SUMMARY
Cloning the first avirulence (avr) gene has led not only to a deeper understanding of gene‐for‐gene interactions in plant disease, but also to fundamental insights into the suppression of basal defences against microbial attack. This article (focusing on Pseudomonas syringae) charts the development of ideas and research progress over the 25 years following the breakthrough achieved by Staskawicz and coworkers. Advances in gene cloning technology underpinned the identification of both avr and hrp genes, the latter being required for the activation of the defensive hypersensitive reaction (HR) and pathogenicity. The delivery of Avr proteins through the type III secretion machinery encoded by hrp gene clusters was demonstrated, and the activity of the proteins inside plant cells as elicitors of the HR was confirmed. Key roles for avr genes in pathogenic fitness have now been established. The rebranding of Avr proteins as effectors, proteins that suppress the HR and cell wall‐based defences, has led to the ongoing search for their targets, and is generating new insights into the co‐ordination of plant resistance against diverse microbes. Bioinformatics‐led analysis of effector gene distribution in genomes has provided a remarkable view of the interchange of effectors and also their functional domains, as the arms race of attack and defence drives the evolution of microbial pathogenicity. The application of our accrued knowledge for the development of disease control strategies is considered.
In the preface to his seminal monograph on Physiological Plant Pathology, R.K.S. Wood (1967) made the simple but profound statement:
. . . there is the frustrating fact that most plants are resistant to colonization by most bacteria and fungi. They are naturally in a state that we still seek to reproduce by the use of fungicides that for the most part have been discovered . . . by empirical methods.
His comments highlight that disease is very much the exceptional outcome of microbe–plant encounters. It is reasonable to suggest that no‐one had the foresight to consider in the 1970s and 1980s that research on gene‐for‐gene interactions might lead to an explanation of how pathogens overcome basal defences using effector proteins, but that is where research has led us. In this article, I retrace, keeping to chronological order where possible and focusing on Pseudomonas syringae, how the breakthrough of the cloning of avirulence (avr) genes has opened up remarkable insights into microbial pathogenicity and plant innate immunity. Many of the topics introduced here are covered in more merited depth in other articles in this volume. Further details on bacterial effectors are given in reviews by Vivian and Arnold (2000), Alfano and Collmer (2004), Mudgett (2005), Grant et al. (2006) and Bent and Mackey (2007).
THE QUEST FOR THE HOLY GRAIL: RACE‐SPECIFIC ELICITORS
In the dark ages before the cloning of the first avr gene (Staskawicz et al., 1984), research had focused on the biochemistry and physiology of plant defence in crop plants. The identification of phytoalexins as strongly antimicrobial compounds that accumulated after infection, and had the potential to restrict microbial growth in plant tissues, led to a search for the elicitors of their biosynthesis (Mansfield, 2000; Mansfield et al., 1974). A number of non‐specific elicitors, including glucans and glycoproteins, were identified and characterized biochemically (Ebel et al., 1976; Hahn et al., 1981). The early emphasis was on the attempted identification of specific elicitors that would activate the hypersensitive reaction (HR) and the associated accumulation of phytoalexins only in resistant crop varieties (Bailey, 1982). Several plant–pathogen systems were developed and examined in great detail, but the real prizes—the elicitors that displayed race and resistance (R) gene specificity—were elusive. The breakthrough came with research by De Wit and colleagues on Cladosporium fulvum, the tomato leaf mould fungus which has an intercellular growth habit much the same as that observed with leaf spotting bacteria, such as pathovars of P. syringae. De Wit and Spikman (1982) demonstrated the presence of R gene‐specific elicitors of HR‐like reactions in intercellular fluids recovered from infected leaves. The direct search for elicitors was therefore encouraged by what we now know to be the rather exceptional release of Avr proteins by C. fulvum (De Wit, 1995). It is interesting to compare the success achieved with the C. fulvum–tomato interaction with the earlier failure of Lyon and Wood (1976) to detect similar activity in apoplastic fluids recovered from leaves colonized by bacteria. Despite completing an impeccably logical series of experiments, quite simply none of the extracts recovered from bacterial infections showed eliciting activity. Research using other approaches ran into the same impasse—a persistent contact between bacteria and plant cells was established as an essential requirement for the transfer of any elicitor (Harper et al., 1987).
ENTER THE WHITE KNIGHTS: BROAD HOST RANGE CLONING VECTORS
The cloning of avr genes would have been near impossible had it not been for the development of efficient broad host range cloning vectors, notably the pRK290 series pioneered by Ditta et al. (1980) and developed as cosmid vectors by Friedman et al. (1982) and Staskawicz et al. (1987). As Brian Staskawicz describes in this volume, the development of new technologies for cloning and the production of cosmid‐based genomic libraries drove new approaches to dissect race‐specific resistance. The new technologies allowed a novel strategy to be adopted and led to the approach that might be summarized as: ‘If you cannot find the elicitor—clone the gene’. The new vectors allowed avr genes to be cloned by function, moving genomic libraries between strains/races of bacteria and searching for changes in virulence. It is important to recognize that, without the pathology and plant genetics that had been used earlier to characterize different races of bacteria and differential varieties of their hosts, the new technologies would have floundered. Collaborative programmes of research between plant breeders and molecular biologists allowed rapid progress to be made with model systems, notably the P. syringae–soybean/bean/pea and Xanthomonas campestris pv. vesicatoria–pepper interactions. An example of the essential hypothetical gene‐for‐gene matrix established that allowed the isolation of avr genes from the bean halo‐blight bacterium P. syringae pv. phaseolicola (Pph) is shown in Fig. 1.
Figure 1.
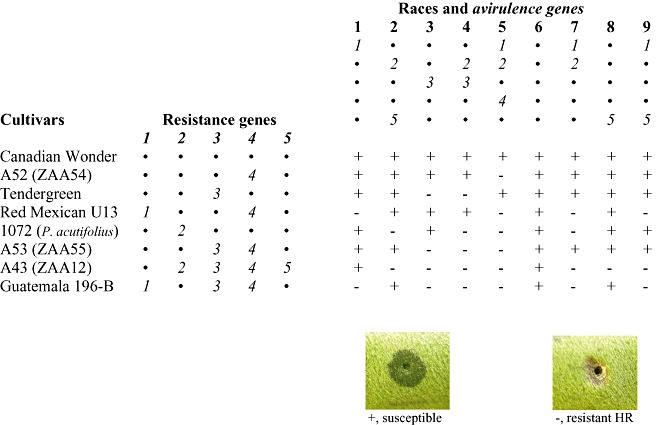
The Phaseolus bean halo‐blight disease gene‐for‐gene matrix proposed by Taylor et al. (1996) to explain race specificity in the Phaseolus syringae pv. phaseolicola (Pph)–Phaseolus vulgaris interaction. The avr genes cloned by function from Pph include avrPphB, avrPphE and avrPphF, which correspond to avr genes 3, 2 and 1 indicated in the matrix. Stab inoculation assays for virulence in bean pods are also illustrated, comparing susceptible water‐soaked lesions with the hypersensitive resistance reaction. Despite the application of targeted cloning and selection strategies, the avr gene matching R4 has not been identified. Other avr genes recovered from Pph by the exchange of genomic libraries between pathovars and assays in soybean and pea include avrPphC and avrPphD, respectively (Arnold et al., 2001a; Yucel et al., 1994).
The efficiency of cosmid libraries prepared with inserts of around 25 kb means that 1000 clones cover the whole genome. Coupled with a pathogenicity test allowing clear qualitative distinction between susceptible and resistant reactions, for example the bean pod assay system (Harper et al., 1987), it was comparatively facile to conjugate individual clones from donor Escherichia coli into the recipient P. syringae strain and search for changes in virulence. In practice, more than one active clone containing overlapping inserts was usually recovered from such experiments. The elegance of the technique was matched by the excitement of finding a clone that worked as expected, changing reactions from disease to the HR. The selection of races as library donors and recipients meant that the functional cloning could have recovered genes that operated as virulence factors, suppressing the HR. For example, referring to Fig. 1, moving the race 3 genomic library into race 5 and screening on bean cv. Tendergreen recovered the avirulence gene avrPphB matching the R3 gene for resistance, but screening in cv. Red Mexican, which was resistant to race 5, did not recover any clones conferring virulence (Hitchin et al., 1989). A second important feature of the screening experiments carried out with P. syringae pathovars and their hosts soybean, pea and bean was that, in all cases, the cloned avr gene was recognized by the activation of a rapid HR. Although the primary role of the HR in genes‐for‐gene interactions has been questioned (Bendahmane et al., 1999; Mansfield et al., 1997), genetic dissection failed to separate resistance from the HR in these bacterial systems.
If functional cloning recovered avr genes that resolved gene‐for‐gene interactions between pathogens and their hosts, what happened when the control of non‐host resistance also conferred by an HR was examined? Moving a P. syringae pv. tomato library into P. syringae pv. glycinea, and testing on soybean, recovered new genes avrD and avrE active in controlling the HR in the non‐host (Kobayashi et al., 1989; Lorang and Keen, 1995). Similarly, reciprocal exchange between P. syringae pv. pisi and P. syringae pv. phaseolicola identified new avr genes, but also demonstrated that some recovered as determinants of cultivar specificity had effects on non‐host plants (Arnold et al., 2001b; Fillingham et al., 1992). Significantly avrPpiA, identified from research on the legumes pea and bean (Vivian et al., 1989), was also found to trigger the HR in Arabidopsis if transferred into the crucifer pathogens P. syringae pv. maculicola and P. syringae pv. tomato (Dangl et al., 1992). avrPpiA was in fact nearly identical to avrRpm1 recovered from P. syringae pv. maculicola. Non‐host resistance to P. syringae appeared to be controlled by multiple avr–R gene interactions rather than a separate set of ‘non‐host super avr genes’ with less specific effects.
The precision allowed in using avr genes to ‘detect’ matching R genes raised some fascinating conundrums, notably the ability of the unrelated avrB and avrRpm1 to activate the HR in Arabidopsis through interaction with the cloned R gene RPM1 (Grant et al., 1995). This has led to interesting debates about genes‐for‐gene interactions which still remain incompletely resolved (Marathe and Dinesh‐Kumar, 2003). The robust strategy of cloning avr genes by function has stood the test of time and advances in genome sequencing. The core set of P. syringae avr genes identified by screening genomic libraries still provides the framework for the more recent bioinformatics‐led characterization of potential effectors (Kvitko et al., 2009).
