Abstract
Background
A growing body of evidence indicates that aberrant expression of miR-107 plays a core role in cancers. This study aims to demonstrate the function of miR-107 and its roles in chemo-drug resistance in breast cancer cells.
Methodology
CCK-8 assays were carried out to test the effect of miR-107 mimics on the proliferation of MCF-7 cells. The apoptosis level of each group was detected by flow cytometry. miR-107 level, mRNA levels of Bcl-2/Bax and TRIAP1 were detected by quantitative real-time Polymerase Chain Reaction (qRT-PCR) analysis. Protein levels of Bcl-2/Bax, p-Akt/Akt in MCF-7 cells were detected by using Western Blot. Lastly, the dual luciferase reporter gene assay system was used to confirm interaction between miR-107 and its target gene TRIAP1.
Results
CCK-8 assays indicated that miR-107 mimics augmented Taxol-induced cell viability inhibition. Flow cytometry showed that miR-107 mimics augmented Taxol-induced elevation of cell apoptosis. qRT-PCR analysis revealed that miR-107 mimics inhibited the mRNA expression of Bcl-2 and induced the mRNA level of Bax. Western Blotting indicated that miR-107 mimics inhibited the expression of proteins Bcl-2 and p-Akt, and induced the expression of Bax, while showing no significant effects on Akt. The relative luciferase activity revealed that oncogene TRIAP1 is a potential target gene of miR-107.
Conclusions
miR-107 plays a role in regulating chemo-drug sensitivity in mammary cancer cell by targeting TRIAP1.
Keywords: miR-107, Breast cancer, TRIAP1
1. Introduction
Breast cancer remains a public-health issue on a global scale in women mainly aged between 20 to 59 years [1]. Although Taxol is a crucial agent in treating breast cancer, the recurrence and metastasis rates of breast cancer are high and the prognosis and survival rate of patients with advanced stage breast cancer are unsatisfactory [2]. The main reason is chemoresistance of cancer cells [3]. In recent years, molecule-targeted treatments were extensively used in clinical settings [4]. As a new method of anti-tumor therapy, the molecule-targeted treatment of tumors has been demonstrated as effective [5]; this treatment has had some visible effects in clinical outcomes. However, the molecular mechanisms underlying the resistance of breast cancer cells to Taxol has not been fully elucidated.
Taxol is a microtubule-stabilizing drug which is widely used in solid tumors such as prostate, lung, sarcoma, and breast [6]. Taxol is reported to contribute to mitotic arrest and inhibits the proliferation of cancer cells [6,7]. Although Taxol is deemed as an efficient anticancer drug, the evolving drug resistance of cancer cells to Taxol may result in treatment failure.
MicroRNAs (miRNAs), a class of small noncoding RNAs, are regulatory, non-coding RNAs about 22 nucleotides in length [8]. Among them, over 200 have been identified in humans [9]. miRNAs can negatively modulate gene expression by targeting mRNAs and trigger either translation repression or RNA degradation [10, 11]. Normally, conserved miRNAs have critical functions across many processes, such as cell proliferation, apoptosis, and metabolism [12, 13, 14, 15]. Most of them play a key role in cancers as oncogenes or tumour suppressor genes [16]. There are studies that have shown that aberrant expression of miRNAs might be involved in tumour formation [17]. In recent years, miR-107 has been extensively investigated in various cancers such as breast, colon, liver, gastric, and bladder cancer [17, 18, 19, 20, 21]. Further evidence has indicated that aberrant expression of miR-107 may lead to cancer [22]. This may be related to the biological behaviors of tumor cell such as proliferation, apoptosis, cell cycle regulation, invasion, and metastasis [23]. miR-107 may also play an important role in the genesis and development of many human malignant tumors [24]. Even though a previous study has explained how miR-107 works in breast cancer [25], it still remains unclear.
In the present study, we report the effect of miR-107 on the chemo-drug sensitivity of breast cancer. Our results indicate that TRIAP1 serves as a novel target gene of miR-107 in breast cancer cells. miR-107 regulates the chemo-drug sensitivity of breast cancer by targeting TRIAP1 via the p-Akt-Bax/Bcl2 signaling pathway.
2. Methods
Cell culture. MCF-7 cells (a breast cancer cell line) were cultured in RPMI-1640 medium supplemented with 10% FBS (Tianhang Biotechnology, Zhejiang, China), 100 U/mL penicillin-G, 100 g/ml streptomycin (Beyotime, Shanghai, China) and 200 ng/mL Taxol [26] at 37 °C in an atmosphere of 5% CO2.
