldehyde gel and blotted onto nylon Hybond‐N+ membrane (GE Healthcare, Buckinghamshire, England). cDNA labelling, hybridization and quantification were carried out as described previously by Ballester et al. (2006). Probes used for Northern hybridization were obtained from different cDNA libraries previously generated in our group (Table S8): (i) RindPdig24 cDNA library, which is derived from ‘Clemenules’ mandarins infected with P. digitatum (Forment et al., 2005; González‐Candelas et al., 2010); (ii) FlavCurFr1, which is derived from heat‐treated ‘Fortune’ mandarins (Forment et al., 2005); and (iii) RindPdigS, a subtractive cDNA library constructed from the peel of ‘Navelina’ oranges infected with P. digitatum (González‐Candelas et al., 2010).
For normalization, filters were hybridized to the 26S rDNA C. sinensis probe (Ballester et al., 2006). With few exceptions, for each gene, a value of 1.0 was assigned to the normalized signal of nontreated flavedo and the expression level of the rest of the samples was referred to it. After stripping the blots, they were hybridized using 28S rDNA P. digitatum probe (Ballester et al., 2006).
Ethylene production
Ethylene production from 10‐mm discs, obtained with a cork borer around the point of inoculation, was determined by incubating the discs in sealed glass tubes at 20 °C. After 15 min of incubation at this temperature, a 1‐mL headspace gas sample was withdrawn from each tube and analysed as described by Lafuente et al. (2001) with a Perkin‐Elmer gas chromatograph (GC) (Norwalk, CT, USA) equipped with a flame ionization detector and an alumina column (1 m × 2 mm diameter, 80/100 mesh) from Supelco (Barcelona, Spain). Nitrogen was used as carrier gas and the temperature of the column was maintained at 140 °C. Ethylene standard was obtained from Abello‐Oxígeno‐Linde S. A. (Valencia, Spain). The results are the means of three replicate samples of nine discs from three different oranges.
Supporting information
Fig. S1 Hierarchical view of gene ontology (GO) biological process categories significantly over‐represented in nontreated flavedo (FNT) compared with nontreated albedo (ANT) using AgriGO.
Fig. S2 Hierarchical view of gene ontology (GO) biological process categories significantly over‐represented in elicited flavedo 7 days after the beginning of the experiment (FIC7) compared with nontreated flavedo (FNT) using AgriGO.
Fig. S3 Hierarchical view of gene ontology (GO) biological process categories significantly over‐represented in elicited albedo 5 days after the beginning of the experiment (AIC5) compared with nontreated albedo (ANT) using AgriGO.
Fig. S4 Hierarchical view of gene ontology (GO) biological process categories significantly over‐represented in elicited albedo 7 days after the beginning of the experiment (AIC7) compared with nontreated albedo (ANT) using AgriGO.
Fig. S5 Graphical representation of the methionine and ethylene biosynthesis pathways showing gene expression values in the flavedo (F) and albedo (A) of elicited oranges. Light blue colour indicates genes with differential expression according to significant analysis of microarrays (SAM). The first number in parentheses indicates the number of genes spotted onto the microarray that have an Arabidopsis thaliana homologue, whereas the second number indicates the total number of A. thaliana genes for each step in the pathway. Numbers in squares indicate the log2 ratio of: a, I1/NT; b, IC4/NT; c, IC5/NT; d, IC7/NT. n.d., not detected. The symbol ‘+’ indicates no expression in NT and the symbol ‘–’ indicates expression in NT fruits but not in the compared treatment. NT, nontreated fruits; I1, infected fruits 1 day after pathogen inoculation; IC, infected and cured fruits at 4, 5 or 7 days after the beginning of the experiment.
Table S1 List of genes showing higher expression in flavedo than in albedo in all five situations analysed according to significant analysis of microarrays (SAM) (P < 0.01).
