Abstract
Woodiness (secondary xylem derived from vascular cambium) has been gained and lost multiple times in the angiosperms, but has been lost ancestrally in all monocots. Here, we investigate the conservation of genes involved in xylogenesis in fully sequenced angiosperm genomes, hypothesizing that monocots have lost some essential orthologs involved in this process. We analyzed the conservation of genes preferentially expressed in the developing secondary xylem of two eudicot trees in the sequenced genomes of 26 eudicot and seven monocot species, and the early diverging angiosperm Amborella trichopoda. We also reconstructed a regulatory model of early vascular cambial cell identity and differentiation and investigated the conservation of orthologs across the angiosperms. Additionally, we analyzed the genome of the aquatic seagrass Zostera marina for additional losses of genes otherwise essential to, especially, secondary cell wall formation. Despite almost complete conservation of orthology within the early cambial differentiation gene network, we show a clear pattern of loss of genes preferentially expressed in secondary xylem in the monocots that are highly conserved across eudicot species. Our study provides candidate genes that may have led to the loss of vascular cambium in the monocots, and, by comparing terrestrial angiosperms to an aquatic monocot, highlights genes essential to vasculature on land.
Keywords: vascular cambium, eudicotyledons, monocotyledons, vasculature, xylogenesis, Zostera marina
Introduction
Plant vasculature from a vascular cambium evolved ∼420 million years ago (Ma), with the oldest recorded fossils displaying secondary growth dated to the early Devonian, 409–394 Ma (Gerrienne et al. 2011; Hoffman and Tomescu 2013). Wood (secondary xylem derived from a bifacial vascular cambium) is thought to be ancestral to the seed plants, as it can be observed in most eudicot lineages as well as in the gymnosperms. Although the angiosperms likely did not originate as tall woody trees but rather as understory shrubs or small trees, early diverging lineages (except for the Nymphaeales) do have a vascular cambium (Spicer and Groover 2010; Cronk and Forest 2017). Wood is characterized by increased deposition of secondary cell wall within specialized cells such as fibers and vessels in angiosperms and tracheids in the case of gymnosperms—with independent evolution of vessels in the gymnospermous Gnetales (Carlquist 2012). With the exception of the monocot lineage, the vascular cambium of the seed plants is responsible for the development of secondary growth and consists of meristematic initials, the daughter cells of which differentiate into secondary phloem and secondary xylem (Larson 1994; De Rybel et al. 2016).
In the eudicots specifically, woodiness is thought to be the ancestral state, whereas all herbaceous species represent a derived adaptation (Spicer and Groover 2010). Indeed, frequent and independent reversion to woodiness are observed in some herbaceous lineages, and have been particularly documented in species that colonize islands, filling the as yet unoccupied niche that woody trees occupy on the mainland. The reversion to woodiness has also been observed in multiple independent members of the Brassicaceae (Lens et al. 2013; Davin et al. 2016). An experimentally derived example of such secondary woodiness is the woody Arabidopsis thaliana phenotype observed when two key flowering genes, SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1) and FRUITFULL (FUL), are simultaneously knocked out, and the typically annual plant becomes a perennial (Melzer et al. 2008).
While most monocots are herbaceous, arborescent monocots exist, as seen in species from the Pandanales, Asparagales, Arecales, and Zingiberales. However, this “wood” is thought to be the result of differentiation from the cambium of the parenchyma of the cortex or pericycle, and may be referred to as secondary thickening meristem (Rudall 1991; Spicer and Groover 2010) or, more recently, the monocot cambium (Zinkgraf et al. 2017). The function of secondary growth, to allow the plant to increase in stature, is comparable between the arborescent monocots and eudicots, but its development is analogous. It is evident that all monocots have had a likely ancestral loss of vascular cambium development (Spicer and Groover 2010), abolishing the possibility of circular vascular bundle patterning, bifacial cambial differentiation, and, eventually, wood formation.
The complete and irreversible absence of a vascular cambium in the monocot lineage likely started with an initial ancestral loss of one or a few essential genes in vascular cambium formation or differentiation in the common ancestor of extant monocots. This loss abolished the developmental program, and, consequently, rendered some downstream genes that were essential exclusively for vascular bundle patterning, vascular cambial development/differentiation, and secondary xylem formation redundant, resulting in further gene loss in all subsequent lineages. Conversely, and as is the case for nonwoody eudicots, the loss or gain of interactions between genes related to xylogenesis resulting in an herbaceous or arborescent growth form is supported by a study of regulatory changes between the woody Arabidopsis mutant from Melzer et al. (2008) and the wild type Arabidopsis (Davin et al. 2016), as well as the recurrent return to a woody phenotype observed in herbaceous eudicots colonizing islands (Lens et al. 2013). Regulatory interactions between genes in developmental programs involved in secondary growth in the eudicots may, therefore, change when new environments are entered or when artificially manipulated, as opposed to new genes being co-opted or evolved for the required functions.
Our understanding of the development of the vascular cambium reinforces this idea; the main genes involved in vascular cambium differentiation are also pleiotropically hardwired to procambium formation, such as meristem development for other nonwoody tissues (Jouannet et al. 2015). Observing the differences in achieving arborescence between eudicots and monocots and considering that a vascular cambium and secondary growth are more likely ancestral to seed plants than independently acquired, we hypothesized that the lack of a vascular cambium in monocots are likely due to the loss of individual genes, while herbaceous eudicots may have merely lost the regulatory interactions between genes essential for woody growth, while the required genes remain intact and conserved. This hypothesis has not been formally tested. Our knowledge on genes related to vascular cambium development and differentiation is largely based on the model herbaceous plant A. thaliana, as well as some eudicot trees, including eucalyptus and poplar. Discovering the potential genes related to the loss of a vascular cambium in monocots could highlight new genes essential specifically for secondary xylem development.
To this end, we compared the genomes of 34 angiosperms (26 eudicots, 7 monocots, and the early diverging Amborella trichopoda) and aimed to identify potential genes responsible for the absence of a vascular cambium in the monocots. First, we identified candidate genes exclusively involved in xylogenesis in the eudicots by searching for genes preferentially expressed in developing xylem in poplar (Populus trichocarpa) and Eucalyptus (E. grandis), and also in early xylogenesis expression clusters from Populustremula, in the 34 angiosperm species with highly conserved orthologs in all eudicot species but absent in the monocots. Next, we constructed a network of early vascular cambial identity, development and differentiation, and secondary cell wall biosynthesis and investigated the conservation of genes involved in these developmental processes across the angiosperms. To further elucidate the possibility that genes involved in other aspects of secondary growth, such as secondary cell wall development and lignification may have been additionally lost after the loss of essential orthologs in vascular cambium development, we investigated the genome of the marine seagrass Zostera marina and inferred further losses of genes otherwise essential to the angiosperms, as this species has undergone losses of many traits associated with a terrestrial habitat (Olsen et al. 2016).
