Abstract
Selenocysteine, the 21st genetically encoded amino acid, is the major form of the antioxidant trace element selenium in the human body. In eukaryotes and archaea its synthesis proceeds through a phosphorylated intermediate in a tRNA-dependent fashion. The final step of selenocysteine formation is catalyzed by (O-phosphoseryl-tRNA:selenocysteinyl-tRNA synthase (SepSecS) that converts phosphoseryl-tRNASec to selenocysteinyl-tRNASec. The human SepSecS protein is also known as soluble liver antigen/liver pancreas (SLA/LP), which represents one of the antigens of autoimmune hepatitis. Here we review the discovery of human SepSecS and the current understanding of the immunogenicity of SLA/LP in autoimmune hepatitis.
Keywords: selenium, selenocysteine tRNA, Sep-tRNA:Sec-tRNA synthase, Soluble Liver Antigen/Liver Pancreas, stop codon recoding, UGA recoding
Introduction
Selenium is an essential antioxidant micronutrient for humans. The 21st genetically encoded amino acid, selenocysteine (Sec), is the principal metabolite of selenium in the human body and the means for exerting its various health benefits (Rayman, 2000). Although chemically similar to cysteine (Cys), Sec is significantly more nucleophilic at physiological pH. The lower pKa of the selenol group (approx. 5.2) ensures that the ionized selenolate form is dominant within the cell. This is in stark contrast to the predominantly reduced thiol group (pKa 8.5) in Cys (Ambrogelly et al., 2007). Similarly to the rest of the 20 canonical amino acids Sec is delivered to the ribosome for protein synthesis by its cognate transfer RNA (tRNA) molecule, tRNASec. Knockout of tRNASec in the mouse is embryonic lethal, which underscores the importance of Sec-containing proteins (or selenoproteins) in mammalian development (Bösl et al., 1997). Mutations and polymorphisms in several of the currently known 25 human selenoproteins have been implicated in cancer and diseases of the muscular, nervous, immune and endocrine systems (reviewed in Bellinger et al., 2009). This is not surprising because most selenoenzymes (i.e., thioredoxin reductase, glutathione peroxidase, thioredoxin/gluta-thione reductase and methionine sulfoxide reductase) safeguard the cell from the detrimental effects of reactive oxygen species and regenerate important antioxidants such as vitamin C, vitamin E and coenzyme Q (Lu and Holmgren, 2009). Sec is also the catalytic residue in iodothyronine deiodinases, the enzymes that enable circulating thyroid hormone to exert its pro-metabolic actions in peripheral tissues. The erroneous replacement of the active site Sec with the chemically similar serine (Ser) or Cys diminishes the activity of several selenoenzymes (Zhong and Holmgren, 2000; Kuiper et al., 2003).
Selenocysteine, an eccentric amino acid
Sec is unique among amino acids in two respects. First, the codon that specifies Sec is one of the stop codons. In the late 1980s and early 1990s, Sec was established as the 21st genetically encoded amino acid specified by UGA that normally serves as a stop codon. Apart from its most common function to signal translational termination and its reassignment to Cys in Euplotes (Meyer et al., 1991) an in-frame UGA codon within the coding region of certain proteins also codes for selenocysteine in organisms from all three domains of life (reviewed in Ambrogelly et al., 2007). An elegant recoding mechanism allows the translation machinery to accurately discriminate between a Sec UGA and a Stop UGA codon. The presence of a stem-loop structure known as the selenocysteine insertion sequence (SECIS) element in the mRNA of selenoproteins signals the insertion of Sec to the translation apparatus. SECIS is present right after the UGA to be recoded in bacterial selenoprotein mRNAs, whereas it is mostly in the 3′ UTR in eukaryotic and archaeal mRNAs, which allows for the insertion of multiple Sec residues in a single eukaryotic polypeptide (Zinoni et al., 1990; Berry et al., 1991; Wilting et al., 1997).
