Abstract
Background
Tumour necrosis factor receptor associated factor 6 (TRAF6) promotes inflammation in response to various cytokines. Aberrant Wnt3a signals promotes cancer progression through accumulation of β-Catenin. Here we investigated a potential role for TRAF6 in Wnt signaling.
Methods
TRAF6 expression was silenced by siRNA in human prostate cancer (PC3U) and human colorectal SW480 cells and by CRISPR/Cas9 in zebrafish. Several biochemical methods and analyses of mutant phenotype in zebrafish were used to analyse the function of TRAF6 in Wnt signaling.
Findings
Wnt3a-treatment promoted binding of TRAF6 to the Wnt co-receptors LRP5/LRP6 in PC3U and LNCaP cells in vitro. TRAF6 positively regulated mRNA expression of β-Catenin and subsequent activation of Wnt target genes in PC3U cells. Wnt3a-induced invasion of PC3U and SW480 cells were significantly reduced when TRAF6 was silenced by siRNA. Database analysis revealed a correlation between TRAF6 mRNA and Wnt target genes in patients with prostate cancer, and high expression of LRP5, TRAF6 and c-Myc correlated with poor prognosis. By using CRISPR/Cas9 to silence TRAF6 in zebrafish, we confirm TRAF6 as a key molecule in Wnt3a signaling for expression of Wnt target genes.
Interpretation
We identify TRAF6 as an important component in Wnt3a signaling to promote activation of Wnt target genes, a finding important for understanding mechanisms driving prostate cancer progression.
Fund
KAW 2012.0090, CAN 2017/544, Swedish Medical Research Council (2016-02513), Prostatacancerförbundet, Konung Gustaf V:s Frimurarestiftelse and Cancerforskningsfonden Norrland. The funders did not play a role in manuscript design, data collection, data analysis, interpretation nor writing of the manuscript.
Keywords: β-Catenin, LRP5, Prostate cancer, TRAF6, Wnt3a, Zebrafish
Highlights
-
•
TRAF6 positively regulated mRNA expression of b-Catenin and subsequent activation of Wnt target genes in prostate cancer cells in vitro.
-
•
High expression of LRP5, TRAF6 and c-Myc correlated with poor prognosis for patients with prostate cancer.
Research in context.
Evidence before this study
Prostate cancer is the second most common cancer form and one of the main leading cancer-related death in men in the world. Aberrant Wnt signaling has recently emerged as a cause of aggressive prostate cancer, but the underlying molecular mechanisms have remained unknown. The E3 ubiquitin ligase tumour necrosis factor receptor associated factor 6 (TRAF6) has recently been reported to act as a tumour promoter in prostate cancer cells and is known to be amplified on the gene as well on the protein level, in several cancers forms, such as lung cancer, breast cancer and head and neck cancer. In this study, we investigated a potential role of TRAF6 in Wnt-signaling in prostate cancer.
Added value of this study
Our study revealed a functional role for TRAF6 in Wnt3a signaling and prostate cancer progression through biochemical studies of prostate cancer cells, colorectal SW480 cells and bioinformatics analyses. We found that TRAF6 forms a complex with the Wnt co-receptor LRP5 and the expression of TRAF6 and Wnt co-receptor LRP5 correlates with aggressive stage i.e. Gleason Score 7 or higher in human prostate cancer tissue sections. Silencing TRAF6 in human prostate and colorectal cancer cells and in zebrafish demonstrated that TRAF6 is required for Wnt induced regulation of β-Catenin and subsequent activation of Wnt target genes and that this mechanism is evolutionary conserved. Since Wnt3a and several cytokines activate TRAF6, our study supports the notion that TRAF6 is an attractive therapeutic target to inhibit prostate cancer progression.
Implications of all the available evidence
We showed that TRAF6 is a key regulatory factor for Wnt3a-induced regulation of β-Catenin and subsequent activation of Wnt3a target genes implicated in prostate cancer progression. TRAF6 may be an important novel target for inhibiting prostate cancer progression and improving prognosis for patients with prostate cancer.
Alt-text: Unlabelled Box
1. Introduction
Wnt signaling plays a crucial role during embryogenesis for the regulation of cell proliferation, cell polarity, axis formation and apoptosis [1]. It is also implicated in tissue homoeostasis, regeneration of tissues in adults along with differentiation and maintenance of stem cells [[2], [3], [4]]. Components of the Wnt pathway participate in gene transcription and cell-cell adhesion [5,6] and abnormalities in these molecules often causes birth defects [7] and several types of cancers [4,[8], [9], [10], [11]]. Wnt proteins are highly conserved cysteine-rich lipid-modified secreted proteins. There are 19 different Wnt ligands through which the signaling is initiated [12]. In the canonical Wnt pathway or Wnt/β-Catenin pathway, the Wnt ligand binds to the Frizzled (FZD) receptors and Low-density lipoprotein receptor-related proteins, 5 and 6 (LRP5/6), which recruits Dishevelled protein to the intracellular domains. This triggers the phosphorylation of cytosolic domains of LRP5/6 at five reiterated PPPSPxS motifs by Glycogen synthase kinase 3-β isoform (GSK3β) and Casein Kinase 1 (CK1), causing subsequent recruitment of Axin to the phosphorylated sites. These events result in the disruption of β-Catenin from destruction complex, consisting of GSK3β, CK1, Axin, Adenomatous polyposis coli protein (APC), Protein Phosphatase 2A (PP2A) and E3 ubiquitin ligase β-TrCP [13]. Stabilisation of β-Catenin leads to its accumulation and translocation into the nucleus, where it associates with the T-cell-specific transcription factor/lymphoid enhancer binding factor I family protein (TCF/LEF-I) transcription factors and drives Wnt target gene expression [8,[14], [15], [16], [17]]. In the absence of Wnt ligands, the destruction complex recruits available β-Catenin, so that active GSK3β and CK1 phosphorylate it, and further, β-TrCP ubiquitinates β-Catenin; which makes it available to proteasomes for proteolytic degradation of β-Catenin [13]. Mutations in the destruction complex components results in increased stabilisation of β-Catenin causing constitutive activation of downstream Wnt target genes, which is commonly seen in many cancer forms [10,[18], [19], [20]]. Aberrant growth factor signaling, including activation of Wnt-β-Catenin signaling occurs in several cancer forms [2,18].
Prostate cancer is the second most common cancer and one of the main leading cancer-related deaths in men in the world [21]. The prostate is a male sex gland, which depends on androgens for its development, maturation and maintenance [22]. Mutations in the androgen receptors and/or increased sensitivity of androgen receptors (AR) towards androgens are related to the progression of prostate cancer [23,24]. Chromosomal translocations juxtaposing the androgen-responsive TMPRSS2 promoter with the ETS-family transcription factor ERG result in aberrant ERG upregulation in approximately 50% of prostate cancers. ERG activates the Wnt/LEF1 signaling cascade by binding to several Wnt-promoters causing accumulation of β-Catenin [25,26]. Certain growth factors might also activate AR and induce AR target genes; such as Insulin-like growth factor-I (IGF-I), Epidermal growth factor (EGF), Keratinocyte growth factor (KGF) and Wnt3a [8,[27], [28], [29], [30]]. High levels of Wnt3a has recently been demonstrated to be produced by the androgen-independent and aggressive human prostate cancer (PC3) cells and Wnt3a was shown to promote generation of bone metastasis in a transgenic mouse prostate cancer model [31]. Inactivation of the Wnt co-receptor LRP5 in prostate cancer cells promotes mesenchymal to epithelial shift and thereby decreases the migration and invasion of cells and also causes a significant decrease of skeletal metastasis [32]. As in mammals, canonical Wnt signaling plays a crucial role during embryonic development in zebrafish [2,33]. The Wnt component β-Catenin is required for the establishment of dorso-ventral axis during early development. During embryogenesis, β-Catenin nuclear accumulation in the dorsal side of the embryo is the earliest hallmark of dorso-ventral axis patterning [34]. The canonical Wnt pathway is also involved in cranial neural crest cell migration [35], epimorphic regeneration of zebrafish tail fin [36] and early development of swim bladder in zebrafish [37].
Tumour necrosis factor receptor associated factor 6 - TRAF6 is a member of TRAF family, which is a class of adaptor protein involved in various signaling pathways such as NF-κB and Mitogen-activated protein kinases (MAPKs) [38]. TRAF6 is important in regulating adaptive immunity and bone homeostasis [39]. TRAF6 is a cytosolic E3 ubiquitin ligase made of highly conserved amino terminal RING domain along with four zinc finger motifs, a central coiled core and TRAF-C domain at the carboxyl terminal [40]. TRAF6 has a unique consensus binding motif Pro-X-Glu-X-X-(aromatic/acidic residue) through which it interacts with other proteins [41]. Earlier studies reported that TRAF6−/− deficient mice develop severe bone defects i.e., osteopetrosis, exencephaly and die at early age [[42], [43], [44]]. TRAF6 has previously been found to play an important role in activation of the AKT pathway [45,46] and to promote prostate cancer growth and invasion in vitro [47] and in in vivo xenograft model [48], suggesting that it can favour cancer progression. TRAF6 has also recently been suggested to act as an oncogene due to amplifications of the TRAF6 gene found in lung carcinoma [49]. TRAF6 is upregulated in colon cancer and promotes proliferation, invasion and metastasis in colon cancer and gastric cancer [50,51].
Our study provide evidence for a functional role of TRAF6 as a positive co-regulator of Wnt3a-induced activation of β-Catenin and Wnt3a classical target genes in human prostate and colorectal cancer cells in vitro through biochemical experiments. TRAF6 was required for Wnt3a-induced invasion of both human prostate and colorectal cancer cells in vitro. Moreover, bioinformatics analysis of patient samples with prostate cancer revealed that the mRNA expression of TRAF6 correlates with mRNA expression of several classical Wnt target genes. Moreover, a TRAF6 consensus-binding motif was found in LRP5, which can explain the herein described association between TRAF6 and the Wnt co-receptor LRP5, in several cell lines and in human prostate cancer tissues. By the use of CRISPR/Cas9 genomic editing to silence TRAF6 expression in zebrafish, we show that TRAF6 plays an important role for positive regulation of Wnt3a target genes. This study identifies TRAF6 as an evolutionary conserved co-regulatory protein in the Wnt pathway that also promotes progression of prostate and colorectal cancer due to its positive effects on Wnt3a signaling.
2. Material and methods
2.1. Cell culture
PC3U (RRID: CVCL_0482), a human prostate cancer androgen-independent cell line which is a clonal derivative of PC3 cells as previously reported [52]. DU145 (RRID: CVCL_0105), a human prostate carcinoma cell line derived from brain metastatic site. LNCaP (RRID: CVCL_0395) were human prostate cancer androgen-dependent cell lines derived from a lymph node metastatic site. PC3U, DU145 and LNCaP cells were cultured in RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO, USA) with 10% fetal bovine serum (FBS), 1% glutamine and 1% penicillin-streptomycin (PEST) supplements at 37 °C in the presence of 5% CO2. RPMI 1640 media complemented with 1% FBS, 1% glutamine and 1% penicillin-streptomycin (PEST) was used to starve the cells. SW480 (RRID: CVCL_0546), a human colorectal adenocarcinoma cell line cultured in DMEM medium (Sigma-Aldrich) with 10% FBS and 1% PEST and was cultured at 37 °C in the presence of 5% CO2. DMEM media complemented with 1% FBS and 1% PEST used to starve the cells. Cells were stimulated with Wnt3a (50 ng/ml) after the cells were starved for 18 h. Human embryonic kidney (HEK293) cells were grown in Eagle's Minimum Essential Medium (EMEM) complemented with 10% FBS and was cultured at 37 °C in the presence of 5% CO2.