THE RETURN TRIP TO ELICITORS VIA AN IN PLANTA EXPRESSION DETOUR
As the numbers of cloned avr genes increased rapidly during the 1980s and 1990s, attention returned to gene function. There were remarkably few clues from the bioinformatics analysis of the encoded proteins. At the time, the only consistent feature was their lack of similarity, apart from some homology between AvrB and AvrC (Tamaki et al., 1988). Intriguingly and exceptionally, strains of E. coli expressing avrD were found to elicit a cultivar‐specific HR in soybean (Keen et al., 1990); these authors also showed that infiltration of the AvrD protein into leaves did not cause the HR. This exciting finding suggested that a bacterial metabolite that was a direct or indirect product of AvrD acted as an elicitor. The syringolides, unusual acyl glycosides, were subsequently identified as the elicitors (Midland et al., 1993). The expression of other avr genes in E. coli (avrA, avrB, avrC, avrPphB, avrRpt2, avrRpm1 or avrPto) failed to demonstrate the similar production of elicitors (Keen, 1997). AvrD and syringolides stood out as a quite different scenario from that operating with other avr genes, whether from Pseudomonas or Xanthomonas. Significantly, E. coli strains expressing other avr genes were later found to be able to elicit HRs following their inoculation into leaves, but only if the E. coli strain also expressed the hrp gene cluster (Pirhonen et al., 1996; Puri et al., 1997; see below).
Vanderplank (1978), in a perceptive but rather eccentric review, argued that direct interaction between the products of avr and R genes was required to activate defence—that Avr proteins should themselves act as elicitors of the HR. His conclusion was based on knowledge of the genetics of gene‐for‐gene interactions accumulated over years of plant breeding for disease resistance in crop plants. In support of this theory, the elicitors from C. fulvum were shown to be small peptides (Scholtens‐Toma and De Wit, 1988), later to be identified as the processed protein products of the fungal avr genes (De Wit, 1995; Van den Ackerveken et al., 1992).
The demonstration that bacterial Avr proteins were capable of eliciting plant cell death when expressed in cells with the matching R gene was reported in landmark papers published in 1996 for avrB/RPM1 (Gopalan et al., 1996), avrB/RPM1 and avrRpt2/RPS2 (Leister et al., 1996), avrBs3/Bs3 (Van den Ackerveken et al., 1996) and avrPto/Pto (Scofield et al., 1996; Tang et al., 1996). Again the experiments were facilitated by the application of new technologies, in this case for plant transformation. The recognition that Avr proteins were themselves active in plant cells explained the significance of the initially puzzling presence of eukaryotic processing and targeting signals in the prokaryotic proteins; for example, myristoylation sites in AvrPhB and AvrRPM1 (Nimchuk et al., 2000) and nuclear targeting signals in the AvrBs3 family from X. campestris pv. vesicatoria (Szurek et al., 2001). We now understand that Avr proteins are often processed and targeted to their sites of action within plant cells.
Although the action of Avr proteins within plant cells was established and widely accepted, it soon became clear that they did not all interact directly with R proteins as proposed by Vanderplank (1978). AvrPto and AvrPtoB certainly bind to the Pto kinase, but the induction of the HR requires the presence of a second protein, Prf, which has the nucleotide‐binding site leucine‐rich repeat motif common to many R proteins. Van der Biezen and Jones (1998) first proposed the guard hypothesis, whereby the R protein does not interact directly with the Avr protein, but acts as a molecular guard to protect the effector target. Strong evidence for the hypothesis has come from the discovery of the RIN4 protein, which appears to regulate the interaction of AvrB and AvrRpm1 with RPM1 in Arabidopsis (Kim et al., 2005). Similarly, AvrPphB targets PBS1 (Zhu et al., 2004). Fascinatingly, AvrBs3‐related effectors have been shown to act as transcription factors, binding not to R proteins, but to the promoters of R and other target genes, to activate or suppress their expression (Gu et al., 2005; Kay et al., 2007; Romer et al., 2007). In certain fungal diseases, however, the Avr and R gene proteins do interact directly (2004, 2006; Jia et al., 2000). Despite the differences in Avr recognition, all of the established bacterial gene‐for‐gene interactions appear to confer resistance through the activation of the HR (Mansfield et al., 1997).
SPECIAL DELIVERY: THE ROLE OF THE hrp CLUSTER
In addition to cloning avr genes, the availability of genomic libraries and conjugation technology allowed the genetic dissection of pathogenicity determinants by mutational screens and complementation assays. Bacterial pathogens were mutagenized using chemicals such as nitrosoguanidine, or through transposon mutagenesis, and screened for the loss of pathogenicity and also the ability to cause the HR in non‐host plants (Bonas et al., 1991; Boucher et al., 1987; Daniels et al., 1984; Niepold et al., 1985; Van Gijsegem et al., 1995). Lindgren et al. (1986), using P. syringae pv. phaseolicola, were the first to confirm the link between pathogenicity and the ability to elicit the HR in resistant plants, whether host or non‐host. Their Tn5‐based mutagenesis strategy allowed a genomic library in pLAFR3 to be probed directly for clones containing hrp genes controlling the HR and pathogenicity. Importantly, the hrp genes were found in a cluster of at least 20 kb.
The full detail of hrp clusters was revealed through marker exchange mutagenesis and sequencing projects. Particularly significant was the work of Huynh et al. (1989), who defined the hrp regulon in P. syringae pv. glycinea and, using reporter gene constructs, demonstrated the key roles for hrpR/S and hrpL in the regulation of hrp gene expression. The link between hrpL and avr gene expression also emerged from this research, and led to the identification of the hrp box within the promoters of genes regulated by HrpL (Innes et al., 1993; Jenner et al., 1991).
The hrp box has subsequently been used as a bioinformatics tool to identify type III secretion system (T3SS)‐secreted Hop proteins (Hrp secreted out proteins) in the analysis of different P. syringae genomes (Fouts et al., 2002; Vencato et al., 2006; Zwiesler‐Vollick et al., 2002). The Hop proteins of P. syringae represent the repertoire of potential effectors within the bacterial pathogen. The most comprehensive analysis has now been completed with P. syringae pv. tomato DC3000. This Arabidopsis and tomato pathogen deploys 28 potential effectors. Many of these would not have been detected based simply on the screening for Avr functions (Cunnac et al., 2009; Kvitko et al., 2009).
The availability of clones containing all hrp genes from P. syringae pv. syringae (Huang et al., 1995), and all except hrpL from P. syringae pv. phaseolicola (Jenner et al., 1991), was exploited to show that non‐pathogenic Pseudomonas fluorescens and, indeed, E. coli could elicit an HR if they expressed both an avr gene and the complete hrp cluster (Pirhonen et al., 1996; Puri et al., 1997). Not all genes within hrp clusters were found to be essential for pathogenicity. Notably, hrpZ emerged as a puzzle because, in P. syringae, it encoded a protein which caused HR‐like symptoms in tobacco and some other plants, but did not seem to be essential for disease development (He et al., 1993; Lee et al., 2001). By contrast, the homologue HrpN from Erwinia amylovora has a clear role in the pathogenicity of the fireblight bacterium (Wei et al., 1992).
The sequencing of hrp clusters in diverse plant pathogenic bacteria revealed several genes containing conserved proteins and prompted a new hrc nomenclature for the conserved elements (Bogdanove et al., 1996). The analysis of protein similarities revealed the striking link between hrp genes and T3SS, first characterized in animal pathogens such as Yersinia pestis (Cornelis and Van Gijsegem, 2000; Fenselau and Bonas, 1995; Huang et al., 1995; Van Gijsegem et al., 1995). I first read about the similarities with the plague pathogen, having inadvertently injected my thumb with an E. coli strain expressing all the hrp genes, avrPphB and avrPphE from P. syringae pv. phaseolicola (Puri et al., 1997). Fortunately, the similarity between secretion systems does not extend significantly to secreted proteins!
How are Avr proteins delivered into plant cells through the T3SS? Clues came from research into pili on the bacterial surface and proteins secreted under conditions allowing hrp gene expression. 1997a, 1997b) brought together the two lines of research and demonstrated that HrpA encoded the subunits that not only formed, but were also capable of the autoassembly of, a 6–8‐nm‐diameter pilus. Similar pili had not, at that time, been directly linked with the T3SS, although filamentous appendages were known to be produced by Salmonella on contact with animal cells (Ginocchio et al., 1994).
The production of polyclonal antisera to the HrpA protein and the development of methods for the incubation of bacteria on grids used for electron microscopy (thereby maintaining delicate surface features intact) allowed the growth of the Hrp pilus to be examined in detail (Brown et al., 2001). The examination of P. syringae pv. tomato DC3000 bacterial infections in Arabidopsis identified gold‐labelled tracks of HrpA crossing the plant cell wall. Remarkably, the HrpA pilus was found to act as the needle of the T3SS syringe. Immunolocalization of HrpZ and AvrPto, which were both known to be secreted through the T3SS, revealed how the proteins accumulated along the growing pilus, as shown in Fig. 2.
Figure 2.
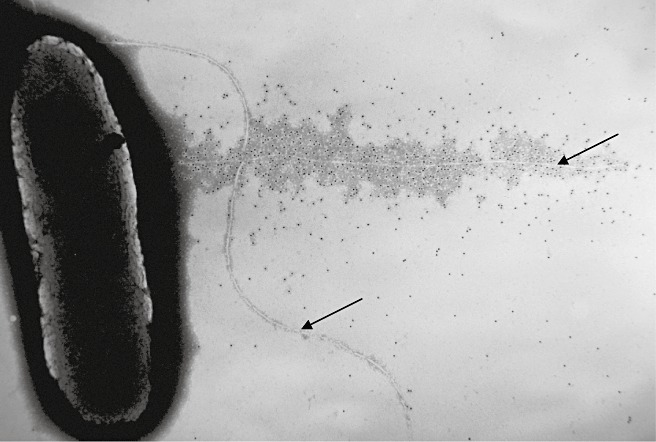
Immunogold localization of HrpZ (viewed as black electron‐dense dots) coating the HrpA pilus (top arrow) of Phaseolus syringae pv. tomato DC3000. A curved flagellum (bottom arrow) containing flagellin, which activates basal defences through the receptor FLS2 (Felix and Boller, 2009), is also shown, but is not associated with HrpZ. Image kindly provided by Ian Brown (see also Li et al., 2002).
