Abstract
Monitoring of antimalarial resistance is important to prevent its further spread, but the available options for assessing resistance are less than ideal for field settings. Although molecular detection is perhaps the most efficient method, it is also the most complex because it requires DNA extraction and PCR instrumentation. To develop a more deployable approach, we designed new probes, which, when used in combination with an inhibitor-tolerant Taq polymerase, enable single-nucleotide polymorphism genotyping directly from whole blood. The probes feature two strategic design elements: locked nucleic acids to enhance specificity and the reporter dyes Cy5 and TEX615, which have less optical overlap with the blood absorbance spectra than other commonly used dyes. Probe performance was validated on a traditional laboratory-based instrument and then further tested on a field-deployable Adaptive PCR instrument to develop a point-of-care platform appropriate for use in malaria settings. The probes discriminated between wild-type Plasmodium falciparum and the chloroquine-resistant CRT PF3D7_0709000:c.227A>C (p.Lys76Thr) mutant in the presence of 2% blood. Additionally, in allelic discrimination plots with the new probes, samples clustered more closely to their respective axes compared with samples using minor groove binder probes with 6-FAM and VIC reporter dyes. Our strategy greatly simplifies single-nucleotide polymorphism detection and provides a more accessible alternative for antimalarial resistance surveillance in the field.
CME Accreditation Statement: This activity (“JMD 2019 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the accreditation requirements and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2019 CME Program in Molecular Diagnostics”) for a maximum of 18.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
The drugs used to treat malaria have followed a pattern typical for many other antimicrobials. Initially, the drug works well, but over time, it becomes less effective through complex mechanisms that lead to the development of drug resistance. This paradigm was observed for chloroquine starting in the 1950s, then later with sulfadoxine-pyrimethamine in the following decade (reviewed by Wongsrichanalai et al1). Indications of a similar pattern are being observed with the current first-line treatment for Plasmodium falciparum malaria.2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 Several reports also suggested that resistance emerged at least in part as independent events.15, 16 However, recently, microsatellite analysis indicates that a mutant lineage carrying the Kelch13 PF3D7_1343700: p.Cys580Tyr single-nucleotide polymorphism (SNP) spread from Cambodia to Thailand, Laos, and Vietnam.17, 18 This is a continuously evolving situation, and it is unclear how and if this parasite lineage will continue to spread.
For previous generations of antimalarials, resistance arose from the development and subsequent spread of one or more parasite SNPs. To monitor and prevent their spread, these SNPs can be detected with molecular assays, like sequencing or real-time PCR. However, the tests are relatively expensive, are labor intensive, and are generally performed in central laboratories. We sought to make SNP genotyping for antimalarial drug resistance more accessible outside of central laboratories by developing an assay that has the potential to be performed directly on blood in a point-of-care setting. To demonstrate proof of principle for the detection of drug resistance–associated SNPs, we tested our design with the well-characterized SNP associated with chloroquine resistance, c.227A>C (p.Lys76Thr) of the P. falciparum chloroquine resistance transporter (CRT) gene.19, 20
A common approach for PCR-based SNP identification uses hydrolysis probes because it can be performed in any laboratory equipped with a real-time PCR instrument. This technique relies on a large differential melting temperature between the probes specific for each polymorphism. The perfectly matched probe has a higher melting temperature for its target compared with the other mismatched probe.21 The difference in melting temperatures can be accomplished by using various probe modifications, like the minor groove binder (MGB).22 In the present study, locked nucleic acids (LNAs) were incorporated in the hydrolysis probes because they, like the MGB, raise the melting temperature of the probes, resulting in preferential binding of the wild-type probe to the wild-type target and the mutant probe to the mutant target.23 In the case of LNAs, this is achieved by a methylene bridge within the ribose sugar of the nucleotide, resulting in a more rigid or locked conformation. This structural modification does not alter its participation in Watson-Crick base pairing, but does enhance the probe's affinity for its complementary target.24 The increase of the oligonucleotide melting temperature by LNAs provides a critical advantage for SNP genotyping and has been exploited for several assays, including Chlamydia pneumoniae genotyping.23, 25, 26, 27, 28
In addition to developing the molecular tools for SNP genotyping, our goal was to apply them within a simplified assay design appropriate for a low-resource setting. To avoid the complexities of sample preparation, a PCR-based assay that can be performed directly on whole blood was designed. Blood poses several obstacles for real-time PCR: it inhibits PCR amplification,29, 30 and its absorption spectra can interfere with the fluorescent reporter dyes.31 Several manufacturers have developed Taq and other DNA polymerases to be highly resistant to PCR inhibitors, which are often referred to as robust polymerases.32, 33, 34 For Taq polymerase, this can be accomplished by an N-terminal deletion that enhances performance, but at the cost of removing the 5′→3′ exonuclease activity required for cleavage of hydrolysis probes.33, 35 In addition, although MGB probes with the reporter dyes 6-FAM (FAM) and VIC have been developed for c.227A>C (p.Lys76Thr) genotyping, they are not ideal for use with blood because of the high overlap between blood absorption and the reporter dyes' excitation and emission wavelengths.36 The limitations associated with blood were overcome by incorporating two key changes to the PCR reagents: a robust Taq polymerase optimized for use with blood and the use of the fluorescent reporter dyes Cy5 and TEX615, which emit at sufficiently long wavelengths to reduce the optical interference from blood. The assay was also evaluated using Adaptive PCR, a previously described real-time PCR platform that uses left-handed DNA (L-DNA) additives to monitor the reaction for more reliable point-of-care performance.37
Materials and Methods
Materials
The P. falciparum reference line 3D7 was cultured, as previously described.38 Genomic DNA from the 3D7 parasite culture was extracted with the Qiagen (Germantown, MD) DNeasy Blood and Tissue kit, according to the manufacturer's protocol. The P. falciparum line 7G8 genomic DNA (MRA-926G, MR4; BEI Resources, Manassas, VA) was obtained through the Malaria Research and Reference Reagent Resource Center. Human whole blood with citrate phosphate dextrose anticoagulant was purchased from Bioreclamation IVT (Westbury, NY). For experiments using packed red blood cells (RBCs), the cells were washed in phosphate-buffered saline and used at a final concentration of 0.8%, which is equivalent to 2% whole blood for a 40% hematocrit.
