Abstract
Light is a key environmental regulator in all photosynthetic organisms. Many studies focused on the physiologic response to changes in light availability of species from the Zygnematophyceae, but the impact of the absence of light and the molecular acclimation process on the other side have been poorly understood. Here we present transcriptomic analyses of Cosmarium crenatum from a polar habitat exposed to darkness. The algae were cultured in dark for one week; cell number and quantum yield of photosystem II (Fv/Fm) were monitored. Cell number was stable, but the Fv/Fm decreased in both groups, darkness-treated and control. Gene expression analysis revealed a strong repression of transcripts associated with photosynthesis, photorespiration and cell wall development. General carbohydrate and lipid metabolism were differentially regulated, but starch is shown to be the primary energy source in these conditions. Additionally, C. crenatum induced mRNA responsible for epigenetic modifications which may be a specific response to an adaption and acclimation to polar conditions. Our study sheds light on the molecular acclimation process to darkness and provides ecological implications for new perspectives in this specialized group of green algae.
Subject terms: Microbiology, Transcriptomics, Light responses
Introduction
Numerous studies showed that light is a key regulator of plant metabolism, development and a central factor for survival. Change of gene expression as reaction to alterations of light availability has been studied in model land plants since decades1,2. Plants fast-react to changes of environmental conditions in order to survive and therefore exhibit a complex remodeling of the transcriptome3. The acclimation to various abiotic conditions of photosynthetic organisms was key for the colonization of terrestrial habitats about 450 Mya4. Land plants evolved from an aquatic ancestor from the group of Charophyta, sharing fundamental phenotypic traits5–7. The genus Cosmarium forms the largest taxonomic entity within the Zygnematophyceae8, the closest living relatives to the Embryophyta9,10. This makes Cosmarium and other genera of the conjugating green algae useful organisms to study early land plant evolution11.
A broad physiological acclimation potential has been described for different Cosmarium species with origins ranging from the tropics to polar regions12. Furthermore, the physiological acclimation to certain environmental regimes points to the origin of the isolate13, but molecular information on the acclimation process is still missing. The species Cosmarium crenatum is distributed worldwide and has been discovered in e.g. Scotland14, the northwest territories in Canada15, New Zealand16 as well as the sub Antarctic Kerguelen islands17. Described as an essentially arctic-alpine species, C. crenatum is predominantly found under harsh climatic conditions of high altitude or latitude, but exhibits a great acclimation potential, and thus, may be encountered even in the lowlands18. In these wide-spread localities environmental factors like temperature and nutrients differ tremendously, but the availability of light may be the most challenging factor for a photoautotroph. Polar freshwater ponds are subjected to continuous winter darkness with ice covers persisting up to 9 months19. Under different climate change scenarios, a rise in temperature is projected to increase precipitation and cloud cover in high Arctic regions20 and, therefore, the ice-covered period may even be extended in the future. Photosynthetic primary production in polar regions is strongly influenced by underwater irradiance which highly depends on prolongation of snow cover in spring, latitudinal position and turbidity21,22. Water temperatures below 0 °C are ubiquitous in winter in Arctic and Subarctic regions, but also above 20 °C may be easily reached in shallow ponds in summer23. The physiological acclimation to naturally occurring abiotic stress scenarios of a number of charophyte green algae has been well studied24 and molecular acclimation studies in these non-model organisms are usually built around specific stressors e.g. desiccation25,26 or the influence of phytohormones27. Although many physiological studies were performed on temperature and light stress in these groups, different stressors are poorly understood and molecular information on the process of acclimation and comprehensive metabolic information is still missing.
This study aims to characterize the molecular response to long-term darkness-induced changes in gene expression in the non-model microalga C. crenatum. Furthermore, we established a comprehensive de novo transcriptome as a basis for future studies on the mechanisms involved in light and temperature acclimation. It is our key hypothesis that an extensive remodeling of the transcriptome is necessary for the alga for survival during elongated darkness as photosynthesis cannot be the energy source.
It is our long-term goal to understand the molecular basis of acclimation to environmental alterations like the influence of polar night. A RNA-seq approach was utilized to construct the transcriptome reference and to investigate differential gene expression. Physiological responses to darkness of one week were determined by cell growth and the maximum quantum yield of photosystem II and compared to gene expression changes.
Material and Methods
Algal material
Liquid cultures of a polar strain of C. crenatum Ralfs 1884 var. boldtianum Gutwinski (Microalgae and Zygnematophyceae Collection Hamburg (MZCH) 561), originating from Franz Josef Land (Russia), were cultured in a synthetic mineral Woods Hole (WH) medium28 at 12 °C and 100 µmol photons m−2 s−1 in a light:dark cycle of 16:8 h. The cultures were acclimated to culture medium and conditions at least 6 months before the experiment was carried out. The algae were pre-cultured in separate 1 L Erlenmeyer bottles filled with 800 mL medium and dispersed with atmospheric air. For gaining high cell concentrations, dense pre-cultures were disconnected from gasification and irradiated 1 h under standard conditions. Algal cells started to form dense conglomerates and rose to the upper surface layer. The algae were transferred with sterile 10 mL pipettes to clean glass bottles and examined prior with an inverted microscope to check vitality and purity of the culture.
Experimental conditions and reference transcriptome RNA sampling
Cultures were transferred from standard growth conditions at 12 °C and 100 µmol photons m−2 s−1 to 2, 12, 22 and 32 (±1) °C for one week at 100 and 500 µmol photons m−2 s−1 (n = 8) in a light:dark cycle of 16:8 in a Multi-Cultivator MC 1000-OD (Photon Systems Instruments, Brno, Czech Republic). Additionally, algae were cultured at 2 °C in darkness for one week. Sampling was performed initially (1 h), short-term (16 h) and long-term (1 week), replicates n = 8. An external cooling aggregate was mounted to the MC for the treatments at 2 and 12 °C. Exposures to UV-radiation (UVR) were performed short-term (16 h) at all mentioned temperatures for high and low light (500 and 100 µmol photons m−2 s−1) exposure in a sun simulator (SonSi, iSiTECGmbH, Bremerhaven, Germany). For a solar-like spectrum including UVR (UVA 8.8 Wm−2, UVB 0.61 W m−2), the cultures were illuminated with a 400 W Metallogen lamp (Philips MSR 400 HR, Germany), as described in literature29,30. After exposure, samples from all treatments were collected. Additionally, samples from standard culture conditions and from one day, one week and one month old cultures (n = 3) were taken. Samples were centrifuged in 2 mL reaction tubes (Sarstedt, Nümbrecht, Germany) for 10 s, the liquid phase was discarded. The tubes were initially flash-frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
Physiology
Cell number was measured with a particle counter (Beckman Coulter GmbH, Krefeld, Germany) with a tube aperture of 100 µm. Chlorophyll fluorescence was determined in vivo with an Imaging Pulse Amplitude fluorometer (Imaging PAM, Heinz Walz GmbH, Effeltrich, Germany) and analyzed with the ImagingWin software (Heinz Walz GmbH, Effeltrich, Germany). Chlorophyll fluorescence was measured as Fv/Fm ((Fm/-Fo)/Fm), which is the optimal quantum yield indicating photosynthetic efficiency of dark adapted reaction centers, i.e. which amount of absorbed photons may be converted into electron transport. The maximum fluorescence (Fm) is the theoretical highest possible fluorescence where all reaction centers are closed, the minimum fluorescence (Fo) is the fluorescence when all reaction centers are open. (for further information see31). Prior to the measurements, the number of cells was adjusted to approximately 110,000 cells mL−1 with fresh culture medium ensuring comparable results of the photosynthetic activity and to avoid fluorescence reabsorption. Photosynthetic data and cell numbers were statistically analyzed with SPSS software version 23 (IBM, Armonk, USA). Based on homogeneity of variance tested with Levene statistic and normality testing with Lilliefors significance correction on Kolmogorov-Smirnov test, either an ANOVA or a non-parametric Friedman’s test was applied (p ≤ 0.05).
RNA isolation and reference transcriptome sequencing
RNA extraction was performed with guanidine thiocyanate phenol chloroform, following the protocol for microalgae samples32. After RNA purity control with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), all samples with high purity and quantity were assessed on a Fragment Analyzer (Advanced Analytical Technologies Inc., Heidelberg, Germany).
Two sequencing approaches were utilized, the first to generate a comprehensive de novo reference transcriptome and, the second to profile the gene expression after one week of darkness exposure. For the reference transcriptome generation, high quality samples with RNA quality number (RQN) above 6 (214 in total) of differentially treated algae from all experimental conditions were pooled in equal amounts to generate a normalized reference transcriptome. The pooling of various RNA samples from different treatments was conducted to ensure a high functional coverage of the reference transcriptome. Reference library sequencing was carried out with a genome sequencer Illumina HiSeq 2500 in rapid run 300 bp paired-end modus.