MiRNA transfection. MCF-7 cells were seeded into 6-well plates and cultured for 24 h at 37°C in an atmosphere of 5% CO2 before transfection. 50 nmol/L miR-107 mimics and negative control sequences were transfected into cells by Lipofectamine 2000 (Invitrogen, USA). Hsa-miR-107 mimics (5’-AGCAGCAUUGUACAGGGCUAUCA-3’) and miR-NA-NC mimics (Ctrl: 5’-UUCUUCGAAGGUGUGACGU-3’), were synthesized by GenePharma (Shanghai, China).
Quantitative real-time polymerase chain reaction (qRT-PCR). MCF-7 cells were collected 24 h after transfection. Trizol (Invitrogen, USA) was used to extract total RNA from cells. Total RNA was polyadenylated with poly (A) polymerase (Thermo Fisher Scientific, USA). Reverse Transcriptase came from TransGen Biotech (Beijing, China). qRT-PCR was carried out using TransStart Top Green qPCR SuperMix (TransGen Biotech, China). GAPDH was used as an internal control for TRIAP1, Bax and Bcl-2.
For miRNA semi-quantification, the RNA was reverse-transcribed using a Mir-X miRNA First Strand Synthesis Kit (TaKaRa, Japan) followed by qRT-PCR with Mir-X™ miRNA qRT-PCR SYBR Kit (TaKaRa, Japan). The relative expression of miR-107 was normalized to internal control U6 using the method of 2-ΔΔCt. The sequences of the primers are listed in Table 1.
Table 1.
Oligonucleotide primers used for Q-PCR
| Name | Primer sequence | |
|---|---|---|
| Forward | 5 ’-ATGATGAGCAGCATTGTACAGG-3’ | |
| miR-107 | Reverse | 5 ’-GCAGGGTCCGAGGTATTC-3’ |
| Forward | 5 ’-CTCGCTTCGGCAGCACA-3’ | |
| U6 | Reverse | 5 ’-AACGCTTCACGAATTTGCGT-3’ |
| Forward | 5 ’-CGACCTCTTCAAGCGCTACC-3’ | |
| TRIAP1 | Reverse | 5 ’-CCCATGAACTCCAGTCCTTCAA-3’ |
| Forward | 5 ’-CACCAGCTCTGAACAGATCATGA-3’ | |
| Bax | Reverse | 5 ’-TCAGCCCATCTTCTTCCAGATGT-3’ |
| Forward | 5 ’-CATCCTCATGGAAGAGATCCGC-3’ | |
| Akt | Reverse | 5 ’-GAGGAAGAACCTGTGCTCCATG-3’ |
| Forward | 5 ’-CACCCCTGGCATCTTCTCCTT-3’ | |
| Bcl-2 | Reverse | 5 ’-AGCGTCTTCAGAGACAGCCAG-3′ |
| Forward | 5 ’-GGAGCGAGATCCCTCCAAAAT-3’ | |
| GAPDH | Reverse | 5 ’-GGCTGTTGTCATACTTCTCATGG-3’ |
Western blot. MCF-7 cells were plated at a density of 5×105 cells/well. Cells were treated with miR-107 mimics and Taxol, Taxol alone and control group (untreated) and were cultured in serum free medium. Cells were then lysed with RIPA lysis buffer (Beyotime, Shanghai, China). Whole extracts were prepared, and the protein concentrations were determined using a BCA protein assay kit (Boster, Wuhan, China). Equal amounts of protein lysates (30 μg) were separated by SDS-PAGE (10%, 80 V for 30 min and then 120 V for 60 min). The proteins were transferred onto PVDF membranes (Millipore Corp, Billerica, MA, USA). Then the PVDF membranes were incubated in TBS/Tween-20 containing 5% non-fat dry milk at 37 °C for 3 h. After blocking, the PVDF membranes were incubated with specific primary antibodies overnight at 4 °C. Rabbit monoclonal antibodies against Bcl-2 (ab32124, Abcam, MA, USA), Bax (ab32503, Abcam, MA, USA), p-Akt (ab106693, Abcam, MA, USA), Akt (ab235958, Abcam, MA, USA) and GAPDH (ab37168, Abcam, MA, USA) were used at the working concentration of 1:1000. Following incubation with primary antibodies, blots were washed three times in TBS/Tween-20 before being incubated at 37°C for about 1 h in goat anti-mouse or goat anti-rabbit horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, USA) conjugate antibody at 1:10000 dilution in TBS/Tween-20 containing 5% non-fat dry milk. After extensive washing in TBS/Tween-20 another three times, the enhanced chemiluminescence system was used to detect proteins on the membranes. Proteins were visualized with an ECL chemiluminescent kit (ECL-plus, Thermo Fisher Scientific, Waltham, MA, USA). Autoradiographs were scanned using the Image-Pro Plus Imaging analysis software (Media Cybernetics, MD Rockville, USA).