Materials and Methods
Gene Family Clustering and GO Enrichment
We first collected 5,825 genes that are primarily expressed in wood forming tissues in P. trichocarpa, E. grandis (Hefer et al. 2015), and P. tremula (Sundell et al. 2017). By using BLASTP (E value <10−5), we searched for the best hits of poplar genes against the protein coding genes from 37 angiosperms, which are used in Li et al. (2016), including 28 eudicots, 8 monocots, as well as the early diverging angiosperm Am. trichopoda. Three genomes, that is, Lotus japonicus, Phoenix dactylifera, and Medicago truncatula, were excluded from our study due to the incomplete nature of their genomes. In short, the gene families were identified by OrthoMCL to group similar protein sequences into gene families after all-against-all BLASTP searches of protein sequences from the 34 angiosperms from Li et al. (2016). The best BLAST results from each P. trichocarpa gene from Hefer et al. (2015) against the 34 angiosperms were examined if they exist in a single orthogroup from Li et al. (2016) by evaluating the percentage calculated as the ratio of the number of species with best hits to the number of species in an orthogroup. This indicated a high consistency between the two data sets, having a mean of 81.5% and a median of 89.2%. We removed best hits that were not in orthogroups, as they were not considered as orthologous by OrthoMCL. This was done to ensure that only orthologs, and not out-paralogs (duplicated genes before a speciation event), were identified in the species investigated (Li et al. 2003). Furthermore, 29 poplar genes were removed, because the best hits of these poplar genes could not reach a cut-off percentage of 50%.
After this filtering process, we applied a modified normalized phylogenetic profile analysis to explore coevolving xylem genes (Tabach et al. 2013; Sadreyev et al. 2015). For each query gene, we normalized the BLASTP bit-scores of its best hits from different species by the bit-score of each query to the target poplar gene itself (to minimize the effects of gene length). We then calculated Z-scores for each species using the population of normalized bit-scores to reduce the effects of evolutionary distance. Next, Pearson correlation coefficients r were calculated between each gene profile with the Z-scores and these r values were used to perform hierarchical clustering with Ward’s minimum variance method. The gene profiles were further divided into three clusters based on the dendrograms obtained from this hierarchical clustering based on Z-scores. A heat map was drawn based on the gene similarity scores (fig. 1), which were sequence identities in the genome of the investigated species using the list of P. trichocarpa orthologs as reference (Tabach et al. 2013; Sadreyev et al. 2015).
Fig. 1.
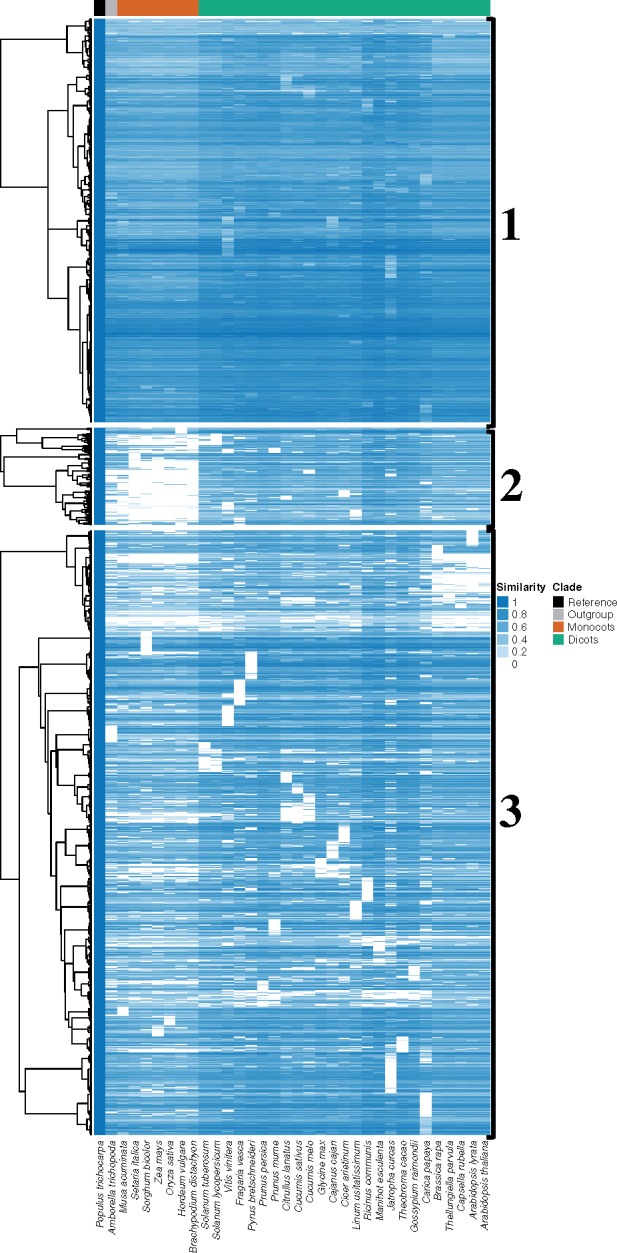
—Heat map representing presence and similarity of the 75 and 5,750 genes preferentially expressed in the xylem tissue of Populus trichocarpa from Hefer et al. (2015). A darker and lighter blue color indicate higher and lower sequence similarity, respectively. Cluster 1 includes genes that are the most conserved across most angiosperm species included in the study. Cluster 2 shows lowest conservation of xylem genes across the monocot species. Cluster 3 represents intermediate conservation across species. The seven monocot species together with the early diverging angiosperm Amborella trichopoda are grouped to the left of the heat map, while the 25 eudicot species are grouped to the right. The three clusters are labeled with a 1, 2, and 3 to the right of the image.
Both the P. trichocarpa and A. thaliana gene IDs of each of the three clusters were extracted into separate gene lists and GO enrichment analysis performed for biological process, molecular function, and cellular component using BLAST2GO, and Fisher’s exact test performed to calculate the enrichments of each one of the three clusters against the genes in all three other clusters (e.g., Cluster 1 against the 5,825 poplar genes preferentially expressed in xylem tissue) as reference (Conesa et al. 2005). While GO term assignment and enrichment have important limitations, it allowed for a starting point in elucidating the potential roles genes of interest might play. The resultantenriched GO terms for the three aspects of each cluster were loaded into REVIGO to reduce redundancy and summarize GO terms. The resultant similarity scores for genes in Cluster 2 (fig. 1) were also compared with the similarity scores of orthologs in Am. trichopoda to compare low similarity monocot orthologs with both the eudicots and the early diverging angiosperm by comparing the average monocot sequence similarity of under 50% with Am. trichopoda sequence similarity >50% (supplementary table S3, Supplementary Material online).
