Abstract
Toxicity challenges by antifungal arsenals and emergence of multidrug resistance scenario has posed a serious threat to global community. To cope up with this alarming situation, phytoactive molecules are richest, safest, and most effective source of broad spectrum antimicrobial compounds. In the present investigation, six phytoactive molecules [cinnamaldehyde (CIN), epigallocatechin, vanillin, eugenol (EUG), furanone, and epigallocatechin gallate] were studied against Candida glabrata and its clinical isolates. Among these, CIN and EUG which are active components of cinnamon and clove essential oils, respectively, exhibited maximum inhibition against planktonic growth of C. glabrata at a concentration of 64 and 128 μg mL–1, respectively. These two molecules effectively inhibited and eradicated approximately 80% biofilm of C. glabrata and its clinical isolates from biomaterials. CIN and EUG increased reactive oxygen species generation, cell lysis, and ergosterol content in plasma membrane and reduced virulence attributes (phospholipase and proteinase) as well as catalase activity of C. glabrata cells. Reduction of mitochondrial membrane potential with increased release of cytochrome c from mitochondria to cytosol indicated initiation of early apoptosis in CIN- and EUG-treated C. glabrata cells. Transcriptional analysis showed that multidrug transporter (CDR1) and ergosterol biosynthesis genes were downregulated in the presence of CIN, while getting upregulated in EUG-treated cells. Interestingly, genes such as 1,3-β-glucan synthase (FKS1), GPI-anchored protein (KRE1), and sterol importer (AUS1) were downregulated upon treatment of CIN/EUG. These results provided molecular-level insights about the antifungal mechanism of CIN and EUG against C. glabrata including its resistant clinical isolate. The current data established that CIN and EUG can be potentially formulated in new antifungal strategies.
Introduction
Mortality and morbidity incidences of infections caused by Candida have increased in the last few decades. This escalating rate of infection depends upon a number of factors including age of the patient, antibiotic therapy, and immune state of patients.1,2 In catheter-associated urinary tract infection, Candida is ranked second, whereas third is the blood-stream infections caused in intensive care unit.3−5 Among Candida species, Candida albicans is the major etiological agent of invasive candidiasis in hospitalized patients. However, non-albicansCandida (NAC) species such as Candida glabrata, Candida tropicalis, and Candida parapsilosis have emerged as a leading cause of systemic candidiasis because of the arbitrary use of antibiotics and increased implanted devices.6 In Australia, the incidence rate of C. glabrata-associated candidemia rose from 16 to 26.7% between 2004 and 2015.7 The distribution of Candida species has changed in last decade resulting in an increase in proportion of C. glabrata in the U.S., Australia, and Europe, whereas C. parapsilosis in Latin America and Africa along with C. albicans.8 In India, a total of 70 Candida isolates were collected in which C. albicans was present in 34 samples, whereas in 36 samples, predominant NAC spp. namely C. tropicalis, Candida haemulonii, C. glabrata, and Candida pelliculosa were found.9 Similar reports related to incidences of candidiasis and dominance of NAC spp.-related infections are available from different parts of India, indicating the severity of fungal infections and their distribution.10−12 Among NAC species, C. glabrata is highly infectious in immunocompromised, diabetic, and hematologic malignant patients.13−17 It is also the major causative agent of vulvo vaginal candidiasis and candiduria.18
The recurrent infections caused by Candida spp. are difficult to treat because of their ability to form biofilm, a three-dimensional, complex architecture of surface-adhered cells encased into extracellular matrix (ECM) where microbes afford protected environment.19 Cell surface hydrophobicity has an important role in cell adherence to substratum and is mediated by cell-surface-attached hydrophobic proteins.20 Biofilm ECM is composed of exopolymeric substances in which the ratio of all macromolecules varies with the environment.21 ECM acts as a barrier to toxic substances and drugs, protects cell from phagocytic cells, and maintains nutrients.22 Also, it offer structural scaffold for cell adherence to different surfaces.23 Extracellular DNA (eDNA) is also one of the important components of ECM and provides structural integrity.24 The presence of hydrolytic enzymes (proteinase and phospholipase) in ECM facilitates tissue penetration and invasion.25 Therefore, all of these characteristic features and components turn biofilm as a source of recalcitrant infections which are undoubtedly difficult to eradicate and hence liable for clinical repercussions.26
Sterol is an important component of eukaryotic cell membrane which is crucial for the structure maintenance and functioning of the cell. Ergosterol is a principal fungal sterol and a well-established target for three major classes of antifungals: azoles, polyenes, and echinocandin.27,28 Any defect in ergosterol biosynthesis or drop in ergosterol content in Candida results in upregulation of ERG genes, AUS1, TIR3 (sterol influx transporter), SUT1, and UPC2 (sterol metabolism regulator).29C. glabrata is inherently resistant to azoles but a partial loss-of-function mutation in MSH2 (DNA mismatch repair gene) is responsible for its unusual high resistance to azoles in clinical isolates.2 Further, recent surveillance data have revealed the development of echinocandin resistance in C. glabrata because of mutations in hotspot regions of the genes FKS1 and FKS2.30 Echinocandin is the latest class of antifungal which was introduced 15 years back and till date a long pause in the discovery of clinically active antifungal reveals the hurdles associated with drug development for eukaryotic pathogens.31
Phytoactive molecules have emerged as a promising antibiofilm candidate which acts by inhibiting synthesis/degrading the signal molecule or blocking the binding site on receptor thereby, inhibiting the signal transduction cascade events.32−34 Occurrence of phytoactive molecules has been reported in a variety of secondary metabolites (flavanoids and catechins) and essential oils (EOs).35−37 EOs are plant-derived concentrated hydrophobic volatile liquids which serve as potential candidates for treating superficial infections.38 They are well-documented antifungal agents and offer an advantage of being used in synergy with conventional antimycotics, even at a lower dose.35,39,40 Earlier, reports on antifungal activity of phytoactive molecules have indicated their curative effect against C. albicans.41 However, the role of phytoactive molecules on C. glabrata is still to be deciphered as it is different from C. albicans in terms of virulence, ploidy, size, phenotypic switching, and antifungal susceptibility.42
The present scenario of antifungal resistance against conventional therapies demands the need for more effective remedy against C. glabrata infections. The naturally occurring bioactive molecules stand out as potential therapeutic candidates against oral and superficial infections.43 This study aimed to highlight the antifungal activity of six different phytoactive molecules, namely eugenol (EUG), epigallocatechin gallate, cinnamaldehyde (CIN), vanillin, furanone, and epigallocatechin (Figure 1), for their biofilm eradication potency and their effect on transcriptional expression, biofilm matrix, and cell wall components against C. glabrata and its clinical isolates. Of the selected phytoactive molecules, CIN and EUG are active components of EOs of cinnamon and clove, respectively, whereas catechins (epigallocatechin gallate and epigallocatechin) are derived from green tea. The phenolic compound vanillin is present in the vanilla pod extract, and furanone presence has been reported in red algae (Delisea pulchra).
Figure 1.
Chemical structure of phytoactive molecules. (a) Vanillin, (b) CIN, (c) EUG, (d) furanone, (e) epigallocatechin, and (f) epigallocatechin gallate. Chemical structures of the molecules were drawn using ChemDoddle software.
Results
Biofilm Formation Ability of C. glabrata and Its Clinical Isolates
The biofilm-forming ability of C. glabrata and its clinical isolates were compared at different time intervals (0–72 h) formed on 96-well microtiter plates (MTPs) and quantified using a 2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide (XTT) reduction assay (Figure 2). Data showed no significant difference in surface adherence by C. glabrata and its clinical isolates on MTP, initially. However, results at 24 h biofilm of the clinical isolates CCG1, CCG3, and CCG4 were significantly higher (20, 33, and 30%, respectively; P < 0.05) to that of the control. Once the mature biofilm was formed after 48 h, again no considerable difference in the optical density (OD) values at 492 nm was recorded by XTT reduction assay (Figure 2).
Figure 2.
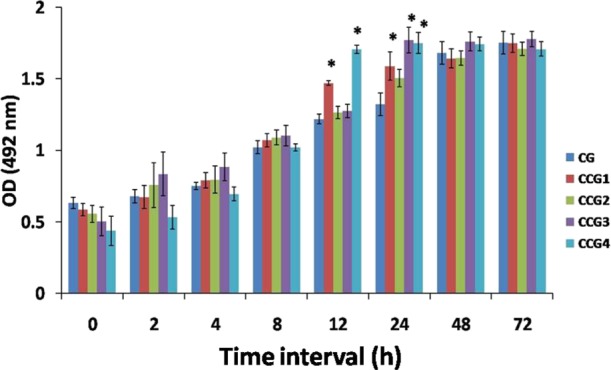
Comparative evaluation of C. glabrata and its clinical isolates biofilm development at different time interval (0−72 h). The value of XTT reduction assay at 492 nm is plotted with different time interval (*P < 0.05).
Fungicidal Activity of Phytoactive Molecules
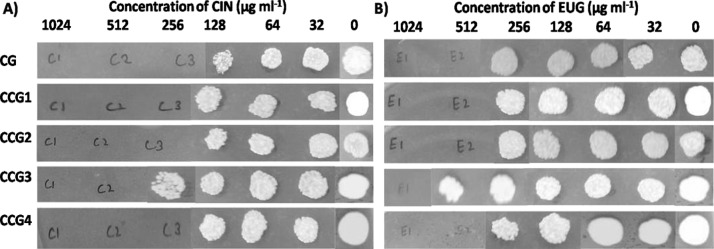
Planktonic growth inhibition of C. glabrata was recorded using six different phytoactive molecules (CIN, EUG, epigallocatechin gallate, vanillin, epigallacatechin, and furanone), as shown in Table 1. Among these, CIN and EUG having an MIC90 value of 64 and 128 μg mL–1, respectively, were selected. These selected molecules were then tested against the growth of C. glabrata and its clinical isolates by performing a spotting assay. Data depicted fungicidal concentration of CIN and EUG to be 256 and 512 μg mL–1, respectively, except CCG3 (Figure 3). Minimum fungicidal concentration (MFC) value of CIN was 512 μg mL–1, whereas that of EUG was 1024 μg mL–1 for CCG3, suggesting this clinical isolate to be the most resistant strain among the chosen isolates (Figure 3).
Table 1. MIC90 Value of Phytoactive Molecules for C. glabrata Planktonic Growth.
| s. no. | phytoactive molecules | source | MIC90 (μg mL–1) |
|---|---|---|---|
| 1 | CIN | EO of cinnamon | 64 |
| 2 | EUG | EO of clove | 128 |
| 3 | EPG | leaf extract of green tea | 512 |
| 4 | vanillin | vanilla pod extract | >1024 |
| 5 | EPC | leaf extract of green tea | 512–1024 |
| 6 | furanone | extract of red algae | 512–1024 |
Figure 3.
MFC of CIN (A) and EUG (B) against planktonic growth of C. glabrata and its isolates on YPD media plates after 48 h of preincubation in RPMI medium with phytoactive molecules.
