Abstract
Background
Zinc finger proteins (ZFPs) containing only a single zinc finger domain play important roles in the regulation of plant growth and development, as well as in biotic and abiotic stress responses. To date, the evolutionary history and functions of the ZFP gene family have not been identified in cotton.
Results
In this paper, we identified 29 ZFP genes in Gossypium hirsutum. This gene family was divided into seven subfamilies, 22 of which were distributed over 17 chromosomes. Bioinformatic analysis revealed that 20 GhZFP genes originated from whole genome duplications and two originated from dispersed duplication events, indicating that whole genome duplication is the main force in the expansion of the GhZFP gene family. Most GhZFP8 subfamily genes, except for GhZFP8–3, were highly expressed during fiber cell growth, and were induced by brassinosteroids in vitro. Furthermore, we found that a large number of GhZFP genes contained gibberellic acid responsive elements, auxin responsive elements, and E-box elements in their promoter regions. Exogenous application of these hormones significantly stimulated the expression of these genes.
Conclusions
Our findings reveal that GhZFP8 genes are involved in cotton fiber development and widely induced by auxin, gibberellin and BR, which provides a foundation for the identification of more downstream genes with potential roles in phytohormone stimuli, and a basis for breeding better cotton varieties in the future.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1932-6) contains supplementary material, which is available to authorized users.
Keywords: Cotton, Zinc finger proteins, Plant hormone, Expression patterns, Fiber development
Background
Cotton is one of the most important crops globally for industrial fiber and oil seed production. Cotton fiber, a primary resource for the textile industry, contributes more than thirteen billion dollars annually to the global economy and creates 330 million jobs in the agricultural or industrial sectors [1]. Gossypium hirsutum (G. hirsutum), the most commonly cultivated cotton species for fiber and oil, accounts for more than 90% of annual global cotton production [2]. Cotton is composed of both diploid and tetraploid species belonging to the Gossypium genus. G. hirsutum is an AADD allotetraploid species, which evolved from A-genome diploids resembling Gossypium arboreum and D-genome diploids resembling Gossypium raimondii, approximately 1–2 million years ago (MYA) [2].
Previous research has shown that phytohormones play critical roles in the process of fiber development. For example, it has been reported that endogenous levels of gibberenllic acid (GA3) are higher in a long staple cotton cultivar compared to medium and short staple cultivars [3]. Exogenous application of GA3 improved fiber elongation, increased fiber cell wall thickness and increased the weight of individual fibers [3, 4]. In vitro ovule culture assay showed that brassinosteroid (BR) significantly promoted fiber elongation, whereas its biosynthesis inhibitor brassinazole (BRZ) abrogated fiber elongation [5]. Treatment with BR and BRZ increased and decreased the expression of cell wall related genes, respectively. In addition, genes involved in the BR biosynthetic pathway were up-regulated during fiber initiation and elongation stages [6, 7]. Exogenous application of indole-3-acetic acid (IAA) significantly increased the total fiber volume [8], while application of IAA transport inhibitor, 1-naphthylphthalamic acid (NPA), dramatically reduced IAA content and the number of fiber cells [9]. Overexpression of the IAA biosynthetic gene iaaM significantly increased fiber cell number, final yield, and overall quality [9].
Zinc-finger proteins (ZFPs) have been shown to act a pivotal part in diverse biological processes, and can be divided into 23 subfamilies based on their structural differences, including A20, AN, Bbox, CDGSH, CHY, DHHC, Dof, FYVE, GATA, LYAR, MSRING, NFX1, PADPP, PHD, RBPO, Ring, TAZ, TDDP, TFIIB, Ubox, UBR, WRKY, and ZK [10–12]. Although ZFPs are abundant in plants, only a few ZFPs, which contain only single zinc finger domain, have been characterized to function in regulation of plant height [13, 14], plant development [15], secondary cell wall thickening [16], anther development [17], root development [18], flower development [19], seed germination [20], and fruit ripening [21].
ZFP genes have been found to participate in various biological processes, including signal transduction, transcriptional regulation, RNA binding and morphogenesis, and stress response [22–24]. In Arabidopsis, ZFP proteins have been devided into 9 groups: C2H2, C8, C6, C3HC4, C2HC, C2HC5, C4, C3H and C4HC3 [25]. AtZFP1, was highly expressed in apical meristem, vascular system, and seedlings at three days post germination. The zfp1 mutant has a dominant phenotype in leaf initiation [26]. ZFP6 encoded a C2H2 zinc finger protein, which are involved in regulating trichome development by integrating gibberellic acid and cytokinin signaling [27]. ZFP5, a gene downstream of ZFP6 signaling, encodes a cell-to-cell mobile mRNA, necessary for regulating trichome development [27]. ZFP10, a DNA binding transcription factor that targets specific sequences, was found to be involved in zinc ion and nucleic acid binding [28]. Overexpression of ZFP11 resulted in mortality and a deformed phenotype in Arabidopsis [29].
In this study, we identified 29 GhZFP genes in G. hirsutum and charactered their evolutionary relationships, chromosomal distribution, gene duplication and gene structure to gain insight into the role of GhZFPs in cotton. Gene expression patterns showed that GhZFP8 subfamily genes, except for GhZFP8–3, were significantly expressed during fiber cell development. Exogenous application of BR enhanced the transcription level of these genes. In addition, we found that a large number of GhZFP genes contained gibberellic acid responsive element (GARE), auxin responsive element (AuxRE), or E-box element in their promoter regions. Our results showed that the majority of GhZFP genes containing these elements in their promoters were significantly up-regulated by exogenous application of gibberellic acid, auxin, and BR, respectively.
Methods
Plant materials and growth conditions
Gossypium hirsutum (Xuzhou 142) was grown in a climate-controlled greenhouse with a 16 h light and 8 h dark cycle at 30 °C, as previously reported [30]. For phytohormone treatment experiments, a total of 30 ovules were used for each treatment and three biological triplicates were performed for each experiment.
Sequence retrieval, multiple sequence alignment, and phylogenetic analysis
The cotton genome sequences were acquired from the CottonGen website (https://www.cottongen.org). The Arabidopsis genome sequences were downloaded from TAIR 10 (http://www.arabidopsis.org). HMMER software with default parameters was used to search for corresponding protein sequences using the conserved ZFP domain as a query. We used the BLAST program to further identify ZFP sequences based on homology. Multiple sequence alignments of all identified ZFPs were performed using Clustal X [31]. A phylogenetic tree of deduced amino acid sequences was constructed using the neighbor-joining algorithm with default parameters and 1000 bootstrap replicates in MEGA 6.0 (https://www.megasoftware.net).
Analysis of chromosomal location, gene structure and conserved motif
The positional information for GhZFP was obtained from the parsed general feature format (GFF) files downloaded from the CottonGen website. For the exon-intron structural analysis of GhZFP genes, the coding sequences were used to align with their genomic DNA sequences and the structure diagrams were drawn using the online Gene Structure Display Server (GSDS) program (http://gsds.cbi.pku.edu.cn/). Conserved motifs of GhZFP proteins were investigated using the online toolkit Multiple Expectation maximization for Motif Elicitation (MEME 3.0.3; http://meme-suite.org/). The optimized parameters of MEME were employed as follows: number of repetitions, any; maximum number of motifs, 50; and the optimum width of each motif, between 6 and 300 residues, and retaining only motifs associated with an E value <e− 5. The identified protein motifs were further annotated with ScanProsite (http://prosite.expasy.org/scanprosite/).
