SUMMARY
Autophagy is a complex degradative process in which cytosolic material, including organelles, is randomly sequestered within double‐membrane vesicles termed autophagosomes. In Saccharomyces cerevisiae, the autophagy genes ATG1 and ATG8 are crucial for autophagy induction and autophagosome assembly, respectively, and their deletion has an impact on the autophagic potential of the corresponding mutant strains. We were interested in the role of autophagy in the development and virulence of U. maydis. Using a reverse genetic approach, we showed that the U. maydis ATG8 orthologue, atg8, is associated with autophagy‐dependent processes. Deletion of atg8 abolished autophagosome accumulation in the vacuoles of carbon‐starved cells and drastically reduced the survival of U. maydisΔatg8 mutant strains during these conditions. In addition, atg8 deletion had an impact on the budding process during saprobic haploid growth. The infection of maize with compatible Δatg8 strains resulted in fewer galled plants, and fungal gall colonization was strongly reduced, as reflected by the very low hyphal density in these tissues. Δatg8 infections resulted in the formation of very few teliospores. To corroborate the role of autophagy in U. maydis development, we also deleted the ATG1 orthologue, atg1. Deletion of atg1 yielded phenotypes similar to the Δatg8 strains during saprobic growth, but of lower magnitude. The Δatg1 strains were only slightly less pathogenic than the wild‐type and teliospore production was not affected. Surprisingly, atg1 deletion in the Δatg8 background exacerbated those phenotypes already observed in the Δatg8 and Δatg1 single‐mutant strains, strongly suggesting an additive phenotype. In particular, the double mutant was completely suppressed for plant gall induction.
INTRODUCTION
Autophagy is a well‐conserved cellular degradative pathway that allows eukaryotic cells to recycle cytoplasmic components as needed and to eliminate obsolete proteins and organelles (Reggiori and Klionsky, 2002). During autophagy, cytoplasmic material is randomly sequestered into double‐membrane vesicles called autophagosomes, which are targeted to the lytic compartment, vacuole (fungi and plants) or lysosome (animals). After degradation by resident hydrolases, the contents are recycled.
Initially described as a cellular response to nutrient stress conditions, the study of autophagy has historically been limited to the descriptive analysis of its morphological aspects. However, in recent years, many of the autophagy genes have been identified, primarily through extensive genetic screening of Saccharomyces cerevisiae autophagy‐deficient mutants. Since then, orthologues of the autophagy (ATG) genes have been identified and characterized in other organisms. Autophagy has now been linked to a great variety of developmental processes, including seed germination, leaf senescence, fungal conidiation, spore germination and appressorium development, among others (Bassham, 2007; 2006, 2007; Liu and Lin, 2008). In humans, the lack of normal autophagic activity is associated with several health disorders, ranging from tumour development and cellular ageing to attenuated defence against pathogen invasion (Huang and Klionsky, 2007; Sachdeva and Thompson, 2008; Vellai, 2009).
The induction of autophagy results in the de novo formation of double‐membrane vesicles called autophagosomes. The first distinguishable step towards autophagosome formation is the appearance of cup‐shaped, double‐membrane structures, referred to as isolating membranes, pre‐autophagosomes or phagophores, in the cytoplasm. These structures expand, randomly engulfing cytoplasmic material, ultimately maturing into double‐membrane vesicles. Autophagosomes are then docked to the lytic compartment and there their outer membrane fuses with the membrane of this organelle, releasing the inner, single‐membrane vesicles, now termed autophagic bodies, into the lumen. Autophagic bodies are degraded by hydrolases present in the lumen of the lytic compartments and the building blocks of the molecules are then recycled (Suzuki and Ohsumi, 2007).
The process of autophagosome formation is complex and requires the orchestrated actions of a subset of autophagic proteins. To date, 32 autophagy‐related genes (ATG genes) have been discovered and described in S. cerevisiae, 18 of which are specifically involved in autophagosome formation (Kanki et al., 2009; Kawamata et al., 2008; Klionsky et al., 2003; Suzuki et al., 2007). ATG1 encodes a serine/threonine protein kinase that functions early during autophagy induction (Kabeya et al., 2005; Kamada et al., 2000). Another key player is ATG8, encoding a ubiquitin‐like protein (Ichimura et al., 2000) required for autophagosome formation (Mizushima et al., 1998). Atg8p is processed post‐transcriptionally by the cysteine protease encoded by ATG4 (Kirisako et al., 2000) and subsequently covalently linked to the phagophore phospholipid phosphatidylethanolamine (PE) in a ubiquitin‐like reaction catalysed by the E1‐like activating enzyme Atg7 and the E2‐like conjugating enzyme Atg3 (Geng and Klionsky, 2008; Ichimura et al., 2000). It is thought that Atg8 bound to PE facilitates membrane hemifusion leading to autophagosome maturation (Nakatogawa et al., 2007).
Within the Kingdom Fungi, several reports have addressed the role of autophagy in species other than S. cerevisiae. In Aspergillus oryzae, deletion of the ATG8 orthologue leads to defects in conidiation and conidial germination (Kikuma et al., 2006). In the rice blast fungus Magnaporthe grisea, deletion of the ATG1 or ATG8 orthologues results in the loss of autophagy induction, normal appressorium development and pathogenicity (Liu et al., 2007; Veneault‐Fourrey et al., 2006). In the basidiomycetous human pathogen Cryptococcus neoformans, RNAi silencing of the ATG8 orthologue affects the ability of the fungus to trigger autophagy and results in a loss of virulence (Hu et al., 2008).
Ustilago maydis is a plant pathogenic fungus that belongs to the phylum Basidiomycota and is responsible for corn smut disease. A central feature of U. maydis biology is its dimorphic switch: from a saprobic haploid yeast phase to a parasitic filamentous dikaryon (Banuett, 1991; Bolker, 2001; Nadal et al., 2008). The first step towards pathogenic development is the mating of two compatible haploid cells, termed sporidia, on the plant surface to form a dikaryotic cell. Mitotic division in the initial dikaryotic cell is arrested while its tip elongates and eventually forms a poorly differentiated appressorium from which it penetrates the plant. Once inside the plant, mitosis resumes and the dikaryotic filament proliferates and branches within the host tissues (Banuett and Herskowitz, 1989). Later, the dikaryotic filaments experience a series of dramatic developmental changes that culminate in the differentiation of teliospores (Banuett and Herskowitz, 1996). We hypothesized that, in U. maydis, autophagy is critical for undergoing the developmental programmes required to complete its life cycle. On the basis of previous reports regarding the critical role of ATG8 in other pathogenic fungi, to test this hypothesis we initially deleted atg8, the ATG8 orthologue in U. maydis, and showed that this gene is required to trigger autophagy during nutritional stress. Incidentally, the ability of Δatg8 strains to survive during carbon starvation conditions was drastically reduced. We also showed that atg8 is required for the wild‐type budding pattern of haploid sporidia and its deletion results in an abnormal lateral budding phenotype. Most importantly, deletion of atg8 results in a substantial decrease in U. maydis virulence, accompanied by a reduction in teliospore production. In an effort to better characterize the process of autophagy in U. maydis, we subsequently deleted the ATG1 orthologue, atg1. The phenotypes of Δatg1 mutants resembled those observed in Δatg8 strains, but were less severe. However, double‐mutant Δatg1Δatg8 strains exhibited a more pronounced attenuation of disease symptoms than did either single‐mutant strain, suggesting additive action.
RESULTS
Identification and deletion of U. maydis ATG8 and ATG1 orthologues
In order to investigate the potential roles of autophagy in U. maydis development, we sought to create mutant strains in which the pathway was no longer functional. Considering that ATG8 and ATG1 orthologues are essential for normal autophagic activity in organisms as distantly related as plants and fungi, we decided to delete the U. maydis atg8 and atg1 genes to generate the desired autophagy‐deficient strains. The U. maydis atg8 and atg1 genes were identified using the S. cerevisiae Atg8 and Atg1 protein sequences, respectively, to search the U. maydis genome using the blast homology search algorithm (http://mips.gsf.de/genre/proj/ustilago).
A single highly related (3e−54) U. maydis gene (um05567), named here atg8, encoding a 118‐amino‐acid protein was identified. Alignment of the U. maydis Atg8 protein with other fungal, plant and human (LC3) Atg8 proteins showed a high degree of amino acid conservation (Fig. S1). Residue G116 of S. cerevisiae Atg8 is of vital importance for protein function. Examination of the U. maydis Atg8 sequence indicates that residue G116 and the sequence context in which it is located have been conserved (Fig. S1, asterisk).
The U. maydis gene identified as most closely related to S. cerevisiae ATG1 was um06363 (2.9e−83), encoding a predicted 990‐amino‐acid serine/threonine protein kinase (Fig. S2).
