The risk of an oil spill accident in pristine regions of the world’s oceans is increasing due to the development and transport of crude oil resources, especially in the Arctic region, as a result of the opening of ice-free transportation routes, and there is currently no consensus regarding the complex interplay among the environmental controls of petroleum hydrocarbon biodegradation for predictive modeling. We examined the hydrocarbon biodegradation potential of bacterioplankton from three representative geographic regions of oil exploration. Our results showed that rates of aerobic respiration coupled to hydrocarbon degradation in surface ocean waters are controlled to a large extent by effects of temperature and nutrient limitation; hydrocarbon exposure history did not appear to have a major impact. Further, the relationship between temperature and biodegradation rates is linked to microbial community structure, which is specific to the geographic origin.
KEYWORDS: biodegradation, microbial communities, hydrocarbons, oil, temperature, nutrients
ABSTRACT
The objective of this study was to quantify the potential for hydrocarbon biodegradation in surface waters of three sites, representing geographic regions of major oil exploration (Beaufort Sea in the Arctic, northern Gulf of Mexico [GOM], and southern GOM), in a systematic experimental design that incorporated gradients in temperature and the availability of major nutrients. Surface seawater was amended in microcosms with Macondo surrogate oil to simulate an oil slick, and microcosms were incubated, with or without nutrient amendment, at temperatures ranging from 4 to 38ºC. Using respiration rate as a proxy, distinct temperature responses were observed in surface seawater microcosms based on geographic origin; biodegradation was nearly always more rapid in the Arctic site samples than in the GOM samples. Nutrient amendment enhanced respiration rates by a factor of approximately 6, stimulated microbial growth, and generally elevated the taxonomic diversity of microbial communities within the optimal temperature range for activity at each site, while diversity remained the same or was lower at temperatures deviating from optimal conditions. Taken together, our results advance the understanding of how bacterioplankton communities from different geographic regions respond to oil perturbation. A pulsed disturbance of oil is proposed to favor copiotrophic r-strategists that are adapted to pointed seasonal inputs of phytoplankton carbon, displaying carbon and nutrient limitations, rather than oil exposure history. Further understanding of the ecological mechanisms underpinning the complex environmental controls of hydrocarbon degradation is required for improvement of predictive models of the fate and transport of spilled oil in marine environments.
IMPORTANCE The risk of an oil spill accident in pristine regions of the world’s oceans is increasing due to the development and transport of crude oil resources, especially in the Arctic region, as a result of the opening of ice-free transportation routes, and there is currently no consensus regarding the complex interplay among the environmental controls of petroleum hydrocarbon biodegradation for predictive modeling. We examined the hydrocarbon biodegradation potential of bacterioplankton from three representative geographic regions of oil exploration. Our results showed that rates of aerobic respiration coupled to hydrocarbon degradation in surface ocean waters are controlled to a large extent by effects of temperature and nutrient limitation; hydrocarbon exposure history did not appear to have a major impact. Further, the relationship between temperature and biodegradation rates is linked to microbial community structure, which is specific to the geographic origin.
INTRODUCTION
The Deepwater Horizon (DWH) catastrophe represents the largest accidental marine oil spill in human history (1, 2). Approximately 3.19 million barrel equivalents of oil were released into the Gulf of Mexico (GOM) (3), and an estimated 10% of the released oil formed slicks on the surface ocean (4). While most of the oil was rapidly dispersed, a persistent oil slick was present at the wellhead during and after the spill (5). From June to November 2010, the integrated surface oil concentrations in the northern GOM varied from nondetectable to over 10,000 ppm, with the highest concentrations being observed near the wellhead and in coastal areas (2).
Microbial biodegradation is considered to be the primary means by which spilled oil is eliminated from the environment (6). Previous studies suggested that petroleum hydrocarbon degradation is site specific and correlated with the hydrocarbon exposure history (7, 8). The GOM is a prolific hydrocarbon basin that receives approximately 604,150 liters of oil per year through natural seeps alone (9, 10). It has been suggested that, because of chronic hydrocarbon exposure, microbial communities in the GOM are primed for hydrocarbon degradation and would rapidly respond to oil input (11–13). Apart from oil exposure history, however, microorganisms are under strong selection pressure from in situ environmental conditions (14). Environmental factors have been shown to be key determinants of microbial community structure and function (15, 16), which suggests that resource limitation may outweigh any effects of oil exposure history on biodegradation. Moreover, relatively few rate measurements are available to support the “priming” hypothesis, and further studies are warranted (17).
Hydrocarbon biodegradation is coupled to aerobic respiration in oceanic surface waters (18). Like all respiration processes, biodegradation is limited by the availability of oxygen and nutrients, temperature, and the physiology of hydrocarbon-degrading microorganisms (19). Since hydrocarbons are often distributed in seawater as liquid droplets, the form and solubility of oil also limit biodegradation. A large body of research, including laboratory and field studies, has shown that the ocean environment dictates the efficiency and capacity of microbial communities to degrade hydrocarbons (13, 20, 21). Despite this extensive knowledge base, however, quantitative understanding is lacking, and we have yet to determine how environmental factors interact to regulate the fate and transport of spilled oil in the oceans. After the DWH disaster, many studies focused on the microbial responses to oil contamination in deepsea oil plumes and on shorelines (22–26). Less information is available on the microbial responses to oil slicks in oligotrophic surface seawater.
As traditional shallow oil reservoirs in temperate regions are being depleted, oil exploration is moving toward regions at high latitudes, including in the Beaufort Sea (27). Moreover, due to global climate change, an elongated ice-free season promotes marine transportation through the Northwest Passage (28). These phenomena may lead to increased risk in the potential for an oil spill in fragile, pristine, polar ecosystems (29). Unlike the GOM, which spans subtropical to tropical climates, cold temperatures in the Arctic region may alter the form/solubility of oil and inhibit biodegradation (30), thereby increasing oil longevity in the environment. A systematic understanding of the rates and controls of hydrocarbon biodegradation, as well as the microbial community response, will be critical for assessing the environmental risks of oil exploration in polar regions.
The objective of this study was to quantify the potential for hydrocarbon biodegradation in simulated oil slicks generated from the surface waters of three sites that represent geographical regions of major oil exploration, based on hydrocarbon exposure history, potential hydrocarbon spill risk, and climatic region (Beaufort Sea in the Arctic region, northern GOM, and southern GOM), in a systematic experimental design that incorporates gradients in temperature and the availability of major nutrients. We hypothesized that hydrocarbon exposure history along with nutrient availability would override temperature as biodegradation controls. In contrast, we observed the highest biodegradation potential in surface seawater from the permanently cold Arctic site, with approximately equal impacts of temperature and nutrient availability on degradation activity at all sites.
RESULTS
Site characteristics.
In situ nutrient concentrations and temperatures of the three sampling sites are provided in Table 1. As expected, the temperature at the time of sampling was highest at the tropical site (IXTOC01), intermediate at the subtropical site (DWH01), and much lower at the polar site (CB2). Major nutrient concentrations, including total dissolved inorganic nitrogen (the sum of nitrate, nitrite, and ammonium) and soluble phosphorus levels, were highest at CB2, but these differences were not statistically significant (P = 0.23 for total inorganic nitrogen levels and P = 0.18 for phosphorus levels) (Table 1).
TABLE 1.
Characteristics and results for each sampling site
| Characteristica | CB2 | DWH01 | IXTOC01 |
|---|---|---|---|
| Latitude | 75°47ʹN | 28°43ʹN | 19°22ʹN |
| Longitude | 129°17ʹW | 88°23ʹW | 92°19ʹW |
| Total inorganic nitrogen level (μM) | 6.16 ± 3.0 | 2.83 ± 3.8 | 2.54 ± 2.1 |
| Phosphate level (μM) | 2.08 ± 1.5 | 0.7 ± 0.1 | 1.72 ± 0.8 |
| In situ temperature (°C) | 0.7 | 31 | 35.9 |
| Topt (°C) | 30 | 38 | 38 |
| Activation energy (kJ/mol) | 54.7 ± 11.3 | 83.1 ± 16.5 | 76.2 ± 12.0 |
| Q10 | 2.1 | 3.1 | 2.8 |
| Rate constant (k) at 20°C (day−1) | |||
| UN treatment | 0.003 ± 0.0004 | 0.001 ± 0.0002 | 0.002 ± 0.0002 |
| NA treatment | 0.01 ± 0.002 | 0.008 ± 0.0006 | 0.009 ± 0.0002 |
| RTopt/R4°C | 5 | 99 | 55 |
Topt indicates the temperature at which maximum degradation occurred. Activation energy indicates the apparent temperature response of the degradation rates. Q10 indicates the degradation rate change with a 10°C increase in temperature at 20°C. RTopt/R4°C is the ratio of degradation rates at the Topt and at 4°C for each site.
Biodegradation rates.
In the current study, surface seawater samples from three geographical regions of oil exploration (Beaufort Sea in the Arctic region, northern GOM in the subtropics, and southern GOM in the tropics) were incubated with amended crude oil in a systematic experimental design that included six temperatures and two nutrient treatments. The temperature responses of microbial respiration were determined in triplicate microcosms, with and without nutrient amendment, for all sites. Microbial respiration, as determined by CO2 accumulation, was used as a proxy for hydrocarbon degradation, as in previous studies (Fig. 1) (31–34). This assumption is supported by the fact that little respiration was detected in control microcosms to which no oil was added (see Fig. S1 in the supplemental material). Dissolved oxygen concentrations were quantified at the final time point, and it was confirmed that at least 50% oxygen saturation remained in all incubations. In addition, known aerobic hydrocarbon-degrading bacteria dominated the seawater microbial communities in all oil-amended treatments.