Cell viability assay. Cell proliferation in different experimental groups was tested by CCK-8 assay and flow cytometry [26]. MCF-7 cells were seeded in 96-well plates. The medium was changed before the assay. After cultivation for 24 h, cells were divided into three groups. CCK-8 was added to the culture medium. After incubation at 37 °C for 4 h, the culture media containing CCK-8 was removed, then DMSO was added into each well and the absorbance at 450 nm was measured by microplate reader (TECAN M1000, Austria GmbH, Austria).
Flow cytometry assay. Cell apoptosis levels in different groups were determined by flow cytometry [19]. Cells were seeded onto 6-well-plates at the concentration of 3 × 105/well and incubated overnight for 24 h. Then, cells were collected according to the manufacturer’s instructions. The cells were incubated with 5 μl Annexin V-FITC for 15 min and 10 μl PI for 5 min, and analyzed using flow cytometry (FCM) (BD Biosciences Clontech, United States) within 30 min.
Dual luciferase activity assay. To examine TRIAP1 and miR-107 binding, pGL3-TRIAP1 WT or pGL3-TRIAP 1 Mut was co-transfected with miR-NC mimics or miR-107 mimics into MCF-7 cells. After 48 h, the MCF-7 cells were subjected to luciferase activity detection by Dual-luciferase reporter system (Promega, USA) according to manufacturer’s protocol. The luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis. Experiments were performed in triplicate. Statistical analysis was thought as significant by comparing mean values (±standard deviation, SD). Differences between two groups were compared by Student’s t test, and differences among three groups were compared using one-way ANOVA followed by Newman Keul’s test. P < 0.05 was considered statistically significant.
3. Results
miR-107 enhanced the chemosensitivity of breast cancer cells in vitro. Treating MCF-7 cells with Taxol significantly increased the apoptosis rate of breast cancer cells, which was more remarkable after treatment with miR-107 mimics and Taxol (Figure 1A and B).
Figure 1.
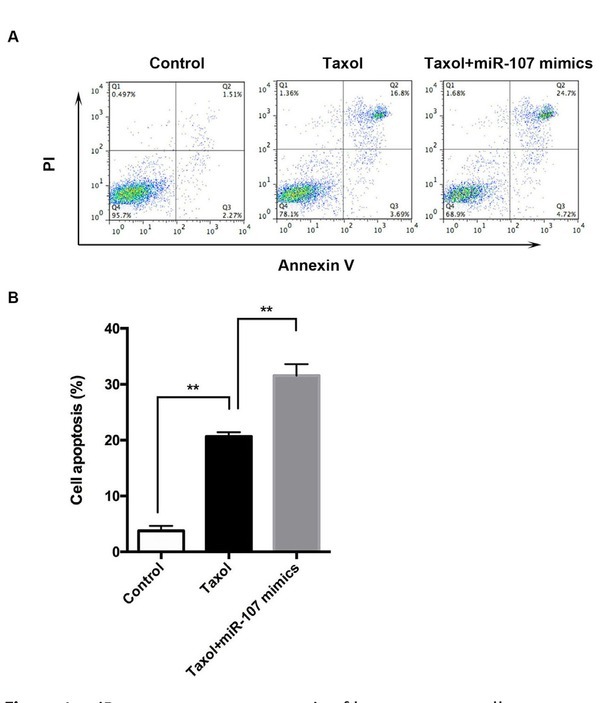
miR-107 promotes apoptosis of breast cancer cells.
A-B: Treating cells with Taxol significantly increased the apoptosis rate of breast cancer cells compared with the control group. The cell apoptosis rate of cells treated with Taxol/ miR-107 mimics was significantly higher compared with that of the breast cancer cell line in Taxol. Data are presented as mean ± SD. **, p < 0.01.