It was not clear from these observations exactly what route was taken by the secreted proteins, for example along or through the pilus (Brown et al., 2001). The similarity between Hrp proteins and some flagellum assembly components was pointed out by He and Jin (2003). The assembly of the flagellum occurs at the tip of the developing appendage by the secretion of FliC subunits through the core basal body (MacNab, 1999). In order to distinguish basal or apical secretion of HrpA subunits, Li et al. (2002) used the ectopic expression of a FLAG‐tagged version of HrpA under the control of a mercury‐inducible promoter. Growing bacteria on electron microscopy grids under hrp‐inducing conditions allowed pilus formation to commence; then, by the addition of HgCl2, the tagged HrpA subunit was also induced and its incorporation into the growing pilus was visualized by immunogold localization using anti‐FLAG antibodies. The results showed, unambiguously, that the pilus grew from its tip, presumably following the secretion of subunits through the central lumen of the appendage (Li et al., 2002). A similar experimental design was used to follow the secretion of AvrPto and HrpZ (Jin and He, 2001; Li et al., 2002). As confirmed with HrpA itself, the other proteins were also secreted from the tip of the pilus. The similarity between flagellum assembly and type III secretion was confirmed. By deconstructing electron microscopic images, Tristan Boureau (University of Angers, France) has developed movies that illustrate the secretion of HrpA and HrpZ through the pilus, and these are provided in Supporting Information (Figs S1 and S2).
The mechanism by which proteins move through the pilus remains unknown, but commonly requires chaperones to support unfolded proteins (Büttner et al., 2002). Romantschuk et al. (2001) put forward several hypotheses, of which the pilus rotation/Archimedes' screw concept seems most likely, given the possible evolutionary links with the flagellum, and would entail rotation of the pilus as it is constructed by the addition of HrpA subunits. The energy source for protein transfer is probably the HrcN ATPase complex found to be located at the base of the T3SS architecture, as demonstrated for the P. syringae pv. phaseolicola hrp cluster expressed in E. coli (Pozidis et al., 2003). Although the secretion route may be similar, a clear difference between the T3SS and flagellum machinery lies in the coordination of expression of genes for assembly. Whereas the various flagellar components are induced in a sequential and apparently logical manner, the whole range of hrp gene operons seem to be induced almost simultaneously, without any pattern that would integrate with the ordered construction of the T3SS (Soutourina and Bertin, 2003; Thwaites et al., 2004).
THE TIME FOR METAMORPHOSIS: FROM AVIRULENCE GENE TO EFFECTOR
The idea that virulence to a wide range of varieties of a crop might carry a fitness penalty was proposed in several early studies of rust and powdery mildew diseases of cereals (Grant and Archer, 1983; Leonard, 1969). The cloning of avr genes allowed the first real test of the hypothesis that a loss of fitness might be a result of the lack of the avr gene product. How else could we explain the paradox of pathogens carrying genes that actually reduced their host range? Studies on the first few avr genes cloned did not support a general role in pathogenicity. For example, in P. syringae pv. phaseolicola, strains of race 6 lacked the three cloned avr genes avrPphB, avrPphE and avrPphF, but were fully virulent on all cultivars of bean tested (Fig. 1). The deletion of avrPpiA from P. syringae pv. pisi allowed the strain to colonize pea cultivars carrying the matching R2 gene, but did not generally reduce fitness (Gibbon et al., 1997). However, deletion of the avrPpiA homologue avrRpm1 from P. syringae pv. maculicola did indeed reduce the ability of the strain to colonize Arabidopsis (Ritter and Dangl, 1995), and fitness functions were attributed to avrA and avrE (Lorang et al., 1994). Although it was only functional in races 2, 4 and 7 of P. syringae pv. phaseolicola, forms of AvrPphE (encoded by avr gene 2 in Fig. 1) were present in all races of the bean pathogen, suggesting some unidentified role for the Avr‐inactive proteins (Stevens et al., 1998). Members of the AvrBs3 family of Avr proteins were shown to cause the symptom of pustule formation in pepper, but individual effectors were not absolutely required for colonization (Marois et al., 2002).
The concept of redundancy of effector functions compensating for the loss of individual avr genes was soon established. To overcome such redundancy required the deletion of several avr or effector genes within a strain. Such an approach has now been achieved through the elegant and rational deletion of effectors from P. syringae pv tomato DC3000, and their analysis in both tomato and Nicotiana benthamiana (Cunnac et al., 2009; Kvitko et al., 2009), but initial experiments relied on the natural clustering of avr genes on plasmids in P. syringae pv. phaseolicola (Jackson et al., 1999) or linked to the hrp cluster as conserved or exchangeable effector loci (CEL and EEL, respectively; Alfano et al., 2000).
The analysis of the role of cryptic plasmids in the pathogenicity of P. syringae pv. phaseolicola led to the discovery that the forced loss of the 154‐kb plasmid from race 7 strain 1449B produced a strain, RW60, that was no longer pathogenic to any bean cultivar, but triggered an HR‐like response on genotypes susceptible to the wild‐type strain (Jackson et al., 1999). Clones identified in a genomic library of 1449B containing the 154‐kb plasmid DNA were mated into RW60 and found to restore virulence. The clone pAV520 was particularly effective, and transposon mutagenesis and subcloning showed that several genes within the 30‐kb insert contributed to complementation. One gene was very active on its own, apparently suppressing the HR induced by RW60 and allowing water‐soaked lesions to develop in pods. Because of the clear virulence phenotype, the gene was named virPphA, as the first gene cloned for virulence function to be isolated from P. syringae pv. phaseolicola. Testing virPphA in soybean showed that it had avr gene activity in the non‐host; dual virulence (effector) and avirulence functions were therefore clearly established. Although VirPphA suppressed the HR‐like reaction caused by RW60 in genotypes normally susceptible to 1449B very effectively, it did not block the HR triggered by the avrPphE–R2 interaction (Jackson et al., 1999).
Several avr genes, including avrPphF, avrD and avrPphC, were also found to be clustered in pAV520 (Fig. 3). The proposal that effector functions were associated with suppression of the HR also received support from the analysis of avrPphF (Tsiamis et al., 2000). The avrPphF gene (homologous to hopPtoF) comprised two open reading frames, the smaller of which has subsequently been identified to encode a chaperone for the functional protein (Singer et al., 2004), and suppressed the HR caused by RW60 particularly effectively on cv. Tendergreen. In cv. Canadian Wonder, however, avrPphF enhanced the HR triggered by RW60 alone and a second gene within the region, avrPphC, was found to suppress the HR activity of avrPphF towards Canadian Wonder (Tsiamis et al., 2000). The gene‐for‐gene framework outlined in Fig. 1 is clearly not as simple as it seemed from the first analyses of cloned avr genes, but represents the outcome of unexplored interactions between several effectors and their targets, including the matching R proteins in bean.
Figure 3.
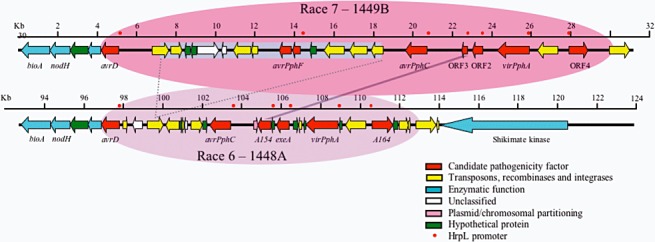
Comparison of the plasmid‐borne pathogenicity island containing effector genes in Phaseolus syringae pv phaseolicola strains 1448A (race 6) and 1449B (race 7). The cosmid clone pAV520, used to detect virulence functions, contains the region from 1449B shaded in dark pink (Jackson et al., 1999). The avrPphF gene conferring avirulence on beans with R1‐based resistance to halo‐blight is absent from 1448A, probably because of a deletion of 9471 nucleotides flanked by transposon fragments. In 1448A, an insertion of 10 nucleotides also generates a deduced 218AA effector, A514 (HopAW1). Kindly provided by Rob Jackson (see also Joardar et al., 2005; Rivas et al., 2005).
Homologues of VirPphA, which suppressed the HR in bean, were detected in numerous pathovars of P. syringae, either as closely similar proteins or more distantly related forms, such as AvrPtoB (Jackson et al., 2002). AvrPtoB, like AvrPto, interacts with the product of the Pto resistance gene, but, unlike AvrPto, AvrPtoB was found to suppress the HR in tomato (Abramovitch and Martin, 2005). Intriguingly, suppression of the avr gene‐triggered response was found to require E3 ubiquitin ligase activity identified for the multidomain AvrPtoB protein (Abramovitch et al., 2006; Janjusevic et al., 2006). The ability of effectors, including some newly identified from bioinformatic screens, to suppress plant cell death was also confirmed in tobacco by Jamir et al. (2004). They described suppressive activity for HopPtoE, HopPtoF (AvrPphF homologue), AvrPphE, AvrPpiB and AvrPtoB.
BEHIND ENEMY LINES, DISABLING SURVEILLANCE AND DEFENSIVE WEAPONRY
Work on the suppression of the HR firmly established a function for Avr proteins as effectors, but the HR is clearly not the only mechanism of defence against bacterial attack. Jakobek et al. (1993) first showed that non‐pathogens and hrp mutants induced defence gene expression without activating the HR. The structural framework of defence against non‐pathogens was investigated at the level of electron microscopy by Bestwick et al. (1995) using the P. syringae pv. phaseolicola–lettuce interaction, and also by Brown et al. (1995) in experiments with the well‐defined X. campestris pv. vesicatoria–pepper model. The reactions of both plants to the hrp mutants was characterized by the deposition of papillae containing callose (β‐1,3‐glucan) in cells next to bacteria in the intercellular space, and structural modification of the adjacent plant cell wall (illustrated in Fig. 4).