2.2. Antibodies and other reagents
Recombinant human Wnt3a (# NP_149122, R&D systems, Minneapolis, MN, USA) was used to stimulate cells. Monoclonal rabbit anti-TRAF6 (#ab40675, RRID: AB_778573) antibody was obtained from Abcam (Cambridge, MA, USA) and used for immunoblotting. Mouse monoclonal anti-TRAF6 (D10; #sc-8409, RRID: AB_628391) from Santa Cruz Biotechnology (Santa Cruz, CA, USA) was used in immunofluorescence. For Proximity Ligation Assay (PLA) anti-TRAF6 rabbit monoclonal (#ab33915, RRID: AB_778572) from Abcam and anti-K63 polyubiquitin mouse monoclonal (HWA4C4; #BML-PW0600, RRID: AB_10540645) from Enzo Life Sciences Inc., (NY, USA) were used. Anti-LRP5 (D80F2) rabbit monoclonal antibody (#5731, RRID: AB_10705602) purchased from Cell Signaling technology (Denvers, MA, USA) was used for immunoblotting and immunofluorescence. Goat polyclonal anti-LRP5 (#ab36121, RRID: AB_776077) purchased from Abcam for PLA and immunofluorescence. Anti-β-Catenin monoclonal mouse antibody (#610154, RRID: AB_397555) was from BD Biosciences (San Jose, CA, USA) and Anti-Non-phospho (Active) β-Catenin (Ser33/37/Thr41) (D13A1) Rabbit monoclonal antibody (#8814, RRID: AB_11127203) was purchased from Cell Signaling Technology. Mouse monoclonal anti-β-actin (#A5441, RRID: AB_476744) was from Sigma-Aldrich (Saint Louis; ML, USA). Anti-c-Myc rabbit monoclonal antibody (#5605S, RRID: AB_1903938) was purchased from Cell Signaling Technology. Anti-HA (12CA5) mouse monoclonal antibody (#11666606001, RRID: AB_514506) was obtained from Roche (Basel, Switzerland). Secondary horseradish peroxide conjugated (HRP) polyclonal goat anti-rabbit (P0448, RRID: AB_2617138) and polyclonal goat anti-mouse (P0447, RRID: AB_2617137) antibodies were obtained from Dako (Denmark). IgG light chain-specific anti-rabbit (211-032-171A, RRID: AB_2339149) and anti-mouse (115-035-174, RRID: AB_2338512) HRP conjugated antibodies were from Jackson Immuno-Research Laboratories, Inc (Cambridgeshire, UK). Secondary Antibodies IRDye® 800CW goat anti-Rabbit (925-32211, RRID: AB_2651127) and IRDye® 680RD goat anti-Mouse (925-68070, RRID: AB_2651128) were obtained from Licor Biosciences (Lincoln, NE, USA). Alexa Flour 555-conjugated donkey anti-rabbit (A-31572, RRID: AB_162543), Alexa Flour 488-conjugated goat anti-mouse (A-10680, RRID: AB_2534062) and Alexa Flour 555 Rabbit anti-goat (A-21431, RRID: AB_2535852) antibodies was purchased from Invitrogen (CA, USA). VECTASHIELD Hardset mounting media Fluorescent dye 4, 6-Diamidino-2-phenylindole dihydrochloride (DAPI) from Vector Laboratories (CA, USA) was used to stain the nuclei for microscopy. Pefabloc (#11429876001) was purchased from Roche (Basel, Switzerland) and Sodium orthovanadate from Sigma. Protein-G Sepharose beads (#17061801) were purchased from GE Healthcare (IL, USA). Prestained PageRuler protein ladder and Spectra Multicolor High Range Protein ladder were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
2.3. Generation of zebrafish TRAF6 specific antibodies
The full-length protein sequence of Danio rerio TRAF6 was subjected to GenScripts Optimun Antigen design tool. Two rabbits were immunized with the peptide spanning from amino acid 528 to 541 to which a cysteine was added to the N-terminus for KLH conjugation (CLRREGVQPRGPEPS). Antibodies were purified from the pooled sera by affinity column purification and tested by limited dilution in a peptide ELISA assay. This antibody was obtained from GenScript HK Ltd. (Nanjing, China).
2.4. Immunoprecipitation and Western blot
Cells were washed with PBS and starved for 16 h in DMEM with 1% FCS. After starvation, cells were treated with Wnt3a for indicated times and thereafter washed once with ice-cold PBS. Lysis of the cells was carried out using ice-cold RIPA lysis buffer (50 mM Tris (pH 8), 150 mM NaCl, 1% TritonX-100, 10% (v/v) glycerol, 1 mM aprotinin, 1 mM Pefabloc and 1 mM Sodium orthovanadate), incubated on ice for 20 min and centrifuged at 13000 rpm for 10 min and supernatant was collected. Protein concentrations were determined by using the BCA protein assay kit from Thermo Scientific. The total cell lysates were immunoprecipitated with corresponding antibodies overnight and incubated with Protein G-Sepharose beads for 1 h at 4 °C. Beads were washed four times with the lysis buffer and then LDS sample buffer (Invitrogen) was added and boiled for 5 min to elute the immunocomplexes [53]. Uniform amounts of protein from total cell lysates and immunoprecipitation were run in a SDS-PAGE using 7% Tris-acetate or 10% Bis-Tris polyacrylamide precast gels (Invitrogen) and blotted onto nitrocellulose membrane with an iBlot apparatus (Invitrogen). Membranes were blocked in 5% bovine serum albumin (BSA) (Roche) in 0.05% Tween-PBS for 1 h at room temperature. The membranes were further incubated with the indicated primary antibodies overnight at 4 °C. Primary antibodies were detected using HRP-conjugated antibodies or light chain specific HRP-antibodies and developed using the enhanced chemiluminescence (ECL) kit from GE Healthcare. The primary antibodies were also detected using either IRDye® 800CW goat anti-rabbit or IRDye® 680RD goat anti-mouse from Licor Biosciences. The membrane blots were visualized using Odyssey CLx Imaging System [54].
2.5. siRNA and plasmids
siGENOME human SMARTpool siRNA was used for the knockdown of human TRAF6 and non-specific control siRNA was purchased from Dharmacon Research (Lafayette, CO, USA). The expression plasmid for 3xHA-tagged wild-type ubiquitin was a gift from V. M. Dixit (Genentech, San Francisco, CA). PC3U cells were transfected with either specific siRNA or control siRNA. siRNA transfections were carried out using Oligofectamine (Invitrogen) according to the manufacturer's protocol. Plasmid transient transfection was carried out using FuGENE® HD (Promega, Madison, WI, USA) according to the manufacturer's protocol. TCF-reporter plasmid TOPflash firefly luciferase contains two sets of three copies of TCF-binding site obtained from Merck-Millipore (#17-285; Darmstadt, Germany). pGL4.73 [hRluc/SV40] renilla luciferase plasmid as internal transfection control was purchased from Promega. A synthetic gene encoding for the intracellular domain of human LRP5 from amino acid 1407 until 1615 was ordered from Eurofinsgenomics (Ebersberg, Germany) as wild type LRP51407–1615WT or with a mutant TRAF6 consensus binding motif LRP51407–1615 E1425A and further cloned into the eukaryotic expression vector pCMVtag5 upstream of the HA and (His)6 tag. Expression of LRP51407–1615HA-(His)6 WT was performed in human embryonic kidney (HEK293) cells and purified by Ni-NTA chromatography. Recombinant human TRAF6-Flag was purchased from Origene, USA (#:TP319528).
2.6. Dual luciferase assay
Wnt signaling was investigated by TCF reporter plasmid TOPflash transiently transfected into PC3U and SW480 cell lines along with either siTRAF6 or siCtrl and renilla plasmid, as an internal transfection control. After starvation of cells for 16 h, the next day the cells were stimulated with Wnt3a for 30 min and 6 h. Cells were harvested after stimulation, and cell extracts were prepared using the Dual-luciferase® reporter assay system (Promega). Firefly luciferase and renilla luciferase activities were assessed using GloMax®96 Luminometer (Promega) according to the manufacturer's instructions.
2.7. Immunofluorescence and confocal microscopy
Immunofluorescence was performed as previously described [48]. PC3U cells were grown on sterile coverslips in 6-well plates. After starving for 16–18 h, cells were stimulated with Wnt3a for 30 min and 6 h. Cells were then washed once with PBS, fixed and permeabilized with 4% formaldehyde and 2% Triton X-100 respectively for 10 min each. Cells were blocked using 5% BSA for at least 1 h. Cells were then incubated for 1 h with the corresponding primary antibodies followed by incubation for 1 h using AlexaFlour® 555 and 488 secondary antibodies. The coverslips were then mounted onto slides using VECTASHIELD Hardset mounting media with DAPI according to the manufacturer's instructions. Confocal images were obtained by Zeiss LSM 710 Meta microscope (Carl Zeiss MicroImaging, Inc. Jena, Germany), equipped with a RET-EXi-F-M-12-C digital camera by using a 63× lens with 1.4 numerical aperture magnification lens. ZEN software was used to capture and analyse the images. Representative results shown were performed from at least three independent experiments.
2.8. In situ proximity ligation assay (PLA)
2.8.1. In situ PLA analysis in PC3U, LNCaP and SW480 cells
PC3U, LNCaP and SW480 cells were grown in 8 well chamber or 6 well dishes on coverslips and stimulated with Wnt3a according to figure legends. Cells were further fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. In situ proximity ligation assay (PLA) was performed according to the manufacturer's protocol using Duo-link detection kit (Sigma). Immunofluorescence images were taken at 40× objective lens (Carl Zeiss MicroImaging, Inc.) as stated above after mounting on to slides with DAPI. Zeiss Zen software was used to acquire images, which were then converted into TIFF format. Duo-link image tool software was used to analyse and quantify the signals.
2.8.2. In situ PLA analysis in tissue sections
Malignant tissue sections from prostate adenocarcinoma with Gleason scores ranging from 6 (3 + 3) to 9 (4 + 5) were obtained from male patients who had undergone through prostatectomy. The use of patient tissues was approved by the Regional Ethical Review Board and with the patients informed consensus. Sections were rehydrated in an ethanol gradient (2 × 5 min Xylen, 2 × 5 min 99% EtOH, 5 min 95% EtOH, 5 min 70% EtOH, 2 × 5 min PBS). Heat-induced retrieval of antigens was performed in a decloaking chamber (4 min at 125 °C followed by 3 min at 90 °C) in Tris-EDTA buffer pH 9 (10 mM Tris Base, 1 mM EDTA, 0.05% Tween 20, pH 9.0), washed with PBS 3 × 5 min, then blocked with Duolink Blocking solution for 1 h 37 °C. Antibodies were diluted in Duolink Antibody dilution buffer (LRP5 (#ab36121) 5 μg/ml, TRAF6 (#ab33915) 2 μg/ml) and incubated on samples overnight 4 °C, followed by 3 × 5 min washes with TBS 0.05% Tween (TBST). Duolink anti goat PLUS and Duolink anti rabbit MINUS probes were diluted 1:5 in Duolink Antibody dilution buffer and incubated on samples for 1 h at 37 °C followed by 3 × 5 min washes with TBST. Ligation was carried out by T4 Ligase diluted 1:40 into Duolink Ligation Buffer 1×, incubated on samples for 30 min 37 °C followed by 3 × 5 min washes with TBST. The tissue sections were further subjected to amplification using Phi29 polymerase, diluted 1:80 into Duolink 1× Amplification buffer Red, and samples were incubated for 100 min 37 °C followed by 3 × 10 min washing with TBST. Counterstaining was performed with Phalloidin-Alexa 488 diluted 1:40 into 1× Hoechst solution for 15 min and slides were finally rinsed with water and mounted using Slow Fade mounting medium, Life Technologies. Images were obtained by using Zeiss Imager Z2, with objectives plan-Apochromat dry NA 0.8 and plan-Apochromat oil immersion NA 1.4. Pictures were processed and exported with Zeiss ZEN software.
2.8.3. In situ PLA analysis in tissue sections by bright field microscope
Paraffin-embedded sections of prostate cancer tissue were rehydrated twice in xylene for 10 min each, and then in 100% ethanol for 10 min followed by 5-min incubations in 95%, 80%, and 70% ethanol and deionized H2O, finally in PBS for 10 min. Thereafter, the sections were treated with Antigen Retrieval Reagents (RD913M, Biocare Medical, CA, USA) at 95 °C for 5 min and rinsed with PBS. The sections were thereafter incubated in 3% H2O2/methanol for 10 min, washed twice with PBS, and blocked in 5% normal goat serum for 1 h at room temperature. The sections were next incubated overnight at 4 °C with 1:500 LRP5 antibody (#ab36121) and PBS (negative control), then treated with PLA Duolink probes (anti-goat PLUS and anti-goat MINUS) for 60 min at 37 °C followed by 90 min amplification, 60 min bright field detection, substrate development, and nuclear staining. At last samples were mounted and digital images were acquired by scanning with bright field microscope (3D Histech, Hungary). From the whole scanning pictures, three different images with at least 300 cells were taken at random and the mean PLA dots (representing products from rolling-circle amplification (RCA) versus total cells (mean ± S.E.M) were counted and analyzed in a Mann-Whitney U test for group difference evaluation.
2.9. Ubiquitination assay
Starved PC3U cells were treated with Wnt3a, and washed once with ice-cold PBS. Cell lysates were heated in the presence of 1% SDS and diluted with lysis buffer containing 0.5% NP-40. The procedure was performed as described previously [48,55].
2.10. RNA isolation and quantitative real time-PCR
Total RNA was isolated from PC3U cells using RNeasy Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's protocol and the RNA concentration, was measured using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific). cDNA was generated from purified total RNA using ThermoScript RT-PCR system (Invitrogen) using Random hexamers according to Manufacturer's protocol. qRT PCR was performed with an Applied Biosystems 7900HT Fast Real-time PCR system (Applied Biosystems, CA, USA) using Power SYBR Green PCR master mix (Applied Biosystems). Primers used are represented in Supplementary Fig. S4. GAPDH was used as internal control.
