Abstract
Climate change is one of the most important threats to biodiversity and crop sustainability. The impact of climate change is often evaluated on the basis of expected changes in species' geographical distributions. Genomic diversity, local adaptation, and migration are seldom integrated into future species projections. Here, we examine how climate change will impact populations of two wild relatives of maize, the teosintes Zea mays ssp. mexicana and Z. mays ssp. parviglumis. Despite high levels of genetic diversity within populations and widespread future habitat suitability, we predict that climate change will alter patterns of local adaptation and decrease migration probabilities in more than two-thirds of present-day teosinte populations. These alterations are geographically heterogeneous and suggest that the possible impacts of climate change will vary considerably among populations. The population-specific effects of climate change are also evident in maize landraces, suggesting that climate change may result in maize landraces becoming maladapted to the climates in which they are currently cultivated. The predicted alterations to habitat distribution, migration potential, and patterns of local adaptation in wild and cultivated maize raise a red flag for the future of populations. The heterogeneous nature of predicted populations’ responses underscores that the selective impact of climate change may vary among populations and that this is affected by different processes, including past adaptation.
Keywords: global warming, local adaptation, migration, genomics, climate change
1. Introduction
Climate change is having detrimental effects on the geographical distribution, local abundance, and levels of genetic diversity of species [1–6] and is a major contributor to the current loss of biodiversity. In cultivated species, these impacts will have significant economic and alimentary repercussions at local and global scales [2,4,7]. The development of strategies to mitigate the impacts of climate change on cultivated species has been a high priority, with recent proposals highlighting the importance of genetic diversity and local adaptation of wild relatives for crop improvement and mitigation [4,8–12].
Cultivated landraces of maize (Zea mays ssp. mays) have had a close association with their wild relatives, the teosintes, throughout their evolutionary history. Maize was domesticated in Mexico from lowland teosinte (Zea mays spp. parviglumis) nearly 9000 years ago [13], with subsequent introgression from highland teosinte (Zea mays spp. mexicana) that allowed cultivated maize to grow at higher elevation environments, particularly in Central Mexico [14,15]. More than 200 Latin-American landraces of maize have been described, which are the result of past and ongoing sociocultural processes that shape and maintain extraordinary biodiversity in this species [16,17]. Teosintes grow wild in Mexico under a wide range of climatic conditions, from very hot and humid coastal environments to temperate and dry inland regions [10,18,19]. Maize landraces have an even wider geographical and environmental range [4,15,20]. Historical (as mentioned above) and current evidence of gene flow from teosintes into maize and the presence of fertile hybrids [21,22] indicates that gene introgression into landraces is possible. Such introgression could be important in enabling maize to continue to evolve in response to new environmental challenges [18,23].
Both the teosintes and several maize landraces are threatened by habitat loss and climate change [4,7,16,20,24], primarily increasing temperature and aridity [4,6]. Genomic analyses in teosintes have identified significant genetic differences between populations that are associated with varying local climates, suggesting local adaptation to contrasting environments (e.g. warm and dry climates) is common in teosintes [18,25–27]. Teosinte populations locally adapted to warm and dry environments are likely to contain alleles that could reduce the negative effects of global warming and increased aridity. Given that teosintes are not cultivated in Mexico, genetic variation should not have been directly affected by the cultural and agronomical practices influencing maize landraces [10,28]. Thus, patterns of gene–environment associations in teosinte populations might prove useful as a blueprint of local adaptation for crop improvement and mitigation in maize landraces.
Ecological niche modelling has become a powerful tool to assess the impacts of climate change on species' geographical distribution and persistence [29–31]. Commonly used ecological niche models assume that populations within a species are identical and share the same environmental tolerance limits [31–34], disregarding differences in the local adaptive response of populations to climate [35,36]. Given that local adaptation is common, among-population variation in climatic tolerances and varying levels of climate–gene relationships across the genome are expected [26,35]. This variation could have important consequences for population and species responses to a changing climate [32,34]. However, there are relatively few studies that have integrated adaptive divergence among subpopulations into ecological niche modelling to predict species’ response to climate change [34,35,37,38].
Here, we examined how climate change is expected to affect the geographical distribution and the frequency of single-nucleotide polymorphisms (SNPs) associated with climate-related traits in two teosinte subspecies: Z. mays ssp. mexicana and Z. mays ssp. parviglumis (hereafter referred to as mexicana and parviglumis, respectively). These teosintes thrive under distinct environmental conditions: mexicana is native to the dryer and colder highland regions of Central Mexico, whereas parviglumis is native to more tropical lowlands of southwestern Mexico [10,18,19,26].
2. Material and methods
(a) Ecological niche models
For each of the two teosintes, we built species distribution models using Maxent v.3.3.3 [39] and present-day climate data from the WorldClim [40] database. We used 254 (mexicana) and 329 (parviglumis) occurrence data points and the 19 bioclimatic variables available in WorldClim [40] at a resolution of 30 arc/sec (approx. 1 km2), with an area extent of 90°–120°W, 10°–30°N and background sampling points randomly chosen across the study area (electronic supplementary material). We examined predicted range shifts under four climate models: two general circulation models (Community Climate System Model (CCSM), Model for Interdisciplinary Research on Climate, (MIROC)) and two greenhouse gas concentration models adopted by the Intergovernmental Panel on Climate Change (Representative Conservation Pathways: RCP 4.5, RCP 8.5) at each of two future projections (2050, 2070). After validating the models, we generated binary presence/absence maps to quantify the degree of change in the geographical distribution of the two teosinte species (electronic supplementary material).
