Abstract
Functional variation in rhodopsin, the dim-light-specialized visual pigment, frequently occurs in species inhabiting light-limited environments. Variation in visual function can arise through two processes: relaxation of selection or adaptive evolution improving photon detection in a given environment. Here, we investigate the molecular evolution of rhodopsin in Gymnotiformes, an order of mostly nocturnal South American fishes that evolved sophisticated electrosensory capabilities. Our initial sequencing revealed a mutation associated with visual disease in humans. As these fishes are thought to have poor vision, this would be consistent with a possible sensory trade-off between the visual system and a novel electrosensory system. To investigate this, we surveyed rhodopsin from 147 gymnotiform species, spanning the order, and analysed patterns of molecular evolution. In contrast with our expectation, we detected strong selective constraint in gymnotiform rhodopsin, with rates of non-synonymous to synonymous substitutions lower in gymnotiforms than in other vertebrate lineages. In addition, we found evidence for positive selection on the branch leading to gymnotiforms and on a branch leading to a clade of deep-channel specialized gymnotiform species. We also found evidence that deleterious effects of a human disease-associated substitution are likely to be masked by epistatic substitutions at nearby sites. Our results suggest that rhodopsin remains an important component of the gymnotiform sensory system alongside electrolocation, and that photosensitivity of rhodopsin is well adapted for vision in dim-light environments.
Keywords: rhodopsin, Gymnotiformes, likelihood-based codon models, clade models of molecular evolution, evolution of fish vision, positive selection
1. Introduction
Animals rely on information from multiple sensory systems to navigate, avoid predators and find mates. However, sensory systems are generally energetically expensive [1], and suboptimal sensory systems are rapidly reduced and eventually lost [2]. For example, the transition from aquatic to terrestrial environments resulted in the loss of the lateral line and electrosensory organs of many fishes because neither sensory system is effective out of water [3]. Also, regressive evolution of eyes is observed in vertebrate lineages inhabiting lightless caves and deep-sea habitats [2,4].
However, regressive evolution in one sensory modality can be correlated with adaptive evolution in an alternative sensory system better suited for the environment [5], an evolutionary pattern described as a sensory trade-off [6]. Reduced visual performance in bats and cetaceans is associated with the evolution of echolocation [7–10], and organisms as diverse as mammals [6] and Drosophila [11] exhibit trade-offs between olfactory and visual systems. Molecular genetic evidence for sensory trade-offs has been reported in bats, the naked mole-rat, the star-nosed mole and the blind Mexican cave fish. In these taxa, the molecular evolution of vision genes is relaxed coincident with an expansion of auditory, mechanosensory or gustatory sensory systems [7,9,10,12,13]. However, interpretation of trade-offs is not always straightforward [7], and in some taxa, such as vipers with infrared-detecting pit organs, trade-offs with vision are not observed [14].
The electric knifefishes of South and Central America (Order Gymnotiformes) are capable of active electrolocation. This sensory system involves the production of stereotypical electric fields by specialized electric organs made up of modified muscle or neuronal tissue [15]. Disturbances in the weak self-generated fields are detected by specialized high-frequency-sensitive tuberous electroreceptors and interpreted by the brain [15]. Active electrolocation allows navigation in the absence of light, and gymnotiforms are mostly nocturnally active and often live in turbid and tannin-stained rivers [16,17]. Gymnotiforms have small eyes and are thought to have poor vision [18]. This suggests a possible sensory trade-off between vision and active electrolocation, with electrolocation providing a clear advantage in highly obscured river environments where downwelling light is rapidly attenuated, especially at night.
Rhodopsin is the key visual pigment for dim-light vision. Because of this, we expected rhodopsin to be the visual pigment most likely to be involved in a trade-off with electrolocation, an adaptation for perception in low light. Rhodopsin mediates the first step in the visual transduction cascade within the rod photoreceptor cells of the eye. Rod photoreceptors show specializations for dim-light activation, including high photosensitivity and signal amplification [19]. Rhodopsin activation has an extremely high signal-to-noise ratio and exhibits highly efficient activation of downstream second messenger molecules [20]. These properties appear to place rhodopsin under considerable functional constraint [21]. In humans, mutations in rhodopsin can cause genetic degenerative eye diseases such as retinitis pigmentosa (RP) [22]. Rhodopsin has long served as a model system for studies of the molecular evolution of sensory systems, and also for investigations of the broader superfamily of pharmacologically important G protein-coupled receptors (GPCRs), resulting in a wealth of related resources, including large disease databases, sequence databases and crystal structures, as well as numerous in vitro mutagenesis studies of rhodopsin and other GPCRs [20,22–24].
In this study, we investigate the molecular evolution of gymnotiform rhodopsin. We ask whether the evolution of active electrolocation in gymnotiforms has reduced selective constraint on rhodopsin as part of a sensory trade-off, or instead whether rhodopsin has undergone adaptive evolution to improve photosensitivity in extremely light-limited environments. Using models that measure site-specific substitution rates across a gene, we investigate rates and patterns of rhodopsin evolution within Gymnotiformes, and we compare patterns of gymnotiform rhodopsin evolution with those of other fishes. Finally, we investigate the relevance of a disease-associated mutation newly discovered in gymnotiforms for the structure and function of rhodopsin.
