Abstract
Eukaryotic genes are regulated by multivalent transcription factor complexes. Through cooperative self-assembly, these complexes perform non-linear regulatory operations involved in cellular decision-making and signal processing. Here, we apply this design principle to synthetic networks, testing whether engineered cooperative assemblies can program non-linear gene circuit behavior in yeast. Using a model-guided approach we show that specifying strength and number of assembly subunits enables predictive tuning between linear and non-linear regulatory response for single- and multi-input circuits. We demonstrate that assemblies can be adjusted to control circuit dynamics. We harness this capability to engineer circuits that perform dynamic filtering, enabling frequency-dependent decoding in cell populations. Programmable cooperative assembly provides a versatile way to tune nonlinearity of network connections, dramatically expanding the engineerable behaviors available to synthetic circuits.
Cooperativity is a widespread biological phenomenon by which coordinated behavior within a molecular system emerges from energetic coupling between its components (1, 2). In eukaryotic gene networks, cooperative assembly occurs when core initiation machinery is recruited to basal promoter regions through multivalent, mutually-reinforcing interactions between transcription factors (TFs) and associated co-factors (3) (Fig 1A). The resulting nucleoprotein complexes play a critical signal processing and decision-making role (4–11); they convert analog TF inputs into switch-like transcriptional outputs (12, 13), or incorporate multiple TFs to carry out decision functions by activating transcription only in the presence of TF combinations (4, 14).
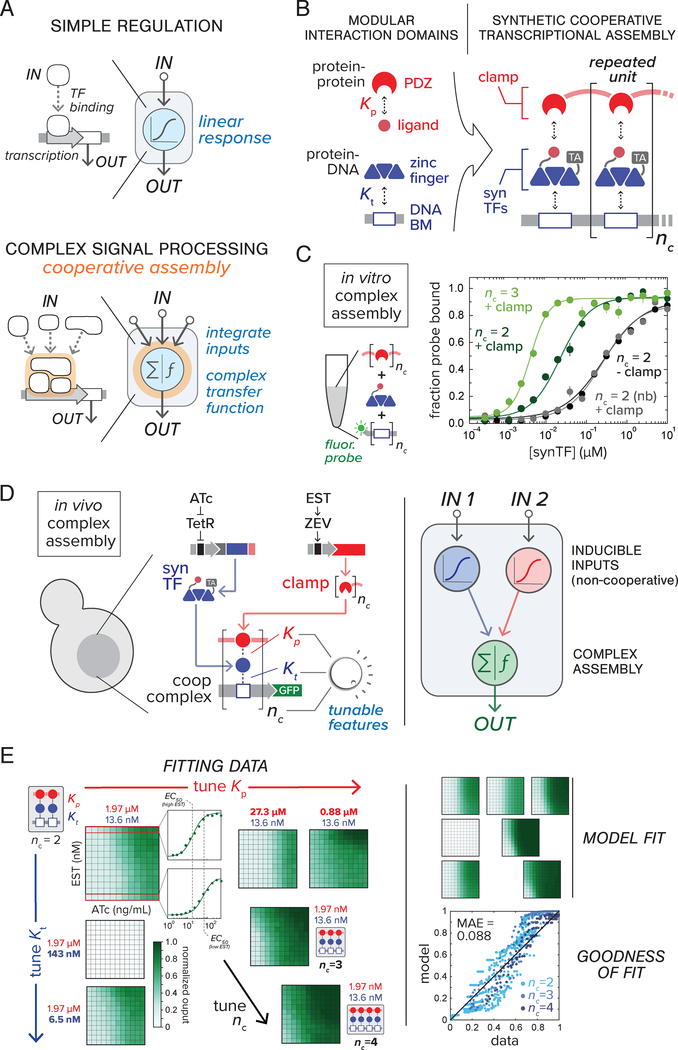
Figure 1.
