In chronic lymphocytic leukemia (CLL), mutations in the NOTCH1 coding region (coding NOTCH1 mutations) have been associated with impaired degradation of NOTCH1 protein1,2 and, clinically, with shorter time to first treatment, shorter overall survival,1–3 and resistance to anti-CD20 immunotherapy in the fludarabine plus cyclophosphamide plus rituximab combination.4,5 In this context, we recently provided evidence that coding NOTCH1 mutations in CLL are associated with reduced CD20 expression, due to a NOTCH1 mutation-driven epigenetic dysregulation involving histone deacetylases.6 More recently, novel recurrent mutations have been identified in the 3′ untranslated region (3′ UTR) of NOTCH1 (3′ UTR NOTCH1 mutations), determining an alternative splicing event within the last NOTCH1 exon,7 again leading to impaired degradation of NOTCH1 protein through a mechanism similar to that occurring in CLL cells bearing coding NOTCH1 mutations. CLL with 3′ UTR NOTCH1 mutations show features of adverse prognosis similar to CLL with coding NOTCH1 mutations in terms of both time to first treatment and overall survival.7 On the other hand, evidence is still lacking regarding the levels of CD20 expression in CLL cases carrying 3′ UTR NOTCH1 mutations. In this study, we provide evidence that 3′ UTR NOTCH1 mutations are associated with low CD20 expression and with relative resistance to anti-CD20 immunotherapy in vitro, thus indicating the need to expand the NOTCH1 mutational analysis to 3′ UTR as a tool to identify anti-CD20 resistant cases.
This study was part of a comprehensive CLL characterization approved by the Internal Review Board of the Aviano Cancer Referral Center (approval n. IRB-05-2010, n. IRB-05-2015) upon informed consent in accordance with the declaration of Helsinki. The study included peripheral blood samples from 662 patients affected by CLL.8 All analyses, including evaluation of CD20 expression, and of NOTCH1 mutational status, were performed on highly purified neoplastic cells (>95% pure). CLL case samples were subjected to purification for negative selection by immunomagnetic beads when required.6,9,10 CD20 expression was evaluated by flow cytometry with a fluorescein isothiocyanate (FITC)-conjugated anti-CD20 antibody (clone L27, BD Biosciences, Milan, Italy), using a FACSCanto II (BD Biosciences).6 NOTCH1 mutational status was assessed by next-generation sequencing covering the whole NOTCH1 exon 34 and part of the 3′ UTR.7 Further details regarding the methods and statistical approaches are provided as Online Supplementary Material and in Online Supplementary Tables S1–S5.
NOTCH1 mutations were detected in 116/662 (17.52%) cases (Online Supplementary Tables S6 and S7), overall accounting for 127 mutations (78 c.7541–7542delCT, 9 other frameshift, 14 nonsense, and 26 3′ UTR mutations) (Online Supplementary Table S7). No missense mutations were detected (Online Supplementary Table S7). Twenty-three of the 26 mutations at the 3′ UTR of NOTCH1 were clonal, i.e. with a variant allele frequency ≥12%,7,11 whereas 55/101 mutations in the NOTCH1 coding region were clonal mutations (Online Supplementary Figure S1A–E).
For the purpose of our analysis, the 116 mutated cases were subdivided into cases with coding NOTCH1 mutations (coding NOTCH1-mut, 90 cases) and cases with 3′ UTR NOTCH1 mutations (3′ UTR NOTCH1-mut, 26 cases). Five cases with concomitant 3′ UTR NOTCH1 mutation and coding NOTCH1 mutation were assigned to the 3′ UTR NOTCH1-mut group according to the substantially higher variant allele frequency detected for the 3′ UTR NOTCH1 mutation.