Early Vascular Cambium Differentiation Gene Network
A network of genes involved in early cambial differentiation to xylem tissue was constructed from literature (Demura and Fukuda 2007; Lucas et al. 2013; Jouannet et al. 2015; De Rybel et al. 2016; Smet and De Rybel 2016) (fig. 2). Genes in the early cambial differentiation network constructed from literature were cross-referenced with the PLAZA 3.0 angiosperm databases, including dicots, monocots and Am. trichopoda (Proost et al. 2015), and the eudicot specific gene list (supplementary table S1, Supplementary Material online). The eudicot specific genes were those present only in eudicots obtained from the gene families from Li et al. (2016), and included those that were present in at least 22 of 26 eudicots to account for putative missing orthologs.
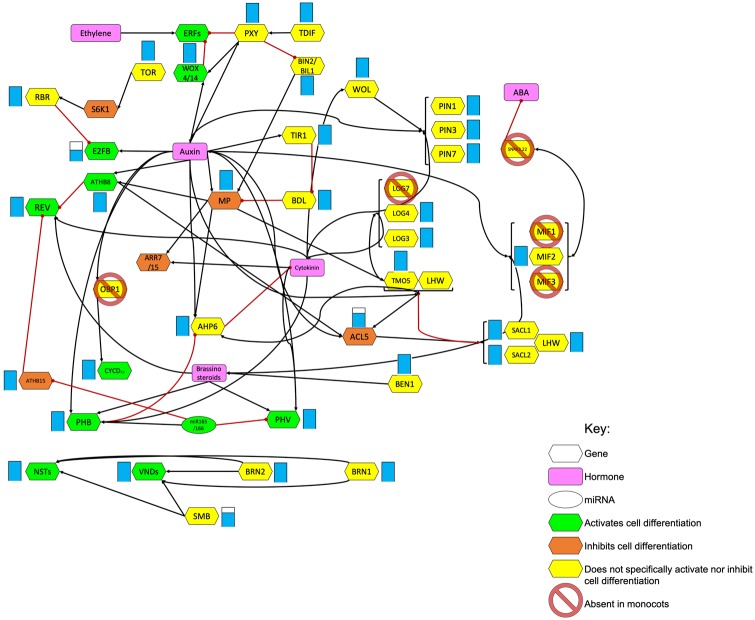
Fig. 2.
—Early cambial differentiation network constructed from literature (see text for details). The network represents the key genes and hormones involved in vascular cambium differentiation. Black arrows indicate positive regulation, while red lines indicate repression or downregulation. Filled blue bars indicate the conservation of genes in the monocots.
Genes Absent in Z. marina
The orthologs from Cluster 1 (fig. 1) generated using the P. trichocarpa gene set from Hefer et al. (2015) as described earlier and representing those genes that are the most conserved across the angiosperms, were searched in the genome of Z. marina by performing a reciprocal BLASTP search. For many-to-many orthologs, OrthoFinder was used to build a gene family set for poplar and Z. marina (Emms and Kelly 2015). Genes absent in Z. marina were identified by removing P. trichocarpa–Z. marina matches from the P. trichocarpa Cluster 1, 2, and 3 gene sets (fig. 1 and supplementary tables S8–S10, Supplementary Material online). GO terms were assigned using BLAST2GO and enrichment of genes absent in Z. marina identified using the Fisher’s exact test and the three respective clusters as reference (Conesa et al. 2005; Supek et al. 2011) (supplementary fig. S1, Supplementary Material online). The top 20 enriched GO terms were manually extracted and is shown in figure 3.
Fig. 3.
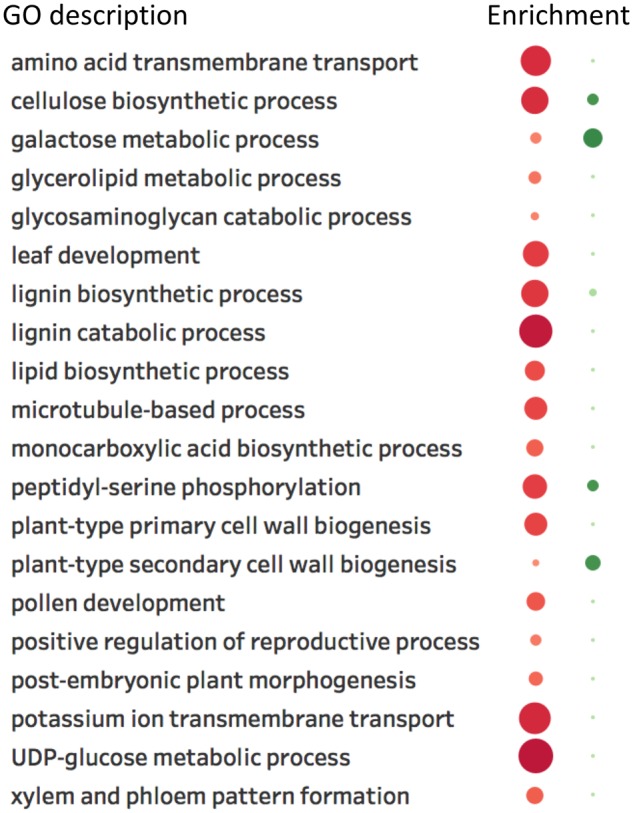
—Selected gene ontology enrichment descriptions of genes from Cluster 1 in figure 1 absent (red) and present (green) in the aquatic monocot Zostera marina. Description of the GO ID is shown in the list on the left, while their corresponding enrichments are indicated in colored circles on the right. Red and green circles represent the enrichment of genes absent and present in Z. marina, respectively. Depth of color and size of the circles indicate the enrichment significance of a particular GO term, with darker color and larger size specifying higher significance, and vice versa.
Results
Conserved Orthologs Preferentially Expressed in Xylem Tissue of P. trichocarpa and E. grandis are Absent in the Monocotyledons
We identified candidate genes involved in xylogenesis in the eudicots by using the gene sets from Hefer et al. (2015) and Sundell et al. (2017) (see Materials and Methods for details). The heat map analysis presented in figure 1 shows the sequence similarity of the preferentially expressed P. trichocarpa genes from Hefer et al. (2015) in 34 angiosperms, and reveals three major clusters (supplementary table S2, Supplementary Material online), with overlap from similar analyses using the vascular cambium and early xylogenesis expression clusters H, E1, and E2 from Sundell et al. (2017) (supplementary fig. S2, Supplementary Material online).