The Δatg8 and Δatg1 deletion strains were generated using DelsGate methodology (Garcia‐Pedrajas et al., 2008) by complete replacement of the corresponding gene open reading frame (ORF) with the plasmid sequences containing the carboxin (cbx)‐ or hygromycin (hyg)‐resistant selectable marker, respectively. Several potential Δatg8 and Δatg1 mutants were identified by polymerase chain reaction (PCR) screening of transformants. We confirmed the deletion of atg8 and atg1 in a subset of these transformants by Southern blot (Fig. S3). Employing the same approach, double‐mutant Δatg1Δatg8 strains were generated by deleting atg1 in a Δatg8 background.
atg8 and atg1 transcripts accumulated during carbon stress conditions
In many organisms, such as S. cerevisiae and Arabidopsis thaliana, the exposure of cells to nutrient stress conditions (low nitrogen or carbon) brings about a rapid accumulation of several ATG gene transcripts (Kirisako et al., 1999; Rose et al., 2006). To investigate whether U. maydis behaves in a similar manner, we examined the levels of atg8 and atg1 transcripts by quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) as cells were starved for carbon. Wild‐type cells grown in potato dextrose broth (PDB) to the exponential phase [optical density (OD) ∼ 0.4, approximately 107 cells/mL] were transferred to minimal medium lacking any carbon source (MM – C) and incubated for 8 h. cDNA was synthesized from RNA samples collected 30 min, 1 h, 2 h, 4 h and 8 h after transfer, and relative transcript abundance was estimated by qRT‐PCR. As expected, an increased accumulation of atg8 transcript was observed in wild‐type U. maydis cells on transfer to MM – C. After 4 and 8 h of carbon starvation, atg8 transcript levels increased more than 13 and 25 times the initial value, respectively. A similar pattern of transcript accumulation, but of reduced amplitude, was observed for atg1 (Fig. 1). These results indicated that, as in other systems, nutrient stress conditions are sufficient to induce the transcript accumulation of U. maydis autophagy genes atg8 and atg1. We used microarray data (Islamovic H., unpublished) to investigate whether these genes were also induced during the pathogenic development of wild‐type U. maydis. The levels of RNA extracted from galled tissue, 14 and 21 days after inoculation (dai), were compared with the corresponding RNA levels of 24 h old PDB liquid cultures. Two biological replicates were considered. At 14 dai, the atg8 and atg1 genes registered an average eight‐ and six fold increase in transcript relative abundance, respectively. At 21 dai, atg8 and atg1 exhibited a five‐ and six fold increase, respectively. In addition to atg8 and atg1, other autophagy‐related genes also showed an increase in RNA levels at 14 and 21 dai, including atg7 (um04880), atg12 (um12234) and atg4 (um05142). These results indicate that, during U. maydis pathogenic development, several elements of the autophagic machinery are induced compared with saprobic growth.
Figure 1.
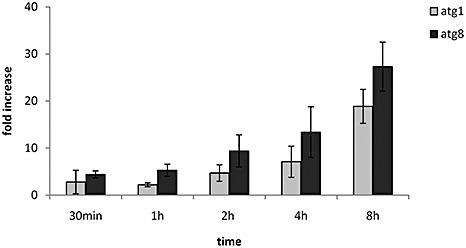
Transcript accumulation of Ustilago maydis atg8 and atg1 genes during carbon starvation. Relative levels of atg8 and atg1 transcripts were calculated by quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) methodology. Wild‐type RNA samples were collected at the time points indicated in the graphs. Expression levels were normalized to the control gene cpr1 (Doehlemann et al., 2009). The indicated values correspond to the means of three biological replicates; the bars represent the standard error of biological variation.
atg8 and atg1 deletion prevents vacuolar accumulation of autophagosomes in U. maydis
When cells undergo autophagy, autophagic bodies tend to accumulate within the vacuole. The monitoring of the accumulation of autophagic bodies is thus a reliable and standard method used to evaluate the autophagic activity within a cell (Baba et al., 1994; Galluzzi et al., 2009). In order to determine whether atg8 and atg1 were important for autophagy in U. maydis, we evaluated the ability of the wild‐type, Δatg8 and Δatg1 strains to accumulate autophagic bodies in the vacuoles when cells were exposed to carbon stress conditions. Wild‐type and Δatg8 cells were grown in PDB until they reached the exponential growth phase (OD ∼ 0.4, approximately 107 cells/mL), and were then transferred to MM – C in the presence of the proteinase inhibitor phenylmethylsulphonylfluoride (PMSF) (1 mm). After 5 h of incubation in MM – C, the cells were collected by centrifugation and prepared for transmission electron microscopy. Transmission electron micrographs showed that wild‐type cells had accumulated numerous autophagic bodies within their vacuoles (Fig. 2A). By contrast, no autophagic bodies were observed in the vacuoles of Δatg8 cells (Fig. 2B). Occasionally (one of 10 cells examined), a few vesicles were observed in the vacuoles of Δatg1 cells, but these were smaller than those observed in the wild‐type cells (Fig. 2C). These results indicate that carbon stress conditions are sufficient to trigger autophagy in U. maydis, and that the atg8 and atg1 genes are necessary for normal autophagosome accumulation under these conditions.
Figure 2.
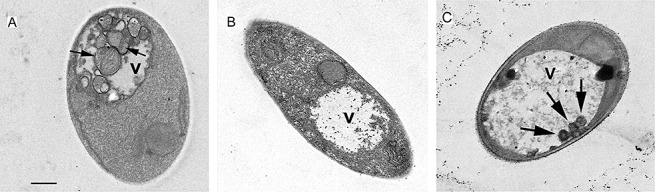
Starvation‐induced autophagosome accumulation in wild‐type, Δatg8 and Δatg1 Ustilago maydis cells. Wild‐type and Δatg8 cells were incubated for 5 h in medium lacking a carbon source (MM – C). Transmission electron images show the accumulation of autophagic bodies (arrows) within the vacuole (v) of wild‐type cells (A). Note the absence of these structures from vacuoles of Δatg8 cells (B). Smaller vesicles occasionally appeared in vacuoles of Δatg1 (C). At least 40 cells were inspected per sample. Scale bar, 1 µm.
atg8 and atg1 deletions affect U. maydis survival during carbon starvation
To determine whether the deletion of atg1 and atg8 affected the ability of U. maydis to survive under carbon starvation conditions, we analysed the capacity of the wild‐type, Δatg8, Δatg1 and Δatg1Δatg8 strains to survive in MM – C for a period of 5 days. The three mutant strains showed a severe loss of viability under carbon starvation when compared with wild‐type cells. After 3 days of incubation in MM – C, on average only 8.5%, 16.5% and 7.5% of Δatg8, Δatg1 and Δatg1Δatg8 cells survived, respectively, whereas more than 50% of wild‐type cells remained alive. Moreover, by day 4, only 3% of cells from Δatg1 and none from Δatg8 and Δatg1Δatg8 strains survived, but approximately 36% of wild‐type cells remained viable (Fig. 3). These results clearly indicate that the autophagy genes atg8 and atg1 are required for survival of U. maydis during long‐term exposure to conditions of carbon stress.
Figure 3.
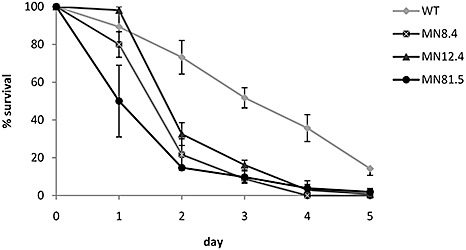
Survival of wild‐type, Δatg8, Δatg1 and Δatg1Δatg8 Ustilago maydis cells during carbon starvation. Wild‐type, MN8.4 (Δatg8), MN12.4 (Δatg1) and MN81.5 (Δatg1Δatg8) cells were grown in liquid medium lacking a carbon source (MM – C). Cell aliquots were diluted and plated on potato dextrose agar (PDA) at the indicated times. Colonies were counted after 2–3 days and the percentage of survival was calculated on the basis of the initial colony number (day 0) for each strain.
atg8 and atg1 are required for wild‐type budding of haploid sporidia
Ustilago maydis wild‐type sporidia divide by budding, with buds emerging at or near the tips of cigar‐shaped cells. During the late exponential growth phase of liquid‐grown wild‐type cells apical budding predominates (Fig. 4A). By the time wild‐type cells reach the stationary phase, they have ceased to divide and virtually no emerging buds are observed (Fig. 4B,C). As in the wild type, the Δatg8, Δatg1 and Δatg1Δatg8 strains also displayed a high degree of budding during exponential growth. However, in addition to buds emerging apically, cells from Δatg8, Δatg1 and Δatg1Δatg8 strains displayed lateral budding (Fig. 4D–F). During the exponential growth phase, the average percentages of Δatg8, Δatg1 and Δatg1Δatg8 cells with lateral buds were 6%, 7% and 12%, respectively, more than 15‐fold higher than the value of 0.4% of the wild‐type. As cultures reached the stationary phase, the percentages of Δatg8, Δatg1 and Δatg1Δatg8 cells bearing lateral buds increased substantially to 25%, 10% and 29%, respectively, with Δatg8 and Δatg1Δatg8 cells exhibiting more frequent lateral budding than Δatg1 strains (Fig. 4B). Moreover, in some of the cells, more than one lateral bud was present or the cell bore multiple apical buds. In addition, at that stage, approximately 1% and 6.5% of Δatg8 and Δatg1 cells, respectively, bore an apical bud. Similar results were obtained when several independent deletion mutant strains were observed. These results indicate that the deletion of either atg8 or atg1 increases the frequency of lateral budding and interferes with the process of cell separation in U. maydis.
Figure 4.
