Abstract
Systemic lupus erythematosus (SLE) is characterized by the expansion of extrafollicular pathogenic B cells derived from newly activated naïve cells. Although these cells express distinct markers, their epigenetic architecture and how it contributes to SLE remains poorly understood. To address this, we determined the DNA methylomes, chromatin accessibility and transcriptomes from five human B cell subsets, including a newly defined effector B cell subset from SLE and healthy subjects. Our data define a differentiation hierarchy between the subsets and elucidate the epigenetic and transcriptional differences between effector and memory B cells. Importantly, an SLE molecular signature was already established in resting naïve cells and was dominated by accessible chromatin enriched in AP-1 and EGR transcription factor motifs. Together, these factors acted in synergy with T-BET to shape the epigenome of expanded SLE effector B cell subsets. Thus, our data define the molecular foundation of pathogenic B cell dysfunction in SLE.
INTRODUCTION
Systemic lupus erythematosus (SLE) is characterized by the production of autoantibodies, placing B cells centrally in SLE etiology. Autoreactive B cells are censored through tolerance checkpoints that can be overcome by the convergence of Toll-like receptor (TLR), cytokine, and/or co-receptor malfunction thereby leading to the expansion of pathogenic B cells1. GWAS SLE studies reveal a striking concentration of disease susceptibility alleles in B cell antigen receptor (BCR) signaling and B cell co-stimulation pathways2, 3. Experimental evidence supports a role for both germinal center reactions and extrafollicular B cell activation and differentiation pathways in the generation of autoantibodies and autoimmunity in mice4. We showed previously that SLE is characterized by the expansion of naïve B cells with an activated phenotype5 and a distinct subset of isotype switched B cells that harbor significant somatic hypermutation but lack CD27, a universal marker of memory B cells, termed DN2 (CD27–CD11c+T-BET+CXCR5–) B cells6. DN2 cells are poised to differentiate into antibody secreting cells (ASCs) through unregulated TLR7 and interleukin 21 (IL-21) stimulation6 and bear resemblance to murine age-associated B cells (ABCs). ABCs require TLR7 signaling for expansion, express CD11c and T-BET, and are enriched in BCR encoding autoantibodies7, 8, 9. TLR signaling can be modulated by type-I interferon (IFN)10, which is prevalent in SLE and may epigenetically influence disease flares11, 12. The strong linkage of DN2 cells and related subsets to TLR-driven pathways suggests that a unique combination of stimuli influences the fate of autoreactive B cells.
Altered epigenetic states, including DNA hypomethylation in B cells11 and chromatin accessibility of naïve B cells13, have been described for SLE and other autoimmune diseases14, 15, suggesting that epigenetic differences may influence B cell responses. Therefore, we examined and compared the DNA methylation epigenome, chromatin accessibility, and transcriptome of human B cells from HC and SLE subjects that represented resting, activated, and memory compartments. A hierarchy in B cell differentiation and multiple SLE disease signatures already manifested in naïve B cells13 were found to persist throughout B cell differentiation. Transcription factor networks altered in SLE converged on signaling networks and revealed external environmental cues that contribute to expansion of pathogenic B cell subsets. Thus, the SLE environment predisposes B cells to a pathogenic phenotype that is epigenetically propagated through B cell differentiation and primes extrafollicular naïve B cell differentiation into ASCs.
RESULTS
Molecular relationships of B cell subsets in SLE and HC
To characterize the epigenetic relationships among human B cell populations in SLE subjects, a cohort of nine SLE and twelve healthy control (HC) African American females were recruited (Supplementary Table 1). Peripheral human B cells can be divided into subtypes that represent naive, activated stages, and isotype-switched memory cells5, 16, 17. Five circulating human B cell subsets were isolated by flow cytometry from each subject using MitoTracker Green dye (MTG) and the following phenotypic markers5, 17: resting naïve (rN, CD19+IgD+CD27–MTG–CD24+CD38+), transitional 3 (T3, CD19+IgD+CD27–MTG+CD24mid/+CD38–), activated naïve (aN, CD19+IgD+CD27–MTG+CD24–CD38–), switched memory (SM, CD19+IgD–CD27+), and double negative (DN2, CD19+IgD–CD27–CXCR5–)(Supplementary Fig. 1). For comparison, circulating ASCs were also isolated (CD19+IgD–CD27+CD38++) from a subset of subjects. The DN2 and aN populations, both characterized by a CXCR5− phenotype, are expanded in SLE subjects experiencing flares and potential disease drivers5, 6. Consistent with results from previous cohorts5, 6, SLE subjects had highly significant expansions of aN and DN2 B cells, smaller but significant increases in T3, and a subtle decrease in rN B cells (Supplementary Fig. 1).
An integrated approach was taken that assayed the DNA methylation status by reduced representation bisulfite sequencing (RRBS)18 and the effect of epigenetic programs to influence the accessibility of chromatin as determined by ATAC-seq13, 19 (Supplementary Fig. 2). The phenotypic readout of the epigenetic programs was assessed by RNA-seq. Differentially methylated loci (DML), differentially accessible regions (DAR), and differentially expressed genes (DEG) were determined between each cell type and SLE disease status. Principal component analysis (PCA) of all DML indicated a linear separation of cell types that progressed from rN to SM B cells in both SLE and HC (Fig. 1a). One major epigenetic feature of B cell differentiation is the targeted and progressive global hypomethylation that occurs as B cells differentiate to ASCs18, 20, suggesting the changes observed by PCA may represent a differentiation hierarchy. Indeed, we observed the same phenomena with a clear progressive loss of DNA methylation across these cell subsets (Fig. 1b). A phylogenetic analysis was performed using the DML between all cell types including ASCs, such that the beginning and differentiation end points were represented. For both SLE and HC, aN and T3 B cells were much more similar to rN; whereas DN2 and SM B cells were closely related to ASCs (Fig. 1c). Thus, DNA methylation changes separate the various cell subsets and is indicative of a hierarchical relationship.
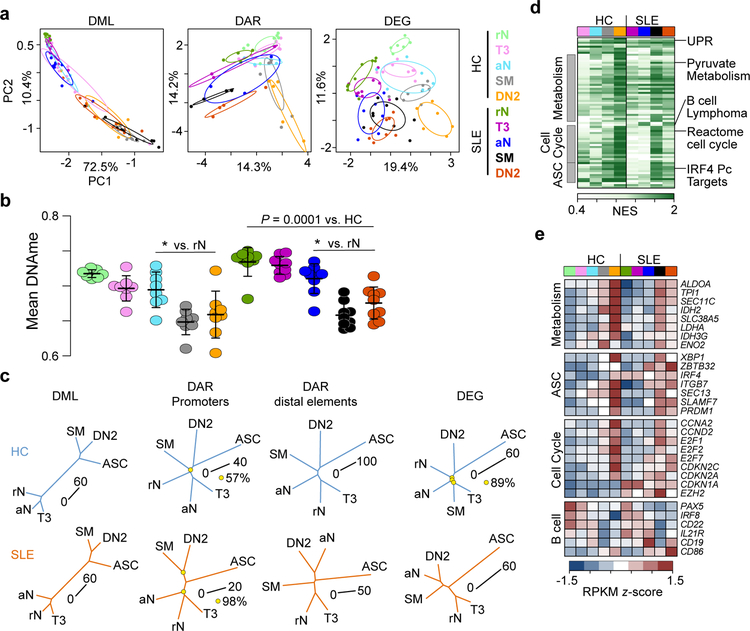
Figure 1. Epigenetic states of B cell subsets identify cell type relationships and differentiation hierarchies.