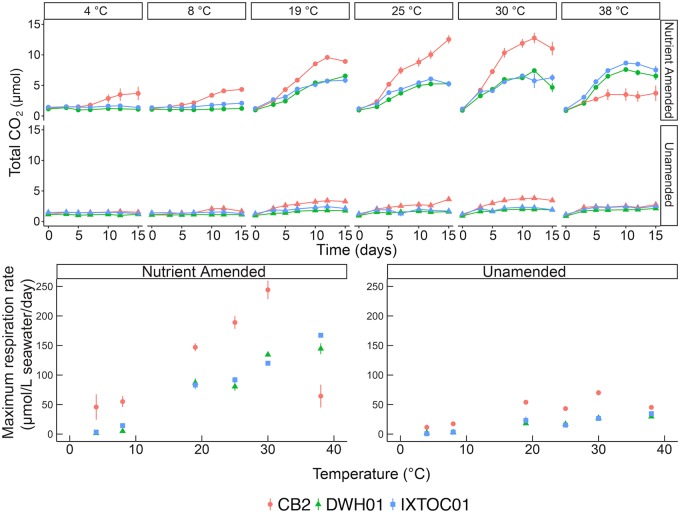
FIG 1.
Biodegradation rates, as determined by carbon dioxide accumulation (upper), and estimated maximum respiration rates (lower) according to temperature and nutrient amendment in microcosms of surface seawater. Scatterplots show average values from triplicate measurements. Error bars indicate standard deviations.
Rapid biodegradation was observed as CO2 production at 25°C, the optimal temperature (Topt) for activity, in oil-amended incubations from all three sites, whereas little to no CO2 production was observed in the control treatments to which no oil was added (representative controls are shown in Fig. S1), indicating that CO2 production could be used an effective proxy for hydrocarbon degradation. Whereas no-oil controls showed little to no activity during the incubation, respiration began immediately in the oil-amended microcosm treatments, and no significant lag phase was observed. Respiration rates were nearly always higher at CB2 for both treatments (244.3 ± 27.0 μmol CO2/liter/day for nutrient-amended [NA] treatment and 45.2 ± 4.8 μmol CO2/liter/day for unamended [UN] treatment), in comparison with the GOM sites, except at the highest temperature studied (38°C), which is well above the ambient range for this permanently cold polar site. Maximum rates at the GOM sites were 144.6 ± 15.8 μmol/liter/day and 167.3 ± 1.6 μmol/liter/day for NA treatment in the DWH01 and IXTOC01 microcosms, respectively. Rates for the UN treatments were 29.6 ± 3.2 μmol CO2/liter/day and 34.6 ± 5 μmol CO2/liter/day for the DWH01 and IXTOC01 microcosms, respectively. At the Topt for activity determined for samples from each site, GOM rates were 32 to 41% and 23 to 35% lower than CB2 rates for the NA and UN treatments, respectively.
According to permutational multivariate analysis of variance (PERMANOVA), nutrient amendment explained the greatest amount of variation in respiration (38%; P = 0.01), followed by temperature (12%; P = 0.03) and site (7%; P = 0.064) (Fig. 1). At most temperatures studied, activity was stimulated substantially by nutrient amendment, with an average 6-fold increase in respiration rates. By comparison, rates at the Arctic site increased an average of 3- to 5-fold across the temperature range studied. Interestingly, at cold temperatures well below the ambient range (4°C and 8°C), activities were suppressed at the GOM sites and the addition of nutrients did not enhance degradation rates. Rates increased with incubation temperature and showed maxima in the mesophilic range (at 30°C for CB2 and 38°C for the GOM sites). Our study might not have captured the Topt for biodegradation for the GOM sites, as rates continued to increase throughout the range studied.
Microbial community analysis.
Overall bacterial abundance was estimated at the end of each incubation using quantitative PCR (qPCR) of small-subunit (SSU) rRNA genes (Fig. S2). Nutrient amendment apparently stimulated microbial growth in all mesocosms, as bacterial abundance was 5- to 10-fold greater with the NA treatments than with the UN treatments. A positive correlation was observed from the linear regression between bacterial abundance and respiration rates at the Topt (R2 = 0.44, P < 0.001). For the GOM samples, bacterial abundance was much greater at the Topt for microbial activity (30°C to 38°C) than at the lowest temperature studied (4°C). Bacterial abundance at 4°C in the CB2 incubations represented approximately one-half of that determined at the Topt for activity. Neither temperature nor nutrient availability significantly affected the cell-specific respiration rates.
The abundance of nifH (nitrogenase) genes, a proxy for the abundance of nitrogen-fixing or diazotrophic microorganisms, represented approximately 0.4% and 5% of the SSU rRNA copies in NA and UN treatments, respectively. Among all treatments for the same temperature and site, the nifH relative abundance increased approximately 32-fold (P = 0.015) (Fig. S2).
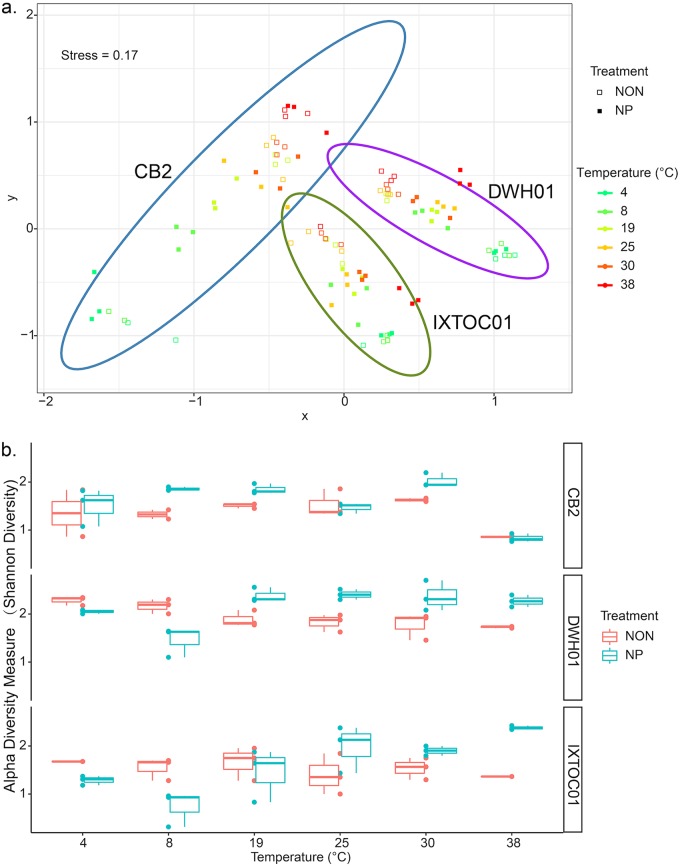
Over 3 million paired-end reads were generated on an Illumina MiSeq platform, and 2.4 million SSU rRNA gene sequences remained after quality control. Operational taxonomic units (OTUs) with a relative abundance of less than 0.05% of the total reads were removed. Samples that contained fewer than 6,000 reads were discarded. Beta diversity, as determined by the Bray-Curtis distance metric, showed a strong selection of microbial communities by sample site (Fig. 2a). Statistical analyses, using PERMANOVA, supported this interpretation. The primary parameter affecting community diversity was site, which accounted for 30% of the variation (P < 0.001), followed by temperature (14%; P < 0.001) and nutrient availability (5%; P < 0.001) (Fig. 2a). Communities were shown to cluster according to site and temperature.
FIG 2.
Beta (a) and alpha (b) diversity, determined for microbial communities in seawater microcosms, according to temperature and nutrient amendment. Beta diversity is displayed as the Bray-Curtis distance metric on a nonmetric multidimensional scaling (NMDS) plot, and alpha diversity is shown as Shannon entropy. Boxplots show average values of triplicate samples. Error bars indicate standard deviations. NON and NP refer to the unamended and nutrient-amended treatments, respectively.
The taxonomic (alpha) diversity of microbial communities in each microcosm was generally elevated with NA treatments within the optimal range of temperatures for activity at each site, while diversity remained the same or was lower at temperatures deviating from optimal conditions (Fig. 2b). The optimal range indicates the temperatures over which significant biodegradation was observed, i.e., 4°C to 30°C for CB2 and 19°C to 38°C for the GOM sites.
Attempts were made to characterize microbial communities in preincubation samples from all sites. However, only small volumes of seawater were available from each site (5 ml) and thus preincubation samples did not yield sufficient DNA for PCR amplification and sequencing of SSU rRNA genes. Because we did not have sufficient sample volume, we were not able to further test extraction methods (35, 36). The focus of the current study was to investigate the complex interplay of the environmental parameters that affect hydrocarbon degradation potential in different oil exploration areas. Therefore, the initial community composition, although important for determining hydrocarbon degradation potential, should not alter our interpretation of relative differences observed between sites.