2. Results
(a). Gymnotiform rhodopsin is under stronger purifying selection than in other vertebrates
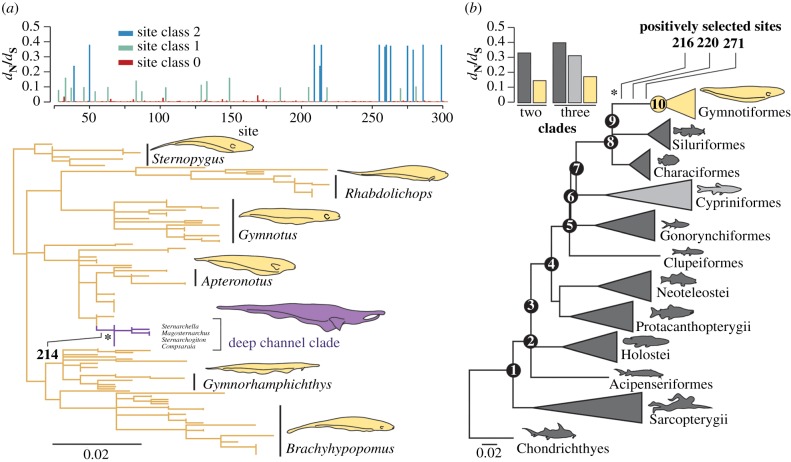
Analysis of the molecular evolution of rhodopsin sequences generated for 147 gymnotiform species reveals no evidence for relaxed selection, in contrast with predictions of a hypothesized sensory trade-off between vision and electrolocation (figure 1a). Instead, estimates of non-synonymous to synonymous substitutions (dN/dS) using the random-sites models in PAML [25] suggest that gymnotiform rhodopsin is evolving under strong purifying selection (figure 1a). We observe among-site rate variation in dN/dS in rhodopsin, but find no evidence for a class of positively selected sites (m1a versus m0: p < 0.0001; m2a versus m1a: p = 1.00; m8 versus m7: p = 1.00; electronic supplementary material, table S1), or support for a class of neutrally evolving sites (M3 versus m2aREL: p < 0.0001; electronic supplementary material, table S1). In PAML, a discrete model with three site classes, each with a dN/dS estimate below 0.5 fits the data best (figure 1a). FUBAR analyses in HYPHY [26] confirm that gymnotiform rhodopsin is evolving under strong purifying selection, with 256 of the 276 sites tested evolving under purifying selection with a posterior probability above 0.8 (electronic supplementary material, figure S1), and no sites found to be positively selected using the same cut-off.
Figure 1.
Patterns of molecular evolution in gymnotiform rhodopsin compared across sites and with other vertebrate lineages. (a) Mean dN/dS estimates for each site in our gymnotiform rhodopsin dataset estimated using the random-sites model M3 in PAML and a maximum-likelihood gymnotiform rhodopsin gene tree (more detailed trees in electronic supplementary material, figures S2 and S3). Clade of deep-channel specialists shown in purple. (b) dN/dS estimates for sites in the divergent site class of clade model D using different partitioning schemes (coloured to match the vertebrate species tree). All branch lengths represent the number of amino acid substitutions per site and positively selected sites identified on branches under positive selection (asterisks) are labelled. (Online version in colour.)
To test if the low dN/dS estimates for gymnotiform rhodopsin are significantly different from other vertebrates, we applied clade models C and D in PAML [27] to a vertebrate rhodopsin dataset (figure 1b). We find a subset of sites in the rhodopsin gene of gymnotiforms have dN/dS estimates that are half that of other vertebrates, indicating stronger purifying selection (figure 1b). Estimating dN/dS separately for the gymnotiform clade is significantly better fitting than nested null models that assume a uniform dN/dS across the entire vertebrate phylogeny (figure 1b, CmC versus M2aREL: p < 0.0001, CmD versus M3: p < 0.0001; electronic supplementary material, table S2). Using RELAX [28] and clade models [27], we directly compared dN/dS in the gymnotiform clade with the Cypriniformes (minnows and their allies), a diverse group of highly visual fishes also belonging to the superorder Ostariophysii. More parameter-rich clade models with both clades set as independent foreground lineages are not better fitting than a model with gymnotiforms set as the sole foreground clade (figure 1b; electronic supplementary material, table S2). Similarly, RELAX reveals an intensification of purifying selection in gymnotiforms relative to Cypriniformes (RELAX alternative versus RELAX null: p = 0.004). These tests indicate that a subset of sites in rhodopsin are evolving under stronger purifying selection in gymnotiforms than in cypriniforms and other vertebrates.