Gene expression sequencing
The cDNA libraries for analysing differential gene expression in darkness were constructed in triplicates. Strand-specific cDNA libraries for analyzing differential gene expression were sequenced in HiSeq 2500 rapid run 100 bp single read modus. All library preparations and sequencing were carried out by GATC Biotech (Konstanz, Germany). The cleaned raw data were deposited in the European Nucleotide Archive (ENA) at the European Molecular Biological Laboratory–European Bioinformatics Institute under study accession number PRJEB30351.
Transcriptome assembly and annotation
Quality of reads were controlled with FastQC33. Adapter sequences were removed with bbduk.sh, from the BBtools suite, version 36.3834 with following parameters: ktrim = r, k = 23, mink = 11, hdist = 1, tpe, tbo. Remaining sequences were searched for rRNA sequences by SortMeRNA version 2.135 which were removed before further processing. To filter the sequences for the common Illumina spikein PhiX, bbduk.sh was used with a kmer size of 31 and a hdist of 1. A final quality trimming was performed with bbduk.sh using Q10 as minimum quality and 36 bases as the minimum length. The quality filtered reads were assembled de novo into transcripts yielding 141,711 transcripts using the Trinity assembler version 2.5.136 with normalized max. read coverage of 100 and a minimum transcript length of 300 bp as parameters. To estimate the completeness of the reference transcriptome Benchmarking Universal Single-Copy Orthologs (BUSCO) was used with the plant datasets 1.1 and Embryophyta odb9 as databases37. The generated transcripts were functional annotated using the Trinotate annotation suite version 3.0.2 (https://trinotate.github.io/). Functional annotation included homology search against known sequence data (BLAST+/UniProtKB Swiss-Prot), protein domain identification (HMMER/PFAM) and retrieving data from additional databases (KEGG/COG/EggNOG). To assess the coverage of metabolic and regulatory pathways, the assigned KEGG, Kegg orthology (KO) numbers and COG were mapped to a KEGG38–40 pathway map (Supplementary Fig. S1).
The reference transcriptome of C. crenatum was compared in a reciprocal best hit analysis with the transcriptome of Zygnema circumcarinatum (Zygnematophyceae)26 and the coding sequences of the genome of Marchantia polymorpha (Embryophyta)41. Using Transdecoder version 3.0.0 (http://transdecoder.github.io), protein sequences were predicted for all three species, which were then used for a reciprocal blast search applying an e-value cut-off of 1e−7.
Differential expression analysis
Short reads of each sample were aligned against the reference de novo transcriptome, using Bowtie2 (version 2.3.3). Low-count transcripts were filtered using a row sum cut-off (>10) on the generated count matrix. Relative abundances were estimated by RSEM (version 1.2.26) and genes were analysed for differential expression using the edgeR Bioconductor package42 with a standard level of p ≤ 0.001 and a log fold change of 2 indicating significance. To detect gene expression changes in darkness, pairwise comparison of the darkness treatment with the control light treatment were conducted.
To identify significantly enriched gene functions among the induced and repressed genes under darkness conditions, a GO enrichment analysis, as implemented in the trinity package (version 2.5.1), was performed. GO terms were classified with CateGOrizer43, using the EGAD2GO classification categories with the additional terms GO:0015979 “photosynthesis” and GO:0009579 “thylakoid”, applied for generating an overview of the GO enrichment analysis (Supplementary Fig. S2).
Results and Discussion
Reference transcriptome and comparative analysis
In this study, we present a comprehensive de novo reference transcriptome from C. crenatum under 18 different temperature and light regimes, covering initial, short-term and long-term acclimation. The Illumina Rapid Run yielded in ~29 million paired end reads with an average length of 300 bp, ~363 million single end reads with an average length of 100 bp, respectively. The de novo assembly yielded in 141,711 transcripts with an average length of 1,231 bp and an N50 of 1,592 bp. In total, 94.34% of the reads could be re-aligned (Bowtie2) to the assembly. Annotation of the assembled transcriptome against sprot BLASTX resulted in 58,826 hits (41.5%). The conducted KEGG mapping resulted in a high coverage, indicating that the established reference represents most parts of the functional genome (Supplementary Fig. S1).
BUSCO analysis using the dataset for plants (Embryophyta odb9) yielded in 39.1% complete, 4.2% fragmented and 56.7% missing BUSCOs, the reference set for Embryophyta odb9 only covers higher plants and orthologous groups that were described within these (n = 1,440). We additionally compared our reference to BUSCO plant v 1.1 (n = 956) and reached 67.2% complete, 10% fragmented and 22.8% missing BUSCOs, indicating a good coverage of orthologous groups37. For Z. circumcarinatum 76% complete BUSCOs, 8% fragmented and 16% were previously shown to be present accordingly26. BUSCO uses the single-copy characteristic of orthologues groups to estimate the contained redundancy of a (functional) genome assembly, but duplications and polyploidy may bias this function in Embryophyta44.
The similarity of the Zygnematophyceae to early land plants is mirrored in the results from reciprocal BLAST analysis between C. crenatum, Z. circumcarinatum26 and M. polymorpha41 (Fig. 1). The three species share 3,327 protein sequences (center of diagram), the second highest number after direct comparison of the close relatives C. crenatum and Z. circumcarinatum with 3,915. Z. circumcarinatum and M. polymorpha share with 3,237 the second highest similarities. These results agree with Timme et al.45 who observed that Spirogyra pratensis, another member of Zygnematales, shares a high number of orthologous groups with land plants45.
Figure 1.
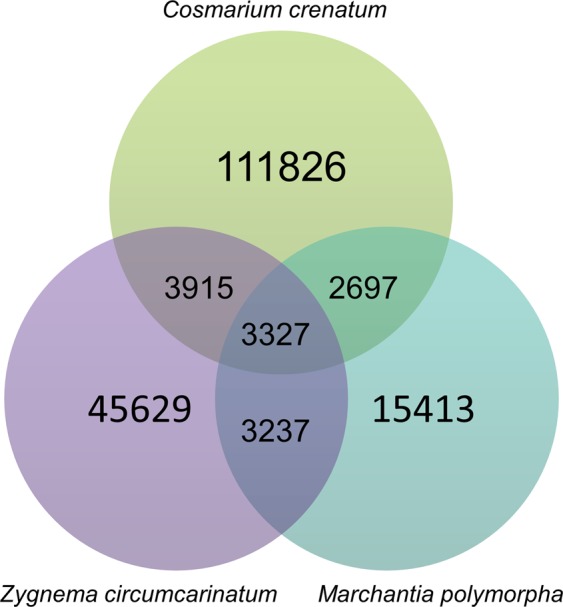
Venn diagram with reciprocal best hits between the reference transcriptomes of Cosmarium crenatum and Zygnema circumcarinatum and the genome of the liverwort Marchantia polymorpha. Larger numbers in circles indicate the number of unique protein sequences, shared sequences are found in overlapping areas. Protein sequences shared between all species tested are in the overlapping center.
More than 100 sequences in the comparison of C. crenatum, Z. circumcarinatum and M. polymorpha are encoding transcription factors, regulators or enhancers. An expansion of the number of transcription factors has been detected in the genome of M. polymorpha41. The expansion is hypothesized to may have allowed a diversification in the response to environmental changes46, which are more prominent and occur more rapidly on land than in an aquatic habitat. Along the shared sequences are plant hormone-responsive transcription factors, e.g. ethylene-responsive transcription factor (AIL1) and auxin response factors (ARFI). Phytohormones participate in a wide range of physiological and developmental processes and have been well studied in angiosperms47. For another streptophytic alga S. pratensis (Zygnematales) was recently shown that phytohormones alter the transcriptomic response of cell wall modification, photosynthesis and abiotic stress27. Our data indicates that phytohormones might also play a role in development and/or metabolism in C. crenatum. However, to confirm this hypothesis further studies should be conducted to investigate in more detail if and how phytohormones affect C. crenatum.
Gene expression analysis of genes responding to darkness
Sequencing yielded in ∼14 million reads per replicate and on average 12 million reads were aligned to the reference (79%). In total, 7,905 differentially expressed genes (DEGs) were found, of which 3,767 transcripts were induced and 4,138 repressed in darkness. About 28% of the up-regulated and 47% of the down-regulated contigs could be successfully assigned an annotation. Furthermore, all differentially regulated genes were annotated using NCBI non-redundant (nr) database with diamond local aligner48 (cut-off 1E−9), which increased the number of annotated functions to about 40% in the up-regulated genes and 65% of the down-regulated genes. A full list of DEGs can be retrieved from the Supplemental Material (Supplemental Table 1).
Functional annotation of darkness-responding transcripts
GO enrichment analysis resulted in 316 enriched GO terms among the in darkness up-regulated DEGs and 858 among the down-regulated transcripts in darkness (p < 0.01). General metabolism and carrier proteins were significantly enriched among both, the up- and down-regulated transcripts in darkness. Enriched GO terms among the down-regulated transcripts belonged mainly to the categories lipid metabolism, cofactor metabolism, energy/tricarboxylic acid (TCA) cycle and photosynthesis.