The effects of miR-107 on MCF-7 cell viability was explored. As depicted in Figure 2, compared with the control group, cell viability of MCF-7 cells was downregulated by Taxol, which was promoted by miR-107 mimics. Therefore, miR-107 expression can promote the drug sensitivity of breast cancer cells to Taxol in vitro.
Figure 2.
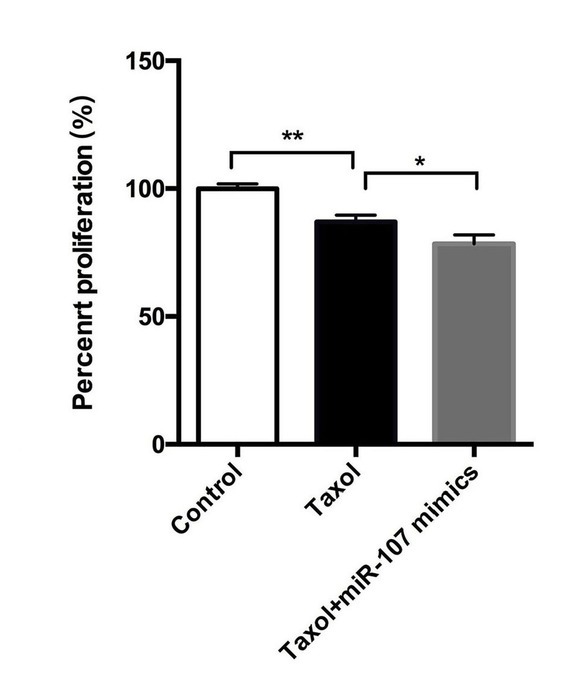
miR-107 inhibits the proliferation of breast cancer
Cell proliferation of the group treated with Taxol+miR-107 mimics is considerably lower than that of the cells in Taxol alone. Therefore, miR-107 expression can increase the drug sensitivity of breast cancer cells in vitro. Data are presented as means ± SD of three independent experiments. *, p < 0.05, **, p < 0.01.
miR-107 regulated the expression of Bcl-2 and Bax. In order to examine the primary mechanism of miR-107-mediated change of drug sensitivity in breast cancer cells, the expression levels of Bcl-2 and Bax were measured. The Bcl-2 family proteins play a role as the key regulators of cell apoptosis and survival; these include the antiapoptotic proteins Bcl-2, Bcl-xL, Mcl-1 and the proapoptotic proteins Bax, Bix etc. [27, 28, 29]. The PI3K/Akt signal transduction pathway was regarded as the chief pathway for the survival of tumor cells [29, 30, 31]. Bax, a proapoptotic member of the Bcl-2 family [32], moves into the mitochondria in response to a wide variety of stimuli for cell apoptosis.
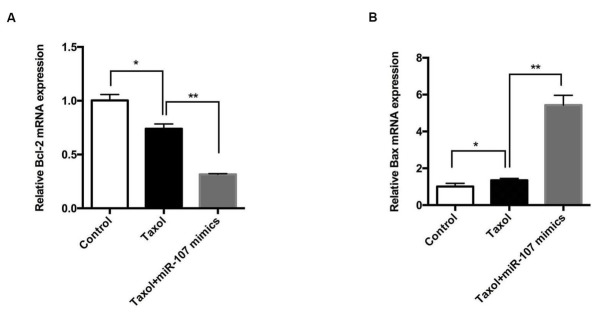
mRNA expression of Bcl-2 and Bax was tested by qPCR assay. As shown in Figure 2A and B, compared with the control group, the mRNA level of Bcl-2 was significantly decreased (P<0.05) and the differences between Taxol and Taxol+miR-107 mimics were more remarkable (P<0.01), while the mRNA level of Bax in cells treated with Taxol was increased (P<0.05), especially in the cells treated with Taxol+miR-107 mimics (P<0.01).
The protein levels of p-Akt, Bcl-2, and Bax were also measured. Results indicated that the protein level changes of Bcl-2 and Bax were similar with that of mRNA levels (Figure 4A-C).Moreover, compared to the control group, Taxol reduced the activation of p-Akt, which was further reduced by the treatment of miR-107 mimics (Fig 4A and D). This preliminarily indicated that the mechanism of miR-107- mediated change of drug sensitivity in breast cancer cells is through up-regulating the expression of Bax and down-regulating the expression of Bcl-2 and p-Akt.
