Abstract
Purpose:
Untargeted metabolomic analysis is increasingly being used in the screening and management of individuals with inborn errors of metabolism (IEM). We aimed to test whether untargeted metabolomic analysis in plasma might be useful for monitoring the disease course and management of urea cycle disorders (UCDs).
Methods:
Untargeted mass spectrometry-based metabolomic analysis was used to generate z-scores for more than 900 metabolites in plasma from 48 individuals with various UCDs. Pathway analysis was used to identify common pathways that were perturbed in each UCD.
Results:
Our metabolomic analysis in plasma identified multiple potentially neurotoxic metabolites of arginine in arginase deficiency and, thus, may have utility in monitoring the efficacy of treatment in arginase deficiency. In addition, we were also able to detect multiple biochemical perturbations in all UCDs that likely reflect clinical management, including metabolite alterations secondary to dietary and medication management.
Conclusions:
In addition to utility in screening for IEM, our results suggest that untargeted metabolomic analysis in plasma may be beneficial for monitoring efficacy of clinical management and off-target effects of medications in UCDs and potentially other IEM.
Keywords: Metabolomics, urea cycle disorder, arginase deficiency, branched-chain amino acids, guanidino compounds
INTRODUCTION
Urea cycle disorders (UCDs) comprise a group of inborn errors of metabolism (IEM) caused by a deficiency of one of the enzymes or transporters involved in the hepatic detoxification of nitrogenous waste. This group of disorders includes N-acetylglutamate synthase (NAGS) deficiency, ornithine transcarbamylase (OTC) deficiency, carbamoyl-phosphate synthase 1 (CPS1) deficiency, argininosuccinate synthase deficiency (citrullinemia type 1), argininosuccinate lyase (ASL) deficiency, arginase deficiency, hyperornithinemia-hyperammonemia-homocitrullinuria (HHH) syndrome, and citrin deficiency. Classically, individuals with UCDs present with neonatal hyperammonemia which, if untreated, can progress to coma and death. However, later onset forms of UCDs can present with hyperammonemia during later childhood or adulthood. Moreover, some disorders, such as arginase deficiency, which typically presents with spastic diplegia and developmental delay with or without seizures, have unique presentations that may reflect perturbations of pathways other than the urea cycle. In many cases, typical biochemical features are suggestive of a particular UCD, and molecular or enzymatic testing can confirm the suspected diagnosis. However, because of the X-linked inheritance pattern, diagnosis using metabolites alone can be difficult in females with partial OTC deficiency. Moreover, in ~10–15% of individuals with suspected OTC deficiency, conventional genetic testing is unable to identify a pathogenic variant1. Treatment for UCDs includes avoidance of catabolic stress, protein-restricted diet with or without essential amino acid formula, supplementation with deficient amino acids, and nitrogen-scavenging medications, such as sodium benzoate, sodium phenylbutyrate, and glycerol phenylbutyrate. Plasma amino acids and metabolites of nitrogen-scavenging medications are used for monitoring treatment efficacy2.
Untargeted metabolomic profiling has increasingly become an important tool in screening for IEMs and in the management of IEMs3, 4. Metabolomic profiling facilitates the simultaneous detection and quantification of potentially thousands of metabolites in plasma3, 4, urine5, and cerebrospinal fluid6. In addition to facilitating screening by detecting known disease-associated metabolites that were previously only detectable in difficult to obtain samples such as cerebrospinal fluid7, metabolomic profiling has identified new disease biomarkers3, biomarkers reflecting treatment3, 8, and perturbations in pathways that reveal new clues to the pathogenesis of various IEM9. In a recent study of plasma metabolomic profiling in various IEM, including a small population of individuals with UCDs, we identified pathognomonic biomarkers for individual disorders such as argininosuccinic acid in ASL deficiency3. In addition, we detected biomarkers for treatment with nitrogen-scavenging agents, and for arginase deficiency, we identified new biomarkers in nitric oxide and nucleoside pathways that had not been previously detectable in routinely available clinical testing platforms8.
In the present study, we performed metabolomic profiling in a larger sample of 48 individuals with various UCDs to assess whether we could identify novel biomarkers for these disorders which might provide clues to alternative pathways impacted by urea cycle dysfunction. Our metabolomic studies demonstrate the identification of several new potential clinical biomarkers for arginase deficiency, metabolite alterations reflecting dietary and medical management, and the impact of urea cycle dysfunction on pyrimidine metabolism.
MATERIALS AND METHODS
Data Collection.
Data for the metabolomic analysis at Baylor Genetics Laboratory came from one of three sources. First, blood samples were collected from individuals with partial OTC deficiency (females), ASL deficiency, and arginase deficiency after informed consent (N=29). Blood samples were collected (EDTA-coated tubes) at routine clinic visits when subjects were at their baseline state of health. Plasma was separated and stored at −80°C. Clinical information including age, diet, nitrogen-scavenging medications, and branched-chain amino acid (BCAA) supplementation was collected. Second, heparinized plasma samples (n=7) received at the Baylor Genetics Laboratory from individuals with citrullinemia type 1 were used for metabolomic analysis. Third, data from plasma samples (from blood collected in EDTA-coated tubes) that were submitted to the Baylor Genetics Laboratory for clinical metabolomic testing were included (n=12). All samples were typically collected during routine clinic visits. Besides clinical diagnosis, no additional clinical information was available for subjects with citrullinemia type 1 and the 12 individuals for whom samples were received for clinical metabolomic analysis. The study procedures were approved by the Institutional Review Board of Baylor College of Medicine. Data from the analysis of samples from individuals with citrullinemia was previously published3. Data from all other individuals has not been previously published. Each sample represents a unique individual.
Metabolomic Profiling.
Metabolomic profiling was performed as previously described3, 5. Briefly, using 200 µl of plasma, small molecules were extracted in an 80% methanol solution containing recovery standards, outlined below, and used to monitor extraction efficiency. The clarified supernatant was divided into five aliquots, one for each of the individual LC/MS analyses, briefly evaporated to remove the organic solvent and stored overnight under nitrogen before analysis.