Oligonucleotides
Ultramer DNA oligonucleotides containing the 166-base wild-type and the c.227A>C (p.Lys76Thr) mutant CRT (PlasmoDB Gene identification: PF3D7_0709000; https://plasmodb.org, last accessed February 1, 2019) amplicons were synthesized by Integrated DNA Technologies (Coralville, IA). All of the primer and probe sequences are shown in Table 1. The custom MGB probes (pfcrt_76K and pfcrt_76T) were supplied in a premixed solution with the primers pfcrt_F and pfcrt_R from Applied Biosystems (Foster City, CA). The MGB probes and their primers have been previously described.36 The 5′ ends of the wild-type and mutant MGB probes were labeled with FAM and VIC fluorescent reporter dyes, respectively. The MGB quencher was located on their 3′ ends.
Table 1.
Sequences of Oligonucleotides Used for Real-Time PCR
| Name | Description | Sequence∗ |
|---|---|---|
| pfcrt_F36 | Forward primer | 5′-TGGTAAATGTGCTCATGTGTTT-3′ |
| pfcrt_R36 | Reverse primer | 5′-AGTTTCGGATGTTACAAAACTATAGT-3′ |
| pfcrt_76K36 | Wild-type probe with MGB | 5′-6-FAM-TGTGTAATGAATAAAATTTTTGCTAA-MGB-3′ |
| pfcrt_76T36 | Mutant probe with MGB | 5′-VIC-TGTGTAATGAATACAATTTTTGCTAA-MGB-3′ |
| LNA_W (ML045) | Wild-type probe with LNAs | 5′-Cy5-AT+G+AA+T+A+A+AAT+TTT+T+GC-IABRQSP-3′ |
| LNA_M (ML047) | Mutant probe with LNAs | 5′-TEX615-GTAATGAA+T+A+C+AA+TT+TTTGCT-IABRQSP-3′ |
| ML040 | Wild-type probe with LNAs (candidate 1) | 5′-Cy5-TGTGTAATGAA+T+A+A+AA+TTTTTG+CT-IABRQSP-3′ |
| ML044 | Wild-type probe with LNAs (candidate 2) | 5′-Cy5-AT+G+AA+T+A+A+AA+TT+TTT+GCT-IABRQSP-3′ |
| ML046 | Wild-type probe with LNAs (candidate 4) | 5′-Cy5-T+G+AA+T+A+A+AAT+TTT+T+GCT-IABRQSP-3′ |
| ML036 | Mutant probe with LNAs (candidate 1) | 5′-HEX-TGTGTAATGAA+T+A+C+AA+TTTTTG+CTA-IABKFQ-3′ |
| ML037 | Mutant probe with LNAs (candidate 2) | 5′-HEX-TGTGTAATGAA+T+A+C+AA+T+TTTTGCTA-IABKFQ-3′ |
IABKFQ, Iowa Black FQ; IABRQSP, Iowa Black RQ; LNA, locked nucleic acid; MGB, minor groove binder.
Locked nucleic acid bases are preceded by a plus sign (+).
A second set of pfcrt_F and pfcrt_R primers as well as the LNA probes were synthesized by Integrated DNA Technologies. Wild-type probes incorporating between six and nine LNA bases were labeled with Cy5 on the 5′ end. The probes for the mutant alleles were labeled with either HEX or TEX615. The FAM and HEX dyes were quenched with a 3′ Iowa Black FQ quencher and the TEX615 and Cy5 dyes with a 3′ Iowa Black RQ quencher. Each was tested for its SNP discrimination potential in a competition assay with each probe from the other allele. The probes with the highest ratio of perfect-match-to-mismatch end point fluorescence, labeled LNA_W and LNA_M in Table 1, were chosen for further study. They contained nine and six locked nucleotides, respectively, and were synthesized with a 5′ reporter dye (Cy5 or TEX615) and a 3′ Iowa Black RQ quencher.
L-DNA enantiomer oligonucleotides were synthesized by Biomers (Ulm, Germany). To monitor primer annealing, L-DNA enantiomers of the pfcrt_F primer and its reverse complement were synthesized. The previously described 77-mer sequence derived from Mycobacterium tuberculosis (labeled as TB77) was synthesized and used to monitor melting of the amplicon.37
PCR
For reactions with the LNA probes, each 25-μL reaction contained 1× Taq Mutant Reaction Buffer (DNA Polymerase Technology, St. Louis, MO), 200 nmol/L of each dNTP (Sigma-Aldrich, St. Louis, MO), 400 nmol/L of each probe, 900 nmol/L of each primer, 0.25 μL of OmniTaq 3 DNA polymerase (DNA Polymerase Technology), and up to 3% whole blood or washed RBCs from the P. falciparum (strain 3D7) parasite culture. The reactions with MGB probes were composed identically with the exception of the probes and primers, which were supplied in a 20× mixture by the manufacturer for a final concentration of 200 and 900 nmol/L, respectively. In addition, the target DNA was added at 107 copies for the synthesized oligonucleotides and 8 ng (approximately 3 × 105 copies) for the genomic DNA.
