Abstract
Background
The molecular mechanism underlying clear cell renal cell carcinoma (ccRCC) metastasis remains unclear. We therefore aimed to elucidate the role of IMPA2 in ccRCC metastatic progression.
Methods
Using the Cancer Genome Atlas (TCGA) database and immunohistochemistry (IHC) staining, we investigated differences in IMPA2 mRNA and protein expression, as well as their clinical relevance, in ccRCC. To investigate the function of IMPA2 in ccRCC metastasis, we performed in vitro migration and in vivo lung colony-forming assays. We further explored the effect of microRNA (miR)-25 on IMPA2 expression by performing a luciferase reporter assay.
Findings
We show that ccRCC expresses relatively lower transcript levels of IMPA2 than normal kidney tissue. IMPA2 downregulation was greater in high-grade ccRCC than in low-grade ccRCC and was correlated with a poor prognosis in ccRCC patients. Importantly, we demonstrate that IMPA2 expression is inversely associated with the metastatic potential of ccRCC cells. We found that IMPA2 knockdown promotes, but overexpression suppresses, the cellular migration and lung colony-forming abilities of ccRCC cells. By using in silico and luciferase reporter assays, we found that IMPA2 expression is primarily influenced by miR-25 in ccRCC cells. Significantly, the inhibition of miR-25 function restored IMPA2 expression, thereby diminishing the metastatic potential of ccRCC cells.
Interpretation
We conclude that miR-25-mediated IMPA2 downregulation constitutes a novel signature for cancer metastasis and poor outcomes in ccRCC. We further postulate that the therapeutic targeting of miR-25 can be useful for preventing the metastatic progression of ccRCC associated with IMPA2 downregulation.
Fund
This study was supported by the Ministry of Science and Technology, Taiwan (MOST 107-2314-B-038-094, MOST 106-2314-B-038-069-MY3, MOST 105-2320-B-038-021-MY3 and MOST 107-2320-B-038-056).
Keywords: Renal cell carcinoma, Metastasis, IMPA2, MicroRNA, Biomarker
Research in context.
Evidence before this study
Clear cell renal cell carcinoma (ccRCC) is the most common type of RCC and named from the deposition of high lipid content in the cytoplasm. Approximately 30% of clear cell renal cell carcinoma (ccRCC) cases develop metastases, and 20 to 40% of patients show recurrence after surgery. Metastatic spread to lungs is extensively found in clinical ccRCC patients; however, molecular mechanism underlying ccRCC metastasis remains largely unknown. Therefore, it is urgently needed to unlock the molecular mechanism for ccRCC metastatic progression and identify a new therapeutic target for combating ccRCC metastasis.
Added value of this study
Here we find that IMPA2 downregulation is commonly detected in high-grade ccRCC and refers to a poor prognosis in ccRCC patients. In addition, IMPA2 knockdown or overexpression negatively modulates the metastatic ability of ccRCC cells. Furthermore, we identified that a dysregulation of miR-25-3p determines the IMPA2-modulated metastatic progression in ccRCC. In summary, this study demonstrated that the miR-25-IMPA2 axis plays a critical role in driving ccRCC metastasis.
Implications of all the available evidence
This study identified a novel miR-25-IMPA2 axis that modulates the metastatic potentials of ccRCC, and suggested that miR-25-IMPA2 axis servers as a clinical biomarker and a promising therapeutic target for ccRCC metastasis.
Alt-text: Unlabelled Box
1. Introduction
Renal cell carcinoma (RCC) is the most common type of malignant adult kidney tumor. Approximately 75% of RCC patients have the clear cell renal cell carcinoma (ccRCC) subtype [1]. Approximately 30% of ccRCC cases develop metastases, and 20 to 40% of patients show recurrence after cancer resection [2]. Metastasizing tumor cells spread out from the original lesions via invading the lymphatics or entering the circulation [3]. Lung metastases are common and are the result of metastatic spread to the lungs from a variety of tumor types, including RCC [4]. Despite the clear importance of metastasis, the process is still incompletely characterized at the molecular and biochemical levels. There are numerous targeted therapy agents approved for clinical use in metastatic RCC. These agents target the vascular epithelial growth factor (VEGF) pathway or are mammalian target of rapamycin (mTOR) inhibitors [5]. Many RCC patients receiving targeted therapy develop acquired resistance and experience subsequent tumor progression. Therefore, there is an urgent need to identify a new therapeutic target to treat RCC [6].
Inositol monophosphatase (IMPase) is an enzyme that dephosphorylates myo-inositol monophosphate to generate free myo-inositol. This enzymatic pathway is critical in cellular functions because myo-inositol is the substrate for the synthesis of the membrane lipid phosphatidylinositol (PI). The dephosphorylation of myo-inositol monophosphate by IMPase is a rate-limiting step for the regeneration of PI [7,8]. When cells receive stimuli, PI 4,5-bisphosphate is hydrolyzed by phospholipase C, producing two second messengers: inositol 1,4,5-trisphosphate (IP3) and diacylglycerol. IP3 triggers the release of Ca2+ from intracellular stores and undergoes multistep dephosphorylation by multiple enzymes to enable the reuse of myo-inositol [9,10]. There are two IMPase genes in mammals, the well-characterized myo-inositol monophosphatase 1 (IMPA1) and myo-inositol monophosphatase 2 (IMPA2) [7]. Until now, myo-inositol has been clinically important in the field of psychiatry, such as bipolar disorder. Lithium, which directly inhibits IMPase, has been used for half a century as a first-line drug in bipolar disorder in vivo and in vitro [11]. Recently, French et al. indicated that the expression of IMPA2 genes accounted for more variation in methotrexate polyglutamates in leukemia cells (46%) than in normal cell lines (20%) [12]. However, there are few published articles describing the relationship between RCC and IMPA2.
MicroRNAs (miRNAs) are small single-stranded noncoding RNAs (21–23 nucleotides long) encoded in the genomes of plants, invertebrates, and vertebrates. miRNAs mainly bind imperfectly to target messenger RNAs (mRNAs) and negatively regulate gene expression posttranscriptionally by inhibiting translation [13]. The accumulated evidence indicates that miRNAs can posttranscriptionally regulate the expression of various oncogenes and tumor suppressor genes. Furthermore, miRNAs have a role in angiogenesis, the epithelial-mesenchymal transition, metastasis, and drug resistance. Loss of one or several miRNAs can have substantial effects or cause tumorigenesis [14]. Numerous studies have reported correlations between miRNAs and tumor type, tumor stage, or survival in ccRCC. For example, miR-338-3p has been found to target the sex-determining region Y-box 4 (SOX4) and inhibit cell proliferation and invasion in renal cell carcinoma [15]. However, miR-543 has been found to promote the proliferation and invasion of ccRCC cells by targeting Krüppel-like factor 6 [16]. Therefore, an improved understanding of miRNA mechanisms in RCC tumorigenesis would provide important information about cancer diagnosis or prognosis. Importantly, this knowledge could be used in the development of anticancer therapies for RCC [17]. Our recent results demonstrated that the expression of IMPA2 is predominantly downregulated in primary tumors compared to normal tissues derived from patients with ccRCC. Therefore, the aims of this study were to evaluate the role of the IMPA2 gene in determining the tumor grade, pathologic metastatic stage and prognosis of ccRCC. Furthermore, we analyzed the correlations of IMPA2 levels with tumor invasion and metastatic progression in ccRCC in vitro and in vivo. Finally, we assessed the relationship between miRNAs and IMPA2 using in silico analysis.
2. Material & methods
2.1. Clinical and molecular data for RCC patients
The clinical information for the patients in the TCGA RCC cohort, including age, gender, cancer grade, cancer stage, TNM stage, and overall survival (OS) time, was collected from the TCGA website (Supplementary Table 1, Supplementary Table 2). The molecular data for the TCGA RCC cohort, which were obtained by RNAseq (polyA þ Illumina HiSeq) analysis, were also downloaded from the UCSC Xena website (http://xena.ucsc.edu/welcome-to-ucsc-xena/).