2.11. In silico gene expression analysis
We used cBioPortal as the repository to study gene expression in prostate cancer. Specifically, we screened the provisional TCGA study of prostate adenocarcinoma (498 samples). We then quantified the gene expression correlation in these samples between TRAF6 and other genes like EGFR, REL, TCF4, β-Catenin, LRP5, c-Myc, DVL1 and AXIN1. We represented these correlations using the logarithm of respective gene expressions. We also obtained the survival analysis data from the Human Protein Atlas using TCGA Pan Cancer Atlas (PRAD) database where Kaplan-Meier plots showing the correlation between patient survival and mRNA expression levels of the Wnt-target gene TRAF6, LRP5 and c-Myc. Survival analysis was made by dividing the patients into two groups with low and high levels of mRNA expression. GENEVESTIGATOR® a public database was used to find the TRAF6 mRNA expression levels among various cancers.
2.12. Invasion assay
The Invasion assay kit was obtained from Cell Biolabs, San Diego, USA (CBA-110-COL). The invasion chambers were coated with Bovine Type I Collagen and the assay procedure was performed according to the manufacturer's protocol. PC3U and SW480 cells were transfected with either siTRAF6 or siCtrl. After 48 h cells were starved for 12 h and then trypsinized. Further, the trypsinized cells were seeded into upper invasion chamber containing media with 2% FBS. The chambers were further placed in 24 well plate containing media with 10% FBS. Cells were stimulated with Wnt3a for 24 h. Images were taken using Zeiss light microscope. The invaded cells were lysed and the optical density (OD) was measured at 560 nm.
2.13. Label-free monitoring of a TRAF6-LRP5 protein interaction
Protein interaction was monitored in real-time by interferometry Bio-Layer detection with the BLItz System from Pall forte BIO, USA. Recombinant LRP51407–1615-HA-(His)6 was immobilized on a Ni-NTA sensor (#18-5101) and excess protein was removed by washing with PBS. For the Kon determination, TRAF6-Flag (Origene, USA #:TP319528) was either added at a concentration of 167 nM or 822 nM, and Koff was determined by washing with PBS until a stable binding curve was achieved.
2.14. Zebrafish strains and maintenance
Embryos and larvae were obtained from wild-type zebrafish (Danio rerio), London wild type (LWT) and tupfel long fin (TL). Zebrafish (ZF) was maintained by standard procedures at the zebrafish facility at Umea University. Ethical permit for generating zebrafish mutants with Crispr/Cas9 and keeping mutant lines is A-13-15 (Umeå Djurförsöksetiska nämnd).
2.15. Generation of Zebrafish TRAF6 mutants using CRISPR/Cas9
Zebrafish TRAF6 mutants were generated using clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR associated protein-9 nuclease (Cas9) CRISPR/Cas9 [56,57]. A customised 20 nucleotide sequence pair targeting TRAF6 exon 2 was designed with the nucleotide sequences 5′-CTGTCTGATGGGTCTCCGCT-3′ (right) and 5′-AGCGGAGACCCATCAGACAG-3′ (left). These nucleotide sequences were annealed and cloned into zebrafish single guide RNA (sgRNA) expression vector pDR274 obtained from Addgene (plasmid # 42250) [56] and the T7 mMESSAGE mMACHINE transcription kit (Ambion) was used to transcribe sgRNAs according to the manufacturer's protocol. Cas9 nuclease, S. pyogenes (New England BioLabs, Inc. MA, USA) along with 300 ng/μl of in-vitro transcribed sgRNA was injected into single cell staged zebrafish embryos. Mutated DNA loci in Fo fish were identified by an incorporated restriction site in each customised target nucleotide sequence as described in [58]; here we used MbiI. The genotyping was further confirmed by sequencing.
2.16. Zebrafish RNA and DNA extraction and genotyping
Heterozygous TRAF6 male and female were incrossed and seven days post fertilization (dpf) embryos were collected for RNA extractions as described earlier [[59], [60], [61]]. Each embryo was collected in an Eppendorf tube, 250 μl of TRIzol (Thermo Fisher Scientific) was added and the samples were homogenised in a Qiagen Tissuelyser II. 50 μl of Chloroform was added to the mixture and vortexed. Samples were centrifuged at 13000 rpm for 15 min, the clear top aqueous layer was rescued carefully and an equal volume of 70% ethanol was added. The mixture was applied to Nucleospin® RNA XS columns (MACHEREY-NAGEL, Duren, Germany) and further procedures were performed according to the manufacturer's protocol. RNA was eluted with 20 μl of RNase-free water and a purity check was performed using Agilent 2100 Bioanalyzer (CA, USA). DNA was extracted from the bottom organic layer by precipitating in 100% ethanol and washing with 0.1 M sodium citrate solution.
2.17. Zebrafish quantitative RT-PCR
cDNA and qRT PCR was performed as described above. β-Actin was used as reference gene [62]. Primers used are represented in Supplementary Fig. S4.
2.18. Whole mount staining for bone
Whole-mount in-situ Alizarin red staining method was used for staining the bone in the adult zebrafish at 50dpf as described earlier [63]. Images were taken after zebrafish were exposed to lethal dose of Tricaine mesylate and then positioned on a 2% agarose plate. Nikon SMZ1500 stereomicroscope and a Nikon D5200 digital camera were used for imaging. Images were edited using Image Composite Editor from Microsoft. Photoshop CS6 was used to improve image quality.
2.19. Statistical analysis
All the experiments were performed independently at least three times unless otherwise stated. Student's t-test and Mann-Whitney U test was used to calculate the significance, unless otherwise stated. P values <.05 were considered statistically significant. Error bars indicate standard error of mean (S.E.M). Kaplan-Meier plots were used to show the survival data in zebrafish. P-values obtained from cBioPortal databases were calculated using bootstrap hypothesis testing as described earlier [64].
3. Results
3.1. TRAF6 associates with the Wnt co-receptors LRP5 and LRP6
TRAF6 is activated by its autoubiquitination [65,66] and acts in concert with specific E2 ligases such as Ubc13 and functions in Lys63-linked polyubiquitination of its target proteins. As we hypothesized that TRAF6 might be implicated in Wnt signaling, we investigated a potential role for TRAF6 in Wnt3a induced responses in androgen-independent aggressive human prostate cancer (PC3U) cells, a derivative of the human PC3 cells, which are known to express high levels of Wnt3a, as previously reported by Nandana et al. [31]. Firstly, we examined the subcellular localization of endogenous LRP5 and β-Catenin upon Wnt3a stimulation of PC3U cells by using confocal imaging (Fig. 1a). Accumulation of nuclear β-Catenin was observed already in unstimulated PC3U cells as a sign of constitutive activation of the canonical Wnt3a-β-Catenin pathway in line with the recent report from Nandana et al. [31]. An association between the carboxy-terminal part of LRP5 and β-Catenin was observed after Wnt3a-stimulation of PC-3U cells after 0.5 h and 6 h (Fig. 1a). We then investigated a possible association between endogenous LRP5 and TRAF6, upon Wnt3a stimulation again by using confocal imaging and observed a colocalization of the two proteins as shown in Fig. 1b. The LRP5 antibody recognizes the extracellular domain of LRP5 in Fig. 1b which explain the difference in localization as seen in Fig. 1a. We examined also the Wnt3a dependent effects on the levels of TRAF6 and β-Catenin in PC3U cells. TRAF6 showed increase in the protein levels upon Wnt3a stimulation with a significant increase by 6 h time point (Supplementary Fig. S1a). However, no significant accumulation of β-Catenin was observed upon Wnt3a stimulation in line with the observation of nuclear β-Catenin in untreated cells (Fig. 1a), suggesting the existence of a constitutive active Wnt3a - β-Catenin pathway in these highly malignant cells.
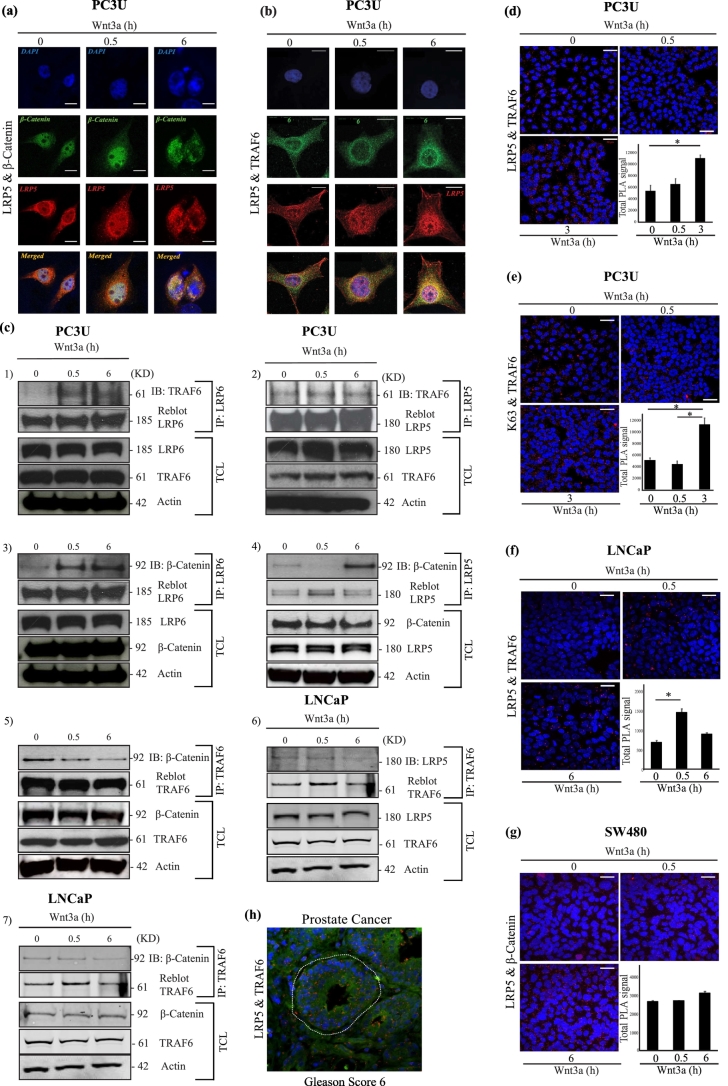
Fig. 1.
Wnt3a promotes interaction between TRAF6 and Wnt Components.
(a and b) Confocal images showing PC3U cells treated with Wnt3a as indicated. (a) Endogenous LRP5 (red) and endogenous β-Catenin (green) in PC3U cells. Nuclei were stained with DAPI (blue). Rabbit Anti-LRP5 (Cell Signaling technologies) antibody used is specific to the carboxyl terminus of human LRP5. Co-localization of these proteins were observed (yellow) in the merged images. Scale bar 20 μM. (b) Immunofluorescence of endogenous LRP5 (red) and endogenous TRAF6 (green) visualized by confocal microscope. DAPI was used to stain the nuclei (blue). Goat Anti-LRP5 (Abcam) antibody used is specific to the extracellular domain of human LRP5. Co-localization of these proteins were observed (yellow) in the merged images. Scale bar 20 μM. (c) Total cell lysates from PC3U cells and LNCaP cells were extracted after treating with Wnt3a as indicated. Lysates were immuno-precipitated with anti-LRP6, LRP5 and TRAF6 antibodies. (c 1–2) Representative immunoblots from PC3U cell lysates were probed to detect TRAF6 associated with LRP6 and LRP5. (c 3–5) Immunoblots from PC3U cell lysates probed to identify β-Catenin associated with LRP6, LRP5 and TRAF6. (c 6–7) Immunoblots from LNCaP cell lysates were probed to detect LRP5 and β-Catenin associated with TRAF6. Corresponding total cell lysates were immunoblotted for LRP6, TRAF6, LRP5, β-Catenin and actin served as internal control for equal loading of proteins. (d-g) In situ PLA analysis of Wnt3a induced formation of the (d) LRP5 and TRAF6 complexes (red dots) in PC3U cells (e) Lys63 and TRAF6 complexes (red dots) in PC3U cells. (f) LRP5 and TRAF6 complexes in LNCaP cells and (g) LRP5 and β-Catenin complexes in SW480 cells were identified. Scale bar 50 μM. Quantification of signal were shown in bar graphs and error bars represent mean ± SEM from three independent experiments. ⁎P < .05 (student t-test). h) Representative PLA analysis of tumour tissue from prostate cancer with Gleason Score 6, which recognizes the co-localization of LRP5 and TRAF6 proteins (red dots). The white dotted line represent tumour glands. Scale bar 20 μM (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Thereafter we examined the possible TRAF6 association with Wnt canonical surface receptors LRP5 and LRP6. Co-immunoprecipitation (Co-IP) experiments demonstrated that endogenous TRAF6 interacts with LRP6 and that the interaction was enhanced by Wnt3a stimulation. Furthermore, the interaction between TRAF6 and LRP5 was also observed (Fig. 1c 1–2). Endogenous β-Catenin was also found to be immunoprecipitated with LRP5, LRP6 and TRAF6, and the intensity of complex formation was enhanced by Wnt3a stimulation in PC3U cells (Fig. 1c 3–5). We next investigated by co-immunoprecipitation if TRAF6 could associate with LRP5, also in the human prostate LNCaP cancer cell line. As shown in Fig. 1c 6–7, an interaction between TRAF6 and LRP5 and also β-Catenin and TRAF6 was observed, in line with the data achieved in PC3U cells. No accumulation of total β-Catenin was observed in PC3U cells upon Wnt3a-stimulation as shown in Fig. 1c 3–5, which together with the observation of nuclear β-Catenin (Fig. 1a) and no significant increase of total β-Catenin (Supplementary Fig. S1a) suggested that the Wnt3a pathway is constitutively active in these aggressive and highly malignant prostate cancer cells. We analyzed also the protein expression of TRAF6, LRP5 and active β-Catenin without Wnt3a stimulation in PC3U, LNCaP, DU145 and SW480 cell lines to check the difference in the levels of protein expression among the cell lines (Supplementary Fig. S1b). We observed low levels of active β-Catenin in PC3U cells, while low levels of TRAF6 and LRP5 were seen in LNCaP and DU145 cells. The colorectal SW480 cells, known to possess constitutive active Wnt-signaling due to a mutation in APC, showed higher levels of active β-Catenin as expected and of TRAF6, while the levels of LRP5 was intermediate. From these data, we decided to include also LNCaP and SW480 cells in our study.