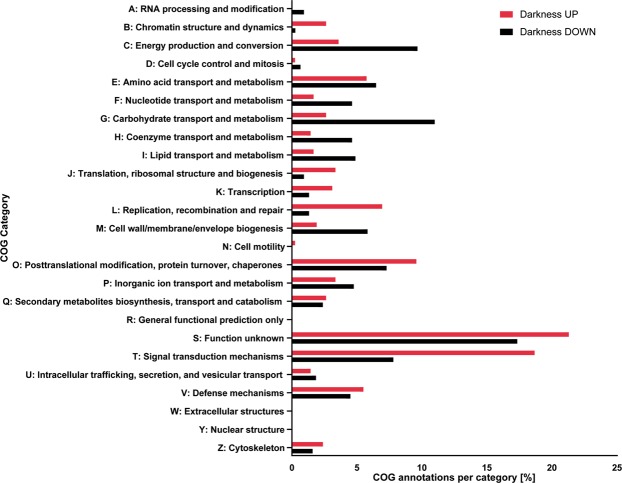
The distribution of annotated COG categories of regulated transcripts indicates a broad response to the darkness treatment regarding cellular functions. Largest percentages of DEGs were found in the category “unknown function” and “signal transduction mechanisms”. Darkness triggered more induction than repression of transcripts in several categories, e.g. “chromatin structure and dynamics”, “translation, ribosomal structure and biogenesis” and “signal transduction mechanisms”. Furthermore, darkness caused more down- then up regulation of transcripts belonging to the classes “energy production and conversion”, “carbohydrate transport and metabolism”. “Carbohydrate transport and metabolism” was the second largest number of COGs found in the down-regulated transcripts with 11.98%, followed by “energy production and conversion” with 9.66% (Fig. 2).
Figure 2.
Categorized (A to Z) Clusters of Orthologous Groups (COG) analysis for darkness-induced and darkness-repressed transcripts, in percent (%) excluding “X: No COG/KOG” per group.
Physiological response and gene expression adjustments to darkness
Under darkness cell density was roughly stable from day 1 (129,347 ± 1,063 cells mL−1) to day 7 (128,890 ± 4,336.15) with a decrease of 0.35%, indicating no cell divisions. Under low light cell density was highest at day 1 (114,803.75 ± 4,582.15), and decreased gradually until day 7 (94,662.5 ± 7,647.77) by 17.5%.
After one week cell density remained the same in darkness (Friedman’s, p < 0.05, n = 8) (Fig. 3), this heterogeneous response is also reflected in the underlying molecular response in cell growth associated mRNAs. Expression of transcripts annotated endoglucanase 1 and 13 were regulated with logFC of 5.87 and −4.24, respectively (Table 1). Up- regulation was found for the probable pectinesterase 53 (logFC of 7.04), down-regulation was observed for pectinesterase 31 (logFC −4.42). Transcripts involved in cell wall growth, like xyloglucan endotransglucosylase/hydrolase and transcripts coding for various expansins, were repressed with logFC from −2.8 to −6.9. Expansins are proteins with diverse classification that induce loosening of wall molecules and subsequently cell extensibility49. Although often theorized about an enzymatic function; so far no catalytic activity has been determined50. In Micrasterias denticulata (Desmidiales), four different expansins are transcriptionally active during reorganization, assembly and selective degradation of the cell wall, two of the genes were active in earlier morphogenetic stages and two being induced in later stages51. Based on these findings, a manual BLASTx analysis was performed and the sequences of transcripts DN11092_c0_g1, DN11092_c0_g2, DN13260_c0_g1 and DN19416_c0_g1 retrieved as best hits the expansins of M. denticulata, which were all down-regulated in the darkness dataset. Furthermore, the involvement of xyloglucan endotransglucosylase/hydrolase (XTH) in cell wall loosening, strengthening and therefore growth is suggested for desmids51. All regulated transcripts encoding XTHs of Cosmarium were manually analyzed in BLASTx and corresponded to the XTH of M. denticulata, except for transcript DN18252_c0_g1, which retrieved a predicted XTH in Brassica rapa. All XTH-encoding genes were down-regulated in darkness, except for the one with the B. rapa annotation. These expression patterns are in accordance with the transcriptomic processes of M. denticulata as described by Vannerum et al.51, where the four expansin genes and the XTH were only significantly regulated during growth or in the initiation of it. Generally, when cells are in a growing stage one or more expansin genes are involved49. Our findings in the gene expression of expansins and XTHs in combination with no significant changes in cell counts in Cosmarium, indicate that neither cell divisions nor substantial modification of the cell wall occurred in darkness.
Figure 3.
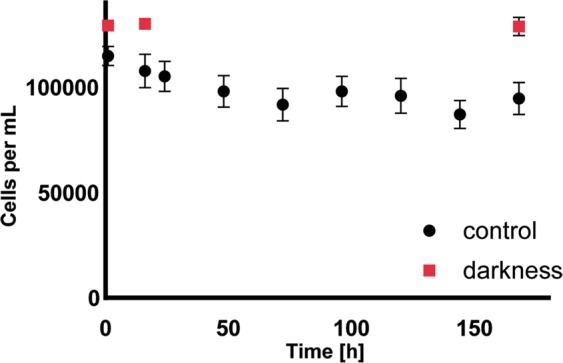
Cell number (cells/mL) of C. crenatum cultivated in 100 µmol photons m−2 s−1 (control) and darkness at 2 °C, error bars indicating standard deviation (n = 8).
Table 1.
Selected list of differentially expressed transcripts in response to darkness (complete list in Supplementary Table S1).
| Contig ID | Putative gene product | E-value | logFC |
|---|---|---|---|
| Cell wall development | |||
| DN15769c0g1 | Endoglucanase 1 | 9.67E-126 | 5.87 |
| DN19491c0g1 | Endoglucanase 13 | 1.62E-63 | −4.24 |
| DN21391c0g1 | Pectinesterase 31 | 1.79E-50 | −4.42 |
| DN14496c0g1 | Putative xyloglucan endotransglucosylase/hydrolase protein 1 | 3.03E-23 | −6.80 |
| DN11092c0g1 | Expansin-A8 | 2.93E-15 | −6.92 |
| DN13260c0g1 | Expansin-A18 | 9.92E-29 | −2.87 |
| DN19416c0g1 | Expansin-A14 | 8.91E-25 | −5.64 |
| DN14554c0g1 | Probable xyloglucan endotransglucosylase/hydrolase protein 18 | 2.36E-16 | −4.93 |
| DN18208c0g2 | Probable xyloglucan endotransglucosylase/hydrolase protein 33 | 4.61E-25 | −4.05 |
| DN18252c0g1 | Xyloglucan endotransglucosylase/hydrolase protein 31 | 5.15E-10 | 2.03 |
| Photosynthesis, photorespiration and accessory pigments | |||
| DN16759c3g1 | Chlorophyll a-b binding protein | 3.66E-94 | −10.05 |
| DN13506c0g2 | Photosystem I reaction center subunit II | 3.3E-89 | −4.00 |
| DN10003c0g1 | Photosystem II 10 kDa polypeptide | 2.18E-22 | −2.79 |
| DN10098c0g1 | Plastocyanin | 6.83E-47 | −5.03 |
| DN11429c0g1 | Cytochrome b6-f complex iron-sulfur subunit 2 | 3.65E-65 | −3.43 |
| DN22938c0g3 | Cytochrome c6 | 8.24E-31 | −2.13 |
| DN22290c0g6 | Ribulose bisphosphate carboxylase large chain | 0 | −2.32 |
| DN12878c0g1 | Ribulose bisphosphate carboxylase small chain C | 6.32E-46 | −2.60 |
| DN16824c1g1 | Glyceraldehyde-3-phosphate dehydrogenase GAPB | 0 | −3.39 |
| DN16824c1g2 | Glyceraldehyde-3-phosphate dehydrogenase A | 0 | −4.34 |
| DN19513c0g2 | Phosphoribulokinase | 0 | −5.48 |
| DN19513c0g1 | Phosphoribulokinase | 3.64E-32 | −5.01 |
| DN16537c0g1 | Tetrapyrrole-binding protein, | 5.68E-37 | −5.12 |
| DN23145c0g1 | Chlorophyll synthase | 2.06E-144 | −2.71 |
| DN19195c1g2 | Magnesium protoporphyrin IX methyltransferase | 1.63E-99 | −5.95 |
| DN21095c1g2 | Pheophytinase | 3.79E-17 | 2.20 |
| DN14118c0g1 | Electron transfer flavoprotein subunit beta | 8.92E-108 | 2.03 |
| DN9942c0g1 | Chlorophyllase-1 | 1.43E-32 | 2.42 |
| DN21727c0g1 | Violaxanthin de-epoxidase | 2.88E-127 | −2.18 |
| DN23964c2g2 | Zeaxanthin epoxidase | 8.8E-61 | 2.05 |
| DN16052c1g2 | ATP-dependent 6-phosphofructokinase 4 | 3.77E-49 | −6.82 |
| DN20695c0g1 | Cytochrome c | 3.3E-63 | −7.01 |