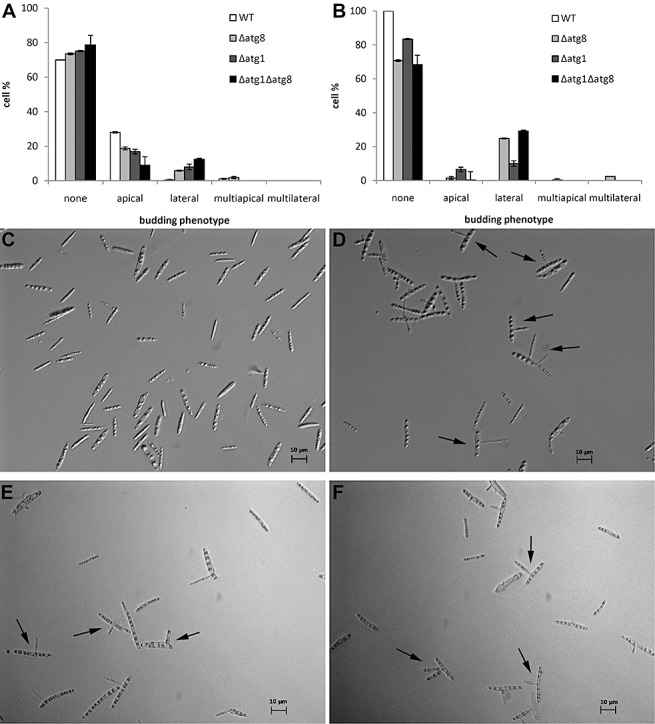
Budding patterns of wild‐type, Δatg8, Δatg1 and Δatg1Δatg8 Ustilago maydis haploid sporidia. The budding phenotype of wild‐type and Δatg8 strains was scored at several time points. The percentage of cells with a particular budding morphology at the exponential (A) and stationary (B) phases of growth. Differential interference contrast images of stationary phase cultures of wild‐type (C), Δatg8 (D), Δatg1 (E) and Δatg1Δatg8 (F). Examples of lateral buds are indicated by arrows.
Mating and filament formation are unaffected by atg8 and atg1 deletion
In U. maydis, the mating of compatible haploid sporidia is a prerequisite for dikaryon establishment and pathogenic development. When compatible sporidia are co‐spotted onto charcoal plates, an initial dikaryotic cell is established that elongates at its tip. This reaction results in a ‘fuzzy’ white colony that is easily distinguished from those in which mating does not occur. Mating reactions of compatible Δatg1Δatg8 strains on charcoal mating plates were indistinguishable from those of compatible wild‐type strains (Fig. 5). These results indicate that Δatg1Δatg8 strains are competent to mate with comparable efficiency to wild‐type strains. Likewise, mating of single Δatg8 or Δatg1 mutants was unaffected (data not shown).
Figure 5.
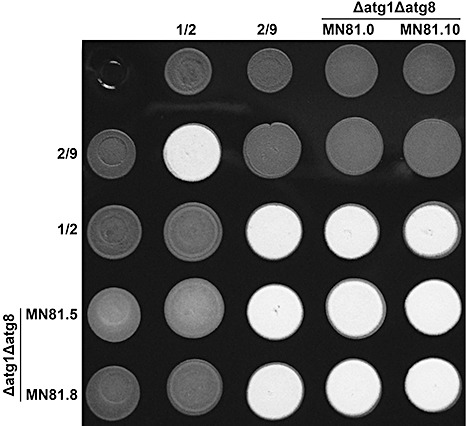
Mating reaction comparison of wild‐type and Δatg1Δatg8 Ustilago maydis strains. Five microlitres of overnight cell cultures of the indicated column and row strains (indicated by the number above and to the left) were spotted onto charcoal‐containing medium and dried in a transfer hood. Mating reactions were incubated at room temperature and photographed 24 h post‐inoculation. The production of white fuzzy colonies is indicative of successful mating reactions.
To investigate whether Δatg1Δatg8 double mutants were also able to sustain filamentous growth on plant inoculation, we examined symptomatic tissue at 5 dai for the presence of U. maydis filaments. At the end of that time period, Δatg1Δatg8 co‐inoculation had resulted in abundant production of filaments on the plant surface (Fig. 6). This result clearly indicates that deletion of atg1 and atg8 does not alter the ability of U. maydis to grow as a filament in the presence of its host. Filaments produced by the single Δatg1 and Δatg8 mutant strains at 5 dai were also indistinguishable from those produced by wild‐type infection (data not shown).
Figure 6.
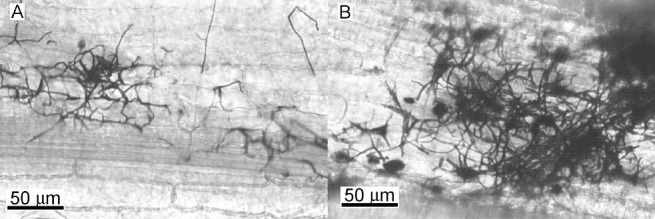
Filamentous development in wild‐type and Δatg1Δatg8 Ustilago maydis strains on maize plants. Wild‐type (A) and Δatg1Δatg8 mutant (B) filaments were detected on the surface of maize plants 5 days after inoculation (dai) after staining with chlorazole black. The Δatg1Δatg8 mutant produced filaments indistinguishable from those of the wild‐type. Scale as indicated.
atg8 and atg1 are required for complete symptom development during pathogenic growth
In order to test the pathogenic capacity of the Δatg8 strains, maize seedlings were co‐inoculated with pairwise combinations of compatible wild‐type and Δatg8 strains, and disease progression was monitored. Two sets of independent compatible mutants were tested for their virulence and, for each pair, three biological replicates of the pathogenicity test were conducted. Results for 10 dai, employing Δatg8 mutant strains MN8.1 and MN8.11, are summarized in Table 1. Similar results were obtained when independent compatible Δatg8 strains MN8.4 and MN8.16 were employed (data not shown). Results at 14 and 21 dai reflected the same trend among the treatments (data not shown). Dikaryons formed between combinations of Δatg8 strains always resulted in less severe disease symptoms, reflected in the lower disease index, than did inoculations with other strain combinations. Moreover, by the end of the experiment (21 dai), plants inoculated with compatible Δatg8 strains had developed considerably fewer galls than those inoculated with any of the other treatments (Fig. 7). In addition, the few galls formed during infection by Δatg8 remained white at 28 dai and did not show mature teliospores. These results clearly indicate that the atg8 gene of U. maydis is required for full symptom development during the infection of maize seedlings.
Table 1.
Δatg8 pathogenicity at 10 days after inoculation (dai).
| Treatment* | Dikaryon | Replicates | Plants per replicate | Disease index† | t‐grouping‡ |
|---|---|---|---|---|---|
| 1 | +/+ | 3 | 20 | 2.8 ± 0.41 | A |
| 2 | +/− | 3 | 20 | 2.7 ± 0.45 | A |
| 3 | −/+ | 3 | 20 | 2.8 ± 0.17 | A |
| 4 | −/− | 3 | 20 | 1.28 ± 0.12 | B |
Treatment: paired strains are as follows (see Table 4 for strain genotypes): 1, (1/2 × 2/9); 2, (1/2 × MN8.11); 3, (MN8.1 × 2/9); 4, (MN8.1 × MN8.11). Inoculation was at 106 cells/mL for all strains.
Mean ± standard errors were calculated for each treatment on the basis of independent biological replicates.
Statistical analysis was performed using a nonparametric test of ordinal data in designed factorial experiments (Shah and Madden, 2004).
Figure 7.

Gall production in wild‐type, Δatg8 and Δatg1Δatg8 Ustilago maydis strains during maize seedling infection. Dikaryon genotype as follows: +/+, (1/2 × 2/9); +/−, (1/2 × either MN8.1 or MN81.5); −/+, (either MN8.11 or MN81.0 × 2/9); −/−, (MN8.1 × MN8.11 or MN8.5 × MMN8.0). The average percentages of plants that developed galls 21 days after inoculation were calculated on the basis of three independent biological replicates; bars represent the standard error of biological variation.
To evaluate the importance of the atg1 gene during U. maydis pathogenic development, we conducted pathogenicity tests with the Δatg1 strains, MN12.1 and MN29.4, in the same manner as for the Δatg8 strains. The pathogenicity test results showed that the Δatg1 mutant strains were slightly less virulent than the wild‐type with no substantial reduction in the number of galled plants observed (Table 2). However, when plants were co‐inoculated with a mixture of compatible Δatg1Δatg8 double‐mutant strains, MN81.5 and MN81.0, the disease symptoms were drastically reduced, to an even greater extent than infection with compatible Δatg8 strains. Remarkably, Δatg1Δatg8 dikaryons completely failed to induce galls (Table 3 and Fig. 7). Similar results were obtained when independent compatible Δatg1Δatg8 strains, MN81.8 and MN81.10, were employed (data not shown).
Table 2.
Δatg1 pathogenicity at 10 days after inoculation (dai).
| Treatment* | Dikaryon | Replicates | Plants per replicate | Disease index† | t‐grouping‡ |
|---|---|---|---|---|---|
| 1 | +/+ | 3 | 20 | 2.56 ± 0.23 | A |
| 2 | +/− | 3 | 20 | 2.55 ± 0.15 | A |
| 3 | −/+ | 3 | 20 | 2.25 ± 0.10 | AB |
| 4 | −/− | 3 | 20 | 1.9 ± 0.20 | B |
Treatment: paired strains are as follows (see Table 4 for strain genotypes): 1, (1/2 × 2/9); 2, (1/2 × MN29.4); 3, (MN12.4 × 2/9); 4, (MN12.4 × MN29.4). Inoculation was at 106 cells/mL for all strains.