(a) Principal component analysis (PCA) of differentially methylated loci (DML)(left), differentially accessible loci (DAR)(middle), and differentially expressed genes (DEG)(right) between cell types. Each cell type is indicated by a point and 99% confidence intervals for each cell subset are denoted by a circle. Sample sizes for each cell type can be found in Supplementary Table 5. (b) Mean CpG methylation for each cell type is plotted. Data represent mean ±SD. Significance determined by two-way ANOVA with Tukey’s post-hoc test. * indicates P < 0.05 for the indicated comparison to rN. (c) Phylogenetic dendrograms for HC (top) and SLE (bottom) subsets for the indicated data. Length of tree branches between cell types represents Euclidean distance, which is denoted for each tree with a scale bar. Reproducibility of tree structure was tested using a bootstrapping analysis and nodes that were less than 100% reproducible are indicated with a yellow circle. Data were averaged for each cell type. (d) Heatmap of normalized enrichment score (NES) calculated by GSEA for pathways upregulated in cell types compared to rN B cells. HC and SLE B cells are separated and a subset of gene sets are grouped into common pathways. ASC, antibody-secreting cell. See also Supplementary Fig. 3. (e) Heatmap of z-score normalized RPKM expression for selected genes from enriched GSEA gene sets. Data represent the mean expression for each cell type.
PCA of DAR and DEG indicated that there was a progression from rN through T3, aN, and SM to DN2 B cells. Whereas the DNA methylation analysis separated the cell types along both principal components, chromatin accessibility and gene expression PCA identified SLE B cells as distinct from HC with PC1 separating disease and PC2 separating cell types (Fig. 1a). For phylogenetic analysis of chromatin accessibility data, promoters and distal elements were separated as the latter has been shown to better separate cell types21, 22. Accessible promoters had a small overall phylogenetic distance between cell types in SLE and HC, indicating a similar promoter accessibility architecture, although the SLE DN2 and SM were closer than in HC (Fig. 1c). In contrast, accessibility at distal elements displayed an expanded phylogenetic distance, indicating cis-elements such as enhancers were distinct between SLE and HC cell types. Similar to previously reported data5, 6, analysis of the RNA-seq, ATAC-seq, and RRBS data indicated that DN2 cells were most closely related to ASCs by phylogenetic tree position and distance (Fig. 1c). As with the accessibility data, the transcriptomes of DN2 and aN cells showed a much closer relationship in SLE than in HC, a finding that was supported by fewer DAR and DEG in SLE than HC (Supplementary Fig. 3). Thus, distinct epigenomes denote critical SLE and HC B cell subsets.
Progressive upregulation of B cell differentiation transcriptional program
To better understand the phenotypic relationships of the cell subsets, the transcriptional programs were analyzed using gene set enrichment analysis (GSEA)23 comparing HC and SLE B cell subsets to their respective rN B cells. Across cell subsets, a progressive enrichment was observed in gene sets associated with metabolism (e.g., ALDOA), cell cycle (e.g., E2F1), unfolded protein response (UPR)(e.g., XBP1), activated B cells (e.g., ZBTB32)24, and genes expressed in plasma cells (e.g., PRDM1, SLAMF7) (Fig. 1d–e, Supplementary Fig. 3). The overall expression levels of the gene sets increased in T3 and aN and peaked in SM and DN2 B cells. For example, consistent with its transcriptional upregulation, the PRDM1 promoter demonstrated gains in accessibility and a decrease in DNA methylation that was most pronounced in DN2 and SM B cells. Additionally, transcriptional activity, as well as promoter and distal element accessibility progressively increased at the SLAMF7 and XBP1 loci. Together, these data provide epigenetic evidence for a progressive upregulation of genes associated with B cell activation and plasma cell differentiation and a differentiation hierarchy among the B cell subsets.
rN B cells are epigenetically distinct in SLE
As expected, both HC and SLE rN B cells express many hallmark B cell genes such as PAX5, IRF8, CD22, IL21R, and CD19, and lacked activation markers such as CD86 (Fig. 1d,e, Supplementary Fig. 3), suggesting that core features of B cell development were not grossly altered. In healthy immune systems, resting naïve cells represent the earliest unperturbed mature B cells available to mount primary immune responses. However, we previously reported that SLE rN B cells contained distinct differences in accessible chromatin compared to HC13, indicating that the unique environment in SLE may alter the molecular machinery and properties of B cells conventionally defined as resting naïve cells. Comparison of the epigenetic and transcriptional states of HC and SLE rN B cells revealed clear differences in each of the assays (Fig. 2a). Correlation of the DNA methylation changes with gene expression revealed a group of genes that were upregulated and demethylated in SLE B cells, including IFI44, PDCD1, and SPRY2 (Fig. 2b). Additionally, a subset of genes was upregulated and contained DAR that increased in accessibility in SLE rN and included IFI44, STAT4, FOSL2, NR4A1, NR4A3, and SPRY2 (Fig. 2b–d, Supplementary Fig. 4). IFI44 is induced in SLE and in response to IFN12. NR4A3 and NR4A1 (Nur77) are induced in response to TLR stimulation25 and signaling through the BCR26 respectively, suggesting that SLE rN B cells may have received stimulation through these pathways. SPRY2 is a negative regulator of receptor tyrosine kinase (RTK) signaling27. Thus, these data indicate that rN B cells in SLE are epigenetically primed and distinct from their HC counterpart.
Figure 2. rN B cells are epigenetically distinct in SLE subjects.
(a) Volcano plot of DML (left), DEG (middle), and DAR (right) between SLE and HC rN B cells. The number of differential features is indicated. DML represent CpG with >20% change and FDR < 0.05 as determined by DSS and DEG and DAR represent features with >=2-fold change and FDR < 0.05 as determined by edgeR. (b) Scatter plot of the correlation of DML vs. DEG (left) and DAR vs. DEG (right) for the data in a. (c) Bar plot of gene expression levels for the indicated gene. Data represent mean ±SD, * indicates DEG between SLE and HC (>=2-fold change and FDR < 0.05) as determined by edgeR. (d) Genome plot showing the average accessibility levels in each cell type at the indicated loci. Data represent the mean for each cell type from one experiment. DAR between HC and SLE rN B cells are highlighted with a box. See also Supplementary Fig. 4.
Intriguingly, differences observed in SLE rN B cells were also present in all the downstream B cell subsets (Fig. 2c,d), suggesting that the SLE epigenetic B cell program is established as early as the rN stage. Notably however, transcriptional upregulation of other gene sets at significant levels was only initiated at the aN stage and did not differ from those of HC. This dichotomy is illustrated by two conventional activation markers, with CD69 already highly overexpressed by SLE rN cells and CD86 becoming upregulated in aN B cells and peaking in DN2 cells (Supplementary Fig. 3). The second pattern was also found for other immunological relevant genes, including TBX21 (T-BET), ITGAX (CD11c), AICDA (Supplementary Fig. 5) and PDCD1 (PD-1) (as described below), with the latter two also suggesting BCR engagement. This pattern reflects a very high degree of overall transcriptional similarity between aN and DN2 cells. Together, these data suggest that there are unique and common disease-specific features that may impact cellular differentiation in SLE.