Secondly, Sec is the universal exception to Crick’s paradigm that there should be one aminoacyl-tRNA synthetase (AARS) for each genetically encoded amino acid. AARSs are the enzymes that ‘read’ the genetic code by selecting the right amino acid and pairing it with its cognate tRNA. A prerequisite for this process is the presence of a cellular pool of free amino acids synthesized before this step. Indeed, biochemical experimental efforts of the last century have identified biosynthetic pathways for all 20 canonical amino acids. Sec is the only exception; there is no biosynthetic pathway for free Sec formation within the cell and a selenocysteinyl-tRNA synthetase does not exist. Instead, Sec synthesis occurs on its cognate tRNA in a route that is based on misacylation by seryl-tRNA synthetase (SerRS) and subsequent tRNA-dependent amino acid transformation.
The route to Sec synthesis
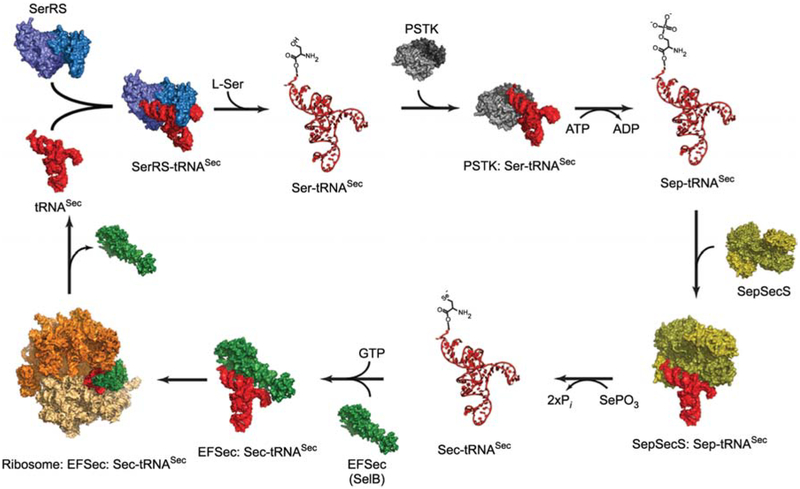
Sec synthesis is interwoven with Sec-tRNA formation in all Sec-encoding bacterial, archaeal and eukaryal organisms. It begins with the misacylation of tRNASec with Ser by SerRS (Figure 1). The structural homology of tRNASer and tRNASec enables SerRS to form Ser-tRNASec, although with only 1% of the efficiency with which it serylates the five tRNASer isoacceptors (Baron et al., 1990). The promiscuity displayed by SerRS is absent from all other Sec-synthetic enzymes. In fact, it is replaced by stringent specificity in which several enzymes recognize only an appropriate ligand that is covalently attached to tRNASec. In bacteria, Ser-tRNASec is directly converted to Sec-tRNASec via the action of the pyridoxal phosphate (PLP)-dependent enzyme selenocysteine synthase (SelA). Selenophosphate made by selenophosphate synthetase (SelD) acts as the selenium donor (reviewed in Böck et al., 2005).
Figure 1.
A schematic diagram of the synthetic cycle of selenocysteine in eukaryotes.
The process begins with serylation of tRNASec (red) by SerRS (light and dark blue). PSTK (light and dark grey) then phosphorylates Ser-tRNASec and releases Sep-tRNASec and ADP. A SepSecS tetramer (gold and olive) subsequently binds Sep-tRNASec and catalyzes a two-step transformation of Sep into Sec using selenophosphate as the selenium donor. The final product, Sec-tRNASec, is delivered to the 80S ribosome (orange and beige) by the specialized elongation factor EFSec (green). Once the Sec residue is inserted into the nascent polypeptide chain, free tRNASec is released for another round of Sec synthesis. All molecules are shown in surface representation, whereas Ser-tRNASec, Sep-tRNASec and Sec-tRNASec are shown as ribbon diagrams. Crystal structures of the bacterial SerRS (Biou et al., 1994), the archaeal PSTK (Araiso et al., 2009), the human SepSecS-tRNASec complex (Palioura et al., 2009), the human tRNASec (Itoh et al., 2009), the archaeal SelB (Leibundgut et al., 2005) and that of the bacterial 70S ribosome in complex with EF-Tu (Schmeing et al., 2009) were used for modeling. Except for the SepSecS-tRNASec complex, all other complexes are proposed models and not true structures.
