Abstract
Complete ammonia oxidation (comammox) to nitrate by certain Nitrospira-lineage bacteria (CMX) could contribute to overall nitrogen cycling in engineered biological nitrogen removal (BNR) processes in addition to the more well-documented nitrogen transformations by ammonia-oxidizing bacteria (AOB), nitrite-oxidizing bacteria (NOB), and anaerobic ammonia-oxidizing (anammox) bacteria (AMX). A metagenomic survey was conducted to quantify the presence and elucidate the potential functionality of CMX in 16 full-scale BNR configurations treating mainstream or sidestream wastewater. CMX proposed to date were combined with previously published AOB, NOB, and AMX genomes to create an expanded database for alignment of metagenomic reads. CMX-assigned metagenomic reads accounted for between 0.28 and 0.64% of total coding DNA sequences in all BNR configurations. Phylogenetic analysis of key nitrification functional genes amoA, encoding the α-subunit of ammonia monooxygenase, haoB, encoding the β-subunit of hydroxylamine oxidoreductase, and nxrB, encoding the β-subunit of nitrite oxidoreductase, confirmed that each BNR system contained coding regions for production of these enzymes by CMX specifically. Ultimately, the ubiquitous presence of CMX bacteria and metabolic functionality in such diverse system configurations emphasizes the need to translate novel bacterial transformations to engineered biological process interrogation, operation, and design.
INTRODUCTION
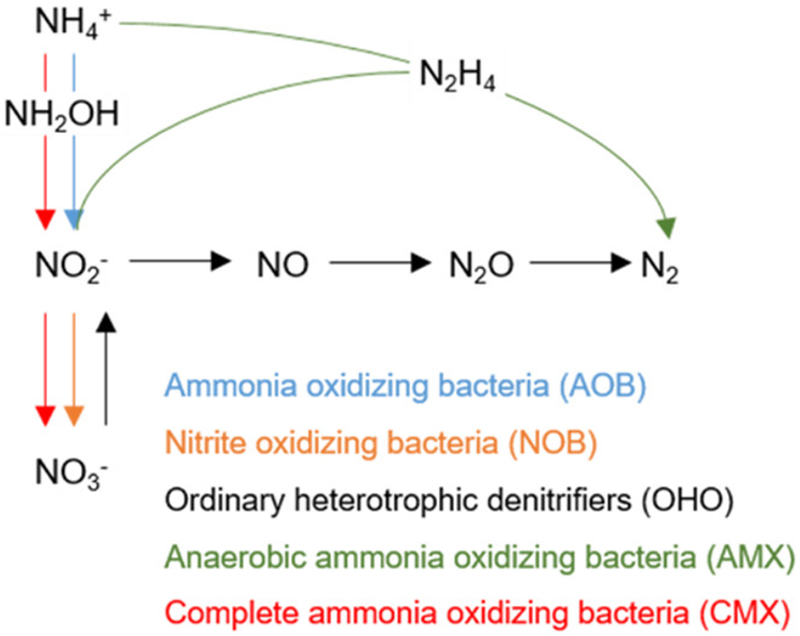
Biological nitrogen removal (BNR) is traditionally accomplished through the concerted action of nitrifying and denitrifying bacteria. During nitrification, the oxidation of ammonia to nitrate is mediated by two distinct groups of chemolithoautotrophs, ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB).1 Nitrate is typically reduced by chemoorganoheterotrophic denitrifying bacteria to dinitrogen gas (N2) (Figure 1).2 However, alternate “shortcut” nitrogen removal pathways allow for potential cost savings of aeration energy for nitrification and external carbon for denitrification and have therefore been applied to mainstream and sidestream wastewater treatment processes in recent years.3–7The coupling of partial nitritation (fractional aerobic oxidation of influent ammonia to nitrite by AOB) with anaerobic ammonia oxidation (anammox) represents one such shortcut process. In such partial nitritation–anammox processes, the selective enrichment of AOB and anammox bacteria (AMX) with the concomitant out-selection of NOB can result in a significant reduction (≤62.5%) of aeration requirements relative to those of conventional BNR approaches, while eliminating the need to supply external organic carbon (Figure 1).8
Figure 1.