DNA methylation stratifies SLE and HC B cells
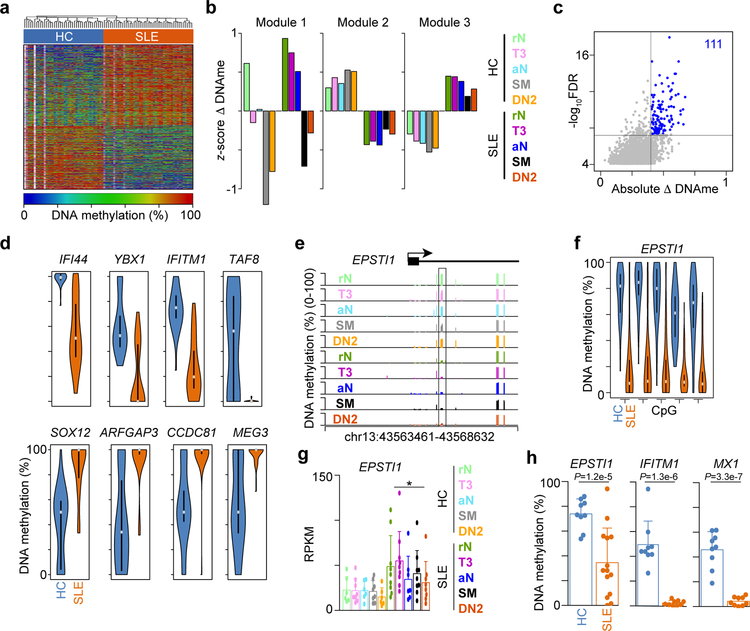
Analysis of DNA methylation profiles identified a distinct molecular SLE disease signature consisting of 6,664 DML that stratified all HC and SLE samples (Fig. 3a). DML were classified into three distinct modules by K-means clustering. CpG Module 1 lost DNA methylation as the cells progressed through differentiation but were hypermethylated in all SLE B cells subsets compared to their HC counterparts (Fig. 3b). The mean CpG methylation for each cell type was quantitated and revealed that SLE B cells were hypermethylated compared to HC. CpG Modules 2 and 3 revealed a subset of DML that were hypo- and hypermethylated, respectively, in all SLE B cells. Because DNA methylation may provide a useful disease biomarker, we identified the CpGs in Modules 2 and 3 that best stratified SLE from HC regardless of a patient’s age, which is known to affect DNA methylation28. Using a minimum DNA methylation change of 40%, we identified 111 CpG that stratified SLE and included hypomethylated sites near the IFI44, IFITM1, YBX1, and TAF8 genes and hypermethylated CpG surrounding SOX12, ARFGAP3, CCDC81, and MEG3 (Fig. 3c,d, Supplementary Table 2). EPSTI1, which encodes a positive regulator of NF-κB signaling29, that is induced by IFN and is associated with SLE susceptibility30, has a highly predictive cluster of five CpGs in its first intron (Fig. 3e,f). Each of the CpGs was hypomethylated and EPSTI1 was upregulated in all of the SLE B cell subsets (Fig. 3g). To confirm these findings, we performed a qPCR quantitation assay18 on rN B cells from an independent cohort and found that CpG at the EPSTI1, IFITM1, and MX1 loci were consistently demethylated in SLE subjects compared to HC (Fig. 3h). Thus, clear differences in the DNA methylation patterns of SLE and HC B cells exist and identify DML that could be used as potential SLE biomarkers.
Figure 3. DNA methylation status stratifies HC and SLE B cell subsets.
(a) Heatmap of percentage change in DNA methylation at 6,664 DML shared across all B cell subsets. White represent CpG with no coverage in a particular sample. (b) DML in a were K-means clustered into three modules. Bar plot showing the average DNA methylation for each cell type in each module. (c) Significance test for DML in modules 2 and 3 from b identifying the DML that best stratify HC (n=43) and SLE (n=49) cell types. Scatter plot showing 111 DML with FDR < 1x10−6 and absolute DNA methylation change greater than 40%. Significance determined by two-sided t-test followed by Benjamini-Hochberg FDR correction. (d) Violin plots of the percent DNA methylation across all B cell subsets for HC (n=43) and SLE (n=49). Grey dots represent mean and filled boxes represent 1st and 3rd quartile ranges. (e) Genome plot of the EPSTI1 locus showing the location of CpG covered by RRBS. Significant DML between HC and SLE are boxed. Data represent the mean for each cell type from one experiment. (f) Violin plot of 5 CpG highlighted in e across all B cell subsets for HC (n=43) and SLE (n=49). Grey dots represent mean and filled boxes represent 1st and 3rd quartile ranges. (g) Bar plot of gene expression levels for EPSTI1. Data represent mean ±SD. * indicates DEG between SLE and HC (>=2-fold change and FDR < 0.05) as determined by edgeR. (h) qPCR quantitation of DNA methylation levels in rN B cells from an independent cohort of HC (n = 9) and SLE (n = 9) subjects. Significance determined by two-tailed Student’s t-test.
SLE DN2 B cells have a distinct chromatin landscape from HC and SM B cells
The DNA methylation phylogenetic analysis revealed that DN2 and SM B cells were in a similar position with respect to their differentiation state/hierarchy. However, the PCA and phylogenetic analyses of accessible chromatin and gene expression datasets indicated that distinct differences existed between DN2 and SM. A comparison of DAR revealed thousands of loci that were distinct between DN2 and SM B cells in both SLE and HC (Fig. 4a). Annotation of DAR to genes revealed that ~50% of accessible chromatin in DN2 cells was shared between HC and SLE subjects with both HC and SLE displaying uniquely accessible gene promoters or regulatory regions (Supplementary Table 3). Irrespective of disease, a highly significant correlation was observed between DAR and DEG for SM and DN2 B cells (Fig. 4b), suggesting that events that generate DAR may influence gene expression. Moreover, direct comparison of SLE and HC DN2 revealed significant numbers of DAR and DEG (Fig. 4c). Therefore, SLE DN2 are distinct from HC DN2 and SM B cells.
Figure 4. SLE DN2 B cells have a chromatin conformation driven by TLR and RTK signaling pathways.
(a) Volcano plots of the DAR between DN2 and SM cell types for HC (top) and SLE (bottom) subjects. DN2 DAR were annotated to the nearest gene and the overlap of HC and SLE is shown as a Venn diagram. DAR represent features with >=2-fold change and FDR < 0.05 as determined by edgeR. (b) Correlation of DAR from a and DEG for the same comparisons for HC (top) and SLE (bottom) subjects. Genes with positive correlation between DAR and DEG (top right quadrant) that overlap between HC and SLE is shown as a Venn diagram. Significance determined by one-way ANOVA. (c) Volcano plot of DAR (top) and DEG (bottom) comparing SLE vs. HC DN2. The number of differential features is indicated. DAR represent features with >=2-fold change and FDR < 0.05 as determined by edgeR. (d) Bar plot of top enriched Gene Ontology Biological Process processes for HC-specific, shared, and SLE-specific genes from Venn diagram in a. Significance determined by Fisher’s exact test. (e) Bar plot of gene expression levels for PDCD1. Data represent mean ±SD. * indicates DEG between SLE and HC (>=2-fold change and FDR < 0.05) as determined by edgeR. (f) Genome plot of the PDCD1 locus showing the accessibility levels for each cell type. DAR between DN2 and SM are highlighted. Data represent the mean for each cell type from one experiment. (g) Flow cytometry analysis of percentage of PD-1+ cells for the indicated cell type. Data represent mean ±SD from 4 SLE subjects performed once. Significance determined by two-tailed Student’s t-test. See also Supplementary Fig. 5.
Compared to SM (HC and SLE), DN2 B cell gene ontology (GO) functional annotation revealed accessibility patterns surrounding genes involved in signaling from the TCR/BCR, costimulatory molecules, as well as response to lipopolysaccharide (Fig. 4d). As described for ABCs7, 8 and consistent with our initial functional and transcriptional results6, these data suggest that TLR stimulation may be unique to the differentiation history of DN2 cells, a contention supported by the significantly lower expression TRAF5, a negative regulator of TLR activation in B cells, including in DN2 cells6, 31. Additional genes associated with ABCs that were unique to both SLE and HC DN2 and aN cells (compared to SM) included TBX21, AICDA, and GAS7 (Supplementary Fig. 5). We further characterized the similarity of DN2 B cells with previously published datasets of ABCs8, 32. Both HC and SLE DN2 B cells displayed a significant enrichment of multiple ABC gene signatures8, 32 (Supplementary Fig. 6). However, no significant enrichment of ABC signatures was observed comparing SLE and HC DN2, indicating the signature was common to DN2 B cells irrespective of disease status. DN2 B cells also contained unique accessibility patterns at the TNF receptor TNFRSF1B and ZAP70, encoding a Src family protein tyrosine kinase that enhances BCR signaling and which is highly expressed in chronic lymphocytic leukemia subjects with poor clinical outcome33. HC DN2 cells were uniquely enriched for distinct signaling pathways, transcriptional repression, and regulation of the cell cycle (Fig. 4d, Supplementary Fig. 3).
