Abstract
Since many oncogenes, including BCR-ABL, may promote the acquisition and maintenance of the glycolytic phenotype, we tested whether treatment of BCR-ABL-driven human leukemia cells with imatinib, a selective BCR-ABL inhibitor, can modulate the expression of key glycolytic enzymes and mitochondrial complex subunits thus causing alterations of glucose metabolism. BCR-ABL-driven K562 and KCL-22 cells were incubated with increasing concentrations of imatinib to preliminarily test drug sensitivity. Then untreated and treated cells were analyzed for levels of BCR-ABL signaling mediators and key proteins of glycolytic cascade and oxidative phosphorylation. Effective inhibition of BCR-ABL caused a concomitant reduction of p-ERK1/2, p-AKT, phosphorylated form of STAT3 (at Tyr705 and Ser727), c-Myc and cyclin D1 along with an increase of cleaved PARP and caspase 3 at 48 h after treatment. Furthermore, a strong reduction of the hexokinase II (HKII), phosphorylated form of PKM2 (at Tyr105 and Ser37) and lactate dehydrogenase A (LDH-A) was observed in response to imatinib along with a strong upregulation of mitochondrial complexes (OXPHOS). According to these findings, a significant reduction of glucose consumption and lactate secretion along with an increase of intracellular ATP levels was observed in response to imatinib. Our findings indicate that imatinib treatment of BCR-ABL-driven human leukemia cells reactivates mitochondrial oxidative phosphorylation thus allowing potential co-targeting of BCR-ABL and OXPHOS.
Keywords: BCR-ABL, OXPHOS, aerobic glycolysis, chronic myeloid leukemia
1. Introduction
Chronic myelogenous leukemia (CML) is a myeloproliferative disorder consistently associated with a t(9;22)(q34;q11) translocation which gives rise to the Philadelphia chromosome [1,2]. This translocation generates the BCR-ABL fusion gene that encodes for the chimeric BCR-ABL protein and drives neoplastic transformation of hematopoietic stem cells [2,3]. When fused with BCR, ABL tyrosine kinase is constitutively activated and retained within the cytoplasm where it can interact with several adaptor molecules creating multiprotein signaling complexes. BCR-ABL is reported to interact with Ras-mitogen-activated protein kinase (MAPK) pathway leading to increased proliferation, the Janus-activated kinase (JAK/STAT) pathway leading to impaired transcriptional activity, and the phosphoinositide 3-kinase (PI3K/AKT) pathway resulting in enhanced survival [4].
Imatinib mesylate is a small-molecule inhibitor of BCR-ABL tyrosine kinase activity that, by competing with ATP for the binding to the kinase domain of ABL, prevents chimeric protein autophosphorylation and downstream signaling leading to growth arrest and apoptosis [5]. Imatinib was approved for treatment of patients with chronic-phase CML and rapidly became the standard first-line therapy for those patients [6,7]. Although the mechanism of action of imatinib and its clinical efficacy have been well established, little is known about mechanisms underlying changes in energy metabolism of BCR-ABL driven CML cells in response to imatinib. Previous studies, using magnetic resonance spectroscopy, showed that imatinib treatment of BCR-ABL positive cells causes a decrease of glucose uptake and lactate production whereas it increases the production of intermediates of the Krebs cycle [8,9].
The aim of the present study was to test whether inhibition of BCR-ABL signaling by imatinib can modulate the expression of key glycolytic enzymes and mitochondrial complex subunits thus causing alterations of glucose metabolism.