Nitrogen cycling in engineered biological nitrogen removal (BNR) systems. In conventional BNR, autotrophic ammonia-oxidizing bacteria (AOB) convert ammonium (NH4+) to nitrite (NO2−) through hydroxylamine (NH2OH), and nitrite-oxidizing bacteria (NOB) convert nitrite to nitrate (NO3−). Ordinary heterotrophic denitrifiers (OHO) then convert nitrate to dinitrogen gas (N2) through nitrite, nitric oxide (NO), and nitrous oxide (N2O). Alternatively, anaerobic ammonia-oxidizing bacteria (AMX) convert ammonium and nitrite directly to dinitrogen gas through hydrazine (N2H4). Complete ammonia-oxidizing bacteria (CMX) have only recently been described and studied, and are capable of converting ammonium to nitrate through hydroxylamine and nitrite in a single organism.
In an exciting expansion of our current knowledge of the microbial nitrogen cycle, recent studies have uncovered the potential for complete ammonia oxidation (comammox) to nitrate by a single organism (CMX) rather than by distinct AOB and NOB (Figure 1).9,10 Three CMX organisms related to Nitrospira spp., Candidatus “Nitrospira inopinata”, Candidatus “Nitrospira nitrificans”, and Candidatus “Nitrospira nitrosa”, have been proposed to date.9,10 However, understanding CMX metabolic activity and their interactions with other bacterial groups is still rather limited in natural and engineered systems, including BNR processes.
In conventional BNR processes, CMX may be a beneficial or equivalent alternative to AOB-mediated nitritation and NOBmediated nitratation depending on the relative kinetics and substrate utilization of CMX-mediated nitritation and nitratation. Additionally, the coupling of ammonia and nitrite oxidation within CMX could potentially decrease production of the greenhouse gas nitrous oxide (N2O), as N2O production by classical AOB has been linked to an imbalance in electron flow due to transient dissolved oxygen (DO) concentrations, accumulation of ammonia or nitrite, or limited inorganic carbon supply.11–14On the other hand, energy- and cost-effective alternates to conventional BNR processes such as those targeting full or partial nitritation (e.g., for coupling with denitrification or anammox) would be less effective as a result of direct conversion of ammonia to nitrate by CMX. Therefore, the abundance of CMX and their contribution to nitrogen turnover across different BNR process configurations need to be characterized to better understand and guide process design and operation. More broadly, with few publications describing the presence of CMX in drinking water15,16 and limited-scale discussion of CMX in wastewater treatment systems,17–20 much work remains to improve our understanding of the potential for CMX capabilities across different natural and engineered systems.
In this study, shotgun metagenomic analyses were conducted on 16 samples from six full-scale wastewater treatment plants in the United States, Denmark, and Singapore. These plants employ conventional and shortcut BNR in mainstream and/or sidestream wastewater using varying reactor configurations (Table 1). This work aimed to identify and quantify the presence of CMX in each of these systems and to explore possible CMX functionality through phylogenetic analysis of metagenomic reads assigned to key nitrogen metabolism enzymes.
Table 1.
Full-Scale Wastewater Treatment Plants Surveyed
| sample | location | reactor type | biomass type | feed type | % CMXg CDS/total CDS |
|---|---|---|---|---|---|
| EB DEMONa | California, USA | DEMON | granule | SSb | 0.46 |
| EB MBBRc | California, USA | MBBR | biofilm | SS | 0.46 |
| SF DEMON | California, USA | DEMON | granule | SS | 0.49 |
| SF MBBR | California, USA | MBBR | biofilm | SS | 0.47 |
| PDR 1 | California, USA | ClearGreend | mixed liquor | MSe | 0.38 |
| PDR 2 | California, USA | ClearGreen | mixed liquor | MS | 0.45 |
| DK overflow | Denmark | hydrocyclone | overflow | MS | 0.49 |
| DK underflow | Denmark | hydrocyclone | underflow | MS | 0.54 |
| DK ALT | Denmark | hydrocyclone | ALT (mixed liquor) | MS | 0.32 |
| DK inoculum | Denmark | – | inoculum | – | 0.64 |
| SG biofilm | Singapore | BNRf | biofilm | MS | 0.46 |
| SG AS | Singapore | BNR | activated sludge | MS | 0.57 |
| VA MBBR 1 | Virginia, USA | MBBR | biofilm | MS | 0.28 |
| VA MBBR 2 | Virginia, USA | MBBR | biofilm | MS | 0.45 |
| VA BNR 1 | Virginia, USA | BNR | mixed liquor | MS | 0.57 |
| VA BNR 2 | Virginia, USA | BNR | mixed liquor | MS | 0.36 |
DEMON, DEamMONification.