SLE DN2 B cells, compared to SM, were specifically enriched for genes associated with tyrosine kinase signaling (Fig. 4d). For example, FGFR1, which is upregulated in SLE DN2, contains accessible chromatin peaks in DN2 B cells compared to SM (Supplementary Fig. 5). The phosphorylation of specific tyrosine residues on TLRs, by growth factor receptors such as EGFR, modulates downstream signaling events34, 35, suggesting that similar activation of FGFR1 or other tyrosine kinases, may enhance TLR signaling in SLE B cells. ITGAX (CD11c), a marker associated with ABCs8, was also highly upregulated in SLE DN2 B cells and contains unique accessible regulatory regions. Additionally, the PDCD1 promoter and cis-regulatory elements were highly accessible in DN2 cells compared to SM and was upregulated at both the mRNA and protein levels in SLE B cell subsets (Fig. 4e–g). Supportive of the phylogenetic analysis, highly similar patterns of gene expression and accessibility were observed for DN2 and aN B cells in SLE. For example, GAS7 is only expressed in aN and DN2 B cells. These data suggest a common epigenetic programming relationship between these cell subsets and a mechanistic link for their prominent expansions in active SLE.
DN2 accessibility is programmed by T-BET, AP-1, and EGR transcription factors
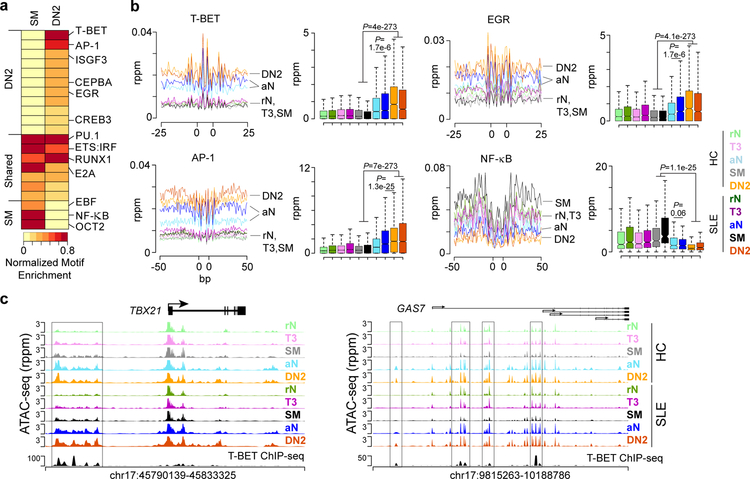
To identify the transcription factor networks driving DN2 and SM accessibility patterns, the DAR were searched for enriched transcription factor binding motifs. Enriched motifs were grouped, revealing common and unique sets of motifs for DN2 and SM accessible loci. Shared motifs included lineage factors important for B cell identity such as PU.1, RUNX1, E2A, and the ETS:IRF composite motif (Fig. 5a). The top enriched motifs in DN2 B cells were T-BET, AP-1, ISGF3, and EGR family factors. T-BET and AP-1 motifs were found to be enriched in the accessible chromatin of ABCs that accumulate in SWEF family-deficient mice that develop systemic autoimmunity9. Conversely, SM loci were uniquely enriched for EBF, NF-κB, and OCT motifs.
Figure 5. DN2 accessibility is driven by T-BET, AP-1 and EGR transcription factors.
(a) Heatmap depicting normalized enrichment of transcription factor motifs in DN2 and SM cell types. Enrichment P-values are normalized to the minimum value for each cell type. Motif grouping is indicated on the left. Significance determined by binomial distribution by HOMER. (b) Histogram (left) and box plot (right) of accessibility at the indicated motif and for the indicated surrounding sequence for each cell types. Data represent the mean for each cell type. The location of cell types is indicated for the histograms. Boxplot center line indicates data median, lower and upper bounds of boxes the 1st and 3rd quartile ranges, and whiskers the upper and lower ranges of the data. Significance determined by two-tailed Student’s t-test. See also Supplementary Fig. 7. (c) Genome plot showing accessibility and T-BET binding in GM12878 B cells36 for the indicated loci. DAR between DN2 and SM with T-BET binding peaks are highlighted. Data represent the mean for each cell type from one experiment.
Compared to all other cell subsets, DN2 B cells had the highest levels of accessibility at T-BET, EGR, and AP-1 motifs (Fig. 5b, Supplementary Fig. 7). Similar to the findings above, aN B cells were also highly accessible at these motifs with SLE aN displaying higher accessibility. For each of the three motifs, minimal accessibility was observed in T3, rN and SM. In contrast, NF-κB motifs displayed the highest levels of accessibility in SM cells and lowest in aN and DN2. T-BET binding profiles from human B cells36 displayed enrichment at loci accessible in aN and DN2 cells at the GAS7, TNFRSF1B, ITGAX (CD11c), and ZAP70 loci (Fig. 5c, Supplementary Fig. 5). Interestingly the TBX21 locus, which encodes T-BET, also contained an upstream distal element that was bound by T-BET and showed enhanced accessibility in aN and DN2 cells. Thus, SM and DN2 cells express distinct transcription factors that correlate with unique accessibility patterns, transcription factor networks, and gene expression programs.
Coordination of SLE transcriptome and transcription factor signatures
For genes such as STAT4, a major SLE risk allele2, and IFI44, the expression and accessibility differences observed in SLE versus HC rN B cells were also shared among the other cell subsets, indicating the presence of a common SLE disease signature. Indeed, 5,090 DEG were identified across all cell types (Fig. 6a). GSEA revealed pathways upregulated in all SLE B cells including inflammatory, IFN-γ, and IFN-α response gene sets previously identified in SLE12 (Fig. 6b, Supplementary Fig. 8). Additionally, we identified the WNT, Notch, and estrogen signaling pathways, cytokine signaling from IL-6 and IL-2 receptors, and the p53 and apoptosis pathways as enriched in SLE B cells. By contrast, SLE DN2 cells showed negative enrichment for several pathways, including those driving UPR and G2M checkpoint.
Figure 6. SLE transcription factor networks correlate with disease-specific transcriptomes.
(a) Heatmap of 5,090 DEG that stratify SLE and HC cell types. DEG represent features with >=2-fold change and FDR < 0.05 as determined by edgeR. (b) GSEA plots for the indicated gene set showing the enrichment score for each SLE cell type compared to HC. See also Supplementary Fig. 8. (c) Volcano plot of DAR between all HC and SLE cell types. DAR represent features with >=2-fold change and FDR < 0.05 as determined by edgeR. (d) Heatmap of the log2 fold change (FC) of PageRank statistic for each cell type between SLE and HC. (e) Bar plot of gene expression levels for selected factors from d. Data for each gene is summarized for all HC and SLE cell types and is displayed as mean ±SD. * indicates DEG between SLE and HC (>=2-fold change and FDR < 0.05) as determined by edgeR. See also Supplementary Fig. 8. (f) qRT-PCR analysis of select genes in rN B cells from an independent cohort of HC (n = 15) and SLE (n = 8). Data represent mean ±SD and are normalized to percentage of ACTB expression. Significance determined by two-tailed Student’s t-test. (g) Network diagram depicting gene sets from b that are regulated by EGR factors. Line thickness is scaled to significance as determined by Fisher’s Exact test. See also Supplementary Fig. 8.
Analysis of the SLE disease signature manifested in the accessible chromatin landscape identified thousands of SLE-specific DAR (Fig. 6c). The PageRank algorithm37 was used to identify transcription factor motifs that correlated with changes in gene expression at target genes and determine the overall importance of each transcription factor to the SLE transcriptional network. PageRank analysis identified 31 transcription factors that were enriched in three or more SLE B cell subsets compared to their HC counterparts (Fig. 6d). Genes encoding these factors were upregulated in SLE cells (e.g., EGR4, AHR, JUN, JUND, FOSL1) (Fig. 6e, Supplementary Fig. 8). However, some factors like BCL6 were expressed at the same levels irrespective of disease, indicating that they may be recruited to unique targets genes in SLE versus HC B cells.