2. Results
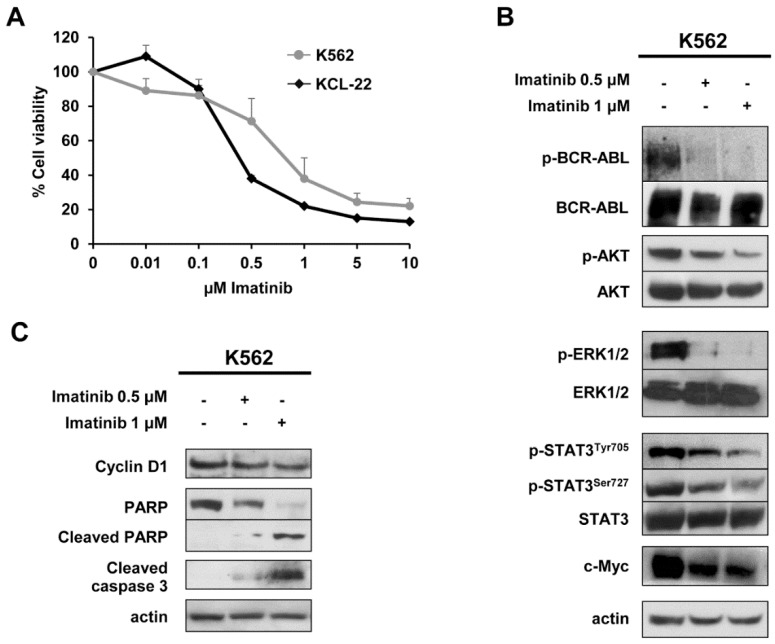
The sensitivity of K562 and KCL-22 cells to increasing concentrations of imatinib was tested by MTS assay and the results are shown in Figure 1A. Viability of K562 and KCL-22 cells decreased in a dose-dependent manner showing an EC50 of approximately 0.7 and 0.3 µM, respectively, indicating sensitivity to imatinib. Furthermore, exposure of K562 cells to imatinib for 48 h caused a dose-dependent decrease of p-BCR-ABL, p-AKT, p-ERK1/2, p-STAT3Tyr705, p-STAT3Ser727 and c-Myc levels (Figure 1B) that lead to a decrease of cyclin D1 and a concomitant enhancement of cleaved PARP and cleaved caspase 3 levels (Figure 1C) indicating a drug-induced growth arrest and apoptosis. Similarly, a dose-dependent decrease of p-BCR-ABL and cyclin D1 was also observed in KCL-22 cells (Figure S1).
Figure 1.
Modulation of BCR-ABL signalling by imatinib in chronic myelogenous leukemia (CML) cells. (A) Cell toxicity assays performed after 72 h of treatment with increasing concentration of imatinib showing an EC50 of 0.7 and 0.3 µM in K562 and KCL-22 cells respectively. Three independent assays were performed and data are expressed as mean ± SD. (B,C) Representative western blot analyses of whole-cell lysates obtained from K562 cells exposed to 0.5 and 1 μM imatinib or vehicle for 48 h. Actin served to ensure equal loading. At least three independent assays were performed.
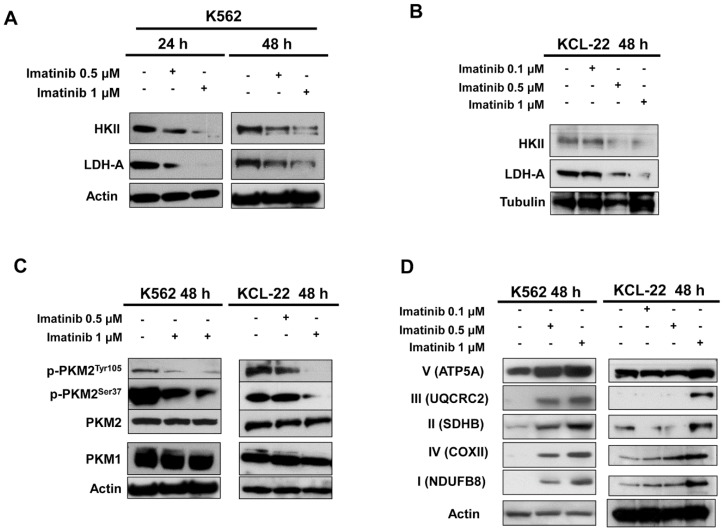
Then levels of key glycolytic enzymes were determined in untreated and treated K562 and KCL-22 cells. A dose-dependent decrease of HKII and LDH-A expression (Figure 2A,B) was observed after 24 and 48 h treatment in K562 cells and confirmed in KCL-22 cells. Similarly, both cell lines treated for 48 h showed a dose-dependent decrease of p-PKM2Tyr105 and p-PKM2Ser37 levels (Figure 2C). In addition, 48 h imatinib treatment of K562 and KCL-22 cells caused a strong up-regulation of mitochondrial complex subunits (OXPHOS) indicating a concomitant reactivation of mitochondrial oxidative phosphorylation (Figure 2D). In contrast to OXPHOS increase, no significant changes of mitochondrial markers were found in response to imatinib as shown in Figure S2.