Inhibitory and Eradication Potency of CIN and EUG
The biofilm inhibitory and eradication potency of CIN and EUG were checked against C. glabrata and its clinical isolates (Figures S1 and S2; Table 2). The results were expressed as biofilm inhibitory concentration (BIC80) and biofilm eradication concentration (BEC80). The inhibition of C. glabrata and its clinical isolate biofilm were gradual with increasing concentration of CIN from 0 to 512 μg mL–1. Enhanced percent inhibition of biofilm was recorded in EUG when the concentration was raised from 32 to 512 μg mL–1 (Figure S1). The BIC80 value of CIN for C. glabrata and its clinical isolate (except CCG3) was 64 μg mL–1, whereas it was 128 μg mL–1 for CCG3, suggesting that clinical isolate CCG3 to be more surface adhering and less susceptible to CIN and EUG (Table 2). EUG also exhibited similar inhibition pattern; BIC80 value of CCG3 (256 μg mL–1) was twice the BIC80 value for all strains (128 μg mL–1). The biofilm susceptibility of C. glabrata and its clinical isolates toward CIN were higher (2× times) than that of EUG (Table 2).
Table 2. BEC80 and BIC80 of CIN and EUG Against C. glabrata and Its Isolates.
| CIN (μg mL–1) | EUG (μg mL–1) | |||
|---|---|---|---|---|
| strain | BIC80 | BEC80 | BIC80 | BEC80 |
| C. glabrata | 64 | 128 | 128 | 512 |
| CCG1 | 64 | 64 | 128 | 512 |
| CCG2 | 64 | 64 | 128 | 256 |
| CCG3 | 128 | 128 | 256 | 512 |
| CCG4 | 64 | 128 | 128 | 256 |
Efficacy of CIN and EUG in eradicating C. glabrata and its clinical isolate biofilm was examined in 96-well MTPs using XTT reduction assay (Table 2; Figure S2). The BEC80 value of CIN for all strains was 128 μg mL–1 except CCG1 and CCG2 for which it was twofold lower (64 μg mL–1), as shown in Table 2. The BEC80 value of CIN for CCG1, CCG2, and CCG3 were similar to their respective BIC80, whereas that of C. glabrata and CCG4, it was twofold higher than their BIC80, indicating less susceptibility of their mature biofilm. The BEC80 value of EUG for C. glabrata, CCG1, and CCG3 was 512 μg mL–1, whereas it was 256 μg mL–1 for CCG2 and CCG4, indicating sensitivity of mature biofilm of CCG2 and CCG4 toward EUG (Table 2; Figure S2).
The biofilm inhibitory effect of CIN and EUG can be correlated with the results of surface hydrophobicity index (HI) of CIN- and EUG-treated C. glabrata and CCG3 which was measured by a two-phase system. The HI value showed that CCG3 (90%) was significantly more hydrophobic than C. glabrata (71%). The hydrophobicity of CIN (68.4%)- and EUG (65.34%)-treated C. glabrata decreased as compared to control. Whereas the HI value of CCG3 treated with CIN (35.0%) decreased but remain unchanged when exposed to EUG (84.3%) (Figure S3).
Effect of CIN and EUG on C. glabrata Extracellular Matrix
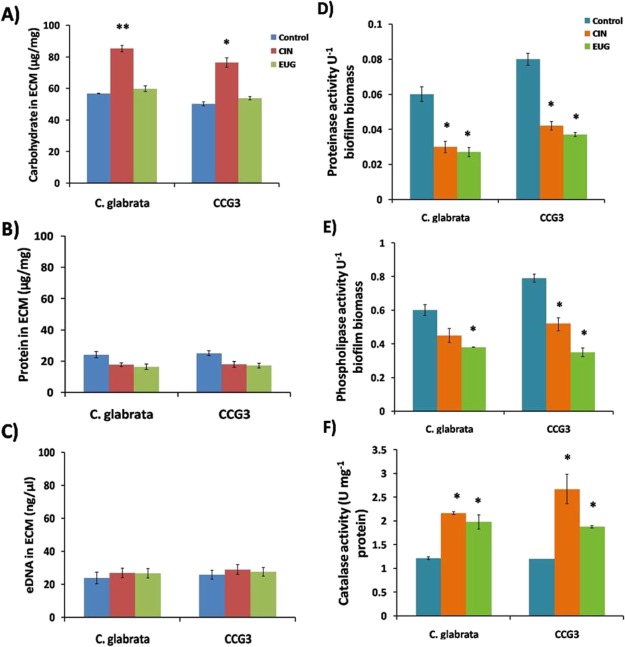
The effect of CIN and EUG treatment on biochemical composition of C. glabrata extracellular matrix (ECM) was studied and compared with that of CCG3. The carbohydrate content of ECM in control, C. glabrata, and CCG3 control were almost similar. An increase in the carbohydrate content of ECM was observed in both C. glabrata (30%) and CCG3 (26%) upon CIN exposure, as compared to their respective control (Figure 4A). However, no change in carbohydrate content was noticed upon EUG treatment. Indeed, no change was observed in the protein content and eDNA content of CIN- and EUG-treated C. glabrata and CCG3 ECM (Figure 4B,C).
Figure 4.
Quantification of biochemical composition (A) carbohydrate; (B) protein and (C) eDNA in ECM and the specific activity of the enzymes (D) proteinase; (E) phospholipase and (F) catalase activity in C. glabrata; and CCG3 biofilm exposed to CIN (128 μg mL–1) and EUG (256 μg mL–1) after 48 h. Data represent means ± SDs of three independent experiments (**P < 0.01 and *P < 0.05).
The enzymatic activity of C. glabrata ECM treated with CIN and EUG was also studied. Proteinase activity was found to be higher in CCG3 as compared to C. glabrata. However, in the presence of CIN and EUG, the proteinase activity decreased in both C. glabrata and CCG3 (Figure 4D). Likewise, the phospholipase activity of CIN- and EUG-treated C. glabrata and CCG3 ECM was reduced (Figure 4E). The catalase activity of untreated C. glabrata and CCG3 was same, whereas in the presence of CIN and EUG, the activity increased 2- and 1.8-fold, respectively, for both C. glabrata and CCG3 (Figure 4F).
Above observations suggested that biochemical composition and enzymatic activity of C. glabrata and CCG3 ECM were affected by CIN and EUG in a similar manner. As expected, in CCG3 control samples, the hydrolytic enzyme activity (phospholipase and proteinase) was more as compared to C. glabrata control cells. However, no noticeable change was observed in catalase activity between control and CCG3.
Assessment of Morphological Changes in C. glabrata Biofilm
Morphological analysis of C. glabrata and CCG3 biofilm samples at BEC80 value of CIN and EUG was performed on polystyrene disc (1 cm2) for 48 h using field-emission scanning electron microscopy (FESEM). The biofilm of untreated C. glabrata and CCG3 retained their structural integrity as healthy yeast cells (Figure 5A,B). CCG3 exhibited substantial cell-rupturing features as compared to C. glabrata upon treatment of CIN. CCG3 cells depicted sunken and shrunk cellular features and are separated from outer cell wall (Figure 5C,D). However, EUG treatment on both C. glabrata and CCG3 biofilm depicted pore formation with wrinkled topology (Figure 5E,F).
Figure 5.
FESEM images of 48 h mature C. glabrata (A) and CCG-3 biofilm (D); after 24 h of treatment with 128 μg mL–1 CIN (B,E); and 256 μg mL–1 EUG (C,F). The scale bar of magnification is 10 μm/1 μm.
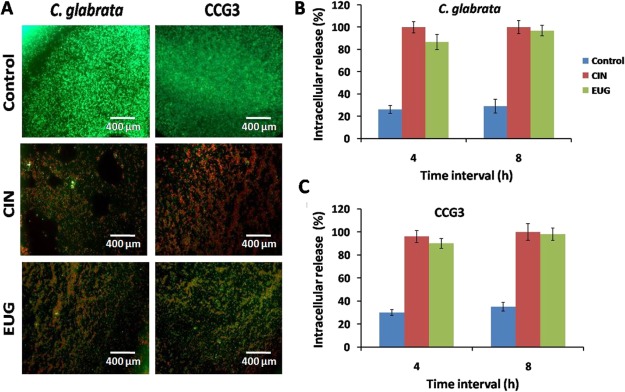
To assess the cellular damage caused by CIN and EUG to C. glabrata biofilm cells, fluorescence microscopy was performed to visualize live–dead cells by FDA–PI. All metabolically active cells emit diffusely distributed green fluorescence, whereas those with damaged membrane showed red fluorescence. In FDA–PI stained biofilm, control emitted only green fluorescence which indicated live cells, whereas CIN- and EUG-treated C. glabrata and CCG3 biofilm emitted red–green fluorescence (Figure 6A).
Figure 6.
(A) Fluorescence microscopy of 48 h mature C. glabrata and CCG3 biofilm after treatment with CIN (128 μg mL–1) and EUG (256 μg mL–1) stained with FDA + PI. Scale bar represents 400 μm. Quantification of intracellular material release in C. glabrata and CCG3 cells treated with (B) CIN (128 μg mL–1) and (C) EUG (256 μg mL–1).
To further co-relate the observations of fluorescence microscopy, the effect of CIN and EUG on cell membrane was determined in terms of rate and amount of released nucleic acid from C. glabrata and CCG3 cells (Figure 6B,C). Both CIN and EUG caused approximately 90% nucleic acid release in C. glabrata and CCG3 after 4 h of incubation. Hundred percent cell lysis was observed after 8 h of incubation of cells with CIN and EUG. Hence, the cell lytic effect of CIN and EUG on C. glabrata and CCG3 can be interpreted.
Reactive Oxygen Species Generated When Cells Exposed to CIN and EUG
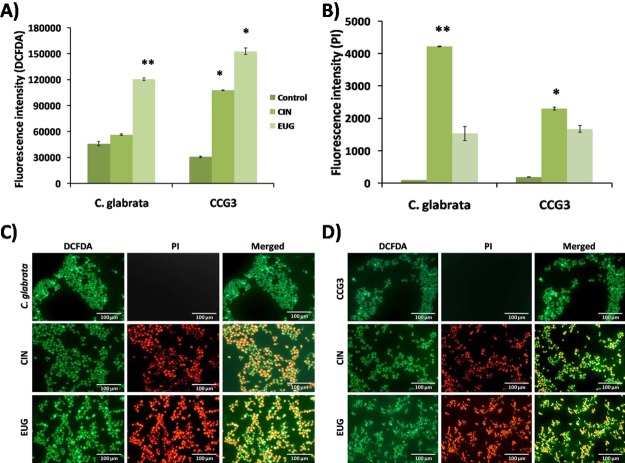
Two fluorogenic dyes, 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA) and PI, were used for reactive oxygen species (ROS) study; DCFDA measure ROS level inside the cell, whereas PI showed cell lysis by binding the DNA. Increased level of intracellular ROS accumulation was recorded in the presence of EUG but not in CIN-treated C. glabrata, whereas the level of ROS accumulation was increased in CCG3 cells upon both, CIN and EUG exposure (Figure 7A). The damaging effect of ROS accumulation on cells was demonstrated in terms of the binding ability of PI to DNA of the lysed cells, and the fluorescence intensity of bounded PI was found to be higher in CIN-exposed cells (Figure 7B). Fluorescence microscopy images of CIN/EUG treated and untreated cells depict red and green fluorescence indicating dead and live cells, respectively (Figure 7C,D).