In archaea and eukarya, Sec-tRNA synthesis involves an additional phosphorylated intermediate. O-phosphoseryl-tRNA kinase (PSTK) phosphorylates Ser-tRNASec to form Sep-tRNASec in the presence of ATP and magnesium (Figure 1). PSTK exhibits a remarkable specificity for Ser-tRNASec. It does not phosphorylate free Ser or Ser attached to its cognate tRNASer. It is thus thought that only the unusual structure of tRNASec can appropriately position Ser in the active site of PSTK for phosphorylation to occur (Carlson et al., 2004). Moreover, in contrast to most AARSs that bind their cognate tRNAs with micromolar affinities, PSTK binds both unacylated and serylated tRNASec with nanomolar affinity (Sherrer et al., 2008). Such tight binding of tRNASec to PSTK might compensate for the significantly lower abundance of tRNASec than tRNASer even in Sec-rich tissues such as liver, kidney and testis (Diamond et al., 1993).
While the discovery of PSTK lent credence to the old finding of Sep-tRNASec in mammalian cell extracts (Mäenpää and Bernfield, 1970), the concomitant discovery of Sep-tRNA:Cys-tRNA synthase (SepCysS) in some methanogenic archaea paved the way for the elucidation of the route to Sec synthesis from Sep-tRNA in archaea and eukarya (Sauerwald et al., 2005). SepCysS uses PLP as a cofactor and a sulfur donor to convert Sep attached to tRNACys to Cys. Given the similar chemistries of Cys and Sec, the presence of Sep-tRNACys as an intermediate of Cys biosynthesis in certain methanogenic archaea suggested Sep-tRNASec as an intermediate in the anabolic cycle of Sec and further supported the quest for an archaeal and eukaryal enzyme that would perform the Sep to Sec conversion (Yuan et al., 2006; Abe et al., 2007; Xu et al., 2007).
Human SepSecS or SLA/LP
The most promising candidate to exhibit such an activity was the human protein Soluble Liver Antigen/Liver Pancreas (SLA/LP). This protein was first identified in the early 1990s as it co-precipitated with tRNASec when mammalian cell extracts were treated with serum from patients with autoimmune hepatitis (Gelpi et al., 1992). Through a computational approach SLA/LP was classified as a PLP-dependent serine hydroxymethyltransferase (Kernebeck et al., 2001) and its archaeal orthologs were only found in known Sec-containing archaea (Yuan et al., 2006). Interestingly, other known tRNA-dependent enzymes that carry out Ser or Sep conversions also require PLP for activity (Leinfelder et al., 1989; Sauerwald et al., 2005). In vivo complementation assays in a heterologous system and in vitro activity assays established the human SLA/LP and its archaeal orthologs as the Sep-tRNA:Sec-tRNA synthase (SepSecS). SepSecS performs the ultimate step in the route to Sec synthesis, the conversion of Sep-tRNASec to Sec-tRNASec (Yuan et al., 2006; Abe et al., 2007; Xu et al., 2007; see Figure 1). However, earlier RNA interference results in mammalian cells could not exclude the existence of an alternative SepSecS-independent pathway of Sec-tRNASec (Xu et al., 2005). Genetic knockouts and in vivo selenoprotein analysis in the parasitic protozoon Trypanosoma brucei showed that the PSTK/SepSecS sequence is the sole route to Sec in T. brucei, and thus possibly in all eukaryotes (Aeby et al., 2009).
Crystal structures of the archaeal (Araiso et al., 2008) and murine apo-SepSecS (Ganichkin et al., 2008) and the most recent of the human SepSecS-tRNASec complex (Palioura et al., 2009) revealed the basis of substrate specificity and the catalytic mechanism of SepSecS. Its homotetrameric structure is distinct from its closest homologue, the dimeric SepCysS, and places SepSecS in its own branch in the family of fold type I PLP-dependent enzymes that stems from the last universal common ancestor (Araiso et al., 2008). Biochemical assays and molecular genetics established a reaction mechanism that proceeds through an external aldimine formed between the bound PLP cofactor and the incoming Sep that is attached to tRNASec (Palioura et al., 2009). SepSecS, like PSTK, exhibits remarkable substrate specificity. The enzyme acts only on Sep-tRNASec and not on free Sep, free Ser or Ser attached to tRNASer (Yuan et al., 2006; Abe et al., 2007; Xu et al., 2007). Structural studies on the human SepSecS-tRNASec complex suggested that tRNASec plays a crucial role in positioning Sep in the active site of SepSecS for catalysis to occur (Palioura et al., 2009).