Table S2 List of genes showing higher expression in albedo than in flavedo in all five situations analysed according to significant analysis of microarrays (SAM) (P < 0.01).
Table S3 List of genes showing higher expression in nontreated flavedo (FNT) than in nontreated albedo (ANT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of the gene ontology (GO) (Fig. S1).
Table S4 List of genes showing higher expression in elicited flavedo 5 days after the beginning of the experiment (FIC5) than in nontreated flavedo (FNT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of gene ontology (GO) (Fig. 4).
Table S5 List of genes showing higher expression in elicited flavedo 7 days after the beginning of the experiment (FIC7) than in nontreated flavedo (FNT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of gene ontology (GO) (Fig. S2).
Table S6 List of genes showing higher expression in elicited albedo 5 days after the beginning of the experiment (AIC5) than in nontreated albedo (ANT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of gene ontology (GO) (Fig. S3).
Table S7 List of genes showing higher expression in elicited albedo 7 days after the beginning of the experiment (AIC7) than in nontreated albedo (ANT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of gene ontology (GO) (Fig. S4).
Table S8 Genes analysed by Northern blot hybridization.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
The technical assistance of Ana Izquierdo (IATA‐CSIC, Valencia‐Spain) is gratefully acknowledged. This work was supported by Research Grants AGL2002‐1227 and AGL2005‐04921‐C02‐01 from the Spanish Ministry of Science and Technology and PROMETEO/2010/010 from the Generalitat Valenciana. A‐RB, RCHdV and AGB acknowledge the Centre for Biosystems Genomics, which is part of the Netherlands Genomics Initiative, for additional funding.
REFERENCES
- Achilea, O. , Chalutz, E. , Fuchs, Y. and Rot, I. (1985a) Ethylene biosynthesis and related physiological changes in Penicillium digitatum infected grapefruit (Citrus paradisi). Physiol. Plant Pathol. 26, 125–134. [Google Scholar]
- Achilea, O. , Fuchs, Y. , Chalutz, E. and Rot, I. (1985b) The contribution of host and pathogen to ethylene biosyntesis in Penicillium digitatum‐infected citrus fruit. Physiol. Plant Pathol. 27, 55–63. [Google Scholar]
- Adato, A. , Mandel, T. , Mintz‐Oron, S. , Venger, I. , Levy, D. , Yativ, M. , Domínguez, E. , Wang, Z. , De Vos, R.C.H. , Jetter, R. , Schreiber, L. , Heredia, A. , Rogachev, I. and Aharoni, A. (2009) Fruit‐surface flavonoid accumulation in tomato is controlled by a SlMYB12‐regulated transcriptional network. PLoS Genet. 5, e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afek, U. , Orenstein, J. , Carmeli, S. , Rodov, V. and Joseph, M.B. (1999) Umbelliferone, a phytoalexin associated with resistance of immature Marsh grapefruit to Penicillium digitatum . Phytochemistry, 50, 1129–1132. [Google Scholar]
- Albrecht, U. and Bowman, K.D. (2008) Gene expression in Citrus sinensis (L.) Osbeck following infection with the bacterial pathogen Candidatus Liberibacter asiaticus causing Huanglongbing in Florida. Plant Sci. 175, 291–306. [Google Scholar]
- Angioni, A. , Cabras, P. , D'Hallewin, G. , Pirisi, F.M. , Reniero, F. and Schirra, M. (1998) Synthesis and inhibitory activity of 7‐geranoxycoumarin against Penicillium species in Citrus fruit. Phytochemistry, 47, 1521–1525. [Google Scholar]
- Arcas, M.C. , Botía, J.M. , Ortuño, A. and Del Río, J.A. (2000) UV irradiation alters the levels of flavonoids involved in the defence mechanism of Citrus aurantium fruits against Penicillium digitatum . Eur. J. Plant Pathol. 106, 617–622. [Google Scholar]