| DN15902c0g3 | NADH dehydrogenase 1 alpha subcomplex subunit 8-A | 8.93E-35 | −3.48 |
| DN13480c1g15 | Cytochrome b | 1.01E-24 | −2.05 |
| DN22923c1g1 | ATP synthase subunit beta | 0 | −2.05 |
| DN15210c0g2 | ATP synthase subunit epsilon | 3.5E-19 | −2.04 |
| Carbohydrate metabolism and associated transport | |||
| DN21236c0g2 | Sucrose synthase 4 | 1.18E-71 | 2.88 |
| DN24074c1g2 | Probable starch synthase 4 | 2.34E-119 | 2.81 |
| DN21996c0g1 | Beta-amylase 3 | 3.04E-153 | 2.47 |
| DN19743c0g2 | Beta-amylase 7 | 8.54E-18 | 2.20 |
| DN23791c2g1 | Soluble starch synthase 3 | 4.15E-17 | 3.43 |
| DN22147c0g1 | Beta-amylase | 3.13E-109 | 2.07 |
| DN23709c2g1 | Sugar transport protein 9 | 4.14E-75 | 6.04 |
| DN23709c1g1 | Sugar transport protein 7 | 1.12E-80 | 4.25 |
| DN23709c2g3 | Sugar transport protein 13 | 3.41E-93 | 2.26 |
| Lipid metabolism | |||
| DN21626c0g2 | Delta(12)-acyl-lipid-desaturase | 1.33E-139 | −10.36 |
| DN23736c0g1 | Oleoyl-acyl carrier protein thioesterase | 3.87E-60 | −6.53 |
| DN16200c0g1 | Acetyl-CoA carboxylase carboxyl transferase subunit alpha | 1.29E-72 | −3.84 |
| DN24419c0g2 | Acetyl-CoA carboxylase 2 | 4.97E-25 | −3.77 |
| DN21155c3g1 | Phospholipase SGR2 | 3.55E-119 | 4.20 |
| DN21919c0g1 | Triacylglycerol lipase 1 | 1.21E-30 | 4.83 |
| DN13622c0g1 | Autophagy-related protein 9 | 1.08E-10 | 2.04 |
| DN15846c2g2 | Autophagy-related protein 11 | 4.84E-10 | 2.07 |
| DN16399c0g1 | Autophagy-related protein 22-1 | 3.5E-21 | 2.37 |
| Epigenetic modifications | |||
| DN23893c0g2 | Histone-lysine N-methyltransferase ATXR3 | 8.39E-114 | 2.19 |
| DN19070c1g3 | Histone-lysine N-methyltransferase family member SUVH2 | 8.14E-26 | 3.62 |
| DN14298c1g2 | Lysine-specific histone demethylase 1B | 2.87E-24 | 3.16 |
| DN20928c2g1 | Histone acetyltransferase GCN5 | 3.45E-77 | 2.38 |
| DN11922c0g1 | Histone-lysine N-methyltransferase SUVR5 | 2.77E-11 | 2.45 |
| DN26242c0g1 | Histone-lysine N-methyltransferase SUVR4 | 5.96E-12 | 3.37 |
| DN19418c0g3 | Histone acetyltransferase GCN5 | 7.60E-11 | 2.57 |
A significant difference in the photosynthetic response was observed between groups in darkness (D) and control conditions (C) for measurement point zero and after one hour (Friedman’s, p ≤ 0.05, n = 8). After one week of experiment, Fv/Fm was 0.449 (±0.008) in D and 0.461 (±0.016) in C (Fig. 4).
Figure 4.
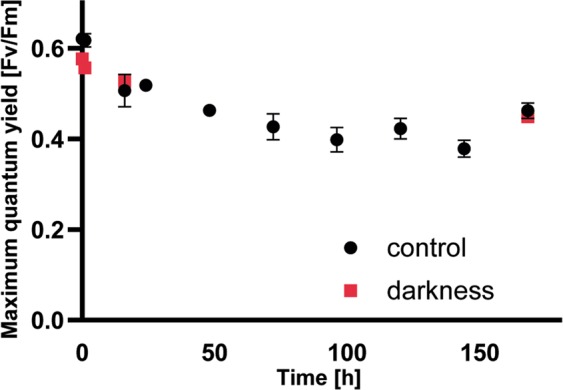
Photosynthetic efficiency of PSII (Fv/Fm) of C. crenatum cultivated in 100 µmol photons m−2 s−1 (control) and darkness at 2 °C, error bars indicating standard deviation (n = 8).
A slight decrease of Fv/Fm was overall observed and had been detected for several Cosmarium strains at low temperatures before13. Our results with 0.449 (78% of t0) in darkness and 0.461 (74% of t0) at 100 µmol photons m−2 s−1 are matching the findings from Stamenkovic and Hanelt (2013), where the Fv/Fm in C. crenatum has decreased to 0.5 after two weeks at 0.6 °C and 30 µmol photons m−2 s−1. A similar response was observed from Micrasterias denticulata, where after nine weeks of darkness the Fv/Fm dropped to 76% of t052. Furthermore, it has been shown that cold temperatures reduce the activity of the Calvin cycle, which leads to a decreased oxidation of electron acceptors (e.g. NADPH) and even low light intensities may damage the photosynthetic apparatus by forming reactive oxygen species (ROS)53. As photoinhibition caused by cold shock, chilling stress and the resulting formation of ROS54 are unlikely to occur under prolonged darkness, other mechanisms, e.g. chlororespiration, might be responsible for the reduction of Fv/Fm. A slight decrease in Fv/Fm after the short-term exposure to darkness had been described for the Antarctic alga Palmaria decipiens (Rhodophyta)55, but these authors concluded that the further reduction of the Fv/Fm was a sign for the start of the degradation of the photosynthetic apparatus and do not provide a theory for the decrease in the first place. The mechanism of chlororespiration has been theorized as an alternative respiratory chain within the thylakoid membrane, enabling electron transport even in the dark56. In theory, the plastoquinone (PQ) pool is oxidized in darkness as light energy is not provided for electron delivery by the oxygen-evolving complex, reaction center II and electron transport to PS I56. In reality, there are indications that parts of the PQ pool remain or become reduced also in darkness e.g. in tomato after 5 hours of darkness57 and in the green alga Chlamydomonas reinhardtii58 causing a decrease of the maximal possible value of Fv/Fm. The specific pattern of a reduced Fv/Fm after one week in darkness exhibited by C. crenatum may reflect a specific adaption to unfavorable conditions of this Arctic opportunist. There are at least six reactions occurring within the chloroplast that can lead to a reduced plastoquinone pool59. A central oxidase located in the thylakoid membrane, the plastid terminal oxidase (PTOX also called IMMUTANS or AOX4), may participate in the electron transfer in the light-independent chlororespiration and already shown to be the major oxidase involved in the process in C. reinhardtii58. Although not significantly regulated in darkness in C. crenatum, there is at least one copy of the gene present and therefore an indication of occurrence of an alternative respiratory mechanism within the chloroplast. Additionally, a significant alteration of the gene expression of photosynthetic components and associated pathways (e.g. chlorophyll metabolism) was shown for C. crenatum. These hidden alterations, not detectable by the Fv/Fm parameter, were observed in the brown macroalga Saccharina latissima60,61. In our experiments, C. crenatum responded to darkness with a down-regulation of nearly all transcripts coding for components of the photosynthetic apparatus. Mainly all transcripts encoding proteins of PSI and PSII were down-regulated, the strongest repression was observed for a chlorophyll a-b binding protein (logFC −10.1). Genes encoding parts of the electron transport chain like cytochrome b6-f complex proteins, plastocyanin and cytochrome c6 were repressed with fold changes of −3.44, −5.0 and −2.1, respectively. The mRNA level of most of the photosynthesis associated genes was reduced in darkness, which is in accordance with findings in A. thaliana for light-controlled metabolic and regulatory pathways62. Furthermore, genes involved in photosynthesis were down-regulated in the Antarctic marine diatom Fragilariopsis cylindrus after one week of prolonged darkness compared to expression under continuous light63, indicating a common response.