tRNASec, an unusual tRNA
Comprised of 90 nucleotides, human tRNASec is among the largest eukaryotic tRNAs, the structure of which was recently determined (Itoh et al., 2009; Palioura et al., 2009). Its acceptor-TΨC ‘helix’ contains an additional base pair resulting in a 9/4 fold, in contrast to the 7/5 fold adopted by all known canonical tRNAs (Sturchler et al., 1993). Except for tRNASec, all tRNAs are transferred to the ribosome bound to either EF-Tu in bacteria or eEFlA in eukaryotes. The atypical 9/4 fold of tRNASec accounts for the evolution of a specialized elongation factor, known as SelB in bacteria (Forchhammer et al., 1989) and EFSec in eukaryotes (Fagegaltier et al., 2000; Tujebajeva et al., 2000), which binds only Sec-tRNASec and not other aminoacyl-tRNAs. Both the variable and D arms of tRNASec are longer than the corresponding elements in canonical tRNAs (Palioura et al., 2009; Itoh et al., 2009), whereas the eighth position in tRNASec is occupied by adenine instead of the highly conserved uridine which is found in all canonical tRNAs. In a striking contrast to U8, the base of A8 does not form any tertiary interactions with the D- and TΨC arms leaving a hole in the core of the tRNA molecule (Yuan et al., 2010). All these features result in a distinct three-dimensional structure of tRNASec, which is likely to be recognized by all Sec-specific enzymes.
Whereas SerRS recognizes common structural features of tRNASer and tRNASec, it is thought that PSTK, SepSecS and EFSec recognize the distinct structural elements of tRNASec instead (Yuan et al., 2010). The human SepSecS-tRNASec complex structure revealed that SepSecS does exactly that. It binds the longer acceptor-TΨC ‘helix’, the long variable arm, the 5′ phosphate and the acceptor-TΨC-variable elbow. Practically, SepSecS measures the length of the acceptor- TΨC ‘helix’ as the distance between the variable arm and the acceptor tip of tRNASec. Modeling of canonical tRNAs onto the human SepSecS showed that the length of their acceptor-TΨC ‘helix’ is too short to reach the active site of the enzyme. Even if productive interaction between the tip of the acceptor arm and the enzyme were forced to form, multiple steric clashes would prevent binding of the canonical tRNA to SepSecS (Palioura et al., 2009).
SLA/LP in autoimmune hepatitis
Autoimmune hepatitis (AIH) is a chronic inflammatory liver disease associated with autoantibodies and liver-infiltrating lymphocytes (Krawitt, 2006; Bogdanos et al., 2009). The pathogenesis of AIH is not understood, but it is assumed that the disease is driven by an inappropriate immune response against self antigens. Indeed, the majority of the patients present with autoantibodies that most commonly recognize nuclear antigens (ANA), filamentous actin (SMA), perinuclear antigen of neutrophils (atypical p-ANCA), the SepSecS molecule (SLA/LP), or cytochrome P450 2D6 (LKM-1) (Krawitt, 2006). SLA/LP auto antibodies are present in approximately 20% of the patients (Manns et al., 1987; Wies et al., 2000; Baeres et al., 2002) In contrast to all other autoantibodies detectable in immune-mediated liver diseases, SLA/LP autoantibodies are highly specific for autoimmune hepatitis (Baeres et al., 2002). SLA/LP auto antibodies seem to react specifically with an immunodominant region of the SLA/LP molecule located near the carboxy-terminus between amino acids 450 and 490 (FINRLDRCLKAVR-KERSKESDDNYDKTEDVDIEEMALKLDN), as identified by analysis of carboxy-terminally truncated proteins (Wies et al., 2000). By analyzing a set of overlapping linear peptides covering this region, the dominant epitope recognized by SLA/LP autoantibodies could be confirmed and further restricted to a linear epitope sequence of 30 amino acids (residues 459–490: KAVRKERSKESDDNYDKTEDVDIE-EMALKLDN) (Herkel et al., 2002). Interestingly, two immunodominant CD4 T cell epitopes have been identified, of which one is situated within the immunodominant region recognized by autoantibodies (residues 452–465: NRLDR-CLKAVRKER) (Mix et al., 2008).