2.2. Immunohistochemistry (IHC) staining analysis
RCC tissue microarrays were purchased from SuperBioChips (Seoul, Korea) and the detailed information on all tumor specimens can be found at http://www.tissue-array.com/main.html and Supplementary Table 3. For IHC staining, paraffin-embedded tumor sections (3 μm thickness) were heated, deparaffinized using xylene, and rehydrated in a graded series of ethanol with a final wash in tap water. Antigen retrieval was performed with Target Retrieval Solution (DAKO) in a Decloaking Chamber (Biocare Medical). Endogenous peroxidase activity was quenched with hydrogen peroxide. The sections were then incubated with anti-IMPA2 antibody (Sigma-Aldrich Cat# HPA029561) at 4 °C overnight. A VECTASTAIN ABC peroxidase system (Vector Laboratories) was used to detect the reaction products.
IHC intensity of the IMPA2 protein was recorded by combining a four-tier scale (0, negative; 1+, week; 2+, moderate; 3+, strong) without (for Kaplan-Meier analysis) or with (for paired normal and tumor tissues) staining percentage (0–100%) in tissues derived from ccRCC patients.
2.3. Cell culture
The human renal adenocarcinoma cell line 769-P was obtained from ATCC; the cells were maintained in RPMI 1640 medium supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100 μg/ml) (Gibco Life Technologies). The human renal adenocarcinoma cell lines ACHN and A498 were purchased from ATCC; the cells were maintained in minimal essential medium (MEM) supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100 μg/ml). The human renal adenocarcinoma cell lines Caki-1 and Caki-2 were purchased from ATCC; the cells were maintained in McCoy's 5A medium supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100 μg/ml). All cells were incubated in 5% CO2 at 37 °C.
2.4. Migration and invasion assays
In vitro migration and invasion was assessed by using a Boyden chamber assay (NeuroProbe, Gaithersburg, MD, USA). Cells (1.5 × 104) in serum-free culture medium were added to the upper chamber of the device on a membrane with an 8.0 μm hole and pre-coated without (for migration) or with (for invasion) matrigel (Sigma-Aldrich), and the lower chamber was filled with 10% FBS culture medium. After a 3 h (for migration) or 16 h (for invasion) incubation, the membrane was soaked in methanol for 10 min and stained with Giemsa, which was diluted 10-fold by double-distilled water, for 1 h. Then, the membrane was attached to slides, and the upper cells of the membrane were carefully removed with a cotton swab. The lower cells were photographed. The migrated cells were quantified by counting the cells in three random areas under a microscope at 400× magnification.
2.5. miRNA-25-3p inhibitor and miR-25-3p/miR-25-5p transfection
Cells were seeded in six-well plates and transfected with 10 nM, 30 nM, or 100 nM miR-25-3p inhibitor or co-transfected with 100 nM miR-25-3p inhibitor with non-silencing or IMPA2 shRNA (0.3 μg of each) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's instructions. The miR-25-3p inhibitor was purchased from Thermo Fisher (Pittsburgh, PA, USA). After 24 h of the transfection, the cells were used for migration, RNA, and protein assays.
Low sensor backbone (LSB) plasmid containing miR-25-3p or miR-25-5p was a gift from Ron Weiss (Addgene plasmid # 103375; http://n2t.net/addgene:103375; RRID: Addgene_103,375) and stably transfected into A498 cells according to the procedure reported previously [18]. The miR-25-3p-expressing cells were further transiently transfected with pLAS3w/Pbsd plasmid containing without or with IMPA2 gene.
2.6. Lentivirus production and transduction
Lentiviral IMPA2 shRNA constructs were purchased from the RNAi Core in Academia Sinica (Taipei, Taiwan). The mature antisense sequences of the IMPA2 shRNAs were as follows: shRNA1 (CDS), 5′-GCTGTTCGACAAGAGCTTGAA-3′; shRNA2 (3’ UTR), 5′-AGAGGGAGTTGTCACGCTACA-3′; shRNA3 (3′ UTR), 5′-ACGTCTCTCTCACCAGGATTT-3′. The gene encoding IMPA2 was amplified from human cDNA (Invitrogen) using the standard polymerase chain reaction (PCR) procedure with paired primers (forward: 5′-TAAGCAGCTAGCATGAAGCCGAGCGGC-3′ reverse: 5′-TGTTTAGAATTCTCACTTCTCATCATCCCGC-3′) and subcloned into the pLAS3w/Pbsd vector with NheI/EcoRI restriction sites. Lentiviruses were produced by cotransfection of shRNA or IMPA2-expressing plasmids, envelope plasmids (pMD.G) and packaging plasmids (pCMV-dR8.91) in 293 T cells. The 293 T cells were incubated for 24 h, and then the culture medium was removed and refreshed. The viral supernatants were harvested and the titers determined at 48 h posttransfection. RCC cells were transduced with the lentiviruses in the presence of polybrene and were selected using puromycin.
2.7. Western blotting assay
Protein was extracted from cells using lysis buffer. After centrifugation, the protein content of the supernatant was quantified at 280 nm. Aliquots of 100 μg of total protein and HR Pre-Stained Protein Marker 10–170 kDa (BIOTOOLS, Taiwan) were loaded onto each lane of a SDS gel and transferred to PVDF membranes. The membranes were incubated with blocking buffer (5% nonfat milk in TBS containing 0.1% Tween-20) for 2 h at room temperature. After that, the samples were probed with primary antibodies against IMPA2/Slug (Cell Signaling) and GAPDH (AbFrontier) overnight at 4 °C. After extensive washing, the membranes were incubated with a secondary peroxidase-labeled IgG for 1 h at room temperature. Immunoreactive bands were visualized by enhanced chemiluminescence.
2.8. Reverse transcription polymerase chain reaction and real-time polymerase chain reaction
Total RNA was isolated from the cells using TRIzol reagent. cDNA was synthesized from 2 μg of total RNA with a Transcriptor First Strand cDNA Synthesis Kit (Roche). Specific primers (IMPA2: forward_5′-GCTTCTGGGGCCAAGTGTGT-3′ and reverse_5′-CTTCAGGGTCGCA GGGTCAC-3′; miR25: forward_5′-GGGGGGCTCCAAAGTGCTGTT-3′ and reverse_5′-GGGGACACCCTTGTTCTGGC-3′; CDH1: forward_5′-GGCACAGATGGTGTGATTACAGTC-3′ and reverse_ 5′-CAGCGTGAGAGAAGAGAGTGTATGTG-3′; CDH2: forward_5′-CTGTAGAGGCTTCTGGTGAAATCG-3′ and reverse: 5′-GCAAGTTGATTGGAGGGATGAC-3′) GAPDH: forward_5′-ATAGTATTCCACCCATGGCAAATTC-3′ and reverse_5′-GGTCATGAGTCCTTCCACGATACC-3′) and 2× PCR reagent were used for the 25–35 cycles of PCR. Power SYBR™ Green PCR Master Mix (Thermo Fisher) was used for real-time PCR. The mRNA levels were normalized to those of GAPDH. Fold changes were calculated using the 2−ΔΔCt method.
2.9. Luciferase reporter assay
The DNA sequence corresponding to the 3′ untranslated region (3’UTR) of IMPA2 transcript at different lengths was amplified from human cDNA (Invitrogen) using the standard polymerase chain reaction (PCR) procedure with paired primers [forward_5′-TGGGCTAGCGCTGTTTCTC TCTTTAATCTCACGTAG-3′; reverse_5′-CCGCTCGAGACACACAGGGCCATTATTTTGA-3′ (for full-length); reverse_5′-CCGCTCGAGAGCAGTTCTAAAGCCTTATTACCTGAGA-3′ (for deletion)]. The PCR products of IMPA2 3’ UTR were inserted into the pmiRGLO plasmid (Promega) using NheI/XhaI restriction sites. The side-directed mutagenesis for the miR-25-3p DNA-binding site was performed by using pmiRGLO plasmid containing the full-length of IMPA2 3′-UTR as template in the standard PCR procedure with PfuUtra high-fidelity DNA polymerase (Agilent) and paired primers (forward_5′- CCCCCCGGAGTACTTCAGAATC -3′; reverse_5′- CCGCTCGAGTCTGAAGTACTCCGGGGGG-3′). For determining luciferase activity, ACHN cells were seeded in 6-well plates and transfected with pmiRGLO or pmiRGLO containing the IMPA2 3′ UTR variants. After 24 h, the luciferase activity was measured using a Dual-Glo® Luciferase Assay System (Promega). Briefly, the cells were lysed in lysis buffer containing luciferase substrate for 10 min. The total lysate was centrifuged at 12,000 rpm for 1 min, the supernatant was divided between 3 wells in a white 96-well plate, and the firefly luminescence was measured. One volume of Dual-Glo® Stop & Glo® reagent was added to each well. After 10 min, Renilla luminescence was measured. The level of firefly luminescence was normalized to that of the Renilla luminescence.