LC/MS/MSn Analyses.
All methods utilized a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution10 and are described in Supplementary Materials and Methods.
BCAA Analysis.
Plasma BCAA levels and prealbumin were collected as part of the Longitudinal Study of Urea Cycle Disorders (ClinicalTrials.gov NCT00237315) conducted by the Urea Cycle Disorders Consortium (UCDC) of the NIH Rare Diseases Clinical Research Network (RDCRN)11. This study is a natural history study conducted at 16 sites across the United States, Canada, and Europe with a total of 730 individuals enrolled at the time of data analysis. A standardized format is used for data collection. Data from each site are entered into an electronic database that is maintained by the RDCRN Data Management and Coordinating Center. Data from the enrollment visit were used for the analysis.
Pathway Analysis Visualization.
Pathway knowledge was curated by Metabolon using MetaboLync Pathway Visualizations (MetaboLync Pathway Visualizations software, version 1.1.2, Copyright 2014 Metabolon, Inc., Research Triangle Park, NC, USA) a Cytoscape12 plugin for pathway visualizations) that incorporates pathway knowledge from pathway databases, such as KEGG. Using the plugin, we exported Node Attributes values for “Compound Type”, “DisplayName” and “ID”, which were associated with the Metabolic Pathways pathway map, containing 1099 unique compounds (depicted as circles) and 597 unique enzymes (depicted as green rectangles). We then manually inspected the Display Names associated with each compound node in the Metabolic Pathways pathway map and compared them to our dataset’s metabolite identifiers. We identified 253 unique metabolites that matched between our dataset and the Metabolic Pathways pathway map. We then reconstructed this pathway with the igraph13 R package using the R programming language. Using igraph, we could resize the nodes based on the strength of the z-score, and color the nodes based on the direction of the perturbation. Mean profiles for our cohort of subjects with the same diagnosis were plotted onto the pathway map.
Statistical Analysis.
For metabolomic data, semi-quantitative analysis was achieved by comparing subject samples to a set of invariant anchor specimens included in each batch, as previously described3, 5, 6. Raw spectral intensity values were normalized to anchor samples, log transformed, and compared to a normal reference population to generate z-score values. The reference population was a pediatric, randomly sampled population that was matched for age, diet and gender (n=181 for heparin samples, n=500 for EDTA samples). Rare compounds are those analytes detected in the patient specimens but only rarely seen in the reference population (<5% of all patients tested). Median raw intensity values were calculated for all analytes identified in 2/3 of the anchor specimens, and these median values were used to normalize corresponding analyte raw intensity values in patient specimens. Analytes not identified in 2/3 or more of the anchor specimens were excluded from statistical analysis. Anchor-adjusted analyte values were scaled to the median anchor-adjusted analyte values of all previous specimens analyzed to date. Z-scores were calculated using the mean and standard deviation of the entire median-scaled log-transformed dataset.
For each disorder, the mean and median z-score for each metabolite were calculated. Metabolites with mean z-scores ≥ 2 or ≤ −2 were identified and used for analysis. In addition, metabolites that were present in study samples but not in the anchor sample or control data set were identified. All box-and-whisker plots show minimum, 25th percentile, median, 75th percentile and maximum values.
All statistical analyses of BCAA and prealbumin from subjects enrolled in the Longitudinal Study of Urea Cycle Disorders were completed using SAS version 9.4 (Cary, NC). One baseline measure for concentrations of leucine, isoleucine, valine, and prealbumin per subject were included in the analyses. Exclusion criteria for this analysis included liver transplantation prior to baseline visit, use of more than one nitrogen-scavenging drug, and use of leucine, isoleucine or valine supplementation (Figure S1). Using PROC GLM (SAS Institute Inc.), ANOVA was performed to separately test for differences in the concentrations of the BCAA and prealbumin due to the type of nitrogen-scavenging drug treatment (sodium benzoate, sodium or glycerol phenylbutyrate, or no use of a nitrogen-scavenging drug) and disorder (OTC deficiency, citrullinemia, ASL deficiency, or arginase deficiency). When data were skewed, log transformations were performed. If data remained skewed after the log transformation, then rank variables from dependent variables were calculated using PROC RANK (SAS Institute, Inc.). Using PROC GLM, ANOVA with the rank values was performed. Statistical significance was set at p < 0.05.
RESULTS
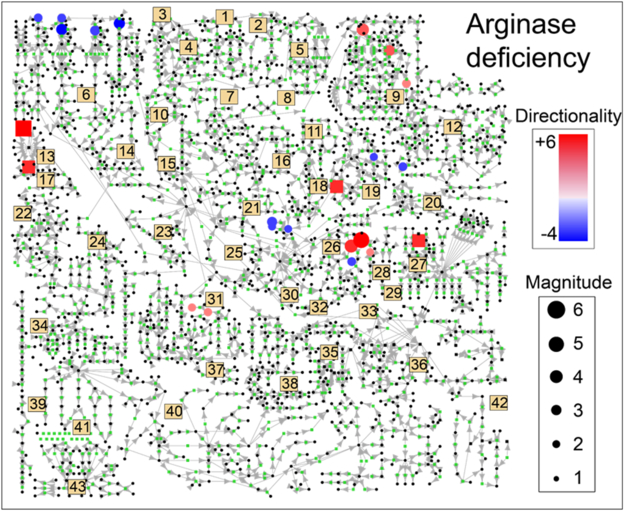
The data set consisted of 48 children and adults with UCDs. Clinical information was available for 29 subjects (Table S1). We identified metabolites with a mean z-score ≥ 2 or ≤ −2 for each UCD (Figure 1 and S2; Table S2 and S3). From these metabolites, xenobiotics, food additives, products derived from anticoagulants used for sample processing, metabolites of nitrogen-scavenging agents, un-identified compounds, and metabolites that were detectable in fewer than 50% of samples for that disorder were removed for the initial analysis. We removed a single metabolite (glucose in arginase deficiency) which had a median that was more than 50% lower than the mean to prevent a single outlier from a single individual from driving any observations. Of all UCDs, subjects with arginase deficiency and citrullinemia had the largest number of metabolites increased or decreased across samples (Figure 1).