(b) Population genomic data
We merged the datasets of [25,26] (available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.8m648 [41] and https://doi.org/10.5061/dryad.tf556 [42]), both obtained using the MaizeSNP50 Illumina Bead chip, to construct a dataset of 33 454 high-quality single-base bi-allelic genetic variants, known as SNPs, distributed along the 10 chromosomes of maize [25,26]. The dataset comprises 23 populations for mexicana and 24 populations for parviglumis, covering the entire geographical and environmental distribution of teosintes [26]. The sets of locally adapted SNPs used here are described in detail by Aguirre-Liguori et al. [26], who used Bayescenv [43] and Bayenv [44] to identify SNPs with significant genetic differentiation among populations (FST) and significantly associated with temperature and precipitation (electronic supplementary material). SNPs that show genetic differentiation within the expected distribution of FST across the genome are presumably affected mainly by neutral processes (hereafter referred to as reference SNPs). On the contrary, the presence of SNPs with significantly high genetic differentiation is presumably affected by divergent selection enhancing allele frequency differences between populations. The SNPs identified through these methods bear statistical signatures consistent with local adaptation to climate. However, we caution that their functional importance has not been experimentally validated and statistical approaches to identify locally adapted genetic variants, such as those used here, are likely to include false positives and may include variants that differ due to non-adaptive processes.
Recent genomic scans in the two teosinte subspecies [26] identified 68 SNPs bearing a signal of local adaptation. We divided these SNPs into two groups based on the results of a discriminant analyses on principal components (DAPC) using climatically defined groups of populations and the 68 putatively locally adapted SNPs. The SNPs with the greatest power to discriminate among populations (the upper third quartile of the distribution of loadings) are hereafter referred to as putatively adaptive SNPs (paSNPs) and the others as candidate SNPs (canSNPs) (electronic supplementary material, figure S1). The DAPC was conducted for each subspecies separately using the adegenet package [45] in R [46] (electronic supplementary material). We estimated levels of genetic diversity (Hs) for reference SNPs within each population using the adegenet package [45] and hierfstat package [47] in R [46]. Genetically poor populations were defined as those with an Hs lower than 0.2 [25].
(c) Gradient Forest
Recently, species ecological distribution modelling techniques have been developed to consider intraspecific genetic variation in the assessment of the impact of climate change [31,35,37,38]. Among these is Gradient Forest [48], a regression tree-based approach that implements nonlinear regressions to estimate patterns of turnover in species (or allelic) composition as a function of environmental variables [35] (electronic supplementary material). The algorithm identifies regions along an environmental gradient associated with a high rate of change in allele frequencies, irrespective of the underlying allele frequencies. Gradient Forest then uses this information to construct allele turnover functions [35], which can be used to model the change in allele frequencies throughout a species’ range as a function of climatic variables.
To examine the potential effect of climate change on the genetic composition of local populations, we applied an extension of Gradient Forest [48] developed by Fitzpatrick & Keller [35] to predict how climate change will alter the frequencies of three sets of SNPs (see above). We used the gradientforest package [48] in R [46] to construct turnover functions for each SNP dataset (i.e. reference, candidate, putatively adaptive) with the 19 bioclimatic variables used for the species distribution models. We used these models to predict within-population genetic turnover throughout the teosintes' geographical range [35,48]. For each SNP dataset, we estimated the allelic frequencies for each subspecies and implemented Gradient Forest using 100 trees and a correlation threshold of 0.5 [35]. Based on the best allele turnover models for each set of SNPs, we constructed landscape models for the present and each of the eight future climate models using the raster package [49] in R [46].
By integrating the present and future allele turnover models, we predict the extent of observed allele frequency changes through time as a function of climate change [48], hereafter referred to as genomic offset [35,37,38]. This provides insights into the disruptive effect of climate change on the frequencies of alleles within contemporary populations. Geographical regions and populations with the greatest genomic offset are those in which climate change is predicted to have the greatest disruptive impact on current gene–environment relationships (electronic supplementary material).
To gauge the robustness of the Gradient Forest models to the use of different sets of SNPs, we ran additional analyses using: (i) a set of reference SNPs with an allele frequency distribution similar to those of candidate and putatively adaptive SNPs, (ii) different sets of outlier SNPs identified by Aguirre-Liguori et al. [26], and (iii) the complete set of 33 454 SNPs (electronic supplementary material).
(d) Allele distribution models
Since the climates of the future are predicted to be warmer, we were interested in modelling the distribution of the putatively warm-adapted alleles at each putatively adaptive SNP (electronic supplementary material). For this, we used Maxent v. 3.3.3 [39] to predict the future geographical distribution of warm-adapted alleles, using the corresponding populations’ geographical coordinates where these alleles were recorded as input. We used the same settings and validation procedures used for the species distribution models (see above). We generated binary presence/absence distribution models for each warm-adapted allele for the present and for future climate models (electronic supplementary material). We estimated the geographical overlap between the present model and each future model to define areas where at least five alleles are predicted to occur in the future, indicating favourable areas where the current gene–environment relationships will remain. We cross-validated the allele distribution models using the Gradient Forest models by inspecting the genomic offset of populations for three sets of regions: present-only, future-only, and overlap.
(e) Barriers to migration
Based on the joint distribution models for the putatively warm-adapted alleles, we approximated the potential capacity of populations to migrate into the new regions predicted under future climate scenarios using circuit theory [50]. This represents a simplified model of population migration based solely on a handful of alleles and assumes that population migration would be mostly limited by current local adaptation. We constructed maps of potential migration using the present and future distribution models for putatively warm-adapted alleles to determine landscape resistance (environmental distances) as a proxy of limitation to successful migration, where increasing resistance indicates decreasing probabilities of allele movement (electronic supplementary material). These maps of potential migration are based on the joint distribution of warm-adapted alleles, for which suitable areas were defined as those predicted with at least five alleles. For each sampled population, we constrained our analyses to a 1° × 1° grid-cell centred on that population. We used the 10-percentile of resistance values as a minimum threshold to estimate migration potential, which can be interpreted as the resistance to successful migration into at least 10% of the future areas of potential settlement (electronic supplementary material).