Mean ± standard errors were calculated for each treatment on the basis of independent biological replicates.
Statistical analysis was performed using a nonparametric test of ordinal data in designed factorial experiments (Shah and Madden, 2004).
Table 3.
Δatg1Δatg8 pathogenicity at 10 days after inoculation (dai).
| Treatment* | Dikaryon | Replicates | Plants per replicate | Disease index† | t‐grouping‡ |
|---|---|---|---|---|---|
| 1 | +/+ | 3 | 20 | 3.05 ± 0.03 | A |
| 2 | +/− | 3 | 20 | 2.6 ± 0.07 | A |
| 3 | −/+ | 3 | 20 | 2.86 ± 0.19 | A |
| 4 | −/− | 3 | 20 | 0.95 ± 0.03 | B |
Treatment: paired strains are as follows (see Table 4 for strain genotypes): 1, = (1/2 × 2/9); 2, (1/2 × MN81.0); 3, (MN81.5 × 2/9); 4, (MN81.5 × MN81.0). Inoculation was at 106 cells/mL for all strains.
Mean ± standard errors were calculated for each treatment on the basis of independent biological replicates.
Statistical analysis was performed using a nonparametric test of ordinal data in designed factorial experiments (Shah and Madden, 2004).
Deletion of atg8 affects gall formation and teliospore production in ears of mature maize
In order to determine whether Δatg8 mutants were affected at a particular stage of teliospore development, we followed microscopically the process of gall formation during maize seedling infection. At 10 dai, many of the plants infected with wild‐type U. maydis contained several large galls with massive fungal proliferation and defined areas in which high hyphal densities were observed (Fig. 8A). Most of the hyphae were thick and undergoing fragmentation or ramification, and many exhibited small bumps at the tips of the short branches (Banuett and Herskowitz, 1996). No galls were observed in the Δatg8 inoculated plants at 10 dai. At 14 dai, galls from wild‐type infected plants contained large pockets of ‘worm stage’ hyphal segments embedded in mucilaginous material, together with some fully formed teliospores (Fig. 8C,E,F). At that time point, the Δatg8 infected plants contained a few stem galls comparable in size to the wild‐type. However, these galls were barely colonized. The hyphae appeared isolated, randomly distributed and there was no evidence of ‘worm stage’ development (Fig. 8D,G,H). These Δatg8 hyphae did, however, show the characteristic thickening and ramification that precedes teliospore formation, but no teliospores were observed at 14 dai. At 21 dai, wild‐type infection resulted in copious amounts of teliospores (Fig. 8I,K,L), whereas Δatg8 hyphae within galled tissue remained scarce and isolated (Fig. 8J). Some Δatg8 hyphae were observed undergoing rounding, but no further development was evident (Fig. 8L). Δatg8 gall samples from 28 and 35 dai were indistinguishable from those at 21 dai (data not shown). After that period, Δatg8 galls were still white and started to rot, probably as a result of secondary infections. These results indicate that the deletion of atg8 severely impairs U. maydis proliferation within the galled plant tissue and the normal process of teliospore development.
Figure 8.
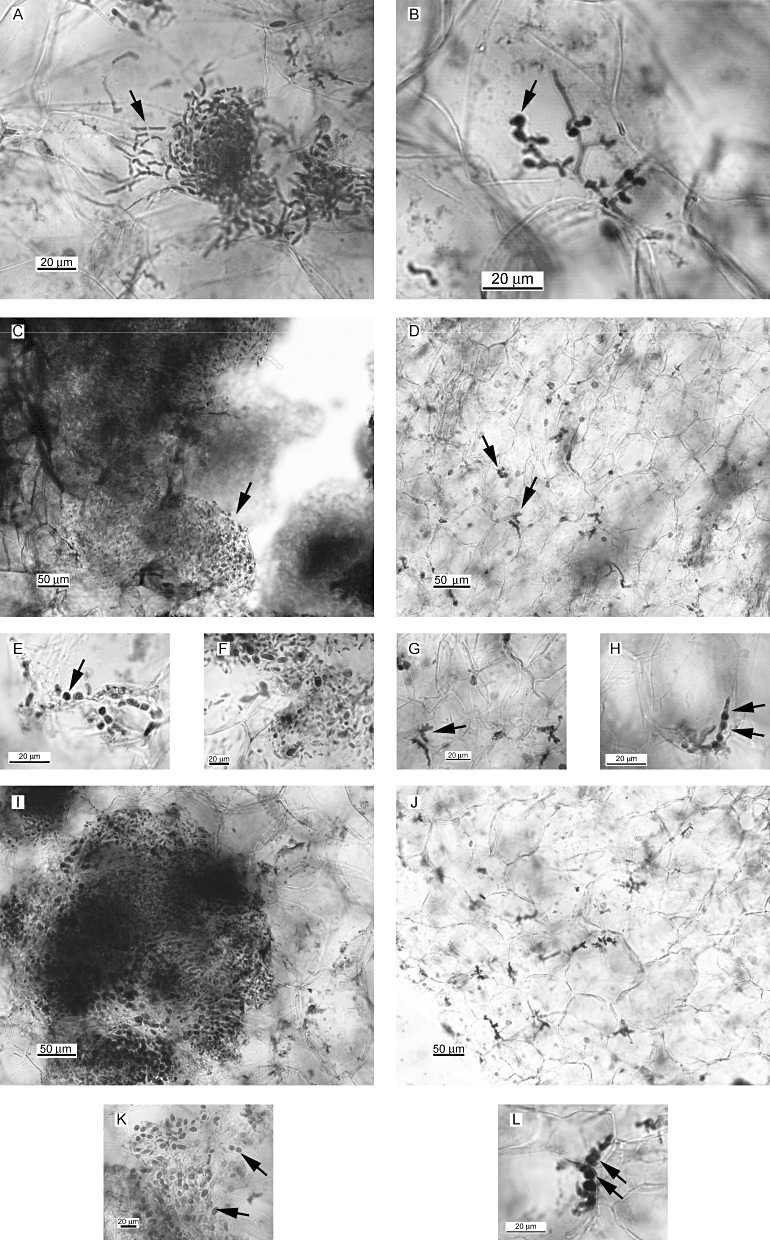
Teliospore formation in wild‐type and Δatg8 Ustilago maydis strains. Teliospore formation was evaluated during wild‐type and Δatg8 maize seedling infections. Wild‐type [10 days after inoculation (dai)] initial accumulation of hyphae showing initial stages of fragmentation and ramification (A). Note the several short branches and bumps (B, arrows). Wild‐type (14 dai) infected galls contained massive accumulation of fragmented hyphae embedded in mucilaginous substance, the ‘worm stage’ (C, arrow). Many hyphae were at the rounding‐up stage (E, arrow) and several teliospores were formed (F). Very few isolated hyphae were present in Δatg8 galled tissue at 14 dai (D, cf. wild‐type C). Several Δatg8 hyphae had branched. Note the presence of several bumps (G, arrow). Some Δatg8 were at the rounding stage (H). Wild‐type infections at 21 dai resulted in a high level of teliospores (I and K, arrow), whereas Δatg8 samples showed little further development (cf. J and G, Δatg8 at 14 dai, and I, wild‐type at 21 dai). Some hyphae were still at the rounding stage (L, arrows). Several samples were examined per strain and the time period and images are representative of each. Scale as indicated.
We inoculated the ears of maturing maize plants with the same strain combinations utilized in the pathogenicity test to better evaluate gall development and teliospore production. We used nine plants per treatment and at least two ears per plant were inoculated. Three weeks after inoculation, numerous galls had formed on the ears of plants co‐inoculated with wild‐type strains or with combinations of compatible wild‐type and Δatg8 strains. Of the nine plants inoculated with compatible Δatg8 strains, one possessed a single ear in which a few very immature galls were present, whereas the remaining eight plants were gall free. The galls in this single ear lacked the characteristic dark coloration indicative of massive teliospore production and very few black teliospores were present (Fig. 9, arrow). Plants co‐inoculated with any other strain combination bore numerous ears in which abundant teliospores developed (Fig. 9). This experiment was repeated twice with similar results. These results corroborate the fact that some Δatg8 hyphae are able to undergo the changes that lead to teliospore formation, but overall production is severely impaired.
Figure 9.

Comparison between wild‐type and Δatg8 gall development in maize ears. Maize ears were inoculated with 106 cells/mL. Paired strains are as follows: 1, (1/2 × 2/9); 2, (1/2 × MN8.11); 3, (MN8.1 × 2/9); 4, (MN8.1 × MN8.11). Nine plants were used for each treatment. Note the reduced number of galls and teliospores in ears co‐inoculated with compatible Δatg8 strains (arrow indicates limited teliospores in galled ear). Experiment was repeated twice with similar results.