2.10. Animal study
NOD/SCID male mice (6–8 weeks old) were obtained from BioLASCO in Taiwan and maintained in compliance with the institutional policy. All animal procedures were approved by the Institutional Animal Care and Use Committee at Taipei Medical University (IACUC Approval No.: LAC-2017-0364). For the in vivo lung metastatic colonization assay, cells were implanted into mice through tail vein injection at a concentration of 1 × 106 cells/0.1 ml of PBS. The mice were sacrificed 15 weeks after cell injection, and the lungs were dissected for histological analyses. The metastatic lung nodules were quantified after H&E staining using a microscope.
2.11. Statistical analysis
Overall survival (OS) was assessed by Kaplan-Meier analysis using a log-rank test. Cox Regression test was used to analyze hazard ratio of variables in the univarible and multivariable modes. The t-test and one-way ANOVA Tukey's post-hoc test were used to analyze the statistical significance of parametric data from 2 and more independent samples, respectively. Non-parametric Wilcoxon Signed-Ranks test, Friedman test, Mann-Whitney U test and Kruskal-Wallis test were used to analyze data from 2 related samples, 3 or more related samples, 2 independent samples and 3 or more independent samples, respectively. Pearson and Spearman correlation tests were used to analyze parametric and non-parametric data, respectively. All statistical calculations were performed in the statistical package SPSS version 22.0 for Windows (SPSS, Inc., Chicago, IL).
3. Results
3.1. IMPA downregulation correlates with tumorigenesis, cancer progression and poor prognosis in ccRCC
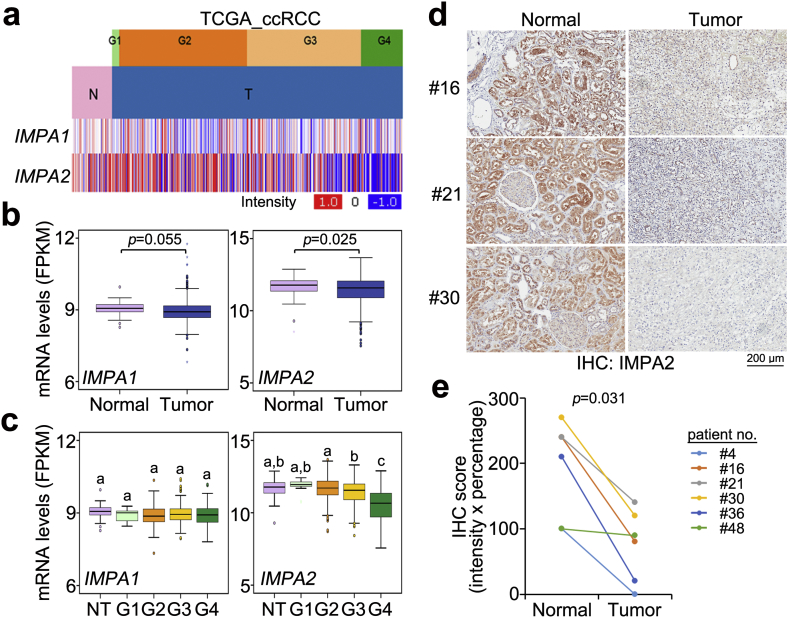
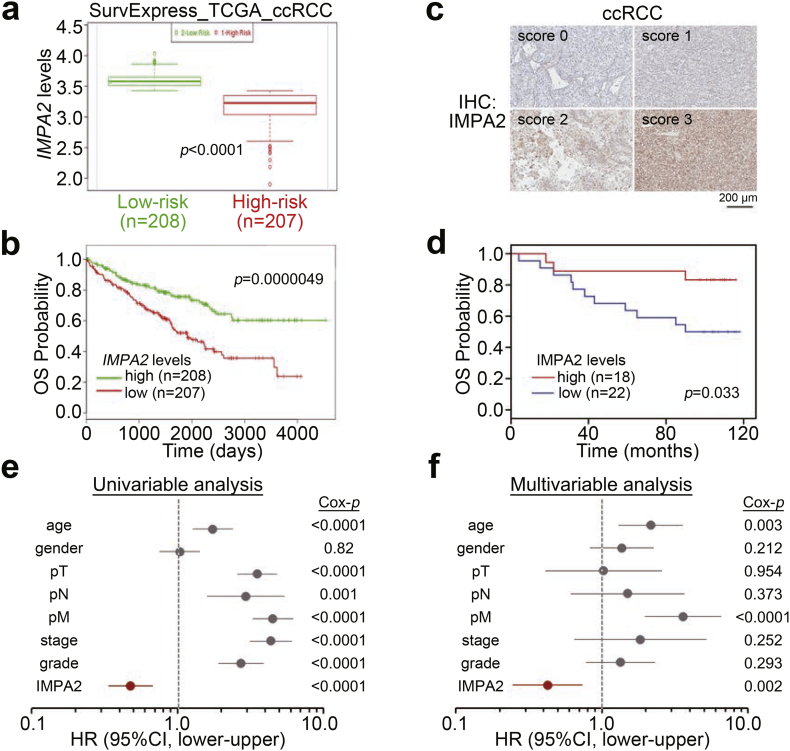
We first analyzed the transcriptional profiles of IMPA1 and IMPA2 in clinical tissues from TCGA ccRCC patients. The data showed that the mRNA levels of IMPA2, but not IMPA1, were significantly (p < .05) downregulated in primary tumors compared to normal renal tissues (Fig. 1a and b). Moreover, in comparison with IMPA1 expression, IMPA2 downregulation in the grade 4 tumors was more predominant than that of normal renal tissues and the grades 1–3 tumors from ccRCC patients (Fig. 1c). Accordingly, immunohistochemistry (IHC) experiments demonstrated that IMPA2 protein levels were dramatically reduced in tumors compared to paired normal renal tissues from 6 ccRCC patients (Fig. 1d and e). Moreover, Kaplan-Meier analysis revealed that low levels of IMPA2 mRNA predicted poor overall survival rates in TCGA ccRCC patients using the SurvExpress program (Fig. 2a and b). Similarly, Kaplan-Meier analysis using IMPA2 protein levels from the IHC experiment showed that IMPA2 downregulation was related to a poor prognosis in ccRCC patients for overall survival probability (Fig. 2c and d) even though the numbers of enrolled patients and clinicopathological results were limited. Importantly, a Cox regression test indicated that high levels of IMPA2 mRNA correlated with good outcomes in the univariate mode and served as an independent factor to predict favorable outcomes in multivariate analyses with other pathologic variables, including age, gender, pT, pN, pM, stage and grade (Fig. 2e and f), as well as other risk factors reported previously, including p53, ki67, CD44, CA9 and PCNA (Figs. S1a and S1b) [19], in ccRCC patients.
Fig. 1.