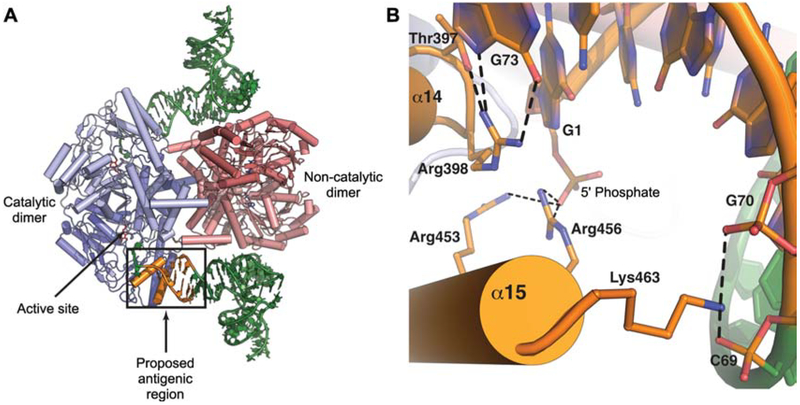
In the structure of the human SepSecS-tRNASec binary complex the first 14 residues of the antigenic region (residues 450–463: FIKRLDRCLKAVRK) form the C-terminal helix α15 (Palioura et al., 2009). Interestingly, helix α15 is spatially located near the entrance to the active-site cleft and it is proximal to helix α14 (Figure 2A). Both α14 and α15 interact with distinct parts of the acceptor arm (Figure 2A). The side chains of Thr397 and Arg398 from helix α14 interact with the discriminator base G73 and, thus, establish the identity of the bound tRNA molecule (Figure 2B). The importance of Thr397 and Arg398 for tRNASec binding has been confirmed by in vivo studies (Palioura et al., 2009). By contrast, the residues Arg453, Arg456 and Lys463 in α15 form the 5′-phosphate binding groove and they interact with the tRNA backbone atoms (Figure 2B). Since autoantibodies from patients with autoimmune hepatitis can precipitate the ribonucleoprotein SepSecS-tRNASec complex, we have proposed that such autoantibodies bind to an interface that lies between the α14 and α15 helices of SepSecS and the tip of the acceptor arm of tRNASec (Figure 2B). The remaining residues of the antigenic region (residues 463–501) form the extreme C-terminal tail of SepSecS and have been disordered in all crystal structures of the enzyme determined to date (Araiso et al., 2008; Ganichkin et al., 2008; Palioura et al., 2009). This more flexible region of the enzyme would be more amenable to the proteolytic cleavage steps required for presentation to the immune system.
Figure 2.
The proposed antigenic region in SepSecS or SLA/LP.
(A) The putative antigenic region of SepSecS is located near the active site of the enzyme (arrow). This region interacts with the tip of the acceptor arm of tRNASec and is crucial for tRNA recognition. The catalytic dimer of SepSecS is in shades of blue, the non-catalytic dimer is colored in shades of pink, two molecules of tRNASec are green, and the antigenic region is orange and demarcated with a box. (B) A close-up view of the interactions at the enzyme-tRNA interface. Residues from helices α14 and α15 interact with the tip of tRNASec. The helix α14 residues interact with the discriminator base G73: Arg398 forms hydrogen bonds with the Hoogsteen face of G73, whereas Thr397 stabilizes this interaction. The C-terminal helix α15 interacts with the tRNA backbone and with the 5′ phosphate. Arg453 and Arg456 are within hydrogen bonding distance from the 5′ phosphate, whereas Lys463 interacts with the non-bridging oxygens of C69 and G70.