In order to investigate whether the few teliospores produced in the plant co‐inoculated with Δatg8 strains were capable of typical germination and meiosis, we observed the segregation of the mating type locus of the progeny. Analysis of 20 teliospore‐derived haploid progeny isolated from the plants co‐inoculated with these strains included descendants with the parental mating types, a1b1 and a2b2 (4 and 3, respectively), as well as the recombinants a1b2 and a2b1 (5 and 8, respectively). The frequency of recombinants was similar to the parents a1b1 and a2b2 and indicated that meiosis did take place. These results indicate that teliospore formation in Δatg8 strains is severely compromised, but that those that do form are functional.
DISCUSSION
Historically recognized as a cellular nutrient stress adaptation mechanism (Levine and Klionsky, 2004), autophagy has in more recent years been described as an integral part of the cellular machinery that facilitates the programmed developmental changes that occur during organ or tissue remodelling in many organisms (Mizushima, 2007). In this work, we have explored the process of autophagy in the plant pathogenic fungus U. maydis. Using a reverse genetic approach, we were able to show that a tight connection exists between autophagy and several aspects of U. maydis biology.
We identified U. maydis autophagy genes, atg8 and atg1, as unique orthologue of the well‐characterized S. cerevisiae ATG8 and ATG1 genes, respectively, based on sequence similarity. In S. cerevisiae, Atg8 is post‐translationally modified at its C‐terminus by cysteine protease Atg4 to generate Atg8G116. The resulting Atg8G116 is then covalently bound to the lipid PE in a ubiquitin‐like reaction catalysed by Atg7 and Atg3 (Ichimura et al., 2000; Kirisako et al., 2000). Examination of the protein sequence indicates that residue G116 and the sequence context in which it is located have been conserved in U. maydis Atg8, reinforcing the notion that atg8 is, indeed, the functional orthologue of ATG8.
When exploring the U. maydis genome in search of the ATG1 orthologue, several genes were identified that encode proteins related to ATG1. On the basis of sequence similarity to S. cerevisiae ATG1, we established that U. maydis atg1 was gene um06363 (2.9e−83). Supporting the hypothesis of um06363 being a unique atg1 orthologue is the fact that the second most similar protein was the well‐characterized protein kinase A (PKA) catalytic subunit, Adr1, encoded by um04456 (1.4e−28). In addition to um06363 and um04456, other U. maydis genes encoding proteins related to serine/threonine kinase showed some degree of similarity to S. cerevisiae ATG1. However, when the blast search was performed using the S. cerevisiae Atg1 protein sequence from which the kinase domain was removed, only the U. maydis gene um06363 was identified as the orthologue. Therefore, these observations strongly suggest that atg1 is the single U. maydis true orthologue of the S. cerevisiae ATG1 gene.
We have demonstrated here that carbon depletion is able to trigger autophagy in haploid U. maydis sporidia, as shown by the accumulation of autophagic bodies within the vacuoles of wild‐type, carbon‐starved cells and the induction of at least two autophagy genes, atg1 and atg8, during these conditions. Importantly, our unpublished microarray data showed that atg1 and atg8, together with other autophagy‐related genes, are induced during teliospore formation, further reinforcing the hypothesis that autophagy operates during U. maydis pathogenic development. The absence of autophagic bodies from vacuoles of carbon‐starved Δatg8 mutant cells strongly supports the hypothesis that autophagy activity is lost. The vacuoles of Δatg1 carbon‐starved cells lacked wild‐type‐like‐appearing autophagic bodies, but did occasionally contain a few smaller vesicles. Whether these vesicles correspond to smaller autophagosomes or to a different type of membrane vesicle formed through an autophagy‐independent manner is yet to be determined. It is possible that deletion of atg1 does not completely shut down autophagy, but affects the size of the resulting autophagosome. A reduced but not completely lost autophagic activity would be consistent with the milder budding and virulence phenotypes observed in Δatg1. Alternatively, the small vesicles might be part of a completely different pathway that acts independently of atg1. In S. cerevisiae, Atg1 forms a complex with Atg13 and Atg17 which, in turn, recruits other Atg proteins to phagophore assembly site (PAS) when stress conditions trigger autophagy (Kabeya et al., 2005). In addition, Atg1 is required to maintain the normal dynamic of PAS component recycling during autophagosome formation (Cebollero and Reggiori, 2009; Cheong and Klionsky, 2008). Nevertheless, Atg1 is associated with the initial step of autophagy induction in other organisms in which no PAS is involved. At present, we do not know whether autophagosome formation in U. maydis proceeds with the assistance of PAS. Notably, no clear orthologues of S. cerevisiae ATG13 and ATG17 are present in the U. maydis genome, thus increasing the uncertainty regarding the modus operandi of Atg1 in this fungus. It is conceivable, however, that a high degree of evolutionary divergence has occurred, impairing the recognition of these proteins as orthologues by simple amino acid sequence analysis. Future research should more precisely address the role of Atg1 and provide further insight into the process of autophagy within U. maydis. In any case, this lack of normal autophagic activity is consistent with the reduced survival capacity of Δatg8, Δatg1 and Δatg1Δatg8 mutants under conditions of nutrient stress. Therefore, it is very likely that these phenotypes observed in Δatg8, Δatg1 and Δatg1Δatg8 cells result from a failure to properly trigger this pathway during saprobic and pathogenic growth.
During exponential growth in liquid culture, U. maydis sporidia actively divide, with new buds typically emerging at or near the tip of the mother cell, and very occasionally cells bud laterally. At the stationary phase, cells cease to divide and complete detachment of the daughter from the mother cells occurs. Thus, in wild‐type cultures, an exponential growth phase of actively budding cells precedes a stationary phase in which cells no longer divide and no buds are observed.
The deletion of autophagy genes, atg8 or atg1, alters the budding process in U. maydis. During the exponential growth phase, Δatg8, Δatg1 and Δatg1Δatg8 mutant strains showed at least 15‐fold higher frequency of lateral buds than did the wild‐type. However, all mutant strains sustained apical budding as well. As cultures reached the stationary phase, the percentages of mutant cells bearing lateral buds increased substantially. The fact that, at that time, many of the Δatg8, Δatg1 and Δatg1Δatg8 cells still carried buds indicated a detachment defect. Thus, the increased number of lateral buds in the mutant cells appears to be the result of a combination of two defects. The atg8 and atg1 deletions not only altered the site of bud emergence, but also negatively affected the process of mother–daughter cell separation. Over the years, a few U. maydis mutants have been reported to exhibit, among other phenotypes, lateral budding. That the budding pattern is influenced by the cyclic adenosine monophosphate (cAMP) and PKA pathway in U. maydis is evident, as exposure of wild‐type cells to exogenous cAMP results in multibudding with an increased level of lateral bud formation, a phenotype that is also mimicked by ubc1 mutations (Gold et al., 1994). Therefore, a high level of PKA activity in U. maydis is associated with lateral budding of haploid sporidia. Most interesting, increased PKA activity has been shown to have an inhibitory effect in autophagy activation through Atg1 in S. cerevisiae (Stephan et al., 2009). The ukb1 gene was identified in the screen for genes encoding PKA catalytic subunits in U. maydis (Durrenberger et al., 1998), and its deletion resulted in increased lateral budding, together with a reduction in virulence (Abramovitch et al., 2002). It was hypothesized that high PKA activity would act to inhibit Ukb1, consequently causing the abnormal budding phenotype. Whether a connection exists between the cAMP pathway and Ukb1 remains a mystery. Future work should clarify whether the lateral budding phenotype produced by high PKA activity in U. maydis is a result of an inhibitory effect on autophagy induction or on Ukb1 kinase. Lateral budding also occurs when entry to mitosis is delayed as a consequence of high Wee1 kinase activity (Sgarlata and Perez‐Martin, 2005). Microtubules and the bidirectional trafficking of early endosomes that they support are major determinants of the polar growth that occurs during bud formation in U. maydis (Steinberg et al., 2001; Wedlich‐Soldner et al., 2002). Mutations in the α‐tubulin gene, tub1, alter the budding pattern, increasing the incidence of lateral budding (Steinberg et al., 2001). At the moment, however, no clear connection exists between cell cycle regulation, tubulin cytoskeleton organization and autophagy to provide a satisfactory hypothesis to explain the observed phenotype. On the other hand, lateral budding has been reported to occur in stressed wild‐type cells (Jacobs et al., 1994). If autophagy is a pathway that allows cells to cope with nutrient stress, it is reasonable to expect that typical behaviours of wild‐type stressed cells would be exacerbated in an autophagy‐deficient mutant. Moreover, the lateral budding of the autophagy mutants reported here increased as cells entered the stationary phase, a stage at which autophagy is known to be induced in S. cerevisiae, most probably as a result of nutrient exhaustion (Wang et al., 2001). Nevertheless, how autophagy specifically influences bud site selection and the completion of cytokinesis in U. maydis will require further exhaustive investigation, and any current attempt at a cohesive hypothesis is speculation.
Because nutrient availability is generally limited in the host plant, the ability of the fungus to trigger autophagy might be crucial for its overall pathogenic development. From the time when two haploid sporidia mate on the plant surface until a competent infection filament reaches the interior of the maize tissue, cells are likely to experience extreme nutrient deprivation and must rely on their stored energy supplies and the recycling of their macromolecules to secure the components needed to continue cellular activities. Even in later stages, the plant tissue itself might represent a relatively nutrient‐poor environment and dikaryotic U. maydis might continue to depend on the recycling of its own cellular material. Therefore, an important point when considering U. maydis pathogenic development is that the ability to trigger autophagy might be crucial during the early and later stages of plant infection when nutrient availability is most likely limiting.