RNA extraction and quantitative RT-PCR (qRT-PCR) analysis
Cotton ovules harvested after phytohormone treatment for each indicated time were frozen in liquid nitrogen and then ground into fine powder with a mortar and pestle using a previously described method [1]. The total RNA was extracted using a PureLink™ RNA mini kit (Invitrogen, Lot no.1687455) according to the manufacturer’s instructions, and cDNA was reverse-transcribed from 1.2 μg total RNA [32]. In the qRT-PCR experiments, each gene was run in three biological replicates and three technical replicates with the reaction parameters as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s and 56 °C for 30 s. A melting curve was generated from 65 to 95 °C. Cotton GhUBQ7 (GenBank no. AY189972) was used as the internal control. Primers for qRT-PCR analysis are listed in Additional file 1: Table S3. SigmaStat software was used for one-way statistical variance analysis.
In vitro ovule culture
The in vitro ovule culture was performed according to a previously published method [26]. Cotton ovules collected at 1 day post anthesis (DPA) were sterilized in 10% sodium hypochlorite and cultured in medium at 30 °C [33]. Five Micrometre 1-Naphthylacetic acid (NAA, Sigma), 1 μM gibberellin acid (GA3, Sigma) and 5 μM BR (Sigma) were added to the culture medium for the indicated time, respectively. After treatment, samples were collected for qRT-PCR experiments.
Identification of cis-elements in GhZFP promoter region
The predicted promoter sequences of GhZFP were downloaded from the CottonGen website (https://www.cottongen.org). The cis-elements in GhZFP promoter regions were predicted using the website Plant Cis-acting Regulatory DNA Elements (PLACE, https://www.dna.affrc.go.jp/PLACE/?action=newplace) [34].
Results
Genome-wide identification of the ZFP gene family in Gossypium
Whole genome sequences of three sequenced cotton species (G. hirsutum, G. arboreum and G. raimondii) were used to identify the ZFP proteins. Arabidopsis ZFP protein sequences were used as queries to search the three reference genomes to screen out candidate ZFP proteins in cotton. Using HMMR software for further selection of ZFP candidates based on conserved domains, we identified 29 ZFPs in G. hirsutum, along with 23 in G. arboreum and 23 in G. raimondii. Among the 29 GhZFP proteins found in G. hirsutum genome, 13 members originated from the At sub-genome and 16 from the Dt sub-genome (Additional file 1: Table S1). The lengths of GhZFP proteins ranged from 170 (GhZFP10–9) to 295 (GhZFP10–1) amino acids (aa), with an average length of 238 aa. The GaZFP proteins ranged from 165 (GaZFP11–3) to 295 (GaZFP10–1) amino acids, with an average length of 226 aa. The GrZFP proteins ranged from 116 (GrZFP4–1) to 490 (GrZFP10–1) amino acids with an average length of 272 aa. The physicochemical parameters analysis showed that the molecular weight of GhZFP proteins ranged from 21.49 (GhZFP8–1) to 33.23 (GhZFP10–2) KDa with an average value of 26.42 KDa and the isoelectric point (pl) of GhZFP proteins ranged from 5.12 (GhZFP4–1) to 9.34 (GhZFP11–1) with an average value of 6.90 (Table 1).
Table 1.
Physicochemical parameters of 29 GhZFP genes in G. hirsutum genome
| Name | Protein length | Protein MW (kD) | Protein pI | Extinction coefficient | Instability index | Aliphatic index |
|---|---|---|---|---|---|---|
| GhZFP1–1 | 250 | 27.65 | 5.45 | 0.49 | 41.23 | 77.36 |
| GhZFP2–1 | 253 | 28.22 | 7.79 | 0.77 | 57.66 | 61.7 |
| GhZFP2–2 | 293 | 32.00 | 8.29 | 1.02 | 44.54 | 59.32 |
| GhZFP3–1 | 253 | 28.15 | 8.28 | 0.77 | 56.71 | 63.24 |
| GhZFP3–2 | 292 | 31.94 | 8.28 | 1.07 | 45.78 | 58.87 |
| GhZFP4–2 | 250 | 27.06 | 5.45 | 0.49 | 41.23 | 77.36 |
| GhZFP5–1 | 239 | 26.56 | 6.11 | 0.67 | 61.39 | 49.04 |
| GhZFP5–2 | 239 | 26.56 | 6.11 | 0.67 | 61.39 | 49.04 |
| GhZFP6–1 | 205 | 21.60 | 8.93 | 0.43 | 53.39 | 61.56 |
| GhZFP7–1 | 250 | 27.05 | 5.96 | 0.43 | 44.25 | 59.4 |
| GhZFP8–1 | 194 | 21.49 | 9.07 | 1.07 | 69.61 | 44.79 |
| GhZFP10–10 | 222 | 24.97 | 6.83 | 1.13 | 56.11 | 53.24 |
| GhZFP10–12 | 270 | 29.22 | 6.29 | 1.07 | 58.02 | 65.78 |
| GhZFP4–1 | 250 | 27.06 | 5.12 | 0.43 | 39.92 | 78.92 |
| GhZFP8–2 | 248 | 27.53 | 6.92 | 0.78 | 59.24 | 49.96 |
| GhZFP8–3 | 235 | 26.28 | 6.19 | 0.77 | 59.79 | 46.13 |
| GhZFP8–4 | 194 | 21.49 | 9.07 | 1.07 | 69.61 | 44.79 |
| GhZFP10–1 | 295 | 33.10 | 6.17 | 1.37 | 56.79 | 56.2 |
| GhZFP10–2 | 295 | 33.23 | 6.12 | 1.46 | 57.52 | 58.85 |
| GhZFP10–3 | 206 | 23.33 | 6.99 | 1.39 | 64.01 | 65.39 |
| GhZFP10–4 | 243 | 27.91 | 7.1 | 1.32 | 60.71 | 46.17 |
| GhZFP10–5 | 206 | 23.33 | 6.99 | 1.39 | 64.01 | 65.39 |
| GhZFP10–6 | 215 | 23.65 | 5.73 | 1.26 | 61.54 | 64.98 |
| GhZFP10–7 | 215 | 23.65 | 5.73 | 1.26 | 61.54 | 64.98 |
| GhZFP10–8 | 206 | 23.33 | 6.99 | 1.39 | 64.01 | 65.39 |
| GhZFP10–9 | 170 | 23.33 | 6.99 | 1.39 | 64.01 | 65.39 |
| GhZFP10–11 | 274 | 29.61 | 6.03 | 1.05 | 57.64 | 69.12 |
| GhZFP10–13 | 215 | 23.65 | 5.73 | 1.26 | 61.54 | 64.98 |
| GhZFP11–1 | 207 | 23.19 | 9.34 | 1.04 | 63.08 | 56.52 |
Phylogenetic analysis, chromosomal location and gene duplication of the ZFP gene family
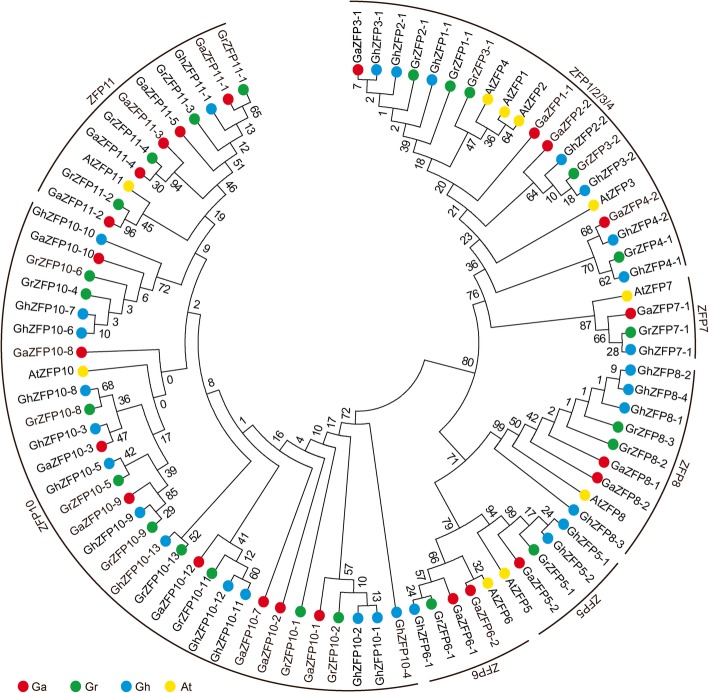
To gain further insights into the evolutionary history and phylogenetic relationships of the ZFP gene family, a neighbor-joining phylogenetic tree was constructed. Our results showed that ZFP family genes were clustered into seven subfamilies and that most of the orthologous genes between the diploids and the corresponding allotetraploid were grouped into the same clade (Fig. 1). As shown in phylogenetic tree (Fig. 1), a similar organization for cotton and Arabidopsis ZFP proteins and some orthologous relationships between both species were identified. Based on this analysis, cotton ZFP proteins were named based on their relationships to known Arabidopsis ZFPs. To validate this result from the neighbor-joining (NJ) method, we reconstructed the phylogenetic tree of ZFP genes using the maximum likelihood method and again found that ZFP genes were divided into seven subfamilies (Additional file 2: Figure S1), similar to the result obtained using the neighbor-joining (NJ) method.