Figure 2.
Protein levels of glycolytic enzymes and OXPHOS in CML cells. (A,B) Protein levels of HKII and LDH-A were assessed by western blot analysis of whole cell lysates from K562 (A) and KCL-22 (B) cells exposed to 0.1, 0.5, 1 μM imatinib or vehicle for 24 or 48 h. (C) Levels of p-PKM2Tyr105, p-PKM2Ser37 along with total form of PKM2 and PKM1 after 48 h treatment with imatinib assessed by western blotting. (D) Levels of OXPHOS in K562 and KCL-22 cells exposed to 0.1, 0.5 or 1 μM imatinib at 48 h. Actin served to ensure equal loading. At least three independent assays were performed.
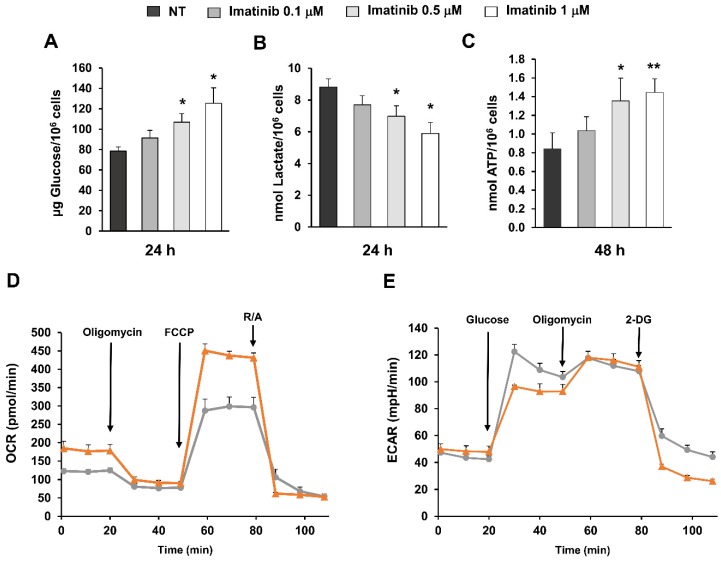
Then we evaluated glucose consumption, lactate secretion and ATP production in untreated and imatinib-treated K562 cells. A significant increase of glucose concentration was observed after 24 h in conditioned media of treated cells as compared to untreated controls (p < 0.05) indicating a lower glucose consumption (Figure 3A). A parallel significant decrease (p < 0.05) of lactate levels was found at 24 h in treated cells (Figure 3B) whereas intracellular ATP levels were significantly increased after 48 h of treatment with 1 µM imatinib (p < 0.01) (Figure 3C). In agreement with western blot analysis, functional assays indicate that imatinib treatment causes an early reduction of glucose consumption and lactate production through glycolysis followed by an increase of ATP indicating reactivation of oxidative phosphorylation. To confirm the results of functional assays, oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured in K562 cells exposed to 0.5 μM imatinib or vehicle for 48 h. An enhancement of both basal and maximal respiration rate was observed in treated K562 cells as compared to untreated controls (Figure 3D). Conversely, a reduction of ECAR was observed in treated cells as compared to untreated controls following the addition of glucose indicating a downregulation of glycolysis (Figure 3E).