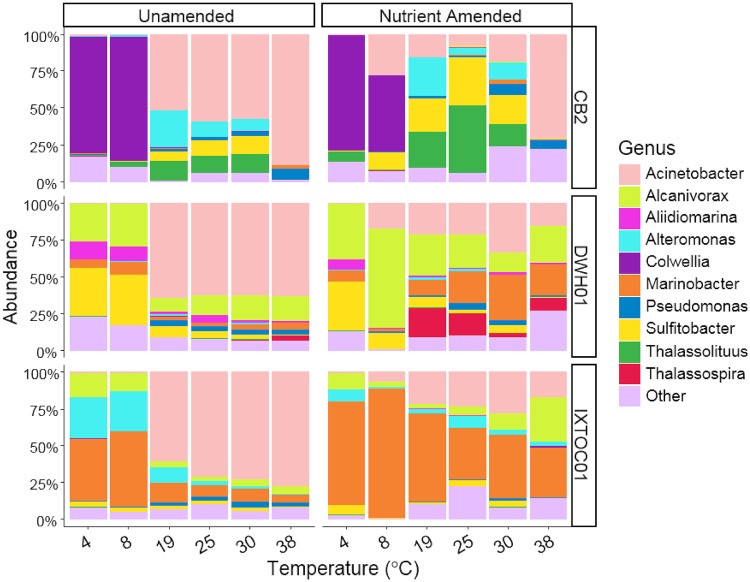
At the class level, Gammaproteobacteria, Alphaproteobacteria, and Betaproteobacteria dominated the microbial communities, constituting up to 98%, 22%, and 38% of the total sequences retrieved, respectively (Fig. 3). Equal or greater alpha diversities were observed in CB2 NA microcosms, compared to UN treatments, across the temperature range, and the Shannon entropy plunged in both treatments at 38°C (Fig. 2b). Colwellia dominated both CB2 treatments at low temperatures (4°C and 8°C) and was elevated, compared to GOM sites, according to linear discriminant analysis effect size (LEfSe) analysis. At mesophilic temperatures, Thalassolituus was strongly enriched in all CB2 microcosms, in comparison with GOM incubations. Both GOM sites demonstrated similar patterns in alpha diversity (Fig. 2b). Low microbial diversity was observed at low temperatures, with significantly greater relative abundances of Alcanivorax, Sulfitobacter, Marinobacter, and Alteromonas being observed at GOM sites. At mesophilic temperatures, the GOM NA treatments were enriched in Alcanivorax and Marinobacter, compared to CB2. The genus Acinetobacter constituted the majority of the microbial communities for all UN treatments at mesophilic temperatures and the NA treatment at 38°C for CB2.
FIG 3.
Microbial community compositions in surface seawater microcosms from CB2 (upper), DWH01 (middle), and IXTOC01 (lower) sites. Barplots show the mean relative abundance of triplicate microcosms. Taxa are grouped at the genus level, and relative abundance was calculated relative to total sequences retrieved.
DISCUSSION
The fate and transport of discharged oil in seawater are determined by a complex interplay among hydrocarbon chemistry, the microbial food web, and ambient oceanographic parameters (21). The complex interactions between environmental factors that regulate the efficiency of microbially mediated hydrocarbon degradation are not completely understood (13).
Environmental controls of biodegradation.
Since the GOM is exposed to a substantial amount of natural oil seepage, it was suggested immediately after the DWH disaster that microbial communities are primed or adapted for an intrinsically high potential for oil biodegradation (22). Conversely, based on analysis of SSU rRNA gene amplicons, numerous studies have shown that hydrocarbon-degrading bacteria are likely to be ubiquitous, albeit rare, in GOM ecosystems, including areas that are not immediately exposed to natural seepage (21, 37). Known petroleum hydrocarbon-degrading bacteria may be separated into two categories, based on their ranges in the utilization of carbon substrates (38). Generalist taxa, such as Acinetobacter and Marinobacter, have the potential to use a wide range of substrates other than hydrocarbons (39, 40). In contrast, specialists, including Alcanivorax (41) and Thalassolituus (42), almost exclusively use hydrocarbons as substrates for growth (18). In the absence of substantial petroleum hydrocarbon input, these organisms may utilize analogous compounds produced by phytoplankton. For the specialists, there are multiple sources of hydrocarbons in the ocean. Indigenous microbial communities in the immediate vicinity of natural hydrocarbon seeps are physiologically adapted to the processing of released hydrocarbons (37). However, natural hydrocarbon seepage occurs in all of the world’s ocean basins. Moreover, phytoplankton represent a widespread source of hydrocarbons, such as alkanes, throughout the world’s oceans (17, 43), and hydrocarbon-degrading bacteria are likely to be equally distributed. Evidence, in the form of potential rates, to support the hypothesis that GOM microbial communities are inordinately primed for hydrocarbon degradation, in comparison to other ocean basins, is lacking.
In this study, the activity and growth of microorganisms in oil-amended microcosms of pristine Arctic seawater, which was collected far from any oil production areas along the coast of northern Canada and Alaska, nearly always exceeded those of seawater sampled from the northern and southern GOM at sites of major oil spills. These data should be verified by direct measurements of petroleum hydrocarbons, and clearly further sampling across ocean basins is needed to assess spatiotemporal variations in hydrocarbon degradation potential. Nonetheless, our results contrast with the paradigm that the GOM is primed for degradation, in comparison to other ocean basins. Rather, this study demonstrates strong selection for hydrocarbon-degrading microbial communities by site, depending on factors other than the history of exposure to high levels of hydrocarbons. Beta diversity analysis showed that microbial communities were most strongly separated by the site of origin, from the polar to tropical sites. Alpha diversity variance reflected microbial adaptation to the respective preferential temperature conditions. Finally, the compositions of major hydrocarbon-degrading communities were clearly distinct among sites, as suggested by LEfSe analysis, especially considering the ranges of activity.
One concern with our experimental design is that samples were stored at 4°C for 1 to 2 months before incubation. The systematic design of this experiment required that large numbers of tubes be incubated in incubators set to different temperatures. Initiation of the experiment was delayed for this period so that all incubations could be conducted in parallel under identical conditions in various incubators. We acknowledge that sample storage should always be performed with caution, especially when refrigeration is employed to slow microbial metabolism. However, comparable studies showed that sample integrity was maintained using the same refrigeration-based storage methods and potential rates were comparable even after 3 months of storage at 4°C (44). In more extreme cases, a 2-month period of frozen storage did not induce permanent damage to microbial activity, with rates comparable to in situ measurements being observed (45). Further, Stenberg et al. reported that, although long-term refrigeration (13 months) reduced organic matter content in samples, substrate-induced microbial activity remained the same after storage (46). In the present study, microbial communities immediately responded to oil amendment without a lag phase, suggesting no suppression of microbial activity. Thus, we contend that sample integrity was maintained to the best of our ability and relative differences in potential rates likely trend with the properties of the different sites. The community composition of the microcosms might have been affected by sample storage, however.
Hydrocarbon-degrading bacteria are ubiquitous in the world’s oceans, as well as in terrestrial environments (47, 48). Since organic matter produced by extant photosynthetic organisms contains compounds analogous to petroleum hydrocarbons, such as alkanes (43), it is likely that microbial communities in any ecosystem exposed to such compounds are primed or adapted to hydrocarbon degradation to some extent. Thus, the potential for oil biodegradation is more likely limited by the complex interplay of environmental parameters unique to each ecosystem. We suggest that the site-specific responses can be explained by the ecological strategy employed by indigenous planktonic microbial communities (49). Previous studies have suggested that marine microorganisms can be separated into two categories according to their responses to resource limitation, namely, copiotrophic r-strategists, which are well adapted to pulsed disturbances of high nutrient and carbon levels interspersed with long periods of starvation, and oligotrophic K-strategists, which are adapted to consistently low inputs of carbon and nutrients, with less ability to respond to disturbances (50, 51). Indeed, r-strategists have been associated with colder ecosystems such as the deepsea and polar regions exposed to pulse disturbances (52–54). We posit that hydrocarbon degraders in Arctic bacterioplankton adopt an r-strategy, which derives from rapid responses to pulses of organic matter over an abbreviated seasonal cycle, whereas communities in the GOM are K-strategists adapted to a more stable, nutrient-poor environment. During the spring bloom in the Arctic Ocean, phytoplankton carbon is injected into surface waters in a short period (55–57), and heterotrophic bacterioplankton respond rapidly to the carbon input through a series of physiological modifications, including shifting kinetic parameters to those more suitable for higher substrate concentrations (52). Despite the significantly increased biomass, community structure appeared to be stable (58), indicating that the majority of the microbial community adopted an r-strategy. Similarly, Arctic zooplankton demonstrated r-strategy-like behavior by sustaining low metabolic rates for much of the year and rapidly becoming more active during spring phytoplankton blooms (59). Thus, it is reasonable to speculate that Arctic surface waters are replete with r-strategist copiotrophs capable of rapidly responding to intense carbon inputs, such as that during an oil spill. In contrast, GOM bacterioplankton experience more consistent conditions of warmer temperatures, sunlight, and organic matter inputs from rivers, leading to more stable resource availability (60, 61). Thus, contrasting ecological strategies to resource utilization may account for the observed differences in biodegradation potentials. Our conclusions are corroborated by previous studies that hypothesized that hydrocarbon degraders employing an r-strategy could respond more quickly during oil spills than those using a K-strategy (37). In studies of in situ microbial communities, it remains challenging to distinguish r-strategists from K-strategists.