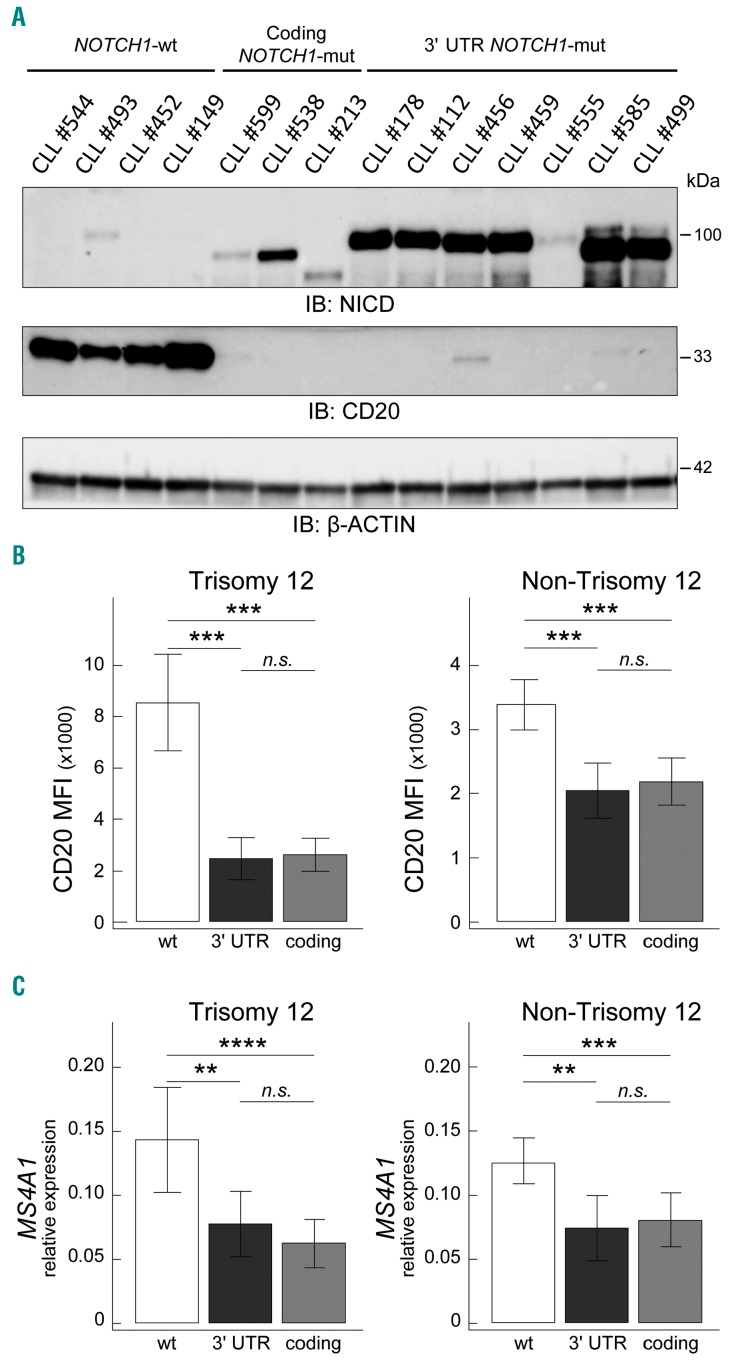
NOTCH1 protein expression was evaluated by western blot in CLL cases carrying either 3′ UTR NOTCH1 mutations or coding NOTCH1 mutations, and, for comparison, in NOTCH1 wild-type (NOTCH1-wt) cases. As shown in Figure 1A, 3′ UTR NOTCH1-mut cases showed high NOTCH1 intracellular domain (NICD) levels, consistent with the presence of an alternative splicing event resulting in a large deletion that disrupts the C-PEST domain causing the subsequent impaired degradation of the NICD.7 In keeping with the presence of coding NOTCH1 mutations that generate a truncated protein with impaired degradation,12 coding NOTCH1-mut cases also showed accumulation of NICD, with molecular weights consistent with the presence of a mutated protein (Figure 1A), both in cases with the c.7541–7542delCT mutation and in cases with other coding NOTCH1 mutations.2,6 In this context, NICD levels were generally consistent with NOTCH1 mutational burden both in 3′ UTR NOTCH1-mut cases and coding NOTCH1-mut cases (Online Supplementary Table S7 and Figure 1A). Conversely, NOTCH1-wt CLL, although expressing discrete amounts of transmembrane NOTCH1 in some instances, usually expressed negligible NICD levels (Figure 1A and Online Supplementary Figure S2A).2,6
Figure 1.