Figure 4.
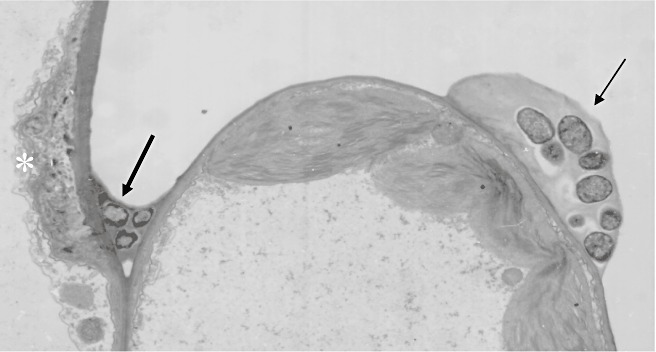
Suppression of basal defences by virulent Xanthomonas campestris pv. vesicatoria in pepper. Mixed inocula of wild‐type and hrp mutant bacteria were infiltrated into leaf mesophyll tissue. The hrp mutant bacteria were tagged with AvrBs3 and identified by immunocytochemistry. The hrp mutants (large arrow) have triggered papilla development (white asterisk) in one cell, but no papilla has formed in the cell in contact with the colony containing wild‐type X. campestris pv. vesicatoria, in which bacteria are embedded in extracellular polysaccharides (small arrow). It is noticeable, however, that chloroplasts have accumulated next to the wild‐type colony. Image kindly provided by Mansureh Keshavarzi; for further details see Keshavarzi et al. (2004).
Callose has emerged as a marker for such basal defence, but it is important to remember that many other modifications occur at reaction sites, including the deposition of phenolics (Bestwick et al., 1995), indolics (in Arabidopsis; Hagemeier et al., 2001) and various glycoproteins (Brown et al., 1998). The mechanisms by which such changes to the cell wall restrict bacterial multiplication within the intercellular space remain unknown, but it seems probable that oxidative cross‐linking of secreted wall components, polymerized through the action of local H2O2 accumulation, may act to agglutinate non‐pathogenic bacteria, generating bacteriostatic, but not strongly bacteriocidal, conditions (Bestwick et al., 1995; Brown et al., 1998; Soylu et al., 2005; K. Mitchell and J.W. Mansfield, unpublished data). In experiments with mixed inocula, using X. campestris pv. vesicatoria hrp mutants tagged with AvrBs3, the presence of wild‐type virulent bacteria was shown to suppress reactions to the hrp mutant, as illustrated in Fig. 4.
The critical link between effectors and the observed suppression of wall alterations was made by Sheng Yang He and colleagues, who found that the expression of AvrPto in Arabidopsis blocked the deposition of callose induced by hrp mutants and by DC3000 with a deletion in CEL (Debroy et al., 2004; Hauck et al., 2003). Importantly, Li et al. (2005) also found that the transient expression of AvrPto blocked the induction of transcription of the non‐host defence gene NHO1 by Arabidopsis protoplasts, normally caused by the conserved flagellin peptide (flg22), which is an example of a pathogen‐associated molecular pattern (PAMP). In addition to AvrPto, other effectors (HopS1, HopT1‐2, HopAA1‐1, HopF2 and HopC1) also suppressed defence gene expression. Finally, we were beginning to address the ability of pathogens to overcome basal defences that are induced by PAMPs.
The attenuated P. syringae pv. phaseolicola strain RW60 fails to cause an HR in Arabidopsis despite its functional T3SS. The strain was used as a donor to deliver members of the VirPphA family of effectors and AvrPto into Arabidopsis (de Torres et al., 2006). Only AvrPtoB significantly promoted bacterial colonization and effects were less marked in Columbia than the Wassilewskija accession that lacks FLS2. Induced expression in planta of avrPtoB, like avrPto, suppressed resistance to a hrpA mutant of P. syringae pv. tomato DC3000 and the callose response of leaf cells to the flg22 peptide (de Torres et al., 2006). The effect on PAMP was only observed if AvrPtoB was induced at least 1 h before treatment with flg22, indicating that, during natural infection, the timing of exposure of cells to PAMPs and the speed of delivery of effectors may significantly influence the outcome of bacterial challenge. Interestingly, deletion of the C‐terminus of AvrPtoB, to remove the ubiquitin ligase domain, did not completely prevent suppression of resistance to RW60, indicating that the enzymatic function attributed to the C‐terminus was not absolutely required for the suppression of PAMP‐induced immunity. A similar conclusion was reached by He et al. (2006) using the transient expression of AvrPtoB in Arabidopsis protoplasts.
Both AvrPto and AvrPtoB have subsequently been found to target the receptor‐like kinases FLS2, BAK1 and CERK, responsible for PAMP perception (Gimenez‐Ibanez et al., 2009; Shan et al., 2008; Xiang et al., 2008). Göhre et al. (2008) found that the ubiquitin ligase function of AvrPtoB was responsible for the observed removal of the FLS2 receptor from the Arabidopsis cell membrane during challenge by DC3000. The idea that a bacterial effector, such as AvrPtoB, may operate behind enemy lines, deactivating the PAMP ‘burglar alarm’ system, is intuitively attractive. However, both AvrPtoB and AvrPto are also known to modify the transcriptional response to challenge by hrp mutants (Hauck et al., 2003; de Torres et al., 2006; de Torres‐Zabala et al., 2007). Expression of AvrPtoB in planta also causes changes in hormone concentrations, notably the accumulation of abscisic acid (de Torres‐Zabala et al., 2007). It therefore seems probable that there are additional targets for AvrPtoB that may impact directly on defence gene expression.
Like the E3 ligase AvrPtoB, several P. syringae effectors have now been shown to have enzymatic function, notably ADP ribosyl transferases (HopU1, AvrPphF) and cysteine proteases (HopC1, HopN1 and AvrPphB), but the link between enzymatic activities and defence suppression, for example callose deposition and the HR, is by no means clear (Bock et al., 2008). As their targets begin to be revealed, however, effectors will become increasingly useful as probes to identify currently unknown components of the innate immune system. There is already good evidence for this in the finding that AvrPtoB (a ‘kinase killer’?) targets CERK, which had previously been thought to be responsible for the recognition of chitin‐containing fungi (Gimenez‐Ibanez et al., 2009), and AvrB cleaves the guardee RIN4, which has an undetermined role in defence (Kim et al., 2005; Lim and Kunkel, 2004). Unravelling how other targets or activities, for example the ribosyl transferase function of HopU1, compromise defence responses should provide new insights into the co‐ordination of immunity (Speth et al., 2007). One fascinating development is the finding that certain effectors, including both AvrPto and AvrPtoB, appear to target the virtually unexplored miRNA regulatory network (Navarro et al., 2008). Current knowledge of effector functions is succinctly summarized by Bock et al. (2008) and Cunnac et al. (2009). An interesting twist to the study of AvrPtoB is that its R gene‐encoded binding partner, the Pto kinase, actually blocks the ubiquitin ligase activity of the effector by phosphorylating the C‐terminal domain (Ntoukakis et al., 2009).
It seems probable, given that similar modes of defence are activated by taxonomically diverse groups of plant pathogens, that effectors from bacteria, oomycetes, fungi and even insects may target the same fundamental plant processes. Evidence is already emerging for common targets, for example the tomato defence protease Rcr3 is inhibited by oomycete and fungal effectors (Song et al., 2009), and defences against P. syringae are suppressed by the oomycete effectors ATR1 and ATR13 (Sohn et al., 2007).
THE CO‐EVOLUTIONARY ARMS RACE: SHUFFLING THE EFFECTOR PACK
Several authors have highlighted the apparent arms race between the development of virulence in microbes and defences in plants (Bergelson et al., 2001; Jones and Dangl, 2006; Tsiamis et al., 2000). Cloning avr genes and the subsequent identification of effector armouries have allowed bioinformatics analysis of the evolution of gene and protein structures, and the generation of genomic insights into the acquisition and loss of clusters of pathogenicity genes. Early experiments demonstrated that homologues of certain avr genes, such as avrPphE, were present in many pathovars of P. syringae, whereas others, such as avrPphB, were much less common (Mansfield et al., 1994). More recently, Stavrinides et al. (2006) have provided fascinating insights into the role of chimera formation, terminal reassortment and transposon rearrangements of individual effectors. The combination of functional domains within an effector is highlighted by the multiple activities reported for AvrPtoB (Abramovitch et al., 2006). Detailed analysis of HopZ1 homologues (including AvrPpiG; Arnold et al., 2001b) has revealed probable evolution through a process termed ‘pathoadaptation’ and also horizontal transfer (Ma et al., 2006).
Bioinformatics analyses present a retrospective view of how genomes have probably evolved. Evidence for the occurrence of avr gene mobility through the transposition of variable sections of chromosomes and plasmids has come from the identification of common flanking regions of DNA associated with different avr genes (Arnold et al., 2001b; Kim et al., 1998). Transposon insertions and deletions affecting the distribution of avrPphF have been characterized in strains of P. syringae pv. phaseolicola (Rivas et al., 2005). avrPphF is a component of the virulence region found on the 154‐kb plasmid in the bean pathogen. Sequencing the region in strains 1448A (race 6, lacking avrPphF) and 1449B (race 7 with avrPphF) reveals that a large deletion has removed avrPphF, allowing virulence on R1‐carrying varieties of bean. A 10‐nucleotide insertion in 1448A actually generates a second potential effector (A154, HopAW1) within the race 6 strain (Fig. 3). The deduced 218AA protein HopAW1 contains the catalytic triad CHD motif, indicating cysteine protease activity, as found in several Pseudomonas and Xanthomonas effectors (Joardar et al., 2005; Shao et al., 2002; Zhu et al., 2004).