Design scheme for assembly-mediated regulatory control of synthetic gene circuits. (A) Nodes in cellular networks use one-to-one regulatory interactions to execute simple computational tasks (top); cooperative interactions within multivalent assemblies enable complex, nonlinear signal processing (bottom). (B) Design of synthetic TF regulatory assemblies. Complexes are built from interaction domains (ZF and PDZ) and their respective binding partners (DBM and PDZ ligand). Clamp-mediated synTF complex-formation (TA, transcriptional activator domain drives coding sequence transcription). Interaction affinities (Kt and Kp) and the number of repeated complex units (nc) determine thermodynamics of assembly. (C) In vitro assembly of purified cooperative complex components. Fluorescence anisotropy for synTF titration of DBM oligo probe (FITC labeled) was measured in the presence or absence of clamp (5 μM) and converted to fraction probe bound (see Supplementary Materials and Methods) (Nb, synTFs with binding-deficient PDZ ligand mutants; nc, number of PDZ domains per clamp and DBMs probe). Points represent mean values for three measurements ± SE. (D) Testing in vivo complex assembly in yeast using a synthetic gene circuit (GFP output). All genes are chromosomally-integrated (left, see Table S3). Small molecule-inducible expression systems (ncTET and ncZEV) control intracellular expression levels (see fig. S4) of synTF (induced by ATc, 2 – 500 ng/mL) and clamp (EST, 0.05 – 12.5 nM), respectively. Adjusting molecular features of assembly (Kt, Kp, nc) tunes overall circuit transfer function (right). (E) Parameterization of thermodynamic complex assembly model. Dose response data for circuit configurations with various Kt, Kp, nc values were fit to a thermodynamic model (fig. S7–8, Supplementary Materials and Methods); regression plot showing residual from fit are shown at the right. MAE, mean absolute error.
To date, most synthetic gene circuits have been constructed using TFs that bind to promoters in one-to-one fashion (15–17), constraining the ability to tune circuit cooperativity and potentially imposing limits on engineerable behavior (18) (Fig. 1A, top). Here, we ask whether circuits with expanded signal processing function can be implemented using engineered multivalent assembly (Fig. 1A, bottom). We establish theoretical and design frameworks for programming cooperative TF assembly based on the configuration and strength of intra-complex interactions, and construct synthetic gene circuits composed of interconnected regulatory assemblies.
In our scheme for engineering cooperative TF assemblies (Fig. 1B), transcription is activated when synthetic zinc-finger (ZF) proteins fused to transcriptional activator domains (synTFs) bind tandem DNA binding motifs (DBMs) located upstream of a core promoter (19, 20). Assembly is mediated by a “clamp” protein: multiple covalently-linked PDZ domains that bind peptide ligands located on the C-termini of adjacent DBM-bound synTFs. Complex free energy can be adjusted by varying either the number of clamp/synTF/DBM repeats (nc), or the affinity of synTF-DBM and PDZ-ligand interactions (Kt and Kp, respectively), which is enabled by affinity variants for both domains (15 ZF-DNA and 13 PDZ-ligand interactions) (fig. S2–3).
In order to directly test whether the synTF/clamp/DBM module can support cooperative assembly, we conducted in vitro fluorescence anisotropy binding experiments on purified complex components (fig. S2–3, Supplementary Materials and Methods). synTF binding to a nc=2 probe showed a non-cooperative dose response profile, while presence of a two-PDZ clamp lowered the synTF binding threshold and steepened the dose response; an effect not observed for a non-binding synTF or for an nc=1 probe (Fig. 1C). Complex formation using an nc=3 probe (and three-PDZ clamp) demonstrated a still sharper, lower-threshold response. Thus, synTF binding cooperativity is enhanced by the clamp and scales in magnitude with complex size.
We next demonstrated in vivo complex assembly in yeast cells by constructing a transcriptional circuit in which a nc=2 synTF assembly drives a GFP reporter, with synTF and clamp levels controlled via non-cooperative, small-molecule inducible expression systems (Fig. 1D, fig. S4): ncTET (for synTF expression) (21), induced with anydrotetracycline (ATc), and ncZEV (for clamp expression) (22), induced by estradiol (EST). A dose response surface was obtained by titrating ATc against EST and recording GFP fluorescence output using flow cytometry (Fig. 1E, upper left). Consistent with clamp-enhanced synTF binding, ATc dose response threshold was reduced in the presence of increasing EST (Fig. 1E).