One of the highest scoring transcription factors was EGR4. EGR factors are induced in response to multiple cellular stimuli and have been implicated in autoimmunity, including SLE38. RNA-seq suggested that all four members of the EGR family were upregulated in SLE B cells (Supplementary Fig. 8). To validate these findings, we performed qRT-PCR for each of the EGR family members in rN B cells from an independent cohort of SLE and HC subjects. Here, we observed significant upregulation of EGR1 and EGR2 in SLE rN B cells and minimal expression of EGR3 and EGR4 (Fig. 6f), findings that were consistent with other cohorts6. Moreover, we analyzed ENCODE ChIP-seq data36 for EGR1 and found a significant overlap of EGR1 peaks with SLE DAR (Supplementary Fig. 7). To demonstrate the importance of EGR factors to the disease transcriptional network, the target genes for each family member were mapped to GSEA gene sets up in the DEG disease network above. Importantly, genes predicted by network analysis to be regulated by EGR factors were enriched in 19 of the 22 (86%) gene sets upregulated in SLE (Fig. 6b,g). The EGR factors also targeted other transcription factors, placing them at the apex of the disease network and further indicating the importance of these factors to the SLE transcriptional state. Analyzing the network for each of the factors showed a remarkable connectivity and overlap between the networks for EGR1 and ERG4 compared to the others, suggesting that the SLE transcriptional differences are, in part, driven by EGR transcription factors.
SLE DN2 B cells have activation of ATF3 response pathways
To specifically identify transcription factors and networks associated with the expanded DN2 cells in SLE, analyses of the PageRank enrichment in SLE DN2 cells versus the change in gene expression was performed and revealed a positive correlation between the datasets (Fig. 7a). As described above, all four EGR family members were upregulated and highly enriched in the SLE DN2 network. The next set of significant regulators included the ATF/CREB family of factors CREM, CHOP (DDIT3), and ATF3. ATF3 is induced in response BCR and TLR stimulation, cellular stress, and can mediate the responses of p53 and various hormone receptors39, 40, 41. ATF3 was upregulated in all SLE B cell subsets with maximal expression in SLE DN2 cells (Fig. 7b). To validate these findings, we quantitated ATF3 mRNA levels in rN B cells by qRT-PCR and found that ATF3 was significantly upregulated in SLE rN B cells compared to HC (Fig. 7c). Consistent with mRNA levels, ATF3 protein was more abundant in SLE rN, aN, SM, and DN2 subsets compared to HC (Fig. 7d,e). Using the predicted target genes from the PageRank analysis, we identified 98 ATF3 target genes that were disease DEG with 87% being upregulated in SLE (Fig. 7f, Supplementary Table 4). DNA footprinting analysis revealed a large difference at ATF3 binding motifs and at the surrounding chromatin in SLE B cells compared to HC (Fig. 7g). Quantitation of ATF3 motif accessibility in each cell subset identified SLE DN2 cells as having the highest levels of accessibility (Fig. 7h). Additionally, ATF3 peaks identified by ChIP-seq36 significantly overlapped SLE DAR (Supplementary Fig. 7). ATF3 heterodimerizes with Jun family members40, of which three (JUN, JUNB, JUND) were enriched in the DN2 networks and upregulated in SLE (Fig. 7a, Supplementary Fig. 8). Additionally, for these ATF3 motifs we did not find any co-occurrence with IRF4 in the AICE composite motif context42 and only 7 of the 98 genes (7%) were predicted to be regulated by IRF4 by PageRank. ATF3 target genes were mapped to functional gene sets enriched in SLE and found to have roles in cell cycle and metabolism through the MTORC1 pathway, G2M checkpoint, TNF signaling, Hypoxia, UPR, and Apoptosis (Fig. 7i). Thus, ATF3 and EGR and their respective family members regulate DN2 specific B cell transcriptional pathways important for SLE epigenetic programming.
Figure 7. SLE DN2 B cells display activation of ATF3-regulated stress response pathways.
(a) Scatter plot of PageRank fold change between SLE DN2 and HC DN2 versus the gene expression changes for the same comparison. (b) Bar plot of gene expression levels for ATF3 Data represent mean ±SD. * indicates DEG between SLE and HC (>=2-fold change and FDR < 0.05) as determined by edgeR. (c) qRT-PCR analysis of ATF3 in rN B cells from an independent cohort of HC (n = 15) and SLE (n = 8). Data represent mean ±SD and are normalized to percentage of ACTB expression. Significance determined by two-tailed Student’s t-test. (d) Flow cytometry of ATF3 expression compared to isotype control in a representative HC and SLE DN2 B cell sample. The experiment was performed once from a cohort of HC (n = 5) and SLE (n = 8) subjects. (e) ATF3 MFI normalized to isotype MFI for the indicated cell type for a cohort of HC and SLE subjects defined in e. Data represent mean ±SD. Significance determined by Wilcoxon rank sum test. (f) Heatmap of gene expression for 98 genes with ATF3 binding motifs determined by PageRank analysis. Data represent the mean for each cell type. (g) Histogram of ATF3 motif accessibility in HC and SLE cells types. The location of the ATF3 motif is indicated. (h) Box plot for each cell type (right) summarizing accessibility at ATF3 motifs enriched in SLE DN2 DAR. * P-value < 0.05 compared to all other cell types as determined by two-tailed Student’s t-test. Boxplot center line indicates data median, lower and upper bounds of boxes the 1st and 3rd quartile ranges, and whiskers the upper and lower ranges of the data. (i) Network diagram depicting gene sets from Fig. 6b that are regulated by ATF3. Line thickness is scaled to significance as determined by Fisher’s Exact test.
DISCUSSION
Here, we used an integrated transcriptional and epigenetic approach to unravel the nature of B cell populations, their differentiation hierarchy, and the molecular programs that underpin their abnormal behavior in SLE. We focused on finely discriminated subsets, representing the human extrafollicular naïve B cell differentiation pathway responsible for the generation of DN2 cells5, 6. Our analysis was performed on an African American cohort with high disease activity. Going forward it will be important to determine how these epigenetic programs evolve during the progression of SLE, in quiescent versus active disease, and whether this epigenetic imprint exists in other ethnic groups.
The data suggest a differentiation order with T3 and aN being derived from activation of rN cells with DN2 and SM cells placed further down the differentiation path to ASCs. T3 cells have been previously defined as transitional cells in human studies of repopulating B cells after Rituximab treatment43. Our study suggests that in active SLE T3 cells represent an early phase of naïve B cell activation. In SLE, consistent with their phenotypic similarity6, aN and DN2 cells shared features of accessible chromatin; were transcriptionally related to each other, yet distinct from the other three B cell subsets; and were closer to ASC than any other subset, including SM B cells. Combined, these data support the notion that DN2 B cells represent a distinct effector subset that is expanded in SLE. Our results are also consistent with separate ASC differentiation pathways derived from either aN-DN2 cells or SM cells.
Consistent with our initial findings13, rN B cells in SLE subjects are distinctly primed at an early differentiation stage. That some of the SLE transcriptional and epigenetic signatures were shared by all SLE B cell subsets demonstrates that SLE B cells are under the control of a unique transmissible epigenome that regulates the impact of the SLE environment on all mature B cells and modulates their activation threshold. Elucidating the earliest stage of development at which B cell precursors may be epigenetically modified in SLE could guide optimization of B cell therapies.
Changes in epigenetic modifications identified programs responsible for the abnormal behavior of individual B cell subsets in SLE and in particular, programs associated with extrafollicular effector cells. The accessible chromatin of DN2 B cells was specifically enriched for T-BET, EGR, and AP-1 transcription factor motifs. Our data provide insight into a T-BET-driven program that is shared by HC and SLE DN2 cells. T-BET was upregulated in DN2 B cells irrespective of disease, was enriched in DN2-specific accessible chromatin, and regulated other markers characteristic of ABCs and DN2, such as CD11c. Moreover, T-BET bound to its own gene, suggesting that it may function in an autoregulatory manner in these cells. This indicates that DN2 B cells are a normal product of B cell differentiation, but in abnormal immune environments, such as SLE, their formation is enhanced and/or their presence sustained, and they may harbor additional properties based on their unique disease specific transcriptome and epigenomes.