(b). Episodic positive selection in a background of purifying selection in gymnotiform rhodopsin
We found significant evidence for positive selection on the branch leading to the gymnotiform clade (figure 1b). An alternative branch-sites model [29] was a significantly better fit than a nested null model without a positively selected site class (branch-sites versus branch-sites null: p = 0.0455; electronic supplementary material, table S2). Bayes empirical Bayes (BEB) analyses [30] most strongly support the inclusion of sites 216, 220 and 271 in the positively selected class of sites, with posterior probabilities of 0.939, 0.956 and 0.837, respectively (figure 1b).
We also find evidence for positive selection within gymnotiforms on a branch leading to a clade of deep-channel specialists within the family Apteronotidae using models that do not specify phylogenetic partitions a priori (branch-sites REL [31], full adaptive model versus baseline model: corrected p-value = 0.0014; figure 1a). This shift in selection pressure corresponds to an ecological transition to deep habitats of large rivers in the Amazon basin, and not differences in the electric organ discharge (electronic supplementary material, figure S2 and table S1). BEB analysis using the branch-sites model identifies site 214 to be under positive selection on this branch (figure 1a). Ancestral reconstruction recovers a threonine to phenylalanine substitution at site 214 along this branch with high posterior probability. The phenylalanine is subsequently conserved throughout the deep-channel clade. Other fishes with phenylalanine at site 214 are typically deeper-dwelling than fishes with a threonine (electronic supplementary material, figure S4).
(c). Structural differences mask the effects of a visual disease-associated mutation found in gymnotiform rhodopsin
We were surprised to observe an amino acid in rhodopsin (cysteine at site 220), found in all 147 gymnotiform species sequenced in this study, that in humans is associated with RP, a genetic disorder characterized by progressive night-blindness, tunnel vision and in some cases the complete loss of sight [22,32]. We investigated if differences in the genetic background of gymnotiform and human rhodopsin might alter the pathogenicity of the F220C substitution. Ancestral reconstructions using the vertebrate rhodopsin dataset identified numerous amino acid residues in close proximity to site 220 in gymnotiforms that differ from those found in human rhodopsin (figure 2; electronic supplementary material, table S3). We hypothesize that substitutions at sites 217 and 258 to phenylalanine (Phe) mitigate the disease-associated loss of Phe at 220. These two substitutions are found in all gymnotiforms and precede the evolution of the disease-associated substitution (figure 2b). In the meta-II active-state structure of rhodopsin [33], the side chain of site 258 is closer to the phenylalanine at site 220 than any other residue in rhodopsin (figure 2a). The F220C mutation has been shown to impair both rhodopsin dimerization [34] and its kinetic properties [35]. These structural differences in helices 5 and 6, critical domains for rhodopsin dimerization and activation [20], might act to mitigate, or even mask the pathogenic effect of the substitutions at site 220.
Figure 2.
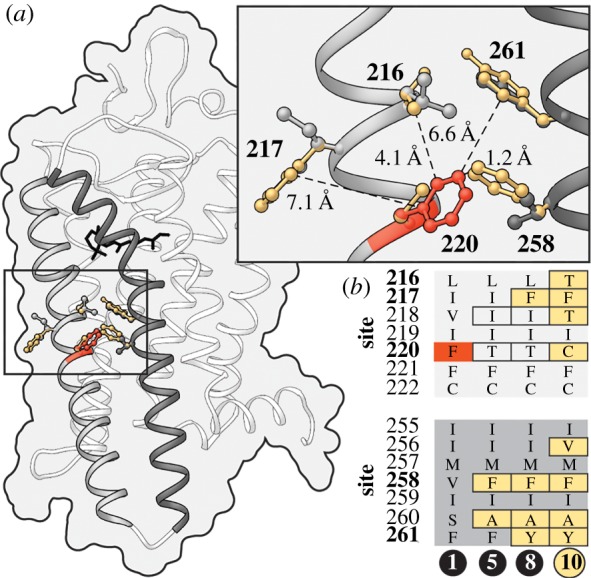
Structural evidence for compensatory changes in rhodopsin allowing the persistence of the disease-associated F220C mutation in gymnotiforms. (a) Amino acid residues on helices 5 and 6 (grey and dark grey, respectively) with side chains in close proximity to site 220 in gymnotiform (gold) and human (grey) rhodopsin are modelled on the active-state structure of rhodopsin [33]. Distances are shown from the side chains of residues that differ in gymnotiform and human rhodopsin to the aromatic ring of the wild-type phenylalanine at site 220 in human rhodopsin (red). (b) Ancestral amino acid sequences of residues near site 220 are shown for nodes between the common ancestor of fishes and humans and gymnotiforms. For corresponding node labels, see figure 1b. (Online version in colour.)