(f). Identifying putative adaptive SNPs in maize landraces
We explored whether the putative adapted SNPs identified for teosintes have been documented in particular landraces of maize [51]. For this, we downloaded the genotype data for maize described by Arteaga et al. [51], consisting of 36 931 SNPs across 46 landraces in Mexico. After identifying warm-adapted alleles across landraces, we estimated a relative ‘adaptive’ score for each landrace by adding up warm-adapted allele frequencies across all putative adapted SNPs (electronic supplementary material).
We evaluated the correlation between the adaptive scores and temperature, genomic offset and the geographical extension of each landrace (electronic supplementary material). For this, we employed simple linear regressions using the adaptive score as the response variable and climatic and geographical data for landraces as the predictors. We used the estimated genomic offset for the two teosintes under the less extreme climate change models (CCSM_2050_RCP4.5) to extract the mean genomic offset values from the geographical occurrences of each landrace. To assess the possibility of uncovering positive regressions between predictors and the adaptive score of landraces irrespective of their potential for adaptation, we calculated linear regressions using adaptive scores estimated from 1000 different sets of randomly sampled reference SNPs (13 per set).
3. Results and discussion
Ecological niche models can be used to predict suitable geographical ranges [29–34]. The predicted shifts and shrinkage in the teosintes' geographical range represent a potential threat to their persistence under climate change [20]. We employed ecological niche modelling and climate averages from 1960 to 1990 [40] to build geographical distribution models for the two teosinte subspecies under eight projections of climate change. As expected due to geographical constraints [6,20,52,53], the highland teosintes (mexicana) are predicted to be more susceptible to climate change than the lowland teosintes (parviglumis) [19,20]. These models predict that the potentially suitable geographical range for mexicana will be reduced by 30–39% in the next 30–50 years, whereas parviglumis range is predicted to be reduced by 4–9% (electronic supplementary material, figure S2 and table S1). Consistent with the predicted contraction in geographical range, 29 and 15% of populations of mexicana and parviglumis, respectively, are predicted to fall outside of suitable habitat by 2050 (45 and 29% by 2070). However, these models do not provide direct information on the potential effects that climate change will have on the genetic composition of local populations [36].
To have a more comprehensive assessment of the potential effects of climate change on wild teosintes, we predicted how climate change would alter the frequencies of three sets of SNPs (see above). The set of putatively adaptive SNPs (paSNPs) is comprised of nine parviglumis and eight mexicana SNPs with frequencies that differ strongly among populations living either in warm or cold environments, a signal that these loci might be contributing to local adaptation to climate. The majority of these SNPs are in or near genes involved in responses to heat, drought, and other environmental stress [26] (electronic supplementary material, table S2). The candidate SNPs (canSNPs) include 24 parviglumis and 25 mexicana SNPs that are also significantly associated with climate, but not as strongly as putatively adapted SNPs (electronic supplementary material). The reference SNPs were randomly selected from the 33 454 SNPs, and showed some, but not particularly strong, population differentiation (FST values within the 10 and 90% percentiles of the distribution of per-SNP population FST).
Our estimates of genomic offset based on Gradient Forest, in combination with projections of future climate change, allowed us to predict the temporal patterns of within-population genetic turnover throughout the teosintes’ geographical range [35,48]. The extent of genetic offset observed for the putatively adaptive and candidate SNPs, relative to reference SNPs, provides insights into the disruptive effect of climate change on patterns of gene–environment associations among contemporary populations. In this case, the higher genomic offset in several populations is indicative of stronger shifts in the frequencies of climate-associated SNPs than populations having a lower genomic offset.
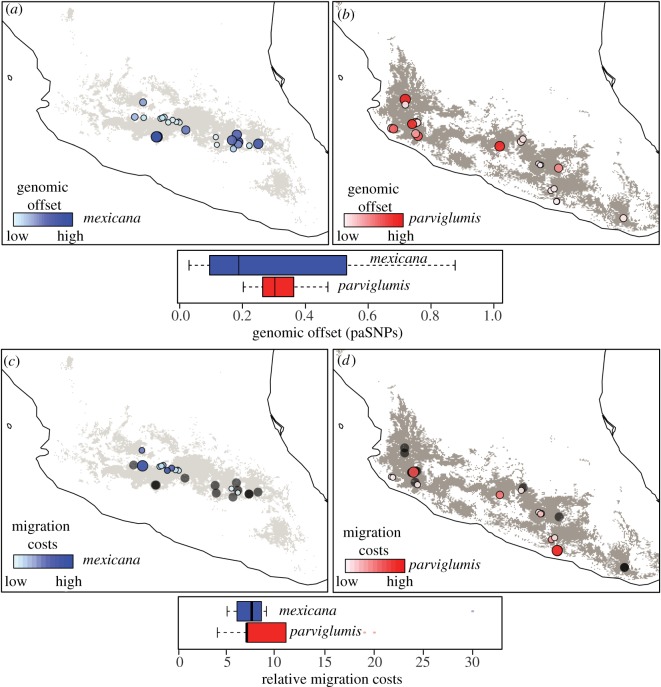
Although a more pronounced geographical contraction is predicted for mexicana than parviglumis, genomic offset at putatively adaptive and candidate SNPs is higher in parviglumis than in mexicana (mean = 0.27 versus 0.06 across all models, respectively; electronic supplementary material, tables S3–S5). The higher genome-wide offset in parviglumis (electronic supplementary material, figures S3 and S4) likely reflects greater population genetic structure (i.e. higher genetic differentiation among populations) in parviglumis than mexicana [26]. The higher genomic offset might also indicate that the genetic composition of parviglumis populations will be more affected by climate change than mexicana populations. The levels of genomic offset are, however, highly variable among populations (figure 1a,b), with genomic offset lower for populations either having higher frequencies (greater than 0.5) of the warm-adapted alleles or having little expected change in climatic conditions (electronic supplementary material, figure S5). The among-population variation underscores that both the selective impact of climate change and the genomic consequences of subsequent evolutionary response may both vary among populations and are affected by past local adaptation.