We previously postulated a developmental link between SLE aN, DN2, and ASCs6. This model is supported by the fact that aN and DN2 cells in SLE share distinct epigenetic and transcriptional profiles and have close relationships in the phylogenetic trees. This close relationship was mirrored for many genes, such as ITGAX, GAS7, and PDCD1. Additionally, the enhanced accessibility of core SLE AP-1 and EGR transcription factor binding motifs was present in aN cells (relative to rN and T3 cells), increased further in DN2 cells, and was largely absent in SM B cells. Thus, these data provide a mechanistic link between dysregulation of central SLE networks and transcription factors and the skewing of B cell differentiation towards the aN-DN2 cell fate.
An informative example of the SLE programs shared across B cell populations that are already engaged in resting naïve cells is illustrated by the NR4A family of nuclear receptors. NR4A1 is induced specifically by BCR stimulation26, while NR4A2 and NR4A3 are induced in response to TLR stimulation25. This is consistent with mouse lupus models induced by BCR and TLR co-stimulation leading to tolerance breakdown44 and our previous data showing that aN and DN2 B cells are hyper responsive to TLR7 stimulation and contain clonally expanded autoreactive B cells5, 6. Of note, DN2 cells demonstrated unique chromatin accessibility patterns surrounding BCR and TLR signaling components, suggesting that initial engagement of both these pathways in naïve B cells may be propagated to later stages of differentiation. These data fit a model of autoimmunity through which BCR and TLR signals can converge in extrafollicular settings to stimulate and expand autoreactive B cells4, 44, 45. Our data indicate that in SLE, rN cells are subjected to positive antigenic stimulation and therefore provide an important window to understand early antigenic triggers in SLE.
The expression of ATF/CREB, AP-1, and EGR family transcription factors was maximally enriched in the SLE DN2 subset. EGR proteins have been linked to autoimmunity, are induced by BCR stimulation46, and regulate plasma cell differentiation47, 48. EGR1 was highly induced in SLE in multiple cohorts and demonstrated connectivity to many dysregulated transcriptional networks. Similarly, ATF3 was highly enriched in the DN2 B cell network. Each of these factors was predicted to impact metabolism, cell cycle, apoptosis, and response to cellular stress. The upregulation of oxidative phosphorylation enhances plasma cell differentiation49. Interestingly, G2M checkpoint and apoptosis pathways were induced in all SLE B cell subsets except DN2 B cells, which might explain their expansion in SLE. These data suggest the balance between pathways that promote or oppose differentiation may be altered in SLE DN2 B cells. Mechanistically, this may occur by switching transcription factor partners. ATF3 for example, functions as a repressor when bound as a homodimer but can heterodimerize with JUN family members to function as an activator40, 41. The coordinated upregulation of JUN family members with ATF3 suggests a shifting equilibrium of ATF3 activity towards activation in SLE, particularly in the DN2 B cell subset. Moreover, the main T-BET inducer, IFN-γ, can synergize with TLR stimulation to enhance the expression of ATF350. Thus, the coordination of stimuli with known pathogenic significance may offer a mechanistic linkage between molecular and cellular abnormalities in SLE.
SLE B cell subsets contained higher overall levels of DNA methylation than their HC counterparts. This may reflect a more recent differentiation from a highly methylated progenitor B cell20. Alternatively, the SLE program may depend on the maintenance of methylation at the loci observed. Nonetheless, while the DNA methylation did not specifically identify disease pathways, the presence/absence of DNA methylation could serve as potential biomarkers of disease and disease progression as illustrated by 111 CpG that discriminated between SLE and HC B cells. While these differential DNA methylation modifications occurred in all B cell subsets, including rN B cells, it is not known when such modifications are first imprinted into the SLE epigenome and may represent an epigenetic record of differentiation history and disease state.
In summary, we have defined the transcriptomes and epigenomes of B cell subsets that are expanded in SLE. Our data indicate a disease signature across all cell subsets, and importantly on mature resting B cells, suggesting that such cells may have been exposed to disease inducing signals. Together, these data define the imprint of SLE on the epigenome of specific B cell populations and highlight transcription factor networks and programming of normal and pathogenic B cells subsets.
METHODS
Human subjects
Four independent cohorts were used for this study and are detailed in Supplementary Table 1. Whole blood samples were obtained with informed consent from all participants in accordance with Emory University School of Medicine Institutional Review Board protocols.
FACS Isolation of human B cell subsets
50–100 ml of whole blood were isolated from each subject via venous puncture utilizing cell preparation tubes (CPT) containing sodium heparin and Ficoll-Hypaque solution (Becton Dickinson). Washed PBMCs were resuspended in 1 ml of pre-warmed complete media (RPMI 1640, 10% FBS, and 1% L-glutamine) containing 20 nM of MitoTracker Green (MTG) and incubated at 37°C in 5% CO2 for 30 min. Next, PBMCs were centrifuged at 1,300 RPM for 10 min at RT, resuspended in 10 ml of pre-warmed complete media, and incubated at 37°C in 5% CO2 for 30 min to allow for excess MTG to be pumped out of B cells that possess the ABCB1 transporter17. PBMCs were centrifuged at 1,300 RPM for 10 min at 4°C and resuspended at 107 PBMCs/100 μl in PBS with 0.5% BSA, 5% normal mouse serum, and 5% normal rat serum containing the following fluorochrome conjugated anti-human monoclonal antibodies: CD3, CD24 (Invitrogen), CD19, IgD, CD27, and CD38 (BD Biosciences). Cells were incubated in the dark for 30 min at 4°C, washed with 5 ml of PBS, and centrifuged at 1,300 RPM for 10 min at 4°C. PBMCs were resuspended in 1 ml of PBS and subjected to LIVE/DEAD Aqua staining (Invitrogen) according to the manufacturer’s protocol. Following LIVE/DEAD incubation, PBMCs were washed in 5 ml of PBS containing 0.5% BSA and centrifuged at 1,300 RPM for 10 min at 4°C. Pelleted PBMCS were resuspended in 1 ml of pre-warmed complete media and filtered using a pre-wetted 35 μm cell strainer FACS tube (Corning). B cell subsets were purified using a FACS Aria II. To ensure sort purity, the FACS Aria II was calibrated to 99.9% purity using fluorescent beads prior to each sort.
Quantitation of PD-1 and ATF3 expression
PBMCs were isolated as described above and stained with the following fluorochrome-conjugated antibodies: CD19, CD27, CD38, IgD (BD Biosciences), CD11c, CD3, CXCR5, and CD279 (PD-1) (BioLegend) in the presence of 10% normal mouse serum. For intracellular staining, cells were fixed, permeabilized and stained using the Tru-nuclear Transcription Buffer set (BioLegend). Cells were blocked with 50 μg/ml Rabbit IgG and stained with 2 μg/ml rabbit anti-human ATF3 (USBIO) or Rabbit IgG isotype control (Invitrogen). Single color stained anti-mouse Ig beads (Bangs) were used as compensation controls. Flow cytometry were collected with BD FACSDiva v6.2 and data analysis was performed FlowJo (TreeStar) v10.4.
RNA-seq
RNA from sorted cell populations was isolated using the AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer’s instructions. 50 pg of total RNA was used as input for the SMART-seq v3 cDNA synthesis kit (Takara) using 10 cycles of PCR amplification. 1 ng of cDNA was used as input for the NexteraXT kit (Illumina) using 9 cycles of PCR amplification. Final libraries were quantitated by qPCR and sized distribution determined by Bioanalyzer prior to pooling and sequencing on a HiSeq2500 using 50bp PE chemistry.
RNA-seq data analysis
Raw fastq files were mapped to the hg19 version of the human genome using Tophat2 v2.0.1351 with the default parameters and the UCSC KnownGene reference transcriptome52. Duplicate reads were removed with PICARD v1.127 (http://broadinstitute.github.io/picard/). For all unique ENTREZ genes, the coverage at each exon was summarized using custom R/Bioconductor scripts and the GenomicRanges v1.22.453 package and normalized to reads per kilobase per million reads (RPKM). Genes were filtered for detection based on being expressed at greater than 3 reads per million (rpm) in all samples for at least one group (i.e., HC rN or SLE DN2). Differential gene expression for detected genes was determined using edgeR v3.18.154 and a generalized linear model. To determine disease differences covariates included patient and cell type, for cell type differences covariates included disease status and patient. Genes with a greater than 2-fold change and FDR < 0.05 between comparisons were termed significant. To generate heatmaps the RPKM values for all detected genes were quantile normalized and then select genes for plotting were z-score normalized. For GSEA all detected genes were ranked by multiplying the direction of the fold change (i.e., positive or negative) by the -log10 of the P-value from edgeR. The resulting list was used as input for the PreRanked analysis in GSEA v3.0.