Two PCR instruments were used for this study: the Rotor-Gene Q (Qiagen), a traditional PCR instrument; and the recently described Adaptive PCR instrument. The latter uses fluorescently labeled L-DNA analogs of the primers and amplicon to identify heating and cooling switch points by optically monitoring the annealing of L-DNA primers and melting of the L-DNA amplicon, respectively.37 For the Rotor-Gene instrument, each reaction was performed with the following conditions: an initial 95°C hold for 3 minutes, followed by 45 cycles at 95°C for 15 seconds and 62°C for 15 seconds. On the Adaptive PCR instrument, the 3-minute initial denaturation was performed by programming LabVIEW software version 2017 (National Instruments, Austin, TX) to monitor the fluorescence signal of labeled L-DNA analogs of the TB77 amplicon while heating and then maintaining the fluorescence signal once the melt plateau was reached. The program maintained the melted state for 3 minutes by turning the heater on if the fluorescence signal fell below the melt state and turning the heater off once the fluorescence reached the melt state. Validation studies with a thermocouple inserted into the reaction tube confirmed that the hold temperature maintained approximately 88°C ± 2°C for the denaturation step. The samples were then cooled until the L-DNA analog of the pfcrt_F primer annealed to its target. The instrument then began heating until the L-DNA TB77 amplicon strands separated. This was repeated for a total of 40 cycles. The end point fluorescence values for the allelic discrimination plots were taken from PCR cycle number 40, regardless of the instrument.
Results
LNA Probes for CRT c.227A>C Genotyping Are Comparable to MGB Probes
Our goal was to streamline SNP genotyping assays to make them more compatible for malaria fieldwork. One of the main bottlenecks is DNA extraction because it is time and labor intensive, and consequently, is usually performed in a traditional laboratory setting. Aside from the inhibitors found in blood, the excitation and emission spectra of commonly used probe reporter dyes are also incompatible with the absorbance spectra of blood. To overcome this problem, the Cy5 and TEX615 reporter dyes were assessed because they excite and emit in the orange-red end of the spectrum, where blood absorbance is lower.
The strategy for evaluating the probes in blood was to increase the experimental complexity one step at a time. First, the LNA probes were compared with previously described MGB genotyping probes36 by using amplicon-length oligonucleotide targets. They were initially validated on the traditional Rotor-Gene PCR instrument. These same probes were further tested on the field-deployable Adaptive PCR device. Once their performance was confirmed on these two instruments, the Adaptive PCR instrument was used to assess the LNA probes' allelic discrimination potential, first in genomic P. falciparum DNA and then with P. falciparum–infected RBCs.
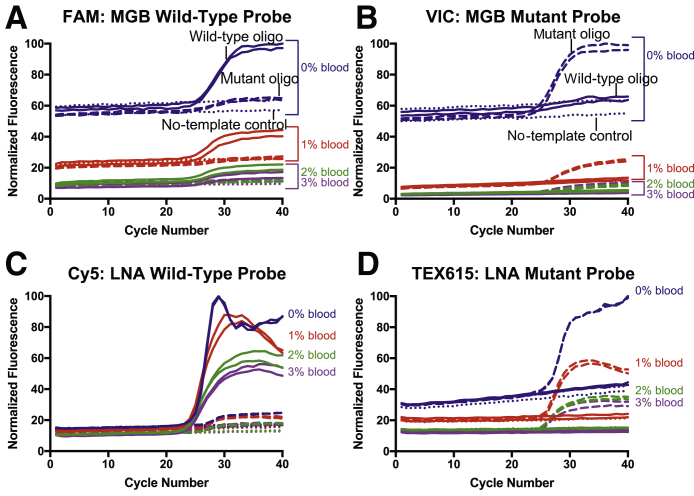
In the first set of experiments, the LNA probes were evaluated side by side with MGB probes in increasing concentrations of blood. Synthesized amplicon-length oligonucleotides were used as targets to simplify the experimental parameters. As expected, the overall fluorescence intensity of the probes is inversely proportional to the percentage of blood in the reaction. The intensities of the FAM and VIC reporter dyes (Figure 1, A and B) decrease more with the addition of blood compared with the Cy5 and TEX615 dyes (Figure 1, C and D), which is presumably caused by the overlap of the blood absorbance spectra with their excitation and emission wavelengths. The absence of amplification of the wild-type sequence in the VIC and TEX615 amplification plots (Figure 1, B and D) suggests that neither of the mutant probes has observable mismatch binding to the wild-type oligonucleotide. However, both the MGB and LNA wild-type probes exhibit leakage (Figure 1, A and C), indicating that the wild-type probes bind to the mutant oligonucleotides at a low level. This is likely a consequence of the middle nucleotide of the CRT codon 76 changing from an adenine to a cytosine in the mutant sequence, which increases the affinity of the mutant probe to the mutant sequence. The wild-type probes, therefore, have inherently lower melting temperatures and, accordingly, lower specificity than their mutant probe counterparts.
Figure 1.
Comparison of the minor groove binder (MGB) and locked nucleic acid (LNA) CRT c.227A>C probes in blood with a traditional PCR instrument. Each panel combines reactions with 0% to 3% whole blood. The results are shown as wild-type (solid line) or mutant (dashed line) oligonucleotide (oligo) of the CRT amplicon or a no-template control (dotted line). Each sample was performed in duplicate on the Rotor-Gene instrument. The normalized fluorescence intensity for the MGB (A and B) and the LNA (C and D) probe pairs is plotted for each cycle. In all cases, as the percentage of blood increases, the fluorescence intensity decreases. The FAM (A) and VIC (B) fluorescence intensities are more dramatically attenuated by the addition of blood compared with the Cy5 (C) and TEX615 (D) reporter dyes.
The LNA probes were further tested for their discriminatory potential in mixed infections. To mimic a low percentage of a drug-resistant isolate compared with a drug-sensitive isolate, 106 copies of the mutant oligonucleotide were combined with 9 × 106 copies of the wild-type oligonucleotide for a total reaction copy number of 107. In addition, a mixture containing 5 × 106 copies of each oligonucleotide was also evaluated. For comparison, 106, 5 × 106, and 9 × 106 copies of each oligonucleotide alone were also included. The wild-type and mutant oligonucleotides clustered near their respective axes on the allelic discrimination plot (Supplemental Figure S1). The mixed reaction samples were located in the zone between the individual oligonucleotides, indicating that the LNA probes can detect the chloroquine-resistance–associated SNP in the context of a mixed infection, as low as 10% of the total copy number.