IMPA2 downregulation significantly correlates with tumorigenesis and cancer progression in ccRCC. (a) Heatmap for the transcriptional profiles of the IMPA1 and IMPA2 genes in normal (N) tissues and primary tumors (T) of different grades from the TCGA ccRCC database. (b and c) Boxplot for the mRNA levels of IMPA1 and IMPA2 in normal tissues (n = 72) and primary tumors (n = 534) (b) or normal tissues (NT, n = 72) and tumors with grade G1 (n = 14), G2 (n = 229), G3 (n = 207) or G4 (n = 76) (c), as shown in a. In b and c, the band inside the box is the second quartile (the median). The upper and lower lines of box are the third and first quartiles, respectively. Box plots have lines extending vertically from the whiskers indicating minimum and maximum of all of the data. The individual points indicate outliers. The significant differences were analyzed by independent sample t-test and Tukey's post-hoc test, respectively. Different letters in the boxplot indicate the statistical significance at p < .05. (d) Immunohistochemistry (IHC) staining for the IMPA2 protein in paired normal and tumor tissues from ccRCC patients #16, #21 and #30. (e) IHC intensity of the IMPA2 protein in paired normal and tumor tissues derived from 6 ccRCC patients. Statistical significance was estimated with a non-parametric Wilcoxon signed rank test.
Fig. 2.
IMPA2 downregulation correlates with a poor prognosis in ccRCC patients. (a and b) Boxplot for the mRNA levels of IMPA2 in the low-risk (green box) and high-risk (red box) groups (a) that were stratified in SurvExpress Kaplan-Meier analysis (b) by the median of the transcriptional profiles in tumors from 415 ccRCC patients from the TCGA database. The band inside the box is the second quartile (the median). The upper and lower lines of box are the third and first quartiles, respectively. Box plots have lines extending vertically from the whiskers indicating minimum and maximum of all of the data. The individual points indicate outliers. In a, significant differences were analyzed by t-test. (c) The IHC results for the IMPA2 protein intensities ranged from 0 to 3. (d) Kaplan-Meier analysis for low (scores 0 and 1) and high (scores 2 and 3) IMPA2 protein levels from the IHC experiment for tumors from 40 ccRCC patients. (E and F) Cox regression test using univariable (e) and multivariable (f) modes for IMPA2 mRNA levels (high vs. low) and other pathological variables including age (median = 60 years, elder vs. younger), gender (male vs. female), pathologic T (pT, T1/T2 vs. T3/T4), pN (N1 vs. N0), pM (M1 vs. M0), stage (III/IV vs. I/II) and grade (III/IV vs. I/II) under the condition of overall survival probability for 415 ccRCC patients from the TCGA database using the SurvExpress program. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. IMPA2 downregulation is associated with metastatic progression in ccRCC
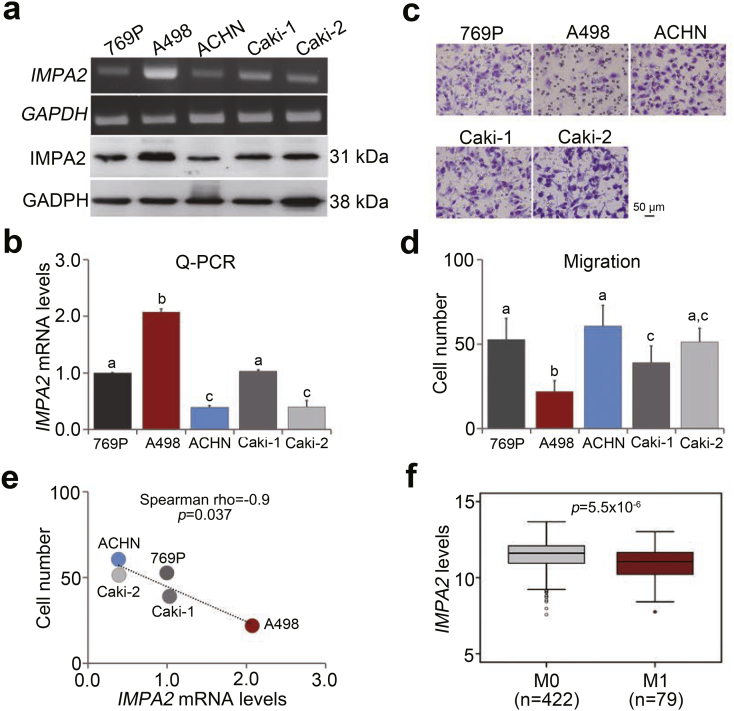
We next examined the endogenous levels of IMPA2 mRNA and protein in a panel of ccRCC cell lines including 769P, A498, ACHN, Caki-1 and Caki-2. The data showed that the endogenous IMPA2 mRNA and protein levels were very elevated in A498 cells but were relatively lower in ACHN cells (Fig. 3a and b). Conversely, the cellular migration abilities were relatively poor in A498 cells but were much stronger in ACHN cells (Fig. 3c and d). The negative correlation between IMPA2 mRNA levels and cellular migration abilities was statistically significant (p = .037) in the tested ccRCC cell lines (Fig. 3e). In clinical specimens, IMPA2 mRNA levels appeared to be significantly (p < .0001) downregulated in metastatic ccRCC tissues (Fig. 3f).
Fig. 3.
IMPA2 expression is inversely associated with metastatic potential in ccRCC cells. (a and b) IMPA2 levels determined by RT-PCR (a, upper), Western blotting (a, bottom) and quantitative PCR (b) in the tested ccRCC cell lines 769P, A498, ACHN, Caki-1 and Caki-2. (c and d) Giemsa staining (c) and data from three independent experiments (d) for the migrated cells in the 3-hour transwell assay for the ccRCC cells. In b and d, error bars represent the mean ± SEM of data obtained from three independent experiments and the statistical differences were analyzed by Kruskal Wallis test. Different letters in each column of b and d indicate the statistical significance at p < .05. (e) Scatchard plot for the endogenous IMPA2 mRNA levels from b and the migrated cell number from d. Spearman's correlation test was used to determine the statistical significance. (f) Transcriptional profile of the IMPA2 gene in primary tumors from TCGA ccRCC patients without (M0) or with (M1) metastatic progression. The band inside the box is the second quartile (the median). The upper and lower lines of box are the third and first quartiles, respectively. Box plots have lines extending vertically from the whiskers indicating minimum and maximum of all of the data. The individual points indicate outliers. The independent sample t-test was used to estimate the statistical significance.