Figure 1.
Genomic offset in climate-related SNPs and limited migratory potential for sampled populations of two teosintes in Mexico: Zea mays spp. mexicana and Zea mays spp. parviglumis. (a–d) Distribution of genomic offset (a,b) and relative migration costs (c,d) for sampled populations of the two species of teosintes estimated under the climate change model CCSM_2050_RCP4.5. Light and dark grey areas represent the species distribution models for mexicana and parviglumis, respectively. Circle size is proportional to the estimated genomic offset and migration costs, with white circles representing populations with the lowest genomic offset and migration costs. Black circles in (c,d) represent populations with no migration potential (infinite migration costs) according to the allele distribution models. (Online version in colour.)
The considerable levels of genomic offset predicted in most populations have the potential to greatly alter patterns of local adaptation to climate, or at least the genetic basis of local adaptation. The genomic offset may also limit the adaptive potential of populations, thereby leading to reduced population sizes and possibly increase the probabilities of local extinction [3,26,31,37,38,52]. This points towards more complex impacts of climate change on teosinte populations than those predicted using geographical distribution models alone [20,32,34,36].
To complement the use of Gradient Forest, we used the same modelling tools employed for species distributions to predict the present and future geographical distribution of the putatively warm-adapted alleles (electronic supplementary material). This modelling showed that by 2050, the predicted geographical distribution of most of these warm-adapted alleles is expected to contract by approximately 45% in mexicana and by 36% parviglumis (electronic supplementary material, table S6). The geographical areas not suitable for the warm-adapted alleles in the future also show higher levels of genomic offset (electronic supplementary material, figure S6), indicating that areas outside the predicted distribution of alleles are expected to have big shifts in climate–gene relationships, either due to a lower starting allele frequency or to a larger change in climate.
The putatively warm-adapted alleles are expected to confer selective advantages to expanding warmer and drier environments, yet populations lacking these alleles might follow a different evolutionary trajectory to adapt to climate change. In this context, standing ‘neutral’ genetic diversity or new mutations could act as an important source of variation for future local adaptation [1,5,54,55] through the generation of novel allele combinations serving as the basis of future adaptive evolution. We found a significantly negative association between genetic diversity at reference SNPs (Hs) and the predicted genomic offset for putatively adaptive SNPs (electronic supplementary material, figure S7). These results suggest that, on average, genetically rich populations will be less vulnerable to climate change, probably because the adaptive potential of populations is maximized with intermediate allele frequencies. This result is consistent with the relevance usually given to standing genetic variation to allow responding to a changing climate [54,55]. However, more than two-thirds of teosinte populations (71% in parviglumis and 91% in mexicana) show high levels of genetic diversity (Hs greater than 0.2, [25]), yet even under the most conservative climate model, many of these populations will experience high levels of genomic offset (figure 1a,b).
While some populations might adapt to the conditions of a changing climate, other populations might migrate to track the climatic conditions to which they are currently adapted [52,53]. With a focus on the alleles with major associations with current climatic conditions, we evaluated the potential for sampled populations to migrate to locations with future climates that are similar to the climates they experience today. To do this, we used the distribution models for putative warm-adapted alleles to estimate environmental costs [50,56] (i.e. lower probabilities of populations successfully migrating) along all potential migration routes radiating from each sampled population.
Depending on the climate model, as the climate changes, 46–54% of parviglumis populations and 22–52% of mexicana populations are expected to have the possibility of migrating into sites in which the alleles of major effect will remain advantageous (electronic supplementary material, table S7). Although more mexicana populations are predicted to have no migration capabilities (i.e. infinite migration costs) into climatically suitable sites, probably due to isolation from areas predicted to have suitable conditions in the future, the remaining populations of mexicana actually showed lower migration costs than populations of parviglumis (figure 1c,d). Thus, although migration costs are expected to be higher for parviglumis than for mexicana, populations living in the Central Highlands of Mexico are expected to have the highest migratory constraints (figure 1c). The variation in migration costs might result from the differing rates of estimated temperature change between highland environments compared to lowland regions [53]. The predicted likelihood of successful migration into a climatically suitable location was positively correlated with within-population frequency of warm-adapted alleles (electronic supplementary material, figure S8).
Under the most conservative model of climate change (CCSM_4.5_2050), our species distribution models predicted 88 and 84% of sampled populations within future suitable areas for parviglumis and mexicana, respectively (electronic supplementary material, table S1). In addition, most of these populations exhibited high levels of genetic diversity [26]. However, the heterogeneous geographical patterns of genomic offset and migration potential suggest that the impacts of climate change will vary considerably among populations. Overall, eastern populations of mexicana and western populations of parviglumis showed high levels of genomic offset and lower migration potential (figure 1), thus we expect that these populations will experience stronger pressures to adapt to climate change than other teosinte populations. From this basic comparison, we conclude that integrating ecological niche modelling and genomic data is an important step forward to better understand the impacts that climate change will have on natural populations.
Locally adapted populations of teosintes can be used as a source of advantageous genetic diversity for landraces of maize [4,19,23], through the introduction of ‘new wild alleles’ into cultivated populations [8,9,11,12]. Experimental evidence has proven that teosintes germplasm can be successfully transferred into maize and can increase crop yields and drought resistance [16,17,22,57]. Alternatively, maize improvement under warmer climates can be achieved more easily by selecting warm-adapted alleles already present in maize landraces. Thus, we estimated the frequency of putatively warm-adapted alleles identified for teosintes in 46 Mexican landraces of maize [51] (electronic supplementary material, table S8).