Direct PCR in Blood with Adaptive PCR Instrument
We recently reported the design of a PCR platform called Adaptive PCR that does not rely on temperature checkpoints. Instead, it monitors the denaturation and annealing of fluorescently labeled, mirror-image L-DNA molecules to switch between heating and cooling steps, which significantly simplifies instrument programming and calibration.37 To develop an SNP genotyping process appropriate for field deployment, next, the probes' discrimination potential was tested in the simpler Adaptive PCR device. Subsequent experiments were performed in the Adaptive PCR instrument.
For the evaluation of the LNA probes in the Adaptive PCR instrument, the L-DNA analog of the 166-bp CRT amplicon could not be synthesized because of the length limitations with synthesis. Therefore, in its place, the previously published M. tuberculosis target sequence and its complement, which were synthesized as L-DNA oligonucleotides, were substituted.37 The GC-rich, 77-bp TB77 sequence has a melting temperature of 87°C, which is approximately 13°C higher than the AT-rich, 166-bp CRT amplicon sequence (data not shown), and ensures that the naturally occurring D-DNA target will be sufficiently denatured during the heating step. The L-DNA analog of the CRT forward primer and its complement were used to monitor annealing of the primers to their target.
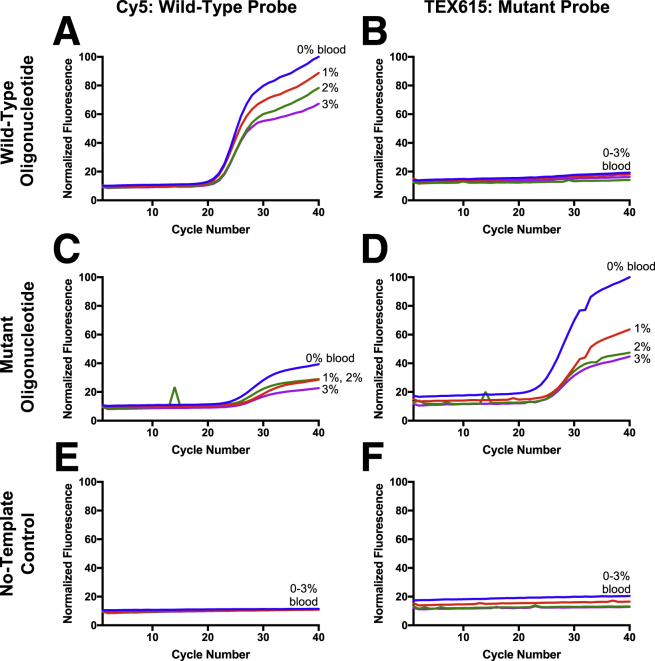
Whole blood at a final concentration of 1%, 2%, or 3% was added to each sample. Phosphate-buffered saline was added to a subset of samples for a 0% blood comparison. Detection of the wild-type and mutant CRT amplicon-length oligonucleotides was compared (Figure 2). Small blips in the amplification curves were occasionally observed for reactions performed on the Adaptive PCR instrument and were attributable to glitches in the early instrument software. The infrequent spikes were isolated to individual cycles and did not change the interpretation of the data.
Figure 2.
Performance of the locked nucleic acid CRT c.227A>C probes in blood with the Adaptive PCR instrument. The wild-type (A and B) or mutant (C and D) oligonucleotide or a no-template control (E and F) was amplified in the presence of 0%, 1%, 2%, or 3% whole blood on the Adaptive PCR device.
The TEX615-labeled mutant probe identified the mutant sequence (Figure 2D) without detectable mismatch binding (Figure 2B). Although the Cy5-labeled wild-type probe detected the wild-type oligonucleotides (Figure 2A), a small, delayed amplification curve was also observed for the mutant sequence (Figure 2C). This corroborates the earlier finding that the wild-type probe has lower specificity than the mutant probe (Figure 1). Amplification curves were not observed for the no-template control samples (Figure 2, E and F).
Comparison of Allelic Discrimination for Each Probe Pair
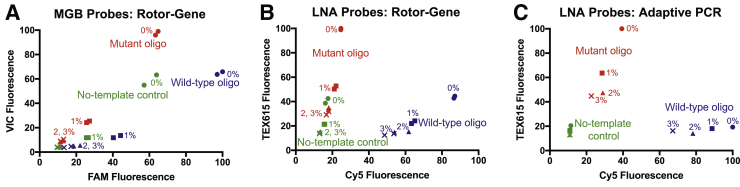
To further assess the probe pairs, the end point fluorescence values from each experiment were plotted in an allelic discrimination plot (Figure 3). Each allele is expected to have high signal from its probe and low signal from the probe for the other allele, and consequently, cluster near its respective axis.39 However, in the Rotor-Gene experiments with the MGB probe pair, the decrease in fluorescence caused by the addition of blood resulted in poor clustering regardless of the allele (Figure 3A). For comparison, in the same instrument with the LNA probes, the wild-type samples locate closer to the Cy5 axis and the mutant samples to the TEX615 axis (Figure 3B). Despite the greater separation between alleles, the 0% blood no-template sample was an exception as it clusters with the mutant samples (Figure 3B). The emission and excitation wavelengths of the TEX615 dye have greater overlap with the absorbance of blood than those of the Cy5 dye. Therefore, the Cy5 initial fluorescence is less impacted by the presence of blood than the TEX615 dye. At 2% and 3% blood, the TEX615 fluorescence starts lower than 0% blood sample, and even after amplification, the end point value remains below the no-template control sample (Figure 1D).
Figure 3.
Allelic discrimination plots for each PCR instrument. The end point fluorescence values are plotted in allelic discrimination plots for the Rotor-Gene (A and B) and Adaptive PCR (C) instruments. For the former, minor groove binder (MGB; A) and locked nucleic acid (LNA; B) probes were tested, whereas LNA probes were evaluated in Adaptive PCR (C).