Figure 3.
Functional assays, oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements in K562 cells exposed to imatinib. K562 cells were treated for 24 or 48 h with imatinib at the indicated concentrations. (A) Residual glucose levels were measured in conditioned media from untreated and treated cells. Data are expressed as μg of glucose normalized to 106 cells. (B) Lactate levels were measured in conditioned media of untreated and treated cells and expressed as nmol of secreted lactate normalized to 106 cells. (C) Intracellular ATP levels were determined in lysates of untreated and treated cells and expressed as nmol of ATP normalized to 106 cells. The symbol * indicates significant differences versus untreated control with * p < 0.05 and ** p < 0.01. At least three independent assays were performed. (D,E) Simultaneous measurements of OCR (D) and ECAR (E) were performed in quintuplicate samples of untreated (grey line) and treated (orange line) K562 cells. All data are expressed as mean ± SE.
3. Discussion
The present study shows that imatinib treatment causes an early downregulation of the expression of glycolytic enzymes and an upregulation of mitochondrial complex subunits in BCR-ABL driven CML cells leading to a reduction of glucose consumption and an increase of intracellular ATP production. In particular, the decrease of HKII reduces the first step of glycolysis and glucose flux through the glycolytic cascade while the reduction of LDH-A caused a decrease of lactate secretion. Pyruvate kinase is a glycolytic enzyme that converts phosphoenolpyruvate to pyruvate [10,11]. Two isoforms of this enzyme, PKM1 and PKM2, are derived from alternative splicing of the primary RNA transcript of the PKM gene. PKM2 was found to be overexpressed in cancer cells and exists in a dimeric and tetrameric form associated with a low and high catalytic activity, respectively [12,13]. The reduction of p-PKM2Tyr105 levels in response to imatinib promotes tetramer formation and a high enzymatic catalytic activity leading to an increase of pyruvate that can be directed toward mitochondrial oxidative phosphorylation [14]. The enhanced pyruvate flux through Krebs cycle and the upregulation of OXPHOS may allow the increase of intracellular ATP levels.
Phosphorylation of PKM2 at Ser37 was reported to induce its translocation into the nucleus where PKM2 serves as a transcriptional factor of several genes including c-Myc [15,16]. Imatinib causing a reduction of p-PKM2Ser37 may modulate the expression of a pool of c-Myc-dependent genes.
In our previous study, we showed the reversal of glycolytic phenotype in EGFR-driven non-small cell lung cancer cells and xenografts in response to EGFR inhibitors [17]. Here we demonstrated that similar results can be obtained by inhibiting BCR-ABL suggesting that different oncogene drivers can share common mechanisms to modulate energy metabolism.
The upregulation of OXPHOS, the increase of intracellular ATP levels and the enhanced OCR in response to imatinib reflect the restoration of mitochondrial function. However, in these BCR-ABL driven cells, imatinib caused also an induction of apoptosis that leads to mitochondrial disruption. It is conceivable that in this context where glycolysis is downregulated and mitochondria are the only source of energy, the addition of OXPHOS inhibitors to imatinib would have a lethal effect on BCR-ABL driven cells. In this respect, the mitochondrial ATP-synthase inhibitor oligomycin A was reported to greatly sensitized leukaemia cells to imatinib predicting a potential benefit for patients treated with a combination of imatinib and OXPHOS inhibitors [18].
In conclusion, our study provided insights into the mechanisms underlying the changes of glucose metabolism in BCR-ABL driven cells treated with imatinib identifying OXPHOS as a potential target for therapy in combination with TKIs.