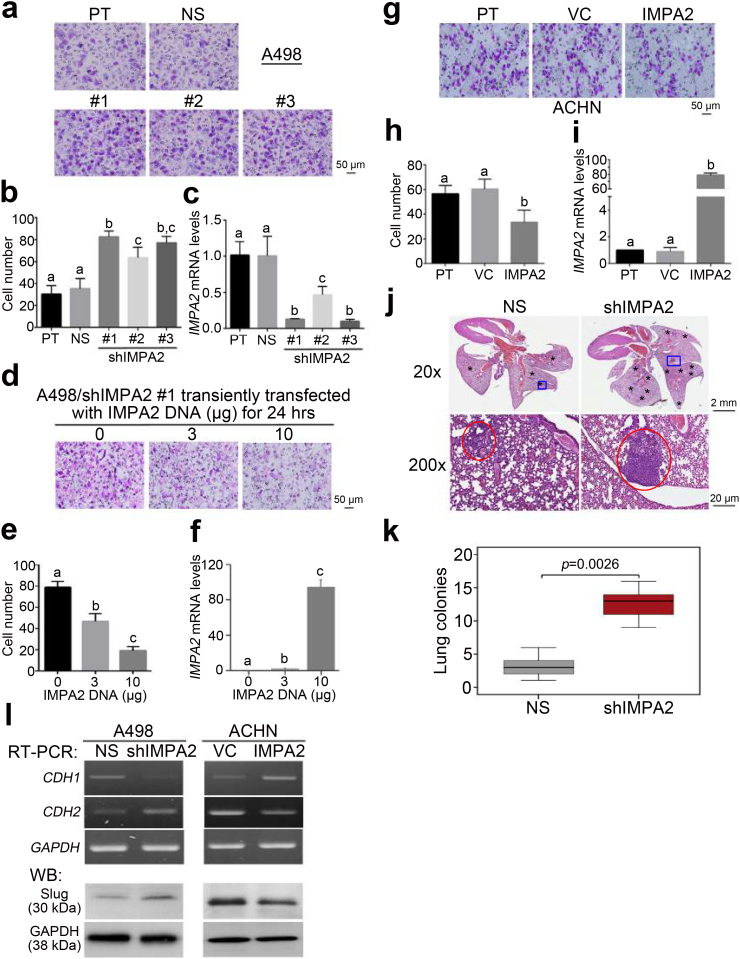
Knockdown (KD) of IMPA expression using 3 independent shRNA clones markedly enhanced the cellular migration ability (Fig. 4a and b) and decreased endogenous IMPA2 mRNA levels (Fig. 4c) in A498 cells. Robustly, the reconstitution of IMPA2 expression in the IMPA2-silenced A498 cells suppressed the IMPA2 KD-enhanced cellular migration ability (Fig. 4d and e) and elevated the IMPA2 mRNA levels (Fig. 4f) in a concentration-dependent manner. Similarly, the forced expression of the exogenous IMPA2 gene predominantly suppressed the cellular migration ability (Fig. 4g and h) but dramatically elevated the endogenous mRNA levels of IMPA2 (Fig. 4i) in ACHN cells with high metastatic potential. Although the negative correlation between the endogenous IMPA2 levels and cellular invasiveness appeared to be not significant, IMPA2 overexpression in ACHN cells (Figs. S2a and S2b) suppressed but IMPA2 knockdown in A498 cells (Figs. S2c and S2d) enhanced the cellular invasion ability dramatically. The lung colony-forming assay demonstrated that IMPA2 knockdown significantly (p = .00022) promoted the formation of tumor foci of A498 cells in the lungs of tumor-bearing mice (Fig. 4j and k). Since epithelial-mesenchymal transition (EMT) plays important role in cancer metastasis, we next examined the expression of E-type marker CDH1 (E-cadherin) and M-type markers CDH2 (N-cadherin) and Slug. Our data showed that IMPA2 knockdown promotes metastatic potentials of A498 cells by triggering EMT as judged by CDH1 downregulation but CDH2/Slug upregulation. Conversely, IMPA2 overexpression suppressed metastatic ability in ACHN cells by reversing EMT as shown by CDH1 upregulation but CDH2/Slug downregulation (Fig. 4l).
Fig. 4.
IMPA2 downregulation promotes metastatic progression in ccRCC cells. (a–c) Giemsa staining (a) and cell number (b) for the migrated cells in 3-hour transwell assays and quantitative PCR for IMPA2 gene expression (c) in A498 cells transfected without (parental, PT) or with a nonsilencing (NS) sequence or one of 3 independent IMPA2 shRNA clones. (d-f) Giemsa staining (d) and cell number (e) for the migrated cells in 3-hour transwell assays and quantitative PCR for IMPA2 gene expression (f) in IMPA2 shRNA clone #1 (targeting 3’ UTR)-overexpressing A498 cells transiently transfected with exogenous IMPA2 DNA in the designated amounts for 24 h. (g-i) Giemsa staining (g) and cell number (h) for the migrated cells in 3-hour transwell assays and quantitative PCR for IMPA2 gene expression (i) in ACHN cells stably transfected without (parental, PT) or with a vector control (VC) or a vector containing the IMPA2 gene. In b, c, e, f, h, and i, error bars represent the mean ± SEM of data obtained from three independent experiments. The different letters in each column of b, c, e, f, h, and i indicate the statistical significance at p < .05 as determined by Friedman test. (j) Hematoxylin/eosin staining for lung tissues derived from mice transplanted with A498 cells transfected with nonsilencing (NS) sequences or IMPA2 shRNA via tail-vein injections. The symbols (*) and red circles indicate the colonies of A498 cells. (k) Boxplot for the lung colonies counted from tumor-bearing mice (n = 5) as shown in j. The band inside the box is the second quartile (the median). The upper and lower lines of box are the third and first quartiles, respectively. Box plots have lines extending vertically from the whiskers indicating minimum and maximum of all of the data. The individual points indicate outliers. Mann-Whitney U test was used to estimate the statistical significance. (l) RT-PCR for CDH1, CDH2 and GAPDH genes and Western blotting (WB) for Slug and GAPDH proteins derived from A498 cells without (NS) or with IMPA2 knockdown (shIMPA2) and ACHN cells without (VC) or with IMPA2 overexpression (IMPA2). GAPDH was used as internal controls for RT-PCR and Western blot analyses. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. MicroRNA-25 posttranscriptionally inhibits IMPA2 expression in highly metastatic ccRCC cells
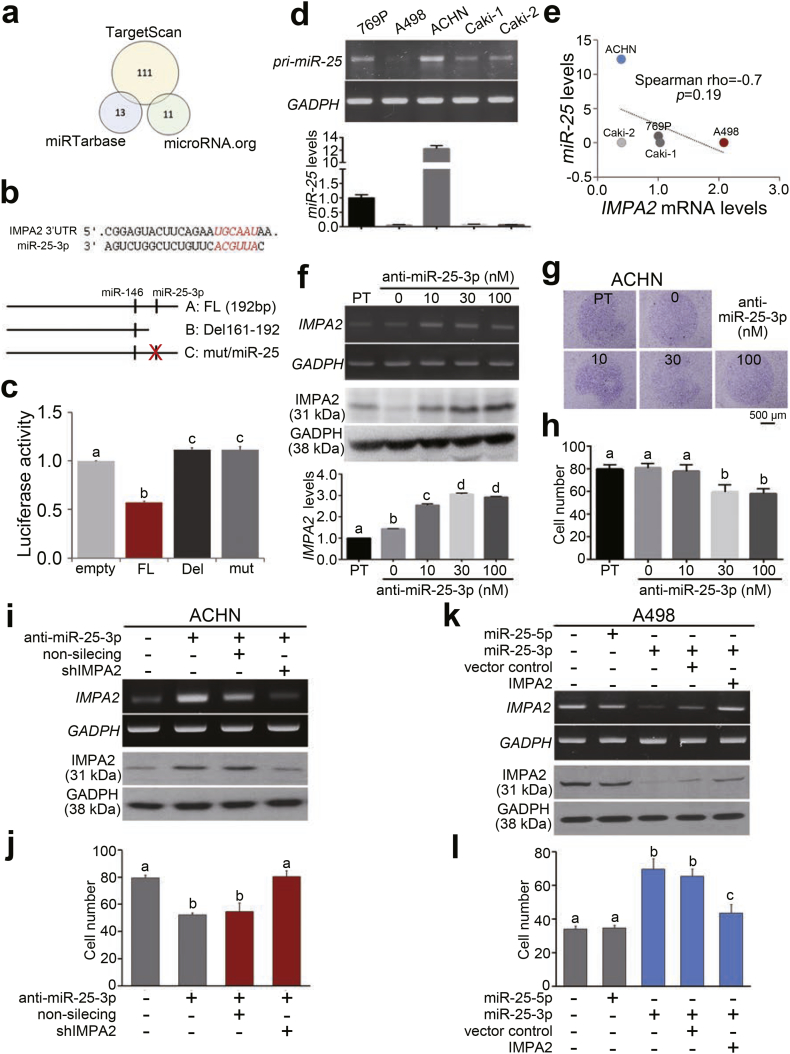
To identify the possible microRNA(s) that binds the 3′ untranslated region (3′ UTR) and thereby posttranscriptionally inhibits the translation of the IMPA2 gene, we performed an in silico analysis using the TargetScan, miRTarbase and microRNA.org databases (Fig. 5a) and then identified 2 miRNA candidates, miR-146 and miR-25-3p (Fig. 5b). We inserted a full-length (192 base pairs) IMPA2 3’ UTR into a luciferase reporter plasmid (Fig. 5b) and then performed a luciferase activity assay in ACHN cells. The data showed that the construct containing the IMPA2 3’ UTR significantly (p < .001) suppressed luciferase activity compared to the empty vector control (Fig. 5c). However, the deletion and mutant variants of the IMPA2 3’ UTR (Fig. 5b) did not affect the luciferase activity in comparison with the vector control (Fig. 5c). Furthermore, we analyzed the endogenous levels of miR-25 in the tested ccRCC cell lines and found that ACHN cells express very high levels of miR-25, but A498 cells exhibit relatively lower miR-25 levels (Fig. 5d). The endogenous levels of miR-25 appeared to be negatively correlated with IMPA2 mRNA levels in the examined ccRCC cell lines (Fig. 5e). The transfection of anti-miR-25-3p oligonucleotides into ACHN cells markedly rescued the endogenous IMPA2 mRNA and protein levels (Fig. 5f) but suppressed the cellular migration ability (Fig. 5g and h). To demonstrate that miR-25-3p promotes ccRCC progression through IMPA2, we further performed IMPA2 knockdown in ACHN cells that were co-transfected with anti-miR-25-3p oligonucleotides. The data showed that IMPA2 knockdown compared to non-silencing control dramatically reduces the RNA and protein levels of IMPA2 (Fig. 5i) and rescues the cellular migration ability in the anti-miR-25-3p oligonucleotide-treated ACHN cells (Fig. 5j and Fig. S3a). On the other hand, the overexpression of miR-25-3p, not miR-25-5p, predominantly decreased the RNA and protein levels of IMPA2 (Fig. 5k) but enhanced cellular migration ability in A498 cells (Fig. 5l and Fig. S3b). In comparison with vector control, the restoration of exogenous IMPA2 gene markedly elevated the RNA and protein levels of IMPA2 (Fig. 5k) but suppressed the cellular migration ability in miR-25-3p-overexpressing A498 cells (Fig. 5l and Fig. S3b). These findings indicate a critical role of miR-25-3p/IMPA2 axis in driving ccRCC progression.