Figure 1. Metabolite Profile in Arginase Deficiency.
Z-scores for metabolites in subjects with arginase deficiency were averaged. All metabolite z-scores that were mapped to the pathway map are plotted. Only metabolites with z-scores ≥ 2 or ≤ - 2 were plotted. Blue nodes indicate negative z-scores, whereas red nodes indicate positive z-scores. Node size reflects the strength of the z-score. Numbers refer to particular pathways and biological processes, described below.
1–Glycosylation 2–Ascorbate Metabolism 3–Glycogen Metabolism
4–Galactose Metabolism 5–Inositol Metabolism 6–BCAA Metabolism
7–Riboflavin Metabolism 8–Pentose Phosphate Pathway 9–Pyrimidine Metabolism
10–Sorbitol/Glycerol Metabolism 11–Benzoate Metabolism 12–Purine Metabolism
13–Protein Degradation 14–Lysine Catabolism 15–Glycolysis
16–Glycine/Serine/Threonine 17–Carnitine Biosynthesis 18–Methionine/Cysteine
Metabolism Metabolism
19–Glutathione Metabolism 20–Histidine Metabolism 21–Ketone Bodies
22–Nicotinate/Nicotinamide 23–Mevalonate Metabolism 24–Tryptophan
Metabolism Metabolism
25–TCA Cycle 26–Arginine Metabolism and 27–Eicosanoids
Urea Cycle
28–Polyamine Metabolism 29–Creatine Metabolism 30–Aspartate/Glutamate
Metabolism
31–Phenylalanine/Tyrosine 32–Proline Metabolism 33–Endocannabinoid
Metabolism Synthesis
34–Fibrinogen Cleavage Peptides 35–Sphingolipid Metabolism 36–Phospholipid
Metabolism
37–Thyroid Hormone Synthesis 38–Fatty Acid Metabolism 39 Hemoglobin/Porphyrin
Metabolism
40–Steroid Hormone Biosynthesis 41–Bile Acid Metabolism 42–Dicarboxylic Acid
Metabolism
43-Secondary Bile Acids
Metabolomic Signature in Arginase Deficiency.
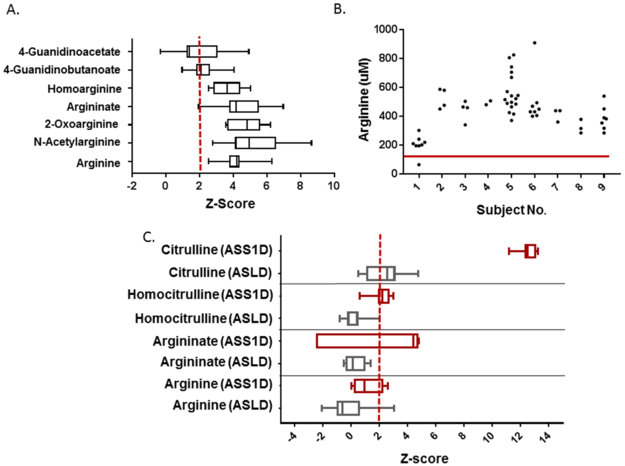
Plasma metabolomic data were available for 13 subjects with arginase deficiency. As expected, the mean z-score for plasma arginine was 4.06 (range: 2.45 to 4.52) (Figure 2). Clinical data were available for 9 subjects (ages 4 – 43 years). Over the five years prior to sample collection, the plasma arginine levels obtained from quantitative plasma amino acids for these subjects was typically above the upper limit of the normal range despite protein-restriction and nitrogen-scavenging medications (Figure 2). Similar findings have been described in other cohorts of individuals with arginase deficiency14, 15. Arginine is a precursor for numerous metabolites including guanidino compounds, and indeed, multiple guanidino compounds including N-acetylarginine, guanidinoacetate, guanidinobutanoate, homoarginine, 4-guanidinobutanal, argininate, and 2-oxoarginine were elevated. The mean z-score for some of these guanidino compounds, such as argininate, 2-oxoarginine, and N-acetylarginine, was higher than arginine, which is the standard biochemical used for newborn screening and clinical diagnosis (Figure 2). Plasma guanidinoacetate has been measured clinically in arginase deficiency and is a proposed target of treatment16. Notably, the z-scores for other guanidino compounds were often higher than guanidinoacetate. These findings suggest that guanidinoacetate may not be the sole guanidino compound contributing to the phenotype.
Figure 2. A. Elevated Guanidino Compounds in Arginase Deficiency.
A. Multiple plasma guanidino compounds are markedly elevated in subjects with arginase deficiency. Data for each metabolite was available for n=7-13 subjects (e.g., total analyte identifications per sample varied by batch and platform used). The red dashed line indicates the z-score of 2. B. Plasma arginine levels over the five years prior to sample collection for metabolomics in subjects with arginase deficiency. The red line represents the upper limit of normal for plasma arginine level. C. Elevated metabolites in citrullinemia (ASS1D, red) and ASL deficiency (ASLD, grey) are shown. The red dashed line indicates the z-score of 2. Data for these metabolites were available for n=7 subjects with ASS1D and n=9-10 subjects with ASLD.
Metabolomic Signature in ASL Deficiency and Citrullinemia Type 1.