Genes associated with photosynthetic carbon fixation of the reductive pentose phosphate pathway were also down-regulated. Transcripts encoding ribulose bisphosphate carboxylase (RuBisCO) small and large chains were repressed with −2.6 and −2.3. Two chloroplast localized phosphoribulokinase (PRK) transcripts were down-regulated with logFC of −5.0 and −5.5. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was repressed with logFC −4.3 and −3.4. Both are essential enzymes in carbon fixation and glycolysis. The two subunits of GAPDH with chloroplast localization, GAPA and GAPB, were repressed in darkness. The GAPB subunit originated from duplication of the GAPA subunit, but differs in 20% of its amino acids from GAPA64. The gene fusion leading to the higher plant chloroplast-specific GAPDH complex took place in the time before the transition from water to land habitat65 and therefore GAPB is found in nearly all groups of Charophyta66. The occurrence of both subunits in C. crenatum supports the hypothesis that the gene fusion happened before entering terrestrial habitats. The small Calvin cycle protein (CP12) is known for its regulatory interaction with the photosynthetic GAPDH67. It forms disulfide bonds with the PRK in darkness and is directly lowering enzyme activity68, therefore enabling metabolic flexibility in carbon fixation, which happens upon irradiation. This is in accordance with the gene expression of C. crenatum as GAPA, GAPB and PRK were all significantly repressed. CP12 is not significantly regulated in darkness in C. crenatum, which is different from A. thaliana, where after five days still an up-regulation of expression was observed. Nevertheless, the data indicates a reduction of the other gene products associated with the Calvin cycle and a reduced carbon fixation in darkness.
Chlorophyll and carotenoid metabolism
Transcripts involved in chlorophyll or porphyrin biosynthesis like tetrapyrrole-binding protein (GUN4C), chlorophyll synthase and magnesium protoporphyrin IX methyltransferase were down-regulated with logFC of −5.1, −2.7 and −5.9, respectively. Transcripts coding for proteins involved in chlorophyll catabolism, e.g. pheophytinase, chlorophyllase-1 and electron transfer flavoprotein subunit beta (ETFB or ETF β) were up-regulated resulting in logFCs of 2.2, 2.4 and 2.0. The electron transfer flavoprotein (ETF)/ETF ubiquinone oxide reductase (ETFQO) electron transfer complex is essential for A. thaliana under extended darkness conditions, and is supposed to be involved in the chlorophyll degradation pathway activated during dark-induced carbohydrate deprivation and catabolism of leucine69. Even though the expression of ETF β, is not affected in response to prolonged darkness in A. thaliana, it has been shown that the ETF re-oxidizing protein ETFQO responds with high expression levels in sucrose-starved conditions in darkness70. As structure and function of ETF and ETFQO are well conserved it can be concluded that C. crenatum did not enter a sugar starvation phase as the expression of ETFQO was not significantly regulated. Adding up to this, the isovaleryl-CoA dehydrogenase (IDV) gene has been shown to be active under sucrose starvation in almost all cell types of A. thaliana71 and is present in the here presented reference transcriptome (DN22252_c1_g1_i5, E:0), but not significantly regulated in darkness.
Central enzymes of the xanthophyll cycle were significantly differentially expressed in darkness. Violaxanthin de-epoxidase showed lower mRNA level in darkness (logFC −2.2) whereas transcripts encoding zeaxanthin epoxidase were induced (logFC of 2.1). In plants and green algae the violaxanthin-cycle is present and serves as a control mechanism against the formation of ROS72. The onset of violaxanthin de-epoxidase under high light as well as contrary the activation of zeaxanthin epoxidase in darkness is an early defined system73 and in accordance to our findings. For C. crenatum it has been shown that the xanthophyll cycle pool strongly reacts to high light with an accumulation of the intermediate antheraxanthin, but remains stable in low light and recovery74. Therefore, the here detected expression of genes of the xanthophyll cycle is in accordance with previously published physiological data of this isolate.
Energy metabolism
Exposure to long-term darkness caused repression of most transcripts involved in primary energy metabolism, e.g. glycolysis or tricarboxylic acid cycle (TCA). Mainly all key components of glycolysis were repressed, starting with the ATP-dependent 6-phosphofructokinase (logFC −6.8) (Table 1). Furthermore, expression of transcripts also coding for enzymes and membrane complexes of the oxidative phosphorylation, like NADH dehydrogenase to cytochrome c, cytochrome b and the subunits beta and epsilon of ATP synthase, were down-regulated with logFCs between −2.05 and −7.0. Contrarily, synthesis of numerous transcripts that are encoding genes of the starch metabolism, like starch synthase; sucrose synthase and beta-amylase were induced after one week of darkness with logFCs between 2.0 and 3.4. Another indication for the use of organic molecules are the expression patterns in carbohydrate transporters e.g. specific sucrose transporters with logFC of 2.9 and hexose transporters with an increase of up to 6-fold and can directly be linked to energy metabolism. Starch is the most abundant carbohydrate storage molecule in land plants and organized in pyrenoids within the chloroplast7, if present. In general, starch is stored during the day inside the chloroplast of plants and metabolized at night75. In the green microalga Acutodesmus obliquus it has been recently proven that starch acts as the major transitory energy storage compound, which was accumulated in the very last part of the illuminated period and consumed in the whole cause of darkness76. For A. thaliana it has been confirmed on the protein level that when facing very short light periods or extended darkness, starch synthesis rate increases during the day and degradation rate decreases rapidly during night77. Furthermore, the content of sugars and amino acids in the end of the dark period were largely independent from the length of the radiation period, presumably plants acclimated quickly to the prolonged dark conditions77. Light regulates transcript abundance of a large number of genes2 and darkness appears to be a trigger vice versa. The exposure of A. thaliana plants to altered conditions of light availability leads to an adjustment of transcript level of starch degradation-related enzymes75. In continuous darkness, starch degradation-related gene transcripts decline within the first 9 h and are barely expressed in the experiments after 9 h of darkness. This is in contrast to the results found in the polar C. crenatum where expression of transcripts of genes encoding beta-amylases (AMYB, BAM3, BAM7) as well as different starch synthases (Soluble starch synthase 3 + 4) and sugar transporters were significantly induced even after one week of darkness. Especially, beta-amylase (BAM) is required for standard starch breakdown in A. thaliana78,79. From the patterns in energy metabolism it is assumed that C. crenatum survives extended darkness periods by the breakdown of starch stored within the chloroplast.
In our dataset the mRNA levels of specific sugar transport proteins 7, 9 and 13 were induced in darkness (log FC 4.2, 6.0 and 2.2), which are localized within the plasma membrane and mediate active uptake of hexoses e.g. glucose80. There are indications that C. crenatum may survive the extended dark period due to heterotrophic utilisation of hexoses derived from the very specific nature of desmids. Desmidiaceae produce a sheath of extracellular polymeric substances (EPS) which are known to function in various matters e.g. movement or adhesion81 and nutrient capture82. The EPS cover of Cosmarium turpinii has been shown to mainly consist of xylose, galactose, glucose and other organic carbohydrates83. Therefore it may be possible to utilize the monosaccharides in the EPS, but further investigations in heterotrophic conditions will be necessary in the future that monitor the uptake of hexoses from the sheath layer. It has been proven for many microalgae lineages to be capable of heterotrophic utilization of different carbon sources84, with most records from the green algae lineage. The ability to increase the heterotrophic potential and the uptake of dissolved organic material or lipids has been reported from polar diatoms and may be a way to thrive within polar night conditions85.
Transcripts annotated with functions in the lipid metabolism were differently regulated in darkness, even though the number of repressed gene transcripts was higher. Strongest repressed gene with a logFC of −10.3 was a delta-(12)-acyl-lipid-desaturase, but amongst the other repressed transcripts many different desaturases were present (Table 1). Transcripts involved in key steps of lipid biogenesis like acetyl-coenzyme A carboxylase (logFC −2.4 and −3.8) and oleoyl-acyl carrier protein thioesterase (log FC −6.5) were also repressed. The phospholipase SGR2 was induced in darkness in C. crenatum with logFC of 4.2. It is known to be involved in gravitropism in A. thaliana and induces leaf movement in darkness86. Therefore, this is an indication of the gene for darkness-specific response, already present early in the Zygnematophyceae. Specific mRNAs of genes encoding long-chain-alcohol-oxidases, like lipase (LIP1) were induced under darkness with logFC 4.8. These are involved in triacylglycerol (TAG) breakdown87 and may be a source of NAD(P)H when cells are low in major carbon reserves like starch59. Recently it has been indicated for a close relative of C. crenatum, Micrasterias denticulata, that classical autophagy and lipid degradation appears during carbon starvation52. In M. denticulata during carbon starvation (e.g. induced by darkness) lipid droplets are transported outside the chloroplast, where they occasionally fuse with the vacuolar compartments and are degraded. Similar shifts of chloroplast lipids to the cytoplasm has been observed in another transmission electron microscope (TEM) study in starved cells of the green alga Chlorella88. This accumulation of lipid bodies by microalgae is a response to store energy in unfavorable environmental conditions, hypothesized to quickly utilize TAG to synthetize metabolites or biomembranes once conditions resume to normal89. Classical autophagy of lipids (or lipophagy) has been indicated for M. denticulata and also an orthologue of the autophaphy-related gene 8 was shown to be present in it52. At least one ATG8 orthologue is present in C. crenatum in our reference, but not significantly regulated. In darkness, different autophagy-related genes (ATG9, ATG11, ATG22-1) are up-regulated and can serve for autophagy-induced lipid mobilization or recycling. Therefore, it may be assumed that storage lipids and TAGs are another possible energy source utilized in C. crenatum in darkness.