Cluster 1 (fig. 1) contained ∼34% of the genes preferentially expressed in xylem (1,949 P. trichocarpa genes) and showed highest conservation across all angiosperms, with high similarity scores in most species of monocots and eudicots investigated, while Cluster 3 (∼51%, 2,920 P. trichocarpa genes) contained sporadic losses across both the monocot and eudicot lineages, possibly indicating that these genes are less essential for xylogenesis in general compared with Cluster 1 (fig. 1). Cluster 2 (∼8%, 466 P. trichocarpa genes), however, showed loss of orthologs in the monocots specifically, with many genes evidently present and conserved in the genomes of eudicot species only. To estimate if the fraction of Cluster 2 genes is similar to the fraction of poplar genes that do not exist in monocots but in eudicots in the poplar genome, we inquired the gene families from Li et al. (2016) and found 933 poplar genes belonging to eudicot specific gene families (in 24 out of 28 eudicot species) and 20,415 poplar genes exist in 32 out of 37 angiosperms investigated. We, therefore, observe a higher fraction of genes in Cluster 2 than eudicot specific poplar genes (Fisher’s exact test, 2.2×10−16), suggesting that the loss of Cluster 2 genes in monocots may be driven by selection. Furthermore, 73 of the 361 Cluster 2 genes with A. thaliana gene IDs (∼20%, supplementary table S5, Supplementary Material online) had specifically low conservation in the monocots compared not only to the eudicots but also to the early diverging angiosperm Am. trichopoda (average monocot sequence similarity of under 50% with Am. trichopoda sequence similarity >50%) (supplementary table S3, Supplementary Material online), indicating that these genes may be specifically associated with the absence of secondary growth in the monocot lineage.
Most of the A. thaliana IDs in all three clusters from figure 1 were annotated, containing detailed gene descriptions when submitted to The Arabidopsis Information Resource (Berardini et al. 2015), but statistical analysis using Fisher’s exact test revealed that Clusters 1 and 3 contained fewer unknown genes than Cluster 2 (P = 0.045 and P = 0.095, respectively). Of the 1,370 Cluster 1 genes with A. thaliana IDs, 15 were of unknown function (∼1%), while for Cluster 3, 28 of the 2,028 genes were of unknown function (∼1.4%) (supplementary tables S4 and S5, Supplementary Material online). Of the genes in Cluster 2 with A. thaliana gene IDs, 9 of 362 had unknown functions (∼2.5%) (supplementary table S6, Supplementary Material online), and included TBL3 (AT5G01360), a DUF231-containing gene involved in xylan acetylation (Yuan et al. 2016; Zhong et al. 2017). The higher prevalence of genes of unknown function in Cluster 2, and the high conservation of orthologs in all eudicots with low conservation across the monocots may signify candidate genes for further investigation that are responsible for the absence of secondary growth from vascular cambium in the monocot lineage.
Although GO term assignment has important limitations that need to be considered, we used it here as a starting point to uncover putative functions of the genes of interest in Clusters 1, 2, and 3. Supplementary figure S3, Supplementary Material online, a summary of the enrichment results for figure 1 Cluster 1, indicated that this cluster was highly enriched for processes including carbohydrate metabolism, cell surface receptor signaling, organ development, and protein phosphorylation. The cytosol and membrane-associated structures were highlighted in cellular component enrichment, while phosphotransferase, ion and protein binding, and hydrolase activity were among enrichment statistics for molecular function. Notable enrichments in Cluster 3, the cluster indicating lower conservation of genes in both monocots and eudicots, included gene expression regulation, and macromolecule metabolic and biosynthesis regulation among biological processes, together with transcription factor complex as the enrichment for cellular component, and additional transcription factor activity for molecular function (supplementary fig. S4, Supplementary Material online). Cluster 3 was, therefore, more inclusive of genes associated with roles in regulatory processes and indicated variation across the angiosperms included in this study, while Cluster 1 showed enrichment for housekeeping activities expected to be present and conserved across the angiosperms. Cluster 2 did not present with enrichment results according to the Fisher’s exact test through BLAST2GO (Conesa et al. 2005) when the same methods as above were used, which may be due to the small number of genes in this cluster (361 genes vs. 3,757 genes in the full gene set).
Genes Involved in Early Vascular Cambial Differentiation are Absent in the Monocotyledons
We constructed an early vascular cambium differentiation gene network for A. thaliana from literature (Demura and Fukuda 2007; Lucas et al. 2013; Jouannet et al. 2015; De Rybel et al. 2016; Smet and De Rybel 2016) and investigated the presence of orthologs of these genes in the eudicots and monocots, as well as in Am. trichopoda (fig. 2 and supplementary table S7, Supplementary Material online). Although the network was constructed using data from A. thaliana, we found that all genes included in the network showed 100% ortholog conservation in all eudicot species investigated, indicating that this network of vascular cambium differentiation is highly conserved in the eudicots. Furthermore, orthologs for most genes in the constructed gene network were also present in all monocot species investigated, indicating that many of these genes are essential to all angiosperms.
Despite the overall high conservation of the network across the angiosperms, with most genes being present across the species investigated, orthologs for some genes in the network were found to be absent in some, but not all, monocot species. These included the transcription factors E2FB (an important regulator of cell division and proliferation; Magyar et al. 2005), SOMBRERO (SMB, important for root cap development; Bennett et al. 2010), and ACAULIS 5 (ACL5), a spermine synthase encoding gene. E2FB, SMB, and ACL5 were not absent from certain species in a monophyletic nor consistent manner. E2FB was absent from Musa acuminata (banana, Order: Zingiberales), Oryza sativa (Asian rice, Order: Poales), and Setaria italica (Foxtail millet, Order: Poales), while SMB was absent from M. acuminata and Hordeum vulgare (barley, Order: Poales), and ACL5 from H. vulgare and Zea mays (maize, Order: Poales), while present in Brachypodium distachyon (Order: Poales) and Sorghum bicolor (Order: Poales). While the variable presence of orthologs in the monocots may be due to lineage-specific losses or the result of missing annotations, the clear presence and high conservation of orthologs of these genes in the eudicots suggest that they may play essential roles in this lineage of plants.