As expected, we observed a reduction in virulence in Δatg8, Δatg1 and Δatg1Δatg8 strains compared with wild‐type infections. Interestingly, the reduced virulence phenotypes displayed by each of these mutants reflected the same trend observed with regard to altered bud site selection and cell separation in budding pattern. Both virulence reduction and altered budding pattern were greater in Δatg8 than in Δatg1 strains. However, when atg1 was deleted in Δatg8 strains, the resulting Δatg1Δatg8 double‐mutant virulence was even lower than Δatg8 single mutants to the point that gall formation was completely suppressed.
A particularly important aspect of U. maydis pathogenic development in the light of our observations is that teliospore production involves drastic remodelling of the fungal hyphae. During wild‐type infections, large areas filled with fungal hyphae occur in or between the plant tumour cells. These hyphae ramify, fragment, round‐up and, eventually, a sculpted, darkly pigmented cell wall is deposited around cytoplasmic regions containing single diploid nuclei. Our microscopy results indicate that the reduced teliospore production observed in Δatg8 infections is most probably a consequence of the lower proliferation of mutant strains within the galled tissue, combined with a delay in the later stages of spore development. Many Δatg8 hyphae were caught at the rounding‐up stage, but mature spores were not observed in maize infected seedlings. It is possible for this particular phenotype to be associated with an overall lower capacity of the autophagy mutant strains to cope with the nutrient‐poor environment of plant growth. Alternatively, it could result from a reduced ability to undergo a determinate developmental change during the later stages of teliospore formation. In many organisms in which developmental changes are triggered by nutrient starvation, autophagy appears to facilitate the nutrient recycling necessary for these events (Mizushima, 2007). In diploid S. cerevisiae autophagy‐deficient strains, nutrient stress‐induced sporulation is blocked (Tsukada and Ohsumi, 1993). In the filamentous ascomycete Aspergillus oryzae, deletion of the Aoatg8 gene results in several developmental defects, including a loss of conidiation and conidial germination (Kikuma et al., 2007). As in these organisms, the sporulation phenotypes of Δatg8 strains could be explained by autophagy playing a critical role in nutrient mobilization during teliospore development in U. maydis. In other organisms, including insects and humans, autophagy is directly involved in the programmed cell death that takes place during developmental changes which are not necessarily triggered by starvation (Baehrecke, 2002, 2003). Therefore, a role of autophagic programmed cell death during teliospore formation cannot be ruled out. Further experiments need to be conducted to determine precisely what role is played by autophagy during gall and teliospore development in U. maydis. In any case, the fact that at least a few Δatg8 teliospores formed in the ears of Tom Thumb maize plants demonstrates that the lack of autophagy does not preclude the hyphae from developing into spores. Therefore, in U. maydis, normal autophagic activity is necessary for the full colonization of galled tissue and normal levels of spore production.
Our interpretation is that the atg8 and atg1 genes are necessary for the full integrity of the autophagy pathway during the budding of haploid wild‐type sporidia and dikaryotic pathogenic development of U. maydis. The atg8 gene plays a crucial role by ensuring full autophagic activity. On the other hand, the atg1 gene appears to play a less significant role, probably because its deletion might not completely impair the pathway. The role of atg1 becomes more apparent in Δatg8 strains, suggesting an additive relationship. The simultaneous deletion of atg8 and atg1 probably affects two, not fully co‐dependent stages of the pathway, and thus resulted in the additive phenotype observed in the double Δatg1Δatg8 mutant. Alternatively, Atg8 or Atg1, or both, could have additional roles in independent pathways other than autophagy, controlling budding and pathogenic development in U. maydis.
An important aspect of autophagy in the context of this work is its connection with glycogen accumulation (Wang et al., 2001). Glycogen is a widespread energy storage molecule in fungi and animals, and autophagy‐deficient mutants of S. cerevisiae experience a reduction in its accumulation. In Ustilago nuda, a species closely related to U. maydis, glycogen, together with lipids, constitutes the main energy reserve of teliospores (Van Laere and Fransen, 1989). In M. grisea, conidial glycogen storage is mobilized during germination to the developing appressorium. This seems to contribute to the production of the high glycerol concentration associated with the turgor pressure needed during this organ's full development (Thines et al., 2000). Moreover, autophagy seems to play a crucial role in the glycogen homeostasis that is associated with normal aerial growth and conidiation in this fungus (Deng et al., 2009). Therefore, the reduced virulence of U. maydis mutant strains could potentially result from a defect in glycogen metabolism associated with a lack of autophagic activity.
In this work, we have presented substantial evidence to support the hypothesis that U. maydis autophagy genes atg8 and atg1 are important for several aspects of U. maydis biology, including normal budding of haploid sporidia, nutrient starvation survival and pathogenic development. Most importantly, atg8 and atg1 are required for U. maydis to exist as an effective pathogen in the relatively low‐nutrient environment of the plant host.
EXPERIMENTAL PROCEDURES
Strains, media and growth conditions
The Ustilago maydis strains utilized in this study are listed in Table 4. Fungal cultures were grown on potato dextrose agar (PDA) or PDB (Difco, Franklin Lakes, NJ, USA). Nitrate minimal medium (Holliday, 1974) without glucose (MM – C) was employed for gene expression experiments, survival assays and autophagy induction. MM – C employed for the vacuolar evaluation of autophagosome accumulation was amended with 1 mM PMSF (Sigma, St. Louis, MO, USA) to inhibit autophagosome degradation by hydrolysis. Ustilago maydis cultures for protoplast production were grown in yeast extract peptone sucrose (YEPS) medium (1% yeast extract, 2% bacto‐peptone, 2% sucrose). Ustilago maydis transformants were selected on YEPS medium amended with 1 m sorbitol (YEPS‐S) and either 3 µg/mL of carboxin (Gustafson, McKinney, TX, USA) or 150 µg/mL of hygromycin B (Calbiochem, San Diego, CA, USA). Fungal cultures were grown at 30 °C and, for liquid cultures, agitation was at 250 rpm. Escherichia coli DH5α cells were used for transformation during deletion construct assembly. Luria–Bertani (LB) medium containing 50 µg/µL kanamycin A (Research Products International Corp., Chicago, IL, USA) was employed for the selection of E. coli transformants. Mating assays were performed on complete medium containing 1% charcoal (Sigma) (Holliday, 1965).
Table 4.
Ustilago maydis strains used in this study.
| Strain | Relevant genotype | Source |
|---|---|---|
| 1/2 | a1b1 (also known as strain 521) | Gold et al. (1997) |
| 2/9 | a2b2 (BX7A22, near‐isogenic to 1/2) | Gold et al. (1997) |
| MN8.1 | a1b1Δatg8::cbx | This study |
| MN8.4 | a1b1Δatg8::cbx | This study |
| MN8.11 | a2b2Δatg8::cbx | This study |
| MN8.16 | a2b2Δatg8::cbx | This study |
| MN12.4 | a1b1Δatg1::hyg | This study |
| MN29.4 | a2b2Δatg1::hyg | This study |
| MN81.5 | a1b1Δatg1::hygΔatg8::cbx | This study |
| MN81.8 | a1b1Δatg1::hygΔatg8::cbx | This study |
| MN81.0 | a2b2Δatg1::hygΔatg8::cbx | This study |
| MN81.10 | a2b2Δatg1::hygΔatg8::cbx | This study |
Gene deletion
All U. maydis deletion mutants were generated using DelsGate methodology (Garcia‐Pedrajas et al., 2008). The atg8 and atg1 ORFs were completely replaced with DelsGate deletion constructs carrying the carboxin (cbx) or hygromycin (hyg) resistance selectable markers, respectively. Gene replacement was assessed by PCR followed by Southern blot hybridization. For each Southern blot hybridization, 5 µg of genomic DNA from wild‐type and either Δatg8 or Δatg1 strains was digested with EcoRI or Eco47III (New England Biolabs, Ipswich, MA, USA), respectively, and resolved on a 0.7% agarose gel. DNA was transferred overnight to Hybond XL (Amersham Pharmacia Biotech, Piscataway, NJ, USA) nylon membrane in 0.4 m NaOH and UV light cross‐linked. For the atg8 and atg1 probes, 1 kb of the corresponding gene 3′ flank was amplified and DIG labelled using a Roche DIG‐High Prime Labeling and Detection kit (Roche, Indianapolis, IN, USA). Probe labelling and hybridization procedures were performed according to the manufacturer's instructions. Double Δatg1Δatg8 mutants were generated by deleting atg1 in a Δatg8 background and confirmed as above.
qRT‐PCR quantification
Total RNA was extracted using a Spectrum Plant Total RNA kit (Sigma). cDNA was synthesized using the SuperScript III First Strand Synthesis System for RT‐PCR (Invitrogen, Carlsbad, CA, USA) using oligo‐dT as primer and according to the manufacturer's recommendations. Transcript abundance was quantified by qRT‐PCR using SYBR‐GREEN methodology (Bio‐Rad, Hercules, CA, USA) with gene‐specific primers designed through the Integrated DNA Technologies (IDT) website (http://www.idtdna.com/Home/Home.aspx). Reactions were performed on a Cepheid SmartCycler I (Cepheid, Sunnyvale, CA, USA). atg1 and atg8 transcript relative expression levels were calculated according to ΔCT calculations (2−ΔΔCT method) (Pfaffl, 2001) with primer pairs atg1f (TCAACACTCTCGCAGAGACCCTTT) and atg1r (TTCCCACCGTCATCTCAAAGAGCA), and atg8f (TCGGATCTCACTGTGGGCCAATTT) and atg8r (AACCATCCTCGTCCTTGTGCTCTT), respectively, and normalized to a reference cyclophilin gene, cpr1 (um03726) using primers ppif (ACGCCGATTCACTTCGTC) and ppir (AACGACGATCCCTCGTAACCGAAA). The means of gene expression fold increase and their corresponding standard error were calculated on the basis of three biological replicates.