3. Discussion
In contrast with our expectations of a possible sensory trade-off between vision and active electrolocation, we find no evidence for relaxed selection in the rhodopsin gene of Gymnotiformes. Instead, we find that the dN/dS estimate for gymnotiform rhodopsin is much lower than values previously reported for other groups of fishes [36–39], indicating that it is even more conserved. However, we do find evidence for positive selection on a branch leading to the gymnotiform clade, as well as on another branch representing a transition into deep-channel habitats, suggesting episodic adaptive evolution in gymnotiforms. This is also supported by the nature of the substitutions observed along the positively selected branches. Finally, we find surprising evidence for a substitution (F220C) that is associated with RP in humans but is conserved throughout gymnotiforms. Analysis of gymnotiform rhodopsin structure and evolution suggests that the effects of this substitution are likely to be compensated or masked by substitutions at sites in close proximity. We discuss below the implications of these results for the ecology and evolution of gymnotiforms, the evolution of their vision and the sensory trade-off hypothesis.
Overall, our analysis of rhodopsin evolution indicates that gymnotiforms have a highly conserved dim-light visual system that appears to be well adapted to the turbid nocturnal environment in which they live. This is counter to the sensory trade-off hypothesis, suggesting that for gymnotiforms, there is an advantage of integrating information from multiple sensory modalities. The integration of sensory information can help recover more robust representations of an environment by minimizing the effects of noise produced by any one sensory modality [40]. Poor vision in gymnotiforms has been primarily suggested on the basis of their small eyes [18]. However, physiological studies have shown that electric fishes use vision and active electrolocation for the respective detection of near and far objects [41]. Other studies show that vision improves the sensory ability of gymnotiform fishes by integrating information received from the visual and electrosensory systems [42], and may be used to corroborate signals from the electrosensory system when hunting [18]. Thus, our results would be consistent with the idea that dim-light vision and electrolocation interact via either integration or a division of labour, rather than a trade-off.
Our results also suggest that adaptations in rhodopsin for improved photosensitivity in dim-light environments have occurred concurrently with the evolution of more specialized electrosensory systems in early gymnotiforms, and may have facilitated transitions into deeper, darker and more turbid environments as well as a more nocturnal life history. We found positively selected substitutions in the lineage leading to gymnotiforms in helices that are known to undergo the largest molecular movements upon rhodopsin photoactivation [33], and where substitutions have been shown in mutagenesis studies to affect the kinetics of chromophore release by rhodopsin [43–45]. Substitutions that affect rhodopsin kinetics are known to have evolved convergently in distantly related species inhabiting dim-light environments [46,47], including other fishes inhabiting the same rivers of the Amazon basin [37]. Interestingly, mutagenesis studies in bovine rhodopsin have also shown that the F220C substitution experimentally decreases the rate of retinal release [35]. Although the link between these shifts in rhodopsin kinetics and effects on visual physiology remains to be investigated, our results suggest that shifts in important aspects of rhodopsin function are coincident with the evolution of electrosensory systems in gymnotiforms.
Reconstructions of protein evolutionary history can be used to infer ancestral visual environments [48,49]. The origination of gymnotiforms pre-dates the bulk of Andean uplift and the formation of the modern Amazonian watersheds [50]. Whether the ancient rivers occupied by ancestral gymnotiforms were similar in optical qualities to the rivers making up the present-day Amazon river basin is unclear [17]. However, some geomorphological evidence does suggest that the major water types characteristic of the contemporary Amazon basin (white water, black water and clear water) have existed since the origination of gymnotiforms [17]. Molecular evidence of positive selection along the ancestral gymnotiform lineage provides additional indication that dimmer black water and white water rivers were present when gymnotiforms originated.
We also found evidence for positive selection on a branch within the gymnotiform clade representing a transition into large-river deep-channel habitats by a lineage within the family Apteronotidae. Members of this lineage primarily inhabit epibenthic habitats of large turbid and tannin-stained rivers of the Amazon and Orinoco drainages where ambient light levels drop within only 10 m of the surface to intensities similar to those observed in the deep sea [16]. In fact, many of these species have only recently been discovered due to the secrecy afforded to them by the depths they inhabit [16]. Visual adaptation has been extensively examined in deep-dwelling fishes, but these studies have focused on spectral tuning in deep-sea species [24]. Like deep-sea species, deep-channel gymnotiforms have multiple morphological adaptations to deep-water habitats, including feeding and diet specializations, and reduced skin pigmentation and eye size [16]. However, the blue-shifting substitutions in rhodopsin often observed in deep-sea fishes are not observed in gymnotiforms. This is probably because of differences in available light spectra in freshwater versus marine environments. Interestingly, we find that members of this deep-channel gymnotiform lineage have a rare rhodopsin amino acid substitution (T214F) found more frequently in fishes that live in marine mesopelagic or twilight zone habitats. In other opsins, substitutions at site 214 have been shown to alter dimer formation [51], a process important for maintaining thermal stability and improving signal amplification following the detection of light [52].