However, when plotting the same LNA probes from the Adaptive PCR experiment, all blood dilutions of the no-template controls clustered tightly near the origin (Figure 3C). The wild-type samples clustered near the Cy5 wild-type axis, and the mutant samples clustered near the TEX615 mutant axis (Figure 3C). As the percentage of blood was increased, the overall fluorescence intensity decreased. The fluorescence decrease was more notable for the TEX615 dye than the Cy5 dye with the Adaptive PCR instrument. The tighter clustering of the Cy5 and TEX615 probes indicated that they are better reporter dyes for use with blood than the FAM and VIC probes (Figure 3A). When compared with the same experiment performed in the traditional Rotor-Gene instrument (Figure 3B), the wild-type and no-template control (Figure 3C) samples clustered more tightly in the experiment was performed in the Adaptive PCR device. These data indicate that the LNA probes work at least as well in the Adaptive PCR instrument.
The LNA Hydrolysis Probes Discriminate between SNPs in Genomic DNA
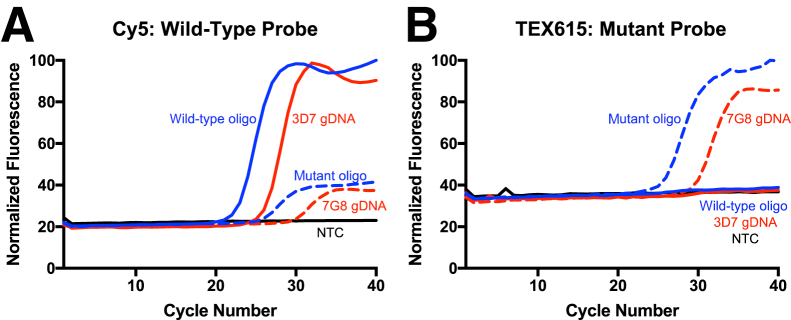
Molecular analyses with short oligonucleotide targets do not accurately reflect the complexity of clinical pathogen testing. Therefore, the CRT c.227A>C (p.Lys76Thr) probes were further tested using genomic DNA isolated from two different strains of P. falciparum parasites: the chloroquine-sensitive 3D7 (wild-type) strain and the chloroquine-resistant 7G8 (mutant) strain. In addition to the canonical chloroquine-resistance c.227A>C (p.Lys76Thr) DNA sequence change, a second nonsynonymous mutation in the 7G8 strain is also present, which results in the c.214T>A (p.Cys72Ser) SNP. However, this SNP is upstream of the probe sequence and was not expected to influence probe binding.
All samples were tested in the presence of a final concentration of 2% blood, which sought to provide a balance between maximizing available targets and maximizing the fluorescence signal. Robust amplification curves were observed with their corresponding SNP genotyping probes (Figure 4). Similar to the previous experiments (Figures 1 and 2), delayed mismatch amplification curves were observed with the wild-type (Figure 4A) but not the mutant probe (Figure 4B), indicating the wild-type probe had some low affinity for the mutant target sequence.
Figure 4.
Single-nucleotide polymorphism genotyping from genomic Plasmodium falciparum DNA in the presence of 2% blood. Each Adaptive PCR sample was spiked with 2% blood and P. falciparum genomic DNA (gDNA) from either a chloroquine-sensitive/reference strain (3D7; solid red line) or a chloroquine-resistant strain (7G8; dashed red line). As a positive control, a subset of samples was spiked with the amplicon-length oligonucleotide (blue lines). The wild-type probe is labeled with Cy5 (A), and the mutant is labeled with TEX615 (B). NTC, no-template control.
The c.227A>C (p.Lys76Thr) SNP was detectable from genomic parasite DNA, even in the presence of 2% blood, indicating that the LNA probes can be used to detect the c.227A>C (p.Lys76Thr) chloroquine-resistance SNP from parasite DNA. Furthermore, the addition of blood to the PCR did not substantially interfere with SNP detection.
c.227A>C SNP Genotyping Directly from Infected Red Blood Cells
In a final test of these probes and the potential point-of-care advantages of the Adaptive PCR instrument's design, PCR amplification was performed directly from blood. Unlike the earlier experiments, which used purified DNA, SNP genotyping was performed without an initial DNA extraction step. As P. falciparum infects RBCs, the potential to perform PCR directly on blood would eliminate the requirement for a DNA extraction step. To this end, infected RBCs were added directly to the Adaptive PCR sample and no DNA extraction step was performed. For optimal amplification, the samples were held at the initial denaturation temperature for 3 minutes for cellular lysis.
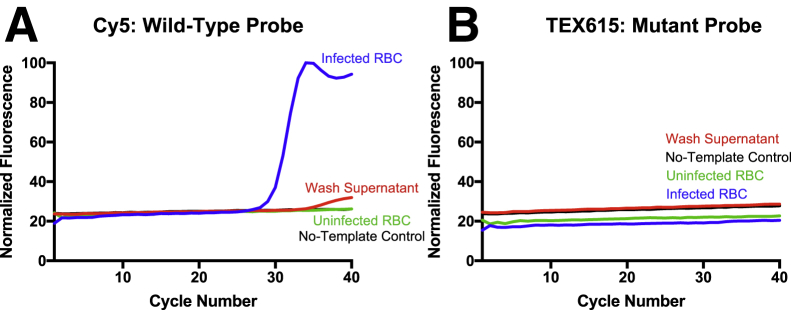
Cultured parasites at a parasitemia of 4% were added to the reaction tube at a concentration equivalent to 2% whole blood. To ensure that the amplified DNA is from the infected RBCs and not the media, the supernatant following the wash was also tested. The wild-type probe (Figure 5A), but not the mutant probe (Figure 5B), detected the drug-sensitive 3D7 strain. These results are similar to those obtained with genomic parasite DNA spiked into blood (Figure 4). Although no experiment could be performed with a chloroquine-resistant parasite culture, it can be expected that the mutant probe would detect the c.227A>C SNP directly from infected red blood cells, similarly to the genomic DNA spike (Figure 4).
Figure 5.