Fewer metabolite perturbations were identified in ASL deficiency and citrullinemia type 1. As expected, citrulline was markedly elevated in citrullinemia (mean z-score: 12.52, range: 11.2 to 13.3) and the related metabolite, homocitrulline (mean z-score: 2.16, range: 0.60 to 2.99), was also elevated (Figure 2). We also noted slight elevations in arginine metabolites likely secondary to arginine supplementation in citrullinemia, but the elevations were lower than the levels observed in arginase deficiency. In particular, the mean z-score for argininate in citrullinemia is 3.29 (range: −2.51 to 4.82) with only two subjects having a z-score for argininate lower than 3.76. In contrast, the mean z-score for arginine was 1.13 (range: 0.04 to 2.61). The levels of arginine (and possibly its metabolites) in citrullinemia likely fluctuate throughout the day depending on arginine dose timing. We also noted elevations of stearate (methyl ester) and stearidonate (18:4n3) which are long chain fatty acids found in dietary oils in citrullinemia, and although the clinical significance is unclear, we suspect that these compounds are derived from the diet.
Likewise, citrulline was mildly elevated (mean z-score: 2.47, range: 0.53 to 4.78) in most samples from individuals with ASL deficiency. Argininosuccinate was detected in all samples from individuals with ASL deficiency, but no z-scores could be calculated because this metabolite is not detected in control samples and was not present in the reference population. Although subjects with ASL deficiency are typically supplemented with arginine, the dose may be lower than in subjects with citrullinemia in this cohort given that many of our subjects with ASL deficiency have mild forms of the disorder without prior history of hyperammonemia (Table S1; Figure 2), and we have previously shown that lower doses of arginine are associated with reduction in markers of liver inflammation in the short term17. Otherwise, metabolite pattern in ASL deficiency was similar to individuals without a UCD.
Biomarkers in OTC Deficiency.
Samples from 17 individuals with OTC deficiency were analyzed. The only metabolite with z- score ≥ 2 or ≤ −2 was orotate. Of the 10 female subjects with partial OTC deficiency who were prospectively enrolled, five subjects had a history of at least one prior episode of hyperammonemia, and five subjects had no history of hyperammonemia. We calculated mean metabolite z-scores for individuals with and without a history of hyperammonemia to test whether any metabolites distinguished the two groups. The only metabolite with a z-score ≥ 2 or ≤ −2 in the subjects without history of hyperammonemia was adenosine monophosphate (AMP). In contrast, in the group with history of hyperammonemia, urea and creatine were reduced whereas orotate, uridine, and glutamine were increased. Of these metabolites, the z-scores for glutamine had the least overlap in the two groups even though the subjects with prior history of hyperammonemia were treated with protein restriction and nitrogen-scavenging agents (Figure S3). Of note, one of the other seven samples was from a male with late-onset OTC deficiency and was a diagnostic sample (pre-treatment). This sample had the highest orotate (z-score=10.2), uracil (z-score=3.02), and uridine (z-score =4.02) and the lowest citrulline (z-score =−3.80), ornithine (z-score =−2.71) and arginine (z-score=3.78) of all the samples from individuals with this diagnosis.
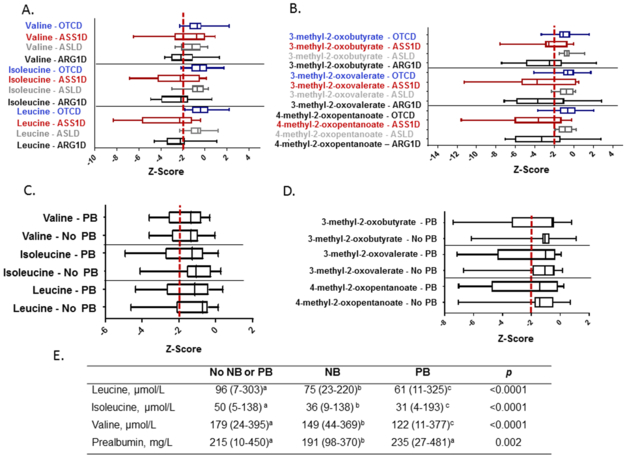
Decreased Branched-Chain Amino Acid (BCAA) Metabolism in UCDs.
BCAA and their branched-chain ketoacids (BCKA) were decreased in arginase deficiency (Figures 3A and 3B), and similar decreases were noted in citrullinemia. Similar trends demonstrating low BCAA in the plasma amino acid analyses collected for routine management over the five years prior to the sample collection were observed in the subjects with arginase deficiency (Figures S4, S5, and S6). Low BCAA are a known off-target effect of sodium and glycerol phenylbutyrate18–20. However, we observed decreased BCAA and BCKA even in subjects who were not taking these medications (Figures 3C, 3D, S4, S5, and S6). Thus, in this study population, the decreased BCAA were likely not solely the result of an off-target effect of sodium or glycerol phenylbutyrate. These reduced levels of BCAA could also be secondary to protein restriction (Table S1). Although most patients with a UCD consume a low-protein diet, a subset of subjects with OTC deficiency and ASL deficiency had no history of hyperammonemia, and thus, the diet in these subjects was likely not as protein-restricted as the subjects with citrullinemia and arginase deficiency. The inclusion of subjects with milder forms of OTC deficiency and ASL deficiency may explain why this low BCAA signature was primarily observed in citrullinemia and arginase deficiency.
Figure 3. Reduced Plasma Levels of Branched-Chain Amino Acids (BCAA) in UCDs.