- Arras, G. (1996) Mode of action of an isolate of Candida famata in biological control of Penicillium digitatum in orange fruits. Postharvest Biol. Technol. 8, 191–198. [Google Scholar]
- Ballester, A.R. , Lafuente, M.T. and González‐Candelas, L. (2006) Spatial study of antioxidant enzymes, peroxidase and phenylalanine ammonia‐lyase in the citrus fruit–Penicillium digitatum interaction. Postharvest Biol. Technol. 39, 115–124. [Google Scholar]
- Ballester, A.R. , Izquierdo, A. , Lafuente, M.T. and González‐Candelas, L. (2010a) Biochemical and molecular characterization of induced resistance against Penicillium digitatum in citrus fruit. Postharvest Biol. Technol. 56, 31–38. [Google Scholar]
- Ballester, A.R. , Molthoff, J. , de Vos, R. , Hekkert, B.t.L. , Orzaez, D. , Fernandez‐Moreno, J.‐P. , Tripodi, P. , Grandillo, S. , Martin, C. , Heldens, J. , Ykema, M. , Granell, A. and Bovy, A. (2010b) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Yehoshua, S. , Rodov, V. , Kim, J.J. and Carmeli, S. (1992) Preformed and induced antifungal materials of citrus fruit in relation to the enhancement of decay resistance by heat and ultraviolet treatment. J. Agric. Food Chem. 40, 1217–1221. [Google Scholar]
- Ben‐Yehoshua, S. , Rodov, V. , Nafussi, B. , Peretz, J. , Porat, R. , D'hallewin, G. and Schirra, M. (2000) Biotic and abiotic induction of resistance against pathogens in citrus fruits In: Proceedings of the International Society of Citriculture, Orlando, FL, 3–7 December 2000 (International Society of Citriculture, publisher), pp. 1107–1112. [Google Scholar]
- Bernardi, J. , Licciardello, C. , Russo, M.P. , Chiusano, M.L. , Carletti, G. , Recupero, G.R. and Marocco, A. (2010) Use of a custom array to study differentially expressed genes during blood orange (Citrus sinensis L. Osbeck) ripening. J. Plant Physiol. 167, 301–310. [DOI] [PubMed] [Google Scholar]
- Bostock, R.M. (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu. Rev. Phytopathol. 43, 545–580. [DOI] [PubMed] [Google Scholar]
- Conrath, U. (2009) Priming of induced plant defense responses In: Advances in Botanical Research, Vol. 51 (Loon L.C.V., ed.), pp. 361–395. Amsterdam, The Netherlands: Academic Press. [Google Scholar]
- De Vos, M. , Van Oosten, V.R. , Van Poecke, R.M.P. , Van Pelt, J.A. , Pozo, M.J. , Mueller, M.J. , Buchala, A.J. , Métraux, J.P. , van Loon, L.C. , Dicke, M. and Pieterse, C. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant–Microbe Interact. 18, 923–937. [DOI] [PubMed] [Google Scholar]
- Dixon, M.T. , Achnine, L. , Kota, P. , Liu, C.J. , Reddy, S. and Wang, L. (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol. Plant Pathol. 3, 371–390. [DOI] [PubMed] [Google Scholar]
- Droby, S. , Chalutz, E. , Horev, B. , Cohen, L. , Gaba, V. , Wilson, C.L. and Wisniewski, M. (1993) Factors affecting UV‐induced resistance in grapefruit against the green mold decay caused by Penicillium digitatum . Plant Pathol. 42, 418–424. [Google Scholar]
- Droby, S. , Vinokur, V. , Weiss, B. , Cohen, L. , Daus, A. , Goldschmidt, E.E. and Porat, R. (2002) Induction of resistance to Penicillium digitatum in grapefruit by the yeast biocontrol agent Candida oleophila . Phytopathology, 92, 393–399. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E. and Dong, X. (2004) Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Ecker, J.R. (1995) The ethylene signal transduction pathway in plants. Science, 268, 667–675. [DOI] [PubMed] [Google Scholar]