SS, sidestream wastewater.
MBBR, moving bed biofilm reactor.
ClearGreen, cyclic low-energy ammonium removal.
MS, mainstream wastewater.
BNR, (conventional) biological nitrogen removal.
CMX, complete ammonia-oxidizing bacteria; total CDS found in Table S1.
MATERIALS AND METHODS
Microbial Sampling and Sequencing.
Sixteen biomass samples from six full-scale wastewater treatment plants (WWTPs) were collected for analysis (Table 1). The following reactor configurations were covered: conventional BNR, moving bed biofilm reactor (MBBR), DEMON deammonification, ClearGreen cyclic low-energy ammonium removal (ClearGreen), and a hydrocyclone. These systems were operated in the United States (California and Virginia), Denmark, and Singapore. Approximately 50 mL was sampled directly from reactor granules, mixed liquor, or biofilm within each BNR process (Table 1) and stored at −80 °C after shipment on dry ice. DNA was extracted from the biomass samples with the MoBio PowerLyzer PowerSoil DNA Isolation Kit per the manufacturer’s instructions (Qiagen) and quantified using Qubit dsDNA HS assay kits and the Qubit 2.0 Fluorometer (Thermo Fisher). Extracted DNA was shipped on dry ice from Columbia University to the Cincinnati Children’s Hospital DNA Core Facility for next-generation sequencing. Shotgun metagenomic libraries were prepared using the Nextera XT Library Preparation Kit (Illumina) with the recommended input of DNA normalized to 5 ng/μL and the default protocol for barcoded whole-genome libraries. An Illumina MiSeq sequencer and pair-ended 2 × 250 bp Illumina MiSeq version 2 sequencing kit were used (Illumina).
Taxonomic Classification.
Pair-ended reads were assembled into contigs and filtered for quality assurance (maximum homopolymeric region length of 10 bp, minimum length of 250 bp, maximum length of 450 bp, maximum number of ambiguous bases of zero) using mothur version 1.36.1.21 The existing NCBI nonredundant protein nr database (version 123) was manually expanded as part of this work to include protein-coding sequences from the three currently published CMX metagenome-derived genomes: Candidatus “Nitrospira inopinata”,9 Candidatus “Nitrospira nitrificans”, and Candidatus “Nitrospira nitrosa”.10 Protein-coding regions from two metagenome-derived AMX genomes that were previously unavailable in nr database version 123, Candidatus “Brocadia caroliniensis”22 and Candidatus “Scalindua profunda”,23 were also included in the custom database. Filtered reads from each sample were then aligned against this custom database using NCBI’s BLASTX. Alignments were curated for minimum identity percentage (80%) and maximum e value (1 × 10−10). Resulting reads in each metagenome were assigned taxonomic classification according to an automated search of NCBI protein databases, with emphasis on the contributions of new and previously indexed CMX and AMX species, as well as AOB and NOB.
Phylogenetic Analysis of Key Nitrification Genes.