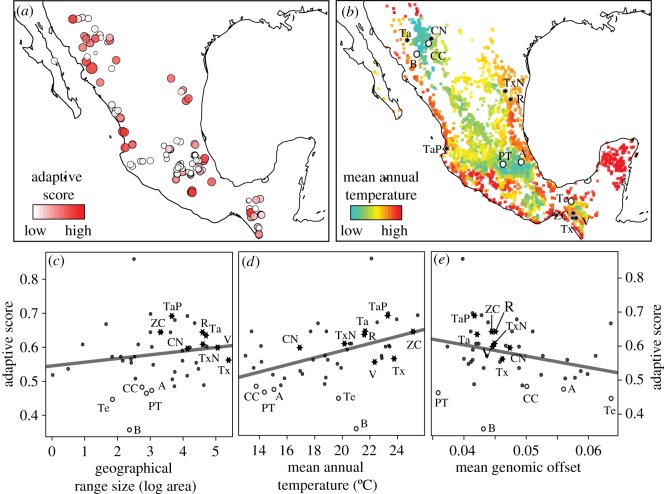
We found an uneven presence of warm-adapted alleles across distinct maize landraces in Mexico (figure 2a), with a positive trend of increasing allele frequency in maize landraces with larger geographical ranges and higher mean annual temperatures (figure 2c,d) (β = 0.01, F = 17.7, d.f. = 45, p < 0.001, R2 = 0.27). Although the positive relationship between the frequency of adaptive alleles and range size was non-significant, several landraces with narrow distribution ranges, such as the almost extinct ‘Tehua’ landrace, and other highly restricted landraces [16] (depicted as open circles in figure 2b–e), showed warm-adapted alleles at fewer loci and at lower frequencies. In turn, we found a negative relationship between adaptive scores and the genomic offset (figure 2e). However, the relationship between warm-adapted allele frequencies and both temperature and genomic offset were greater than those estimated between reference SNPs and these factors, for all but 0.5% of the subsamples (electronic supplementary material, figure S9).
Figure 2.
Adaptive scores and genomic offset in climate-related SNPs among landraces of maize accessions in Mexico. (a) Distribution of adaptive scores for accessions of 46 maize landraces in Mexico estimated from the climate-SNP models for the two teosinte species under the climate change model CCSM_2050_RCP4.5. Circle size is proportional to the estimated adaptive scores, with white circles representing accessions with the low adaptive scores. (b) Known geographical occurrences for the 46 maize landraces in Mexico coloured by the mean annual temperature. Black stars represent accessions for landraces with high adaptive scores that have been used for maize improvement and white circles represent accessions for restricted landraces with low adaptive potential. (c–e) Association of adaptive scores versus the geographical range size, mean annual temperature, and genomic offset estimated for 46 maize landraces. Regression coefficients for range size: β = 0.02, F = 1.1, d.f. = 45, p = 0.31; for temperature: β = 0.01, F = 17.7, d.f. = 45, p < 0.001, R2 = 0.27; for genomic offset: β = −3.31, F = 3.76, d.f. = 45, p = 0.06. ‘Conico Norteño’ (CN), ‘Ratón’ (R), ‘Tabloncillo’ (Ta), ‘Tabloncillo Perla’ (TaP), ‘Tuxpeño’ (Tx), ‘Tuxpeño Norteño’ (TxN), ‘Vandeño’ (V), ‘Zapolote Chico’ (ZC), ‘Arrocillo’ (A), ‘Bofo’ (B), ‘Cristalino de Chihuahua’ (CC), ‘Palomero Toluqueño’ (PT), and ‘Tehua’ (Te). (Online version in colour.)
The statistically significant relationships between warm-adapted alleles identified in teosintes and the climatic environment in which maize landraces are grown, could result from past introgression of adaptive genes into maize via gene flow from teosintes [14,15,18,23,58]. However, hybridization between maize and teosintes appears to be limited to areas of sympatry [23,58] and thus, it seem more likely that the higher adaptive scores for landraces growing in warmer climates reflect past adaptation to these conditions [4]. The non-significant regression observed for genomic offset (figure 2e) may partially result from the transferability of models of teosinte into maize or the quantitative nature of phenotypic traits associated with adaptation to climate [59–62]. Nonetheless, to the extent that the warm-adapted alleles identified in teosinte contribute to local adaptation in maize, our results suggest that climate change will alter the genetic basis of local adaptation in maize landraces. This disruption might result in maize landraces becoming locally maladapted to the climatic conditions in which they have been grown historically [4]. Such maladaptation may lead to yield declines in many of these landraces as temperatures increase [7].
Based on our analyses, we identified several maize landraces (depicted as black stars in figure 2b–e) with moderate to high adaptive scores and known to be drought resistant [16] (electronic supplementary material, table S9). Indeed, several of these landraces have been used to improve maize lines with enhanced heat and drought resistance, with increased yields [16,57]. We acknowledge that adaptation to climate very likely involves quantitative traits [54,59–62], yet identifying alleles with small effects on polygenic traits remains a major challenge [62], especially in species with complex genetic structures, such as teosintes. Nonetheless, the putatively adaptive and candidate SNPs we identified are promising candidates to test the response of maize and teosintes to changing climate under experimental settings. The convergence between the adaptive score of landraces, together with historical agronomical practices, opens up the opportunity to fully integrate the vast cultural knowledge on maize landraces with analyses of genomic data in an ecological context.
In an era in which population-level genomic data are accessible for an increasing number of non-model species, approaches integrating genomic and environmental data provide powerful tools to better understand the impacts of climate change on natural populations [5,35–38]. Predictions of the impact of climate change on species based solely on projected shifts in species' ranges, although valuable, only provide partial information [30–34]. By building spatio-temporal models of climate–gene relationships, we show that the future of wild populations of teosintes and possibly of several landraces of maize appears to be much harsher than previously estimated [20].
Although it is tempting to directly relate our results to future declines in population fitness [38], the association between climate and fitness should be properly validated using experimental or at least simulation approaches [37,63]. Nevertheless, the fact that future climate change predicts significant alterations to habitat distribution, migration potential, and patterns of local adaptation raises a red flag for the future of wild and cultivated populations of maize.