The remarkable uniformity in epitope recognition among the patients suggests that autoimmunity to SLA/LP is antigen-driven and induced by a common mechanism (Herkel et al., 2002). Most probably, autoimmunity to SepSecS is driven by the human SepSecS antigen itself and not by molecular mimicry (Wang et al., 2006). Thus, it is likely that SLA/LP autoimmunity is related to the pathogenesis of autoimmune hepatitis, at least in the subgroup of patients who display anti-SLA/LP reactivity. However, it is currently not clear how the biosynthesis of selenocysteine could be related to autoimmune hepatitis. Selenoproteins are synthesized in various organs, but nutritional selenium is mainly metabolized in the liver, from where selenium is distributed to other organs in the form of selenoprotein P (Gromer et al., 2005). One could thus speculate that nutritional selenium compounds or their metabolites or, alternatively, selenium deficiency might alter hepatic SepSecS in such ways that it becomes an immunogenic neoantigen. Such alterations could include dysfunction of the SepSecS enzyme, aberrant sub-cellular localization of SepSecS molecules in hepatocytes, or the formation of immunogenic molecular complexes of SepSecS with other proteins or non-protein molecules.
However, it is conceivable that modifications of SepSecS molecules by metabolites in the liver may initiate an immune response to SepSecS. A similar scenario has been suggested to initiate autoimmunity to the pyruvate dehydrogenase complex in primary biliary cirrhosis, which seems to be related to lipoic acid and neoantigenic xenobiotic lipoic acid analogs (Bruggraber et al., 2003; Walden et al., 2008). Of note, the catalytic domain of pyruvate dehydrogenase seems to carry a dominant target epitope of antimitochondrial antibodies in primary biliary cirrhosis patients (Braun et al., 2010). Moreover, such antibodies to pyruvate dehydrogenase have been reported to inhibit its enzymatic activity (Van de Water et al., 1988; Teoh et al., 1994). Given that autoantibodies to the SepSecS molecule also seem to target the catalytic domain, it is thus possible that SLA/LP autoantibodies could inhibit the enzymatic function of SepSecS and thereby contribute to the pathogenesis of autoimmune hepatitis.
Acknowledgments
Work in the D.S. laboratory was supported by grants from the Department of Energy, the National Institute for General Medical Sciences, and the National Science Foundation. S.P. holds a fellowship of the MD/PhD Program of Yale University School of Medicine.
References
- Abe K, Mihara H, Tobe R, and Esaki N (2007). Characterization of human selenocysteine synthase involved in selenoprotein biosynthesis. Biomed. Res. Trace Elements 19, 80–83. [Google Scholar]
- Aeby E, Palioura S, Pusnik M, Marazzi J, Lieberman A, Ullu E, Söll D, and Schneider A (2009). The canonical pathway for selenocysteine insertion is dispensable in Trypanosomes. Proc. Natl. Acad. Sci. USA 106, 5088–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrogelly A, Palioura S, and Söll D (2007). Natural expansion of the genetic code. Nat. Chem. Biol 3, 29–35. [DOI] [PubMed] [Google Scholar]
- Araiso Y, Palioura S, Ishitani R, Sherrer RL, O’Donoghue P, Yuan J, Oshikane H, Domae N, Defranco J, Söll D, et al. (2008). Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation. Nucleic Acids Res. 36, 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araiso Y, Sherrer RL, Ishitani R, Ho JM, Söll D, and Nureki O (2009). Structure of a tRNA-dependent kinase essential for selenocysteine decoding. Proc. Natl. Acad. Sci. USA 106, 16215–16220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeres M, Herkel J, Czaja AJ, Wies I, Kanzler S, Cancado EL, Porta G, Nishioka M, Simon T, Daehnrich C, et al. (2002). Establishment of standardized SLA/LP immunoassays: specificity for autoimmune hepatitis, worldwide occurrence, and clinical characteristics. Gut 51, 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C, Heider J, and Böck A (1990). Mutagenesis of selC, the gene for the selenocysteine-inserting tRNA-species in E. coli: effects on in vivo function. Nucleic Acids Res. 18, 6761–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger FP, Raman AV, Reeves MA, and Berry MJ (2009). Regulation and function of selenoproteins in human disease. Biochem. J 422, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Chen Y, Mandel SJ, Kieffer JD, Harney JW, and Larson PR (1991). Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353, 273–276. [DOI] [PubMed] [Google Scholar]