Figure 7.
Amount of ROS accumulation in terms of fluorescence intensity of (A) DCFDA and (B) PI in C. glabrata and CCG3 biofilms exposed to CIN (128 μg mL–1) and EUG (256 μg mL–1) after 48 h. Fluorescence microscopy images of ROS generation in (C) C. glabrata and (D) CCG3 biofilm for the detection of ROS (green), cell lysis (bright red), and eDNA (diffuse red). The scale bar represents 100 μm.
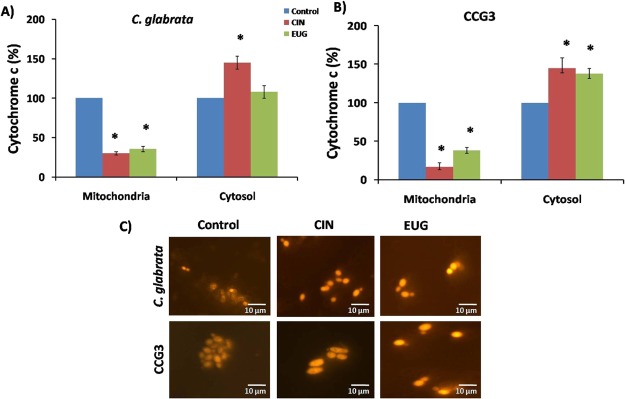
Cytochrome c Release into Cytosol and Mitochondrial Membrane Potential
Cytochrome c content was measured in mitochondria and cytosol of CIN and EUG treated C. glabrata and CCG3 because it gives an indirect evidence of apoptosis. In C. glabrata cells, cyt c release was increased from 25 to 145 and 38 to 108% in the presence of CIN and EUG, respectively, from mitochondria to cytosol (Figure 8A). Similarly, in CIN- and EUG-treated CCG3 cells, cyt c was released from 14 to 140 and 38 to 133%, respectively (Figure 8B).
Figure 8.
Release of cytochrome c from mitochondria to cytoplasm in response to CIN (128 μg mL–1) and EUG (256 μg mL–1) treatment in (A) C. glabrata and (B) CCG3. (C) Fluorescence microscopy of C. glabrata and CCG3 cells stained with rhodamine B to study the effect of CIN (128 μg mL–1) and EUG (256 μg mL–1) on MMP. Images were taken at 60× magnification. The scale bar represents 10 μm.
The change in MMP of CIN (128 μg mL–1) and EUG (256 μg mL–1) treated log phase C. glabrata, and CCG3 cells was analyzed by fluorescent cationic rhodamine B dye using a fluorescent microscope. The effect of CIN and EUG treatment on ATP production in C. glabrata cells was determined indirectly by measuring MMP using rhodamine B (Figure 8C). Rhodamine B, a hexyl ester, emits red fluorescence which in response to transmembrane potential distributes itself across biological membrane. CIN and EUG treatment increased the MMP, making the membrane more negatively charged which resulted in more accumulation of rhodamine B as compared to untreated control C. glabrata and CCG3. Collectively, cyt c quantification and MMP data depict the role of CIN and EUG in mediating early apoptosis.
CIN and EUG Differentially Modulate Transcriptional Expression
The ergosterol content in plasma membrane of CIN-and EUG-treated C. glabrata and CCG3 was quantified spectrophotometrically. No noticeable change in the ergosterol content was observed on CIN-treated C. glabrata cells. A significant increase in the ergosterol content was observed upon EUG treatment in both C. glabrata and its clinical isolate CCG3 (Figure 9). Furthermore, the enhancement of ergosterol in CCG3 is higher to that of the reference strain.
Figure 9.
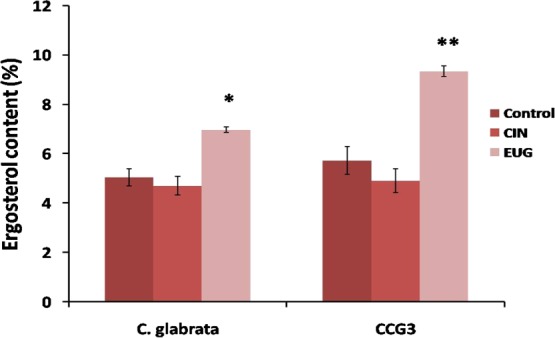
Ergosterol content as percent wet weight of C. glabrata and CCG3 in CIN (32 μg mL–1) and EUG (64 μg mL–1) untreated and treated cells (**P < 0.01 and *P < 0.05).
To gain insights into the mechanism of action of CIN and EUG against C. glabrata and CCG3 growth, transcriptional analysis of ergosterol synthesis genes (ERG2, ERG3, ERG4, ERG10, and ERG11), sterol importer (AUS1), GPI-anchored cell wall protein (KRE1), 1,3-β-glucan synthase (FKS1), and multidrug transporter (CDR1) genes were investigated by qRT-PCR, as summarized in Table S1. Expression levels of AUS1, KRE1, and FKS1 were significantly downregulated upon treatment of both CIN/EUG. However, the expression levels of ergosterol synthesis genes showed differential behavior upon treatment of CIN/EUG. Upon CIN treatment, ERG2, ERG4, and ERG11 were moderately downregulated, whereas ERG10 was moderately upregulated. In case the of EUG upregulation of ERG2, ERG3, ERG10, and ERG11, CDR1 was observed (Table 3).
Table 3. CG and CCG3 Genes Up- and Downregulated in Response to Subinhibitory Concentration of CIN (32 μg mL–1) and EUG (64 μg mL–1) and EUGa.
| fold expression |
||||||
|---|---|---|---|---|---|---|
| C. glabrata | CCG3 | |||||
| C. glabrata (ORF status) | S. cerevisiae homologue | CIN | EUG | CIN | EUG | description |
| CAGL0M01760g (verified) | CDR1 | –1.4 | 1.84 | –1.13 | 2.4 | ABC multidrug transporter regulated by Pdr1p |
| CAGL0L10714g (uncharacterized) | ERG2 | –1.67 | 1.3 | –1.4 | 2.1 | C-8 sterol isomerase |
| CAGL0F01793g (verified) | ERG3 | –1.4 | 2.7 | –2.12 | 4.2 | C-5 sterol desaturase |
| CAGL0A00429g (uncharacterized) | ERG4 | –3.34 | –1.11 | –2.0 | –1.25 | C-24 sterol reductase |
| CAGL0L12364g (uncharacterized) | ERG10 | 2.3 | 2.0 | 3.4 | 3.7 | acetyl-CoA C-acetyltransferase have role in sterol biosynthesis |
| CAGL0E04334g (verified) | ERG11 | –1.64 | 1.7 | –1.4 | 2.6 | cytochrome P-450 lanosterol 14-alpha-demethylase role in ergosterol synthesis |
| CAGL0F01419g (verified) | AUS1 | –52 | –20.0 | –7.14 | –5.55 | ABC transporter involved in sterol uptake |
| CAGL0M04169g (uncharacterized) | KRE1 | –16.39 | –20.4 | –18.86 | –12.19 | role in cell wall biogenesis and organization |
| CAGL0G01034g (verified) | FKS1 | –50.0 | –7.24 | –25.0 | –4.76 | component of 1,3-beta-glucan synthase |
Genes showing a fold expression ≥1.5 were only considered to assess the changes. Fold expression of 1.5–5.0 are considered as moderate expression, and those showing a change ≥5.0 are considered to be significant.
Biofilm Eradication from Clinically Relevant Biomaterials
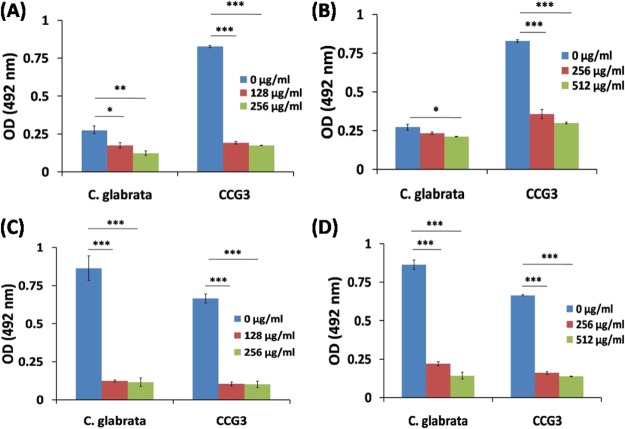
To investigate the biofilm eradication of C. glabrata and CCG3 by CIN and EUG formed on the surface of clinically relevant biomaterials (silicone urinary catheter and contact eye lens), XTT reduction assay was performed. The absorbance values of XTT reduction assay at 492 nm indicated that the clinical isolate CCG3 formed three times more biofilm as compared to C. glabrata on urinary catheters indicating thereby that the CCG3 strain was more pathogenic/virulent because of its stronger adhering properties on biomaterial devices (Figure 10). Both CIN and EUG showed eradication of C. glabrata and CCG3 biofilm from the urinary catheter (Figure 10A,B). CIN at a concentration of 256 μg mL–1 eradicated ∼55% of C. glabrata biofilm from the urinary catheter, whereas EUG showed ∼23% eradication of C. glabrata biofilm at 512 μg mL–1 (Table S2). However, CIN has eradicated more than 75% of CCG3 biofilm at a concentration of 256 μg mL–1, whereas EUG eradicated 64% CCG3 biofilm from the urinary catheter (Table S2). This suggests that CIN and EUG to be a potent antifungal against clinical isolate CCG3 biofilm. The differences in the eradication of CIN and EUG between C. glabrata and CCG3 can be attributed to their differential adherence properties on silicone urinary catheters.
Figure 10.
Quantification of C. glabrata and CCG3 biofilm developed on urinary catheter (A,B) and contact eye lens (C,D) after treatment at different concentrations of CIN (A,C) and EUG (B,D).
C. glabrata and CCG3 formed almost similar amount of biofilm on contact eye lens with OD values 0.86 (CG) and 0.67 (CCG3), suggesting the contact lens to be good surface-adhering biomaterial. CIN and EUG have eradicated a biofilm of C. glabrata and CCG3 to a significant extent from the contact eye lens (Figure 10C,D). CIN (256 μg mL–1) and EUG (512 μg mL–1) showed a maximum eradication of C. glabrata biofilm from the eye lens (86.6 ± 2.5 and 83.7 ± 2.8%) at their highest concentrations (Figure 10C,D; Table S2).