Epigenetic modifications
We observed enhanced mRNA levels in response to darkness for several transcripts correlated to histone modifications. Histone modifications influence chromatin structure and gene activity by changing DNA–histone interaction and accessibility of transcription factors90. Transcripts encoding proteins involved in H3 K4 methylation/demethylation processes were induced under darkness, e.g. histone-lysine N-methyltransferase ATXR3 (logFC 2.2), histone-lysine N-methyltransferase family member SUVH2 (logFC 3.6) and lysine-specific histone demethylase 1B (logFC 3.2). Genes connected to methylation of H3 K9 showed significantly higher transcript abundances, i.e. histone-lysine N-methyltransferase SUVR4 (logFC 3.4) and SUVR5 (logFC 2.5). Additional, we observed induction of 2 transcripts involved in histone acetylation, namely histone acetyltransferase GCN5 and increased DNA methylation, which were up-regulated with log FC 2.4 and 2.6. Even though the presence of histone methyltransferases in algae have been reported91 up to now little is known on changes in histone modification status under abiotic stresses in algae. A study conducted on the unicellular alga C. reinhardtii showed that copper starvation and heat stress causes histone acetylation of H3/4 as well as changes in the methylation state of H3 K492. Histone modifications are involved in plant stress response, hypoxia in rice seedlings caused dynamic and reversible changes of histone H3 K4 methylation and H3 acetylation93. Heat stress response in A. thaliana causes enrichment of H3 K4 trimethylation and H3 K9 acetylation90. Furthermore, it was observed that dehydration stress in A. thaliana triggers dynamic changes in genome-wide histone H3K4 methylation patterns94. Our results indicate that adaptation/acclimation to darkness in C. crenatum involves a sophisticated network of histone modifications. In general, H3 K4 and H3 K36 methylation are correlated with gene activation, whereas H3K9 and H3K27 methylation as well as H3 acetylation are associated with gene silencing95. However, as the same histone mark can have different functions in different organisms96, more detailed studies should focus on how gene expression adjustments in response to abiotic changes in algae are associated with certain histone modification. As a recent study on microevolution in C. reinhardtii showed that transgenerational epigenetic effects play an important role in adaptive evolution97, future studies on histone modifications in C. crenatum should investigate whether these are passed through mitotic cell division.
Ecological implications and darkness acclimation
In our experiments, the switch to absence of light was the required trigger for expression changes in the transcriptome and acclimation to winter conditions. According to a communication by Mix (1970), Cosmarium species have developed gelatinous sheaths outside the cells or incorporate storage molecules, especially starch and lipids, to protect the vegetative cells from damage caused by freezing98. The mucilage sheaths also have an influence on the sinking rate, which ensures that certain species (e.g. C. botrytis) rapidly sink to the ground in a water body99. Usually freezing damage does not occur to benthic Cosmarium species in ponds and lakes in the high Arctic as, depending on depth, water bodies do not freeze solid to the ground (e.g. Greenland with lowest temperature between 0.01–0.1 °C in 2 m depth)100. In accordance with our findings, laboratory experiments on C. botrytis showed a survival of vegetative cells in darkness101.
C. crenatum maintains a vegetative cell which is non-dormant and holds the ability to photosynthesize when exposed to light. Polar winter laboratory experiments on the endemic Antarctic red alga Iridaea cordata over six months of darkness show a similar pattern102. The maximum photosynthetic rate is slightly decreased within the first months, but afterwards maintained at a certain level. I. cordata uses floridean starch as energy source in darkness and maintains the structure of the photosynthetic apparatus without structural changes. As C. crenatum is exhibiting gene expression associated with starch catabolism but no growth occurs it can be hypothesized that the species has a comparable survival strategy. Further experiments with longer darkness periods will be necessary to verify its ecological adaption to polar winter.
Conclusion
It is demonstrated that darkness was associated with down-regulation of genes of the primary metabolism, photosynthesis as well as cell divisions and photosynthetic efficiency in C. crenatum as ecophysiological acclimation to enhanced dark periods. As gene expression of transcripts encoding starch catabolism and to some extent sugar transport proteins is clearly induced in darkness, we assume that these pathways ensure main energy supply in dark periods, and therefore phases of heterotrophic energy supply when photosynthesis is not efficient or does not occur at all. Chlororespiration may play a role in electron transport within the chloroplast, but needs to be further investigated. We were able to show that cell division stops immediately and after one week of darkness no significant changes are detectable in the number of cells, resulting in a mechanism to save storage molecules. Significant changes in gene expression were observed under conditions where the Fv/Fm remained unchanged. Moreover, genes of the primary energy metabolism which involve glycolysis, TCA and oxidative phosphorylation are repressed. During a long dark period under polar conditions it could be beneficial for this organism to avoid extensive usage of storage compounds. Considering the polar origin of C. crenatum MZCH 561, its tolerance to low temperatures in combination with total darkness was shown as the algae were able to cope with the conditions and specifically react. A period of prolonged darkness (up to 9 months) and low temperatures seem to be key altering factor in this environment during polar winter.
Artificial laboratory conditions are hardly mimicking the exact conditions of a long polar winter where gradual acclimation in response to seasonal change is acquired. Therefore, future studies should include year-round in situ sampling in the environment to further shed light on dark acclimation and recovery of these photoautotrophs.
Supplementary information
Acknowledgements
Funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC 2037 ‘CLICCS - Climate, Climatic Change, and Society’ – Project Number: 390683824, contribution to the Center for Earth System Research and Sustainability (CEN) of Universität Hamburg.
Author Contributions
Florian Mundt (F.M.), Sandra Heinrich (S.H.) and Dieter Hanelt (D.H.) designed the experimental set-up. F.M. did the practical work, manual gene expression interpretation and wrote the first draft of the manuscript. S.H. coordinated the sequencing strategies and bioinformatics. L.H. performed bioinformatics including de novo transcriptome assembly, quality evaluation, annotation and mapping. All authors worked on data interpretation and approved the final manuscript.
Data Availability
The cleaned raw sequencing data are deposited in the European Nucleotide Archive (ENA) at the European Molecular Biological Laboratory–European Bioinformatics Institute under study accession number PRJEB30351 https://www.ebi.ac.uk/ena/data/view/PRJEB30351.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47041-7.
References
- 1.Thompson WF, White MJ. Physiological and molecular studies of light-regulated nuclear genes in higher-plants. Annu Rev Plant Phys. 1991;42:423–466. doi: 10.1146/annurev.pp.42.060191.002231. [DOI] [Google Scholar]
- 2.Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Phys. 1995;46:445–474. doi: 10.1146/annurev.arplant.46.1.445. [DOI] [Google Scholar]
- 3.Jiao YL, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 4.Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature. 1997;389:33–39. doi: 10.1038/37918. [DOI] [Google Scholar]
- 5.Graham LE, Cook ME, Busse JS. The origin of plants: Body plan changes contributing to a major evolutionary radiation. P Natl Acad Sci USA. 2000;97:4535–4540. doi: 10.1073/pnas.97.9.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker B, Marin B. Streptophyte algae and the origin of embryophytes. Ann Bot-London. 2009;103:999–1004. doi: 10.1093/aob/mcp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leliaert F, et al. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31:1–46. doi: 10.1080/07352689.2011.615705. [DOI] [Google Scholar]
- 8.Gontcharov AA, Melkonian M. A study of conflict between molecular phylogeny and taxonomy in the Desmidiaceae (Streptophyta, Viridiplantae): analyses of 291 rbcL sequences. Protist. 2011;162:253–267. doi: 10.1016/j.protis.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Timme, R. E., Bachvaroff, T. R. & Delwiche, C. F. Broad phylogenomic sampling and the sister lineage of land plants. Plos One7, 10.1371/journal.pone.0029696 (2012). [DOI] [PMC free article] [PubMed]
- 10.Wickett NJ, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. P Natl Acad Sci USA. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwiche, C. F. & Cooper, E. D. The evolutionary origin of a terrestrial flora. Curr Biol25, 10.1016/j.cub.2015.08.029 (2015). [DOI] [PubMed]
- 12.Stamenkovic M, Hanelt D. Growth and photosynthetic characteristics of several Cosmarium strains (Zygnematophyceae, Streptophyta) isolated from various geographic regions under a constant light-temperature regime. Aquat Ecol. 2011;45:455–472. doi: 10.1007/s10452-011-9367-7. [DOI] [Google Scholar]
- 13.Stamenkovic M, Hanelt D. Adaptation of growth and photosynthesis to certain temperature regimes is an indicator for the geographical distribution of Cosmarium strains (Zygnematophyceae, Streptophyta) European Journal of Phycology. 2013;48:116–127. doi: 10.1080/09670262.2013.772657. [DOI] [Google Scholar]
- 14.West, W. & West, G. S. A monograph of the British Desmidiaceae. Vol. 3 (Ray society, 1908).