ATAC-seq
ATAC-seq13 was performed by resuspending 1,000 to 20,000 FACS sorted cells in 50 μl nuclei lysis buffer (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and centrifuged at 500g for 30 min at 4°C. Next, the supernatant was removed and cells resuspended in 25 μl tagmentation mix (300 mM NaCl, 100 mM EDTA, 0.6% SDS, 1.6 μg Proteinase-K) and placed in a thermocycler at 37°C for 1 h. Cells were lysed and high molecular weight DNA removed using a 0.6× SPRI-bead negative selection and low molecular weight DNA purified by 1.2× SPRI-bead positive selection. The resulting tagmented DNA was PCR amplified and indexed using Nextera Indexing primers (Illumina, Inc) and 2× HiFi Hotstart Ready Mix (Roche, Inc). Following PCR, high molecular weight DNA was removed using a 0.3× SPRI-bead negative selection and low molecular weight DNA purified by 1× SPRI-bead positive selection. Final libraries were checked for ATAC-seq specific patterning on a Bioanlyzer and quantitated by qPCR prior to pooling at equimolar ratios and sequenced on a HiSeq2500 using 50bp PE chemistry.
ATAC-seq data analysis
Raw fastq files were mapped to the hg19 version of the human genome using Bowtie v1.1.155 with the default parameters and duplicate reads removed with PICARD v1.127 (http://broadinstitute.github.io/picard/). Accessible peaks of enrichment were called using MACS2 v2.156 with the default parameters. A unified set of peaks found in 1 or more samples was created using the mergePeaks.pl function in HOMER v4.8.257 and the raw read count for each peak annotated for each sample using the GenomicRanges v1.22.453 package. The resulting data was reads per peak per million (RPPM) normalized for the fraction of reads in peaks (FRiP)58 and sequencing depth as previously described13. Differentially accessible regions were identified using edgeR v3.18.154 and a generalized linear model. To determine disease differences covariates included patient and cell type, for cell type differences covariates included disease status and patient. Peaks with a greater than 2-fold change and FDR < 0.05 between comparisons were termed significant. To identify motifs enriched in DAR, the findMotifsGenome.pl function of HOMER v4.8.257 and the ‘de novo’ output used for downstream analysis. For the motif heatmap analysis the enrichment P-value for each motif was normalized to the maximum P-value in each cell type. To generate motif footprints the motifs occurring in peaks were annotated with the HOMER v4.8.257 annotatePeaks.pl function using the options ‘-size given’. Next, the read depth at the motif and surrounding sequence was computed using the GenomicRanges v1.22.453 package and custom scripts in R/Bioconductor and the coverage at each base plotted as a histogram or summarized as a boxplot. GO Biological Process enrichment analysis of DAR was performed using DAVID59. The resulting list of GO terms was filtered using REVIGO60 to generate a list of terms containing unique sets of genes.
Reduced representation bisulfite sequencing (RRBS)
DNA was isolated from the same fraction as RNA using the AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer’s instructions. A dual restriction enzyme RRBS assay18 was performed by splitting the DNA into two aliquots and digesting over night with either MspI or TaqI (NEB). The resulting DNA was purified, and digestion efficiency quantitated by qPCR using primers spanning sequences that contained one, both, or no restriction site. For bisulfite conversion controls, ΦX174 phage DNA (NEB) was in vitro methylated with M.SssI methylase (NEB). Methylated ΦX174 phage DNA and unmethylated lambda phage DNA (NEB) were sonicated to generate random 200–400 bp fragments. 10 pg (1:10,000 ratio of phage to genomic DNA) of each DNA were spiked into the purified digested genomic DNA to use as bisulfite conversion controls. Sequencing adapters were ligated on using the HyperPrep kit (KAPA Biosystems) and fully methylated short adapters61. Adapter ligated DNA was bisulfite converted using the EpiTect Bisulfite kit (Qiagen) according to the manufacturer’s protocol except the denaturation temperature was increased to 99°C for 10 min. Final libraries were PCR amplified with custom indexed primers and HiFi Uracil+ polymerase (KAPA Biosystems). The resulting RRBS library was quality checked by bioanalyzer, quantitated by qPCR, pooled at equimolar ratio, and sequenced on a HiSeq2500 using 50bp PE chemistry.
RRBS data analysis
Raw fastq files were mapped to a composite genome index containing hg19, ΦX174, and lambda phage genomes using Bismark v0.16.362. CpG calls were determined using the Rsamtools v1.22.0 and data.table v1.10.4.3 packages and custom R scripts as previously described18. All CpGs with at least 10× coverage per group (9,803,151) were determined and differential methylation analysis performed using the DSS v2.10.063 package and a generalized linear model. To determine disease differences, covariates included patient and cell type, for cell type differences covariates included disease status and patient. CpG with an FDR < 0.05 and a difference in methylation greater than 20% between comparisons were called significant. K-means clustering of the disease DML was performed using the Biganalytics package (https://CRAN.R-project.org/package=biganalytics). To determine predictive CpG from modules 2 and 3, we built a linear regression model at each CpG to find associations between SLE status and DNA methylation using a two-sided t-test with age as a covariate. The resulting P values were FDR corrected using the Benjamini-Hochberg method and CpG with a minimum change of 40% between HC and SLE and FDR < 5×10−8 were considered highly predictive.
Meta-analysis
For PCA and phylogenetic analysis, all significant differences (DML, DEG, DAR) between all cell types pairwise were selected and the data was z-score normalized. PCA was performed using the vegan v2.4–3 package in R/Bioconductor. Phylogenetic analysis was performed by computing a Euclidean distance matrix between samples which was clustered using the ‘hclust’ function in R/Bioconductor. Phylogenetic trees were plotted with APE v3.464 as ‘unrooted’ trees. The reproducibility of each phylogenetic tree was assessed by bootstrapping using the ‘boot.phylo’ function with 10,000 permutations. To correlate epigenetic and transcriptome datasets, DML and DAR were annotated to the nearest transcription start site using the annotatePeaks.pl function in HOMER v4.8.257. To cross reference data the ENTREZ ID for each gene was used. T-BET ChIP-seq data was downloaded from the ENCODE Consortium website under accession ENCFF483RCB36. The hg38 bigWig file was converted to hg19 using the ‘lift’ function of the bwtool v1.065. For GSEA analysis of ABC datasets the raw microarray data from GSE288878 and GSE8165032 was processed using oligo v1.40.266. Genes displaying a FDR < 0.05 and fold change > 4 for the indicated comparisons were used as genes sets. PageRank37 analysis of ATAC-seq and RNA-seq data was performed using the raw ATAC-seq data and normalized differential expression files comparing each SLE and HC cell type. The significance of transcription factor target genes within GSEA gene sets were calculated by Fisher’s exact test and network diagrams plotted using Cytoscape v3.4.0 scaling the network connections to the -log10 of the P-value.
qPCR DNA Methylation Analysis
DNA from sorted rN B cells was split into three equal aliquots and digested with the methyl-sensitive HpaII, the methyl-insensitive isoschizomer MspI, or mock digested. Equal portions of each reaction were subject to qPCR using primers covering target CpG that are detailed in Supplementary Table 6. DNA methylation was determined as the ratio of HpaII to mock digested signal and quantitated using a standard curve.