To confirm the potential for ammonia and nitrite oxidation by CMX rather than AOB, AMX, or NOB, unique representative sequences from each BNR system assigned through BLASTX to CMX amoA, haoB, and nxrB with at least 30× coverage were selected. Single-nucleotide variants (SNVs) were calculated, and a distance matrix was generated using these unique sequences with MAFFT version 7.24 CMX (Candidatus “Nitrospira inopinata”, Candidatus “Nitrospira nitrificans”, and Candidatus “Nitrospira nitrosa”), AOB (Nitrosomonas europaea, Nitrosomonas eutropha, Nitrosospira multiformis, and Nitrosococcus halophilus), NOB (Candidatus “Nitrospira defluvii”, Nitrobacter hamburgensis, and Nitrobacter winogradskyii), and AMX (Candidatus “Brocadia caroliniensis”, Candidatus “Brocadia fulgida”, Candidatus “Brocadia sinica”, and Candidatus “Kuenenia stuttgartiensis”) reference genomes were also included in the MAFFT alignment to identify likely phylogenetic origins of sequences in the BNR systems studied here (see Table S1 for a complete list of accession numbers used). RAxML version 8.225 was used to generate a maximum likelihood tree and calculate branch node support over 100 bootstrap iterations under the GAMMA GTR substitution model.25 AOB (for amoA and haoB), NOB (for nxrB), and AMX (for haoB and nxrB) sequences were also included for the sake of completeness and to confirm whether the representative sequences from the 16 BNR systems clustered with, and therefore likely traced to, CMX in each engineered process.
RESULTS AND DISCUSSION
Varying Degrees of Comammox Capability Detected in All Systems.
Even prior to the discovery of comammoxcapable bacteria, complete oxidation of ammonia to nitrate in a single organism had been previously theorized.26 Within the framework of engineered BNR and biological wastewater treatment systems, the potential for complex interactions among CMX, AOB, NOB, and AMX could be particularly interesting given the utilization of common substrates (ammonia, nitrite, and even inorganic carbon).9,10 While targeted 16S rRNA gene analyses can describe only the microbial community structure of a system, shotgun metagenomics can reveal both the community structure and functional potential of engineered bioreactors.
The results reported here include a discussion of the relative abundance [in reads per kilobase mapped per million reads (RPKM)] of key genes involved in nitrification, related to the capability of each BNR process to produce key nitrifying enzymes encoded by these genes. This approach serves as a reference blueprint for similarly broad meta-transcriptomics or meta-proteomics studies as well as more focused targeted studies. Additionally, phylogenetic analyses are used here to complement the metagenomics approach and provide improved resolution for taxonomic assignment of sequences from organisms with limited genomic reference availability.
Previous work has identified CMX in drinking water or wastewater systems through a combination of next-generation sequencing and phylogenetics.15,16,18,19 However, unique to this study is the characterization of the contribution of CMX to the overall protein-producing capability of full-scale wastewater treatment plant metagenomes in a wide variety of reactor configurations. The systems assessed here have been designed for conventional BNR or partial nitritation by AOB followed by deammonification by anammox (Table 1). Conventional BNR systems included those from Singapore (SG Biofilm, SG AS) and Virginia (VA BNR 1 and 2). DEMON and ClearGreen systems from California (EB DEMON, SF DEMON, and PDR 1 and 2) were designed and operated for partial nitritation, and the partial oxidization of ammonia to nitrite by AOB, followed by anammox. These systems used granules enriched with AMX due to resultant substrate and oxygen gradients formed therein and treated both sidestream (EB DEMON and SF DEMON) and mainstream (PDR 1 and 2) wastewater.27–29 The nitritation–anammox MBBRs from California and Virginia (EB MBBR, SF MBBR, and VA MBBR 1 and 2) similarly contained biofilms enriched with AMX and consisted of either single-stage simultaneous partial nitritation–anammox treating sidestream wastewater (as in EB or SF) or two-stage decoupled operation treating mainstream wastewater (as in VA), in which an aerated nitritation process was followed by the anaerobic deammonification MBBR.30 The hydrocyclone-based process in Denmark (samples denoted DK ALT, overflow, and underflow) was utilized to selectively retain granular biomass in a mainstream BNR system.28
In total, 59.65 million filtered contigs were available for this study, with an average of 3.73 million sequences and a 393 bp contig length across the 16 samples (Table S1). For each sample, the number of total coding DNA sequences (CDS) was calculated as the number of contigs aligned with the expanded protein database above the described score thresholds. According to this definition, the 16 samples contained on average 3.14 ± 0.48 million total CDS (Table S1).
Interestingly, despite high variability in microbial community profiles across the different samples [0.59–14.35% AMX, 2.25–7.98% AOB, and 2.39–3.47% NOB (Table 1 and Table S1)], CMX-assigned coding regions remained consistently between 0.28 and 0.64% of total CDS.