Fig. 5.
IMPA2 downregulation is mediated by the miR-25 posttranscriptional inhibition in ccRCC cells. (a) An in silico analysis of IMPA2-targeting microRNAs using the TargetScan, miRTarbase and microRNA.org databases. The numbers indicate the microRNAs that are computationally predicted to bind the 3′ untranslated region (3′ UTR) of the IMPA2 transcript. (b) Experimental designs for the cloning of the IMPA2 3′ UTR with no alteration (FL, full-length), with a deletion (Del) among base pairs (bp) 161–192 (numbered from the bp next to the stop codon) or with a mutation (mut) at the miR-25 binding sequences into the luciferase-expressing pmirGLO plasmid. (c) Luciferase activity in ACHN cells transiently transfected with the empty pmirGLO plasmid or the plasmid containing the IMPA2 3’ UTR without alterations (FL) or with a deletion (Del 161–192) or miR-25 binding site mutation. One-way ANOVA with Tukey's test was used to estimate the statistical significance. (d) RT-PCR (upper) and quantitative PCR (bottom) analyses for miR-25 and GAPDH expression in a panel of ccRCC cell lines. GAPDH was used as an internal control and was utilized to normalize the miR-25 levels in quantitative PCR. (e) Scatchard plot for the endogenous IMPA2 mRNA levels from Fig. 3b and the miR-25 levels from d. Spearman's correlation test was used to determine the statistical significance. (f) RT-PCR (upper), Western blot (middle) and quantitative PCR (Q-PCR, bottom) analyses for IMPA2 and GAPDH expression in ACHN cells transiently transfected with anti-miR-25-3p oligonucleotides at the designated concentrations for 24 h. GAPDH was used as an internal control. (g and h) Giemsa staining (g) and cell number (h) for the migrated cells in 3-h transwell assays for ACHN cells transiently transfected with anti-miR-25 oligonucleotides at the designated concentrations for 24 h. (i) RT-PCR (upper) and Western blot (bottom) analyses for IMPA2 and GAPDH expression in ACHN cells transiently transfected without or with anti-miR-25-3p oligonucleotides combining with non-silencing or IMPA2 shRNA for 24 h. GAPDH was used as an internal control. (j) Migrated cell number of ACHN cells without or with the designed transfections as shown in i. (k) RT-PCR (upper) and Western blot (bottom) analyses for IMPA2 and GAPDH expression in A498 cells without or with the overexpression of miR-25-5p or miR-25-3p combining without (vector control) or with the restoration of exogenous IMPA2 gene. GAPDH was used as an internal control. (l) Migrated cell number of A498 cells without or with the designed transfections as shown in k. In c, f, h, j and l, error bars represent the mean ± SEM of data obtained from three independent experiments. The different letters in each column of c, f, h, j and l indicate the statistical significance at p < .05 as determined by Friedman test.
3.4. The combined signature of high miR-25 levels and low IMPA2 levels predicts a worse outcome in ccRCC patients
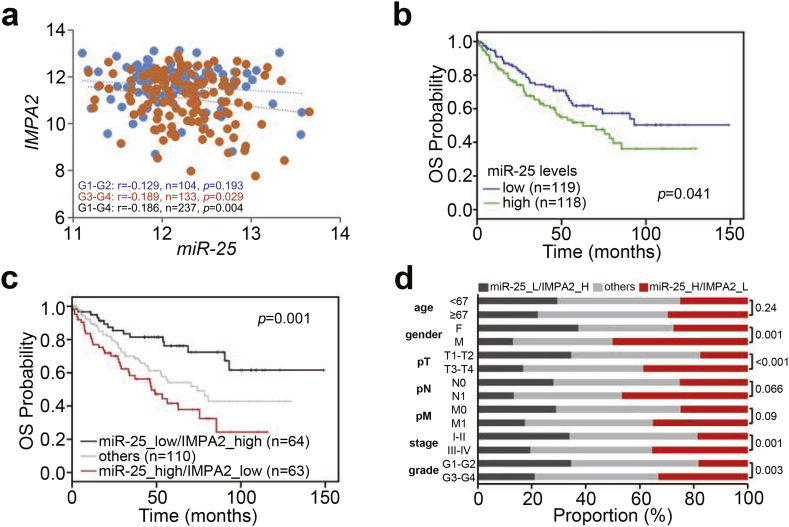
We next analyzed the transcriptional profiles of miR-25 and IMPA2 in primary tumors from TCGA ccRCC patients. We found that the expression of miR-25 was negatively correlated with IMPA2 transcript levels in the analyzed ccRCC tissues (Fig. 6a). Moreover, Kaplan-Meier analysis demonstrated that high miR-25 levels and the combination of high miR-25 levels and low IMPA2 levels are associated with a poorer prognosis in TCGA ccRCC patients (Fig. 6b and c). In addition, the combined signature of high miR-25 levels and low IMPA2 levels was not associated with age, was relatively associated with pathologic N and M status, and was significantly correlated with gender, pathologic T status, stage and grade in TCGA ccRCC patients (Fig. 6d). Importantly, the Cox regression test revealed that the combined signature of high miR-25 levels and low IMPA2 levels predicted an increased hazard ratio in univariate mode and served as an independent risk factor in multivariate mode under the condition of overall survival probability in TCGA ccRCC patients (Table 1).
Fig. 6.