The c.227A>C probes enable single-nucleotide polymorphism genotyping directly from Plasmodium falciparum–infected red blood cells (RBCs). Amplification curves for the wild-type (A) and mutant (B) probes. Washed, packed RBCs from a 3D7 (wild-type) culture were added to an Adaptive PCR sample at a final concentration of 0.8%. The same concentration of the supernatant after washing was also evaluated for the presence of parasite DNA. Uninfected packed RBCs and no-template controls served as negative controls.
A small amplification curve was observed for the wash supernatant as well; however, this curve was at a much lower level (Figure 5A). Unlike the delayed amplification curve representing mismatch binding, observed in Figure 4A, the supernatant curve indicates that, even after washing, either free DNA or parasites are present in the supernatant at a lower concentration, ie, a small portion of the DNA detected in the infected RBC sample may be attributable to DNA that is not associated with infected RBCs (Figure 5A). No amplification was observed for the uninfected and no-template negative controls. As expected, amplification curves were not observed for the mutant probe (Figure 5B). The fluorescence intensity of the mutant probe was normalized to a positive control, which contained the mutant oligonucleotide.
Together, the data indicate that SNP genotyping can be performed directly from P. falciparum–infected RBCs. The initial preheating step is sufficient to lyse the RBCs and parasite cells, resulting in the release of genomic DNA. Despite the presence of blood in the reaction, the sample composition does not prevent amplification for SNP detection.
Discussion
The LNA probes designed for this study enable SNP genotyping directly from blood (Figure 5). As a consequence, drug-resistance testing for malaria can be performed without an additional DNA extraction step, which significantly decreases the test time and complexity. Although the World Health Organization no longer recommends chloroquine as a first-line treatment for P. falciparum infection,40 this study provides the framework for the general detection of SNPs in blood. It is particularly valuable for malaria investigations because malaria parasites infect red blood cells, but the approach can be further extended to studies of genetic markers in other organisms.
Because of the demonstrated effect of blood on fluorescence intensity, the choice of the reporter dyes is critical. Several of the most common reporter dyes (ie, SYBR Green, FAM, HEX, and VIC) fall into the green-yellow range of the spectrum, which overlaps with the absorbance spectra of blood. Cy5 and TEX615 were selected because they are orange-red dyes, which were found to be more compatible with the blood absorbance spectra than those traditionally used in MGB genotyping probes (Figure 1). In addition, the selection of a more inhibitor-tolerant Taq polymerase was the second essential feature of this work. OmniTaq 3 was chosen for our study on the basis of its performance in blood and its retention of 5′→3′ exonuclease activity. The field-deployable Adaptive PCR instrument further complements the molecular biology reagents. Together, these tools can be used to perform SNP genotyping directly from blood in the field.
Historically, monitoring of resistance to artemisinin, the current first-line therapy for P. falciparum infection, has been a tedious process. The standard approaches are based on parasite phenotype after drug treatment either clinically or in a ring-stage survival assay.41, 42, 43 In addition to a delayed-clearance phenotype, artemisinin-resistant parasites generally also harbor 1 of 20 different nonsynonymous mutations in the propeller domain of the gene that encodes for the Kelch13 protein.44 A potential approach would be to assess several SNPs simultaneously with all drug-resistance markers in the same fluorescence channel and a positive control in a separate channel. Although it would not be possible to determine the exact position if an SNP was found, the test would quickly provide information regarding the patient's resistance status. One recent study used the amplification-refractory mutation system to detect the five most prevalent SNPs associated with resistance.45 Their strategy and ours offer field-deployable approaches for complex molecular analyses. They have the potential to serve as a quick preliminary assessment of artemisinin resistance, thereby replacing sequencing as a screening tool.
Although the reagents outlined in this study could, in theory, also be used as a molecular detection test, they are likely less sensitive than is required to detect low copy numbers. PCR-based detection of malaria infection usually relies on amplification of targets that appear multiple times in the genome, thereby increasing the copies of the starting material.46, 47 Molecular markers of drug resistance, like those targeted in our assay, are usually found only once within each genome and, therefore, have inherently lower starting copy numbers. As such, the test described herein has lower sensitivity for the detection of malaria. It is better suited for assessing antimalarial resistance status from patients with well-established malaria infections, in whom it is expected that a high level of sensitivity is not required.
Although monitoring and containment are important to prevent the further spread of drug-resistant parasites, designing field-deployable methods to rapidly detect artemisinin resistance has been challenging. Our strategy eliminates the most time- and labor-intensive step: DNA extraction. By generating a procedure that overcomes the obstacles presented by blood, we have developed a simple and straightforward method to quickly identify SNPs associated with drug resistance. As a consequence, higher-throughput testing and more rapid sample-to-result turnaround will be possible, which will enable more comprehensive studies. The approach outlined herein could be optimized for assessing resistance to artemisinin or future drugs, and it could provide informative clues regarding the drug's efficacy, thereby preventing its further spread.
Acknowledgment
We thank BEI Resources (National Institute of Allergy and Infectious Diseases, NIH) for kindly providing the genomic DNA from Plasmodium falciparum, strain 7G8, MRA-926G, which was deposited by Thomas E. Wellems.
Footnotes
Supported by NIH Small Business Technology Transfer (STTR) grant R42 HG009470 (M.L., N.M.A., W.E.G. and F.R.H.) and the NIH-funded Vanderbilt-Zambia Network for Innovation in Global Health Technologies grant D43TW009348 (M.L., N.M.A., D.W.W. and F.R.H.).
Disclosures: None declared.
Current address of M.L., Department of Biomedical Engineering, Vanderbilt University, Nashville, TN.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2019.02.004.
Contributor Information
David W. Wright, Email: david.wright@vanderbilt.edu.
Frederick R. Haselton, Email: rick.haselton@vanderbilt.edu.
Supplemental Data
Effect of concentration and ratio on c.227A>C single-nucleotide polymorphism detection with locked nucleic acid probes. Allelic discrimination plot for two percentages of mutant oligonucleotide (oligo): 10% (9 × 106 copies of wild type to 106 copies of mutant; purple circles) and 50% (5 × 106 copies of each; purple squares). The equivalent copy numbers for the wild-type (blue symbols) or mutant (red symbols) oligonucleotides alone are included for comparison. The data were acquired on the Rotor-Gene Q instrument.