- Fajardo, J.E. , McCollum, T.G. , McDonald, R.E. and Mayer, R.T. (1998) Differential induction of proteins in orange flavedo by biologically based elicitors and challenge by Penicillium digitatum . Sacc. Biol. Control. 13, 143–151. [Google Scholar]
- Ferreira, R.B. , Monteiro, S. , Freitas, R. , Santos, C.N. , Chen, Z. , Batista, L.M. , Duarte, J. , Borges, A. and Teixeira, A. (2006) Fungal pathogens: the battle for plant infection. Crit. Rev. Plant Sci. 25, 505–524. [Google Scholar]
- Forment, J. , Gadea, J. , Huerta, L. , Abizanda, L. , Agusti, J. , Alamar, S. , Alos, E. , Andres, F. , Arribas, R. , Beltrán, J.P. , Berbel, A. , Blazquez, M.A. , Brumos, J. , Canas, L.A. , Cercos, M. , Colmenero‐Flores, J.M. , Conesa, A. , Establés, B. , Gandia, M. , García‐Martínez, J.L. , Gimeno, J. , Gisbert, A. , Gomez, G. , González‐Candelas, L. , Granell, A. , Guerri, J. , Lafuente, M.T. , Madueno, F. , Marcos, J.F. , Marques, C. , Martinez, F. , Martínez‐Godoy, M.A. , Miralles, S. , Moreno, P. , Navarro, L. , Pallás, V. , Perez‐Amador, M.A. , Perez‐Valle, J. , Pons, C. , Rodrigo, I. , Rodriguez, P.L. , Royo, C. , Serrano, R. , Soler, G. , Tadeo, F. , Talón, M. , Terol, J. , Trenor, M. , Vaello, L. , Vicente, O. , Vidal, C. , Zacarías, L. and Conejero, V. (2005) Development of a citrus genome‐wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol. Biol. 57, 375–391. [DOI] [PubMed] [Google Scholar]
- Fujii, H. , Shimada, T. , Sugiyama, A. , Nishikawa, F. , Endo, T. , Nakano, M. , Ikoma, Y. , Shimizu, T. and Omura, M. (2007) Profiling ethylene‐responsive genes in mature mandarin fruit using a citrus 22K oligoarray. Plant Sci. 173, 340–348. [Google Scholar]
- Gandía, M. , Conesa, A. , Ancillo, G. , Gadea, J. , Forment, J. , Pallás, V. , Flores, R. , Duran‐Vila, N. , Moreno, P. and Guerri, J. (2007) Transcriptional response of Citrus aurantifolia to infection by Citrus tristeza virus. Virology, 367, 298–306. [DOI] [PubMed] [Google Scholar]
- Gimeno, J. , Gadea, J. , Forment, J. , Pérez‐Valle, J. , Santiago, J. , Martinez‐Godoy, A.M. , Yenush, L. , Belles, J.M. , Brumós, J. , Colmenero‐Flores, J. , Talon, M. and Serrano, R. (2009) Shared and novel molecular responses of mandarin to drought. Plant Mol. Biol. 70, 403–420. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- González‐Candelas, L. , Alamar, S. , Sanchez‐Torres, P. , Zacarias, L. and Marcos, J. (2010) A transcriptomic approach highlights induction of secondary metabolism in citrus fruit in response to Penicillium digitatum infection. BMC Plant Biol. 10, 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, J. , Díaz‐Uriarte, R. and Dopazo, J. (2003) Gene expression data preprocessing. Bioinformatics, 19, 655–656. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kavanagh, J.A. and Wood, R.K.S. (1967) The role of wounds in the infection of oranges by Penicillium digitatum . Sacc. Ann. Appl. Biol. 60, 375–383. [Google Scholar]
- Kim, J.J. , Ben Yehoshua, S. , Shapiro, B. , Henis, Y. and Carmeli, S. (1991) Accumulation of scoparone in heat‐treated lemon fruit inoculated with Penicillium digitatum Sacc. Plant Physiol. 97, 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.‐S. , Sagaram, U.S. , Burns, J.K. , Li, J.‐L. and Wang, N. (2009) Response of sweet orange (Citrus sinensis) to ‘Candidatus Liberibacter asiaticus’ infection: microscopy and microarray analyses. Phytopathology, 99, 50–57. [DOI] [PubMed] [Google Scholar]
- Király, L. , Barna, B. and Király, Z. (2007) Plant resistance to pathogen infection: forms and mechanisms of innate and acquired resistance. J. Phytopathol. 155, 385–396. [Google Scholar]