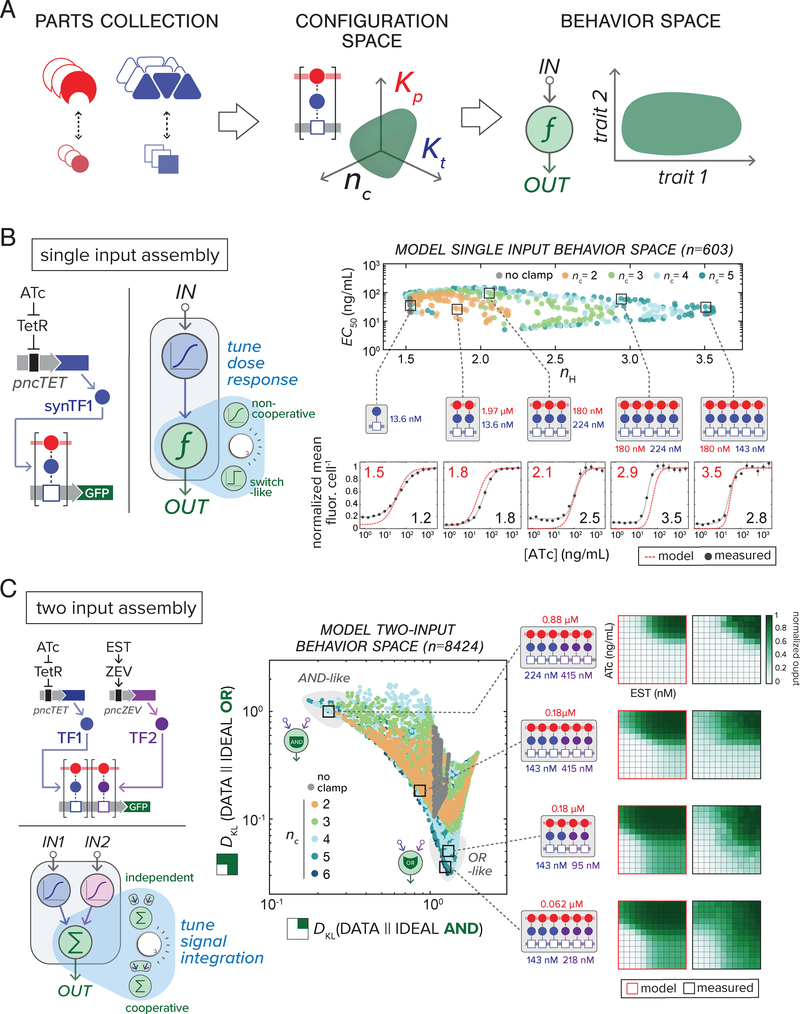
To quantitatively describe complex formation, we formulated a simple statistical thermodynamic model (14, 23, 24) relating intracellular synTF and clamp expression to promoter occupancy and resulting GFP output (fig. S7–10, see Supplementary Text for full model description). The model was fit to experimental two-input dose response data for seven complex configurations, where Kt, Kp, and nc were experimentally adjusted (Fig. 1E). Parameter fitting was constrained by in vitro-measured PDZ and ZF interaction affinities (fig. S2–3) and inducible component expression data (fig. S4). The fitted model can be used to guide circuit engineering, enumerating potential circuit behaviors (behavior space) based on input-output functions calculated for available configurations of parts (configuration space) (Fig 2A, fig. S2–4).
Figure 2.