Correlation between 3′ UTR NOTCH1 mutations and CD20 expression. (A) Western blot showing NICD (upper panel) and CD20 (L26 epitope, with short exposure, lower panel) protein expression in representative CLL cases, i.e. four NOTCH1-wt cases, three coding NOTCH1-mut cases (2 cases with the g.139390648CAG>C, c.7541-7542delCT, p.P2514Rfs*4; 1 case with the g.139390929AC>A, c.7261delG, p.V2421*), and seven 3′ UTR NOTCH1-mut cases (5 cases with the g.139390152T>C, c.*7668+371A>G; 2 cases with the g.139390145T>C, c.*7668+378A>G). β-actin was used as loading control. Identification (ID) number according to Online Supplementary Table S1 is also reported. (B) Bar graphs showing CD20 protein expression levels evaluated by flow cytometry in 112 trisomy 12 CLL cases (9 3′ UTR NOTCH1-mut cases, 35 coding NOTCH1-mut cases, 68 NOTCH1-wt cases) and 550 non-trisomy 12 CLL cases (17 3′ UTR NOTCH1-mut cases, 55 coding NOTCH1-mut cases, 478 NOTCH1-wt cases). Data were analyzed using the t test. *: P<0.05; **, P<0.01; ***, P<0.001; n.s.= not significant. Bar graphs represent mean values, error bars represent the 95% confidence interval (CI). Abbreviations: NOTCH1-wt: wt; 3′ UTR NOTCH1-mut: 3′ UTR; coding NOTCH1-mut: coding. (C) Bar graphs showing MS4A1 transcript expression levels, as evaluated by quantitative real-time polymerase chain reaction, in 662 CLL cases subdivided according to NOTCH1 mutational status as reported in (B). Data were analyzed using the t test. *P<0.05; **P<0.01; ***P<0.001; n.s.= not significant. Bar graphs represent mean values, error bars represent the 95% confidence interval. NOTCH1-wt: wt; 3′ UTR NOTCH1-mut: 3′ UTR; coding NOTCH1-mut: coding.
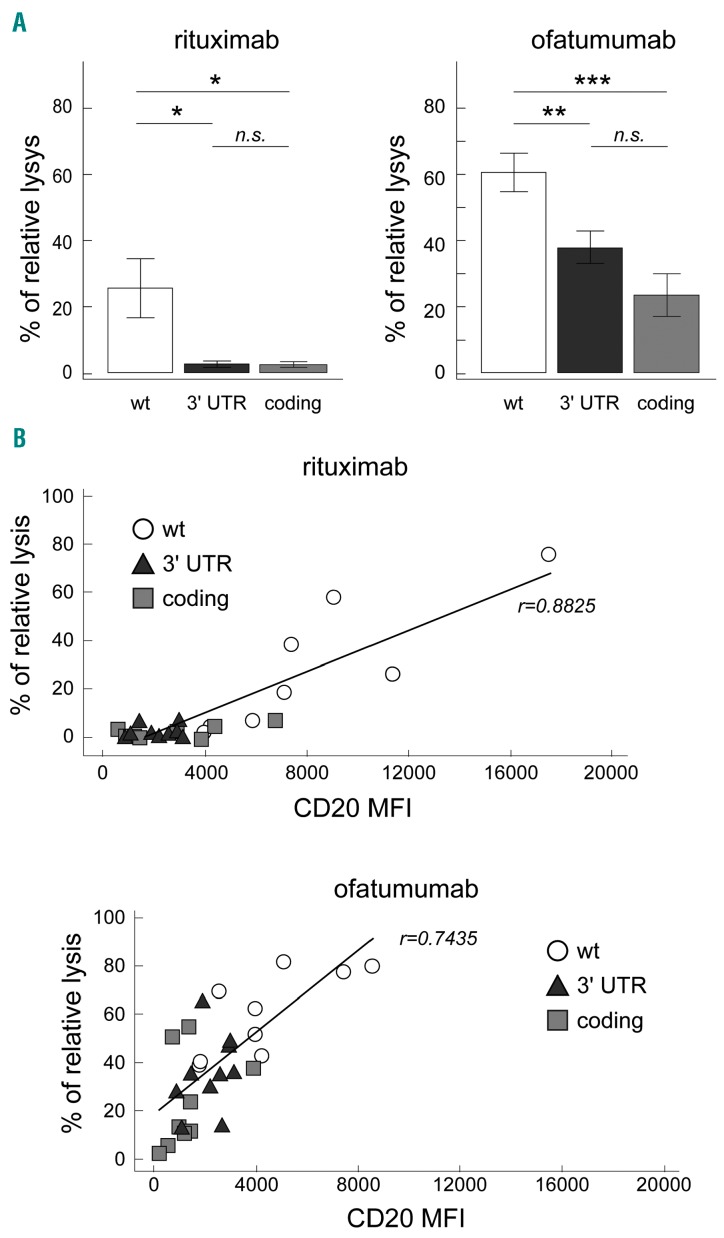
Figure 3.