We further confirmed the association of LRP5 and TRAF6 in PC3U cells by using in situ proximity ligation assay (PLA) (Fig. 1d). The in situ PLA assay showed a significant higher amount of the endogenous LRP5 and TRAF6 protein complex at 3 h of Wnt3a stimulation, than observed by co-immunoprecipitation at 6 h of Wnt3-stimulation of PC3U cells (Fig. 1c-2). The difference between results shown in Fig. 1c-2 and d is probably explained by that the in situ PLA assay is more sensitive than the co-immunoprecipitation method used in Fig. 1c-2.
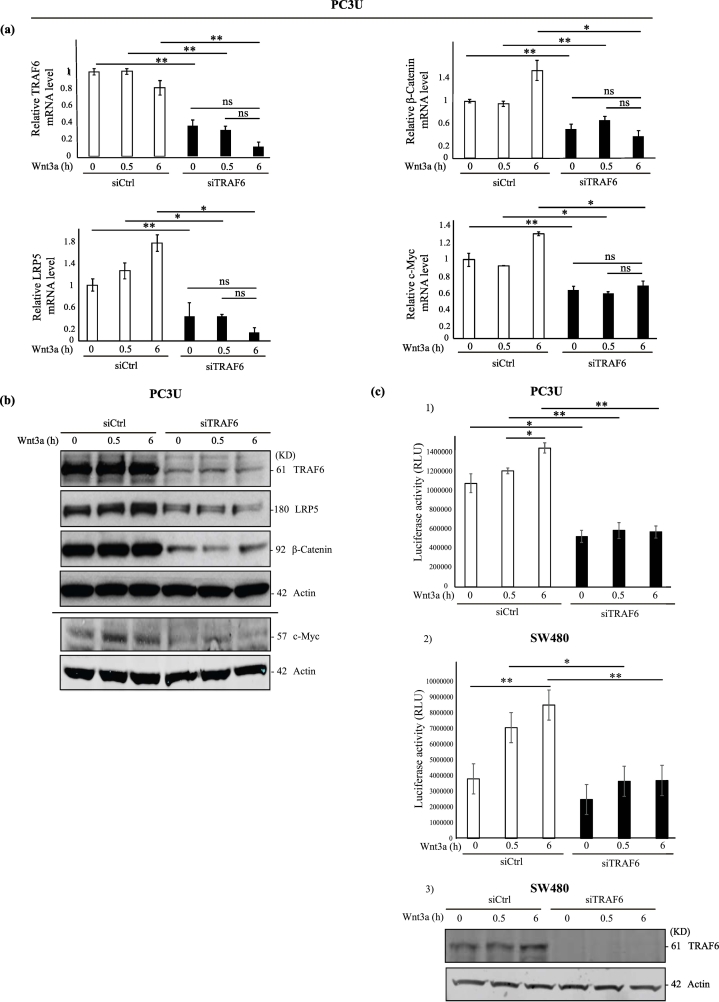
Fig. 2.
TRAF6 is required for Wnt3a-induced activation of β-Catenin and Wnt target genes.
(a-b) PC3U cells were transiently transfected with control siRNA (siCtrl) and TRAF6 siRNA (siTRAF6) and treated with Wnt3a as indicated. Total cell lysates and total RNA were isolated from these cells and subjected to immunoblot and qRT-PCR. (a) Relative mRNA levels of TRAF6, β-Catenin, LRP5 and c-Myc in PC3U cells, which were treated as indicated. GAPDH served as an internal control (n = 4 independent experiments). Bar graphs show the means ± SEM from three independent experiments. ⁎P < .05, ⁎⁎P ≤ .005, ns-not significant (student t-test). (b) Total cell lysates from PC3U cells were treated as indicated and subjected to immunoblot for TRAF6, LRP5, β-Catenin and c-Myc. Actin served as internal control. (c) Dual luciferase assay performed to measure the transcriptional activity of nuclear β-Catenin in siTRAF6 treated PC3U and SW480 cells. (c 1–2) PC3U and SW480 cells were transiently transfected with TOP Flash along with control siRNA or TRAF6 siRNA. Renilla acted as internal control (n = 3 independent experiments). The values shown are luciferase activity in relative luciferase units (RLU). Bar graphs show the means ± SEM from three to four independent experiments. ⁎P < .05, ⁎⁎P ≤ .005 (student t-test). (c 3) Representative immunoblot of total cell lysates of SW480 cells used for luciferase assay showing the knock down of TRAF6 and actin control.
The enzymatic activity of the E3-ligase TRAF6 was found to be activated and auto-ubiquitinated upon Wnt3a stimulation, revealing the association between Lys63-linked ubiquitination and TRAF6 (Fig. 1e, Supplementary Fig. S1c). Earlier studies suggest that auto-ubiquitination of TRAF6 serves as activation marker for TRAF6 [66]. A Wnt3a-enhanced association between endogenous LRP5 and TRAF6 was found also in LNCaP cells by an in situ PLA assay at 30 min (Fig. 1f.). In human colorectal SW480 cells, known to possess constitutive active Wnt-signaling due to a mutation in APC, a constitutive association between LRP5 and β-Catenin with a slight increase in 6 h with Wnt stimulation was demonstrated by in situ PLA assay (Fig. 1g). These results showed that TRAF6 interacts with the Wnt components LRP5, LRP6 and β-Catenin, upon Wnt stimulation. By in silico analyses we identified an approximate consensus binding motif Pro-X-Glu-X-X-(aromatic/acidic residue) for TRAF6 in LRP5 (Supplementary Fig. S1d). We speculate that a phospho-Serine in last position of consensus sequence in LRP5 may substitute the properties of acidic amino acid, making it a binding site for TRAF6. By introducing a point mutation in the TRAF6 consensus binding site in LRP5 (HA-LRP51407–1615HA-E1425A), we investigated in transient transfections of PC3U cells, if this motif was important for the observed association between TRAF6 and LRP5. We observed a reduced binding of endogenous TRAF6 to HA-tagged mutant LRP51407–1615HA-E1425A, compared to HA-tagged wild type LRP51407–1615HA-WT (Supplementary Fig. S1e), supporting our hypothesis that the identified TRAF6 consensus motif in LRP5 could associate with TRAF6. An increased stability of the mutant LRP51407–1615HA-E1425A, compared to wild type LRP51407–1615HA-WT was observed (Supplementary Fig. S1e), but the reason behind this is currently not known. We also generated plasmids for recombinant expression of the intracellular domain of wild type LRP51407–1615HA-(His)6-WT to investigate a possible direct interaction between recombinant TRAF6 and the intracellular domain of LRP5. However, no direct interaction between the recombinant proteins was observed (Supplementary Fig. S1f). Based on this observation, we find it likely that both TRAF6 and LRP5 undergo posttranslational modifications in vivo in cancer cells, which contribute to regulate their binding to each other. Another possibility is that LRP5, as a transmembrane protein has to be inserted in the cell membrane in order to fold properly to make a complex with TRAF6.
We observe an association between LRP5 and β-Catenin as shown in Fig. 1a and c, however no association after 0.5 h (Fig. 1a 4). The reason behind the potential disruption of the protein association 0.5 h post Wnt3a-treatment as observed by co-immunoprecipitation, is not known at this moment, but might be due to movement of the LRP5 - β-Catenin complex to a subcellular fraction from where proteins might not be as soluble, or by other technical reasons. Since LRP5 and β-Catenin is co-localized at 0.5 h after Wnt3a-stimulation (Fig. 1a), we conclude that TRAF6, LRP5/LRP6 and β-Catenin are associated in a protein complex upon Wnt3a-stimulation in PC3U cells, although these cells showed signs of a constitutive active Wnt3a-β-Catenin signaling pathway due to accumulation of nuclear β-Catenin already in non-stimulated PC3U cells (shown in Fig. 1a).
We further analyzed if the TRAF6-LRP5 protein complex could be detected by in situ PLA in human prostate cancer tissues and found the complex to be formed in high degree in human prostate cancer tissues with Gleason Score 6, a commonly used histopathological marker for aggressive disease (Fig. 1h). The data derived from PLA in situ assay, visualized the LRP5-TRAF6 complex also in prostate cancer tissues, thereby providing further support for a functional role of the protein complex in prostate cancer.
3.2. TRAF6 is required for expression and regulation of Wnt co-regulators and Wnt target genes in prostate cancer tissues
In order to understand the potential role of TRAF6 in context with its observed association with the Wnt components, we knocked down TRAF6 using siRNA in PC3U cells and analyzed the expression of Wnt target genes by qRT-PCR. We found that silencing of TRAF6 leads to down-regulation of LRP5 and its downstream target β-Catenin mRNA and the classical Wnt-target genes c-Myc upon Wnt3a stimulation (Fig. 2a). No significant increase of the c-Myc mRNA level was observed in siCtrl transfected PC3U cells upon Wnt3a-stimulation, which is likely due to a constitutive activation of the Wnt3a-pathway in these cells as demonstrated by accumulation of nuclear β-Catenin (shown in Fig. 1a), and consistent with the report from Nandana et al., [31]. We also observe in siRNA TRAF6 groups that TRAF6 seems to be required both for maintenance of basal levels of TRAF6 and LRP5 mRNA expression and for positive regulation of TRAF6 mRNA and LRP5 mRNA, as a result of Wnt3a-induced expression of these genes. Furthermore, immunoblotting of cell lysates from siTRAF6 treated PC3U cells further confirmed that TRAF6 downregulation inhibits the protein expression of LRP5, β-Catenin and c-Myc, which corresponds to the mRNA levels (Fig. 2b). To further confirm these findings at the transcriptional level, we performed a dual luciferase assay in PC3U and SW480 cells to investigate the transcriptional activity of nuclear β-Catenin, using TCF/LEF wild type binding sites in the presence and absence of TRAF6. These results show a significant difference in the relative levels of luciferase activity between the control and TRAF6 siRNA treatment groups upon Wnt3a stimulation in both PC3U and SW480 cells, suggesting that the amount of nuclear β-Catenin that could bind to the TCF/LEF transcription factors was remarkably decreased in TRAF6 silenced cells (Fig. 2c 1–3). Moreover, a significant increase of luciferase activity upon Wnt3a-stimulation of PC3U cells was observed in the siCtrl group between cells stimulated at 0.5 h and 6 h (Fig. 2c-1). The reason why no significant increase was observed between unstimulated siCtrl group at 0 h and between the 0.5 h and 6 h, likely is due to the constitutive activation of Wnt3a-β-Catenin observed in the cells (as shown by nuclear accumulation of β-Catenin in unstimulated cells; Fig. 1a). Taken together these results demonstrated that TRAF6 contributed to maintenance of basal levels of LRP5, β-catenin and c-Myc mRNA expression in PC3U cells and Wnt3a induced activation of TopFlash luciferase activity in both PC3U and SW480 cells.