Significantly, among the known genes involved in regulation of early cambial differentiation, the orthologs of five genes were completely absent from all monocot species while showing 100% conservation in all eudicots (see Materials and Methods for criteria used). These included the transcription factor OBF BINDING PROTEIN 1 (OBP1), SNF1-RELATED PROTEIN KINASE 3.22 (SNRK3.22), Mini Zinc Finger (MIF) gene family members 1 and 3 (MIF1 and MIF3), and the lysine decarboxylase family gene LONELY GUY 7 (LOG7). Of these, OBP1, SNRK3.22, and LOG7 had orthologs in Am. trichopoda, while the orthogroup containing MIF1 (ORTHO03D033600) and MIF3 (ORTHO03D011418) were Brassicaceae- and dicot-specific, respectively, according to the PLAZA 3.0 database (Proost et al. 2015). The A. thaliana Dof protein OBP1, a DNA binding protein that contains a single 51-amino acid zinc finger domain, likely plays an important role in the development and growth of plants (Yanagisawa 2002). It belongs to the DOF zinc finger family which is responsible for regulating transcription due to the presence of a highly conserved DNA-binding domain (Shimofurutani et al. 1998). SNRK3.22 (also known as PROTEIN KINASE SOS2-LIKE 5 [PKS5]), a member of the calcineurin B-like-interacting protein kinase family, is closely involved in the abscisic acid signal transduction through phosphorylation of ABSCISIC ACID-INSENSITIVE5 (ABI5) and has been shown to play an essential role in seed germination in A. thaliana (Zhou et al. 2015). The MIF genes encode small zinc finger proteins important in various aspects of plant development through hormonal regulation. Overexpression of MIF1 results in dramatic developmental defects, inhibiting cell division and elongation which are important characteristics of cambial differentiation, emphasizing its importance in xylogenesis (Hu and Ma 2006), while overexpressed MIF3 induces ectopic meristem on leaf margins (Hu et al. 2011). LOG7 belongs to the lysine decarboxylase family proteins, and is most prominently involved in plant development through its interacting role with the hormone cytokinin. Cytokinin is particularly important in maintaining plant stem cell activity, a vital characteristic of the vascular cambium. The protein encoded by LOG7 promotes cytokinin activation as has been shown by the significant inhibition of cytokinin activation in LOG7 knock-out experiments (Tokunaga et al. 2012).
The observed absence of the orthologs of these genes, especially those also present in Am. trichopoda, may underpin, wholly or in part, the absence of the specific vascular bundle arrangement and subsequent vascular cambial differentiation present in both nonmonocot angiosperms and gymnosperms. Whether a single gene loss event led to the absence of this specific growth form in the monocots or whether a combination of losses is responsible for the phenotype is difficult to ascertain. In addition, the possibility remains that losses of other genes involved in early vascular cambial development that have not yet been associated with this process are the central reason for the absence of a vascular cambium in the monocots, and that the gene losses observed here are downstream effects of that single loss event. Nevertheless, the high conservation of the network presented here indicates that most of the known genes involved in early cambial differentiation are essential and conserved to at least some aspects of vascular growth in all angiosperms, and that certain elements of the network are ubiquitously absent in the monocots.
Zostera m arina has Undergone Additional Losses of Secondary Cell Wall Genes Essential to Terrestrial Angiosperms
Genes involved in cell wall synthesis are expected to be present and conserved across the angiosperms, because although the monocots lack growth from a vascular cambium, they do have secondary cell walls. Indeed, genes involved in secondary cell wall development were found to be highly conserved across the angiosperms (supplementary fig. S5, Supplementary Material online), with only seven genes belonging to the leucine-rich repeat family being eudicot specific and indicating absence in the monocots. Certain species, however, have undergone extreme habitat shifts that may alter the need for these otherwise vital genes, such as the aquatic monocot Z.marina.
Zostera marina has undergone major gene losses associated with the transition from terrestrial to aquatic environments, including genes associated with stomatal developmental, as well as genes involved in pathways such as terpenoid synthesis and ethanol signaling (Olsen et al. 2016). Notable alterations have also occurred in the cell wall composition of Z. marina, transitioning convergently to that similar to aquatic algae (Olsen et al. 2016). Since Cluster 1 (fig. 1) represented genes conserved almost ubiquitously across all angiosperms, we specifically queried genes in this cluster for additional losses in the genome of Z. marina, hypothesizing that transitioning to an almost exclusively submerged aquatic environment should result in further losses of orthologs of genes associated with secondary cell wall development due to the reduced need for mechanical support. Indeed, after querying these genes against the Z. marina genome, we found that Z. marina has undergone additional losses of these otherwise “essential” genes. A total of 437 genes (∼32% of Cluster 1 genes with A. thaliana gene IDs) were lost from the genome of Z. marina (supplementary table S8, Supplementary Material online), compared with 21% of Z. marina genes being absent from Cluster 2, and 20% from Cluster 3 (supplementary tables S9 and S10, Supplementary Material online, respectively). Fisher’s exact test revealed that Cluster 1 had significantly more loss in Z. marina than Cluster 2 (P = 3.25E-10) and Cluster 3 (P = 2.2E-16), while there was no significant difference in loss between Clusters 2 and 3. An enrichment analysis of genes lost in Cluster 1 indicated that these genes were primarily associated with, among others, anatomical structure development, ion transmembrane transport, and functions such as xylem and phloem pattern formation and cellular response to abiotic stress (fig. 3).
Several genes potentially associated with normal secondary cell wall formation and lignin biosynthesis have also been lost from the genome of Z. marina, including known secondary cell wall genes IRREGULAR XYLEM 1, 3, 6, 10, and 12 (IRX1, IRX3, IRX6, IRX10, and IRX12), together with ESKIMO 1 (ESK1), NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1 (NST1), and XYLEM CYSTEINE PEPTIDASE 1 (XCP1). NST1 is partially redundant with NST3, and together they play important roles in eudicot fiber secondary cell wall formation (Mitsuda et al. 2007). NST3 was not found to be absent in Z. marina, therefore, the loss of NST1 might not have such a dramatic effect as the loss of both would have had. Furthermore, three genes highly expressed in the xylem tissue of E. grandis were also absent from the genome of Z. marina. These included ETHYLENE-FORMING ENZYME (EFE)/ACO4, GALACTINOL SYNTHASE 4 (GOLS4), and MICROTUBULE-ASSOCIATED PROTEIN 65-8 (MAP65-8). EFE/ACO4 is highly upregulated during tension wood formation in poplar (Andersson-Gunnerås et al. 2006) and Eucalyptus (Mizrachi et al. 2015). Interestingly, REVOLUTA (REV), a positive regulator of xylogenesis (Prigge et al. 2005), and PIN-FORMED 1 (PIN1), were also absent from Z. marina, and together with MAP65-8 (Oda and Fukuda 2012), are genes potentially involved in xylem and phloem pattern formation and various aspects of early plant development.