Figure 4.
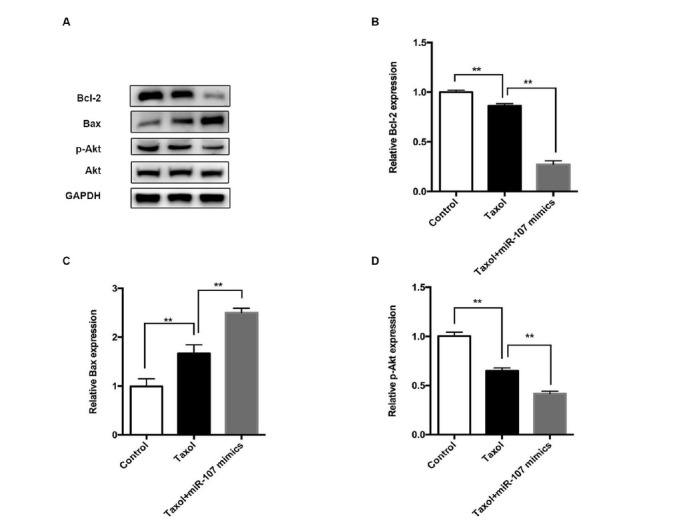
Treating cells with miR-107 and Taxol regulated the protein level of Bax, Bcl-2, Akt, and p-Akt.
The protein expression of Bcl-2 and p-Akt was significantly reduced after treatment with Taxol; this was more remarkable in the Taxol+miR-107 mimics group. There is no significant difference in Akt. Data are presented as mean ± SD. **, p < 0.01.
Figure 3.
miR-107 combined with Taxol regulates the mRNA level of Bax and Bcl-2.
A-B: The expression of Bax in cells treated with Taxol was significantly increased, and was higher in cells treated with Taxol+miR-107 mimics, while Bcl-2 was expressed in low amounts. Data are presented as mean ± SD. *, p < 0.05, **, p < 0.01.
Relative Dual-Luciferase activity. miRNA mainly targets the 3’ untranslated region (UTR) of genes, and in the plasmid construct it is behind the luciferase of the carrier reporter gene [33]. Compared with the change of carrier reporter gene expression, overexpression of interfering miRNA reflects the inhibitory effects of miRNA on target genes. In combination with site-directed mutagenesis of target genes, this can confirm the interaction site of miRNA with the 3’UTR of the target gene. As depicted in Figure 5A, miR-107 targeted the 3’UTR of TRIAP1. hsa-miR-107 mimics inhibited the expression of TRIAP1 via targeting the 3’UTR. Luciferase activity was significantly decreased after treatment with miR-107 mimics in the TRIAP1 WT group, while there was no significant difference in the TRIAP1 MUT group (Figure 5B). After mutation of the binding site, it showed no regulatory role. This result confirmed that after treatment of Taxol, miR-107 enhanced chemosensitivity of breast cancer, suppressed cell viability, and induced cell apoptosis by targeting TRIAP1.
Figure 5.
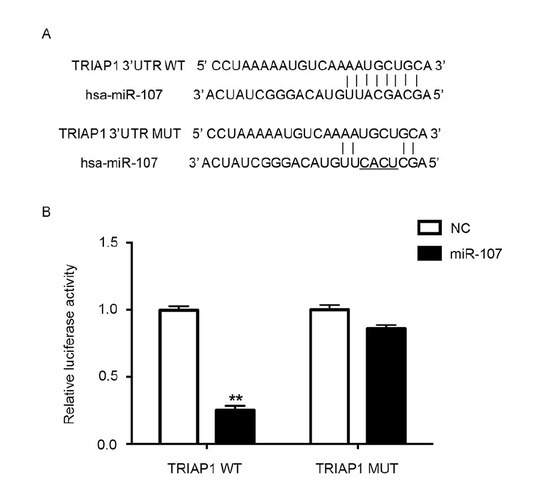
Relative dual-luciferase activity.
A: hsa-miR-107 regulated the expression of TRIAP1 via the binding site on 3’UTR.
B: Transfecting cells with miR-107 significantly downregulated the luciferase activity in TRIAP1 group, while the difference in TRIAP1 MUT is of no statistical significance.. **, p < 0.01.
4. Discussion
In our study, the relationship between miR-107 and chemo-drug sensitivity in breast cancer was evaluated in the MCF-7 cell line. We found that overexpression of miR-107 could enhance the drug sensitivity of breast cancer cells in vitro.