Future investigations of sensory trade-offs between vision and electrolocation in gymnotiforms could consider the role of other vision genes. Interestingly, opsins and other genes involved in sensory systems have exceptionally high rates of gene birth and death [53]. This dynamic evolutionary process suggests that factors more typically associated with population genetics, such as the strength of selection, effective population size and mutational load [54], may play a significant role in setting the threshold for gene loss in sensory trade-off scenarios. Some gymnotiforms lack genes encoding cone opsins SWS1 and SWS2 that are sensitive to the short-wavelengths of light most rapidly attenuated in turbid and tannin-stained waters, and express only LWS, the long-wavelength sensitive opsin gene and rhodopsin in their eyes [55]. A reduced opsin complement is also observed in the other electrogenic clade of fishes, the mormyroids [56]. Thus, while the evolution of rhodopsin in gymnotiforms fails to match the predictions of the sensory trade-off hypothesis, the evolution of opsins that are less well adapted to turbid underwater habitats may better reflect the trade-offs observed in other taxa with alternative sensory modalities for light-limited environments [8–10,12].
Surprisingly, one of the positively selected substitutions in gymnotiforms involves an amino acid that is associated with visual disease in humans. This substitution (F220C) is conserved throughout the gymnotiform clade, but in humans is associated with the degenerative eye disease RP [32]. Disease-associated mutations are generally quite rare in nature, and usually occur at highly conserved sites that are typically characterized by lower rates of dN/dS [21]. Given the evidence for functional vision in gymnotiforms [18,41,42], this would suggest that the persistence of this disease-associated substitution is probably compensated, or masked by substitutions at nearby sites. Substitutions at sites 217 and 258 might allow for the re-establishment of the dimerization interface [34] or restore normal membrane integration in the photoreceptor outer segment [35]. Similar compensatory effects have been proposed in other systems, including mammals and insects [57,58]. The approach of mining natural variation for disease-associated substitutions can improve understanding of the molecular underpinnings of genetic disease by illuminating the compensatory mechanisms required to establish normal function. Rhodopsin represents an ideal system for such studies, as expansive disease databases already exist for the gene, and rhodopsin sequences are available from a wide array of vertebrate species that have evolved unique solutions to a diverse set of selective pressures. The functional flexibility of rhodopsin has allowed its adaptation to many different environments, and appears to have cemented its place in vertebrate vision and in the overall integration of sensory information, even in taxa with novel sensory systems like the gymnotiforms.
4. Methods
(a). Rhodopsin datasets
We amplified and sequenced an 828 bp fragment, spanning all transmembrane helices, of rhodopsin from 147 gymnotiform species using PCR and Sanger sequencing (detailed methods in electronic supplementary material). Sequences are deposited in GenBank (electronic supplementary material, table S4). A rhodopsin gene tree was generated in PhyML [59] using the best-fitting nucleotide substitution model, HKY + I + G (see electronic supplementary material, figure S2). We also assembled a vertebrate rhodopsin sequence dataset for analyses of molecular evolution, including only species with sequence lengths longer than 700 bp, and with robust phylogenetic placement in a multi-gene species tree of vertebrates (electronic supplementary material, table S5).
(b). Analyses of molecular evolution
Codon-based models of evolution were used to investigate patterns of selection. Random sites, branch-sites and clade models as implemented in the codeml program in the PAML 4 software package [60] were used to investigate changes in selective constraint in both the gymnotiform and vertebrate rhodopsin datasets (see electronic supplementary material for detailed descriptions). We also conducted analyses, including FUBAR, RELAX and branch-sites REL, in the HyPhy package [61]. We tested for positive selection in gymnotiforms using random-sites models in PAML [25] and FUBAR [26]. Clade models C and D [27], as well as RELAX [28], were used to compare dN/dS in gymnotiforms with other vertebrates. The fit of the clade models were compared with nested null models M2aREL and M3, where the dN/dS estimate for the divergent site class in CmC and CmD is collapsed to uniformity across the phylogeny. We used branch-sites models to test for positive selection on the branch leading to gymnotiforms [29]. Branch-sites REL models were also used to identify positively selected sites on a branch detected to be under positive selection within the gymnotiform clade, composed of deep-channel specialist species [31]. Support for sites under positive selection was inferred using BEB [30]. Ancestral sequences were reconstructed in PAML using the best-fitting empirical amino acid substitution model, mtZOA + I+G4, and codon model, m3, for both the gymnotiform and vertebrate datasets.
Supplementary Material
Acknowledgements
For assistance with tissues and specimens, we thank Mark Sabaj (ANSP), Hernán López-Fernández (ROM), Erling Holm (ROM), Margaret Zur (ROM), Tiago Carvalho (UFRGS), John Armbruster (AUM), Victor Tagliacollo (UL), James Albert (UL) and the curators of all of the following institutions for also providing fish tissue samples: UF, MCP, INPA, MNRJ, UNELLEZ, LBP, IAvH-BT, MUSM, MZUSP, IAvHP, ZOO.A.V.Pe, IMCN, AMNH, CBF, UMSS and MHNG. We thank Fangyu Ren for assistance with gymnotiform silhouettes in figure 1.
Data accessibility
New sequence data are available from GenBank (accession nos MN031442–MN031588) and listed in electronic supplementary material, table S4.