Discussion
The sequencing of genomes from early diverging land plant lineages such as Selaginella moellendorffii (a representative of the earliest diverging vascular plants; Banks et al. 2011), Physcomitrella patens (a representative of the earliest diverging land plants; Rensing et al. 2008), and a variety of charophytic and chlorophytic algae (Derelle et al. 2006; Merchant et al. 2007; Prochnik et al. 2010) has revealed that many of the pathways involved in wood formation, particularly those involved in the synthesis of polysaccharide and phenolic biopolymers deposited in the secondary cell wall, have an ancient ancestry often preceding land plants themselves (Bowman et al. 2007; Lang et al. 2008; Zhong et al. 2010). Over 2,000 genes have been implicated in the development of the plant primary and secondary cell wall, with more likely to be discovered (Arabidopsis Genome Initiative 2000; Carpita et al. 2001; Carpita and McCann 2002), and comparative genomics and transcriptomics within and between woody species have revealed specifically conserved orthologs differentially expressed during xylogenesis (Nystedt et al. 2013; Hefer et al. 2015; Jokipii-Lukkari et al. 2017). Although the conservation of the cell wall is high enough for early diverging species such as Physcomitrella to be used as a model organism for primary cell wall structure and function in angiosperms, much is still to be resolved about our understanding of xylem development, vascular cambium differentiation, and patterning which manifests not only in the case of wood in trees but also in a variety of specialized growth patterns that afford high plasticity in adaptation to environments (Popper 2008; Spicer and Groover 2010). Additionally, many proteins implicated in xylogenesis have unknown functions (Mewalal et al. 2014).
The vascular cambium likely had a single origin early in seed plant evolution, as most angiosperm lineages as well as the gymnosperms contain a vascular cambial layer that differentiates into secondary xylem and secondary phloem. However, secondary growth from vascular cambium is completely absent in the monocot lineage, likely due to a loss of one or multiple crucial genes required for vascular cambium identity, patterning, differentiation, or development early in their evolutionary history. Another possibility, in addition to, or independent from, gene loss, is that the monocots could have acquired functions including closed and dispersed vascular bundles as opposed to the peripherally arranged and open vascular bundles of woody species. Vascular cambial differentiation is tightly controlled by key hormones and highly conserved across plant lineages. Auxin, a phytohormone, plays a crucial role in vascular patterning (fig. 2). Expression of an auxin-responsive transcription factor MONOPTEROS (MP) is increased after accumulation of the phytohormone in procambial cells (Campbell and Turner 2017). MP interacts with the dimer formed from the transcription factors TARGET OF MONOPTEROS 5 (TMO5) and LONESOME HIGHWAY (LHW), forming a heterodimer that promotes periclinal divisions (fig. 2) (Ohashi-Ito and Bergmann 2007; Ohashi-Ito et al. 2014). Class III homeodomain leucine-zipper (HD-ZIP) transcription factors play a key role in determining the polarity of vascular bundles arranged radially in stems. The phytohormones cytokinin and brassinosteroids, together with auxin, coordinate early procambium formation, affecting the number and size of vascular bundles (Campbell and Turner 2017). Auxin regulates the position and patterning of vascular bundles, and cytokinin and brassinosteroids affect the number of vascular bundles in Arabidopsis by stimulating procambial divisions (Matsumoto-Kitano et al. 2008; Ibañes et al. 2009).
In this work, we show that, despite high conservation of genes potentially involved in downstream xylogenesis (genes preferentially expressed in developing xylem tissue from E. grandis and P. trichocarpa) across the angiosperms, many were found to be absent from or poorly conserved in the monocot genomes investigated (fig. 1). Cluster 2 from figure 1 included 361 genes with A. thaliana gene IDs that indicated least conservation to the corresponding orthologs in monocots. From these, 73 genes showed especially low sequence similarity in monocots when not only compared with the eudicots but also to Am. trichopoda (supplementary table S3, Supplementary Material online). Among these were HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (HP6), the overexpression of which results in an increased number of xylem cells (Jang et al. 2017), and FRUCTOKINASE 1 (FRK1), which has been associated with vascular development in knock-out experiments by (Stein et al. 2016), although the smaller xylem cells and cambial necrosis phenotype was apparent in quadruple fpk1 fpk4 fpk6 fpk7 and quintuple fpk1 fpk3 fpk4 fpk6 fpk7 mutants (Stein et al. 2016, 2017). The involvement of MICROTUBULE-ASSOCIATED PROTEINS 70-5 (MAP70-5) in secondary cell wall patterning has been established in A. thaliana (Pesquet et al. 2010), as has the role of VASCULAR-RELATED NAC-DOMAIN PROTEIN 4 (VND4) in developing xylem in vessel cells (Zhou et al. 2014) and both of these appear to be lost in the monocot genomes investigated here. It is evident that none of these genes was found to be individually important for their investigated roles, but that all their products functioned closely with others, and that often the manipulation of both was required to achieve the reported phenotype. HP6 is tightly regulated by the jasmonic acid-responsive transcription factor MYELOCYTOMATOSIS ONCOGENE HOMOLOG 2 (MYC2) (Jang et al. 2017), the knock-down of multiple FRK genes resulted in underdeveloped vasculature (Stein et al. 2016, 2017), MAP70-5 functions with MAP70-1, a close binding partner (Pesquet et al. 2010), and VND1 to VND5 are important for secondary cell wall biosynthesis in vessels (Zhou et al. 2014). These genes, individually or taken together, are, therefore, not necessarily solely responsible for the absolute loss of secondary growth from vascular cambium in the monocot lineage, but are potential candidates for knock-in and knock-out studies to specifically investigate the effects of their absence in the monocots.
From our constructed network of early vascular cambial development (fig. 2), it is evident that this network is highly conserved across the angiosperms. We observed 100% conservation of orthologs in this network across the eudicots, supporting our hypothesis that herbaceous eudicots have never lost orthologs of these genes, but that changes in gene interactions likely led to the absence of secondary xylem and an herbaceous growth habit in nonarborescent species. This observation also supports our understanding of vascular development; that many genes involved in vascular cambial differentiation are also pleiotropically linked to other processes in eudicots such as procambial formation and meristem development (Jouannet et al. 2015). An example involves the homeobox genes REV and PHABULOSA (PHB) that are not only involved in shoot apical meristem differentiation but also in the development of leaf polarity, flower development as well as xylem differentiation in eudicots (McConnell and Barton 1998). Furthermore, although the entire monocot lineage completely lacks secondary growth from a vascular cambium, most orthologs of genes associated with early cambial differentiation in the eudicots were present in the monocots, and are likely pleiotropically linked to processes present in and vital to the monocots, or indeed involved with monocot cambium formation, and would, therefore, be under selective pressure to remain conserved in their genomes. Despite the high conservation of most orthologs, some components (E2FB, SMB, and ACL5) appear to be auxiliary or redundant in the monocots compared with the ubiquitous conservation in the eudicots, while others were completely absent in the monocot lineage.