Studies of miRNA functions have confirmed that miRNAs are associated with various functions including embryonic development, metabolism, viral infections, and human malignancies [34]. Different expressions of miR-107 are found in cell cycle arrest, angiogenesis and hypoxia signal pathways [35]. Previous evidence has indicated that miR-107 is overexpressed in colon, pancreatic, and stomach cancers [13, 36]. miR-107 enhanced the chemosensitivity of ovarian cancer to Taxol [36]. A recent study has also shown that miR-107 is expressed in low amounts in breast cancer patients [37]. Our findings further confirm these reports. These findings suggest that treating cells with miR-107and Taxol enhanced the chemosensitivity of breast cancer cells which can be seen in the promoted apoptosis and inhibited proliferation of breast cancer cells.
To explore potential mechanisms, we evaluated the expression level of Bax/Bcl-2 and p-Akt. We found that miR-107 regulated mRNA levels of Bax/Bcl-2 and protein expression of p-Akt and Bax/Bcl-2. Notably, we showed that miR-107 targets TRIAP1, which participates in breast cancer progression.
TP53-regulated inhibitor of apoptosis 1 (TRIAP1), also known as P53CSV, is involved in programmed cell death [38]. TRIAP1 has been shown to be upregulated in many types of cancers [39]. TRIAP1 is deemed as a novel biomarker for chemoresistance in prostate cancer and breast cancer [39, 40]. Evidence has indicated that the expression of TRIAP1 mRNA correlates with myelomas and breast cancer [40, 41]. TRIAP1 is up-regulated in drug-resistant breast cancer cells, and regulates breast cancer cells’ sensitivity to doxorubicin [10, 39, 40]. In the present study, it was demonstrated that TRIAP1 is the direct target of miR-107. Furthermore, western blotting results indicated that miR-107 regulated the expression of the downstream targets of TRIAP1, including Bax, Bcl-2, Akt and p-Akt, which further enhanced the sensitivity of breast cancer cells to Taxol [42]. The results here indicate that miR-107 changes the drug sensitivity of the breast cancer cell line MCF-7 in vitro through targeting TRIAP1.
In summary, miR-107 directly targets TRIAP1 to regulate the apoptosis of breast cancer cells, and increases the sensitivity of cancer cells to Taxol. Manipulating miR-107 expression may have the potential to reverse chemo-resistance in breast cancer patients.
Footnotes
Conflict of interest: Authors state no conflict of interest.
References
- [1].van ‘t Veer L.J., Dai H., van de Vijver M.J.. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530. doi: 10.1038/415530a. et al. –. [DOI] [PubMed] [Google Scholar]
- [2].Ao X., Nie P.P., Wu B.Y., Xu W., Zhang T., Wang S.M., Chang H.C., Zou Z.Z. Decreased expression of microRNA-17 and microRNA-20b promotes breast cancer resistance to Taxol therapy by upregulation of NCOA3. Cell Death Dis. 2016;7:e2463. doi: 10.1038/cddis.2016.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yang F., Zhao N., Wu N., J.. TNFR2 promotes adriamycin resistance in breast cancer cells by repairing DNA damage. Mol Med Rep. 2017;16:2962. doi: 10.3892/mmr.2017.6898. –. [DOI] [PubMed] [Google Scholar]
- [4].Simon M., Stefan N., Plückthun A., Zangemeister W.U. Epithelial cell adhesion molecule-targeted drug delivery for cancer therapy. Expert Opin Drug Deliv. 2013;10:451. doi: 10.1517/17425247.2013.759938. –. [DOI] [PubMed] [Google Scholar]
- [5].Jaferian S., Soleymaninejad M., Negahdari B., Eatemadi A. Stem cell, biomaterials and growth factors therapy for hepatocellular carcinoma. Biomed Pharmacother. 2017;88:1046. doi: 10.1016/j.biopha.2017.01.154. –. [DOI] [PubMed] [Google Scholar]