3′ UTR NOTCH1 mutations and susceptibility to anti-CD20 antibodies in CLL. (A) Left panel: bar graphs showing the percentage of relative lysis of CLL cells, from nine 3′ UTR NOTCH1-mut, nine coding NOTCH1-mut and nine NOTCH1-wt CLL cases, treated with rituximab in a standard complement-dependent cytotoxicity assay. Right panel: bar graph showing the percentage of relative lysis of CLL cells, from nine 3′ UTR NOTCH1-mut, nine coding NOTCH1-mut and nine NOTCH1-wt CLL cases, treated with ofatumumab in a standard complement-dependent cytotoxicity assay. Data were analyzed using a t test. *: P<0.05; **, P<0.01; ***, P<0.001; n.s.= not significant. Bars represent mean values, error bars represent standard error of the mean (SEM). (B) Correlation plots showing CD20 expression versus percentage of relative lysis (r= Pearson correlation coefficient) in 3′ UTR NOTCH1-mut, coding NOTCH1-mut and NOTCH1-wt CLL cases, as evaluated by a complement-dependent cytotoxicity assay using rituximab (upper panel) or ofatumumab (lower panel). NOTCH1-wt: wt; 3′ UTR NOTCH1-mut: 3′ UTR; coding NOTCH1-mut: coding.
Using flow cytometry to evaluate CD20 expression by mean fluorescence intensity (MFI) in the 662 cases classified according to the main cytogenetic aberrations,13 variable CD20 levels were found, the highest levels being detected in trisomy 12 CLL (Online Supplementary Figure S3A).6,14 According to NOTCH1 mutational status, 3′ UTR NOTCH1-mut cases expressed lower levels of CD20 than did NOTCH1-wt cases in both trisomy 12 CLL (mean MFI in 9 3′ UTR NOTCH1-mut cases = 2446 versus mean MFI in 68 NOTCH1-wt cases = 8504; P<0.0001) and non-trisomy 12 CLL (mean MFI in 17 3′ UTR NOTCH1-mut cases = 2049 versus mean MFI in 478 NOTCH1-wt cases = 3389; P<0.0001) (Figure 1B and Online Supplementary Figure S3B). The CD20 levels found in 3′ UTR NOTCH1-mut cases were similar to those detected in coding NOTCH1-mut cases (trisomy 12 CLL: mean MFI in 35 coding NOTCH1-mut cases = 2601; P=0.7470; non-trisomy 12 CLL: mean MFI in 55 coding NOTCH1-mut cases = 2181; P=0.6275). As expected,6 coding NOTCH1-mut cases had lower levels of CD20 expression than NOTCH1-wt cases in both trisomy 12 CLL (P<0.0001) and non-trisomy 12 CLL (P<0.0001) (Figure 1B and Online Supplementary Figure S3B).