Discussion
Candida is an opportunistic commensal fungal pathogen known to cause superficial to systemic infections. The aptitude of these pathogens to form biofilm is a prime virulence trait responsible for their multidrug resistance which often leads to failure of therapeutic strategies.44 Biofilm is a structured community of harmonically communicating sessile cells encapsulated in ECM.34 Disintegration of this irreversible structure is a powerful target for therapeutic intervention. Screening of traditional medicine which can reduce biofilm is a promising approach in modern era.45 The antimicrobial activity and molecular target of CIN in combination with citral has been elucidated against Penicillium expansum growth.46 Besides this, many researchers have highlighted the potential of phytoactive molecules against bacterial and fungal species.47−49
In this investigation, the antifungal activity of two effective phytoactive molecules (CIN and EUG) was studied against two different growth forms (planktonic and sessile) of C. glabrata and its clinical isolates. The effect of CIN and EUG on C. glabrata cell wall, ROS generation, ECM, transcription of selected genes, and hydrophobicity was also explored and compared with that of CCG3. The results indicated C. glabrata biofilm eradicating potency of CIN and EUG from clinically relevant biomaterials (contact eye lens and urinary catheter). Data suggested that clinical isolate (CCG3) is more resistant, more surface adhering, and hydrophobic, as compared to C. glabrata, although no significant changes were observed in their ECM biochemical compositions. Similar variation in biofilm-forming ability of clinical isolates as compared to Candida laboratory strain has been reported by other researchers.50,51 The plausible mechanisms observed for such a variation is the ability of pathogens to modulate their biochemical composition/differential adherence/hydrophobicity ability, which is the resultant of exposure to antifungal agents, close contact with host, and timely mutations.20,25,51
Molecular Insights into C. glabrata Biofilm Eradication Mechanism by CIN and EUG
The antimicrobial property of natural compounds or molecules depends on the functional group present in them.52 CIN is a naturally occurring organic compound in cinnamon oil, whereas EUG is a phenylpropanoid present in aromatic plants and a major component of clove oil.53,54 The antimicrobial activity of both compounds is due to their lipophilic nature they interact with hydrophobic components (ergosterol) of cell membrane to generate pores. This eventually reduces cell membrane integrity and permeability, thus resulting in cell lysis and leakage of intracellular contents (nucleic acid, protein, and ATP) from the cell.46,53,55−59 CIN exposure might mediate damage to C. glabrata cell membrane in a similar way, as evident from results which depicts increased release of nucleic acid followed by cell death, without any change in ergosterol content (Figures 6 and 9). EUG also causes cell death as well as release of nucleic acid (Figure 6); and unlike CIN, EUG increased the cell membrane ergosterol content (Figure 9), indicating that EUG and CIN interacts differently with cell membrane ergosterol.
Previously, researchers have reported strong inhibitory effect of CIN on plasma membrane ATPase which has a potential role in the secretion of hydrolytic enzymes.60 However, upsurge of this ATPase-dependent transportation of hydrolytic enzymes enhances secreted aspartyl proteases activity in C. albicans.61 Therefore, the observed decrease in the proteinase and phospholipase activity in this study can be attributed to the de-escalation of CIN/EUG-mediated ATPase, which decreased the efflux of hydrolytic enzymes across cell membrane and ultimately results in less enzymatic activity (Figure 4D,E). In line with our observations, CIN reduced the proteinase and phospholipase activities in C. albicans.62
Many studies have also proposed that the antifungal activity of CIN is due to its inhibitory effect on cell wall and membrane synthesizing enzymes, mainly 1,3-β-glucan, chitin, and ergosterol.63−66 It is worth noting that cells cannot overcome the stress of ergosterol deficiency and were more susceptible to stress conditions as ergosterol is a crucial ingredient of fungal cell membrane in terms of membrane rigidity, fluidity, and permeability.29,67 CIN induced moderate transcriptional downregulation of ERG2, ERG3, ERG4, and ERG11 and was in congruence with the data of ergosterol content which showed no significant changes although the ergosterol importer gene AUS1 was significantly downregulated (Figure 9, Table 3). However, RT-PCR results of EUG-treated C. glabrata showed upregulation of ERG genes and an increase in ergosterol content. These results are in sharp contrast with the previous findings because of a drop in ergosterol content in C. albicans at fungicidal concentrations of EUG.68 Moreover, in Trichophyton rubrum, also EUG did affect ergosterol content.69 The differences in the ergosterol content observed in the present case with respect to other studies can be attributed to: (a) the differential composition of cell membrane of C. glabrata to that of C. albicans and T. rubrum and (b) difference in EUG concentrations because this study used subinhibitory concentration (64 μg mL–1).
CIN is also a known noncompetitive inhibitor of 1,3-β-glucan synthase and mixed inhibitor of chitin synthase in Saccharomyces cerevisiae and thus acts as cell-wall-active antifungal molecule.63,69 Interestingly, a significant decrease in the expression of FKS1 (1,3-β-glucan synthase) and KRE1 (cell wall biogenesis) was observed in CIN/EUG-treated cells. These data are in support of previous observations suggesting the importance of using CIN and EUG as antifungal molecule against C. glabrata.
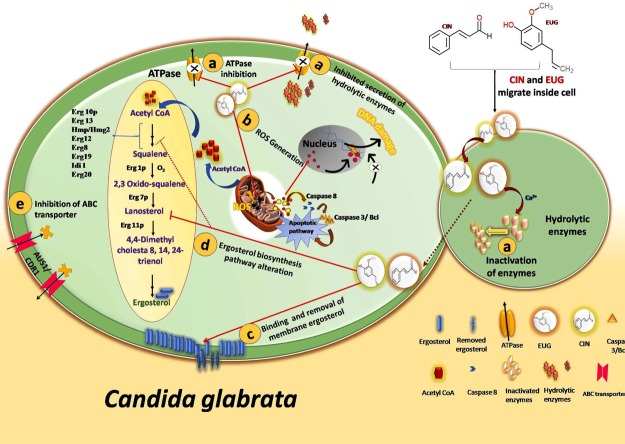
In addition to cell wall and membrane, CIN is also responsible for oxidative stress generation and apoptosis.70 CIN increases the MMP that leads to ROS generation and release of cyt c from mitochondria to cytoplasm.70 In C. albicans, CIN induces apoptosis via metacaspase-dependent pathway activated by cytochrome c release and ROS generation. Increased MMP is an early event of apoptosis, whereas ROS and cytochrome c are known to activate the proapoptotic pathway.71 Similarly, EUG is also known to induce oxidative stress which causes lipid peroxidation of cytoplasmic membrane lipids and finally cell death.72 Earlier researchers mentioned that EUG induces apoptosis in C. albicans as a consequence of the inhibition of cell cycle progression at G1–S and G2–M phases.73 ROS, MMP, and cytochrome c results of CIN/EUG-exposed C. glabrata cells were in agreement with the prior indications of CIN/EUG-induced apoptosis. The activity of catalase enzyme, which is known to protect cell from ROS, was found to be reduced in CIN/EUG-treated C. glabrata biofilm cells which is in resemblance with the increased catalase activity in CIN-exposed C. albicans.46 Considering these evidence, the mechanism of action through which CIN and EUG exhibit antifungal activity can be highlighted by the following cellular features (Figure 11). They include: (a) inhibition of plasma membrane ATPase which has a role in secretion of hydrolytic enzyme, (b) ROS generation and apoptosis, (c) damaging cell structure by binding and removing membrane ergosterol, (d) disturbing functionality of genes involved in membrane biosynthesis such as ERG genes, FKS1 and KRE1, and (e) inhibiting membrane ATP binding cassette (ABC) sterol importer and drug transporter.
Figure 11.
Schematic diagram representing possible antifungal mechanism (a–e) of CIN and EUG in C. glabrata. (a) By inhibition of membrane ATPase pump which mediates secretion of hydrolytic enzyme or by degradation of hydrolytic enzymes; (b) by ROS generation which results in release of cyt c and activates proapoptotic pathways; (c) by disturbing cell membrane integrity via removal of membrane ergosterol; (d) by inhibiting/altering the functionality of biosynthetic pathway enzymes similar to ergosterol; and (e) by blocking import of extracellular sterol or downregulating the expression of ABC sterol importer and drug transporter.
Concluding Remarks
The current study highlights the antibiofilm activity of phytoactive molecules (CIN and EUG) in C. glabrata and its clinical isolates. These two compounds are mediating the antifungal activity via deactivating its hydrolytic enzymes, ROS generation, apoptosis, and selectively modulating the ergosterol content. The study established that these compounds are highly effective in their biofilm eradication properties even on clinically relevant biomaterials such as urinary catheter and eye lens. The benefit of using phytoactive molecules in antifungal therapy is that no new formulation development is needed for their therapeutic application as they are naturally present in EOs. A recent study reported that EO components, such as CIN, EUG, thymol, and carvacrol, exhibited excellent antimicrobial activity against bacterial biofilms and shown better cytotoxicity values against fibroblasts, macrophages, and keratinocytes cell lines compared to the traditional antiseptic drug chlorhexidine.74 Moreover, these EOs are kept under GRAS category by FDA (U.S.).75 Indeed, CIN is an active component of cinnamon oil, which has passed clinical trial phase 1 for treatment of oral candidiasis.76 Furthermore, EUG containing clove oil has also been widely used in dentistry for treating dental caries and periodontal diseases.77,78 The study demonstrates that coating of medical implant devices with CIN and EUG will help in preventing implant-associated fungal infections. With these perspectives, we believe that naturally occurring phytoactive molecules stand out as a potential source of bioactive molecule with immense therapeutic applications in treating dental, oral, and superficial fungal infections.
Materials and Methods
Strain Collection and Culturing of C. glabrata and Its Clinical Isolates
C. glabrata (CG; MTCC 3019) used in the study was procured from IMTECH (Microbial Type Culture Collection), Chandigarh, India. C. glabrata clinical isolates (CCG1, CCG2, CCG3, and CCG4) used in this study were a kind gift from Dr. Navin Kumar, Graphic Era Deemed to be University, Dehradun, India.79 Clinical isolates of C. glabrata were classified as susceptible (CCG4), susceptible dose-dependent (CCG2), and resistant (CCG1 and CCG3) according to interpretive guidelines for in vitro susceptibility testing of Candida species for fluconazole.79 All strains were routinely maintained and cultured in YPD media (1% yeast extract, 2% dextrose, and 2% peptone; 2% agar for solid media) at 37 °C. The urinary catheter and eye lens (EtafilconA; 14.0 DIA) were purchased locally. All chemicals used in the study were procured from Sigma-Aldrich chemicals Ltd and Himedia, India. RNeasy kit and RT-PCR reagents were purchased from QIAGEN and Applied Biosystems, respectively. Stock solutions of CIN and EUG were prepared in dimethyl sulfoxide.