Fig. 1.
Phylogenetic analysis of the ZFP gene family. The phylogenetic tree was constructed using the full length ZFP protein amino acid sequences from G. arboreum, G. raimondii, G. hirsutum and A. thaliana. MEGA 6.0 software was used with the neighbor-joining method and bootstrapping with 1,000 iterations. At, Arabidopsis thaliana; Ga, Gossypium arboreum; Gr, Gossypium raimondii; Gh, Gossypium hirsutum
In contrast to Arabidopsis, two GaZFP8 and eight GaZFP10 genes were found in G. arboreum, as well as two GrZFP8 and nine GrZFP10 genes in G. raimondii. Furthermore, four TcZFP8 genes and two TcZFP10 genes were found in T. cacao (Additional file 3: Figure S2). To validate the evolutionary relationship of ZFPs, we introduced the ZFP proteins from O. sativa and G. max. As shown in Additional file 4: Figure S3, ZFP proteins were also clustered into seven subfamilies, consistent with our previous results (Additional file 4: Figure S3). Comparison of gene number of ZFP8 and ZFP10 genes in three Gossypium species and Arabidopsis suggests that the expansion of these gene subfamilies may have occurred in Gossypium after divergence from the common ancestor of Arabidopsis and Gossypium.
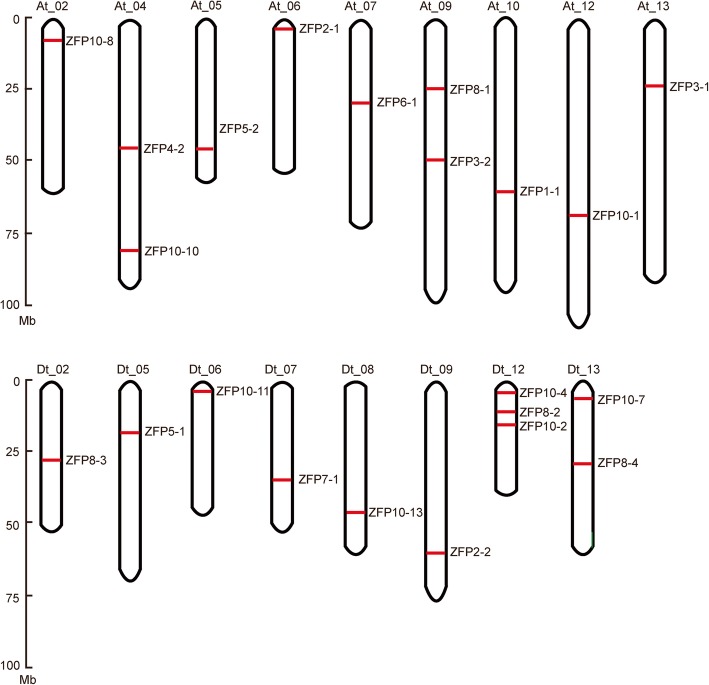
To determine the chromosomal location of GhZFP genes in G. hirsutum, the physical distribution of GhZFP genes along the chromosomes was performed using positional information files downloaded from the CottonGen website. Among the 29 GhZFP genes, 22 genes deposited on 17 chromosomes, including nine chromosomes from the At subgenome and eight from the Dt subgenome (Fig. 2). Most of the chromosomes possessed only one GhZFP gene, except for the At_04 and At_09 chromosomes. Among the GaZFP and GrZFP genes, 23 members from G. arboreum were distributed on 10 chromosomes and 17 members from G. rainomdii were distributed on nine chromosomes (Additional file 5: Figure S4).
Fig. 2.
Chromosomal location of GhZFP genes on 17 chromosomes. The genome visualization tool CIRCOS was used to display the chromosomal location of GhZFP genes. The chromosome number is shown on the top of each chromosome. The scale bar indicates the length in megabases (Mb)
We further investigated possible GhZFP genes created by whole-genome duplications (WGD). Among all of the GhZFP genes, 25 were produced from WGD and 4 from dispersed duplication events (Additional file 1: Table S2), indicating that WGD events played an important role in the expansion of GhZFP gene. Additionally, in order to investigate the evolutionary history of GhZFP genes, the KaKs_calculator 2.0 program was used to calculate synonymous and non-synonymous substitution rates. The Ks ratios of GhZFP gene pairs ranged from 0.016 to 0.555. The approximate time of duplication events of GhZFPs ranged from 0.53 million years ago (MYA) to 18.5 MYA (Table 2).
Table 2.