Loss of the avrPphB gene from P. syringae pv. phaseolicola has provided a model system for the analysis of the evolution of virulence in real time. Race change towards the ability to colonize bean cultivars with the R3 gene for resistance was found to be a result of the loss of a genomic island containing avrPphB (Jackson et al., 2002; Pitman et al., 2005). Loss of the genomic island from the bacterial population was monitored following repeated passage of P. syringae pv. phaseolicola through the resistant bean cv. Tendergreen. The excised island was found to circularize after deletion from the bacterial chromosome, and recent data have shown that it transfers by transformation into other strains of P. syringae pv. phaseolicola, inserting at specific att sites in the chromosome (H. Lovell and D. L. Arnold, UWE Bristol, unpublished data). Both excision of the island and transformation competence appear to be enhanced by the stress to bacteria generated by the defensive HR (Arnold et al., 2007; Pitman et al., 2005). Exposure to plant innate immunity drives the evolution of more virulent strains of P. syringae pv. phaseolicola, first by activating genome rearrangements, and second by providing the selective pressure of an antimicrobial environment. An intriguing feature of the island is why it is retained by many African strains of P. syringae pv. phaseolicola, despite the presence of the R3 gene for resistance in local varieties (Taylor et al., 1996). Other genes on the island may improve fitness outside the plant.
CONCLUDING REMARKS: avr GENES AND DISEASE CONTROL?
Integrated and experimentally corroborated models of diverse effector functions and co‐ordinated defence suppression should soon emerge. The current state of play is well summarized by Bock et al. (2008). It should be borne in mind, however, that parasitism is a mode of microbial nutrition. Pathovars of Pseudomonas are biotrophic during the early phase of their colonization and must establish an intimate nutritional relationship with the living cells of their hosts. Many proteinaceous effectors may be found to have primary roles in the modification of plant metabolism to promote the release of bacterial nutrients from host cells rather than, or in addition to, the more easily assayed suppression of defences.
Although the future looks bright for research on effector cell biology and evolution, I am drawn back to the second part of Wood's preface which continued to state that . . .
It is a sobering but also a challenging thought that we know very little indeed . . . about the nature of this resistance, but we can be confident that anything significant we do learn . . . will help, perhaps considerably, in the development of better methods of controlling plant disease.
We now have remarkable insight into the molecular biology of attack and defence. However, his statement provides a strong reminder that, despite our improved understanding of innate immunity in plants, we still have to dig deep to find any examples of the new knowledge achieving an impact in agriculture and food production. Will we be able to design rationally new chemotherapeutics that target effector delivery or, indeed, restrict the transfer of pathogenicity islands between pathogens? Will the manipulation of genes regulating basal defences be able to confer broad spectrum resistance without reducing plant growth potential? Will it be possible to generate designer R genes that specifically interact with the effectors most needed for full pathogenicity? Whatever the answers to these questions, the next 25 years of work on avr genes should be strongly focused on bringing the application of our new knowledge to fruition.
Supporting information
Fig. S1 Growth of the Hrp pilus by the addition of HrpA subunits to the tip of the appendage in Pseudomonas syringae pv. tomato. The deconstructed electron micrograph shows the immunogold localization of Flag‐tagged HrpA subunits produced after an initial period of growth of the pilus in hrp gene‐inducing medium, followed by the induction of tagged HrpA by the addition of mercury. The black dots of gold label demonstrate the secretion of HrpA and its inclusion into the elongating pilus. Movie kindly provided by Tristan Boureau, University of Angers, France. For further details, see Li et al. (2002).
Fig. S2 Secretion of HrpZ through the Hrp pilus of Pseudomonas syringae pv. tomato. The deconstructed electron micrograph shows the immunogold localization of HrpZ produced after an initial period of growth of the pilus in hrp gene‐inducing medium, followed by induction of hrpZ by the addition of mercury. The black dots of gold label demonstrate the secretion of HrpZ from the tip of the elongating pilus. Movie kindly provided by Tristan Boureau, University of Angers, France. For further details, see Li et al. (2002).
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
ACKNOWLEDGEMENTS
This article includes part of the British Society for Plant Pathology Presidential Address presented in December 2000. I wish to acknowledge support from the Biotechnology and Biological Sciences Research Council (BBSRC) and the European Union (EU) for research on plant–microbe interactions in my laboratory. Many thanks are also due to numerous colleagues for synergistic interactions and, in particular, early collaborations with Mike Daniels, John Taylor and Alan Vivian.
REFERENCES
- Abramovitch, R.B. and Martin, G.B. (2005) Avrptob: a bacterial type III effector that both elicits and suppresses programmed cell death associated with plant immunity. FEMS Microbiol. Lett. 245, 1–8. [DOI] [PubMed] [Google Scholar]
- Abramovitch, R.B. , Janjusevic, R. , Stebbins, C.E. and Martin, G.B. (2006) Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. USA, 103, 2851–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Alfano, J.R. , Charkowski, A.O. , Deng, W.L. , Badel, J.L. , Petnicki‐Ocwieja, T. , Van Dijk, K. and Collmer, A. (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA, 97, 4856–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, D.L. , Gibbon, M.J. , Jackson, R.W. , Wood, J.R. , Brown, J. , Mansfield, J.W. , Taylor, J.D. and Vivian, A. (2001a) Molecular characterization of avrPphD, a widely‐distributed gene from Pseudomonas syringae pv. phaseolicola involved in non‐host recognition by pea (Pisum sativum). Physiol. Mol. Plant Pathol. 58, 55–62. [Google Scholar]
- Arnold, D.L. , Jackson, R.W. , Fillingham, A.J. , Goss, S.C. , Taylor, J.D. , Mansfield, J.W. and Vivian, A. (2001b) Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv. pisi . Microbiology, 147, 1171–1182. [DOI] [PubMed] [Google Scholar]
- Arnold, D.L. , Jackson, R.W. , Waterfield, N.R. and Mansfield, J.W. (2007) Evolution of microbial virulence: the benefits of stress. Trends Genet. 23, 293–300. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A. (1982) Mechanisms of phytoalexin accumulation In: Phytoalexins (Bailey J.A. and Mansfield J.W., eds), pp. 289–318. Glasgow: Blackie and Sons. [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Bergelson, J. , Dwyer, G. and Emerson, J.J. (2001) Models and data on plant–enemy coevolution. Annu. Rev. Genet. 35, 469–499. [DOI] [PubMed] [Google Scholar]
- Bestwick, C.S. , Bennett, M.H. and Mansfield, J.W. (1995) Hrp mutant of Pseudomonas syringae pv. phaseolicola induces cell wall alterations but not membrane damage leading to the HR in lettuce (Lactuca sativa). Plant Physiol. 108, 503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, A. , Li, G. , Fu, Z.Q. and Alfano, J. (2008) Phytopathogen type III weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Beer, S.V. , Bonas, U. , Boucher, C.A. , Collmer, A. , Coplin, D.L. , Cornelis, G.R. , Huang, H.C. , Hutcheson, S.W. , Panopoulos, N.J. and Van Gijsegem, F. (1996) Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20, 681–683. [DOI] [PubMed] [Google Scholar]
- Bonas, U. , Schulte, R. , Fenselau, S. , Minsavage, G.V. , Staskawicz, B.J. and Stall, R.E. (1991) Isolation of a gene‐cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant–Microbe Interact. 4, 81–88. [Google Scholar]
- Boucher, C.A. , Van Gijsegem, F. , Barberis, P.A. , Arlat, M. and Zischek, C. (1987) Pseudomonas solanacearum genes controlling both pathogenicity on tomato and hypersensitivity on tobacco are clustered. J. Bacteriol. 169, 5626–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, I.R. , Mansfield, J.W. and Bonas, U. (1995) hrp genes in Xanthomonas campestris pv. vesicatoria determine ability to suppress papilla deposition in pepper mesophyll cells. Mol. Plant–Microbe Interact. 8, 825–836. [Google Scholar]
- Brown, I. , Trethowan, J. , Kerry, M. , Mansfield, J.W. and Bolwell, G. (1998) Localisation of components of the oxidative cross‐linking of glycoproteins and of callose synthesis in papillae formed during the interaction between non‐pathogenic strains of Xanthomonas campestris and French bean mesophyll cells. Plant J. 15, 333–343. [Google Scholar]
- Brown, I. , Mansfield, J. , Taira, S. , Roine, E. and Romantschuk, M. (2001) Immunocytochemical localization of HrpA and HrpZ supports a role for the Hrp pilus in the transfer of effector proteins from Pseudomonas syringae pv. tomato across the host plant cell wall. Mol Plant–Microbe Interact. 14, 394–404. [DOI] [PubMed] [Google Scholar]