Phylogenetic analyses confirmed that each BNR process contained sequences aligned with key nitrification genes originating from CMX for amoA encoding the ammonia monooxygenase α-subunit, haoB encoding the hydroxylamine oxidoreductase β-subunit, and nxrB encoding the β-subunit of nitrite oxidoreductase. Each system also contained AOB, NOB, and AMX functional gene sequences, but only contigs aligned with CMX through BLASTX are included here for evidence of potential CMX functionality. SF MBBR, SG BNR AS and Biofilm, and VA MBBR 2 contained amoA sequences clustered with Candidatus “Nitrospira inopinata” [P = 0.71 (Figure 2A)].
Figure 2.
Phylogenetic relationships between published amoA, haoB, and nxrB sequences and CMX-assigned sequences from each BNR process. Unique merged contigs from each BNR process that were assigned with ≥30× coverage as CMX amoA, haoB, and nxrB were included in the phylogenetic analysis, along with amoA, haoB, and nxrB sequences from representative published sequences (see Table S2 for a complete list of accession numbers). (A) CMX (red) and AOB (blue) reference amoA sequences compared through phylogenetics to CMX-assigned amoA in each BNR process. (B) CMX (red), AOB (blue), and AMX (green) reference haoB sequences compared to CMX-assigned haoB from each BNR system. (C) CMX (red), NOB (orange), and AMX (green) reference nxrB sequences compared to CMX-assigned nxrB from each BNR system. Tree-scale units are average amino acid substitutions per site, and circles at each node are scaled to reflect P values at nodes with ≥50% support. Each BNR process contained at least one amoA, haoB, and nxrB sequence that clustered with CMX rather than AOB, NOB, or AMX, providing evidence of potential CMX functionality in each system.
EB DEMON and MBBR, SF DEMON and MBBR, DK overflow and inoculum, and VA BNR 2 samples contained amoA sequences that clustered with Candidatus “Nitrospira nitrosa” (P = 0.99). Interestingly, many samples (EB DEMON and MBBR, SF DEMON and MBBR, PDR 2, DK overflow, ALT, and inoculum, and VA MBBR 1 and 2) contained amoA sequences clustered closely with Candidatus “Nitrospira nitrificans” (P = 1). EB DEMON, SF DEMON and MBBR, DK ALT and inoculum, and VA BNR 1 contained haoB sequences that clustered with Candidatus “Nitrospira inopinata” (P = 0.97) (Figure 2B). EB DEMON, SF DEMON, DK overflow and inoculum, SG BNR AS and biofilm, and VA MBBR 2 haoB clustered with Candidatus “Nitrospira nitrosa”, while EB MBBR, SF DEMON, PDR 1, DK underflow and inoculum, and VA MBBR 2 sequences clustered with Candidatus “Nitrospira nitrificans” haoB 1 and 2 (P = 0.83).
Previously, it was theorized that CMX have high growth yields rather than high specific growth rates associated with AOB and NOB, leading to a competitive advantage at low substrate concentrations and conditions favoring microbial aggregation.26 Additionally, previous work by van Kessel et al. suggested functional interplay between CMX and AMX due to tight clustering of functional gene sequences,10 which was also observed here to an extent (Figure 2). The data presented here may support these hypotheses, as the processes promoting biofilm growth, including nitritation–anammox, biofilm, and anammox inoculum samples, contained more unique CMX amoA and haoB sequences than the conventional BNR and hydrocyclone systems did (Figure 2A,B). Future analysis will be necessary to quantify the abundance of CMX amoA and haoB, aided partly by CMX amoA-targeted polymerase chain reaction primers.31
Each BNR process also contained contigs that clustered with CMX nxrB sequences rather than NOB or AMX (Figure 2C). EB DEMON and MBBR, SF DEMON and MBBR, DK overflow and inoculum, and VA BNR2 samples had nxrB contigs that clustered closely with Candidatus “Nitrospira nitrosa” nxrB 1 and 2 (P = 0.97). SF MBBR, SG BNR AS and Biofilm, VA BNR 1, and VA MBBR 1 and 2 contained contigs related to Candidatus “Nitrospira inopinata” nxrB (P = 0.82).