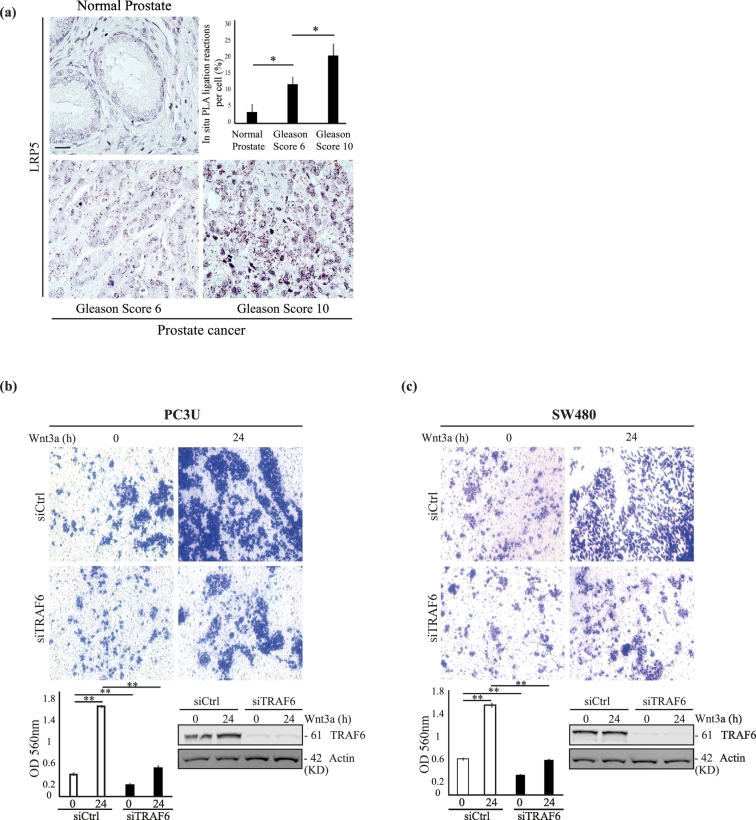
We also investigated the expression of LRP5 by employing a sensitive in situ PLA assay in human tissue sections, including normal prostate, as well as prostate cancer tissues with Gleason Score 6 and Gleason Score 10 (most aggressive). We observed a significantly higher amount of PLA signals; i.e. products of the rolling-circle amplification (RCA), as visualized as “blobs” in the cytoplasm of the majority of prostate cancer cells, with an appearance typical for the in situ PLA technique [67,68] demonstrating higher expression of LRP5 in Gleason Score 10 cancer tissue sections than in prostate cancer with lower Gleason Score 6, and importantly the lowest expression of LRP5 was found in normal prostate tissues (Fig. 3a). No signal was observed in the negative control (Supplementary Fig. S2a). To avoid false positive signals from the sensitive in situ PLA assay, we selected a specific LRP5 antibody and optimized the protocol, so that normal tissue in sections from human tissues containing tumour cells, still were negative. The different techniques used for detection of endogenous LRP5 protein expression by immunofluorescence performed on PC3U cells in culture, shown in Fig. 1b and by in situ PLA in human tissues shown in Fig. 3a which visualizes the RCA as “blobs”, as a results from the in situ PLA assay, explains why the pattern for endogenous LRP5 is not looking the same in the two images.
Fig. 3.
Levels of LRP5 is increased in prostate cancer and TRAF6 is required for Wnt3a-induced invasion of cancer cells.
(a) In situ proximity ligation analysis of LRP5 in human normal prostate tissue sections (n = 10) along with prostate cancer tissue sections with Gleason Score 6 (n = 20) and Gleason Score 10 (n = 18). Brown dots represents LRP5 protein visualized from in situ PLA ligation reactions i.e. products of rolling-circle amplification. Data shown as mean in situ PLA ligation reactions of LRP5 in normal prostate and in tumour cells (number of in situ PLA reactions per cell; %) ± S.E.M, and ⁎P < .05 (Mann-Whitney U test). Scale bar 50 μM. (b and c) Invasion assay showing the invasion of PC3U and SW480 cells respectively. PC3U and SW480 cells were transfected with either siCtrl or siTRAF6 and were treated with Wnt3a for 24 h. Bar graphs represent optical density (OD) of invaded cells measured at 560 nm. Error bars represent means ± SEM from three independent experiments, ⁎⁎P ≤ .005 (student t-test). Representative immunoblot of total cell lysates of PC3U and SW480 cells showing the knock down of TRAF6 and actin control. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further investigate a functional role for TRAF6 in Wnt3a-induced cancer responses, we next silenced TRAF6 expression in PC3U and SW480 cells and investigated the role for TRAF6 in Wnt3a-induced cancer cell invasion. We observed that expression of TRAF6 was required in both cancer cell lines to promote Wnt3a-induced invasion of cancer cells (Fig. 3b and c).
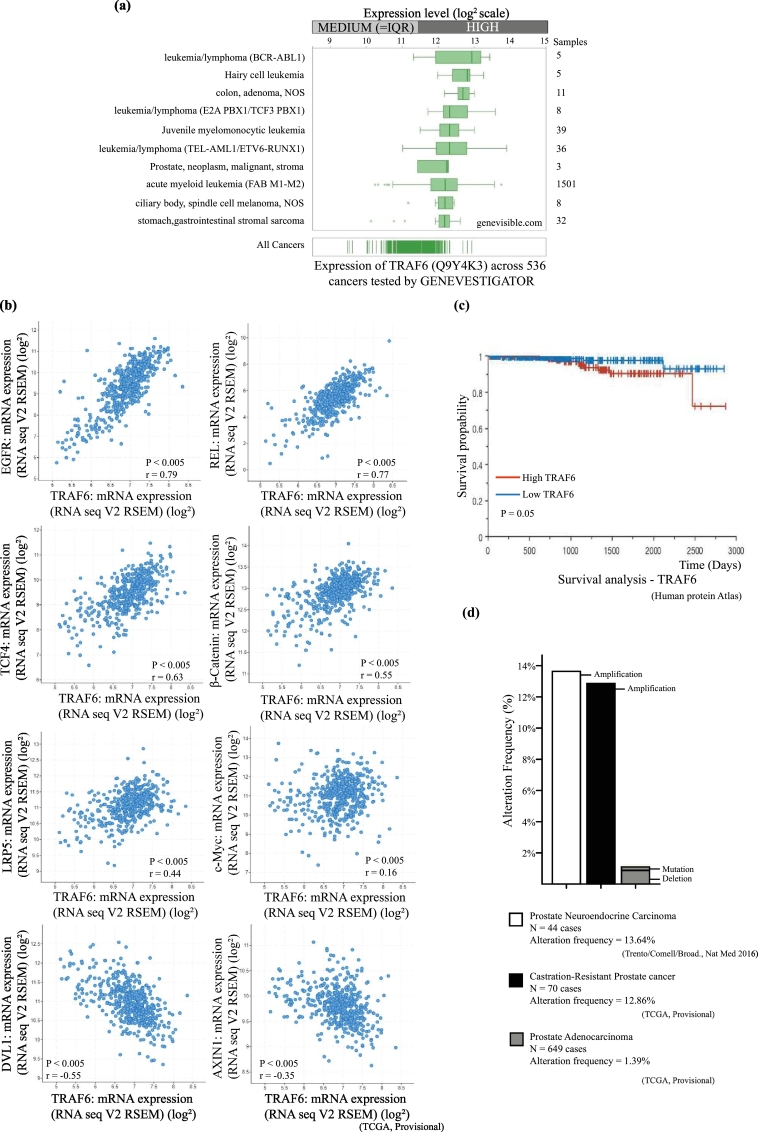
Further by using GENEVISGATOR database we have observed that TRAF6 is highly expressed in prostate neoplasm, stroma and in malignant prostate tissues. High expression of TRAF6 was also observed in several cancers such as lymphoma, hairy cell leukaemia and gastrointestinal sarcoma (Fig. 4a). Using the cBioPortal database from TCGA provisional, we next investigated the clinical relevance of our findings for TRAF6 and observed a significant positive correlation between the TRAF6 mRNA expression and two Wnt target genes: the epidermal growth factor receptor (EGFR); localized on cytoband 7p11.2 (Pearson coefficient r = 0.79, P < .005) and the REL proto-oncogene (r = 0.77, P < .005), NF-kB subunit; localized on cytoband 2p16.1 [[69], [70], [71]]. A positive correlation between TRAF6 mRNA and TCF4 mRNA was also observed (r = 0.63, P < .005) in a study containing 498 patients with prostate adenocarcinoma. Using the same dataset, a positive correlation between the TRAF6 mRNA and β-Catenin mRNA (r = 0.55, P < .005) along with LRP5 mRNA (r = 0.44, P < .005) and c-Myc (r = 0.16, P < .005) was observed. While a negative correlation between TRAF6 mRNA expression and DVL1 mRNA (r = −0.5, P = .005) and AXIN1 mRNA (r = −0.35, P < .005) expression was found (Fig. 4b). Further analysis from Human Protein atlas using TCGA Pan Cancer Atlas (PRAD) database also demonstrated that high expression of TRAF6 mRNA, correlated with poor prognosis (Fig. 4c). High expression of TRAF6 is correlated with reduced ~10-year survival of men with prostate cancer when compared with low expression of TRAF6 with significant P value (P = .05). Moreover, high expression of LRP5 is correlated with reduced survival of men with prostate cancer (96%, n = 327) when compared with low expression of LRP5 (99%, n = 167) (Supplementary Fig. S2b) [72]. High expression of c-Myc is correlated with reduced survival of men with prostate cancer (95%, n = 138) when compared with low expression of c-Myc (99%, n = 356) [73,74] (Supplementary Fig. S2c).
Fig. 4.
TRAF6 positively regulates Wnt signaling and its aberrant expression correlates with occurrence of prostate cancer.
(a) Bar graphs represent log2 expression levels of TRAF6 across 536 cancers. The analysis was performed using GENEVESTIGATOR database in which TRAF6 show medium to high expression levels in prostate neoplasm, stroma and in malignant prostate tissues. (b) The dot plots show the correlations between mRNA expression of TRAF6 with Wnt components and its target genes, in which TRAF6 mRNA positively correlates with EGFR, REL, TCF4, β-Catenin, LRP5 and c-Myc mRNA and negatively correlates DVL1 and AXIN1 mRNA. Data obtained from a study containing 498 patients with prostate adenocarcinoma obtained from the cBioPortal TCGA Provisional databases in which log2 fold change (RNA seq V2 RSEM) was represented. P value-Bootstrap hypothesis and Pearson correlation coefficient (r) were represented in the corresponding images. (c) Kaplan Meier Plot showing the survival probability of patients categorized based on high and low expression of TRAF6 mRNA from disease onset until 3000 days (~10 years). P-value (Kaplan Meier analysis) was represented. Representative image obtained from Human Protein atlas using data from TCGA Pan Cancer Atlas (PRAD) database. (d) Representative Bar graphs shows the alteration frequency (%) of TRAF6 gene loci among neuroendocrine prostate cancer patients, castration resistant prostate cancer and prostate adenocarcinoma patients. The alteration frequencies are 13.64% amplification, 12.86% amplification and 1.39% deletion and mutation respectively.
During recent years it has been suggested that castration-resistant prostate cancer is developing due to trans-differentiation of cancer cells, to instead show signs of a neuroendocrine phenotype [75]. Intriguingly TRAF6 has been found to be amplified in 18% of patients with neuroendocrine prostate cancer [76]. Data from Neuroendocrine Prostate Cancer database by Trento/Cornell/Broad, Nat Med 2016 [76] and Prostate adenocarcinoma TCGA Provisional in cBioPortal databases illustrate that 13.6% Neuroendocrine Prostate Cancer patients and 12.9% castration resistant prostate cancer patients have amplification of TRAF6 gene loci (Fig. 4d). Our observations manifest that TRAF6 positively regulates Wnt signaling and its components, is consistent with published data from public TCGA and Human Protein Atlas databases [73], supporting the biological relevance for a role for TRAF6 in aberrant Wnt signals in prostate and colorectal cancer.