Constructing gene circuits using cooperative TF assemblies enables expanded steady-state signal processing behavior. (A) Computational model-driven approach for exploring engineerable circuit behavior. Our parts collection (left) defines available configuration space (middle: PDZ-ligand variants, Kp; synTF-DBM variants, Kt; clamp/synTF/DBM units, nc). Our model (fig. S7–9) computes input-output functions for this space, mapping the potential circuit behavior range (behavior space, right). (B) Programmed complex assembly enables tuning of single-input circuit dose response. For a single-input (2-node) circuit, synTF is induced by ATc addition (input node) and assembles with constitutively-expressed clamp to regulate GFP transcription (reporter node, left). In model-computed circuit behavior space (right) colors indicate different complex sizes (nc). Five circuits with different assemblies (parameters: Kt affinity in blue, Kp affinity in red, nc), were constructed and tested by inducing with ATc and measuring GFP by FACS after 16 h (below). Points represent mean values for three experiments ± SE. (C) Programmed complex assembly enables tuning of two-input dose response between linear and non-linear computations. In a two-input circuit, synTFI and synTF2 are induced by ATc and EST respectively (input nodes), assembling with constitutively-expressed clamp to regulate a downstream reporter node (left). Behavior space for the full set of available circuit configurations (center) are plotted as Kullback-Leibler divergence (DKL): “similarity” between model-computed output surfaces and archetypal Boolean AND and OR surfaces (see fig. S13). Grey areas in the plot indicate regions of AND- and OR-like behavior. Selected circuits, with corresponding reporter complex parameters (Kt affinity, blue; Kp affinity, red; nc), were constructed and their 2D output surfaces experimentally measured (right) by inducing with ATc and EST and measuring GFP by FACS after 16 h (see Supplementary Materials and Methods).
We examined dose response behavior for a single-input (2-node) circuit motif consisting of an inducible upstream input node (driving synTF production) and a downstream reporter node where the synTF assembles with constitutively expressed clamp (Fig. 2B). We asked whether features of the circuit dose response—half-maximal dose response (EC50) and Hill coefficient (nH)—could be systematically tuned by adjusting complex Kt, Kp and nc (Fig. 2B, fig. S11). Dose responses for all part-allotted configurations (603) were computed, and EC50 and nH values plotted as a two-dimensional behavior space (Fig. 2B, fig. S12). Low valency configurations conferred linear (lower nH) dose responses, while those with higher nc were broadly distributed, including configurations exhibiting the most switch-like (nH > 3.0) behavior (fig. S12). Circuit configurations with different predicted nH values were tested experimentally (fig. S12), and showed good correspondence with the model (Fig. 2B, fig. S12C).
We next assessed the relationship between assembly and Boolean computation for complexes integrating multiple synTF inputs (TF1 and TF2 respectively controlled by ncTET and ncZEV) (Fig. 2C). We calculated dose response surfaces for two-input circuits containing complexes ranging from nc = 2 to 6, for all possible combinations of TF1 and TF2 occupancy and interaction affinity (8,424 in total) (fig. S13A). We used Kullback-Leibler (K-L) divergence to compare idealized digital AND- and OR-like logic functions and simulated circuit behavior (fig. S13B, C). Plotting K-L divergences as a scatter revealed that regions of behavior space containing the most non-linear, Boolean-like circuits generally contained higher-order (nc > 3) assemblies (Fig 2C, fig. S14). Circuits were constructed and experimentally tested for configurations representing different regions of configuration space, with most performing in model-predicted fashion (fig. S15), including those from AND- and OR-like regions (Fig. 2C).