Temperature has long been recognized as a critical parameter that regulates hydrocarbon biodegradation (6). It influences biodegradation through effects on the physicochemical properties of oil and on microbial structure/function (62). Many studies, primarily conducted in the laboratory, indicate that temperature strongly regulates the capacity and efficiency of petroleum hydrocarbon degradation in seawater (63). Results are equivocal, however, and kinetic constraints may not be as important as previously thought (64–68).
In this study, distinct temperature responses were observed in surface seawater microcosms based on geographical origin, from the subtropical and tropical GOM sites (DWH01 and IXTOC01) to the Arctic Beaufort Sea (CB2). We observed more rapid degradation in surface seawater samples between 4°C and 30°C in the Arctic samples, compared to subtropical and tropical samples, in corroboration of the findings by Bagi et al., who found faster hydrocarbon degradation in Arctic Ocean seawater than in temperate Atlantic Ocean seawater (32). Results from CB2 microcosms indicated that the in situ microbial community from a permanently cold Arctic environment could maintain relatively rapid degradation across a large range in temperatures, indicating cold adaptation (28, 63, 69–72). In contrast, little degradation activity was observed at lower temperatures (4°C and 8°C) in microcosms from the GOM sites, suggesting adaptation to mesophilic temperatures (73). Our observations are corroborated by the fact that the CB2 site had slightly elevated nutrient concentrations, relative to the GOM sites (74). Generally, nitrogen and phosphorus levels are depleted at the surface by extensive photosynthetic production in low-latitude tropical oceans (74), while low temperatures and light deficiencies lead to higher nutrient levels in the polar region. Thus, higher inorganic nutrient levels may support a greater potential for hydrocarbon degradation in polar waters (75).
Multiple lines of evidence were provided for temperature adaptation. A comparison of both apparent activation energy values and the ratios of the degradation rate at the Topt for activity (RTopt) to the rate at the lowest temperature (4°C) (R4°C) revealed large differences between the Arctic and GOM sites. Lower activation energies and RTopt/R4°C ratios for CB2 microcosms pointed to less change in microbial activity across the range in temperatures, while higher values for both GOM sites indicated more variation with temperature (76). These results reflect the differences in temperatures at the sampling sites. Whereas the average annual sea surface temperature in the GOM is approximately 25°C (77), the temperature at CB2 in the Beaufort Sea permanently remains at <1°C (78). The change in rates with a 10°C increase (Q10) values also diverged between Arctic and GOM sites and fell within the range of findings from previous work (63). In corroboration of evidence from rate measurements, endpoint biomass, as indicated by the abundance of rRNA genes, in Arctic seawater incubations equaled or exceeded biomass measured in GOM incubations, and cell-specific respiration rates did not differ substantially between sites. Differences in the degradation potential observed across temperatures indicate a critical role of microbial adaptation to the indigenous conditions. Microorganisms native to the cold environment showed greater activity at low temperatures, compared to microorganisms from warm environments. The consensus appears to be that it is not possible to interpret rates of oil biodegradation as first order with respect to temperature, due to confounding factors such as nutrient limitations, the solubility of hydrocarbons, and microbial community compositions in various studies (63). Variations in methodology and experimental conditions also likely contribute to uncertainty.
Our interpretation of nutrient limitation in surface waters is supported by the quantification of nitrogen-fixing or diazotrophic microbial communities in the microcosms. The abundance of diazotrophs was estimated by qPCR of the most commonly used molecular marker for nitrogen fixation, nifH (nitrogenase). Nitrogen fixation transforms nitrogen gas into bioavailable ammonia, which provides a competitive advantage for a large diversity of prokaryotes under nutrient-limited conditions (79, 80). In the current study, diazotroph abundance was 32 times higher for UN treatments than for NA treatments, suggesting that nitrogen fixers were responding to limited nutrient availability in the presence of a simulated oil slick. These results are corroborated by field observations during the DWH oil spill. Previous studies reported elevated nitrogen fixation potential in response to oil contamination in GOM waters and sediments (81, 82). Nitrogen fixation was suggested to be a primary nitrogen source in oiled environments during the DWH spill (83), and the findings described above indicate that nutrient limitation likely inhibits oil degradation.
Microbial community dynamics across site, temperature, and nutrient availability.
The dominance of the class Gammaproteobacteria was expected, as most well-known marine hydrocarbon-degrading bacteria reside in this group (22, 25, 28, 84). Although both generalists (Acinetobacter [39], Colwellia [85], and Marinobacter [40]) and specialists (Alcanivorax [41] and Thalassolituus [42]) were present in the microcosms, the microbial populations were likely shaped by the petroleum hydrocarbon amendment rather than other natural carbon sources. Petroleum hydrocarbons represented the only carbon source available in the incubations, as indicated by comparison to control treatments. Previous work indicated that members of the Gammaproteobacteria are well adapted to changing environmental conditions and respond rapidly to pulses in organic matter input (50, 86). At higher taxonomic resolution, shifts in community composition reflected adaptation in response to environmental conditions. At cold temperatures closest to ambient conditions in polar surface waters, the genus Colwellia was selected in all microcosms. The genus Colwellia was often found in oil-contaminated seawater under cold conditions (22, 32, 48, 87). In the current study, abundant Colwellia OTUs showed the highest sequence identity to the psychrophilic strains Colwellia maris and Colwellia rossensis, both of which were isolated from cold environments (88, 89); this may explain the rapid degradation that occurred in CB2 microcosms at 4°C and 8°C. At higher temperatures in incubations of Arctic waters, other genera (Thalassolituus and Sulfitobacter) dominated the communities with nutrient addition. While the genus Thalassolituus is an obligate hydrocarbon-degrading group (42), members of Sulfitobacter have not yet been shown to degrade hydrocarbons. Both microbial groups were often associated with oil-contaminated sites under various conditions (22, 33, 90–92).
Unlike the Arctic microcosms, no distinct microbial group at the genus level was selected by low temperature in incubations of GOM seawater, and the dominant OTUs were all most closely affiliated with mesophilic microorganisms isolated at warm temperatures (93–96). This finding suggests that the minimum degradation observed was due to dysbiosis and a lack of cold-adapted microorganisms in GOM microcosms pushed to temperature extremes. The enrichment of known hydrocarbon degraders, including Alcanivorax, Alteromonas, Marinobacter, and Thalassospira, indicated that these groups are adapted to high oil concentrations and elevated nutrient concentrations. These findings are corroborated by previous studies of planktonic ecosystems in the GOM, where these genera were enriched under heavily oiled conditions either in situ (44, 97) or ex situ (98).
At mesophilic temperatures, all UN microcosms were dominated by the genus Acinetobacter, regardless of the site sampled, and the group persisted in all NA microcosms. Acinetobacter species are well known to degrade a range of hydrocarbons, including long-chain alkanes (99) and polyaromatic hydrocarbons such as phenanthrene and pyrene (100). The high relative abundance of Acinetobacter in UN treatments suggests that it outcompetes other hydrocarbon degraders in heavily oiled and nutrient-depleted environments. In agreement with this finding, Acinetobacter was observed at high abundance in oil slicks and heavily oiled beach and saltmarsh sediments (25, 101, 102) but not in the dispersed oil plumes resulting from the DWH oil spill in May 2010 (44).
Here we demonstrate that the microbial community responses to oil contamination in surface waters of major oil exploration regions are site specific and dependent on ambient conditions, temperature, and nutrients. The activity, diversity, composition, and growth of microbial communities were all strongly selected by site in seawater microcosms, suggesting adaptation to deterministic environmental parameters. Surprisingly, the highest potential hydrocarbon degradation rates were observed in pristine polar waters, with approximately equal effects of temperature and nutrient availability on degradation activity at all sites. The results call into question the role of chronic oil pollution in the priming of GOM waters for oil biodegradation. We hypothesize that the adoption of different ecological strategies for resource utilization provides a basis for hydrocarbon degradation potential in surface seawater. It is important to note that this study represents only a snapshot of the likely spatiotemporal variation of the three sites and should not be extrapolated to the entire geographic region. Future studies should incorporate direct measurements of hydrocarbons and should focus on further verification of the complex interplay among environmental controls of biodegradation.
MATERIALS AND METHODS
Sample collection.
Surface water samples were collected in Niskin bottles at 10 m below the sea surface, from research cruises aboard the Canadian Coast Guard icebreaker Amundsen (10 September 2015), the R/V Weatherbird II (20 August 2015), and the R/V Justo Sierra (2 August 2015) for the polar site CB2, the subtropical site DWH01, and the tropical site IXTOC01, respectively. Site characteristics are presented in Table 1. DWH01 and IXTOC01 represent subtropical and tropical sites, respectively, where the largest accidental marine oil spills in history occurred in the GOM (11). The Beaufort Sea represents a pristine, permanently cold, polar site that is being considered for offshore drilling platforms on the continental slope. Site CB2 was chosen because it had many oceanographic similarities to DWH01. Like DWH01, CB2 is a pelagic site located on the continental slope, remote from shore. In addition, the region surrounding CB2 is socioeconomically and ecologically relevant as a highly productive marine ecosystem in the path of the Northwest Passage (28). Samples for nutrient analysis were immediately filtered through 0.2-μm polycarbonate filters (MoBio Laboratories, Carlsbad, CA, USA) and stored at −20°C. Samples for incubation were stored in Nalgene bottles at 4°C until use. Major inorganic nutrient (nitrite/nitrate, ammonia, and soluble phosphate) levels were determined using established methods (103–105).