- Lafuente, M.T. and Sala, J.M. (2002) Abscisic acid levels and the influence of ethylene, humidity and storage temperature on the incidence of postharvest rindstaning of ‘Navelina’ orange (Citrus sinensis L. Osbeck) fruit. Postharvest Biol. Technol. 25, 49–57. [Google Scholar]
- Lafuente, M.T. , Zacarías, L. , Martínez‐Téllez, M.A. , Sánchez‐Ballesta, M.T. and Dupille, E. (2001) Phenylalanine ammonia‐lyase as related to ethylene in the development of chilling symptoms during cold storage of citrus fruits. J. Agric. Food Chem. 49, 6020–6025. [DOI] [PubMed] [Google Scholar]
- Lers, A. , Burd, S. , Lomaniec, E. , Droby, S. and Chalutz, E. (1998) The expression of a grapefruit gene encoding an isoflavone reductase‐like protein is induced in response to UV irradiation. Plant Mol. Biol. 36, 847–856. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- López‐García, B. , González‐Candelas, L. , Pérez‐Payá, E. and Marcos, J.F. (2000) Identification and characterization of a hexapeptide with activity against phytopathogenic fungi that cause postharvest decay in fruits. Mol. Plant–Microbe Interact. 13, 837–846. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O. , Piqueras, R. , Sánchez‐Serrano, J.J. and Solano, R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell, 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos, J.F. , González‐Candelas, L. and Zacarías, L. (2005) Involvement of ethylene biosynthesis and perception in the susceptibility of citrus fruit to Penicillium digitatum infection and the accumulation of defense‐related mRNAs. J. Exp. Bot. 56, 2183–2193. [DOI] [PubMed] [Google Scholar]
- Martinez‐Godoy, A.M. , Mauri, N. , Juarez, J. , Marques, C.M. , Santiago, J. , Forment, J. and Gadea, J. (2008) A genome‐wide 20 K citrus microarray for gene expression analysis. BMC Genomics, 9, 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayuoni, L. , Sharabi‐Schwager, M. , Feldmesser, E. and Porat, R. (2011) Effects of ethylene degreening on the transcriptome of mandarin flesh. Postharvest Biol. Technol. 60, 75–82. [Google Scholar]
- McCollum, G. (2000) Defensive proteins in grapefruit flavedo In: Proceedings of the International Society of Citriculture, Orlando, FL, 3–7 December 2000 (International Society of Citriculture, publisher), pp. 1113–1116. [Google Scholar]
- Montaner, D. , Tárraga, J. , Huerta‐Cepas, J. , Burguet, J. , Vaquerizas, J.M. , Conde, L. , Minguez, P. , Vera, J. , Mukherjee, S. , Valls, J. , Pujana, M.A.G. , Alloza, E. , Herrero, J. , Al‐Shahrour, F. and Dopazo, J. (2006) Next station in microarray data analysis: GEPAS. Nucleic Acids Res. 34, W486–W491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, L.A. , Zhang, P. and Rhee, S.Y. (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 132, 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore, K.S. and Ryu, C.M. (2004) Nonhost resistance: how much do we know? Trends Plant Sci. 9, 97–104. [DOI] [PubMed] [Google Scholar]
- Ortuño, A. , Báidez, A. , Gómez, P. , Arcas, M.C. , Porras, I. , García‐Lidón, A. and Del Río, J.A. (2006) Citrus paradisi and Citrus sinensis flavonoids: their influence in the defence mechanism against Penicillium digitatum . Food Chem. 98, 351–358. [Google Scholar]
- Pavoncello, D. , Lurie, S. , Droby, S. and Porat, R. (2001) A hot water treatment induces resistance to Penicillium digitatum and promotes the accumulation of heat shock and pathogenesis‐related proteins in grapefruit flavedo. Physiol. Plant. 111, 17–22. [Google Scholar]
- Porat, R. , Lers, A. , Dori, S. , Cohen, L. , Weiss, B. , Daus, A. , Wilson, C.L. and Droby, S. (1999a) Induction of chitinase and beta‐1,3‐endoglucanase proteins by UV irradiation and wounding in grapefruit peel tissue. Phytoparasitica, 27, 1–6. [Google Scholar]