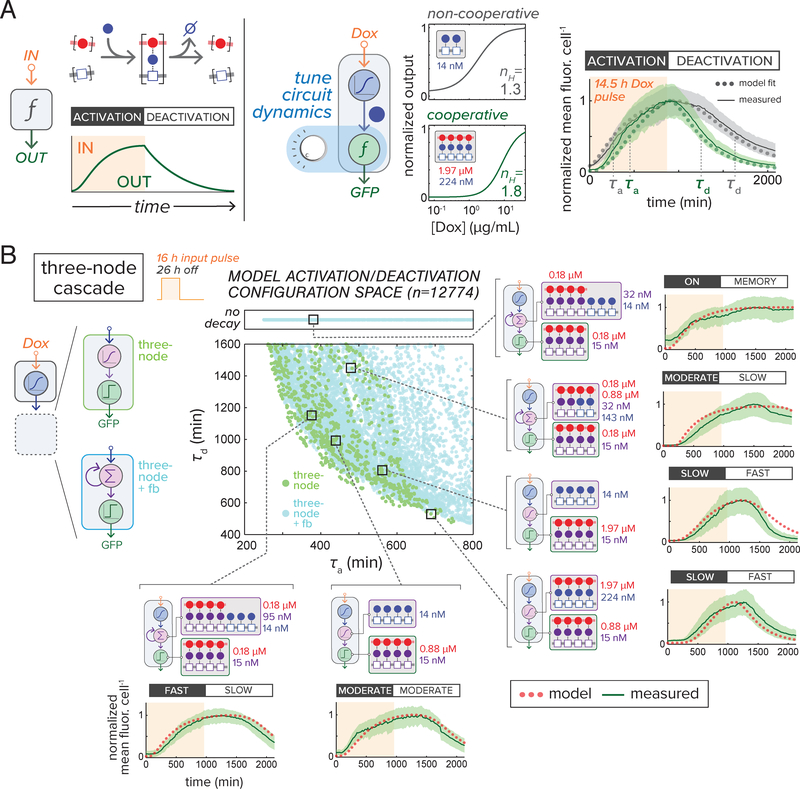
Since the timing of complex assembly is dependent on the rate of synTF accumulation, we hypothesized that assembly configuration could be adjusted to control circuit dynamics (Fig. 3A). Using time-lapse fluorescence microscopy of microfluidic device-cultured yeast (Supplementary Materials and Methods), circuit GFP expression was measured in response to transient doxycycline (Dox) inducer pulses (fig. S16). We extended our model to account for temporal circuit behavior by fitting the model to time course experiments (fig. S17A, B, Supplementary Text). We highlight two fitted circuits: a nc=2 complex that exhibits a non-cooperative steady-state dose response, and one with a more cooperative nc=4 complex (Fig. 3A). The later exhibits a delayed activation onset and rapid decay upon Dox removal, demonstrating that the timing of circuit activation and deactivation phases can be tuned by adjusting complex cooperativity.
Figure 3.
Controlling gene circuit dynamics using programmed complex assembly. (A) Tuning assembly cooperativity to control phases of circuit activation. synTF complex formation (left) determines circuit activation and deactivation kinetics (green) in response to transient inducer input (orange). Model-generated dose response profiles for two-node cascades regulated by non-cooperative two-TF (grey) or cooperative four-TF (green) assemblies are shown. Data from time course experiments using time-lapse microscopy in a microfluidic device (lines = mean fluor. Cell−1, shaded boundaries = ± 1 SD of population mean; τa, activation half-time; τd, deactivation half-time) were compared to model-fitted behavior (dots) (see Supplementary Materials and Methods). (B) Cooperative assemblies enable activation and decay phases to be broadly and independently tuned for a three-node cascade motif. Model-predicted dynamic behavior space compares τa and τd for two-node (orange) and three-node motifs with (light blue) and without (light green) feedback (‘No decay’, configurations did not return to basal activity upon input removal). Highlighted circuits were tested by time-lapse microscopy/microfluidics (16 h Dox induction, light orange; lines = mean fluor. Cell−1 normalized to maximum output, shaded boundaries = ± 1 SD) and compared to model simulations (dots). See movies S1 and S2 for time-lapse videos.
We analyzed circuit dynamics behavior space for three-node cascades in two distinct synTF assemblies connected in series (Fig. 3B), examining the basis of apparent half-time for circuit activation (τa) and decay (τd). We consider circuits with two middle node configurations: one with a single-synTF assembly and the other where the promoter is also under positive auto-regulation by its own output synTF. Plotting of simulated dynamics for part-permitted three-node behavior space (12,774) revealed a broad distribution of temporal behaviors (Fig. 3B, fig. S18), with circuits composed of cooperative, higher nc assemblies demonstrating the broadest τa and τd tuning (fig. S19). Only positive feedback configurations could access both slow activation/slow decay behavior and stable memory (no decay), suggesting circuit sensitivity to TF integration at the middle node of the cascade (Fig. 3B). We selected circuit configurations located throughout the behavior space to test experimentally; performance of these circuits confirmed the model’s ability to describe the range of temporal behaviors that can be achieved (Fig. 3B, fig. S20, movies S1–2).