A. The levels of leucine, isoleucine, and valine in each UCD are shown. B. The levels of the branched-chain ketoacids (BCKA) in each UCD are shown. C. Leucine, isoleucine, and valine levels in subjects taking phenylbutyrate-containing nitrogen-scavenging agents vs. subjects not taking these agents. D. The BCKA levels in subjects taking phenylbutyrate-containing nitrogen scavenging agents vs. subjects not taking these agents. The red dashed line indicates the z-score of −2. OTCD = OTC deficiency, ASS1D = citrullinemia, ASLD = ASL deficiency, ARG1D = arginase deficiency. PB=phenylbutyrate. E. BCAA and prealbumin concentrations in subjects from the UCDC Longitudinal Study are provided based on type of nitrogen-scavenging drug treatment. Statistical analysis includes one-way ANOVA for type of nitrogen-scavenging drug treatment. Values are medians (minimum – maximum). Sample sizes for the branched-chain analyses included: No NB or PB, n = 269-270; NB, n = 61; PB, n = 193-195. Sample sizes for prealbumin included: No NB or PB, n = 211; NB, n = 50; PB, n = 152. a,b,cMedians with different superscripted letters are significantly different (p<0.05). NB, sodium benzoate; PB, sodium or glycerol phenylbutyrate.
To further investigate whether reduced BCAA are specific to individuals taking phenylbutyrate-containing medications, we analyzed BCAA levels in individuals enrolled in the Longitudinal Study of Urea Cycle Disorders (n=730) performed by the Urea Cycle Disorders Consortium (Figure S1). We previously demonstrated that subjects taking sodium phenylbutyrate had lower plasma BCAA levels as compared to subjects who were not taking this medication18. In this study, we expanded our previous analysis to reflect current enrollment (n=730 subjects) and included one additional comparison group (individuals taking sodium benzoate) (Figures 3E and S7; Table S4 and S5). Interestingly, the plasma BCAA levels in subjects taking sodium benzoate were also significantly lower than in subjects not requiring a nitrogen-scavenging agent (Figure 3). Since the prealbumin levels were significantly lower in subjects taking sodium benzoate (Figure 3), we suspect that the reduction in plasma BCAA in subjects taking sodium benzoate may be due to dietary protein restriction.
Urea Cycle Dysfunction and Pyrimidine Metabolism.
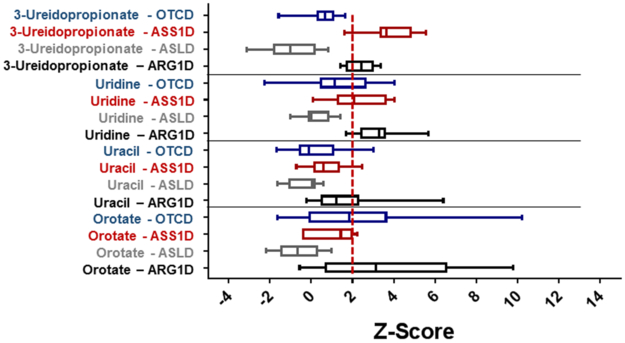
Metabolites involved in pyrimidine metabolism were significantly elevated in arginase deficiency, citrullinemia, and OTC deficiency (Figure 4). Individuals with arginase deficiency had elevated orotate, uridine, 5,6-dihydrouracil, and ß-ureidopropionate, which likely results from intrahepatic deficiency of ornithine. In citrullinemia, elevated ß-ureidopropionate and uridine were elevated whereas in OTC deficiency, orotate was elevated. It is unclear whether this alteration in pyrimidine metabolism impacts purine/pyrimidine ratios or has downstream impact on cellular processes.
Figure 4. Pyrimidine Metabolites are Elevated in UCDs.
The levels of elevated pyrimidine metabolites in subjects with UCDs are shown. The red dashed line indicates the z-score of 2. OTCD = OTC deficiency, ASS1D = citrullinemia, ASLD = ASL deficiency, ARG1D = arginase deficiency.
Metabolomic Profile Reflects Treatment in UCDs.
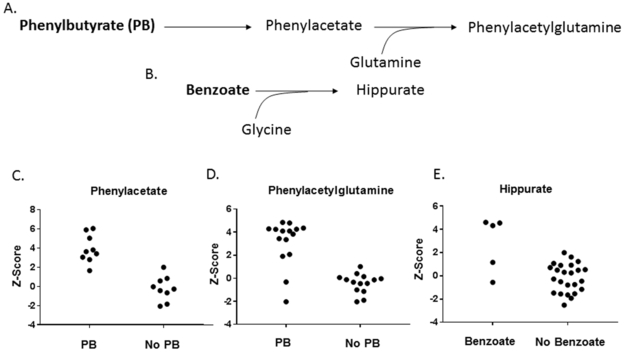
Nitrogen-scavenging agents (sodium benzoate, sodium phenylbutyrate, glycerol phenylbutyrate) are typically used to divert nitrogen away from the urea cycle in UCDs to reduce risk for hyperammonemia. Metabolites of these agents were detectable by the metabolomic platform. For example, hippurate was elevated in 3 of 5 individuals taking sodium benzoate, and phenylacetate and phenylacetylglutamine were elevated in individuals taking sodium or glycerol phenylbutyrate (Figure 5).
Figure 5. Metabolites of the Nitrogen Scavenging Agents are Detected in Plasma Metabolomic Analysis.
A. Phenylbutyrate is metabolized to phenylacetate and combines with glutamine to form phenylacetylglutamine which is excreted in the urine. B. Benzoate combines with glycine to form hippurate which is excreted in the urine. C. Phenylacetate is elevated in subjects taking sodium phenylbutyrate or glycerol phenylbutyrate. D. Phenylacetylglutamine is elevated in subjects taking sodium phenylbutyrate or glycerol phenylbutyrate. E. Hippurate is elevated in most subjects taking sodium benzoate. PB = taking sodium phenylbutyrate or glycerol phenylbutyrate;
DISCUSSION
Untargeted metabolomic profiling has great potential as a tool in screening and management of individuals with IEM3, 4. Although future studies in larger populations of individuals with UCDs are needed, our analysis is a first step toward demonstrating the potential utility of this technology in screening and management of individuals with UCDs. Metabolites, enzymatic assays, and/or molecular testing often facilitate a UCD diagnosis. However, in ~10–15% of individuals with OTC deficiency, standard clinical DNA testing does not reveal a pathogenic variant21, and thus, in the absence of a classic metabolite pattern in plasma amino acids, diagnosis of at-risk relatives of individuals can be challenging. Although elevated urine orotic acid is classically used in diagnosis, plasma levels of uridine and other pyrimidine metabolites were noted to be elevated in at least one female with OTC deficiency22. Here, we demonstrate that plasma uridine and orotate may be supportive of a diagnosis of partial OTC deficiency in some individuals and are markedly high in one male at diagnosis. For instance, of 17 individuals with OTC deficiency (males and females, varying severity), 10 (including all five females with known history of hyperammonemia) had a z-score > 2 for either orotate or uridine. Of the five females without a known history of hyperammonemia, one also had elevated uridine. Although elevated plasma uridine and orotate might support a diagnosis of OTC deficiency, our data demonstrate that normal levels of these metabolites are not sufficient to rule out a diagnosis. Future studies using a larger population of individuals with OTC deficiency are needed to determine the specificity and sensitivity of these metabolites. Other biochemical hallmarks in UCDs, such as citrulline elevation in citrullinemia and in ASL deficiency, increased arginine in arginase deficiency, and argininosuccinate in ASL deficiency, were all detected by metabolomic analysis.