- Biou V, Yaremchuk A, Tukalo M, and Cusack S (1994). The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263, 1404–1410. [DOI] [PubMed] [Google Scholar]
- Böck A, Thanbichler M, Rother M, and Resch A (2005). Selenocysteine In: Aminoacyl-tRNA Synthetases, Ibba M, Francklyn CS and Cusack S, eds. (Georgetown, TX, USA: Landes Bioscience; ), pp. 320–327. [Google Scholar]
- Bogdanos DP, Mieli-Vergani G, and Vergani D (2009). Autoantibodies and their antigens in autoimmune hepatitis. Semin. Liver Dis. 29, 241–253. [DOI] [PubMed] [Google Scholar]
- Bösl MR, Takaku K, Oshima M, Nishimura S, and Taketo MM (1997). Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc. Natl. Acad. Sci. USA 94, 5531–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Berg C, Buck S, Gregor M, and Klein R (2010). Catalytic domain of PDC-E2 contains epitopes recognized by antimitochondrial antibodies in primary biliary cirrhosis. World J. Gastroenterol 16, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggraber SF, Leung PS, Amano K, Quan C, Kurth MJ, Nantz MH, Benson GD, Van de Water J, Luketic V, Roche TE, et al. (2003). Autoreactivity to lipoate and a conjugated form of lipoate in primary biliary cirrhosis. Gastroenterology 125, 1705–1713. [DOI] [PubMed] [Google Scholar]
- Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, and Hatfield DL (2004). Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl. Acad. Sci. USA 101, 12848–12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond AM, Choi IS, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, McCloskey JA, et al. (1993). Dietary selenium affects méthylation of the wobble nucleoside in the anticodon of selenocysteine tRNA[Ser]Sec. J. Biol. Chem 268, 14215–14223. [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, and Krol A (2000). Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19, 4796–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K, Leinfelder W, and Böck A (1989). Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature 342, 453–456. [DOI] [PubMed] [Google Scholar]
- Ganichkin OM, Xu XM, Carlson BA, Mix H, Hatfield DL, Gladyshev VN, and Wahl MC (2008). Structure and catalytic mechanism of eukaryotic selenocysteine synthase. J. Biol. Chem 283, 5849–5865. [DOI] [PubMed] [Google Scholar]
- Gelpi C, Sontheimer EJ, and Rodriguez-Sanchez JL (1992). Autoantibodies against a serine tRNA-protein complex implicated in cotranslational selenocysteine insertion. Proc. Natl. Acad. Sci. USA 89, 9739–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromer S, Eubel JK, Lee BL, and Jacob J (2005). Human selenoproteins at a glance. Cell. Mol. Life Sci. 62, 2414–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkel J, Heidrich B, Nieraad N, Wies I, Rother M, and Lohse AW (2002) Fine specificity of autoantibodies to soluble liver antigen and liver/pancreas. Hepatology 35, 403–408. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Chiba S, Sekine S, and Yokoyama S (2009). Crystal structure of human selenocysteine tRNA. Nucleic Acids Res. 37, 6259–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernebeck T, Lohse AW, and Grötzinger J (2001). A bioinfor-matical approach suggests the function of the autoimmune hepatitis target antigen soluble liver antigen/liver pancreas. Hepatology 34, 230–233. [DOI] [PubMed] [Google Scholar]
- Krawitt EL (2006). Autoimmune hepatitis. N. Engl. J. Med 354, 54–66. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Klootwijk W, and Visser TJ (2003). Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase reduces catalytic efficiency and alters substrate preference. Endocrinology 144, 2505–2513. [DOI] [PubMed] [Google Scholar]
- Leibundgut M, Frick C, Thanbichler M, Böck A, and Ban N (2005). Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 24, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinfelder W, Stadtman TC, and Böck A (1989). Occurrence in vivo of selenocysteyl-tRNA(SERUCA) in Escherichia coli. Effect of sel mutations. J. Biol. Chem 264, 9720–9723. [PubMed] [Google Scholar]
- Lu J and Holmgren A (2009). Selenoproteins. J. Biol. Chem 284, 723–727. [DOI] [PubMed] [Google Scholar]
- Mäenpää PH and Bernfield MR (1970). A specific hepatic transfer RNA for phosphoserine. Proc. Natl. Acad. Sci. USA 67, 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns M, Gerken G, Kyriatsoulis A, Staritz M, and Meyer zum Buschenfelde KH (1987). Characterization of a new subgroup of autoimmune chronic active hepatitis by autoantibodies against a soluble liver antigen. Lancet 1, 292–294. [DOI] [PubMed] [Google Scholar]