Electron microscopy
Ustilago maydis cells were initially fixed in 2.5% glutaraldehyde in buffer (50 mm potassium phosphate buffer, pH 7.0) at 4 °C overnight, and then post‐fixed in 1% OsO4 in buffer for 2 h at 4 °C and left overnight for en‐bloc staining with 0.5% aqueous uranyl acetate. Cells were dehydrated using a graded ethanol series (25–100%) and rinsed twice in 100% acetone for 10 min each time. Cells were infiltrated with Spurr's resin (Electron Microscopy Sciences, Philadelphia, PA, USA) by serial gradient replacements (33%, 66% Spurr's in acetone and 100% Spurr's) and placed in an oven for 48 h. Polymerization occurred after 48 h at 60 °C. Thin sections (80 nm) were cut using an ultramicrotome (Reichert‐Jung, Wien, Austria) equipped with a Diatome Diamond knife (Diatome AG, Biel, Switzerland), picked up using slot grids and allowed to dry down onto Formvar films according to the procedure of Rowley and Moran (1975). Samples were post‐stained in uranyl acetate and lead citrate and viewed, and microphotographs were taken using an EM 902A transmission electron microscope (Zeiss, Oberkochen, Germany). A minimum of 40 cells was examined per sample for their vacuolar vesicle contents.
Starvation survival assay
Cultures were grown on PDB to stationary phase, harvested by centrifugation, washed with H2O and resuspended in carbon starvation medium (MM – C). Samples were taken at the indicated times, diluted and plated on PDA. Colonies were counted after 3 days at 30 °C. The mean and standard error for each strain were calculated on the basis of two biological replicates.
Mating and pathogenicity analyses
Mating plate assays were used to determine mutant mating ability. The indicated strains were grown overnight in PDB, and equal volumes of each of the mating strains were co‐spotted onto 1% charcoal‐containing complete medium plates that were sealed with parafilm and incubated at room temperature in the dark for 24 h. White dikaryotic filaments indicated a successful mating reaction.
For pathogenicity tests, 7‐day‐old Golden Bantam maize seedlings were co‐inoculated with strain mixtures of 106 cells/mL. Plants were kept in a growth chamber with a cycle of a 16‐h day at 28°C and an 8‐h night at 20°C. Symptom development was scored 7, 10 and 14 dai, and each plant was individually assigned a disease rating based on the following disease scale: 0, no symptoms; 1, anthocyanin production and/or chlorosis; 2, small leaf galls; 3, small stem galls; 4, large stem galls; 5, plant death (Gold and Kronstad, 1994). The disease index is calculated as the summation of the disease ratings divided by the number of plants. For each mutant strain considered, three independent biological replicates of pathogenicity tests were conducted. A nonparametric statistical test was performed to simultaneously evaluate the differences among treatments (Shah and Madden, 2004).
Staining of fungal structures during in planta growth
Symptomatic plant tissue was collected 5, 10, 14, 21, 28 and 35 dai, and stained with chlorazole black E as described previously (Brundrett et al. 1996). Leaf tissue at 5 dai was cut into smaller samples and destained for 48 h in ethanol, washed twice in water and stained with a solution of 0.03% chlorazole black E (Sigma) in a 1:1:1 mixture of water–glycerol–lactic acid overnight at 65 °C. Free hand sections of galled tissue were made with a razor blade and directly stained with chlorazole black as described for the 5 dai samples without the ethanol destaining step. Samples were stored in 50% glycerol until microscopic observation.
Supporting information
Fig. S1 Ustilago maydis Atg8 shares a high degree of sequence similarity with other fungal orthologues.
Fig. S2 Ustilago maydis Atg1 and Saccharomyces cerevisiae Atg1 share a high degree of sequence similarity.
Fig. S3 Deletion of atg8 and atg1 in Ustilago maydis.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
REFERENCES
- Abramovitch, R.B. , Yang, G. and Kronstad, J.W. (2002) The ukb1 gene encodes a putative protein kinase required for bud site selection and pathogenicity in Ustilago maydis . Fungal Genet. Biol. 37, 98–108. [DOI] [PubMed] [Google Scholar]
- Baba, M. , Takeshige, K. , Baba, N. and Ohsumi, Y. (1994) Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke, E.H. (2002) How death shapes life during development. Nat. Rev. Mol. Cell Biol. 3, 779–787. [DOI] [PubMed] [Google Scholar]
- Baehrecke, E.H. (2003) Autophagic programmed cell death in Drosophila . Cell Death Differ. 10, 940–945. [DOI] [PubMed] [Google Scholar]
- Banuett, F. (1991) Identification of genes governing filamentous growth and tumor induction by the plant pathogen Ustilago maydis . Proc. Natl. Acad. Sci. USA, 88, 3922–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F. and Herskowitz, I. (1989) Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA, 86, 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F. and Herskowitz, I. (1996) Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis . Development, 122, 2965–2976. [DOI] [PubMed] [Google Scholar]
- Bassham, D.C. (2007) Plant autophagy—more than a starvation response. Curr. Opin. Plant Biol. 10, 587–593. [DOI] [PubMed] [Google Scholar]
- Bolker, M. (2001) Ustilago maydis—a valuable model system for the study of fungal dimorphism and virulence. Microbiology, 147, 1395–1401. [DOI] [PubMed] [Google Scholar]
- Brundrett, M. , Bougher, N. , Dell, B. , Grove, T. and Malajczuk, N. (1996) Working with Mycorrhizas in Forestry and Agriculture. Australian Centre for International Agricultural Research, Canberra, Australia, Monograph 32.
- Cebollero, E. and Reggiori, F. (2009) Regulation of autophagy in yeast Saccharomyces cerevisiae . Biochim. Biophys. Acta, 1793, 1413–1421. [DOI] [PubMed] [Google Scholar]
- Cheong, H. and Klionsky, D.J. (2008) Dual role of Atg1 in regulation of autophagy‐specific PAS assembly in Saccharomyces cerevisiae . Autophagy, 4, 724–726. [DOI] [PubMed] [Google Scholar]
- Deng, Y.Z. , Ramos‐Pamplona, M. and Naqvi, N.I. (2009) Autophagy‐assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae . Autophagy, 5, 33–43. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. , Van Der Linde, K. , Assmann, D. , Schwammbach, D. , Hof, A. , Mohanty, A. , Jackson, D. and Kahmann, R. (2009) Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5, e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrenberger, F. , Wong, K. and Kronstad, J.W. (1998) Identification of a cAMP‐dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis . Proc. Natl. Acad. Sci. USA, 95, 5684–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi, L. , Aaronson, S.A. , Abrams, J. , Alnemri, E.S. , Andrews, D.W. , Baehrecke, E.H. , Bazan, N.G. , Blagosklonny, M.V. , Blomgren, K. , Borner, C. , Bredesen, D.E. , Brenner, C. , Castedo, M. , Cidlowski, J.A. , Ciechanover, A. , Cohen, G.M. , De Laurenzi, V. , De Maria, R. , Deshmukh, M. , Dynlacht, B.D. , El‐Deiry, W.S. , Flavell, R.A. , Fulda, S. , Garrido, C. , Golstein, P. , Gougeon, M.L. , Green, D.R. , Gronemeyer, H. , Hajnoczky, G. , Hardwick, J.M. , Hengartner, M.O. , Ichijo, H. , Jaattela, M. , Kepp, O. , Kimchi, A. , Klionsky, D.J. , Knight, R.A. , Kornbluth, S. , Kumar, S. , Levine, B. , Lipton, S.A. , Lugli, E. , Madeo, F. , Malorni, W. , Marine, J.C. , Martin, S.J. , Medema, J.P. , Mehlen, P. , Melino, G. , Moll, U.M. , Morselli, E. , Nagata, S. , Nicholson, D.W. , Nicotera, P. , Nunez, G. , Oren, M. , Penninger, J. , Pervaiz, S. , Peter, M.E. , Piacentini, M. , Prehn, J.H. , Puthalakath, H. , Rabinovich, G.A. , Rizzuto, R. , Rodrigues, C.M. , Rubinsztein, D.C. , Rudel, T. , Scorrano, L. , Simon, H.U. , Steller, H. , Tschopp, J. , Tsujimoto, Y. , Vandenabeele, P. , Vitale, I. , Vousden, K.H. , Youle, R.J. , Yuan, J. , Zhivotovsky, B. and Kroemer, G. (2009) Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 16, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Pedrajas, M.D. , Nadal, M. , Kapa, L.B. , Perlin, M.H. , Andrews, D.L. and Gold, S.E. (2008) DelsGate, a robust and rapid gene deletion construction method. Fungal Genet. Biol. 45, 379–388. [DOI] [PubMed] [Google Scholar]