Authors' contributions
A.V.N., N.R.L. and B.S.W.C. conceived and designed the study. K.B., F.H.J., J.A.M-O., N.R.L. and W.G.R.C. collected specimens in the field. K.B., F.H.J. and A.V.N. carried out the molecular laboratory work. A.V.N. analysed experimental data, conducted computational and statistical analyses and drafted the manuscript. A.V.N., N.R.L. and B.S.W.C. interpreted the results and revised the manuscript, with assistance from W.G.R.C.
Competing interests
We declare we have no competing interests.
Funding
Funding was provided by NSERC Discovery grants (to B.S.W.C. and N.R.L.) and a Vision Science Research Fellowship (to A.V.N.).
References
- 1.Niven JE, Laughlin SB. 2008. Energy limitation as a selective pressure on the evolution of sensory systems. J. Exp. Biol. 211, 1792–1804. ( 10.1242/jeb.017574) [DOI] [PubMed] [Google Scholar]
- 2.Niemiller ML, Fitzpatrick BM, Shah P, Schmitz L, Near TJ. 2013. Evidence for repeated loss of selective constraint in rhodopsin of amblyopsid cavefishes (Teleostei: Amblyopsidae). Evolution 67, 732–748. ( 10.1111/j.1558-5646.2012.01822.x) [DOI] [PubMed] [Google Scholar]
- 3.Hodos W, Butler AB. 1997. Evolution of sensory pathways in vertebrates. Brain Behav. Evol. 50, 189–197. ( 10.1159/000113333) [DOI] [PubMed] [Google Scholar]
- 4.Fernholm B, Holmberg K. 1975. The eyes in three genera of hagfish (Eptatretus, Paramyxine and Myxine)—a case of degenerative evolution. Vision Res. 15, 253–259, IN1–IN4. [DOI] [PubMed] [Google Scholar]
- 5.Müller J, Bickelmann C, Sobral G. 2018. The evolution and fossil history of sensory perception in amniote vertebrates. Annu. Rev. Earth Planet. Sci. 46, 495–519. ( 10.1146/annurev-earth-082517-010120) [DOI] [Google Scholar]
- 6.Nummela S, Pihlström H, Puolamäki K, Fortelius M, Hemilä S, Reuter T. 2013. Exploring the mammalian sensory space: co-operations and trade-offs among senses. J. Comp. Physiol. 199, 1077–1092. ( 10.1007/s00359-013-0846-2) [DOI] [PubMed] [Google Scholar]
- 7.Sadier A, Davies KT, Yohe LR, Yun K, Donat P, Hedrick BP et al. . 2018. Multifactorial processes underlie parallel opsin loss in neotropical bats. eLife 7, 58 ( 10.7554/eLife.37412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS. 2013. Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9, e1003432 ( 10.1371/journal.pgen.1003432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez EA, Schott RK, Preston MW, Loureiro LO, Lim BK, Chang BSW. 2018. The role of ecological factors in shaping bat cone opsin evolution. Proc. R. Soc. B 285, 20172835 ( 10.1098/rspb.2017.2835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Jiao H, Simmons NB, Lu Q, Zhao H. 2018. Testing the sensory trade-off hypothesis in New World bats. Proc. R. Soc. B 285, 20181523 ( 10.1098/rspb.2018.1523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keesey IW, et al. 2019. Inverse resource allocation between vision and olfaction across the genus Drosophila. Nat. Commun. 10, 698 ( 10.1038/s41467-019-09087-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerling CA, Springer MS. 2014. Eyes underground: regression of visual protein networks in subterranean mammals. Mol. Phylogenetics Evol. 78, 260–270. ( 10.1016/j.ympev.2014.05.016) [DOI] [PubMed] [Google Scholar]
- 13.Shiriagin V, Korsching SI. 2018. Massive expansion of bitter taste receptors in blind cavefish, Astyanax mexicanus. Chem. Senses 44, 23–32. ( 10.1093/chemse/bjy062) [DOI] [PubMed] [Google Scholar]
- 14.Gower DJ, et al. 2019. Evolution of the eyes of vipers with and without infrared-sensing pit organs. Biol. J. Linn. Soc. Lond. 126, 796–823. ( 10.1093/biolinnean/blz003) [DOI] [Google Scholar]
- 15.Crampton WGR. 2019. Electroreception, electrogenesis and electric signal evolution. J. Fish Biol. 204, 185 ( 10.1111/jfb.13922) [DOI] [PubMed] [Google Scholar]
- 16.Crampton WGR. 2008. Diversity and adaptation in deep channel Neotropical electric fishes. In Fish life in special environments (eds Sebert P, Onyango DW, Kapoor BG), pp. 283–339. Enfield, NH: Science Publishers. [Google Scholar]
- 17.Crampton WGR. 2011. An ecological perspective on diversity and distributions. In Historical biogeography of neotropical freshwater fishes (ed. Albert JS.), pp. 165–189. Berkeley, CA: University of California Press. [Google Scholar]
- 18.Takiyama T, Luna da Silva V, Moura Silva D, Hamasaki S, Yoshida M. 2015. Visual capability of the weakly electric fish Apteronotus albifrons as revealed by a modified retinal flat-mount method. Brain Behav. Evol. 86, 122–130. ( 10.1159/000438448) [DOI] [PubMed] [Google Scholar]