Microcosm experiments.
For each site, microcosms were constructed by amending 5 ml of seawater with 5 μl of surrogate MC252 oil (106) in 30-ml sealed glass tubes. The surrogate oil is a sweet light crude with analytical and toxicological properties similar to those of the MC252 oil discharged during the DWH disaster; it was set aside by BP as a tractable model oil for experimentation. Experimental treatments included UN microcosms, to which no nutrient was added, and NA microcosms, which received 32 μM ammonium (NH4Cl) and 2 μM phosphate (K2HPO4) (final concentrations) (31). Microcosms with no oil addition were constructed and incubated at 25°C to indicate the amount of respiration supported by recently produced natural organic matter present in seawater at the time of sampling. Triplicate microcosms were incubated in the dark for 15 days at six different temperatures, spanning the temperature range of polar to tropical climates (4°C, 8°C, 19°C, 25°C, 30°C, and 38°C). Further reduction to subzero temperatures could not be achieved with the available laboratory equipment. Microcosms were sampled at regular intervals for respiration rate measurements.
Respiration was used as a proxy for oil biodegradation, as established in previous work (24). Respiration rates were quantified as CO2 accumulation by sampling the microcosm headspace with a gastight syringe and immediately injecting the sample into a gas chromatograph with a flame ionization detector that was equipped with a methanizer (Shimadzu Scientific, Kyoto, Japan). Total carbon dioxide production was calculated as the sum of the gas phase and the dissolved phase using Henry’s law, with temperature compensation (107). Maximum respiration rates of the linear growth stage were calculated using the R package grofit, with default settings (108). Calculation of activation energy and temperature coefficient (Q10) values was carried out following the methods described by Bagi et al. (32). Rate coefficients (k) were calculated using the pseudo-first-order equation dC/dt = −kt, where C is the residual carbon concentration (in micromolar) and t is the incubation time (in days). Dissolved oxygen concentrations were determined using a Presens Microx 4 optode system with a PSt7 needle-type sensor (Presens, Regensburg, Germany).
DNA extraction and sequence analysis.
After incubation, all of the seawater volume was pelleted and extracted with the Quick-DNA fungal/bacterial microprep kit (Zymo Research, Irvine, CA). Extracted DNA was quantified with the Qubit HS assay kit (Invitrogen, Carlsbad, CA, USA), and 10 ng per reaction was used to generate SSU rRNA amplicons. Established next-generation sequencing protocols were applied for profile prokaryotic community compositions. V4 variable regions of the SSU rRNA gene were amplified using the primers CS1_515F and CS2_806R (109, 110). The amplicons were further barcoded with commercial 10-base barcodes (Fluidigm Corp., South San Francisco, CA, USA), concentrations were normalized, and sequencing was performed on an Illumina MiSeq 2000 platform at the DNA services facility of the University of Chicago (111–113).
Bioinformatic tools were used for downstream analysis of the sequence libraries. PEAR was used for merging of pair-end reads. The merged sequences were processed through vsearch and mothur for demultiplexing and trimming, respectively. Chimeras were eliminated from the libraries using vsearch. Dereplicated sequences were grouped into OTUs using SWARM (with d = 1) (114). Clustered OTUs were then assigned against the SILVA database (115, 116). Downstream analyses were conducted with R packages, including phyloseq (117), vegan, DESeq2, and ggplot2 (117). Multivariate analysis was performed for statistical analysis of microbial community dissimilarity. LEfSe analysis was performed to identify significantly affected taxa (by uploading the abundance table to http://huttenhower.sph.harvard.edu/galaxy).
Quantitative PCR was employed for quantification of SSU rRNA genes and dinitrogenase (nifH) genes on a StepOnePlus platform (Applied Biosystems, Foster City, CA, USA), using PolF/PolR (118) and 331F/518R (119) primers, respectively. Standard curves were obtained with standard plasmids containing target Escherichia coli K12 SSU rRNA genes (2.76 × 103 to 2.76 × 108 copies). The running conditions were as follows: 2 min at 50°C, 2 min at 95°C, and 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 1 min. The nifH genes were amplified using established methods (120). Standard curves were obtained with standard plasmids containing target Azotobacter vinelandii nifH gene fragments as the insert (3.2 × 102 to 3.2 × 107 copies). The running conditions were as follows: 2 min at 50°C, 2 min at 95°C, and 45 cycles of 95°C for 15 s and 63°C for 1 min. In all experiments, negative controls containing no template DNA were subjected to the same qPCR procedure, to exclude or to detect any possible DNA contamination.
Data availability.
Data are publicly available through the Gulf of Mexico Research Initiative Information and Data Cooperative (https://data.gulfresearchinitiative.org/data/R4.x267.179:0009 [DOI: https://doi.org/10.7266/n7-df3b-bq71]) (121). The generated sequence data are available at the NCBI under BioProject no. PRJNA434326.
Supplementary Material
ACKNOWLEDGMENTS
This research was made possible by grants from The Gulf of Mexico Research Initiative to the C-IMAGE II, C-IMAGE III, and Deep-C consortia.
All data are publicly available through the Gulf of Mexico Research Initiative information & Data Cooperative (GRIIDC) at https://data.gulfresearchinitiative.org/data/R4.x267.179:0009 (DOI: https://doi.org/10.7266/n7-df3b-bq71).
We thank the crews of the R/V Weatherbird II, the R/V Justo Sierra, and the icebreaker CCGS Amundsen for their assistance during the field program. We thank Casey Hubert (University of Calgary) for the opportunity to participate in the icebreaker cruise.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00443-19.
REFERENCES
- 1.Farrington JW. 2013. Oil pollution in the marine environment. I. Inputs, big spills, small spills, and dribbles. Environ Sci Policy Sustain Dev 55:3–13. doi: 10.1080/00139157.2013.843980. [DOI] [Google Scholar]
- 2.Sammarco PW, Kolian SR, Warby RAF, Bouldin JL, Subra WA, Porter SA. 2013. Distribution and concentrations of petroleum hydrocarbons associated with the BP/Deepwater Horizon oil spill, Gulf of Mexico. Mar Pollut Bull 73:129–143. doi: 10.1016/j.marpolbul.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 3.Barbier CJ. 2015. MDL 2179 oil spill by the oil rig “Deepwater Horizon.” http://www.laed.uscourts.gov/sites/default/files/OilSpill/Orders/1152015FindingsPhaseTwo.pdf.
- 4.Ryerson TB, Camilli R, Kessler JD, Kujawinski EB, Reddy CM, Valentine DL, Atlas E, Blake DR, de Gouw J, Meinardi S, Parrish DD, Peischl J, Seewald JS, Warneke C. 2012. Chemical data quantify Deepwater Horizon hydrocarbon flow rate and environmental distribution. Proc Natl Acad Sci U S A 109:20246–20253. doi: 10.1073/pnas.1110564109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Hénaff M, Kourafalou VH, Paris CB, Helgers J, Aman ZM, Hogan PJ, Srinivasan A. 2012. Surface evolution of the Deepwater Horizon oil spill patch: combined effects of circulation and wind-induced drift. Environ Sci Technol 46:7267–7273. doi: 10.1021/es301570w. [DOI] [PubMed] [Google Scholar]
- 6.Atlas RM. 1981. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol Rev 45:180–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caparello DM, Larock PA. 1975. A radioisotope assay for the quantification of hydrocarbon biodegradation potential in environmental samples. Microb Ecol 2:28–42. doi: 10.1007/BF02010379. [DOI] [PubMed] [Google Scholar]
- 8.Powell SM, Harvey PM, Stark JS, Snape I, Riddle MJ. 2007. Biodegradation of petroleum products in experimental plots in Antarctic marine sediments is location dependent. Mar Pollut Bull 54:434–440. doi: 10.1016/j.marpolbul.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Joye SB, Teske AP, Kostka JE. 2014. Microbial dynamics following the Macondo oil well blowout across Gulf of Mexico environments. Bioscience 64:766–777. doi: 10.1093/biosci/biu121. [DOI] [Google Scholar]