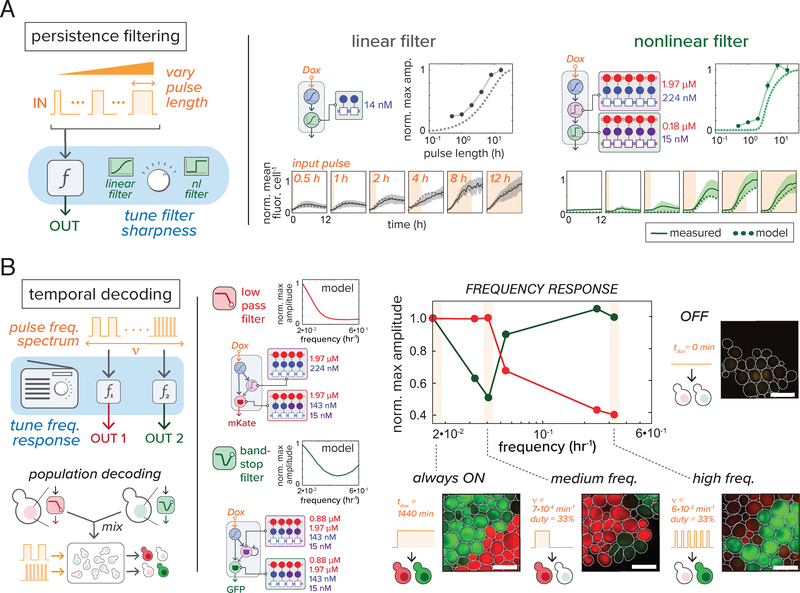
Cellular networks are capable of responding to information encoded in the dynamics of an input signal (25, 26), responding to inputs of a specific duration (27), or decoding features of an input time series (e.g., frequency) (28). To demonstrate that cooperative assemblies could facilitate engineering of dynamic filtering behavior, we computationally identified two- and three-node motifs (fig. S21A) capable of persistence filtering—activation only in the presence of a sufficiently long duration input (Fig. 4A). Filtering behavior with steep duration-dependent threshold was exhibited by three-node cascades with cooperative, high nc complexes (fig. S21B). Results of Dox pulse titration experiments were consistent with these predictions: a two-node circuit with a low valency assembly demonstrated linear filtering, while a three-node circuit with highly co-operative assemblies showed sharp filtering (Fig. 4A).
Figure 4.
Circuits engineered with cooperative assemblies perform temporal signal processing behavior. (A) Persistence filtering in cooperative assembly circuits. Computationally identified two- and three-node motifs reveal persistence filtering behavior – activation in response to an input of a sufficient duration (left, fig. S21). Circuits predicted to show linear (shallow, grey) and nonlinear (sharp, green) filtering were tested in microfluidic time course experiments (lines = mean fluor. Cell−1, shaded region = ± 1 SD of cell population) by inducing with increasing Dox pulse lengths (light orange). “temporal dose response” curves (line = measured data, dots = model) were generated from normalized time course output maxima (see Supplementary Materials and Methods). (B) Population-level temporal decoding of frequency input. Computationally identified (fig. S22) two- and three-node network motifs (left) were tested for frequency filtering behavior in microfluidic experiments. Mixed populations of yeast harboring low-pass filtering (LPF) and band-stop filtering (BSF) feed-forward circuits (driving mKate and GFP, respectively) were tested for their ability to decode a frequency-modulated square wave Dox pulses (fig. S23). Model-predicted frequency responses are shown next to each circuit. Experimentally measured frequency response is shown to the right. Maximum reporter output for each input frequency was normalized to maximum output for constitutive Dox. (fig. S22). Representative mixed population fluorescence images are shown for frequencies highlighted in orange: constitutive Dox (‘always ON’), medium frequency treatment (~10−5 Hz, 33%), and high frequency (~10−4 Hz, 33%). Cells without Dox treatment (‘OFF’) are shown to the right. In images, cells harboring LPF and BSF circuits are false-colored red and green, respectively; cell boundaries were determined by segmentation software (see Supplementary Materials and Methods). Scale bar, 10 μM.