Dates of duplication for the duplicated gene pairs
| Gene 1 | Gene 2 | Length | Ks | T = Ks/2λ |
|---|---|---|---|---|
| GhZFP3–4 | GhZFP33 | 876 | 0.026 | 0.87 |
| GhZFP3–2 | GhZFP3–1 | 759 | 0.016 | 0.53 |
| GhZFP10–8 | GhZFP10–3 | 684 | 0.041 | 1.37 |
| GhZFP10–11 | GhZFP1012 | 804 | 0.022 | 0.73 |
| GhZFP10–7 | GhZFP10–6 | 645 | 0.019 | 0.63 |
| GhZFP10–2 | GhZFP10–1 | 885 | 0.024 | 0.8 |
| GhZFP8–2 | GhZFP84 | 717 | 0.555 | 18.5 |
| GhZFP5–1 | GhZFP4–1 | 717 | 0.046 | 1.53 |
Gene structure and conserved motifs of GhZFP proteins
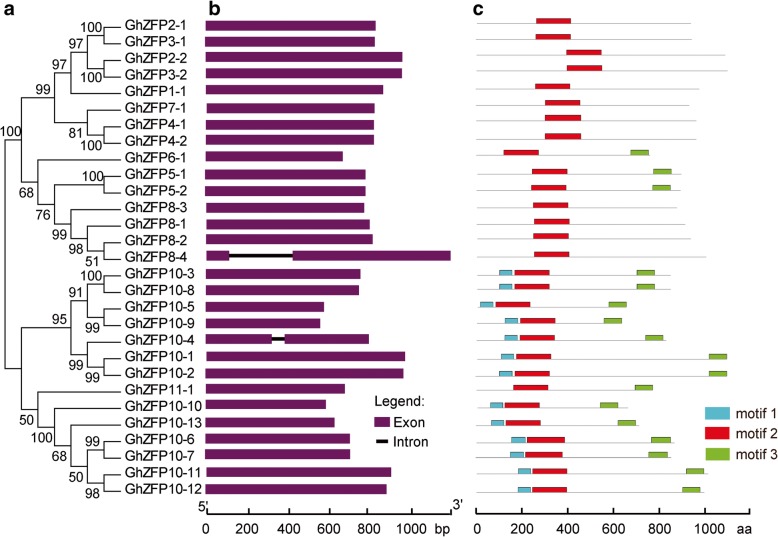
With the aim of gaining better understanding of the similarity and diversity between different members of GhZFP proteins, we generated an unrooted phylogenetic tree with the deduced GhZFP amino acid sequences (Fig. 3a) and performed a comparative analysis of exon-intron structure. We found that the gene length of GhZFP was relatively conserved, with GhZFP8–4 having the longest (1.2 Kb) and GhZFP10–9 having the shortest (0.6 Kb) genomic sequence (Fig. 3b). Twenty seven out of 29 GhZFP genes had no introns, except for GhZFP8–4 and GhZFP10–4, which contained one intron. These results were quite similar to the structure of ZFP genes in Populus trichocarpa [35]. Most GhZFPs within the same subclades exhibited similar gene structure in terms of numbers and lengths of introns and exons, which was consistent with subfamilies in the phylogenetic tree (Fig. 3a).
Fig. 3.
Phylogenetic relationships, gene structures and protein domain architecture of GhZFP genes. a Phylogenetic relationships between GhZFPs. A phylogenetic tree was generated using the maximum likelihood method with 1,000 bootstrap iterations in MEGA 6.0 software. b Gene structure (exon-intron organization) analysis of GhZFPs. Exons and introns are represented by purple boxes and black lines, respectively. The scale bar is shown at the bottom. c The protein domain architecture of GhZFP genes. The number and order of motifs in each GhZFP genes are shown. Motif 2 in (c) is the ZFP domain
Furthermore, we investigated the conserved motifs in GhZFP proteins to understand the diversity of motif compositions among GhZFP proteins. A total of three conserved motifs, named motif 1 to motif 3, were identified in GhZFP proteins. The number of conserved motifs in each GhZFP varied from 2 to 3 and most GhZFPs within the same subfamily exhibited similar motif compositions (Fig. 3c). In addition, most GhZFP proteins possessed three conserved motifs, except for GhZFP1, GhZFP2, GhZFP3, GhZFP4 and GhZFP8 subfamilies, which contained two conserved motifs, indicating that GhZFP proteins showed functional divergence.
GhZFP8 subfamily genes were highly expressed during fiber cell development
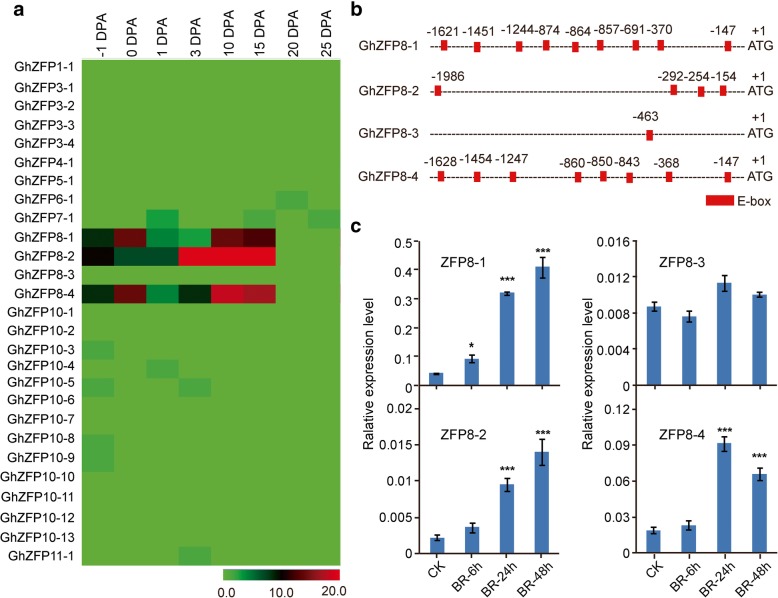
To determine which GhZFP genes potentially function in fiber cell development, the expression profiles of individual genes were investigated using transcriptome data from different developmental stages of fiber cells, including fibers at 0, 3, 10, and 15 DPA.
We found that most of GhZFP genes from the same subfamily shared similar expression patterns. Notably, all members of the GhZFP8 subfamily, except for GhZFP8–3, were highly expressed during fiber cell initiation and elongation development (Fig. 4a), suggesting that these genes may be involved in cotton fiber cell development. We extracted 2000 bp sequences upstream of transcription start codon (ATG) as promoter region. When analyzing the promoter regions of GhZFP8 genes, a large number of E-box elements (CANNTG, where N can be any nucleotide) were found in GhZFP8 promoter regions (Fig. 4b). E-box elements can be recognized by BES1, a core transcription factor in the BR signaling pathway [36].
Fig. 4.
Expression profiles of GhZFP genes during fiber development. a RNA-seq data heat map of GhZFP gene expression levels during different stages of fiber growth. The differences in gene expression are shown in different colors. b Cis-element analysis of GhZFP8 promoters. E-box (CANNTG) motifs (red) were predicted using the online toolkit, PLACE. A total of 2,000 bp promoter sequence for each GhZFP8 were used for the analysis. c Exogenous BR promotes the transcription of GhZFP8. Statistical significance was determined using one-way analysis of variance combined with Tukey’s test. *P < 0.05; **P < 0.01; ***P < 0.001
Three GhZFP8 genes, GhZFP8–1, GhZFP8–2 and GhZFP8–4, contained at least four E-box elements in their promoter regions, while GhZFP8–3 contained only one E-box (Fig. 4b). These results indicate that the expression of GhZFP8 genes may be induced by BR. To further confirm this, we performed qRT-PCR experiments to assess the transcription levels of GhZFP8 genes with or without BR treatment in vitro (Fig. 4c). Our results showed that the transcripts of GhZFP8–1, GhZFP8–2 and GhZFP8–4 were significantly induced after BR application for 24 h (Fig. 4c). However, BR did not induce GhZFP8–3 transcripts even when the ovules were treated with BR for 48 h. These results imply that GhZFP8 may be required for cotton fiber cell development.