When CD20 expression was investigated by western blotting, both 3′ UTR NOTCH1-mut and coding NOTCH1-mut cases showed negligible CD20 levels (Figure 1A) that, in the majority of cases, became detectable only with very high antibody concentrations and long exposure time (Online Supplementary Figure S2B). On the other hand, NOTCH1-wt cases had relevant amounts of CD20 protein (Figure 1A and Online Supplementary Figure S2B). Western blotting also confirmed the inverse correlation between NICD and CD20 levels in both 3′ UTR NOTCH1-mut and coding NOTCH1-mut cases (Figure 1A). Thus, although with discrepancies allegedly due to the different detection method (western blotting versus flow cytometry) and anti-CD20 clone employed (clone L26 in western blotting versus clone L27 in flow cytometry), the western blot experiments corroborated the observation of lower CD20 protein expression in both 3′ UTR NOTCH1-mut and coding NOTCH1-mut cases compared to NOTCH1-wt cases, as determined by flow cytometry.
In keeping with western blot and flow cytometry results, transcript levels of the MS4A1 gene, encoding the CD20 protein, were lower in 3′ UTR NOTCH1-mut cases than in NOTCH1-wt cases in both trisomy 12 and non-trisomy 12 categories (P=0.0053 and P=0.0013, respectively), and similar to those of coding NOTCH1-mut cases (P=0.3294 and P=0.6990, respectively) (Figure 1C). Again, coding NOTCH1-mut cases showed lower MS4A1 transcript levels than NOTCH1-wt cases in both trisomy 12 CLL (P=0.0004) and non-trisomy 12 CLL (P=0.0009) (Figure 1C).6
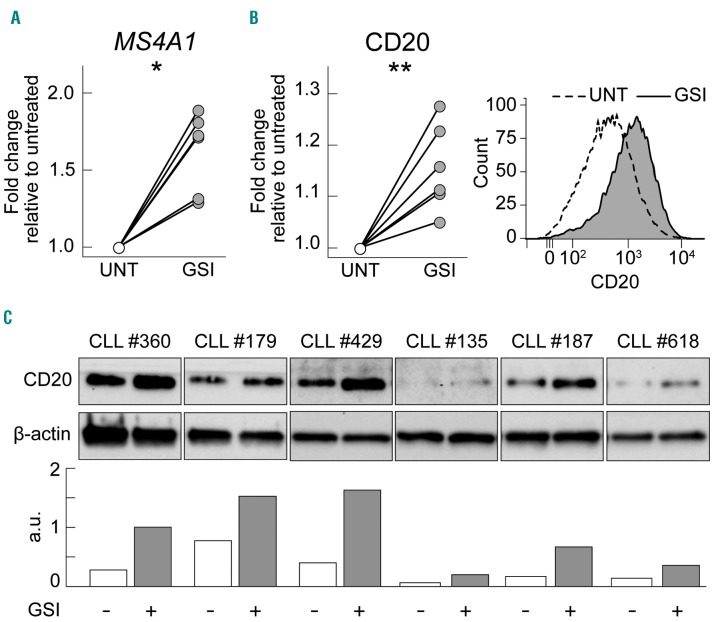
To confirm the direct correlation between NOTCH1 signaling and CD20 expression levels also in 3′ UTR NOTCH1-mut cases,6 CLL cells from 3′ UTR NOTCH1-mut cases were exposed to a gamma-secretase inhibitor and evaluated for CD20 expression.6 Treatment with a gamma-secretase inhibitor, performed in six 3′ UTR NOTCH1-mut cases, increased both MS4A1 transcript levels, at 6 h (P=0.0138) (Figure 2A), and CD20 protein levels, at 24 h, as shown by flow cytometry (mean MFI in untreated samples = 1939 versus mean MFI in gammasecretase inhibitor-treated samples = 2147; P=0.0011) (Figure 2B) and by western blotting (Figure 2C).6
Figure 2.