References
- 1.Wongsrichanalai C., Pickard A.L., Wernsdorfer W.H., Meshnick S.R. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 2.Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 3.Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S.S., Yeung S., Singhasivanon P., Day N.P., Lindegardh N., Socheat D., White N.J. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S. Tracking resistance to artemisinin C: spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phyo A.P., Nkhoma S., Stepniewska K., Ashley E.A., Nair S., McGready R., ler Moo C., Al-Saai S., Dondorp A.M., Lwin K.M., Singhasivanon P., Day N.P., White N.J., Anderson T.J., Nosten F. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thanh N.V., Thuy-Nhien N., Tuyen N.T., Tong N.T., Nha-Ca N.T., Dong L.T., Quang H.H., Farrar J., Thwaites G., White N.J., Wolbers M., Hien T.T. Rapid decline in the susceptibility of Plasmodium falciparum to dihydroartemisinin-piperaquine in the south of Vietnam. Malar J. 2017;16:27. doi: 10.1186/s12936-017-1680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., Shrestha S., Li X., Miao J., Yuan L., Cabrera M., Grube C., Yang Z., Cui L. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007-2012. Malar J. 2015;14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thriemer K., Hong N.V., Rosanas-Urgell A., Phuc B.Q., Ha do M., Pockele E., Guetens P., Van N.V., Duong T.T., Amambua-Ngwa A., D'Alessandro U., Erhart A. Delayed parasite clearance after treatment with dihydroartemisinin-piperaquine in Plasmodium falciparum malaria patients in central Vietnam. Antimicrobial Agents Chemother. 2014;58:7049–7055. doi: 10.1128/AAC.02746-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye R., Hu D., Zhang Y., Huang Y., Sun X., Wang J., Chen X., Zhou H., Zhang D., Mungthin M., Pan W. Distinctive origin of artemisinin-resistant Plasmodium falciparum on the China-Myanmar border. Sci Rep. 2016;6:20100. doi: 10.1038/srep20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulle M., Witkowski B., Duru V., Sriprawat K., Nair S.K., McDew-White M., Anderson T.J., Phyo A.P., Menard D., Nosten F. Artemisinin-resistant Plasmodium falciparum K13 mutant alleles, Thailand-Myanmar border. Emerg Infect Dis. 2016;22:1503–1505. doi: 10.3201/eid2208.160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang F., Takala-Harrison S., Jacob C.G., Liu H., Sun X., Yang H., Nyunt M.M., Adams M., Zhou S., Xia Z., Ringwald P., Bustos M.D., Tang L., Plowe C.V. A single mutation in K13 predominates in Southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis. 2015;212:1629–1635. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Menard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2013;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tun K.M., Imwong M., Lwin K.M., Win A.A., Hlaing T.M., Hlaing T., Lin K., Kyaw M.P., Plewes K., Faiz M.A., Dhorda M., Cheah P.Y., Pukrittayakamee S., Ashley E.A., Anderson T.J., Nair S., McDew-White M., Flegg J.A., Grist E.P., Guerin P., Maude R.J., Smithuis F., Dondorp A.M., Day N.P., Nosten F., White N.J., Woodrow C.J. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menard D., Khim N., Beghain J., Adegnika A.A., Shafiul-Alam M., Amodu O. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takala-Harrison S., Jacob C.G., Arze C., Cummings M.P., Silva J.C., Dondorp A.M. Independent emergence of Plasmodium falciparum artemisinin resistance mutations in Southeast Asia. J Infect Dis. 2014;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imwong M., Suwannasin K., Kunasol C., Sutawong K., Mayxay M., Rekol H., Smithuis F.M., Hlaing T.M., Tun K.M., van der Pluijm R.W., Tripura R., Miotto O., Menard D., Dhorda M., Day N.P.J., White N.J., Dondorp A.M. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imwong M., Hien T.T., Thuy-Nhien N.T., Dondorp A.M., White N.J. Spread of a single multidrug resistant malaria parasite lineage (PfPailin) to Vietnam. Lancet Infect Dis. 2017;17:1022–1023. doi: 10.1016/S1473-3099(17)30524-8. [DOI] [PubMed] [Google Scholar]
- 19.Fidock D.A., Nomura T., Talley A.K., Cooper R.A., Dzekunov S.M., Ferdig M.T., Ursos L.M., Sidhu A.B., Naude B., Deitsch K.W., Su X.Z., Wootton J.C., Roepe P.D., Wellems T.E. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basco L.K., Ringwald P. Analysis of the key pfcrt point mutation and in vitro and in vivo response to chloroquine in Yaounde, Cameroon. J Infect Dis. 2001;183:1828–1831. doi: 10.1086/320726. [DOI] [PubMed] [Google Scholar]
- 21.Kim S., Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng. 2007;9:289–320. doi: 10.1146/annurev.bioeng.9.060906.152037. [DOI] [PubMed] [Google Scholar]
- 22.Kutyavin I.V., Afonina I.A., Mills A., Gorn V.V., Lukhtanov E.A., Belousov E.S., Singer M.J., Walburger D.K., Lokhov S.G., Gall A.A., Dempcy R., Reed M.W., Meyer R.B., Hedgpeth J. 3'-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouritzen P., Nielsen A.T., Pfundheller H.M., Choleva Y., Kongsbak L., Moller S. Single nucleotide polymorphism genotyping using locked nucleic acid (LNA) Expert Rev Mol Diagn. 2003;3:27–38. doi: 10.1586/14737159.3.1.27. [DOI] [PubMed] [Google Scholar]
- 24.Braasch D.A., Corey D.R. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 25.Rupp J., Solbach W., Gieffers J. Single-nucleotide-polymorphism-specific PCR for quantification and discrimination of Chlamydia pneumoniae genotypes by use of a “locked” nucleic acid. Appl Environ Microbiol. 2006;72:3785–3787. doi: 10.1128/AEM.72.5.3785-3787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson M.P., Haupt L.M., Griffiths L.R. Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. Nucleic Acids Res. 2004;32:e55. doi: 10.1093/nar/gnh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simeonov A., Nikiforov T.T. Single nucleotide polymorphism genotyping using short, fluorescently labeled locked nucleic acid (LNA) probes and fluorescence polarization detection. Nucleic Acids Res. 2002;30:e91. doi: 10.1093/nar/gnf090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latorra D., Campbell K., Wolter A., Hurley J.M. Enhanced allele-specific PCR discrimination in SNP genotyping using 3' locked nucleic acid (LNA) primers. Hum Mutat. 2003;22:79–85. doi: 10.1002/humu.10228. [DOI] [PubMed] [Google Scholar]