We also detected metabolites that could contribute to long-term complications of UCDs. For instance, multiple guanidino compounds, which are derived from arginine, were elevated in arginase deficiency, as described previously23, 24. Guanidinoacetate, a metabolite known to be associated with intellectual disability and seizures in guanidinoacetate methyltransferase deficiency,25 has been suggested to cause GABA receptor dysfunction26, 27, inhibition of Na+, K+-ATPase and creatine kinase activities in the brain28, and increased oxidative stress in the brain28, 29. It has been suggested that guanidinoacetate should be a therapeutic target in arginase deficiency16. However, the z-scores for other guanidino compounds including argininate, N-acetylarginine, α-keto-δ-guanidinovaleric acid (also known as 2-oxoarginine), homoarginine, and 4-guanidinobutanoate were even higher than guanidinoacetate. Several of these other guanidino compounds have been implicated in the pathogenic mechanism of spastic diplegia, seizures, and intellectual disability in arginase deficiency by mediating increased oxidative stress30, disrupting Na+, K+-ATPase activity31, and other potential mechanisms of neurotoxicity32, 33. The elevations of these potentially neurotoxic compounds derived from arginine are especially concerning given the challenges in normalizing plasma arginine in our subjects and in published cohorts14, 34. These guanidino compounds may prove to be useful biomarkers for monitoring efficacy of existing therapies including liver transplantation35 and may be useful biomarkers for future interventional studies, such as enzyme therapy for arginase deficiency14.
Likewise, in a subset of subjects with arginase deficiency, orotate and other pyrimidine metabolites were elevated. Marked elevations of urinary orotic acid and other pyrimidine metabolites have been observed in individuals with arginase deficiency and have been attributed to hepatic ornithine deficiency36. Similarly, plasma elevations of a subset of these metabolites were also identified in OTC deficiency (especially subjects with history of hyperammonemia) and in citrullinemia. The impact of this elevation in pyrimidine metabolites is unclear, and it is unclear if this finding impacts purine/pyrimidine ratios. Interestingly, orotic acid induces lipogenesis in rat liver and in human hepatoma cell lines37. However, it is unclear if orotic acid has any impact on liver disease in individuals with UCDs.
The various therapeutic strategies used in UCDs impact the plasma metabolomic profile. For instance, metabolites of sodium benzoate and sodium or glycerol phenylbutyrate were readily detectable in plasma metabolomic profiles and may be useful markers for monitoring therapy adherence. Likewise, arginine supplementation in citrullinemia was associated with elevated argininate, a guanidino compound derived from arginine that is also markedly elevated in arginase deficiency. Thus, untargeted metabolomic profiling may have utility for optimizing arginine dose in citrullinemia and ASL deficiency if high doses of arginine are associated with elevations of guanidino compounds. Further metabolomics studies in individuals on various doses of arginine would demonstrate whether elevations in guanidino compounds are a consistent finding in individuals on high doses of this amino acid supplement. In addition, we hypothesize that the low BCAA and BCKA observed in arginase deficiency and citrullinemia are likely the consequence of protein restriction and the use of phenylbutyrate-containing medications18–20. The impact of chronically low BCAA and BCKA is unclear. However, at least two genetic forms of autism are associated with reduced BCAA transport in the brain38, 39, but it is unclear whether low circulating plasma BCAA levels impact the levels in cerebrospinal fluid or brain tissue.
The limitations of this study include the small sample sizes and lack of comprehensive clinical data, including data regarding sample collection, for subjects who were not prospectively enrolled. Future studies utilizing larger sample sizes and standard collection procedures are an important next step to demonstrate the potential utility of metabolomics in the clinical setting. One potential limitation of the current clinical metabolomic platform used in this study is the turnaround time (21 days), which is longer than other standard monitoring laboratory analyses used in UCDs, such as plasma amino acids. This longer turnaround time reflects the complex methodology and large number of metabolites analyzed. Perhaps, as technology advances, a shorter turnaround time will be feasible. A second limitation of the platform is the use of z-scores which are calculated using a reference population. As a result, z-scores for rare compounds, which are not present in the reference population, cannot be calculated. However, in many cases, the detection of these “rare molecules”, such as argininosuccinic acid, is diagnostic regardless of the absolute level.