- [6].Weaver B.A. How Taxol/paclitaxel kills cancer cells. Mol Biol Cell. 2014;25:2677. doi: 10.1091/mbc.E14-04-0916. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li W., Liu J., Jackson K., Shi R., Zhao Y. Sensitizing the therapeutic efficacy of Taxol with shikonin in human breast cancer cells. Plos One. 2014;9:e94079. doi: 10.1371/journal.pone.0094079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ye X.M., Bai W.D., Zhu Z.Y., Zhang X., Chen Y., Wang L., Yang A.G., Zhao J., Jia L. miR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 2014;47:268. doi: 10.5483/BMBRep.2014.47.5.165. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang B., Zuo Z., Lv F.. miR-107 inhibits proliferation of lung cancer cells through regulating TP53 regulated inhibitor of apoptosis 1 (TRIAP1) Open Life Sciences. 2017;12:200. et al. –. [Google Scholar]
- [11].Lee K.H., Lotterman C., Karikari C.. Epigenetic silencing of microRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293. doi: 10.1159/000186051. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Takahashi Y., Forrest A.R., Maeno E.. miR-107 and miR-185 can induce cell cycle arrest in human non-small cell lung cancer cell lines. Plos One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen P.S., Su J.L., Cha S.T.. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J Clin Invest. 2017;127:1116. doi: 10.1172/JCI92099. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Feng L., Xie Y., Zhang H., Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Medical Oncology. 2012;29:856. doi: 10.1007/s12032-011-9823-1. –. [DOI] [PubMed] [Google Scholar]
- [15].Yuan W., Chen F., Man Z.. miR-107 suppresses proliferation of hepatoma cells through targeting HMGA2 mRNA 3′UTR. Biochem Biophys Res Commun. 2016;480:455. doi: 10.1016/j.bbrc.2016.10.070. et al. –. [DOI] [PubMed] [Google Scholar]
- [16].Sharma P., Saraya A., Gupta P., Sharma R. Decreased levels of circulating and tissue miR-107 in human esophageal cancer. Biomarkers. 2013;18:322. doi: 10.3109/1354750X.2013.781677. –. [DOI] [PubMed] [Google Scholar]
- [17].Zhou C., Li G., Zhou J.. miR-107 activates ATR/Chk1 pathway and suppress cervical cancer invasion by targeting MCL1. Plos One. 2014;9:e111860. doi: 10.1371/journal.pone.0111860. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cui J., Mo J., Luo M.. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:12400. et al. –. [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang L., Ma P., Sun L.M.. miR-107 down-regulates SIAH1 expression in human breast cancer cells and silencing of miR-107 inhibits tumor growth in a nude mouse model of triple-negative breast cancer. Mol Carcinog. 2016;55:768. doi: 10.1002/mc.22320. et al. –. [DOI] [PubMed] [Google Scholar]
- [20].Wang S., Ma G., Zhu H.. miR-107 regulates tumor progression by targeting NF1 in gastric cancer. Sci Rep. 2016;6:36531. doi: 10.1038/srep36531. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ayremlou N., Mozdarani H., Mowla S.J., Delavari A. Increased levels of serum and tissue miR-107 in human gastric cancer: correlation with tumor hypoxia. Cancer Biomark. 2015;15:851. doi: 10.3233/CBM-150529. [DOI] [PubMed] [Google Scholar]
- [22].Taisuke I, Shuhei K, Daisuke I,. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep. 2017;7:5708. doi: 10.1038/s41598-017-06137-8. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li X.H., Zhang Y., Shi Y.Q.. microRNA-107, an oncogene microRNA that regulates tumour invasion and metastasis by targeting DICER1 in gastric cancer. J Cell Mol Med. 2011;15:1887. doi: 10.1111/j.1582-4934.2010.01194.x. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang J.J., Wang C.Y., Hua L., Yao K.H., Chen J.T., Hu J.L. miR-107 promotes hepatocellular carcinoma cell proliferation by targeting Axin2. Int J Clin Exp Pathol. 2015;8:5168. –. [PMC free article] [PubMed] [Google Scholar]
- [25].Isabel S., Brigitte R., Wolfgang J., Bernadette J., Klaus P., Heidi S. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget. 2015;6:13387. doi: 10.18632/oncotarget.3874. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yun H., Yi S., Tingting L.. Cell viability, HUVEC proliferation and apoptosis on different doses of netrin-4 was detected by using the CCK-8 assay and flow cytometry. Plos One. 2015 et al. [Google Scholar]