Biofilm Formation Studies on C. glabrata and Its Clinical Isolates
Biofilm forming ability of C. glabrata and its clinical isolates were studied in 96-well flat bottom MTPs in RPMI medium (Roswell Park Memorial Institute). Log-phase cultures of all strains were diluted individually in phosphate-buffered saline (PBS) (pH 7.0) to a concentration of 1 × 107 cells mL–1. The suspension (100 μL) of these cells was separately added to wells of MTP and incubated for 90 min. After incubation, PBS was replaced with RPMI, and MTP was again incubated at 37 °C. For quantification of biofilm formed at different time intervals (2, 4, 8, 10, 12, 24, and 48 h), XTT reduction assay was used.80
For studying the effect of CIN and EUG on C. glabrata and its clinical isolate biofilm, log phase cells were diluted in sterile PBS to a concentration of 1 × 107 cells mL–1, and 100 μL of suspension was added into each well of MTP.81 Plates were then incubated at 37 °C for 90 min (adhesion phase), and 200 μL of RPMI containing CIN and EUG in the concentration range of 512–2 μg mL–1 was added after washing wells thrice with PBS. The plates were again incubated for 48 h, and the inhibitory effect of CIN and EUG was evaluated in terms of metabolic activity by XTT reduction assay.
For mature biofilm, after 90 min incubation of adhesion phase, wells were washed with PBS and 200 μL of RPMI was added. Then, plates were reincubated for 48 h followed by washing with PBS, and 200 μL of RPMI containing CIN and EUG in the concentration range of 512–2 μg mL–1 was added. The eradicating effect of CIN and EUG on biofilm was evaluated by XTT reduction assay after 24 h of incubation.
Minimum Inhibitory Concentration Measurements
The inhibitory effect of six different phytoactive molecules was initially screened against C. glabrata in MTP as per M27-A2 guidelines for yeast broth microdilution.82 The concentration of log phase C. glabrata cells was adjusted to 2.5 × 103 cells mL–1 in RPMI medium. Briefly, 100 μL of cell suspension was added into each well of MTP and 100 μL of twofold diluted phytoactive molecule in RPMI media (range 1024–32 μg mL–1) was also added. The plates were incubated for 48 h at 37 °C and visualized for growth in the form of turbidity. The results are represented as MIC90 values; a concentration where 90% growth was inhibited as compared to control.
MFC Measurements
MFC of phytoactive molecules was determined against C. glabrata and its clinical isolates on YPD agar media plates. The 48 h-treated cells from MTP of MIC were spotted on to solid media plates. The drop was then air dried and incubated at 37 °C for 18 h. After incubation, plates were photographed on a black background.79
Determination of Ergosterol Content
Ergosterol content in the cell membrane of C. glabrata and CCG3 cells was measured by incubating log phase cells in sabouraud dextrose broth with or without CIN (32 μg mL–1) and EUG (64 μg mL–1) for 24 h at 37 °C.69 The cells were centrifuged at 6000 rpm for 5 min, and the pellet was washed with sterile water. The wet weight of the cell pellet was measured, and the pellet was suspended in 3 mL of lysing agent (25% alcoholic KOH) and vortexed for 1 min. Cell suspension was incubated for 1 h at 85 °C in water bath. Then, sterols were extracted by adding a mixture of distilled water and n-heptane in a 1:3 ratio, followed by vigorous vortexing. The heptane layer was collected carefully in glass tubes and stored for 24 h at −20 °C. The sterol extracts were studied by mixing 20 μL of sample and 100 μL of absolute ethanol and scanning from 230 to 300 nm using a UV–visible spectrophotometer. The amount of ergosterol was quantified by using the following equation
where F is the dilution factor; 290 and 518 are the E values for crystalline ergosterol and 24(28) dehydroergosterol, respectively.
ROS Generation
ROS level in C. glabrata and CCG3 biofilm cells on exposure to lethal concn of CIN (128 μg mL–1) and EUG (256 μg mL–1) was studied by adding a mixture of DCFDA (10 μM) and propidium iodide (1 mg mL–1 PI) in MTP, as described earlier.83 The MTP was incubated in dark for 30 min at 37 °C. For measurement of ROS generation in biofilm cells, DCFDA fluorescence was recorded at an emission wavelength of 520 nm and excitation wavelength of 485 nm. Whereas the emission and excitation wavelength for PI was taken at 617 and 543 nm, respectively. Fluorescence microscopic images of cells were taken at 40× magnification in fluorescence microscope.
Biochemical Composition of ECM
ECM was isolated from C. glabrata and CCG3 biofilm developed in the presence of CIN and EUG by scrapping it from the MTP well surface using sterile scrapper and sterile PBS (pH 7.0). The scrapped biofilm was sonicated (Q700 sonicator, QSonica) at 35 W in an ice bath for five cycles of 30 s each, as described by Fonseca et al.84 The suspension was centrifuged at 12 000 rpm for 5 min. The supernatant was then used for biochemical and enzymatic analysis of ECM, whereas the cell pellet was used for catalase activity measurement. The ECM was examined for carbohydrate, protein, and eDNA quantification. Phenol–sulfuric acid method was used for total carbohydrate estimation with glucose as a standard. Briefly, 1 mL of H2SO4, 200 μL of phenol (5% w/v), and 100 μL of sample were mixed in glass tubes. The tubes were then incubated at 30 °C for 30 min. Then, tubes were cooled and the absorbance was measured at 485 nm. For total protein measurement, BCA kit was used with bovine serum albumin (BSA) as a standard. The absorbance was measured at 562 nm. The quantity of eDNA in sample was measured by precipitating the eDNA. Precipitation was done by adding one-tenth the volume of sodium acetate (3 M) in a sample, followed by adding phenol, chloroform, and isoamyl alcohol (25:24:1). The aqueous layer was collected in fresh tube, and 2.5 volume of ethanol was added to precipitate the eDNA. The purity of eDNA was checked by using nanodrop by 260/280 ratio.
Phospholipase and Proteinase Activity
At neutral pH, the activity was measured by preparing the substrate consisting of 50 mM Tris-HCl buffer (pH 7.5), 1.6 mM phosphatidylcholine (min. 41%), 0.25% Triton X-100, 20 mM AlCl3, and 0.124% bromothymol blue. The solution was then filtered and stored at 4 °C. Before absorbance was measured, the pH of ECM was adjusted to 7.5 with 10 mM NaOH. The phospholipasic activities were determined by mixing 100 μL of ECM with 900 μL of substrate and the reading was taken at OD630 nm. The specific phospholipase activity was recorded as the absorbance shift per minute of the reaction. The proteinase activity was determined by mixing 1% w/v azocasein substrate with the supernatants and incubating it at 37 °C for 1 h. Trichloroacetic acid (10%) was used to stop the reaction, and the mixture was centrifuged for 5 min at 12 000 rpm. The supernatant obtained was then mixed with 0.5 M NaOH and incubated for 15 min. The proteinase activity was measured at 440 nm. The specific proteinase activity was the amount of enzyme that elicited an increase of 0.001 units in absorbance per minute of reaction.83
Catalase Activity
The catalase activity of CIN- and EUG-treated C. glabrata and CCG3 sessile cells collected after ECM isolation were lysed with glass beads.85 The samples were then centrifuged at 10 000 rpm, and the supernatant was collected. For determining the catalase activity, 333 μL of 50 mM H2O2, 567 μL of PBS (pH 7.0), and 100 μL of supernatant were mixed, followed by measuring absorbance of sample at 240 nm. One unit of catalase activity corresponded to the amount of enzyme that decomposes 1 μmol of H2O2 per minute of reaction at 37 °C. The catalase activity (U/mg) was calculated using the following formula
where, A0 – A2 is the difference in absorbance; Vt is the total volume of reaction; Vs is the volume of sample; Σ240 is the molar extinction coefficient for H2O2; d is the optical path length of cuvette; and Ct is the protein concentration in sample.
Hydrophobicity Assay
To determine hydrophobicity, overnight grown C. glabrata and CCG3 cell at 0.1 OD600 were exposed to subinhibitory concentration of CIN (64 μg mL–1) and EUG (128 μg mL–1) and then incubated for 24 h at 37 °C. Cells were harvested after incubation, washed with sterile PBS, and suspended in 3 mL of 50 mM sodium phosphate buffer (pH 7.2) at a concentration of 2 × 106 cells mL–1. Octane (500 μL) was then added to cell suspension and vortexed for 1 min. Cells present in aqueous phase were calculated by taking absorbance at OD600.86 The HI was calculated by the following equation
where A1 is the absorbance of inoculum and A2 is the absorbance of aqueous phase.
Cytochrome c Determination
Briefly, early stationary phase cells were diluted in PBS to a concentration of 1 × 107 cells mL–1 and further incubated in 3 mL of YPD broth containing CIN (128 μg mL–1) and EUG (256 μg mL–1) separately at 30 °C for 24 h. The cells were washed with PBS, and the pellet was suspended in the homogenization medium [50 mM Tris (pH 7.5) 2 mM EDTA, 2% glucose, and 1 mM phenylmethylsulfonyl fluoride]. The suspension was vortexed and centrifuged at 5000 rpm for 10 min. The supernatant so obtained was again centrifuged at 20 000 rpm for 45 min. After centrifugation, the supernatant was quantified for cyt c released from mitochondria to cytoplasm. Whereas the pellet was suspended in 50 mM Tris (pH 5.0) having 2 mM EDTA and incubated for 5 min at 37 °C. The dissolved pellet again centrifuged at 10 000 rpm for 30 s was used for the determination of cyt c remaining in mitochondria. Protein content in the supernatant and the pellet was estimated by Bradford reagent kit, using BSA as the standard. Before quantifying cyt c quantity in supernatant and pellet, both were reduced with 500 mg L–1 ascorbic acid at room temperature for 5 min, and the absorbance was measured at 550 nm.70
Mitochondrial Membrane Potential
For studying the change in MMP, log phase C. glabrata cells were incubated in SDB medium with or without CIN and EUG for 6 h at 37 °C in shaking. After incubation, cells were collected by centrifugation and washed with PBS (pH 7.0). Then, cells were suspended in 10 mM HEPES buffer (pH 7.4) containing 5% glucose and 100 nM rhodamine B. The suspension was incubated for 30 min in dark at 37 °C. After incubation, cells were washed and MMP was visualized by fluorescence microscope at the excitation wavelength of 555 nm and emission wavelength of 579 nm.87
FESEM Measurements
FESEM was used to monitor the morphological changes in C. glabrata and CCG3 (resistant isolate) biofilm cells before and after treatment with CIN and EUG. Briefly, 1 cm2 polystyrene discs were incubated in fetal calf serum for 24 h. After incubation, polystyrene discs were placed in 24-well plates, and 1 mL of cell suspension prepared in RPMI medium was added. Plates were incubated for 48 h, and then medium was replaced with CIN- and EUG-containing media. Plates were again incubated for 24 h after which discs were washed with PBS and fixed overnight in 2.5% gluteraldehyde solution. Discs were then again washed with PBS and treated with ethanol gradient. Air dried samples were mounted on stubs, and gold sputtering was done. Finally, samples were visualized under FESEM at a voltage of 15 kV and magnification from 1000 to 5000×.88
Release of Nucleic Acid Content
The rate of release of nucleic acid from C. glabrata and CCG3 cells exposed to CIN (128 μg mL–1) and EUG (256 μg mL–1) for 2, 4, and 8 h at 37 °C was measured by taking absorbance at 260 nm.89 Cells exposed to alcoholic KOH which causes 100% cellular leakage were used as a reference sample. The sample was centrifuged at 5000 rpm for 5 min, and the supernatant collected was used for measuring the intracellular leakage in terms of rate of release of DNA from C. glabrata and CCG3 cells.