Induction of CD20 expression by NOTCH1 signaling inhibition. (A) Dot-and-line plots showing fold change increases of MS4A1 transcript expression levels between untreated (UNT) samples and samples treated with a gamma-secretase inhibitor (GSI) for 6 h, of six 3′ UTR NOTCH1-mut CLL cases, as evaluated by quantitative real-time polymerase chain reaction. Data were analyzed using a paired t-test. *: P<0.05; **, P<0.01; ***, P<0.001; n.s.= not significant. (B) Dot-and-line plots showing fold-change increases of CD20 protein expression levels between untreated (UNT) samples and samples treated with GSI (GSI) for 24 h, of six 3′ UTR NOTCH1-mut CLL cases, as evaluated by flow cytometry; a representative overlay histogram of CD20 expression by flow cytometry of CLL cell samples left untreated or GSI treated of a 3′ UTR NOTCH1-mut case is also shown. Data were analyzed using a paired t-test. *: P<0.05; **P<0.01; *** P<0.001; n.s.= not significant. (C) Upper panel: western blot showing CD20 protein expression of CLL cell samples left untreated (−) or GSI treated (+) of six 3′ UTR NOTCH1-mut cases. Lower panel: bar graphs representing the relative densitometric analysis of the same western blot. β-actin was used as loading control. For evaluation of CD20 expression high sensitivity conditions were applied (Online Supplementary Table S5).
Finally, we investigated whether the presence of 3′ UTR NOTCH1 mutations could effectively influence susceptibility to anti-CD20 immunotherapy,6 by evaluating the capability of rituximab and ofatumumab to kill in vitro CLL cells in a standard complement-dependent cytotoxicity assay.15 Consistent with CD20 expression levels, 3′ UTR NOTCH1-mut cases showed lower relative lysis induced by rituximab than did NOTCH1-wt cases (9 3′ UTR NOTCH1-mut cases, mean relative lysis upon rituximab: 2.09% versus 9 NOTCH1-wt cases, mean relative lysis upon rituximab: 25.57%; P=0.0314) (Figure 3A), and similar to those of coding NOTCH1-mut cases (9 coding NOTCH1-mut cases; mean relative lysis upon rituximab: 2.36%; P=0.8159). In the same manner, 3′ UTR NOTCH1-mut cases showed lower relative lysis induced by ofatumumab than did NOTCH1-wt cases (9 3′ UTR NOTCH1-mut cases, mean relative lysis upon ofatumumab: 37.97% versus 9 NOTCH1-wt cases, mean relative lysis upon ofatumumab: 60.64%; P=0.0095), and again similar to those of coding NOTCH1-mut cases (9 coding NOTCH1-mut cases; mean relative lysis upon ofatumumab: 23.40%; P=0.0970). As expected,6 coding NOTCH1-mut cases showed lower relative lysis induced by rituximab and ofatumumab than did NOTCH1-wt cases (P=0.0330 and P=0.0006, respectively) (Figure 3A). Moreover, CD20 levels directly correlated with the killing capacity of both rituximab and ofatumumab, as expressed by percentage of relative lysis (r=0.8825 and r=0.7435, respectively) (Figure 3B). In this context, ofatumumab appeared more efficient than rituximab (Figure 3B).6,15 These results remain to be confirmed by considering another effector function such as antibody-dependent cell-mediated cytotoxicity in a specific in vitro assay.