Quantitative RT-PCR Analysis
RNA from 1000 rN B cells was isolated using the Quick-RNA MicroPrep kit (Zymo Research) and the resulting RNA was used as input for the SMART-seq v4 cDNA synthesis kit (Takara) using 10 cycles of PCR amplification. cDNA was diluted 1:25 and Real-time PCR performed to quantitate the target gene expression using SybrGreen incorporation on a CFX96 Instrument (BioRad) with CFX Manager v3.1 software. Data were normalized to percentage of ACTB. All primers are detailed in Supplementary Table 6.
Statistics and Reproducibility
All statistical analyses were performed with R or Excel and P-values less than 0.05 were considered significant. Exact statistical tests are noted in the figure legends and Methods section for specific experiments and analyses. The statistical analysis of RNA-seq and ATAC-seq data was performed using edgeR54 and DSS for RRBS63. The sample sizes for each HC and SLE cell type profiled by RRBS, ATAC-seq, and RNA-seq can be found in Supplementary Fig. 2 and Supplementary Table 5. The genomics data represent one experiment from a single cohort. The expression of select genes and DNA methylation status at select CpG was validated in independent cohorts as noted.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Boss and Sanz laboratories for critical reading of the manuscript, the New York University Genome Technology Center for Illumina sequencing, the Yerkes Genomics Core for RNA-seq library preparation, the Emory Pediatrics Flow Cytometry core for FACS isolation of cell subsets, and the Emory Integrated Genetics and Computational Core for Bioanalyzer and sequencing library QC. This work was supported by NIH U19 AI110483 to J.M.B. and I.S., P01 AI125180 to I.S., E.H.L., and J.M.B., RO1 AI113021 to J.M.B., F31 AI112261 to B.G.B., and T32 GM008490 to J.M.B.
Footnotes
Data Availability
The data that support the findings of this study are available from the NCBI Gene Expression Omnibus (GEO) under accession GSE118256 and are detailed in Supplementary Table 5. Code and data processing scripts are available from the corresponding author upon request and at https://github.com/cdschar.
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Rawlings DJ, Metzler G, Wray-Dutra M & Jackson SW Altered B cell signalling in autoimmunity. Nat Rev Immunol 17, 421–436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langefeld CD et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun 8, 16021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung SA et al. Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS genetics 7, e1001323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.William J, Euler C, Christensen S & Shlomchik MJ Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science 297, 2066–2070 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Tipton CM et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol 16, 755–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenks SA et al. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 49, 725–739 e726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao Y, O’Neill P, Naradikian MS, Scholz JL & Cancro MP A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood 118, 1294–1304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubtsov AV et al. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c(+) B-cell population is important for the development of autoimmunity. Blood 118, 1305–1315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manni M et al. Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat Immunol 19, 407–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poovassery JS & Bishop GA Type I IFN receptor and the B cell antigen receptor regulate TLR7 responses via distinct molecular mechanisms. J Immunol 189, 1757–1764 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Ulff-Moller CJ et al. Twin DNA Methylation Profiling Reveals Flare-Dependent Interferon Signature and B Cell Promoter Hypermethylation in Systemic Lupus Erythematosus. Arthritis Rheumatol 70, 878–890 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Banchereau R et al. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell 165, 551–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scharer CD et al. ATAC-seq on biobanked specimens defines a unique chromatin accessibility structure in naive SLE B cells. Sci Rep 6, 27030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javierre BM et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 20, 170–179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewagama A & Richardson B The genetics and epigenetics of autoimmune diseases. J Autoimmun 33, 3–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanz I, Wei C, Lee FE & Anolik J Phenotypic and functional heterogeneity of human memory B cells. Semin Immunol 20, 67–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wirths S & Lanzavecchia A ABCB1 transporter discriminates human resting naive B cells from cycling transitional and memory B cells. Eur J Immunol 35, 3433–3441 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Barwick BG, Scharer CD, Bally APR & Boss JM Plasma cell differentiation is coupled to division-dependent DNA hypomethylation and gene regulation. Nat Immunol 17, 1216–1225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulis M et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet 47, 746–756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corces MR et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scharer CD, Barwick BG, Guo M, Bally APR & Boss JM Plasma cell differentiation is controlled by multiple cell division-coupled epigenetic programs. Nat Commun 9, 1698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon HS et al. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J Immunol 189, 2393–2403 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaoka M et al. The Orphan Nuclear Receptor NR4A3 Is Involved in the Function of Dendritic Cells. J Immunol 199, 2958–2967 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Ashouri JF & Weiss A Endogenous Nur77 Is a Specific Indicator of Antigen Receptor Signaling in Human T and B Cells. J Immunol 198, 657–668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross I, Bassit B, Benezra M & Licht JD Mammalian sprouty proteins inhibit cell growth and differentiation by preventing ras activation. J Biol Chem 276, 46460–46468 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Otin C, Blasco MA, Partridge L, Serrano M & Kroemer G The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T et al. Identification of epithelial stromal interaction 1 as a novel effector downstream of Kruppel-like factor 8 in breast cancer invasion and metastasis. Oncogene 33, 4746–4755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo J-S, Chae S-CJ G. & Genomics. EPSTI1 polymorphisms are associated with systemic lupus erythematosus. 39, 445–451 (2017). [Google Scholar]
- 31.Buchta CM & Bishop GA TRAF5 negatively regulates TLR signaling in B lymphocytes. J Immunol 192, 145–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell Knode LM et al. Age-Associated B Cells Express a Diverse Repertoire of VH and Vkappa Genes with Somatic Hypermutation. J Immunol 198, 1921–1927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiestner A et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood 101, 4944–4951 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Veleeparambil M et al. Constitutively Bound EGFR-Mediated Tyrosine Phosphorylation of TLR9 Is Required for Its Ability To Signal. J Immunol 200, 2809–2818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chattopadhyay S et al. EGFR kinase activity is required for TLR4 signaling and the septic shock response. EMBO Rep 16, 1535–1547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu B et al. Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat Immunol 18, 573–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myouzen K et al. Regulatory polymorphisms in EGR2 are associated with susceptibility to systemic lupus erythematosus. Hum Mol Genet 19, 2313–2320 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Jadhav K & Zhang Y Activating transcription factor 3 in immune response and metabolic regulation. Liver Res 1, 96–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Juilland M et al. CARMA1- and MyD88-dependent activation of Jun/ATF-type AP-1 complexes is a hallmark of ABC diffuse large B-cell lymphomas. Blood 127, 1780–1789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilchrist M et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441, 173–178 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Glasmacher E et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338, 975–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palanichamy A et al. Novel human transitional B cell populations revealed by B cell depletion therapy. J Immunol 182, 5982–5993 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leadbetter EA et al. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 416, 603–607 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Lau CM et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med 202, 1171–1177 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Martin D, Diaz-Zamudio M, Galindo-Campos M & Alcocer-Varela J Early growth response transcription factors and the modulation of immune response: implications towards autoimmunity. Autoimmun Rev 9, 454–458 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Oh YK, Jang E, Paik DJ & Youn J Early Growth Response-1 Plays a Non-redundant Role in the Differentiation of B Cells into Plasma Cells. Immune Netw 15, 161–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gururajan M et al. Early growth response genes regulate B cell development, proliferation, and immune response. J Immunol 181, 4590–4602 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MJ, Patterson DG, Scharer CD & Boss JM Progressive Upregulation of Oxidative Metabolism Facilitates Plasmablast Differentiation to a T-Independent Antigen. Cell Rep 23, 3152–3159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho HH, Antoniv TT, Ji JD & Ivashkiv LB Lipopolysaccharide-induced expression of matrix metalloproteinases in human monocytes is suppressed by IFN-gamma via superinduction of ATF-3 and suppression of AP-1. J Immunol 181, 5089–5097 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 51.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu F et al. The UCSC Known Genes. Bioinformatics 22, 1036–1046 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Lawrence M et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landt SG et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Supek F, Bosnjak M, Skunca N & Smuc T REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barwick BG et al. B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat Commun 9, 1900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krueger F & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng H, Conneely KN & Wu H A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res 42, e69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paradis E, Claude J & Strimmer K APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Pohl A & Beato M bwtool: a tool for bigWig files. Bioinformatics 30, 1618–1619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carvalho BS & Irizarry RA A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.