Our results show the relevance of moving beyond the standard species distribution models to assess climate change impacts [30–38] and prove how the analysis of genomic data can identify important genetic resources to aid wildlife conservation and crop sustainability under a rapidly changing climate. However, our approach remains an oversimplification of the complex evolutionary and ecological processes affecting populations [36,52,54,62,63]. This underscores the need for continued integration of agronomical practices, genomic data, and climate models to better understand the impacts of a rapidly changing climate on cultivated and wild species.
Supplementary Material
Acknowledgements
We thank Eria A. Rebollar, Erika Aguirre Planter, and Laura Espinosa Asuar for technical and logistic support during this project. This article was written during a sabbatical leave of L.E.E. in the Department of Plant and Microbial Biology at the University of Minnesota, with support from the program PASPA-DGAPA, UNAM.
Data accessibility
Genomic data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.8m648 [41], https://doi.org/10.5061/dryad.tf556 [42] and https://doi.org/10.5061/dryad.4t20n [64]. The list of candidate SNPs at https://doi.org/10.1111/mec.14203. Geographical occurrences of teosintes and maize landraces at http://www.biodiversidad.gob.mx. R data and code at https://github.com/spiritu-santi/teosintes.
Authors' contributions
J.A.A.-L., S.R.-B., and L.E.E. conceived and designed the framework for the analyses. J.A.A.-L. and S.R.-B. collected the data, conducted analyses, and drafted the manuscript. L.E.E. secured the funding. All authors discussed results and wrote the manuscript.
Competing interests
The authors declare no competing financial interests.
Funding
This study was funded by CONACYT Investigación Científica Básica CB2011-167826 and SEP-CONACYT-ANUIES-ECOS France M12-A03-CONACYT-ANUIES 207571 granted to L.E.E.; and postdoctoral fellowship from DGAPA-UNAM granted to S.R.-B.
References
- 1.Allendorf FW, Hohenlohe PA, Luikart G. 2010. Genomics and the future of conservation genetics. Nat. Rev. Genet. 11, 697–709. ( 10.1038/nrg2844) [DOI] [PubMed] [Google Scholar]
- 2.Dai A. 2012. Increasing drought under global warming in observations and models. Nat. Clim. Change 1633, 52–57. [Google Scholar]
- 3.Pauls SU, Nowak C, Bálint M, Pfenninger M. 2012. The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 22, 925–946. ( 10.1111/mec.12152) [DOI] [PubMed] [Google Scholar]
- 4.Hellin J, Bellon MR, Hearne SJ. 2014. Maize landraces and adaptation to climate change in Mexico. J. Crop Improv. 28, 482–501. ( 10.1080/15427528.2014.921800) [DOI] [Google Scholar]
- 5.Schafer ABA, et al. 2015. Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 30, 78–87. ( 10.1016/j.tree.2014.11.009) [DOI] [PubMed] [Google Scholar]
- 6.Pacifici M, et al. 2017. Species’ traits influenced their response to recent climate change. Nat. Clim. Change 3223, 1–4. [Google Scholar]
- 7.Tigcheelar M, Battisti DS, Naylor RL, Ray DK. 2018. Future warming increases probability of globally synchronized maize production shocks. Proc. Natl Acad. Sci. USA 115, 6644–6649. ( 10.1073/pnas.1718031115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmgren MG, et al. 2014. Are we ready for back-to-nature crop breeding? Trends Plant Sci. 20, 155–164. ( 10.1016/j.tplants.2014.11.003) [DOI] [PubMed] [Google Scholar]
- 9.Warschefsky E, Penmetsa RV, Cook DR, Wettberg EJ. 2014. Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am. J. Bot. 101, 1791–1800. ( 10.3732/ajb.1400116) [DOI] [PubMed] [Google Scholar]
- 10.Aguirre-Liguori JA, Aguirre-Planter E, Eguiarte LE. 2016. Genetics and ecology of wild and cultivated maize: domestication and introgression. In Ethnobotany of Mexico (eds R Lira, A Casas, J Blancas), pp. 403–416. New York, NY: Springer. [Google Scholar]
- 11.Zhang H, Mittal N, Leamy LJ, Barazani O, Song BH. 2016. Back into the wild—apply untapped genetic diversity of wild relatives for crop improvement. Evol. Appl. 10, 5–24. ( 10.1111/eva.12434) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dempewolf H, et al. 2017. Past and future use of wild relatives in crop breeding. Crop Sci. 57, 1070–1082. ( 10.2135/cropsci2016.10.0885) [DOI] [Google Scholar]
- 13.Matsuoka Y, et al. 2002. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl Acad. Sci. USA 99, 6080–6084. ( 10.1073/pnas.052125199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hufford MB, Lubinksy P, Pyhäjärvi T, Devengenzo MT, Ellstrand NC, Ross-Ibarra J. 2013. The genomic signature of crop-wild introgression in maize. PLoS Genet. 9, e1003477 ( 10.1371/journal.pgen.1003477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Heerwaarden J, et al. 2011. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl Acad. Sci. USA 108, 1088–1092. ( 10.1073/pnas.1013011108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CONABIO. 2011. Base de datos del proyecto global ‘Recopilación, generación, actualización y análisis de información acerca de la diversidad genética de maíces y sus parientes silvestres en méxico’. México, DF: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad. [Google Scholar]
- 17.Sánchez González JJ. 2011. Diversidad del maíz y teocintle. Informe preparado para el proyecto global ‘Recopilación, generación, actualización y análisis de información acerca de la diversidad genética de maíces y sus parientes silvestres en México’ de la Comisión Nacional para el Conocimiento y Uso de la Biodiversidad. México, DF: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad. [Google Scholar]
- 18.Hufford MB, Martínez-Meyer E, Gaut BS, Eguiarte LE, Tenaillon MI. 2012. Inferences from the historical distribution of wild and domesticated maize provide ecological and evolutionary insight. PLoS ONE 11, e47659 ( 10.1371/journal.pone.0047659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González JJS, et al. 2018. Ecogeography of teosinte. PLoS ONE 13, e0192676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ureta C, Martínez-Meyer E, Perales HR, Álvarez-Buylla ER. 2012. Projecting the effects of climate change on the distribution of maize races and their wild relatives in Mexico. Glob. Change Biol. 18, 1073–1108. ( 10.1111/j.1365-2486.2011.02607.x) [DOI] [Google Scholar]