4. Materials and Methods
4.1. Cell Line and Treatment
The CML cell line, K562, was purchased from American Type Culture Collection whereas KCL-22 cells were kindly provided by Prof. B. Izzo and Dr. F. Quarantelli. K562 and KCL-22 cells were grown in IMDM and RPMI 1640 (Gibco, Thermo Fisher, Waltham, MA, USA) culture media, respectively, containing 10% fetal bovine serum (FBS), 100 IU/mL penicillin and 50 μg/mL streptomycin in a humidified incubator with 5% CO2 at 37 °C. Drug-induced toxicity was assessed by MTS assay (Promega, Madison, WI, USA) as previously described [19]. Briefly, cells were plated at a density of 5000/well in 96-well plates and then treated with increasing concentration (0.1–10 μM) of imatinib (Biaffin GmbH & Co KG, Germany) or vehicle for 72 h. The optical density (OD) was measured at 490 nm using microplate spectrophotometer, after 2 h incubation with MTS at 37 °C. At least three independent assays were performed and data are expressed as percentage of viable cells, considering the untreated control cells as 100%. EC50 values were calculated using GraphPad.
4.2. Immunoblotting Analysis
Whole cell lysates were prepared as previously described [20] after treatment with imatinib at 0.1, 0.5 or 1 μM for 24 or 48 h. Then untreated and treated cells were lysed with RIPA buffer (Sigma Aldrich, St. Louis, MO, USA), homogenized and centrifuged at 13,000× g at 4 °C for 20 min. Western blot analysis was performed using a standard procedure.
Antibodies used for western blotting included mouse monoclonal antibodies recognizing BCR-ABL (1 µg/µl, Thermo scientific), STAT3 (1:1000), p42/44 MAP kinase (ERK1/2) (0.1 mg/mL) (Cell Signaling), HSP70 (2 μg/mL) (Santa Cruz Biotechnology), cytochrome C (1 μg/mL, BD Pharmingen, San Jose, CA, USA), α-tubulin (1 μg/mL), actin (1 μg/mL) (Sigma), PARP (1:1000, BD Pharmingen), the OXPHOS cocktail of 5 mAbs (Mitoscience, Eugene, OR; 1:1000) recognizes the following proteins: 20-kD subunit of Complex I (20 kD), COX II of Complex IV (22 kD), 30-kD Ip subunit of Complex II (30 kD), core 2 of Complex III (~50 kD), and F1α (ATP synthase) of Complex V (~60 kD); rabbit monoclonal antibodies against p-c-AblTyr412 (1:1000), Hexokinase II (1:1000), PKM2 (1:1000), LDH-A (1:1000), p-STAT3Tyr705 (1:1000) (Cell Signaling); rabbit polyclonal antibodies against PKM1 (1:1000, Abgent), p-AKTSer473 (1:1000), PGC-1α (1:1000) (Santa Cruz Biotechnology), p-PKM2Ser37 (1:1000) (Signalway Antibody), p-PKM2Tyr105 (1:1000), phospho-p42/44 MAP kinase (ERK1/2) (1:1000), AKT (1:1000), cyclin D1 (1:1000), cleaved Caspase-3 (1:1000), c-Myc (1:1000), p-STAT3Ser727 (1:1000), (Cell Signaling). A commercially available ECL kit (GE Healthcare, UK) was used to reveal the reaction. At least three independent assays were performed.
4.3. Glucose Consumption, Lactate Secretion and Intracellular ATP
Cells were seeded in six-well flat-bottomed plates at a density of 3 × 105 cells per well and then treated with imatinib at the indicated concentration for 24 or 48 h. Untreated and treated cells were analyzed for glucose and lactate levels in the conditioned media and for intracellular ATP levels. Briefly, after treatment, conditioned media were removed, centrifuged at 13,000× g at 4 °C for 10 min and then assayed for glucose and lactate concentrations using the Glucose Assay Kit (Sigma-Aldrich) and the Lactate Assay Kit (Sigma-Aldrich) following manufacturers’ instructions. Cells were simultaneously subjected to intracellular ATP determination using the ATPlite Luminescence Assay (Perkin Elmer; Waltham, MA, USA) following manufacturer’s instructions. Briefly, cells were lysed, incubated with the ATP reaction mixture for 5 min and then subjected to luminescence measurements. Absolute glucose, lactate and ATP levels were calculated from the corresponding standard curve and normalized to 106 cells. At least three independent assays were performed.