- 10.Macdonald IR. 1998. Natural oil spills. Sci Am 279:56–61. doi: 10.1038/scientificamerican1198-56. [DOI] [Google Scholar]
- 11.Atlas RM, Hazen TC. 2011. Oil biodegradation and bioremediation: a tale of the two worst spills in U.S. history. Environ Sci Technol 45:6709–6715. doi: 10.1021/es2013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleindienst S, Paul JH, Joye SB. 2015. Using dispersants after oil spills: impacts on the composition and activity of microbial communities. Nat Rev Microbiol 13:388–396. doi: 10.1038/nrmicro3452. [DOI] [PubMed] [Google Scholar]
- 13.Hazen TC, Prince RC, Mahmoudi N. 2016. Marine oil biodegradation. Environ Sci Technol 50:2121–2129. doi: 10.1021/acs.est.5b03333. [DOI] [PubMed] [Google Scholar]
- 14.Baldwin AJ, Moss JA, Pakulski JD, Catala P, Joux F, Jeffrey WH. 2005. Microbial diversity in a Pacific Ocean transect from the Arctic to Antarctic circles. Aquat Microb Ecol 41:91–102. doi: 10.3354/ame041091. [DOI] [Google Scholar]
- 15.Rodríguez-Blanco A, Duval A, Pelletier E, Delille D, Ghiglione JF. 2013. Effects of temperature and fertilization on the structure of total versus active bacterial communities from sub-Antarctic seawater exposed to crude oil and diesel fuel. Polar Res 32:1. doi: 10.3402/polar.v32i0.18521. [DOI] [Google Scholar]
- 16.Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, Gonzalez JM, Luo H, Wright JJ, Landry ZC, Hanson NW, Thompson BP, Poulton NJ, Schwientek P, Acinas SG, Giovannoni SJ, Moran MA, Hallam SJ, Cavicchioli R, Woyke T, Stepanauskas R. 2013. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci U S A 110:11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentine DL, Reddy CM. 2015. Latent hydrocarbons from cyanobacteria. Proc Natl Acad Sci U S A 112:13434–13435. doi: 10.1073/pnas.1518485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Head IM, Jones DM, Röling W. 2006. Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 19.Joye S, Kleindienst S, Gilbert J, Handley K, Weisenhorn P, Overholt W, Kostka J. 2016. Responses of microbial communities to hydrocarbon exposures. Oceanography 29:136–149. doi: 10.5670/oceanog.2016.78. [DOI] [Google Scholar]
- 20.Prince RC. 2010. Bioremediation of marine oil spills, p 2617–2630. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 21.Joye SB, Bracco A, Özgökmen TM, Chanton JP, Grosell M, MacDonald IR, Cordes EE, Montoya JP, Passow U. 2016. The Gulf of Mexico ecosystem, six years after the Macondo oil well blowout. Deep Sea Res Part II 129:4–19. doi: 10.1016/j.dsr2.2016.04.018. [DOI] [Google Scholar]
- 22.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D’haeseleer P, Holman H-Y, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 23.Valentine DL, Kessler JD, Redmond MC, Mendes SD, Heintz MB, Farwell C, Hu L, Kinnaman FS, Yvon-Lewis S, Du M, Chan EW, Garcia Tigreros F, Villanueva CJ. 2010. Propane respiration jump-starts microbial response to a deep oil spill. Science 330:208–211. doi: 10.1126/science.1196830. [DOI] [PubMed] [Google Scholar]
- 24.Baelum J, Borglin S, Chakraborty R, Fortney JL, Lamendella R, Mason OU, Auer M, Zemla M, Bill M, Conrad ME, Malfatti SA, Tringe SG, Holman H-Y, Hazen TC, Jansson JK. 2012. Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon spill. Environ Microbiol 14:2405–2416. doi: 10.1111/j.1462-2920.2012.02780.x. [DOI] [PubMed] [Google Scholar]
- 25.Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M. 2011. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl Environ Microbiol 77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kappell AD, Wei Y, Newton RJ, Van Nostrand JD, Zhou J, McLellan SL, Hristova KR. 2014. The polycyclic aromatic hydrocarbon degradation potential of Gulf of Mexico native coastal microbial communities after the Deepwater Horizon oil spill. Front Microbiol 5:205. doi: 10.3389/fmicb.2014.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arctic Monitoring and Assessment Programme. 2007. Arctic oil and gas 2007, p 13–40. Arctic Monitoring and Assessment Programme, Oslo, Norway. [Google Scholar]
- 28.Yergeau E, Michel C, Tremblay J, Niemi A, King TL, Wyglinski J, Lee K, Greer CW. 2017. Metagenomic survey of the taxonomic and functional microbial communities of seawater and sea ice from the Canadian Arctic. Sci Rep 7:42242. doi: 10.1038/srep42242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crisafi F, Giuliano L, Yakimov MM, Azzaro M, Denaro R. 2016. Isolation and degradation potential of a cold-adapted oil/PAH-degrading marine bacterial consortium from Kongsfjorden (Arctic region). Rend Fis Acc Lincei 27:261–270. doi: 10.1007/s12210-016-0550-6. [DOI] [Google Scholar]
- 30.Karhu K, Auffret MD, Dungait JAJ, Hopkins DW, Prosser JI, Singh BK, Subke J-A, Wookey PA, Agren GI, Sebastià M-T, Gouriveau F, Bergkvist G, Meir P, Nottingham AT, Salinas N, Hartley IP. 2014. Temperature sensitivity of soil respiration rates enhanced by microbial community response. Nature 513:81–84. doi: 10.1038/nature13604. [DOI] [PubMed] [Google Scholar]
- 31.Edwards BR, Reddy CM, Camilli R, Carmichael CA, Longnecker K, Van Mooy B. 2011. Rapid microbial respiration of oil from the Deepwater Horizon spill in offshore surface waters of the Gulf of Mexico. Environ Res Lett 6:035301. doi: 10.1088/1748-9326/6/3/035301. [DOI] [Google Scholar]
- 32.Bagi A, Pampanin DM, Lanzén A, Bilstad T, Kommedal R. 2014. Naphthalene biodegradation in temperate and arctic marine microcosms. Biodegradation 25:111–125. doi: 10.1007/s10532-013-9644-3. [DOI] [PubMed] [Google Scholar]
- 33.Röling WFM, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJP, Head IM. 2002. Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol 68:5537–5548. doi: 10.1128/AEM.68.11.5537-5548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortazavi B, Horel A, Beazley MJ, Sobecky PA. 2013. Intrinsic rates of petroleum hydrocarbon biodegradation in Gulf of Mexico intertidal sandy sediments and its enhancement by organic substrates. J Hazard Mater 244–245:537–544. doi: 10.1016/j.jhazmat.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 35.Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, Catroux G. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl Environ Microbiol 67:2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Lipthay JR, Enzinger C, Johnsen K, Aamand J, Sørensen SJ. 2004. Impact of DNA extraction method on bacterial community composition measured by denaturing gradient gel electrophoresis. Soil Biol Biochem 36:1607–1614. doi: 10.1016/j.soilbio.2004.03.011. [DOI] [Google Scholar]
- 37.Kleindienst S, Grim S, Sogin M, Bracco A, Crespo-Medina M, Joye SB. 2016. Diverse, rare microbial taxa responded to the Deepwater Horizon deep-sea hydrocarbon plume. ISME J 10:400–415. doi: 10.1038/ismej.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutierrez T. 2017. Marine, aerobic hydrocarbon-degrading gammaproteobacteria: overview In McGenity TJ. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 39.Overholt WA, Green SJ, Marks KP, Venkatraman R, Prakash O, Kostka E. 2013. Draft genome sequences for oil-degrading bacterial strains from beach sands impacted by the Deepwater Horizon oil spill. Genome Announc 1:e01015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mounier J, Camus A, Mitteau I, Vaysse P-J, Goulas P, Grimaud R, Sivadon P. 2014. The marine bacterium Marinobacter hydrocarbonoclasticus SP17 degrades a wide range of lipids and hydrocarbons through the formation of oleolytic biofilms with distinct gene expression profiles. FEMS Microbiol Ecol 90:816–831. doi: 10.1111/1574-6941.12439. [DOI] [PubMed] [Google Scholar]
- 41.Yakimov MM, Golyshin PN, Lang S, Moore ERB, Abraham W, Lunsdorf H, Timmis KN. 1998. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int J Syst Bacteriol 48:339–348. doi: 10.1099/00207713-48-2-339. [DOI] [PubMed] [Google Scholar]
- 42.Yakimov MM, Giuliano L, Denaro R, Crisafi E, Chernikova TN, Abraham WR, Luensdorf H, Timmis KN, Golyshin PN. 2004. Thalassolituus oleivorans gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int J Syst Evol Microbiol 54:141–148. doi: 10.1099/ijs.0.02424-0. [DOI] [PubMed] [Google Scholar]