As a final demonstration, we identified sets of circuits capable of differentially responding to distinct input frequencies (Fig. 4B). We computationally identified a pair of circuits, both coherent feed-forward loops (CFFL), activated by distinct square-wave frequencies (fig. S22). This included low-pass filters (LPF), which only respond to low frequency input, and band-stop filters (BSF), which filter medium frequency while activating at high frequency (both with 33% duty cycle). For both circuits, cooperative, high nc assemblies were critical for sharp filtering (fig. S22C). To test filtering function, we created two strains: a BSF circuit driving a GFP reporter, and an LPF with mKate reporter. As predicted, at different input frequencies, strains differentially activated within a mixed population (BSF at ~10−4; LPF at ~10−5 Hz) (Fig. 4B, fig. S23).
Our work demonstrates that cooperative assembly is a powerful, highly flexible design strategy for engineering non-linear circuit behavior, and offers clues as to why TF assemblies evolved as a dominant mode of transcriptional regulatory control (29, 30). Adjusting promoter assemblies may have provided networks with a simple way to interpolate between diverse regions of functional space (31, 32). Use of engineering approaches that incorporate cooperative assembly could facilitate creation of signal processing circuitry (33), enabling precision control in applications where non-linear temporal and spatial signal processing are critical, such as circuit-directed cell differentiation or dynamic regulation of homeostasis in engineered tissues.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. K. Joung and members of the Collins and Khalil laboratories for helpful discussions.
Funding: This work was supported by NSF Expeditions in Computing grant CCF-1522074 (A.S.K.) and DARPA grant W911NF-11–2-0056 (J.J.C. and A.S.K.). A.S.K. also acknowledges funding from the NIH Director’s New Innovator Award (1DP2AI131083–01), NSF CAREER Award (MCB-1350949), and DARPA Young Faculty Award (D16AP00142).
Footnotes
Competing interests: C.J.B, N.P., J.J.C. and A.S.K. are inventors on U.S. Provisional Patent Application 62/691,187 filed June 28, 2018 by Boston University.
Data and materials availability: All DNA constructs and cell strains are available from A.S.K. The datasets generated and analyzed and the computer code used during the current study are available upon request from the corresponding author.
References and Notes
- 1.Hill AV, The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol. 40, iv–vii (1910). [Google Scholar]
- 2.Whitty A, Cooperativity and biological complexity. Nat Chem Biol. 4, 435–9 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Ptashne M, How eukaryotic transcriptional activators work. Nature. 335, 683–9 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Antebi YE, et al. , Combinatorial Signal Perception in the BMP Pathway. Cell. 170, 1184–1196 e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhalla US and Iyengar R, Emergent properties of networks of biological signaling pathways. Science. 283, 381–7 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Ferrell JE Jr. and Ha SH, Ultrasensitivity part III: cascades, bistable switches, and oscillators. Trends Biochem Sci. 39, 612–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine M, Transcriptional enhancers in animal development and evolution. Curr Biol. 20, R754–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spitz F and Furlong EE, Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 13, 613–26 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Struhl K, Mechanisms for diversity in gene expression patterns. Neuron. 7, 177–81 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Williamson JR, Cooperativity in macromolecular assembly. Nat Chem Biol. 4, 458–65 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Bhattacharya S, and Andersen ME, Ultrasensitive response motifs: basic amplifiers in molecular signalling networks. Open Biol. 3, 130031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carey M, The enhanceosome and transcriptional synergy. Cell. 92, 5–8 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Veitia RA, A sigmoidal transcriptional response: cooperativity, synergy and dosage effects. Biol Rev Camb Philos Soc. 78, 149–70 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Buchler NE, Gerland U, and Hwa T, On schemes of combinatorial transcription logic. Proc Natl Acad Sci U S A. 100, 5136–41 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashor CJ, et al. , Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annu Rev Biophys. 39, 515–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brophy JA and Voigt CA, Principles of genetic circuit design. Nat Methods. 11, 508–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purnick PE and Weiss R, The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 10, 410–22 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Buchler NE and Cross FR, Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol Syst Biol. 5, 272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keung AJ, et al. , Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell. 158, 110–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalil AS, et al. , A Synthetic Biology Framework for Programming Eukaryotic Transcription Functions. Cell. 150, 647–658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nevozhay D, et al. , Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci U S A. 106, 5123–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mclsaac RS, et al. , Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 41, e57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia HG, et al. , Thermodynamics of biological processes. Methods Enzymol. 