- 15.Sheath RG, Steinman AD. A checklist of freshwater algae of the Northwest Territories, Canada. Canadian Journal of Botany. 1982;60:1964–1997. doi: 10.1139/b82-245. [DOI] [Google Scholar]
- 16.Croasdale, H. & Flint, E. (Botany Division, DSIR: New Zealand, 1988).
- 17.Thérézien, Y. & Couté, A. Algues d’eau douce des Iles Kerguelen et Crozet: à l’exclusion des diatomées. Vol. 43 (Comité national français des recherches antarctiques, 1977).
- 18.Coesel PFM. Biogeography of desmids. Hydrobiologia. 1996;336:41–53. doi: 10.1007/Bf00010818. [DOI] [Google Scholar]
- 19.Rautio M, et al. Shallow freshwater ecosystems of the circumpolar. Arctic. Ecoscience. 2011;18:204–222. doi: 10.2980/18-3-3463. [DOI] [Google Scholar]
- 20.IPCC. Climate change 2014: synthesis report. Intergovernmental panel on climate change. 151 (IPCC, 2014).
- 21.Priddle J. Production ecology of benthic plants in some Antarctic lakes. 1. In situ production studies. J Ecol. 1980;68:141–153. doi: 10.2307/2259248. [DOI] [Google Scholar]
- 22.Riis T, Christoffersen KS, Baattrup-Pedersen A. Effects of warming on annual production and nutrient-use efficiency of aquatic mosses in a high Arctic lake. Freshwater Biol. 2014;59:1622–1632. doi: 10.1111/fwb.12368. [DOI] [Google Scholar]
- 23.Laurion I, et al. Variability in greenhouse gas emissions from permafrost thaw ponds. Limnol Oceanogr. 2010;55:115–133. doi: 10.4319/lo.2010.55.1.0115. [DOI] [Google Scholar]
- 24.Holzinger, A. & Pichrtova, M. Abiotic stress tolerance of charophyte green algae: new challenges for omics techniques. Frontiers in Plant Science7, 10.3389/Fpls.2016.00678 (2016). [DOI] [PMC free article] [PubMed]
- 25.Holzinger, A. et al. Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense reaction. Plos One9, 10.1371/journal.pone.0110630 (2014). [DOI] [PMC free article] [PubMed]
- 26.Rippin M, Becker B, Holzinger A. Enhanced desiccation tolerance in mature cultures of the streptophytic green alga Zygnema circumcarinatum revealed by transcriptomics. Plant Cell Physiol. 2017;58:2067–2084. doi: 10.1093/pcp/pcx136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Poel B, Cooper ED, Van der Straeten D, Chang CR, Delwiche CF. Transcriptome profiling of the green alga Spirogyra pratensis (Charophyta) suggests an ancestral role for ethylene in cell wall metabolism, photosynthesis, and abiotic stress responses. Plant Physiol. 2016;172:533–545. doi: 10.1104/pp.16.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols HW. In Handbook of phycological methods: culture methods and growth measurements. 1973;1:7–24. [Google Scholar]
- 29.Dethlefsen V, von Westernhagen H, Tug H, Hansen PD, Dizer H. Influence of solar ultraviolet-B on pelagic fish embryos: osmolality, mortality and viable hatch. Helgoland Mar Res. 2001;55:45–55. doi: 10.1007/s101520000062. [DOI] [Google Scholar]
- 30.Hanelt D, Hawes I, Rae R. Reduction of UV-B radiation causes an enhancement of photoinhibition in high light stressed aquatic plants from New Zealand lakes. J Photoch Photobio B. 2006;84:89–102. doi: 10.1016/j.jphotobiol.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Hanelt, D. In Bioassays: Advanced Methods and Applications Vol. 1 (ed. Donat-P.; Erzinger, Häder, Gilmar, S.) 169–198 (Elsevier, 2018).
- 32.Mundt, F., Heinrich, S. & Hanelt, D. RNA isolation from taxonomically diverse photosynthetic protists. Limnology and Oceanography: Methods, 10.1002/lom3.10299 (in press).
- 33.Andrews, S. FastQC: A quality control tool for high throughput sequence data. Available from, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 34.Bushnell, B. BBMap Short Read Aligner, and Other Bioinformatics Tools. Available online at, http://www.osti.gov/scitech/biblio/1241166 (2015).
- 35.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 36.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–U130. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 38.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/Nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowman JL, et al. Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell. 2017;171:287–304.e215. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 42.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Z-L, Bao J, Reecy JM. CateGOrizer: a web-based program to batch analyze gene on-tology classification categories. Online J Bioinform. 2008;9:108–112. [Google Scholar]
- 44.Veeckman E, Ruttink T, Vandepoele K. Are we there yet? reliably estimating the completeness of plant genome sequences. Plant Cell. 2016;28:1759–1768. doi: 10.1105/tpc.16.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timme, R. E. & Delwiche, C. F. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. Bmc Plant Biol10, 10.1186/1471-2229-10-96 (2010). [DOI] [PMC free article] [PubMed]
- 46.Delwiche CF, Goodman CA, Chang C. Land plant model systems branch out. Cell. 2017;171:265–266. doi: 10.1016/j.cell.2017.09.036. [DOI] [PubMed] [Google Scholar]
- 47.Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14:290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/Nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 49.Cosgrove DJ. Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol. 2015;25:162–172. doi: 10.1016/j.pbi.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Bio. 2005;6:850–861. doi: 10.1038/Nrm1746. [DOI] [PubMed] [Google Scholar]
- 51.Vannerum, K. et al. Transcriptional analysis of cell growth and morphogenesis in the unicellular green alga Micrasterias (Streptophyta), with emphasis on the role of expansin. Bmc Plant Biol11, 10.1186/1471-2229-11-128 (2011). [DOI] [PMC free article] [PubMed]
- 52.Schwarz V, Andosch A, Geretschlager A, Affenzeller M, Lutz-Meindl U. Carbon starvation induces lipid degradation via autophagy in the model alga Micrasterias. J Plant Physiol. 2017;208:115–127. doi: 10.1016/j.jptph.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Pfannschmidt T. Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci. 2003;8:33–41. doi: 10.1016/S1360-1385(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 54.Ruelland E, Vaultier MN, Zachowski A, Hurry V. Cold signalling and cold acclimation in plants. Advances in Botanical Research. 2009;49:35–150. doi: 10.1016/S0065-2296(08)00602-2. [DOI] [Google Scholar]
- 55.Luder UH, Wiencke C, Knoetzel J. Acclimation of photosynthesis and pigments during and after six months of darkness in Palmaria decipiens (Rhodophyta): A study to simulate Antarctic winter sea ice cover. J Phycol. 2002;38:904–913. doi: 10.1046/j.1529-8817.2002.t01-1-01071.x. [DOI] [Google Scholar]
- 56.Bennoun P. Evidence for a respiratory-chain in the chloroplast. P Natl Acad Sci-Biol. 1982;79:4352–4356. doi: 10.1073/pnas.79.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trouillard M, et al. Kinetic properties and physiological role of the plastoquinone terminal oxidase (PTOX) in a vascular plant. Bba-Bioenergetics. 2012;1817:2140–2148. doi: 10.1016/j.bbabio.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Houille-Vernes L, Rappaport F, Wollman FA, Alric J, Johnson X. Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. P Natl Acad Sci USA. 2011;108:20820–20825. doi: 10.1073/pnas.1110518109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson X, Alric J. Interaction between starch breakdown, acetate assimilation, and photosynthetic cyclic electron flow in Chlamydomonas reinhardtii. J Biol Chem. 2012;287:26445–26452. doi: 10.1074/jbc.M112.370205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinrich S, Frickenhaus S, Glockner G, Valentin K. A comprehensive cDNA library of light- and temperature-stressed Saccharina latissima (Phaeophyceae) European Journal of Phycology. 2012;47:83–94. doi: 10.1080/09670262.2012.660639. [DOI] [Google Scholar]
- 61.Heinrich S, Valentin K, Frickenhaus S, Wiencke C. Temperature and light interactively modulate gene expression in Saccharina latissima (Phaeophyceae) J Phycol. 2015;51:93–108. doi: 10.1111/jpy.12255. [DOI] [PubMed] [Google Scholar]
- 62.Ma LG, et al. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.13.12.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mock, T. et al. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature541, 10.1038/nature20803 (2017). [DOI] [PubMed]