- Meyer F, Schmidt HJ, Plümper E, Hasilik A, Mersmann G, Meyer HE, Engström A, and Heckmann K (1991). UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc. Natl. Acad. Sci. USA 88, 3758–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mix H, Weiler-Normann C, Thimme R, Ahlenstiel G, Shin EC, Herkel J, David CS, Lohse AW, and Rehermann B (2008). Identification of CD4 T cell epitopes within Soluble Liver Antigen/Liver Pancreas autoantigen in autoimmune hepatitis. Gastroenterology 135, 2107–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palioura S, Sherrer RL, Steitz TA, Söll D, and Simonović M (2009). The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science 325, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman MP (2000). The importance of selenium to human health. Lancet 356, 233–241. [DOI] [PubMed] [Google Scholar]
- Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR 3rd, Ibba M, and Söll D (2005). RNA-dependent cysteine biosynthesis in archaea. Science 307, 1969–1972. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy F.V.t., Weir JR, and Ramakrishnan V (2009). The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrer RL, O’Donoghue P, and Söll D (2008). Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation. Nucleic Acids Res. 36, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturchler C, Westhof E, Carbon P, and Krol A (1993). Unique secondary and tertiary structural features of the eukaryotic selenocysteine tRNASec. Nucleic Acids Res. 21, 1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh KL, Mackay IR, Rowley MJ, and Fussey SP (1994). Enzyme inhibitory autoantibodies to pyruvate dehydrogenase complex in primary biliary cirrhosis differ for mammalian, yeast and bacterial enzymes: implications for molecular mimicry. Hepatology 19,1029–1033. [PubMed] [Google Scholar]
- Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, and Berry MJ (2000). Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 1, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden HR, Kirby JA, Yeaman SJ, Gray J, Jones DE, and Palmer JM (2008). Xenobiotic incorporation into pyruvate dehydrogenase complex can occur via the exogenous lipoylation pathway. Hepatology 48, 1874–84. [DOI] [PubMed] [Google Scholar]
- Van de Water J, Fregeau D, Davis P, Ansari A, Danner D, Leung P, Coppel R, and Gershwin ME (1988). Autoantibodies of primary biliary cirrhosis recognize dihydrolipoamide acetyltransferase and inhibit enzyme function. J. Immunol 141, 2321–2324. [PubMed] [Google Scholar]
- Wang C, Teufel A, Cheruti U, Grötzinger J, Galle PR, Lohse AW, and Herkel J (2006). Characterisation of human gene encoding SLA/LP autoantigen and its conserved homologues in mouse, fish, fly and worm. World J. Gastroenterol 12, 902–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wies I, Brunner S, Henninger J, Herkel J, Kanzler S, Meyer zum Büschenfelde KH, and Lohse AW (2000). Identification of target antigen for SLA/LP autoantibodies in autoimmune hepatitis. Lancet 355, 1510–1515. [DOI] [PubMed] [Google Scholar]
- Wilting R, Schorling S, Persson BC, and Böck A (1997). Selenoprotein synthesis in Archaea: identification of an mRNA element of Methanococcus jannaschii probably directing selenocysteine insertion. J. Mol. Biol 266, 637–641. [DOI] [PubMed] [Google Scholar]
- Xu XM, Mix H, Carlson BA, Grabowski PJ, Gladyshev VN, Berry MJ, and Hatfield DL (2005). Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen, in the selenoprotein synthesis machinery. J. Biol. Chem 280, 41568–41575. [DOI] [PubMed] [Google Scholar]
- Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, and Hatfield DL (2007). Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 5, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Palioura S, Salazar JC, Su D, O’Donoghue P, Hohn MJ, Cardoso AM, Whitman WB, and Söll D (2006). RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. USA 103, 18923–18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, O’Donoghue P, Ambrogelly A, Gundllapalli S, Lynn Sherrer R, Palioura S, Simonović M, and Söll D (2010). Distinct genetic code expansion strategies for selenocysteine and pyrrolysine are reflected in different aminoacyl-tRNA formation systems. FEBS Lett. 584, 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L and Holmgren A (2000). Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J. Biol. Chem 275, 18121–18128. [DOI] [PubMed] [Google Scholar]
- Zinoni F, Heider J, and Böck A (1990). Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl. Acad. Sci. USA 87, 4660–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]