We infer that the genes showing complete absence of orthologs in the monocots likely resulted from an original single gene loss early in the evolutionary history of the monocot lineage. Loss of one or two genes previously regarded as essential to early vascular cambial differentiation, which did not adversely affect the plant’s ability to thrive, likely led to a cascade of subsequent losses of genes that were no longer regarded as essential or had no function when upstream genes were lost. These losses could also have led to the absence of vascular bundle patterning and vascular cambial development, rendering the lineage incapable of undergoing secondary growth through the formation of secondary xylem. Furthermore, certain orthologs that were preferentially expressed in xylem tissue of E. grandis and P. trichocarpa from Hefer et al. (2015) and particularly involved in secondary cell wall formation and xylogenesis, were also absent from the monocots.
In addition to vascular cambium specific gene losses in the monocot lineage, we also show further losses of genes regarded as essential across the angiosperms (in early diverging angiosperms, monocots, and eudicots), particularly those associated with cell wall formation, in the seagrass species, Z. marina. The shift from a terrestrial to a marine environment was possibly the most severe habitat shift achieved by any angiosperm, and resulted in fundamental land plant innovations to be lost (Olsen et al. 2016). This analysis highlighted genes with known roles in xylogenesis, including IRXs, ESK1, NST1, and REV, as well as other candidates that have not previously been investigated for their role in secondary growth. These include GIBBERELLIC ACID INSENSITIVE (GAI) involved in cell proliferation and expansion, TREHALOSE-6-PHOSPHATE SYNTHASE (TPS1) involved in cell wall deposition and cell division, and others of unknown function (supplementary table S8, Supplementary Material online) which will be valuable targets for subsequent investigations.
In conclusion, despite high conservation of genes involved in early xylogenesis in all angiosperms, monocots have lost important genes associated with vascular cambial and secondary xylem development. Due to the limited availability of noneudicot plant genomes, fewer monocot than eudicot species were included in this study. Naturally, more species, particularly eudicots such as Ceratophyllum, which secondarily lost xylem formation (Schneider and Carlquist 1996), and monocots from a wider range in the lineage, will undoubtedly supplement these findings, but we must acknowledge that there could be multiple independent mechanisms through which xylem formation could have been lost. Nevertheless, this study provides many candidate genes for further investigation to elucidate and better understand their holistic roles in vascular cambial differentiation and secondary growth in the angiosperms. We also highlight genes essential to vasculature on land by comparing terrestrial angiosperms to an almost permanently submerged aquatic species, and further studies focusing on other independent aquatic angiosperms will contribute more to our knowledge of the evolution of xylogenesis in the land plants. In addition, it is not known whether all monocots with secondary thickening meristem obtained this from a shared common ancestor, or whether it was obtained independently, as differences between the monocot cambium has been reported between species (Zinkgraf et al. 2017). More studies focusing on these areas will aid greatly in understanding the loss of vascular cambium in the monocots and the genes that could have been co-opted for use in developing alternative plant tissues.
Availability of Data and Material
Data generated and analyzed during this study are included in this article and its supplementary information files. Additional data are available from PLAZA 3.0, http://bioinformatics.psb.ugent.be/plaza/ Last accessed December 2018.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of South Africa (grant number 102186 to D.R.), the University of Pretoria Research Development Programme (RDP) to E.M., and the European Union Seventh Framework Programme (FP7/2007-2013) under European Research Council Advanced Grant Agreement 322739—DOUBLEUP to Y.V.d.P. Z.L. is funded by a postdoctoral fellowship from the research fund of UGent with number BOFPDO2018001701.
Author Contributions
D.R. Y.V.d.P., and E.M. planned and designed the research. D.R. and Z.L. performed the experiments. D.R., Z.L., Y.V.d.P., and E.M. analyzed the data. D.R. wrote the article.
Literature Cited
- Andersson-Gunnerås S, et al. 2006. Biosynthesis of cellulose‐enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 45(2):144–165. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. [DOI] [PubMed] [Google Scholar]
- Banks JA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332(6032):960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, et al. 2010. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22(3):640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, et al. 2015. The Arabidopsis Information Resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53(8):474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K.. 2007. Green genes—comparative genomics of the green branch of life. Cell 129(2):229–234. [DOI] [PubMed] [Google Scholar]
- Campbell L, Turner S.. 2017. Regulation of vascular cell division. J Exp Bot. 68(1):27–43. [DOI] [PubMed] [Google Scholar]
- Carlquist S. 2012. Wood anatomy of Gnetales in a functional, ecological, and evolutionary context. Aliso 30:33–47. [Google Scholar]
- Carpita N, Tierney M, Campbell M.. 2001. Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant Mol Biol. 47(1–2):1–5. [PubMed] [Google Scholar]
- Carpita NC, McCann MC.. 2002. The functions of cell wall polysaccharides in composition and architecture revealed through mutations. Plant Soil. 247(1):71–80. [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Cronk QCB, Forest F.. 2017. The evolution of angiosperm trees: from palaeobotany to genomics In: Groover A, Cronk Q, editors. Comparative and evolutionary genomics of angiosperm trees. New York: Springer International Publishing. [Google Scholar]
- Davin N, et al. 2016. Functional network analysis of genes differentially expressed during xylogenesis in soc1ful woody Arabidopsis plants. Plant J. 86(5): 376–390. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Mähönen AP, Helariutta Y, Weijers D.. 2016. Plant vascular development: from early specification to differentiation. Nat Rev Mol Cell Biol. 17(1):30–40. [DOI] [PubMed] [Google Scholar]
- Demura T, Fukuda H.. 2007. Transcriptional regulation in wood formation. Trends Plant Sci. 12(2):64–70. [DOI] [PubMed] [Google Scholar]
- Derelle E, et al. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A. 103(31):11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrienne P, et al. 2011. A simple type of wood in two early Devonian plants. Science 333(6044):837.. [DOI] [PubMed] [Google Scholar]
- Hefer CA, Mizrachi E, Myburg AA, Douglas CJ, Mansfield SD.. 2015. Comparative interrogation of the developing xylem transcriptomes of two wood‐forming species: Populus trichocarpa and Eucalyptus grandis. New Phytol. 206(4):1391–1405. [DOI] [PubMed] [Google Scholar]
- Hoffman LA, Tomescu AM.. 2013. An early origin of secondary growth: Franhueberia gerriennei gen. et sp. nov. from the Lower Devonian of Gaspe (Quebec, Canada). Am J Bot. 100(4):754–763. [DOI] [PubMed] [Google Scholar]
- Hu W, Feng B, Ma H.. 2011. Ectopic expression of the Arabidopsis MINI ZINC FINGER1 and MIF3 genes induces shoot meristems on leaf margins. Plant Mol Biol. 76(1–2):57–68. [DOI] [PubMed] [Google Scholar]