- 21.Baltazar BM, Sanchez-Gonzalez JJ, de la Cruz-Larios L, Schoper J B. 2005. Pollination between maize and teosinte: an important determinant of gene flow in Mexico. Theor. Appl. Genet. 110, 519–526. ( 10.1007/s00122-004-1859-6) [DOI] [PubMed] [Google Scholar]
- 22.Kato TAY, Sánchez JJG. 2002. Introgression of chromosome knobs from Zea diploperennis into maize. Maydica 47, 33–50. [Google Scholar]
- 23.Warburton ML, et al. 2011. Gene flow among different teosinte taxa and into the domesticated maize gene pool. Genet. Resour. Crop Evol. 58, 1243–1261. ( 10.1007/s10722-010-9658-1) [DOI] [Google Scholar]
- 24.Wilkes HG. 2006. Urgent notice to all maize researchers: disappearance and extinction of the last wild teosinte populations is more than half completed. A modest proposal for teosinte evolution and conservation in situ: the Balsas, Guerrero, Mexico. Maydica 52, 49–58. [Google Scholar]
- 25.Pyhäjärvi T, Hufford MB, Mezmouk S, Ross-Ibarra J. 2013. Complex patterns of local adaptation in teosinte. Genome Biol. Evol. 5, 1594–1609. ( 10.1093/gbe/evt109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguirre-Liguori JA, et al. 2017. Connecting genomic patterns of local adaptation and niche suitability in teosintes. Mol. Ecol. 26, 4226–4240. ( 10.1111/mec.14203) [DOI] [PubMed] [Google Scholar]
- 27.Fustier M-A, et al. 2017. Signatures of local adaptation in lowland and highland teosintes from whole-genome sequencing of pooled samples. Mol. Ecol. 26, 2738–2756. ( 10.1111/mec.14082) [DOI] [PubMed] [Google Scholar]
- 28.Hufford MB, Bilinski P, Pyhäjärvi T, Ross-Ibarra J. 2012. Teosinte as a model system for population and ecological genomics. Trends Genet. 28, 606–615. ( 10.1016/j.tig.2012.08.004) [DOI] [PubMed] [Google Scholar]
- 29.Peterson AT, et al. 2011. Ecological niches and geographic distributions. Princeton, NJ: Princeton University Press. [Google Scholar]
- 30.Pagel J, Schurr FM. 2012. Forecasting species ranges by statistical estimation of ecological niches and spatial populations dynamics. Glob. Ecol. Biogeogr. 21, 293–304. ( 10.1111/j.1466-8238.2011.00663.x) [DOI] [Google Scholar]
- 31.Gotelli NJ, Stanton-Geddes J. 2015. Climate change, genetic markers and species distribution modelling. J. Biogeogr. 42, 1577–1585. ( 10.1111/jbi.12562) [DOI] [Google Scholar]
- 32.Pelini S, et al. 2009. Translocation experiments with butterflies reveal limits to enhancement of poleward populations under climate change. Proc. Natl Acad. Sci. USA 106, 11 160–11 165. ( 10.1073/pnas.0900284106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urban MC, et al. 2016. Improving the forecast for biodiversity under climate change. Science 353, aad8466 ( 10.1126/science.aad8466) [DOI] [PubMed] [Google Scholar]
- 34.Hällfors MH, et al. 2016. Addressing potential local adaptation in species distribution models: implications for conservation under climate change. Ecol. Appl. 26, 1154–1169. ( 10.1890/15-0926) [DOI] [PubMed] [Google Scholar]
- 35.Fitzpatrick MC, Keller SR. 2015. Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 18, 1–16. ( 10.1111/ele.12376) [DOI] [PubMed] [Google Scholar]
- 36.Peterson ML, Doak DF, Morris WF. 2019. Incorporating local adaptation into forecasts of species' distribution and abundance under climate change. Global Change Biol. 25, 775–793. ( 10.1111/gcb.14562) [DOI] [PubMed] [Google Scholar]
- 37.Exposito-Alonso M, et al. 2017. Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana. Nat. Ecol. Evol. 2, 352–358. ( 10.1038/s41559-017-0423-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bay RA, et al. 2018. Genomic signals of selection predict climate-driven populations declines in a migratory bird. Science 359, 83–86. ( 10.1126/science.aan4380) [DOI] [PubMed] [Google Scholar]
- 39.Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. ( 10.1016/j.ecolmodel.2005.03.026) [DOI] [Google Scholar]
- 40.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high-resolution interpolated climate surfaces for global land areas. Int. J. Climatology 25, 1965–1978. ( 10.1002/joc.1276) [DOI] [Google Scholar]
- 41.Pyhäjärvi T, Hufford MB, Mezmouk S, Ross-Ibarra J. 2013. Data from: Complex patterns of local adaptation in Teosinte Dryad Digital Repository. ( 10.5061/dryad.8m648) [DOI] [PMC free article] [PubMed]
- 42.Aguirre-Liguori J, Tenaillon M, Vázquez-Lobo A, Gaut B, Jaramillo-Correa J, Montes-Hernandez S, Souza V, Eguiarte L. 2017. Data from: Connecting genomic patterns of local adaptation and niche suitability in teosintes Dryad Digital Repository. ( 10.5061/dryad.tf556) [DOI] [PubMed]
- 43.Villemereuil P, Gaggiotti OE. 2015. A new FST-based method to uncover local adaptation using environmental variables. Methods Ecol. Evol. 6, 1248–1258. ( 10.1111/2041-210X.12418) [DOI] [Google Scholar]
- 44.Coop G, Witonsky D, Di Rienzo A, Pritchard JK. 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185, 1411–1423. ( 10.1534/genetics.110.114819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jombart T. 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. ( 10.1093/bioinformatics/btn129) [DOI] [PubMed] [Google Scholar]
- 46.Core Team R. 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 47.Goudet J. 2005. Hierfstat, a package for R to compute and test variance components and F-statistics. Mol. Ecol. Notes 5, 184–186. ( 10.1111/j.1471-8286.2004.00828.x) [DOI] [Google Scholar]
- 48.Ellis N, Smith SJ, Pitcher CR. 2012. Calculating importance gradients on physical predictors. Ecology 93, 156–168. ( 10.1890/11-0252.1) [DOI] [PubMed] [Google Scholar]
- 49.Hijmans RJ.2017. raster: geographic data analysis and modeling. R package version 2.6-7. See https://CRAN.R-project.org/package=raster .