Overall, our results demonstrate the potential utility of plasma metabolomics as a tool for monitoring the clinical management of arginase deficiency given that metabolomics facilitates the detection of potentially neurotoxic metabolites of arginine. Moreover, untargeted metabolomics facilitates the detection of off-target effects of medical management and arginine supplementation in UCDs and, in the future, may have utility in assessing the impact of new therapeutic strategies on the UCD metabolome. Importantly, metabolomics has the potential to provide insights into multiple metabolic pathway dysregulation observed in UCDs, thereby offering an opportunity for a broader and more integrated view of patient status beyond other modalities such as independent measures of orotic acid/orotidine, amino acids, and phenylbutyrate metabolite analysis. While not quantitative per se compared to these targeted assays, untargeted metabolomic profiling provides a holistic and integrated view that can be used in the context of screening and management of UCDs and other IEM.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alyssa Tran and Mercedes Alejandro for technical assistance, and Dr. William Wilson, Dr. Kirk Pappan and Jacob Wulff for helpful discussions. We thank the patients and families for their participation. LCB is supported by NIH K08DK106453 and holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. LA is supported by a training fellowship from the Gulf Coast Consortia, on the NLM Biomedical Informatics Training Program (Grant No. T15 LM007093). BS is supported by NIH T32DK007664–28 (BS) and the US Public Health Service grant P30DK56338, which funds the Texas Medical Center Digestive Disease Center. This work was supported by NIH R01 DK102641 (BL) and the Clinical Translational Core of Baylor College of Medicine (BCM) Intellectual and Developmental Disabilities Research Center (HD024064) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The Longitudinal Study of UCDs was supported by the Urea Cycle Disorders Consortium (UCDC) (U54HD061221). UCDC is a part of Rare Disease Clinical Research Network (RDCRN), an initiative of the Office of Rare Disease Research (ORDR), NCATS. This consortium is funded through collaboration between NCATS, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the NIH.
Footnotes
Competing interests
L.C.B., B M.S., Q.S., MJ.M., S.C.S.N., W.C., F.S., V.R.S., B.H.L., and S.H.E. are employees of Baylor College of Medicine, which has a partnership with Baylor Genetics and derives revenue from genetic testing. The other authors declare no conflict of interest.
REFERENCES
- [1].Jang YJ, LaBella AL, Feeney TP, Braverman N, Tuchman M, Morizono H, et al. Disease-causing mutations in the promoter and enhancer of the ornithine transcarbamylase gene. Hum Mutat. 2018;39:527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Haberle J, Boddaert N, Burlina A, Chakrapani A, Dixon M, Huemer M, et al. Suggested guidelines for the diagnosis and management of urea cycle disorders. Orphanet J Rare Dis. 2012;7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Miller MJ, Kennedy AD, Eckhart AD, Burrage LC, Wulff JE, Miller LA, et al. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. J Inherit Metab Dis. 2015;38:1029–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Coene KLM, Kluijtmans LAJ, van der Heeft E, Engelke UFH, de Boer S, Hoegen B, et al. Next-generation metabolic screening: targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients. J Inherit Metab Dis. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kennedy AD, Miller MJ, Beebe K, Wulff JE, Evans AM, Miller LA, et al. Metabolomic Profiling of Human Urine as a Screen for Multiple Inborn Errors of Metabolism. Genet Test Mol Biomarkers. 2016;20:485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kennedy AD, Pappan KL, Donti TR, Evans AM, Wulff JE, Miller LAD, et al. Elucidation of the complex metabolic profile of cerebrospinal fluid using an untargeted biochemical profiling assay. Mol Genet Metab. 2017;121:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Atwal PS, Donti TR, Cardon AL, Bacino CA, Sun Q, Emrick L, et al. Aromatic L-amino acid decarboxylase deficiency diagnosed by clinical metabolomic profiling of plasma. Mol Genet Metab. 2015;115:91–4. [DOI] [PubMed] [Google Scholar]
- [8].Cappuccio G, Pinelli M, Alagia M, Donti T, Day-Salvatore DL, Veggiotti P, et al. Biochemical phenotyping unravels novel metabolic abnormalities and potential biomarkers associated with treatment of GLUT1 deficiency with ketogenic diet. PLoS One. 2017;12:e0184022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sandlers Y, Mercier K, Pathmasiri W, Carlson J, McRitchie S, Sumner S, et al. Metabolomics Reveals New Mechanisms for Pathogenesis in Barth Syndrome and Introduces Novel Roles for Cardiolipin in Cellular Function. PLoS One. 2016;11:e0151802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Evans AMBB, Liu Q, Mitchell MW, Robinson RJ, Dai H, Stewart SJ, DeHaven CD, Miller LAD. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics. 2014;4:132. [Google Scholar]
- [11].Batshaw ML, Tuchman M, Summar M, Seminara J, Members of the Urea Cycle Disorders C. A longitudinal study of urea cycle disorders. Mol Genet Metab. 2014;113:127–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].G C, T N. The igraph software package for complex network research, . InterJournal Complex Systems. 2006:1695. [Google Scholar]
- [14].Burrage LC, Sun Q, Elsea SH, Jiang MM, Nagamani SC, Frankel AE, et al. Human recombinant arginase enzyme reduces plasma arginine in mouse models of arginase deficiency. Hum Mol Genet. 2015;24:6417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Diez-Fernandez C, Rufenacht V, Gemperle C, Fingerhut R, Haberle J. Mutations and common variants in the human arginase 1 (ARG1) gene: Impact on patients, diagnostics, and protein structure considerations. Hum Mutat. 2018;39:1029–50. [DOI] [PubMed] [Google Scholar]