- 19.Baylor DA. 1996. How photons start vision. Proc. Natl Acad. Sci. USA 93, 560–565. ( 10.1073/pnas.93.2.560) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. 2013. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163. ( 10.1021/cr4003769) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauser FE, Schott RK, Castiglione GM, Van Nynatten A, Kosyakov A, Tang PL, Gow DA, Chang BSW. 2016. Comparative sequence analyses of rhodopsin and RPE65 reveal patterns of selective constraint across hereditary retinal disease mutations. Vis. Neurosci. 33, 183 ( 10.1017/S0952523815000322) [DOI] [PubMed] [Google Scholar]
- 22.Daiger SP, Sullivan LS, Bowne SJ. 2013. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 84, 132–141. ( 10.1111/cge.12203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erlandson SC, McMahon C, Kruse AC. 2018. Structural basis for G protein-coupled receptor signaling. Annu. Rev. Biophys. 47, 1–18. ( 10.1146/annurev-biophys-070317-032931) [DOI] [PubMed] [Google Scholar]
- 24.Bowmaker JK. 2008. Evolution of vertebrate visual pigments. Vision Res. 48, 2022–2041. ( 10.1016/j.visres.2008.03.025) [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Nielsen R, Goldman N, Pedersen A-MK. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol. Biol. Evol. 30, 1196–1205. ( 10.1093/molbev/mst030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bielawski JP, Yang Z. 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evol. 59, 121–132. ( 10.1007/s00239-004-2597-8) [DOI] [PubMed] [Google Scholar]
- 28.Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2014. RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 32, 820–832. ( 10.1093/molbev/msu400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917. ( 10.1093/oxfordjournals.molbev.a004148) [DOI] [PubMed] [Google Scholar]
- 30.Yang Z. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. ( 10.1093/molbev/msi097) [DOI] [PubMed] [Google Scholar]
- 31.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evol. 28, 3033–3043. ( 10.1093/molbev/msr125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunge S, Wedemann H, David D, Terwilliger DJ, Van Den Born LI, Aulehla-Scholz C et al. . 1993. Molecular analysis and genetic mapping of the rhodopsin gene in families with autosomal dominant retinitis pigmentosa. Genomics 17, 230–233. ( 10.1006/geno.1993.1309) [DOI] [PubMed] [Google Scholar]
- 33.Choe H-W, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. 2011. Crystal structure of metarhodopsin II. Nature 471, 651–655. ( 10.1038/nature09789) [DOI] [PubMed] [Google Scholar]
- 34.Ploier B, Caro LN, Morizumi T, Pandey K, Pearring JN, Goren MA et al. . 2016. Dimerization deficiency of enigmatic retinitis pigmentosa-linked rhodopsin mutants. Nat. Commun. 7, 177 ( 10.1038/ncomms12832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallory DP, Gutierrez E, Pinkevitch M, Klinginsmith C, Comar WD, Roushar FJ, Schlebach JP, Smith AW, Jastrzebska B. 2018. The retinitis pigmentosa-linked mutations in transmembrane helix 5 of rhodopsin disrupt cellular trafficking regardless of oligomerization state. Biochemistry 57, 5188–5201. ( 10.1021/acs.biochem.8b00403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stieb SM, Cortesi F, Sueess L, Carleton KL, Salzburger W, Marshall NJ. 2017. Why UV vision and red vision are important for damselfish (Pomacentridae): structural and expression variation in opsin genes. Mol. Ecol. 26, 1323–1342. ( 10.1111/mec.13968) [DOI] [PubMed] [Google Scholar]
- 37.Hauser FE, Ilves KL, Schott RK, Castiglione GM, López-Fernández H, Chang BSW. 2017. Accelerated evolution and functional divergence of the dim light visual pigment accompanies cichlid colonization of Central America. Mol. Biol. Evol. 34, 2650–2664. ( 10.1093/molbev/msx192.) [DOI] [PubMed] [Google Scholar]
- 38.Van Nynatten A, Bloom DD, Chang BSW, Lovejoy NR. 2015. Out of the blue: adaptive visual pigment evolution accompanies Amazon invasion. Biol. Lett. 11, 20150349 ( 10.1098/rsbl.2015.0349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rolland J, Silvestro D, Litsios G, Faye L, Salamin N. 2018. Clownfishes evolution below and above the species level. Proc. R. Soc. B 285, 20171796 ( 10.1098/rspb.2017.1796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munoz NE, Blumstein DT. 2012. Multisensory perception in uncertain environments. Behav. Ecol. 23, 457–462. ( 10.1093/beheco/arr220) [DOI] [Google Scholar]
- 41.Bastian J. 1982. Vision and electroreception: integration of sensory information in the optic tectum of the weakly electric fish Apteronotus albifrons. J. Comp. Physiol. 147, 287–297. ( 10.1007/BF00609662) [DOI] [Google Scholar]