- Büttner, D. , Nennstiel, D. , Klusener, B. and Bonas, U. (2002) Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas pv. vesicatoria . J. Bacteriol. 184, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis, G.R. and Van Gijsegem, F. (2000) Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54, 735–774. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Lindeberg, M. and Collmer, A. (2009) Pseudomonas syringae type III effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. , Ritter, C. , Gibbon, M.J. , Mur, L.A. , Wood, J.R. , Goss, S. , Mansfield, J. , Taylor, J.D. and Vivian, A. (1992) Functional homologs of the Arabidopsis RPM1 disease resistance gene in bean and pea. Plant Cell, 4, 1359–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, M.J. , Barber, C.E. , Turner, P.C. , Sawczyc, M.K. , Byrde, R.J.W. and Fielding, A.H. (1984) Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J. 3, 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy, S. , Thilmony, R. , Kwack, Y.B. , Nomura, K. and He, S.Y. (2004) A family of conserved bacterial effectors inhibits salicylic acid‐mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA, 101, 9927–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit, P.J.G.M. (1995) Fungal avirulence genes and plant resistance genes: unraveling the molecular basis of gene‐for‐gene interactions. Adv. Bot. Res. 21, 148–185. [Google Scholar]
- De Wit, P.J.G.M. , Spikman, G. (1982) Evidence for the occurrence of race‐ and cultivar‐specific elicitors of necrosis in intercellular fluids of compatible interactions between Cladosporium fulvum and tomato. Physiol. Plant Pathol. 21, 1–11. [Google Scholar]
- Ditta, G.S. , Stanfield, S. , Corbin, D. and Helinski, D.R. (1980) Broad host‐range DNA cloning system for Gram‐negative bacteria: construction of a gene bank of Rhizobium meliloti . Proc. Natl. Acad. Sci. USA, 77, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Ayliffe, M.A. and Ellis, J.G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Teh, T. , Wang, C.I.A. , Ayliffe, M.A. , Kobe, B. and Ellis, J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel, J. , Ayers, A.R. , Albersheim, P. (1976) Host–pathogen interactions XII. Response of suspension‐cultured soybean cells to the elicitor isolated from Phytophthora megasperma var. sojae, a fungal pathogen of soybeans. Plant Physiol. 57, 775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G. and Boller, T. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Fenselau, S. and Bonas, U. (1995) Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol. Plant–Microbe Interact. 8, 845–854. [DOI] [PubMed] [Google Scholar]
- Fillingham, A.J. , Wood, J. , Bevan, J.R. , Crute, I.R. , Mansfield, J.W. , Taylor, J.D. and Vivian, A. (1992) Avirulence genes from Pseudomonas syringae pathovars phaseolicola and pisi confer specificity towards both host and non‐host species. Physiol. Mol. Plant Pathol. 40, 1–15. [Google Scholar]
- Fouts, D.E. , Abramovitch, R.B. , Alfano, J.R. , Baldo, A.M. , Buell, C.R. , Cartinhour, S. , Chatterjee, A.K. , D'Ascenzo, M. , Gwinn, M.L. , Lazarowitz, S.G. , Lin, N.C. , Martin, G.B. , Rehm, A.H. , Schneider, D.J. , Van Dijk, K. , Tang, X. and Collmer, A. (2002) Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA, 99, 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, A.M. , Long, S.R. , Brown, S.E. , Buikema, W.J. and Ausubel, F.M. (1982) Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene, 18, 289–296. [DOI] [PubMed] [Google Scholar]
- Gibbon, M.J. , Jenner, C. , Mur, L.A.J. , Puri, N. , Mansfield, J.W. , Taylor, J.D. and Vivian, A. (1997) Avirulence gene avrPpiA from Pseudomonas syringae pv. pisi is not required for full virulence on pea. Physiol. Mol. Plant Pathol. 50, 219–236. [Google Scholar]
- Gimenez‐Ibanez, S. , Hann, D.R. , Ntoukakis, V. , Petutschnig, E. , Lipka, V. and Rathjen, J.P. (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. [DOI] [PubMed] [Google Scholar]
- Ginocchio, C.C. , Olmsted, S.B. , Wells, C.L. and Galan, J.E. (1994) Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium . Cell, 76, 717–724. [DOI] [PubMed] [Google Scholar]
- Göhre, V. , Spallek, T. , Haweker, H. , Mersmann, S. , Mentzel, T. , Boller, T. , De Torres, M. , Mansfield, J.W. and Robatzek, S. (2008) Plant pattern‐recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Gopalan, S. , Bauer, D.W. , Alfano, J.R. , Loniello, A.O. , He, S.Y. and Collmer, A. (1996) Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype‐specific hypersensitive cell death. Plant Cell, 8, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M.R. , Godiard, L. , Straube, E. , Ashfield, T. , Lewald, J. , Sattler, A. , Innes, R.W. and Dangl, J.L. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science, 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Grant, M.W. and Archer, S.A. (1983) Calculation of selection coefficients against unnecessary genes for virulence from field data. Phytopathology, 73, 547–551. [Google Scholar]
- Grant, S.R. , Fisher, E.J. , Chang, J.H. , Mole, B.M. and Dangl, J.L. (2006) Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60, 425–449. [DOI] [PubMed] [Google Scholar]
- Gu, K. , Yang, B. , Tian, D. , Wu, L. , Wang, D. , Sreekala, C. , Yang, F. , Chu, Z. , Wang, G.L. , White, F.F. and Yin, Z. (2005) R gene expression induced by a type‐III effector triggers disease resistance in rice. Nature, 435, 1122–1125. [DOI] [PubMed] [Google Scholar]
- Hagemeier, J. , Schneider, B. , Oldham, N.J. and Hahlbrock, K. (2001) Accumulation of soluble and wall‐bound indolic metabolites in Arabidopsis thaliana leaves infected with virulent or avirulent Pseudomonas syringae pathovar tomato strains. Proc. Natl. Acad. Sci. USA, 98, 753–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M.G. , Darvill, A.G. and Albersheim, P. (1981) Host–pathogen interactions: XIX. The endogenous elicitor, a fragment of a plant cell wall polysaccharide that elicits phytoalexin accumulation in soybeans. Plant Physiol. 68, 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, S. , Zewdie, N. , Brown, I.R. and Mansfield, J.W. (1987) Histological, physiological and genetic studies of the responses of leaves and pods of Phaseolus vulgaris to three races of Pseudomonas syringae pv. phaseolicola and to Pseudomonas syringae pv. coronafaciens . Physiol. Mol. Plant Pathol. 31, 153–172. [Google Scholar]
- Hauck, P. , Thilmony, R. and He, S.Y. (2003) A Pseudomonas syringae type III effector suppresses cell wall‐based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA, 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P. , Shan, L. , Lin, N.C. , Martin, G.B. , Kemmerling, B. , Nürnberger, T. and Sheen, J. (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell, 125, 563–575. [DOI] [PubMed] [Google Scholar]
- He, S.Y. and Jin, Q. (2003) The Hrp pilus: learning from flagella. Curr. Opin. Microbiol. 6, 15–19. [DOI] [PubMed] [Google Scholar]
- He, S.Y. , Huang, H.C. and Collmer, A. (1993) Pseudomonas syringae pv. syringae HarpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell, 73, 1–20. [DOI] [PubMed] [Google Scholar]
- Hitchin, F.E. , Jenner, C.E. , Harper, S. , Mansfield, J.W. , Barber, C.E. and Daniels, M.J. (1989) Determinant of cultivar specific avirulence cloned from Pseudomonas syringae pv. phaseolicola race 3. Physiol. Plant Pathol. 34, 309–322. [Google Scholar]
- Huang, H.C. , Lin, R.H. , Chang, C.J. , Collmer, A. and Deng, W.L. (1995) The complete hrp gene cluster of Pseudomonas syringae pv. syringae 61 includes two blocks of genes required for HarpinPss secretion that are arranged colinearly with Yersinia ysc homologs. Mol. Plant–Microbe Interact. 8, 733–746. [DOI] [PubMed] [Google Scholar]
- Huynh, T.V. , Dahlbeck, D. and Staskawicz, B.J. (1989) Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science, 245, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Innes, R.W. , Bent, A.F. , Kunkel, B.N. , Bisgrove, S.R. and Staskawicz, B.J. (1993) Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J. Bacteriol. 175, 4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.W. , Athanassopoulos, E. , Tsiamis, G. , Mansfield, J.W. , Sesma, A. , Arnold, D.L. , Gibbon, M.J. , Murillo, J. , Taylor, J.D. and Vivian, A. (1999) Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola . Proc. Natl. Acad. Sci. USA, 96, 10 875–10 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.W. , Mansfield, J.W. , Ammouneh, H. , Dutton, L.C. , Wharton, B. , Barredo, A. , Arnold, D.L. , Tsiamis, G. , Sesma, A. , Butcher, D. , Boch, J. , Kim, Y.J. , Martin, G.B. , Tegli, S. , Murillo, J. and Vivian, A. (2002) Location and activity of members of a family of virPphA homologues in pathovars of Pseudomonas syringae and P. savastanoi . Mol. Plant Pathol. 3, 205–216. [DOI] [PubMed] [Google Scholar]
- Jakobek, J.L. , Smith, J.A. and Lindgren, P.B. (1993) Suppression of bean defense responses by Pseudomonas syringae . Plant Cell, 5, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamir, Y. , Guo, M. , Oh, H.‐S. , Petnicki‐Ocwieja, T. , Chen, S. , Tang, X. , Dickman, M.B. , Collmer, A. and Alfano, J.R. (2004) Identification of Pseudomonas syringae type III secreted effectors that suppress programmed cell death in plants and yeast. Plant J. 37, 554–565. [DOI] [PubMed] [Google Scholar]
- Janjusevic, R. , Abramovitch, R.B. , Martin, G.B. and Stebbins, C.E. (2006) A bacterial inhibitor of host programmed cell death is an E3 ubiquitin ligase. Science, 311, 221–226. [DOI] [PubMed] [Google Scholar]
- Jenner, C. , Hitchin, E. , Mansfield, J. , Walters, K. , Betteridge, P. and Taylor, J. (1991) Gene‐for‐gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus . Mol. Plant–Microbe Interact. 4, 553–562. [PubMed] [Google Scholar]
- Jia, Y. , McAdams, S.A. , Bryan, G.T. , Hershey, H.P. and Valent, B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q.L. and He, S.Y. (2001) Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae . Science, 294, 2556–2258. [DOI] [PubMed] [Google Scholar]
- Joardar, V. , Lindeberg, M. , Jackson, R.W. , Selengut, J. , Dodson, R. , Brinkac, L.M. , Daugherty, S.C. , DeBoy, R. , Durkin, A.S. , Giglio, M.G. , Madupu, R. , Nelson, W.C. , Rosovitz, M.J. , Sullivan, S. , Crabtree, J. , Creasy, T. , Davidsen, T. , Haft, D.H. , Zafar, N. , Zhou, L.W. , Halpin, R. , Holley, T. , Khouri, H. , Feldblyum, T. , White, O. , Fraser, C.M. , Chatterjee, A.K. , Cartinhour, S. , Schneider, D.J. , Mansfield, J. , Collmer, A. and Buell, C.R. (2005) Whole‐genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187, 6488–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.F. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Hause, G. and Bonas, U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science, 318, 648–651. [DOI] [PubMed] [Google Scholar]