- 43.Lea-Smith DJ, Biller SJ, Davey MP, Cotton CAR, Perez Sepulveda BM, Turchyn AV, Scanlan DJ, Smith AG, Chisholm SW, Howe CJ. 2015. Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc Natl Acad Sci U S A 112:13591–13596. doi: 10.1073/pnas.1507274112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redmond M, Valentine D. 2012. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci U S A 109:20292–20297. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Techtmann SM, Zhuang M, Campo P, Holder E, Elk M, Hazen TC, Conmy R, Domingo JWS. 2017. Corexit 9500 enhances oil biodegradation and changes active bacterial community structure of oil-enriched microcosms. Appl Environ Microbiol 83:e03462-16. doi: 10.1128/AEM.03462-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenberg B, Johansson M, Pell M, Sjödahl-Svensson K, Stenström J, Torstensson L. 1998. Microbial biomass and activities in soil as affected by frozen and cold storage. Soil Biol Biochem 30:393–402. doi: 10.1016/S0038-0717(97)00125-9. [DOI] [Google Scholar]
- 47.Prince RC, Gramain A, McGenity TJ. 2010. Prokaryotic hydrocarbon degraders, p 1669–1692. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 48.Kleindienst S, Seidel M, Ziervogel K, Grim S, Loftis K, Harrison S, Malkin SY, Perkins MJ, Field J, Sogin ML, Dittmar T, Passow U, Medeiros PM, Joye SB. 2015. Chemical dispersants can suppress the activity of natural oil-degrading microorganisms. Proc Natl Acad Sci U S A 112:14900–14905. doi: 10.1073/pnas.1507380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews JH, Harris RF. 1986. r- and K-selection and microbial ecology. Adv Microb Ecol 9:99–147. doi: 10.1007/978-1-4757-0611-6_3. [DOI] [Google Scholar]
- 50.Eilers H, Pernthaler J, Amann R. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl Environ Microbiol 66:4634–4640. doi: 10.1128/AEM.66.11.4634-4640.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M, Johnson J, Montgomery R, Ferriera S, Beeson K, Williamson SJ, Tovchigrechko A, Allen AE, Zeigler LA, Sutton G, Eisenstadt E, Rogers YH, Friedman R, Frazier M, Venter JC. 2010. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468:60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 52.Yager PL, Connelly TL, Mortazavi B, Wommack KE, Bano N, Bauer JE, Opsahl S, Hollibaugh JT. 2001. Dynamic bacterial and viral response to an algal bloom at subzero temperatures. Limnol Oceanogr 46:790–801. doi: 10.4319/lo.2001.46.4.0790. [DOI] [Google Scholar]
- 53.Eloe EA, Fadrosh DW, Novotny M, Zeigler Allen L, Kim M, Lombardo MJ, Yee-Greenbaum J, Yooseph S, Allen EE, Lasken R, Williamson SJ, Bartlett DH. 2011. Going deeper: metagenome of a hadopelagic microbial community. PLoS One 6:e20388. doi: 10.1371/journal.pone.0020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kvíderová J, Elster J, Komárek J. 2019. Ecophysiology of cyanobacteria in the polar regions, p 277–302. In Mishra AK, Tiwari DN, Rai AN (ed), Cyanobacteria: from basic science to applications. Academic Press, New York, NY. [Google Scholar]
- 55.Mundy CJ, Gosselin M, Gratton Y, Brown K, Galindo V, Campbell K, Levasseur M, Barber D, Papakyriakou T, Bélanger S. 2014. Role of environmental factors on phytoplankton bloom initiation under landfast sea ice in Resolute Passage, Canada. Mar Ecol Prog Ser 497:39–49. doi: 10.3354/meps10587. [DOI] [Google Scholar]
- 56.Pomeroy LR, Wiebe WJ, Deibel D, Thompson RJ, Rowe GT, Pakulski JD. 1991. Bacterial responses to temperature and substrate concentration during the Newfoundland spring bloom. Mar Ecol Prog Ser 75:143–159. doi: 10.3354/meps075143. [DOI] [Google Scholar]
- 57.Smith REH, Clement P, Cota GF. 1989. Population dynamics of bacteria in Arctic Sea ice. Microb Ecol 17:63–76. doi: 10.1007/BF02025594. [DOI] [PubMed] [Google Scholar]
- 58.Kirchman DL, Cottrell MT, Lovejoy C. 2010. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol 12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- 59.Conover RJ, Siferd TD. 1993. Dark-season survival strategies of coastal zone zooplankton in the Canadian Arctic. Arctic 46:303–311. doi: 10.14430/arctic1357. [DOI] [Google Scholar]
- 60.Galicia M, Salazar M, Lopez-Veneroni D. 2009. Prospective and potential impact of biological communities in Deepwater metallic structures in the southern Gulf of Mexico In NACE Corrosion 2009 conference papers. NACE International, Houston, TX. [Google Scholar]
- 61.Fredericks AD, Sackett WM. 1970. Organic carbon in the Gulf of Mexico. J Geophys Res 75:2199–2206. doi: 10.1029/JC075i012p02199. [DOI] [Google Scholar]
- 62.Leahy JG, Colwell RR. 1990. Microbial degradation of hydrocarbons in the environment. Microbiol Rev 54:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bagi A, Pampanin DM, Brakstad OG, Kommedal R. 2013. Estimation of hydrocarbon biodegradation rates in marine environments: a critical review of the Q10 approach. Mar Environ Res 89:83–90. doi: 10.1016/j.marenvres.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 64.Nedwell D. 1999. Effect of low temperature on microbial growth: lowered affinity for substrates limits growth at low temperature. FEMS Microbiol Ecol 30:101–111. doi: 10.1111/j.1574-6941.1999.tb00639.x. [DOI] [PubMed] [Google Scholar]
- 65.Garneau MÉ, Michel C, Meisterhans G, Fortin N, King TL, Greer CW, Lee K. 2016. Hydrocarbon biodegradation by Arctic sea-ice and sub-ice microbial communities during microcosm experiments, Northwest Passage (Nunavut, Canada). FEMS Microbiol Ecol 92:fiw130. doi: 10.1093/femsec/fiw130. [DOI] [PubMed] [Google Scholar]
- 66.Brakstad OG, Nonstad I, Faksness LG, Brandvik PJ. 2008. Responses of microbial communities in Arctic sea ice after contamination by crude petroleum oil. Microb Ecol 55:540–552. doi: 10.1007/s00248-007-9299-x. [DOI] [PubMed] [Google Scholar]
- 67.Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Greer CW. 2013. Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7:1200–1210. doi: 10.1038/ismej.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee K, Nedwed T, Prince RC, Palandro D. 2013. Lab tests on the biodegradation of chemically dispersed oil should consider the rapid dilution that occurs at sea. Mar Pollut Bull 73:314–318. doi: 10.1016/j.marpolbul.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 69.Horowitz A, Atlas RM. 1977. Continuous open flow-through system as a model for oil degradation in the Arctic Ocean. Appl Environ Microbiol 33:647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McFarlin KM, Prince RC, Perkins R, Leigh MB. 2014. Biodegradation of dispersed oil in Arctic seawater at −1°C. PLoS One 9:e84297. doi: 10.1371/journal.pone.0084297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brakstad OG, Ribicic D, Winkler A, Netzer R. 2018. Biodegradation of dispersed oil in seawater is not inhibited by a commercial oil spill dispersant. Mar Pollut Bull 129:555–561. doi: 10.1016/j.marpolbul.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 72.Brakstad OG, Farooq U, Ribicic D, Netzer R. 2018. Dispersibility and biotransformation of oils with different properties in seawater. Chemosphere 191:44–53. doi: 10.1016/j.chemosphere.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 73.Shiah FK, Ducklow HW. 1994. Temperature regulation of heterotrophic bacterioplankton production, and specific growth rate in Chesapeake Bay. Limnol Oceanogr 39:1243–1258. doi: 10.4319/lo.1994.39.6.1243. [DOI] [Google Scholar]
- 74.Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, Galbraith ED, Geider RJ, Guieu C, Jaccard SL, Jickells TD, La Roche J, Lenton TM, Mahowald NM, Marañón E, Marinov I, Moore JK, Nakatsuka T, Oschlies A, Saito MA, Thingstad TF, Tsuda A, Ulloa O. 2013. Processes and patterns of oceanic nutrient limitation. Nat Geosci 6:701–710. doi: 10.1038/ngeo1765. [DOI] [Google Scholar]
- 75.Atlas RM, Bartha R. 1972. Degradation and mineralization of petroleum in sea water: limitation by nitrogen and phosphorous. Biotechnol Bioeng 14:309–318. doi: 10.1002/bit.260140304. [DOI] [PubMed] [Google Scholar]
- 76.Canion A, Kostka JE, Gihring TM, Huettel M, Van Beusekom JEE, Gao H, Lavik G, Kuypers MM. 2014. Temperature response of denitrification and anammox reveals the adaptation of microbial communities to in situ temperatures in permeable marine sediments that span 50° in latitude. Biogeosciences 11:309–320. doi: 10.5194/bg-11-309-2014. [DOI] [Google Scholar]
- 77.National Oceanic and Atmospheric Administration. Sea surface temperature (SST) contour charts. http://www.ospo.noaa.gov/Products/ocean/sst/contour.