Fluorescence Microscopy Studies
For studying the effect of CIN and EUG on the biofilm cell viability, biofilm of C. glabrata and CCG3 developed was washed with PBS (pH 7.0) after CIN and EUG treatment. Then, biofilm was stained with FDA (2 μg mL–1 fluorescein diacetate) and PI (0.6 μg mL–1 propidium iodide).90 Plate was incubated for 20 min at room temperature in dark, and wells were washed before visualization under fluorescence microscope. All images were captured at 40× magnification.
Transcriptional Analysis
Effect of CIN (32 μg mL–1) and EUG (64 μg mL–1) subinhibitory concentration on the expression of selected C. glabrata and CCG3 was studied by qRT-PCR. Log-phase cells were treated with CIN and EUG in YPD broth. Total RNA was extracted from cells by using RNeasy kit, QIAGEN by following manufacturer’s instructions. The RNA was quantified by a nanodrop (Thermo Fisher Scientific), which was used for cDNA synthesis using Verso cDNA synthesis kit, Thermo. Primers for RT-PCR of the above-mentioned genes and housekeeping gene (ACT1) were designed by using Primer Quest, Integrated DNA Technologies (IDT) and were synthesized by IDT (Table 3). SYBR green mix (Applied Biosystems) was used in Mastercycler for qRT-PCR. cDNA template (100 ng) and gene-specific forward and reverse primers (200 nM) were used in the reaction. The following parameters were used for qRT-PCR: an initial denaturation at 95 °C (3 min), followed by 40 cycles of denaturation (95 °C/30 s), annealing (52 °C/30 s), and extension (72 °C/30 s), melting-curve analysis starting from initial temperature 45 to 95 °C, with a gradual increase in 0.5 °C/15 s. Specificity of the primers was confirmed by the melting curve analysis. The generated CT values of target genes were normalized to the CT value of housekeeping ACT1 gene. Relative expression fold changes were evaluated by ΔΔCT method using 2–ΔΔCT formula.91
Biofilm Study on Biomaterials
Commercially available urinary catheter (silicone) and contact eye lens were procured from a local medical store and were used as biomaterials for biofilm study. Contact lens were used as a whole, whereas 1 cm long pieces of urinary catheter coupons were cut out and sterilized with 100% ethylene oxide and kept under UV for 4 h.92−94 Sterile lens and catheter coupons were incubated in RPMI medium containing 1 × 107 cells mL–1 for 48 h. Biomaterials with biofilms were treated for 24 h with CIN and EUG containing RPMI and then was quantified by XTT reduction assay.
Statistical Analysis
All experiments were performed in triplicates, and the values presented the mean with standard deviation, obtained from three different observations for each assay. Student’s t-test was used for the statistical analysis, and a value of P < 0.05 was considered statistically significant (*), P < 0.01 as highly significant (**) and P < 0.001 as extremely significant (***).
.
Acknowledgments
P.G. acknowledges the financial support of NPDF Grant PDF/2016/001670, from Science and Engineering Research Board (SERB), Government of India. K.M.P. acknowledges the receipt of DBT-IYBA fellowship and SERB-LS young scientist award. Authors are thankful to the Institute Instrumentation Centre, IIT Roorkee, for the microscopy analysis. Authors are grateful to Dr. V. K. Kateria, Department of Microbiology, Shri Guru Ram Rai Institute of Medical and Health Sciences, Dehradun, for the clinical isolates of C. glabrata.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01856.
Candida glabrata biofilm inhibition, mature biofilm eradication, cell hydrophobicity, primer sequence, and metabolic activity of C. glabrata biofilm on urinary catheter and contact eye lens (PDF)
Author Contributions
P.G., V.P., and K.M.P. participated in the study design and preparation of the manuscript. P.G. and N.K. procured the clinical isolates of C. glabrata and performed the experiments. P.G., S.G., and M.S. analyzed the data. All authors have read and approved the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Dadar M.; Tiwari R.; Karthik K.; Chakraborty S.; Shahali Y.; Dhama K. Candida Albicans-Biology, Molecular Characterization, Pathogenicity, and Advances in Diagnosis and Control–An Update. Microb. Pathog. 2018, 117, 128–138. 10.1016/j.micpath.2018.02.028. [DOI] [PubMed] [Google Scholar]
- Healey K. R.; Nagasaki Y.; Zimmerman M.; Kordalewska M.; Park S.; Zhao Y.; Perlin D. S. The Gastrointestinal Tract Is a Major Source of Echinocandin Drug Resistance in a Murine Model of Candida Glabrata Colonization and Systemic Dissemination. Antimicrob. Agents Chemother. 2017, 61, e01412-17 10.1128/aac.01412-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J. Global Trends in Candidemia: Review of Reports from 1995–2005. Curr. Infect. Dis. Rep. 2005, 7, 429–439. 10.1007/s11908-005-0044-7. [DOI] [PubMed] [Google Scholar]
- Pfaller M. A.; Diekema D. J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. 10.1128/cmr.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapar N. Epidemiology and Risk Factors for Invasive Candidiasis. Ther. Clin. Risk Manage. 2014, 10, 95. 10.2147/tcrm.s40160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushi M. F.; Mtemisika C. I.; Bader O.; Bii C.; Mirambo M. M.; Groß U.; Mshana S. E. International Journal of Infectious Diseases High Oral Carriage of Non- albicans Candida Spp. among HIV-Infected Individuals. Int. J. Infect. Dis. 2016, 49, 185–188. 10.1016/j.ijid.2016.07.001. [DOI] [PubMed] [Google Scholar]
- Chapman B.; Slavin M.; Marriott D.; Halliday C.; Kidd S.; Arthur I.; Bak N.; Heath C. H.; Kennedy K.; Morrissey C. O. Changing Epidemiology of Candidaemia in Australia. J. Antimicrob. Chemother. 2017, 72, 1103–1108. 10.1093/jac/dkx047. [DOI] [PubMed] [Google Scholar]
- Lamoth F.; Lockhart S. R.; Berkow E. L.; Calandra T. Changes in the Epidemiological Landscape of Invasive Candidiasis. J. Antimicrob. Chemother. 2018, 73, i4–i13. 10.1093/jac/dkx444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee P. Epidemiology and Antifungal Susceptibility of Candida Species in a Tertiary Care Hospital, Kolkata, India. Curr. Med. Mycol. 2016, 2, 20. 10.18869/acadpub.cmm.2.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma Y.; Chumber S.; Kaur M. Studying the Prevalence, Species Distribution, and Detection of in Vitro Production of Phospholipase from Candida Isolated from Cases of Invasive Candidiasis. J. Global Infect. Dis. 2017, 9, 8. 10.4103/0974-777x.199995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi V.; Patel M. Prevalence of Candida Species and Its Antifungal Susceptibility Isolated from Blood Culture at Tertiary Care Hospital, Ahmedabad, India. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 884–892. 10.20546/ijcmas.2017.606.104. [DOI] [Google Scholar]
- Kalaiarasan K.; Singh R.; Chaturvedula L. Fungal Profile of Vulvovaginal Candidiasis in a Tertiary Care Hospital. J. Clin. Diagn. Res. 2017, 11, DC06. 10.7860/JCDR/2017/23578.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi G.; Ebrahimi-Rad M.; Mousavi S. F.; Shams-Ghahfarokhi M.; Razzaghi-Abyaneh M. Emergence of Non-Candida Albicans Species: Epidemiology, Phylogeny and Fluconazole Susceptibility Profile. J. Mycol. Med. 2018, 28, 51–58. 10.1016/j.mycmed.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Goswami D.; Goswami R.; Banerjee U.; Dadhwal V.; Miglani S.; Lattif A. A.; Kochupillai N. Pattern of Candida Species Isolated from Patients with Diabetes Mellitus and Vulvovaginal Candidiasis and Their Response to Single Dose Oral Fluconazole Therapy. J. Infect. 2006, 52, 111–117. 10.1016/j.jinf.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Ray D.; Goswami R.; Banerjee U.; Dadhwal V.; Goswami D.; Mandal P.; Sreenivas V.; Kochupillai N. Prevalence of Candida Glabrata and Its Response to Boric Acid Vaginal Suppositories in Comparison with Oral Fluconazole in Patients with Diabetes and Vulvovaginal Candidiasis. Diabetes Care 2007, 30, 312–317. 10.2337/dc06-1469. [DOI] [PubMed] [Google Scholar]
- Hachem R.; Hanna H.; Kontoyiannis D.; Jiang Y.; Raad I. The Changing Epidemiology of Invasive Candidiasis. Cancer 2008, 112, 2493–2499. 10.1002/cncr.23466. [DOI] [PubMed] [Google Scholar]
- Silva S.; Hooper S. J.; Henriques M.; Oliveira R.; Azeredo J.; Williams D. W. The Role of Secreted Aspartyl Proteinases in Candida Tropicalis Invasion and Damage of Oral Mucosa. Clin. Microbiol. Infect. 2011, 17, 264–272. 10.1111/j.1469-0691.2010.03248.x. [DOI] [PubMed] [Google Scholar]
- Whaley S. G.; Berkow E. L.; Rybak J. M.; Nishimoto A. T.; Barker K. S.; Rogers P. D. Azole Antifungal Resistance in Candida Albicans and Emerging Non-Albicans Candida Species. Front. Microbiol. 2017, 7, 2173. 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalheiro M.; Teixeira M. C. Candida Biofilms: Threats, Challenges, and Promising Strategies. Front. Med. 2018, 5, 28. 10.3389/fmed.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali A.; Vavala E.; Marzano V.; Leone C.; Castagnola M.; Iavarone F.; Angiolella L. Cell Wall Composition and Biofilm Formation of Azoles-Susceptible and-Resistant Candida Glabrata Strains. J. Chemother. 2017, 29, 164–172. 10.1080/1120009x.2016.1199507. [DOI] [PubMed] [Google Scholar]