3.3. TRAF6 is required for expression and regulation of Wnt co-regulators and Wnt target genes in zebrafish
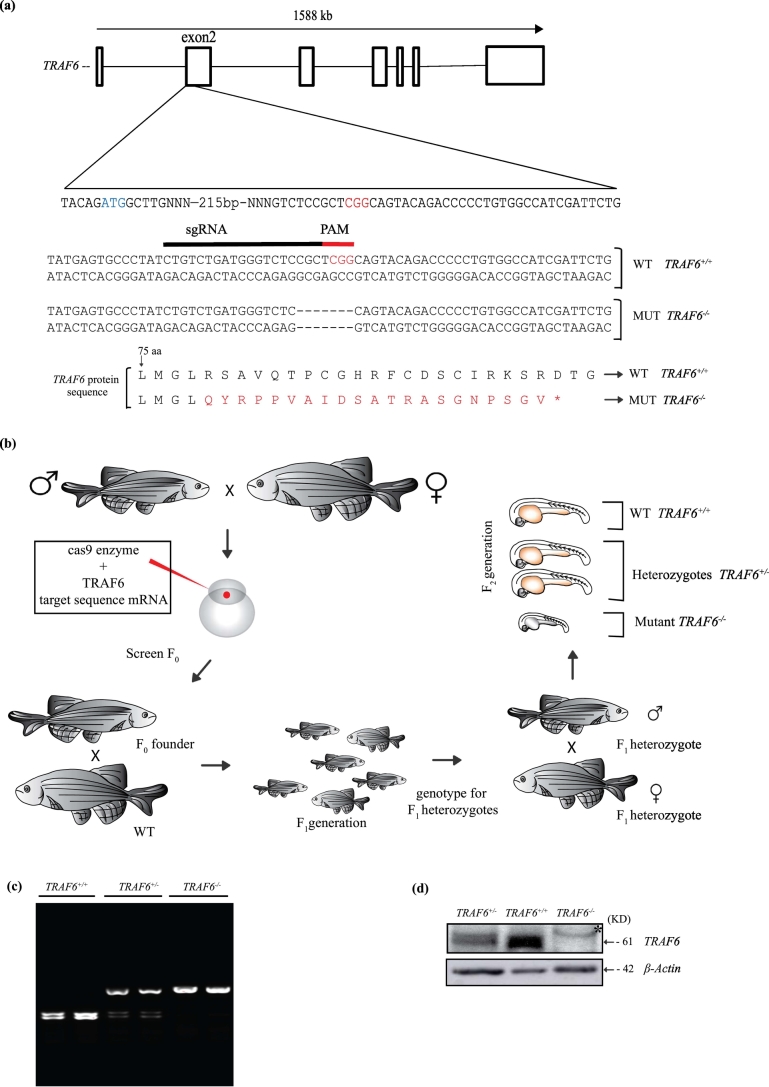
As we observed that TRAF6 was required for several Wnt3a-induced cellular responses in several human cancer cell lines as described above, we next wanted to understand if this role for TRAF6 is evolutionary conserved, as that could support the human in vitro and cell culture based data. Thus to further explore the importance of TRAF6 in Wnt signaling in vivo, we generated a TRAF6 deficient zebrafish line, using (CRISPR/Cas9). The zebrafish TRAF6 gene exon 2 (Fig. 5a and b) was targeted, which resulted in genetic deletions leading to frame shift mutations and introduction of a premature stop codon in the TRAF6 mRNA resulting in a non-functional aberrant protein. Genotyping was performed by restriction digestion of the mutated loci by restriction endonuclease MbiI that has an endogenous restriction site, which is destroyed in the mutant allele (Fig. 5c). Immunoblotting with an antibody specific for zebrafish TRAF6 on F2 embryos 25 days post fertilization confirmed the lack of TRAF6 protein in TRAF6−/− mutants (Fig. 5d).
Fig. 5.
CRISPR/Cas9 mediated knockout of TRAF6 in Zebrafish.
(a) Schematic overview of CRISPR/Cas9 mediated deletion of nucleotides resulted in a frameshift of TRAF6 gene in zebrafish. Deletion induced in TRAF6 exon 2 using complementary sgRNA (black line) next to the PAM sequence (indicated in red). Represented the nucleotide sequences of TRAF6+/+ and TRAF6−/− containing the seven base deletion in TRAF6 exon 2. Protein sequences (from 75 amino acid) of TRAF6+/+ and TRAF6−/− representing the mutated sequence (in red) due to frame shift and resulted in premature stop codon (*) (b) Schematic strategy for developing TRAF6 knockout zebrafish model using CRISPR/Cas9. (c) Representative Agarose gel image showing TRAF6+/+, TRAF6+/− and TRAF6−/− identified by their mutated loci using restriction enzyme MbiI (CCG^CTC) against restriction site situated in the PAM and sgRNA sequence. d) Western blot analysis of TRAF6 protein. Image showing the western blot of total cell lysate obtained from whole zebrafish at 25dpf stage, indicates the presence of TRAF6 (marked by an arrow) in TRAF6+/− and TRAF6+/+ but not in TRAF6−/− lysates. * indicates an unspecific band. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
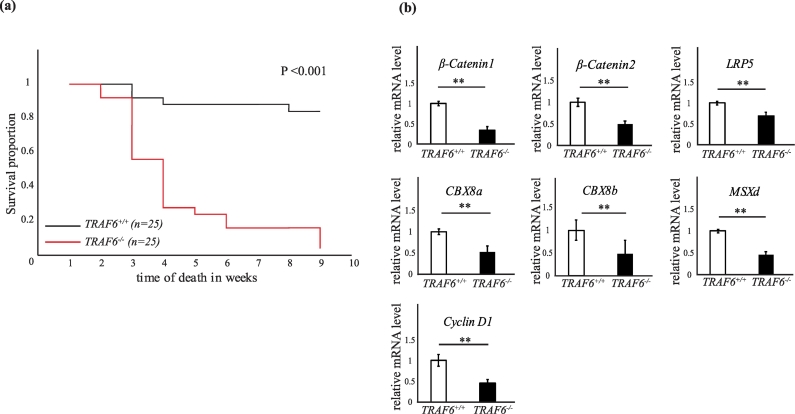
TRAF6 heterozygous fish carrying a 7-base pair deletion were incrossed to obtain TRAF6−/− mutants. These mutants showed a high incidence of mortality with age compared to their wild type or heterozygous siblings. TRAF6−/− fish appeared normal at birth but failed to survive during the development. >95% of mutant fish died and very few TRAF6−/− fish survived during the observation time of 10 weeks (Fig. 6a). The mortality of TRAF6−/− fish at week 3 post fertilization was 44% compared to 8% of TRAF6+/+ siblings and >95% of TRAF6−/− died by week 9 (Table 1). Earlier studies in TRAF6 knock out mice have shown similar results, as these mice failed to thrive normally [42,43].
Fig. 6.
TRAF6 positively regulates Wnt-target genes and is crucial for the survival in zebrafish.
(a) Kaplan-Meier Plot showing the survival curves of wild type (TRAF6+/+) n = 25 and mutant (TRAF6−/−) n = 25 fish. P < .001 (Kaplan-Meier analysis) (b) 7 dpf TRAF6+/+ and TRAF6−/− embryos were identified by genotyping and RNA extracted for qRT-PCR analysis. Bar graphs showing relative mRNA expression levels of β-Catenin1, β-Catenin2, LRP5, CBX8a, CBX8b, MSXd and Cyclin D1. β-Actin was used as internal control. All the qRT-PCR experiments were repeated three times (n = 3 TRAF6+/+ and 3 TRAF6−/−). ⁎⁎P ≤ .005 (student t-test).
Table 1.
Representative data showing the percentage of mortality rate in TRAF6+/+ and TRAF6−/− fishes.
| n = 25 | Total No. of deaths by week |
|||
|---|---|---|---|---|
| TRAF6+/+ | % | TRAF6−/− | % | |
| Week 3 | 2 | 8% | 11 | 44% |
| Week 6 | 3 | 12% | 21 | 84% |
| Week 9 | 4 | 16% | 24 | 96% |
Here n = 25. Data obtained from three independent experiments/observations.
Since we observed that TRAF6 influenced Wnt3a-induced expression of β-Catenin, transcription of its co-receptor LRP5, and its target gene c-Myc, in PC3U cells, we investigated whether silencing of TRAF6 in zebrafish, could influence mRNA expression of β-Catenin in zebrafish. qRT-PCR analyses of mRNA extracted from wild type and mutant TRAF6 zebrafish embryos at 7dpf, confirmed that TRAF6 expression regulated the β-Catenin1 and β-Catenin2 mRNA expression (Fig. 6b). This observation is in line with the previous report from Naito et al. [77] who reported that knock down of TRAF6, reduced the expression of β-Catenin in epidermis of the mice [77]. Furthermore, evidence for transcriptional regulation of β-Catenin has previously been reported in the invasive front of colorectal liver metastasis [78]. In addition, the mRNA expression of LRP5 and Cyclin D1 was found to be regulated in zebrafish embryos in a TRAF6-dependent manner, consistent with our data observed in PC3U cells (Fig. 2). Furthermore, mRNA derived from TRAF6+/+ and TRAF6−/− zebrafish embryos, demonstrated TRAF6-dependent down-regulation of homeobox genes such as msxd. We also observed TRAF6-dependent down-regulation of cbx8a and cbx8b genes, which are encoding proteins in the polycomb repressing complex 1 (PRC1), all implicated in breast cancer progression [79].
The few TRAF6−/− fish that developed beyond juvenile stages and continued to adulthood, showed signs of defective bone formation in the vertebrae of tail region (Supplementary Fig. S3a, 1–2). Whole-mount alizarin red staining show fusion of the most posterior vertebrae in TRAF6−/− zebrafish. Six to seven vertebral discs were fused which resulted in abnormal tail formation and a malformed body axis (Supplementary Fig. S3a, 3–4). This fusion of vertebrae might be due to high bone density with hard and denser bones, a typical trait of osteopetrosis where the bone reabsorption by osteoclasts are significantly reduced [80]. These findings concur with earlier findings in which TRAF6−/− mice show severe osteopetrosis due to osteoclasts being disorganised from bone surfaces and resulting in reduced areas for bone resorption [43,44]. To validate these findings, we confirmed TRAF6 mRNA expression in the bone of wild-type zebrafish (Supplementary Fig. S3b). The identification of LRP5, β-Catenin and msxd as TRAF6 target genes, might correlate to the effects observed in this study on bone formation and loss of survival of the TRAF6−/− mutant zebrafish.
From the studies performed in TRAF6−/− deficient zebrafish we conclude that TRAF6 is required for regulation of a subset of previously described targets for Wnt3a. This is in line with our human prostate cancer cell data and our findings in clinical patient-material with activated Wnt3a signaling.
4. Discussion
Activation of Wnt pathways are recognized as key regulatory mechanisms identified in cancer due to the potent effects exerted by these signals, on cell renewal and cell growth [81]. In this study, we have elucidated intriguing functions for TRAF6 as a coregulatory factor for expression of Wnt-target genes in human prostate and colorectal cancer cells, which was confirmed in vivo by using CRISPR/Cas9 genomic editing, in zebrafish. Our data suggest that TRAF6 is required for gene expression of β-Catenin as well as for the Wnt-ligand co-receptor LRP5 itself. The observed association of TRAF6 with LRP5 both in vivo and in vitro seems therefore to be positively regulated upon a Wnt3a induced responses in prostate cancer as well as in the development of zebrafish. The majority of TRAF6 mutant zebrafish did not survive longer than 9 weeks post fertilization a finding that is consistent with the important role for TRAF6 in survival of mice [43,44]. Since both LRP5 and TRAF6 has been reported to be crucial for bone development and bone formation [43,[81], [82], [83], [84], [85]], the fused vertebras observed in zebrafish is likely related to reduced levels of LRP5 and TRAF6, as we also observed a decrease of Cyclin D1 mRNA levels in zebrafish embryos in TRAF6 deficient zebrafish. Earlier studies in colon cancer have shown that knockdown of TRAF6, caused a decreased Cyclin D1 expression and that TRAF6 regulates the proliferation of colon cancer cells through its induction of Cyclin D1 [50]. High expression of TRAF6 mRNA was found to be correlated with high mRNA expression of Wnt-target genes such as EGFR, REL, as well as with c-Myc, LRP5 and β-Catenin mRNA expression [[69], [70], [71],73,86].
In siTRAF6 PC3U cells we observed a tendency of a decrease of TRAF6 and LRP5 mRNA 6 h after Wnt3a stimulation (Fig. 2a), suggesting that TRAF6 is might be required for a positive regulation of these genes also in these cells. However, since this difference was not statistically significant this finding needs to be further elucidated in future studies as we for the moment have no clear molecular explanation for this observation. The PC3U cells used in our study shows sign of a constitutive activation of the Wnt3a-β-Catenin signaling pathway as nuclear accumulation of β-Catenin was observed in unstimulated cells (Fig. 1a). Moreover, as no robust accumulation of total β-Catenin (Fig. 2b and Supplementary Fig. S1a) and no significant increase of the relative c-Myc mRNA levels was observed in PC3U cells in response to Wnt3a stimulation, as shown in Fig. 2a, these data provides support of constitutive active Wnt3a-β-Catenin signaling in PC3U cells. These data are consistent with a recent report from Nandana et al., [31] which demonstrated that the highly aggressive and metastatic PC3 cells, from which PC3U cells are derived [87] produces high levels of Wnt3a, which promoted invasion of PC3 cells. In this study, we observed a slight increased level of β-Catenin protein after 0.5 h and 6 h of Wnt3a-stimulation of PC3U cells (Fig. 2b), while the increase of β-Catenin mRNA expression levels is observed after 6 h (Fig. 2a), consistent with the observed increase of TOPFlash luciferase activity between Wnt3a stimulation of PC3U cells at 0.5 h and 6 h (Fig. 2c). We did so far not find a mechanism that explains this phenomenon and future studies are required for elucidating this potential post-transcriptional regulation of β-Catenin in prostate cancer cells in response to Wnt3a stimulation. Previous studies have however showed that accumulation of β-Catenin occurs as a result of that Wnt signaling is triggering the sequestration of GSK3 from the cytosol into multivesicular bodies (MVBs), so that this enzyme becomes separated from its many cytosolic substrates including β-Catenin [17]. As we observe that LRP5 and β-Catenin is colocalized 6 h after Wnt3a-stimulation in PC3U cells (Fig. 1a), it is possible that the protein complex is localized in specific subcellular compartments such as multivesicular bodies in line with the report from Taelman and collegues [17]. Since TRAF6 was required for Wnt3a induced invasion of both the castration-resistant prostate cancer PC3U cells and the SW480 colorectal cancer cells, these findings give further support for that TRAF6 can promote cancer progression of certain forms of cancer as presented in this report. The observations from invasion assay explains a potential role of TRAF6 in cancer cells for their invasion. As we observed a clear reduction of Wnt3a induced invasion in TRAF6 knockout cells, our data suggest that TRAF6 and Wnt may work concomitantly for the cancer cell invasion. These observations were identical to the previous studies in gastric cancer [51]. Furthermore, high expression of both TRAF6 and c-Myc is correlated with poor survival in patients with prostate cancer [73]. Castration-resistant prostate cancer has during the recent years been proposed to develop due to trans-differentiation of prostate cancer cells to a neuroendocrine phenotype with a poor prognosis [75]. Intriguingly has TRAF6 recently been found to be amplified in neuroendocrine prostate cancer patients [76], as also reported here in Fig. 4d. We also observed a significantly higher expression of LRP5 in aggressive prostate cancer tissues (with Gleason Score 10) compared to less aggressive prostate cancer tissues (Gleason Score 6) and tissues derived from normal prostate (Fig. 3a) consistent with the report from Rabbani et al., demonstrating a role for LRP5 in prostate cancer invasion and skeletal metastasis [32]. Identification of TRAF6 as a key regulatory adaptor protein for transduction of Wnt3a/β-Catenin pathway and regulation of Wnt3a-induced invasion of cancer cells and Wnt-target genes provides an important knowledge for future therapeutic strategies to interfere with oncogenic signals in cancers dependent on Wnt-signals.