The combined signature of high miR-25 transcript levels and low IMPA2 transcript levels predicts a worse prognosis in ccRCC patients. (a) Scatchard plot for IMPA2 and miR-25 mRNA levels from the TCGA ccRCC database. Pearson's correlation test was used to determine the statistical significance of IMPA2 and miR-25 coexpression in primary tumors derived from patients diagnosed with G1-G2 (blue dots), G3-G4 (orange dots) or G1-G4 (blue and orange dots) ccRCC. (b and c) Kaplan-Meier analyses for miR-25 mRNA levels (b) or miR-25 and IMPA2 mRNA levels (c) in TCGA ccRCC patients (n = 237). The high and low expression levels of miR-25 and IMPA2 were determined by the median of their mRNA levels in primary tumors from the TCGA ccRCC database. (d) Chi-square test for the combined signature of miR-25 and IMPA2 mRNA levels and pathologic variables including age, gender, pT, pN, pM, stage and grade from the TCGA ccRCC database. In c and d, the high (H) and low (L) expression levels of miR-25 and IMPA2 are presented as the signatures miR-25_L/IMPA2_H, others (miR-25_L/IMPA2_L and miR-25_H/IMPA2_H) and miR-25_H/IMPA2_L. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Cox univariable and multivariable analyses under the condition of overall survival probability in association with miR-25/IMPA2 mRNA expression levels and pathological stage derived TCGA cohort with clear-cell renal cell carcinoma.
| Overall survival (n = 237) | ||||
|---|---|---|---|---|
| Variables | Univariable (HR, 95%CI) | P | Multivariable (HR, 95%CI) | P |
| Age | ||||
| <67 | 1 | NA | 1 | NA |
| ≥67 | 1.47 (0.97–2.23) | 0.069 | 1.43 (0.92–2.19) | 0.111 |
| Gender | ||||
| Female | 1 | NA | 1 | NA |
| Male | 1.05 (0.69–1.60) | 0.827 | 1.74 (1.06–2.86) | 0.029 |
| pT | ||||
| T1-T2 | 1 | NA | 1 | NA |
| T3-T4 | 2.74 (1.79–4.17) | <0.001 | 1.08 (0.46–2.55) | 0.853 |
| pN | ||||
| N0 | 1 | NA | 1 | NA |
| N1 | 2.95 (1.52–5.71) | <0.001 | 1.33 (0.65–2.71) | 0.437 |
| pM | ||||
| M0 | 1 | NA | 1 | NA |
| M1 | 3.81 (2.46–5.92) | <0.001 | 2.55 (1.50–4.32) | <0.001 |
| Stage | ||||
| I-II | 1 | NA | 1 | NA |
| III-IV | 3.10 (1.99–4.81) | <0.001 | 1.53 (0.59–3.95) | 0.377 |
| Grade | ||||
| I-II | 1 | NA | 1 | NA |
| III-IV | 2.47 (1.56–3.92) | <0.001 | 1.74 (1.06–2.86) | 0.029 |
| miR-25/IMPA2 levels | ||||
| Low/high | 1 | NA | 1 | NA |
| High/low | 3.22 (1.75–5.95) | <0.001 | 2.43 (1.21–4.91) | 0.013 |
HR and CI denote hazard ratio and confident interval, respectively.
4. Discussion
IMPA2 is gene that encodes a catalytic protein that converts inositol monophosphate into free inositol through dephosphorylation [20]. Inositol regulates >712 genes and is involved in many intracellular signaling pathways [21]. Previous studies have suggested that a promoter polymorphism of IMPA2 contributes to the risk of schizophrenia by elevating transcription activity in Han Chinese individuals [22]. Another recent study concluded that a potential association exists between single nucleotide polymorphisms in the IMPA2 gene and increased risk of ischemic stroke [23]. Lithium, which inhibits IMPase, is a major therapeutic agent for bipolar disorder and inhibits two important biochemical systems: the inositol signaling pathway and GSK-3β function [24]. However, there are few published articles describing the relationship between IMPA2 and cancer. French et al. indicated that the expression of the IMPA2 gene accounts for more variation in methotrexate polyglutamates in leukemia cells than in normal cell lines [12]. Our data showed that the expression of IMPA2 is predominantly downregulated in primary tumors compared to normal tissues derived from patients with ccRCC (Fig. 1b, d and e). Moreover, IMPA2 expression levels were significantly correlated with disease grade (Fig. 1a and c). Patients with low IMPA2 expression had significantly worse OS than those with high IMPA2 expression (Figs. 2a-d). In addition, comparing IMPA2 expression with clinicopathological factors in affecting the survival of ccRCC patients, lower IMPA2 expression was significantly correlated with poor OS in univariate and multivariate analysis (Fig. 2e and f).
ccRCC is the most common and aggressive type of RCC. Metastatic RCC is often already present due to the asymptomatic nature of RCC and relapse after nephrectomy [25]. However, RCC metastases are surgically difficult to remove and are resistant to radiotherapy and systemic therapies [26]. Previous studies have demonstrated that IMPase increases free inositol and IP3 [9]. Recent evidence has shown that IP3 receptors (IP3Rs) are calcium (Ca2+) channels and play important roles in tumor growth, aggressiveness and drug resistance via the modulation of different signaling pathways [27,28]. Shibao et al. indicated that knockdown of IP3R3 in colorectal carcinoma cells increased apoptosis, while overexpression of IP3R3 decreased apoptosis [29]. Another recent study concluded that high levels of IP3R3 were associated with increased metastasis in the lymph nodes and liver and with decreased 5-year survival [30]. Therefore, IP3Rs may be regulated by proto-oncogenes and tumor-suppressor proteins [28,31]. In our in vitro study, endogenous IMPA2 levels were inversely correlated with metastatic capability in a panel of ccRCC cell lines (Fig. 3). Silencing IMPA2 expression appeared to effectively increase the metastatic colonization of A498 cells in vitro and in the lungs of tumor-bearing mice (Fig. 4). Furthermore, IMPA2 overexpression inhibited the metastatic ability of ACHN cells (Fig. 4). These findings demonstrate that IMPA2 is likely correlated with the metastatic progression of ccRCC.
Our results revealed that miR-25 levels are negatively associated with IMPA2 expression in ccRCC tissues and that miR-25 probably inhibits IMPA2 expression by targeting the 3′ untranslated region of IMPA2 transcripts in ccRCC cells (Figs. 5a-e and 6a). Moreover, inhibiting miR-25 suppressed the metastatic ability of ccRCC cells (Figs. 5f-h). To the best of our knowledge, this study is the first to describe the relationship between miR-25 and IMPA2. MiR-25, which is a member of the miR-106b-25 cluster, has been reported to be involved in the progression and development of many types of cancers [32]. Recent evidence has shown the upregulation of miR-25 in gastric cancer patients and patients with positive lymph node metastasis [33]. Conversely, miR-25 inhibits the proliferation, migration, and invasion of osteosarcoma by targeting SRY-related high-mobility group box 4 (SOX4). We found that patients with high miR-25 expression had significantly poorer OS than those with low miR-25 expression (Fig. 6b). Furthermore, both low miR-25 expression and high IMPA2 expression were good prognostic markers in ccRCC (Fig. 6c and d).
We have shown that downregulation of IMPA2 expression maintains the metastatic potential of human ccRCC tumor cells in vitro and in vivo. Furthermore, IMPA2 likely serves as a good prognostic marker. In addition, the dysregulation of miR-25 may result in IMPA2 downregulation and thereby promote metastatic progression in ccRCC. However, the complete mechanism by which IMPA2 disrupts cellular signaling in metastatic RCC is unknown and requires further investigation.
The following are the supplementary data related to this article.
Clinical information of TCGA RCC patients and IMPA2 mRNA levels.
Clinical information of RCC patients enrolled in Superbiochips tissue microarray and IMPA2 IHC intensity.
Clinical information of TCGA RCC patients and mRNA levels of IMPA2 and miR-25.
Supplementary material
Acknowledgments
Acknowledgements
The authors would like to thank Dr. Michael Hsiao at the Genomics Research Center, Academia Sinica, Taiwan for kindly providing the ccRCC cell lines.
Funding sources
This study was supported by the Ministry of Science and Technology, Taiwan (MOST 107-2314-B-038-094, MOST 106-2314-B-038-069-MY3, MOST 105-2320-B-038-021-MY3 and MOST 107-2320-B-038-056).
Declaration of competing interests
The authors have declared no potential conflicts of interest.
Author contributions
Y-F Lin, J-L Chou, H-W Chiu and Y-F Lin performed and analyzed most of the experiments. J-S Chang and I-J Chiu provided intellectual and technical support for the in vitro and in vivo experiments. Y-F Lin, H-W Chiu and Y-F Lin oversaw and designed the study and experiments, analyzed the data, and cowrote the manuscript.