- 42.Rose GJ, Canfield JG. 1993. Longitudinal tracking responses of the weakly electric fish, Sternopygus. J. Comp. Physiol. 171, 791–798. ( 10.1007/bf00213075) [DOI] [Google Scholar]
- 43.Goncalves JA, South K, Ahuja S, Zaitseva E, Opefi CA, Eilers M, Vogel R, Reeves PJ, Smith SO. 2010. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc. Natl Acad. Sci. USA 107, 19 861–19 866. ( 10.1073/pnas.1009405107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piechnick R, Ritter E, Hildebrand PW, Ernst OP, Scheerer P, Hofmann KP, Heck M. 2012. Effect of channel mutations on the uptake and release of the retinal ligand in opsin. Proc. Natl Acad. Sci. USA 109, 5247–5252. ( 10.1073/pnas.1117268109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrow JM, Chang BSW. 2015. Comparative mutagenesis studies of retinal release in light-activated zebrafish rhodopsin using fluorescence spectroscopy. Biochemistry 54, 4507–4518. ( 10.1021/bi501377b) [DOI] [PubMed] [Google Scholar]
- 46.Castiglione GM, Chang BSW. 2018. Functional trade-offs and environmental variation shaped ancient trajectories in the evolution of dim-light vision. eLife 7, 348 ( 10.7554/eLife.35957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugawara T, Imai H, Nikaido M, Imamoto Y, Okada N. 2010. Vertebrate rhodopsin adaptation to dim light via rapid meta-II intermediate formation. Mol. Biol. Evol. 27, 506–519. ( 10.1093/molbev/msp252) [DOI] [PubMed] [Google Scholar]
- 48.Gaucher EA, Thomson JM, Burgan MF, Benner SA. 2003. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425, 285–288. ( 10.1038/nature01977) [DOI] [PubMed] [Google Scholar]
- 49.Chang BSW, Jönsson K, Kazmi MA, Donoghue MJ, Sakmar TP. 2002. Recreating a functional ancestral archosaur visual pigment. Mol. Biol. Evol. 19, 1483–1489. ( 10.1093/oxfordjournals.molbev.a004211) [DOI] [PubMed] [Google Scholar]
- 50.Lavoué S, Miya M, Arnegard ME, Sullivan JP, Hopkins CD, Nishida M. 2012. Comparable ages for the independent origins of electrogenesis in African and South American weakly electric fishes. PLoS ONE 7, e36287 ( 10.1371/journal.pone.0036287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jastrzebska B, Comar WD, Kaliszewski MJ, Skinner KC, Torcasio MH, Esway AS, Jin H, Palczewski K, Smith AW. 2016. A G protein-coupled receptor dimerization interface in human cone opsins. Biochemistry 56, 61–72. ( 10.1021/acs.biochem.6b00877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gunkel M, Schöneberg J, Alkhaldi W, Irsen S, Noé F, Kaupp UB, Al-Amoudi A. 2015. Higher-order architecture of rhodopsin in intact photoreceptors and its implication for phototransduction kinetics. Structure 23, 628–638. ( 10.1016/j.str.2015.01.015) [DOI] [PubMed] [Google Scholar]
- 53.Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. ( 10.1016/S0169-5347(03)00033-8) [DOI] [Google Scholar]
- 54.Platt A, Weber CC, Liberles DA. 2018. Protein evolution depends on multiple distinct population size parameters. BMC Evol. Biol. 18, 9541 ( 10.1186/s12862-017-1085-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu D-W, Lu Y, Yan HY, Zakon HH. 2017. South American weakly electric fish (Gymnotiformes) are long-wavelength-sensitive cone monochromats. Brain Behav. Evol. 88, 204–212. ( 10.1159/000450746) [DOI] [PubMed] [Google Scholar]
- 56.Liu D-W, Wang F-Y, Lin J-J, Thompson A, Lu Y, Vo D, Yan HY, Zakon HH. 2018. The cone opsin repertoire of osteoglossomorph fishes: gene loss in mormyrid electric fish and a long wavelength-sensitive cone opsin that survived 3R. Mol. Biol. Evol. 282, 1711 ( 10.1093/molbev/msy241) [DOI] [PubMed] [Google Scholar]
- 57.Kondrashov AS, Sunyaev S, Kondrashov FA. 2002. Dobzhansky-Muller incompatibilities in protein evolution. Proc. Natl Acad. Sci. USA 99, 14 878–14 883. ( 10.1073/pnas.232565499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulathinal RJ. 2004. Compensated deleterious mutations in insect genomes. Science 306, 1553–1554. ( 10.1126/science.1100522) [DOI] [PubMed] [Google Scholar]
- 59.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. ( 10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 60.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 61.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. ( 10.1093/bioinformatics/bti079) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New sequence data are available from GenBank (accession nos MN031442–MN031588) and listed in electronic supplementary material, table S4.