Gene expression analysis of GhZFP genes in response to plant hormone treatment
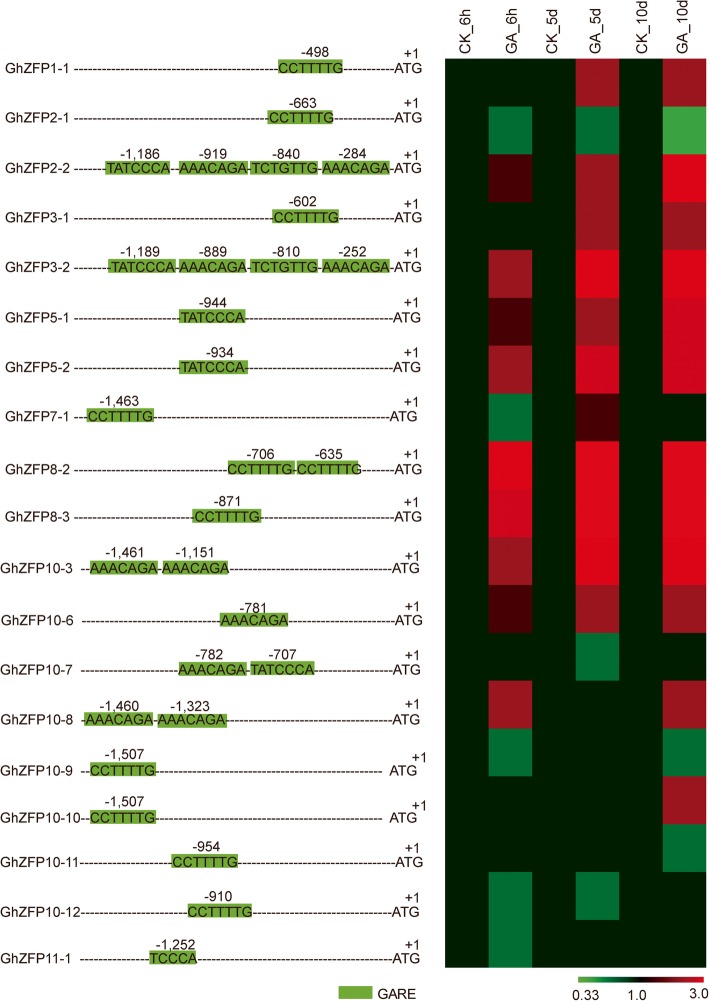
Phytohormones play pivotal roles in plant development. Considerable evidence suggests that gibberellic acid, auxin and BR are required for cotton fiber cell development [34, 37–39]. In order to explore the relationship between GhZFP genes and gibberellin, we analyzed the cis-elements in GhZFP promoter regions. A large number of GARE elements (CCTTTG or TATCCCA or AAACAGA or TCTGTTG) were found within GhZFP promoter regions (Fig. 5). Specifically, 19 out of 29 GhZFP genes possessed at least one GARE element. These results strongly suggest that the expression of GhZFP genes may be regulated by gibberellic acid. To confirm this finding, expression analysis of GhZFP genes was carried out after treatment with gibberellin. Our results showed that a total of 15 GhZFP genes were responsive to gibberellin treatment (Fig. 5), except for GhZFP2–1, GhZFP7–1, GhZFP10–7 and GhZFP11–1 genes.
Fig. 5.
Expression profiles of 19 GhZFP genes under GA3 treatment. Analysis of 19 GhZFP genes with GARE elements present in their promoter regions (left). GAREs (green) were predicted using the PLACE website (http://www.dna.affrc.go.jp/htdocs/PLACE/). Fold change of 19 GhZFP genes under GA3 treatment (right). All treatments were performed with three biological and three technical replicates. The relative gene expression levels were determined using cotton GhUBQ7 as a control
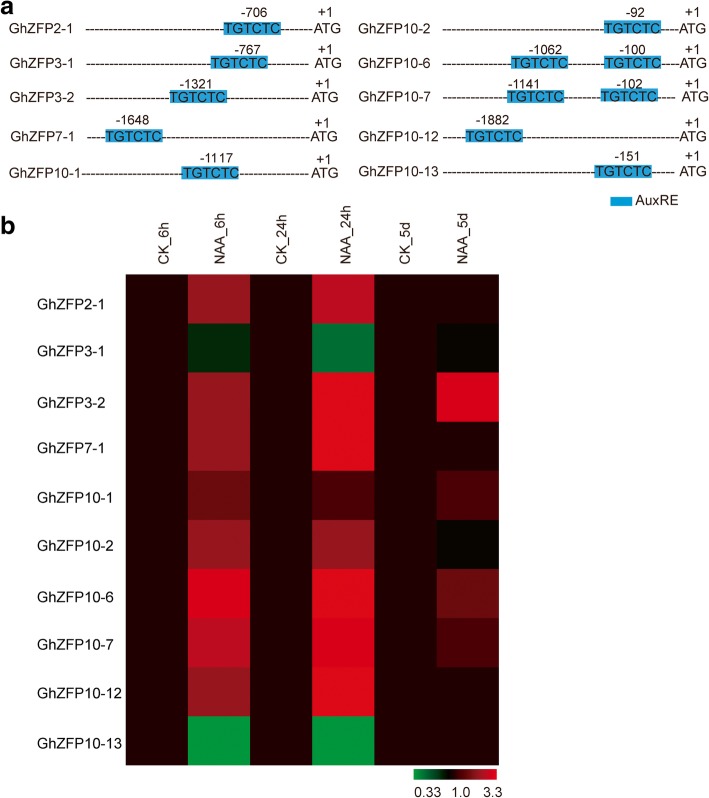
Auxin is also known to play important roles in promoting cotton fiber cell development. Overexpression of the auxin biosynthetic gene iaaM significantly increased fiber cell initiation [38]. We then identified the GhZFP genes responsive to auxin. To this end, we used a similar method to identify genes that were responsive to gibberellic acid. Auxin response factors (ARF), key components in the auxin signaling pathway, specifically recognize and bind to auxin responsive elements (AuxRE, TGTCTC) to regulate downstream auxin responsive genes [40]. Our data showed that a total of 10 GhZFP genes were found to contain AuxRE elements in their promoter regions (Fig. 6a). qRT-PCR analysis showed that the transcript levels of seven GhZFP genes significantly increased after application of 5 μM NAA, an auxin analog, for 24 h (Fig. 6b). These results suggest that auxin may promote the expression of GhZFP genes.
Fig. 6.
Expression profiles of 10 GhZFP genes under NAA treatment. a Analysis of 10 GhZFP with AuxRE elements in their promoter regions. AuxREs (blue) were predicted using the PLACE website. b NAA activated transcription of most GhZFP genes with AuxRE elements in their promoter regions. Relative expression levels of each gene were determined after normalizing to the expression level in CK (no chemical added) ovules, which was set to 1.0
BR is also involved in fiber growth and exogenous application of BR in vitro promoted fiber cell elongation [33, 37]. E-box elements are specifically recognized by the BES1 transcription factor, which plays a key role in BR-regulated gene expression [36]. We analyzed the distribution of E-box elements in GhZFP promoter regions and assayed the transcription levels of the GhZFP genes with or without BR treatment. Surprisingly, 25 out of 29 GhZFP genes contained an E-box element in their promoter regions (Additional file 6: Figure S5), indicating that GhZFP transcripts may be induced by BR. Further evidence was found in significantly stimulated transcription levels for the majority of GhZFP genes with an E-box element in their promoter regions when they were treated with 5 μM BR (Fig. 7).
Fig. 7.