EB DEMON and MBBR, SF DEMON and MBBR, PDR 1 and 2, DK overflow, underflow, and inoculum, SG BNR AS and biofilm, and VA MBBR 1 and 2 all contained nxrB contigs that clustered with Candidatus “Nitrospira nitrificans” nxrB 1, 2, and 3 (P = 0.98). Of note, CMX-assigned nxrB contigs from EB MBBR, DK inoculum, VA BNR 1, and VA MBBR 1 and 2 were revealed through phylogenetic analysis to be more closely related to the non-CMX, canonical NOB Candidatus “Nitrosomonas defluvii” (P = 1). Thus, relying solely on BLASTbased alignment could lead to overestimation of CMX, highlighting the need for integrated metagenomics approaches that include phylogenetic interrogation for closely related organisms or those without published references. NOB outselection is important in deammonification systems primarily because of the competition between NOB and AMX for nitrite.4 Similarly, CMX utilization of nitrite could impact deammonification by AMX. Therefore, it is critical to note the potential for each nitritation–anammox process to produce nxrB by CMX to develop and apply more accurate out-selection strategies.
Nitrification by CMX Is Possible in Engineered Water Systems.
The emergence of newly discovered microorganisms capable of full nitrification is understandably exciting. Detection of comammox in such a wide array of systems does reveal the need for re-evaluation and adjustment of current BNR models given these newly available metabolic pathways. Metagenomic analysis alone, though, cannot determine if comammox is active within engineered nitrogen removal systems, given the operation conditions and competitive microbial communities involved. Lab-scale enrichment of CMX under appropriate conditions could improve our understanding of CMX thermodynamics, biokinetic parameters, and electron flow.32 Subsequently, calculation of theoretical relative abundances of CMX, AOB, and NOB required for nitrification33,34 and improved CMX-inclusive BNR modeling would be possible. Such directed studies will be required to compare the likelihood of comammox to that of conventional two-step nitrification or anammox under different conditions, link the presence of CMX and functionality to reactor operational parameters, and ultimately quantify the contribution of CMX to overall nitrogen turnover in BNR systems. These insights will be key in both the diagnosis of existing systems and the design of new nitrogen removal processes.
Supplementary Material
ACKNOWLEDGMENTS
This study was made possible with support from the Water Environment Research Foundation (WERF) STAR_N2R14 project. M.K.A. was additionally supported by a Presidential Fellowship through the Columbia University Fu Foundation School of Engineering & Applied Sciences. V.K. was supported by the U.S. Environmental Protection Agency (EPA) via a postdoctoral appointment administered by the Oak Ridge Institute for Science and Education through an Interagency agreement between the U.S. Department of Energy and the U.S. EPA. The authors thank Michael Elk for critical review of the manuscript. The manuscript has been subjected to the EPA’s peer review and has been approved as an EPA publication. Mention of trade names or commercial products does not constitute endorsement or recommendation by the EPA for use. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.estlett.7b00577.