- [27].Tsujimoto Y., Shimizu S. VDAC regulation by the Bcl-2 family of proteins. Cell Death Differ. 2000;7:1174. doi: 10.1038/sj.cdd.4400780. –. [DOI] [PubMed] [Google Scholar]
- [28].Trudel S., Stewart A.K., Li Z.. The Bcl-2 family protein inhibitor, ABT-737, has substantial antimyeloma activity and shows synergistic effect with dexamethasone and melphalan. Clin Cancer Res. 2007;13:621. doi: 10.1158/1078-0432.CCR-06-1526. et al. –. [DOI] [PubMed] [Google Scholar]
- [29].Soini Y., Kinnula V., Kaarteenaho-Wiik R.. Apoptosis and expression of apoptosis regulating proteins bcl-2, mcl-1, bcl-X, and bax in malignant mesothelioma. Clin Cancer Res. 1999;5:3508. et al. –. [PubMed] [Google Scholar]
- [30].Xi G.M., Niu R.F. Effects of blocking P13K-Akt signal pathway in breast cancer therapy. Chinese Journal of Cancer Prevention and Treatment. 2007;14:230. –. [Google Scholar]
- [31].Ma X., Bai Y. IGF-1 activates the P13K/AKT signaling pathway via upregulation of secretory clusterin. Mol Med Rep. 2012;6:1433. doi: 10.3892/mmr.2012.1110. [DOI] [PubMed] [Google Scholar]
- [32].Brady H.J.M., Gil-Gómez G. Molecules in focus Bax, The pro-apoptotic Bcl-2 family member, Bax. Int J Biochem Cell Biol. 1998;30:647. doi: 10.1016/s1357-2725(98)00006-5. –. [DOI] [PubMed] [Google Scholar]
- [33].Zou H., Li Y., Liu X., Wang X., Zou H., Li Y., Liu X., Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549. doi: 10.1074/jbc.274.17.11549. –. [DOI] [PubMed] [Google Scholar]
- [34].Tenev T., Maran i M., Mcneish I., Lemoine N.R.. Pro-caspase-3 overexpression sensitises ovarian cancer cells to proteasome inhibitors. Cell Death Differ. 2001;8:256. doi: 10.1038/sj.cdd.4400808. –. [DOI] [PubMed] [Google Scholar]
- [35].Zhou P., Xu W., Peng X.. Large-scale screens of miRNA-mRNA interactions unveiled that the 3’UTR of a gene is targeted by multiple miRNAs. Plos One. 2013;8:e68204. doi: 10.1371/journal.pone.0068204. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim Y.W., Kim E.Y., Jeon D., Liu J.L., Kim H.S., Choi J.W., Ahn W.S. Differential microRNA expression signatures and cell type-specific association with Taxol resistance in ovarian cancer cells. Drug Des Devel Ther. 2014;8:293. doi: 10.2147/DDDT.S51969. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Martello G., Rosato A., Ferrari F.. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195. doi: 10.1016/j.cell.2010.05.017. et al. –. [DOI] [PubMed] [Google Scholar]
- [38].Li Y.Q., Tang X.R., He Q.G. Overexpression of mitochondria mediator gene TRIAP1 by miR-320b loss is associated with progression in nasopharyngeal carcinoma, J. PLoS Genet. 2016;12:e1006183. doi: 10.1371/journal.pgen.1006183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Adams C., Cazzanelli G., Rasul S.. Apoptosis inhibitor TRIAP1 is a novel effector of drug resistance. Oncol Rep. 2015;34:415. doi: 10.3892/or.2015.3988. et al. –. [DOI] [PubMed] [Google Scholar]
- [40].Siu M.K., Abou-Kheir W., Yin J.J.. Loss of EGFR signaling regulated miR-203 promotes prostate cancer bone metastasis and tyrosine kinase inhibitors resistance. Oncotarget. 2014;5:3770. doi: 10.18632/oncotarget.1994. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fook-Alve s V.L., de Oliveira MB., Zanatta D.B., Strauss B.E., Colleoni G.W.. TP53 Regulated Inhibitor of Apoptosis 1 (TRIAP1) stable silencing increases late apoptosis by upregulation of caspase 9 and APAF1 in RPMI8226 multiple myeloma cell line. Biochim Biophys Acta. 2016;1862:1105. doi: 10.1016/j.bbadis.2016.03.011. –. [DOI] [PubMed] [Google Scholar]
- [42].Andrysik Z., Kim J., Tan A.C., Espinosa J.M. A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell Rep. 2013;3:1346. doi: 10.1016/j.celrep.2013.04.014. –. [DOI] [PMC free article] [PubMed] [Google Scholar]