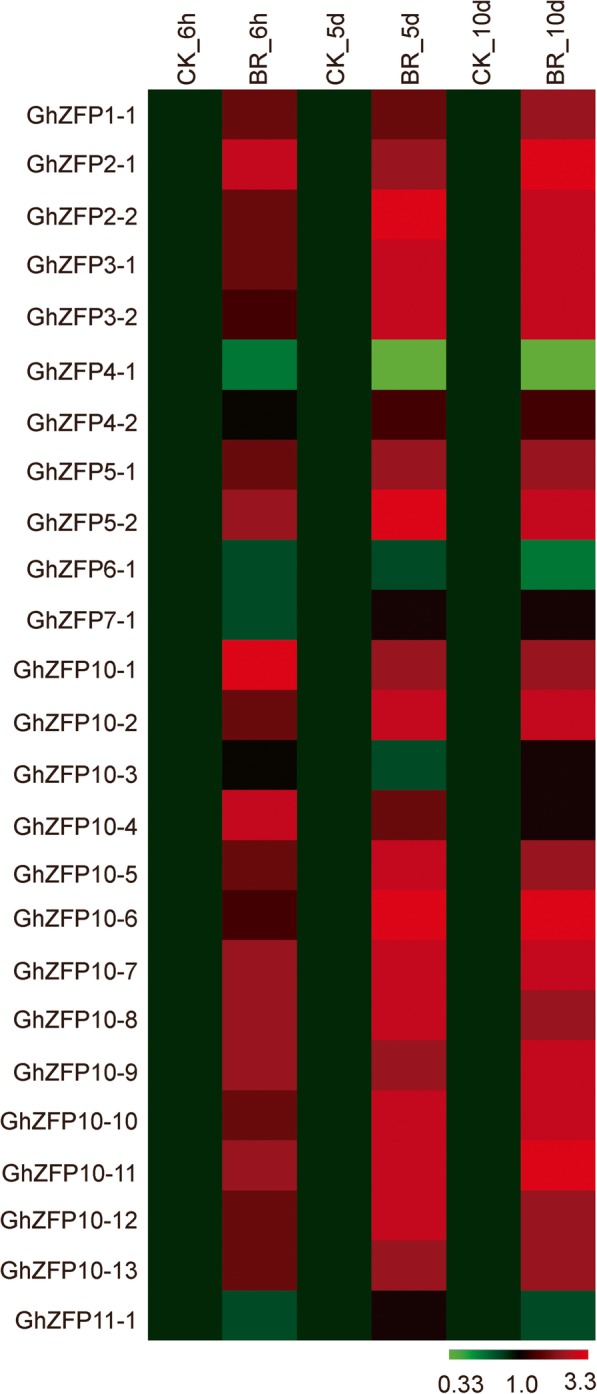
Heat map of expression levels of GhZFP genes with the BES1 element in their promoter regions under BR treatment. Differences in gene expression are shown in different colors indicated in the scale. All treatments were performed with three biological and three technical replicates
Taken together, transcription of GhZFP genes was widely induced by gibberellic acid, auxin and BR, suggesting that these genes play important roles in phytohormone regulation of cotton development.
Discussion
The ZFP gene family is one of the largest gene families and its members are involved in a wide range of functions in plant growth and development [41, 42]. In Arabidopsis, there are 211 zinc finger proteins, which constitute the most abundant family of putative transcriptional regulators in plants [42]. Among them, only 10 members contain just a single zinc finger domain, for which they are named ZFP proteins. ZFP family genes have been studied in Arabidopsis, rice and petunia [42–44]. However, the ZFP gene family has not been investigated in G. hirsutum. In this study, we identified 29 GhZFP genes in G. hirsutum, including 13 genes from the At subgenome and 16 from the Dt subgenome (Additional file 1: Table S1). Notably, 23 ZFP genes were found in G. arboreum and G. raimondii, each, suggesting that gene loss events have occurred in the ZFP gene family after the polyploidization in G. hirsutum, which also confirmed prior work that a large number of gene loss events occurred in allotetraploid cotton [45, 46].
Gene duplication events are involved in gene expansion and genomic realignments [47]. Gene duplication contributed to functional innovation and expansion of genes, especially for transcription factor gene families in plants [35]. To investigate the origin of ZFP genes in G. hirsutum, we analyzed their distribution along the chromosomes and gene duplication events. Among the 29 GhZFP genes, 22 members were located across 17 chromosomes (Fig. 2). Exon-intron organization, or the genomic structure of genes, plays an important role in the evolution of gene families and gene splicing [48]. Our results showed that all of the GhZFP genes had only one exon, except for GhZFP8–4 and GhZFP10–4, which had one intron between two exons, respectively (Fig. 3). This may be due to incomplete cDNA molecules recombining with their genomic copies and horizontal transfer, which are found to lead to the loss of introns within genomes [49–51]. In Populus trichocarpa, most of the C2H2-ZF group I genes had no introns [35], consistent with the exon-intron arrangement in G. hirsutum.
To better understand the role of GhZFP genes in G. hirsutum, we performed a systematic analysis on gene expression patterns throughout several developmental stages of cotton fiber cells. The transcriptome data were used to analyze the expression levels of GhZFP genes. Interestingly, most GhZFP8 subfamily genes were significantly expressed in fiber cells (Fig. 4a), strongly suggesting the involvement of GhZFP8 genes in fiber development. We also performed cis-element prediction on GhZFP8 promoter regions to explore the mechanisms of GhZFP8 regulating fiber growth. We found abundant E-box elements in GhZFP8 promoter regions. Since BES1 is reportedly part of the BR response pathway, this finding suggests that transcription of GhZFP8 genes could be activated by BR. The phytohormone, BR, plays an important role in regulating fiber development [33, 52]. Our findings suggest that BR could induce GhZFP8 expression to regulate fiber development.
Auxin, gibberellic acids and BR play important roles in fiber development [33, 37–39]. In order to explore the link between GhZFP genes and these hormones in detail, we investigated the transcriptional expression levels of GhZFP genes before and after treatment with the individual hormones. When analyzing the promoter regions of all GhZFP genes, 19 of them were found to possess GARE elements. Exogenous application of GA3 significantly induced the transcription of most GhZFP genes with GARE elements in their promoter regions (Fig. 5). Applications of NAA, an auxin analog, resulted in significant transcriptional up-regulation of seven genes which contained AuxRE elements in their promoters (Fig. 6). Among the 29 GhZFP genes, 25 members contained an E-box element in their promoter region, and most of which were up-regulated after treatment with BR for 5 days (Fig. 7). Our findings suggest that GhZFP genes were widely induced by auxin, gibberellin and BR, which provides a foundation for the identification of more downstream genes with potential roles in phytohormone stimuli, and a basis for breeding better cotton varieties in the future.
Conclusions
Our study provides a comprehensive analysis of the GhZFP gene family. We revealed that GhZFP8 genes were involved in cotton fiber development. This study will expand our understand the precise role of GhZFP genes in cotton fiber development and in adaption to phytohormone stimuli. Our findings will also further provide clues for breeding better cotton varieties in the future.
Additional files
Table S1. Analysis of G. hirsutum ZFP gene family and its orthologs in AA and DD cotton genomes. Table S2. Analysis of duplication events in G. hirsutum ZFP genes located in chromosomes. Table S3. A list of primers used in this study. (PDF 112 kb)
Figure S1. Phylogenetic analysis of the ZFP gene family. MEGA 6.0 software was used with the maximum likelihood method and bootstrapping with 1,000 iterations. At, Arabidopsis thaliana; Ga, Gossypium arboreum; Gr, Gossypium raimondii; Gh, Gossypium hirsutum. (PDF 165 kb)
Figure S2. Phylogenetic analysis of the ZFP gene family. MEGA 6.0 software was used with the Neighbor-Joining method and bootstrapping with 1,000 iterations. (PDF 253 kb)
Figure S3. Phylogenetic analysis of the ZFP gene family. The phylogenetic tree was constructed using the full length ZFP protein amino acid sequences. At, Arabidopsis thaliana; Ga, Gossypium arboreum; Gr, Gossypium raimondii; Gh, Gossypium hirsutum; Tc, Theobroma cacao; BGIOSGA, Oryza sativa; Glyma, Glycine max. (PDF 515 kb)
Figure S4. Chromosomal location of GaZFP (a) and GrZFP (b) genes on chromosomes. The chromosome number is shown on the top of each chromosome. The scale bar indicates the length in megabases (Mb). (PDF 199 kb)
Figure S5. Analysis of 25 GhZFP with BES1 element present in their promoter regions. BES1 elements (red) were predicted using the online PLACE website. (PDF 424 kb)
Acknowledgements
Not applicable.