Details regarding contig assembly, filtering, and database alignment (Table S1), reference NCBI Genome Assembly and GenBank accession numbers used in the phylogenetic analyses (Table S2), raw sequencing reads that have been submitted to the NCBI Sequencing Read Archive (SRA) (BioProject PRJNA377913), and all phylogenetic trees that have been visualized through the Interative Tree of Life (iTOL) viewer (https://itol.embl.de/)35 (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Grady CPL; Daigger GT; Love NG; Filipe CDM Biological Wastewater Treatment; IWA Publishing, 2011. [Google Scholar]
- (2).Lu H; Chandran K; Stensel D Microbial ecology of denitrification in biological wastewater treatment. Water Res. 2014, 64, 237–254. [DOI] [PubMed] [Google Scholar]
- (3).Hu B.-l.; Zheng P; Tang C.-j.; Chen J.-w.; van der Biezen E; Zhang L; Ni B.-j.; Jetten MS; Yan J; Yu H-Q; Kartal B. Identification and quantification of anammox bacteria in eight nitrogen removal reactors. Water Res. 2010, 44, 5014–5020. [DOI] [PubMed] [Google Scholar]
- (4).Lackner S; Gilbert EM; Vlaeminck SE; Joss A; Horn H; van Loosdrecht MC Full-scale partial nitritation/anammox experiences–an application survey. Water Res. 2014, 55, 292–303. [DOI] [PubMed] [Google Scholar]
- (5).Laureni M; Falås P; Robin O; Wick A; Weissbrodt DG; Nielsen JL; Ternes TA; Morgenroth E; Joss A Mainstream partial nitritation and anammox: long-term process stability and effluent quality at low temperatures. Water Res. 2016, 101, 628–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).van Dongen L; Jetten M; van Loosdrecht MC The combined SHARON/Anammox process; IWA Publishing, 2001. [PubMed] [Google Scholar]
- (7).Winkler M-K; Kleerebezem R; Van Loosdrecht M Integration of anammox into the aerobic granular sludge process for main stream wastewater treatment at ambient temperatures. Water Res. 2012, 46, 136–144. [DOI] [PubMed] [Google Scholar]
- (8).Kartal B; Maalcke WJ; De Almeida NM; Cirpus I; Gloerich J; Geerts W; Op den Camp HJM; Harhangi HR; Janssen-Megens EM; Francoijs KJ; Stunnenberg HG; Keltjens JT; Jetten MSM; Strous M Molecular mechanism of anaerobic ammonium oxidation. Nature 2011, 479, 127–130. [DOI] [PubMed] [Google Scholar]
- (9).Daims H; Lebedeva EV; Pjevac P; Han P; Herbold C; Albertsen M; Jehmlich N; Palatinszky M; Vierheilig J; Bulaev A; Kirkegaard RH; von Bergen M; Rattei T; Bendinger B; Nielsen PH; Wagner M Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).van Kessel MA; Speth DR; Albertsen M; Nielsen PH; Op den Camp HJM; Kartal B; Jetten MS; cker S. Complete nitrification by a single microorganism. Nature 2015, 528, 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yu R; Kampschreur MJ; van Loosdrecht MCM; Chandran K Mechanisms and specific directionality of autotrophic nitrous oxide and nitric oxide generation during transient anoxia. Environ. Sci. Technol. 2010, 44, 1313–1319. [DOI] [PubMed] [Google Scholar]
- (12).Chandran K; Stein LY; Klotz MG; van Loosdrecht MCM Nitrous oxide production by lithotrophic ammonia-oxidizing bacteria and implications for engineered nitrogen-removal systems. Biochem. Soc. Trans. 2011, 39, 1832–1837. [DOI] [PubMed] [Google Scholar]
- (13).Jiang D; Khunjar WO; Wett B; Murthy SN; Chandran K Characterizing the metabolic trade-off in Nitrosomonas europaea in response to changes in inorganic carbon supply. Environ. Sci. Technol. 2015, 49, 2523–2531. [DOI] [PubMed] [Google Scholar]
- (14).Kim YM; Park H; Chandran K Nitrification inhibition by hexavalent chromium Cr(VI) – Microbial ecology, gene expression and off-gas emissions. Water Res. 2016, 92, 254–261. [DOI] [PubMed] [Google Scholar]
- (15).Wang Y; Ma L; Mao Y; Jiang X; Xia Y; Yu K; Li B; Zhang T Comammox in drinking water systems. Water Res. 2017, 116, 332–341. [DOI] [PubMed] [Google Scholar]