- Hu W, Ma H.. 2006. Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J. 45(3):399–422. [DOI] [PubMed] [Google Scholar]
- Ibañes M, Fàbregas N, Chory J, Caño-Delgado AI.. 2009. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci U S A. 106:13630–13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G, et al. 2017. Antagonistic interaction between jasmonic acid and cytokinin in xylem development. Sci Rep. 7(1):10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokipii-Lukkari S, et al. 2017. NorWood: a gene expression resource for evo-devo studies of conifer wood development. New Phytol. 216(2): 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet V, Brackmann K, Greb T.. 2015. (Pro)cambium formation and proliferation: two sides of the same coin? Curr Opin Plant Biol. 23:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Zimmer AD, Rensing SA, Reski R.. 2008. Exploring plant biodiversity: the Physcomitrella genome and beyond. Trends Plant Sci. 13(10):542–549. [DOI] [PubMed] [Google Scholar]
- Larson PR. 1994. The vascular cambium: development and structure. Berlin Heidelberg:Springer-Verlag. [Google Scholar]
- Lens F, Davin N, Smets E, del Arco M.. 2013. Insular woodiness on the Canary Islands: a remarkable case of convergent evolution. Int J Plant Sci. 174(7):992–1013. [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. 2016. Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell 28(2):326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, et al. 2013. The plant vascular system: evolution, development and functions. J Integr Plant Biol. 55(4):294–388. [DOI] [PubMed] [Google Scholar]
- Magyar Z, et al. 2005. The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17(9):2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Kitano M, et al. 2008. Cytokinins are central regulators of cambial activity. Proc Natl Acad Sci U S A. 105(50):20027–20031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Barton MK.. 1998. Leaf polarity and meristem formation in Arabidopsis. Development 125(15):2935–2942. [DOI] [PubMed] [Google Scholar]
- Melzer S, et al. 2008. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet. 40(12):1489–1492. [DOI] [PubMed] [Google Scholar]
- Merchant SS, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318(5848):245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewalal R, Mizrachi E, Mansfield SD, Myburg AA.. 2014. Cell wall-related proteins of unknown function: missing links in plant cell wall development. Plant Cell Physiol. 55(6):1031–1043. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, et al. 2007. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19(1):270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrachi E, et al. 2015. Investigating the molecular underpinnings underlying morphology and changes in carbon partitioning during tension wood formation in Eucalyptus. New Phytol. 206(4):1351–1363. [DOI] [PubMed] [Google Scholar]
- Nystedt B, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497(7451):579–584. [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H.. 2012. Secondary cell wall patterning during xylem differentiation. Curr Opin Plant Biol. 15(1):38–44. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC.. 2007. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 134(16):2959–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, et al. 2014. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr Biol. 24(17):2053–2058. [DOI] [PubMed] [Google Scholar]
- Olsen JL, et al. 2016. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530(7590):331–335. [DOI] [PubMed] [Google Scholar]
- Pesquet E, Korolev AV, Calder G, Lloyd CW.. 2010. The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr Biol. 20(8):744–749. [DOI] [PubMed] [Google Scholar]
- Popper ZA. 2008. Evolution and diversity of green plant cell walls. Curr Opin Plant Biol. 11(3):286–292. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, et al. 2005. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17(1):61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, et al. 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329(5988):223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, et al. 2015. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43(Database issue):D974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319(5859):64–69. [DOI] [PubMed] [Google Scholar]
- Rudall P. 1991. Lateral meristems and stem thickening growth in monocotyledons. Bot Rev. 57(2):150–163. [Google Scholar]
- Sadreyev IR, Ji F, Cohen E, Ruvkun G, Tabach Y.. 2015. PhyloGene server for identification and visualization of co-evolving proteins using normalized phylogenetic profiles. Nucleic Acids Res. 43(W1): W154–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider EL, Carlquist FS.. 1996. Conductive tissue in Ceratophyllum demersum (Ceratophyllaceae). SIDA Contrib Bot. 437–443. [Google Scholar]
- Shimofurutani N, Kisu Y, Suzuki M, Esaka M.. 1998. Functional analyses of the Dof domain, a zinc finger DNA-binding domain, in a pumpkin DNA-binding protein AOBP. FEBS Lett. 430(3):251–256. [DOI] [PubMed] [Google Scholar]
- Smet W, De Rybel B.. 2016. Genetic and hormonal control of vascular tissue proliferation. Curr Opin Plant Biol. 29:50–56. [DOI] [PubMed] [Google Scholar]
- Spicer R, Groover A.. 2010. Evolution of development of vascular cambia and secondary growth. New Phytol. 186(3):577–592. [DOI] [PubMed] [Google Scholar]
- Stein O, et al. 2016. Arabidopsis fructokinases are important for seed oil accumulation and vascular development. Front Plant Sci. 7:2047.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein O, et al. 2017. Corrigendum: Arabidopsis fructokinases are important for seed oil accumulation and vascular development. Front Plant Sci. 8:303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell D, et al. 2017. AspWood: high-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29(7):1585–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T.. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, et al. 2013. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature 493(7434):694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga H, et al. 2012. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 69(2):355–365. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. 2002. The Dof family of plant transcription factors. Trends Plant Sci. 7(12):555–560. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Teng Q, Zhong R, Ye ZH.. 2016. TBL3 and TBL31, two Arabidopsis DUF231 domain proteins, are required for 3-O-monoacetylation of xylan. Plant Cell Physiol. 57(1):35–45. [DOI] [PubMed] [Google Scholar]
- Zhong R, Cui D, Ye ZH.. 2017. Regiospecific acetylation of xylan is mediated by a group of DUF231-containing O-acetyltransferases. Plant Cell Physiol. 58(12):2126–2138. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye Z-H.. 2010. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 15(11):625–632. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhong R, Ye ZH.. 2014. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9(8):e105726.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2015. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 168(2):659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkgraf M, Gerttula S, Groover A.. 2017. Transcript profiling of a novel plant meristem, the monocot cambium. J Integr Plant Biol. 59(6): 436–449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated and analyzed during this study are included in this article and its supplementary information files. Additional data are available from PLAZA 3.0, http://bioinformatics.psb.ugent.be/plaza/ Last accessed December 2018.