492, 27–59 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gertz J, Siggia ED, and Cohen BA, Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 457, 215–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine JH, Lin Y, and Elowitz MB, Functional roles of pulsing in genetic circuits. Science. 342, 1193–200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purvis JE and Lahav G, Encoding and decoding cellular information through signaling dynamics. Cell. 152, 945–56 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangan S and Alon U, Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 100, 11980–5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell A, Wei P, and Lim WA, Oscillatory stress stimulation uncovers an Achilles’ heel of the yeast MAPK signaling network. Science. 350, 1379–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hnisz D, et al. , A Phase Separation Model for Transcriptional Control. Cell. 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J, Shang Y, and Zhang M, Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat Rev Neurosci. 17, 209–23 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Baker CR, et al. , Protein modularity, cooperative binding, and hybrid regulatory states underlie transcriptional network diversification. Cell. 151, 80–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuch BB, Li H, and Johnson AD, Evolution of eukaryotic transcription circuits. Science. 319, 1797–9 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Novershtern N, et al. , Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144, 296–309 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jantz D and Berg JM, Probing the DNA-binding affinity and specificity of designed zinc finger proteins. Biophys J 98, 852–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang ZX, An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 360, 111–4 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Cookson S, et al. , Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol Syst Biol 1, 2005 0024 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vega NM, et al. , Signaling-mediated bacterial persister formation. Nature chemical biology. 8, 431–33 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duffy DC, et al. Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane). Anal Chem 70, 4974–84 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Unger MA, et al. , Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 288, 113–6 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Wang Q, et al. , Image segmentation and dynamic lineage analysis in single-cell fluorescence microscopy. Cytometry A 77, 101–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris BZ, Hillier BJ, and Lim WA, Energetic determinants of internal motif recognition by PDZ domains. Biochemistry. 40, 5921–30 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Wiedemann U, et al. , Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J Mol Biol 343, 703–18 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Bintu L, et al. , Transcriptional regulation by the numbers: models. Current opinion in genetics & development. 15, 116–24 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirny LA, Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci U S A. 107, 22534–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen AS and O’Shea EK, Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol Syst Biol 9, 704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao C, et al. , Quantitative analysis of the transcription control mechanism. Mol Syst Biol 6, 431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aymoz D, et al. , Timing of gene expression in a cell-fate decision system. Mol Syst Biol 14, e8024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guarente L, et al. , Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 36, 503–11 (1984). [DOI] [PubMed] [Google Scholar]
- 49.Estrada J, et al. , Information Integration and Energy Expenditure in Gene Regulation. Cell. 166, 234–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bintu L, et al. , Transcriptional regulation by the numbers: applications. Current opinion in genetics & development. 15, 125–35 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang JS, Correlation between functional and binding activities of designer zinc-finger proteins. Biochem J 403, 177–82 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrell JE Jr., Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol 14, 140–8 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Bennett MR, et al. , Metabolic gene regulation in a dynamically changing environment. Nature. 454, 1119–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bialek W, et al. , Reading a neural code. Science. 252, 1854–7 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.