References
- 1.Simard E.P., Ward E.M., Siegel R., Jemal A. Cancers with increasing incidence trends in the United States: 1999 through 2008. CA Cancer J Clin. 2012;62(2):118–128. doi: 10.3322/caac.20141. [DOI] [PubMed] [Google Scholar]
- 2.Leibovich B.C., Lohse C.M., Crispen P.L., Boorjian S.A., Thompson R.H., Blute M.L. Histological subtype is an independent predictor of outcome for patients with renal cell carcinoma. J Urol. 2010;183(4):1309–1315. doi: 10.1016/j.juro.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Steeg P.S. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3(1):55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 4.Schroeder A., Heller D.A., Winslow M.M., Dahlman J.E., Pratt G.W., Langer R. Treating metastatic cancer with nanotechnology. Nat Rev Cancer. 2011;12(1):39–50. doi: 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- 5.Choueiri T.K., Motzer R.J. Systemic therapy for metastatic renal-cell carcinoma. N Engl J Med. 2017;376(4):354–366. doi: 10.1056/NEJMra1601333. [DOI] [PubMed] [Google Scholar]
- 6.Small J., Washburn E., Millington K., Zhu J., Holder S.L. The addition of abemaciclib to sunitinib induces regression of renal cell carcinoma xenograft tumors. Oncotarget. 2017;8(56):95116–95134. doi: 10.18632/oncotarget.19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohnishi T., Ohba H., Seo K.C., Im J., Sato Y., Iwayama Y. Spatial expression patterns and biochemical properties distinguish a second myo-inositol monophosphatase IMPA2 from IMPA1. J Biol Chem. 2007;282(1):637–646. doi: 10.1074/jbc.M604474200. [DOI] [PubMed] [Google Scholar]
- 8.Harwood A.J. Lithium and bipolar mood disorder: the inositol-depletion hypothesis revisited. Mol Psychiatry. 2005;10(1):117–126. doi: 10.1038/sj.mp.4001618. [DOI] [PubMed] [Google Scholar]
- 9.Fisher S.K., Novak J.E., Agranoff B.W. Inositol and higher inositol phosphates in neural tissues: homeostasis, metabolism and functional significance. J Neurochem. 2002;82(4):736–754. doi: 10.1046/j.1471-4159.2002.01041.x. [DOI] [PubMed] [Google Scholar]
- 10.Rapoport S.I., Primiani C.T., Chen C.T., Ahn K., Ryan V.H. Coordinated expression of phosphoinositide metabolic genes during development and aging of human dorsolateral prefrontal cortex. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0132675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berridge M.J., Downes C.P., Hanley M.R. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59(3):411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- 12.French D., Yang W., Cheng C., Raimondi S.C., Mullighan C.G., Downing J.R. Acquired variation outweighs inherited variation in whole genome analysis of methotrexate polyglutamate accumulation in leukemia. Blood. 2009;113(19):4512–4520. doi: 10.1182/blood-2008-07-172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Kong Y.W., Ferland-McCollough D., Jackson T.J., Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 15.Tong Z., Meng X., Wang J., Wang L. MicroRNA-338-3p targets SOX4 and inhibits cell proliferation and invasion of renal cell carcinoma. Exp Ther Med. 2017;14(5):5200–5206. doi: 10.3892/etm.2017.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F., Ma J., Tang Q., Zhang W., Fu Q., Sun J. MicroRNA-543 promotes the proliferation and invasion of clear cell renal cell carcinoma cells by targeting Kruppel-like factor 6. Biomed Pharmacother. 2017;97:616–623. doi: 10.1016/j.biopha.2017.10.136. [DOI] [PubMed] [Google Scholar]
- 17.He Y.H., Chen C., Shi Z. The biological roles and clinical implications of MicroRNAs in clear cell renal cell carcinoma. J Cell Physiol. 2018;233(6):4458–4465. doi: 10.1002/jcp.26347. [DOI] [PubMed] [Google Scholar]
- 18.Gam J.J., Babb J., Weiss R. A mixed antagonistic/synergistic miRNA repression model enables accurate predictions of multi-input miRNA sensor activity. Nat Commun. 2018;9(1):2430. doi: 10.1038/s41467-018-04575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroeze S.G., Bijenhof A.M., Bosch J.L., Jans J.J. Diagnostic and prognostic tissuemarkers in clear cell and papillary renal cell carcinoma. Cancer Biomark. 2010;7(6):261–268. doi: 10.3233/CBM-2010-0195. [DOI] [PubMed] [Google Scholar]
- 20.Bloch P.J., Weller A.E., Doyle G.A., Ferraro T.N., Berrettini W.H., Hodge R. Association analysis between polymorphisms in the myo-inositol monophosphatase 2 (IMPA2) gene and bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34(8):1515–1519. doi: 10.1016/j.pnpbp.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Jesch S.A., Zhao X., Wells M.T., Henry S.A. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J Biol Chem. 2005;280(10):9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J., Huang S., Dai H.R., Wang J., Lin L.H., Xiao H. A promoter polymorphism rs2075824 within IMPA2 gene affecting the transcription activity: possible relationship with schizophrenia. J Cell Mol Med. 2017;21(4):658–664. doi: 10.1111/jcmm.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Q., Yang Y., Na Y., Jin T., Xue Y., Shi Y. IMPA2 polymorphisms and risk of ischemic stroke in a Northwest Han Chinese population. Oncotarget. 2016;7(46):75273–75278. doi: 10.18632/oncotarget.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams R.S., Cheng L., Mudge A.W., Harwood A.J. A common mechanism of action for three mood-stabilizing drugs. Nature. 2002;417(6886):292–295. doi: 10.1038/417292a. [DOI] [PubMed] [Google Scholar]
- 25.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 26.Escudier B., Porta C., Bono P., Powles T., Eisen T., Sternberg C.N. Randomized, controlled, double-blind, cross-over trial assessing treatment preference for pazopanib versus sunitinib in patients with metastatic renal cell carcinoma: PISCES study. J Clin Oncol. 2014;32(14):1412–1418. doi: 10.1200/JCO.2013.50.8267. [DOI] [PubMed] [Google Scholar]
- 27.Parys J.B., Decuypere J.P., Bultynck G. Role of the inositol 1,4,5-trisphosphate receptor/Ca2+−release channel in autophagy. Cell Commun Signal. 2012;10(1):17. doi: 10.1186/1478-811X-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akl H., Bultynck G. Altered ca(2+) signaling in cancer cells: proto-oncogenes and tumor suppressors targeting IP3 receptors. Biochim Biophys Acta. 2013;1835(2):180–193. doi: 10.1016/j.bbcan.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Shibao K., Fiedler M.J., Nagata J., Minagawa N., Hirata K., Nakayama Y. The type III inositol 1,4,5-trisphosphate receptor is associated with aggressiveness of colorectal carcinoma. Cell Calcium. 2010;48(6):315–323. doi: 10.1016/j.ceca.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang S.S., Han K.S., Ku B.M., Lee Y.K., Hong J., Shin H.Y. Caffeine-mediated inhibition of calcium release channel inositol 1,4,5-trisphosphate receptor subtype 3 blocks glioblastoma invasion and extends survival. Cancer Res. 2010;70(3):1173–1183. doi: 10.1158/0008-5472.CAN-09-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R., Valencia I., Zhong F., McColl K.S., Roderick H.L., Bootman M.D. Bcl-2 functionally interacts with inositol 1,4,5-trisphosphate receptors to regulate calcium release from the ER in response to inositol 1,4,5-trisphosphate. J Cell Biol. 2004;166(2):193–203. doi: 10.1083/jcb.200309146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caiazza C., Mallardo M. The roles of miR-25 and its targeted genes in development of human cancer. Microrna. 2016;5(2):113–119. doi: 10.2174/2211536605666160905093429. [DOI] [PubMed] [Google Scholar]
- 33.P L.A., Ahadi A., Zare A., Tarighi S., Zaheri M., Souri M. Up-regulation of miR-21, miR-25, miR-93, and miR-106b in gastric cancer. Iran Biomed J. 2018;22(6):367–373. doi: 10.29252/.22.6.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical information of TCGA RCC patients and IMPA2 mRNA levels.
Clinical information of RCC patients enrolled in Superbiochips tissue microarray and IMPA2 IHC intensity.
Clinical information of TCGA RCC patients and mRNA levels of IMPA2 and miR-25.
Supplementary material