- 50.McRae BH, Dickson BG, Keitt T. 2008. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 89, 2712–2724. ( 10.1890/07-1861.1) [DOI] [PubMed] [Google Scholar]
- 51.Arteaga MC, et al. 2016. Genomic variation in recently collected maize landraces from Mexico. Genomics Data 7, 38–45. ( 10.1016/j.gdata.2015.11.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann AA, Sgrò CM. 2011. Climate change and evolutionary adaptation. Nature 470, 479–485. ( 10.1038/nature09670) [DOI] [PubMed] [Google Scholar]
- 53.Loarie SR, et al. 2009. The velocity of climate change. Nature 462, 1052–1055. ( 10.1038/nature08649) [DOI] [PubMed] [Google Scholar]
- 54.Siol M, Wright SI, Barrett SC. 2010. The population genomics of plant adaptation. New Phytol. 188, 313–332. ( 10.1111/j.1469-8137.2010.03401.x) [DOI] [PubMed] [Google Scholar]
- 55.Frankham R. 2010. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 143, 1919–1927. ( 10.1016/j.biocon.2010.05.011) [DOI] [Google Scholar]
- 56.Manel S, Schwartz MK, Luikart G, Taberlet P. 2003. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18, 189–197. ( 10.1016/S0169-5347(03)00008-9) [DOI] [Google Scholar]
- 57.Wang L, Yang A, He C, Qu M, Zhang J. 2008. Creation of new maize germplasm using alien introgression from Zea mays ssp. mexicana. Euphytica 164, 789–801. ( 10.1007/s10681-008-9730-5) [DOI] [Google Scholar]
- 58.Moreno-Letelier A, et al. 2017. Was maize domesticated in the Balsas Basin? Complex patterns of genetic divergence, gene flow and ancestral introgressions among Zea subspecies suggest an alternative scenario. bioRxiv 239707. [Google Scholar]
- 59.Yoder JB, et al. 2014. Genomic signature of adaptation to climate in Medicago truncatula. Genetics 196, 1263–1275. ( 10.1534/genetics.113.159319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiffin P, Ross-Ibarra J. 2014. Advances and limits of using population genetics to understand local adaptation. Trends Ecol. Evol. 29, 673–680. ( 10.1016/j.tree.2014.10.004) [DOI] [PubMed] [Google Scholar]
- 61.Kawecki TJ, Ebert D. 2004. Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241. ( 10.1111/j.1461-0248.2004.00684.x) [DOI] [Google Scholar]
- 62.Yeaman S. 2015. Local adaptation by alleles of small effect. Am. Nat. 186, S74–S89. ( 10.1086/682405) [DOI] [PubMed] [Google Scholar]
- 63.Fitzpatrick MC, Keller SR, Lotterhos KE. 2018. Comment on ‘Genomic signals of selection predict climate-driven population declines in a migratory bird’. Science 361, eaat7279. [DOI] [PubMed] [Google Scholar]
- 64.Arteaga MC, Moreno-Letelier A, Mastretta-Yanes A, Vázquez-Lobo A, Breña-Ochoa A, Moreno-Estrada A, Eguiarte LE, Piñero D. 2015. Data from: Genomic variation in recently collected maize landraces from Mexico Dryad Digital Repository. ( 10.5061/dryad.4t20n) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Pyhäjärvi T, Hufford MB, Mezmouk S, Ross-Ibarra J. 2013. Data from: Complex patterns of local adaptation in Teosinte Dryad Digital Repository. ( 10.5061/dryad.8m648) [DOI] [PMC free article] [PubMed]
- Aguirre-Liguori J, Tenaillon M, Vázquez-Lobo A, Gaut B, Jaramillo-Correa J, Montes-Hernandez S, Souza V, Eguiarte L. 2017. Data from: Connecting genomic patterns of local adaptation and niche suitability in teosintes Dryad Digital Repository. ( 10.5061/dryad.tf556) [DOI] [PubMed]
- Arteaga MC, Moreno-Letelier A, Mastretta-Yanes A, Vázquez-Lobo A, Breña-Ochoa A, Moreno-Estrada A, Eguiarte LE, Piñero D. 2015. Data from: Genomic variation in recently collected maize landraces from Mexico Dryad Digital Repository. ( 10.5061/dryad.4t20n) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Genomic data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.8m648 [41], https://doi.org/10.5061/dryad.tf556 [42] and https://doi.org/10.5061/dryad.4t20n [64]. The list of candidate SNPs at https://doi.org/10.1111/mec.14203. Geographical occurrences of teosintes and maize landraces at http://www.biodiversidad.gob.mx. R data and code at https://github.com/spiritu-santi/teosintes.