- Keen, N.T. (1997) Elicitor generation and receipt—the mail gets through but how? In: The Gene‐for‐Gene Relationship in Host–Parasite Interactions (Crute I.R., Holub E.B. and Burdon J.J., eds), pp. 379–388. London: CAB International. [Google Scholar]
- Keen, N.T. , Tamaki, S. , Kobayashi, D. , Gerhold, D. , Stayton, M. , Shen, H. , Gold, S. , Lorang, J. , Thordal‐Christensen, H. , Dahlbeck, D. and Staskawicz, B. (1990) Bacteria expressing avirulence gene D produce a specific elicitor of the soybean hypersensitive reaction. Mol. Plant–Microbe Interact. 3, 112–121. [Google Scholar]
- Keshavarzi, M. , Soylu, S. , Brown, I. , Bonas, U. , Nicole, M. , Rossiter, J. and Mansfield, J. (2004) Basal defenses induced in pepper by lipopolysaccharides are suppressed by Xanthomonas campestris pv. vesicatoria . Mol. Plant–Microbe Interact. 7, 805–815. [DOI] [PubMed] [Google Scholar]
- Kim, J.F. , Charkowski, A.O. , Alfano, J.A. , Collmer, A. and Beer, S.V. (1998) Sequences related to transposable elements and bacteriophages flank avirulence genes of Pseudomonas syringae . Mol. Plant–Microbe Interact. 11, 1247–1252. [Google Scholar]
- Kim, M.G. , Da Cunha, L. , McFall, A.J. , Belkhadir, Y. , DebRoy, S. , Dangl, J.L. and Mackey, D. (2005) Two Pseudomonas syringae type III effectors inhibit RIN4‐regulated basal defense in Arabidopsis. Cell, 121, 749–759. [DOI] [PubMed] [Google Scholar]
- Kobayashi, D.A. , Tamaki, S.J. and Keen, N.T. (1989) Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer cultivar specificity on soybean. Proc. Natl. Acad. Sci. USA, 86, 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko, B.H. , Park, D.H. , Velásquez, A.C. , Wei, C.F. , Russell, A.B. , Martin, G.B. , Schneider, D.J. and Collmer, A. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388. Epub 2009 April 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Klüsener, B. , Tsiamis, G. , Stevens, C. , Neyt, C. , Cornelis, G.R. , Panopoulos, N.J. , Weiler, E.W. , Mansfield, J.W. and Nürnberger, T. (2001) HrpZPsph from the plant pathogen Pseudomonas syringae pv. phaseolicola is exported by the type III secretion pathway and forms an ion‐conducting pore in vitro. Proc. Natl. Acad. Sci. USA, 98, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister, R.T. , Ausubel, F.M. and Katagiri, F. (1996) Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc. Natl. Acad. Sci. USA, 93, 15 497–15 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, K.J. (1969) Selection in heterogeneous populations of Puccinia graminis f. sp. tritici . Phytopathology, 59, 1845–1850. [Google Scholar]
- Li, C.M. , Brown, I. , Mansfield, J. , Stevens, C. , Boureau, T. , Romantschuk, M. and Taira, S. (2002) The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J. 21, 1909–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Lin, H. , Zhang, W. , Zou, Y. , Zhang, J. , Tang, X. and Zhou, J. (2005) Flagellin induces innate immunity in non‐host interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA, 102, 12 990–12 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, M.T. and Kunkel, B.N. (2004) The Pseudomonas syringae type III effector AvrRpt2 promotes virulence independently of RIN4, a predicted virulence target in Arabidopsis thaliana . Plant J. 40, 790–798. [DOI] [PubMed] [Google Scholar]
- Lindgren, P.B. , Peet, R.C. and Panopoulos, N.J. (1986) Gene‐cluster of Pseudomonas syringae pv. phaseolicola controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J. Bacteriol. 168, 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang, J.M. and Keen, N.T. (1995) Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp‐linked avirulence locus consisting of at least two transcriptional units. Mol. Plant–Microbe Interact. 8, 49–57. [DOI] [PubMed] [Google Scholar]
- Lorang, J.M. , Shen, H. , Kobayashi, D. , Cooksey, D. and Keen, N.T. (1994) avrA and avrE in Pseudomonas syringae pv. tomato PT23 play a role in virulence on tomato plants. Mol. Plant–Microbe Interact. 7, 508–515. [Google Scholar]
- Lyon, F. and Wood, R.K.S. (1976) The hypersensitive reaction and other responses of bean leaves to bacteria. Ann. Bot. 40, 479–491. [Google Scholar]
- Ma, W. , Dong, F.F. , Stavrinides, J. and Guttman, D.S. (2006) Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2, e209. Epub 2006 October 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNab, R.M. (1999) The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181, 7149–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, J. , Jenner, C. , Hockenhull, R. , Bennett, M.A. and Stewart, R. (1994) Characterization of avrPphE, a gene for cultivar‐specific avirulence from Pseudomonas syringae pv. phaseolicola which is physically linked to hrpY, a new hrp gene identified in the halo blight bacterium. Mol. Plant–Microbe Interact. 7, 726–739. [DOI] [PubMed] [Google Scholar]
- Mansfield, J.W. (2000) Antimicrobial compounds and disease resistance In: Mechanisms of Disease Resistance in Plants (Slusarenko A., ed.), pp. 325–370. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Mansfield, J.W. , Hargreaves, J.A. and Boyle, F.C. (1974) Phytoalexin production by live cells in broad bean‐leaves infected with Botrytis cinerea . Nature, 252, 316–317. [Google Scholar]
- Mansfield, J.W. , Bennett, M.H. , Bestwick, C.S. and Woods‐Tor, A.M. (1997) Phenotypic expression of gene‐for‐gene interactions involving fungal and bacterial pathogens: variation from recognition to response In: The Gene‐for‐Gene Relationship in Host–Parasite Interactions (Crute I.R., Holub E.B. and Burdon J.J., eds), pp. 265–291. London: CAB International. [Google Scholar]
- Marathe, R. and Dinesh‐Kumar, S.P. (2003) Plant defense: one post, multiple guards?! Mol. Cell. 11, 284–286. [DOI] [PubMed] [Google Scholar]
- Marois, E. , Van den Ackerveken, G. and Bonas, U. (2002) The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant–Microbe Interact. 15, 637–646. [DOI] [PubMed] [Google Scholar]
- Midland, S.L. , Keen, N.T. , Sims, J.J. , Midland, M.M. , Stayton, M.M. , Burton, V. , Smith, M.J. , Mazzola, E.P. , Graham, J.K. and Clardy, J. (1993) The structure of syringolides 1 and 2, novel C glycosidic elicitors from Pseudomonas syringae pv. tomato . J. Org. Chem. 58, 2940–2945. [Google Scholar]
- Mudgett, M.B. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56, 509–531. [DOI] [PubMed] [Google Scholar]
- Navarro, L. , Jay, F. , Nomura, K. , He, S.Y. and Voinnet, O. (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science, 321, 964–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepold, F. , Anderson, D. and Mills, D. (1985) Cloning determinants of pathogenesis from Pseudomonas syringae pathovar syringae . Proc. Natl. Acad. Sci. USA, 82, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z. , Marois, E. , Kjemtrup, S. , Leister, R.T. , Katagiri, F. and Dangl, J.L. (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae . Cell, 101, 353–363. [DOI] [PubMed] [Google Scholar]
- Ntoukakis, V. , Mucyn, T.S. , Gimenez‐Ibanez, S. , Chapman, H.C. , Gutierrez, J.R. , Balmuth, A.L. , Jones, A.M. and Rathjen, J.P. (2009) Host inhibition of a bacterial virulence effector triggers immunity to infection. Science, 324, 784–787. [DOI] [PubMed] [Google Scholar]
- Pirhonen, M.U. , Lidell, M.C. , Rowley, D.L. , Lee, S.W. , Jin, S. , Liang, Y. , Silverstone, S. , Keen, N.T. and Hutcheson, S.W. (1996) Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp‐encoded secretion system. Mol. Plant–Microbe Interact. 9, 252–260. [DOI] [PubMed] [Google Scholar]
- Pitman, A. , Jackson, R.W. , Mansfield, J.M. , Kaitell, V. , Thwaites, R. and Arnold, D.L. (2005) Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr. Biol. 15, 2230–2235. [DOI] [PubMed] [Google Scholar]
- Pozidis, C. , Chalkiadaki, A. , Gomez‐Serrano, A. , Stahlberg, H. , Brown, I. , Tabakaki, A.P. , Lustig, A. , Sianidis, G. , Politou, S.A. , Engel, A. , Panopoulos, N.J. , Mansfield, J. , Pugsley, A.P. , Karamanou, S. and Economou, A. (2003) Type III protein translocase: HrcN is a peripheral membrane ATPase that is activated by oligomerization. J. Biol. Chem. 278, 25 816–25 824. [DOI] [PubMed] [Google Scholar]
- Puri, N. , Jenner, C. , Bennett, M.A. , Stewart, R. , Mansfield, J.W. , Lyons, N. and Taylor, J. (1997) Expression of avrPphB, an avirulence gene from Pseudomonas syringae pv. phaseolicola, and the delivery of signals causing the hypersensitive reaction in bean. Mol. Plant–Microbe Interact. 10, 247–256. [DOI] [PubMed] [Google Scholar]
- Ritter, C. and Dangl, J.L. (1995) The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol. Plant–Microbe Interact. 8, 444–453. [DOI] [PubMed] [Google Scholar]
- Rivas, L.A. , Mansfield, J.W. , Tsiamis, G. , Jackson, R.W. and Murillo, J. (2005) Changes in race‐specific virulence in Pseudomonas syringae pv. phaseolicola are associated with a chimeric transposable element and rare deletion events in a plasmid‐borne pathogenicity island. Appl. Environ. Microbiol. 71, 3778–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine, E. , Saarinen, J. , Kalkkinen, N. and Romantschuk, M. (1997a) Purified HrpA of Pseudomonas syringae pv. tomato DC3000 reassembles into pili. FEBS Lett. 417, 168–172. [DOI] [PubMed] [Google Scholar]
- Roine, E. , Wei, W. , Yuan, J. , Nurmiaho‐Lassila, E.L. , Kalkkinen, N. , Romantschuk, M. and He, S.Y. (1997b) Hrp pilus: an hrp‐dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 94, 3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantschuk, M. , Roine, E. and Taira, S. (2001) Hrp pilus—reaching through the plant cell wall. Eur. J. Plant Pathol. 107, 153–160. [Google Scholar]