- Pierce C.; Vila T.; Romo J.; Montelongo-Jauregui D.; Wall G.; Ramasubramanian A.; Lopez-Ribot J. The Candida Albicans Biofilm Matrix: Composition, Structure and Function. J. Fungi 2017, 3, 14. 10.3390/jof3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panariello B. H. D.; Klein M. I.; Mima E. G. D. O.; Pavarina A. C. Fluconazole Impacts the Extracellular Matrix of Fluconazole-Susceptible and-Resistant Candida Albicans and Candida Glabrata Biofilms. J. Oral Microbiol. 2018, 10, 1476644. 10.1080/20002297.2018.1476644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K. F.; Zarnowski R.; Andes D. R. Fungal Super Glue: The Biofilm Matrix and Its Composition, Assembly, and Functions. PLoS Pathog. 2016, 12, e1005828 10.1371/journal.ppat.1005828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M.; Henriques M.; Lopez-Ribot J. L.; Oliveira R. Addition of DNase Improves the in Vitro Activity of Antifungal Drugs against Candida Albicans Biofilms. Mycoses 2012, 55, 80–85. 10.1111/j.1439-0507.2011.02047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri S.; Shams-Ghahfarokhi M.; Razzaghi-Abyaneh M. Comparative Analysis of Proteinase, Phospholipase, Hydrophobicity and Biofilm Forming Ability in Candida Species Isolated from Clinical Specimens. J. Mycol. Med. 2018, 28, 437–442. 10.1016/j.mycmed.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Kong E. F.; Tsui C.; Kucharíková S.; Van Dijck P.; Jabra-Rizk M. A. Modulation of Staphylococcus Aureus Response to Antimicrobials by the Candida Albicans Quorum Sensing Molecule Farnesol. Antimicrob. Agents Chemother. 2017, 61, e01573-17 10.1128/aac.01573-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S.; Lemetais G.; Ferreira T.; Cayot P.; Gervais P.; Beney L. Ergosterol Biosynthesis: A Fungal Pathway for Life on Land?. Evolution Int. J. Org. Evolution 2012, 66, 2961–2968. 10.1111/j.1558-5646.2012.01667.x. [DOI] [PubMed] [Google Scholar]
- Stylianou M.; Kulesskiy E.; Lopes J. P.; Granlund M.; Wennerberg K.; Urban C. F. Antifungal Application of Non-Antifungal Drugs. Antimicrob. Agents Chemother. 2013, 58, 1055. 10.1128/aac.01087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Tsai H.-F.; Mandal A.; Walker B.; Noble J.; Fukuda Y.; Bennett J. Sterol Uptake and Sterol Biosynthesis Act Coordinately to Mediate Antifungal Resistance in Candida Glabrata under Azole and Hypoxic Stress. Mol. Med. Rep. 2018, 17, 6585–6597. 10.3892/mmr.2018.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo-Cardona M. Á.; Marcos-Zambrano L. J.; Sánchez-Carrillo C.; de la Pedrosa E. G. G.; Cantón R.; Bouza E.; Escribano P.; Guinea J. Mutant Prevention Concentration and Mutant Selection Window of Micafungin and Anidulafungin in Clinical Candida Glabrata Isolates. Antimicrob. Agents Chemother. 2018, 62, e01982-17 10.1128/aac.01982-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T.; Krysan D. J. Antifungal Drug Development: Challenges, Unmet Clinical Needs, and New Approaches. Cold Spring Harbor Perspect. Med. 2014, 4, a019703. 10.1101/cshperspect.a019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackman G.; Coenye T. Quorum Sensing Inhibitors as Anti-Biofilm Agents. Curr. Pharm. Des. 2015, 21, 5–11. 10.2174/1381612820666140905114627. [DOI] [PubMed] [Google Scholar]
- Tang K.; Zhang X.-H. Quorum Quenching Agents: Resources for Antivirulence Therapy. Mar. Drugs 2014, 12, 3245–3282. 10.3390/md12063245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham W.-R. Going beyond the Control of Quorum-Sensing to Combat Biofilm Infections. Antibiotics 2016, 5, 3. 10.3390/antibiotics5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh C.; Sam C.; Yin W.; Tan L. Y.; Krishnan T.; Chong Y. M.; Chan K. Plant-Derived Natural Products as Sources of Anti-Quorum Sensing Compounds. Sensors 2013, 13, 6217–6228. 10.3390/s130506217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A.; Singh B. N.; Deepak D.; Rawat A. K. S.; Singh B. R. Colostrum Hexasaccharide, a Novel Staphylococcus Aureus Quorum- Sensing Inhibitor. Antimicrob. Agents Chemother. 2015, 59, 2169–2178. 10.1128/aac.03722-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetzner S. Quorum Quenching Enzymes. J. Biotechnol. 2015, 201, 2–14. 10.1016/j.jbiotec.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Lade H.; Paul D.; Kweon J. H. Quorum Quenching Mediated Approaches for Control of Membrane Biofouling. Int. J. Biol. Sci. 2014, 10, 550–565. 10.7150/ijbs.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guleria S.; Tiku A. K.; Koul A.; Gupta S.; Singh G.; Razdan V. K. Antioxidant and Antimicrobial Properties of the Essential Oil and Extracts of Zanthoxylum Alatum Grown in North-Western Himalaya. Sci. World J. 2013, 2013, 1–9. 10.1155/2013/790580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A.; Khan A.; Khan L. A.; Manzoor N. Vitro Synergy of Eugenol and Methyleugenol with Fluconazole against Clinical Candida Isolates. J. Med. Microbiol. 2010, 59, 1178–1184. 10.1099/jmm.0.020693-0. [DOI] [PubMed] [Google Scholar]
- Bacha K.; Tariku Y.; Gebreyesus F.; Zerihun S.; Mohammed A.; Weiland-Bräuer N.; Schmitz R. A.; Mulat M. Antimicrobial and Anti-Quorum Sensing Activities of Selected Medicinal Plants of Ethiopia: Implication for Development of Potent Antimicrobial Agents. BMC Microbiol. 2016, 16, 139. 10.1186/s12866-016-0765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunke S.; Hube B. Two Unlike Cousins: Candida Albicans and C. Glabrata Infection Strategies. Cell. Microbiol. 2013, 15, 701–708. 10.1111/cmi.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J.; Cragg G. M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- McCall A.; Edgerton M. Real-Time Approach to Flow Cell Imaging of Candida Albicans Biofilm Development. J. Fungi 2017, 3, 13. 10.3390/jof3010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos G. C. d O.; Vasconcelos C. C.; Lopes A. J. O.; Cartagenes M. D. S. D. S.; Filho A. K. D. B.; do Nascimento F. R. F.; Ramos R. M.; Pires E. R. R. B.; de Andrade M. S.; Rocha F. M. G. Candida Infections and Therapeutic Strategies: Mechanisms of Action for Traditional and Alternative Agents. Front. Microbiol. 2018, 9, 1351. 10.3389/fmicb.2018.01351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Feng K.; Yang H.; Zhang Z.; Yuan Y.; Yue T. Effect of Cinnamaldehyde and Citral Combination on Transcriptional Profile, Growth, Oxidative Damage and Patulin Biosynthesis of Penicillium Expansum. Front. Microbiol. 2018, 9, 597. 10.3389/fmicb.2018.00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.; Gautam P.; Rai N.; Kumar N. An Emerging Hope to Combat Candida Albicans: Plant Based Therapeutics. Biotechnol. Int. 2012, 5, 85–114. [Google Scholar]
- Bacha K.; Tariku Y.; Gebreyesus F.; Zerihun S.; Mohammed A.; Weiland-Bräuer N.; Schmitz R. A.; Mulat M. Antimicrobial and Anti-Quorum Sensing Activities of Selected Medicinal Plants of Ethiopia: Implication for Development of Potent Antimicrobial Agents. BMC Microbiol. 2016, 16, 139. 10.1186/s12866-016-0765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alalwan H.; Rajendran R.; Lappin D. F.; Combet E.; Shahzad M.; Robertson D.; Nile C. J.; Williams C.; Ramage G. The Anti-Adhesive Effect of Curcumin on Candida Albicans Biofilms on Denture Materials. Front. Microbiol. 2017, 8, 659. 10.3389/fmicb.2017.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanloo S.; Shams-Ghahfarokhi M.; Ghahri M.; Razzaghi-Abyaneh M. Drug Susceptibility Profile of Candida Glabrata Clinical Isolates from Iran and Genetic Resistant Mechanisms to Caspofungin. Rev. Iberoam. Micol. 2018, 35, 88–91. 10.1016/j.riam.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Alnuaimi A. D.; O’Brien-Simpson N. M.; Reynolds E. C.; McCullough M. J. Clinical Isolates and Laboratory Reference Candida Species and Strains Have Varying Abilities to Form Biofilms. FEMS Yeast Res. 2013, 13, 689–699. 10.1111/1567-1364.12068. [DOI] [PubMed] [Google Scholar]
- Dorman H. J. D.; Deans S. G. Antimicrobial Agents from Plants: Antibacterial Activity of Plant Volatile Oils. J. Appl. Microbiol. 2000, 88, 308–316. 10.1046/j.1365-2672.2000.00969.x. [DOI] [PubMed] [Google Scholar]
- Abdelwahab S. I.; Mariod A. A.; Taha M. M. E.; Zaman F. Q.; Abdelmageed A. H. A.; Khamis S.; Sivasothy Y.; Awang K. Chemical Composition and Antioxidant Properties of the Essential Oil of Cinnamomum Altissimum Kosterm. (Lauraceae). Arabian J. Chem. 2017, 10, 131–135. 10.1016/j.arabjc.2014.02.001. [DOI] [Google Scholar]
- Marchese A.; Barbieri R.; Coppo E.; Orhan I. E.; Daglia M.; Nabavi S. F.; Izadi M.; Abdollahi M.; Nabavi S. M.; Ajami M. Antimicrobial Activity of Eugenol and Essential Oils Containing Eugenol: A Mechanistic Viewpoint. Crit. Rev. Microbiol. 2017, 43, 668–689. 10.1080/1040841x.2017.1295225. [DOI] [PubMed] [Google Scholar]
- Pinto E.; Vale-Silva L.; Cavaleiro C.; Salgueiro L. Antifungal Activity of the Clove Essential Oil from Syzygium Aromaticum on Candida, Aspergillus and Dermatophyte Species. J. Med. Microbiol. 2009, 58, 1454–1462. 10.1099/jmm.0.010538-0. [DOI] [PubMed] [Google Scholar]
- Latifah-Munirah B.; Himratul-Aznita W. H.; Zain N. M. Eugenol, an Essential Oil of Clove, Causes Disruption to the Cell Wall of Candida Albicans (ATCC 14053). Front. Life Sci. 2015, 8, 231. 10.1080/21553769.2015.1045628. [DOI] [Google Scholar]
- Devi K. P.; Nisha S. A.; Sakthivel R.; Pandian S. K. Eugenol (an Essential Oil of Clove) Acts as an Antibacterial Agent against Salmonella Typhi by Disrupting the Cellular Membrane. J. Ethnopharmacol. 2010, 130, 107–115. 10.1016/j.jep.2010.04.025. [DOI] [PubMed] [Google Scholar]
- de Souza E. L.; Almeida E. T. d. C.; Guedes J. P. d. S. The Potential of the Incorporation of Essential Oils and Their Individual Constituents to Improve Microbial Safety in Juices: A Review. Compr. Rev. Food Sci. Food Saf. 2016, 15, 753–772. 10.1111/1541-4337.12208. [DOI] [PubMed] [Google Scholar]