- 64.Brinkmann H, Cerff R, Salomon M, Soll J. Cloning and sequence-analysis of cDNAs encoding the cytosolic precursors of subunits Gapa and Gapb of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinach. Plant Mol Biol. 1989;13:81–94. doi: 10.1007/Bf00027337. [DOI] [PubMed] [Google Scholar]
- 65.Wedel N, Soll J. Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. P Natl Acad Sci USA. 1998;95:9699–9704. doi: 10.1073/pnas.95.16.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petersen J, Teich R, Becker B, Cerff R, Brinkmann H. The GapA/B gene duplication marks the origin of streptophyta (Charophytes and land plants) Mol Biol Evol. 2006;23:1109–1118. doi: 10.1093/molbev/msj123. [DOI] [PubMed] [Google Scholar]
- 67.Pohlmeyer K, Paap BK, Soll J, Wedel N. CP12: A small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol Biol. 1996;32:969–978. doi: 10.1007/Bf00020493. [DOI] [PubMed] [Google Scholar]
- 68.Howard TP, Metodiev M, Lloyd JC, Raines CA. Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. P Natl Acad Sci USA. 2008;105:4056–4061. doi: 10.1073/pnas.0710518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ishizaki K, et al. The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 2006;47:751–760. doi: 10.1111/j.1365-313X.2006.02826.x. [DOI] [PubMed] [Google Scholar]
- 70.Ishizaki K, et al. The critical role of Arabidopsis electron-transfer flavoprotein: ubiquinone oxidoreductase during dark-induced starvation. The Plant Cell. 2005;17:2587–2600. doi: 10.1105/tpc.105.035162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daschner K, Couee I, Binder S. The mitochondrial isovaleryl-coenzyme A dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol. 2001;126:601–612. doi: 10.1104/Pp.126.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siefermann-Harms D. Carotenoids in photosynthesis. 1. Location in photosynthetic membranes and light-harvesting function. Biochim Biophys Acta. 1985;811:325–355. doi: 10.1016/0304-4173(85)90006-0. [DOI] [Google Scholar]
- 73.Hager A. Light dependent decrease of the pH-value in a chloroplast compartment causing the enzymatic interconversion of violaxanthin to zeaxanthin; relations to photophosphorylation. Planta. 1969;89:224–243. doi: 10.1007/BF00385028. [DOI] [PubMed] [Google Scholar]
- 74.Stamenkovic M, Bischof K, Hanelt D. Xanthophyll cycle pool size and composition in several Cosmarium strains (Zygnematophyceae, Streptophyta) are related to their geographic distribution patterns. Protist. 2014;165:14–30. doi: 10.1016/j.protis.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 75.Lu Y, Gehan JP, Sharkey TD. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005;138:2280–2291. doi: 10.1104/pp.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leon-Saiki GM, et al. The role of starch as transient energy buffer in synchronized microalgal growth in Acutodesmus obliquus. Algal Res. 2017;25:160–167. doi: 10.1016/j.algal.2017.05.018. [DOI] [Google Scholar]
- 77.Gibon Y, et al. Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ. 2009;32:859–874. doi: 10.1111/j.1365-3040.2009.01965.x. [DOI] [PubMed] [Google Scholar]
- 78.Fulton DC, et al. beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell. 2008;20:1040–1058. doi: 10.1105/tpc.107.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith SM, et al. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004;136:2687–2699. doi: 10.1104/pp.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schneidereit A, Scholz-Starke J, Buttner M. Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiol. 2003;133:182–190. doi: 10.1104/pp.103.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Domozych D, Kort S, Benton S, Yu T. The extracellular polymeric substance of the green alga Penium margaritaceum and its role in biofilm formation. Biofilms. 2005;2:129–144. doi: 10.1017/S147905050500181X. [DOI] [Google Scholar]
- 82.Coesel, P. F. On the ecological significance of a cellular mucilaginous envelope in planktic desmids. Algological Studies/Archiv für Hydrobiologie, Supplement Volumes, 65–74 (1994).
- 83.Ha Y, Dyck L, Thomas R. Hydrocolloids from the freshwater microalgae, Palmella texensis and Cosmarium turpinii. Journal of Food Science. 1988;53:841–844. doi: 10.1111/j.1365-2621.1988.tb08967.x. [DOI] [Google Scholar]
- 84.Morales-Sanchez D, Martinez-Rodriguez OA, Kyndt J, Martinez A. Heterotrophic growth of microalgae: metabolic aspects. World J Microb Biot. 2015;31:1–9. doi: 10.1007/s11274-014-1773-2. [DOI] [PubMed] [Google Scholar]
- 85.Palmisano AC, Sullivan CW. Physiology of sea ice diatoms. I. Response of three polar diatoms to a simulated summer‐winter transition. J Phycol. 1982;18:489–498. doi: 10.1111/j.1529-8817.1982.tb03215.x. [DOI] [Google Scholar]
- 86.Mano E, Horiguchi G, Tsukaya H. Gravitropism in leaves of Arabidopsis thaliana (L.) Heynh. Plant Cell Physiol. 2006;47:217–223. doi: 10.1093/pcp/pci237. [DOI] [PubMed] [Google Scholar]
- 87.Karim EK, et al. Identification and characterization of a triacylglycerol lipase in Arabidopsis homologous to mammalian acid lipases. Febs Lett. 2005;579:6067–6073. doi: 10.1016/j.febslet.2005.09.072. [DOI] [PubMed] [Google Scholar]
- 88.Goncalves EC, Johnson JV, Rathinasabapathi B. Conversion of membrane lipid acyl groups to triacylglycerol and formation of lipid bodies upon nitrogen starvation in biofuel green algae Chlorella UTEX29. Planta. 2013;238:895–906. doi: 10.1007/s00425-013-1946-5. [DOI] [PubMed] [Google Scholar]
- 89.Murphy DJ. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma. 2012;249:541–585. doi: 10.1007/s00709-011-0329-7. [DOI] [PubMed] [Google Scholar]
- 90.Kim JM, et al. Alterations of lysine modifications on the Histone H3 N-Tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1580–1588. doi: 10.1093/pcp/pcn133. [DOI] [PubMed] [Google Scholar]
- 91.Montsant A, et al. Identification and comparative genomic analysis of signaling and regulatory components in the diatom Thalassiosira pseudonana. J Phycol. 2007;43:585–604. doi: 10.1111/j.1529-8817.2007.00342.x. [DOI] [Google Scholar]
- 92.Strenkert D, Schmollinger S, Sommer F, Schulz-Raffelt M, Schroda M. Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell. 2011;23:2285–2301. doi: 10.1105/tpc.111.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsuji H, Saika H, Tsutsumi N, Hirai A, Nakazono M. Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol. 2006;47:995–1003. doi: 10.1093/pcp/pcj072. [DOI] [PubMed] [Google Scholar]
- 94.van Dijk, K. et al. Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. Bmc Plant Biol10, 10.1186/1471-2229-10-238 (2010). [DOI] [PMC free article] [PubMed]
- 95.Luo M, et al. Chromatin modifications and remodeling in plant abiotic stress responses. Bba-Gene Regul Mech. 2012;1819:129–136. doi: 10.1016/j.bbagrm.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 96.Feng SH, Jacobsen SE. Epigenetic modifications in plants: an evolutionary perspective. Curr Opin Plant Biol. 2011;14:179–186. doi: 10.1016/j.pbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kronholm I, Bassett A, Baulcombe D, Collins S. Epigenetic and genetic contributions to adaptation in Chlamydomonas. Mol Biol Evol. 2017;34:2285–2306. doi: 10.1093/molbev/msx166. [DOI] [PubMed] [Google Scholar]
- 98.Mix, M. Die Desmidiaceen des Zeller Lochs bei Fulda. Vol. 13 71–92 (Staatsinstitut für Allgemeine Botanik Hamburg, 1970).
- 99.Duthie, H. C. Some observations on the ecology of desmids. J Ecol53, 10.2307/2257628 (1965).
- 100.Riis T, Christoffersen KS, Baattrup-Pedersen A. Mosses in High-Arctic lakes: in situ measurements of annual primary production and decomposition. Polar Biol. 2016;39:543–552. doi: 10.1007/s00300-015-1806-9. [DOI] [Google Scholar]
- 101.Duthie H. The survival of desmids in ice. British Phycological Bulletin. 1964;2:376–377. [Google Scholar]
- 102.Weykam G, Thomas DN, Wiencke C. Growth and photosynthesis of the Antarctic red algae Palmaria decipiens (Palmariales) and Iridaea cordata (Gigartinales) during and following extended periods of darkness. Phycologia. 1997;36:395–405. doi: 10.2216/i0031-8884-36-5-395.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cleaned raw sequencing data are deposited in the European Nucleotide Archive (ENA) at the European Molecular Biological Laboratory–European Bioinformatics Institute under study accession number PRJEB30351 https://www.ebi.ac.uk/ena/data/view/PRJEB30351.