- Geng, J. and Klionsky, D.J. (2008) The Atg8 and Atg12 ubiquitin‐like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9, 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, S.E. , Brogdon, S.E. , Mayorga, M.E. and Kronstad, J.W. (1997) The Ustilago maydis regulatory subunit of a cAMP‐dependent protein kinase is required for gall formation in maize. Plant Cell, 9, 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, S. , Duncan, G. , Barrett, K. and Kronstad, J. (1994) cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis . Genes Dev. 8, 2805–2816. [DOI] [PubMed] [Google Scholar]
- Gold, S.E. and Kronstad, J.W. (1994) Disruption of two genes for chitin synthase in the phytopathogenic fungus Ustilago maydis . Mol. Microbiol. 11, 897–902. [DOI] [PubMed] [Google Scholar]
- Holliday, R. (1965) Induced mitotic crossing‐over in relation to genetic replication in synchronously dividing cells of Ustilago maydis . Genet. Res. 10, 104–120. [DOI] [PubMed] [Google Scholar]
- Holliday, R. (1974) Ustilago maydis In: Handbook of Genetics (King R. C. ed.), pp. 575–595. New York: Plenum Press. [Google Scholar]
- Hu, G. , Hacham, M. , Waterman, S.R. , Panepinto, J. , Shin, S. , Liu, X. , Gibbons, J. , Valyi‐Nagy, T. , Obara, K. , Jaffe, H.A. , Ohsumi, Y. and Williamson, P.R. (2008) PI3K signaling of autophagy is required for starvation tolerance and virulence of Cryptococcus neoformans . J. Clin. Invest. 118, 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. and Klionsky, D.J. (2007) Autophagy and human disease. Cell Cycle, 6, 1837–1849. [DOI] [PubMed] [Google Scholar]
- Ichimura, Y. , Kirisako, T. , Takao, T. , Satomi, Y. , Shimonishi, Y. , Ishihara, N. , Mizushima, N. , Tanida, I. , Kominami, E. , Ohsumi, M. , Noda, T. and Ohsumi, Y. (2000) A ubiquitin‐like system mediates protein lipidation. Nature, 408, 488–492. [DOI] [PubMed] [Google Scholar]
- Jacobs, C.W. , Mattichak, S.J. and Knowles, J.F. (1994) Budding patterns during the cell cycle of the maize smut pathogen Ustilago maydis . Can. J. Bot. 72, 1675–1680. [Google Scholar]
- Kabeya, Y. , Kamada, Y. , Baba, M. , Takikawa, H. , Sasaki, M. and Ohsumi, Y. (2005) Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell, 16, 2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, Y. , Funakoshi, T. , Shintani, T. , Nagano, K. , Ohsumi, M. and Ohsumi, Y. (2000) Tor‐mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki, T. , Wang, K. , Cao, Y. , Baba, M. and Klionsky, D.J. (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell, 17, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata, T. , Kamada, Y. , Kabeya, Y. , Sekito, T. and Ohsumi, Y. (2008) Organization of the pre‐autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell, 19, 2039–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuma, T. , Ohneda, M. , Arioka, M. and Kitamoto, K. (2006) Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae . Eukaryot. Cell, 5, 1328–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuma, T. , Arioka, M. and Kitamoto, K. (2007) Autophagy during conidiation and conidial germination in filamentous fungi. Autophagy, 3, 128–129. [DOI] [PubMed] [Google Scholar]
- Kirisako, T. , Baba, M. , Ishihara, N. , Miyazawa, K. , Ohsumi, M. , Yoshimori, T. , Noda, T. and Ohsumi, Y. (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T. , Ichimura, Y. , Okada, H. , Kabeya, Y. , Mizushima, N. , Yoshimori, T. , Ohsumi, M. , Takao, T. , Noda, T. and Ohsumi, Y. (2000) The reversible modification regulates the membrane‐binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J. , Cregg, J.M. , Dunn, W.A., Jr , Emr, S.D. , Sakai, Y. , Sandoval, I.V. , Sibirny, A. , Subramani, S. , Thumm, M. , Veenhuis, M. and Ohsumi, Y. (2003) A unified nomenclature for yeast autophagy‐related genes. Dev. Cell, 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Levine, B. and Klionsky, D.J. (2004) Development by self‐digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell, 6, 463–477. [DOI] [PubMed] [Google Scholar]
- Liu, X.H. and Lin, F.C. (2008) Investigation of the biological roles of autophagy in appressorium morphogenesis in Magnaporthe oryzae . J. Zhejiang Univ. Sci. B, 9, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.H. , Lu, J.P. , Zhang, L. , Dong, B. , Min, H. and Lin, F.C. (2007) Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell, 6, 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N. (2007) Autophagy: process and function. Genes Dev. 21, 2861–2873. [DOI] [PubMed] [Google Scholar]
- Mizushima, N. , Noda, T. , Yoshimori, T. , Tanaka, Y. , Ishii, T. , George, M.D. , Klionsky, D.J. , Ohsumi, M. and Ohsumi, Y. (1998) A protein conjugation system essential for autophagy. Nature, 395, 395–398. [DOI] [PubMed] [Google Scholar]
- Nadal, M. , Garcia‐Pedrajas, M.D. and Gold, S.E. (2008) Dimorphism in fungal plant pathogens. FEMS Microbiol. Lett. 284, 127–134. [DOI] [PubMed] [Google Scholar]
- Nakatogawa, H. , Ichimura, Y. and Ohsumi, Y. (2007) Atg8, a ubiquitin‐like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell, 130, 165–178. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F. and Klionsky, D.J. (2002) Autophagy in the eukaryotic cell. Eukaryot. Cell, 1, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, T.L. , Bonneau, L. , Der, C. , Marty‐Mazars, D. and Marty, F. (2006) Starvation‐induced expression of autophagy‐related genes in Arabidopsis . Biol. Cell, 98, 53–67. [DOI] [PubMed] [Google Scholar]
- Rowley, J.C., 3rd and Moran, D.T. (1975) A simple procedure for mounting wrinkle‐free sections on formvar‐coated slot grids. Ultramicroscopy, 1, 151–155. [DOI] [PubMed] [Google Scholar]
- Sachdeva, U.M. and Thompson, C.B. (2008) Diurnal rhythms of autophagy: implications for cell biology and human disease. Autophagy, 4, 581–589. [DOI] [PubMed] [Google Scholar]
- Sgarlata, C. and Perez‐Martin, J. (2005) Inhibitory phosphorylation of a mitotic cyclin‐dependent kinase regulates the morphogenesis, cell size and virulence of the smut fungus Ustilago maydis . J. Cell Sci. 118, 3607–3622. [DOI] [PubMed] [Google Scholar]
- Shah, D.A. and Madden, L.V. (2004) Nonparametric analysis of ordinal data in designed factorial experiments. Phytopathology, 94, 33–43. [DOI] [PubMed] [Google Scholar]
- Steinberg, G. , Wedlich‐Soldner, R. , Brill, M. and Schulz, I. (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell. Sci. 114, 609–622. [DOI] [PubMed] [Google Scholar]
- Stephan, J.S. , Yeh, Y.Y. , Ramachandran, V. , Deminoff, S.J. and Herman, P.K. (2009) The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA, 106, 17049–17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K. and Ohsumi, Y. (2007) Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae . FEBS Lett. 581, 2156–2161. [DOI] [PubMed] [Google Scholar]
- Suzuki, K. , Kubota, Y. , Sekito, T. and Ohsumi, Y. (2007) Hierarchy of Atg proteins in pre‐autophagosomal structure organization. Genes Cells, 12, 209–218. [DOI] [PubMed] [Google Scholar]
- Thines, E. , Weber, R.W. and Talbot, N.J. (2000) MAP kinase and protein kinase A‐dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea . Plant Cell, 12, 1703–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M. and Ohsumi, Y. (1993) Isolation and characterization of autophagy‐defective mutants of Saccharomyces cerevisiae . FEBS Lett. 333, 169–174. [DOI] [PubMed] [Google Scholar]
- Van Laere, A. and Fransen, M. (1989) Metabolism of germinating teliospores of Ustilago nuda . Arch. Microbiol. 153, 33–37. [Google Scholar]
- Vellai, T. (2009) Autophagy genes and ageing. Cell Death Differ. 16, 94–102. [DOI] [PubMed] [Google Scholar]
- Veneault‐Fourrey, C. , Barooah, M. , Egan, M. , Wakley, G. and Talbot, N.J. (2006) Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science, 312, 580–583. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Wilson, W.A. , Fujino, M.A. and Roach, P.J. (2001) Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP‐activated protein kinase, and the cyclin‐dependent kinase Pho85p. Mol. Cell Biol. 21, 5742–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich‐Soldner, R. , Straube, A. , Friedrich, M.W. and Steinberg, G. (2002) A balance of KIF1A‐like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis . EMBO J. 21, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Ustilago maydis Atg8 shares a high degree of sequence similarity with other fungal orthologues.
Fig. S2 Ustilago maydis Atg1 and Saccharomyces cerevisiae Atg1 share a high degree of sequence similarity.
Fig. S3 Deletion of atg8 and atg1 in Ustilago maydis.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item