Funding sources
This work was supported by grants from the Knut and Alice Wallenberg Foundation (KAW 2012.0090), Swedish Cancer Society (CAN 2017/544), Swedish Medical Research Council (2016-02513), Prostatacancerförbundet, Konung Gustaf den V:s och Drottning Victorias Frimurarestiftelse and Cancerforskningsfonden Norrland.
Author contributions
K.A, S.K.G., G.Z, A.S., S.S.Å., and L.Ö. performed the experiments; A.B. provided the human samples; K.A., J.v.H. and M.L. planned the project; K.A., J.v.H. and M.L. prepared the manuscript.
Declaration of Competing Interest
This work was supported by grants to Marene Landström from the Knut and Alice Wallenberg Foundation (KAW 2012.0090), Swedish Cancer Society (CAN 2017/544), Swedish Medical Research Council (2016-02513), Prostatacancerförbundet, Konung Gustaf den V:s och Drottning Victorias Frimurarestiftelse and Lion's Cancer Research Foundation LP 18-2197, Umeå University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The other authors have declared no conflict of interest. The other authors have declared no conflict of interest.
Acknowledgements
We thank our colleagues at the Department of Medical Biosciences, Umeå University, Sweden for valuable discussions. We specially thank Pernilla Andersson, Reshma Sundar and Anders Wallenius for technical support and advices. Radiosa Gallini at the PLA Proteomics platform, a national infrastructure facility at SciLifeLab at Uppsala University, Sweden for performing in situ PLA. Hanna Nord and Nils Dennhag for support in Zebrafish experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.06.046.
Appendix A. Supplementary data
Supplementary material Fig. S1. (a) Total cell lysates from PC3U cells were extracted after treating with Wnt3a as indicated. Lysates were immunoblotted to detect TRAF6 and β-Catenin proteins. Actin served as internal control. Bar graphs represent the quantification of TRAF6 and β-Catenin bands against actin control. n = 4 independent experiments. *P ≤ .05 (student t-test). (b) Representative immunoblot shows the protein levels of TRAF6, LRP5 and active β-Catenin across various cell lines. Total cell lysates from PC3U, LNCaP, DU145 and SW480 cell lines were extracted without any treatment and immunoblotted for corresponding protein. Actin served as internal control. (c) Auto-ubiquitination of TRAF6 upon Wnt stimulation. PC3U cells were transfected with HA-tagged ubiquitin. Total cell lysates were immuno-precipitated with rabbit anti-TRAF6 antibody and lysine-63 (K63) ubiquitination visualized by immunoblotting with anti-K63 antibody. The total cell lysates were subjected to immunoblotting with HA antibody to verify the equal expression levels of ubiquitin. (d) Representation of a TRAF6 consensus-binding motif in LRP5 intracellular domain identified in silico. (e) PC3U cells were transfected with LRP51407-1615HA-WT and mutant LRP51407-1615HA-E1425A plasmids. Total cell lysates from these cells were extracted after treating with Wnt3a as indicated. Lysates were immuno-precipitated with HA antibody. Representative immunoblots were probed to detect TRAF6 associated with LRP5-HA. Bar graphs represents the normalization of TRAF6 with immuno-precipitated LRP5-HA. (f) Recombinant LRP51407-1615-HA-(His)6-WT was immobilized on a Ni-NTA sensor and binding against human TRAF6-Flag (Origene, USA Cat#:TP319528) at a concentration of 167 nM (green line) and 822 nM (pink line) respectively was analyzed with a BLitz (Forte BIO, Pall Life Sciences). No binding between recombinant LRP5-HA-(His)6-WT and TRAF6-Flag could be detected. The intracellular domain of human LRP5 spanning from the end of the transmembrane domain to the very C-terminal end (amino acid 1407 to 1615) was expressed recombinantly as an HA- and (His)6-tagged fusion protein in human HEK293 cells and purified by Ni-NTA chromatography. The protein LRP51407-1615-HA-(His)6-WT with a theoretical mass of 25 kDa was analyzed in an immunoblot by using mouse anti-HA monoclonal antibody followed by HRP-conjugated anti-mouse IgG.
Fig. S2: a) In situ PLA negative control (PBS control) of paraffin-embedded sections of prostate cancer tissue corresponding to the Fig. 3a. Scale bar 50 μM (b) Kaplan-Meier Plot showing the survival probability of patients categorized based on high and low expression of LRP5 and (c) c-Myc mRNA. P-values (Kaplan-Meier analysis) were represented. Representative image obtained from Human Protein atlas using data from TCGA Pan Cancer Atlas (PRAD) database.
Fig. S3: (a) (1-2) Images representing the phenotypic differences between TRAF6+/+ and TRAF6−/− fishes at 50dpf. (3-4) Whole-mount in-situ Alizarin red staining of TRAF6+/+ and TRAF6−/− fishes at 50dpf. Boxes indicate area of enlargement on the right. White arrows indicate normal areas and black arrows indicate affected areas in vertebrae. Images were taken using a Nikon SMZ1500 stereomicroscope and a Nikon D5200 digital camera. Scale bar 2 mm. (b) mRNA expression levels of TRAF6 in bone tissue in 60 dpf wild-type zebrafish (n = 4 WT fish). Bar graphs show the means ± SEM.
Fig. S4: Data represents the list of primers used for the qRT-PCR mRNA analysis of human prostate cancer cell line (PC3U) and zebrafish experiments.
References
- 1.Moon R.T., Bowerman B., Boutros M., Perrimon N. The promise and perils of Wnt signaling through β-catenin. Science. 2002;296(5573):1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H., Loh K.M., Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346(6205) doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 3.Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 4.Duchartre Y., Kim Y.-M., Kahn M. The Wnt signaling pathway in cancer. Crit Rev Oncol Hematol. 2016;99:141–149. doi: 10.1016/j.critrevonc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heuberger J., Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2(2) doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson W.J., Nusse R. Convergence of Wnt, ß-catenin, and cadherin pathways. Science. 2004;303(5663):1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y. Wnts and wing: Wnt signaling in vertebrate limb development and musculoskeletal morphogenesis. Birth Defects Res C Embryo Today. 2003;69(4):305–317. doi: 10.1002/bdrc.10026. [DOI] [PubMed] [Google Scholar]
- 8.Kypta R.M., Waxman J. Wnt/beta-catenin signalling in prostate cancer. Nat Rev Urol. 2012;9(8):418–428. doi: 10.1038/nrurol.2012.116. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Giles R.H., van Es J.H., Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta Rev Biomembr Cancer. 2003;1653(1):1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 11.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17(1):45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamos J.L., Weis W.I. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5(1) doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14(15):1837–1851. [PubMed] [Google Scholar]
- 15.Angers S., Moon R.T. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10(7):468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald B.T., He X. Frizzled and LRP5/6 receptors for Wnt/beta-catenin signaling. Cold Spring Harb Perspect Biol. 2012;4(12) doi: 10.1101/cshperspect.a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taelman V.F., Dobrowolski R., Plouhinec J.-L., Fuentealba L.C., Vorwald P.P., Gumper I. Wnt Signaling requires the sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143(7):1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clevers H., Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 20.Salahshor S., Woodgett J.R. The links between axin and carcinogenesis. J Clin Pathol. 2005;58(3):225–236. doi: 10.1136/jcp.2003.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou C.K., Check D.P., Lortet-Tieulent J., Laversanne M., Jemal A., Ferlay J. Prostate cancer incidence in 43 populations worldwide: an analysis of time trends overall and by age group. Int J Cancer. 2016;138(6):1388–1400. doi: 10.1002/ijc.29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper L.A., Page S.T. Androgens and prostate disease. Asian J Androl. 2014;16(2):248–255. doi: 10.4103/1008-682X.122361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pienta K.J., Bradley D. Mechanisms underlying the development of androgen-independent prostate Cancer. Clin Cancer Res. 2006;12(6):1665–1671. doi: 10.1158/1078-0432.CCR-06-0067. [DOI] [PubMed] [Google Scholar]
- 24.Feldman B.J., Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 25.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.-W. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 26.Wu L., Zhao J.C., Kim J., Jin H.J., Wang C.Y., Yu J. ERG is a critical regulator of Wnt/LEF1 signaling in prostate cancer. Cancer Res. 2013;73(19):6068–6079. doi: 10.1158/0008-5472.CAN-13-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Culig Z., Hobisch A., Cronauer M.V., Radmayr C., Trapman J., Hittmair A. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res. 1994;54(20):5474–5478. [PubMed] [Google Scholar]
- 28.Terry S., Yang X., Chen M.-W., Vacherot F., Buttyan R. Multifaceted interaction between the androgen and Wnt signaling pathways and the implication for prostate cancer. J Cell Biochem. 2006;99(2):402–410. doi: 10.1002/jcb.20983. [DOI] [PubMed] [Google Scholar]
- 29.Verras M., Sun Z. Roles and regulation of Wnt signaling and β-catenin in prostate cancer. Cancer Lett. 2006;237(1):22–32. doi: 10.1016/j.canlet.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Verras M., Brown J., Li X., Nusse R., Sun Z. Wnt3a growth factor induces androgen receptor-mediated transcription and enhances cell growth in human prostate cancer cells. Cancer Res. 2004;64(24):8860–8866. doi: 10.1158/0008-5472.CAN-04-2370. [DOI] [PubMed] [Google Scholar]
- 31.Nandana S., Tripathi M., Duan P., Chu C.-Y., Mishra R., Liu C. Bone metastasis of prostate cancer can be therapeutically targeted at the TBX2–WNT signaling axis. Cancer Res. 2017;77(6):1331–1344. doi: 10.1158/0008-5472.CAN-16-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabbani S.A., Arakelian A., Farookhi R. LRP5 knockdown: effect on prostate cancer invasion growth and skeletal metastasis in vitro and in vivo. Cancer Med. 2013;2(5):625–635. doi: 10.1002/cam4.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu N., Kawakami K., Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Dev Biol. 2012;370(1):71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Schneider S., Steinbeisser H., Warga R.M., Hausen P. β-Catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos. Mech Dev. 1996;57(2):191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- 35.Willems B., Tao S., Yu T., Huysseune A., Witten P.E., Winkler C. The Wnt co-receptor Lrp5 is required for cranial neural crest cell migration in zebrafish. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0131768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoick-Cooper C.L., Weidinger G., Riehle K.J., Hubbert C., Major M.B., Fausto N. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134(3):479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 37.Yin A., Korzh S., Winata C.L., Korzh V., Gong Z. Wnt signaling is required for early development of zebrafish swimbladder. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0018431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi N., Kadono Y., Naito A., Matsumoto K., Yamamoto T., Tanaka S. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 2001;20(6):1271–1280. doi: 10.1093/emboj/20.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H., Arron J.R. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays. 2003;25(11):1096–1105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- 40.Yin Q., Lin S.-C., Lamothe B., Lu M., Lo Y.-C., Hura G. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16(6):658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye H., Arron J.R., Lamothe B., Cirilli M., Kobayashi T., Shevde N.K. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418(6896):443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 42.Lomaga M.A., Henderson J.T., Elia A.J., Robertson J., Noyce R.S., Yeh W.-C. Tumor necrosis factor receptor-associated factor 6 (TRAF6) deficiency results in exencephaly and is required for apoptosis within the developing CNS. J Neurosci. 2000;20(19):7384–7393. doi: 10.1523/JNEUROSCI.20-19-07384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lomaga M.A., Yeh W.C., Sarosi I., Duncan G.S., Furlonger C., Ho A. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13(8):1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naito A., Azuma S., Tanaka S., Miyazaki T., Takaki S., Takatsu K. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells. 1999;4(6):353–362. doi: 10.1046/j.1365-2443.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 45.Yang W.-L., Wang J., Chan C.-H., Lee S.-W., Campos A.D., Lamothe B. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamidi A., Song J., Thakur N., Itoh S., Marcusson A., Bergh A. TGF-β promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85α. Sci Signal. 2017;10(486) doi: 10.1126/scisignal.aal4186. [DOI] [PubMed] [Google Scholar]