Abbreviations
- ARF
Auxin responsive factors
- BR
Brassinosteroids
- DPA
Day post anthesis
- GFF
General Feature Format
- GSDS
Gene Structure Display Server
- MARD1
The mediator of ABA-regulated dormancy1
- MEME
Multiple Expectation maximization for Motif Elicitation
- MYA
Million years ago
- RD21A
Responsive to dehydration 21A
- WGD
Whole genome duplication
Authors’ contributions
GX and PH conceived the original research plan, YYang, ZW and PZ performed experiments, YYuan, LZ, YM and CP carried out the data analysis; GX wrote the article, JY and PH revised the manuscript. YYuan would correspond to the author furthest up on the author list. All authors contributed to the research and read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31600223), Hong Kong Scholars Program (Grant No. XJ2017017), Natural Science Basic Research Plan in Shaanxi Province of China (Grant No. 2018JZ3006 and 2019JM-491), Shaanxi Youth Entrusted Talents Program (Grant No. 20190205), Shaanxi Postdoctoral Project (Grant No. 2018BSHYDZZ76), Fundamental Research Funds for the Central Universities (Grant No. GK201803041 and GK201901004), State Key Laboratory of Cotton Biology Open Fund (Grant No. CB2018A03 and CB2019A09), National Undergraduate Innovation and Entrepreneurship Training Program (Grant No. cx2018143), and Diligent Research and Innovation Fund Project (Grant No. KY2018YB003). The funding bodies provided the financial support to the research projects, but didn’t involved in study design, data collection, analysis, or preparation of the manuscript.
Availability of data and materials
The cotton genome data analyzed during this article is available from CottonGen (https://www.cottongen.org) and the A. thaliana genome sequence is accessible at TAIR 10 (http://www.arabidopsis.org).
Ethics approval and consent to participate
All the cotton lines used and analyzed for this study were collected from the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences, which were public and available for non-commercial purpose. This article did not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Peng He, Email: phe@snnu.edu.cn.
Yan Yang, Email: 360226919@qq.com.
Zihua Wang, Email: wangzi@snnu.edu.cn.
Peng Zhao, Email: zhaopeng1817@163.com.
Yi Yuan, Email: 1208404121@qq.com.
Li Zhang, Email: 1012884880@qq.com.
Yueqin Ma, Email: 1010426519@qq.com.
Chaoyou Pang, Email: chypang@163.com.
Jianing Yu, Email: jnyu@snnu.edu.cn.
Guanghui Xiao, Email: guanghuix@snnu.edu.cn.
References
- 1.Zhang Y, He P, Yang Z, Huang G, Wang L, Pang C, Xiao H, Zhao P, Yu J, Xiao G. A genome-scale analysis of the PIN gene family reveals its functions in cotton fiber development. Front Plant Sci. 2017;8:461. doi: 10.3389/fpls.2017.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. doi: 10.1016/S0065-2113(02)78004-8. [DOI] [Google Scholar]
- 3.Gokani SJ, Thaker VS. Role of gibberellic acid in cotton fibre development. J Agric Sci. 2002;138:255–260. doi: 10.1017/S0021859602001934. [DOI] [Google Scholar]
- 4.Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Xing H, Ashikari M, Kitano H, Yamaguchi I, Matsuoka M. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell. 2007;19:2140–2155. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, Yoshida S, Allen RD. Brassinosteroid regulates fiber development on cultured cotton ovules. Plant Cell Physiol. 2005;46:1384–1391. doi: 10.1093/pcp/pci150. [DOI] [PubMed] [Google Scholar]
- 6.Xiao G, Zhao P, Zhang Y. A pivotal role of hormones in regulating cotton fiber development. Front Plant Sci. 2019;10:87. [DOI] [PMC free article] [PubMed]
- 7.Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HJ, Triplett BA. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 2001;27:1361–1366. doi: 10.1104/pp.010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M, Xiao Y, Luo X, Zhang J, Xiang C, Pei Y. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol. 2011;29:453–458. doi: 10.1038/nbt.1843. [DOI] [PubMed] [Google Scholar]
- 10.Luo X, Bai X, Zhu D, Li Y, Ji W, Cai H, Wu J, Liu B, Zhu Y. GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta. 2012;235:1141–1155. doi: 10.1007/s00425-011-1563-0. [DOI] [PubMed] [Google Scholar]
- 11.Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot. 2010;61:2807–2818. doi: 10.1093/jxb/erq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pradhan S, Kant C, Verma S, Bhatia S. Genome-wide analysis of the CCCH zinc finger family identifies tissue specific and stress responsive candidates in chichpea (Cier arietinum L.) PLoS One. 2017;12:e0180469. doi: 10.1371/journal.pone.0180469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Xu Y, Xiao J, Ma Q, Li D, Xue Z, Chong K. OsDOG, a gibberellin-induced A20/AN1 zinc-finger protein, negatively regulates gibberellin-mediated cell elongation in rice. J Plant Physiol. 2011;168:1098–1105. doi: 10.1016/j.jplph.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Joseph MP, Papdi C, Kozma-Bognár L, Nagy I, López-Carbonell M, Rigó G, Koncz C, Szabados L. The Arabidopsis ZINC FINGER PROTEIN3 interferes with abscisic acid and light signaling in seed germination and plant development. Plant Physiol. 2014;165:1203–1220. doi: 10.1104/pp.113.234294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Wang J, Sun Q, Li W, Yu Y, Zhao M, Meng Z. Expression analysis of genes encoding double B-box zinc finger proteins in maize. Funct Integr Genomics. 2017;17:653–666. doi: 10.1007/s10142-017-0562-z. [DOI] [PubMed] [Google Scholar]
- 16.Seok HY, Woo DH, Park HY, Lee SY, Tran HT, Lee EH, Vu Nguyen L, Moon YH. AtC3H17, a non-tandem CCCH zinc finger protein, functions as a nuclear transcriptional activator and has pleiotropic effects on vegetative development, flowering and seed development in Arabidopsis. Plant Cell Physiol. 2016;57:603–615. doi: 10.1093/pcp/pcw013. [DOI] [PubMed] [Google Scholar]
- 17.Chai G, Kong Y, Zhu M, Yu L, Qi G, Tang X, Wang Z, Cao Y, Yu C, Zhou G. Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J Exp Bot. 2015;66:2595–2609. doi: 10.1093/jxb/erv060. [DOI] [PubMed] [Google Scholar]
- 18.Liu XM, Nguyen XC, Kim KE, Han HJ, Yoo J, Lee K, Kim MC, Yun DJ, Chung WS. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem Biophys Res Commun. 2013;430:1054–1059. doi: 10.1016/j.bbrc.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Ma C, Xu Y, Wei Q, Imtiaz M, Lan H, Gao S, Cheng L, Wang M, Fei Z, Hong B, Gao J. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell. 2014;26:2038–2054. doi: 10.1105/tpc.114.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baek D, Cha JY, Kang S, Park B, Lee HJ, Hong H, Kim DH, Kim MC, Lee SY, Yun DJ. The Arabidopsis a zinc finger domain protein ARS1 is essential for seed germination and ROS homeostasis in response to ABA and oxidative stress. Front Plant Sci. 2015;6:963. doi: 10.3389/fpls.2015.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng L, Zhao F, Li R, Xu C, Chen K, Xiao H. The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol. 2015;167:931–949. doi: 10.1104/pp.114.255174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai G, Hu R, Zhang D, Qi G, Zuo R, Cao Y, Chen P, Kong Y, Zhou G. Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa) BMC Genomics. 2012;13:253. doi: 10.1186/1471-2164-13-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng X, Zhao Y, Cao J, Zhang W, Jiang H, Li X, Ma Q, Zhu S, Cheng B. CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS One. 2012;7:e40120. doi: 10.1371/journal.pone.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta SK, Rai AK, Kanwar SS, Sharma TR. Comparative analysis of zinc finger proteins involved in plant disease resistance. PLoS One. 2012;7:e42578. doi: 10.1371/journal.pone.0042578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zang D, Li H, Xu H, Zhang W, Zhang Y, Shi X, Wang Y. An Arabidopsis zinc finger protein increases abiotic stress tolerance by regulating sodium and potassium homeostasis, reactive oxygen species scavenging and osmotic potential. Front Plant Sci. 2016;7:1272. doi: 10.3389/fpls.2016.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chrispeels HE, Oettinger H, Janvier N, Tague BW. AtZFP1, encoding Arabidopsis thaliana C2H2 zinc-finger protein 1, is expressed downstream of photomorphogenic activation. Plant Mol Biol. 2000;42:279–290. doi: 10.1023/A:1006352809700. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z, Sun L, Zhao Y, An L, Yan A, Meng X, Gan Y. Zinc finger protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol. 2013;198:699–708. doi: 10.1111/nph.12211. [DOI] [PubMed] [Google Scholar]
- 28.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 29.Dinkins RD, Pflipsen C, Collins GB. Expression and deletion analysis of an Arabidopsis SUPERMAN-like zinc finger gene. Plant Sci. 2003;165:33–38. doi: 10.1016/S0168-9452(03)00123-7. [DOI] [Google Scholar]
- 30.He P, Zhao P, Wang L, Zhang Y, Wang X, Xiao H, Yu J, Xiao G. The PIN gene family in cotton (Gossypium hirsutum): genome-wide identification and gene expression analyses during root development and abiotic stress responses. BMC Genomics. 2017;18:507. doi: 10.1186/s12864-017-3901-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao GH, Wang K, Huang G, Zhu YX. Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin a regulated fiber growth. J Integr Plant Biol. 2016;58:577–589. doi: 10.1111/jipb.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higo K, Ugawa Y, Iwamoto M, Higo H. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Wang Z, Xu X, Zhang H, Li C. Genome-wide analysis of C2H2 zinc-finger family transcription factors and their responses to abiotic stress in poplar (Populus trichocarpa) PLoS One. 2015;10:e0134753. doi: 10.1371/journal.pone.0134753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulated gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 37.Beasley CA. Hormonal regulation of growth in unfertilized cotton ovules. Science. 1973;179:1003–1005. doi: 10.1126/science.179.4077.1003. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M, et al. Spationtemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol. 2011;29:453–458. doi: 10.1038/nbt.1843. [DOI] [PubMed] [Google Scholar]
- 39.Shan CM, Shangguan XX, Zhao B, Zhang XF, Chao LM, Yang CQ, Wang LJ, Zhu HY, Zeng YD, Guo WZ, et al. Control of cotton fiber elongation by a homeodomain transcription factor GhHOX3. Nat Commun. 2014;5:5519. doi: 10.1038/ncomms6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guilfoyle TJ, Hagen G. Auxin response factor. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Meissner R, Michael AJ. Isolation and characterization of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger proteins gene and cDNAs. Plant Mol Biol. 1997;33:615–624. doi: 10.1023/A:1005746803089. [DOI] [PubMed] [Google Scholar]
- 42.Englbrecht CC, Schoof H, Böhm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics. 2004;5:39. doi: 10.1186/1471-2164-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, Kapoor S, Tyagi AK. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol. 2007;65:467–485. doi: 10.1007/s11103-007-9199-y. [DOI] [PubMed] [Google Scholar]
- 44.Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Takatsuji H. Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 1998;26:608–615. doi: 10.1093/nar/26.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33:524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 46.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 47.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 48.Xu G, Guo C, Shan H, Kong H. Divergence of duplication genes in exon-intron structure. Proc Natl Acad Sci U S A. 2012;109:1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaughn JC, Mason MT, Sper-Whitis GL, Kuhlman P, Palmer JD. Fungal origin by horizontal gene transfer of a plant mitochondrial group I intron in the chimeric coxI gene of Peperomia. J Mol Evol. 1995;41:563–572. doi: 10.1007/BF00175814. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD. Frequent, phylogenetically local horizontal transfer of the cox1 group I intron in flowering plant mitochondria. Mol Biol Evol. 2008;25:1762–1777. doi: 10.1093/molbev/msn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson H. M. The Large srh Family of Chemoreceptor Genes in Caenorhabditis Nematodes Reveals Processes of Genome Evolution Involving Large Duplications and Deletions and Intron Gains and Losses. Genome Research. 2000;10(2):192–203. doi: 10.1101/gr.10.2.192. [DOI] [PubMed] [Google Scholar]
- 52.Yang Z, Zhang C, Yang X, Liu K, Wu Z, Zhang X, Zheng W, Xun Q, Liu C, Lu L, et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 2014;203:437–448. doi: 10.1111/nph.12824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Analysis of G. hirsutum ZFP gene family and its orthologs in AA and DD cotton genomes. Table S2. Analysis of duplication events in G. hirsutum ZFP genes located in chromosomes. Table S3. A list of primers used in this study. (PDF 112 kb)
Figure S1. Phylogenetic analysis of the ZFP gene family. MEGA 6.0 software was used with the maximum likelihood method and bootstrapping with 1,000 iterations. At, Arabidopsis thaliana; Ga, Gossypium arboreum; Gr, Gossypium raimondii; Gh, Gossypium hirsutum. (PDF 165 kb)
Figure S2. Phylogenetic analysis of the ZFP gene family. MEGA 6.0 software was used with the Neighbor-Joining method and bootstrapping with 1,000 iterations. (PDF 253 kb)
Figure S3. Phylogenetic analysis of the ZFP gene family. The phylogenetic tree was constructed using the full length ZFP protein amino acid sequences. At, Arabidopsis thaliana; Ga, Gossypium arboreum; Gr, Gossypium raimondii; Gh, Gossypium hirsutum; Tc, Theobroma cacao; BGIOSGA, Oryza sativa; Glyma, Glycine max. (PDF 515 kb)
Figure S4. Chromosomal location of GaZFP (a) and GrZFP (b) genes on chromosomes. The chromosome number is shown on the top of each chromosome. The scale bar indicates the length in megabases (Mb). (PDF 199 kb)
Figure S5. Analysis of 25 GhZFP with BES1 element present in their promoter regions. BES1 elements (red) were predicted using the online PLACE website. (PDF 424 kb)
Data Availability Statement
The cotton genome data analyzed during this article is available from CottonGen (https://www.cottongen.org) and the A. thaliana genome sequence is accessible at TAIR 10 (http://www.arabidopsis.org).