- Porat, R. , Weiss, B. , Cohen, L. , Daus, A. , Goren, R. and Droby, S. (1999b) Effects of ethylene and 1‐methylcyclopropene on the postharvest qualities of ‘Shamouti’ oranges. Postharvest Biol. Technol. 15, 155–163. [Google Scholar]
- Porat, R. , Vinokur, V. , Holland, D. , McCollum, T.G. and Droby, S. (2001) Isolation of a citrus chitinase cDNA and characterization of its expression in response to elicitation of fruit pathogen resistance. J. Plant Physiol. 158, 1585–1590. [Google Scholar]
- Porat, R. , McCollum, T.G. , Vinokur, V. and Droby, S. (2002) Effects of various elicitors on the transcription of a β‐1,3‐endoglucanase gene in citrus fruit. J. Phytopathol. 150, 70–75. [Google Scholar]
- Rodov, V. , Ben‐Yehoshua, S. , Kim, J.J. , Shapiro, B. and Ittah, Y. (1992) Ultraviolet illumination induces scoparone production in kumquat and orange fruit and improves decay resistance. J. Am. Soc. Hortic. Sci. 117, 788–792. [Google Scholar]
- Saeed, A.I. , Sharov, V. , White, J. , Li, J. , Liang, W. , Bhagabati, N. , Braisted, J. , Klapa, M. , Currier, T. , Thiagarajan, M. , Sturn, A. , Snuffin, M. , Rezantsev, A. , Popov, D. , Ryltsov, A. , Kostukovich, E. , Borisovsky, I. , Liu, Z. , Vinsavich, A. , Trush, V. and Quackenbush, J. (2003) TM4: a free, open‐source system for microarray data management and analysis. BioTechniques, 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Ballesta, M.T. , Zacarías, L. , Granell, A. and Lafuente, M.T. (2000) Accumulation of Pal transcript and Pal activity as affected by heat‐conditioning and low‐temperature storage and its relation to chilling sensitivity in Mandarin fruits. J. Agric. Food Chem. 48, 2726–2731. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M. , Kazan, K. , Wilson, I. , Anderson, J.P. , Richmond, T. , Somerville, S.C. and Manners, J.M. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA, 97, 11 655–11 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, T. , Fuiii, H. , Endo, T. , Yazaki, J. , Kishimoto, N. , Shimbo, K. , Kikuchi, S. and Omura, M. (2005) Toward comprehensive expression profiling by microarray analysis in citrus: monitoring the expression profiles of 2213 genes during fruit development. Plant Sci. 168, 1383–1385. [Google Scholar]
- Stracke, R. , Werber, M. and Weisshaar, B. (2001) The R2R3‐MYB gene family in Arabidopsis thaliana . Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Talon, M. and Gmitter, F.G. , Jr (2008) Citrus genomics. Int. J. Plant Genomics, 2008, Article ID 528361, 17 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V.G. , Tibshirani, R. and Chu, G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas, J.M. , Dopazo, J. and Díaz‐Uriarte, R. (2004) DNMAD: web‐based diagnosis and normalization for microarray data. Bioinformatics, 20, 3656–3658. [DOI] [PubMed] [Google Scholar]
- Venditti, T. , Molinu, M.G. , Dore, A. , Agabbio, M. and D'Hallewin, G. (2005) Sodium carbonate treatment induces scoparone accumulation, structural changes, and alkalinization in the albedo of wounded Citrus fruits. J. Agric. Food Chem. 53, 3510–3518. [DOI] [PubMed] [Google Scholar]
- Zhou, X. and Su, Z. (2007) EasyGO: Gene Ontology‐based annotation and functional enrichment analysis tool for agronomical species. BMC Genomics, 8, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Hierarchical view of gene ontology (GO) biological process categories significantly over‐represented in nontreated flavedo (FNT) compared with nontreated albedo (ANT) using AgriGO.