- Tsuchiya H.; Sato M.; Miyazaki T.; Fujiwara S.; Tanigaki S.; Ohyama M.; Tanaka T.; Iinuma M. Comparative Study on the Antibacterial Activity of Phytochemical Flavanones against Methicillin-Resistant Staphylococcus Aureus. J. Ethnopharmacol. 1996, 50, 27–34. 10.1016/0378-8741(96)85514-0. [DOI] [PubMed] [Google Scholar]
- Shreaz S.; Sheikh R. A.; Bhatia R.; Neelofar K.; Imran S.; Hashmi A. A.; Manzoor N.; Basir S. F.; Khan L. A. Antifungal Activity of α-Methyl Trans Cinnamaldehyde, Its Ligand and Metal Complexes: Promising Growth and Ergosterol Inhibitors. BioMetals 2011, 24, 923–933. 10.1007/s10534-011-9447-0. [DOI] [PubMed] [Google Scholar]
- Wu T.; Wright K.; Hurst S. F.; Morrison C. J. Enhanced Extracellular Production of Aspartyl Proteinase, a Virulence Factor, by Candida Albicans Isolates Following Growth in Subinhibitory Concentrations of Fluconazole. Antimicrob. Agents Chemother. 2000, 44, 1200–1208. 10.1128/aac.44.5.1200-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootong A.; Norrapong B.; Cowawintaweewat S. Antifungal Activity of Cinnamaldehyde against Candida Albicans. Southeast Asian J. Trop. Med. Public Health 2017, 48, 150–158. [PubMed] [Google Scholar]
- Bang K.-H.; Lee D.-W.; Park H.-M.; Rhee Y.-H. Inhibition of Fungal Cell Wall Synthesizing Enzymes by Trans-Cinnamaldehyde. Biosci., Biotechnol., Biochem. 2000, 64, 1061–1063. 10.1271/bbb.64.1061. [DOI] [PubMed] [Google Scholar]
- Yen T.-B.; Chang S.-T. Synergistic Effects of Cinnamaldehyde in Combination with Eugenol against Wood Decay Fungi. Bioresour. Technol. 2008, 99, 232–236. 10.1016/j.biortech.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Thakre A. D.; Mulange S. V.; Kodgire S. S.; Zore G. B.; Karuppayil S. M. Effects of Cinnamaldehyde, Ocimene, Camphene, Curcumin and Farnesene on Candida Albicans. Adv. Microbiol. 2016, 06, 627–643. 10.4236/aim.2016.69062. [DOI] [Google Scholar]
- Khan M. S.; Ahmad I.; Cameotra S. Phenyl Aldehyde and Propanoids Exert Multiple Sites of Action towards Cell Membrane and Cell Wall Targeting Ergosterol in Candida Albicans. AMB Express 2013, 3, 54. 10.1186/2191-0855-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks L. W.; Casey W. M. Physiological Implications of Sterol Biosynthesis in Yeast. Annu. Rev. Microbiol. 1995, 49, 95–116. 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- de Paula S. B.; Bartelli T. F.; Di Raimo V.; Santos J. P.; Morey A. T.; Bosini M. A.; Nakamura C. V.; Yamauchi L. M.; Yamada-Ogatta S. F. Effect of Eugenol on Cell Surface Hydrophobicity, Adhesion, and Biofilm of Candida Tropicalis and Candida Dubliniensis Isolated from Oral Cavity of HIV-Infected Patients. J. Evidence-Based Complementary Altern. Med. 2014, 2014, 1–8. 10.1155/2014/505204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F. d. O.; Mendes J. M.; Lima E. d. O. Investigation on Mechanism of Antifungal Activity of Eugenol against Trichophyton rubrum. Med. Mycol. 2013, 51, 507–513. 10.3109/13693786.2012.742966. [DOI] [PubMed] [Google Scholar]
- Khan S. N.; Khan S.; Iqbal J.; Khan R.; Khan A. U. Enhanced Killing and Antibiofilm Activity of Encapsulated Cinnamaldehyde against Candida Albicans. Front. Microbiol. 2017, 8, 1641. 10.3389/fmicb.2017.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreaz S.; Bhatia R.; Khan N.; Muralidhar S.; Manzoor N.; Khan L. A. Influences of Cinnamic Aldehydes on H+ Extrusion Activity and Ultrastructure of Candida. J. Med. Microbiol. 2013, 62, 232–240. 10.1099/jmm.0.036145-0. [DOI] [PubMed] [Google Scholar]
- Khan A.; Ahmad A.; Akhtar F.; Yousuf S.; Xess I.; Khan L. A.; Manzoor N. Induction of Oxidative Stress as a Possible Mechanism of the Antifungal Action of Three Phenylpropanoids. FEMS Yeast Res. 2011, 11, 114–122. 10.1111/j.1567-1364.2010.00697.x. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Huang S.; Dai B.; Zhu Z.; Lu H.; Dong L.; Cao Y.; Wang Y.; Gao P.; Chai Y.; Jiang Y. Candidaalbicans Cells Lacking CaMCA1-Encoded Metacaspase Show Resistance to Oxidative Stress-Induced Death and Change in Energy Metabolism. Fungal Genet. Biol. 2009, 46, 183–189. 10.1016/j.fgb.2008.11.001. [DOI] [PubMed] [Google Scholar]
- García-Salinas S.; Elizondo-Castillo H.; Arruebo M.; Mendoza G.; Irusta S. Evaluation of the Antimicrobial Activity and Cytotoxicity of Different Components of Natural Origin Present in Essential Oils. Molecules 2018, 23, 1399. 10.3390/molecules23061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyldgaard M.; Mygind T.; Meyer R. L. Essential Oils in Food Preservation: Mode of Action, Synergies, and Interactions with Food Matrix Components. Front. Microbiol. 2012, 3, 12. 10.3389/fmicb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranasinghe P.; Pigera S.; Premakumara G. A. S.; Galappaththy P.; Constantine G. R.; Katulanda P. Medicinal Properties of “true” Cinnamon (Cinnamomum Zeylanicum): A Systematic Review. BMC Complementary Altern. Med. 2013, 13, 275. 10.1186/1472-6882-13-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanomaru-Filho M.; Tanomaru J. M. G.; Barros D. B.; Watanabe E.; Ito I. Y. In Vitro Antimicrobial Activity of Endodontic Sealers, MTA-Based Cements and Portland Cement. J. Oral Sci. 2007, 49, 41–45. 10.2334/josnusd.49.41. [DOI] [PubMed] [Google Scholar]
- Elaissi A.; Rouis Z.; Salem N. A. B.; Mabrouk S.; ben Salem Y.; Salah K. B. H.; Aouni M.; Farhat F.; Chemli R.; Harzallah-Skhiri F. Chemical Composition of 8 Eucalyptus Species’ Essential Oils and the Evaluation of Their Antibacterial, Antifungal and Antiviral Activities. BMC Complementary Altern. Med. 2012, 12, 81. 10.1186/1472-6882-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.; Chanda R.; Rai N.; Kataria V. K.; Kumar N. Antihypertensive, Amlodipine Besilate Inhibits Growth and Biofilm of Human Fungal Pathogen Candida. Assay Drug Dev. Technol. 2016, 14, 291–297. 10.1089/adt.2016.714. [DOI] [PubMed] [Google Scholar]
- Gupta P.; Nath S.; Meena R. C.; Kumar N. Comparative Effects of Hypoxia and Hypoxia Mimetic Cobalt Chloride on in Vitro Adhesion, Biofilm Formation and Susceptibility to Amphotericin B of Candida Glabrata. Journal Med. Mycol. 2014, 24, e169–e177. 10.1016/j.mycmed.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Gupta P.; Meena R. C.; Kumar N. Functional Analysis of Selected Deletion Mutants in Candida Glabrata under Hypoxia. 3 Biotech 2017, 7, 193. 10.1007/s13205-017-0821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex J. H.Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard, 2nd ed.; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Pemmaraju S. C.; Padmapriya K.; Pruthi P. A.; Prasad R.; Pruthi V. Impact of Oxidative and Osmotic Stresses on Candida Albicans Biofilm Formation. Biofouling 2016, 32, 897–909. 10.1080/08927014.2016.1212021. [DOI] [PubMed] [Google Scholar]
- Fonseca E.; Silva S.; Rodrigues C. F.; Alves C. T.; Azeredo J.; Henriques M. Effects of Fluconazole on Candida Glabrata Biofilms and Its Relationship with ABC Transporter Gene Expression. Biofouling 2014, 30, 447–457. 10.1080/08927014.2014.886108. [DOI] [PubMed] [Google Scholar]
- Orta-Zavalza E.; Briones-Martin-del-Campo M.; Castano I.; De Las Penas A. Catalase Activity Assay in Candida Glabrata. Bio-Protoc. 2014, 4, e1072 10.21769/bioprotoc.1072. [DOI] [Google Scholar]
- Rajkowska K.; Kunicka-Styczyńska A.; Maroszyńska M. Selected Essential Oils as Antifungal Agents against Antibiotic-Resistant Candida Spp.: In Vitro Study on Clinical and Food-Borne Isolates. Microb. Drug Resist. 2017, 23, 18–24. 10.1089/mdr.2016.0001. [DOI] [PubMed] [Google Scholar]
- Kwolek-Mirek M.; Zadrag-Tecza R. Comparison of Methods Used for Assessing the Viability and Vitality of Yeast Cells. FEMS Yeast Res. 2014, 14, 1068–1079. 10.1111/1567-1364.12202. [DOI] [PubMed] [Google Scholar]
- Panwar R.; Pemmaraju S. C.; Sharma A. K.; Pruthi V. Efficacy of Ferulic Acid Encapsulated Chitosan Nanoparticles against Candida Albicans Biofilm. Microb. Pathog. 2016, 95, 21–31. 10.1016/j.micpath.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Latifah-Munirah B.; Himratul-Aznita W. H.; Zain N. M. Eugenol, an Essential Oil of Clove, Causes Disruption to the Cell Wall of Candida Albicans (ATCC 14053). Front. Life Sci. 2015, 8, 231–240. 10.1080/21553769.2015.1045628. [DOI] [Google Scholar]
- Kwolek-Mirek M.; Zadrag-Tecza R. Comparison of Methods Used for Assessing the Viability and Vitality of Yeast Cells. FEMS Yeast Res. 2014, 14, 1068–1079. 10.1111/1567-1364.12202. [DOI] [PubMed] [Google Scholar]
- Haque F.; Alfatah M.; Ganesan K.; Bhattacharyya M. S. Inhibitory Effect of Sophorolipid on Candida Albicans Biofilm Formation and Hyphal Growth. Sci. Rep. 2016, 6, 23575. 10.1038/srep23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J.; Kuhn D. M.; Mukherjee P. K.; Hoyer L. L.; McCormick T.; Ghannoum M. A. Biofilm Formation by the Fungal PathogenCandida Albicans: Development, Architecture, and Drug Resistance. J. Bacteriol. 2001, 183, 5385–5394. 10.1128/jb.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y.; Chandra J.; Mukherjee P. K.; Lattif A. A.; Szczotka-Flynn L. B.; Pearlman E.; Lass J. H.; O’Donnell K.; Ghannoum M. A. Fusarium and Candida Albicans Biofilms on Soft Contact Lenses: Model Development, Influence of Lens Type, and Susceptibility to Lens Care Solutions. Antimicrob. Agents Chemother. 2008, 52, 171–182. 10.1128/aac.00387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.; Agrawal V.; Pemmaraju S. C.; Panwar R.; Pruthi V. Impact of Infectious Candida Albicans Biofilm on Biomaterials. Indian J. Biotechnol. 2011, 10, 417–422. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.