- 78.Carmack EC, Aagaard K, Swift JH, MacDonald RW, McLaughlin FA, Jones EP, Perkin RG, Smith JN, Ellis KM, Killius LR. 1997. Changes in temperature and tracer distributions within the Arctic Ocean: results from the 1994 Arctic Ocean section. Deep Sea Res Part II 44:1487–1502. doi: 10.1016/S0967-0645(97)00056-8. [DOI] [Google Scholar]
- 79.Gaby JC, Rishishwar L, Valderrama-Aguirre LC, Green SJ, Valderrama-Aguirre A, Jordan IK, Kostka JE. 2018. Diazotroph community characterization via a high-throughput nifH amplicon sequencing and analysis pipeline. Appl Environ Microbiol 84:e01512-17. doi: 10.1128/AEM.01512-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuypers MMM, Marchant HK, Kartal B. 2018. The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 81.Rodriguez-R LM, Overholt WA, Hagan C, Huettel M, Kostka JE, Konstantinidis KT. 2015. Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J 9:1928–1940. doi: 10.1038/ismej.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A, Lee Y-J, Mason OU, Dubinsky EA, Chavarria KL, Tom LM, Fortney JL, Lamendella R, Jansson JK, D'haeseleer P, Hazen TC, Zhou J. 2012. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J 6:451–460. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mason OU, Scott NM, Gonzalez A, Robbins-Pianka A, Bælum J, Kimbrel J, Bouskill NJ, Prestat E, Borglin S, Joyner DC, Fortney JL, Jurelevicius D, Stringfellow WT, Alvarez-Cohen L, Hazen TC, Knight R, Gilbert JA, Jansson JK. 2014. Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J 8:1464–1475. doi: 10.1038/ismej.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brakstad OG, Throne-Holst M, Netzer R, Stoeckel DM, Atlas RM. 2015. Microbial communities related to biodegradation of dispersed Macondo oil at low seawater temperature with Norwegian coastal seawater. Microb Biotechnol 8:989–998. doi: 10.1111/1751-7915.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang T, Nigro LM, Gutierrez T, Ambrosio LD, Joye SB, Highsmith R, Teske A. 2016. Pulsed blooms and persistent oil-degrading bacterial populations in the water column during and after the Deepwater Horizon blowout. Deep Sea Res Part II 129:282–291. doi: 10.1016/j.dsr2.2014.01.014. [DOI] [Google Scholar]
- 86.Gihring TM, Humphrys M, Mills HJ, Huette M, Kostka JE. 2009. Identification of phytodetritus-degrading microbial communities in sublittoral Gulf of Mexico sands. Limnol Oceanogr 54:1073–1083. doi: 10.4319/lo.2009.54.4.1073. [DOI] [Google Scholar]
- 87.Brakstad OG, Nordtug T, Throne-Holst M. 2015. Biodegradation of dispersed Macondo oil in seawater at low temperature and different oil droplet sizes. Mar Pollut Bull 93:144–152. doi: 10.1016/j.marpolbul.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 88.Yumoto I, Kawasaki K, Iwata H, Matsuyama H, Okuyama H. 1998. Assignment of Vibrio sp. strain ABE-1 to Colwellia maris sp. nov., a new psychrophilic bacterium. Int J Syst Evol Microbiol 48:1357–1362. doi: 10.1099/00207713-48-4-1357. [DOI] [PubMed] [Google Scholar]
- 89.Gosink JJ, Staley JT. 1995. Biodiversity of gas vacuolate bacteria from Antarctic Sea ice and water. Appl Environ Microbiol 61:3486–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu P, Dubinsky EA, Probst AJ, Wang J, Sieber CMK, Tom LM, Gardinali PR, Banfield JF, Atlas RM, Andersen GL. 2017. Simulation of Deepwater Horizon oil plume reveals substrate specialization within a complex community of hydrocarbon degraders. Proc Natl Acad Sci U S A 114:7432–7437. doi: 10.1073/pnas.1703424114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Coulon F, McKew BA, Osborn AM, McGenity TJ, Timmis KN. 2007. Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ Microbiol 9:177–186. doi: 10.1111/j.1462-2920.2006.01126.x. [DOI] [PubMed] [Google Scholar]
- 92.Brakstad OG, Lødeng A. 2005. Microbial diversity during biodegradation of crude oil in seawater from the North Sea. Microb Ecol 49:94–103. doi: 10.1007/s00248-003-0225-6. [DOI] [PubMed] [Google Scholar]
- 93.Green DH, Llewellyn LE, Negri AP, Blackburn SI, Bolch C. 2004. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol Ecol 47:345–357. doi: 10.1016/S0168-6496(03)00298-8. [DOI] [PubMed] [Google Scholar]
- 94.Kwon KK, Oh JH, Yang SH, Seo HS, Lee JH. 2015. Alcanivorax gelatiniphagus sp. nov., a marine bacterium isolated from tidal flat sediments enriched with crude oil. Int J Syst Evol Microbiol 65:2204–2208. doi: 10.1099/ijs.0.000244. [DOI] [PubMed] [Google Scholar]
- 95.Gonzalez JM, Mayer F, Moran MA, Hodson RE, Whitman WB. 1997. Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int J Syst Bacteriol 47:773–780. doi: 10.1099/00207713-47-3-773. [DOI] [PubMed] [Google Scholar]
- 96.Jin HM, Kim KH, Jeon CO. 2015. Alteromonas naphthalenivorans sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from tidal-flat sediment. Int J Syst Evol Microbiol 65:4208–4214. doi: 10.1099/ijsem.0.000563. [DOI] [PubMed] [Google Scholar]
- 97.Liu Z, Liu J. 2013. Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the Deepwater Horizon oil spill. Microbiologyopen 2:492–504. doi: 10.1002/mbo3.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kasai Y, Kishira H, Sasaki T, Syutsubo K, Watanabe K, Harayama S. 2002. Predominant growth of Alcanivorax strains in oil- contaminated and nutrient-supplemented sea water. Environ Microbiol 4:141–147. doi: 10.1046/j.1462-2920.2002.00275.x. [DOI] [PubMed] [Google Scholar]
- 99.Throne-Holst M, Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB. 2007. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl Environ Microbiol 73:3327–3332. doi: 10.1128/AEM.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao Y, Yu XZ, Wu SC, Cheung KC, Tam NFY, Qian PY, Wong MH. 2006. Interactions of rice (Oryza sativa L.) and PAH-degrading bacteria (Acinetobacter sp.) on enhanced dissipation of spiked phenanthrene and pyrene in waterlogged soil. Sci Total Environ 372:1–11. doi: 10.1016/j.scitotenv.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 101.Hamdan LJ, Fulmer PA. 2011. Effects of COREXIT EC9500A on bacteria from a beach oiled by the Deepwater Horizon spill. Aquat Microb Ecol 63:101–109. doi: 10.3354/ame01482. [DOI] [Google Scholar]
- 102.Atlas RM, Stoeckel DM, Faith SA, Minard-Smith A, Thorn JR, Benotti MJ. 2015. Oil biodegradation and oil-degrading microbial populations in marsh sediments impacted by oil from the Deepwater Horizon well blowout. Environ Sci Technol 49:8356–8366. doi: 10.1021/acs.est.5b00413. [DOI] [PubMed] [Google Scholar]
- 103.García-Robledo E, Corzo A, Papaspyrou S. 2014. A fast and direct spectrophotometric method for the sequential determination of nitrate and nitrite at low concentrations in small volumes. Mar Chem 162:30–36. doi: 10.1016/j.marchem.2014.03.002. [DOI] [Google Scholar]
- 104.Strickland JDH, Parsons TR. 1972. A practical handbook of seawater analysis. Fisheries Research Board of Canada, Ottawa, Canada. [Google Scholar]
- 105.Murphy J, Riley JP. 1962. A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36. doi: 10.1016/S0003-2670(00)88444-5. [DOI] [Google Scholar]
- 106.Pelz O, Brown J, Huddleston M, Rand G, Gardinali P, Stubblefield W, Benkinney MT, Ahnell A. Selection of a surrogate MC252 oil as a reference material for future aquatic toxicity tests and other studies. http://gulfresearchinitiative.org/wp-content/uploads/2012/05/Surrogate-Oil-selection-Paper-at-SETAC.pdf.
- 107.Weiss RF. 1974. Carbon dioxide in water and seawater: the solubility of a non-ideal gas. Mar Chem 2:203–215. doi: 10.1016/0304-4203(74)90015-2. [DOI] [Google Scholar]
- 108.Kahm M, Hasenbrink G, Lichtenberg-Frate H, Ludwig J, Kschischo M. 2010. Grofit: fitting biological growth curves. Nat Preced doi: 10.1038/npre.2010.4508.1. [DOI] [Google Scholar]
- 109.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, Mcdonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moonsamy PV, Williams T, Bonella P, Holcomb CL, Höglund BN, Hillman G, Goodridge D, Turenchalk GS, Blake LA, Daigle DA, Simen BB, Hamilton A, May AP, Erlich HA. 2013. High throughput HLA genotyping using 454 sequencing and the Fluidigm Access Array system for simplified amplicon library preparation. Tissue Antigens 81:141–149. doi: 10.1111/tan.12071. [DOI] [PubMed] [Google Scholar]
- 111.Herbold CW, Pelikan C, Kuzyk O, Hausmann B, Angel R, Berry D, Loy A. 2015. A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Front Microbiol 6:731. doi: 10.3389/fmicb.2015.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Green SJ, Venkatramanan R, Naqib A. 2015. Deconstructing the polymerase chain reaction: understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS One 10:e0128122. doi: 10.1371/journal.pone.0128122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bybee SM, Bracken-Grissom H, Haynes BD, Hermansen RA, Byers RL, Clement MJ, Udall JA, Wilcox ER, Crandall KA. 2011. Targeted amplicon sequencing (TAS): a scalable next-gen approach to multilocus, multitaxa phylogenetics. Genome Biol Evol 3:1312–1323. doi: 10.1093/gbe/evr106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. 2014. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ 2:e593. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Poly F, Monrozier LJL, Bally R. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103. doi: 10.1016/S0923-2508(00)01172-4. [DOI] [PubMed] [Google Scholar]
- 119.Muyzer G, De Waal E, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gaby JC, Buckley DH. 2017. The use of degenerate primers in qPCR analysis of functional genes can cause dramatic quantification bias as revealed by investigation of nifH primer performance. Microb Ecol 74:701–708. doi: 10.1007/s00248-017-0968-0. [DOI] [PubMed] [Google Scholar]
- 121.Sun X, Kostka JE. 2018. Hydrocarbon-degrading microbial communities are site-specific and their activity is limited by synergies in temperature and nutrient availability in surface ocean waters. Gulf of Mexico Research Initiative Information and Data Cooperative (GRIIDC), Harte Research Institute, Texas A&M University-Corpus Christi, Corpus Christi, TX. doi: 10.7266/n7-df3b-bq71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are publicly available through the Gulf of Mexico Research Initiative Information and Data Cooperative (https://data.gulfresearchinitiative.org/data/R4.x267.179:0009 [DOI: https://doi.org/10.7266/n7-df3b-bq71]) (121). The generated sequence data are available at the NCBI under BioProject no. PRJNA434326.