- Romer, P. , Hahn, S. , Jordan, T. , Strauss, T. , Bonas, U. and Lahaye, T. (2007) Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science, 318, 645–648. [DOI] [PubMed] [Google Scholar]
- Scholtens‐Toma, I.M.J. and De Wit, P.J.G.M. (1988) Purification and primary structure of a necrosis‐inducing peptide from the apoplastic fluids of tomato infected with Cladosporium fulvum (syn. Fulvia fulva). Physiol. Mol. Plant Pathol. 33, 59–67. [Google Scholar]
- Scofield, S.R. , Tobias, C.M. , Rathjen, J.P. , Chang, J.H. , Lavelle, D.T. , Michelmore, R.W. and Staskawicz, B.J. (1996) Molecular basis of gene‐for‐gene specificity in bacterial speck disease of tomato. Science, 274, 2063–2065. [DOI] [PubMed] [Google Scholar]
- Shan, L. , He, P. , Li, J. , Heese, A. , Peck, S.C. , Nürnberger, T. , Martin, G.B. , and Sheen, J. (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor‐signaling complexes and impede plant immunity. Cell Host & Microbe, 4, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F. , Merritt, P.M. , Bao, Z. , Innes, R.W. and Dixon, J.E. (2002) A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell, 109, 575–588. [DOI] [PubMed] [Google Scholar]
- Singer, A.U. , Desveaux, D. , Betts, L. , Chang, J.H. , Nimchuk, Z. , Grant, S.R. , Dangl, J.L. and Sondek, J. (2004) Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure, 12, 1669–16681. [DOI] [PubMed] [Google Scholar]
- Sohn, K.H. , Lei, R. , Nemri, A. and Jones, J.D. (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana . Plant Cell, 19, 4077–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. , Win, J. , Tian, M. , Schornack, S. , Kaschani, F. , Ilyas, M. , Van Der Hoorn, R.A. and Kamoun, S. (2009) Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA, 106, 1654–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina, O.A. and Bertin, P.N. (2003) Regulation cascade of flagellar expression in Gram‐negative bacteria. FEMS Microbiol. Rev. 27, 505–523. [DOI] [PubMed] [Google Scholar]
- Soylu, S. , Brown, I.R. and Mansfield, J.W. (2005) Cellular reactions in Arabidopsis following challenge by strains of Pseudomonas syringae: from basal resistance to compatibility. Physiol. Mol. Plant Pathol. 66, 232–243. [Google Scholar]
- Speth, E.B. , Lee, Y.N. and He, S.Y. (2007) Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr. Opin. Plant Biol. 10, 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J. , Dahlbeck, D. and Keen, N.T. (1984) Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race‐specific incompatibility on Glycine max (L.) Merr. Proc. Natl. Acad. Sci. USA, 81, 6024–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J. , Dahlbeck, D. , Keen, N.T. and Napoli, C. (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea . J. Bacteriol. 169, 5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides, J. , Ma, W. and Guttman, D.S. (2006) Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathog. 2, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, C. , Bennett, M.A. , Athanassopoulos, E. , Tsiamis, G. , Taylor, J.D. and Mansfield, J.W. (1998) Sequence variations in alleles of the avirulence gene avrPphE.R2 from Pseudomonas syringae pv. phaseolicola lead to loss of recognition of the AvrPphE protein within bean cells and a gain in cultivar‐specific virulence. Mol. Microbiol. 29, 165–177. [DOI] [PubMed] [Google Scholar]
- Szurek, B. , Marois, E. , Bonas, U. and Van den Ackerveken, G. (2001) Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 26, 523–534. [DOI] [PubMed] [Google Scholar]
- Tamaki, S. , Dahlbeck, D. , Staskawicz, B.J. and Keen, N.T. (1988) Characterisation and expression of two avirulence genes cloned from Pseudomonas syringae pv. glycinea . J. Bacteriol. 170, 4846–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. , Frederick, R.D. , Zhou, J. , Halterman, D.A. , Jia, Y. and Martin, G.B. (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science, 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Taylor, J.D. , Teverson, D.M. , Allen, D.J. and Pastor‐Corrales, M.A. (1996) Identification and origin of races of Pseudomonas syringae pv. phaseolicola from Africa and other bean growing areas. Plant Pathol. 45, 469–478. [Google Scholar]
- Thwaites, R. , Spanu, P.D. , Panopoulos, N.J. , Stevens, C. and Mansfield, J.W. (2004) Transcriptional regulation of components of the type III secretion system and effectors in Pseudomonas syringae pv. phaseolicola . Mol. Plant–Microbe Interact. 17, 1250–1258. [DOI] [PubMed] [Google Scholar]
- De Torres, M. , Mansfield, J.W. , Grabov, N. , Brown, I.R. , Ammouneh, H. , Tsiamis, G. , Forsyth, A. , Robatzek, S. , Grant, M. and Boch, J. (2006) Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis . Plant J. 47, 368–382. [DOI] [PubMed] [Google Scholar]
- De Torres‐Zabala, M. , Truman, W. , Bennett, M.H. , Lafforgue, G. , Mansfield, J.W. , Egea, P.R. , Bogre, L. and Grant, M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 26, 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiamis, G. , Mansfield, J.W. , Hockenhull, R. , Jackson, R.W. , Sesma, A. , Athanassopoulos, E. , Bennett, M.A. , Stevens, C. , Vivian, A. , Taylor, J.D. and Murillo, J. (2000) Cultivar‐specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo‐blight disease. EMBO J. 19, 3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ackerveken, G. , Marois, E. and Bonas, U. (1996) Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell, 87, 1307–1316. [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken, G.F.J.M. , Van Kan, J.A.L. and De Wit, P.J.G.M. (1992) Molecular analysis of the avirulence gene avr9 of the fungal tomato pathogen Cladosporium fulvum fully supports the gene‐for‐gene hypothesis. Plant J. 2, 359–366. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A. and Jones, J.D.G. (1998) Plant disease resistance proteins and the gene‐for‐gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Van Gijsegem, F. , Gough, C. , Zischek, C. , Niqueux, E. , Arlat, M. , Genin, S. , Barberis, P. , German, S. , Castello, P. and Boucher, C. (1995) The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol. Microbiol. 15, 1095–1114. [DOI] [PubMed] [Google Scholar]
- Vanderplank, J.E. (1978) Genetic and Molecular Basis of Plant Pathogenesis. Berlin‐Heidelberg‐New York: Springer‐Verlag. [Google Scholar]
- Vencato, M. , Tian, F. , Alfano, J.R. , Buell, C.R. , Cartinhour, S. , DeClerck, G.A. , Guttman, D.S. , Stavrinides, J. , Joardar, V. , Lindeberg, M. , Bronstein, P.A. , Mansfield, J.W. , Myers, C.R. , Collmer, A. and Schneider, D.J. (2006) Bioinformatics‐enabled identification of the HrpL regulon and type III secretion system: effector proteins of Pseudomonas syringae pv. phaseolicola 1448A (2006) Mol. Plant–Microbe Interact. 19, 1193–1206. [DOI] [PubMed] [Google Scholar]
- Vivian, A. and Arnold, D. (2000) Bacterial effector genes and their role in host–pathogen interactions. J. Plant Pathol. 82, 163–178. [Google Scholar]
- Vivian, A. , Atherton, G.T. , Bevan, J.R. , Crute, I.R. , Mur, L.A.J. and Taylor, J.D. (1989) Isolation and characterization of cloned DNA conferring specific avirulence in Pseudomonas syringae pv. pisi to pea (Pisum sativum) cultivars, which possess the resistance allele, R2 . Physiol. Mol. Plant Pathol. 34, 335–344. [Google Scholar]
- Wei, Z.M. , Laby, R.J. , Zumoff, C.H. , Bauer, D.W. , He, S.Y. , Collmer, A. and Beer, S.V. (1992) Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora . Science, 257, 85–88. [DOI] [PubMed] [Google Scholar]
- Wood, R.K.S. (1967) Physiological Plant Pathology. Oxford and Edinburgh: Blackwell Scientific Publications. [Google Scholar]
- Xiang, T. , Zong, N. , Zou, Y. , Wu, Y. , Zhang, J. , Xing, W. , Li, Y. , Tang, X. , Zhu, L. , Chai, J. and Zhou, J.‐M. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80. [DOI] [PubMed] [Google Scholar]
- Yucel, I. , Slaymaker, D. , Boyd, C. , Murillo, J. , Buzzell, R.I. and Keen, N.T. (1994) Avirulence gene avrPphC from Pseudomonas syringae pv. phaseolicola 3121: a plasmid‐borne homologue of avrC closely linked to an avrD allele. Mol. Plant–Microbe Interact. 7, 677–679. [DOI] [PubMed] [Google Scholar]
- Zhu, M. , Shao, F. , Innes, R.W. , Dixon, J.E. and Xu, Z. (2004) The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain‐like fold with a distinct substrate‐binding site. Proc. Natl. Acad. Sci. USA, 10, 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiesler‐Vollick, J. , Plovanich‐Jones, A. , Nomura, K. , Bandyopadhyay, S. , Joardar, V. , Kunkel, B.N. and He, S.Y. (2002) Identification of novel hrp‐regulated genes through functional genomic analysis of the Pseudomonas syringae pv. syringae DC3000 genome. Mol. Microbiol. 45, 1207–1218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Growth of the Hrp pilus by the addition of HrpA subunits to the tip of the appendage in Pseudomonas syringae pv. tomato. The deconstructed electron micrograph shows the immunogold localization of Flag‐tagged HrpA subunits produced after an initial period of growth of the pilus in hrp gene‐inducing medium, followed by the induction of tagged HrpA by the addition of mercury. The black dots of gold label demonstrate the secretion of HrpA and its inclusion into the elongating pilus. Movie kindly provided by Tristan Boureau, University of Angers, France. For further details, see Li et al. (2002).
Fig. S2 Secretion of HrpZ through the Hrp pilus of Pseudomonas syringae pv. tomato. The deconstructed electron micrograph shows the immunogold localization of HrpZ produced after an initial period of growth of the pilus in hrp gene‐inducing medium, followed by induction of hrpZ by the addition of mercury. The black dots of gold label demonstrate the secretion of HrpZ from the tip of the elongating pilus. Movie kindly provided by Tristan Boureau, University of Angers, France. For further details, see Li et al. (2002).
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