- (16).Pinto AJ; Marcus DN; Ijaz UZ; Bautista-de lose Santos QM; Dick GJ; Raskin L Metagenomic evidence for the presence of comammox Nitrospira-Like Bacteria in a Drinking Water System. mSphere 2016, 1, e00054–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Bartelme RP; McLellan SL; Newton RJ Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammoniaoxidizing Archaea and comammox Nitrospira. Front. Microbiol. 2017, 8, n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chao Y; Mao Y; Yu K; Zhang T Novel nitrifiers and comammox in a full-scale hybrid biofilm and activated sludge reactor revealed by metagenomic approach. Appl. Microbiol. Biotechnol. 2016, 100, 8225–8237. [DOI] [PubMed] [Google Scholar]
- (19).Gonzalez-Martinez A; Rodriguez-Sanchez A; van Loosdrecht MCM; Gonzalez-Lopez J; Vahala R Detection of comammox bacteria in full-scale wastewater treatment bioreactors using tag-454pyrosequencing. Environ. Sci. Pollut. Res. 2016, 23, 25501–25511. [DOI] [PubMed] [Google Scholar]
- (20).Palomo A; Jane Fowler S; Gülay A; Rasmussen S; SicheritzPonten T; Smets BF Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J. 2016, 10, 2569–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Schloss PD; Westcott SL; Ryabin T; Hall JR; Hartmann M; Hollister EB; Lesniewski RA; Oakley BB; Parks DH; Robinson CJ; Sahl JW; Stres B; Thallinger GG; Van Horn DJ; Weber CF Introducing mothur: open-source, platformindependent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Park H; Brotto AC; van Loosdrecht MCM; Chandran K Discovery and metagenomic analysis of an anammox bacterial enrichment related to Candidatus “Brocadia caroliniensis” in a fullscale glycerol-fed nitritation-denitritation separate centrate treatment process. Water Res. 2017, 111, 265–273. [DOI] [PubMed] [Google Scholar]
- (23).van de Vossenberg J; Woebken D; Maalcke WJ; Wessels HJ; Dutilh BE; Kartal B; Janssen-Megens EM; Roeselers G; Yan J; Speth D; et al. The metagenome of the marine anammox bacterium ‘Candidatus Scalindua profunda’ illustrates the versatility of this globally important nitrogen cycle bacterium. Environ. Microbiol. 2013, 15, 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Katoh K; Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Costa E; Perez J; Kreft JU Why is metabolic labour divided in nitrification? Trends Microbiol. 2006, 14, 213–219. [DOI] [PubMed] [Google Scholar]
- (27).Vlaeminck SE; Terada A; Smets BF; De Clippeleir H; Schaubroeck T; Bolca S; Demeestere L; Mast J; Boon N; Carballa M; Verstraete W Aggregate size and architecture determine microbial activity balance for one-stage partial nitritation and anammox. Appl. Environ. Microbiol. 2010, 76, 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Winkler MK; Kleerebezem R; Kuenen JG; Yang J; van Loosdrecht MC Segregation of biomass in cyclic anaerobic/aerobic granular sludge allows the enrichment of anaerobic ammonium oxidizing bacteria at low temperatures. Environ. Sci. Technol. 2011, 45, 7330–7337. [DOI] [PubMed] [Google Scholar]
- (29).Degremont I Cleargreen Low Energy Deammonification System: Pilot Study Report; Suez Environment: Paris, 2015. [Google Scholar]
- (30).Mehrdad M; Park H; Ramalingam K; Fillos J; Beckmann K; Deur A; Chandran K Anammox moving bed biofilm reactor pilot at the 26th Ward wastewater treatment plants in Brooklyn, New York: start-up, biofilm population diversity and performance optimization. Water Sci. Technol. 2014, 70, 1448–1455. [DOI] [PubMed] [Google Scholar]
- (31).Pjevac P; Schauberger C; Poghosyan L; Herbold CW; van Kessel MAHJ; Daebeler A; Steinberger M; Jetten MSM; cker S; Wagner M; Daims H AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Front. Microbiol. 2017, 8, 1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Chandran K; Smets BF Estimating biomass yield coefficients for autotrophic ammonia and nitrite oxidation from batch respirograms. Water Res. 2001, 35, 3153–3156. [DOI] [PubMed] [Google Scholar]
- (33).Winkler MK; Bassin JP; Kleerebezem R; Sorokin DY; van Loosdrecht MC Unravelling the reasons for disproportion in the ratio of AOB and NOB in aerobic granular sludge. Appl. Microbiol. Biotechnol. 2012, 94, 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Hooper AB; Vannelli T; Bergmann DJ; Arciero DM Enzymology of the oxidation of ammonia to nitrite by bacteria. Antonie van Leeuwenhoek 1997, 71, 59–67. [DOI] [PubMed] [Google Scholar]
- (35).Letunic I; Bork P Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.