4.4. Oxygen Consumption and Extracellular Acidification Rates
The oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) were determined using the Seahorse Extracellular Flux Analyzer (XF-96, Seahorse Bioscience, North Billerica, MA, USA). Briefly, K562 cells were plated at a density of 5 × 106 cells in 10 cm petri dishes and then treated with 0.5 µM imatinib for 48 h. Then cells were plated on XF 96-well microplates and allowed to equilibrate. OCR was measured in basal conditions and after the subsequent addition of 5 μM oligomycin, 1.5 μM carbonylcyanide-4-(trifluoromethoxy)-phenylhydrazone (FCCP) and 1 μM rotenone/antimycin A. ECAR was simultaneously measured in basal conditions and after the subsequent addition of 10 mM glucose, 5 μM oligomycin and 100 mM 2-deoxyglucose.
4.5. Statistical Analysis
Statistical analysis was performed using the software MedCalc for Windows, version 12.7.0.0, (MedCalc Software, Mariakerke, Belgium). Unpaired Student’s t-test was used to compare means. Differences between means were considered statistically significant for p < 0.05.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/13/3134/s1.
Author Contributions
Conception and design, V.D.R, F.I. and S.D.V.; Development of methodology, M.M., C.T. and V.D.R.; Acquisition of data, M.M. and C.T.; Analysis and interpretation of data, V.D.R., R.F. and F.I.; Writing the manuscript, V.D.R., F.I. and S.D.V.; Revision of the manuscript, V.D.R., F.I. and S.D.V.; Study supervision, S.D.V.
Funding
This work was partly supported by AIRC, Associazione Italiana per la Ricerca sul Cancro (project No. IG-17249) and a grant from Regione Campania SATIN Project 2018–2020.
Conflicts of Interest
The authors declare that there are no conflict of interest.
References
- 1.Nowell P.C., Hungerford D.A. A minute chromosome in human chronic granulocotic leukemia. Science. 1960;132:1497–1501. [Google Scholar]
- 2.Ren R. Mechanisms of bcr-abl in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 3.Melo J.V., Barnes D.J. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat. Rev. Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 4.Cilloni D., Saglio G. Molecular pathways: Bcr-abl. Clin. Cancer Res. 2012;18:930–937. doi: 10.1158/1078-0432.CCR-10-1613. [DOI] [PubMed] [Google Scholar]
- 5.Marcucci G., Perrotti D., Caligiuri M.A. Understanding the molecular basis of imatinib mesylate therapy in chronic myelogenous leukemia and the related mechanisms of resistance. Commentary re: A. N. Mohamed et al. The effect of imatinib mesylate on patients with philadelphia chromosome-positive chronic myeloid leukemia with secondary chromosomal aberrations. Clin. Cancer res., 9: 1333–1337, 2003. Clin. Cancer Res. 2003;9:1248–1252. [PubMed] [Google Scholar]
- 6.Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantarjian H., Capdeville R., Ohno-Jones S., et al. Efficacy and safety of a specific inhibitor of the bcr-abl tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 7.Rea D., Etienne G., Nicolini F., Cony-Makhoul P., Johnson-Ansah H., Legros L., Huguet F., Tulliez M., Gardembas M., Bouabdallah K., et al. First-line imatinib mesylate in patients with newly diagnosed accelerated phase-chronic myeloid leukemia. Leukemia. 2012;26:2254–2259. doi: 10.1038/leu.2012.92. [DOI] [PubMed] [Google Scholar]