In conclusion, we showed here that 3′ UTR NOTCH1 mutations are associated with low CD20 expression and with relative resistance to anti-CD20 immunotherapy in vitro, as previously demonstrated for CLL carrying coding NOTCH1 mutations.6 This suggests that it would be useful to introduce a comprehensive diagnostic evaluation of NOTCH1 mutational status, including 3′ UTR NOTCH1 mutations, in CLL patients undergoing anti-CD20 immunochemotherapy.
Supplementary Material
Footnotes
Funding: supported in part by the Associazione Italiana Ricerca Cancro (AIRC), Investigator Grants IG-17622, and Special Program Molecular Clinical Oncology 5 × 1000 N. 10007; Progetto Ricerca Finalizzata I.R.C.C.S. n. RF-2010-2307262, RF-2011-02349712, Progetto Giovani Ricercatori n. GR-2011-02347441, n. GR-2011-02346826, n. GR-2011-02351370, Ministero della Salute, Rome, Italy; Associazione Italiana contro le Leucemie, linfomi e mielomi (AIL), Venezia Section, Pramaggiore Group, Italy; Fondazione per la Vita di Pordenone, Italy; 5×1000 Intramural Program, Centro di Riferimento Oncologico, Aviano, Italy.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Fabbri G, Rasi S, Rossi D, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208(7):1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabbri G, la-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16(3):145–162. [DOI] [PubMed] [Google Scholar]
- 4.Stilgenbauer S, Schnaiter A, Paschka P, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123(21):3247–3254. [DOI] [PubMed] [Google Scholar]
- 5.Bo MD, Del Principe MI, Pozzo F, et al. NOTCH1 mutations identify a chronic lymphocytic leukemia patient subset with worse prognosis in the setting of a rituximab-based induction and consolidation treatment. Ann Hematol. 2014;93(10):1765–1774. [DOI] [PubMed] [Google Scholar]
- 6.Pozzo F, Bittolo T, Arruga F, et al. NOTCH1 mutations associate with low CD20 level in chronic lymphocytic leukemia: evidence for a NOTCH1 mutation-driven epigenetic dysregulation. Leukemia. 2016;30(1):182–189. [DOI] [PubMed] [Google Scholar]
- 7.Puente XS, Bea S, Valdes-Mas R, et al. Non-coding recurrent mutations ichronic lymphocytic leukaemia. Nature. 2015; 526(7574):519–524. [DOI] [PubMed] [Google Scholar]
- 8.Matutes E, Owusu-Ankomah K, Morilla R, et al. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia. 1994;8(10):1640–1645. [PubMed] [Google Scholar]
- 9.Gattei V, Bulian P, Del Principe MI, et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111(2):865–873. [DOI] [PubMed] [Google Scholar]
- 10.Dal Bo M, Bulian P, Bomben R, et al. CD49d prevails over the novel recurrent mutations as independent prognosticator of overall survival in chronic lymphocytic leukemia. Leukemia. 2016;30(10):2011–2018. [DOI] [PubMed] [Google Scholar]
- 11.Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and sub-clonal TP53, SF3B1, BIRC3, NOTCH1 and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127(17):2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. [DOI] [PubMed] [Google Scholar]
- 13.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000; 343(26):1910–1916. [DOI] [PubMed] [Google Scholar]
- 14.Tam CS, Otero-Palacios J, Abruzzo LV, et al. Chronic lymphocytic leukaemia CD20 expression is dependent on the genetic subtype: a study of quantitative flow cytometry and fluorescent in-situ hybridization in 510 patients. Br J Haematol. 2008;141(1):36–40. [DOI] [PubMed] [Google Scholar]
- 15.Golay J, Lazzari M, Facchinetti V, et al. CD20 levels determine the in vitro susceptibility to rituximab and complement of B-cell chronic lymphocytic leukemia: further regulation by CD55 and CD59. Blood. 2001;98(12):3383–3389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.