- 47.Mu Y., Sundar R., Thakur N., Ekman M., Gudey S.K., Yakymovych M. TRAF6 ubiquitinates TGFbeta type I receptor to promote its cleavage and nuclear translocation in cancer. Nat Commun. 2011;2:330. doi: 10.1038/ncomms1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gudey S.K., Sundar R., Mu Y., Wallenius A., Zang G., Bergh A. TRAF6 stimulates the tumor-promoting effects of TGFb type I receptor through polyubiquitination and activation of Presenilin 1. Sci Signal. Jan 7, 2014;7(307) doi: 10.1126/scisignal.2004207. [DOI] [PubMed] [Google Scholar]
- 49.Starczynowski D.T., Lockwood W.W., Deléhouzée S., Chari R., Wegrzyn J., Fuller M. TRAF6 is an amplified oncogene bridging the RAS and NF-κB pathways in human lung cancer. J Clin Invest. 2011;121(10):4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun H., Li X., Fan L., Wu G., Li M., Fang J. TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int J Biochem Cell Biol. 2014;53:195–201. doi: 10.1016/j.biocel.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 51.Han F., Zhang L., Qiu W., Yi X. TRAF6 promotes the invasion and metastasis and predicts a poor prognosis in gastric cancer. Pathol Res Pract. 2016;212(1):31–37. doi: 10.1016/j.prp.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Franzén P., Ichijo H., Miyazono K. Different signals mediate transforming growth factor-β1-induced growth inhibition and extracellular matrix production in prostatic carcinoma cells. Exp Cell Res. 1993;207(1):1–7. doi: 10.1006/excr.1993.1156. [DOI] [PubMed] [Google Scholar]
- 53.Nakao A., Imamura T., Souchelnytskyi S., Kawabata M., Ishisaki A., Oeda E. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16(17):5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sitaram R.T., Mallikarjuna P., Landström M., Ljungberg B. Transforming growth factor-β promotes aggressiveness and invasion of clear cell renal cell carcinoma. Oncotarget. 2016;7(24) doi: 10.18632/oncotarget.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorrentino A., Thakur N., Grimsby S., Marcusson A., von Bulow V., Schuster N. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10(10):1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 56.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D. Efficient in vivo genome editing using RNA-guided nucleases. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140(24):4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- 58.Nord H., Dennhag N., Muck J., von Hofsten J. Pax7 is required for establishment of the xanthophore lineage in zebrafish embryos. Mol Biol Cell. 2016;27(11):1853–1862. doi: 10.1091/mbc.E15-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Jong M., Rauwerda H., Bruning O., Verkooijen J., Spaink H.P., Breit T.M. RNA isolation method for single embryo transcriptome analysis in zebrafish. BMC Res Notes. 2010;3:73. doi: 10.1186/1756-0500-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lan C.C., Tang R., Un San Leong I., Love D.R. Quantitative real-time RT-PCR (qRT-PCR) of zebrafish transcripts: optimization of RNA extraction, quality control considerations, and data analysis. Cold Spring Harb Protoc. 2009;2009(10) doi: 10.1101/pdb.prot5314. [pdb prot5314] [DOI] [PubMed] [Google Scholar]
- 61.Leung Y.F., Ma P., Dowling J.E. Gene expression profiling of zebrafish embryonic retinal pigment epithelium in vivo. Invest Ophthalmol Vis Sci. 2007;48(2):881–890. doi: 10.1167/iovs.06-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang R., Dodd A., Lai D., McNabb W.C., Love D.R. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim Biophys Sin. 2007;39(5):384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Connolly M.H., Yelick P.C. High-throughput methods for visualizing the teleost skeleton: capturing autofluorescence of alizarin red. J Appl Ichthyol. 2010;26(2):274–277. [Google Scholar]
- 64.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinforma. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng L., Wang C., Spencer E., Yang L., Braun A., You J. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 66.Walsh M.C., Kim G.K., Maurizio P.L., Molnar E.E., Choi Y. TRAF6 autoubiquitination-independent activation of the NFκB and MAPK pathways in response to IL-1 and RANKL. PLoS One. 2008;3(12) doi: 10.1371/journal.pone.0004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Söderberg O., Leuchowius K.-J., Gullberg M., Jarvius M., Weibrecht I., Larsson L.-G. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45(3):227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 68.Borroto Escuela D., Hagman B., Woolfenden M., Pinton L., Jiménez-Beristain A., Oflijan J. 2016. In Situ Proximity Ligation Assay to Study and Understand the Distribution and Balance of GPCR Homo- and Heteroreceptor Complexes in the Brain; pp. 109–124. [Google Scholar]
- 69.Ma B., Hottiger M.O. Crosstalk between Wnt/β-catenin and NF-κB signaling pathway during inflammation. Front Immunol. 2016;7:378. doi: 10.3389/fimmu.2016.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwitalla S., Fingerle A.A., Cammareri P., Nebelsiek T., Göktuna S.I., Ziegler P.K. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1):25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 71.Li D., Beisswenger C., Herr C., Hellberg J., Han G., Zakharkina T. Myeloid cell RelA/p65 promotes lung cancer proliferation through Wnt/β-catenin signaling in murine and human tumor cells. Oncogene. 2013;33:1239. doi: 10.1038/onc.2013.75. [DOI] [PubMed] [Google Scholar]
- 72.Human Protein Atlas. 18 Aug, 2017. https://www.proteinatlas.org/ENSG00000162337-LRP5/pathology/tissue/prostate+cancer#imid_3743383
- 73.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352) doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 74.Human Protein Atlas. 2019. https://www.proteinatlas.org/ENSG00000136997-MYC/pathology/tissue/prostate+cancer#img
- 75.Tsaur I., Heidegger I., Kretschmer A., Borgmann H., Gandaglia G., Briganti A. Aggressive variants of prostate cancer – are we ready to apply specific treatment right now? Cancer Treat Rev. 2019;75:20–26. doi: 10.1016/j.ctrv.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 76.Beltran H., Prandi D., Mosquera J.M., Benelli M., Puca L., Cyrta J. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Naito A., Yoshida H., Nishioka E., Satoh M., Azuma S., Yamamoto T. TRAF6-deficient mice display hypohidrotic ectodermal dysplasia. Proc Natl Acad Sci. 2002;99(13):8766. doi: 10.1073/pnas.132636999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bandapalli O.R., Dihlmann S., Helwa R., Macher-Goeppinger S., Weitz J., Schirmacher P. Transcriptional activation of the β-catenin gene at the invasion front of colorectal liver metastases. J Pathol. 2009;218(3):370–379. doi: 10.1002/path.2539. [DOI] [PubMed] [Google Scholar]
- 79.Chung C.-Y., Sun Z., Mullokandov G., Bosch A., Qadeer Z.A., Cihan E. Cbx8 acts non-canonically with Wdr5 to promote mammary tumorigenesis. Cell Rep. 2016;16(2):472–486. doi: 10.1016/j.celrep.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Popoff S.N., Marks S.C. The heterogeneity of the osteopetroses reflects the diversity of cellular influences during skeletal development. Bone. 1995;17(5):437–445. doi: 10.1016/8756-3282(95)00347-4. [DOI] [PubMed] [Google Scholar]
- 81.Nusse R., Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 82.Johnson M.L., Harnish K., Nusse R., Van Hul W. LRP5 and Wnt signaling: a union made for bone. J Bone Miner Res. 2004;19(11):1749–1757. doi: 10.1359/JBMR.040816. [DOI] [PubMed] [Google Scholar]
- 83.Moon R.T., Kohn A.D., De Ferrari G.V., Kaykas A. WNT and [beta]-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5(9):691. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 84.Holmen S.L., Zylstra C.R., Mukherjee A., Sigler R.E., Faugere M.-C., Bouxsein M.L. Essential role of β-catenin in postnatal bone acquisition. J Biol Chem. 2005;280(22):21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- 85.Gong Y., Slee R.B., Fukai N., Rawadi G., Roman-Roman S., Reginato A.M. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107(4):513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 86.Tan X., Apte U., Micsenyi A., Kotsagrelos E., Luo J.-H., Ranganathan S. Epidermal growth factor receptor: a novel target of the Wnt/β-catenin pathway in liver. Gastroenterology. 2005;129(1):285–302. doi: 10.1053/j.gastro.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zang G., Mu Y., Gao L., Bergh A., Landström M. PKCζ facilitates lymphatic metastatic spread of prostate cancer cells in a mice xenograft model. Oncogene. 2019;38(22):4215–4231. doi: 10.1038/s41388-019-0722-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Fig. S1. (a) Total cell lysates from PC3U cells were extracted after treating with Wnt3a as indicated. Lysates were immunoblotted to detect TRAF6 and β-Catenin proteins. Actin served as internal control. Bar graphs represent the quantification of TRAF6 and β-Catenin bands against actin control. n = 4 independent experiments. *P ≤ .05 (student t-test). (b) Representative immunoblot shows the protein levels of TRAF6, LRP5 and active β-Catenin across various cell lines. Total cell lysates from PC3U, LNCaP, DU145 and SW480 cell lines were extracted without any treatment and immunoblotted for corresponding protein. Actin served as internal control. (c) Auto-ubiquitination of TRAF6 upon Wnt stimulation. PC3U cells were transfected with HA-tagged ubiquitin. Total cell lysates were immuno-precipitated with rabbit anti-TRAF6 antibody and lysine-63 (K63) ubiquitination visualized by immunoblotting with anti-K63 antibody. The total cell lysates were subjected to immunoblotting with HA antibody to verify the equal expression levels of ubiquitin. (d) Representation of a TRAF6 consensus-binding motif in LRP5 intracellular domain identified in silico. (e) PC3U cells were transfected with LRP51407-1615HA-WT and mutant LRP51407-1615HA-E1425A plasmids. Total cell lysates from these cells were extracted after treating with Wnt3a as indicated. Lysates were immuno-precipitated with HA antibody. Representative immunoblots were probed to detect TRAF6 associated with LRP5-HA. Bar graphs represents the normalization of TRAF6 with immuno-precipitated LRP5-HA. (f) Recombinant LRP51407-1615-HA-(His)6-WT was immobilized on a Ni-NTA sensor and binding against human TRAF6-Flag (Origene, USA Cat#:TP319528) at a concentration of 167 nM (green line) and 822 nM (pink line) respectively was analyzed with a BLitz (Forte BIO, Pall Life Sciences). No binding between recombinant LRP5-HA-(His)6-WT and TRAF6-Flag could be detected. The intracellular domain of human LRP5 spanning from the end of the transmembrane domain to the very C-terminal end (amino acid 1407 to 1615) was expressed recombinantly as an HA- and (His)6-tagged fusion protein in human HEK293 cells and purified by Ni-NTA chromatography. The protein LRP51407-1615-HA-(His)6-WT with a theoretical mass of 25 kDa was analyzed in an immunoblot by using mouse anti-HA monoclonal antibody followed by HRP-conjugated anti-mouse IgG.
Fig. S2: a) In situ PLA negative control (PBS control) of paraffin-embedded sections of prostate cancer tissue corresponding to the Fig. 3a. Scale bar 50 μM (b) Kaplan-Meier Plot showing the survival probability of patients categorized based on high and low expression of LRP5 and (c) c-Myc mRNA. P-values (Kaplan-Meier analysis) were represented. Representative image obtained from Human Protein atlas using data from TCGA Pan Cancer Atlas (PRAD) database.
Fig. S3: (a) (1-2) Images representing the phenotypic differences between TRAF6+/+ and TRAF6−/− fishes at 50dpf. (3-4) Whole-mount in-situ Alizarin red staining of TRAF6+/+ and TRAF6−/− fishes at 50dpf. Boxes indicate area of enlargement on the right. White arrows indicate normal areas and black arrows indicate affected areas in vertebrae. Images were taken using a Nikon SMZ1500 stereomicroscope and a Nikon D5200 digital camera. Scale bar 2 mm. (b) mRNA expression levels of TRAF6 in bone tissue in 60 dpf wild-type zebrafish (n = 4 WT fish). Bar graphs show the means ± SEM.
Fig. S4: Data represents the list of primers used for the qRT-PCR mRNA analysis of human prostate cancer cell line (PC3U) and zebrafish experiments.