- 8.Gottschalk S., Anderson N., Hainz C., Eckhardt S.G., Serkova N.J. Imatinib (sti571)-mediated changes in glucose metabolism in human leukemia bcr-abl-positive cells. Clin. Cancer Res. 2004;10:6661–6668. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- 9.Kominsky D.J., Klawitter J., Brown J.L., Boros L.G., Melo J.V., Eckhardt S.G., Serkova N.J. Abnormalities in glucose uptake and metabolism in imatinib-resistant human bcr-abl-positive cells. Clin. Cancer Res. 2009;15:3442–3450. doi: 10.1158/1078-0432.CCR-08-3291. [DOI] [PubMed] [Google Scholar]
- 10.Hitosugi T., Chen J. Post-translational modifications and the warburg effect. Oncogene. 2014;33:4279–4285. doi: 10.1038/onc.2013.406. [DOI] [PubMed] [Google Scholar]
- 11.Hitosugi T., Kang S., Vander Heiden M.G., Chung T.W., Elf S., Lythgoe K., Dong S., Lonial S., Wang X., Chen G.Z., et al. Tyrosine phosphorylation inhibits pkm2 to promote the warburg effect and tumor growth. Sci. Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dayton T.L., Jacks T., Vander Heiden M.G. Pkm2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christofk H.R., Vander Heiden M.G., Harris M.H., Ramanathan A., Gerszten R.E., Wei R., Fleming M.D., Schreiber S.L., Cantley L.C. The m2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 14.Christofk H.R., Vander Heiden M.G., Wu N., Asara J.M., Cantley L.C. Pyruvate kinase m2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 15.Yang W., Zheng Y., Xia Y., Ji H., Chen X., Guo F., Lyssiotis C.A., Aldape K., Cantley L.C., Lu Z. Erk1/2-dependent phosphorylation and nuclear translocation of pkm2 promotes the warburg effect. Nat. Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakasam G., Iqbal M.A., Bamezai R.N.K., Mazurek S. Posttranslational modifications of pyruvate kinase m2: Tweaks that benefit cancer. Front Oncol. 2018;8:22. doi: 10.3389/fonc.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Rosa V., Iommelli F., Monti M., Fonti R., Votta G., Stoppelli M.P., Del Vecchio S. Reversal of warburg effect and reactivation of oxidative phosphorylation by differential inhibition of egfr signaling pathways in non-small cell lung cancer. Clin. Cancer Res. 2015;21:5110–5120. doi: 10.1158/1078-0432.CCR-15-0375. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez-Calderon F., Gregory M.A., Pham-Danis C., DeRyckere D., Stevens B.M., Zaberezhnyy V., Hill A.A., Gemta L., Kumar A., Kumar V., et al. Tyrosine kinase inhibition in leukemia induces an altered metabolic state sensitive to mitochondrial perturbations. Clin. Cancer Res. 2015;21:1360–1372. doi: 10.1158/1078-0432.CCR-14-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iommelli F., De Rosa V., Gargiulo S., Panico M., Monti M., Greco A., Gramanzini M., Ortosecco G., Fonti R., Brunetti A., et al. Monitoring reversal of met-mediated resistance to egfr tyrosine kinase inhibitors in non-small cell lung cancer using 3’-deoxy-3’-[18f]-fluorothymidine positron emission tomography. Clin. Cancer Res. 2014;20:4806–4815. doi: 10.1158/1078-0432.CCR-14-0264. [DOI] [PubMed] [Google Scholar]
- 20.Monti M., Iommelli F., De Rosa V., Carriero M.V., Miceli R., Camerlingo R., Di Minno G., Del Vecchio S. Integrin-dependent cell adhesion to neutrophil extracellular traps through engagement of fibronectin in neutrophil-like cells. PLoS ONE. 2017;12:e0171362. doi: 10.1371/journal.pone.0171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.