- [16].Amayreh W, Meyer U, Das AM. Treatment of arginase deficiency revisited: guanidinoacetate as a therapeutic target and biomarker for therapeutic monitoring. Dev Med Child Neurol. 2014;56:1021–4. [DOI] [PubMed] [Google Scholar]
- [17].Nagamani SC, Shchelochkov OA, Mullins MA, Carter S, Lanpher BC, Sun Q, et al. A randomized controlled trial to evaluate the effects of high-dose versus low-dose of arginine therapy on hepatic function tests in argininosuccinic aciduria. Mol Genet Metab. 2012;107:315–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Burrage LC, Jain M, Gandolfo L, Lee BH, Members of the Urea Cycle Disorders C, Nagamani SC. Sodium phenylbutyrate decreases plasma branched-chain amino acids in patients with urea cycle disorders. Mol Genet Metab. 2014;113:131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Scaglia F, Carter S, O’Brien WE, Lee B. Effect of alternative pathway therapy on branched chain amino acid metabolism in urea cycle disorder patients. Mol Genet Metab. 2004;81 Suppl 1:S79–85. [DOI] [PubMed] [Google Scholar]
- [20].Brunetti-Pierri N, Lanpher B, Erez A, Ananieva EA, Islam M, Marini JC, et al. Phenylbutyrate therapy for maple syrup urine disease. Hum Mol Genet. 2011;20:631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamaguchi S, Brailey LL, Morizono H, Bale AE, Tuchman M. Mutations and polymorphisms in the human ornithine transcarbamylase (OTC) gene. Hum Mutat. 2006;27:626–32. [DOI] [PubMed] [Google Scholar]
- [22].Webster DR, Simmonds HA, Barry DM, Becroft DM. Pyrimidine and purine metabolites in ornithine carbamoyl transferase deficiency. J Inherit Metab Dis. 1981;4:27–31. [DOI] [PubMed] [Google Scholar]
- [23].Marescau B, Qureshi IA, De Deyn P, Letarte J, Ryba R, Lowenthal A. Guanidino compounds in plasma, urine and cerebrospinal fluid of hyperargininemic patients during therapy. Clin Chim Acta. 1985;146:21–7. [DOI] [PubMed] [Google Scholar]
- [24].Deignan JL, De Deyn PP, Cederbaum SD, Fuchshuber A, Roth B, Gsell W, et al. Guanidino compound levels in blood, cerebrospinal fluid, and post-mortem brain material of patients with argininemia. Mol Genet Metab. 2010;100 Suppl 1:S31–6. [DOI] [PubMed] [Google Scholar]
- [25].Stockler S, Isbrandt D, Hanefeld F, Schmidt B, von Figura K. Guanidinoacetate methyltransferase deficiency: the first inborn error of creatine metabolism in man. Am J Hum Genet. 1996;58:914–22. [PMC free article] [PubMed] [Google Scholar]
- [26].Neu A, Neuhoff H, Trube G, Fehr S, Ullrich K, Roeper J, et al. Activation of GABA(A) receptors by guanidinoacetate: a novel pathophysiological mechanism. Neurobiol Dis. 2002;11:298–307. [DOI] [PubMed] [Google Scholar]
- [27].Schulze A, Tran C, Levandovskiy V, Patel V, Cortez MA. Systemic availability of guanidinoacetate affects GABAA receptor function and seizure threshold in GAMT deficient mice. Amino Acids. 2016;48:2041–7. [DOI] [PubMed] [Google Scholar]
- [28].Zugno AI, Scherer EB, Schuck PF, Oliveira DL, Wofchuk S, Wannmacher CM, et al. Intrastriatal administration of guanidinoacetate inhibits Na+, K+-ATPase and creatine kinase activities in rat striatum. Metab Brain Dis. 2006;21:41–50. [DOI] [PubMed] [Google Scholar]
- [29].Mori A, Kohno M, Masumizu T, Noda Y, Packer L. Guanidino compounds generate reactive oxygen species. Biochem Mol Biol Int. 1996;40:135–43. [DOI] [PubMed] [Google Scholar]
- [30].Wyse AT, Bavaresco CS, Hagen ME, Delwing D, Wannmacher CM, Severo Dutra-Filho C, et al. In vitro stimulation of oxidative stress in cerebral cortex of rats by the guanidino compounds accumulating in hyperargininemia. Brain Res. 2001;923:50–7. [DOI] [PubMed] [Google Scholar]
- [31].da Silva CG, Parolo E, Streck EL, Wajner M, Wannmacher CM, Wyse AT. In vitro inhibition of Na+,K(+)-ATPase activity from rat cerebral cortex by guanidino compounds accumulating in hyperargininemia. Brain Res. 1999;838:78–84. [DOI] [PubMed] [Google Scholar]
- [32].Balz D, de Souza Wyse AT, Morsch VM, da Silva AC, Vieira VL, Morsch AL, et al. In vitro effects of L-arginine and guanidino compounds on NTPDase1 and 5’-nucleotidase activities from rat brain synaptosomes. Int J Dev Neurosci. 2003;21:75–82. [DOI] [PubMed] [Google Scholar]
- [33].Delwing-de Lima D, Wollinger LF, Casagrande AC, Delwing F, da Cruz JG, Wyse AT, et al. Guanidino compounds inhibit acetylcholinesterase and butyrylcholinesterase activities: effect neuroprotector of vitamins E plus C. Int J Dev Neurosci. 2010;28:465–73. [DOI] [PubMed] [Google Scholar]
- [34].Diez-Fernandez C, Rufenacht V, Gemperle C, Fingerhut R, Haberle J. Mutations and common variants in the human arginase 1 (ARG1) gene: impact on patients, diagnostic and protein structure considerations. Hum Mutat. 2018. [DOI] [PubMed] [Google Scholar]
- [35].Silva ES, Cardoso ML, Vilarinho L, Medina M, Barbot C, Martins E. Liver transplantation prevents progressive neurological impairment in argininemia. JIMD Rep. 2013;11:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Naylor EW, Cederbaum SD. Urinary pyrimidine excretion in arginase deficiency. J Inherit Metab Dis. 1981;4:207–10. [DOI] [PubMed] [Google Scholar]
- [37].Jung EJ, Kwon SW, Jung BH, Oh SH, Lee BH. Role of the AMPK/SREBP-1 pathway in the development of orotic acid-induced fatty liver. J Lipid Res. 2011;52:1617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tarlungeanu DC, Deliu E, Dotter CP, Kara M, Janiesch PC, Scalise M, et al. Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder. Cell. 2016;167:1481–94 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Novarino G, El-Fishawy P, Kayserili H, Meguid NA, Scott EM, Schroth J, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.