- 29.Al-Soud W.A., Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abu Al-Soud W., Radstrom P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J Clin Microbiol. 2000;38:4463–4470. doi: 10.1128/jcm.38.12.4463-4470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosschaart N., Edelman G.J., Aalders M.C., van Leeuwen T.G., Faber D.J. A literature review and novel theoretical approach on the optical properties of whole blood. Lasers Med Sci. 2014;29:453–479. doi: 10.1007/s10103-013-1446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abu Al-Soud W., Radstrom P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kermekchiev M.B., Kirilova L.I., Vail E.E., Barnes W.M. Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res. 2009;37:e40. doi: 10.1093/nar/gkn1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trombley Hall A., McKay Zovanyi A., Christensen D.R., Koehler J.W., Devins Minogue T. Evaluation of inhibitor-resistant real-time PCR methods for diagnostics in clinical and environmental samples. PLoS One. 2013;8:e73845. doi: 10.1371/journal.pone.0073845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawyer F.C., Stoffel S., Saiki R.K., Myambo K., Drummond R., Gelfand D.H. Isolation, characterization, and expression in Escherichia coli of the DNA polymerase gene from Thermus aquaticus. J Biol Chem. 1989;264:6427–6437. [PubMed] [Google Scholar]
- 36.Wilson P.E., Kazadi W., Kamwendo D.D., Mwapasa V., Purfield A., Meshnick S.R. Prevalence of pfcrt mutations in Congolese and Malawian Plasmodium falciparum isolates as determined by a new Taqman assay. Acta Trop. 2005;93:97–106. doi: 10.1016/j.actatropica.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Adams N.M., Gabella W.E., Hardcastle A.N., Haselton F.R. Adaptive PCR based on hybridization sensing of mirror-image l-DNA. Anal Chem. 2017;89:728–735. doi: 10.1021/acs.analchem.6b03291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandlin R.D., Fong K.Y., Wicht K.J., Carrell H.M., Egan T.J., Wright D.W. Identification of beta-hematin inhibitors in a high-throughput screening effort reveals scaffolds with in vitro antimalarial activity. Int J Parasitol Drugs Drug Resist. 2014;4:316–325. doi: 10.1016/j.ijpddr.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak K.J. Allelic discrimination using fluorogenic probes and the 5' nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 40.World Health Organization . ed 3. Geneva, Switzerland; 2015. Global malaria programme: guidelines for the treatment of malaria. Available at https://apps.who.int/iris/bitstream/handle/10665/162441/9789241549127_eng.pdf;jsessionid=2A183FF8182287D534335EACAC9AB1B8?sequence=1 (accessed May 9, 2019) [Google Scholar]
- 41.Witkowski B., Amaratunga C., Khim N., Sreng S., Chim P., Kim S., Lim P., Mao S., Sopha C., Sam B., Anderson J.M., Duong S., Chuor C.M., Taylor W.R., Suon S., Mercereau-Puijalon O., Fairhurst R.M., Menard D. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flegg J.A., Guerin P.J., White N.J., Stepniewska K. Standardizing the measurement of parasite clearance in falciparum malaria: the parasite clearance estimator. Malar J. 2011;10:339. doi: 10.1186/1475-2875-10-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.World Health Organization . Geneva, Switzerland; 2017. Status report: artemisinin and artemisinin-based combination therapy resistance. Available at https://apps.who.int/iris/bitstream/handle/10665/255213/WHO-HTM-GMP-2017.9-eng.pdf?sequence=1&isAllowed=y (accessed May 9, 2019) [Google Scholar]
- 44.Fairhurst R.M., Dondorp A.M. Artemisinin-resistant Plasmodium falciparum malaria. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.EI10-0013-2016. EI10-0013-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vachot-Ganee L., Khim N., Iannello A., Legrand E., Kim S., Eam R., Khean C., Ken M., Ashley E., Tun K.M., Dhorda M., Nosten F., Souleymane I.M., Blein S., Pachot A., Ariey F., Kaiser K., Menard D. A novel field-based molecular assay to detect validated artemisinin-resistant k13 mutants. Malar J. 2018;17:175. doi: 10.1186/s12936-018-2329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demas A., Oberstaller J., DeBarry J., Lucchi N.W., Srinivasamoorthy G., Sumari D., Kabanywanyi A.M., Villegas L., Escalante A.A., Kachur S.P., Barnwell J.W., Peterson D.S., Udhayakumar V., Kissinger J.C. Applied genomics: data mining reveals species-specific malaria diagnostic targets more sensitive than 18S rRNA. J Clin Microbiol. 2011;49:2411–2418. doi: 10.1128/JCM.02603-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rougemont M., Van Saanen M., Sahli R., Hinrikson H.P., Bille J., Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–5643. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of concentration and ratio on c.227A>C single-nucleotide polymorphism detection with locked nucleic acid probes. Allelic discrimination plot for two percentages of mutant oligonucleotide (oligo): 10% (9 × 106 copies of wild type to 106 copies of mutant; purple circles) and 50% (5 × 106 copies of each; purple squares). The equivalent copy numbers for the wild-type (blue symbols) or mutant (red symbols) oligonucleotides alone are included for comparison. The data were acquired on the Rotor-Gene Q instrument.