Fig. S2 Hierarchical view of gene ontology (GO) biological process categories significantly over‐represented in elicited flavedo 7 days after the beginning of the experiment (FIC7) compared with nontreated flavedo (FNT) using AgriGO.
Fig. S3 Hierarchical view of gene ontology (GO) biological process categories significantly over‐represented in elicited albedo 5 days after the beginning of the experiment (AIC5) compared with nontreated albedo (ANT) using AgriGO.
Fig. S4 Hierarchical view of gene ontology (GO) biological process categories significantly over‐represented in elicited albedo 7 days after the beginning of the experiment (AIC7) compared with nontreated albedo (ANT) using AgriGO.
Fig. S5 Graphical representation of the methionine and ethylene biosynthesis pathways showing gene expression values in the flavedo (F) and albedo (A) of elicited oranges. Light blue colour indicates genes with differential expression according to significant analysis of microarrays (SAM). The first number in parentheses indicates the number of genes spotted onto the microarray that have an Arabidopsis thaliana homologue, whereas the second number indicates the total number of A. thaliana genes for each step in the pathway. Numbers in squares indicate the log2 ratio of: a, I1/NT; b, IC4/NT; c, IC5/NT; d, IC7/NT. n.d., not detected. The symbol ‘+’ indicates no expression in NT and the symbol ‘–’ indicates expression in NT fruits but not in the compared treatment. NT, nontreated fruits; I1, infected fruits 1 day after pathogen inoculation; IC, infected and cured fruits at 4, 5 or 7 days after the beginning of the experiment.
Table S1 List of genes showing higher expression in flavedo than in albedo in all five situations analysed according to significant analysis of microarrays (SAM) (P < 0.01).
Table S2 List of genes showing higher expression in albedo than in flavedo in all five situations analysed according to significant analysis of microarrays (SAM) (P < 0.01).
Table S3 List of genes showing higher expression in nontreated flavedo (FNT) than in nontreated albedo (ANT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of the gene ontology (GO) (Fig. S1).
Table S4 List of genes showing higher expression in elicited flavedo 5 days after the beginning of the experiment (FIC5) than in nontreated flavedo (FNT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of gene ontology (GO) (Fig. 4).
Table S5 List of genes showing higher expression in elicited flavedo 7 days after the beginning of the experiment (FIC7) than in nontreated flavedo (FNT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of gene ontology (GO) (Fig. S2).
Table S6 List of genes showing higher expression in elicited albedo 5 days after the beginning of the experiment (AIC5) than in nontreated albedo (ANT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of gene ontology (GO) (Fig. S3).
Table S7 List of genes showing higher expression in elicited albedo 7 days after the beginning of the experiment (AIC7) than in nontreated albedo (ANT). Those genes with an Arabidopsis thaliana homologue were used to generate the hierarchical view of gene ontology (GO) (Fig. S4).
Table S8 Genes analysed by Northern blot hybridization.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


